Abstract
Natural transformation is the main means of horizontal genetic exchange in the obligate human pathogen Neisseria gonorrhoeae. Neisseria spp. have been shown to preferentially take up and transform their own DNA by recognizing the nonpalindromic 10- or 12-nucleotide sequence 5′-ATGCCGTCTGAA-3′ (additional semiconserved nucleotides are underlined), termed the DNA uptake sequence (DUS10 or DUS12). Here we investigated the effects of the DUS on transformation and DNA uptake for several laboratory strains of N. gonorrhoeae. We found that all strains showed efficient transformation of DUS containing DNA (DUS10 and DUS12) but that the level of transformation with DNA lacking a DUS (DUS0) was variable in different strains. The DUS-enhanced transformation was 20-fold in two strains, FA1090 and FA19, but was approximately 150-fold in strains MS11 and 1291. All strains tested provide some level of DUS0 transformation, and DUS0 transformation was type IV pilus dependent. Competition with plasmid DNA revealed that transformation of MS11 was enhanced by the addition of excess plasmid DNA containing a DUS while FA1090 transformation was competitively inhibited. Although FA1090 was able to mediate much more efficient transformation of DNA lacking a DUS than was MS11, DNA uptake experiments showed similar levels of uptake of DNA containing and lacking a DUS in FA1090 and MS11. Finally, DNA uptake was competitively inhibited in both FA1090 and MS11. Taken together, our data indicate that the role of the DUS during DNA transformation is variable between strains of N. gonorrhoeae and may influence multiple steps during transformation.
Natural transformation is a widespread mechanism of horizontal gene transfer in bacteria, with at least 66 bacterial species identified as naturally competent (reviewed in reference 26). Competence is defined as the ability to take up exogenous DNA from the environment (10). Both chromosomal and plasmid DNA can be transformed, resulting in homologous recombination into the bacterial chromosome and plasmid reconstitution, respectively. DNA uptake systems may have evolved as a means to disseminate genetic information or as a mechanism to use DNA as a source of food. The ability to transport DNA into the cell is tightly regulated by quorum sensing, nutrient starvation, and/or growth phase in many naturally competent bacteria, and DNA uptake into bacterial cells for transformation requires specialized systems for competence which may not be expressed at all times (reviewed in reference 41). With the exception of Helicobacter pylori, which uses a distinct apparatus related to type IV secretion, all other currently identified DNA uptake systems resemble either type IV pili (TFP) or type II secretion systems (reviewed in reference 10). TFP are long, thin appendages about 50 to 80 Å wide, consisting of thousands of repeating subunits of pilin (12), and function in bacterial adherence, twitching motility, and transformation (43, 45). Twitching motility is mediated by the depolymerization of pilus oligomers to promote pilus retraction, which is directly correlated with DNA transformation (45).
Natural transformation is the main means of horizontal genetic exchange in the obligate human pathogen Neisseria gonorrhoeae and the related pathogen Neisseria meningitidis (30, 42). N. gonorrhoeae and N. meningitidis are constitutively competent and undergo high-frequency DNA transformation during all phases of growth (43). A lack of stable clonal lineages of N. gonorrhoeae indicates that exchange of chromosomal DNA during mixed infections is common (40), similar to H. pylori (32). Furthermore, intergeneric transfer of DNA has been identified between Haemophilus and Neisseria (31). Frequent transformation between lineages of N. gonorrhoeae may aid the spread of antibiotic resistance determinants. Although antibiotic therapy has historically been effective for treatment of gonorrhea, only cephalosporin drugs are presently recommended for treatment of gonorrhea in the United States (4).
N. gonorrhoeae and N. meningitidis preferentially take up and transform their own DNA by virtue of the DNA uptake sequence (DUS) (5′-GCCGTCTGAA-3′), which is found in abundance in Neisseria genomes (3, 17, 22). DUSs are often found as inverted repeats in transcriptional terminators, although efficient transformation requires only one DUS. Haemophilus influenzae has a similar constraint on DNA uptake, although the uptake sequence is distinct (14, 15). The Neisseria DUS was initially identified by the ability of cloned gonococcal genomic DNA containing the DUS to competitively inhibit transformation (22). Furthermore, the DUS was shown to positively increase DNA uptake and transformation, since adding the DUS to nontransformable plasmid DNA allowed transformation and protected the DNA from exogenous DNase I added to the medium (17). Genomic scanning revealed that a majority of DUSs in both the gonococcal and meningococcal genomes contain two additional semiconserved nucleotides at the −2 (A) and −1 (T) positions, and this extended 12-mer DUS (5′-ATGCCGTCTGAA-3′) allowed slightly higher transformation efficiencies than the 10-mer DUS (3). The molecular basis of DUS recognition leading to the selective uptake and transformation of DNA from Neisseria species or Haemophilus species is not known.
Natural transformation can be divided into three basic steps: (i) DNA binding, (ii) DNA uptake (comprising transport through the outer membrane, periplasm, and inner membrane), and (iii) DNA recombination into the chromosome. Many factors have been shown to be involved in these steps of transformation in N. gonorrhoeae; these factors can be divided between proteins involved in DNA transport and proteins involved in DNA recombination. Double-stranded DNA binds nonspecifically to the cell surface via an unknown mechanism, although Opa proteins and the minor pilus protein ComP can affect DNA binding to the cell surface and transformation efficiency (1, 2, 25). Although the molecular basis of DUS recognition is unknown, the DUS is thought to bind a surface-localized, sequence-specific, DNA-binding protein, and this interaction is required for efficient DNA uptake into the periplasm. DNA uptake requires all of the gene products required to produce functional TFP and mediate twitching motility, including the secretin PilQ, the major pilus subunit PilE, and the ATPase PilT. The proteins Tpc and ComL are localized to the periplasm and may aid DNA transport through the peptidoglycan layer (18, 19). One strand of the incoming DNA is degraded, while the remaining strand is delivered to the cytoplasm through the inner membrane protein ComA (8). Finally, DNA recombination into the chromosome is mediated by RecA (29) and requires the helicase PriA (28). RecBCD recombination appears to play a partial role in transformation, since null mutants show intermediate levels of transformation (36).
Here we characterized the effect of the DUS on transformation in several different N. gonorrhoeae strains. Different DNA substrates carrying point mutations or an insertion all transformed very efficiently when carrying a DUS, but matched DNA substrates without a DUS (DUS0) transformed less efficiently. Although all tested strains transformed DUS0 substrates, the efficiency of DUS0 transformation varied between strains. Contrary to previous reports, DUS containing DNA failed to competitively inhibit transformation in all strains and surprisingly enhanced transformation in one strain. These observations prompted us to reexamine the effect of the DUS on DNA uptake. We found that DUS0-dependent DNA uptake did not fully correlate with differences in transformation efficiencies. Taken together, these data show strain variability in the requirement for the DUS, are inconsistent with the accepted DUS receptor hypothesis in all strains, and may indicate a dual role for the DUS during transformation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Transformation and DNA uptake were investigated in the laboratory strains FA1090 (11), FA19 (34), MS11 (37), and 1291 (5). Strain FA1090 1-81-S2nv (38), which contains a kanamycin (Kan) resistance cassette upstream of the pilE locus that completely abrogates antigenic variation, was used to investigate the effects of pilin antigenic variation on transformation. We constructed MS11 1-81-S2nv by transforming chromosomal DNA from the FA1090 1-81-S2nv strain into MS11, selecting for Kanr and identifying transformants that had crossed in the 1-81-S2 pilE sequence by DNA sequencing. This yielded an MS11 strain which cannot undergo antigenic variation and has a pilE gene identical to that of FA1090 1-81-S2nv. All N. gonorrhoeae strains were grown on GC medium base (GCB) (Difco) plates with Kellogg's supplements I and II (27) at 37°C in a humidified 5% CO2 atmosphere. The concentrations of antibiotics in GCB were as follows: Kan, 50 μg/ml; kasugamycin (Ksg), 150 μg/ml; chloramphenicol (Cam), 0.75 μg/ml for strains FA1090 and FA19, 10 μg/ml for strain MS11, and 3 μg/ml for strain 1291; and nalidixic acid (Nal), 1 μg/ml for strains FA1090 and FA19 and 3 μg/ml for strains MS11 and 1291.
Construction of ksgA1 DUS10, DUS12, and DUS0 plasmids.
PCR was used to amplify a mutant ksgA gene (G305A, referred to as ksgA1) which confers Ksg resistance in N. gonorrhoeae (16). Taq DNA polymerase (Promega) was used as per the manufacturer's instructions with the primers ksgtopDUS0 and ksgbot3 for the DUS0 plasmid, primers ksgtop1DUS12 and ksgbot3 for the DUS12 plasmid, and primers ksgDUS10top2 and ksgbot3 for the DUS10 plasmid (Table 1). Following initial denaturing at 94°C for 2 min, PCR amplification consisted of 24 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min. The resulting PCR products were blunted by adding 10 mM deoxynucleoside triphosphates (dNTPs) and 0.625 U of Pfu DNA polymerase (Stratagene) and incubating at 72°C for 30 min. The blunted products were cloned into the pCR-Blunt vector (Invitrogen) as per the manufacturer's instructions and transformed into TOP10 Escherichia coli cells (Invitrogen). Positive clones were verified by DNA sequencing (SeqWright) using the primers M13 forward and M13 reverse.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′-3′)a |
|---|---|
| ksgDUS12top1 | TCGTATGCCGTCTGAAAACG |
| ksgDUS0top2 | CTGCCGTTTGCGGATAAC |
| ksgDUS10top2 | GCCGTCTGAAAACGCTGC |
| ksgbot3 | CCAGATAATTGCTCAACGCC |
| gyrB2top | GCCATCGACGAAGCACTC |
| gyrB6bot | GCGGCCGTCTGAAACGATT |
| gyrB12bot | GGCTTTTTCCAAGGCAAGG |
| gytB6botDUS12 | ATGCCGTCTGAAACGATTTTGG |
| comAfor | TTACGGCGTATTCAGAACGGAAG |
| comArev | AGAATTTATACACTCTCAAACGTTGA |
DUS10 sequences are underlined. The additional two nucleotides in the DUS12 are in bold.
Identification of Nalr mutation in gyrB.
The plasmid pSY6 contains a mutant gyrB gene and has been used to measure transformation efficiency in Neisseria species, but the mutation conferring Nalr has not been identified (23, 44). We sequenced the mutant gyrB gene cloned in the plasmid pSY6 and identified three mutations, D429N, R652Q, and A750T. Fragments carrying each mutation were cloned individually into pCR-Blunt (Invitrogen) and transformed into N. gonorrhoeae. Only the clone containing the D429N mutation yielded Nalr; we have designated this mutation gyrB1.
Construction of gyrB1 DUS10, DUS12, and DUS0 plasmids.
PCR was used to amplify the mutant gyrB1 gene (see above), which confers nalidixic acid resistance in N. gonorrhoeae. KOD Hot Start DNA polymerase was used per the manufacturer's instructions with the plasmid pSY6 as the template and primers gyrB2top and gyrB12bot for the DUS0 plasmid, primers gyrB2top and gyrB6botDUS12 for the DUS12 plasmid, and primers gyrB2top and gyrB6bot for the DUS10 plasmid. The resulting PCR products were cloned into the pCR-Blunt vector (Invitrogen) as per the manufacturer's instructions and transformed into TOP10 E. coli cells (Invitrogen). Positive clones were verified by DNA sequencing (SeqWright) using the primers M13 forward and M13 reverse.
Construction of mutants.
The comA::kan mutant was created by PCR amplifying the predicted comA gene from strain FA1090 with the primers comAfor and comArev and cloning the product into the pCR2.1-TOPO plasmid (Invitrogen). The resulting plasmid DNA was isolated, and in vitro transposition with the Ez:Tn Kan-2 transposon (Epicentre) was performed according the manufacturer's instructions to identify a comA::kan mutant plasmid construct with the EZ-Tn Kan transposon disrupting comA. The construct was transformed into FA1090, and PCR was used to verify the insertion of the mutation comA::kan. The pilQ mutant contains a chloramphenicol resistance cassette inserted in the pilQ locus, and the pilT mutant contains an erythromycin resistance cassette inserted in the pilT locus; both have been previously described (33). Both the pilQ::cat and pilT::erm mutants were crossed into wild-type (WT) FA1090, and the pilQ and pilT loci were checked for the presence of cat and erm, respectively. The FA1090 and MS11 recB and recD mutants contain an erythromycin resistance cassette inserted into each locus, and both have been previously described (24). The FA1090 (RM11.2) ΔpilE strain was generated by selecting nonpiliated colonies and has been previously described (9).
Transformation and competition of transformation assays.
Strains were grown for 18 h on GCB plates and resuspended in liquid transformation medium (1.5% protease peptone no. 3 (Difco), 0.1% NaCl, 200 mM HEPES (Sigma), 5 mM MgSO4, and Kellogg supplements I and II, pH 7.2) to an optical density (OD) at 600 nm of approximately 1.5. Thirty μl of the cell suspension was added to tubes containing 150 ng transforming DNA and 200 μl transformation medium. For competition of transformation experiments, 100 ng of transforming DNA (gyrB1-pBlunt) (see Fig. 1) was mixed with no or a 10-, 100-, or 1,000-fold molar excess of pHSS6 or pUP1 DNA. DUS10/12- and DUS0-containing plasmids of ksgA1, gyrB1, and iga::cat (pJKD1314 and pJKD1502 [20]) (see Fig. 1) were used as transforming DNA. Following incubation at 37°C for 20 min, transformation mixtures were added to prewarmed 2 ml transformation medium and incubated at 37°C in the presence of 5% CO2 for 4 h. For competition experiments and where indicated, transformations were stopped after 15 min by the addition of 5 μg of DNase I (Worthington) before being added to the prewarmed transformation medium. The mixtures were serially diluted 10-fold in transformation medium lacking MgSO4 and Kellogg supplements, and 20 μl serial 10-fold dilutions were spotted on GCB plates in the presence and absence of the appropriate antibiotic. Transformation efficiencies are reported as antibiotic-resistant CFU divided by total CFU and are the means of data for at least three replicates.
FIG. 1.
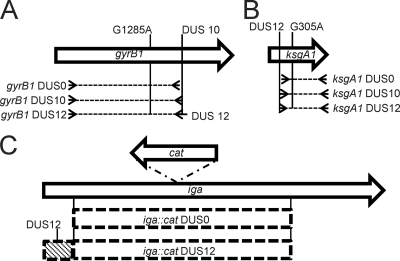
Schematic scale cartoon of transforming constructs. (A) The gyrB1 allele contains a G1285A mutation which confers resistance to Nal and contains a DUS10. Three constructs were created containing gyrB1 (dotted lines); all were cloned with the same 5′ primer but differing 3′ primers. One construct lacks a DUS (3′ primer anneals immediately upstream of DUS), one construct contains the DUS10 (3′ primer anneals to DUS10), and one construct contains the DUS12 (3′ primer anneals to DUS10 and contains “AT” nucleotides on 5′end). (B) The ksgA1 allele contains a G305A mutation which confers resistance to Ksg and contains a DUS12. Three constructs were created containing ksgA1 (dotted lines); all were cloned with the same 3′ primer but differing 5′ primers. One construct lacks a DUS (5′ primer anneals immediately downstream of DUS12), one construct contains the DUS10 (5′ primer anneals to the DUS10), and one construct contains the DUS12 (5′ primer anneals to the DUS12). (C) Schematic cartoon of iga::cat DUS0 and DUS12 constructs (dashed boxes). The DUS0 iga::cat construct contains the indicated region of the iga locus with the cat insertion. The iga::cat DUS12 construct contains an additional 382-bp region of N. gonorrhoeae DNA which contains the DUS12 (diagonal line-filled box).
Radiolabeling of DNA substrates.
The plasmids pHSS6 (DUS0) and pUP1 (DUS12) were radiolabeled with [α-32P]dATP/dCTP, and the DUS10 and DUS0 gyrB1-pBlunt plasmids were labeled with [α-33P]dATP/dCTP to take advantage of the longer half-life of 33P. The plasmids pHSS6 and pUP1 were linearized with NheI prior to labeling. To label only the gyrB1 region in gyrB1-pBlunt, the DUS10 and DUS0 gyrB1-pBlunt plasmids were doubly digested with KpnI and SpeI, leaving an ExoIII-sensitive 3′ end and an ExoIII-resistant 5′ end. Linearized plasmids were purified using Qiaquick columns (Qiagen), and 2 μg DNA was treated with 1 U ExoIII for 10 min and subsequently heat inactivated at 70°C for 20 min. This time was empirically determined to be the longest digestion that allowed quantitative recovering of the double-stranded form after fill-in (data not shown). A fill-in reaction with 10 U of the Klenow fragment of polymerase I was performed in the presence of 1 μM α-32P-or α-33P-labeled dATP/dCTP (3,000 Ci mmol−1; Perkin Elmer) and 25 μM dGTP/dTTP. After incubation for 90 min at 37°C, 25 μM unlabeled dATP/dCTP was added, followed by incubation for another 15 min at 37°C. Klenow was heat inactivated by incubation at 70°C for 20 min. Labeled DNA was purified using Qiaquick columns, and the specific activity was measured by scintillation counting.
DNA binding, uptake, and competition of uptake assays.
Strains were grown for 18 to 20 h on GCB plates and resuspended in 3 ml transformation medium minus Kellogg's supplements to an OD at 600 nm of approximately 1.5. One ml of the cell suspension was added to approximately 200 ng 32P- or 33P-labeled DNA (specific activity of approximately 5 × 106 cpm) and rotated end over end on a rotisserie at 37°C for 30 min. The samples were split into 450-μl fractions, with one fraction receiving 5 μg DNase I (Worthington) to measure DNase I-resistant (DNaser) uptake while the other fraction received no DNase I to measure total cell-associated DNA. Both fractions were incubated at 37°C for 10 min and then placed on ice for 10 min to stop the action of the DNase I. The cells were pelleted at 15,000 × g at 4°C for 10 min and washed three times in 1 ml ice-cold wash buffer (1.5% proteose peptone no. 3, 0.1% NaCl, 200 mM HEPES, pH 7.2). The samples were resuspended in 300 μl Tris-EDTA (TE), and 100 μl was added to 4 ml Ultimate Gold scintillation cocktail (Perkin Elmer) and counted in a liquid scintillation counter (Beckman-Coulter). DNA uptake and cell-associated DNA are reported as the percentages of DNA added and are the means of results for 3 to 6 replicates. Competition of DNA uptake was performed essentially as in DNA binding and uptake assays described above, with 7 plates of cells resuspended into 22 ml transformation medium minus Kellogg's supplements. One ml of the cell suspension was added to 100 ng 33P-labeled gyrB1-pBlunt, along with zero or a 10-, 100-, or 1,000-fold molar excess of either pHSS6 or pUP1. Results of competition of DNA uptake experiments are reported as percentages of DNA added and are the means of results for at least three replicates.
RESULTS
The magnitude of DUS enhancement of transformation is strain dependent.
Natural transformation in several strains of N. gonorrhoeae, including MS11, F62, and FA19, has been extensively studied (1, 3, 17, 22, 43). It has been established that N. gonorrhoeae preferentially transforms its own DNA by the ability of the Neisseria-specific nonpalindromic 10- or 12-nucleotide sequence 5′-ATGCCGTCTGAA-3′ (DUS10 or -12) to mediate DNA uptake leading to transformation. It was also found that transforming DNA must carry at least one DUS10 for full transformation efficiency and that the presence of the DUS10 protects labeled DNA from degradation by exogenous DNase.
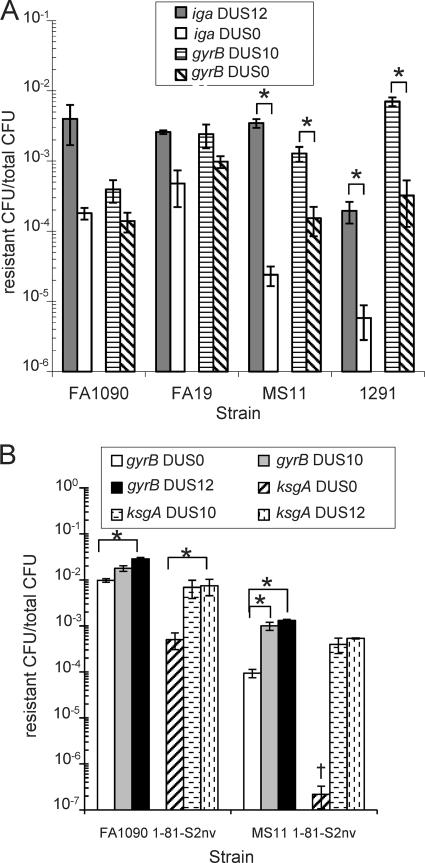
During investigations into the basis of kasugamycin resistance in strain FA1090, we observed substantial levels of transformation in cloned fragments devoid of a DUS (16). Quantitative transformation of strains FA1090, FA19, MS11, and 1291 with matched DNA substrates with or without a DUS10 or DUS12 (Fig. 1) containing two different antibiotic resistance determinants demonstrated that all the strains provided some level of transformation for DUS0 constructs but that the efficiencies for DUS0 transformation were greater for FA1090 and FA19 than those recorded for MS11 or 1291 (Fig. 2 A). The two genes yielded differing absolute transformation efficiencies within each strain, which could be a function of the nature of the resistance mechanisms, differing lengths of homologous DNA, or the differences in degenerate or DUS-like sequences. All strains transformed the DUS10/12 DNA more efficiently than the cognate DUS0 DNA, but the magnitude of the difference between the DUS0 and DUS10/12 transformation efficiencies was dependent on the N. gonorrhoeae strain used (Fig. 2A). A larger difference in transformation efficiency was seen between DUS0 and DUS12 DNA with the iga::cat insertion versus that with the gyrB1 point mutation in all strains. Strains FA1090 and FA19 exhibited similar DUS0 and DUS10/12 transformation phenotypes, with smaller differences between DUS0 and DUS10/12 transformation efficiencies that were not statistically significant. Strains MS11 and 1291 showed similar transformation frequencies, with DUS0 DNA transforming much less efficiently than DUS10/12 DNA (P < 0.05). Strikingly, the fold difference between the DUS0 and DUS12 transformation efficiencies with the iga::cat insertion was about 22-fold for FA1090 but 144-fold for MS11 (Fig. 2A). The relatively high efficiency of the DUS0 DNA transformation in FA1090 and FA19 (10−3 to 10−4 resistance CFU/total CFU) was surprising, since a previous report indicated that the DUS0 DNA transformation occurs at very low efficiency in MS11 (10−7 resistant CFU/total CFU) (3) and inefficient DUS-independent transformation may be pilus independent (7). One possibility was that this efficient DUS0 transformation was utilizing a novel pathway for efficient entry into cells. However, transformation of chromosomal DNA and both DUS10 and DUS0 cloned DNA into strain FA1090 required functional pilT, pilE, pilQ, and comA genes (data not shown), indicating that DUS0 transformation also requires functional components of the TFP. To determine whether an alternative uptake sequence might be responsible for the efficient DUS0 transformation, bioinformatic analysis of the different transforming DNA constructs was performed but failed to reveal any short (≤15 nucleotides) shared repeat sequences in the three fragments (data not shown) (Fig. 1). The two DUS phenotypes (large and small difference between DUS10/12 and DUS0 transformation efficiencies) do not correlate with the presence of the type IV secretion system, since strains FA1090 and 1291 lack the secretion system and strains FA19 and MS11 contain the system. Additionally, the two DUS phenotypes are unlikely to be explained by differences in the minor pilus proteins ComP and PilV, which have been shown to affect transformation efficiency, since ComP is 100% conserved and PilV is 93% identical and 100% similar in all four strains, or differences in the DNA processing chain protein DprA/Smf, which is 98.5% identical and 100% similar in all four strains (data not shown).
FIG. 2.
The magnitude of DUS enhancement of transformation is strain dependent (A), and DUS phenotypes are not due to pilin sequence or pilin antigenic variation (B). (A) Strains FA1090, FA19, MS11, and 1291 were quantitatively transformed with iga::cat DUS0 and DUS12 and gyrB1 DUS10 and DUS0 plasmid DNA. Transformation efficiencies are plotted as resistant CFU/total CFU. Gray bars indicate iga::cat DUS12, white bars indicate DUS0 iga::cat, horizontal dashed lines indicate gyrB1 DUS10, and diagonal lines indicate gyrB1 DUS0 DNA. Error bars are SEM. *, P < 0.05 by Student's t test. (B) Strains FA1090 1-81-S2nv and MS11 1-81-S2, which cannot undergo pilin antigenic variation and are isogenic for pilE, were quantitatively transformed with gyrB1 and ksgA1 DUS0, DUS10, and DUS12 plasmid DNA. Transformation efficiencies are plotted as resistant CFU/total CFU. White bars indicate gyrB1 DUS0, gray bars indicate gyrB1 DUS10, black bars indicate gyrB1 DUS12, diagonal stripes indicate ksgA1 DUS0, horizontal dashed lines indicate ksgA1 DUS10, and vertical dashed lines indicate ksgA1 DUS12 plasmid DNA. Error bars are SEM. †, 2 repeats below limit of detection. *, P < 0.05 by Student's t test.
It was possible that the strain differences in DUS0 transformation were mediated by the recombination systems. As expected, both DUS10 and DUS0 transformations were absolutely recA dependent in all strains (data not shown). A role for the RecBCD nuclease in the DUS0 transformation differences was tested by making FA1090 and MS11 recB, recC, and recD mutants. These strains all showed identical reductions in transformation efficiencies with both DUS0 and DUS10 DNA (data not shown), as has been previously reported for DUS10 transformation (36). These studies genetically show that both DUS10 and DUS0 transformation utilize the same transport and recombination machinery.
DUS phenotypes are not due to variant pilins.
Pilin, the major component of TFP, undergoes high-frequency pilin antigenic variation, resulting in distinct pilin proteins (13). Since different pilin variants have been shown to affect DNA uptake and transformation efficiencies (2), we tested whether the strain-dependent DUS phenotypes might be explained by differences in pilin sequence. In order to test if different pilin sequences and antigenic variation were responsible for the DUS phenotypes, we utilized a transposon insertion located upstream of the pilE gene of FA1090 pilin variant 1-81-S2 (39), which completely abrogates pilin antigenic variation (38), to introduce the 1-81-S2 pilE sequence into MS11. The MS11 1-81-S2nv strain transformed the DUS0 gyrB1 construct with an efficiency similar to that of the parental strain, expressing the original pilin variant (Fig. 2B). As before, the difference in transformation efficiency between DUS0 and DUS10 DNA was not statistically different in FA1090 but was statistically different in MS11 (Fig. 2B, white and gray bars) (P < 0.05), suggesting that neither pilin antigenic variation nor the sequence of pilE affects the DUS0-mediated enhancement of transformation. Interestingly, FA1090 1-81-S2nv transformation efficiencies for both the DUS0 and DUS10 gyrB1 plasmids were statistically increased over WT FA1090 levels (P < 0.05) (Fig. 2B and 2A, respectively), whereas MS11 1-81-S2nv transformation efficiencies were similar to WT MS11 levels (Fig. 2B and A, respectively). These data show that although the pilin sequence affects transformation efficiencies, differences in pilin sequence do not account for the strain-dependent DUS transformation phenotypes.
DUS12 transformation enhancement is modestly increased over that of DUS10.
A previous study demonstrated that 77% of the DUS sequences in the genome of FA1090 contain two semiconserved nucleotides and that this extended 12-mer DUS enhanced transformation efficiencies compared to results with the DUS 10-mer (3). To determine how the DUS0-, -10-, and -12-mer affect transformation efficiencies in FA1090 and MS11, an identical gyrB1 construct carrying a DUS12 sequence was constructed (Fig. 1) and the frequencies of transformation of FA1090 1-81-S2nv and MS11 1-81-S2nv were measured (Fig. 2B). In both strains, the DUS12 gyrB1 plasmids consistently transformed at higher efficiencies than the DUS0 plasmids (P < 0.05) but not statistically differently from the DUS10 plasmids. These results confirm previous findings that the DUS12 modestly enhances transformation over that with the DUS10 (3). We confirmed these results with the mutant ksgA1 gene, carrying both DUS10 and DUS12 sequences (Fig. 1 and 2B). As before, DUS0 transformation of FA1090 was relatively high (10−4 resistance CFU/total CFU) and was not statistically different from the efficiency of the DUS10 plasmid (Fig. 2B), although the DUS12-mer transformation of FA1090 ksgA1 was statistically higher than that of the DUS0 plasmid (P < 0.05). As before, MS11 DUS0 transformation was relatively inefficient (below 10−6 resistant CFU/total CFU) and was below the limit of detection in two repeats (Fig. 2B). This result was in contrast to the transformation efficiency of either the DUS10 or DUS12 ksgA1 plasmid, which mediated high levels of transformation in MS11. Although statistical analysis was unavailable since levels of ksgA1 DUS0 transformation were below the limit of detection, the DUS10 and DUS12 plasmids exhibited at least 1,800-fold and 2,400-fold increases in transformation enhancement from DUS0 levels, respectively, in MS11 (Fig. 2B).
The DUS competitively inhibits transformation of FA1090 but not MS11.
The DUS was initially identified as a sequence which could competitively inhibit transformation in strain FA19 when added in excess, while DNA lacking a DUS had no effect on transformation (22). These results were interpreted as suggesting that the DUS functions by binding to a cell surface receptor. Elkins et al. established that DNA must contain a DUS to be transported into cells using two nonreplicating plasmids, pHSS6, which lacks a DUS, and pUP1, in which an inverted repeat of the DUS12 was cloned into pHSS6 (17).
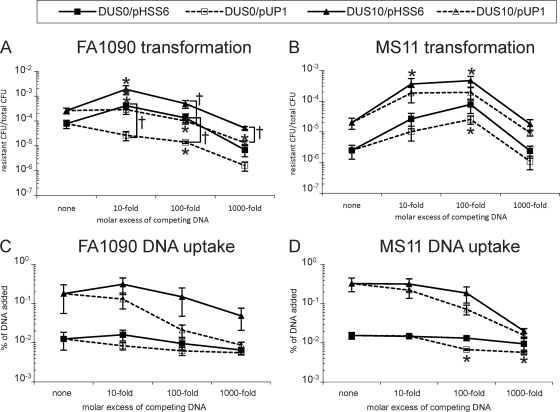
To determine whether a DUS carrying DNA would inhibit transformation similarly in FA1090 and MS11, pHSS6 and pUP1 were mixed with DUS10 and DUS0 transforming DNA at 0-, 10-, 100-, and 1,000-fold molar excess (Fig. 3 A and B). In strain FA1090, pHSS6 had a positive effect on transformation of both DUS0 and DUS10, transforming DNA at a 10-fold molar excess (5-fold and 7-fold, respectively; P < 0.05 compared to results with no competitor), but had no effect at a 100-fold molar excess (Fig. 3A, solid lines). In contrast, excess pUP1 DNA inhibited FA1090 transformation, and this inhibition was dose dependent, with approximately a 50-fold reduction with DUS0 transforming DNA (P < 0.05 compared to results with no competitor) and a 20-fold reduction with DUS10 transforming DNA at a 1,000-fold excess (Fig. 3A). At some levels of competitor (Fig. 3), the effects of pHSS6 were statistically distinct from the effects of pUP1 (†, P < 0.05) in FA1090, suggesting that FA1090 can recognize the DUS carried on competitor DNA.
FIG. 3.
Competition of DUS0 and DUS10 gyrB1 transformation (A and B) or DNA uptake (C and D) by pHSS6 and pUP1. FA1090 (A) or MS11 (B) was transformed with gyrB1 DUS10 (triangles) or DUS0 (squares) in the presence of zero or a 10-, 100-, or 1,000-fold molar excess of pHSS6 (solid lines and shapes) or pUP1 (dashed lines and open shapes). Transformation efficiency is plotted as resistant CFU/total CFU. DNA uptake of 33P-labeled gyrB1 DUS10 (triangles) or DUS0 (squares) was measured in FA1090 (C) or MS11 (D) in the presence of zero or 10-, 100-, or 1,000-fold molar excess of pHSS6 (solid lines and shapes) or pUP1 (dashed lines and open shapes). DNA uptake is plotted as the percentage of DNA added. Error bars are SEM. *, P < 0.05 compared to results with no competitor by Student's t test. †, P < 0.05 by Student's t test.
The results with competitors in MS11 were very different from those in FA1090. Addition of pHSS6 or pUP1 strongly stimulated both DUS0 and DUS10 transformation of MS11 at 10- and 100-fold excess (Fig. 3B). While both pHSS6 and pUP1 enhanced transformation in MS11, pHSS6 exhibited a slightly greater effect (23-fold and 31-fold for DUS0 and DUS10 transforming DNA, respectively) than pUP1 (10-fold for both DUS0 and DUS10 transforming DNA). The effect of pHSS6 compared to that of pUP1 was consistently different but not statistically different in MS11 at any level of competitor (Fig. 3B). Interestingly, while both the pHSS6 and pUP1 plasmids enhanced transformation when added at 10- and 100-fold excess, addition of these plasmids at a 1,000-fold excess returned the transformation levels to within 2-fold of no-competitor levels (Fig. 3B). These results were surprising given previous reports of inhibition of transformation by DUS containing DNA (22).
Strain differences in transformation efficiencies are not due solely to DNA uptake.
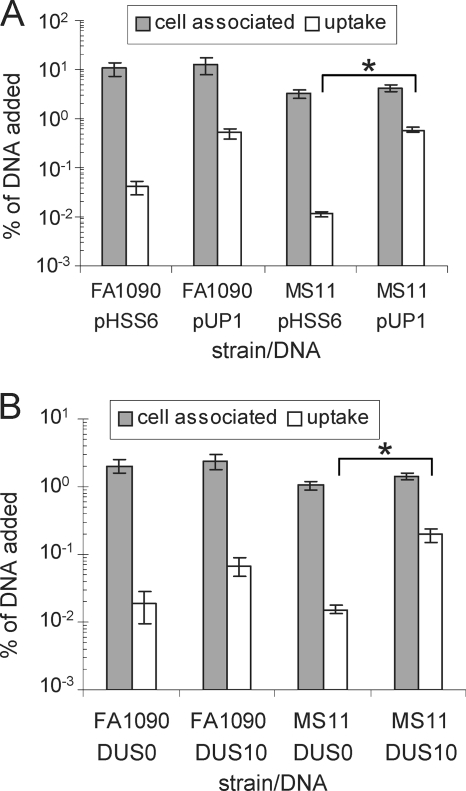
Previous studies have shown that DNA must contain a DUS to be protected from exogenous DNase in the medium when incubated with strains MS11 (1) and FA19 (17). These studies utilized radiolabeled pHSS6 and pUP1 to measure DNA binding and uptake. We revisited these studies to determine whether the increased DUS0 transformation seen in FA1090 relative to that in MS11 would correlate with an increase in DUS0 uptake. Radiolabeled pHSS6 and pUP1 were used to measure both the total cell-associated and DNaser transported DNA in both FA1090 and MS11 (Fig. 4 A). These data confirm previous reports that the DUS is required for nonhomologous DNA to be transported into a DNase-resistant state in MS11, since less than 0.01% of DUS0 DNA was resistant to exogenous DNase (Fig. 4A). Although strain FA1090 bound more DNA and transported 3-fold more DUS0 DNA than MS11, the amounts of DUS12 DNA transported were identical between the strains (Fig. 4A). The difference between transported DUS0 and DUS12 DNA was less in FA1090 than in MS11, confirming a less-stringent requirement for the DUS in FA1090. The small differences in uptake of DUS0 DNA between FA1090 and MS11 do not correlate with the larger difference in transformation. However, since pHSS6 and pUP1 are nonreplicating in N. gonorrhoeae and cannot be recombined into the chromosome since they lack any homology to Neisseria DNA, uptake of these plasmids may not accurately represent the uptake of bona fide transforming DNA.
FIG. 4.
DNA uptake of DUS10/12 and DUS0 DNA correlates with transformation efficiencies in MS11 but not FA1090. (A) Binding and uptake of 32P-labeled pHSS6 (DUS0) and pUP1 (DUS12) DNA in strains FA1090 and MS11. Cell-associated (bound plus taken up) DNA is shown in gray, and DNaser DNA is shown in white; data are graphed as a percentage of DNA added. Error bars are SEM. *, P < 0.05 by Student's t test. (B) Binding and uptake of 33P-labeled gyrB1 DUS0 and DUS10 DNA in strains FA1090 and MS11. Total cellular DNA is shown in gray, and DNaser DNA is shown in white; data are graphed as a percentage of DNA added. Error bars are SEM. *, P < 0.05 by Student's t test.
To directly measure the uptake of homologous DNA, gyrB1 DUS10 and gyrB1 DUS0 plasmids were selectively radiolabeled in the N. gonorrhoeae DNA and used in uptake assays (Fig. 4B). The relative magnitudes of binding and uptake of homologous gyrB1 DNA (Fig. 4B) were similar to the binding and uptake of nonhomologous DNA (Fig. 4A) in both FA1090 and MS11, with about a 3-fold difference between DUS0 and DUS10 uptake for FA1090 and about a 10-fold difference for MS11. Since the iga::cat insertion showed a much larger difference in DUS0 and DUS12 transformation than the gyrB1 point mutation (Fig. 2A), we repeated the uptake assay with radiolabeled iga::cat-DUS12 and iga::cat-DUS0 plasmids to test whether uptake matched the transformation (data not shown). Both the relative and absolute magnitudes of DNA binding and uptake of iga::cat plasmids were essentially the same as those seen with the gyrB1 plasmids in both strains, indicating that the larger difference in transformation of DUS0 compared to that of DUS12 is not due to differences in DNA uptake.
The DUS competitively inhibits uptake of DUS10 DNA.
The unexpected enhancement of transformation efficiency in FA1090 and MS11 at a 10-fold excess of pHSS6 and at a 10- and 100-fold excess of pHSS6 and pUP1 in MS11 prompted us to determine how these competitors affected DNA uptake. Therefore, DNA uptake experiments were performed with radiolabeled gyrB1 DUS0/10 DNA in the presence of unlabeled pHSS6 or pUP1 at a 0-, 10-, 100-, or 1,000-fold molar excess (Fig. 3C and D). Similar to the competition of transformation results, pUP1 competitively inhibited uptake of DUS10 DNA in FA1090 and was dose dependent, with a 20-fold inhibition at a 1,000-fold excess (Fig. 3C, open triangles). In contrast, the DUS0 competitor pHSS6 had little effect on DUS10 DNA uptake, inhibiting only 3-fold at a 1,000-fold excess. As before, very little DUS0 DNA was transported into cells, and neither pHSS6 nor pUP1 had any effect on DUS0 DNA transport in FA1090. In contrast to the enhancing effect of competitor DNA in transformation competition experiments in MS11, no stimulation of DNA uptake was observed with competing pHSS6 or pUP1 in MS11 (Fig. 3D). Interestingly, both pHSS6 and pUP1 inhibited DUS10 DNA uptake and were dose dependent, with 16-fold and 21-fold inhibition at a 1,000-fold excess, respectively (Fig. 3D). Very little DUS0 DNA was transported in MS11; however, 100- and 1,000-fold excesses of pUP1 inhibited this transport 2-fold and 3-fold, respectively (P < 0.05).
DISCUSSION
The accepted models for the action of the DUS in DNA transformation invoke DUS recognition at the bacterial surface by a sequence-specific DNA binding protein to allow efficient DNA transport into the periplasm by the pilus apparatus. Here we show that the magnitude of the effect of the DUS on transformation efficiency and the ability of excess DUS to inhibit transformation are both strain dependent and that this difference is not due solely to differential DNA uptake. Taken together, these data suggest that action on the DUS cannot be explained solely by surface binding.
In all the strains, DUS10/12 transformation was more efficient than DUS0 transformation of the same DNA, and while the absolute level of DUS10/12 transformation was different for each transforming construct, the frequencies were all within approximately 36-fold of one another (Fig. 2A and B). It is not surprising that the three transformation substrates utilized have different DUS10/12 transformation efficiencies, since they have different total sizes, the position of the mutation in each fragment is different, and two carry point mutations that provide antibiotic resistance while the other carries an entire antibiotic resistance gene. However, there was a larger difference measured between DUS0 and DUS12 DNA with the cat insertion and the transforming DNA carrying point mutations (Fig. 2A). It is possible that the additional difficulty in recombining insertions into the chromosome creates additional problems for DUS0 DNA.
The magnitude of DUS enhancement of transformation was much greater than the magnitude of DNA uptake differences between the strains (Fig. 4A and B). Also, similar levels of radiolabeled, gyrB1 or iga::cat DUS10 or DUS0 DNA were transported into the strains (Fig. 4B and data not shown) even though their transformation efficiencies were quite distinct (Fig. 2A). The transformation assays reported allow for multiple rounds of transformation, which may have accentuated differences in transformation efficiencies between the strains. Up to 1,000-fold differences in DNA uptake and transformation in different H. influenzae strains have been reported (35), although the influence of the uptake signal sequence (USS) on these differences was not examined. Similar to our results, the differences seen in H. influenzae transformation were not completely explained by the amount of DNA transported into cells (35). There are two major hypotheses that explain these observations. Either the DUS and USS function only to allow efficient transport of the DNA and measuring bulk transport through the outer membrane does not measure the transported DNA that is able to be used for transformation, or alternatively, the DUS and USS function during transport through the outer membrane but can also influence steps subsequent to transport.
Consistent with the idea that the DUS has more-complicated roles than binding a surface receptor are the differences observed when competing transformation with excess competitor DNA in FA1090 and MS11 was compared. The dose-dependent enhancement of MS11 transformation by pHSS6 and pUP1 at 10- and 100-fold excess (Fig. 3B) demonstrates that addition of excess DNA to this strain significantly boosts transformation efficiency. This is unlikely to be a transcriptional response, since the competitors were added at the same time as the transforming DNA and transformation was limited to 15 min. It is unclear why transformation efficiency with a 1,000-fold excess of competitor returns to no-competitor levels in MS11, but it is possible that at this level of excess competitor, the stimulation of transformation is competed back to noncompetitor levels (Fig. 3B). If this explanation is correct, then there must be a high-affinity interaction that stimulates transformation at lower levels of competitor and a low-affinity interaction which is inhibitory. The enhancement of transformation efficiency was more robust for pHSS6 versus that for pUP1 in MS11, indicating that a DUS on the competitor DNA decreases the stimulatory activity. FA1090 also showed a small enhancement of transformation with the DUS0 competitor at a 10-fold molar excess but was mostly inhibited by both DUS12 and DUS0 competitors at higher levels of competitor. We do not know the mechanism for the enhancement of transformation, but these results confirm that these strains react differently during transformation and show differential recognition of the DUS.
Direct measurement of nonhomologous radiolabeled-DNA uptake into cells recapitulated previous reports of work using MS11 showing that small amounts of DUS0 DNA enter a DNase-protected state (Fig. 4A). The modest increase in transported nonhomologous DUS0 DNA in FA1090 compared to that in MS11 does not fully explain the differences seen in transformation efficiencies (Fig. 4A). To directly measure DNA uptake of transforming substrates, we also utilized homologous radiolabeled DNA (Fig. 4B). The relative magnitudes of DNA binding and DNA uptake for homologous DNA were similar to those for nonhomologous DNA; again the modest increase in transported DUS0 DNA in FA1090 versus that in MS11 does not correlate with the differences seen in transformation efficiencies. The mechanism for increased DUS0 transformation in FA1090 is likely at a step after uptake, since MS11 and FA1090 transported similar levels of DUS0 DNA (Fig. 4A and B).
The fact that DUS10 DNA uptake was competitively inhibited by pUP1 in both FA1090 and MS11 (Fig. 3C and D) suggests that the DUS action during DNA uptake is similar between the strains, a fact supported by the relatively similar levels of uptake observed with no competitors (Fig. 4A and B). In both competition of uptake and transformation assays, pHSS6 and pUP1 differentially affected these processes in FA1090 but not MS11, while in transformation assays, MS11 discerned DUS0 DNA from DUS10/12 DNA. The competition experiments do support a role for the DUS in DNA uptake (DNA uptake is competitively inhibited by pUP1); however, the enhancement of MS11 transformation by the addition of competitor DNA supports an additional activity for the DUS in a process important for transformation that is distinct from DNA uptake.
Our data confirm the findings of previous studies that show that the DUS enhances the ability of heterologous or homologous transforming DNA to become resistant to exogenously added DNase I, but they lend doubt to the hypothesis that DUS activity is due solely to binding to a surface exposed DNA binding protein. There are several models that are consistent with the action of the DUS during transformation. The large differences in the transformation efficiencies of DUS0 DNA between the strains suggest that FA1090 and FA19 recognize DUS0 DNA more efficiently than MS11 or 1291 or that MS11 and 1291 possess mechanisms for limiting the transformation of DUS0 DNA that are less protective in FA1090 and FA19. It is possible that the DUS functions to protect DNA from a periplasmic or cytoplasmic nuclease that is more active in MS11 than in FA1090. We ruled out the RecBCD exonuclease as being involved, since both DUS0 and DUS10 DNA transformation was reduced in recBCD null mutants (data not shown). Nucleases have been shown to be involved during DNA uptake in several bacteria (6, 21), and Chausse and Hill have demonstrated that single-stranded DNA is formed in the periplasm during transformation of strain MS11 and P9; however, they did not identify the responsible nuclease or test the DUS dependence for the single-stranded DNA formation (8). The enhancement of transformation seen in MS11 when either DUS0 or DUS12 DNA is added to the transformation reactions may be consistent with a putative nuclease in MS11, since the extra DNA could sop up or overwhelm the nuclease, resulting in more transforming DNA being available for recombination. Clearly, the lack of an identified mechanism for DUS recognition in Neisseria and the related USS recognition in Haemophilus during transformation makes interpreting the complex DUS DNA uptake and transformation phenotypes difficult.
Acknowledgments
We are grateful to Adrienne Chen and Paul Schook for critical reading and editing of the manuscript. The comA::kan mutant was constructed by Melissa Rohrer. We thank Pamela Shaw and Michael Grobe for their help with bioinformatics analysis and John Davies for the plasmids pJKD1314 and pJKD1502.
This work was supported by National Institutes of Health grants R01 AI055977, R01 AI044239, and R37 AI033493 to H.S.S.
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Aas, F. E., C. Lovold, and M. Koomey. 2002. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to type IV pilus expression. Mol. Microbiol. 46:1441-1450. [DOI] [PubMed] [Google Scholar]
- 2.Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Lovold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. [DOI] [PubMed] [Google Scholar]
- 3.Ambur, O. H., S. A. Frye, and T. Tonjum. 2007. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J. Bacteriol. 189:2077-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 2007. Update to CDC's Sexually Transmitted Diseases Treatment Guidelines, 2006: fluoroquinolones no longer recommended for treatment of gonococcal infections. MMWR Morb. Mortal. Wkly. Rep. 56:332-336. [PubMed] [Google Scholar]
- 5.Apicella, M. A., J. F. Breen, and N. C. Gagliardi. 1978. Degradation of the polysaccharide component of gonococcal lipopolysaccharide by gonococcal and meningococcal sonic extracts. Infect. Immun. 20:228-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blokesch, M., and G. K. Schoolnik. 2008. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J. Bacteriol. 190:7232-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle-Vavra, S., and H. S. Seifert. 1996. Uptake-sequence-independent DNA transformation exists in Neisseria gonorrhoeae. Microbiology 142(Pt. 10):2839-2845. [DOI] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., and S. A. Hill. 1998. Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae. J. Bacteriol. 180:5117-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. J., D. M. Tobiason, C. E. Thomas, W. M. Shafer, H. S. Seifert, and P. F. Sparling. 2004. A mutant form of the Neisseria gonorrhoeae pilus secretin protein PilQ allows increased entry of heme and antimicrobial compounds. J. Bacteriol. 186:730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 11.Connell, T. D., W. J. Black, T. H. Kawula, D. S. Barritt, J. A. Dempsey, K. Kverneland, Jr., A. Stephenson, B. S. Schepart, G. L. Murphy, and J. G. Cannon. 1988. Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Mol. Microbiol. 2:227-236. [DOI] [PubMed] [Google Scholar]
- 12.Craig, L., M. E. Pique, and J. A. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363-378. [DOI] [PubMed] [Google Scholar]
- 13.Criss, A. K., K. A. Kline, and H. S. Seifert. 2005. The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol. Microbiol. 58:510-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danner, D. B., R. A. Deich, K. L. Sisco, and H. O. Smith. 1980. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11:311-318. [DOI] [PubMed] [Google Scholar]
- 15.Danner, D. B., H. O. Smith, and S. A. Narang. 1982. Construction of DNA recognition sites active in Haemophilus transformation. Proc. Natl. Acad. Sci. U. S. A. 79:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duffin, P. M., and H. S. Seifert. 2009. ksgA mutations confer resistance to kasugamycin in Neisseria gonorrhoeae. Int. J. Antimicrob. Agents 33:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins, C., C. E. Thomas, H. S. Seifert, and P. F. Sparling. 1991. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J. Bacteriol. 173:3911-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fussenegger, M., D. Facius, J. Meier, and T. F. Meyer. 1996. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol. Microbiol. 19:1095-1105. [DOI] [PubMed] [Google Scholar]
- 19.Fussenegger, M., A. F. Kahrs, D. Facius, and T. F. Meyer. 1996. Tetrapac (tpc), a novel genotype of Neisseria gonorrhoeae affecting epithelial cell invasion, natural transformation competence and cell separation. Mol. Microbiol. 19:1357-1372. [DOI] [PubMed] [Google Scholar]
- 20.Fyfe, J. A., C. S. Carrick, and J. K. Davies. 1995. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a sigma 70 promoter during growth in vitro. J. Bacteriol. 177:3781-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaasbeek, E. J., J. A. Wagenaar, M. R. Guilhabert, M. M. Wosten, J. P. van Putten, L. van der Graaf-van Bloois, C. T. Parker, and F. J. van der Wal. 2009. A DNase encoded by integrated element CJIE1 inhibits natural transformation of Campylobacter jejuni. J. Bacteriol. 191:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman, S. D., and J. J. Scocca. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 85:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helm, R. A., M. M. Barnhart, and H. S. Seifert. 2007. pilQ missense mutations have diverse effects on PilQ multimer formation, piliation, and pilus function in Neisseria gonorrhoeae. J. Bacteriol. 189:3198-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helm, R. A., and H. S. Seifert. 2009. Pilin antigenic variation occurs independently of the RecBCD pathway in Neisseria gonorrhoeae. J. Bacteriol. 191:5613-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill, S. A. 2000. Opa expression correlates with elevated transformation rates in Neisseria gonorrhoeae. J. Bacteriol. 182:171-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnsborg, O., V. Eldholm, and L. S. Havarstein. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767-778. [DOI] [PubMed] [Google Scholar]
- 27.Kellogg, D. S., Jr., I. R. Cohen, L. C. Norins, A. L. Schroeter, and G. Reising. 1968. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J. Bacteriol. 96:596-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kline, K. A., and H. S. Seifert. 2005. Mutation of the priA gene of Neisseria gonorrhoeae affects DNA transformation and DNA repair. J. Bacteriol. 187:5347-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koomey, J. M., and S. Falkow. 1987. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J. Bacteriol. 169:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koomey, M. 1998. Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl. 84:56-61. [DOI] [PubMed] [Google Scholar]
- 31.Kroll, J. S., K. E. Wilks, J. L. Farrant, and P. R. Langford. 1998. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. U. S. A. 95:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine, S. M., E. A. Lin, W. Emara, J. Kang, M. DiBenedetto, T. Ando, D. Falush, and M. J. Blaser. 2007. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J. 21:3458-3467. [DOI] [PubMed] [Google Scholar]
- 33.Long, C. D., D. M. Tobiason, M. P. Lazio, K. A. Kline, and H. S. Seifert. 2003. Low-level pilin expression allows for substantial DNA transformation competence in Neisseria gonorrhoeae. Infect. Immun. 71:6279-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maness, M. J., and P. F. Sparling. 1973. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J. Infect. Dis. 128:321-330. [DOI] [PubMed] [Google Scholar]
- 35.Maughan, H., and R. J. Redfield. 2009. Extensive variation in natural competence in Haemophilus influenzae. Evolution 63:1852-1866. [DOI] [PubMed] [Google Scholar]
- 36.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, T. F., N. Mlawer, and M. So. 1982. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell 30:45-52. [DOI] [PubMed] [Google Scholar]
- 38.Sechman, E. V., M. S. Rohrer, and H. S. Seifert. 2005. A genetic screen identifies genes and sites involved in pilin antigenic variation in Neisseria gonorrhoeae. Mol. Microbiol. 57:468-483. [DOI] [PubMed] [Google Scholar]
- 39.Seifert, H. S., C. J. Wright, A. E. Jerse, M. S. Cohen, and J. G. Cannon. 1994. Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J. Clin. Invest. 93:2744-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solomon, J. M., and A. D. Grossman. 1996. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 12:150-155. [DOI] [PubMed] [Google Scholar]
- 42.Sox, T. E., W. Mohammed, E. Blackman, G. Biswas, and P. F. Sparling. 1978. Conjugative plasmids in Neisseria gonorrhoeae. J. Bacteriol. 134:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sparling, P. F. 1966. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J. Bacteriol. 92:1364-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein, D. C., R. J. Danaher, and T. M. Cook. 1991. Characterization of a gyrB mutation responsible for low-level nalidixic acid resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 35:622-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321-330. [DOI] [PubMed] [Google Scholar]