Abstract
A Gram-positive polychlorinated-biphenyl (PCB) degrader, Rhodococcus jostii RHA1, degrades PCBs by cometabolism with biphenyl. A two-component BphS1T1 system encoded by bphS1 and bphT1 (formerly bphS and bphT) is responsible for the transcription induction of the five gene clusters, bphAaAbAcAdC1B1, etbAa1Ab1CbphD1, etbAa2Ab2AcD2, etbAdbphB2, and etbD1, which constitute multiple enzyme systems for biphenyl/PCB degradation. The bphS2 and bphT2 genes, which encode BphS2 and BphT2, virtually identical to BphS1 (92%) and BphT1 (97%), respectively, were characterized. BphS2T2 induced the activation of the bphAa promoter in a host, Rhodococcus erythropolis IAM1399, in the presence of a variety of aromatics, including benzene, toluene, ethylbenzene, xylenes, isopropylbenzene, and chlorinated benzenes, as effectively as BphS1T1. The substrate spectrum of BphS2T2 was the same as that of BphS1T1, except for biphenyl, which is a substrate only for BphS1T1. BphS2T2 activated transcription from the five promoters of biphenyl/PCB degradation enzyme gene clusters as effectively as BphS1T1. The targeted disruptions of the bphS1, bphS2, bphT1, and bphT2 genes indicated that all these genes are involved in the growth of RHA1 on aromatic compounds. The hybrid system with bphS1 and bphT2 and that with bphS2 and bphT1 were constructed, and both systems conducted induced activation of the bphAa promoter, indicating cross-communication. These results indicated that RHA1 employs not only multiple enzyme systems, but also dual regulatory systems for biphenyl/PCB degradation. Comparison of the sequences, including bphS2T2, with the bphS1T1-containing sequences and the corresponding sequences in other rhodococcal degraders suggests that bphS2T2 might have originated from bphS1T1.
Polychlorinated biphenyls (PCBs) have been widely used for industrial purposes because of their excellent stability, insulation properties, and resistance to combustion. In recent years, however, the use and synthesis of PCBs has been prohibited because of their toxicity and their contamination of and recalcitrance in the environment. Nonetheless, PCBs are still widely present as environmental contaminants and continue to cause serious problems throughout the world. Degradation of PCBs by microorganisms has been regarded as a promising procedure for their removal from the environment. Many kinds of bacteria that can aerobically degrade PCBs have been isolated, and their degradation pathway enzymes and genes have been characterized (1, 6, 15, 23). Most of these bacteria cometabolize PCBs through the metabolic pathway of biphenyl.
A PCB degrader, Rhodococcus jostii RHA1, was isolated from γ-hexachlorocyclohexane-contaminated upland soil and has a great capacity to degrade mono- to octachlorobiphenyls by cometabolism with biphenyl and ethylbenzene (32, 33). In the strain RHA1, multiple isozymes are often involved in each degradation step of the upper biphenyl pathway. Biphenyl is transformed to 2-hydroxypenta-2,4-dienoate (HPD) and benzoate through the sequential reactions of a multicomponent biphenyl dioxygenase encoded by bphAaAbAcAd and etbAaAbAcAd (12, 13), a dihydrodiol dehydrogenase encoded by bphB1 and bphB2 (30), a 2,3-dihydroxybiphenyl dioxygenase encoded by bphC1 and etbC (7, 11, 29), and a 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase encoded by bphD1 (7, 10, 46). The resulting HPD is metabolized to pyruvate and acetyl-coenzyme A (CoA) through the lower biphenyl pathway (31), and benzoate is metabolized to succinate and acetyl-CoA through the benzoate pathway (16).
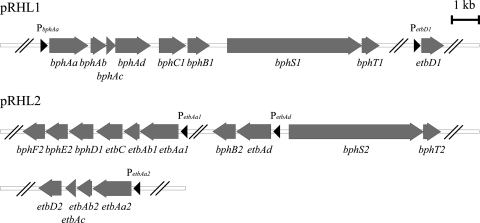
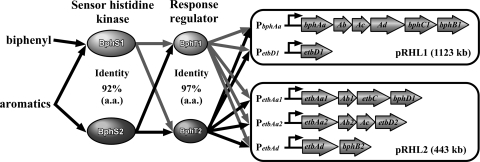
These degradation enzyme genes of the upper biphenyl pathway are carried in two linear plasmids, pRHL1 (1,123 kb) and pRHL2 (443 kb), of RHA1, which has a linear chromosome (7.80 Mb), and by three large linear plasmids comprising a genome with a total size of 9.70 Mb (22). They are distributed as gene clusters of bphAaAbAcAdC1B1 and etbD1 in pRHL1 and gene clusters of etbAa1Ab1CbphD1, etbAa2Ab2AcD2, and etbAdbphB2 in pRHL2 (Fig. 1). The genes etbD1 and etbD2 encode 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase, which has little activity toward 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate, a meta-cleavage intermediate product of biphenyl (10, 46), and the genes etbAa1Ab1 and etbAa2Ab2 are duplicated genes with the same amino acid sequences. The expression of these genes is simultaneously induced from the promoters of bphAa, etbAa1, etbAa2, etbAd, and etbD1 (bphAap, etbAa1p, etbAa2p, etbAdp, and etbD1p, respectively) in the presence of aromatic compounds, including biphenyl and ethylbenzene (37, 38, 46). All these promoters are under the control of a two-component system encoded by bphS and bphT, which encode the sensor histidine kinase and response regulator, respectively, since the heterologous expression of bphST in Rhodococcus erythropolis IAM1399 has been shown to promote transcriptional activation of bphAap, etbAa1p, etbAa2p, etbAdp, and etbD1p connected to the luxAB luciferase reporter genes in the presence of biphenyl (37, 38). Disruption of the bphS gene in RHA1 resulted in growth deficiency on biphenyl, suggesting that the bphS-dependent two-component system is solely responsible for the induction of biphenyl degradation gene promoters by biphenyl.
FIG. 1.
Genetic organization of the biphenyl degradation gene clusters in RHA1 under the control of gene products of bphS1 and bphT1 (BphS1T1), which were previously reported as the bphS and bphT genes, respectively. All the gene clusters are located on either the linear plasmid pRHL1 or pRHL2, as illustrated. The BphS1T1-inducible promoters are indicated by black triangles. Double slashes indicate a space separating two independent gene clusters.
In this study, bphS and bphT homologs, whose presence in RHA1 had previously been suggested by Southern hybridization of a bphS mutant strain using a bphS probe (38), were isolated and characterized. The results revealed the duplicated regulatory systems governing biphenyl degradation genes in the strain RHA1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. R. jostii RHA1 was grown at 30°C in LB (Bacto tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 5 g/liter), 1/5 LB (Bacto tryptone, 2 g/liter; yeast extract, 1 g/liter; NaCl, 5 g/liter), or W minimal medium (32) containing one of the following carbon sources: biphenyl (1.54 or 2 g/liter); sodium benzoate (2 g/liter); sodium succinate (2 g/liter); sodium terephthalate (10 mM); ethylbenzene; toluene; benzene; isopropylbenzene; and o-, m-, and p-xylenes. Volatile compounds were supplied as vapor. To determine growth characteristics, cells precultured in 1/5 LB for 2 days were inoculated in 10 ml W minimal medium at an optical density at 600 nm (OD600) of 0.2 and incubated at 30°C with shaking for 5 days after the addition of each substrate. If necessary, cells were precultured in 1/5 LB in the presence of an appropriate antibiotic(s), including kanamycin (50 μg/ml), chloramphenicol (20 μg/ml), and thiostreptone (20 μg/ml). The host strains, R. erythropolis IAM1399 (ATCC 15963) and Escherichia coli JM109, were grown in LB.
DNA manipulations and analysis.
All of the DNA techniques used, including gene cloning, nucleotide sequencing, and electrotransformation (electroporation), have been described previously (20, 21). Southern hybridization and colony hybridization were performed using the DIG system (Boehringer Mannheim Biochemicals, Indianapolis, IN) according to the manufacture's protocol. Sequence analysis was performed using the GeneWorks (Intelligenetics, Inc., MountainView, CA), BioEdit (9), and in silico MolecularCloning (in silico Biology, Inc., Yokohama, Japan) programs. BLAST (2) was used for homology searches. Sequence alignments were done with ClustalW (39). Genome alignments were done with GenomeMatcher (http://www.ige.tohoku.ac.jp/joho/gmProject/gmdownload.html) (25).
Deletion analysis of bphS2T2.
Deletion of pUSH15 was carried out using a Deletion Kit for Kilo-sequence (Takara Bio Inc., Otsu, Japan) as described in the manufacturer's protocol. The deleted fragments were made blunt ended and cloned into the blunt-ended KpnI site of pKLAF1. The transcription of the genes in a deleted fragment was expected to be promoted by the promoter of an upstream kanamycin resistance gene in pKLAF1.
Luciferase assay.
Cells of a recombinant strain were grown in LB containing kanamycin (50 μg/ml), washed with 50 mM sodium phosphate buffer (pH 7.0), and suspended in 10 ml of 1/5 LB containing kanamycin (50 μg/ml) at an OD600 of 1.0. Each cell suspension was incubated at 30°C for 5 h in the absence or presence of an inducer compound. Solid compounds were supplied as powder at a final concentration of 0.2% (wt/vol), and volatile compounds were supplied in vapor. After the final OD600 was measured, the cell suspension was diluted 1:10 in lux buffer (sodium phosphate, 50 mM, pH 7.0; 2-mercaptoethanol, 50 mM; bovine serum albumin, 1% [wt/vol]), and a 100-μl aliquot was mixed with 390 μl of lux buffer. Then, 10 μl of 0.1% (vol/vol) 1-decanal in lux buffer was added to the resulting 490 μl of diluted cell suspension, and the luciferase activity was measured using a luminometer, Lumitester K-100 (Kikkoman, Noda, Japan). The total light generated during the initial 15 s was recorded, and the activity (relative light units [RLU]/OD600) was expressed as RLU per milliliter per final OD600 unit.
RT-PCR.
RT-PCR was performed using ReverTra Dash (Toyobo Co., Ltd., Osaka, Japan) as described in the manufacturer's protocol. One or two micrograms of total RNA was reverse transcribed with random primers and PCR amplified using bphS2_intF and bphS2_intR (see Table S1 in the supplemental material) for 30 cycles at the annealing temperature of 55°C. RNA samples were analyzed concurrently in PCR mixtures without reverse transcriptase to verify the absence of contaminating genomic DNA. PCR mixtures were analyzed by electrophoresis using a 2% agarose gel.
Gene disruption and complementation.
A disruption mutant of bphS2, SDR2, was obtained, together with SDR1, in an experiment in bphS1 gene disruption using pUKSD (38). A double mutant of bphS1 and bphS2, SDR21, was constructed from SDR2 using pBCSD. Insertions of pBCSD and pUKSD into the bphS1 and bphS2 genes, respectively, were confirmed by Southern hybridization analysis. To perform bphS2 gene complementation in SDR21, pKTS2 was introduced into SDR21 by electroporation, and a transformant was isolated on an LB agar plate containing thiostreptone (20 μg/ml). pK4Ts was used as a control plasmid. The growth characteristics of SDR21/pKTS2 and SDR21/pK4Ts were examined as described above.
The bphT1 and bphT2 genes of RHA1 were deleted using unmarked gene deletion mutagenesis as described previously (42). To make a deletion of bphT1, the 1.6-kb EcoRI-BamHI and 1.5-kb XhoI fragments containing the N-terminal and C-terminal sequences of bphT1, respectively, were inserted in tandem into pK18mobsacB. The 1.5-kb XhoI fragment was inserted into the SalI site of pK18mobsacB. The resulting plasmid, pK18mobT1, was introduced into RHA1, and the deletion mutant strain of bphT1 obtained was designated TDR1. To make a deletion of bphT2, the 1.2-kb SphI-BamHI and 1.9-kb SmaI-PvuII fragments containing the N-terminal and C-terminal sequences of bphT2, respectively, were inserted in tandem into pK18mobsacB. The 1.9-kb SmaI-PvuII fragment was inserted into the SmaI site of pK18mobsacB. The resulting plasmid, designated pK18mobT2, was introduced into RHA1, and the deletion mutant strain of bphT2 obtained was designated TDR2. The double-deletion mutant strain of bphT1 and bphT2 was constructed from TDR1 using pK18mobT2 and designated TDR12. To perform gene complementation in TDR12, pKPA1TKE containing bphT1 or pKPA1T2KE containing bphT2 was introduced into TDR12 by electroporation. pKPA1 was used as a control plasmid. Each transformant was isolated on a 1/5 LB agar plate containing kanamycin (50 μg/ml) and subjected to growth characteristic determination as described above.
Construction of hybrid systems.
Hybrid systems between heterologous bphS and bphT were constructed as follows. (i) The 4.8-kb ApaI fragment carrying bphS1 and the 1.4-kb ApaI-PstI fragment containing bphT2 were inserted into pBSL to obtain pBS1T2. The 6.2-kb SpeI fragment of pBS1T2 carrying bphS1 and bphT2 was cloned into pKLAF1Spe to obtain pKLAS1T2. (ii) The 3.0-kb SmaI fragment of pBAL73R carrying the N-terminal sequence of bphS2 and the 4.2-kb BglII fragment containing the C-terminal sequence of bphS1 and the entire bphT1 gene were cloned into the SmaI and BamHI sites of pU18EH, respectively, to yield pUEHT1. The 2.4-kb ApaI fragment of pUEHT1 containing internal portions of bphS2 and bphS1 was replaced by the 4.6-kb ApaI fragment carrying the internal portion of bphS2 to obtain pUEHS2T1. Because the 89-bp C-terminal sequence of bphS1 remaining in pUEHS2T1 was completely identical to that of bphS2, pUEHS2T1 contained the entire bphS2 and bphT1 genes. The 7.3-kb HindIII fragment of pUEHS2T1 carrying bphS2 and bphT1 was blunt ended and cloned into the blunt-ended KpnI site of pKLAF1 to obtain pKLAS2T1.
Each resultant plasmid, pKLAS1T2 and pKLAS2T1, was introduced into an IAM1399 cell to perform the luciferase assay. The transcription of each hybrid system was expected to originate from the kanamycin resistance gene promoter of the vector.
Nucleotide sequence accession numbers.
The DNA sequences of the bphS2, bphT2, and ro10148 genes have been deposited in the NCBI and GenBank databases as part of the genome sequence of strain RHA1 (NC008270 in NCBI and CP000433 in GenBank).
RESULTS
Isolation of a bphST homolog.
The presence of a bphS homolog in the pRHL2 linear plasmid was suggested by Southern hybridization analysis of RHA1 and its plasmid deletion mutants, RCA1 and RCD1, with deletions of pRHL1 and pRHL2, respectively (data not shown), and this notion was confirmed by a sequence search in the RHA1 genome sequence database (http://www.rhodococcus.ca/index.jsp), which indicated that the second bphS is located in the vicinity of etbAd, encoding a reductase component of a biphenyl dioxygenase, and followed by the second bphT. The 14.1-kb HindIII fragment containing the bphST homolog was isolated, and the deduced amino acid sequences of the second bphST showed identity with those of bphS (92%) and bphT (97%) of RHA1 (38), akbS (100%) and akbT (100%) of Rhodococcus sp. strain DK17 (GenBank accession number AY502075) (14), and bpdS (54%) and bpdT (63%) of a PCB degrader, Rhodococcus sp. strain M5 (GenBank accession number, U85412) (17). Thus, the second bphS and bphT were designated bphS2 and bphT2, respectively, and the original bphS and bphT were renamed bphS1 and bphT1, respectively. In addition to these, another bphS homolog, ro10148, was found in pRHL2 of RHA1 by a sequence search in the RHA1 genome sequence database. The deduced amino acid sequence of ro10148 was not as close to those of bphS1 (65%) and bphS2 (64%) as the identity between bphS1 and bphS2 (92%). No additional bphT homologs were found in RHA1, and no response regulator genes were distributed in the vicinity of ro10148.
The N-terminal amino acid sequence of BphS2 contained a serine/threonine protein kinase catalytic motif. The C-terminal amino acid sequence contained H, N, D, and G boxes, which are highly conserved among sensor histidine kinases, and His-1411 in the H box was estimated to be the residue for self-phosphorylation (8, 35, 38, 45). BphT2 contained Asp-9, Asp-54, and Lys-104, which are highly conserved among response regulators (27, 36, 38). Thus, BphS2 and BphT2 appeared to constitute a functional two-component regulatory system, in addition to the BphS1T1 system.
Expression of bphS2T2 in R. erythropolis IAM1399.
The significant identity between bphS1T1 and bphS2T2 suggested that bphS2T2 was involved in the regulation of biphenyl degradation enzymes, in addition to bphS1T1. To examine the involvement of bphS2T2 in the regulation of transcription from a biphenyl degradation gene promoter, bphAap, the 14.1-kb HindIII fragment including bphS2T2 was inserted into a reporter plasmid, pKLAF1, that consisted of bphAap followed by the luxAB luciferase genes (38) (Fig. 2 A). The resultant plasmid, pKLAH15, was introduced into a rhodococcal host strain, R. erythropolis IAM1399, and the transformants were subjected to a luciferase assay (26). In the cells harboring a vector, pKLAF1, very little bphAap activity was observed (Fig. 2B). bphAap activity was elevated by the introduction of a 14.1-kb HindIII fragment into pKLAH15 and was further elevated in the presence of ethylbenzene. Such further elevation of bphAap activity was not observed in the presence of biphenyl (data not shown). These results suggested that bphS2T2 conducts the inducible activation of bphAap not by biphenyl, but by ethylbenzene. In the absence of ethylbenzene, the cells carrying pKLAH15 exhibited obviously higher luciferase activity than those carrying pKLAF1, suggesting that the basal activation of bphAap is promoted by bphS2T2 products.
FIG. 2.
Deletion analysis of the 14.1-kb HindIII fragment containing bphS2T2. (A) Physical maps of deletion derivatives. The segments represented by solid bars were inserted into pKLAF1, which is a reporter plasmid that does not contain any bphST segment. (B) Luciferase assay of the deletion derivatives. The IAM1399 cells harboring the plasmids were grown in 1/5 LB in the presence or absence of ethylbenzene and were subjected to the luciferase assay. The activity was expressed as RLU measured by a luminometer per milliliter of culture per OD600 unit, as described in Materials and Methods. The data are means ± standard deviations from at least three independent experiments.
To address the functions of bphS2T2 products, deletion analysis of the 14.1-kb HindIII fragment was performed (Fig. 2A). The deleted fragments were inserted into pKLAF1. The resulting plasmids were introduced into R. erythropolis IAM1399, and the transformants were subjected to a luciferase assay (Fig. 2B). IAM1399 with pKAP73 containing the entire bphS2T2 gene showed both the basal activation and the ethylbenzene-dependent inducible activation of bphAap, as was the case with pKLAH15. IAM1399 harboring pKMX1, which contained active bphT2 and inactive bphS2 lacking its N-terminal part, showed only the basal activation of bphAap irrespective of the presence of ethylbenzene. The transformant harboring pKLD9, which contained active bphS2 and inactive bphT2 lacking its C-terminal half, exhibited no activation of bphAap. These results suggest that the bphS2 and bphT2 genes are responsible for the ethylbenzene-dependent inducible activation and the basal activation of transcription from bphAap, respectively, as expected for the sensor histidine kinase and the response regulator of a two-component system.
The inducing-substrate spectrum of BphS2T2.
To examine the inducing-substrate spectrum of the BphS2T2 regulatory system in comparison with that of the BphS1T1 system, a 6.3-kb ApaI-BglII fragment containing bphS1T1 was inserted into pKLAF1 and a 7.3-kb SpeI fragment of pBAL73F containing bphS2T2 was inserted into pKLAF1Spe to generate pKLAST1 and pKLAST2, respectively. These plasmids were then introduced into IAM1399. The significant induction of luciferase activity by a broad range of aromatic compounds, including ethylbenzene, benzene, toluene, xylenes, isopropylbenzene, and chlorinated benzenes, was observed in IAM1399 with pKLAST2 (Table 1). Induction was not observed with benzoate, succinate, 4-chlorobiphenyl, and biphenyl. These results indicate that BphS2T2, like BphS1T1, has a significantly broad spectrum of inducing substrates. Biphenyl is the only substrate that differentiates the two systems, being an inducing substrate only for BphS1T1.
TABLE 1.
Inducing-substrate spectra of BphS1T1 and BphS2T2
a IAM1399 cells harboring pKLAST1 or pKLAST2 were grown in 1/5 LB in the presence or absence (None) of each substrate compound. The activity was expressed as in Fig. 2. The data are means ± standard deviations from at least three independent experiments.
The promoters under the control of BphS2T2.
The involvement of BphS2T2 in the transcriptional induction of bphAap, etbAa1p, etbAa2p, etbAdp, and etbD1p was examined, because these five promoters are under the control of bphS1T1 (37). The bphS2T2 gene fragment was inserted into a recombinant plasmid containing each fragment of these promoters in a promoter probe vector, pKLA1 (46). The resultant plasmids, pKMBPH, pKMETBA, pKMEBDA, pKMETA4, and pKMETBD, containing bphAap, etbAa1p, etbAa2p, etbAdp, and etbD1p, respectively, were introduced into IAM1399. Each transformant was subjected to a luciferase assay using ethylbenzene and biphenyl as inducing substrates (Table 2 ). The luciferase activity was elevated in the presence of ethylbenzene in a series of transformants harboring each of these plasmids. Such elevation was not observed in the absence of ethylbenzene. No elevation was observed in the presence of biphenyl. In contrast, such induction was not observed in transformants harboring pKLAF1, pKLABD1, pKLAED2, pKLAEA4, and pKLAED1, which did not contain bphS2T2 genes. These results indicate that the BphS2T2 regulatory system controls the transcriptional induction of the same five promoters as the BphS1T1 system does.
TABLE 2.
Induction of BphS1T1-governing promoters by BphS2T2
| Promoter | Plasmid | bphS2T2b | Luciferase activitya (105) |
||
|---|---|---|---|---|---|
| None | BPH | ETB | |||
| bphAap | pKLAF1 | − | 3 ± 0.1 | 3 ± 0.1 | 3 ± 0.2 |
| pKMBPH | + | 39 ± 1 | 36 ± 2 | 381 ± 9 | |
| etbAa1p | pKLABD1 | − | 3 ± 0.2 | 3 ± 0.01 | 2 ± 0.2 |
| pKMETBA | + | 39 ± 2 | 36 ± 1 | 419 ± 9 | |
| etbAa2p | pKLAED2 | − | 3 ± 0.1 | 2 ± 0.2 | 2 ± 0.1 |
| pKMEBDA | + | 22 ± 1 | 19 ± 0.4 | 426 ± 10 | |
| etbAdp | pKLAEA4 | − | 2 ± 0.2 | 3 ± 0.2 | 3 ± 0.2 |
| pKMETA4 | + | 13 ± 2 | 19 ± 0.2 | 304 ± 12 | |
| etbD1p | pKLAED1 | − | 1 ± 0.2 | 1 ± 0.2 | 1 ± 0.2 |
| pKMETBD | + | 9 ± 0.1 | 11 ± 0.7 | 271 ± 8 | |
The plasmids were separately introduced into IAM1399, and the resultant transformants were grown in 1/5 LB in the presence or absence (None) of biphenyl (BPH) or ethylbenzene (ETB), and were subjected to a luciferase assay. The activity was expressed as in Fig. 2. The data are means ± standard deviations from at least three independent experiments.
The presence (+) or absence (−) of bphS2T2 is indicated.
Constitutive transcription of bphS2.
To examine the transcription of bphS2, total RNAs from RHA1 cells grown on LB, biphenyl, or ethylbenzene were subjected to reverse transcription-PCR (RT-PCR) analysis using a primer set designed not to amplify the internal segment of bphS1, but to amplify that of bphS2 (Fig. 3 A). Amplified products with the expected size of 323 bp were observed using RNAs grown on LB, biphenyl, and ethylbenzene (Fig. 3B). The promoter activity of the region upstream from bphS2 was also examined. The 2.1-kb KpnI-EcoRV fragment containing the region upstream from bphS2 and the etbAdp region that is in the reverse orientation to bphS2 was blunt ended and inserted just in front of the luxAB luciferase genes in a promoter probe vector, pKLA1. A plasmid, pKLS2F, which contained a 2.1-kb bphS2 upstream fragment in the forward orientation to the luxAB luciferase genes, was introduced into RHA1, and the transformant was subjected to a luciferase assay. The transformant, irrespective of the presence of biphenyl or ethylbenzene, exhibited 20-fold-higher luciferase activity than RHA1 containing a control vector, pKLA1 (Fig. 3C). In contrast, RHA1 harboring a plasmid, pKLS2R, that contained a 2.1-kb bphS2-upstream fragment in the reverse orientation to luxAB exhibited high luciferase activity only in the presence of biphenyl or ethylbenzene, representing the BphST-dependent inducible transcription from etbAdp. These results indicate that bphS2 is transcribed constitutively from a promoter in the adjacent region upstream from bphS2 in RHA1.
FIG. 3.
Constitutive transcription of bphS2. (A) Physical map of the 9.2-kb ApaLI-HindIII fragment containing bphS2T2. The thick bar indicates the position of the 323-bp segment amplified in RT-PCR analysis. The 2.1-kb KpnI-EcoRV fragment, indicated by a two-headed arrow, was cloned in a reporter plasmid, pKLA1, for the luciferase assay. (B) RT-PCR analysis of bphS2 transcription. Total RNA of RHA1 grown in LB (lanes 1 and 2) or on biphenyl (lanes 3 and 4) or ethylbenzene (lanes 5 and 6) was used as a template in the presence (+) or absence (−) of reverse transcriptase. The DNA sizes of molecular size markers (lane M) are indicated on the left. The position and the expected size of the amplified fragment are indicated by a horizontal arrowhead on the right. (C) Luciferase assay of the bphS2 promoter region. The reporter plasmids pKLS2F and pKLS2R containing the 2.1-kb bphS2 upstream region in the forward and reverse orientations to luxAB genes, respectively, and a vector control, pKLA1, were independently introduced into RHA1, and the transformants grown in 1/5 LB in the presence or absence of biphenyl or ethylbenzene were subjected to a luciferase assay. The activity was expressed as in Fig. 2. The data are means ± standard deviations from at least three independent experiments.
Disruption of bphS1 and bphS2 in RHA1.
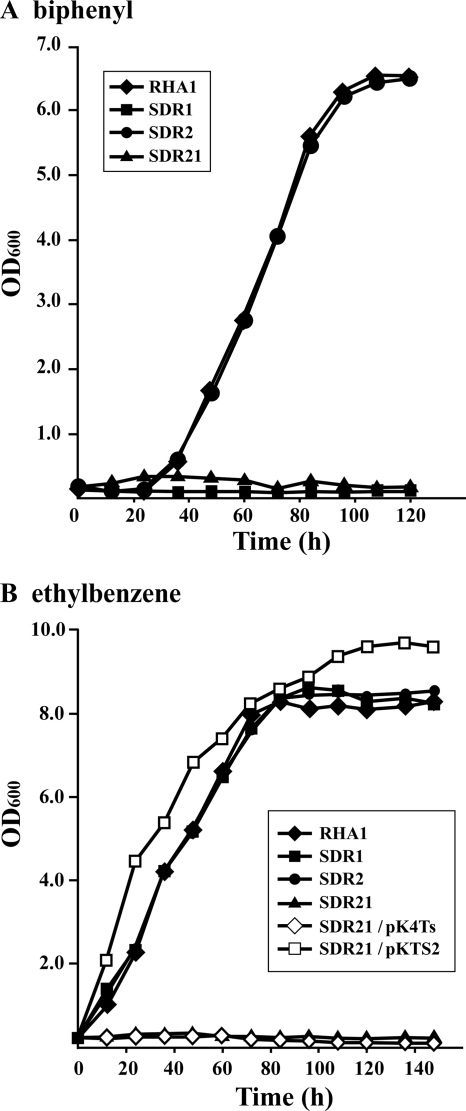
To determine whether bphS2 is really involved in the growth of RHA1 on aromatic compounds, bphS1 and bphS2 were inactivated by homologous recombination. The disruption mutant of bphS1 in strain SDR1 has been described previously (38). The disruption of bphS2 in the mutant strain SDR2 and the additional disruption of bphS1 in SDR21, derived from SDR2, were confirmed by Southern hybridization (data not shown). The growth of RHA1, SDR1, SDR2, and SDR21 on biphenyl (Fig. 4 A) or ethylbenzene (Fig. 4B) was examined. SDR1 (ΔbphS1) and SDR21 (ΔbphS1 ΔbphS2) did not grow on biphenyl. RHA1 and SDR2 (ΔbphS2) grew well on biphenyl. On the other hand, SDR21 did not grow on ethylbenzene, while RHA1, SDR1, and SDR2 grew well on ethylbenzene. When pKTS2 containing an intact bphS2 gene was constructed and introduced into SDR21, the resulting transformant grew well on ethylbenzene. These results indicate that bphS1 is solely responsible for growth on biphenyl and that either bphS1 or bphS2 is required for growth on ethylbenzene. These notions agree with the results on the inducing substrates for BphS1T1 and BphS2T2, since biphenyl is an inducing substrate only for BphS1T1 and ethylbenzene is an inducing substrate for both BphS1T1 and BphS2T2.
FIG. 4.
Growth on biphenyl (A) and ethylbenzene (B) of the disruption mutants of bphS1 or bphS2. The bphS1 mutant, SDR1; the bphS2 mutant, SDR2; the bphS1/S2 double mutant, SDR21; SDR21 harboring pKTS2 containing bphS2; and SDR21 containing a vector control, pK4Ts, were grown in W minimal medium containing biphenyl (2 g/liter) or ethylbenzene. Ethylbenzene was supplied as vapor. Growth was monitored by measuring the OD600.
Deletion of bphT1 and bphT2 in RHA1.
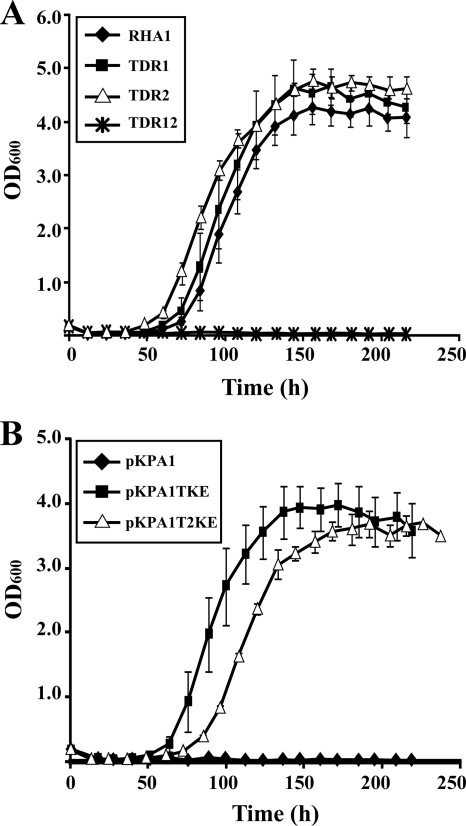
In order to examine the participation of bphT1 and bphT2 in the growth of RHA1 on aromatic compounds, the deletion mutants of bphT1, bphT2, and bphT1T2 were constructed and designated TDR1, TDR2, and TDR12, respectively. The growth of RHA1, TDR1, TDR2, and TDR12 on biphenyl was examined. TDR12 (ΔbphT1T2) did not grow on biphenyl, although RHA1, TDR1 (ΔbphT1), and TDR2 (ΔbphT2) grew well on biphenyl (Fig. 5 A). When the deletion of either bphT1 or bphT2 in TDR12 was complemented by introducing pKPA1TKE or pKPA1T2KE containing bphT1 or bphT2, respectively, the resultant transformant of TDR12 harboring either pKPA1TKE or pKPA1T2KE grew on biphenyl (Fig. 5B). Thus, bphT1 and bphT2 appeared to be equally involved in the growth of RHA1 on biphenyl, and the introduction of either bphT1 or bphT2 was sufficient to restore the growth deficiency of TDR12 on biphenyl.
FIG. 5.
Growth on biphenyl of the disruption mutants of bphT1 and bphT2. (A) The bphT1 mutant, TDR1; the bphT2 mutant, TDR2; and the bphT1/T2 double mutant, TDR12, were grown in W minimal medium containing biphenyl (1.54 g/liter). (B) TDR12 harboring pKPA1TKE containing bphT1 or pKPA1T2KE containing bphT2 was grown in W minimal medium containing biphenyl (1.54 g/liter). pKPA1 was used as a vector control. Growth was monitored by measuring the OD600. Data are means ± standard deviations from at least three independent experiments.
The BphST-dependent growth substrate spectrum.
The growth on a variety of aromatic compounds of mutant strains of bphS and bphT was examined to determine substrates for bphST-dependent growth (Table 3). Neither SDR21 nor TDR12 grew on ethylbenzene, toluene, isopropylbenzene, o-xylene, or benzene, although RHA1, SDR1, SDR2, TDR1, and TDR2 grew on these substrates. None of the strains grew on m- or p-xylene. All of them grew on terephthalate. Although m- and p-xylene are inducers for BphS1T1 and BphS2T2, as shown in the inducing-substrate spectrum (Table 1), neither RHA1 nor any of its mutants grew on these substrates. These results indicate the bphST dependence of the induction of the catabolic pathways for ethylbenzene, toluene, isopropylbenzene, o-xylene, and benzene and that either bphS1 or bphS2 and either bphT1 or bphT2 are also required for induction.
TABLE 3.
Growth characteristics of bphS mutant derivatives
| Substrate | Growth in straina: |
||||||
|---|---|---|---|---|---|---|---|
| RHA1 | SDR1 | SDR2 | SDR21 | TDR1 | TDR2 | TDR12 | |
| Ethylbenzene | + | + | + | − | + | + | − |
| Toluene | + | + | + | − | + | + | − |
| Benzene | + | + | + | − | + | + | − |
| Isopropyl- benzene | + | + | + | − | + | + | − |
| o-Xylene | + | + | + | − | + | + | − |
| m-Xylene | − | − | − | − | − | − | − |
| p-Xylene | − | − | − | − | − | − | − |
| Terephthalate | + | + | + | + | + | + | + |
−, no growth; +, growth.
Cross-communication between heterologous BphS and BphT.
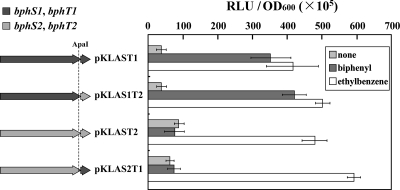
Although biphenyl is the inducing substrate only for BphS1T1, strain TDR1, which has bphT1 deleted and maintains bphT2, grew on biphenyl. Considering that sensor histidine kinases are responsible for inducing the substrate specificity of two-component systems, these results appeared to suggest cross-communication between BphS1 and BphT2. In order to examine the cross-communication between BphS1 and BphT2 or between BphS2 and BphT1, the fragment containing bphS1 and bphT2 or bphS2 and bphT1 was inserted into a luxAB reporter plasmid containing bphAap to construct pKLAS1T2 or pKLAS2T1, respectively. In the presence of ethylbenzene, the transformants of IAM1399 harboring pKLAS1T2 or pKLAS2T1 exhibited luciferase activity as high as that of the IAM1399 transformants harboring pKLAST1 or pKLAST2, which contained bphS1T1 or bphS2T2, respectively (Fig. 6). In the presence of biphenyl, only the transformants of IAM1399 harboring pKLAS1T2 or pKLAST1 exhibited high luciferase activity. These results indicate efficient cross-communication between BphS1 and BphT2 or between BphS2 and BphT1 and suggest that BphS is a key component for the determination of substrate specificity, as expected from its role as a sensor histidine kinase, and that BphS1 is solely responsible for the induction by biphenyl. The luciferase activities of pKLAST1 were 84% and 83% of those of pKLAS1T2 in the presence of biphenyl and ethylbenzene, respectively. That of pKLAST2 was 81% of that of pKLAS2T1 in the presence of ethylbenzene. Thus, the efficiency of the cross-communication between heterologous BphS and BphT components appears to be similar to that of the original communication between cognate BphS and BphT components.
FIG. 6.
Cross-communication between heterologous BphS and BphT. An original bphT was replaced by a heterologous bphT at the common ApaI site to obtain pKLAS1T2 and pKLAS2T1 containing bphS1 plus bphT2 and bphS2 plus bphT1, respectively. Shown is the luciferase activity of IAM1399 harboring pKLAS1T2 or pKLAS2T1. The cells were incubated in 1/5 LB in the presence or absence of biphenyl or ethylbenzene for 5 h and subjected to a luciferase assay. IAM1399 harboring pKLAST1 or pKLAST2 was used as a control. The activity was expressed as in Fig. 2. The data are means ± standard deviations from at least three independent experiments.
Comparison of the sequences surrounding bphST genes.
To obtain insight into the mechanism of bphST gene duplication, we compared the DNA sequences of the region surrounding bphST genes with the genome sequences of RHA1 and its related Rhodococcus strains (Fig. 7). Comparison between the sequences of RHA1 including bphS2T2 and bphS1T1 indicated an 8,374-bp duplication. This duplication started from 370 or 382 bp upstream from bphS2/S1, and ended at 158 or 168 bp downstream from ro10119/ro08049, respectively. This shared sequence of pRHL2 contained bphS2, bphT2, ro10120, and ro10119, and that of pRHL1 contained bphS1, bphT1, ro08050, and ro08049. The open reading frames ro10120 and ro08050 were annotated as medium-chain acyl-CoA ligase, and ro10119 and ro08049 were annotated as enoyl-CoA hydratase. Transposase genes and repetitive sequences were not found in either of the borders of any of the duplicated sequences. Thus, it is difficult to estimate the duplication mechanism of this region. A comparison of the sequences, including the additional bphS homolog, ro10148, and bphS2/S1, did not indicate any duplication of significance, although there were some local similarities between them (data not shown).
FIG. 7.
Comparison of the DNA sequences surrounding the bphST genes in RHA1 and the Rhodococcus strains, including bphST homologs. pROB02 (GenBank accession number AP011117) and pBD2 (GenBank accession number AY223810) are linear plasmids of R. opacus B4 and R. erythropolis BD2, respectively. The gray-shaded sections represent homologous DNA regions with more than 70% identity between two sequences. The exact percentages of identity are indicated by their shades of gray, which are shown in the gray scale bar at the top. The 24-bp direct repeats, including the bphST-dependent promoter consensus of RHA1, are connected by black lines and a gray line between ro10124 and etbD1 of RHA1.
The 20,158-bp region containing bphS1T1 in the center is shared with the corresponding regions in linear plasmids, pROB02 of Rhodococcus opacus B4 (GenBank accession number AP011117) and pBD2 of R. erythropolis BD2 (34). These regions are more than 90% similar to each other. This common region contains mainly degradation enzyme genes, including bphAa to -Ad, bphB1, bphC1, and etbD1. In B4 and BD2, it was sandwiched between two putative transposase genes, but not in RHA1. In addition, two transposase genes in tandem were inserted just below the counterparts of RHA1 bphT1, ROP_pROB02_01660 in B4 and the putative response regulator gene ipbT in BD2. In B4, the transposase genes of this insertion were disrupted by a small deletion including their intergenic sequences. In BD2, an extra transposase gene was inserted between the tandem transposase genes and ipbT. An additional insertion of putative tandem transposase genes was found between PBD2.166 and PBD2.169 in BD2. A further insertion of tandem transposase genes was found just below ipbD in BD2. Thus, this 20,158-bp bphST region in B4 and BD2 appears to be mobilizable with the aid of transposable elements. The region in RHA1 may be an ancestral sequence, because it contains only a fragment of a transposase gene between ro08046 and etbD1, which is shared by B4 and BD2.
The 24-bp direct repeats, TTCCGTAGTTTTCCCGGATGTTCG, were identified in the bphST-containing regions of RHA1, B4, and BD2, which are indicated in Fig. 7 by black lines between ro08061 of RHA1 pRHL1 and ROP_pROB02_01690 of B4, between ro10120 of RHA1 pRHL2 and ro08061 of RHA1 pRHL1, between ro08050 of RHA1 pRHL1 and ROP_pROB02_01570 of B4, between ROP_pROB02_01570 of B4 and alkK of BD2, and between ROP_pROB02_01690 of B4 and PBD2.152 of BD2. However, they appear not to be involved in the rearrangements generating the shared sequences between RHA1, B4, and BD2, because none of them are located within the borders of the shared sequences. Identical repeats with a single-base alteration were also conserved and were indicated by a gray line between ro10124 of RHA1 pRHL2 and etbD1 of RHA1 pRHL1 in Fig. 7. These repeats contained the bphST-dependent promoter consensus sequence of RHA1, CcGTAgTTTttcCGGATG (lowercase letters indicate that the consensus is not unanimous), which was proposed previously (37).
DISCUSSION
In this study, the bphS2 and bphT2 genes, located in the pRHL2 linear plasmid, were isolated and characterized. Their deduced amino acid sequences are nearly identical to those of bphS1 (92%) and bphT1 (97%), respectively, which are located in the pRHL1 plasmid. Along with bphS1 and bphT1, they are responsible for the activated transcription from bphAap, etbAa1p, etbAa2p, etbAdp, and etbD1p in the presence of a variety of aromatic compounds, except for biphenyl (Fig. 8).
FIG. 8.
Transcriptional regulation mechanisms of BphS1T1 and BphS2T2 two-component systems in RHA1. BphS1T1 and BphS2T2 share most characteristics. Both BphS1 and BphS2 are estimated to phosphorylate BphT1 and BphT2, which are both responsible for the transcriptional activation of bphAap, etbAa1p, etbAa2p, etbAdp, and etbD1p in the presence of a variety of aromatic compounds, including ethylbenzene, benzene, toluene, xylenes, isopropylbenzene, and chlorinated benzenes (Table 1 shows details). Biphenyl is an inducing substrate for BphS1, but not for BphS2.
Both BphS1 and BphS2 have wide ranges of inducing substrates, which are broader than the growth substrate range. Such an unnecessarily broad range of inducing substrates was also reported with TodS of Pseudomonas putida DOT-T1E (18), which is a sensor histidine kinase involved in the degradation of toluene. Similar broader ranges of inducers than pathway substrates were often found in other types of regulatory systems for aromatic degradation pathways (41). A broader substrate spectrum of inducing substrates than of growth substrates appears to be advantageous for the acquisition of new metabolic traits. The limitation of the growth substrate range by the restricted range of inducing substrates, which was indicated by TodS of P. putida F1 (4), DmpR of Pseudomonas sp. strain CF600 (5, 28), and PhhR of P. putida P35X (24), appears to support this notion.
Double inactivation of bphT1 and bphT2 or of bphS1 and bphS2 resulted in an inability to grow on aromatic compounds, including biphenyl, ethylbenzene, toluene, benzene, isopropylbenzene, and o-xylene. Although RHA1 is estimated to have 33 sets of two-component system genes (22), these results suggest that these four genes are the only members of two-component systems responsible for the growth of RHA1 on the aromatic compounds mentioned above. These results also exclude the possible involvement of the additional bphS homolog, ro10148, in the growth of RHA1 on the above-mentioned aromatic compounds. The cross-communication between the gene products of ro10148 and bphT1 was not observed in preliminary experiments (data not shown), supporting this notion.
The growth of TDR1 (ΔbphT1) on biphenyl indicated cross-communication between BphS1 and BphT2, and heterologous cross-communication between BphS1 and BphT2 and between BphS2 and BphT1 was also indicated when they were expressed in a rhodococcal host strain. Signal transduction between a heterologous BphS-BphT pair was mostly similar to that between a cognate pair. It is estimated that a single bacterium has on average around 50 species of two-component systems (43). Although some systems in the same family have good sequence identity to each other, they barely cross-communicate in vivo (3, 19). When these systems were reproduced by gene duplication, they should have cross-communicated, and subsequently, they lost the ability to cross-communicate during evolution into divergent systems (3, 44). Cross-communication between a heterologous BphS-BphT pair, which is as efficient as the original communication between a cognate pair, and significant sequence identity between the respective components suggest that these two systems have been duplicated recently. On the other hand, the nucleotide sequences of bphS1 and bphS2 differ by 317 nucleotides among the total 4,797 nucleotides, suggesting that duplication between the bphS1T1 and bphS2T2 regions did not occur very recently. Such duplication appears to confer an advantage for the acquisition of new phenotypes, because the cell can maintain the original phenotype, in addition to the changing (mutating) one. The inducing-substrate spectrum of BphS1 includes biphenyl, in addition to the substrates of BphS2. BphS1 appears to have acquired an expanded substrate spectrum from BphS2, which seems to maintain the original substrate spectrum.
The only difference between them is the response to biphenyl, which is an inducing substrate for the bphS1T1 system, but not for the bphS2T2 system. The following results suggested that in the presence of a single-ring aromatic compound, such as ethylbenzene, both bphS1 and bphS2 are responsible for the transcriptional activation of degradation genes in RHA1. (i) The transcription of the bphS1 and bphS2 genes is constitutive (38). (ii) SDR1 (ΔbphS1) and SDR2 (ΔbphS2) grew well, and SDR21 (ΔbphS1ΔbphS2) did not grow, on ethylbenzene, toluene, benzene, isopropylbenzene, and o-xylene. It is expected that the transcriptional activation promoted by both bphS1 and bphS2 in the presence of a single-ring aromatic compound is superior to that promoted by bphS1 only in the presence of biphenyl. However, the amounts of degradation gene transcripts in RHA1, including bphAa, etbC, and etbAc, were reported to be the same in the cells grown on biphenyl and ethylbenzene (7). Thus, the activation promoted by bphS1 only in the presence of biphenyl appears to be enough to bring out the full promoter activities of degradation genes.
The regions containing bphS1T1 and bphS2T2 share 8,374-bp sequences starting from 382 or 370 bp upstream from bphS1/S2 and ending 168 or 158 bp downstream from ro08049/ro10119, annotated as enoyl-CoA hydratase. Considering that the bphS1T1-containing regions are conserved between RHA1, B4, and BD2 and that bphS1T1 constitutes an operon with bphAaAbAcAdC1B1 genes, the 8,374-bp bphS2T2-containing sequence might have originated from a bphS1T1-containing region.
Two 24-bp sequences (TTCCGTAGTTTTCCCGGATGTTCG) were completely conserved in the bphST-containing regions of RHA1, B4, and BD2. Two identical sequences with a single-base alteration were also found in the bphST-containing regions of RHA1. These sequences contained CcGTAgTTTttcCGGATG, which was previously proposed as the bphST-dependent promoter consensus sequence of RHA1 located in the bphAa, etbD1, etbAa1, etaAa2, and etbAd promoters (37). Recently, it was reported that the mutation of this consensus sequence in the thnA1 promoter sequence in a tetralin-degrading bacterium, Rhodococcus sp. strain TFB, affected the promoter activity (40). These results suggest that the common mechanism of bphST regulation governs the degradation genes located in the conserved bphST-containing regions of RHA1, B4, and BD2.
The results of the genome sequencing of RHA1 indicated that RHA1 has numerous gene duplications among aromatic degradation genes. RHA1 is thought to have evolved its versatile degradation activity on aromatics via gene duplications of not only degradation enzyme genes, but also regulatory genes, such as bphS1T1 and bphS2T2.
Supplementary Material
Acknowledgments
We thank René De Mot for the kind gift of plasmid pFAJ2574. We are grateful to R. van der Geize for his helpful suggestions regarding the unmarked gene deletion mutagenesis of rhodococcal genes.
This work was supported in part by a grant-in-aid for Scientific Research (B) (22380050) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation; QTL-GMB0005).
Footnotes
Published ahead of print on 9 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahmad, D., R. Massé, and M. Sylvestre. 1990. Cloning and expression of genes involved in 4-chlorobiphenyl transformation by Pseudomonas testosteroni: homology to polychlorobiphenyl-degrading genes in other bacteria. Gene 86:53-61. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bijlsma, J. J., and E. A. Groisman. 2003. Making informed decisions: regulatory interactions between two-component systems. Trends. Microbiol. 11:359-366. [DOI] [PubMed] [Google Scholar]
- 4.Choi, E. N., M. C. Cho, Y. Kim, C. K. Kim, and K. Lee. 2003. Expansion of growth substrate range in Pseudomonas putida F1 by mutations in both cymR and todS, which recruit a ring-fission hydrolase CmtE and induce the tod catabolic operon, respectively. Microbiology 149:795-805. [DOI] [PubMed] [Google Scholar]
- 5.Fernández, S., V. Shingler, and V. De Lorenzo. 1994. Cross-regulation by XylR and DmpR activators of Pseudomonas putida suggests that transcriptional control of biodegradative operons evolves independently of catabolic genes. J. Bacteriol. 176:5052-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa, K., and A. M. Chakrabarty. 1982. Involvement of plasmids in total degradation of chlorinated biphenyls. Appl. Environ. Microbiol. 44:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonçalves, E. R., H. Hara, D. Miyazawa, J. E. Davies, L. D. Eltis, and W. W. Mohn. 2006. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 72:6183-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 9.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 10.Hatta, T., T. Shimada, T. Yoshihara, A. Yamada, E. Masai, M. Fukuda, and H. Kiyohara. 1998. Meta-fission product hydrolases from a strong PCB degrader Rhodococcus sp. RHA1. J. Ferment. Bioeng. 85:174-179. [Google Scholar]
- 11.Hauschild, J. E., E. Masai, K. Sugiyama, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1996. Identification of an alternative 2,3-dihydroxybiphenyl 1,2-dioxygenase in Rhodococcus sp. strain RHA1 and cloning of the gene. Appl. Environ. Microbiol. 62:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki, T., K. Miyauchi, E. Masai, and M. Fukuda. 2006. Multiple-subunit genes of the aromatic-ring-hydroxylating dioxygenase play an active role in biphenyl and polychlorinated biphenyl degradation in Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 72:5396-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwasaki, T., H. Takeda, K. Miyauchi, T. Yamada, E. Masai, and M. Fukuda. 2007. Characterization of two biphenyl dioxygenases for biphenyl/PCB degradation in a PCB degrader, Rhodococcus sp. strain RHA1. Biosci. Biotechnol. Biochem. 71:993-1002. [DOI] [PubMed] [Google Scholar]
- 14.Kim, D., J. C. Chae, G. J. Zylstra, H. Y. Sohn, G. S. Kwon, and E. Kim. 2005. Identification of two-component regulatory genes involved in o-xylene degradation by Rhodococcus sp. strain DK17. J. Microbiol. 43:49-53. [PubMed] [Google Scholar]
- 15.Kimbara, K., T. Hashimoto, M. Fukuda, T. Koana, M. Takagi, M. Oishi, and K. Yano. 1989. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitagawa, W., K. Miyauchi, E. Masai, and M. Fukuda. 2001. Cloning and characterization of benzoate catabolic genes in the gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. J. Bacteriol. 183:6598-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labbé, D., J. Garnon, and P. C. Lau. 1997. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J. Bacteriol. 179:2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacal, J., A. Busch, M. E. Guazzaroni, T. Krell, and J. L. Ramos. 2006. The TodS-TodT two-component regulatory system recognizes a wide range of effectors and works with DNA-bending proteins. Proc. Natl. Acad. Sci. U. S. A. 103:8191-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laub, M. T., and M. Goulian. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121-145. [DOI] [PubMed] [Google Scholar]
- 20.Masai, E., K. Sugiyama, N. Iwashita, S. Shimizu, J. E. Hauschild, T. Hatta, K. Kimbara, K. Yano, and M. Fukuda. 1997. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene 187:141-149. [DOI] [PubMed] [Google Scholar]
- 21.Masai, E., A. Yamada, J. M. Healy, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLeod, M. P., R. L. Warren, W. W. Hsiao, N. Araki, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. Jones, R. Holt, F. S. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mondello, F. J. 1989. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J. Bacteriol. 171:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng, L. C., C. L. Poh, and V. Shingler. 1995. Aromatic effector activation of the NtrC-like transcriptional regulator PhhR limits the catabolic potential of the (methyl)phenol degradative pathway it controls. J. Bacteriol. 177:1485-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtsubo, Y., W. Ikeda-Ohtsubo, Y. Nagata, and M. Tsuda. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsson, O., C. Koncz, and A. A. Szalay. 1988. The use of the luxA gene of the bacterial luciferase operon as a reporter gene. Mol. Gen. Genet. 215:1-9. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 28.Pavel, H., M. Forsman, and V. Shingler. 1994. An aromatic effector specificity mutant of the transcriptional regulator DmpR overcomes the growth constraints of Pseudomonas sp. strain CF600 on para-substituted methylphenols. J. Bacteriol. 176:7550-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai, M., E. Masai, H. Asami, K. Sugiyama, K. Kimbara, and M. Fukuda. 2002. Diversity of 2,3-dihydroxybiphenyl dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1. J. Biosci. Bioeng. 93:421-427. [DOI] [PubMed] [Google Scholar]
- 30.Sakai, M., E. Masai, K. Sugiyama, C. Takahashi, A. Yamada, K. Miyauchi, and M. Fukuda. 2004. Characterization of a second 2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase gene of the polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. J. Environ. Biotechnol. 4:41-47. [Google Scholar]
- 31.Sakai, M., K. Miyauchi, N. Kato, E. Masai, and M. Fukuda. 2003. 2-Hydroxypenta-2,4-dienoate metabolic pathway genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 69:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seto, M., K. Kimbara, M. Shimura, T. Hatta, M. Fukuda, and K. Yano. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seto, M., E. Masai, M. Ida, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Multiple polychlorinated biphenyl transformation systems in the gram-positive bacterium Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:4510-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stecker, C., A. Johann, C. Herzberg, B. Averhoff, and G. Gottschalk. 2003. Complete nucleotide sequence and genetic organization of the 210-kilobase linear plasmid of Rhodococcus erythropolis BD2. J. Bacteriol. 185:5269-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 36.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeda, H., N. Hara, M. Sakai, A. Yamada, K. Miyauchi, E. Masai, and M. Fukuda. 2004. Biphenyl-inducible promoters in a polychlorinated biphenyl-degrading bacterium, Rhodococcus sp. RHA1. Biosci. Biotechnol. Biochem. 68:1249-1258. [DOI] [PubMed] [Google Scholar]
- 38.Takeda, H., A. Yamada, K. Miyauchi, E. Masai, and M. Fukuda. 2004. Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J. Bacteriol. 186:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomás-Gallardo, L., E. Santero, E. Camafeita, E. Calvo, M. Schlömann, and B. Floriano. 2009. Molecular and biochemical characterization of the tetralin degradation pathway in Rhodococcus sp. strain TFB. Microb. Biotechnol. 2:262-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tropel, D., and J. R. van der Meer. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68:474-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Geize, R., G. I. Hessels, R. van Gerwen, P. van der Meijden, and L. Dijkhuizen. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Delta1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197-202. [DOI] [PubMed] [Google Scholar]
- 43.Whitworth, D. E. 2008. Genomes and knowledge—a questionable relationship? Trends Microbiol. 16:512-519. [DOI] [PubMed] [Google Scholar]
- 44.Whitworth, D. E., and P. J. Cock. 2009. Evolution of prokaryotic two-component systems: insights from comparative genomics. Amino Acids 37:459-466. [DOI] [PubMed] [Google Scholar]
- 45.Wolanin, P. M., P. A. Thomason, and J. B. Stock. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3:REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.