Abstract
The filamentous cyanobacterium Anabaena sp. strain PCC 7120 forms a periodic pattern of nitrogen-fixing heterocysts when grown in the absence of combined nitrogen. PatA is necessary for proper patterning of heterocysts along filaments. In this study, apparent transcriptional start points (tsps) were identified at nucleotides −305, −614, and −645 relative to the translational start site (−305, −614, and −645 tsps). Transcriptional reporter fusions were used to show that transcription from the −305 tsp was induced in all cells of filaments in response to nitrogen deprivation, required hetR for induction, and increased in a patA mutant. Transcription from −614/−645 tsp reporter fusions was spatially regulated and occurred primarily in cells that would become heterocysts. Complementation of a patA mutant strain by alleles encoding substitutions in, or deletion of, the putative phosphoacceptor C-terminal domain indicates that the PATAN domain can function independently of the C-terminal domain of PatA. Localization of a ring of PatA-GFP at sites of cell division, as well as the formation of enlarged cells with altered cell morphology when patA was overexpressed, suggests that PatA may participate in cell division.
In the filamentous cyanobacterium Anabaena sp. strain PCC 7120, the mutually exclusive metabolic processes of oxygen-evolving photosynthesis and nitrogen fixation occur simultaneously during aerobic growth. The bacterium spatially separates these processes into two distinct cell types. Oxygenic photosynthesis occurs in vegetative cells, while heterocysts, which contain the oxygen-labile nitrogenase complex, are the sites of nitrogen fixation. Heterocysts differentiate from vegetative cells in response to deprivation of combined nitrogen. Vegetative cells provide the heterocyst with fixed carbon to compensate for the latter's lack of PSII, and the heterocysts provide vegetative cells with fixed nitrogen (12, 23). Heterocysts undergo numerous metabolic and morphological modifications to create the microaerobic interior essential for the function of nitrogenase (for a review, see reference 24). They are terminally differentiated cells that do not divide. Approximately every tenth cell of filaments differentiates into a heterocyst when fixed-nitrogen levels are low enough to limit growth. The resulting periodic pattern is one of the oldest known biological patterns, providing an ideal model system for studying the minimal genetic requirements for formation and maintenance of a periodic cell pattern.
PatA is necessary for the generation of a wild-type pattern of heterocysts. In mutant strains that lack a functional patA gene in an otherwise wild-type genetic background, heterocyst differentiation is limited primarily to cells at the filament termini, although rare intercalary heterocysts have been observed (10). Based on its sequence, PatA could be the response regulator of a two-component system or a member of a signaling cascade regulated by phosphorylation. It is unusual in that it contains the CheY-like phosphoacceptor domain at its C-terminal end rather than at the N terminus. PatA is also predicted to contain a putative disrupted helix-turn-helix DNA-binding domain and an N-terminal PATAN (PatA N terminus) domain, which was proposed in a bioinformatics study to mediate protein-protein interactions (11). Although there are many predicted histidine kinases in strain PCC 7120, to date none is known to phosphorylate PatA. Predicted PATAN domains are encoded in the genomes of a variety of environmental bacteria and archaea, where they are associated in proteins with predicted REC, Roadblock, and other signal transduction domains or with helix-turn-helix domains. Genes that encode proteins with PATAN domains are predicted to be involved in processes such as chemotaxis, cell development, and differentiation (11).
PatA appears to promote the activity of HetR, which is considered the master regulator of heterocyst differentiation, while also limiting its accumulation in cells. Deletion of the patA gene suppresses the approximately 3-fold increase in the number of heterocysts formed as a result of ectopic overexpression of hetR (4). It also results in a substantial increase in the level of HetR protein in filaments (17), but as mentioned, fewer cells of the mutant filament differentiate. Epistasis studies suggest additionally that PatA attenuates the negative effects of the two inhibitors of differentiation, PatS and HetN (15).
The patA mutant phenotype can be bypassed by inactivation of two inhibitors of differentiation, PatS (15) and PatU3 (27), or ectopic overexpression of the positive regulator of differentiation, HetF (17). We have proposed that HetF and PatA work together to promote the activity of HetR. HetF is a putative CHF protease (1). As is the case for PatA, inactivation of HetF suppresses the multiple contiguous heterocyst (Mch) phenotype produced by ectopic overexpression of hetR in a wild-type genetic background. Deletion of the hetF gene also results in an increase in the level of HetR in filaments, while decreasing the number of cells that differentiate into heterocysts, in this case preventing it entirely (17, 25). A hetF mutant also has significantly larger cells than those of the wild type, a phenotype that is dependent on functional patA and hetR genes (17). Ectopic overexpression of hetF results in reduced cell size, suggesting that HetF affects the timing of cell division (17).
In the work reported here, we further elucidate the interaction between PatA and HetR, HetN, and PatS in the regulation of heterocyst differentiation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type strain, Anabaena sp. strain PCC 7120, and its derivatives (Table 1) were grown in BG-11 medium as previously described (18). Media were supplemented with neomycin at a concentration of 45 μg ml−1 for selection of replicating plasmids. To induce heterocyst formation, cultures grown to exponential phase (optical density at 750 nm [OD750] of 0.3 to 0.7) were washed twice and resuspended in media lacking combined nitrogen (BG-110) and antibiotics. Anabaena cultures with a plasmid bearing PpetE-patA were created and maintained under copper-deficient conditions as described previously (3). Transcription from the petE promoter was induced by transferring filaments to BG-11 or BG-110, both of which contain copper at a concentration of 0.3 μM. All plasmids were conjugated from Escherichia coli into Anabaena strains as previously described (7). To determine vegetative cell intervals and heterocyst frequencies, the number of vegetative cells between heterocysts and number of each cell type were recorded for 1,500 cells. Reported results represent an average of three replicates. Bright-field and fluorescence microscopy were performed as described previously (3). Confocal micrographs of PatA-GFP localization were obtained using an Olympus FV-1000 confocal system on an IX-81 inverted microscope with FV10ASW1.7 software for 3D reconstruction with appropriate filters.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Anabaena sp. strains | ||
| PCC 7120 | Wild type | Pasteur Culture Collection |
| 7120 PN | PpetE-hetN | 5 |
| UHM101 | ΔpatA mutant | 15 |
| UHM103 | ΔhetR mutant | 3 |
| UHM109 | ΔhetR ΔpatA mutant | 15 |
| UHM114 | ΔpatS mutant | 3 |
| UHM115 | ΔhetN mutant | 3 |
| UHM162 | patA-N270 | This study |
| Plasmids | ||
| pAM504 | Shuttle vector for replication in E. coli and Anabaena; Kmr Nmr | 22 |
| pAM505 | Shuttle vector pAM504 with an inverted multiple cloning site | 22 |
| pAM1956 | Shuttle vector pAM504 with promoterless gfp | 26 |
| pNC101 | Shuttle vector pAM504 with promoterless lacZ | 17 |
| pRL277 | Suicide vector; Smr/Spr | 2 |
| pPpatA-gfp | pAM504 with PpatA-gfp | This study |
| pSMC232 | pAM504 with promoterless gfp; used to make translational fusions | 17 |
| pSSY125 | pAM505 with PpetE-patA | This study |
| pSSY102 | pNC101 with patA promoter fused to lacZ | This study |
| pSSY101 | pNC101 with +25 tsp region fused to lacZ | This study |
| pSSY103 | pNC101 with −164 tsp region fused to lacZ | This study |
| pSSY104 | pNC101 with −305 tsp region fused to lacZ | This study |
| pSSY105 | pNC101 with −614 tsp region fused to lacZ | This study |
| pSSY100 | pAM1956 with +25 tsp region fused to gfp | This study |
| pSSY106 | pAM1956 with −164 tsp region fused to gfp | This study |
| pSSY107 | pAM1956 with −305 tsp region fused to gfp | This study |
| pSSY108 | pAM1956 with −614 tsp region fused to gfp | This study |
| pDR314 | pAM504 with PpatA-patA D292N | This study |
| pDR315 | pAM504 with PpatA-patA D306N | This study |
| pDR316 | pAM504 with PpatA-patA D313N | This study |
| pDR370 | pSMC232 with PpetE-patA | This study |
| pSSY109 | pAM504 with patA-N270 | This study |
| pSSY110 | pRL277 with patA-N270 | This study |
Kmr, kanamycin resistance; Nmr, neomycin resistance; Smr, streptomycin resistance; Spr, spectinomycin resistance.
Construction of plasmids.
The strains and plasmids used in this study are described in Table 1, and primers are listed and referred to here according to the numbering in Table 2. Fusion of the copper-inducible promoter of petE to patA to give PpetE-patA was accomplished by overlap extension PCR (8) with primers 1 to 4. The PpetE-patA PCR product was cloned into pGEM-T (Promega) and subsequently moved into the mobilizable shuttle vector pAM505 (22) as a BamHI-SacI fragment to give plasmid pSSY125. The translational patA-gfp reporter fusion with expression from the petE promoter was created by using primers 39 and 40 to amplify PpetE-patA from pSSY125 and moved as a BamHI-SmaI fragment to pSMC232 to give pDR370. pSMC232 is a replicating plasmid used to make translational fusions to gfp (17).
TABLE 2.
Oligonucleotides used in this study
| Primer no. | Primer name | Sequencea |
|---|---|---|
| 1 | PpetE F | GCTGAGGTACTGAGTACACAGCTAAT |
| 2 | petE-patA F | TAGGAGAACGCATGAAAACACTTCCGATTACTAGAT |
| 3 | patA-petE R | CGGAAGTGTTTTCATGGCGTTCTCCTAACCTGTAG |
| 4 | patA R-BamHI | GGATCCTTACGTAATGTGTTTAAAAATTAC |
| 6 | Race 1 Mut 2 | CTCGCAAAGCATTGAAGACCATACGCG |
| 7 | Race 2 Mut 2 | CAGGTAGTTTTCCAGTAGTGCAAAT |
| 8 | Race 3 Mut 2 | GAAAATTTGTGCCCATTAACATCACC |
| 9 | Region 3 cDNA F | GATTTAGTAGTTACCCAGGTTTGAG |
| 10 | Region 3 RACE | GAGATGTTGTTTGCCAGATTT |
| 11 | PpatA −100 P1 | CAGCAATTTACAGTTAAGTCAACACAGTTAGTC |
| 12 | PpatA −144 P2 | CCAATCTTTCCACTCAGGAGAG |
| 13 | PpatA −173 P3 | CTACACCGTCAGTGCTACTCTCCTGAGT |
| 14 | PpatA −538 P1 | GTCAATAGCAGATTTTCACTAT |
| 15 | −164 Sac1F | ATATATGAGCTCGCCAAGTTAATGAAAGATACAG |
| 16 | −164 KpnI R | TATATAGGTACCCACGCCAATCTTTCCACTCAGGAGAGTT |
| 17 | −305 Sac1F | ATATATGAGCTCGAGCCATTACTTGCGTATCTACC |
| 18 | −305 KpnI R | TATATAGGTACCCTGTATCTTTCATTAACTTGGC |
| 19 | −614 Sac1F | ATATATGAGCTCTCAGGATTGGGGACAAGTG |
| 20 | −614 KpnI R | TATATAGGTACCGGTAGATACGCAAGTAATGG |
| 21 | +25 Sac1F | TATAGAGCTCGTGGAAAGATTGGCGTG |
| 22 | +25 KpnI R | TATATAGGTACCTTACGATAGGGGTTGTATTTTCTG |
| 23 | PpatABamH1F | GAACGGGATCCATCTGGTAATTCTTCTGC |
| 24 | PpatA SmaI R | GAAGTGCCCGGGTGGCGATCGCTCTGTA |
| 25 | +25 BamH1F | TATAGGATCCGTGGAAAGATTGGCGTG |
| 26 | +25 SmaI R | TATATACCCGGGTTACGATAGGGGTTGTATTTTCTG |
| 27 | −164 BamH1F | ATATATGGATCCGCCAAGTTAATGAAAGATACAG |
| 28 | −164 SmaI R | TATATACCCGGGCACGCCAATCTTTCCACTCAGGAGAGTT |
| 29 | −305 BamH1F | ATATATGGATCCGAGCCATTACTTGCGTATCTACC |
| 30 | −305 SmaI R | TATATACCCGGGCTGTATCTTTCATTAACTTGGC |
| 31 | −614 BamH1F | ATATATGGATCCTCAGGATTGGGGACAAGTG |
| 32 | −614 SmaI R | TATATACCCGGGGGTAGATACGCAAGTAATTGG |
| 33 | PpatA BamHI F | ATATAGGATCCCGTTCAACATTCAGGATTGG |
| 34 | PatA Tru SacI | GAGCTCTTATGGATTCTCATCAATACAAAAGATTGTATAG |
| 35 | ATA patA F bgl II | TATATAGATCTCAGAGCGATCGCCATGAAAACAC |
| 36 | ATA patA R | GGTAATAGTTGATATGGATTCTCATCAATACAAAAG |
| 37 | UP ATA F | GAGAATCCATATCCACTATTACCAATTAC |
| 38 | UP ATA R SacI | TATATGAGCTCCAATCAGGGGCAAAAGGGTTTGGGTAAG |
| 39 | PpetE BamH 1 F | ATATAGGATCCCTGAGGTACTGAGTACACAG |
| 40 | patA SmaI R | ATATACCCGGGACGTAATGTGTTTAAAAATTACTTTTAGC |
| 41 | D313IIF | GGTACCTGGTCTATATACGTAGAAGAGGG |
| 42 | D313R | GCATCCCAGGACGAAACAACTCAAC |
| 43 | D292NNF | GCAGTAATTGGTGTTACAAATTCTCTAAAAGCA |
| 44 | D292NNR | GCTTTTAGAGAATTTGTAACACCAATTACTGCA |
| 45 | D306NNF | TACAAAGCCAAATATCATTTTGATCAATGTTG |
| 46 | D306NNR | TTTTACTATAAACCGAAACATGTCTCTTAAA |
| 47 | D313N IIF | GGTACCTGGTCTATATACGTAGAAGAGGG |
| 48 | D313N R | AGTCCGTATAATTGTAACTAGTTTTACTATA |
| 49 | TNL-GFP-F | TTTGGATCCAATCCCGGGGATCGCGTCAGCTAGTAAAGGAGAAGAACTTTTCACTGGA |
| 50 | TNL-GFP-R | ACAGAGCTCTTATTTGTATAGTTCATCCATGCCATG |
| 51 | PatA promoterUP | CCGGACCCGGGCAGGATTGGGGACAAGTGAA |
| 52 | PatA promoterDN | GCGGCCCCGGGCGATCGCTCTGTTATTAAATC |
Oligonucleotides read in the 5′-to-3′ direction.
The promoter of patA was divided into four fragments, each containing one transcriptional start point, and individual fragments were cloned upstream of gfp (primers 15 to 22) or lacZ (primers 25 to 32). The nucleotides included in each fragment contain the promoter region up to the preceding tsp and between 16 and 23 nucleotides downstream of the tsp under investigation. For creation of the gfp transcriptional reporters, the fragments were cloned into pAM1956 as SacI-KpnI fragments to create plasmids pSSY100 and pSSY106 to pSSY108. The lacZ fusions were created by moving the PCR products to pNC101 (17) as a BamHI-SmaI fragment to create plasmids pSSY101 and pSSY103 to pSSY105. The full-length patA promoter was amplified by PCR using primers 23 and 24 and moved to pNC101 as a BamHI-SmaI fragment to make a transcriptional fusion with lacZ and create plasmid pSSY102. A corresponding transcriptional fusion between the full-length promoter region of patA and gfp was made by amplifying the promoter region with primers 51 and 52 and moving the resulting PCR product as a SmaI fragment to pAM1956 to create the plasmid pPpatA-gfp.
Overlap extension PCR was used to create alleles of patA coding for proteins with individual aspartate-to-asparagine substitutions at position 292, 306, or 313. The internal primers (primers 43 to 48) that introduced the point mutations are listed in Table 2 as DXXXN, where XXX designates the position of the residue that was changed. The amplicons were defined with primers D313IIF/D313R (primers 41 and 42) and moved into pAM505 as BamHI-SacI fragments to create plasmids pDR314 to -316. An allele of patA coding for a truncated form of PatA, patA-270N was created by PCR with primers 33 and 34 and cloned onto the replicating plasmid pAM504 to create pSSY109. Plasmid pSSY110 was used to replace the chromosomal copy of patA with patA-N270 by allelic replacement (15). Primers 35 to 38 were used to create the truncated allele of patA by overlap extension PCR, which was moved to pRL277 (2) as a BglII-SacI fragment to create pSSY110.
RNA isolation and RACE.
Filaments of PCC 7120 harvested under nitrogen-replete conditions or 12 h after nitrogen step-down were flash-frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted as previously described (17). Rapid amplification of cDNA ends (RACE) was performed using 3 μg of total RNA and the 5′/3′ RACE kit, 2nd generation, from Roche. The synthesized cDNAs were amplified using primers 6, 11, and 9. Nested primers 7, 12, and 10 were used for RACE PCR, and primers 8, 14, and 13 were used for subsequent reamplification. The products of the RACE procedure were visualized on a 3% agarose gel stained with ethidium bromide and sequenced following gel purification.
β-Galactosidase assays.
Cells were prepared as described by Schaefer and Golden (20) except that the cells were lysed using a Branson1510 cavitator in lieu of treatment with chloroform. β-Galactosidase activity was measured according to Miller (13), and protein concentrations were determined with the Bio-Rad DC protein assay. β-Galactosidase activity was reported as nanomoles of o-nitrophenol min−1 mg protein−1. Assays were conducted in triplicate and reported as an average with one standard deviation.
RESULTS
Induction of patA transcription requires a functional hetR gene.
Transcription of many genes involved in the regulation of heterocyst formation has been shown to be upregulated in developing heterocysts. Wang and Xu recently used a transcriptional GFP fusion to show that transcription from the promoter of patA occurs at a low level in vegetative cells and is induced in proheterocysts (21). To determine how HetR and PatA itself might affect expression of patA, transcription from the native promoter of patA was examined in different genetic backgrounds. Consistent with the previous report, wild-type filaments containing a plasmid with a transcriptional PpatA-gfp fusion displayed a low level of GFP fluorescence in the vegetative cells, and a moderate increase in signal was observed with some proheterocysts 18 h after the removal of combined nitrogen (Fig. 1A). β-Galactosidase activity from whole filaments of the same strain carrying a PpatA-lacZ transcriptional fusion on a plasmid indicated that expression 18 h after removal of combined nitrogen was approximately double that of filaments grown with combined nitrogen (Fig. 2). In contrast, the fluorescence signals from the PpatA-gfp fusion in the ΔhetR strain UHM103 or 7120PN, which has expression of hetR blocked by overexpression of hetN in BG-11 (5), were comparable to those of the wild-type strain with or without a promoterless version of the reporter on a plasmid (data not shown). The low background is presumably due to autofluorescence. There was no evidence of induction of expression in individual cells, in contrast to the wild-type strain. Consistent with the lack of induction of expression indicated by the GFP-reporter fusion, β-galactosidase activity from whole filaments of the same strains carrying the PpatA-lacZ transcriptional fusion on a plasmid was similar to that of negative controls carrying a promoterless copy of the reporter (Fig. 2). We conclude that HetR is necessary for normal levels of expression of patA in vegetative cells and induction of patA transcription in developing heterocysts.
FIG. 1.
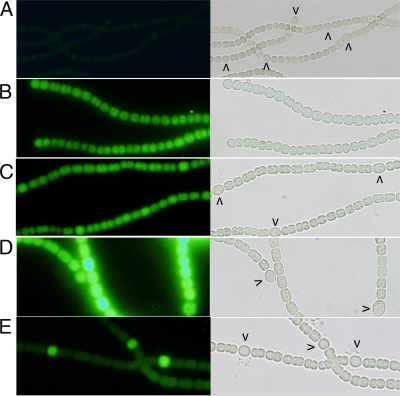
Spatial regulation of the promoter of patA. (A) PCC 7120 with the plasmid pPpatA-gfp. (B) The patA deletion strain UHM101 with plasmid pPpatA-gfp. (C) PCC 7120 with −305 tsp-gfp on plasmid pSSY107. (D) The patA deletion strain UHM101 with −305 tsp-gfp on plasmid pSSY107. (E) PCC 7120 with −614/−645 tsp-gfp fusion on plasmid pSSY108. Fluorescence (left) and bright-field (right) micrographs were taken 18 h after removal of combined nitrogen. Microscope and camera settings were identical for all fluorescence micrographs. Carets indicate proheterocysts.
FIG. 2.
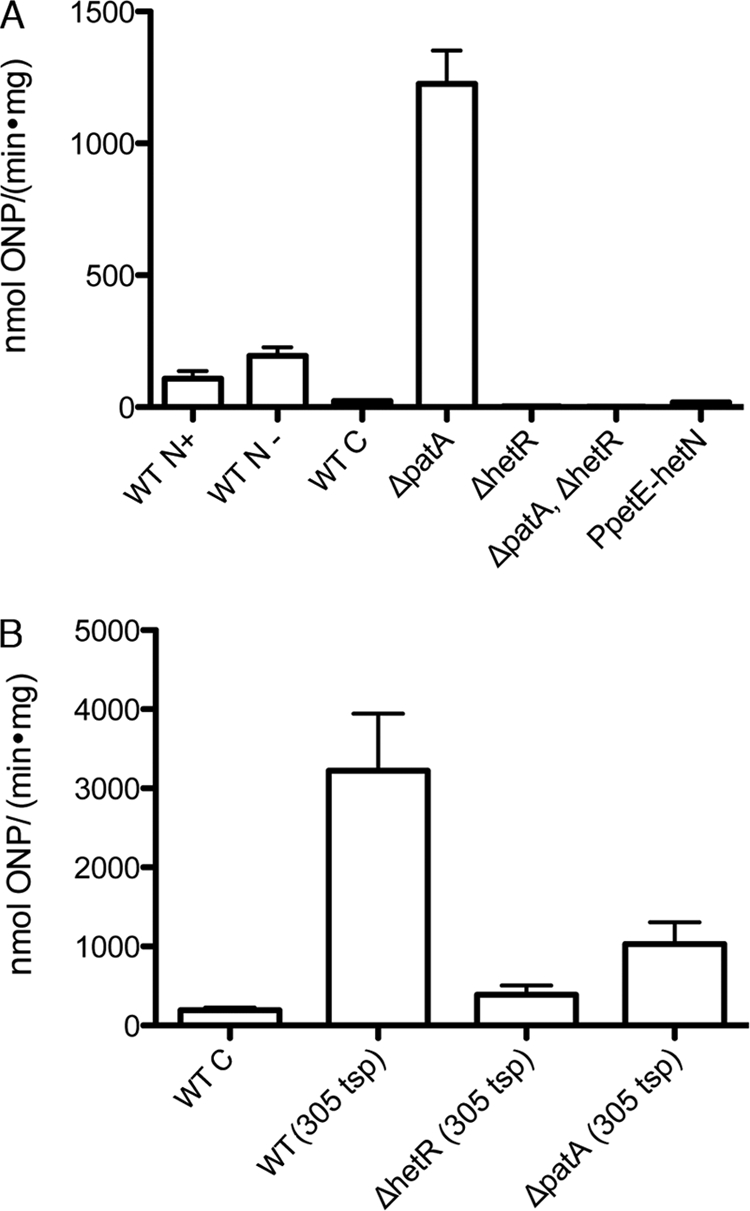
Levels of transcription from the promoter of patA. (A) β-Galactosidase activity from strains with the indicated genetic backgrounds (WT indicates wild type) carrying either pNC101 (WT C), which serves as a control and has a promoterless copy of lacZ, or pSSY102 (the remaining strains), which has a transcriptional fusion between the full promoter region of patA and lacZ. ONP, o-nitrophenol. (B) β-Galactosidase activity from strains with the indicated genetic backgrounds carrying a transcriptional fusion between the indicated tsp region and lacZ. The wild-type strain PCC 7120 carrying pNC101 (WT C) serves as a negative control. Both sets of assays were conducted with filaments grown either in BG-11 (WT N+), which contains nitrate, or 18 h after removal of combined nitrogen (the remaining strains).
In contrast to the hetR deletion strain, in the patA deletion strain UHM101, the fluorescence signal from the PpatA-gfp reporter was markedly increased compared to that of the wild type in both heterocysts and vegetative cells, which had equal levels of fluorescence (Fig. 1B). Consistent with the GFP results, β-galactosidase activity from whole filaments of the ΔpatA strain carrying the PpatA-lacZ transcriptional fusion was more than 10-fold higher than that of the wild-type strain. After an initial decline, activity increased about 5-fold 24 h after removal of combined nitrogen, when mature heterocysts are present, suggesting that transcription from the patA promoter is negatively autoregulated (directly or indirectly) (Fig. 2). Deletion of patA has been reported to markedly increase levels of HetR in most cells of filaments (17). To determine if increased transcriptional activity in the patA mutant is the direct result of a lack of PatA, or if HetR is required, β-galactosidase activity from filaments of the ΔpatA ΔhetR double mutant strain UHM109 carrying the PpatA-lacZ transcriptional fusion was measured. Levels of β-galactosidase activity in the double mutant strain resembled those of the ΔhetR single mutant, indicating that HetR is required for the increased transcription observed with the promoter of patA in a patA mutant (Fig. 2).
Transcriptional start sites of patA.
To characterize the promoter region of patA, RACE was used to identify putative transcriptional start points (tsps). cDNA corresponding to mRNA starting at positions +25, −305, −614, and −645 relative to the putative translational start site of patA were detected. The cDNAs starting at positions +25, −614, and −645 were detected in cultures grown in both the presence or absence of combined nitrogen, while cDNA corresponding to a putative −305 tsp was detected only in filaments 18 h after the removal of combined nitrogen (Fig. 3). cDNA starting at position −164 was also observed with some cultures, in both the presence and absence of combined nitrogen, but not in other replicate cultures.
FIG. 3.
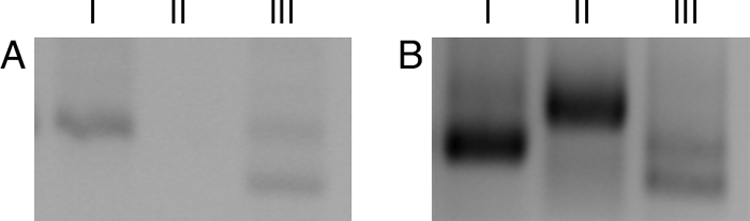
Transcription start points of patA. RACE products from filaments grown either in BG-11 (A), which contains nitrate, or 12 h after removal of combined nitrogen (B). Lane I, RACE product corresponding to cDNA starting at position +25 relative to the translational start site of patA generated with primers 6, 7, and 8; lane II, RACE product corresponding to cDNA starting at position −305 generated with primers 11, 12, and 13; lane III, RACE products corresponding to cDNA starting at position −614 (bottom) and −645 (top) generated with primers 9, 10, and 14.
To examine the spatial and temporal regulation of patA transcription and determine which of the cDNAs generated during RACE is likely to correspond to start points of transcription, regions of DNA upstream of the 5′ end of a given cDNA and extending to the third nucleotide downstream of that cDNA were fused to either gfp or lacZ as indicated in Fig. 4. Each fusion was introduced on a plasmid into the wild type, the patA deletion strain, and the hetR deletion strain. Levels of fluorescence from GFP or β-galactosidase activity were similar to those of the negative controls for both sets of +25 and −164-gfp and -lacZ fusions, respectively, under all conditions of growth and in all genetic backgrounds, suggesting that the cDNAs detected during RACE may not correspond to tsps but instead may have been generated from processed mRNAs (data not shown).
FIG. 4.
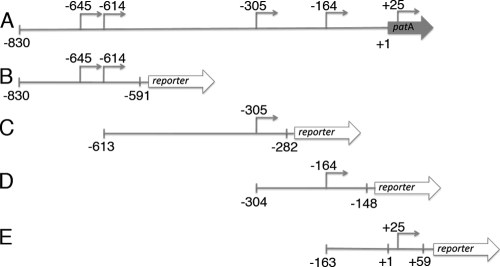
Schematics of reporter fusions made with 5′-end regions of patA transcripts. (A) The patA promoter and coding region with locations of the cDNA start sites indicated by RACE. (B) The −645/−614 tsp-reporter fusion. (C) The −305 tsp-reporter fusion. (D) The −164 tsp-reporter fusion. (E) The +25 tsp-reporter fusion. Arrows indicate potential tsps, and vertical hash marks indicate the boundaries of sequences from the promoter region of patA that were included in each of the fusions. Numbers are relative to the translational start site of patA.
The RACE product corresponding to a putative −305 tsp was detected only from RNA isolated after the removal of combined nitrogen. Fluorescence from the −305 tsp-gfp fusion was detected in both vegetative cells and heterocysts at equal levels in the wild type, was markedly increased in the patA deletion strain, and was similar to background levels in the hetR deletion strain (Fig. 1C and D and data not shown). Levels of fluorescence from −305 tsp-gfp in the wild type were similar to that from the full-length patA-promoter fusion in the patA deletion strain UHM101. β-Galactosidase activity from the wild-type strain carrying the −305 tsp-lacZ fusion in the absence of combined nitrogen was over 10-fold higher than that of the full promoter fusion in the same strain under the same conditions (Fig. 2B). β-Galactosidase activity from the −305 tsp-lacZ fusion in the patA deletion strain, although markedly higher than that from the wild-type strain carrying the full-length promoter region, was less than that from the same fusion in the wild-type strain, which is inconsistent with the GFP results (Fig. 2B). This discrepancy may be explained by the apparent lethality or severe retardation of growth caused by high levels of expression of lacZ in PCC 7120 and its derivatives and the consequent isolation of strains with putative mutations that downregulate expression (16). β-Galactosidase activity from the −305 tsp-lacZ fusion was markedly decreased in the hetR deletion strain (Fig. 2B). The tsp located at −305 thus appears to account for the HetR-dependent induction of transcription in the full-length promoter region of patA and increased levels of transcription in a patA deletion strain.
Fluorescence from the −614/−645 tsp-gfp fusion was observed in both vegetative cells and heterocysts, but fluorescence from heterocysts was noticeably brighter (Fig. 1E). Levels of fluorescence from −614/−645 tsp-gfp fusions in patA and hetR deletion strains were comparable to that from the same cell type in the wild-type strain carrying the same fusion (data not shown). The tsps located at −614/−645 appear to account for the upregulation of transcription from the full-length promoter region of patA in proheterocysts and heterocysts. In addition, transcriptional reporter activity from both the −305 and −614/−645 tsps was higher than that for the full-length promoter, suggesting that in each case an element that limits transcript levels or translation from the full-length promoter is not present in these reporter constructs.
Overexpression of patA causes aberrant vegetative cell morphology in a hetR-dependent manner.
It has been proposed that PatA promotes heterocyst differentiation by attenuating the activity of inhibitors of differentiation, PatS and HetN (15). Thus, it was predicted that the phenotype of a strain overexpressing patA would resemble the phenotype of a strain overexpressing hetR or the phenotype of strains with either patS or hetN deleted, e.g., multiple contiguous heterocysts. However, overexpression of patA from a copper-inducible promoter (PpetE) on a replicating plasmid produced dramatic changes in the morphology of vegetative cells in an otherwise wild-type genetic background, in both complete media and media lacking combined nitrogen; the cells were enlarged, and the cell morphology was aberrant (Fig. 5A and B). Disruptions in the division plane were evident in several cells along the filament, suggesting that patA overexpression affects cell polarity. To determine what role HetR, PatS, or HetN may have in the aberrant morphology caused by overexpression of patA, the replicating plasmid containing PpetE-patA was conjugated into strains with these genes individually deleted. The aberrant vegetative cell morphology was observed with strains UHM114 and UHM115, which lack the patS and hetN gene, respectively, in media with and without combined nitrogen (data not shown). However, when patA was overexpressed in the hetR deletion strain, vegetative cell morphology was similar to that of the parent strain (Fig. 5C). Interestingly, a change in cell size and shape was also seen in a hetF deletion strain, a phenotype that is dependent on functional copies of both hetR and patA (17).
FIG. 5.
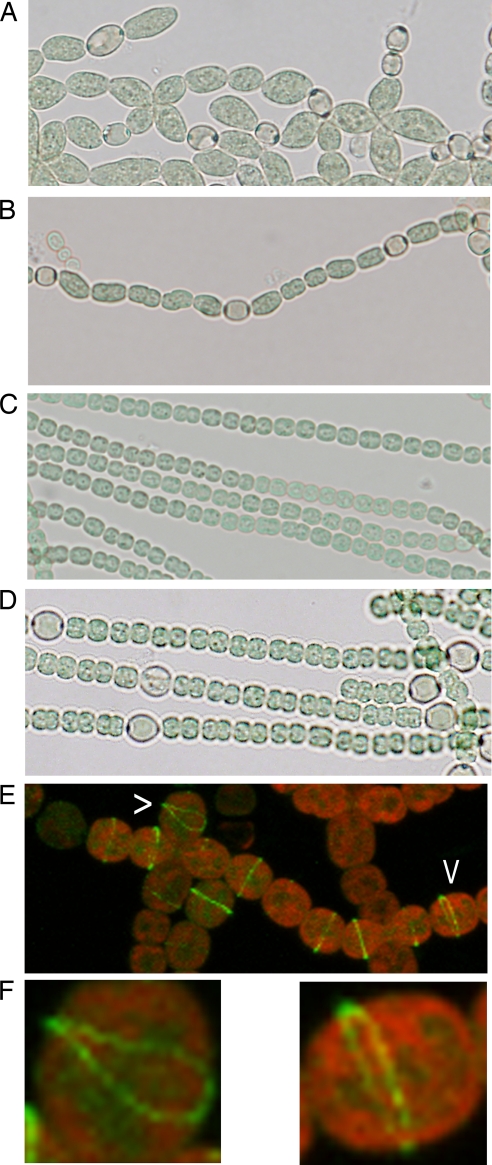
Ectopic expression of patA and localization of PatA-GFP. Strain PCC 7120 (A and B) or the hetR deletion strain UHM103 (C) with the PpetE-patA on plasmid pSSY125 showing altered vegetative cell morphology (A), a reduced number of vegetative cells between heterocysts (B), or the requirement of hetR for either effect (C). (D) Strain UHM162, which has the normal copy of patA replaced by patA-N270. (E) Confocal microscopy of PCC 7120 carrying a patA-gfp translational fusion on plasmid pDR370. (F) Enlargements of cells indicated by carets in panel E. Photomicrographs were taken 48 h after removal of combined nitrogen.
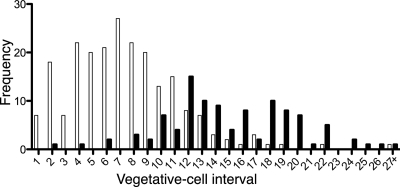
In addition to the change in vegetative cell morphology, overexpression of patA in the wild type results in a reduction in the number of vegetative cells between heterocysts (vegetative cell interval) and an increase in the number of heterocysts that differentiate adjacent to another heterocyst, even in a medium containing nitrate. The strain overexpressing PatA in a wild-type genetic background had an average of 15.5% heterocysts compared to 7.6% for the wild type and an average of 7.4 vegetative cells between heterocysts compared to 15.0 for the wild type 48 h after removal of combined nitrogen (Fig. 6). In medium containing nitrate, the strain overexpressing patA in a wild-type genetic background had an average of 5.5% heterocysts compared to 1.7% for the wild type with a control plasmid under the same conditions.
FIG. 6.
Heterocyst patterning with ectopic expression of patA. The frequency of the indicated vegetative cell intervals, defined as the number of vegetative cells between two heterocysts, is shown for PCC 7120 with the PpetE-patA fusion on plasmid pSSY125 (empty bars) and PCC 7120 with the control plasmid pAM505 (filled bars) 48 h after transfer to BG-110.
PATAN domain of PatA alone is sufficient for heterocyst differentiation.
The amino acid sequence of PatA suggests that it functions as the response regulator of a two-component system with an N-terminal PATAN domain and a C-terminal phosphoacceptor domain, but it has been suggested that the phosphoacceptor domain may not be functional (11). Alignment of PatA to CheY revealed three aspartate residues most likely to act as sites of phosphorylation: D292, D306, and D313. To investigate whether any of these aspartic acids is required for the formation of intercalary heterocysts, alleles of patA encoding conservative substitutions to asparagine were constructed on a replicating plasmid and tested separately for their abilities to restore the formation of intercalary heterocysts to a patA deletion strain. Strains with each of the three substitution alleles under the control of the native patA promoter complemented the patA deletion mutant to the same extent as the wild-type patA gene to give a normal pattern of heterocysts (data not shown). These results indicate that none of the three aspartates of PatA is necessary for promotion of heterocyst formation.
To determine whether the PATAN domain alone is sufficient for promotion of differentiation, the N-terminal portion of patA encoding only the 270-amino acid effector domain of PatA (patA-N270) was tested for its ability to complement a ΔpatA strain when on a replicating plasmid. Morphologically distinct heterocysts were observed with this strain, but they did not appear until 72 h after removal of combined nitrogen, compared to 24 h for the wild type (Fig. 5D). Although vegetative cell intervals were relatively consistent, the intervals were typically 18 to 20 cells, nearly twice that in the wild type. These results indicate that the PATAN domain alone is capable of inducing heterocyst formation but not with the wild-type frequency of heterocysts. To avoid the possible effects of having a regulatory gene on a multicopy plasmid, the native copy of patA in the chromosome of strain PCC 7120 was replaced with patA-N270. The frequency and vegetative cell intervals in the resulting strain were similar to those when patA-N270 was on a plasmid. However, the chromosomal construct also promoted the formation of heterocysts when filaments were grown in BG-11, which contains the combined-nitrogen source, nitrate.
PatA localizes to the division septum.
To examine localization of PatA within cells, a patA-gfp translational fusion expressed from the copper-inducible promoter of petE was introduced into the wild-type strain. Fluorescence from PatA-GFP, although present at a low level throughout most cells, was concentrated in a ring at the center of dividing cells (Fig. 5E). The ring structure resembled that observed for an FtsZ-GFP reporter in strain PCC 7120 (19).
DISCUSSION
The pattern of heterocysts in Anabaena sp. strain PCC 7120 presumably optimizes growth of the organism under conditions of low fixed nitrogen. Heterocysts do not divide, so differentiation of an excessive number of heterocysts would remove dividing and CO2-fixing cells from the population, whereas differentiation of an insufficient number of cells would not supply adequate amounts of fixed nitrogen and would limit the growth of vegetative cells. HetR is thought to be the master regulator of heterocyst differentiation, and several proteins, such as NtcA, NrrA, HetN, and PatS, influence the production or activity of HetR (5, 6, 9, 14). Regulation by each of these proteins is part of a feedback loop, either positive or negative, and either direct or indirect. Here we demonstrate the existence of another feedback loop involving HetR, a positive one with PatA. Induction of expression of hetR in intercalary cells of filaments requires a functional patA gene (4), and as shown here, induction of expression of patA in a pattern that anticipates that of the heterocysts requires a functional hetR gene. The existence of multiple feedback loops that intersect at hetR reinforces the importance of HetR activity in both heterocyst patterning and the commitment to differentiation.
The −271 tsp of hetR requires a functional patA gene for full induction of expression, and expression from this tsp is regulated spatially to give a pattern of expression that anticipates the pattern of heterocysts (16). Conversely, the patA tsp-gfp fusions used here indicate that expression from the hetR-dependent −305 tsp of patA is not regulated spatially in filaments. Instead, hetR was necessary for upregulation of patA from this tsp in all cells of the filament, suggesting that the mutual positive interaction of hetR and patA for induction of transcription is more complex than simple reinforcement to amplify an initial increase in expression of one of the two genes.
In contrast to transcription from the −305 tsp of patA, combined expression from the −645 and −614 tsps was spatially regulated. The spacing between these two tsps is similar to that between the −728 and −696 tsps of hetR, 31 versus 32 bp, respectively (and some variability was observed with these two tsps of patA). The −728 and −696 tsps of hetR have been shown to be regulated directly by NrrA. Expression of nrrA has been shown to be regulated spatially, so regulation of the −645/−614 tsps of patA by NrrA could account for a pattern of transcription from these tsps. However, a search of the sequence upstream of the −645 tsp of patA failed to identify a potential binding site for NrrA, which consists of an 8-bp direct repeat separated by 8 additional nucleotides (6).
For each of the −645/−614 and −305 tsp fusions, reporter activity was substantially greater than that of corresponding fusions using the full-length promoter region of patA, suggesting that sequences downstream of these tsps are involved in either limiting initiation of transcription from the full-length promoter or limiting the half-life or translation of the full-length transcript. We can only speculate what sequences may be involved but suspect that cDNAs beginning at positions −164 and +25 identified by RACE may be informative in this regard. Position +25 is immediately after a 12-bp sequence that comprises the downstream half of an inverted repeat, and position −164 occurs within a 16-bp sequence that forms the downstream half of a second inverted repeat (10). The cDNAs starting at these positions may represent truncated mRNAs, indicative of targeted mRNA degradation. If so, the lack of these sequences in the −645/−614 and −305 tsp fusions, which contain only three bp from patA downstream of the tsp, would be consistent with the increased reporter activity of the tsp fusions relative to fusions with the full-length promoter.
PatA is predicted to be comprised of two functional domains, a CheY-like phosphoacceptor domain typical of the response regulators in two-component systems and a recently described PATAN domain, for which PatA is the namesake (10, 11). It was speculated that phosphorylation at aspartate 313 may be involved in the regulation of PatA activity. However, alleles of patA on a plasmid-encoding protein with asparagine in place of D292, D306, or D313 all complemented a patA mutation, suggesting that either phosphorylation of one or more of these aspartates is not required for PatA function or that having the alleles on a multicopy plasmid overcame the need for activation by phosphorylation. Consistent with the former possibility, an allele of patA encoding only the N-terminal 270 amino acids was capable of promoting intercalary heterocyst formation in a patA mutant. However, it did not fully complement the mutation. The number and pattern of heterocysts differed from that of the same strain carrying the full-length allele, so the possibility remains that phosphorylation is required for full PatA activity.
Overexpression of patA resulted in both the enlargement of individual vegetative cells and a decrease in the number of vegetative cells between heterocysts. Changes in the expression of several genes are known to alter heterocyst number and spacing, so it is difficult to attribute the effect of overexpression of patA to an effect on any one of them. Enlarged cells are observed in a hetF mutant, but the cells have a more regular, consistent shape in the hetF mutant (17), compared to the irregular shape of cells in the strain overexpressing patA, which varied considerably in the degree of aberrant morphology of individual cells. This variation may reflect variation in expression levels of patA from cell to cell, whereas loss of HetF activity would be consistent in all cells of a hetF mutant. The enlarged cell phenotype and alterations in the division plane suggest that overexpression of patA delays cell division and affects its orientation. Localization of PatA-GFP to sites of division in a ring structure reminiscent of that formed by FtsZ suggests that extra PatA in the cell may have a direct effect on the timing and location of cell division. However, the need for a functional hetR gene in the aberrant cell morphology phenotype suggests that the mechanism behind the phenotype is more complex than the activity of PatA alone. In addition, caution should be used in interpreting phenotypes generated by overexpression of alleles. The observed effects on cell division may be artifactual and instead be indicative of regulation of PatA by cell division proteins. Inhibition of cell division prior to induction of differentiation prevents the formation of heterocysts (19). Localization of PatA to division septa may constitute a molecular link between the two processes.
Acknowledgments
We thank Guntram Christiansen, Hongwei Li, and members of the Li lab for help with RNA work.
This work was supported by grants MCB-0343998 and IOS-0919878 from the National Science Foundation.
Footnotes
Published ahead of print on 9 July 2010.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2002. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins 46:355-367. [DOI] [PubMed] [Google Scholar]
- 2.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 3.Borthakur, P. B., C. C. Orozco, S. S. Young-Robbins, R. Haselkorn, and S. M. Callahan. 2005. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 57:111-123. [DOI] [PubMed] [Google Scholar]
- 4.Buikema, W. J., and R. Haselkorn. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. U. S. A. 98:2729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan, S. M., and W. J. Buikema. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40:941-950. [DOI] [PubMed] [Google Scholar]
- 6.Ehira, S., and M. Ohmori. 2006. NrrA directly regulates expression of hetR during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 188:8520-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. U. S. A. 101:4848-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang, J., L. Scappino, and R. Haselkorn. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. U. S. A. 89:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova, K. S., E. V. Koonin, R. Haselkorn, and M. Y. Galperin. 2006. Cyanobacterial response regulator PatA contains a conserved N-terminal domain (PATAN) with an alpha-helical insertion. Bioinformatics 22:1297-1301. [DOI] [PubMed] [Google Scholar]
- 12.Meeks, J. C., C. P. Wolk, W. Lockau, N. Schilling, P. W. Shaffer, and W.-S. Chien. 1978. Pathways of assimilation of [13N]N2 and 13NH4+ by cyanobacteria with and without heterocysts. J. Bacteriol. 134:125-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 14.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 44:1377-1385. [DOI] [PubMed] [Google Scholar]
- 15.Orozco, C. C., D. D. Risser, and S. M. Callahan. 2006. Epistasis analysis of four genes from Anabaena sp. strain PCC 7120 suggests a connection between PatA and PatS in heterocyst pattern formation. J. Bacteriol. 188:1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajagopalan, R., and S. M. Callahan. 2010. Temporal and spatial regulation of the four transcription start sites of hetR from Anabaena sp. strain PCC 7120. J. Bacteriol. 192:1088-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risser, D. D., and S. M. Callahan. 2008. HetF and PatA control levels of HetR in Anabaena sp. strain PCC 7120. J. Bacteriol. 190:7645-7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Risser, D. D., and S. M. Callahan. 2007. Mutagenesis of hetR reveals amino acids necessary for HetR function in the heterocystous cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:2460-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakr, S., R. Jeanjean, C.-C. Zhang, and T. Arcondeguy. 2006. Inhibition of cell division suppresses heterocyst development in Anabaena sp. strain PCC 7120. J. Bacteriol. 188:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer, M. R., and S. S. Golden. 1989. Differential expression of members of a cyanobacterial psbA gene family in response to light. J. Bacteriol. 171:3973-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, Y., and X. Xu. 2005. Regulation of hetC of genes required for heterocyst differentiation and cell division in Anabaena sp. strain PCC 7120. J. Bacteriol. 187:8489-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei, T.-F., R. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 176:4473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolk, C. P. 1968. Movement of carbon from vegetative cells to heterocysts in Anabaena cylindrica. J. Bacteriol. 96:2138-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria, vol. 1. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 25.Wong, F. C. Y., and J. C. Meeks. 2001. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J. Bacteriol. 183:2654-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon, H.-S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, W., Y. Du, I. Khudyakov, Q. Fan, H. Gao, D. Ning, C. P. Wolk, and X. Xu. 2007. A gene cluster that regulates both heterocyst differentiation and pattern formation in Anabaena sp. strain PCC 7120. Mol. Microbiol. 66:1429-1443. [DOI] [PubMed] [Google Scholar]