Abstract
Studying the biogenesis of the Thermus thermophilus cytochrome ba3 oxidase, we analyze heme a cofactor insertion into this membrane protein complex. Only three proteins linked to oxidase maturation have been described for this extreme thermophile, and in particular, no evidence for a canonical Surf1 homologue, required for heme a insertion, is available from genome sequence data. Here, we characterize the product of an open reading frame, cbaX, in the operon encoding subunits of the ba3-type cytochrome c oxidase. CbaX shares no sequence identity with any known oxidase biogenesis factor, and CbaX homologues are found only in the Thermaceae group. In a series of cbaX deletion and complementation experiments, we demonstrate that the resulting ba3 oxidase complexes, affinity purified via an internally inserted His tag located in subunit I, are severely affected in their enzymatic activities and heme compositions in both the low- and high-spin sites. Thus, CbaX displays typical features of a generic Surf1 factor essential for binding and positioning the heme a moiety for correct assembly into the protein scaffold of oxidase subunit I.
Terminal oxidases catalyze the reduction of molecular oxygen to water, coupling the available redox energy to proton translocation across the cytoplasmic (or mitochondrial inner) membrane. Most oxidases belong to the heme/copper superfamily, which is quite diverse in terms of electron donors, subunit compositions, and heme types (30). Two different terminal cytochrome c oxidase complexes have been described for the extremely thermophilic eubacterium Thermus thermophilus, both members of this superfamily. In contrast to the caa3 oxidase, the ba3 oxidase is expressed under limited oxygen supply (20) and is composed of three protein subunits, I, II, and IIa. The most prominent features are associated with subunit I, the core element of the complex, where a protein scaffold consisting of 13 transmembrane helices provides the ligands for hemes b and as3 (23). Heme as3, together with CuB, forms the active site, and in Thermus is characterized by the presence of a hydrophobic hydroxyethyl geranylgeranyl moiety instead of the hydroxyethyl farnesyl side chain found in conventional a-type hemes. All of the N termini of the three subunits are located on the cytoplasmic (negative) side of the membrane, whereas the hydrophilic domain of subunit II containing the primary electron acceptor, a mixed-valence binuclear CuA center, faces the periplasmic space (32).
The biogenesis of cytochrome c oxidases involves numerous steps in which specific chaperones assist the translation of subunits, their insertion or translocation across the membrane, the integration of essential cofactors, and the assembly and final maturation of the enzyme complex. This assembly process, and in particular the exact order of events, is still not very well understood, and most effort has been directed to the mitochondrial enzyme (14, 17, 21, 36).
In this work, we show that the ba3 locus comprising the three structural genes for the complex (cbaDBA) also encodes two additional putative proteins (CbaX and CbaY) downstream from the subunit I gene on the same transcript. One of them, CbaX, is obviously required for proper maturation of the enzyme, because its deletion results in a severe deficit in the heme a content and consequently a drastic decrease in ba3 oxidase activity. From the analysis of the cytochrome ba3 transcription unit and from complementation studies, we conclude that CbaX is critically involved in heme a incorporation into the high-spin site of subunit I of the ba3 cytochrome c oxidase. Even though it lacks any sequence homology, it shares essential functional similarities with the previously identified Surf1 protein (5, 6, 25, 31, 35), which is required for the biogenesis of the mitochondrial oxidase and many bacterial oxidases. Furthermore, we demonstrate that an unbalanced expression of cbaX and the oxidase subunit genes leads to incorporation of heme a even into the low-spin site of the enzyme.
MATERIALS AND METHODS
Construction of the ba3 deletion strains.
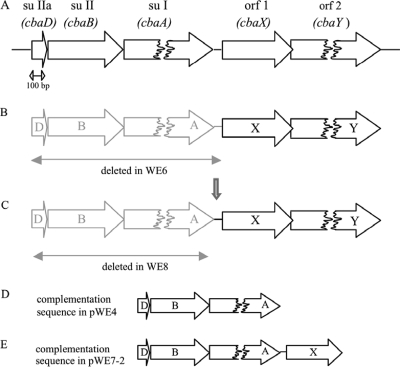
All deletion strains (Fig. 1) were obtained by double homologous recombination using suicide plasmids, each a derivative of pUC18. The structural genes of the ba3 oxidase alone (see i below), as well as together with a 56-bp fragment downstream of subunit I (see ii below), were replaced by the bleomycin resistance gene under the control of the surface layer protein A gene promoter (slAp) (13). The bleomycin resistance gene, including slAp, was amplified via PCR using the plasmid pWUR-bleo (4) and the primer pair 17/18 (primer sequences are listed in Table 1). The forward primer (primer 17) introduced an XbaI site, and the reverse primer (primer 18) introduced a PstI site. (i) Upstream and downstream regions flanking the ba3 locus (flank A and flank B) were amplified via PCR using T. thermophilus HB27 genomic DNA. For flank A (genomic positions 747813 to 748315) the forward primer (primer 1) introduced an EcoRI site, and the reverse primer (primer 2) introduced an XbaI site. For flank B (genomic positions 750807 to 751361), the forward primer (primer 5) gave rise to a PstI site, and the reverse primer (primer 4) introduced a HindIII site. The resulting products (506 bp for flank A and 558 bp for flank B) were digested and cloned via a four-fragment ligation, together with the digested bleomycin resistance gene, into pUC18, leading to pWE8 (Fig. 1C). (ii) Flank A was produced as described above; for flank B′ (genomic positions 750861 to 751361), the forward primer (primer 3) gave rise to a PstI site, and the reverse primer (primer 4) introduced a HindIII site. The resulting products (506 bp for flank A and 502 bp for flank B′) were digested and again cloned via a four-fragment ligation, together with the digested bleomycin resistance gene, into pUC18, leading to pWE6 (Fig. 1B).
FIG. 1.
Schematic drawing of the cytochrome ba3 operon and its deletion and complementation constructs. (A) The ba3 locus (GeneID, 2775625, 2775353, and 2775333) contains the structural genes for ba3 subunits (su) IIa, II, and I, as well as two ORFs encoding proteins with previously unknown functions. The distance between the stop codon of su I and the start codon of ORF 1 is 56 bp. (B to E) In the deletion strain WE6, the structural genes of the oxidase, as well as the stretch of 56 bp downstream, are deleted (B) and exchanged for a bleomycin resistance gene, while in WE8 (C), the 56 bp (vertical arrow) are maintained in the genome. For complementations, pWE4 (D) contains only the structural genes for subunits IIa, II, and I, while pWE7-2 (E) in addition carries the 56-bp intergenic sequence and the cbaX gene.
TABLE 1.
Sequences of primers used in this worka
| No. | Sequence (5′→3′) |
|---|---|
| 1 | CCGGAATTCGGATCTACAGCAAGGTGCC |
| 2 | GCTCTAGATTAAGACCCGCCCGCCAC |
| 3 | AACCAACTGCAGATGCTCCTCGCCCGCGC |
| 4 | CCCCAAGCTTGCCGAGGAGGTAGACCAG |
| 5 | GGTAACTCTGCAGCCTAGGCCCTTTGTCCCTTGACC |
| 6 | ATACTAGCAGCATATGGAAGAAAAGCCCAAAGGCGC |
| 7 | GTGATGGTGATGGTGATGGTGATGGTGATGCCCGGCGGGGTTCGCCGCCTTCCAGC |
| 8 | CATCACCATCACCATCACCATCACCATCACGCCGGGGCCCCAGGGAAGGTGACCC |
| 9 | TGATTATCTGCAGCTACCAGAGCCGCCACCCCGGC |
| 10 | CCTTCGTCCAGTCCTACTACC |
| 11 | GAGCTATCTGCAGCCTTACCCAGGATAGG |
| 12 | GCAGCCCGGGGGATCCACTAGAGTCGAGG |
| 13 | TCACGGCCGGGCCCTCCAGAG |
| 14 | ATCTCCGACGCGCAAAG |
| 15 | TTAGCATTGATCTGCAGTCACGGCCGGGCCCTCCAGAGGAGC |
| 16 | ATGGCCAAGTTGACCAGTGCCGTTC |
| 17 | CTGAATCTAGAGGCGGCGCATGCATTCCTGG |
| 18 | ATAGTCTGCAGCTTCCGGCTCAGATCTTGTGTGG |
See Materials and Methods.
Each suicide plasmid was electroporated (10) into T. thermophilus wild-type strain HB27 (10), and bleomycin-resistant colonies grown at 70°C on plates in the presence of 100 μg/ml bleomycin were checked by dot blot analysis. The deletion of interest was verified via PCR with genomic DNA and by Western blot analysis using antibodies directed against subunit II of the ba3 oxidase.
Cloning of ba3 oxidase complementation plasmids.
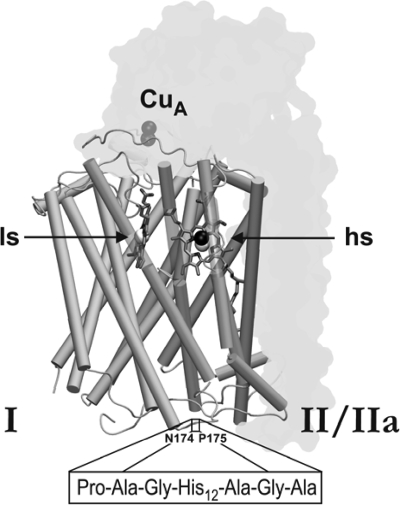
Introduction of a polyhistidine coding region into subunit I followed the gene splicing by overlap extension (SOEing) method of Horton (18). The 12-histidine tag was placed between amino acids N174 and P175 of subunit I into a cytoplasmically located loop using the primer pairs 6/7 and 8/9, respectively. These primers introduce an NdeI and a PstI site and also contain a “linker sequence” coding for three additional amino acids on each side of the histidine coding sequence (Pro-Ala-Gly-His12-Ala-Gly-Ala) (see Fig. 3). The PCR product was digested and cloned into pDM12 (27), an Escherichia coli/Thermus shuttle vector, which contains the Thermus cytochrome bc complex promoter, resulting in pWE4 (Fig. 1D). pWE7 was obtained by restriction of pWE4 with NdeI and BamHI. The 6,526-bp fragment was ligated with the digested PCR product, obtained with primers 10/11 (introducing a PstI site) and T. thermophilus HB27 genomic DNA as a template. pWE7-2 (Fig. 1E) was generated by an inverse-PCR technique (26) using primers 12/13, with pWE7 as a template. These complementation plasmids were electroporated (10) into T. thermophilus WE6 and WE8, as before.
FIG. 3.
Schematic representation of the T. thermophilus ba3 oxidase. Subunits II (harboring the CuA center) and IIa are shown as grey transparent outlines, while subunit I is displayed mainly with its 13 transmembrane helices, partly presented as transparent segments to increase cofactor visibility. The low-spin heme b and the high-spin heme a3/CuB site of subunit I are denoted by ls and hs, respectively. The boxed amino acid sequence corresponds to the internal His tag in subunit I, inserted into the cytoplasmic loop connecting transmembrane helices IV and V between residues N174 and P175, embedded by three additional linker amino acids on either side, as indicated (Protein Data Bank [PDB] code, 1EHK; graphics generated by VMD [http://www.ks.uiuc.edu/Research/vmd/]).
cDNA synthesis with isolated total RNA.
Total RNAs from different Thermus strains were isolated using the SV total RNA isolation system kit (Promega, Germany), and cDNA synthesis was performed with a RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) using primer 15, each according to the manuals. PCR on synthesized cDNA was done using the primer pair 14/15 or 16/15. The isolated RNA without cDNA synthesis was used as a negative control and pWE7 and pWE8 plasmids as positive controls; identical template concentrations were employed whenever possible.
Growth conditions and membrane preparation.
All Thermus strains were grown at 70°C in LB medium containing 25 μg/ml kanamycin and 100 μg/ml bleomycin, where appropriate, with rotary shaking in unbaffled flasks. After 15 to 20 h of growth, cells were collected and membranes were prepared by established methods (28). The protein concentration was determined using a modified Lowry protocol (22, 24).
Membrane solubilization and protein purification.
Membranes were solubilized in the presence of 3% (wt/vol) Triton X-100 in buffer (50 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole, pH 8.0) at a final protein concentration of 10 mg/ml. The solubilized material was loaded on an Ni-nitrilotriacetic acid (NTA) column (Qiagen), and Triton X-100 was exchanged by washing with 10 column volumes (CV) of buffer (as described above) containing 0.02% (wt/vol) n-dodecyl-β-d-maltoside (DDM) and 20 mM imidazole. After the column was washed with 5 CV of detergent buffer containing 60 mM imidazole, His-tagged ba3 was eluted from the column with buffer containing 150 mM imidazole. To remove the imidazole, the protein was immediately loaded on a Sephacryl S-200 column and eluted with buffer (150 mM Tris-HCl, 150 mM NaCl, 0.02% [wt/vol] DDM, pH 8.0).
Spectral analysis.
Redox difference spectra were recorded in the visible range on a Hitachi U-3000 spectrophotometer using potassium ferricyanide for oxidation and sodium dithionite for reduction. For pyridine hemochromogen redox difference spectra, samples were diluted in 20% (vol/vol) pyridine, 0.1 M NaOH. Native (see i below) and pyridine hemochromogen (see ii below) redox spectra were recorded, and the heme/protein concentrations were determined using the Lowry determination (see above) and the following extinction coefficients: (i) Δɛ560-690, 26 cm−1 mM−1 for heme b (8), and Δɛ613-658 nm, 6.3 cm−1 mM−1 for heme a (37); (ii) Δɛ557-540 nm, 22.1 cm−1 mM−1 for heme b (33), and Δɛ587-620 nm, 21.7 cm−1 mM−1 for heme a (33). For cyanide binding spectra, samples were reduced with dithionite, and potassium cyanide (neutralized) was added to the samples to a final concentration of 100 mM.
Enzymatic-activity measurements.
Activity measurements were recorded at room temperature on a Hitachi U-3000 spectrophotometer using 20 μM reduced T. thermophilus cytochrome c552 (50 mM Tris-HCl, 0.02% [wt/vol] DDM, pH 8.0; Δɛ552 nm = 21 cm−1 mM−1).
RESULTS
The cytochrome ba3 oxidase locus contains additional ORFs in its transcription unit.
The aim of the project presented here was to establish the organization of the cytochrome ba3 (cba) locus, since the genomic sequence identifies two additional open reading frames (ORFs) only 56 bp downstream of the three structural genes of the ba3 oxidase itself (Fig. 1A). A BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of ORF 1 (here termed cbaX) did not reveal any specific motif for the putative protein (CbaX) and indicated similar sequences only in Thermus aquaticus (67% sequence identity) and two Meiothermus strains (Meiothermus silvanus, 34% sequence identity; Meiothermus ruber, 35% sequence identity). A topology search with TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) identified the gene product as a membrane protein of 156 amino acids (16.8 kDa) and predicted three transmembrane helices; with respect to its functional role, we note that the protein lacks any histidine residues as potential heme ligands (see Discussion). The gene product of ORF 2 (cbaY, overlapping with cbaX by 4 nucleotides) aligned with a number of sequences, mostly annotated as permeases, with a conserved major facilitator superfamily (MFS) domain.
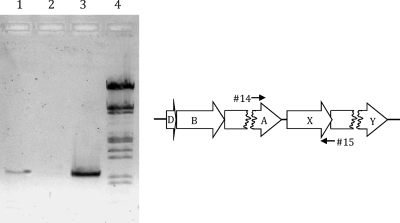
The intergenic distance between cbaA (encoding subunit I [Fig. 1A]) and cbaX is 56 bp and therefore long enough to potentially accommodate its own promoter element. To address this issue, total RNA was isolated from T. thermophilus HB27 wild-type cells, and cDNA was synthesized using a gene-specific primer that binds at the 3′ end of cbaX (see Materials and Methods). The primers used for PCR with this synthesized cDNA (Fig. 2) give rise to a product only if cbaX is part of the same transcription unit as the preceding ba3 structural genes. The PCR product obtained had a size identical to that of the 1,030-bp positive control. This clearly shows that the ba3 transcription unit includes at least cbaX, so that the 56 bp of intervening DNA most likely contains a ribosome binding site (RBS) but no separate promoter acting on cbaX. Due to the low degree of consensus between different RBSs in T. thermophilus (11), its exact position remains unclear. The cbaX and cbaY reading frames overlap by 4 nucleotides, strongly suggesting that the downstream cbaY is part of the cba transcript, as well.
FIG. 2.
Transcriptional analysis of cbaX. Total RNA was isolated from T. thermophilus HB27, and cDNA was produced using the gene-specific primer 15. After cDNA synthesis, PCR was performed using the primer pair 14 and 15 on different templates. Lane 1, cDNA as a template; lane 2, RNA as a template (negative control); lane 3, plasmid DNA as a template (positive control; pWE7) (see Materials and Methods); lane 4, λ DNA marker (restricted with EcoRI and HindIII; relevant fragment sizes, from bottom: 0.83, 0.95, 1.38, and 1.58 kb).
Deletion of cbaX leads to severe loss of heme a in the purified ba3 oxidase.
To assign the cbaX gene product a functional role, a genomic deletion of the structural genes (as well as the promoter region) of the ba3 oxidase, including the downstream stretch of 56 bp, was generated (strain WE6) (Fig. 1 B), preserving, on the other hand, as much of the ORFs in question as possible. Assuming an RBS was present within the 56 bp downstream of cbaA, this deletion strain should no longer have been able to express the CbaX protein. Strain WE6 was then complemented by a plasmid encoding only the ba3 subunits (pWE4) (Fig. 1 D). For ease of purification, subunit I was provided with an internal 12-mer His tag located on a cytoplasmically oriented loop (Fig. 3) between transmembrane helices IV and V, thus avoiding any interference with cytochrome c552 binding on the periplasmic side of the enzyme complex. A C-terminal tag position resulted in a nonfunctional enzyme (not shown), possibly due to steric hindrance, while tag lengths beyond 7 residues on the N terminus (8) apparently are also not tolerated (C. Werner, unpublished data). The internally tagged enzyme, when expressed and purified from HB27 wild-type cells, allowed a single-step purification leading to an oxidase preparation (estimated to have >95% purity) (not shown) that was fully competent in its enzymatic and spectral properties (Fig. 4, pWE4/HB27) and provided a convenient tool to quickly assess the putative effects of a cbaX deletion.
FIG. 4.
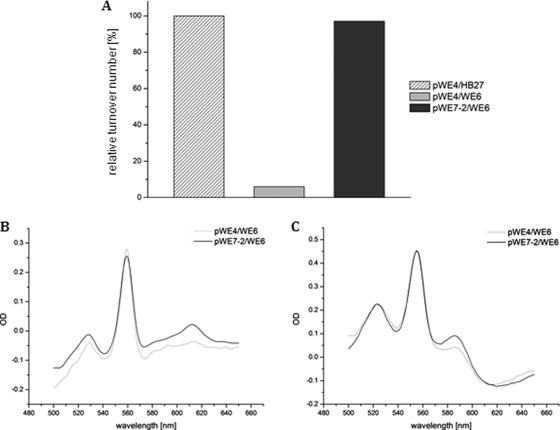
Spectral and enzymatic properties of the affinity-purified cytochrome ba3 oxidase from different strains, recording relative turnover activity (A), as well as native (B) and pyridine (C) difference spectra. (A) pWE4/HB27 served as the recombinant, internally his-tagged wild type in the activity test, using 20 μM of reduced T. thermophilus cytochrome c552 at room temperature. The 100% value corresponded to a molar activity of 70 s−1. (B and C) Complementation of WE6 with pWE4 leads to a heme a/b ratio of less than 0.1:1, as determined by the native spectrum (B, gray line). The pyridine spectrum (C, gray line) displayed a heme a/b ratio of about 0.57:1, while the enzymatic activity (A, gray) was decreased to 7% of the wild type. The complementation of WE6 with pWE7-2 led to a heme a/b ratio of 1:1, as determined by native and pyridine spectra (B and C, black lines), and the enzymatic activity was comparable to the wild type (A, dark gray). The protein concentration for all spectra was 10 μM. OD, optical density.
The loss of the putative cbaX gene product drastically lowered the ba3 activity to about 7% of that of the wild type (Fig. 4A), and less than 10% of the expected heme a content was deduced from the typical peak position at 613 nm in the native redox spectrum of the purified oxidase (Fig. 4B). However, we note that the trace of the redox spectrum appeared rather “noisy,” with small spectral features present on either side of this peak. In contrast to the native spectrum with its severe loss of the heme a peak at 613 nm, the pyridine hemochromogen spectrum revealed that around 60% of the wild-type heme a level was detectable in the enzyme complex (see Discussion). Heme b was not affected by the deletion of cbaX, as the specific heme b content, based on native and denaturating spectra (Fig. 4C), matched the wild-type value.
To prove that the diminished oxidase activity, as well as the loss of heme a3, was indeed caused by the deletion of cbaX, a second complementation plasmid was generated, comprising the ba3 structural genes, the 56-bp intergenic region, and cbaX (pWE7-2) (Fig. 1E). After complementation, the resulting ba3 enzyme was again affinity purified and examined (Fig. 4): both the activity and the heme a content of the purified ba3 oxidase were fully restored in this strain.
A genomic deletion of cbaY was also generated, but it apparently had no deleterious effects on the spectral and catalytic properties of the enzyme (data not shown); however, the total yield of purified enzyme was more than 10 times lower in this case.
Expression of cbaX under the control of a strong promoter leads to partial incorporation of heme a into the low-spin site of the ba3 cytochrome c oxidase.
To separate the expression of the three structural genes of the cytochrome ba3 oxidase from that of the cbaX gene, we constructed a second deletion strain in which the structural genes of the ba3 oxidase were deleted while the intergenic stretch of 56 bp was still present (WE8) (Fig. 1C, vertical arrow). Here, the structural genes of the ba3 oxidase were exchanged for the bleomycin resistance gene placed under the control of the slpA promoter. To confirm a transcriptional read-through into the cbaX gene, thereby mimicking the wild-type situation, we ascertained that no transcriptional terminator was present at the 3′ end of the bleomycin resistance gene. For experimental confirmation, total RNA was isolated from this deletion strain, and again, cDNA was produced with the same gene-specific primer binding at the 3′ end of cbaX. The primer pair used for this PCR with the synthesized cDNA as a template can result in a product only if cbaX is cotranscribed, this time with the bleomycin resistance gene. Figure 5 shows that indeed a PCR product with a length identical to the 1 kb of the positive control was obtained, clearly demonstrating cotranscription of both genes.
FIG. 5.
Transcriptional analysis of the bleomycin transcript in WE8. Total RNA was isolated from the deletion strain WE8 (Fig 1C), and cDNA was produced using a gene-specific primer (primer 15). After cDNA synthesis, PCR was performed using the primer pair 16 and 15 on different templates. In the case of cDNA as the template (lane 2), the presence of a product of 1,000 bp confirmed that cbaX is part of the same transcript as the bleomycin resistance gene. Lane 1, λ DNA marker (restricted with EcoRI and HindIII); lane 2, cDNA as the template; lane 3, RNA as the template (negative control); lane 4, plasmid DNA as the template (positive control; pWE8) (see Materials and Methods).
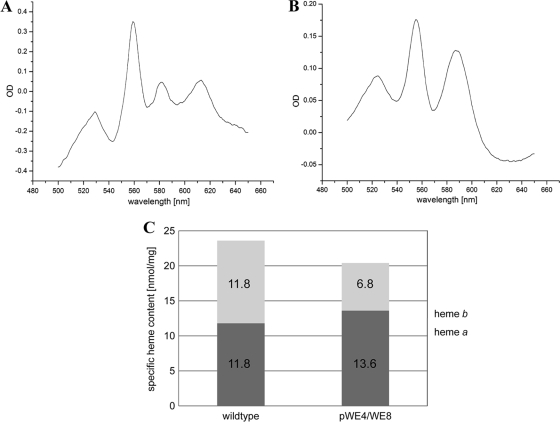
To check the phenotype of the ba3 oxidase, strain WE8 was complemented with a plasmid carrying only the ba3 subunit genes with the internal His tag in subunit I (pWE4) (Fig. 1D). The resulting enzyme was purified and assayed for its enzymatic, as well as spectral, properties. The native spectrum (Fig. 6 A) showed the typical heme b (560-nm) and heme a (613-nm) peaks, but unexpectedly, one additional peak at 582 nm arose. To reveal its identity, heme extraction and separation (data not shown) were performed and a pyridine hemochromogen spectrum (Fig. 6B) was recorded, both confirming the presence of no heme type other than a and b. The expressed oxidase in this strain showed an increase of the heme a/b ratio to 2:1 and a decrease in the turnover number to about 65% of that of the wild-type ba3 enzyme. Determination of the specific heme content (Fig. 6C) verified that the general heme (a + b)-to-protein ratio was not influenced, i.e., the purified enzyme did not carry any surplus heme.
FIG. 6.
Redox spectra and specific heme content of the affinity-purified oxidase from strain pWE4/WE8. (A) Besides the typical heme b peak at 560 nm and the heme a peak at 613 nm, an additional peak at 582 nm was visible in the native spectrum. (B) The pyridine spectrum, however, revealed only heme b at 557 nm and heme a at 587 nm. (C) While the absolute value for the specific heme content of the ba3 oxidase purified from pWE4/WE8 was 20.4 nmol/mg (hemes a and b), close to the value for the wild type (23.6 nmol/mg), the heme a/b ratio had been shifted to 2:1 in the ba3 oxidase from the complemented strain relative to the wild type.
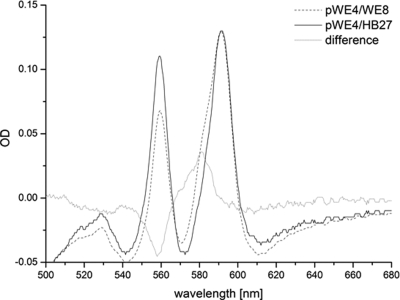
Our finding that the complementation of WE8 leads to a distorted heme a/b ratio raised the question of how the high- and low-spin sites of the enzyme are populated by both heme types. To determine this, a spectrum of the isolated, reduced, and cyanide-liganded enzyme was recorded (Fig. 7). In contrast to the oxidized form of the ba3 oxidase (32), the reduced form readily reacts with cyanide, and a shift of the heme a peak in the α-region from 613 nm to 592 nm has been reported for the five-coordinate a3 site (29), while hemes present in the six-coordinate low-spin site were not affected. Accordingly, the recorded wild-type spectrum (Fig. 7, solid line) showed two peaks at 560 nm (low-spin heme b) and at 592 nm (high-spin heme a3). When the enzyme isolated from strain pWE4/WE8 was analyzed (Fig. 7, dotted line), an additional shoulder at around 580 nm was observed. The difference spectrum (Fig. 7, gray line) revealed a loss of signal for the heme b peak at 560 nm and a distinct extra peak at 582 nm. A comparison of Fig. 6 and 7 clearly shows that neither the position of the 560-nm heme b peak nor that of the extra 582-nm heme a peak was shifted by cyanide treatment. Due to the observed inaccessibility for external ligands, we therefore conclude that both heme types, a and b, are found in the low-spin site in contrast to the wild-type situation: heme a populating this low-spin site replaces one-third of the regular heme b moiety (Fig. 7).
FIG. 7.
Cyanide spectra of the affinity-purified cytochrome ba3 oxidase from the wild type and from strain WE8 complemented with pWE4 (Fig. 1). The high-spin heme a peak (pWE4/HB27) shifted from 613 nm to 592 nm on cyanide incubation, while the low-spin heme b peak remained at 560 nm. In the case of pWE4/WE8, the spectrum displayed a shoulder near 582 nm (dotted line), clearly visualized in the difference spectrum (pWE4/WE8 minus pWE4/HB27). The spectra were normalized for heme a at 592 nm.
DISCUSSION
The mitochondrial cytochrome c oxidase complex is composed of up to 13 subunits encoded by both the mitochondrial and the nuclear genomes. Defects in the assembly pathways for its subunits and redox centers are frequently associated with severe respiratory deficiencies. From extensive work, mostly with yeast and mammalian oxidases, a considerable body of information on various chaperones and biogenesis factors has been established (for reviews, see, e.g., references 7, 21, and 36).
The cytochrome ba3 oxidase from the extremely thermophilic eubacterium T. thermophilus, on the other hand, consists of only three subunits. This organism not only provides a much simpler biogenesis system, but, given its evolutionary location in the deepest branch of the phylogenetic tree (34), is also an interesting object for the study of assembly events of an evolutionarily ancient and thermophilic oxidase.
Only three types of specific auxiliary proteins associated with cytochrome c oxidase assembly have been identified so far in T. thermophilus: a prenyltransferase and a heme a synthase, catalyzing the last two steps of heme a biosynthesis, and Sco1, previously linked to the formation of the CuA center in subunit II (2, 9; see also reference 1, which presents different results). Two further assembly factors of critical importance found almost ubiquitously in all aerobic organisms have not yet been identified in T. thermophilus: CtaG, thought to be responsible for the formation of the CuB center (2, 9, 15), and Surf1, necessary for heme a insertion into subunit I (5, 6, 25, 35).
Here, we show by transcriptional analysis that a novel protein, CbaX, is encoded in the same operon with the structural genes of the ba3 oxidase. Our present data suggest that the 56-bp sequence preceding the cbaX gene (Fig. 1A) harbors an RBS. While the cbaX gene was shown to be cotranscribed (Fig. 2), loss of its presumed translational start signal within the 56-bp stretch (deleted in strain WE6) (Fig. 1B) is responsible for the observed phenotype. Absence of CbaX caused a drastic decrease in the activity of the ba3 oxidase complex, due to a dramatically lowered content of correctly inserted heme a. Even though there is no sequence homology to any known oxidase assembly protein, the reported phenotype suggests this protein is a prime candidate in the biogenesis pathway of the ba3 oxidase, possibly responsible for the correct insertion of heme a into subunit I.
As shown in Fig. 4, the most notable effect of the cbaX deletion on the cytochrome ba3 oxidase consists of a decrease in its enzymatic activity to 7% of that of the wild type, and this observation is paralleled by an apparent loss of heme a of about 90%, as judged from the canonical 613-nm peak in the native spectrum (Fig. 4B, strain pWE4/WE6). However, when the total amount of heme a was assessed for this oxidase preparation under denaturing conditions (Fig. 4C), this loss amounted to only 40% of the heme a of the wild type. This obvious discrepancy may be explained by only a small fraction of correctly assembled heme a contributing to the 613-nm spectral peak, indicative of a normal ligand environment in the high-spin a3 site of subunit I; the majority of heme a present in the oxidase from this assembly mutant strain may be liganded in a nonnative environment, giving rise to spectral “noise” observed on either side of the 613-nm redox spectrum peak (Fig. 4B).
Whereas the deletion of cbaX caused a loss of correctly assembled heme a in the cytochrome ba3 oxidase and dramatically diminished its turnover number, the use of a strong promoter for the expression of CbaX, detached from that of the subunit genes in WE8, led to a distorted heme a/b ratio of about 2:1 (Fig. 6C). Compared to the wild-type situation, where the heme a/b ratio is 1:1, a disturbed ratio of heme a was reflected in pWE4/WE8 by an additional peak at 582 nm (Fig. 6A). As shown in Fig. 6C, the specific heme content (the sum of heme a and heme b) was not changed in this strain; therefore, the presence of any surplus heme sticking to the enzyme is excluded.
The unusual spectral position of the 582-nm redox peak may be explained by fortuitous side chains surrounding the low-spin site, now liganding the heme a artificially. This assumption is supported by the cyanide spectrum (Fig. 7): as expected for the wild type, the high-spin heme a peak was shifted to 592 nm by the cyanide binding, while the position of the low-spin heme b peak remained unchanged (Fig. 7, solid line). In pWE4/WE8 (dotted line), the 592-nm heme a peak displayed an unexpected shoulder with a maximum at 582 nm; this spectral assignment became more obvious in the difference spectrum (gray line). Thus, this wavelength was identical to that of the additional peak in the native redox spectrum (Fig. 6A) and therefore represented heme a occupying the low-spin site. The measured enzymatic activity, two-thirds that of the wild type, supports this notion, where one out of three oxidases carries a heme a moiety instead of heme b in its low-spin site. Because of the bulky geranylgeranyl side chain of heme a, it is highly unlikely that this heme a group would fit into the low-spin site in an orientation identical to that of heme b and be properly positioned between the two histidine ligands, as found in the native structure (32). We conclude that these severe oxidase defects are caused by overexpression of CbaX due to the use of the strong slpA promoter; since S-layer proteins represent up to 15% of the total cellular protein of an S-layer-carrying cell, the promoters of surface layer genes are among the strongest found in prokaryotes (3, 19). We hypothesize that in the wild type, where the transcription of cbaX is linked to that of the subunit genes, a balanced protein level, not necessarily stoichiometric with subunit I, is assured; such a regulated expression may be necessary for proper ba3 assembly and function. Whether the role of CbaX is a truly catalytic one or rather that of a generic chaperone maintaining subunit I in a state competent to receive the heme a moiety remains unclear. However, the low yield for CbaX, even when purified from overexpressing strains, may rather argue for a catalytic function. We note that the CbaX protein is devoid of any histidine residues typically involved in liganding heme groups, thus not favoring direct heme transport.
The lowered activity, the distorted heme a content, and the location of the cbaX gene within the ba3 operon (6, 31) clearly suggest a Surf1-like function for CbaX. This assumption, on the other hand, is not substantiated on the sequence level, where CbaX and Surf1 share no meaningful sequence identity in either of the two T. thermophilus genome sequences (16).
Data on a surf1 deletion in Paracoccus denitrificans or Rhodobacter sphaeroides show that the activities of the resulting oxidases are much less affected (6, 31) than in the results presented here. Also at variance in the case of P. denitrificans, where two heme a-carrying terminal oxidases require two independent surf1 gene variants with noncomplementary functions encoded in each particular operon (5, 6), no further operon-specific homologue was found within the caa3 oxidase operon (12) or elsewhere in the genome of T. thermophilus. At present, we consider it very unlikely that assembly of the cytochrome caa3 oxidase is also dependent on proper CbaX function, as T. thermophilus HB27 completely relies on oxidative phosphorylation and lacks fermentative pathways (16). Therefore, a severe defect in the assembly of the cytochrome caa3 oxidase in the CbaX deletion strain would result in serious growth rate retardation, which was not observed here. The role of the second gene, cbaY, which is most likely also part of the cbaDBAX transcription unit, remains unclear, because its deletion had no effect upon proper function of the cytochrome ba3 oxidase (data not shown), apart from a sharp drop in the yield of the enzyme.
A BLAST search found homologues of CbaX only in members of the Thermaceae group: in the few cases with genomic information available, this gene was located in a position either next to the ba3 structural genes (in T. thermophilus, as well as in T. aquaticus) or next to sco1 (as in either of the two Meiothermus strains). However, in all the above-mentioned bacteria with a CbaX protein present, a Surf1 homologue was missing. The existence of an alternative heme a insertion factor may well reflect an adaption of this biogenesis pathway to hyperthermophilic conditions.
In conclusion, we identified a novel membrane protein, CbaX, that is vital for proper maturation of a thermophilic enzyme, the ba3 cytochrome c oxidase of T. thermophilus. It is encoded, together with the structural genes of the enzyme, in one transcription unit, and its absence causes a severe loss of correctly assembled heme a and therefore of enzymatic activity. We conclude that this protein, devoid of homologues outside the Thermaceae group, plays a specific role in heme a insertion into the high-spin site of subunit I of the ba3 oxidase by an as-yet-unknown mechanism. It may act as a chaperone, keeping the transmembrane helices of newly synthesized subunit I polypeptides in a state competent to sterically position the heme a group, thus facilitating proper formation of the active site. Furthermore, balanced expression seems to be important, as overexpression of CbaX leads to misincorporation of heme a into the low-spin site of the ba3 enzyme, thereby once again emphasizing the specificity of CbaX in heme a incorporation.
Acknowledgments
The Collaborative Research Center “Molecular Bioenergetics” (SFB 472), the Center for Membrane Proteomics (CMP), and the Cluster of Excellence Frankfurt (Macromolecular Complexes, DFG Project EXC 115) are acknowledged for financial support.
We thank B. Averhoff for kindly providing the pWUR-bleo vector and H.-W. Müller for excellent technical assistance.
Footnotes
Published ahead of print on 9 July 2010.
REFERENCES
- 1.Abriata, L. A., L. Banci, I. Bertini, S. Ciofi-Baffoni, P. Gkazonis, G. A. Spyroulias, A. J. Vila, and S. Wang. 2008. Mechanism of Cu(A) assembly. Nat. Chem. Biol. 4:599-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertini, I., and G. Cavallaro. 2008. Metals in the “omics” world: copper homeostasis and cytochrome c oxidase assembly in a new light. J. Biol. Inorg. Chem. 13:3-14. [DOI] [PubMed] [Google Scholar]
- 3.Boot, H. J., C. P. Kolen, F. J. Andreadaki, R. J. Leer, and P. H. Pouwels. 1996. The Lactobacillus acidophilus S-layer protein gene expression site comprises two consensus promoter sequences, one of which directs transcription of stable mRNA. J. Bacteriol. 178:5388-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouns, S. J., H. Wu, J. Akerboom, A. P. Turnbull, W. M. de Vos, and J. van der Oost. 2005. Engineering a selectable marker for hyperthermophiles. J. Biol. Chem. 280:11422-11431. [DOI] [PubMed] [Google Scholar]
- 5.Bundschuh, F. A., A. Hannappel, O. Anderka, and B. Ludwig. 2009. Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. J. Biol. Chem. 284:25735-25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bundschuh, F. A., K. Hoffmeier, and B. Ludwig. 2008. Two variants of the assembly factor Surf1 target specific terminal oxidases in Paracoccus denitrificans. Biochim. Biophys. Acta 1777:1336-1343. [DOI] [PubMed] [Google Scholar]
- 7.Carr, H. S., and D. R. Winge. 2003. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 36:309-316. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., L. Hunsicker-Wang, R. L. Pacoma, E. Luna, and J. A. Fee. 2005. A homologous expression system for obtaining engineered cytochrome ba3 from Thermus thermophilus HB8. Protein Expr. Purif. 40:299-318. [DOI] [PubMed] [Google Scholar]
- 9.Cobine, P. A., F. Pierrel, and D. R. Winge. 2006. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta 1763:759-772. [DOI] [PubMed] [Google Scholar]
- 10.de Grado, M., P. Castan, and J. Berenguer. 1999. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42:241-245. [DOI] [PubMed] [Google Scholar]
- 11.Faraldo, M. M., M. A. de Pedro, and J. Berenguer. 1992. Sequence of the S-layer gene of Thermus thermophilus HB8 and functionality of its promoter in Escherichia coli. J. Bacteriol. 174:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fee, J. A., T. Yoshida, K. K. Surerus, and M. W. Mather. 1993. Cytochrome caa3 from the thermophilic bacterium Thermus thermophilus: a member of the heme-copper oxidase superfamily. J. Bioenerg Biomembr. 25:103-114. [DOI] [PubMed] [Google Scholar]
- 13.Fernández-Herrero, L. A., G. Olabarria, and J. Berenguer. 1997. Surface proteins and a novel transcription factor regulate the expression of the S-layer gene in Thermus thermophilus HB8. Mol. Microbiol. 24:61-72. [DOI] [PubMed] [Google Scholar]
- 14.Fontanesi, F., I. C. Soto, D. Horn, and A. Barrientos. 2006. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 291:C1129-C1147. [DOI] [PubMed] [Google Scholar]
- 15.Greiner, P., A. Hannappel, C. Werner, and B. Ludwig. 2008. Biogenesis of cytochrome c oxidase—in vitro approaches to study cofactor insertion into a bacterial subunit I. Biochim. Biophys. Acta 1777:904-911. [DOI] [PubMed] [Google Scholar]
- 16.Henne, A., H. Bruggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Dencker, R. Huber, H. P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H. J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 17.Herrmann, J. M., and S. Funes. 2005. Biogenesis of cytochrome oxidase—sophisticated assembly lines in the mitochondrial inner membrane. Gene 354:43-52. [DOI] [PubMed] [Google Scholar]
- 18.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 19.Kahala, M., and A. Palva. 1999. The expression signals of the Lactobacillus brevis slpA gene direct efficient heterologous protein production in lactic acid bacteria. Appl. Microbiol. Biotechnol. 51:71-78. [DOI] [PubMed] [Google Scholar]
- 20.Keightley, J. A., B. H. Zimmermann, M. W. Mather, P. Springer, A. Pastuszyn, D. M. Lawrence, and J. A. Fee. 1995. Molecular genetic and protein chemical characterization of the cytochrome ba3 from Thermus thermophilus HB8. J. Biol. Chem. 270:20345-20358. [DOI] [PubMed] [Google Scholar]
- 21.Khalimonchuk, O., and G. Rödel. 2005. Biogenesis of cytochrome c oxidase. Mitochondrion 5:363-388. [DOI] [PubMed] [Google Scholar]
- 22.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 23.Lubben, M., and K. Morand. 1994. Novel prenylated hemes as cofactors of cytochrome oxidases. Archaea have modified hemes A and O. J. Biol. Chem. 269:21473-21479. [PubMed] [Google Scholar]
- 24.Markwell, M. A., S. M. Haas, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 25.Mashkevich, G., B. Repetto, D. M. Glerum, C. Jin, and A. Tzagoloff. 1997. SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J. Biol. Chem. 272:14356-14364. [DOI] [PubMed] [Google Scholar]
- 26.McPherson, M. J., P. Quirke, and G. R. Taylor. 1991. PCR: a practical approach. IRL Press, Oxford, United Kingdom.
- 27.Mooser, D. 2007. Ph.D. dissertation, University of Frankfurt, Frankfurt, Germany.
- 28.Mooser, D., O. Maneg, F. MacMillan, F. Malatesta, T. Soulimane, and B. Ludwig. 2006. The menaquinol-oxidizing cytochrome bc complex from Thermus thermophilus: protein domains and subunits. Biochim. Biophys. Acta 1757:1084-1095. [DOI] [PubMed] [Google Scholar]
- 29.Nicholls, P., and T. Soulimane. 2004. The mixed valence state of the oxidase binuclear centre: how Thermus thermophilus cytochrome ba3 differs from classical aa3 in the aerobic steady state and when inhibited by cyanide. Biochim. Biophys. Acta 1655:381-387. [DOI] [PubMed] [Google Scholar]
- 30.Pereira, M. M., M. Santana, and M. Teixeira. 2001. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta 1505:185-208. [DOI] [PubMed] [Google Scholar]
- 31.Smith, D., J. Gray, L. Mitchell, W. E. Antholine, and J. P. Hosler. 2005. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J. Biol. Chem. 280:17652-17656. [DOI] [PubMed] [Google Scholar]
- 32.Soulimane, T., G. Buse, G. P. Bourenkov, H. D. Bartunik, R. Huber, and M. E. Than. 2000. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus. EMBO J. 19:1766-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams, J. N., Jr. 1964. A method for the simultaneous quantitative estimation of cytochromes a, B, C1, and C in mitochondria. Arch. Biochem. Biophys. 107:537-543. [DOI] [PubMed] [Google Scholar]
- 34.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao, J., and E. A. Shoubridge. 1999. Expression and functional analysis of SURF1 in Leigh syndrome patients with cytochrome c oxidase deficiency. Hum. Mol. Genet. 8:2541-2549. [DOI] [PubMed] [Google Scholar]
- 36.Zee, J. M., and D. M. Glerum. 2006. Defects in cytochrome oxidase assembly in humans: lessons from yeast. Biochem. Cell Biol. 84:859-869. [DOI] [PubMed] [Google Scholar]
- 37.Zimmermann, B. H., C. I. Nitsche, J. A. Fee, F. Rusnak, and E. Munck. 1988. Properties of a copper-containing cytochrome ba3: a second terminal oxidase from the extreme thermophile Thermus thermophilus. Proc. Natl. Acad. Sci. U. S. A. 85:5779-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]