Abstract
Vibrio cholerae switches between free-living motile and surface-attached sessile lifestyles. Cyclic diguanylate (c-di-GMP) is a signaling molecule controlling such lifestyle changes. C-di-GMP is synthesized by diguanylate cyclases (DGCs) that contain a GGDEF domain and is degraded by phosphodiesterases (PDEs) that contain an EAL or HD-GYP domain. We constructed in-frame deletions of all V. cholerae genes encoding proteins with GGDEF and/or EAL domains and screened mutants for altered motility phenotypes. Of 52 mutants tested, four mutants exhibited an increase in motility, while three mutants exhibited a decrease in motility. We further characterized one mutant lacking VC0137 (cdgJ), which encodes an EAL domain protein. Cellular c-di-GMP quantifications and in vitro enzymatic activity assays revealed that CdgJ functions as a PDE. The cdgJ mutant had reduced motility and exhibited a small decrease in flaA expression; however, it was able to produce a flagellum. This mutant had enhanced biofilm formation and vps gene expression compared to that of the wild type, indicating that CdgJ inversely regulates motility and biofilm formation. Genetic interaction analysis revealed that at least four DGCs, together with CdgJ, control motility in V. cholerae.
Cyclic diguanylate (c-di-GMP) is a ubiquitous second messenger in bacteria. It is synthesized by diguanylate cyclases (DGCs) that contain a GGDEF domain and is degraded by phosphodiesterases (PDEs) that contain an EAL or HD-GYP domain (46, 48, 50). The receptors of c-di-GMP, which can be proteins or RNAs (riboswitches), bind to c-di-GMP and subsequently transmit the signal to downstream targets (22). C-di-GMP signaling is predicted to occur via a common or localized c-di-GMP pool(s) through so-called c-di-GMP signaling modules harboring DGCs and PDEs, receptors, and targets that affect cellular function (22).
C-di-GMP controls various cellular functions, including the transition between a planktonic lifestyle and biofilm lifestyle. In general, high concentrations of c-di-GMP promote the expression of adhesive matrix components and result in biofilm formation, while low concentrations of c-di-GMP result in altered motility upon changes in flagellar or pili function and/or production (reviewed in reference 25). C-di-GMP inversely regulates motility and biofilm formation by implementing control at different levels through gene expression or through posttranslational mechanisms (reviewed in reference 25).
Vibrio cholerae, the causative agent of the disease cholera, uses c-di-GMP signaling to undergo a motile-to-sessile lifestyle switch that is important for both environmental and in vivo stages of the V. cholerae life cycle. The survival of the pathogen in both natural aquatic environments and during infection depends on the appropriate regulation of motility, surface attachment, and colonization factors (26). The V. cholerae genome encodes a total of 62 putative c-di-GMP metabolic enzymes: 31 with a GGDEF domain, 12 with an EAL domain, 10 with both GGDEF and EAL domains, and 9 with an HD-GYP domain (21). V. cholerae contains a few known or predicted c-di-GMP receptors: two riboswitches (53), five PilZ domain proteins (43), VpsT (31), and CdgG (6). C-di-GMP regulates virulence, motility, biofilm formation, and the smooth-to-rugose phase variation in V. cholerae (6, 8, 9, 12, 30, 33, 43, 45, 54, 56, 57). However, particular sets of proteins have not been matched to discrete cellular processes.
Some of the DGCs and PDEs involved in regulating motility in V. cholerae have been identified: rocS and cdgG mutants exhibit a decrease in motility (45), while cdgD and cdgH mutants exhibit an increase in motility (6). In addition, VieA (PDE) positively regulates motility in the V. cholerae classical biotype but not in the El Tor biotype (7). AcgA (PDE) positively regulates motility at low concentrations of inorganic phosphate (42). In this study, we investigated the role of each putative gene encoding DGCs and PDEs in controlling cell motility. In addition to the already-characterized proteins CdgD, CdgH, and RocS, we identified two putative DGCs (CdgK and CdgL) that negatively control motility and a putative PDE (CdgJ) that positively controls motility. We further characterized CdgJ and showed that it functions as a PDE and inversely regulates motility and biofilm formation. Genetic interaction studies revealed that DGCs CdgD, CdgH, CdgL, and CdgK and PDE CdgJ form a c-di-GMP signaling network to control motility in V. cholerae.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The wild-type V. cholerae strain used in this work was a smooth variant of strain O1 El Tor A1552 (62). Escherichia coli DH5α was used for standard DNA manipulations, and E. coli S17-1λpir was used for conjugation with V. cholerae. All V. cholerae strains and E. coli strains were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl; pH 7.5) at 30 and 37°C, respectively, unless otherwise noted. When needed, ampicillin and rifampin were used at 100 μg/ml and gentamicin was used at 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F′ endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 (Φ80lacΔM15) | Promega |
| S17-1 λpir | Tpr Smr, recA thi pro rK− mK+RP4:2-Tc:MuKm Tn7 λpir Tpr Smr λpir | 17 |
| DH10B F− | mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 galUgalK λ−rpsL (StrR) nupG ΔlacX74recA1endA1araΔ139 Δ(ara, leu)7697 | Invitrogen |
| V. cholerae | ||
| Fy_Vc_1 | V. cholerae O1 El Tor A1552, smooth variant, Rifr | 61 |
| Fy_Vc_2 | V. cholerae O1 El Tor A1552, rugose variant, Rifr | 61 |
| Fy_Vc_3 | Fy_Vc_1 ΔlacZ, Rifr | 13 |
| Fy_Vc_352 | Fy_Vc_1 ΔcdgD, Rifr | 33 |
| Fy_Vc_970 | Fy_Vc_1 ΔcdgG, Rifr | 6 |
| Fy_Vc_1592 | Fy_Vc_1 ΔcdgH, Rifr | 6 |
| Fy_Vc_99 | Fy_Vc_1 ΔvpsT, Rifr | This study |
| Fy_Vc_1663 | Fy_Vc_1 ΔplzA, Rifr | This study |
| Fy_Vc_1667 | Fy_Vc_1 ΔplzB, Rifr | This study |
| Fy_Vc_1671 | Fy_Vc_1 ΔplzC, Rifr | This study |
| Fy_Vc_1641 | Fy_Vc_1 ΔplzD, Rifr | This study |
| Fy_Vc_1659 | Fy_Vc_1 ΔplzE, Rifr | This study |
| Fy_Vc_4601 | Fy_Vc_3 mTn7-GFP, Rifr Gmr | This study |
| Fy_Vc_4571 | Fy_Vc_3 ΔVC0136, Rifr | This study |
| Fy_Vc_4592 | Fy_Vc_3 ΔcdgJ, Rifr | This study |
| Fy_Vc_4574 | Fy_Vc_3 ΔVC0136-7, Rifr | This study |
| Fy_Vc_4577 | Fy_Vc_3 cdgJ-AAA, Rifr | This study |
| Fy_Vc_4607 | Fy_Vc_3 ΔVC0136 mTn7-GFP, Rifr Gmr | This study |
| Fy_Vc_4604 | Fy_Vc_3 ΔVC0137 mTn7-GFP, Rifr Gmr | This study |
| Fy_Vc_4610 | Fy_Vc_3 ΔVC0136-7 mTn7-GFP, Rifr Gmr | This study |
| Fy_Vc_5399 | Fy_Vc_1 ΔcdgJ ΔplzA, Rifr | This study |
| Fy_Vc_5401 | Fy_Vc_1 ΔcdgJ ΔplzB, Rifr | This study |
| Fy_Vc_5402 | Fy_Vc_1 ΔcdgJ ΔplzC, Rifr | This study |
| Fy_Vc_5405 | Fy_Vc_1 ΔcdgJ ΔplzD, Rifr | This study |
| Fy_Vc_5408 | Fy_Vc_1 ΔcdgJ ΔplzE, Rifr | This study |
| Fy_Vc_5467 | Fy_Vc_1 ΔplzA ΔplzB ΔplzC ΔplzD ΔplzE, Rifr | This study |
| Fy_Vc_5471 | Fy_Vc_1 ΔcdgJ ΔplzA ΔplzB ΔplzC ΔplzD ΔplzE, Rifr | This study |
| Fy_Vc_5392 | Fy_Vc_3 ΔcdgJ ΔvpsT, Rifr | This study |
| Fy_Vc_5413 | Fy_Vc_1 ΔcdgJ ΔcdgG, Rifr | This study |
| Fy_Vc_5390 | Fy_Vc_3 ΔcdgJ ΔcdgD, Rifr | This study |
| Fy_Vc_5411 | Fy_Vc_1 ΔcdgJ ΔcdgH, Rifr | This study |
| Fy_Vc_5474 | Fy_Vc_1 ΔcdgJ ΔcdgD ΔcdgH, Rifr | This study |
| Fy_Vc_5477 | Fy_Vc_1 ΔcdgJ ΔcdgK, Rifr | This study |
| Fy_Vc_5480 | Fy_Vc_1 ΔcdgJ ΔcdgL, Rifr | This study |
| Fy_Vc_5450 | Fy_Vc_1 ΔcdgD ΔcdgH, Rifr | This study |
| Fy_Vc_6325 | Fy_Vc_1 ΔcdgD ΔcdgH ΔcdgK, Rifr | This study |
| Fy_Vc_6347 | Fy_Vc_1 ΔcdgD ΔcdgH ΔcdgK ΔcdgL, Rifr | This study |
| Fy_Vc_6328 | Fy_Vc_1 ΔcdgJ ΔcdgD ΔcdgH ΔcdgK, Rifr | This study |
| Fy_Vc_6350 | Fy_Vc_1 ΔcdgJ ΔcdgD ΔcdgH ΔcdgK ΔcdgL, Rifr | This study |
| Plasmids | ||
| pGP704-sacB28 | pGP704 derivative; sacB, Apr | G. Schoolnik |
| pCC2 | pGP704-sacB28::ΔlacZ, Apr | 13 |
| pFY-824 | pGP704-sacB28::ΔVC0137, Apr | This study |
| pFY-681 | pGP704-sacB28::ΔVC0136, Apr | This study |
| pFY-682 | pGP704-sacB28::ΔVC0136-7, Apr | This study |
| pFY-683 | pGP704-sacB28::VC0137-AAA, Apr | This study |
| pACYC177 | Multicopy-number cloning vector, Apr | New England Biolabs |
| pVC0136 | pACYC177::VC0136, Apr | This study |
| pVC0136-7 | pACYC177::VC0136-7, Apr | This study |
| pcdgJ | pACYC177::VC0137, Apr | This study |
| pBAD/myc-His-B | Arabinose inducible expression vector with C-terminal myc epitope and six-His tags | Invitrogen |
| pBAD::cdgJ | pBAD/myc-His-B::cdgJ, Apr | This study |
| pBAD::cdgJ-AAA | pBAD/myc-His-B::cdgJ-AAA, Apr | This study |
| pMCM11 | pGP704::mTn7-GFP, Gmr, Apr | G. Schoolnik |
| pUX-BF13 | oriR6K helper plasmid, provides Tn7 transposition function in trans; Apr | 3 |
| pRS415 | lacZ-based cloning vector for transcriptional fusion studies; Apr | 52 |
| pCC9 | pGP704-sacB28:: ΔvpsT, Apr | 13 |
| pCC12 | pRS415 vpsL promoter, Apr | 13 |
| pCC10 | pRS415 vpsR promoter, Apr | 13 |
| pCC25 | pRS415 vpsT promoter, Apr | 13 |
| pFY-352 | pRS415 flaA promoter, Apr | 34 |
| pFY-835 | pRS415 VC0136-7 promoter, Apr | This study |
| pFY-640 | pGP704-sacB28::ΔplzA, Apr | 6 |
| pFY-641 | pGP704-sacB28::ΔplzB, Apr | 6 |
| pFY-642 | pGP704-sacB28::ΔplzC, Apr | 6 |
| pFY-643 | pGP704-sacB28::ΔplzD, Apr | 6 |
| pFY-644 | pGP704-sacB28::ΔplzE, Apr | 6 |
| pFY-157 | pGP704-sacB28::ΔcdgD, Apr | 33 |
Plasmid and strain construction.
DNA oligonucleotides used in this study were purchased from Operon Technologies (Alameda, CA) and are listed in Table 2. Green fluorescent protein (GFP)-tagged V. cholerae strains were generated as described previously (20, 33). Plasmids to generate in-frame deletions were constructed using a protocol described previously (19, 24, 32). Briefly, del-A and del-B primers were used to amplify a 500- to 600-bp 5′-flanking sequence of the gene, including several nucleotides of the coding region, and del-C and del-D primers were used to amplify several nucleotides from the 3′ region of the gene plus the downstream flanking region. The two PCR products were joined using the splicing overlap extension (SOE) technique (24, 32), and the resulting PCR product was digested and ligated into a pGP704-sacB28 suicide plasmid. For the chromosomal replacement of the cdgJ wild-type copy with cdgJ-AAA point mutations, a mutated version of cdgJ was cloned into pGP704-sacB28 plasmid as described above with the following alterations: VC0137del-A and VC0137-AAA_B were used to amplify 151 bp 5′ of the region of the VC0137 locus and 541 bp of upstream flanking region, and VC0137-AAA_C and VC0137-AAA_D were used to amplify the coding region of the VC0137 locus from bp 132 to 746.
TABLE 2.
DNA sequences of oligonucleotides used in this studya
| Primer name | DNA sequence (5′-3′) |
|---|---|
| VC0136_del_A | GCAAAGCCCATGGCCTTAAATACC |
| VC0136_del_B | CGAATATAGCGACCCATTGCAATTCCTCATTCCTGAC |
| VC0136_del_C | GCAATGGGTCGCTATATTCGTTCACCTAACATAATG |
| VC0136_del_D | CTAGTCTAGACGACTCGTTCTGCCAAAA |
| VC0137_del_A | ATCGCCATGGTATCTACTGACCGCGGTG |
| VC0137_del_B | TATCACTAAATTAATCGTTGCTTACGCTTTGGCGGTA |
| VC0137_del_C | CAACGATTAATTTAGTGATAAGTTAATGCACCTTTTC |
| VC0137_del_D | CGATTCTAGACCACTGGAATATTCAAGG |
| VC0137op_del_B | ATCACTAAATTAATCCCATTGCAATTCCTCATTCCTGAC |
| VC0137op_del_C | AATGGGATTAATTTAGTGATAAGTTAATGCACCTTTTC |
| VC0137-AAA_B | CTCGAAAAGCAGCAGCATAACCTAGCGTGTGTCGCTTTG |
| VC0137-AAA_C | TTATGCTGCTGCTTTTCGAGATGGAGAAAAAAACGC |
| VC0137-AAA_D | GCTCTAGACGCACATAATCGACTTCAGGCTG |
| VC0137_comp_A | CCGCTCGAGTTCAATCCTGCCATTCACA |
| VC0137_comp_D | CGCGGATCCAATCGATGAACTGGCAGAG |
| VC0136_comp_D | CGGGATCCATAACCTAGCGTGTGTCGCTTTGC |
| VC0137op_prom_Fwd | GGAATTCGGCCTATTTGTCTATCTTCAGTCAGACG |
| VC0137op_prom_Rev | CAGGATCCTGCAATTCCTCATTCCTGACTTATGC |
| VC0137_pBAD_F_NcoI | GTCACCATGGTTCGATGTTTATGGGCTGC |
| VC0137_pBAD_R | CGAATTCCAATTAATCGATTGATGTCCTGGCTCC |
| operon_F | GTCATCTAGAAAGTTATGGTACACGCCACTCATTAGGG |
| operon_R | GTCACCATGGACGCACAAATTCACACGCTTTATCG |
| gyrA_qF | TGTGGGTGCAGTACAAGTGG |
| gyrA_qR | AAGTACGGATCAGGGTCACG |
| VC0137_qF | ACGTCAGTCCAGAGCATGTGA |
| VC0137_qR | ATGAGGAGATGGAGACCGAGA |
Underlined bases indicate the positions of restriction sites.
To complement the mutations, VC0136 and the entire operon with the native promoter were amplified by PCR using wild-type genomic DNA as a template with comp-A and comp-D primers (Table 2). cdgJ with the promoter region upstream of VC0136 was PCR amplified using ΔVC0136 genomic DNA as a template. The resulting PCR products were digested and ligated into vector pACYC177 to generate pVC0136, pcdgJ, and pVC0136-7. To overexpress cdgJ and cdgJ-AAA, cdgJ and cdgJ-AAA were PCR amplified with primers VC0137_pBAD_F_NcoI and VC0137_pBAD_R, using the wild type or the cdgJ-AAA strain as a template. The PCR products were cloned into pBAD/myc-His-B to generate pBAD::cdgJ and pBAD::cdgJ-AAA. The plasmids were sequenced to verify that no PCR errors were present and then introduced into corresponding mutant strains by electroporation.
RNA isolation, RT-PCR, and qRT-PCR.
Total RNA was isolated from 2 ml of mid-exponential-phase V. cholerae wild-type and ΔVC0136 cells grown in LB (optical density at 600 nm [OD600], 0.3 to 0.4) as described previously (61). The Turbo DNA-free kit (Ambion) was used to remove any contaminating genomic DNA from the total RNA samples, and the total RNA yield was calculated using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). cDNA was synthesized from 1 μg of each RNA sample using an iScript cDNA synthesis kit (Bio-Rad). The product was diluted 1:4 with water. Reverse transcription-PCR (RT-PCR) was carried out with cDNA generated with RNA from the wild type as a template and the operon_F/operon_R primer pair. Reaction mixtures lacking reverse transcriptase or RNA template were used as negative controls. A reaction mixture with genomic DNA as the template was used as a positive control. For quantitative PCRs (qRT-PCR), cDNA template was further diluted 10- and 100-fold. The reaction mixture contained 10 μl of the diluted cDNA, 12.5 μl of SYBR green PCR master mix (Applied Biosystems), and 300 nM each primer. Reactions were carried out in a DNA Engine Opticon 2 thermocycler (MJ Research) using the following cycle parameters: 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 1 min, and 72°C for 30 s; and a melting-curve analysis from 60 to 95°C. The results were analyzed using Opticon 2 software. Standard curves were prepared from genomic DNA of wild-type V. cholerae (1, 0.1, 0.01, 0.001, and 0.0001 ng) to correlate the amount of DNA to cycle thresholds. An R2 of 0.99 was obtained for all of the primer pairs that were used in this study (Table 2). The gyrA gene encoding DNA gyrase subunit A was used to normalize expression values. Three biological replicates and three technical replicates were used for each strain.
Motility assays.
LB soft-agar motility plates (0.3% agar) were used to determine the motility of bacterial strains (59). Briefly, single colonies were stabbed into the LB motility plates. The diameter of the migration zone was measured after 16 h of incubation at 30°C. Two-tailed Student's t tests were used to compare motility differences between strains.
Biofilm assays.
Biofilm formation assays using static cultures were carried out as described previously (38), with slight modifications. Each strain was inoculated from overnight cultures into LB broth to an OD600 of about 0.02. Culture (100 μl) was pipetted into each well of a sterile 96-well microtiter plate (polyvinyl chloride) and incubated statically at room temperature. After 2 and 6 h, planktonic bacteria were removed from each microtiter plate by inverting the plate, and the plate was washed gently once with water. A volume of 150 μl of 1% crystal violet solution was added into each well. The plates were incubated at room temperature for 15 min, rinsed thoroughly, and dried at room temperature. The stain was dissolved by adding 150 μl of ethanol and then quantified by measuring absorbance at 595 nm. The assay was repeated two times independently (two biological replicates), each with eight technical replicates.
For flow cell experiments, overnight cultures were diluted to an OD600 of 0.02 in 2% LB (0.02% tryptone, 0.01% yeast extract, 1% NaCl; pH 7.5) and used to inoculate flow chambers. The flow cell system was assembled and prepared as described previously (15). Biofilms were grown at room temperature in flow chambers (with individual channel dimensions of 1 by 4 by 40 mm) supplied with 2% LB at a flow rate of 4.5 ml h−1. Biofilms were analyzed using confocal laser-scanning microscopy (CLSM) with an LSM 5 Pascal laser-scanning microscope (Zeiss). The three-dimensional images were reconstructed using Imaris software (Bitplane) and quantified using COMSTAT (23). The experiments were performed two times independently, and three images were taken for each strain in each run.
β-Galactosidase assays.
Cells were grown to exponential phase (OD600 of 0.3 to 0.4) and stationary phase (grown overnight, OD600 of about 4.2) as described previously (51). β-Galactosidase assays were performed using a previously described protocol (19) similar to that described by Miller (36).
In vitro PDE activity assays.
V. cholerae strains carrying pBAD::cdgJ, pBAD::cdgJ-AAA overexpression plasmids, or empty vector were grown under inducing conditions (0.1% arabinose) at 30°C overnight. Cultures were harvested by centrifugation at 3,220 × g for 15 min and resuspended in phosphate-buffered saline (PBS) with 1 mM phenylmethylsulfonyl fluoride. Cells were lysed by sonication and then centrifuged at 16,100 × g for 10 min at 4°C. The supernatant (crude cell extract) was used in PDE activity assays. The protein concentration in crude cell extract was determined with a standard bicinchoninic acid (BCA) assay using BCA protein assay reagent (Thermo Fisher Scientific Inc., Rockford, IL). PDE activity was assessed using the synthetic substrate bis(p-nitrophenyl) phosphate (bis-pNPP) with a method similar to that previously described (10). Briefly, the 100-μl reaction mixture contains 50 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 5 mM bis-pNPP, and 20 μl of crude cell extract. The reaction mixture was incubated for 10 to 60 min at 37°C. The release of p-nitrophenol was quantified at OD410 in a spectrophotometer.
Determination of intracellular c-di-GMP levels.
C-di-GMP concentrations were measured in V. cholerae wild type (smooth variant), ΔcdgJ, cdgJ-AAA, rugose variant (R), R(pBAD/myc-His-B), R(pBAD::cdgJ), and R(pBAD::cdgJ-AAA). Each strain was inoculated from frozen stock into LB broth with or without ampicillin and grown for 16 h at 30°C with shaking (58). To determine the amount of protein, 100 μl of each culture was centrifuged to pellet cells. The supernatant was removed, and cells were lysed with 0.5 ml of 2% sodium dodecyl sulfate. The samples then were assayed with a standard BCA assay as described above.
To extract c-di-GMP, 2 ml of each culture was harvested by centrifugation at 16,100 × g for 2 min. The cell pellets were resuspended in 400 μl of cold extraction solution (40% acetonitrile, 40% methanol, 0.1% formic acid, 19.9% water), vortexed for 30 s, and then incubated on ice for 15 min (58). The samples then were centrifuged at 16,100 × g for 2 min, and the supernatants were collected and dried under vacuum by centrifugal evaporation and lyophilization. The resulting samples were resuspended in 50 μl high-performance liquid chromatography (HPLC)-grade water and stored at −20°C until analyzed by liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) on a ThermoElectron Finnigan LTQ mass spectrometer (Thermo, San Jose, CA) coupled with a surveyor HPLC. C-di-GMP was detected by selected ion-monitoring analyses (m/z = 687.7 to 690.70) in negative mode and identified by the fragmentation pattern in the second MS. The electrospray voltage was set to 5.0 kV, and a data-dependent acquisition MS mode (over the m/z range of 400 to 2,000) was used, followed by the five MS-MS scans for the five most abundant precursor ions in the MS survey scan. The collision-induced dissociation (CID) at a normalized collision energy of 35% was used for MS-MS.
Reverse-phase liquid chromatography was done with a Synergi Hydro 4u Fusion-RP 80A column (150 by 2.00 mm; 4-μm particle size) (Phenomenex, Torrance, CA). Solvent A was 0.1% formic acid in water. Solvent B was acetonitrile. The gradient was as follows: time (t) = 0, 2% solvent B; t = 3 min, 2% solvent B; t = 15 min, 30% solvent B; t = 15.01 min, 95% solvent B; and t = 20 min, 95% solvent B. Other chromatography parameters were as follows: autosampler, 4°C; injection volume, 20 μl; and flow rate, 200 μl/min.
The amount of c-di-GMP in samples was estimated using a standard curve generated from pure c-di-GMP (Biolog Life Science Institute, Bremen, Germany) with concentrations of 50 nM, 100 nM, 500 nM, 2 μM, 3.5 μM, 5 μM, 7.5 μM, and 10 μM. The assay is linear from 50 nM to 10 μM with an R2 of 0.999. C-di-GMP levels were normalized to total protein per ml of culture. C-di-GMP quantification was performed with at least three biological replicates. Two-tailed Student's t tests were used to compare c-di-GMP levels between strains.
Transmission electron microscopy (TEM).
Bacterial strains were grown overnight on LB plates at 30°C. Ten μl of PBS was dropped on a colony, and a 400-mesh carbon-coated copper grid (Electron Microscopy Sciences) was placed on top of the drop. Grids with cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4), rinsed with water, and then stained with 2% (wt/vol) uranyl acetate solution. Samples were examined with a JEOL JEM-1230 electron microscope by using an accelerating voltage of 120 kV. Micrographs were taken with a charge-coupled device (CCD) camera.
RESULTS
Identification of genes encoding GGDEF and EAL domain proteins modulating motility.
To characterize the functions of GGDEF and EAL domain proteins and their role in the c-di-GMP signaling network of V. cholerae, we constructed in-frame deletions of each individual gene (except VC0515, as it is absent from our prototype strain) in the wild-type genetic background (6). The growth rates of all mutants were similar to that of the wild type when grown in LB medium at 30°C (data not shown). To understand the effect of each individual gene on cell motility, we analyzed the motility phenotype of each mutant on LB motility plates compared to that of the wild type. Of the 52 mutants tested, cdgD (VCA0697), cdgH (VC1067), VC1104, and VC2285 mutants exhibited increased motility compared to that of the wild type (Fig. 1). CdgD and CdgH previously were shown to negatively regulate motility, and it has been shown that CdgH has DGC activity and CdgD is predicted to be a DGC (6, 33). VC1104 and VC2285 both have a conserved GGDEF domain; we designated VC1104 and VC2285 cdgK and cdgL, respectively. In contrast, the rocS, cdgG, and VC0137 mutants exhibited decreased motility compared to that of the wild type (Fig. 1), which is consistent with previously reported phenotypes for rocS and cdgG (6, 33). Our previous studies showed that CdgG is likely to be a c-di-GMP receptor protein with an RXXD motif (6). RocS is a dual GGDEF/EAL domain protein and is predicted to function as a PDE under the experimental conditions utilized in this study (33). VC0137 contains a conserved EAL domain and is predicted to be a PDE. We designated VC0137 cdgJ. We further characterized cdgJ, as its motility phenotype was the most severe among the mutants tested.
FIG. 1.

Motility phenotypes of selected mutants. The diameters of migration zones of the wild type and mutants were measured after 16 h of incubation at 30°C on LB motility plates. Only the mutants with altered motility phenotypes are shown. The results are based on three replicates; error bars indicate standard deviations. The shaded region represents the mean level of the wild type (WT) ± standard deviations.
CdgJ functions as a PDE in vitro and in vivo.
The putative protein product of cdgJ is 432 amino acids long and predicted to be cytoplasmic. CdgJ has a substitution at the active site of the EAL domain and has ELL residues. A known PDE in E. coli, YhjH, also has ELL residues at the active site (41). In addition, CdgJ possesses 13 out of 14 conserved polar residues at the EAL domain, and the only nonconserved residue (L116) appears to play a minor or nonessential role in catalysis (44). Therefore, we hypothesized that CdgJ functions as a PDE.
To test this hypothesis, we overexpressed cdgJ under the control of an arabinose-inducible promoter (pBAD::cdgJ) in wild-type V. cholerae. PDE activity in crude extracts was assayed by a colorimetric assay involving the PDE-mediated cleavage of the synthetic substrate, bis-pNPP (10). We showed that bis-pNPP is cleaved in crude extract containing CdgJ protein, and cleavage occurs in a concentration-dependent manner (Fig. 2A). We also observed background PDE activity in the wild type with empty vector. However, PDE activity observed in the crude extract containing overproduced CdgJ was significantly higher, at all concentrations tested, than PDE activity in the crude extract of cells with empty vector (Fig. 2A), suggesting that CdgJ functions as a PDE. To test whether the PDE activity is dependent on a conserved EAL domain, we generated an overexpression plasmid harboring cdgJ with point mutations converting ELL residues to AAA. The crude extract from cells carrying pBAD::cdgJ-AAA exhibited the same background activity as that observed with empty vector (Fig. 2A), indicating that the ELL residues are required for CdgJ function.
FIG. 2.
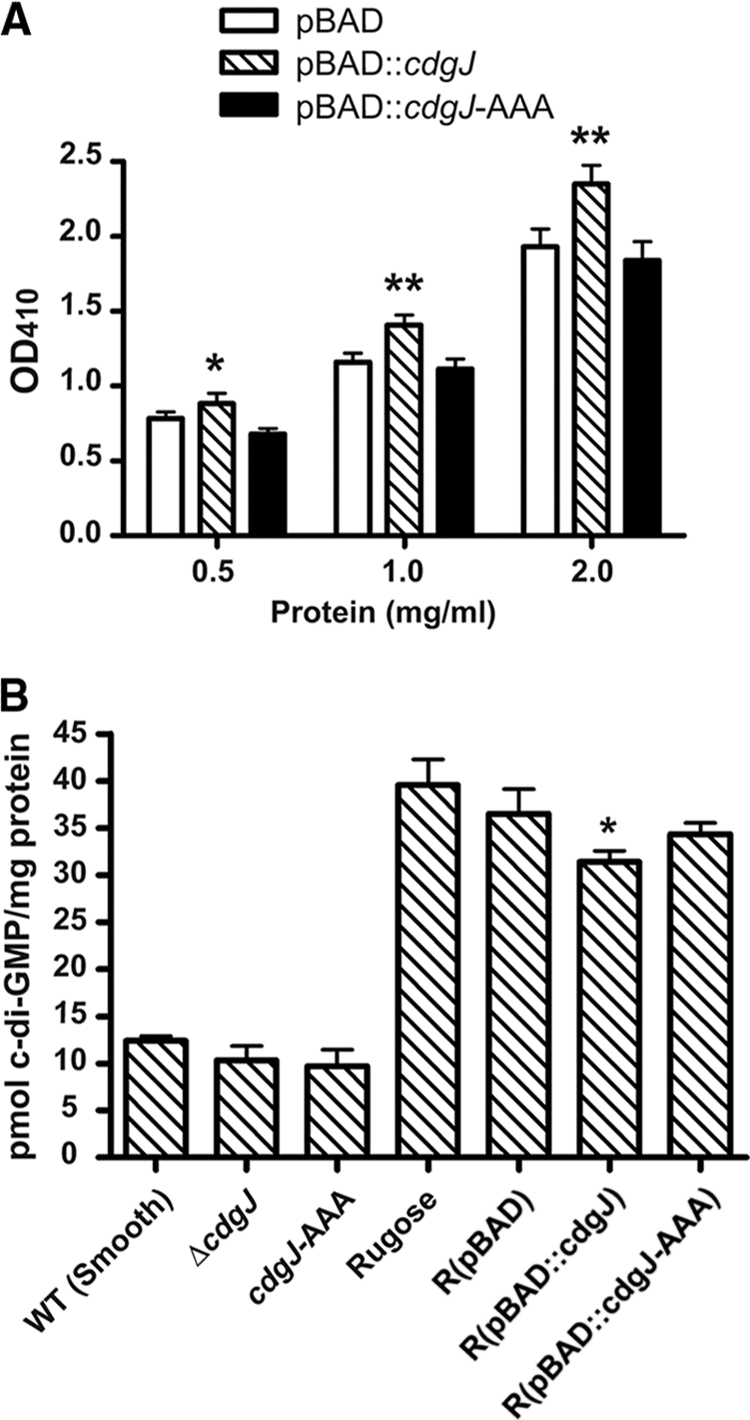
Analysis of PDE activity of CdgJ in vitro and in vivo. (A) Assessment of PDE activity in the crude cell extracts compared to that of control extract, prepared from wild-type V. cholerae carrying the vector only, using the substrate bis-pNPP. The graph shows the dose-response curve generated by 2-fold serial dilutions of crude extract incubated in the presence of bis-pNPP for 10 min at 37°C. The release of p-nitrophenol was quantified at OD410. One asterisk indicates P < 0.05 (statistical significance in two-tailed t test). Two asterisks indicate P < 0.001. The experiments were performed twice, and three biological replicates and two technical replicates were included in each experiment. (B) C-di-GMP levels in V. cholerae strains. C-di-GMP was extracted from whole cells, and levels were measured using HPLC/MS-MS. Data represent averages from at least three replicates; error bars indicate standard deviations. The asterisk indicates P < 0.05.
To confirm that CdgJ functions as a PDE, we also measured cellular c-di-GMP concentrations in wild-type, cdgJ deletion, and overexpression strains using HPLC/MS-MS. The c-di-GMP concentrations in ΔcdgJ and cdgJ-AAA were not significantly different from that of the wild type (Fig. 2B). When CdgJ was overexpressed in a strain with high c-di-GMP background (rugose), the cellular c-di-GMP level was significantly decreased compared to that of a rugose strain carrying vector only. In contrast, the overexpression of cdgJ-AAA does not alter the c-di-GMP level. These data suggest that CdgJ functions as a PDE in vivo. We did not observe a difference in c-di-GMP levels between the wild type (smooth variant) and ΔcdgJ, suggesting either that our method is not sensitive enough to detect the small c-di-GMP concentration differences or that a lack of cdgJ does not significantly affect global c-di-GMP levels.
Proteins encoded by the cdgJ operon positively regulate cell motility.
cdgJ is located in a putative operon with a second gene, VC0136, which is immediately upstream of and overlaps the cdgJ open reading frame (ORF) by 53 nucleotides (Fig. 3A). Database analyses predict that the protein encoded by VC0136 has six transmembrane regions and belongs to the RhtB family of efflux proteins (1). We determined that VC0136 and cdgJ constitute an operon by RT-PCR. The RT-PCRs produced the expected 1,021-bp product (confirmed by sequencing) in the sample and positive control (Fig. 3B). No products were amplified from the no-reverse-transcriptase control (−RT) and no-template control (NTC). These results indicate that VC0136 and cdgJ are transcribed as a single operon.
FIG. 3.
CdgJ regulates motility in V. cholerae. (A) The genomic context of cdgJ. The cdgJ gene (VC0137) is located downstream of VC0136. The two genes overlap and comprise an operon. Individual genes are indicated by block arrows, and the locations of primers used in RT-PCR are indicated by thin arrows. (B) RT-PCR analysis of the VC0136-VC0137 locus shows transcriptional coupling. RT, reverse transcriptase; NTC, no-template control. (C) cdgJ transcription in WT and ΔVC0136 strains measured with qRT-PCR analysis. Two-tailed t test assuming unequal variance reports a P value of 0.258 (no significant difference). (D) Motility phenotypes of WT (I), ΔVC0136 (II), ΔcdgJ (III), cdgJ-AAA (IV), ΔVC0136-7 (V), and ΔflaA (VI) strains. On the left is a picture of a representative motility assay. On the right, the bar graph shows the mean diameters of migration zones for each strain and standard deviations for three replicates. The motility between the WT and all of the mutants was significantly different based on a two-tailed t test (P < 0.01). (E) Motility phenotypes of WT (vector) (I), ΔVC0136 (vector) (II), ΔVC0136 (pVC0136) (III), ΔcdgJ (vector) (IV), ΔcdgJ (pcdgJ) (V), ΔVC0136-7 (vector) (VI), and ΔVC0136-7 (pVC0136-7) (VII) strains. The motility between the complemented strains and the corresponding vector-only controls was significantly different based on a two-tailed t test (P < 0.01).
Since VC0136 and cdgJ are cotranscribed, we hypothesized that the product of VC0136 played a role in motility. To test this hypothesis, we generated an in-frame deletion mutant of VC0136 without interrupting cdgJ. We further verified that the transcript level of cdgJ did not change in the strain ΔVC0136 by qRT-PCR analysis (Fig. 3C). We also constructed a mutant with the operon deletion (ΔVC0136-7) and a mutant harboring point mutations converting ELL residues to AAA at the chromosomal locus (cdgJ-AAA). All mutants were tested for motility on LB motility plates. The VC0136 mutant exhibited a 20% decrease in migration compared to that of the wild type (Fig. 3D). The operon deletion mutant and cdgJ-AAA mutant have phenotypes similar to that of the cdgJ mutant: all of them have about 60% decreases in migration compared to that of the wild type. All of these mutants had a larger migration zone than the nonmotile flaA mutant (Fig. 3D), indicating that they are still motile. These findings suggest that the protein products of VC0136 and cdgJ both positively regulate motility, that the ELL residues are required for CdgJ function, and that the cdgJ mutant phenotype is more dominant than that of VC0136.
To further confirm that the motility defects of the mutants are due to a lack of VC0136 and/or cdgJ genes, we generated three complementation constructs (pVC0136, pcdgJ, and pVC0136-7) and expressed VC0136, cdgJ, and the VC0136-7 operon from the native promoter on the pACYC177 plasmid. The introduction of pVC0136 to ΔVC0136 in trans restored motility to the wild-type level (Fig. 3E). pVC0136 was unable to complement ΔcdgJ or ΔVC0136-7 (data not shown). In contrast, pcdgJ was able to complement ΔVC0136, ΔcdgJ, cdgJ-AAA, and ΔVC0136-7 (Fig. 3E and data not shown), suggesting that CdgJ function is not dependent on the VC0136 gene product. Furthermore, the strains harboring pcdgJ or pVC0136-7 were more motile than the wild type (Fig. 3E), and this finding is consistent with our data showing that CdgJ functions as a PDE, since the increased cdgJ gene copy number is expected to result in a decrease in c-di-GMP concentration and, thus, increased motility.
Mutations in cdgJ operon affect flaA expression but do not affect flagellar morphology.
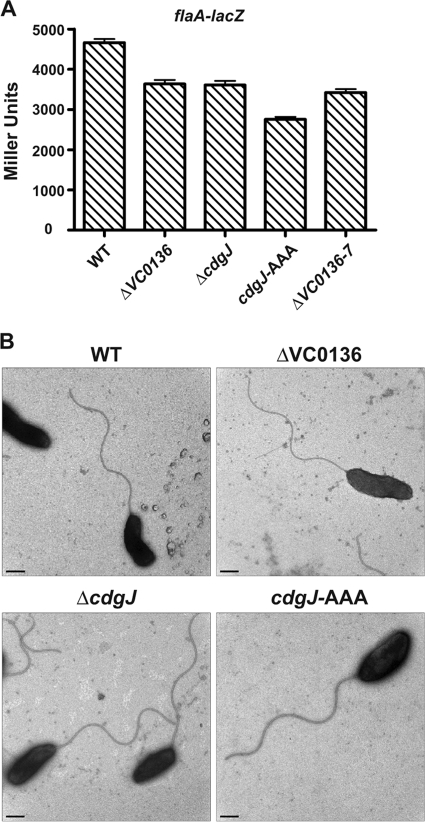
Because the VC0136 and cdgJ mutants exhibited motility defects, we wondered if mutations in the cdgJ operon affect flagellar gene expression in V. cholerae. The core flagellin of V. cholerae is encoded by flaA, which belongs to the class III genes in the flagellar gene transcriptional hierarchy (16, 28). We analyzed the transcription of flaA in the wild type and mutants discussed above using a flaA-lacZ transcriptional fusion. We found that ΔVC0136, ΔcdgJ, cdgJ-AAA, and ΔVC0136-7 mutants exhibited a slight decrease in the transcription of flaA compared to that of the wild type at both exponential phase (Fig. 4A) and stationary phase (data not shown). We also analyzed flagellum production using TEM. There was no significant difference in the presence and the length of flagella between the mutant and wild-type strains (Fig. 4B). These results indicate that a slight decrease in flaA expression does not affect flagellar morphology and cannot account for the dramatic motility defect of ΔcdgJ. Therefore, changes in flagellar function are likely to be contributing to the reduced motility of ΔcdgJ.
FIG. 4.
Mutations in cdgJ and VC0136 affect flaA expression but do not affect flagellar morphology. (A) CdgJ affects flaA expression. The expression of the flaA-lacZ fusion was determined in WT and mutant strains by measuring β-galactosidase activity in cells that were grown to mid-exponential phase in LB broth at 30°C. The results shown are from one representative experiment of at least three independent experiments. The error bars indicate standard deviations from eight technical replicates. The difference between the WT and all of the mutants was significantly different based on two-tailed t tests (P < 0.01). (B) Flagellation of V. cholerae wild-type and mutant strains. TEM was performed on at least 50 cells for each strain, and one representative electron micrograph is shown for each strain. Scale bars = 0.5 μm.
CdgJ negatively regulates biofilm formation.
Since motility and biofilm formation usually are inversely regulated via c-di-GMP regulatory networks in bacteria, we hypothesized that CdgJ negatively regulates biofilm formation. To test this hypothesis, we first compared the biofilm formation capabilities of wild-type, ΔVC0136, ΔcdgJ, cdgJ-AAA, and ΔVC0136-7 strains in static cultures using a crystal violet staining assay. At 2 h, the biofilm phenotypes were similar between the wild type and all mutants, suggesting that the mutations did not affect the initial attachment of cells to the substratum (Fig. 5A). At 6 h, all of the mutants significantly increased biofilm formation compared to that of the wild type, suggesting that protein products of VC0136 and cdgJ both inhibit biofilm development (Fig. 5A). Again, the cdgJ-AAA strain had the same biofilm phenotype as the ΔcdgJ strain, indicating that the ELL residues are necessary for CdgJ function. ΔcdgJ, cdgJ-AAA, and ΔVC0136-7 mutants produced significantly higher levels of biofilm than ΔVC0136.
FIG. 5.
CdgJ regulates biofilm formation in V. cholerae. (A) V. cholerae strains were grown in LB broth under static conditions at room temperature for 2 and 6 h. Biofilms were stained with crystal violet and quantified. The results shown are from one representative experiment of two independent experiments. The error bars indicate standard deviations from eight technical replicates. The biofilm data differed significantly between the WT and all of the mutants based on a two-tailed t test (P < 0.01). (B) Three-dimensional biofilm structures of the V. cholerae strains formed after 48 h postinoculation in a flow cell system. Images were acquired by CLSM, and top-down views (large panes) and orthogonal views (side panes) of biofilms are shown. The results shown are from one representative experiment of two independent experiments. Scale bars = 40 μm.
We also evaluated the roles of VC0136 and CdgJ in biofilm formation in a flow cell system using GFP-labeled strains and CLSM. There was no difference between biofilm phenotypes of the wild type and mutants in the initial attachment stage at 2 h postinoculation (data not shown). However, all of the mutants formed thicker and more structured biofilm than the wild type at 24 (data not shown) and 48 h (Fig. 5B). Quantitative analysis with COMSTAT revealed that the total biomass, average and maximum thickness, roughness, and surface-to-volume ratio of biofilms formed by ΔVC0136, ΔcdgJ, and ΔVC0136-7 mutants were higher than those of the wild type at 48 h (Table 3). These biofilm characteristics were more pronounced in the ΔcdgJ and ΔVC0136-7 strains than in the ΔVC0136 strain (Table 3). These results suggest that the protein products of VC0136 and cdgJ negatively regulate biofilm formation.
TABLE 3.
COMSTAT analysis of biofilmsa
| Strain | Biomass (um3/um2) | Thickness (um) |
Substratum coverage | Roughness | Surface area to vol (um2/um3) | |
|---|---|---|---|---|---|---|
| Avg | Max | |||||
| Wild type | 14.38 ± 0.76 | 16.26 ± 0.68 | 29.33 ± 1.02 | 0.96 ± 0.01 | 0.15 ± 0.01 | 0.96 ± 0.12 |
| ΔVC0136 | 14.93 ± 1.10 | 17.37 ± 0.82 | 34.32 ± 0.88 | 0.96 ± 0.03 | 0.20 ± 0.02 | 1.50 ± 0.25 |
| ΔcdgJ | 15.71 ± 1.68 | 18.37 ± 0.97 | 42.53 ± 5.08 | 0.99 ± 0.01 | 0.23 ± 0.01 | 1.72 ± 0.22 |
| ΔVC0136-7 | 17.45 ± 0.44 | 19.40 ± 0.24 | 44.88 ± 4.90 | 0.99 ± 0.01 | 0.23 ± 0.03 | 1.78 ± 0.24 |
All values are means ± standard deviations derived from at least three z-series image stacks.
CdgJ negatively regulates expression of vps genes.
Because of the increased biofilm formation capabilities observed in the VC0136 and cdgJ mutants, we hypothesized that vps gene expression is elevated in these mutants. We determined the transcription of vpsL, vpsR, and vpsT by measuring β-galactosidase activities of vpsL-lacZ, vpsR-lacZ, and vpsT-lacZ transcriptional fusions in wild-type, ΔVC0136, ΔcdgJ, cdgJ-AAA, and ΔVC0136-7 strains. In all mutants, the expression levels of all three vps genes were elevated compared to those of the wild type during both exponential (Fig. 6) and stationary phases (data not shown). This suggests that VC0136 and CdgJ negatively regulate vps gene expression. The expression levels of vpsL and vpsT (but not vpsR) in ΔcdgJ, cdgJ-AAA, and ΔVC0136-7 were higher than those observed in ΔVC136. This result is consistent with the intermediate biofilm phenotype of ΔVC0136.
FIG. 6.

CdgJ inhibits vps gene expression. Expression of vpsL-lacZ (A), vpsR-lacZ (B), and vpsT-lacZ (C) fusions were determined in WT and mutant strains by measuring β-galactosidase activity in cells that were grown to mid-exponential phase in LB broth with ampicillin at 30°C. The results shown are from one representative experiment of at least three independent experiments. The error bars indicate standard deviations from eight technical replicates. The difference between the WT and all of the mutants was significantly different based on two-tailed t tests (P < 0.01).
Genetic interaction analysis of CdgJ with c-di-GMP receptors.
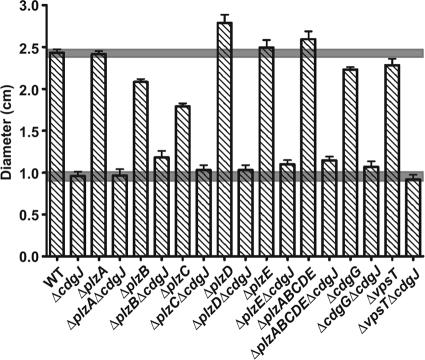
C-di-GMP receptors bind to c-di-GMP and transmit signals to downstream targets (22). Previous studies showed that PilZ domain proteins can bind to c-di-GMP (14, 35, 43, 47). The V. cholerae genome encodes five proteins with the PilZ domain: PlzA, PlzB, PlzC, PlzD, and PlzE (2). Of these, only PlzC and PlzD were shown to bind to c-di-GMP (43). The phenotypic analysis of strains lacking plzB, plzC, and plzD revealed that their protein products regulate biofilm formation, motility, and intestinal colonization in the V. cholerae O1 classical phenotype (43). We previously showed that none of the five Plz proteins are essential for rugosity (6). We constructed and analyzed motility phenotypes of all five single plz deletion mutants, as well as the quintuple mutant, in our prototype V. cholerae O1 El Tor strain. Consistently with the results by Pratt et al. (43), ΔplzB and ΔplzC showed a decrease in motility, while ΔplzD showed an increase in motility compared to that of the wild type in an LB motility plate assay (Fig. 7). The quintuple mutant ΔplzABCDE also was more motile than the wild type, suggesting that plzD is the most dominant gene among the five plz genes with respect to motility (Fig. 7). To determine if one or several of the Plz proteins functions in a signaling module with CdgJ, we constructed double mutants of cdgJ and each of the plz genes and compared their motility phenotypes to those of the single mutants. All of the double mutants had phenotypes similar to that of ΔcdgJ, except that ΔplzB ΔcdgJ and ΔplzE ΔcdgJ mutants had a slight increase in motility compared to that of ΔcdgJ (Fig. 7). In addition, ΔcdgJ ΔplzABCDE exhibited a slight increase in motility compared to that of ΔcdgJ (Fig. 7). These data suggest that plzB and/or plzE is involved in the same signaling module as cdgJ, and that the other plz genes are not involved in this signaling module.
FIG. 7.
Genetic interaction analysis of cdgJ and genes encoding c-di-GMP binding proteins. The diameters of migration zones of each strain were measured in LB motility plates after 16 h of incubation at 30°C. The results shown are from one representative experiment of at least two independent experiments. The error bars indicate standard deviations from three replicates. Based on a two-tailed t test comparing WT and ΔplzB, ΔplzC, ΔplzD, ΔplzABCDE, ΔcdgG, or ΔvpsT, P < 0.05. The motility between the WT and ΔplzA or ΔplzE strain was not significantly different (P > 0.05). P < 0.05 for comparisons of ΔcdgJ and ΔcdgJ ΔplzB, ΔcdgJ ΔplzE, or ΔcdgJ ΔplzABCDE. The motility between ΔcdgJ and other double mutants was not significantly different (P > 0.05). The shaded region represents the mean level of WT or ΔcdgJ ± standard deviations.
Our recent studies revealed that CdgG is likely to be an RXXD-type c-di-GMP binding protein (6) and that VpsT can bind to c-di-GMP (31). We constructed the double mutants of cdgJ with cdgG and vpsT and analyzed their motility phenotypes. ΔcdgJ ΔcdgG and ΔcdgJ ΔvpsT exhibited a motility phenotype similar to that of ΔcdgJ, suggesting that cdgG and vpsT are not involved in the cdgJ signaling module (Fig. 7).
Genetic interaction analysis of CdgJ with DGCs.
Mutations in cdgD, cdgH, cdgK, and cdgL resulted in an increase in motility (Fig. 1 and 8), and we wanted to evaluate the impacts of these mutations on CdgJ-controlled motility. To this end, we constructed single, double, triple, and quadruple mutants of the DGC genes in both wild-type and ΔcdgJ genetic backgrounds and analyzed the motility phenotypes of these mutants on LB motility plates. ΔcdgD ΔcdgH showed an increase in motility compared to that of the single mutants ΔcdgD and ΔcdgH. The triple mutant ΔcdgD ΔcdgH ΔcdgK and the quadruple mutant ΔcdgD ΔcdgH ΔcdgK ΔcdgL showed a further increase in motility (Fig. 8). The deletion of each of the four DGC genes in the ΔcdgJ background led to the partial restoration of the ΔcdgJ motility defect (Fig. 8). The accumulation of deletions of the four DGC genes in ΔcdgJ further increased the motility of ΔcdgJ, and the strain with all four DGC gene deletions restored the motility of the ΔcdgJ mutant to almost-wild-type levels (Fig. 8). These data suggest that CdgD, CdgH, CdgK, and CdgL function as DGCs and synergistically adjust the c-di-GMP pool for motility control. These four DGCs, together with the PDE CdgJ (and possibly other PDEs), set the concentration of c-di-GMP to control motility.
FIG. 8.
Genetic interaction analysis of cdgJ and DGCs. The diameters of migration zones of each strain were measured in LB motility plates after 16 h of incubation at 30°C. The results shown are from one representative experiment of at least three independent experiments. The error bars indicate standard deviations from three replicates. The motility between the WT and all of the single DGC mutants was significantly different (P < 0.05). The motility between the ΔcdgJ mutant and all of the Δdgc ΔcdgJ double mutants also was significantly different (P < 0.05). The shaded region represents the mean level of WT or ΔcdgJ ± standard deviations.
DISCUSSION
The second messenger, c-di-GMP, controls diverse cellular processes, including biofilm formation, motility, and virulence. The V. cholerae genome encodes 62 proteins with GGDEF, EAL, or HD-GYP domains, which are predicted to produce or degrade c-di-GMP (21). However, relatively little is known about what the players are and how they function mechanistically in each cellular process. In previous studies, nine of the genes encoding c-di-GMP-metabolizing enzymes were shown to have motility-associated phenotypes in V. cholerae (6, 7, 9, 33, 42). These studies have revealed that the c-di-GMP regulation of motility is complex. In the rugose genetic background (cells with high cellular c-di-GMP levels), the rocS, mbaA, cdgC, and cdgG mutants exhibit a decrease in motility (6, 33), while the vpvC, cdgD, and cdgH mutants exhibit an increase in motility (6, 9, 33). In the smooth genetic background, the rocS and cdgG mutants exhibit a decrease in motility (45), while the cdgD and cdgH mutants exhibit an increase in motility (6). Interestingly, the two colony morphotypes of V. cholerae differ in that the rugose variant has elevated intracellular c-di-GMP concentrations compared to that of the smooth variant (33 and this study). Therefore, the mbaA, cdgC, and vpvC mutants show their motility phenotype only in a strain with increased c-di-GMP levels (rugose variants). In addition, VieA (PDE) positively regulates motility in the V. cholerae classical biotype but not in the El Tor biotype (7). AcgA (PDE) positively regulates motility at low concentrations of inorganic phosphate (42).
In this study, our objective was to draft a blueprint of the c-di-GMP motility control module of V. cholerae. We determined that seven genes were involved in modulating motility in the V. cholerae O1 El Tor smooth variant under standard growth conditions (growth in LB medium at 30°C). Besides the already-identified genes (cdgD, cdgG, cdgH, and rocS [6, 33, 45]), we identified three additional genes, cdgJ, cdgK, and cdgL, involved in regulating motility. The cdgJ mutant exhibited decreased motility compared to that of the wild type and is predicted to be a PDE. The cdgK and cdgL mutants exhibited increased motility, and CdgK and CdgL are predicted to be DGCs. Other GGDEF and EAL domain proteins may (i) not have enzymatic activity; (ii) not be active under our experimental conditions; (iii) be involved in other cellular processes; or (iv) affect cell motility under different conditions or in different genetic backgrounds (e.g., MbaA, CdgC, VpvC, VieA, and AcgA) (5, 7, 33, 42).
We showed that CdgJ inversely regulates motility and biofilm formation and functions as a PDE. The deletion of cdgJ led to reduced motility, while the overexpression of cdgJ in trans led to increased motility. The LB soft-agar motility assay measures growth, motility, and chemotaxis. Since the cdgJ mutant exhibits a wild-type growth rate in LB, CdgJ likely regulates motility, chemotaxis, or both. C-di-GMP regulates motility at the level of transcription, posttranscription function, flagellar function, and flagellar ejection (reviewed in reference 60). We observed only a slight decrease in flaA expression in cdgJ mutants, and cdgJ mutants have a wild-type flagellar morphology. These data suggest that an increase in c-di-GMP concentration in cdgJ mutants interferes with flagellar function but not with the assembly of flagellar components. Two known c-di-GMP binding proteins, YcgR in E. coli and DgrA in Caulobacter crescentus, interfere with the motor function of fully assembled flagella by directly interacting with flagellar motor proteins (11, 14, 18, 29, 39, 47). The c-di-GMP binding partner of CdgJ could function in a similar way to modulate motility. We are in the process of determining whether the cdgJ mutant phenotype is due to reduced flagellar speed or altered flagellar rotation bias (i.e., clockwise and counter-clockwise rotation).
The operon structure of cdgJ and VC0136 is conserved among a number of Vibrio species, including V. angustum, V. alginolyticus, V. campbellii, V. cholerae, V. harveyi, V. parahaemolyticus, V. shilonii, V. splendidus, and V. vulnificus. Outside of the Vibrio genus, it is present in a couple of Photobacterium species, which also belong to the family Vibrionaceae. Interestingly, V. fischeri genomes do not contain VC0136, although cdgJ is present. A VC0136 mutant also had decreased motility and increased biofilm formation compared to those of the wild type. However, both motility and biofilm phenotypes were less severe than those of the cdgJ mutant. In addition, the double mutant (ΔVC0136-7) had the same phenotype as the cdgJ mutant, suggesting that the cdgJ phenotype is dominant over that of VC0136. VC0136 is predicted to encode a membrane-bound protein belonging to the RhtB family (1), which functions as an exporter of homoserine, homoserine lactone, and some other amino acids (63). Its conservation in operon structure with cdgJ suggests that the two genes function in the same cellular process, which is supported by our data.
Certain environmental signals can affect bacterial physiology via c-di-GMP signaling systems. These include oxygen, blue light, nutrient starvation, antibiotics, bile salts, intercellular signaling molecules, and mucin (reviewed in reference 25). In V. cholerae, polyamine norspermidine (intercellular signaling molecule) enhances biofilm formation via an NspS/MbaA signaling system (27). NspS, a predicted periplasmic sensor for norspermidine (27), is hypothesized to form a complex with a membrane-bound protein, MbaA, a putative c-di-GMP phosphodiesterase, and alter its activity. It will be of interest to determine whether the activity of CdgJ is regulated under certain environmental stress or cellular conditions and whether the protein encoded by VC0136 has any role in this process.
It has been shown that the disruption of a single c-di-GMP-metabolizing enzyme can alter the intracellular c-di-GMP concentration. For example, the deletion of vpvC (DGC) in the V. cholerae rugose variant reduces the intracellular c-di-GMP concentration; in addition, the deletion of binA in V. fischeri increases the intracellular c-di-GMP concentration (4, 9). Our HPLC/MS-MS analysis revealed that the overexpression of cdgJ reduced the cellular concentration of c-di-GMP, indicating that CdgJ functions as a PDE. In addition, the results of the in vitro enzyme assay also suggested that CdgJ functions as a PDE. However, we detected no significant difference between the cellular concentration of c-di-GMP of the cdgJ mutant and that of the wild type. It is possible that the disruption of cdgJ alters levels of a localized c-di-GMP pool without affecting global levels of c-di-GMP. Alternatively, the changes in the c-di-GMP pool were too small to be detected by the methods we used. Previous studies of other bacteria suggest microcompartmentalized c-di-GMP activity. In C. crescentus, PleD was shown to localize to the stalked-cell pole in a phosphorylation-dependent manner, and its DGC activity also is stimulated by phosphorylation (40). If this is also the case in V. cholerae, some DGCs and PDEs may modulate cell processes by fine-tuning c-di-GMP concentrations at small, localized pools without much disturbance in the global concentration of c-di-GMP. It is possible that CdgJ acts in a localized control module containing DGC protein(s) and c-di-GMP receptor(s). CdgJ would act to eliminate the c-di-GMP signal to control the homeostasis of the cellular process. Of the 52 mutants studied in this work, we identified 4 potential GGDEF-containing proteins (CdgD, CdgH, CdgK, and CdgL) that regulated motility in a manner that is the opposite of that of CdgJ. Genetic interaction analysis revealed that CdgD, CdgH, CdgK, and CdgL function as DGCs and work synergistically to regulate motility. These four DGCs, together with PDE CdgJ (and possibly other PDEs, such as RocS), form a network to set the concentration of c-di-GMP for motility control.
C-di-GMP receptors are considered to be central players in the c-di-GMP network. They bind to c-di-GMP and convey the signal to downstream targets. In E. coli, the inactivation of YcgR, which binds to c-di-GMP via its PilZ domain, did not affect motility (47), while disruptions of yhjH, which encodes a PDE, led to decreased motility (29, 47). However, the disruption of ycgR in the yhjH background greatly improved the motility of the yhjH mutant, indicating that YcgR functions downstream of YhjH (47). Recent studies reported that the c-di-GMP-YcgR complex interacts with flagellar switch complex proteins FliG and FliM and motor protein MotA, affecting rotation bias and swimming speed (11, 18, 39). V. cholerae has seven known or putative c-di-GMP binding proteins: PlzA, PlzB, PlzC, PlzD, PlzE, CdgG, and VpsT (6, 31, 43). We sought to determine which c-di-GMP receptor(s) belonged to the c-di-GMP motility control module with CdgJ. Of the ones tested, only mutations in plzB and plzE partially rescued the ΔcdgJ motility phenotype (Fig. 7). This suggests that, in addition to PlzB and PlzE, other c-di-GMP receptor(s) are involved in the c-di-GMP motility control module.
C-di-GMP-regulated cellular processes play important roles in V. cholerae's ability to cause epidemics. VieA, a PDE that positively regulates motility and virulence genes, is upregulated during acute infection and is required for virulence in the infant mouse model of V. cholerae colonization (57). During the late stage of infection, V. cholerae detaches from the epithelial surface and becomes motile (37). Motility and chemotaxis genes are upregulated in the rabbit ileal loop during detachment, a process termed mucosal escape response (37). It also has been shown that some of the genes encoding c-di-GMP-metabolizing enzymes are induced during the late infection stage and help bacteria to become motile and prepare for the aquatic life cycle (42, 49, 55). A better understanding of the molecular mechanisms involved in c-di-GMP-regulated motility therefore is critical for understanding the pathogenesis and environmental survival of V. cholerae. This work constitutes a blueprint for the identification of the c-di-GMP motility control module in V. cholerae and provides a foundation for understanding the molecular mechanisms of c-di-GMP-regulated motility.
Acknowledgments
This research was supported by NIH grant AI055987. C-di-GMP quantification was done at the UCSC Mass Spectrometry Facility, which is funded by NIH grant S10-RR20939 (MS equipment grant).
We thank Qiangli Zhang for help in HPLC/MS-MS analysis, Yu-Chen Hwang (UCSC Life Sciences Microscopy Center) for technical support with TEM, and Karen Ottemann, Holger Sondermann, and members of the Yildiz laboratory for their suggestions on the manuscript.
Footnotes
Published ahead of print on 9 July 2010.
REFERENCES
- 1.Aleshin, V. V., N. P. Zakataeva, and V. A. Livshits. 1999. A new family of amino-acid-efflux proteins. Trends Biochem. Sci. 24:133-135. [DOI] [PubMed] [Google Scholar]
- 2.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3-6. [DOI] [PubMed] [Google Scholar]
- 3.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 4.Bassis, C. M., and K. L. Visick. 2010. The cyclic-di-GMP phosphodiesterase BinA negatively regulates cellulose-containing biofilms in Vibrio fischeri. J. Bacteriol. 192:1269-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyhan, S., K. Bilecen, S. R. Salama, C. Casper-Lindley, and F. H. Yildiz. 2007. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189:388-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyhan, S., L. S. Odell, and F. H. Yildiz. 2008. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J. Bacteriol. 190:7392-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Differences in Gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect. Immun. 74:3633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beyhan, S., and F. H. Yildiz. 2007. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 63:995-1007. [DOI] [PubMed] [Google Scholar]
- 10.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123-130. [DOI] [PubMed] [Google Scholar]
- 11.Boehm, A., M. Kaiser, H. Li, C. Spangler, C. A. Kasper, M. Ackermann, V. Kaever, V. Sourjik, V. Roth, and U. Jenal. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107-116. [DOI] [PubMed] [Google Scholar]
- 12.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christen, M., B. Christen, M. G. Allan, M. Folcher, P. Jeno, S. Grzesiek, and U. Jenal. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 104:4112-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 16.Correa, N. E., and K. E. Klose. 2005. Characterization of enhancer binding by the Vibrio cholerae flagellar regulatory protein FlrC. J. Bacteriol. 187:3158-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 18.Fang, X., and M. Gomelsky. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76:1295-1305. [DOI] [PubMed] [Google Scholar]
- 19.Fong, J. C., K. Karplus, G. K. Schoolnik, and F. H. Yildiz. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bacteriol. 188:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 67:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 22.Hengge, R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263-273. [DOI] [PubMed] [Google Scholar]
- 23.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 24.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 25.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385-407. [DOI] [PubMed] [Google Scholar]
- 26.Jude, B. A., and R. K. Taylor. 2008. Genetics of Vibrio cholerae colonization and motility, p. 67-99. In S. M. Faruque and G. B. Nair (ed.), Vibrio cholerae: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 27.Karatan, E., T. R. Duncan, and P. I. Watnick. 2005. NspS, a predicted polyamine pensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 187:7434-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko, M., and C. Park. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 303:371-382. [DOI] [PubMed] [Google Scholar]
- 30.Kovacikova, G., W. Lin, and K. Skorupski. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol. Microbiol. 57:420-433. [DOI] [PubMed] [Google Scholar]
- 31.Krasteva, P. V., J. C. Fong, N. J. Shikuma, S. Beyhan, M. V. Navarro, F. H. Yildiz, and H. Sondermann. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefebvre, B., P. Formstecher, and P. Lefebvre. 1995. Improvement of the gene splicing overlap (SOE) method. Biotechniques 19:186-188. [PubMed] [Google Scholar]
- 33.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331-348. [DOI] [PubMed] [Google Scholar]
- 34.Lim, B., S. Beyhan, and F. H. Yildiz. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876-895. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Assay of β-galactosidase. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 37.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 39.Paul, K., V. Nieto, W. C. Carlquist, D. F. Blair, and R. M. Harshey. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38:128-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pesavento, C., G. Becker, N. Sommerfeldt, A. Possling, N. Tschowri, A. Mehlis, and R. Hengge. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pratt, J. T., E. McDonough, and A. Camilli. 2009. PhoB regulates motility, biofilms, and cyclic di-GMP in Vibrio cholerae. J. Bacteriol. 191:6632-6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282:12860-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao, F., Y. Yang, Y. Qi, and Z. X. Liang. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190:3622-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rashid, M. H., C. Rajanna, A. Ali, and D. K. Karaolis. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227:113-119. [DOI] [PubMed] [Google Scholar]
- 46.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 103:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Ryjenkov, D. A., R. Simm, U. Romling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310-30314. [DOI] [PubMed] [Google Scholar]
- 48.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schild, S., R. Tamayo, E. J. Nelson, F. Qadri, S. B. Calderwood, and A. Camilli. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shikuma, N. J., J. C. Fong, L. S. Odell, B. S. Perchuk, M. T. Laub, and F. H. Yildiz. 2009. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J. Bacteriol. 191:5147-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 53.Sudarsan, N., E. R. Lee, Z. Weinberg, R. H. Moy, J. N. Kim, K. H. Link, and R. R. Breaker. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamayo, R., J. T. Pratt, and A. Camilli. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamayo, R., S. Schild, J. T. Pratt, and A. Camilli. 2008. Role of cyclic di-GMP during El Tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic di-GMP phosphodiesterase CdpA. Infect. Immun. 76:1617-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waters, C. M., W. Lu, J. D. Rabinowitz, and B. L. Bassler. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. U. S. A. 86:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolfe, A. J., and K. L. Visick. 2008. Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 190:463-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 62.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zakataeva, N. P., V. V. Aleshin, I. L. Tokmakova, P. V. Troshin, and V. A. Livshits. 1999. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 452:228-232. [DOI] [PubMed] [Google Scholar]