Abstract
Advances in bacterial cell biology have demonstrated the importance of protein localization for protein function. In general, proteins are thought to localize to the sites where they are active. Here we demonstrate that in Escherichia coli, MurG, the enzyme that mediates the last step in peptidoglycan subunit biosynthesis, becomes polarly localized when expressed at high cellular concentrations. MurG only becomes polarly localized at levels that saturate MurG's cellular requirement for growth, and E. coli cells do not insert peptidoglycan at the cell poles, indicating that the polar MurG is not active. Fluorescence recovery after photobleaching (FRAP) and single-cell biochemistry experiments demonstrate that polar MurG is dynamic. Polar MurG foci are distinct from inclusion body aggregates, and polar MurG can be remobilized when MurG levels drop. These results suggest that polar MurG represents a temporary storage mechanism for excess protein that can later be remobilized into the active pool. We investigated and ruled out several candidate pathways for polar MurG localization, including peptidoglycan biosynthesis, the MreB cytoskeleton, and polar cardiolipin, as well as MurG enzymatic activity and lipid binding, suggesting that polar MurG is localized by a novel mechanism. Together, our results imply that inactive MurG is dynamically sequestered at the cell poles and that prokaryotes can thus utilize subcellular localization as a mechanism for negatively regulating enzymatic activity.
Cells need ways to deal with having more of a specific protein than they need. Left unchecked, excess protein can be toxic to the cell and interfere with essential processes. In prokaryotes, a common mechanism for dealing with excess protein is degradation (30). Bacterial proteases can break down proteins, salvaging amino acids to produce new protein. This process costs time and energy, especially if the protein being degraded is essential and will need to be resynthesized later. Excess protein can also aggregate into insoluble inclusion bodies. In inclusion bodies, proteins are generally misfolded, and though in some cases these proteins can be refolded (24, 35), inclusion body proteins are not readily accessible for use by the cell (11). A potential alternative strategy for dealing with excess protein is to temporarily store the protein in an inactive form that can later be dynamically remobilized when needed. Here we propose that Escherichia coli uses subcellular localization of MurG to accomplish such dynamic storage.
MurG is an essential, membrane-associated N-acetylglucosaminyl transferase involved in catalyzing the final step of peptidoglycan subunit biosynthesis (4, 21). In E. coli, the peptidoglycan cell wall determines both cell shape and growth rate (17). During growth and division, E. coli cells add new peptidoglycan both along the lateral cylindrical portion of the cell and at the division plane, but no new peptidoglycan is added at the cell poles (10). Previous efforts to study the localization of MurG have found that MurG localizes to the cell periphery and division plane in E. coli (25). In this study, we demonstrate that E. coli MurG also localizes to the cell poles in a concentration-dependent manner. We find that the polar MurG represents a dynamic pool of excess protein, suggesting that polar accumulation represents an accessible form of temporary storage.
MATERIALS AND METHODS
Media.
E. coli strains were grown in Gutnick minimal medium containing 4.7 g KH2PO4, 13.5 g K2HPO4, 1 g K2SO4, 0.1 g MgSO4·7H2O, 0.5 g NH4Cl, and 4 g glucose per liter. Antibiotics were used at the following concentrations: 50 μg carbenicillin/ml and 30 μg kanamycin/ml.
Strains.
The strains used in this study are listed in Table 1. E. coli wild-type strain NCM3722 was a gift from the laboratory of J. Rabinowitz (Princeton University). The temperature-sensitive lambda red recombination-expressing strain DY378 and the Keio collection strains JW6464 (cls〈〉kan) and JW5952 (ybhO〈〉kan) were gifts from the laboratory of T. Silhavy (Princeton University). Removal of antibiotic cassettes using pCP20 (6), lambda red recombineering (9, 38), and P1-mediated transduction (31) were done using previously described methods. Strain ZG249 (DY378 murG〈〉kan/pAM2) was constructed by first using electroporation to introduce pAM2 into DY378 and then using recombineering to replace the chromosomal murG locus with a kanamycin resistance cassette obtained by PCR from plasmid pKD4. Strains ZG250 (NCM3722 murG〈〉kan/pAM2), ZG294 (NCM3722 murG〈〉kan/pAM8), ZG295 (NCM3722 murG〈〉kan/pAM9), ZG296 (NCM3722 murG〈〉kan/pAM10), and ZG297 (NCM3722 murG〈〉kan/pAM11) were constructed using electroporation to introduce pAM2, pAM8, pAM9, pAM10, or pAM11, respectively, into NCM3722 and then using P1 transduction to introduce murG〈〉kan from ZG249. Strain ZG251 (BW25113 Δcls ybhO〈〉kan) was constructed using pCP20 to remove kan from JW6464 and then using P1 transduction to introduce ybhO〈〉kan from JW5952. Strain ZG252 (BW25113 Δcls ΔybhO) was constructed using pCP20 to remove kan from ZG251.
TABLE 1.
Strains used in this study
| Strain | Relevant feature(s) | Source or reference |
|---|---|---|
| NCM3722 | E. coli K-12 wild type | 32 |
| DY378 | W3110 λcI857 Δ(cro-bioA) | 38 |
| ZG249 | DY378 murG〈〉kan/pAM2 | This study |
| ZG250 | NCM3722 murG〈〉kan/pAM2 | This study |
| JW6464 | BW25113 cls〈〉kan | 3 |
| JW5952 | BW25113 ybhO〈〉kan | 3 |
| ZG251 | BW25113 Δcls ybhO〈〉kan | This study |
| ZG252 | BW25113 Δcls ΔybhO | This study |
| ZG294 | NCM3722 murG〈〉kan/pAM8 | This study |
| ZG295 | NCM3722 murG〈〉kan/pAM9 | This study |
| ZG296 | NCM3722 murG〈〉kan/pAM10 | This study |
| ZG297 | NCM3722 murG〈〉kan/pAM11 | This study |
Plasmids.
The plasmids used in this study are listed in Table 2, and the oligonucleotide sequences used are listed in Table S1 in the supplemental material. Plasmids pTrc99a, pKD4, and pCP20 were gifts from the laboratory of T. Silhavy (Princeton University). pAM1 was constructed by amplifying the Gateway cassette with C-terminal mCherry fusion from plasmid gXRC using the primers GWrfpFORSpe and GWrfpREVSpe. pTrc99a was amplified using the primers ptrc99aFwdSpeIMCS and *ptrc99aRevSpeI. Both PCR products were digested with SpeI and ligated together. pAM2 was constructed by amplifying the murG gene from chromosomal NCM3722 DNA using the primers MurG FWD GWY EC and MurG REV GWY EC. The PCR product was moved into pAM1 using Gateway cloning (Invitrogen). pAM4 was constructed using the QuikChange site-directed mutagenesis kit (Stratagene) to modify pAM2 with the primer pair MurG-E268A and Rev-MurG-E268A. pAM7 was constructed by amplifying otsA from chromosomal NCM3722 DNA using the primers OtsA FWD GWY EC and OtsA REV GWY EC. The PCR product was moved into pAM1 using Gateway cloning. pAM8 was constructed by amplifying the N-terminal part of murG out of pAM2 using the primers MurG FWD GWY EC and MurG L79E F82E REV and the C-terminal part of murG out of pAM2 using the primers MurG REV GWY EC and MurG L79E F82E FWD. The two products were used in a SOEing PCR with primers MurG FWD GWY EC and MurG REV GWY EC. This full-length murG product containing the desired mutation was moved into pAM1 using Gateway cloning. pAM9 and pAM10 were constructed by amplifying otsA and ibpA from chromosomal NCM3722 DNA using the primer pairs OtsA FWD GWY EC and OtsA REV GWY EC and IbpA FWD GWY and IbpA REV GWY. The PCR products were moved into plasmid gXGC using Gateway cloning. otsA-GFP and ibpA-GFP were amplified from the resulting Gateway clones using the primers OtsA Fwd SacI/IbpA Fwd SacI and GFP Rev XbaI. The otsA-GFP and ibpA-GFP PCR products and pAM2 were digested with SacI and XbaI and ligated together. pAM11 was constructed by amplifying mraY from chromosomal NCM3722 DNA using the primers MraY Fwd SacI and MraY Rev STOP XbaI. The PCR product and pAM2 were digested with SacI and XbaI and ligated together.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| pKD4 | Template plasmid for gene disruption | 9 |
| pTrc99a | Expression vector; AprPtrc rrnB TtT2ori(pBR322) lacIq | 1 |
| pCP20 | FLP-recombination plasmid: flp bla cat rep101(Ts) | 6 |
| gXRC | Destination vector-cloned GW cassette and mCherry into pXGFP4 | 36 |
| gXGC | Same as gXRC, except mCherry is replaced by GFP | This study |
| pAM1 | rfp fusion vector (pTrc99a-Gateway cassette-mCherry) | This study |
| pAM2 | Ptrc::murG-mCherry bla | This study |
| pAM4 | Ptrc::murG(E268A)-mCherry bla | This study |
| pAM7 | Ptrc::otsA-mCherry bla | This study |
| pAM8 | Ptrc::murG(L79E F82E)-mCherry bla | This study |
| pAM9 | Ptrc::murG-mCherry otsA-GFP bla | This study |
| pAM10 | Ptrc::murG-mCherry ibpA-GFP bla | This study |
| pAM11 | Ptrc::murG-mCherry mraY bla | This study |
Microscopy.
Images were collected on a Nikon 90i microscope equipped with a Nikon Plan Apo 100×/1.4 phase-contrast objective, a Rolera XR cooled charge-coupled device (CCD) camera, and NIS Elements software.
Protein localization.
Strains expressing MurG-mCherry were grown overnight in minimal medium containing carbenicillin for plasmid maintenance. In the case of strains carrying murG〈〉kan, antibiotic selection for the plasmid was not necessary. Overnight cultures were diluted 1:50 into minimal medium containing 0 to 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and grown at 37°C for 2 h, at which point they were spotted onto 1% agarose pads containing minimal medium and IPTG inducer.
Growth curves.
Overnight cultures grown in minimal medium were diluted 1:50 into minimal medium containing 0 to 1 mM IPTG and transferred into a shallow-well 96-well plate. The plate was grown at 37°C with shaking, and at 1-h time intervals, optical density at 600 nm (OD600) and fluorescence intensity were measured using a Perkin-Elmer Wallac EnVision 2103 multilabel reader (Waltham, MA).
FRAP.
For fluorescence recovery after photobleaching (FRAP), photobleaching was performed using a MicroPoint laser system (Photonic Instruments, St. Charles, IL) equipped with an NL100 nitrogen laser (Stanford Research Systems, Sunnyvale, CA). Recovery was measured over 2 min following photobleaching. Images were analyzed using PSICIC (14; http://www.molbio1.princeton.edu/labs/gitai/psicic/psicic.html) and ImageJ (http://rsbweb.nih.gov/ij/). Mean fluorescence intensity was normalized to the maximum intensity for each cell.
Drug treatments.
Overnight cultures grown in minimal medium were diluted 1:50 into minimal medium with or without 0.1 mM IPTG inducer. The cultures were grown at 37°C for 2 h. After 1 h, the cultures without inducer were treated with various concentrations of the drugs. When the 2-h incubation was complete, the cells were spotted onto 1% agarose pads containing minimal medium, 0.1 mM IPTG, and various concentrations of the drugs. The highest drug concentrations tested are as follows: 50 μg/ml cephalexin, 100 μg/ml amdinocillin, 10 μg/ml A22, and 100 μg/ml fosfomycin.
Single-cell biochemical assay for inclusion bodies.
Overnight cultures of strains expressing MurG-mCherry or OtsA-mCherry grown in minimal medium containing carbenicillin were diluted 1:50 into minimal medium containing 0.1 mM IPTG or 1 mM IPTG and grown at 37°C for 2 h. The cells were spotted onto 1% agarose pads containing minimal medium, 0.1 mM IPTG, 1 mg/ml lysozyme, 0.1% Triton X-100, and 10 mM EDTA.
Protein localization in prefilamented cells.
Overnight cultures grown in minimal medium were diluted 1:50 into minimal medium and grown for 1 h at 37°C. At this point, cephalexin was added to a final concentration of 50 μg/ml. The cells were grown for 2 h at 37°C before imaging on 1% agarose pads containing minimal medium, 50 μg/ml cephalexin, and 0.1 mM IPTG.
Flow chamber experiment.
The flow chamber used is model RC-31 (Warner Instruments, Hamden, CT). The cells were adhered to the coverslip using 0.5% polyethylenimine.
NAO staining.
Overnight cultures grown in minimal medium were diluted 1:50 into minimal medium with 0 to 1 mM IPTG inducer and grown at 37°C for 1 h. At this point, nonyl acridine orange (NAO) was added directly to the medium to a final concentration of 200 nM. The cells were then incubated at 37°C for 1 h before imaging on 1% agarose pads containing minimal medium and inducer.
RESULTS
MurG localizes to the poles in a concentration-dependent manner in E. coli.
In the course of investigating the localization dynamics of proteins involved in E. coli cell shape determination, we constructed a plasmid-borne MurG-mCherry fusion that is under the control of the Ptrc IPTG-inducible promoter. We were surprised to discover that the localization pattern of MurG-mCherry changed depending on the level of IPTG inducer used (similar to the pattern shown in Fig. 1). Specifically, this fusion localized diffusely throughout the cell at low induction levels (see the discussion in the supplemental material) but became enriched at both poles at higher induction levels. The cells with bipolar MurG-mCherry still retained some fluorescence along the entire cell, suggesting that MurG was preferentially accumulating at the poles but not excluded from the cell body. Western blot analysis confirmed that increasing IPTG inducer levels correlated well with increased MurG-mCherry protein concentration (data not shown), indicating that the change in MurG localization is a concentration-dependent phenomenon.
FIG. 1.
MurG localizes to the poles when present at high cellular concentrations. Shown are phase-contrast (top) and mCherry fluorescence (bottom) images of ZG250, a strain expressing a functional MurG-mCherry fusion in the absence of the endogenous MurG. The strain was grown for 2 h in minimal medium with the indicated levels of IPTG inducer. Bar, 2 μm.
To investigate whether the MurG-mCherry bipolar localization might be an artifact of the mCherry fusion, we determined if the fusion is functional. We replaced the chromosomal murG with a kanamycin resistance gene, thereby deleting the endogenous murG in the presence of the Ptrc::murG-mCherry. The endogenous murG could only be deleted in the presence of MurG-mCherry, indicating that murG-mCherry is indeed functional for complementing the viability defect of murG. The cells that retained MurG-mCherry as their only copy of MurG also exhibited wild-type morphologies, further confirming the functionality of the fusion protein (Fig. 1). Although the MurG-mCherry was expressed from the Ptrc promoter, the strain was viable even in the absence of inducer, likely due to leaky expression. When the IPTG inducer was added at concentrations of 0.01 mM or lower, MurG-mCherry was diffusely localized (Fig. 1). When the IPTG concentration was increased to 0.1 mM or higher, MurG-mCherry was enriched at the cell poles. At intermediate concentrations (0.02 mM and 0.05 mM), MurG-mCherry was bipolar in some cells and diffusely localized in others (Fig. 1; see the discussion in the supplemental material). These results suggest that MurG accumulates at the poles when present at a high concentration in the cell.
The appearance of polar MurG corresponds to the accumulation of inactive excess protein.
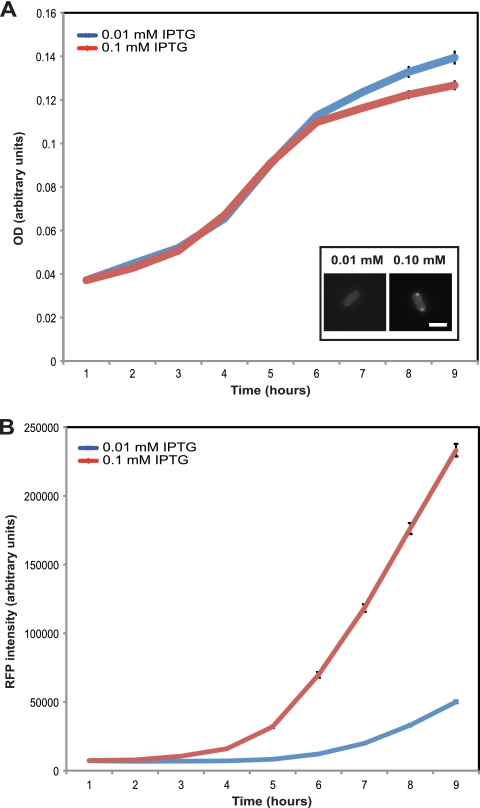
We next sought to determine whether polarly localized MurG represents an active or inactive pool of enzyme. We reasoned that if polar MurG contributes to growth, then cells with increased polar accumulation of MurG would have an increased growth rate. To test this hypothesis, we measured growth curves of cells grown with a range of inducer levels from 0 to 1 mM IPTG. We also measured the fluorescence intensity of these cells to monitor MurG-mCherry levels. Fluorescence intensity and OD600 were measured using a 96-well plate reader at 1-h intervals. As expected, fluorescence intensity increased with increasing level of induction (Fig. 2 B; see Fig. S1B in the supplemental material). The growth rate increased as IPTG levels were raised from 0 to 0.01 mM IPTG (Fig. 2A; see Fig. S1A in the supplemental material). At all IPTG concentrations 0.01 mM and higher, the growth rate remained similar, with perhaps a slight reduction at extremely high induction levels (0.5 and 1 mM). The fact that raising IPTG levels above 0.01 mM increased MurG-mCherry levels but did not increase growth rate indicates that the cellular requirement for MurG is saturated at 0.01 mM IPTG under these growth conditions. Notably, cells with bipolar MurG were only observed at IPTG levels higher than 0.01 mM, suggesting that these cells contain excess MurG. Moreover, the cells with bipolar MurG-mCherry did not grow any faster than cells without polar MurG-mCherry, indicating that the polar protein does not contribute to cell growth and is not actively synthesizing peptidoglycan. Together, these results demonstrate that polar MurG appears only when MurG is in excess and that this polar pool does not actively promote growth.
FIG. 2.
Increased polar accumulation of MurG does not lead to increased growth rate. Growth curves were performed on strain ZG250 grown in minimal medium and 0.01 mM (blue) or 0.1 mM (red) IPTG inducer. A 96-well plate reader was used to measure OD (A) and fluorescence intensity (B) at 1-h time intervals. Error bars represent standard error based on n = 7 measurements. The inset (A, lower right) shows fluorescence micrographs from Fig. 1 for reference. Bar, 2 μm.
MurG polar foci are distinct from inclusion bodies.
The appearance of polar MurG foci at excess MurG concentrations could represent either a transient storage mechanism or a waste removal system, such as inclusion bodies. Consistent with this latter possibility, inclusion bodies have been previously shown to accumulate at the cell poles (19, 28, 37). We thus sought to compare MurG-mCherry to an inclusion body. As a control inclusion body, we focused on a C-terminal fusion of GFP to otsA, which encodes the E. coli homolog of trehalose-6-phosphate synthase. OtsA is typically not expressed during the conditions used for these experiments (12, 16), and its overexpression leads to rapid inclusion body formation, as evidenced by the accumulation of phase-bright foci.
To examine both OtsA and MurG in the same cells, we constructed a double-labeled strain in which OtsA-GFP and MurG-mCherry are coexpressed by the same promoter in a synthetic operon. If MurG accumulates in inclusion bodies, it would be expected to colocalize with OtsA. Instead, when imaged in the presence of 0.1 mM IPTG, MurG-mCherry formed bipolar foci, whereas the cells expressing OtsA-GFP exhibited unipolar foci (Fig. 3 A). Moreover, at the poles that exhibited both OtsA-GFP and MurG-mCherry foci, the two proteins were often found to not colocalize, rather occupying distinct regions of the cell pole (Fig. 3A). As a second marker for inclusion bodies, we also colocalized MurG-mCherry with a GFP fusion to the small heat shock protein (sHsp) IbpA. IbpA has been previously found to colocalize with inclusion bodies in E. coli (19). As was seen with OtsA-GFP, IbpA-GFP formed unipolar foci that did not colocalize with MurG-mCherry when the two fusions were coexpressed (Fig. 3B).
FIG. 3.
Polar MurG foci are distinct from inclusion bodies. (A and B) MurG does not colocalize with inclusion bodies. Cells coexpressing MurG-mCherry and either OtsA-GFP (A) or IbpA-GFP (B) were grown for 2 h in minimal medium with 0.1 mM IPTG inducer before imaging on agarose pads. Shown are an overlay of fluorescence and phase contrast (left), mCherry fluorescence (middle), and GFP fluorescence (right) images. Bar, 2 μm. (C) MurG localizes exclusively to the poles in filamentous cells. Cells coexpressing MurG-mCherry and IbpA-GFP were prefilamented for 2 h in 50 μg/ml cephalexin before imaging on agarose pads containing 0.1 mM IPTG inducer and 50 μg/ml cephalexin. Shown are an overlay (left), mCherry fluorescence (middle), and GFP fluorescence (right) images. The images shown were taken 100 min after the start of induction. The arrows point to nonpolar foci of IbpA-GFP. Bar, 2 μm. (D to G) Polar MurG remains soluble upon cell lysis, while inclusion bodies remain aggregated. Cells expressing MurG-mCherry (D and E) or OtsA-mCherry (F and G) were grown for 2 h in minimal medium with 0.1 mM IPTG inducer before imaging on agarose pads containing minimal medium with lysis buffer. mCherry fluorescence (top) and phase-contrast (bottom) images are shown for cells before (D and F) and after (E and G) lysis. Bar, 2 μm.
It has been previously suggested that polar aggregates such as inclusion bodies accumulate at the poles due to displacement by the nucleoid in a process termed nucleoid occlusion (37). In filamentous cells, nucleiods are spaced at intervals throughout the cell such that aggregates that are affected by nucleoid occlusion tend to localize both at the poles and in foci scattered through the length of the cell (37). We therefore generated long filamentous cells by pretreatment with the division inhibitor cephalexin (27) and subsequently coexpressed MurG-mCherry and IbpA-GFP. We found that MurG-mCherry still localized exclusively to the cell poles, whereas IbpA-GFP formed both polar and nonpolar foci (Fig. 3C). These results confirm that MurG and IbpA do not colocalize and moreover suggest that MurG polar localization is not the result of nucleoid occlusion.
As an additional method to distinguish MurG foci from inclusion bodies, we turned to a single-cell biochemical assay. Inclusion bodies are insoluble protein aggregates which remain aggregated upon cell lysis (11). In contrast, if MurG is being dynamically localized to the poles and not simply aggregating there, the polar MurG foci should dissolve upon lysis. To test this hypothesis, we put unfixed E. coli cells expressing either OtsA-mCherry or MurG-mCherry onto agarose pads made from minimal medium with lysozyme to promote cell lysis. Cells were followed over 3 h by time-lapse microscopy with images taken every 15 min. As expected, OtsA-mCherry fluorescent inclusion bodies remained visible on the pads as insoluble aggregates after the cells had lysed and for the duration of the experiment (Fig. 3F and G). Meanwhile, as soon as the MurG-mCherry cells lysed, they completely lost their fluorescent signal, suggesting that MurG-mCherry remained soluble (Fig. 3D and E). Taken together, the results of the colocalization, cephalexin pretreatment, and cell lysis experiments suggest that MurG polar foci form structures that are distinct from inclusion bodies.
Polar MurG is dynamic and may be remobilized from the poles.
Since MurG polar foci are distinct from inclusion bodies, it is possible that they represent a temporary storage mechanism for excess protein. In that case, MurG would be predicted to be able to dynamically enter and exit the polar foci. To see if MurG protein can be exchanged between existing polar foci and the cytoplasmic pool, we performed FRAP experiments on cells with bipolar MurG foci. We bleached one polar focus of MurG-mCherry and then followed the fluorescence intensities of both the bleached and unbleached foci for 2 min (Fig. 4 A and B). In all cells tested (n = 13), the fluorescence intensity of the bleached pole increased over the 2-min recovery period. This was the case whether the bleached or unbleached focus was brighter initially. By the end of the experiment, the unbleached and bleached foci equilibrated to almost the same level of fluorescence.
FIG. 4.
MurG polar foci are dynamic, while inclusion body polar foci are static. Cells expressing either MurG-mCherry (A and B) or OtsA-GFP (C and D) were grown for 2 h in minimal medium with 0.1 mM IPTG inducer before imaging on agarose pads containing minimal medium and 0.1 mM IPTG. For MurG, cells were bleached at one pole and fluorescence intensity was monitored at both poles. For OtsA, which is unipolar, the focus in a neighboring cell was used as the unbleached pole. (A and C) Shown are mCherry (A) or GFP (C) fluorescence images of a representative cell before bleaching (top), immediately after bleaching (middle), and 2 min after bleaching (bottom). The arrows indicate the bleached pole. Bar, 2 μm. (B and D) Normalized fluorescence intensity was averaged among all cells tested for the unbleached (red) and bleached (blue) pole over the course of the experiment. Error bars represent standard error for n = 13 measurements for MurG and n = 6 measurements for OtsA.
We also performed FRAP experiments on OtsA-GFP. Since OtsA-GFP is unipolar, we could not use a second focus in the same cell for normalization and therefore used an OtsA focus in a neighboring cell as an unbleached control. As with MurG, we bleached one polar focus of OtsA and then followed fluorescence intensity at the bleached focus and the unbleached control for 2 min (Fig. 4C and D). Unlike MurG, the OtsA fluorescence did not recover at the bleached pole in any of the cells examined (n = 6). In every case, the bleached foci always remained well below the fluorescence level of unbleached foci. Thus, MurG fluorescence can recover after photobleaching but OtsA cannot. These FRAP results demonstrate that there is dynamic exchange of MurG protein between the polar foci and the cytoplasmic pool, whereas OtsA polar accumulation is not reversible over the time scales examined.
The ability of MurG to dynamically enter and exit polar foci without aggregating is consistent with the polar foci acting as a dynamic temporary storage mechanism. To test the storage hypothesis more directly, we used a flow chamber to shift cells from conditions under which they grew rapidly and formed polar MurG foci (0.1 mM IPTG) to conditions under which they grew more slowly and lacked polar foci at steady state (0 mM IPTG). The MurG foci did not completely disappear in the period during which we could track these cells, but their fluorescence could be seen to decrease (Fig. 5 A). To account for photobleaching, we normalized the decrease in the fluorescence intensity of the polar foci to the decrease in intensity of the diffuse MurG in the central regions of the cell. We found that the decrease in polar MurG fluorescence was significantly greater than that of photobleaching, suggesting that cells remobilize polar MurG when it is needed (Fig. 5). This result supports our hypothesis that the MurG protein present in polar foci can be remobilized when MurG levels drop.
FIG. 5.
Cells may remobilize polar MurG. Cells expressing MurG-mCherry were grown in minimal medium for 2 h with 0.1 mM IPTG before being transferred to a flow chamber. Medium without inducer was washed into the chamber, and images were taken every 10 min. (A) Fluorescence images of a cell at the indicated time points. Bar, 2 μm. (B) For each cell, the peak fluorescence of the MurG foci (blue) and the average fluorescence of the diffuse midcell portion (green) were determined at each time point and normalized to the value at the first time point (see schematic, lower left). The peak fluorescence (“polar foci”) and diffuse midcell fluorescence (“midcell”) were then averaged over all cells. Error bars represent standard errors of the means for 21 cells.
MurG may be localized to the poles via a novel targeting mechanism.
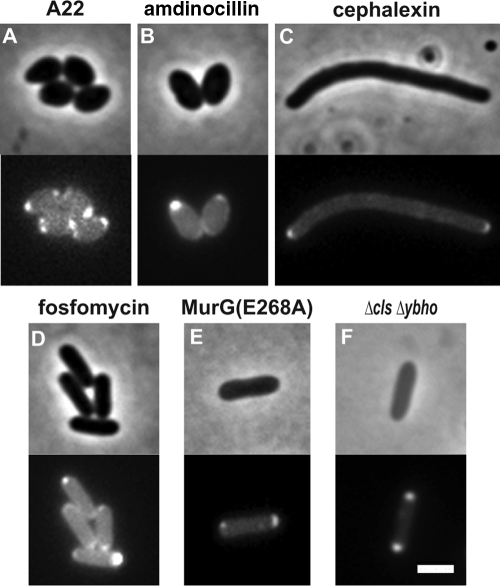
We attempted to address the question of how MurG becomes localized to the cell poles at excess concentration levels. To this end, we imaged MurG-mCherry in the presence of chemical inhibitors of candidate localization proteins. We used inhibitors of the MreB actin cytoskeleton (A22 [13]) and the peptidoglycan biosynthesis enzymes PBP2, PBP3, and MurA (amdinocillin [34], cephalexin [27], and fosfomycin [20], respectively). None of these compounds affected MurG-mCherry localization, regardless of the concentration used or whether treatment was performed before or after MurG-mCherry induction (Fig. 6 A to D). In addition, we examined the possibility that MurG localization is coupled to its enzymatic activity by constructing an E268A mutation in MurG that disrupts its catalytic activity (8). Unlike the mCherry fusion to wild-type MurG, this mutant failed to complement a chromosomal murG deletion, confirming that it is enzymatically inactive. In a wild-type background, this mutant still formed polar foci (Fig. 6E). The fact that an inactive MurG mutant protein localizes to the poles rules out enzymatic activity as the mechanism for MurG localization and is also consistent with our hypothesis that polar MurG is inactive.
FIG. 6.
MurG localization is not mediated by candidate localization pathways. Phase-contrast (top) and mCherry fluorescence (bottom) images are shown for cells expressing wild-type (A to D and F) or mutant (E) MurG-mCherry. Cells were grown for 2 h in minimal medium with 0.1 mM IPTG prior to imaging. (A to D) Cells expressing wild-type MurG-mCherry were spotted on agarose pads containing minimal medium, 0.1 mM IPTG, and either 10 μg/ml A22 (A), 100 μg/ml amdinocillin (B), 50 μg/ml cephalexin (C), or 100 μg/ml fosfomycin (D). The images are representative of the localization of MurG 2 h after exposure to the relevant compound. (E) A cell expressing MurG-mCherry with an engineered E268A point mutation that disrupts enzymatic activity. (F) A cell expressing wild-type MurG-mCherry in a Δcls ΔybhO background. Bar, 2 μm.
Another candidate polar localization determinant is the polarly localized anionic phospholipid, cardiolipin (22, 29). This mechanism was particularly attractive because MurG can bind cardiolipin through an amphipathic helix. Moreover, a previous study found that overexpression of MurG protein correlates with accumulation of phospholipid vesicles at the poles, although this study did not examine the localization of MurG itself (33). We thus examined MurG-mCherry localization in two cardiolipin synthetase mutants (cls and ybhO), as well as in the cls ybhO double mutant. None of these strains had any effect on MurG localization (Fig. 6F). However, it is possible that MurG is still localized by anionic phospholipids that persist in the cls ybhO double mutant. We therefore constructed an L79E F82E double mutant in the lipid binding domain of MurG (15). This MurG lipid interaction double mutant can complement a MurG deletion but had to be expressed at higher levels than wild-type MurG in order to achieve a comparable growth rate. We found that this mutant also had to be expressed at a higher level than wild type before we began to see MurG polar foci (Fig. 7). The lipid binding mutant thus appears to be less active than wild-type MurG, and therefore more of the mutant protein is needed to reach the threshold at which increased levels of MurG will no longer increase the growth rate of the cell. This finding supports our hypothesis that MurG accumulates at the poles when present in excess.
FIG. 7.
MurG may recruit phospholipids to the cell poles. Cells expressing mCherry fusions to either wild-type (WT) MurG or the L79E F82E MurG predicted lipid-binding domain double mutant were grown in minimal medium for 2 h with the indicated level of IPTG inducer. For the second hour, NAO dye was added to a final concentration of 200 nM. Cells were spotted onto agarose pads containing IPTG. Shown are mCherry red fluorescence (top) and NAO green fluorescence (bottom) images. Bar, 2 μm.
We noticed that in contrast to wild-type MurG, the MurG lipid interaction mutant often localized to a single pole in a manner resembling IbpA and OtsA. To confirm that these unipolar foci were not inclusion bodies, we repeated the single-cell lysis experiment with the L79E F82E mutant. Upon cell lysis, MurG(L79E F82E)-mCherry foci disappeared, suggesting that the protein remained soluble and had not formed inclusion bodies (see Fig. S2 in the supplemental material). Thus, MurG localization does not appear to be mediated by any of the major candidate localization pathways, and future unbiased screens will be needed to dissect the mechanism of MurG localization.
Polar MurG can promote polar accumulation of anionic phospholipids.
Since MurG(L79E F82E)-mCherry does not bind anionic phospholipids but can localize to the cell poles, we used this mutant to determine if MurG normally functions to recruit anionic phospholipids to the poles. To label anionic phospholipids such as cardiolipin, we used the NAO dye (23). Cells overexpressing wild-type MurG-mCherry exhibited increased polar NAO staining that colocalized with the polar MurG (Fig. 7). However, cells overexpressing MurG(L79E F82E)-mCherry did not exhibit increased polar NAO staining, even when polar MurG foci were clearly present (Fig. 7). This result suggests that while lipid interaction is not necessary for MurG polar localization, MurG localization may serve to recruit anionic phospholipids to the poles.
DISCUSSION
Sequestration of MurG as a posttranslational regulatory mechanism in E. coli.
We show here that when an E. coli cell has a higher concentration of MurG than is needed for growth, excess MurG is actively sequestered to the poles (Fig. 1 and 2). Why is this pool of MurG retained in the cell? Why is it not degraded to provide raw materials to produce other needed proteins in the cell? It is energetically wasteful for a cell to degrade proteins that then later need to be resynthesized. MurG is an essential protein that is needed for growth. When the cell has enough MurG to grow at its maximal growth rate for the environmental conditions given (temperature, nutrient availability, etc.), the remaining pool of MurG is localized to the poles. Our observations suggest that this polar MurG pool is not active because increased polar MurG does not lead to increased growth rate (Fig. 2). Polar localization might inhibit MurG function by either sequestering MurG from its substrate or by catalytically inactivating the enzyme. We propose that the cell retains excess MurG in an inactive state instead of degrading it so that it can become available for use when environmental conditions become more favorable. Our data support this storage hypothesis (Fig. 5). In addition to saving energy, this dynamic storage mechanism could also allow the cell to respond more quickly to changes in environmental conditions. In this scenario, the cell would be in a primed state, able to rapidly remobilize MurG when growth conditions improve.
Why localize MurG specifically to the cell poles? Having excess MurG diffusing throughout the cell may be toxic to the cell and impair growth rate by interfering with other essential processes. Previous research in E. coli has shown that the poles are inert and excluded from active insertion of new peptidoglycan (10). This would make the poles of the cell an ideal place to localize the inactive pool of MurG. Since there is not active insertion of peptidoglycan at the poles, the inactive MurG would not interfere with this process. Moreover, the MurG substrate may be excluded from the poles, thereby providing a mechanism for rendering the polar MurG inactive. This hypothesis relies on the assumption that the substrate for MurG cannot diffuse to the poles. Alternatively, when MurG is present in excess, it could be using up all of its substrate. However, we found that co-overexpression of MurG with MraY, the upstream enzyme that synthesizes the MurG substrate lipid I (18), did not change MurG localization (data not shown).
Protein sequestration is a well-documented, although perhaps underappreciated, mechanism for protein regulation. In eukaryotes, spatial separation of proteins from their substrates is often used, as in the case of keeping transcription factors out of the nucleus (7). Here we demonstrate that even in the absence of intracellular compartments, bacterial cells may use a similar strategy to spatially sequester MurG. To our knowledge, this work represents the first suggestion of bacteria using subcellularly localized protein sequestration as a transient regulatory mechanism. In future studies, it will prove interesting to determine if this represents a more widespread mechanism.
What is the mechanism of MurG localization?
Our observation that polar MurG is dynamic (Fig. 4) suggests that MurG is actively localized to the poles by either directed transport or a diffusion-capture-type mechanism. Further supporting the active localization of MurG, we found that MurG differs from inclusion bodies in multiple assays of localization and dynamics (Fig. 3 and 4). But how is MurG getting to the poles, and how is it then retained there? Our studies have ruled out most of the obvious candidates, including the peptidoglycan biosynthesis machinery, the MreB actin cytoskeleton, and polar anionic phospholipids like cardiolipin (Fig. 6). Although cardiolipin binding is not necessary for polar MurG localization, polar MurG localization may itself serve to enrich cardiolipin at the poles (Fig. 7). Interestingly, in the specific strain background and under the growth conditions used for this study, NAO staining only revealed polar cardiolipin upon MurG overexpression. Using different E. coli strains and growth conditions, previous reports have found polar cardiolipin without MurG overexpression (23, 29), such that the generality of the role of MurG in cardiolipin localization will require further studies. Thus, while cardiolipin has been previously proposed to localize to the poles in a self-organizing manner (26), additional factors such as MurG may serve to enhance this polarized distribution.
One attractive mechanism for MurG localization is for MurG to diffuse along the cytoplasmic face of the inner membrane and then be captured by a protein that is normally found at the poles of E. coli. This protein could be cytoplasmic, or it could be an inner membrane protein. Large-scale studies of protein interactions in E. coli (2, 5) have not identified any known polar proteins as potential interaction partners for MurG. However, this result does not rule out the possibility that an interaction may have been missed, possibly due to complications associated with studying protein-protein interactions with a membrane-associated protein like MurG. Nevertheless, even in this scenario the question would remain as to how the MurG localization factor itself becomes localized independently of the mechanisms we tested here. Thus, our results suggest that MurG may be directly or indirectly localized by a previously unappreciated mechanism. This discovery will motivate future screens geared toward uncovering this elusive mechanism.
In summary, we find that excess MurG is localized to the poles in an inactive yet dynamic form. We propose here that sequestration of proteins in an inactive pool is a mechanism that prokaryotic cells can use to posttranslationally regulate protein activity. This would be more time and energy efficient than degrading the proteins and resynthesizing them again when needed. Now that this phenomenon has been observed, future studies will help understand its mechanism and utility for both MurG in particular and other proteins in general.
Supplementary Material
Acknowledgments
We are grateful to members of the Silhavy and Rabinowitz labs for reagents and helpful discussions and to Eric Chen for generating the gXGC plasmid. We also thank the members of the Gitai lab, and in particular Kim Cowles Eric Klein, and Sven van Teeffelen, for helpful discussions and critical reading of the manuscript.
Z.G. is supported by funding from grant DE-FG02-05ER64136 from the U.S. Department of Energy Office of Science (Biological and Environmental Research), NIH grant 1DP2OD004389-01, the Human Frontiers Science Program, and the Beckman Foundation.
Footnotes
Published ahead of print on 19 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 2.Arifuzzaman, M., M. Maeda, A. Itoh, K. Nishikata, C. Takita, R. Saito, T. Ara, K. Nakahigashi, H. C. Huang, A. Hirai, K. Tsuzuki, S. Nakamura, M. Altaf-Ul-Amin, T. Oshima, T. Baba, N. Yamamoto, T. Kawamura, T. Ioka-Nakamichi, M. Kitagawa, M. Tomita, S. Kanaya, C. Wada, and H. Mori. 2006. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 16:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bupp, K., and J. van Heijenoort. 1993. The final step of peptidoglycan subunit assembly in Escherichia coli occurs in the cytoplasm. J. Bacteriol. 175:1841-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 6.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 7.Chu, C. T., E. D. Plowey, Y. Wang, V. Patel, and K. L. Jordan-Sciutto. 2007. Location, location, location: altered transcription factor trafficking in neurodegeneration. J. Neuropathol. Exp. Neurol. 66:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crouvoisier, M., G. Auger, D. Blanot, and D. Mengin-Lecreulx. 2007. Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis. Biochimie 89:1498-1508. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Pedro, M. A., J. C. Quintela, J. V. Holtje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahnert, B., H. Lilie, and P. Neubauer. 2004. Inclusion bodies: formation and utilisation. Adv. Biochem. Eng. Biotechnol. 89:93-142. [DOI] [PubMed] [Google Scholar]
- 12.Giaever, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strom. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 14.Guberman, J. M., A. Fay, J. Dworkin, N. S. Wingreen, and Z. Gitai. 2008. PSICIC: noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput. Biol. 4:e1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha, S., D. Walker, Y. Shi, and S. Walker. 2000. The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 9:1045-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengge-Aronis, R., W. Klein, R. Lange, M. Rimmele, and W. Boos. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holtje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda, M., M. Wachi, H. K. Jung, F. Ishino, and M. Matsuhashi. 1991. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J. Bacteriol. 173:1021-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindner, A. B., R. Madden, A. Demarez, E. J. Stewart, and F. Taddei. 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 105:3076-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy, A. J., R. C. Sandlin, and A. T. Maurelli. 2003. In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J. Bacteriol. 185:1218-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mengin-Lecreulx, D., L. Texier, M. Rousseau, and J. van Heijenoort. 1991. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J. Bacteriol. 173:4625-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mileykovskaya, E. 2007. Subcellular localization of Escherichia coli osmosensory transporter ProP: focus on cardiolipin membrane domains. Mol. Microbiol. 64:1419-1422. [DOI] [PubMed] [Google Scholar]
- 23.Mileykovskaya, E., and W. Dowhan. 2000. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182:1172-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogk, A., E. Deuerling, S. Vorderwulbecke, E. Vierling, and B. Bukau. 2003. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50:585-595. [DOI] [PubMed] [Google Scholar]
- 25.Mohammadi, T., A. Karczmarek, M. Crouvoisier, A. Bouhss, D. Mengin-Lecreulx, and T. den Blaauwen. 2007. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol. Microbiol. 65:1106-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhopadhyay, R., K. C. Huang, and N. S. Wingreen. 2008. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys. J. 95:1034-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogliano, J., K. Pogliano, D. S. Weiss, R. Losick, and J. Beckwith. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. U. S. A. 94:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rokney, A., M. Shagan, M. Kessel, Y. Smith, I. Rosenshine, and A. B. Oppenheim. 2009. E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J. Mol. Biol. 392:589-601. [DOI] [PubMed] [Google Scholar]
- 29.Romantsov, T., S. Helbig, D. E. Culham, C. Gill, L. Stalker, and J. M. Wood. 2007. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 64:1455-1465. [DOI] [PubMed] [Google Scholar]
- 30.Rozkov, A., and S. O. Enfors. 2004. Analysis and control of proteolysis of recombinant proteins in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 89:163-195. [DOI] [PubMed] [Google Scholar]
- 31.Silhavy, T. J., M. L. Berman, L. W. Enquist, and Cold Spring Harbor Laboratory. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 32.Soupene, E., W. C. van Heeswijk, J. Plumbridge, V. Stewart, D. Bertenthal, H. Lee, G. Prasad, O. Paliy, P. Charernnoppakul, and S. Kustu. 2003. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185:5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van den Brink-van der Laan, E., J. W. Boots, R. E. Spelbrink, G. M. Kool, E. Breukink, J. A. Killian, and B. de Kruijff. 2003. Membrane interaction of the glycosyltransferase MurG: a special role for cardiolipin. J. Bacteriol. 185:3773-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinella, D., D. Joseleau-Petit, D. Thevenet, P. Bouloc, and R. D'Ari. 1993. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J. Bacteriol. 175:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weibezahn, J., P. Tessarz, C. Schlieker, R. Zahn, Z. Maglica, S. Lee, H. Zentgraf, E. U. Weber-Ban, D. A. Dougan, F. T. Tsai, A. Mogk, and B. Bukau. 2004. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119:653-665. [DOI] [PubMed] [Google Scholar]
- 36.Werner, J. N., E. Y. Chen, J. M. Guberman, A. R. Zippilli, J. J. Irgon, and Z. Gitai. 2009. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. U. S. A. 106:7858-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler, J., A. Seybert, L. Konig, S. Pruggnaller, U. Haselmann, V. Sourjik, M. Weiss, A. S. Frangakis, A. Mogk, and B. Bukau. 2010. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29:910-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.