Abstract
The Lia system, a cell envelope stress response module of Bacillus subtilis, is comprised of the LiaRS two-component system and a membrane-anchored inhibitor protein, LiaF. It is highly conserved in the Firmicutes bacteria, and all orthologs investigated so far are activated by cell wall antibiotics. In response to envelope stress, the systems in Firmicutes cocci induce the expression of a number of genes that are involved in conferring resistance against its inducers. In contrast, a complete picture of the LiaR regulon of B. subtilis is still missing and no phenotypes could be associated with mutants lacking LiaRS. Here, we performed genome-wide transcriptomic, proteomic, and in-depth phenotypic profiling of constitutive “Lia ON” and “Lia OFF” mutants to obtain a comprehensive picture of the Lia response of Bacillus subtilis. In addition to the known targets liaIH and yhcYZ-yhdA, we identified ydhE as a novel gene affected by LiaR-dependent regulation. The results of detailed follow-up gene expression studies, together with proteomic analysis, demonstrate that the liaIH operon represents the only relevant LiaR target locus in vivo. It encodes a small membrane protein (LiaI) and a phage shock protein homolog (LiaH). LiaH forms large oligomeric rings reminiscent of those described for Escherichia coli PspA or Arabidopsis thaliana Vipp1. The results of comprehensive phenotype studies demonstrated that the gene products of the liaIH operon are involved in protecting the cell against oxidative stress and some cell wall antibiotics. Our data suggest that the LiaFSR system of B. subtilis and, presumably, other Firmicutes bacilli coordinates a phage shock protein-like response.
The cell envelope is an essential structure of the bacterial cell, and its integrity is crucial for survival. Accordingly, it is closely monitored in both Gram-negative and Gram-positive bacteria to sense and counteract damages before they can disrupt its functionality. The regulatory networks orchestrating the cell envelope stress response (CESR) have been characterized in detail in the two model organisms Escherichia coli and Bacillus subtilis (28, 41).
In both organisms, the major regulatory principles coordinating the CESR are two-component systems (TCS) and extracytoplasmic function (ECF) σ factors. In E. coli, the key components to counteract damages to the cell envelope are the CpxAR TCS and the ECF σ factor σE. Moreover, the BaeRS TCS and the Rcs phosphorelay also contribute to the E. coli CESR (41). Another system that has been associated with the CESR of E. coli and closely related bacteria is the PspA-dependent phage shock response. But despite two decades of research, the physiological role of this system is still poorly understood (14, 48). In B. subtilis, the regulatory network is even more complex. At least three ECF σ factors (σM, σW, and σX) and four TCS (BceRS, LiaRS, YvcPQ, and YxdJK) have been described to be involved in the response of this organism to the presence of cell wall antibiotics and other envelope-perturbing agents (28).
A unique three-component system orchestrates the cell envelope stress response in most Firmicutes (low-G+C, Gram-positive) bacteria. In B. subtilis, it is comprised of the TCS LiaRS and a membrane-anchored negative regulator, LiaF. The latter is invariantly linked to its cognate TCS by function and genomic context in all species harboring LiaRS homologs (28, 29, 44). While the role of LiaF has so far only been investigated in B. subtilis and Streptococcus mutans (29, 30, 55), LiaRS-like systems have also been described in a number of other Firmicutes species. The best-understood LiaRS homolog is the VraSR system of Staphylococcus aureus. This system has initially been identified and described for its contribution to antibiotic resistance in clinical isolates of S. aureus (9, 19, 36, 37). More recently, a number of detailed biochemical studies have addressed both the phosphotransfer between the sensor kinase VraS and the response regulator VraR and the phosphorylation-dependent DNA-binding specificity of VraR (5, 6, 15, 39). Other Lia-like systems that have been genetically and physiologically investigated include CesSR of Lactococcus lactis and the recently described LiaFSR system of Streptococcus pneumoniae (16, 43).
In all species but B. subtilis, the LiaRS-like TCS regulate a number of target loci, including those that directly contribute to the resistance against envelope-perturbing conditions that function as stimuli. Moreover, the collective data seem to indicate that these systems are the major (and often the only) CESR systems of these bacteria. In contrast, the Lia system of B. subtilis (and presumably also other species of the genera Bacillus and Listeria) is embedded in a complex regulatory network that contains not only other TCS but also ECF σ factors. Interestingly, regulon comparison and physiological data indicate that the Lia systems in Firmicutes cocci regulate genes and confer phenotypes that are subject to ECF-dependent regulation in B. subtilis (28). In contrast, a direct physiological contribution of the LiaRS system to the CESR of B. subtilis has not yet been established. This is especially surprising since the response of the B. subtilis LiaFSR system to the presence of lipid II-interacting antibiotics is much stronger and more specific than that of any of the other Lia-homologous systems investigated so far: the expression of the LiaR-dependent liaIH operon is induced 100- to 1,000-fold in the presence of sublethal amounts of its specific inducers (46, 59), in contrast to a mere 2- to 5-fold induction of the respective target genes in L. lactis, S. aureus, and S. mutans. Homologs of the liaIH operon are absent in all Firmicutes cocci, indicating a functional diversification between the two groups of LiaFSR-like systems. These functional differences are also supported by the LiaR binding site of the Bacillus/Listeria group, which differs significantly from the consensus sequence determined for the Firmicutes cocci (16).
Taken together, the data derived from studies of Lia-like systems in different Firmicutes cocci clearly establish their role as the most important CESR system in these bacteria. In contrast, there is still a very limited understanding of the physiological role of LiaFSR in B. subtilis and other bacilli. In order to gain a comprehensive overview of the Lia response of B. subtilis, we decided to apply a range of “omics” technologies to identify all LiaR-dependent genes and proteins and the associated phenotypes. Using DNA microarrays, we first compared the genome-wide expression profile of a “Lia ON” mutant to that of a “Lia OFF” mutant. We identified three LiaR target loci, including the two known operons liaIH and yhcYZ-yhdA (29, 45). The promoter upstream from the third target, ydhE, was further analyzed and a putative LiaR binding site could be identified. The results of the subsequent expression analysis and proteomic studies strongly suggest that the liaIH operon is the only relevant target of LiaR-dependent gene expression in wild-type cells. We therefore performed in-depth phenotyping by comparing a Lia ON mutant to a liaIH deletion strain. No influence of LiaR-dependent gene expression on any cellular differentiation could be observed. In contrast, a phenotype microarray analysis combined with follow-up phenotypic profiling established a protective role of the liaIH gene products against envelope and oxidative stress. These data, together with the results from oligomerization studies of the primary target protein LiaH, indicate that the LiaFSR system of B. subtilis (and, presumably, other Firmicutes bacilli) coordinates a phage shock protein-like response, reminiscent of PspA in E. coli.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacillus subtilis was grown in LB medium or Mueller-Hinton broth at 37°C with aeration. All strains used in this study are derivates of the laboratory wild-type strains W168 and CU1065 (W168 trpC attSPβ) and are listed in Table 1. The antibiotics kanamycin (10 μg ml−1), chloramphenicol (5 μg ml−1), tetracycline (10 μg ml−1), and erythromycin (1 μg ml−1), plus lincomycin (25 μg ml−1) for macrolide-lincosamide-streptogramin (MLS) resistance, were used for selection of the B. subtilis mutants. Transformation was carried out as described previously (24).
TABLE 1.
Strains used in this study
| Strain | Relevant genotypea | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | recA1 endA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 φ80ΔlacZΔM15 Δ(lacZYA-argF)U169 | Laboratory stock |
| BL21(DE3)/pLysS | F−lon ompT rB mBhsdS gal (cIts857 ind1 Sam7 nin5 lacUV5-T7 gene1) | Laboratory stock |
| TME139 | BL21(DE3)/pLysS, pDW1604 | This study |
| Bacillus subtilis strains | ||
| W168 | trpC2 | Laboratory stock |
| CU1065 | W168 att SPβ2Δ2 trpC2 | Laboratory stock |
| HB0920 | CU1065 liaH::kan | 45 |
| HB0933 | CU1065 liaR::kan | 45 |
| HB0961 | CU1065 liaI::pMUTIN | 45 |
| TMB002 | CU1065 liaF::kan | 29 |
| TMB011 | CU1065 liaI::pMUTIN liaR::kan | 30 |
| TMB016 | CU1065 amyE::pTM1 (PliaI-lacZ) | 29 |
| TMB018 | CU1065 amyE::pTM1 (PliaI-lacZ) liaF::kan | 29 |
| TMB071 | W168 amyE::pAJ603 (PyhcY-lacZ) | 29 |
| TMB095 | W168 amyE::pAJ603 (PyhcY-lacZ) liaF::kan | 29 |
| TMB211 | CU1065 liaH::kan pspA::cat | This study |
| TMB222 | W168 amyE::pSK602 (PydhE-lacZ) | This study |
| TMB224 | W168 amyE::pSK602 (PyhcY-lacZ) liaF::kan | This study |
| TMB421 | W168 amyE::pSK602 (PydhE-lacZ) liaF clean deletion | This study |
| TMB329 | W168 ΔliaF (clean deletion) | This study |
| TMB419 | W168 liaF::tet amyE::pJR601 (PydhE(-57)-lacZ) | This study |
| TMB420 | W168 liaF::tet amyE::pJR602 (PydhE(-34)-lacZ) | This study |
| TMB604 | W168 ΔPliaI-liaIH (clean deletion) | This study |
| TMB611 | W168 liaR-terminator-gerAC (termination sequence inserted) liaF::kan | This study |
| TMB630 | W168 ΔPliaI-liaIH (clean deletion) pspA::cat | This study |
| TMB676 | W168 ΔliaF (clean deletion) amyE::pAJu604 (PydhE(-74)-lacZ) | This study |
| TMB721 | W168 ΔliaF clean deletion, amyE::pDW602 (PydhE(-96)-lacZ) | This study |
| TMB769 | W168 ΔliaF clean deletion, amyE::pDW604 (PydhE(-71)-lacZ) | This study |
Resistance cassettes: kan, kanamycin; cat, chloramphenicol; tet, tetracycline.
Allelic replacement mutagenesis using LFH-PCR.
The long flanking homology PCR (LFH-PCR) technique is derived from a published procedure (58) and was performed as described previously (45). The strains constructed are listed in Table 1, and the corresponding primers are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Primer name and purpose | Sequencea |
|---|---|
| Northern hybridization | |
| TM0227 yhcY-probe fwd | GAGTTGCTGAGTCTGACAAACC |
| TM0294 yhcY-probe-T7-rev | CTAATACGACTCACTATAGGGAGATCGTGAAGCTCCTGAGCGAGGC |
| TM0360 liaIH-probe fwd | TTTGGAGGAGGAAGCTTCG |
| TM0361 liaIH-probe-T7-rev | CTAATACGACTCACTATAGGGAGAGATAGGCAATCGTGTGCTG |
| TM0758 ydhE-probe-fwd | AGCAACGGTCCATCTTCAC |
| TM0759 ydhE-probe-T7-rev | CTAATACGACTCACTATAGGGAGACTCTCTTTGGAAAGCTCGGC |
| Overexpression of LiaH | |
| TM0118 liaH-fwd (XhoI) | ATATCTCGAGATGGTATTAAAAAGAATCAGAGACATG |
| TM0120 liaH-rev (BamHI) | AGCCGGATCCTTATTCATTTGCCGCTTTTGTCTGG |
| Overexpression of LiaR | |
| TM1045 liaR-fwd (NdeI) | TCGTCATATGGTGATTCGAGTATTATTGATTGATGATC |
| TM0124 liaR-rev (BamHI) | AGCCGGATCCCTAATTCACGAGATGATTTCGG |
| Check primers | |
| TM0151 T7-fwd | TAATACGACTCACTATAGGG |
| TM0152 T7-rev | GCTAGTTATTGCTCAGCGG |
| LFH-PCR and clean deletions | |
| 1315 pspA-up fwd | GGACGCTGTACATGTCGATACCTC |
| 1316 pspA-up rev (cat) | CTTGATAATAAGGGTAACTATTGCCGGCTAATTCGGTAACCCTTG |
| 1317 pspA-do fwd (cat) | GGGTAACTAGCCTCGCCGGTCCACGCATACATAGGAGGCCGCAGC |
| 1318 pspA-do rev | CCGTTCATCGCAAAGATATGCTCCGC |
| TM0457 liaF-upfwd (BamHI) | AGCCGGATCCAAGGATTTGCGGTCAAGTCC |
| TM0458 liaF-dorev (NcoI) | AGCTCCATGGTTCAAGCCGTATGAGGAGGC |
| TM0574 liaFclean-uprev (XhoI) | GACTCTCGAGTCCTGGTGTCCGCCTCCTTTC |
| TM0575 liaFclean-dofwd (XhoI) | GACTCTCGAGCGGTGATGTGGATGTGAAGTACG |
| TM1055 PliaI-liaIHclean-dofwd | CCAGACAAAAGCGGCAAATG |
| TM1056 PliaI-liaIHclean-uprev | CATTTGCCGCTTTTGTCTGGCTCGCACCGGACCCATTGGC |
| TM1057 PliaI-liaIH-upfwd (BamHI) | AGCCGGATCCGCGGGCTTCTCTCCGCTGTG |
| TM1058 PliaI-liaIH-dorev (EcoRI) | CCATGAATTCGAATGCGGACGTCCGTCACGC |
| TM1184 liaR-fwd (Term.-up-BamHI) | ACGTGGATCCGGACGGCAGCGAAGGTGTTCGG |
| TM1185 liaR-rev (Term.-do-SmaI) | AGCTCCCGGGAAAAAAAAGCCGTTTCAGGGAAAGGGCTTTTTTTTCTAATTCACGAGATGATTTCGG |
| TM1186 gerAC-rev (Term.-up-SmaI) | AGCTCCCGGGCTATTTGTTTGCGCCTTTCG |
| TM1187 gerAC-fwd (Term.-do-NcoI) | ACGTCCATGGGGAGGGCTCTTCATCTGATCCG |
| Analysis of the ydhE promoter | |
| TM0480 PydhE-rev (BamHI) | CGATGGATCCATCCCGGTGAAGATGGACCG |
| TM0481 PydhE-fwd (EcoRI) | CGATGAATTCGCGAATGTGACAGCTGAGGG |
| TM0682 PydhE-57-fwd (EcoRI) | CGATGAATTCAATAGTTCATTCATTTGTACCT |
| TM0683 PydhE-34-fwd (EcoRI) | CGATGAATTCTGCATGGGTTGTGTTCCCA |
| TM1395 PydhE-74-fwd (EcoRI) | CCATGAATTCTAAGTCCAGAGCAGCAGAATAG |
| TM1559 PydhE-96-fwd (EcoRI) | ATCGGAATTCGCTTGGGTGTCTTTTTTTTG |
| TM1583 PydhE-71-fwd (EcoRI) | ATCGGAATTCGTCCAGAGCAGCAGAATAGTTC |
| Gel mobility shift assay | |
| TM0632 bceA-fwd | GAAGTGCTGAAGGGCATCG |
| TM0633 bceA-rev | CATAGCCGTCATGTCATTTCC |
| TM0099 PliaI-fwd (EcoRI) | CCATGAATTCCCGGTGCGAGATACGACTCC |
| TM0100 PliaI-rev (BamHI) | CGATGGATCCTCCTCCAAAAAAGACGGAGATCCC |
| TM1490 PydhE-fwd (EcoRI) | CCATGAATTCAGACACCAGAGCTTGGGTGTC |
| TM0480 PydhE-rev (BamHI) | CGATGGATCCATCCCGGTGAAGATGGACCG |
| TM0166 PyhcY-fwd (EcoRI) | CGATGAATTCGACAGTGAAAAGCGACTTGCC |
| TM0165 PyhcY-rev (BamHI) | CGATGGATCCGTGTTGCTTTGATATCGTGCC |
The sequences that represent the T7 promoter necessary for in vitro transcription are underlined. Restriction sites are in boldface. Linker sequences used for joining reactions are in italic and boldface.
Construction of markerless deletion and insertion mutants.
Markerless deletions of the liaIH operon (including its promoter) and liaF, as well as an insertion mutant introducing an artificial terminator sequence between the 3′ ends of liaR and gerAC (liaR-terminator-gerAC), were constructed using the vector pMAD (2). The genomic regions approximately 1 kb up- and downstream of the genes were amplified using the primers listed in Table 2, thereby introducing a 26-bp extension to the 3′ end of the upstream fragment which is complementary to the 5′ end of the downstream fragment. The two fragments were fused in a second PCR, and the resulting fragment was cloned into pMAD via BamHI and EcoRI, generating pSJ102 (liaF), pDW103 (PliaI-liaIH), and pDW105 (liaR-terminator-gerAC). The generation of the mutants basically followed the established procedure (2). In brief, B. subtilis W168 was transformed with pSJ102 or pDW105 (Table 3) and incubated at 30°C with MLS selection on LB agar plates supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 100 μg ml−1). Blue colonies were selected and incubated for 6 to 8 h at 42°C in LB medium with MLS selection, resulting in the integration of the plasmids into the chromosome. Blue colonies were again picked from LB (X-Gal) plates and incubated at 30°C for 6 h in LB medium without selection. Subsequently, the liquid culture was shifted to 42°C for 3 h, and the cells were then plated on LB (X-Gal) plates, this time without selective pressure. White colonies that had lost the plasmids were picked and checked for MLS sensitivity. The resulting strains, TMB329 (ΔliaF), TMB604 (ΔPliaI-liaIH), and TMB611 (liaR-terminator-gerAC), were subsequently analyzed by PCR and sequencing for the integrity of the desired genetic modifications.
TABLE 3.
Vectors and plasmids
| Plasmid | Genotype | Primer pair(s) used for cloning | Reference or source |
|---|---|---|---|
| pAC6 | bla lacZ cat amyE′…′amyE | 54 | |
| pGEMcat | ori(f1) lacZ′(MCS) bla cat | 63 | |
| pET-16b | T7 lac promoter, His10 tag, MCS, T7 terminator; lacI pBR322-Ori bla | Novagen | |
| pMAD | erm ori(pE194-Ts) MCS-PclpB-bgaB ori(pBR322) bla | 2 | |
| pAJu604 | pAC6 PydhE(−74)-lacZ | TM1395/TM0480 | This study |
| pDW103 | pMAD PliaI-liaIH up/down overlap | TM1056/TM1057, TM1055/TM1058 | This study |
| pDW105 | pMAD liaR-terminator-gerAC overlap | TM1184/TM1185, TM1186/TM1187 | This study |
| pDW602 | pAC6 PydhE(−96)-lacZ | TM1559/TM0480 | This study |
| pDW604 | pAC6 PydhE(−71)-lacZ | TM1583/TM0480 | This study |
| pDW1604 | pET16b-liaR | TM1045/TM0124 | This study |
| pFK1601 | pET16b-liaH | TM0118/TM0120 | This study |
| pJR601 | pAC6 PydhE(-57)-lacZ | TM0682/TM0480 | This study |
| pJR602 | pAC6 PydhE(-34)-lacZ | TM0683/TM0480 | This study |
| pSJ102 | pMAD liaF up/down overlap | TM0457/TM0574, TM0575/TM0458 | This study |
| pSK602 | pAC6 PydhE-lacZ | TM0480/TM0481 | This study |
Measurement of induction by β-galactosidase assay.
Cells were inoculated from fresh overnight cultures and grown in LB medium at 37°C with aeration until they reached an optical density at 600 nm (OD600) of ∼0.4. The culture was split; bacitracin (50 μg ml−1 final concentration) was added to one-half (induced sample), and the other half was left untreated (uninduced control). After incubation for an additional 30 min at 37°C with aeration, 2 ml of each culture was harvested and the cell pellets were stored at −20°C. The pellets were resuspended in 1 ml working buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol) and assayed for β-galactosidase activity, with normalization to cell density, as described in reference 47.
Preparation of total RNA.
B. subtilis CU1065, TMB002 (liaF::kan), and HB0933 (liaR::kan) were grown aerobically at 37°C in LB medium to late-log phase (OD600 of 0.8; growth rate of the wild type was 0.031 ± 0.001, of the liaR mutant was 0.030 ± 0.001, and of the liaF mutant was 0.026 ± 0.001 [min−1]). Thirty milliliters of each sample was mixed with 15 ml cold killing buffer (20 mM Tris-HCl, pH 7.0, 5 mM MgCl2, 20 mM NaN3), harvested by centrifugation, and frozen in liquid nitrogen. For cell disruption, the pellet was resuspended in 200 μl killing buffer, immediately transferred to a Teflon vessel (filled and precooled with liquid nitrogen), and then disrupted with a Mikro-Dismembrator U (Sartorius). The resulting cell powder was resuspended in 3 ml of lysis solution (4 M guanidine thiocyanate, 0.025 M Na acetate, pH 5.2, 0.5% N-lauroyl sarcosinate), and the RNA was extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1), followed by chloroform-isoamyl alcohol (24:1) extraction and ethanol precipitation. RNA samples were treated with an RNase-free DNase kit (Qiagen) according to the manufacturer's instructions and purified using RNeasy mini columns (Qiagen). Quality control of the RNA preparations was performed with an RNA 6000 Nano LabChip kit (Agilent Technologies) on an Agilent 2100 Bioanalyzer according to the manufacturer's instructions.
DNA microarray analysis.
The RNA samples obtained from three independent cultivations were used for independent cDNA synthesis and DNA microarray hybridization. The generation of the Cy3/Cy5-labeled cDNAs and hybridization to B. subtilis whole-genome DNA microarrays (Eurogentec) were performed as described previously (32). The slides were scanned with a ScanArray Express scanner (PerkinElmer). Quantification of the signal and background intensities was carried out using the ScanArray Express image analysis software.
Transcriptome data analysis.
Data were analyzed using GeneSpring software (Agilent Technologies). The raw signal intensities were first transformed by intensity-dependent LOWESS normalization. The normalized array data were subjected to statistical analysis using Cyber-T, a program based on a t test combined with a Bayesian statistical framework (4). The software is accessible through a web interface at http://cybert.microarray.ics.uci.edu. The mRNA abundance was considered to be significantly different between samples obtained from wild-type and mutant strains if (i) the Cyber-T Bayesian P value was <0.001 and (ii) the averaged fold change was at least 3.
Northern hybridization analysis.
Northern blot analysis was carried out as described previously (25) using equal amounts of total RNA (5 μg) separated under denaturing conditions on a 1.2% formaldehyde agarose gel. Chemiluminescence was detected with a Lumi-Imager (Roche, Germany) using CDP-Star (Roche, Germany) as the substrate. The transcript sizes were determined by comparison with an RNA size marker (Invitrogen). The digoxigenin-labeled specific-RNA probes were synthesized by in vitro transcription using T7 RNA polymerase from the T7 promoter-containing internal PCR products of liaIH, ydhE, and yhcY using the primers listed in Table 2.
Cloning, expression, and purification of recombinant His10-tagged LiaR and LiaH.
The liaR and liaH genes were amplified from B. subtilis W168 genomic DNA by PCR using oligonucleotide pairs TM1045/-0124 and TM0118/-0120, respectively. The PCR product was cloned into the pET-16b expression vector (Novagen, Germany) using the added restriction sites and confirmed by sequencing. For overexpression, E. coli BL21(DE3)/pLysS (Novagen, Germany) was transformed with pDW1604 (His10-LiaR) or pFK1601 (His10-LiaH) and grown in LB medium. In mid-log phase (OD600 of 0.6 to 0.8), protein expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cultures were harvested 3 h after induction and washed in ZAP (10 mM Tris, 200 mM NaCl, pH 7.4). Due to low accessibility of the His tag under native protein folding, His10-LiaH was purified under denaturing conditions in the presence of 2 M urea in the ZAP buffer. Cell pellets were resuspended in 10 ml ZAP, and cells were disrupted by 3 passes through a French press at 1,000 lb/in2. The lysate was centrifuged at 11,000 × g and 4°C for 30 min. The supernatant was loaded on a gravity flow column with 1 ml Ni2+-nitrilotriacetic acid (NTA) Superflow matrix (Qiagen, Germany). After washing steps with ZAP and ZAP containing 5 mM and 10 mM imidazole, His10-LiaR was eluted from the column using ZAP with imidazole concentrations of 25 mM, 50 mM, 100 mM, 200 mM, 300 mM, and 500 mM. All fractions were analyzed using SDS-PAGE. Fractions containing purified His10-LiaR were used for gel mobility shift experiments. Fractions containing purified His-tagged LiaH were dialyzed against ZAP to remove imidazole and urea. After this treatment, His10-LiaH was again unable to bind Ni2+-NTA, indicating proper refolding (data not shown). This protein was then used for oligomerization studies.
Gel mobility shift assays.
DNA fragments of the promoter regions upstream of liaI (180 bp), yhcY (313 bp), ydhE (323 bp), and bceA (161 bp, control) were amplified by PCR (primers are listed in Table 2). The products were purified using a PCR purification kit (SLG, Germany) and diluted to a final concentration of 2.5 ng ml−1 with band shift buffer [20 mM HEPES, 10 mM (NH4)2SO4, 1 mM dithiothreitol (DTT), 30 mM KCl, 10 mM MgCl2, pH 7.9]. Phosphorylation of LiaR was carried out by incubation for 30 min at 37°C in band shift buffer containing 12.5 mM acetyl phosphate (40). Binding of His10-LiaR was carried out in band shift buffer in a 15-μl reaction mixture containing 5 ng PCR product and various amounts of phosphorylated LiaR. The binding reaction mixtures were incubated at 25°C for 20 min. The samples were loaded onto native 8% polyacrylamide gels in 1× TGE buffer (50 mM Tris, 400 mM glycine, 1.73 mM EDTA). Electrophoresis was carried out at 25 mA and room temperature for 1 h. Gels were stained with SYBR green I (Molecular Probes) for 15 min, and DNA was detected by scanning with a Bio-Vision 3000 fluorescence imager (Peqlab, Germany).
Phenotype microarray analysis.
Phenotype microarray assays were performed by Biolog (Hayward, CA) according to the published procedure (www.biolog.com/pmTechDesOver.html), using a tetrazolium dye (8). Incubation and recording of phenotypic data were performed in the OmniLog station by capturing digital images of the microarray and storing quantitative color change values in a computer file displayed as a kinetic graph. The OmniLog-PM software generates time course curves for the redox state of the tetrazolium dye and calculates differences in the areas for mutant and control cells. The units are arbitrary. Positive values indicate that the mutant showed greater rates of growth than the control. The differences are the averages of the values reported for two or more clones of each mutant compared to the average values for the corresponding control strains. All significant hits (as defined by Biolog) are listed in Table S1 in the supplemental material both for the comparison of strain TMB329 (markerless liaF deletion; hereinafter ΔliaF) and of strain TMB211 (liaH::kan pspA::cat) to the wild type (W168).
Determination of the MIC.
For determination of MICs, strains W168, TMB329 (ΔliaF), and TMB604 (ΔPliaI-liaIH markerless deletion) were grown in LB medium overnight at 37°C with aeration. One hundred microliters of each overnight culture was then added to 5 ml Mueller-Hinton broth with different concentrations of the tested substances and a positive control (Mueller-Hinton broth only), respectively. The cultures were incubated for 6 h with aeration at 37°C, and the final OD600 values were determined. The concentration of each compound at which no growth was detectable was defined as the MIC.
Serial dilution spot tests.
The strains W168, TMB329, and TMB604 (Table 1) were incubated in 3 ml LB medium overnight at 37°C with aeration. One hundred microliters of each strain was added to 3 ml of fresh LB medium. After 3 to 4 h at 37°C with aeration, the OD600 was determined and the cultures were diluted to an OD600 of 0.5 (undiluted sample, 100). Five microliters of each dilution (10−1 to 10−6) of every strain was spotted onto Mueller-Hinton agar plates containing the compounds of interest at different concentrations and incubated at 37°C overnight.
Preparation of cytoplasmic proteins and 2-D PAGE.
B. subtilis strains CU1065 (wild type), HB0920 (liaH::kan), and TMB002 (liaF::kan) were grown in LB medium to mid-log phase (OD600 of 0.6 to 0.8). The preparation of cytoplasmic proteins and subsequent two-dimensional (2-D) PAGE were performed as described previously (12). The 2-D gels were stained with colloidal Coomassie brilliant blue and analyzed using Delta-2D software (Decodon, Germany). Proteins of interest were excised and identified by matrix-assisted laser desorption-ionization-tandem time-of-flight (MALDI-TOF-TOF) mass spectrometry.
Oligomerization studies of His10-LiaH by transmission electron microscopy.
Twenty microliters of purified His10-LiaH (diluted 10-fold in ZAP buffer [pH 8.5] supplemented with 100 μg μl−1 PEG 2000) was applied to a piece of parafilm. A carbon-coated Formvar-nickel grid was incubated for 30 s on the drop. After a brief wash with water, the grid was incubated for 10 s in 4% uranyl acetate and briefly dried on filter paper. Electron microscopy was carried out with a Philips EM 301 instrument at calibrated magnifications. Images were recorded on IMAGO electron-sensitive films (Plano, Germany). Magnifications were calibrated with a cross-lined grating replica. Markham rotational analysis of images of LiaH rings was performed as described previously (42).
Size exclusion chromatography.
A Superose 6 column was used on an Äkta fast protein liquid chromatography system (GE Healthcare). The column was equilibrated with running buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5 mM MgCl2, and 0.5 mM DTT). LiaH was used at a final concentration of 10 μM, and a sample of 100 μl was applied to the column. Gel filtration experiments were performed at room temperature with a flow rate of 0.5 ml min−1. The eluted fractions were collected, precipitated with acetone, and subsequently analyzed using SDS-PAGE.
RESULTS
LiaFSR-like three-component systems are highly conserved in Firmicutes bacteria. In contrast to the situation in low G+C Gram-positive cocci, where LiaFSR-like three-component systems function as the primary CESR systems, the physiological role of LiaFSR homologs in the Bacillus group is less well understood (28). Therefore, the aim of this study was to perform in-depth analyses to identify the LiaR regulon and phenotypes associated with LiaR-dependent gene expression. For this purpose, we used two mutants that were previously described to represent the constitutive Lia ON and Lia OFF states of the system (29). In a liaR mutant which lacks the response regulator, no LiaR-dependent gene expression can occur (Lia OFF). In contrast, liaF encodes an inhibitor of the LiaRS TCS; a mutant with a mutation in this gene, therefore, represents the constitutive Lia ON state of the system.
Identification of LiaR-dependent genes.
Previously, we had identified two LiaR target promoters, PliaI and PyhcY (29, 45), but a comprehensive picture of the LiaR regulon was still missing. We therefore performed a DNA microarray analysis by comparing the transcriptome patterns of the Lia ON and Lia OFF mutants with that of the wild-type strain to gain specific information on LiaR-dependent gene expression. The corresponding strains W168 (wild type), TMB002 (liaF::kan), and HB0933 (liaR::kan) (Table 1) were grown in LB medium to mid-logarithmic growth phase (OD600 of 0.5). Cells were harvested and snap-frozen. Subsequently, total RNA was prepared, reverse transcribed, labeled, and hybridized to B. subtilis microarrays representing the complete set of annotated genes (see Materials and Methods for details). The results from competitive hybridization are shown in Table 4. The genomic organization of putative LiaR target genes is illustrated in Fig. 1A. The complete microarray data set can be found in Table S1 in the supplemental material.
TABLE 4.
Results of microarray analysis showing LiaR-dependent marker genes that are induced/repressed more than 3-fold
| Gene(s) | Fold change in:a |
Function, homology, remarkb,c | Reference | |
|---|---|---|---|---|
| ΔliaF mutant | ΔliaR mutant | |||
| Induced | ||||
| liaIH | 310 ± 49 | Membrane protein, phage-shock protein homolog | 22 | |
| liaGFSR | 75 ± 12 | Unknown (liaG), negative regulator (liaF), TCS (liaSR) | 29 | |
| gerAC-gerAB-gerAA | 18 ± 3.2 | 6.7 ± 1.8 | Spore germination, polar effect | 49 |
| yhcYZ-yhdA | 24 ± 2.3 | Putative TCS and azoreductase | 45 | |
| yhdB-yhdC | 3.6 ± 0.3 | Unknown, polar effect | ||
| ydhE | 15 ± 0.8 | Putative macrolide glycosyltransferase | ||
| yqjU | 6.7 ± 1.5 | Unknown | ||
| wapA-yxxG | 6.3 ± 0.9 | Cell wall-associated protein | 13 | |
| yxiF-yxzC-yxiG-yxiH-yxzG-yxiI-yxiJ-yxiK-yxiL | 6.4 ± 1.1 | Unknown | ||
| yxiM-deaD | 4.5 ± 0.6 | Unknown, ATP-dependent RNA helicase | 34 | |
| ytrABCDEF | 4.1 ± 0.9 | Acetoin utilization, stress inducible | 62 | |
| Repressed | ||||
| hag | 3.3 ± 0.05 | Flagellin protein | 18 | |
| yvzB | 3.3 ± 0.04 | Similar to flagellin | 21 | |
| srfAA-srfAB-comS-comAC-comAD | 5.3 ± 0.03 | Surfactin synthetase, regulation of competence | 50, 52 | |
| yxkC | 4 ± 0.02 | Unknown | ||
| pel | 5.8 ± 0.03 | Pectate lyase | 51 | |
| catD-catE | 4.8 ± 0.04 | Catechol catabolism, dioxygenase | 56 | |
Values (relative to wild-type levels) are averages and standard deviations of the results from three independent biological replicates. All genes listed had a Cyber-T Bayesian P value of <0.001 (see Materials and Methods for experimental details).
TCS, two-component system that includes histidine kinase and response regulator.
Positive polar effect was indicated by strong expression of upstream genes.
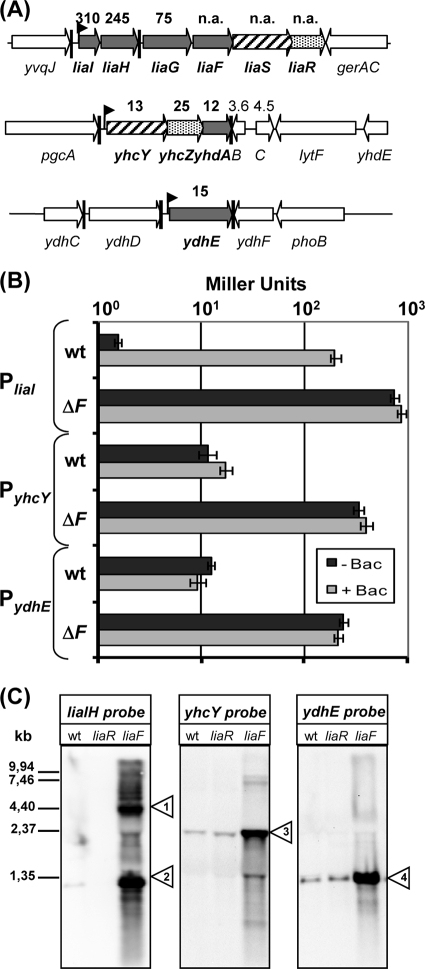
FIG. 1.
Identification of LiaR-dependent target genes. (A) Genomic organization of potential LiaR target genes as identified by DNA microarray analysis. Expression of the liaIHGFSR operon, yhcYZ-ydhA operon, and the ydhE gene are strongly induced in a Lia ON mutant relative to their expression in the wild-type strain (see fold-change values above the arrows; n.a., values derived for these genes are an artifact due to replacing the liaF gene with the kanamycin resistance cassette). (B) Measurement of the β-galactosidase activity of the PliaI, PyhcY, and PydhE promoters in the wild type (wt) and the corresponding liaF::kan background. Cultures of strains TMB016 (PliaI-lacZ), TMB018 (PliaI-lacZ liaF::kan), TMB071 (PyhcY-lacZ), TMB095 (PyhcY-lacZ liaF::kan), TMB222 (PydhE-lacZ), and TMB224 (PydhE-lacZ liaF::kan) were grown in LB medium to mid-log phase and then split. One half was induced by the addition of bacitracin (+ Bac; 50 μg ml−1 final concentration), and the other half served as an uninduced control (− Bac). Cells were harvested after 30 min of incubation and assayed as described previously (46). β-Galactosidase activity is expressed in Miller units (47). Error bars show standard deviations. (C) Northern blot analysis. Expression of liaIHGFSR, yhcYZ-yhdA, and ydhE was measured using 5 μg of total RNA from each strain sample (wild type, TMB002 [liaF::kan], and HB0933 [liaR::kan]) separated on a 1.2% formaldehyde gel. RNA was transferred to a nylon membrane and hybridized with labeled gene-specific RNA probes (see Materials and Methods for details). Small triangles indicate the major transcript(s) for each gene; their sizes correspond to transcripts liaIHG-kan-liaSR (1), liaIH (2), yhcYZ-ydhA (3), and ydhE (4).
The strongest effects relative to the results for the wild type were observed in the liaF mutant. This was to be expected, since deletion of the inhibitor gene liaF results in a constitutively active (i.e., phosphorylated) response regulator LiaR. Therefore, all genes that are affected (directly or indirectly) by LiaR-dependent gene expression should be positively or negatively modulated. Only three loci were significantly upregulated (Fig. 1A and B). The strongest effect was observed for the primary target of LiaR-dependent regulation. Compared to the results for the wild type, the liaIH operon was induced 200- to 300-fold in the liaF mutant, while its expression was reduced about 2.5-fold in the liaR mutant (data not shown; also see Table S1 in the supplemental material). Moreover, the expression of liaG was also strongly induced. The significantly weaker values were caused by partial termination of transcription after liaH (29). While the expression of the complete liaIHGFSR operon in antibiotic-induced wild-type cells is well documented, the increased expression values for liaSR from this microarray work need to be interpreted with caution, since they are most likely influenced by the strong expression of the kanamycin resistance cassette replacing liaF. We also observed a polar effect on the divergently expressed operon downstream of the lia operon, gerAB-gerAC. This expression pattern was verified by Northern blot analysis using a liaIH-specific probe (Fig. 1C). Two major transcripts of about 1.3 kb (liaIH) and 5 kb (liaIHG-kan-liaRS) can be detected. Moreover, a number of larger but weaker transcripts can also be seen, supporting the read-through countertranscription into the ger operon, as indicated by the microarray data. A similar, albeit weaker effect is also observed in the liaR::kan mutant, indicating positive polar downstream effects based on the expression of the resistance cassette from its own strong constitutive promoter. The physiological consequence of this effect is further described below.
The second already-known LiaR target, the yhcYZ-yhdA operon, was also identified in this study as significantly (12- to 25-fold) induced in the liaF mutant. Here again, weakly increased values for two genes further downstream indicate read-through transcription. In addition, one novel potential target gene was identified. The monocistronic ydhE gene (Fig. 1A) was induced 15-fold in the Lia ON mutant, while its expression was slightly reduced in the Lia OFF strain. The LiaR-dependent expression of this gene was analyzed in more detail (see below for details). In both cases, transcripts of the expected sizes were also detected in the corresponding Northern blots (Fig. 1C). It should be noted that these two loci were not induced by bacitracin in the wild type, in contrast to the lia locus (Fig. 1B). The significance of this observation will be addressed below.
In addition, three genes (hag, yvzB, and yxkC) showed a >3-fold reduced expression level in the liaF mutant but also an ∼2-fold reduction in the liaR mutant relative to the level in the wild-type strain. Moreover, the expression of the srf operon was repressed more than 5-fold in the liaR mutant and about 2-fold in the liaF mutant.
Without external stimuli, the Lia system is inactive during mid-logarithmic growth phase (29). Hence, we did not expect to see strong effects in the microarray comparison of the liaR mutant (Lia OFF) versus the wild-type strain. Nevertheless, a number of loci were significantly affected in their expression. More than 12 consecutive genes downstream from and including wapA were induced 4- to 6-fold, while the expression of this locus was unaffected in the liaF mutant (Table 4). Moreover, the expression of the stress-inducible ytrABCDEF operon is also increased about 4-fold. The expression of this operon is a very sensitive, albeit nonspecific marker for a diverse set of stress conditions (62), indicating that the inactivation of the Lia system causes some kind of cellular stress. Two loci (pel and catDE) showed a >5-fold-reduced expression level in the liaR mutant without being affected in the Lia ON mutant.
Leaving these minor and presumably nonspecific effects aside, the results of our microarray analysis strongly suggest that an activated Lia system seems to specifically affect the expression of only three target loci, liaIH-liaGFSR, yhcYZ-yhdA, and the ydhE gene.
The ydhE gene is a novel target of LiaR-dependent gene regulation.
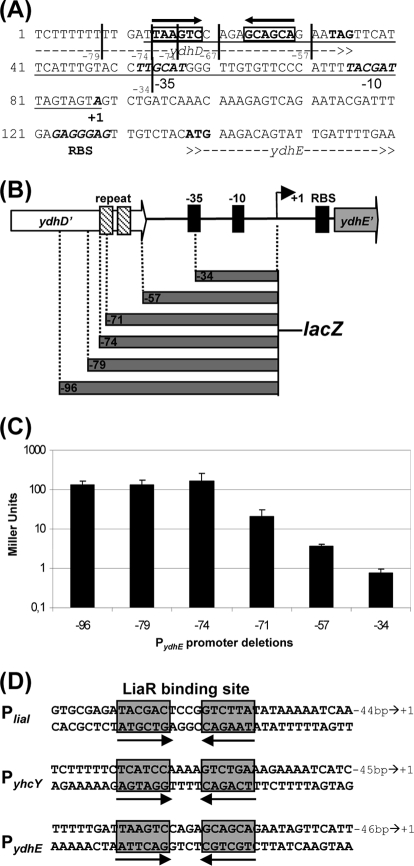
Based on the microarray data, we next analyzed the LiaR-dependent ydhE expression in more detail. The initial bioinformatic analysis of the short intergenic region upstream of ydhE revealed a suitable ribosome binding site but neither a strong promoter nor an obvious LiaR binding site. Using 5′ rapid amplification of cDNA ends (RACE), we mapped the transcriptional start site to an A 50 nucleotides upstream from the start codon, which allowed us to identify a potential but poorly conserved σA-type promoter sequence (Fig. 2A).
FIG. 2.
Characterization of the ydhE promoter and determination of LiaR binding sites. (A) Sequence of the ydhE promoter region, including the postulated LiaR binding site (black arrows, boldface, and bold boxes), the −35 and −10 promoter regions, transcription start site, and ribosome binding site (RBS) (all in boldface italics), and the start and stop codons (ATG and TAG, respectively; in boldface). The minimal LiaR-dependent promoter fragment is underlined. Note that the labeling of the 5′ end of the fragments for the promoter deletion analysis in panels A and B is given relative to the transcription start (+1) for reasons of clarity. This nomenclature differs thereby from the labeling of the fragments for cloning (Tables 1 and 2), which are normalized relative to the start codon of ydhE. (B) Overview of the promoter deletion analysis of PydhE. A graphical representation of the intergenic region and outline of the fragments used for promoter deletion are shown. The −35 region, −10 region, and RBS are highlighted. The black arrow indicates the transcription start site (+1). The inverted repeat representing the LiaR binding site is symbolized by hatched boxes. The sizes of the cloned fragments are illustrated by the vertical gray bars below. (C) The corresponding promoter activities in a liaF deletion mutant, as determined by β-galactosidase assay as described in the Fig. 1B legend (but without the addition of bacitracin), using strains TMB419-421, TMB676, TMB721, and TMB769. Error bars show standard deviations. (D) Summary of the LiaR binding sites upstream from the promoter regions of the liaI, yhcY, and ydhE genes. The corresponding inverted repeats are boxed in gray and underlined with black arrows. The bp number represents the distance to the transcription start site.
Based on this information, we then performed a promoter deletion analysis. Promoter fragments of decreasing length at the 5′ end were cloned into the promoter probe vector pAC6 (54). All fragments had the same 3′ end, extending 169 nucleotides into the ydhE gene (Fig. 2A shows the exact 5′ end of all relevant constructs, and Fig. 2B is a schematic representation). The resulting plasmids, pAJu604, pDW602, pDW604, pJR601-pJR602, and pSK602, were used to transform the liaF mutant, resulting in strains TMB419-421 and TMB676/-721/-769 (see Table 1 for details). Subsequent β-galactosidase assays of the initial promoter set showed that all larger fragments down to position −79 showed a strong Lia-dependent expression in the liaF mutant (Fig. 2C) but only a basal expression level in the wild type (about 10 Miller units; data not shown). The −57 fragment showed a strongly reduced basal activity, while the −34 fragment, which already lacked parts of the −35 promoter regions, only showed background activity of less than one Miller unit (Fig. 2C).
A closer inspection of the sequence between the −79 and the −57 fragment revealed one potential LiaR binding signature (TAaGtC----GcaGcA) at the 3′ end of ydhD (Fig. 2A and D). While the four core residues at the ends of the two repeats are conserved (boldface), only two more residues fit to the position weight matrix (capital letters) (16), and no additional complementary bases could be detected. To exactly map the LiaR binding site upstream of ydhE, two additional fragments (−74 and −71) were cloned and corresponding reporter strains (TMB676 and TMB769) constructed, and β-galactosidase activities were measured in the liaF mutant background. The data support this motif, since the −74 fragment still has the maximum activity, while a further 5′ truncation of only three more nucleotides in the −71 fragment already leads to a 10-fold-reduced level of activity (Fig. 2C). Therefore, we conclude that the inverted repeat described above indeed represents the LiaR binding site upstream of ydhE (Fig. 2D).
The liaIH operon is the only relevant LiaR target in vivo.
Studies of LiaFSR-like systems in four different Firmicutes bacteria allowed the identification of a conserved inverted repeat element as the binding sequence for LiaR-like response regulators (16, 29, 43, 55). While there is some variation between binding sites from Bacillus/Listeria species (group I) and Firmicutes cocci (group II), a minimal and almost invariant core 6-4-6 motif, TNNNNC----GNNNNA, can be extracted, with normally at least two additional residues having complementary partners in each side of the repeat (16). While this rule certainly applies for the binding site of the liaI promoter, the inverted repeats identified upstream of yhcY and ydhE are poorly conserved (Fig. 2D). Moreover, only PliaI can be induced by bacitracin, a stimulus known to strongly activate the LiaFSR system (46). In contrast, the two other promoters only respond in a LiaR-dependent manner in the absence of the inhibitor protein LiaF but cannot be induced by bacitracin (Fig. 1B).
To investigate the relevance of these two potential LiaR binding sites, we performed gel mobility shift assays with the purified response regulator LiaR to directly study its interaction with the three promoter regions containing the identified LiaR boxes (Fig. 2D) in vitro. In the presence of acetyl phosphate as a small-molecule phosphodonor, a clear shift was observed for the liaI promoter region, whereas neither the unspecific DNA fragment (bceA) nor either of the other two promoters showed any shift (Fig. 3). The shift of the liaI promoter was dependent on the phosphorylation of LiaR, since no shift was observed in the absence of acetyl phosphate (Fig. 3). Taken together, these three observations raised doubts about the in vivo relevance of the yhcY and ydhE promoter regions as LiaR target loci of B. subtilis.
FIG. 3.
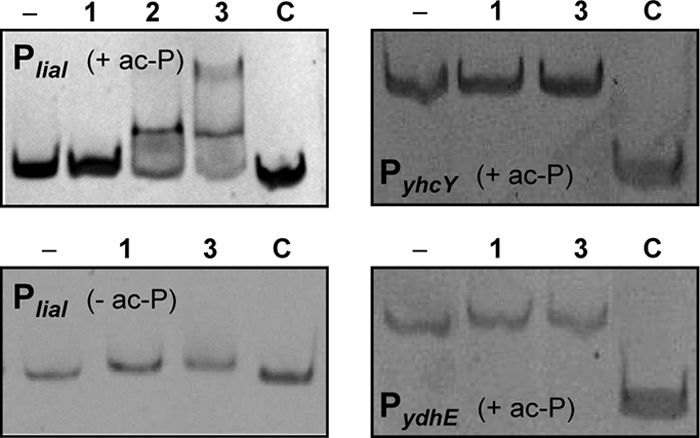
Gel mobility shift assays with purified response regulator LiaR. For the reactions, LiaR was phosphorylated by acetyl phosphate (ac-P), with the exception of the assay whose results are shown in the lower left panel, as described in Materials and Methods. PCR products of PliaI (180 bp), PyhcY, and PydhE were incubated with an increasing molar excess of phosphorylated LiaR (lane 1, 50:1; lane 2, 75:1; lane 3, 100:1) followed by gel electrophoresis on native 8% polyacrylamide gels and detection of the DNA with SYBR green I. The first lane in each panel corresponds to free DNA (−). An internal fragment of bceA (161 bp) was always included as a negative control (C).
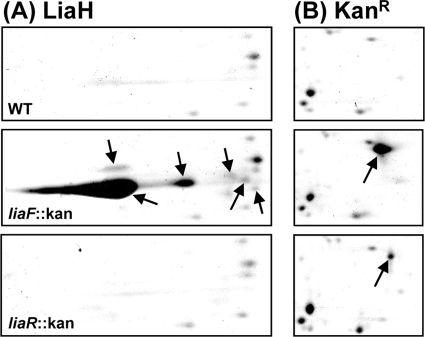
To further substantiate our findings and to analyze whether the significantly increased mRNA levels for all three targets translate into increased amounts of the corresponding proteins, we next performed a comprehensive proteomics study. Since all gene products but one (LiaI) encoded by these three loci are predicted to be soluble proteins, we only investigated the cytoplasmic proteome, again by comparing the protein signature of a wild-type strain with those of the isogenic liaF::kan and liaR::kan mutants. The results are shown in Fig. 4 and Fig. S1 in the supplemental material. Overall, the proteome signatures show very few significant differences in spot intensity. The most drastic changes were observed for LiaH, which was found to be the predominant cytoplasmic protein in the liaF mutant (see Fig. S1 in the supplemental material). It could be identified in six independent spots, indicative of posttranslational modifications (Fig. 4A). A comparably complex pattern of LiaH expression has recently been observed in daptomycin-treated wild-type cells of B. subtilis (59), underlining the relevance of our finding. The nature of these posttranslational modifications remains to be identified. But mass spectroscopy analyses of LiaH protein bands isolated from SDS-PAGE gels identified one LiaH-specific peptide with a mass increase corresponding to one phosphate group (data not shown). This observation, together with the clear pI shift of the spots in the 2-D gels (Fig. 4A), indicates that phosphorylation seems to be at least one type of posttranslational modification for LiaH.
FIG. 4.
Analysis of the cytoplasmic proteome by two-dimensional gel electrophoresis of the wild type (WT) and of otherwise isogenic liaF::kan and liaR::kan mutants. Only selected gel regions are shown. See Materials and Methods for experimental details. (A) The LiaH protein was identified in six independent spots (black arrows) by mass spectroscopy in the Lia ON mutant, indicative of posttranslational modifications. (B) A spot representing the product of the kanamycin resistance cassette (a type III aminoglycoside phosphotransferase derived from the Streptococcus faecalis plasmid pJH1; black arrow) was identified in the gels of the liaF::kan and liaR::kan mutant but not in the gel of the wild type. The larger amount in the liaF mutant relative to the amount in the liaR mutant strain originates from the LiaR-dependent autoregulation in the Lia ON mutant. See text for details.
The only other protein that showed a significant difference in its cellular amounts did not match any of the mapped proteins of the B. subtilis proteome. It could be detected in both mutants but was completely absent in the wild type. Mass spectroscopy revealed that the spot is comprised of the product of the kanamycin resistance gene. Compared to the basal expression level in the liaR mutant, its amount was further increased in the liaF mutant because of the autoregulatory feedback loop (Fig. 4B).
Despite significant promoter activity of both PyhcY and PydhE in the liaF mutant (Fig. 1B) and strong signals for the corresponding transcripts (Fig. 1C), we were unable to detect any of the gene products encoded by these two loci. Taken together, our collective data suggest that the liaIH operon is the only LiaR target of relevance under natural conditions. Hence, we decided to focus our attention on this operon to understand the physiological role of the Lia response of B. subtilis.
The Lia system is not involved in cellular differentiation in B. subtilis.
We had previously shown that the expression of the liaIH operon is embedded in the AbrB-/Spo0A-dependent differentiation cascade, which ultimately leads to the formation of dormant endospores (30). Therefore, we decided to investigate the influence of the Lia system on complex phenotypes associated with this regulatory cascade by comparing the behavior of liaIH and liaF mutants to that of the wild-type strain. No effects on the development of competence and endospore formation were observed. Moreover, we also moved both alleles into the nondomesticated ancestor strain NCIB3610 by SPP1 transduction to investigate multicellular phenotypes, such as colony differentiation, pellicle formation, and swarming motility, which were lost in the course of propagating the soil isolate in the laboratory environment (10). But again, no differences were observed for the lia mutants compared to the isogenic wild type (data not shown). The only identified difference with regard to complex phenotypes was delayed spore germination in the liaF::kan mutant relative to that in the wild type. But this turned out to be an experimental artifact, based on the strong read-through transcription from the lia locus into the divergently expressed ger operon that encodes spore germination proteins (see Fig. S2 in the supplemental material for details).
LiaIH contributes to the protection of cells against envelope and oxidative stress.
The initial phenotypic screens failed to provide evidence for a role of LiaIH in resistance against cell wall antibiotics that interfere with the lipid II cycle and which also function as strong inducers of the LiaFSR system (data not shown). Since the genome of B. subtilis encodes a LiaH paralog, PspA, we wondered if the lack of phenotypes associated with a liaH mutant could perhaps be the result of a functional redundancy of these phage shock proteins, as has been observed for a number of other paralogous protein pairs in B. subtilis (57). We therefore constructed a liaH::kan pspA::cat double mutant (TMB211) which was used in the subsequent physiological screens.
In order to get a comprehensive picture of phenotypes associated with LiaH/PspA of B. subtilis, we performed in-depth profiling by using phenotype microarrays. This technique allows the screening of hundreds of stress conditions in parallel, thereby enabling the fast and accurate identification of novel traits linked to genetic alterations (8), here, between the wild type and the isogenic liaH pspA and liaF mutant strains. The results of this study are summarized in Table S2 in the supplemental material. Overall, a relatively small number of rather diverse phenotypes could be associated with either the Lia ON mutant or the liaH pspA double mutant, and many effects were weak (data not shown). Only two types of stresses were represented by a number of compounds each. β-Lactam (cephalexin and cefotaxime) and additional cell wall antibiotics from our own screens (daptomycin, enduracidin, nisin, and fosfomycin) pointed toward a role of the phage shock proteins from B. subtilis in maintaining envelope integrity. Moreover, the sensitivity of lia mutants was also affected in the case of some compounds causing oxidative stress, such as menadione, plumbagin, sodium selenite, and hydrogen peroxide.
To verify and investigate these phenotypes in more detail, we next performed detailed follow-up studies to determine the MICs for larger panels of cell wall antibiotics and oxidative stress agents. We also performed serial dilution spot tests, using the individual liaH and pspA mutants to analyze the contribution of each of the two phage shock protein homologs to the observed phenotypes. Importantly, any increased sensitivity in the liaH pspA double mutant could be attributed to LiaH alone (data not shown). Therefore, only the results for the liaH mutant are shown. The results are summarized in Table 5, as well as in Fig. S3 in the supplemental material. In general, the MIC differences were moderate, rarely exceeding a 2- to 4-fold increased sensitivity of the liaH mutants relative to that of the wild type. The effects were more obvious in the serial dilution spot tests: at critical stressor concentrations, the survival rate of the liaH mutant was reduced between one and four orders of magnitude, depending on the compound tested (Table 5; also see Fig. S3 in the supplemental material). Upregulation of LiaIH expression in the liaF mutant hardly ever affected the stress sensitivity, with the exception of sensitivity to the β-lactam antibiotic cephalexin. Here, the wild type and the liaH mutant showed comparable behavior, while 10 to 100 times more cells of the liaF mutant survived in the presence of 0.25 μg/ml cephalexin (see Fig. S3 in the supplemental material).
TABLE 5.
Determination of the MICs and survival rates
| Type of compound | Compounds | MIC,a,b survival ratec for: |
||
|---|---|---|---|---|
| Wild type | ΔliaF mutant | ΔliaIH mutant | ||
| Cell wall antibiotics | Bacitracin | 300 | 300 | 300 |
| Cefotaxime | 0.1, 100 | 0.1, 100 | 0.05, 10−2 | |
| Cephalexin | 0.25, 100 | 0.25, 102 | 0.125, 101 | |
| Daptomycin | 1.25, 100 | 1.25, 100 | 0.6, 10−1 to 10−2 | |
| Enduracidin | 25, 100 | 12.5, 100 | 12.5, 10−3 | |
| Nisin | 10 | 2.5 | 10 | |
| Fosfomycin | 100, 100 | 200, 100 | 50, 10−3 | |
| Vancomycin | 0.5 | 0.5 | 0.5 | |
| Oxidative stress reagents | Cumene hydroperoxide | 0.006, 100 | 0.006, 100 | 0.006, 10−2 |
| Hydrogen peroxide | 1.5 | 0.5 | 1.0 | |
| Menadione | 5, 100 | 1.25, 100 | 2.5, 10−1 | |
| Plumbagin | 5, 100 | 2.5, 100 | 1.25, 10−1 | |
| Sodium selenite | 500, 100 | 500, 100 | 125, 10−2 | |
| t-Butyl hydroperoxide | 1, 100 | 1, 100 | 0.5, 10−1 | |
| Others | Cetylpyridinium chloride | 0.5 | 0.5 | 0.5 |
| Domiphen bromide | 0.125 | 0.03 | 0.125 | |
| Phenylarsine oxide | 0.1 | 0.1 | 0.05 | |
MICs are in μg/ml with the exception of cumene hydroperoxide (%), t-butylhydroperoxide (mM) and hydrogen peroxide (%).
Values indicating explicit sensitivity or resistance of the ΔliaF and ΔliaIH mutant strains versus that of the wild-type are highlighted in bold.
Survival rates were determined by serial dilution spot tests on Mueller-Hinton agar plates at the following “critical” compound concentrations: fosfomycin, 250 μg ml−1; cefotaxime, 0.1 μg ml−1; enduracidin, 0.01 μg ml−1; daptomycin, 0.15 μg ml−1; cephalexin, 0.2 μg ml−1; plumbagin, 2.5 μg ml−1; menadione, 1.25 μg ml−1; t-butyl hydroperoxide, 0.3 mM; cumene hydroperoxide, 0.003%; sodium selenite, 125 μg ml−1. All values are given relative to the result for the wild type as a reference, which was therefore set to 100. See Fig. S3 in the supplemental material for the original data and experimental details.
Previous studies had identified a number of cell wall antibiotics as strong inducers of the Lia response, most of which interfered with the membrane-anchored lipid II cycle (i.e., bacitracin, daptomycin, enduracidin, ramoplanin, nisin, and vancomycin). In contrast, antibiotics that inhibit early (cytoplasmic; i.e., fosfomycin and d-cycloserine) or late (extracellular; i.e., β-lactams) steps of cell wall biosynthesis did not induce the liaI promoter (28, 46). Remarkably, LiaH does not confer any resistance against most of its inducers, with the exception of enduracidin and daptomycin (Table 5). The role of LiaH in counteracting daptomycin damage was recently reported in an independent study (20). On the other hand, LiaH had a significant effect on the viability of B. subtilis in the presence of some cell wall antibiotics that do not function as inducers of the Lia response, such as fosfomycin and some β-lactams (Table 5). The significance of this finding remains to be investigated.
LiaH forms large oligomeric rings with a 9-fold rotational symmetry.
LiaH and PspA both belong to the phage shock protein family, which is widely conserved in the microbial world. A phylogenomic analysis revealed that members can be found in the Actinobacteria and Firmicutes (Gram positive) and Proteo- and Cyanobacteria (Gram negative) and also in the Archaea and even in the chloroplasts of plants (data not shown). While the exact physiological role of phage shock proteins is still a matter of ongoing studies, one structural feature that seems to be conserved even in distantly related phage shock proteins is the formation of large oligomeric ring-like structures (3, 22).
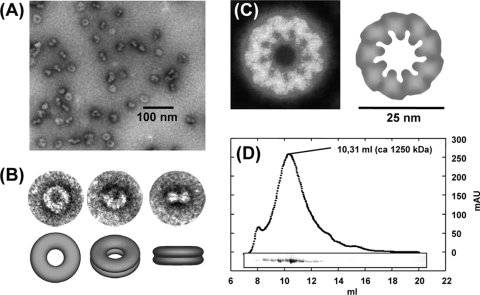
To verify a functional relationship of LiaH to better-investigated phage shock proteins, we overexpressed and purified LiaH to near homogeneity as a His-tagged fusion protein (see Materials and Methods for details). Oligomerization was studied both by transmission electron microscopy and by gel filtration (Fig. 5). In the presence of 100 μg μl−1 PEG-2000 (to increase the viscosity of the protein solution in order to simulate the cytoplasmic environment), a LiaH solution readily formed large ring-like complexes of homogenous sizes (Fig. 5A). Close-up views of individual complexes indicate the formation of bagel-shaped oligomers of about 25 nm in diameter, which can be seen at different angles (Fig. 5B). From 400 particles randomly selected and analyzed, 46.2% showed the top-view projection form, 23.8% the tilted-view form at different angles, and 16.0% the side-view form. Fourteen percent of the particles could not be assigned (mostly smaller fragments of the 25-nm complex). The images of the side-view form clearly show an accumulation of negative staining solution along the long axes of the protein complexes, separating the ring into two substructures. This is indicative for an indentation (as shown in the schematic models) but not necessarily for two separate rings. Side-view forms consisting of just one single ring could not be observed in the preparation.
FIG. 5.
Oligomeric structure of LiaH (see Materials and Methods for experimental details). (A) Transmission electron micrographs of LiaH rings. (B) Details of individual rings at different angles. Schematic models are shown below for illustrative purposes. (C) Markham rotation reveals a 9-fold rotational symmetry. (D) Size exclusion chromatography reveals a homogenous population of LiaH oligomers with a size of at least 1,250 kDa. The inset shows the respective SDS-PAGE analyses of the collected fractions corresponding to the chromatogram above. mAU, milli-absorbance unit.
We performed Markham rotations of top-view forms to analyze the symmetry of these complexes (42). This analysis clearly revealed a 9-fold rotational symmetry of the LiaH rings (Fig. 5C). The stability and homogeneous size distribution of the LiaH complexes was verified by analytical gel filtration experiments (Fig. 5D). Moreover, these studies also demonstrated that the LiaH complexes have a molecular mass of at least 1,250 kDa, which is in good agreement with the formation of a 36-mer (i.e., a nonamer of tetramers), as has also been suggested for E. coli PspA (22).
DISCUSSION
In this work, we have combined transcriptomics, proteomics, and in-depth phenotypic profiling to define the LiaR response of B. subtilis. We could demonstrate that three loci, liaIH-liaGFSR, yhcYZ-yhdA, and ydhE, which are preceded by a putative LiaR binding site, are also expressed in a Lia-dependent manner (Fig. 1 and 2). However, subsequent analyses suggested that only the lia operon itself is a relevant target locus of LiaR in vivo (Fig. 3 and 4). We hypothesize that an important factor contributing to these discrepancies between the wild-type situation and the response in a Lia ON mutant is the autoregulatory feedback loop: in the absence of the inhibitor protein LiaF, the TCS LiaRS is not only fully active, it also strongly (and constitutively) induces its own expression (29). Under these conditions, the accumulated cellular pool of phosphorylated LiaR obviously also mediates enhanced expression from promoters containing weakly conserved LiaR boxes that do not play any role as target loci in the wild-type situation. While the putative LiaR binding sites of PyhcY and PydhE are located at about the same distance relative to the transcriptional start site, they are significantly more weakly conserved (Fig. 2D) and presumably have a LiaR binding affinity that excludes recognition by this response regulator under normal physiological conditions. This hypothesis is supported by the results from the in vitro gel retardation assays (Fig. 3).
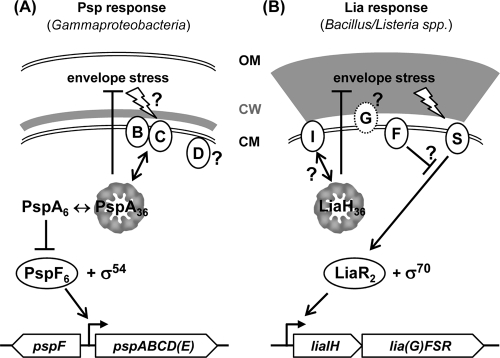
Our collective data argue for the liaIH-liaGFSR operon as the only relevant target of LiaFSR-dependent signal transduction. This hypothesis is also supported by the results of a comparative genomics analysis indicating that in the Bacillus/Listeria group, homologs of the liaIH operon are the primary target(s) of Lia-dependent gene expression (29). This operon encodes a small putative membrane protein with little sequence conservation and a member of the PspA/IM30 protein family, respectively. PspA is the core component of the phage shock response of E. coli and other Gammaproteobacteria, as mentioned above. It is therefore attractive to hypothesize that the LiaFSR system of B. subtilis and other bacilli has adopted the function of the proteobacterial phage shock response.
Comparison of the Psp response of E. coli and the Lia response of B. subtilis.
The core of the Psp response in E. coli consists of the stress-inducible pspABCD operon, which is regulated by PspF, encoded by a constitutive gene located upstream and inversely oriented from the psp operon (Fig. 6A) (31). As a homohexamer, this AAA(+) enhancer protein mediates strong induction from the σ54-dependent pspA promoter in the presence of envelope stress conditions (see below for details). The activity of PspF6 is strictly regulated by PspA. Importantly, recent biochemical analyses have demonstrated that PspA undergoes a shift in its oligomeric structure which affects its activity. Under nonstress conditions, PspA exists in a lower-order oligomer and interacts with PspF to form heterooligomers with a 6:6 stoichiometry (27). In this form, PspA inhibits PspF, resulting in only a very low basal expression level of the psp operon. Stress induction somehow affects the PspF-PspA interaction, resulting in a dissociation of the complex. Thereby, PspF is released to activate the expression of the psp operon, which strongly induces pspA expression. The resulting increase in cellular PspA amounts leads to the formation of large 36-mer PspA complexes, which then directly interact with the cytoplasmic membrane to counteract proton leakage (33). A signaling cascade that perceives envelope damage and transduces this information to the PspA-PspF complex presumably exists but has not been characterized so far. Likely candidates for this function would be the two membrane proteins PspB and PspC (Fig. 6A). Their genes are coexpressed with pspA, and a protein-protein interaction of both with PspA has been demonstrated (1). Joly et al. therefore proposed that part of the signal transduction mechanism is the decay of the PspABC complex in the absence of stress, which ultimately allows the formation of the PspA6-PspF6 repressor complex (27). The role of the fourth gene product of the psp operon, PspD, has not been elucidated so far, and PspE, while coexpressed, is not part of the Psp response (14).
FIG. 6.
Comparison between the Psp response of gammaproteobacteria and the Lia response of Firmicutes bacilli. All proteins are represented by circles at their known or predicted cellular location, with the letters corresponding to the respective genes, shown below. The phage shock proteins PspA and LiaH are depicted in their oligomeric form, using the model from Fig. 5C for illustrative purposes only. LiaG is represented by a dotted circle because homologs are only present in B. subtilis and its closest relatives. Double-ended arrows indicate interactions, normal arrows activation, and T-shaped lines inhibition. CM, cytoplasmic membrane; CW, cell wall; OM, outer membrane. See Discussion for details.
The Lia response, which is conserved in all Firmicutes Bacillus and Listeria species, also consists of five genes organized in two transcriptional units (Fig. 6B). While the expression of the regulatory genes lia(G)FSR is ensured by a weak constitutive promoter, the inducible and strongly LiaR-dependent promoter upstream of the liaIH operon leads to a significant amount of read-through transcription and, therefore, a positive-feedback loop on regulator gene expression (29), as also demonstrated by the results of the microarray analysis (Table 4). LiaF, together with LiaRS, constitutes a cell envelope stress-sensing three-component system that is highly conserved in Firmicutes bacteria (44). A LiaG homolog is only found in bacilli closely related to B. subtilis; its function is still unclear.
The Psp and Lia responses clearly differ from each other with regard to the mechanism of signal transduction and gene regulation (Fig. 6). Most importantly, there is no indication that LiaH plays any regulatory role comparable to that of E. coli PspA. A potential modulator function of LiaH on LiaR-dependent gene expression, as has been discussed previously (29), could be attributed to a genetic artifact, based again on strong positive polar effects from the inserted kanamycin resistance cassette used to replace liaH in the original mutant (data not shown).
The only direct homology between proteins of the Psp and Lia response is of PspA/LiaH, which both belong to the PspA/IM30 protein family. While no additional sequence similarity exists in the remaining proteins involved in both responses, it is noteworthy that pspB and liaI both encode small putative membrane proteins that are coexpressed with the PspA homologs. It is therefore tempting to postulate an interaction between LiaI and LiaH, similar to that described for PspA and PspB (Fig. 6B), but this needs to be further investigated.
A similar physiological role of PspA and LiaH is supported by the overlap of the inducer spectra of B. subtilis LiaH and PspA from E. coli. In addition to the strong induction by envelope stress, expression of the liaIH operon is induced by alkaline shock, secretion stress, and organic solvents (26, 46, 61). In comparison, expression of the pspA operon of E. coli is induced by filamentous phage infection (hence the name), misfolded and oversecreted envelope proteins, and other stress conditions, including heat and osmotic shock, exposure to organic solvents, protonophores, and antibiotics that interfere with phospholipid biosynthesis (7, 11, 14, 23, 48). Moreover, both genes are also induced without external stresses at the onset of or during stationary phase (30, 60). The similarity in inducing conditions is also reflected by the relatively diverse but weak phenotypes associated with mutations in the liaIH operon (Table 5; also see Fig. S3 in the supplemental material), which is again reminiscent of the Psp response of E. coli and related bacteria (3, 14, 22, 48).
The most obvious similarity between E. coli PspA and B. subtilis LiaH is the formation of large oligomeric ring-like structures with a 9-fold rotational symmetry, possibly consisting of 36-mers (Fig. 5) (22). Comparable bagel-shaped structures have also been observed for the PspA homologs, VIPP1 from Synechocystis (Cyanobacteria), and chloroplasts from Chlamydomonas and Arabidopsis thaliana (3, 35, 38). This conservation in both sequence and overall oligomeric structure indicates an evolutionary ancient mechanism of ensuring membrane and envelope integrity which is present in the Archaea, many eubacterial phyla, and plants. More recently, structural studies by Standar et al. (53) with detergent-free PspA preparations from membrane fractions demonstrated the formation of even larger clathrin-like so-called “PspA scaffolds” in E. coli. The exact function of these super structures is still unclear, but these large scaffolds are thought to contribute to the maintenance of membrane integrity in stressed cells, possibly through multiple PspA-membrane interactions over large surface areas (53). This assumption of an overall PspA layer to support membrane integrity is somewhat contradicted by recent findings that PspA localizes in discrete regions of the cell, such as midcell and the lateral cell wall (17). The localization of these PspA complexes, which are highly dynamic structures, depends on the bacterial cytoskeleton element MreB. In the absence of MreB, the Psp response is still induced, but the cells are no longer protected against membrane stress. This finding strongly suggests that the protective function of PspA complexes requires the bacterial cytoskeleton (17). These observations demonstrate that despite a number of groundbreaking recent studies and our progress in understanding the Psp response of gammaproteobacteria, the exact physiological role of PspA/IM30 proteins is still not fully understood.
Conclusions and outlook.
Taken together, this is the first report on the Psp response in low-G+C Gram-positive bacteria. Based on the clear conservation in the genomic context of the lia locus and the identification of putative LiaR binding sites, the overall organization (with the noteworthy exception of liaG, which is only present in Bacillus spp. closely related to B. subtilis) and regulation seem to be conserved in other Firmicutes bacilli and, presumably, also in Listeria spp. (29). For the latter, the liaIH and liaFSR operons are in separate genomic locations. However, both are preceded by a LiaR box (29). The work presented in this study was aimed at defining the Lia response of B. subtilis. Our data strongly suggest that, in contrast to the large LiaRS regulons identified in Firmicutes cocci (16, 36, 43, 55), the Lia response in Bacillus and Listeria spp. only orchestrates a phage shock protein-like response, encoded by the liaIH operon. Future studies now need to unravel the physiological role of the two corresponding proteins. Based on the data from E. coli PspA, phage shock proteins seem to exhibit their function through protein-protein and protein-membrane interactions. Accordingly, we first need to investigate the localization and identify potential protein interaction partners of LiaH. The latter will also help to answer the question of whether LiaH, like PspA, also functions as an AAA(+) adaptor protein, despite the lack of a PspF homolog in B. subtilis.
Supplementary Material
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (MA2837/1-3) and the Fonds der Chemischen Industrie (to T.M.) and the Bundesministerium für Bildung und Forschung (project 03ZIK012, to G.H. and U.M., and project 0313807A, to M.H. and B.V.). T.W. is the recipient of an FCI Chemiefonds Ph.D. stipend.
We thank Susanne Gebhard for critical reading of the manuscript.
Footnotes
Published ahead of print on 16 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, H., W. Teertstra, J. Demmers, R. Boesten, and J. Tommassen. 2003. Interactions between phage shock proteins in Escherichia coli. J. Bacteriol. 185:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aseeva, E., F. Ossenbuhl, L. A. Eichacker, G. Wanner, J. Soll, and U. C. Vothknecht. 2004. Complex formation of Vipp1 depends on its α-helical PspA-like domain. J. Biol. Chem. 279:35535-35541. [DOI] [PubMed] [Google Scholar]
- 4.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 5.Belcheva, A., and D. Golemi-Kotra. 2008. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283:12354-12364. [DOI] [PubMed] [Google Scholar]
- 6.Belcheva, A., V. Verma, and D. Golemi-Kotra. 2009. DNA-binding activity of the vancomycin resistance associated regulator protein VraR and the role of phosphorylation in transcriptional regulation of the vraSR operon. Biochemistry 48:5592-5601. [DOI] [PubMed] [Google Scholar]
- 7.Bergler, H., D. Abraham, H. Aschauer, and F. Turnowsky. 1994. Inhibition of lipid biosynthesis induces the expression of the pspA gene. Microbiology 140(Pt. 8):1937-1944. [DOI] [PubMed] [Google Scholar]
- 8.Bochner, B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4:309-314. [DOI] [PubMed] [Google Scholar]
- 9.Boyle-Vavra, S., S. Yin, and R. S. Daum. 2006. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 262:163-171. [DOI] [PubMed] [Google Scholar]
- 10.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brissette, J. L., M. Russel, L. Weiner, and P. Model. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mäder, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 13.Dartois, V., M. Debarbouille, F. Kunst, and G. Rapoport. 1998. Characterization of a novel member of the DegS-DegU regulon affected by salt stress in Bacillus subtilis. J. Bacteriol. 180:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621-628. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson, L. W. 2008. The NMR structure of the Staphylococcus aureus response regulator VraR DNA binding domain reveals a dynamic relationship between it and its associated receiver domain. Biochemistry 47:3379-3388. [DOI] [PubMed] [Google Scholar]
- 16.Eldholm, V., B. Gutt, O. Johnsborg, R. Bruckner, P. Maurer, R. Hakenbeck, T. Mascher, and L. S. Havarstein. 2010. The pneumococcal cell envelope stress-sensing system LiaFSR is activated by murein hydrolases and lipid II-interacting antibiotics. J. Bacteriol. 192:1761-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engl, C., G. Jovanovic, L. J. Lloyd, H. Murray, M. Spitaler, L. Ying, J. Errington, and M. Buck. 2009. In vivo localizations of membrane stress controllers PspA and PspG in Escherichia coli. Mol. Microbiol. 73:382-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estacio, W., S. S. Anna-Arriola, M. Adedipe, and L. M. Marquez-Magana. 1998. Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis. J. Bacteriol. 180:3548-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardete, S., S. W. Wu, S. Gill, and A. Tomasz. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3424-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hachmann, A.-B., E. R. Angert, and J. D. Helmann. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53:1598-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamze, K., D. Julkowska, S. Autret, K. Hinc, K. Nagorska, A. Sekowska, I. B. Holland, and S. J. Seror. 2009. Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC. Microbiology 155:398-412. [DOI] [PubMed] [Google Scholar]
- 22.Hankamer, B. D., S. L. Elderkin, M. Buck, and J. Nield. 2004. Organization of the AAA(+) adaptor protein PspA is an oligomeric ring. J. Biol. Chem. 279:8862-8866. [DOI] [PubMed] [Google Scholar]
- 23.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 24.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, England.
- 25.Homuth, G., S. Masuda, A. Mogk, Y. Kobayashi, and W. Schumann. 1997. The dnaK operon of Bacillus subtilis is heptacistronic. J. Bacteriol. 179:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyyryläinen, H. L., M. Sarvas, and V. P. Kontinen. 2005. Transcriptome analysis of the secretion stress response of Bacillus subtilis. Appl. Microbiol. Biotechnol. 67:389-396. [DOI] [PubMed] [Google Scholar]
- 27.Joly, N., P. C. Burrows, C. Engl., G. Jovanovic, and M. Buck. 2009. A lower-order oligomer form of phage shock protein A (PspA) stably associates with the hexameric AAA(+) transcription activator protein PspF for negative regulation. J. Mol. Biol. 394:764-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 29.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan, S., E. Rietkötter, M. A. Strauch, F. Kalamorz, B. G. Butcher, J. D. Helmann, and T. Mascher. 2007. LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology 153:2530-2540. [DOI] [PubMed] [Google Scholar]
- 31.Jovanovic, G., L. Weiner, and P. Model. 1996. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J. Bacteriol. 178:1936-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jürgen, B., S. Tobisch, M. Wümpelmann, D. Gördes, A. Koch, K. Thurow, D. Albrecht, M. Hecker, and T. Schweder. 2005. Global expression profiling of Bacillus subtilis cells during industrial-close fed-batch fermentations with different nitrogen sources. Biotechnol. Bioeng. 92:277-298. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi, R., T. Suzuki, and M. Yoshida. 2007. Escherichia coli phage shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66:100-109. [DOI] [PubMed] [Google Scholar]
- 34.Kossen, K., and O. C. Uhlenbeck. 1999. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 27:3811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroll, D., K. Meierhoff, N. Bechtold, M. Kinoshita, S. Westphal, U. C. Vothknecht, J. Soll, and P. Westhoff. 2001. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. U. S. A. 98:4238-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 37.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 38.Liu, C., F. Willmund, J. R. Golecki, S. Cacace, B. Hess, C. Markert, and M. Schroda. 2007. The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 50:265-277. [DOI] [PubMed] [Google Scholar]
- 39.Liu, Y. H., A. Belcheva, L. Konermann, and D. Golemi-Kotra. 2009. Phosphorylation-induced activation of the response regulator VraR from Staphylococcus aureus: insights from hydrogen exchange mass spectrometry. J. Mol. Biol. 391:149-163. [DOI] [PubMed] [Google Scholar]
- 40.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U. S. A. 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacRitchie, D. M., D. R. Buelow, N. L. Price, and T. L. Raivio. 2008. Two-component signaling and gram negative envelope stress response systems. Adv. Exp. Med. Biol. 631:80-110. [DOI] [PubMed] [Google Scholar]
- 42.Markham, R., S. Frey, and G. J. Hills. 1963. Methods for the enhancement of image detail and accentuation of structure in electron microscopy. Virology 20:88-102. [Google Scholar]
- 43.Martinez, B., A. L. Zomer, A. Rodriguez, J. Kok, and O. P. Kuipers. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473-486. [DOI] [PubMed] [Google Scholar]
- 44.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133-144. [DOI] [PubMed] [Google Scholar]
- 45.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 46.Mascher, T., S. L. Zimmer, T. A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 48.Model, P., G. Jovanovic, and J. Dworkin. 1997. The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24:255-261. [DOI] [PubMed] [Google Scholar]
- 49.Moir, A., and D. A. Smith. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44:531-553. [DOI] [PubMed] [Google Scholar]
- 50.Nakano, M. M., M. A. Marahiel, and P. Zuber. 1988. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nasser, W., A. C. Awade, S. Reverchon, and J. Robert-Baudouy. 1993. Pectate lyase from Bacillus subtilis: molecular characterization of the gene, and properties of the cloned enzyme. FEBS Lett. 335:319-326. [DOI] [PubMed] [Google Scholar]
- 52.Ogura, M., L. Liu, M. Lacelle, M. M. Nakano, and P. Zuber. 1999. Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis. Mol. Microbiol. 32:799-812. [DOI] [PubMed] [Google Scholar]
- 53.Standar, K., D. Mehner, H. Osadnik, F. Berthelmann, G. Hause, H. Lunsdorf, and T. Bruser. 2008. PspA can form large scaffolds in Escherichia coli. FEBS Lett. 582:3585-3589. [DOI] [PubMed] [Google Scholar]
- 54.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 55.Suntharalingam, P., M. D. Senadheera, R. W. Mair, C. M. Levesque, and D. G. Cvitkovitch. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 191:2973-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tam, L. T., C. Eymann, D. Albrecht, R. Sietmann, F. Schauer, M. Hecker, and H. Antelmann. 2006. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ. Microbiol. 8:1408-1427. [DOI] [PubMed] [Google Scholar]
- 57.Thomaides, H. B., E. J. Davison, L. Burston, H. Johnson, D. R. Brown, A. C. Hunt, J. Errington, and L. Czaplewski. 2007. Essential bacterial functions encoded by gene pairs. J. Bacteriol. 189:591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 59.Wecke, T., D. Zühlke, U. Mäder, S. Jordan, B. Voigt, S. Pelzer, H. Labischinski, G. Homuth, M. Hecker, and T. Mascher. 2009. Daptomycin versus friulimicin B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob. Agents Chemother. 53:1619-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiner, L., and P. Model. 1994. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. U. S. A. 91:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiegert, T., G. Homuth, S. Versteeg, and W. Schumann. 2001. Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 41:59-71. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida, K. I., Y. Fujita, and S. D. Ehrlich. 2000. An operon for a putative ATP-binding cassette transport system involved in acetoin utilization of Bacillus subtilis. J. Bacteriol. 182:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Youngman, P. 1990. Use of transposons and integrational vectors for mutagenesis and construction of gene fusions in Bacillus subtilis, p. 221-266. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, England.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.