Abstract
The WalRK two-component regulatory system coordinates gene expression that maintains cell wall homeostasis and responds to antibiotic stress in low-GC Gram-positive bacteria. Phosphorylated WalR (VicR) of the major human respiratory pathogen Streptococcus pneumoniae (WalRSpn) positively regulates transcription of several surface virulence genes and, most critically, pcsB, which encodes an essential cell division protein. Despite numerous studies of several species, little is known about the signals sensed by the WalK histidine kinase or the function of the WalJ ancillary protein encoded in the walRKSpn operon. To better understand the functions of the WalRKJSpn proteins in S. pneumoniae, we performed experiments to determine their cellular localization and amounts. In contrast to WalK from Bacillus subtilis (WalKBsu), which is localized at division septa, immunofluorescence microscopy showed that WalKSpn is distributed throughout the cell periphery. WalJSpn is also localized to the cell surface periphery, whereas WalRSpn was found to be localized in the cytoplasm around the nucleoid. In fractionation experiments, WalRSpn was recovered from the cytoplasmic fraction, while WalKSpn and the majority of WalJSpn were recovered from the cell membrane fraction. This fractionation is consistent with the localization patterns observed. Lastly, we determined the cellular amounts of WalRKJSpn by quantitative Western blotting. The WalRSpn response regulator is relatively abundant and present at levels of ≈6,200 monomers per cell, which are ≈14-fold greater than the amount of the WalKSpn histidine kinase, which is present at ≈460 dimers (920 monomers) per cell. We detected ≈1,200 monomers per cell of WalJSpn ancillary protein, similar to the amount of WalKSpn.
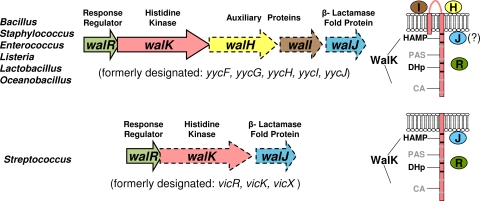
The WalRK (YycFG) two-component regulatory system (TCS) plays critical roles in maintaining cell wall and surface homeostasis and in responding to cell wall stresses in low-GC Gram-positive bacteria (reviewed in references 8, 22, and 43). In Bacillus, Staphylococcus, and many other species, both the WalR (YycF) response regulator and WalK (YycG) histidine kinase are essential, and they cannot be depleted (Fig. 1) (11, 14, 27). In contrast, the WalR (VicR) response regulator of Streptococcus species is essential, whereas the WalK (VicK) histidine kinase is not essential under standard growth conditions (10, 30, 34, 40). WalRK regulons include different sets of genes that mediate peptidoglycan biosynthesis, cell division, and the cell surface in different Gram-positive species (1, 4, 9, 14, 21, 26, 28, 30, 31, 34). The WalK histidine kinases of Streptococcus species have sensing domains that are structurally different from those of Bacillus, Staphylococcus, and most other species (Fig. 1) (8, 32, 43). WalK from Bacillus subtilis (WalKBsu), which is typical of one class, contains two transmembrane domains flanking an extracytoplasmic domain (35). The transmembrane domains of WalKBsu interact with the membrane domains of the auxiliary WalHI (YycHI) proteins to negatively regulate phosphorylation levels of the WalRBsu response regulator (36-38). In contrast, WalK from Streptococcus pneumoniae (WalKSpn), which exemplifies the other class, contains only a single transmembrane domain connected to an extracellular peptide of only 12 amino acids (Fig. 1) (24, 32, 40). Streptococcus species also lack homologues of WalHI (32). On the other hand, the cytoplasmic domains of the two classes of WalK histidine kinases are highly similar and contain a PAS domain (Fig. 1) (8, 16, 32, 43). The signals that are sensed directly by WalK histidine kinases are not yet known in any species (8, 19, 43). To understand better the functions and signaling of WalRKJSpn in S. pneumoniae, we determined their localization and cellular amounts.
FIG. 1.
Organization of the WalRKSpn TCS genes and WalK histidine kinases in low-GC Gram-positive bacteria. In most species (upper row), the operon consists of five genes encoding the essential (solid lines) WalR response regulator and WalK histidine kinase and three nonessential (dashed lines) proteins. In these species, the membrane-bound auxiliary WalHI proteins regulate WalK activity, but the functions and localization of the ancillary WalJ protein, which contains a metallo-β-lactamase fold, are unknown. The WalK histidine kinases of these species possess two transmembrane domains and an extracellular loop. In Streptococcus species, the operon contains only three genes encoding essential WalR and nonessential WalK and WalJ. In addition, the streptococcal WalK histidine kinases contain a single transmembrane domain and no extracellular domain, the WalHI proteins are absent, and WalJ is mostly membrane associated as reported here. The WalK histidine kinases of all species contain the HAMP and PAS sensing domains, the DHp dimerization and histidine phosphorylation domain, and the CA (catalytic) ATPase domain. See text for references and additional details.
MATERIALS AND METHODS
Localization of WalRKJSpn and FtsZ by immunofluorescence microscopy (IFM).
Unencapsulated S. pneumoniae strains IU1824 (D39 non-FLAG negative control), IU1876 [R6 ftsZ-(C)-FLAG3], IU3816 [D39 walJSpn-(C)-L-FLAG3], IU4052 [D39 walRSpn-(C)-L-FLAG3], IU4062 [D39 walKSpn-(C)-FLAG], and IU4099 [D39 ΔwalKSpn walJSpn-(C)-L-FLAG3] (Table 1 ) were cultured statically in 5 ml of BD brain heart infusion (BHI) broth in 16-mm glass tubes at 37°C in an atmosphere of 5% CO2 to an optical density of 0.12 to 0.2 as described previously (39). Four milliliters of cultures was centrifuged at 16,000 × g at 4°C for 10 min, resuspended in 1 ml of phosphate-buffered saline (PBS; Ambion; AM9625), and centrifuged at 16,000 × g at 4°C for 5 min. The pellets were resuspended in 1 ml of 4% paraformaldehyde (EMS; 157-4) and incubated for 15 min at room temperature and 45 min on ice. Fixed cells were centrifuged, and pellets were washed three times with PBS at 4°C, resuspended in 0.3 ml of prechilled (4°C) GTE (50 mM glucose, 1 mM EDTA, 20 mM Tris HCl, pH 7.5), and stored at 4°C for up to 20 h. Fifty microliters of cell suspensions was deposited in 1-cm2 wells made by a Liquid Blocker Super Pap Pen (EMS; 71310) on precleaned microscope slides and incubated for 5 min at room temperature. Unattached cells were aspirated, and attached cells were treated with 50 μl of 0.2% Triton X-100 (Mallinckrodt; H282) in PBS (PBS-T) for 10 s. PBS-T was aspirated from the wells, and the slides were immediately immersed in prechilled −20°C methanol and kept at −20°C for 10 min and then removed from methanol and air dried completely. The following incubations were carried out with 50 μl of reagents at room temperature: PBS-T for 5 min; 5% skim milk powder (BBL; 11915), 0.2% Triton X-100 in PBS (PBS-T-M) for 1 h; PBS for 10 s twice; 1:200 dilution of primary anti-FLAG polyclonal antibody (Sigma; F7425) in 5% skim milk powder in PBS (PBS-M) for 1 h; PBS twice for 10 s and once for 5 min; 1:100 dilution of secondary anti-rabbit-Alexa 488 (Molecular Probes; A11034), 4′,6-diamidino-2-phenylindole (DAPI; final concentration, 0.2 μg/ml) in PBS-M for 1 h; PBS twice for 10 s and once for 5 min. After the wells were air dried, 7 μl of Slowfade Gold antifade reagent (Invitrogen; S36936) was applied to each well and coverslips were applied and sealed. Cells were examined using a Nikon E-400 epifluorescence phase-contrast microscope equipped with a mercury lamp, a 100× Nikon Plan Apo oil-immersion objective (numerical aperture, 1.40), and filter blocks for fluorescence (DAPI, EX 330 to 380, DM 400, and BA 435 to 485; Alexa 488, EX 460 to 500, DM 505, and BA 510 to 560). Images were captured using a CoolSNAP HQ2 charge-coupled device (CCD) camera (Photometrics) and processed with NIS-Elements AR imaging software (Nikon).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype (description)a | Antibiotic resistanceb | Reference or source |
|---|---|---|---|
| Bacterial strains | |||
| S. pneumoniae | |||
| EL59 | R6 (unencapsulated laboratory strain R6 derived from intermediates of strain D39) | None | 25 |
| IU1473 | R6 ΔvicR<>ermAM kan-t1t2-Pc-pcsB | Kanr | 31 |
| IU1690 | D39 (single-colony isolate of serotype 2 strain encapsulated D39 NCTC 7466) | None | 25 |
| IU1781 | D39 rpsL1 | Strr | 23 |
| IU1824 | D39 rpsL1 Δcps2A′-cps2H′ | Strr | 25 |
| IU1876 | R6 ftsZ-(C)-FLAG3 intergenic::Pc-ermAM (EL59 transformed with fusion amplicon) | Ermr | This study |
| IU1885 | D39 rpsL1 ΔwalKSpn::Pc-[Kanr-rpsL+] | Strs Kanr | 19 |
| IU1888 | D39 rpsL1 ΔwalJSpn::Pc-[Kanr-rpsL+] (IU1781 transformed with fusion amplicon) | Strs Kanr | This study |
| IU1896 | D39 rpsL1 ΔwalKSpn | Strr | 19 |
| IU3126 | R6 pcsB-(C)-L-FLAG3 intergenic::Pc-ermAM (EL59 transformed with fusion amplicon) | Ermr | This study |
| IU3150 | D39 ΔbgaA′::Pc-kant1t2-Pfcsk-pcsB (IU1690 transformed with fusion amplicon) | Kanr | This study |
| IU3348 | R6 ΔbgaA′::Pc-kant1t2-Pfcsk-pcsB-(C)-L-FLAG3 (EL59 transformed with fusion amplicon) | Kanr | This study |
| IU3477 | R6 ΔbgaA′::Pc-kant1t2-Pfcsk-walJSpn-(C)-L-FLAG3 (EL59 transformed with fusion amplicon) | Kanr | This study |
| IU3479 | R6 ΔbgaA′::Pc-kant1t2-Pfcsk-walRSpn-(C)-L-FLAG3 (EL59 transformed with fusion amplicon) | Kanr | This study |
| IU3689 | D39 Δcps2A′-cps2H′ rpsL1 walJSpn+ intergenic::Pc-[Kanr-rpsL+] (IU1824 transformed with fusion amplicon) | Strs Kanr | This study |
| IU3816 | D39 Δcps2A′-cps2H′ rpsL1 walJSpn-(C)-L-FLAG3 (IU3689 transformed with fusion amplicon) | Strr | This study |
| IU3953 | D39 Δcps2A′-cps2H′ rpsL1 ΔwalJSpn (IU3689 transformed with fusion amplicon) | Strr | This study |
| IU4021 | D39 Δcps2A′-cps2H′ rpsL1 ΔwalKSpn::Pc-[Kanr-rpsL+] (IU1824 transformed with ΔwalKSpn::Pc-[Kanr-rpsL+] amplicon from IU1885) | Kanr | This study |
| IU4052 | D39 Δcps2A′-cps2H′ rpsL1 walRSpn-(C)-L-FLAG3 (IU4021 transformed with fusion amplicon) | Strr | This study |
| IU4062 | D39 Δcps2A′-cps2H′ rpsL1 walKSpn-(C)-FLAG (IU4021 transformed with fusion amplicon) | Strr | This study |
| IU4092 | D39 Δcps2A′-cps2H′ rpsL1 ΔwalKSpn::Pc-[Kanr-rpsL+] walJSpn-(C)-L-FLAG3 (IU3816 transformed with ΔwalKSpn::Pc-[Kanr-rpsL+] from IU1885) | Strs Kanr | This study |
| IU4099 | D39 Δcps2A′-cps2H′ rpsL1 ΔwalKSpn walJSpn-(C)-L-FLAG3 (IU4092 transformed with ΔwalKSpn amplicon from IU1896) | Strr | This study |
| E. coli | |||
| EL27 | BL21(DE3)(pLysS)(pSP001) | Ampr Cmr | 31 |
| IU4019 | BL21(DE3) Rosetta(pLysS)(pIU251) | Kanr Cmr | This study |
| IU4223 | BL21(DE3) Rosetta(pLysS)(pIU259) | Kanr Cmr | This study |
| Plasmids (E. coli) | |||
| pSP001 | pET16b [expression of (N)-His10-WalRSpn] | Ampr Cmr | 31 |
| pIU251 | pSumo [expression of (N)-Sumo-WalKSpn ΔN35-(C)-FLAG, where ΔN35 removed the transmembrane domain of WalKSpn] | Kanr Cmr | This study |
| pIU259 | pET28a [expression of WalJSpn-(C)-L-FLAG3] | Kanr Cmr | This study |
Primers used to synthesize fusion amplicons are listed in Table S1 in the supplemental material. (N)- or (C)- indicates that fusions were made to the amino- or carboxyl-terminal ends of reading frames, respectively. L refers to a 10-amino-acid spacer linker (GSAGSAAGSG) based on reference 42. FLAG3 indicates three tandem copies of the FLAG epitope (DYKDDDDK) (20). Intergenic means that the resistance gene was cloned into the intergenic region following the indicated gene. BL21(DE3)(pLysS), BL21(DE3) Rosetta(pLysS), pET16b, and pET28a were purchased from Novagen, and pSumo was purchased from LifeSensors.
Antibiotic resistance markers: Kanr, kanamycin; Strr, streptomycin, Ermr, erythromycin; Ampr, ampicillin; Cmr, chloramphenicol. Concentrations of antibiotics used for S. pneumoniae strains: 250 μg Str per ml (Sigma S6501), 250 μg Kan per ml (Sigma K0254), and 0.3 μg Erm per ml (Sigma E6376); for E. coli strains, 34 μg Cm per ml (Sigma C0378) and 30 μg Kan per ml and 100 μg Amp per ml (Sigma A5354).
Fractionation of S. pneumoniae cells.
Biochemical fractionation of IU3816 [D39 walJSpn-(C)-L-FLAG3], IU4052 [D39 walRSpn-(C)-L-FLAG3], and IU4062 [D39 walKSpn-(C)-FLAG] was performed with modifications from references 6 and 44. Briefly, cells were grown in BHI broth to an optical density at 620 nm (OD620) of 0.2 to 0.4 as described above. One milliliter of cultures was sampled to prepare fractions of culture supernatants (S), cell pellets (P), digested cell wall (CW), protoplasts (Prot), cytoplasm (Cyto), and cell membrane (Memb). Cultures were centrifuged at 13,000 × g for 10 min at room temperature to obtain culture supernatants (S), which were stored on ice, and cell pellet fractions (P). Pellets were briefly washed with 0.5 ml 1× SMM buffer (0.5 M sucrose, 20 mM MgCl2, 20 mM morpholineethanesulfonic acid [MES], pH 6.5) at room temperature, collected again by centrifugation, and resuspended in 0.5 ml 1× SMM buffer. To obtain protoplast (Prot) and digested cell wall fractions (CW), 25 μl of 10 mg lysozyme (Sigma) per ml, 2 μl of 1 mg mutanolysin (Sigma; M9901) per ml, and 5 μl of protease inhibitor cocktail III (Calbiochem) were added, and the mixture was incubated at 37°C for 1 h. The protoplast (Prot, pellet) and digested cell wall (CW, supernatant) fractions were separated by centrifugation at 3,000 × g for 10 min at room temperature. The CW fraction was stored on ice, and the protoplast pellet was frozen on dry ice for 10 min and resuspended in 0.5 ml of cold buffer H (20 mM HEPES, pH 8.0, 200 mM NaCl, 1 mM dithiothreitol [DTT], and protease inhibitor cocktail III). Five microliters of 0.1 M MgCl2, 5 μl of 0.1 M CaCl2, 1 μl of 5 mg DNase (Sigma; D4527) per ml, and 1 μl of 10 mg RNase (Sigma; R5500) per ml were added, and the mixture was incubated on ice for 1 h. After centrifugation at 16,100 × g for 30 min at 4°C, the pellet was collected as the membrane fraction (Memb) and the supernatant was collected as the cytoplasmic fraction (Cyto). The supernatants from each spin (S, CW, and Cyto fractions) were precipitated with cold 10% (wt/vol) trichloroacetic acid (TCA; Sigma; T4885) as described in the work of Barendt et al. (2), and pellets were resuspended in 100 μl lysis buffer (1% [wt/vol] SDS, 0.1% [vol/vol] Triton X-100) and 100 μl 2× SDS sample buffer (Bio-Rad; 161-0737) containing 5% (vol/vol) β-mercaptoethanol (Sigma). The pellets (P, Prot, and Memb fractions) were resuspended in 100 μl lysis buffer (1% [wt/vol] SDS, 0.1% [vol/vol] Triton X-100) and 100 μl 2× SDS sample buffer (Bio-Rad; 161-0737) containing 5% (vol/vol) β-mercaptoethanol (Sigma). Samples were boiled for 5 min, and 40 μl of each sample was resolved by SDS-PAGE (4% stacking, 10% Tris-glycine gels) as described in the work of Barendt et al. (2). FLAG-tagged proteins were detected by Western blotting with primary anti-FLAG polyclonal antibody (1:1,000 dilution; Sigma; F7425), secondary donkey anti-rabbit antibody conjugated with horseradish peroxidase (HRP; diluted 1:10,000; GE Healthcare; NA934), and ECL detection kits (Amersham) as described in reference 2. Autoradiography and quantitation using an IVIS imaging system were done as described in reference 2.
To distinguish protein aggregates from membrane-bound proteins, membrane fractions (Memb) were centrifuged on a sucrose cushion (7). The membrane fraction was resuspended in 0.5 ml buffer H. The suspension was carefully overlaid on 0.5 ml of buffer H containing 2 M sucrose, followed by centrifugation at 150,000 × g for 16 h at 4°C. Five equal-volume fractions were collected by removing solution gently along the side of the centrifuge tube from the top. Protein in each fraction was precipitated with 10% (wt/vol) TCA on ice, and the FLAG-tagged proteins were detected by Western blotting as described above.
Determination of cellular amounts of WalRKJSpn by quantitative Western blotting.
Cellular amounts of WalRSpn and WalKSpn-(C)-FLAG or WalJSpn-(C)-L-FLAG3 were determined in strain IU4062 or IU3816, respectively (Table 1). Lysates were prepared from unencapsulated strains IU1473 (R6 Pc-pcsB ΔwalRspn), IU1824 (D39 non-FLAG negative control), IU3816 [D39 walJSpn-(C)-L-FLAG3], and IU4062 [D39 walRSpn+ walKSpn-(C)-FLAG], which had been cultured in BHI broth to an OD620 of 0.35 to 0.45 as described above. One milliliter of cultures was centrifuged at 16,100 × g for 10 min at room temperature, and cell pellets were resuspended in 100 μl of lysis buffer as described in reference 41 with the addition of 40 μg mutanolysin (Sigma; M9901) per ml to digest S. pneumoniae cell walls. Mixtures were incubated at 37°C for 10 min. One hundred microliters of 2× SDS sample buffer (Bio-Rad; 161-0737) containing 5% (vol/vol) β-mercaptoethanol (Sigma) was then added to sample preparations, and the mixtures were incubated at 95°C for 10 min. Lysates were centrifuged at 3,300 × g for 4 min at room temperature, and 30 μl of the supernatants was resolved by 10% SDS-PAGE as described above. Protein standards (N)-His10-WalRSpn, (N)-Sumo-WalKSpn ΔN35-(C)-FLAG, and WalJSpn-(C)-L-FLAG3 were purified as described in Table S2 in the supplemental material. Standard curves for WalRSpn quantitation were generated by adding 0.03 to 0.5 pmol of (N)-His10-WalRSpn to 30-μl samples of IU1473 lysates before SDS-PAGE. Standard curves for WalKSpn-(C)-FLAG and WalJSpn-(C)-L-FLAG3 quantitation were generated by adding 0.016 to 0.45 pmol of (N)-Sumo-WalKSpn ΔN35-(C)-FLAG or 0.03 to 0.45 pmol of WalJSpn-(C)-L-FLAG3, respectively, to 30-μl samples of IU1824 lysate before SDS-PAGE. Detection of the WalRSpn protein on Western blots was achieved using 1:1,000 dilutions of primary anti-WalRSpn rabbit polyclonal antibody (Cocalico Biologicals) (30). FLAG-tagged proteins were detected using primary anti-FLAG polyclonal antibody as described above. Autoradiography and quantitation using an IVIS imaging system were done as described in reference 2.
CFU were determined by serial dilution of cultures used for lysate preparations and plating on Trypticase soy agar II (modified) containing 5% (vol/vol) defibrinated sheep blood agar plates (Becton Dickinson; BBL 221261). Molecules per cell were calculated from CFU, taking into account that these unencapsulated strains form primarily diplococcal cells, which means that there are two cells per CFU (2).
RESULTS AND DISCUSSION
Localization of WalRKJSpn by immunofluorescence microscopy.
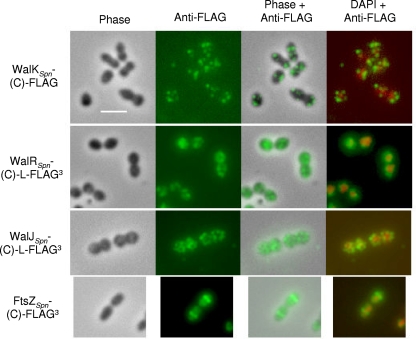
WalKBsu colocalizes with FtsZ at the septa of dividing B. subtilis cells, and it has been postulated that this interaction allows the WalRKBsu to sense cell division directly (15). To determine whether WalRKSpn localized similarly in S. pneumoniae, we performed immunofluorescence microscopy (IFM) as described in Materials and Methods. Attempts to locate the WalKSpn histidine kinase with purified polyclonal antibodies prepared against the entire polypeptide or a synthetic PAS domain peptide were not successful. Consequently, we fused a FLAG epitope tag (20) to the carboxyl terminus of WalKSpn expressed from its native chromosomal locus (Table 1). The resulting strain (IU4062) did not show several characteristic phenotypes of mutants defective in WalKSpn function (data not shown), indicating that the FLAG tag did not significantly interfere with WalKSpn function. IFM using purified polyclonal anti-FLAG antibody (Sigma Corp.) (Fig. 2) showed that WalKSpn-(C)-FLAG was distributed in a random pattern around the periphery of S. pneumoniae cells growing exponentially in brain heart infusion (BHI) broth in an atmosphere of 5% CO2 (see references 33 and 39) and was not localized to division septa, as reported for WalKBsu (15). Control experiments show no labeling of cells lacking a protein fused to the FLAG tag (data not shown). As another control, FtsZ fused at its carboxyl terminus to three tandem copies of the FLAG tag (FLAG3) localized normally to cell division sites at equators and septa (Fig. 2; Table 1) (12, 29, 45). No defects in cell division or morphology were observed for this construct (Fig. 2). The tandem FLAG3 tag amplified detection of FtsZ expressed from its native chromosomal locus, since a single FtsZ-FLAG construct could not be detected.
FIG. 2.
Localization of WalKSpn-(C)-FLAG, WalRSpn-(C)-L-FLAG3, and WalJSpn-(C)-L-FLAG3 by immunofluorescence microscopy (IMF) in exponentially growing unencapsulated strain D39. Representative phase images of fixed cells and IMF of the indicated FLAG-tagged proteins using anti-FLAG antibody are shown in the first and second columns, respectively. Carboxyl-terminal (C) fusion of the chromosomally expressed proteins to the linker (L) and FLAG epitope tags (Table 1) did not cause any observable changes in cell growth or morphology compared to parent strains (first column; data not shown). IMF of cells lacking a FLAG-tag fusion protein did not show any labeling (data not shown). The third and fourth columns show overlays of the first two columns or overlays of the second column and images stained with DAPI to locate nucleoids, respectively. For comparison and to validate the IMF methods, the bottom row shows localization of FtsZSpn-(C)-FLAG3 at the equators of exponentially growing cells of R6, which is an unencapsulated laboratory strain originally derived from strain D39. At least three biological replicates were performed for each strain, and multiple fields of cells were observed and analyzed for each replicate. See Materials and Methods for the IMF staining procedure used and the text for additional details.
We also localized WalRSpn and WalJSpn fused at their carboxyl termini to a 10-amino-acid linker (L) based on reference 42 followed by a FLAG3 tandem tag (Table 1; Fig. 2). WalJSpn is a nonessential protein containing a metallo-β-lactamase fold that may play an ancillary role in WalRKSpn TCS signaling (Fig. 1) (30). The resulting constructs (IU4052 and IU3816; Table 1) showed wild-type diplococcal morphology and cell division, consistent with normal WalRSpn and WalJSpn function (Fig. 2; data not shown). WalRBsu localized to the nucleoid in growing cells and surrounded the nucleoid in nondividing cells (15). In contrast, WalRSpn-(C)-L-FLAG3 was distributed in the cytoplasm around nucleoids in exponentially growing S. pneumoniae (Fig. 2). WalJSpn-(C)-L-FLAG3 was located throughout pneumococcal cells (Fig. 2) and appeared to be localized near cell surfaces, similarly to WalKSpn-(C)-FLAG (Fig. 2). WalJSpn localization was not affected by the absence of WalKSpn in a ΔwalKSpn mutant (IU4099, Table 1; data not shown), although ΔwalKSpn mutants form longer chains (data not shown). On the basis of these data, we conclude that localization of WalKSpn and WalRSpn (Fig. 2) was fundamentally different from that of WalKBsu and WalRBsu in growing cells. This difference may reflect the different structures of WalKSpn and WalKBsu (Fig. 1), and it implies that sensing by WalKSpn of cell division likely does not occur through a direct interaction with FtsZ, as proposed for WalKBsu (15).
Fractionation of WalRKJSpn correlates with their localization.
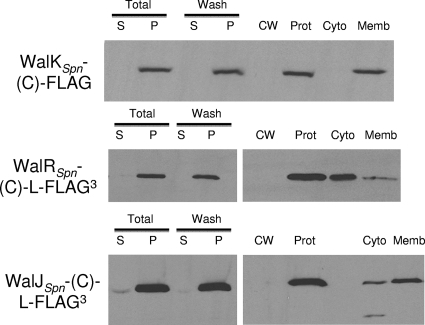
To gain additional information about these WalRKJSpn localization patterns, we fractionated cells using methods described in references 6 and 44 with the modifications described in Materials and Methods. As expected, the vast majority of the recovered WalRSpn-(C)-L-FLAG3 response regulator or WalKSpn-(C)-FLAG histidine kinase was in the cytoplasmic (88%) or membrane (94%) fractions, respectively (Fig. 3; data not shown). Consistent with its apparent localization to the cell surface (Fig. 2), 79% of the recovered WalJSpn-(C)-L-FLAG3 was associated with the membrane fraction (Fig. 3). Some of the small amount of WalJSpn-(C)-L-FLAG3 in the cytoplasmic fraction was degraded at its amino terminus as indicated by a lower-molecular-mass band that retained the FLAG3 tag. WalJSpn contains a metallo-β-lactamase fold (Pfam [13]) and at least one extended amphipathic helix predicted by the Heliquest program (17). WalJSpn does not contain a predicted signal peptide or transmembrane domain, and it seems unlikely that the negatively charged FLAG3 tag would promote membrane association. Moreover, WalJSpn and WalJBsu have been difficult to purify because of aggregation (data not shown; Bill Burkholder, personal communication). To confirm that WalJSpn was associated with the cell membrane and not aggregating during the fractionation, we centrifuged membrane fractions on sucrose cushion gradients as described in reference 7. Significant amounts of both WalKSpn-(C)-FLAG and WalJSpn-(C)-L-FLAG3 were found in fractions near the sucrose interface, indicative of membrane association (data not shown). Thus, the majority of WalJSpn is associated with the cytoplasmic side of the cell membrane in exponentially growing S. pneumoniae (Fig. 1 and 3).
FIG. 3.
Biochemical fractionation of WalKSpn-(C)-FLAG, WalRSpn-(C)-L-FLAG3, and WalJSpn-(C)-L-FLAG3 in an exponentially growing unencapsulated derivative of strain D39. Fractionation and Western immunoblot assays using anti-FLAG antibody were performed as described in Materials and Methods. Total, supernatant (S) and pellet (P) of starting cultures. Wash, supernatant (S) and pellet (P) of cells washed with sucrose-containing buffer. Washed pellets were further separated into solubilized cell wall (CW) and protoplast (Prot) fractions. Lysed protoplasts were separated into cytoplasmic (Cyto) and membrane (Memb) fractions. The image shown is a semiquantitative autoradiogram. Quantitation in the text is based on three independent biological replicate experiments in which band luminescence was determined using an IVIS imaging system. See text for additional details.
WalRSpn is a relatively abundant protein compared to WalKSpn and WalJSpn.
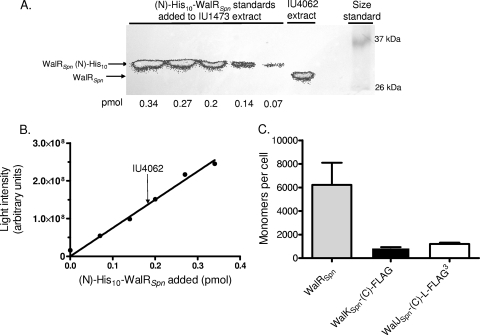
We determined the cellular amounts of WalRKJSpn by quantitative Western blotting as described in references 2 and 41 with the modifications in Materials and Methods. Cellular amounts of cognate histidine kinases and response regulators have not been determined for many TCSs. For the archetypical EnvZ/OmpR TCS, the OmpR response regulator is present in a significant excess over the EnvZ histidine kinase (3,500 monomers versus 59 dimers per cell, respectively) (5). This ratio has important implications for the robustness and response of the system (3, 18). In determinations of WalRSpn amounts, we used polyclonal anti-WalRSpn antibody (30) in the Western blot assays (Fig. 4A). As a standard, we spiked various concentrations of purified (N)-His10-WalRSpn protein (19, 31) (see Table S2 in the supplemental material) into an extract of a ΔwalRSpn Pc-pcsB+ mutant (IU1473, Table 1) containing the same amount of total protein as that used for the walRSpn+ strain IU4062 (Table 1). Unencapsulated mutants that are predominantly diplococci instead of chains of cells were used in these experiments to simplify estimates of cellular amounts (2). Control experiments showed that similar amounts were present in unencapsulated and encapsulated D39 strains (data not shown). The amount of WalRSpn detected in the walRSpn+ strain fell within the linear range of the standard curves (Fig. 4B). We found that WalRSpn was relatively abundant at 6,200 ± 1,900 (standard error of the mean [SEM], n = 3) monomers per cell in cultures growing exponentially in BHI broth in an atmosphere of 5% CO2 (Fig. 4C). Parallel determinations of cellular amounts of WalRSpn-L-FLAG3 using anti-FLAG antibodies gave similar results (data not shown) and provided validation for the use of FLAG-tagged proteins for quantitation. Similar amounts of WalRSpn were detected in cells sampled early or later in exponential phase (OD620 of 0.1, 0.3, and 0.6). Therefore, as with WalRBsu (37), there was no growth phase regulation of WalRSpn amount. The abundance of WalRSpn was consistent with its presence throughout the cytoplasm (Fig. 2). Since the shape of an unencapsulated pneumococcus cell approximates a prolate spheroid (a = 0.62 μm and b = 0.82 μm [2]) with a volume of 1.3 × 10−15 liters, the cellular concentration of WalRSpn in the cytoplasm is ≈8 μM.
FIG. 4.
Quantitative Western immunoblot assays of WalRSpn, WalKSpn-(C)-FLAG, and WalJSpn-(C)-L-FLAG3 cellular amounts in exponentially growing unencapsulated derivatives of strain D39. The quantitation procedure is included in Materials and Methods. (A) Representative quantitative Western immunoblot assay used to determine the cellular amount of WalRSpn. (Left lanes) The indicated amounts of a purified (N)-His10-WalRSpn standard were added to extracts of ΔwalRSpn Pc-pcsB+ mutant IU1473. (Right lanes) Amount of native WalRSpn detected in an extract of WalRSpn+ strain IU4062 and protein molecular mass markers. The same amounts of total protein of the IU1473 and IU4062 extracts were loaded into each lane. Band luminescence was determined using an IVIS imaging system. (B) Standard curve (circles) generated from panel A. The arrow shows the light intensity of the IU4062 band from which the cellular amount of WalRSpn was read from the standard curve. (C) Cumulative data showing the amounts in monomers per pneumococcal cell of WalRSpn, WalKSpn-(C)-FLAG, and WalJSpn-(C)-L-FLAG3. The amount of WalRSpn in strain IU4062 was from three biological replicates, including the one in panels A and B. The amounts of WalKSpn-(C)-FLAG in strain IU4062 and WalJSpn-(C)-L-FLAG3 in strain IU3816 were from two biological replicates each using anti-FLAG antibody with purified (N)-Sumo-WalKSpn ΔN35-(C)-FLAG and partially purified WalJSpn-(C)-L-FLAG3, respectively, as standards. See Materials and Methods and the text for additional details.
We were unable reliably to detect WalKSpn in pneumococcal extracts by using three different polyclonal anti-WalKSpn antibody preparations that did bind to purified WalKSpn protein in Western blots (data not shown). However, we could detect WalKSpn-(C)-FLAG used for localization studies (Fig. 2), probably because the commercial anti-FLAG antibody had a higher titer than did the anti-WalKSpn antibody preparations. In this case, a purified (N)-Sumo-WalKSpn ΔN35-(C)-FLAG standard (Table 1; see also Table S2 in the supplemental material) was added to extracts of strain IU1824 (Table 1) in quantitative Western blot assays (data not shown). We found that the cellular amount of the WalKSpn histidine kinase was 920 ± 20 (SEM, n = 2) monomers (460 dimers) per cell (Fig. 4C). Thus, similarly to the EnvZ/OmpR TCS, the WalRSpn response regulator monomer was present in a sizable molar excess (≈14-fold) over the WalKSpn histidine kinase dimer in exponentially growing pneumococcal cells. Since WalKSpn has both autokinase and phosphorylated WalRSpn (WalRSpn∼P) phosphatase activities (19), this excess implies that WalRKSpn may be a robust signaling system (3, 18). The impact of these cellular amounts and ratios to signal transduction by the WalRKSpn TCS awaits determinations of binding constants, kinetic properties, and cellular phosphorylation levels.
Finally, we approximated the cellular amount of WalJSpn-(C)-L-FLAG3 used for localization (Fig. 2), where the standard was partially purified WalJSpn-(C)-L-FLAG3 added to extracts of strain IU1824 (Table 1; see also Table S2 in the supplemental material). Because of a tendency to aggregate, the WalJSpn-(C)-L-FLAG3 standard could be purified only to ≈40% purity, in contrast to the highly purified (>95%) WalRSpn and WalKSpn standards (Table S2). With this limitation, membrane-associated WalJSpn was approximated at 1,200 ± 110 (SEM, n = 2) monomers per exponentially growing cell, close to the amount of the WalKSpn histidine kinase (Fig. 4C). The functions of WalJSpn and its relationship to WalRKSpn TCS signal transduction remain to be determined, as does the mechanism that reduces cellular expression of cotranscribed WalKSpn and WalJSpn compared to WalRSpn.
Supplementary Material
Acknowledgments
We thank Andre Zapun (CNRS, Grenoble, France) for advice on IMF and Daniel Kearns and members of his laboratory (Indiana University) for advice on microscopy.
This project was supported by grant number AI060744 (to M.E.W.) from the National Institute of Allergy and Infectious Diseases. S.M.B. was a predoctoral trainee on NIH grant number F31FM082090, and K.J.W. was a predoctoral trainee on training grant T32GM007757.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print on 9 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J. Bacteriol. 189:8519-8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barendt, S. M., A. D. Land, L. T. Sham, W. L. Ng, H. C. Tsui, R. J. Arnold, and M. E. Winkler. 2009. Influences of capsule on the cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J. Bacteriol. 191:3024-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor, E., and M. Goulian. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. U. S. A. 100:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisicchia, P., D. Noone, E. Lioliou, A. Howell, S. Quigley, T. Jensen, H. Jarmer, and K. M. Devine. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65:180-200. [DOI] [PubMed] [Google Scholar]
- 5.Cai, S. J., and M. Inouye. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277:24155-24161. [DOI] [PubMed] [Google Scholar]
- 6.Campo, N., K. A. Marquis, and D. Z. Rudner. 2008. SpoIIQ anchors membrane proteins on both sides of the sporulation septum in Bacillus subtilis. J. Biol. Chem. 283:4975-4982. [DOI] [PubMed] [Google Scholar]
- 7.Caulfield, M. P., P. C. Tai, and B. D. Davis. 1983. Association of penicillin-binding proteins and other enzymes with the ribosome-free membrane fraction of Bacillus subtilis. J. Bacteriol. 156:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrac, S., P. Bisicchia, K. M. Devine, and T. Msadek. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307-1322. [DOI] [PubMed] [Google Scholar]
- 9.Dubrac, S., I. G. Boneca, O. Poupel, and T. Msadek. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189:8257-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echenique, J. R., and M. C. Trombe. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fadda, D., A. Santona, V. D'Ulisse, P. Ghelardini, M. G. Ennas, M. B. Whalen, and O. Massidda. 2007. Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD-free context. J. Bacteriol. 189:1288-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn, R. D., J. Mistry, J. Tate, P. Coggill, A. Heger, J. E. Pollington, O. L. Gavin, P. Gunasekaran, G. Ceric, K. Forslund, L. Holm, E. L. Sonnhammer, S. R. Eddy, and A. Bateman. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211-D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 15.Fukushima, T., H. Szurmant, E. J. Kim, M. Perego, and J. A. Hoch. 2008. A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol. Microbiol. 69:621-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, R., and A. M. Stock. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautier, R., D. Douguet, B. Antonny, and G. Drin. 2008. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24:2101-2102. [DOI] [PubMed] [Google Scholar]
- 18.Goulian, M. 2004. Robust control in bacterial regulatory circuits. Curr. Opin. Microbiol. 7:198-202. [DOI] [PubMed] [Google Scholar]
- 19.Gutu, A. D., K. J. Wayne, L. T. Sham, and M. E. Winkler. 2010. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J. Bacteriol. 192:2346-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopp, T. P., K. S. Prickett, V. L. Price, R. T. Libby, C. J. March, D. P. Cerretti, D. L. Urdal, and P. J. Conlon. 1988. A short polypeptide marker sequence useful for recombinant protein identification and purification. Biotechnology (NY) 6:1204-1210. [Google Scholar]
- 21.Howell, A., S. Dubrac, K. K. Andersen, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639-1655. [DOI] [PubMed] [Google Scholar]
- 22.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 23.Kazmierczak, K. M., K. J. Wayne, A. Rechtsteiner, and M. E. Winkler. 2009. Roles of rel in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol. Microbiol. 72:590-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 25.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, M., T. S. Hanks, J. Zhang, M. J. McClure, D. W. Siemsen, J. L. Elser, M. T. Quinn, and B. Lei. 2006. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiology 152:967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohedano, M. L., K. Overweg, A. de la Fuente, M. Reuter, S. Altabe, F. Mulholland, D. de Mendoza, P. Lopez, and J. M. Wells. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morlot, C., A. Zapun, O. Dideberg, and T. Vernet. 2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845-855. [DOI] [PubMed] [Google Scholar]
- 30.Ng, W. L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 31.Ng, W. L., H. C. Tsui, and M. E. Winkler. 2005. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 187:7444-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng, W. L., and M. E. Winkler. 2004. Singular structures and operon organizations of essential two-component systems in species of Streptococcus. Microbiology 150:3096-3098. [DOI] [PubMed] [Google Scholar]
- 33.Ramos-Montanez, S., H. C. Tsui, K. J. Wayne, J. L. Morris, L. E. Peters, F. Zhang, K. M. Kazmierczak, L. T. Sham, and M. E. Winkler. 2008. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol. Microbiol. 67:729-746. [DOI] [PubMed] [Google Scholar]
- 34.Senadheera, M. D., B. Guggenheim, G. A. Spatafora, Y. C. Huang, J. Choi, D. C. Hung, J. S. Treglown, S. D. Goodman, R. P. Ellen, and D. G. Cvitkovitch. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187:4064-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szurmant, H., L. Bu, C. L. Brooks III, and J. A. Hoch. 2008. An essential sensor histidine kinase controlled by transmembrane helix interactions with its auxiliary proteins. Proc. Natl. Acad. Sci. U. S. A. 105:5891-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szurmant, H., M. A. Mohan, P. M. Imus, and J. A. Hoch. 2007. YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 189:3280-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szurmant, H., K. Nelson, E. J. Kim, M. Perego, and J. A. Hoch. 2005. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 187:5419-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szurmant, H., R. A. White, and J. A. Hoch. 2007. Sensor complexes regulating two-component signal transduction. Curr. Opin. Struct. Biol. 17:706-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsui, H. C., D. Mukherjee, V. A. Ray, L. T. Sham, A. L. Feig, and M. E. Winkler. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192:264-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner, C., A. de Saizieu, H. J. Schonfeld, M. Kamber, R. Lange, C. J. Thompson, and M. G. Page. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 70:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner, J. K., K. A. Marquis, and D. Z. Rudner. 2009. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol. Microbiol. 73:963-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldo, G. S., B. M. Standish, J. Berendzen, and T. C. Terwilliger. 1999. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 17:691-695. [DOI] [PubMed] [Google Scholar]
- 43.Winkler, M. E., and J. A. Hoch. 2008. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190:2645-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zapun, A., T. Vernet, and M. G. Pinho. 2008. The different shapes of cocci. FEMS Microbiol. Rev. 32:345-360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.