Abstract
Corynebacterium diphtheriae, the causative agent of the severe respiratory disease diphtheria, utilizes hemin and hemoglobin as iron sources for growth in iron-depleted environments. Because of the toxicity of high levels of hemin and iron, these compounds are often tightly regulated in bacterial systems. In this report, we identify and characterize the C. diphtheriae hrtAB genes, which encode a putative ABC type transporter involved in conferring resistance to the toxic effects of hemin. Deletion of the hrtAB genes in C. diphtheriae produced increased sensitivity to hemin, which was complemented by a plasmid harboring the cloned hrtAB locus. The HrtAB system was not involved in the uptake and use of hemin as an iron source. The hrtAB genes are located on the C. diphtheriae genome upstream from the chrSA operon, which encodes a previously characterized two-component signal transduction system that regulates gene expression in a heme-dependent manner. The hrtB promoter is activated by the ChrAS system in the presence of hemin or hemoglobin, and mutations in the chrSA genes abolish heme-activated expression from the hrtB promoter. It was also observed that transcription from the hrtB promoter is reduced in a dtxR deletion mutant, suggesting that DtxR is required for optimal expression of hrtAB. Previous studies proposed that the ChrS sensor kinase may be responsive to an environmental signal, such as hemin. We show that specific point mutations in the ChrS N-terminal transmembrane domain result in a reduced ability to activate the hrtB promoter in the presence of a heme source, suggesting that this putative sensor region is essential for the detection of a signal produced in response to hemin exposure. This study shows that the HrtAB system is required for protection from hemin toxicity and that expression of the hrtAB genes is regulated by the ChrAS two-component system. This study demonstrates a direct correlation between the detection of heme or a heme-associated signal by the N-terminal sensor domain of ChrS and the transcriptional activation of the hrtAB genes.
Corynebacterium diphtheriae is a Gram-positive bacterium and a well-known human pathogen that causes the severe upper respiratory tract disease diphtheria. C. diphtheriae colonizes the upper respiratory tract and is rarely found outside this region during infection in humans. Much of the severe morbidity and mortality associated with diphtheria is due to the production and secretion of diphtheria toxin, which disseminates throughout the host (12). Numerous studies with C. diphtheriae have focused on the genetic regulation of diphtheria toxin, and it is known that transcription of the structural gene tox is repressed by iron and the diphtheria toxin repressor protein, DtxR, a global iron-dependent regulator (6, 10, 18, 37, 39). While it is well established that iron levels can serve as a signal to regulate expression of bacterial virulence factors, the ability to acquire iron as a nutrient source by pathogenic bacteria is also important for virulence (7, 9). Numerous bacterial pathogens, including C. diphtheriae, utilize various host iron sources for growth, including transferrin, lactoferrin, and heme sources such as hemoglobin (7, 26, 35). Bacteria have developed a variety of mechanisms for the uptake and utilization of iron, including siderophore uptake systems (9, 19, 50) and binding protein-dependent transporters that extract iron from various host sources (7, 50). Hemin is thought to be one of the primary host iron sources used by bacterial pathogens, and hemin iron transport and utilization systems in numerous bacterial species have been described (21, 45). Uptake and utilization of hemin as an iron source in C. diphtheriae involve the iron- and DtxR-regulated hmu hemin transport locus, which contains the hmuTUV genes, which encode an ABC hemin transporter, and the htaA, htaB, and htaC genes (1, 13, 18). HtaA and HtaB were recently shown to be surface-anchored hemin binding proteins, and mutations in htaA and in the hmuTUV genes result in reduced ability to use hemoglobin as an iron source, suggesting that factors encoded by these genes are important in hemin transport (1).
The use of heme as an iron source in C. diphtheriae also involves the hmuO gene, which encodes a heme oxygenase that degrades intracellular heme, which results in the release of the heme-associated iron (31, 49). Transcription of the hmuO gene in C. diphtheriae is repressed in high-iron conditions by DtxR and is activated by heme or hemoglobin (32). Heme-dependent activation of hmuO is primarily mediated by the ChrA-ChrS (ChrAS) two component signal transduction system, which is composed of the ChrS sensor kinase and the response regulator ChrA (4, 33). The HrrS-HrrA (HrrAS) two-component system is also involved in the heme-dependent transcriptional activation of the hmuO promoter and contributes approximately 20% of the activity observed at this promoter (5). Moreover, HrrAS, as well as the ChrAS regulatory system, is involved in the heme-dependent repression of the hemA gene, the promoter-proximal gene in a heme biosynthetic operon. Mutations in either chrA or chrS result in reduced ability to activate expression of hmuO and in an increased sensitivity to hemin (4). The hemin sensitivity observed in the chrSA mutants was not due to the reduced expression of hmuO, since an hmuO mutant shows no increased sensitivity to hemin (4). It was proposed that the heightened hemin sensitivity observed in the chrSA mutants was likely due to reduced expression of an as yet identified gene(s) that is regulated by ChrAS (4).
Recent studies with Staphylococcus aureus and Bacillus anthracis identified an ABC type transporter, designated HrtAB, which is proposed to protect these bacteria from the toxic effects of high levels of hemin (15, 40, 41, 42). Mutations in the S. aureus hrtA and hrtB genes result in an increased sensitivity to hemin (40). The HrtAB system, which is composed of an ATPase (HrtA) and a membrane permease (HrtB), is hypothesized to either directly export excess heme from the cytosol or remove toxic metabolites that accumulate in response to hemin exposure (40, 42). The mechanism by which the HrtAB system reduces heme stress has not been determined (40). The S. aureus hrtAB genes are found adjacent to and transcribed divergently from the recently described hssR-hssS genes, which encode a two-component signal transduction system that controls the expression of the hrtAB genes in a heme-dependent manner (40, 41, 42).
In this study, we have identified a putative ABC type transporter in C. diphtheriae that is involved in protection from the toxic effects of hemin. We have designated this ABC transporter HrtAB to be consistent with previously described orthologous systems (15, 40, 41). A C. diphtheriae hrtAB deletion mutant exhibited an increased sensitivity to hemin; expression of the hrtAB genes is increased in the presence of heme or hemoglobin, and this regulation is mediated by the ChrAS two-component signal transduction system. Optimal expression from the hrtB promoter also requires DtxR. Mutations in the sensor region of ChrS were shown to affect heme-dependent activation of hrtAB transcription, demonstrating a direct association between detection of a heme-associated signal by ChrS and transcription of the hrtAB genes.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli and C. diphtheriae strains used in this study are listed in Table 1. Luria-Bertani (LB) medium was used for culturing E. coli, and heart infusion broth (Difco, Detroit, MI) containing 0.2% Tween 80 (HIBTW) was used for routine growth of C. diphtheriae strains. Bacterial stocks were maintained in 20% glycerol at −80°C. Antibiotics were added to LB medium at 34 μg/ml for chloramphenicol, 50 μg/ml for kanamycin, and 100 μg/ml for spectinomycin; for C. diphtheriae cultures in HIBTW medium antibiotic concentrations were 2 μg/ml for chloramphenicol, 100 μg/ml for spectinomycin, and 50 μg/ml for kanamycin. HIBTW was made low iron by the addition of ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) at 12 μg/ml (unless indicated otherwise). Modified PGT (mPGT) is a semidefined low-iron medium that has been previously described (46). Antibiotics, EDDA, Tween 80, hemin (bovine), and hemoglobin (human) were obtained from Sigma Chemical Co. (St. Louis, MO).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics or use | Reference or source |
|---|---|---|
| Corynebacterium strains | ||
| C7 (−) | C. diphtheriae wild type | 3 |
| 1737 | C. diphtheriae wild type, Gravis biotype | 29 |
| C7chrSΔ | chrS deletion mutant of C7 (−) | 4 |
| C7ChrSAΔ | chrSA deletion mutant of C7 (−) | 5 |
| C7hrrSAΔ | hrrAS deletion mutant of C7 (−) | 5 |
| C7hrtABΔ | hrtAB deletion mutant of C7 (−) | This study |
| C7dtxRΔ | dtxR deletion mutant of C7 (−) | This study |
| CU44 | C. ulcerans hmuU mutant | 31 |
| E. coli strains | ||
| DH5α | Cloning strain | Invitrogen |
| S17-1 | RP4 mobilization functions | 47 |
| TOP10 | Cloning of PCR fragments | Invitrogen |
| XL1-Gold | QuikChange cloning | Stratagene |
| Plasmids | ||
| pCR-Blunt-II | Cloning of PCR fragments, Kanr | Invitrogen |
| pCM502 | Promoter probe vector, Cmr | 32 |
| pSPZ | Promoter probe vector, Spcr | 24 |
| phrtB-lacZ | hrtB promoter-lacZ fusion on pSPZ | This study |
| pCMtox | tox promoter-lacZ fusion on pCM502 | 32 |
| pKN2.6 | C. diphtheriae shuttle vector, Knr | 37 |
| pKN2.6Z | C. diphtheriae shuttle vector, Knr | 13 |
| pCM2.6 | C. diphtheriae shuttle vector, Cmr | 37 |
| pECK18mob2 | Corynebacterium cloning vector, Knr | 30 |
| pECK-chrSA | chrSA genes cloned into pECK18mob2 | 5 |
| pTrc99A | E. coli cloning and expression vector, Ampr | Pharmacia |
| pKN-chrS | chrS cloned into pKN2.6 | This study |
| pKN-hrtAB | hrtAB cloned into pKN2.6Z | This study |
| pMnt-hrtAB | Plasmid pKN-hrtAB with mntA promoter | This study |
| p2.6-dtxR | dtxR gene cloned into pKN2.6Z | This study |
Mutant construction.
A previously described allelic replacement technique was used to construct in-frame deletions of dtxR and the hrtAB locus in C. diphtheriae strain C7 (47). Mutant construction utilized PCR to clone DNA fragments located upstream and downstream of the region targeted for deletion. The deleted region in the hrtAB mutant is predicted to encode a peptide of approximately 21 amino acids that contains residues derived from the N terminus of HrtB and the C terminus of HrtA. The dtxR mutant is predicted to produce a truncated peptide of 42 amino acids. Primers used for the construction of the mutants are listed in Table 2. PCR was used to confirm the mutation in all of the deletion mutants (not shown).
TABLE 2.
Primers used in this study
| Primer | Amplified region | Sequencea (5′-3′) |
|---|---|---|
| Deletion mutants | ||
| 23/24KO-Ds-F | Downstream of hrtAB | GCATTCTAGATCATCACATTCAGCAATGC |
| 23/24KO-Ds-R | TAGCGTCGACATGGCCGACGGAAAAGCC | |
| 23/24KO-Us-F | Upstream of hrtAB | GACTGTCGACACGAATGTCGCGGATTCC |
| 23/24KO-Us-F | GACTGTCGACACGAATGTCGCGGATTCC | |
| dtxR-UP1 | Upstream of dtxR | GGCAGTACTTCAATGCTGAAGATGAGG |
| dtxR-UP2 | GGCGTCGACGATCCTAGCGCGAAGAGG | |
| dtxR-DN1 | Downstream of dtxR | GGCGTCGACGCCCACACTATTCGTATC |
| dtxR-DN2 | GGCACTAGTAGTACTGACCGAATATCCTTCACC | |
| hrtB promoter fusion | ||
| DIP2324-PO-R | hrtB | GGATCCGGCCGGTCAGCATAACG |
| DIP2324-PO-F | Promoter | GTCGACGGGTTCCTACGCTGACC −3′ |
| hrtAB cloning | ||
| DIP2323-R | CTGAGGATCCGACTAGCACACCAAATTCC | |
| DIP2324-PO-F(RV) | GGCGATATCGGGTTCCTACGCTGACC | |
| 2324P | GGCGATATCCCTTACTTTCACAACAAT | |
| mntP-Rev | GGCGATATCTCCTGTTTGACAAACTTG | |
| chrS cloning | ||
| chrS-kin-R | GTCGACGGATCATGAAGTCTCCCG | |
| chrS-wt-F | GGATCCGCAGGAGATCTTTTATGC | |
| ChrS C-terminal expression construct | ||
| chrS-kin-ab-F2 | CTACCTCTGAACATATGGCAGGCCGG | |
| chrS-kin-R | GTCGACGGATCATGAAGTCTCCCG | |
| dtxR cloning | ||
| DTRB | AAAGACATAGGATCCTATTAAAAGCAATC | |
| DTRS | ATCTAATTTGTCGACTTTAGTATTTAGAG |
Restriction sites are underlined.
Growth of C. diphtheriae cultures in the presence of hemin.
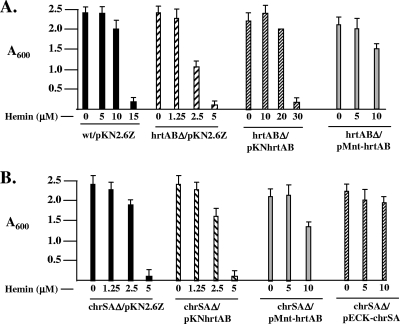
The effect of hemin on the growth of various C. diphtheriae strains was determined by inoculating overnight cultures, at an optical density at 600 nm (OD600) of 0.1, into fresh HIBTW medium that contained various hemin concentrations, as shown in Fig. 4. Cultures were grown aerobically at 37°C, and growth was assessed by measuring the optical density of cultures after 48 h.
FIG. 4.
(A) Deletion of the C. diphtheriae hrtAB genes results in increased sensitivity to hemin. Strains were grown in HIBTW medium containing various amounts of hemin as indicated. Growth of bacterial cultures was measured after 48 h (optical density at 600 nm [A600]). (B) The heme sensitivity of C7chrSAΔ is complemented by the cloned hrtAB genes on plasmid pMnt-hrtAB. Growth conditions are the same as for panel A. A chrS deletion mutant showed a hemin-sensitive phenotype similar to that of C7chrSAΔ (5) (data not shown). Results are the averages of three independent experiments ± standard deviations.
Hemoglobin-iron utilization assays.
The hemoglobin-iron utilization assay has been described previously (20). Briefly, Corynebacterium strains were grown overnight (20 to 22 h at 37°C) in HIBTW and then inoculated to an OD600 of 0.2 into fresh HIBTW that contained 12 μg/ml of the iron chelator EDDA. Strains were grown for several hours at 37°C until log phase, at which time bacteria were recovered by centrifugation, resuspended in mPGT medium, and then inoculated at an OD600 of 0.03 into fresh mPGT medium that contained various supplements. After 20 to 22 h of growth at 37°C, the OD600 of the cultures was determined.
Plasmid construction.
Plasmids used in this study are listed in Table 1. PCR-derived DNA fragments were initially cloned into the pCR-Blunt II-TOPO vector (Invitrogen), and genomic DNA derived from C. diphtheriae strain C7 or 1737 was used as a template for PCRs. Promoter probe vectors pCM502 (31) and pSPZ (24) were used for the construction of lacZ promoter fusions; these plasmids contain a promoterless lacZ gene and replicate at low copy number in C. diphtheriae. To construct plasmid phrtB-lacZ, which contains the hrtAB promoter region fused to a promoterless lacZ gene, a 334-bp fragment encompassing the entire hrtB-cmrA intergenic region plus flanking sequence was amplified with primers DIP2324-PO-F and DIP2324-PO-R and cloned into pSPZ with a BamHI-SalI digest.
Plasmid pKN2.6 is an E. coli-Corynebacterium shuttle vector with a kanamycin resistance cassette that replicates at low copy number in Corynebacterium and has been previously described (37). Plasmid pKN2.6Z is a derivative of pKN2.6 that has the multiple cloning sequence from pBluescript KS and has been previously described (13). Plasmid pKN2.6(−EcoRI) is a derivative of pKN2.6 in which the EcoRI site has been blunt ended using standard techniques. To create plasmid pKN-chrS, a DNA fragment was amplified from either C7 and 1737 using primers chrS-wt-F and chrS-kin-R and ligated into the BamHI-SalI sites in pKN2.6(−EcoRI). Plasmid vector pECK18mob2 (30) was used to construct plasmids utilized in the chrS complementation studies. Plasmid pECK-chrS has a copy of the chrS gene from C7 or 1737 which was derived from plasmid pKN-chrS constructs.
To construct a plasmid with hrtA and hrtB for complementation, primers DIP2324-PO-F(RV) and DIP2323-R were used to amplify hrtAB and the upstream region containing the putative promoter region. This fragment was moved into plasmid pKN2.6Z with a BamHI-EcoRV digest to make pKN-hrtAB. To construct pMnt-hrtAB, the mntA promoter region from plasmid pPO1 (34) was amplified by PCR using primers mntP-Rev and PO1 to generate an approximately 260-bp fragment, which was subsequently ligated upstream of the hrtB gene on plasmid pKN-hrtAB2. The DNA insert in plasmid pKN-hrtAB2 contains the complete coding region and translational signals for hrtAB but lacks the ChrAS-regulated promoter. The insert in plasmid pKN-hrtAB2 was derived from a PCR product using primers DIP2323R and 2324P and C7 chromosomal DNA as the template. The insert was ligated into pKN2.6Z at the BamHI-EcoRV sites.
Plasmid p2.6-dtxR, which carries the C. diphtheriae C7 dtxR gene, was constructed by using PCR and primers DTRB and DTRS to amplify a 901-bp fragment that contains the dtxR gene and upstream elements needed for expression of the gene product. The 901-bp DNA fragment was first cloned into pCR-Blunt II-TOPO and then moved into pKN2.6Z at the SalI-BamHI sites.
To construct PhoA fusions to the sensor region of ChrS, the phoA gene was obtained from plasmid pSS1324 (43) with a SalI-HindIII digest. The DNA fragment containing phoA was ligated into pUC18. Fragments containing various segments of the chrS gene were amplified using various primer sets. Fragments were first cloned into pCR-Blunt II-TOPO and then moved into pUC18 as XbaI-SalI fragments. The chrS-phoA fusions were then cloned into the pTrc99A vector (Pharmacia) with an XbaI-HindIII digest. To construct LacZ fusions to the sensor region of ChrS, a DNA fragment harboring the lacZ gene was first amplified by PCR from plasmid pSS1462 (43) and subsequently ligated into the pTrc99A vector at the HindIII-SalI sites (pTrc99A-lacZ). Fragments containing various segments of the chrS gene were amplified using various primer sets and subsequently ligated into pTrc99A-lacZ at the XbaI-SalI sites.
The plasmid used to express the ChrS C-terminal kinase domain was constructed as follows. PCR was used to amplify a 670-bp product using primers chrS-kin-ab-F2 and chrS-kin-R and C7 chromosomal DNA as the template. This DNA fragment, which encodes the ChrS C-terminal kinase domain, was moved into the expression vector pET28a to allow production of a His-tagged fusion protein. The His-ChrS-C terminus fusion protein was expressed at high levels in E. coli by following standard techniques and purified by nickel affinity chromatography as previously described (1). Polyclonal antiserum against the ChrS-His fusion was obtained from guinea pigs by following standard procedures (Cocalico Inc.). Immunoblot analysis of C. diphtheriae whole-cell lysates was performed as described previously (1). The DNA inserts in all plasmid constructs used in this study were confirmed by DNA sequence analysis.
LacZ assays.
C. diphtheriae strains containing promoter-lacZ fusion constructs were grown overnight in HIBTW medium and then inoculated at an OD600 of 0.2 into fresh HIBTW medium in the presence of various supplements. Cultures were grown for several hours until they reached log phase, and LacZ activity was determined as previously described (38).
PhoA and LacZ fusion analysis.
The various ChrS-PhoA and -LacZ fusion proteins were constructed to map the membrane topography of the ChrS N-terminal domain. DH5α cells carrying the pTrc-ChrS-PhoA or -LacZ fusion construct were inoculated from frozen stocks into 1-ml (LB medium with ampicillin) overnight cultures. Overnight cultures were then inoculated into LB medium with ampicillin at an OD600 of 0.05. Cultures were grown for 2 to 2.5 h at 37°C (for PhoA fusions only, isopropyl-β-d-thiogalactopyranoside [IPTG] was added at a final concentration of 0.1 mM and the cultures were incubated at 37°C for an additional 30 min). Alkaline phosphatase and LacZ activities were determined as previously described (8, 23).
Site-directed mutagenesis.
Site-directed mutants of C7 ChrS were made using the QuikChange or QuikChange Lightning kit (Stratagene) according to the manufacturer's instructions. Briefly, 125 ng of each primer containing the targeted base change (designed using the QuikChange primer design program at http://www.stratagene.com/sdmdesigner/default.aspx) and 50 ng of plasmid template pKN-chrS were used in the QuikChange reaction mixture (50-μl total volume). Methylated template DNA was removed from the reaction mixture by digestion with DpnI restriction endonuclease. Mutagenized DNA was recovered by transformation into XL1-Blue (QuikChange) or XL1-Gold (QuikChange Lightning) competent cells (2 μl of DpnI-treated reaction mixture in 45 μl of cells). The presence of the base changes was confirmed by sequencing. The mutated chrS genes on pKN-chrS were moved into pECK18mob2 for the complementation studies with C7chrSΔ, which harbors phrtB-lacZ.
Computer analysis.
Amino acid sequence similarity searches were done using the BLAST program (2) at the National Center for Biotechnology Information and also using the BLAST server provided at the online site for the Sanger Institute http://www.sanger.ac.uk/Projects/C_diphtheriae. The annotated genome sequence for C. diphtheriae strain NCTC13129 (11) is accessible in the EMBL/GenBank database at accession number BX248353. The online transmembrane programs TMPred (http://www.ch.embnet.org/software/TMPRED_form.html) and DAS (http://www.sbc.su.se/∼miklos/DAS/) were used in the analysis of the ChrS N-terminal region, and sequence alignments were done with ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html/).
RESULTS
Identification of a C. diphtheriae ABC transporter that confers protection from hemin toxicity.
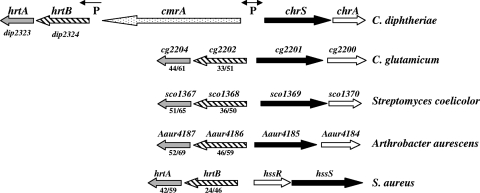
Upstream of the chrSA operon is the cmrA gene, which is predicted to encode a large zinc-regulated surface-anchored protein that is divergently transcribed from chrSA (Fig. 1) (4). Downstream of cmrA are two genes, dip2324 and dip2323, which are predicted to encode the permease and ATPase components, respectively, of an ABC transporter (Fig. 1). The predicted gene products of dip2323 and dip2324 have sequence similarity to the previously described Staphylococcus aureus gene products HrtA (DIP2323, 42% identity/59% similarity) and HrtB (DIP2324, 24% identity/46% similarity), respectively (15, 48). In S. aureus, the hrtAB genes are located adjacent to and divergently transcribed from the hssS-hssR genes, which encode a two-component signal transduction system that is required for the heme-dependent expression of hrtAB (Fig. 1) (42). A similar genetic locus is present in Bacillus anthracis (41). Because of the sequence similarities between DIP2324 and DIP2323 and the orthologous proteins in S. aureus, we have renamed these C. diphtheriae genes hrtB and hrtA, respectively.
FIG. 1.
Genetic map of the hrtAB-chrSA loci in C. diphtheriae and in related organisms. A genetic map of the S. aureus hrtAB-hssRS region is also shown for comparison (40). The percentages of amino acid similarity/identity to the C. diphtheriae HrtA and HrtB proteins are indicated below the gene designations. Genes with homology to hrtA are indicated by gray arrows, and homologs of hrtB are shown with hatched arrows. P indicates the locations of known promoters in C. diphtheriae and the directions of transcription. Genes encoding proteins with homology to sensor kinases are indicated in black, and response regulator homologs are shown with open arrows.
Genomic analysis of various Gram-positive species, some of which are closely related to C. diphtheriae, revealed the presence of putative gene products that share sequence similarity to HrtAB (Fig. 1). All of these hrtAB-related genes were genetically linked to a two-component signal transduction system (Fig. 1). The S. aureus HssS sensor kinase showed no significant sequence similarity with ChrS in either the C-terminal kinase domain or in the N-terminal sensor region, which anchors these proteins to the membrane and is proposed to be critical for recognition of an environmental signal. The amino acid sequence of HssR showed approximately 40% similarity and 23% identity with that of ChrA. Although the genetic organizations of the hrt-chr loci among the species that are closely related to C. diphtheriae are similar, only C. diphtheriae contains the cmrA gene, which resides between the coding sequences for the two-component system and the ABC transporter (Fig. 1). No gene products with significant amino acid similarity to CmrA are predicted to be encoded in any of these organisms, and a function for CmrA is not known (M. P. Schmitt, unpublished observation).
Heme-dependent regulation of hrtAB expression is mediated by ChrAS.
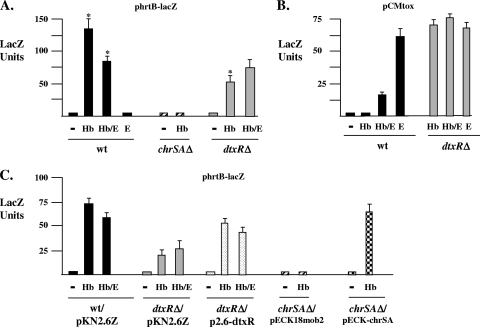
Previous studies in our laboratory demonstrated that the ChrAS two-component signal transduction system was critical for the control of hemin-dependent transcription at promoters upstream of the hmuO and hemA genes (4, 5). To determine if ChrAS also regulates transcription of hrtAB in C. diphtheriae, an hrtB promoter-lacZ fusion construct, phrtB-lacZ, was generated and promoter activity in various C. diphtheriae strains was assessed. These studies revealed that hrtB promoter activity is increased almost 50-fold in the presence of hemoglobin and that only minimal levels of activity are observed in the absence of a heme source (Fig. 2A). Hemin gave results similar to those for hemoglobin in the activation of the hrtB promoter (data not shown); however, because of the toxicity associated with hemin, hemoglobin was used for all experiments shown. Hemoglobin-dependent transcription at the hrtB promoter is abolished in strains carrying deletions in the chrSA genes, which indicates that the hemin-responsive activation of hrtAB requires the ChrAS regulatory system (Fig. 2A). Hemin-responsive expression at the hrtB promoter is restored in the chrSA deletion mutant in the presence of the cloned chrSA genes (Fig. 2C). The C. diphtheriae HrrAS two-component system, which is an additional heme-dependent activator of hmuO expression (5), is not involved in the regulation of the hrtB promoter (not shown).
FIG. 2.
(A) The hrtB promoter is regulated by ChrSA in the presence of hemoglobin. Plasmid phrtB-lacZ was used to measure hrtB promoter activity in C7 (wild type [wt]), C7chrSAΔ, and C7dtxRΔ. Strains were grown in HIBTW medium containing no supplement (−), hemoglobin (Hb; 200 μg/ml), EDDA (E; 12 μg/ml), or EDDA and hemoglobin (Hb/E). The results are the means of at least three independent experiments (±standard deviations). Asterisks indicate that results with wt grown in Hb are significantly different from results with wt grown in Hb/E (P < 0.05) and with C7dtxRΔ grown in Hb (P < 0.02). (B) Expression from the iron- and DtxR-regulated tox promoter was assessed using plasmid pCMtox. Conditions were the same as those for panel A. The results are the means of at least three independent experiments (±standard deviations). (C) Expression of the hrtB promoter in the C. diphtheriae mutants C7dtxRΔ and C7chrSAΔ in the presence of the cloned dtxR and chrSA genes. All strains carried plasmid phrtB-lacZ and a second plasmid, which was either the vector control or a plasmid containing the dtxR gene or the chrSA genes. Conditions were the same as those describe for panel A. wt/pKN2.6Z indicates the C7 wild type harboring vector pKN2.6Z (C7 wt with vector pECK18mob2 gave similar results [data not shown]). The results are the means of at least three independent experiments (±standard deviations).
Expression of the hrtB promoter is reduced approximately 40% in medium containing the iron chelator EDDA and hemoglobin (Fig. 2A), which suggests that reduced iron levels affect hrtB promoter activity. No activity is seen with medium supplemented only with EDDA, since a heme source is required for expression from the hrtB promoter. Since it was previously shown that the ChrAS-regulated hmuO promoter was affected by iron and dtxR, we examined the expression of the hrtB promoter in a dtxR deletion mutant. Expression of the hrtB promoter in the presence of hemoglobin was reduced 2- to 3-fold in the dtxR deletion mutant compared to expression under these conditions in the wild-type strain (Fig. 2A), suggesting that DtxR is required for wild-type levels of transcription at the hrtB promoter. There was no significant difference in expression at the hrtB promoter between the wild type and the dtxR mutant in the presence of EDDA and hemoglobin. Expression from the hrtB promoter in a dtxR mutant was increased to nearly wild-type levels in the presence of the cloned dtxR gene, which suggests that the dtxR mutation is the reason for reduced hrtB expression (Fig. 2C). The inability of the cloned dtxR gene to fully restore hrtB promoter activity to wild-type levels may be due to low expression of the cloned dtxR gene on plasmid pKN2.6Z. In Fig. 2C, it was noted that expression of the hrtB promoter in the wild-type strain and in the dtxR mutant was approximately one-half the levels observed in these same strains in Fig. 2A. The reason for the difference in expression is likely the presence of two plasmids residing in the strains used in the studies in Fig. 2C. The presence of two plasmids in these strains may lower the copy number of phrtB-lacZ, which would be expected to result in reduced levels of LacZ activity. Appropriate antibiotics were present at all times during the expression studies to ensure maintenance of all plasmids.
Expression at the hrtB promoter cannot be assessed under the low-iron conditions that our laboratory has used previously to measure iron-regulated gene expression in C. diphtheriae (32). This is because heme functions as not only a heme source for hrt expression but also an iron source for C. diphtheriae, which results in increased intracellular iron levels. In previous studies, HIBTW medium supplemented only with EDDA was used to generate an optimal low-iron condition for assessing gene expression in C. diphtheriae (32). In order to assess the biologically relevant iron levels under the conditions used in Fig. 2A, expression from the iron-responsive and DtxR-regulated tox promoter on plasmid pCMtox was examined (32). As expected, expression from the tox promoter in a dtxR deletion mutant was derepressed under all conditions tested (Fig. 2B). In the wild-type strain, expression of tox was fully repressed in high-iron medium (HIBTW medium that contained hemoglobin or no supplement); however, in medium supplemented with hemoglobin and EDDA (Hb/E medium), tox expression was approximately 25% of the activity seen under the optimal low-iron conditions (EDDA alone [E medium]) (Fig. 2B, compare wild-type E with Hb/E). Although these results indicate that optimal low-iron conditions are not achieved in the Hb/E medium, a reduction in biologically relevant iron levels is obtained in Hb/E medium, as demonstrated by an increase in expression at the tox promoter compared to the level of expression observed in high-iron conditions (Fig. 2B, compare wild-type Hb with Hb/E). This finding supports the observation that hrtB expression is affected by iron levels and further suggests that optimal expression of hrtAB requires DtxR, hemoglobin, and an iron-rich medium.
Sequence analysis of the promoter regions for ChrA-regulated genes.
Sequence analysis of the region upstream of the ChrA-regulated genes, hrtB, hmuO, and hemA, revealed the presence of a conserved 10-bp region with the consensus sequence 5′-GGTTGATGTG-3′ (Fig. 3). Upstream of the hmuO promoter are two invariant copies of this 10-bp sequence separated by 40 bases, while upstream of hrtB are two copies that match 9 of 10 bases of the consensus sequence and are positioned on opposite strands to create a potential stem-loop structure. A single copy of this conserved sequence, which matches 8 of 10 bases of the consensus, is also present upstream of hemA (Fig. 3). This conserved sequence in the hemA promoter region overlaps putative −10 promoter elements, which is consistent with a regulatory protein, such as ChrA, functioning as a repressor at this promoter. While it has not been determined whether this conserved 10-bp sequence functions as a binding site for ChrA or another regulatory factor, the presence of this sequence upstream of the three known ChrA-regulated genes strongly suggests that this sequence has a role in transcriptional regulation.
FIG. 3.
DNA sequence of the region upstream of the hmuO, hrtB, and hemA genes. Boxed sequences indicates the 10-bp conserved region present in all three upstream sequences. The underlined region in the hmuO sequence indicates the location of the DtxR binding site (DtxR b.s.), and the circled A residue denotes the start site of transcription. The solid bar above the sequence indicates the location of putative −10 promoter elements, and arrows identify start codons.
Deletion of the hrtAB genes results in increased hemin sensitivity.
To determine the function of the HrtAB system, a nonpolar deletion of the hrtAB genes was constructed in C. diphtheriae strain C7, and the mutant is designated C7hrtABΔ. C7hrtABΔ exhibited growth similar to that of the wild-type strain in HIBTW medium, indicating that the deletion strain is not defective for growth in nutrient-rich broth (Fig. 4A). However, growth of C7hrtABΔ was restricted in the presence of elevated levels of hemin. Hemin concentrations above 2.5 μM strongly limited the growth of the mutant, whereas growth of the wild-type strain was not affected by hemin levels as high as 10 μM (Fig. 4A). The hemin sensitivity of C7hrtABΔ was complemented by a plasmid carrying the cloned hrtAB genes on plasmid pKNhrtAB, confirming that the hemin-sensitive phenotype observed in this mutant is specific to hrtAB (Fig. 4A). It was further noted that the presence of the hrtAB cloned genes in C7hrtABΔ and in the wild-type strain resulted in a resistance to hemin toxicity that was greater than that observed in the wild-type strain carrying only the pKN2.6Z vector, suggesting that multiple copies of the hrtAB genes (or increased expression or both) result in enhanced tolerance to hemin (Fig. 4A; data not shown for the wild type harboring the cloned hrtAB genes). C7hrtABΔ exhibited a sensitivity to hemin similar to that of the C. diphtheriae chrSA mutant C7chrSAΔ, which was previously shown to be sensitive to elevated levels of hemin (5) (Fig. 4B). Because hrtAB expression is positively regulated by ChrAS in a heme-dependent manner, we sought to determine if the hemin sensitivity seen in chrA and chrS mutants was caused by reduced expression of hrtAB. The pKN-hrtAB plasmid, which carries the hrtAB genes, did not restore hemin tolerance in C7chrSAΔ (Fig. 4B); however, the hrtAB genes on this clone are under the control of the native hrtB promoter, which is poorly expressed in chrSA mutants (Fig. 2A). Plasmid pMnt-hrtAB contains the hrtAB genes under transcriptional control of the mntA promoter, a manganese-repressible promoter that is independent of ChrAS regulation (34). When plasmid pMnt-hrtAB is moved into C7hrtABΔ and C7chrSAΔ, both of these strains are able to grow at hemin levels as high as 10 μM, which is comparable to the growth observed in the wild-type strain at this hemin concentration (Fig. 4A and B). This finding strongly suggests that the hemin sensitivity observed in the chrSA mutant is due to reduced expression of the ChrAS-regulated hrtAB genes. The hemin-sensitive phenotype was not complemented in strains C7hrtABΔ and C7chrSAΔ, harboring pMnt-hrtAB, in the presence of 20 μM MnCl2 (data not shown). The absence of complementation in the presence of MnCl2 suggests that the Mn2+-repressible mntA promoter drives transcription of the hrtAB genes on plasmid pMnt-hrtAB. Hemin tolerance is fully restored in C7chrSAΔ when the cloned chrSA genes are provided in trans on plasmid pECK-chrSA (5) (Fig. 4B).
HrtAB does not use hemoglobin as an iron source.
The C. diphtheriae C7 hrtAB deletion mutant exhibited no defect in the ability to use hemoglobin as an iron source (data not shown), which suggests that the HrtAB system does not function as a heme uptake and heme-iron utilization system like the previously described HmuTUV ABC transporter in C. diphtheriae and Corynebacterium ulcerans (13, 36). Furthermore, the cloned hrtAB genes on plasmid pKN-hrtAB were unable to complement the heme utilization defect in C. ulcerans strain CU44, which contains a defective permease component (HmuU) of the heme transport system (data not shown) (36).
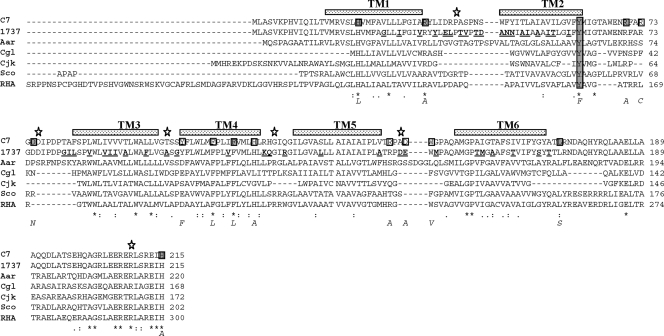
Analysis of the ChrS N-terminal region and effect of ChrS mutations on hrtB promoter activity. (i) Topology mapping of the ChrS N-terminal domain.
The N-terminal region of ChrS is proposed to respond to an environmental signal, which is presumed to be hemin or factors produced in response to hemin (33). It is further postulated that, upon interaction with external stimuli, ChrS autophosphorylates at the conserved histidine residue H215. The phosphate moiety on ChrS is transferred to an aspartate residue on the cognate response regulator ChrA, which then affects transcription of regulated promoters such as the hrtB promoter. A more complete description of the membrane architecture at the N terminus of ChrS is critical for understanding the mechanism of signal transduction and the interaction of ChrS with an activating signal. Analysis of the structure of the ChrS protein reveals that it is composed of a conserved C-terminal kinase domain that is linked to an N-terminal membrane region that exhibits minimal sequence conservation with other proteins (4, 5, 33). The precise number and location of membrane-spanning regions in ChrS have not been reported, and transmembrane prediction programs (TMpred and DAS) indicate that there are either 5 or 6 regions in the N-terminal domain that may traverse the membrane (5). We used PhoA and LacZ translational fusions to facilitate topology mapping of the ChrS N-terminal region. These constructs contained various N-terminal segments of the ChrS protein fused in frame to either PhoA or LacZ. The fusion points were located at specific ChrS sequences that are predicted to reside between membrane-spanning regions, which would result in the C-terminal PhoA or LacZ portion of the fusion protein being inserted either into the cytosol or outside the cytoplasmic membrane. The rationale for membrane mapping with PhoA and LacZ fusions is based on the observation that higher levels of PhoA activity are detected when PhoA is inserted outside the cytoplasmic membrane, while increased LacZ activity is observed for fusions in which LacZ is cytosolic (43). The results of the mapping studies shown in Fig. 5 and Table 3 indicate that six transmembrane regions (TM 1 to 6) are located in the N terminus of ChrS. The even number of transmembrane segments observed with ChrS is consistent with that reported for many related sensor kinases; this predicts that the N terminus (sensor domain) and C terminus (kinase domain) are both located in the cytosol (27).
FIG. 5.
Amino acid sequence of the N-terminal region of ChrS from C. diphtheriae aligned with orthologous proteins from related species. Aar, Arthrobacter aurescens TC1 Aaur4185; Cgl, Corynebacterium glutamicum 13032 cg2201; Cjk, Corynebacterium jeikeium jk1162; Sco, Streptomyces coelicolor A3, SCO1369; RHA, Rhodococcus species RHA1, RoO3970 (portions of the N terminus of the RHA ortholog are not shown). Stippled bars above the aligned sequences indicate predicted locations of the six transmembrane (TM) regions in C. diphtheriae C7, and stars denotes fusion points for PhoA and LacZ translational fusion constructs. Boxed amino acids in the C7 sequence indicate residues that were subjected to site-directed mutagenesis, and the resulting amino acid changes are indicated below the alignment. Substituted resides that resulted in a phenotype different from the wild type are shaded, and the conserved tyrosine, Y61, is shaded in all lanes. Amino acid differences between the C7 and 1737 C. diphtheriae strains are underlined and in boldface in the 1737 sequence.
TABLE 3.
PhoA and LacZ activity observed with ChrS translational fusions
| Fusion sitea | Activityb (avg ± SD) of: |
Locationc | |
|---|---|---|---|
| PhoA | LacZ | ||
| Vector | 7.7 (1.2) | 1.5 (0.43) | NAd |
| P40 | 387.7 (46.6) | 145.0 (19.5) | O |
| D76 | 64.7 (15.6) | 673.3 (143.8) | I |
| T101 | 2,121.0 (377) | 110.7 (36.5) | O |
| G122 | 11.4 (0.9) | 330.7 (53.5) | I |
| A146 | 1,275.4 (133.5) | 61.1 (6.6) | O |
| R209 | 23.6 (4.0) | 479.7 (182.2) | I |
Site of fusion junction in ChrS-PhoA and -LacZ fusions (vector or amino acid).
PhoA and LacZ activities were determined as described previously (8, 23). Results are the averages of at least three independent experiments.
Predicted cellular location of fusion junction. O, outside membrane; I, intracellular.
NA, not applicable.
(ii) Mutations in the ChrS N-terminal domain affect hrtB promoter activity.
Promoters upstream of hmuO and hemA were previously shown to be regulated by ChrAS in a heme-dependent manner; however, heme regulation at both of these promoters also required the activity of the HrrAS system (5). In this report, we have shown that deletion of the chrS gene abolishes heme-dependent activation at the hrtB promoter, which demonstrates that only the ChrAS system is required for heme regulation of hrtAB. Mutations in chrS that affect signal transduction are predicted to directly impact heme-dependent expression from the hrtB promoter. To identify specific sequences important for the function of the N-terminal sensor region of ChrS, point mutations within this region were constructed. To assist in the identification of residues to target for mutagenesis, the ChrS sensor region was aligned with the N-terminal domains from proteins similar in sequence and genetic organization to ChrS (Fig. 1 and 5). It is not known if any of these orthologous proteins have a function similar to that of ChrS. Several highly conserved residues were identified from this alignment, including H21 and Y61; histidine and tyrosine residues are know to be associated with the coordination of hemin in other proteins (28). These two residues, as well as several other conserved or charged amino acids located primarily in putative loop regions, were targeted for site-directed mutagenesis (Fig. 5). Clones carrying the mutated chrS genes were moved into the C7 chrS deletion mutant, and the effect of these mutations on ChrS function was assessed by measuring heme-dependent activation at the hrtB promoter on phrtB-lacZ. Immunoblotting, which used a polyclonal antiserum raised against the C-terminal kinase domain of ChrS, showed that all of the mutated chrS genes expressed the ChrS protein at levels similar to that at which the wild-type gene was expressed (data not shown). Several of the site-directed mutations in the sensor domain strongly reduced heme-dependent hrtB promoter activity, with mutations in Y61 and R70 producing the strongest defect in ChrS function (Table 4). All of the site-directed mutants that were constructed in this study are indicated in Fig. 5, but only data for those mutants that exhibited hrtB promoter activity that was different from the wild type are presented in Table 4. It should be noted that 48 of the approximately 170 amino acids in the N-terminal transmembrane region of ChrS are different between the C7 and 1737 strains (changes in 1737 are indicated in boldface and underlined in Fig. 5). It is assumed that none of the changes in 1737 significantly affect heme-dependent signaling in ChrS, since the cloned 1737 chrS gene is able to restore heme-dependent activation at the hrtB promoter in the C7 chrS mutant (Table 4). These findings suggest that amino acid substitutions that reduced heme-responsive promoter activity disrupt the recognition of a signaling molecule or affect transmission of the signal. No highly conserved residues were present in the loop regions after TM 3, 4, or 5, and none of the constructed mutations (or the 8 changes between C7 and 1737 in this region) had any effect on hrtB promoter activity (Fig. 5). A mutation in the highly conserved H21 residue (H21L) resulted in high-level, hemoglobin-independent activity, suggesting that this mutation causes a conformational change rather than having a specific effect on signal interaction. Additionally, a mutation in the partially conserved L173 residue, which is located after TM 6 (L173S), maintained heme-dependent regulation but resulted in a significant increase in promoter activity in the presence of hemoglobin. The H215 residue is proposed to be the site of phosphorylation and is assumed to be essential for the kinase activity (33), and as expected, the H215A mutation abolished activity (Table 4).
TABLE 4.
Amino acid substitutions in the ChrS sensor region affect hrtB promoter activity
| chrS version or amino acid substitutiona | LacZ activity |
|
|---|---|---|
| +Hb | −Hb | |
| Vector pECK18mob2 | 1.8 (0.6) | 2.1 (0.3) |
| Wild type (C7) | 37.3 (8.4) | 1.9 (0.1) |
| Wild type (1737) | 32.5 (4.4) | 2.9 (0.5) |
| H21L | 162.6 (34.7) | 175.4 (28.4) |
| R34A | 6.9 (2.2) | 3.0 (1.2) |
| Y61F | 2.9 (0.7) | 2.2 (0.4) |
| R70A | 4.2 (0.8) | 2.5 (0.3) |
| D75N | 10.5 (3.1) | 2.3 (0.6) |
| F114L | 9.0 (1.8) | 2.9 (0.7) |
| L173S | 131.6 (14.0) | 2.4 (0.4) |
| H215A | 2.0 (0.2) | 2.2 (0.3) |
chrS wild-type and mutant alleles in pECK18mob2 were tested for LacZ activity in C7chrSΔ/phrtB-lacZ in HIBTW medium in the presence and absence of hemoglobin (Hb).
DISCUSSION
High levels of heme or iron are toxic to many bacteria, including C. diphtheriae, and the expression of genetic systems involved in the uptake and utilization of iron or iron-containing compounds such as heme is often tightly regulated in bacteria (7, 9). Iron and hemin transport systems in C. diphtheriae are regulated by the DtxR repressor in an iron-dependent manner (18, 19). While high iron levels repress the expression of numerous genes in C. diphtheriae, the presence of extracellular hemin or heme sources like hemoglobin results in both activation and repression of gene expression, which are mediated by the ChrAS and HrrAS two-component systems (4, 5, 18). In this study, we have identified in C. diphtheriae an ABC transport system, HrtAB, that is required for resistance to the toxic effects of high levels of hemin and whose genes are regulated by the ChrAS two-component system in a heme-dependent manner. The recent report that identified the hrtAB genes in S. aureus showed that mutations in these genes resulted in hemin sensitivity, that expression of these genes is hemin activated, and that hemin activation is mediated by the HssRS system (40, 48). The hrtAB genes in C. diphtheriae appear to have a function similar to those reported in S. aureus and B. anthracis, which is the detoxification of high levels of hemin. Pfam analysis (14) of the C. diphtheriae HrtB permease indicates that it shares similarity with a family of proteins involved in the export of compounds through the cytoplasmic membrane, suggesting that the heme-sensitive phenotype observed in an hrtAB mutant is associated with a defect in the ability to export excess hemin or compounds produced in response to hemin exposure. Recent studies with S. aureus using mass spectrometry-based tracking did not confirm a function for HrtAB in heme secretion, suggesting that hemin metabolites or other toxic compounds may be the substrate for the HrtAB system in that species (40).
It was recently reported that proteins orthologous to HrtAB were present in various bacterial species within the phylum Firmicutes, including important human pathogens within the genera Bacillus, Staphylococcus, and Listeria (40, 42). To determine the prevalence of the HrtAB genes in other bacterial phyla, we performed a BLAST search using the C. diphtheriae HrtB sequence to identify orthologous proteins in the 1,200 completed bacterial genomes available in the NCBI database (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). The results identified two predominant phyla that contained organisms encoding orthologs of HrtB: the above-mentioned Firmicutes and also the Actinobacteria, which include genera related to Corynebacterium, Streptomyces, Arthrobacter, and Mycobacterium; however, no HrtB orthologs were identified in any Mycobacterium species. No organisms outside these two groups of Gram-positive bacteria and no Gram-negative bacteria encoded proteins with significant sequence similarity to HrtB. In addition to the HrtAB system, other factors are known to protect bacteria from hemin toxicity, including the unusual HbpS protein in Streptomyces reticuli, which has been proposed to degrade hemin at the cell surface (25). Various systems in Gram-negative bacteria have also been identified, including ShuS and HemS in Shigella dysenteriae and Yersinia enterocolitica, respectively (44, 51). The mechanisms of protection conferred by these proteins have not been determined, although it is proposed that these intracellular proteins bind hemin and function as either hemin storage or shuttle proteins.
We previously reported that C. diphtheriae strains with mutations in the chrSA genes are more sensitive to hemin than the wild-type strain (4), and it was further noted that this enhanced sensitivity was not associated with decreased expression of HmuO, an enzyme involved in heme degradation and previously shown to be activated by the ChrAS two-component system (4, 33). It was presumed that the increase in heme sensitivity in a ChrAS mutant was due to reduced expression of an as yet identified gene(s) whose transcription is elevated by the ChrAS regulatory proteins. In this study, we provide strong evidence that the increased hemin sensitivity observed in chrSA mutants is due to reduced expression of the ChrAS-regulated hrtAB genes. The hemin sensitivity in the chrSA mutant C7chrSAΔ was alleviated in the presence of plasmid pMnt-hrtAB, which contains the cloned hrtAB genes under transcriptional control of the mntA promoter, a promoter that is independent of ChrAS regulation (34).
The heme-activated hrtB promoter is the third promoter identified that is regulated by the ChrAS system and the only promoter in the ChrA regulon that is not also affected by the response regulator HrrA. Sequence analysis of the three promoter regions regulated by ChrA, which include hmuO, hrtB, and hemA, revealed the presence of the 10-bp sequence 5′-GGTTGATGTG-3′, which may serve as a possible binding site for ChrA. The hrtB promoter contained two copies of this sequence (9/10 match with consensus), which form a potential stem-loop structure located upstream of the putative −10 element. No sequences matching a strong −35 element were identified at an appropriate distance from the putative −10 region within the hrtB promoter, which is consistent with the requirement for an activator involved in the expression of these genes. Two copies of 5′-GGTTGATGTG-3′ separated by 40 bp are present upstream of the −10 element for the hmuO promoter, and a single copy that matches 8 of 10 residues for this sequence overlaps putative −10 sequences for the hemA promoter (Fig. 3). While additional studies are needed to confirm that these regions are important recognition sequences for the ChrA response regulator, the different relative locations and arrangements of these sequences suggest differences in the ChrA binding configurations at these various promoters. It is possible that the interaction of DtxR and HrrA influences ChrA binding at the hmuO promoter in a manner that is not observed with ChrA binding at the hrtB promoter, where HrrA and DtxR do not appear to have any direct role in regulation.
In C. diphtheriae, it has been well established that DtxR functions in an iron-dependent manner to repress expression of tox and numerous other genes involved in iron uptake and metabolism (6, 18, 37). While DtxR functions as an iron-dependent repressor at the ChrAS-regulated hmuO promoter, this regulatory protein appears to function as an activator at the hrtB promoter. It was observed that expression of the hrtB promoter is reduced in a dtxR mutant, and it was further noted that the presence of the iron chelator EDDA resulted in a 40% decrease in expression of hrtB in the wild-type strain. Since hemoglobin or hemin is required to activate expression of hrtB, the presence of the chelator resulted in only a modest reduction in biologically relevant iron levels, as measured by expression of the tox gene, so the difference between expression under high iron levels and that under very low levels is not possible to determine. Nevertheless, these findings suggest that DtxR functions as an activator at hrtB and that optimal expression occurs under iron-replete conditions. Analysis of the region upstream of the hrtB gene failed to identify sequences with significant similarity to the consensus DtxR binding site, which suggests that DtxR may affect expression of the hrtB promoter through indirect mechanisms. DtxR has not been shown to function as an activator of gene expression in C. diphtheriae. However, orthologs of DtxR in Mycobacterium tuberculosis (16) and in Corynebacterium glutamicum (10) are reported to function as both repressors and activators. IdeR, the DtxR ortholog in M. tuberculosis, activates transcription by directly binding to regions upstream of regulated promoters (16).
A comparison of C. diphtheriae ChrAS with S. aureus and B. anthracis HssRS suggests that these signal transduction systems have similar roles in the regulation of gene expression through a heme-responsive mechanism. However, several notable differences exist between these systems. The HssRS systems have been shown to regulate only the hrtAB promoter, while ChrAS controls the expression of at least three promoters, all of which are associated with various aspects of heme metabolism. Although the response regulators ChrA and HrrR exhibit some sequence homology, the similarity is primarily in regions that are commonly conserved in numerous other response regulators within this group of related regulatory proteins (27). Surprisingly, the cognate sensor kinases for these systems, ChrS and HssS, share no significant sequence similarities (data not shown) (40). The N-terminal sensor domains of these proteins, which are thought to be critical in the detection of the environmental signal and in anchoring the proteins to the membrane, are remarkably different not only in sequence but also in their predicted structures. We show in this study that ChrS contains 6 transmembrane helices in its N-terminal domain, joined by short loop regions, whereas the sequence of HssS is predicted to contain only two transmembrane domains and one large extracellular loop region. A recent analysis of point mutations in conserved residues in the B. anthracis HssS N-terminal region identified mutants that had reduced activity, suggesting that this region is important in signal transduction and in responding to hemin exposure; however, in an expanded analysis of all HssS orthologs, no conserved residues that are known to coordinate with the axial iron atom of heme were identified (40). We have also identified conserved residues in the ChrS N-terminal region that are important for activity; the most notable finding from this analysis is that a mutation in a highly conserved tyrosine (Y61) results in virtually complete loss of ChrS function. Tyrosine residues are known to be important in the coordination of heme in numerous proteins (28). While it is possible that the Y61 residue may be critical in recognition of hemin or a hemin metabolite, additional studies will be needed to confirm the function of this specific residue in signal transduction.
A recent report described the expression and purification of the ChrS protein in E. coli and showed that autophosphorylation of ChrS was enhanced in the presence of hemin (17). Purified ChrS that was incorporated into proteoliposomes gave an absorbance peak of 405 nm in the presence of hemin, suggesting that ChrS directly binds hemin, although the specific nature of the binding and location of the binding site were not determined. It was also noted that the proteoliposomes in the absence of ChrS also bound hemin and gave significant background absorbance in these studies. Numerous attempts in our laboratory to overexpress and purify ChrS have not been successful, even when using techniques very similar to those previously reported (17). Even moderate expression of ChrS appears to be detrimental to E. coli strains used in expression studies, presumably due to the presence of the multiple membrane-spanning regions. Small amounts of ChrS that were expressed in our studies could not be purified as glutathione S-transferase (GST) fusions, presumably due to unusual structural conformations. These fusion proteins also failed to bind to hemin agarose, a technique we previously employed to demonstrate hemin binding to the HmuT protein (13; M. P. Schmitt, unpublished results).
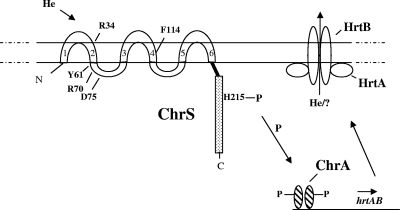
In this study, we show that transcriptional activation of the hrtB promoter is mediated by the ChrAS signal transduction system in response to hemin. As depicted schematically in Fig. 6, it is postulated that hemin or heme-associated factors interact with ChrS, resulting in autophosphorylation at H215, which is followed by the transphosphorylation of ChrA. Phosphorylated ChrA is presumed to undergo a conformational change allowing it to interact with sequences upstream of the hrtB promoter, resulting in activation of transcription. The products of hrtAB are predicted to function as an efflux pump to remove excess levels of hemin or factors produced in response to hemin exposure. If hemin directly interacts with ChrS, as proposed in a recent study (17), then amino acid residues that we have identified that are critical for activity may be associated with hemin binding. Interestingly, 3 of the 5 residues that reduce heme-responsive ChrS function, including the highly conserved Y61, are clustered in a loop region that is predicted to be intracellular, suggesting that the sensing of the signal may occur in the cytosol. Membrane-anchored histidine kinases that detect cytosolic signals are unusual but have been previously described (22). Confirmation that ChrS detects a signal in the cytosol at its N terminus will require additional studies, since other resides that are important for function, such as the conserved R34, are predicted to be outside the membrane. A sensor that is responsive to intracellular heme levels would be consistent with the proposed function of the various ChrAS-regulated systems, which either interact directly with intracellular heme (HmuO and HrtAB) or are involved in the maintenance of heme homeostasis (HemA and associated heme biosynthetic enzymes).
FIG. 6.
Schematic representation of ChrSA regulation of hrtAB and proposed function of HrtAB. In the presence of a heme source, hemin or heme-associated factors are proposed to interact with the membrane-anchored N terminus of ChrS, which results in autophosphorylation at H215 and the subsequent transphosphorylation of ChrA. Phosphorylated ChrA is proposed to bind upstream of the hrtB promoter to activate transcription. The products of hrtAB are predicted to be components of an ABC transporter that may function as an efflux pump to export hemin or factors produced in response to hemin exposure. The locations of amino acids that were identified from mutagenesis studies and are critical for ChrS function are indicated in the ChrS sensor domain. The six transmembrane regions in the N-terminal of ChrS are numbered 1 to 6.
Acknowledgments
We thank Scott Stibitz and Jon Burgos for helpful comments on the manuscript.
Footnotes
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Allen, C. E., and M. P. Schmitt. 2009. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J. Bacteriol. 191:2638-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., G. Warren, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Barksdale, W. L., and A. M. Pappenheimer, Jr. 1954. Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J. Bacteriol. 67:220-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibb, L. A., N. D. King, C. A. Kunkle, and M. P. Schmitt. 2005. Analysis of a heme-dependent signal transduction system in Corynebacterium diphtheriae: deletion of the chrAS genes results in heme sensitivity and diminished heme-dependent activation of the hmuO promoter. Infect. Immun. 73:7406-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibb, L. A., C. A. Kunkle, and M. P. Schmitt. 2007. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun. 75:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, J. M., O. N. Manish, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. U. S. A. 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, V. 2005. Bacterial iron transport related to virulence. Contrib. Microbiol. 12:210-233. [DOI] [PubMed] [Google Scholar]
- 8.Brickman, E., and J. Beckwith. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J. Mol. Biol. 96:307-316. [DOI] [PubMed] [Google Scholar]
- 9.Brown, J. S., and D. W. Holden. 2002. Iron acquisition by gram-positive bacterial pathogens. Microbes Infect. 4:1149-1156. [DOI] [PubMed] [Google Scholar]
- 10.Brune, I., H. Werner, A. T. Huser, J. Kalinowski, A. Puhler, and A. Tauch. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerdeno-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier, R. J. 2001. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon 39:1793-1803. [DOI] [PubMed] [Google Scholar]
- 13.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from hemin and hemoglobin are homologous to ABC hemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 14.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, J. S. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman, D. B., D. L. Stauff, G. Pishchany, C. W. Whitwell, V. J. Torres, and E. P. Skaar. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PloS Pathog. 2:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gold, B., G. M. Rodriguez, S. A. Marras, M. Pentecost, and I. Smith. 2001. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42:851-865. [DOI] [PubMed] [Google Scholar]
- 17.Ito, Y., S. Nakagawa, A. Komagata, M. Ikeda-Saito, Y. Shiro, and H. Nakamura. 2009. Heme-dependent autophophorylation of a heme sensor kinase, ChrS, from Corynebacterium diphtheriae reconstituted in proteoliposomes. FEBS Lett. 583:2244-2248. [DOI] [PubMed] [Google Scholar]
- 18.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkle, C. A., and M. P. Schmitt. 2005. Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J. Bacteriol. 187:422-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkle, C. A., and M. P. Schmitt. 2007. Comparative analysis of hmuO function and expression in Corynebacterium species. J. Bacteriol. 189:3650-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, C. B. 1995. Quelling the red menace: hemin capture by bacteria. Mol. Microbiol. 18:383-390. [DOI] [PubMed] [Google Scholar]
- 22.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70:910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 24.Oram, D. M., A. D. Jacobson, and R. K. Holmes. 2006. Transcription of the contiguous sigB, dtxR, and galE genes in Corynebacterium diphtheriae: evidence for multiple transcripts and regulation by environmental factors. J. Bacteriol. 188:2959-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortiz de Orué Lucana, D., and M. R. Groves. 2009. The three-component signaling system HbpS-SenS-SenR as an example of a redox sensing pathway in bacteria. Amino Acids 37:479-486. [DOI] [PubMed] [Google Scholar]
- 26.Otto, B. R., V. V. Vught, A. M. Verweij-van Vught, and D. M. MacLaren. 1992. Transferrins and heme compounds as iron sources for pathogenic bacteria. Crit. Rev. Microbiol. 18:217-233. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 28.Pilpa, R. M., S. A. Robson, V. A. Villareal, M. L. Wong, M. Phillips, and R. T. Clubb. 2009. Functionally distinct NEAT (near transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J. Biol. Chem. 284:1166-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popovic, T., S. Y. Kombarova, M. W. Reeves, H. Nakao, I. K. Mazurova, M. Wharton, I. K. Wachsmuth, and J. D. Wenger. 1996. Molecular epidemiology of diphtheria in Russia, 1985-1994. J. Infect. Dis. 174:1064-1072. [DOI] [PubMed] [Google Scholar]
- 30.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt, M. P. 1997. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenase and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt, M. P. 1997. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 65:4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt, M. P. 1999. Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J. Bacteriol. 181:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt, M. P. 2002. Analysis of a DtxR-like metalloregulatory protein, MntR, from Corynebacterium diphtheriae that controls expression of an ABC metal transporter by an Mn2+-dependent mechanism. J. Bacteriol. 184:6882-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt, M. P. 2004. Corynebacterium diphtheriae, p. 344-359. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 36.Schmitt, M. P., and E. S. Drazek. 2001. Construction and consequences of directed mutations affecting the hemin receptor in pathogenic Corynebacterium species. J. Bacteriol. 183:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt, M. P., and R. K. Holmes. 1991. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect. Immun. 59:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt, M. P., E. M. Twiddy, and R. K. Holmes. 1992. Purification and characterization of the diphtheria toxin repressor. Proc. Natl. Acad. Sci. U. S. A. 89:7576-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stauff, D. L., and E. P. Skaar. 2009. The heme sensor system of Staphylococcus aureus. Contrib. Microbiol. 16:120-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stauff, D. L., and E. P. Skaar. 2009. Bacillus anthracis HssRS signaling to HrtAB regulates heme resistance during infection. Mol. Microbiol. 72:763-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stauff, D. L., V. J. Torres, and E. P. Skaar. 2007. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J. Biol. Chem. 282:26111-26121. [DOI] [PubMed] [Google Scholar]
- 43.Stibitz, S., and M. Yang. 1991. Subcellular localization and immunological detection of proteins encoded by the vir locus of Bordetella pertussis. J. Bacteriol. 173:4288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stojiljkovic, I., and K. Hantke. 1994. Transport of hemin across the cytoplasmic membrane through a hemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 45.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281-295. [DOI] [PubMed] [Google Scholar]
- 46.Tai, S.-P. S., A. E. Krafft, P. Nootheti, and R. K. Holmes. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb. Pathog. 9:267-273. [DOI] [PubMed] [Google Scholar]
- 47.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 48.Torres, V. J., D. L. Stauff, G. Pishchany, J. S. Bezbradica, L. E. Gordy, J. Iturregui, K. L. Anderson, P. M. Dunman, S. Joyce, and E. P. Skaar. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 19:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilks, A., and M. P. Schmitt. 1998. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]
- 50.Winkelmann, G. 2002. Microbial siderophore-mediated transport. Biochem. Soc Trans. 30:691-696. [DOI] [PubMed] [Google Scholar]
- 51.Wyckoff, E. E., G. F. Lopreato, K. A. Tipton, and S. M. Payne. 2005. Shigella dysenteriae ShuS promotes utilization of heme as an iron source and protects against heme toxicity. J. Bacteriol. 187:5658-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]