Abstract
Adhesive pili on the surface of the serotype M1 Streptococcus pyogenes strain SF370 are composed of a major backbone subunit (Spy0128) and two minor subunits (Spy0125 and Spy0130), joined covalently by a pilin polymerase (Spy0129). Previous studies using recombinant proteins showed that both minor subunits bind to human pharyngeal (Detroit) cells (A. G. Manetti et al., Mol. Microbiol. 64:968-983, 2007), suggesting both may act as pilus-presented adhesins. While confirming these binding properties, studies described here indicate that Spy0125 is the pilus-presented adhesin and that Spy0130 has a distinct role as a wall linker. Pili were localized predominantly to cell wall fractions of the wild-type S. pyogenes parent strain and a spy0125 deletion mutant. In contrast, they were found almost exclusively in culture supernatants in both spy0130 and srtA deletion mutants, indicating that the housekeeping sortase (SrtA) attaches pili to the cell wall by using Spy0130 as a linker protein. Adhesion assays with antisera specific for individual subunits showed that only anti-rSpy0125 serum inhibited adhesion of wild-type S. pyogenes to human keratinocytes and tonsil epithelium to a significant extent. Spy0125 was localized to the tip of pili, based on a combination of mutant analysis and liquid chromatography-tandem mass spectrometry analysis of purified pili. Assays comparing parent and mutant strains confirmed its role as the adhesin. Unexpectedly, apparent spontaneous cleavage of a labile, proline-rich (8 of 14 residues) sequence separating the N-terminal ∼1/3 and C-terminal ∼2/3 of Spy0125 leads to loss of the N-terminal region, but analysis of internal spy0125 deletion mutants confirmed that this has no significant effect on adhesion.
The group A Streptococcus (S. pyogenes) is an exclusively human pathogen that commonly colonizes either the pharynx or skin, where local spread can give rise to various inflammatory conditions such as pharyngitis, tonsillitis, sinusitis, or erysipelas. Although often mild and self-limiting, GAS infections are occasionally very severe and sometimes lead to life-threatening diseases, such as necrotizing fasciitis or streptococcal toxic shock syndrome. A wide variety of cell surface components and extracellular products have been shown or suggested to play important roles in S. pyogenes virulence, including cell surface pili (1, 6, 32). Pili expressed by the serotype M1 S. pyogenes strain SF370 mediate specific adhesion to intact human tonsil epithelia and to primary human keratinocytes, as well as cultured keratinocyte-derived HaCaT cells, but not to Hep-2 or A549 cells (1). They also contribute to adhesion to a human pharyngeal cell line (Detroit cells) and to biofilm formation (29).
Over the past 5 years, pili have been discovered on an increasing number of important Gram-positive bacterial pathogens, including Bacillus cereus (4), Bacillus anthracis (4, 5), Corynebacterium diphtheriae (13, 14, 19, 26, 27, 44, 46, 47), Streptococcus agalactiae (7, 23, 38), and Streptococcus pneumoniae (2, 3, 24, 25, 34), as well as S. pyogenes (1, 29, 32). All these species produce pili that are composed of a single major subunit plus either one or two minor subunits. During assembly, the individual subunits are covalently linked to each other via intermolecular isopeptide bonds, catalyzed by specialized membrane-associated transpeptidases that may be described as pilin polymerases (4, 7, 25, 41, 44, 46). These are related to the classical housekeeping sortase (usually, but not always, designated SrtA) that is responsible for anchoring many proteins to Gram-positive bacterial cell walls (30, 31, 33). The C-terminal ends of sortase target proteins include a cell wall sorting (CWS) motif consisting, in most cases, of Leu-Pro-X-Thr-Gly (LPXTG, where X can be any amino acid) (11, 40). Sortases cleave this substrate between the Thr and Gly residues and produce an intermolecular isopeptide bond linking the Thr to a free amino group provided by a specific target. In attaching proteins to the cell wall, the target amino group is provided by the lipid II peptidoglycan precursor (30, 36, 40). In joining pilus subunits, the target is the ɛ-amino group in the side chain of a specific Lys residue in the second subunit (14, 18, 19). Current models of pilus biogenesis envisage repeated transpeptidation reactions adding additional subunits to the base of the growing pilus, until the terminal subunit is eventually linked covalently via an intermolecular isopeptide bond to the cell wall (28, 41, 45).
The major subunit (sometimes called the backbone or shaft subunit) extends along the length of the pilus and appears to play a structural role, while minor subunits have been detected either at the tip, the base, and/or at occasional intervals along the shaft, depending on the species (4, 23, 24, 32, 47). In S. pneumoniae and S. agalactiae one of the minor subunits acts as an adhesin, while the second appears to act as a linker between the base of the assembled pilus and the cell wall (7, 15, 22, 34, 35). It was originally suggested that both minor subunits of C. diphtheriae pili could act as adhesins (27). However, recent data showed one of these has a wall linker role (26, 44) and may therefore not function as an adhesin.
S. pyogenes strain SF370 pili are composed of a major (backbone) subunit, termed Spy0128, plus two minor subunits, called Spy0125 and Spy0130 (1, 32). All three are required for efficient adhesion to target cells (1). Studies employing purified recombinant proteins have shown that both of the minor subunits, but not the major subunit, bind to Detroit cells (29), suggesting both might act as pilus-presented adhesins. Here we report studies employing a combination of recombinant proteins, specific antisera, and allelic replacement mutants which show that only Spy0125 is the pilus-presented adhesin and that Spy0130 has a distinct role in linking pili to the cell wall.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The serotype M1 S. pyogenes strain SF370 was obtained from the ATCC and has been described by Ferretti et al. (10). Construction of the S. pyogenes SF370 allele replacement mutants Δspy0125, Δspy0128, and Δspy0130 has been described previously (1), and mutants constructed during this study are described in the text. Escherichia coli strains TG1and BL21(DE3) have been described by Sambrook et al. (39). The allele replacement vector pG+host 9 and its E. coli host strain TG1-dev were generously provided by E. Maguin (INRA, Jouy en Josas, France) and have been described previously (12). The expression plasmid pET28a-D is a derivative of pET28a (Novagen, Merck Chemicals Ltd., Nottingham, United Kingdom), where 45 bp immediately upstream of the BamHI site (pET28a nucleotides 203 to 248) have been deleted. Unless otherwise specified, E. coli strains were grown in LB medium and S. pyogenes strains were grown in THY medium as described previously (12). Erythromycin was added to THY medium at a final concentration of 2 μg/ml to select pG+host 9 in S. pyogenes and to LB medium at a final concentration of 300 μg/ml to select pG+host 9 in E. coli TG1-dev. For selection of pET28a or pET28a-D in E. coli, 50 μg/ml of kanamycin was added to LB medium.
General DNA and protein procedures.
Restriction endonucleases, T4 DNA ligase, Taq polymerase, and Pfu polymerase were purchased from various standard commercial sources and used according to the manufacturers' instructions. Purification of plasmid DNA from E. coli, extraction of DNA fragments from agarose gels, and purification of PCR-amplified products employed appropriate kits from Qiagen Ltd. (Crawley, West Sussex, United Kingdom). S. pyogenes chromosomal DNA was purified using Puregene DNA isolation kits (Gentra Systems Inc., Minneapolis, MN), adapted as described previously (12). DNA primers were produced to order by VH Bio Ltd. (Newcastle, United Kingdom) and are described in Table 1. DNA sequencing was performed to order by Eurofins MWG Operon (Ebersberg, Germany). Other DNA manipulations and analyses employed standard procedures, as described previously (12, 39, 48). Protein separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with specific antisera employed standard procedures, as described previously (12, 48).
TABLE 1.
Oligonucleotide primers used in constructing expression plasmids and mutants
| Constructa | Primer sequence (5′-3′)b |
|---|---|
| spy0125 in pET28a | ttacatacatatgAAGACTGTTTTTGGTTTAGTAG |
| aagttctcgagTTAAACTCCTGTAGGAACAAC | |
| spy0125-N in pET28a-D | ctaggatccAAGACTGTTTTTGGTTTAGTAG |
| atgttctcgagTTAACCACCACTCAAAAGATT | |
| spy0125-C in pET28a-D | ctaggatccAATCAACCTCAAACGACTTCAG |
| atgttctcgagTTATGTAGGAACAACAGGCTC | |
| spy0128 in pET28a | ttacatacatatgGAGACTGTTGTAAACGGAGC |
| aagttctcgagTTATACTCCTGTTGGCACTTC | |
| spy0130 in pET28a | ttacatacatatgGAGAACCTCACTGCAAGCATT |
| aagttctcgagTTAACCAGTTGATGGCAAAATACC | |
| Δspy0125-N in pG+Host 9 | atgttctcgagTTATGTAGGAACAACAGGCTC |
| ttagtcgacACCAAAAACAGTCTTAGCAC | |
| attagtcgacAATCAACCTCAAACGACTT | |
| atgttctcgagAATCTGTTTTCCATCAATAA | |
| Δspy0125-C in pG+Host 9 | ttacatactgcagGTTatgtataatggacatccacaa |
| attagtcgacTGGAGGCATTGGTGGGTC | |
| attagtcgaccagctaactgaccttgatttc | |
| aagttctcgagATAGCCTTCAGAATCTGTTTC | |
| spy0128 in pG+Host 9 | attgaattccgatatggctcctagtgtaa |
| aagttctcgagGATGTGCATAGATTAATGAG | |
| spy0128K161A site-directed mutagenesis | TATAAAGAAGGTAGTgcGGTGCCAATTCAGTTC |
| GAACTGAATTGGCACCgcACTACCTTCTTTATA |
The expression plasmid or mutant constructed using the primer shown.
Lowercase letters indicate bases added to facilitate cloning or that were changed for mutagenesis. Restriction endonuclease sites used for cloning are underlined.
Construction of S. pyogenes mutants.
In-frame deletion mutants lacking defined segments of the spy0125 gene were constructed by allele replacement mutagenesis using vector pG+host 9, essentially as described previously (12). For site-directed mutagenesis, spy0128 plus ca. 500 bp of flanking sequences were first cloned into pG+host9, and site-directed mutants of the cloned gene were produced using the QuikChange kit (Stratagene, United Kingdom), essentially as described by the manufacturer. Mutants were confirmed by sequencing the entire cloned region, before transforming the S. pyogenes Δspy0128 mutant and replacing the deleted wild-type gene with the spy0128K161A mutant allele by allele replacement mutagenesis. All S. pyogenes mutant structures were confirmed by DNA sequencing of PCR-amplified chromosomal fragments, ensuring that these products included sequences flanking those that had been cloned and manipulated in vitro during mutant construction. Growth rates of all mutants were confirmed to be the same as the parent wild-type strain.
Production of recombinant proteins and specific antisera.
For production of recombinant Spy0125 (rSpy0125), rSpy0128, and rSpy0130 proteins, PCR primers were designed to amplify sequences encoding the mature cell surface forms of these proteins, i.e., lacking the N-terminal secretion signal peptides and the sequences downstream from the C-terminal threonine after sortase cleavage of the CWS motifs VVPTG, EVPTG, and LPSTG in Spy0125, Spy0128, and Spy0130, respectively. Primers included appropriate restriction endonuclease site tags to allow the resulting PCR products to be cloned in the desired reading frame between the NdeI and XhoI sites of pET28a. Similarly, plasmids expressing the individual N- and C-terminal domains of Spy0125 described here (rSpy0125-N and rSpy0125-C, respectively) were constructed using PCR primers designed to amplify the corresponding subsections of the spy0125 gene, which were then cloned between the BamHI and XhoI sites of pET28a-D. After transformation into E. coli strain BL21(DE3) the recombinant plasmids were induced to express the desired proteins with N-terminal His tag fusions, enabling purification on nickel-chelating affinity His tag columns, essentially as described in the QIAexpress handbook (Qiagen Ltd., Crawley, West Sussex, United Kingdom). The desired protein fractions were identified by SDS-PAGE, pooled, and concentrated by ultrafiltration. Depending on protein size, a Hi-Load 16/60 Superdex 75 or 16/60 Superdex 200 column (GE Healthcare, Little Chalfont, Bucks, United Kingdom), preequilibrated in running buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.5), was used to purify the concentrated protein further by gel filtration. Fractions containing protein purified to apparent homogeneity, as judged by overloading SDS-PAGE gels, were concentrated to ∼10 mg/ml, and 1-ml aliquots were stored at either 4°C for no more than 2 weeks or −80°C for longer periods.
For production of specific antisera, purified protein was dialyzed into phosphate-buffered saline (PBS; Sigma-Aldrich, Dorset, United Kingdom), mixed with Freund's adjuvant, and used to immunize rabbits (New Zealand Whites) following standard procedures. Control (preimmune) serum samples were recovered from each individual animal prior to immunization. Experiments employed purified IgG fractions, which were obtained by affinity chromatography of crude antisera on protein A columns (Sigma-Aldrich, United Kingdom).
Preparation of cell wall and culture supernatant fractions.
To ensure equitable comparisons, all cultures were grown to an optical density at 600 of 0.68 to 0.70, and the recovered bacterial cells and cell-free culture supernatants were concentrated to the same extent (50-fold). Routinely, cells from 50-ml cultures were recovered by centrifugation at 5,000 × g, 4°C, for 10 min, and washed twice in cold Tris-EDTA (TE) buffer (50 mM Tris-Cl [pH 8.0], 1 mM EDTA), before being resuspended in 1 ml of freshly prepared wall extraction buffer consisting of TE buffer containing 20% (wt/vol) sucrose, 100 mg/ml lysozyme (Sigma), 100 U mutanolysin (Sigma), and 400 U protease inhibitor cocktail set II (Calbiochem-Merck, Nottingham, United Kingdom). Cell suspensions were incubated at 37°C with gentle rotation for 2 h to facilitate wall digestion. Following incubation, insoluble material, including protoplasts, was removed by centrifugation at 16,000 × g for 5 min. The 1 ml of supernatant was carefully recovered and stored at −80°C until required. In parallel, the cell-free culture supernatants were filter sterilized (0.2-μm filter; Anachem, Luton, United Kingdom) to remove any residual bacteria before being concentrated 50-fold by ultrafiltration through a 10,000 MWCO Vivaspin 20 filter (Sartorius, Goettingen, Germany). The 1 ml of concentrated supernatant was stored at −80°C until required. For comparisons, equal volumes of wall and supernatant fractions were loaded in adjacent tracks on 4 to 15% (wt/vol) precast gradient SDS-PAGE gels (Bio-Rad, Hemel Hempstead, United Kingdom), and separated proteins were immunoblotted as described above.
Purification of S. pyogenes pili and LC-MS/MS analysis of proteolytic digestion products.
Pili were extracted from S. pyogenes SF370, purified by immunoaffinity chromatography, concentrated, and run on an SDS-PAGE gel, as described previously (18). Bands containing purified pili were recovered, destained, and diced into small segments. The resulting mix was subject to five rounds of trypsin digestion as described previously (18) with the incubation conditions modified to submerging diced gel pieces in 50 ng/μl trypsin (Promega) in 25 mM NH4HCO3 and 10% acetonitrile for 4 h at 37°C. Pooled trypsin digestion products were concentrated to ∼10 μl and analyzed by mass spectrometry using liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described previously (18). The resulting MS/MS data were first searched for matches against the MSDB protein sequence database (ftp://ftp.ncbi.nih.gov/repository/MSDB/msdb.nam), which identified linear peptides only. Searches for a peptide cross-linked by the predicted Spy0125-Spy0128 intermolecular isopeptide bond were made from the peptide peaks that had no match in the sequence database search. This involved first estimating the molecular masses of potential cross-linked peptides, taking into account the loss of H2O (18 Da). These peptides were then sought from among the unmatched peptides, together with searches for corresponding fragment ions. The MS/MS spectra from these manual searches were inspected to confirm the presence of two noncontiguous peptides.
Bacterial adhesion and protein binding assays.
Adhesion of S. pyogenes to cultured HaCaT cells and to fresh ex vivo samples of intact human tonsil epithelium was assayed essentially as described previously (1), with the following minor modification to the fixing and staining conditions in the case of HaCaT cells. Following washing to remove nonadherent bacteria, cells were fixed by incubation at 4°C for 15 min in 2% (vol/vol) paraformaldehyde in PBS. All subsequent steps were at room temperature. After washing three times in PBS, cells were permeabilized by incubation for 15 min in 0.1% (vol/vol) Triton X-100 in PBS, and residual Triton was removed by washing three times in PBS. Cells were then blocked by incubation for 1 h in 10% (vol/vol) rabbit serum in PBS. To stain HaCaT cells and adherent S. pyogenes, respectively, 50 μg ml−1 of rhodamine-labeled phalloidin (Sigma) and 5 μg/ml fluorescein isothiocyanate (FITC)-labeled polyclonal antibody to group A streptococcus (Acris Antibodies) were added and incubation was continued in the dark for a further 1 h. Following three washes in PBS, samples were mounted and analyzed by confocal laser microscopy, as described previously (1). For antibody inhibition assays, 20 μg of affinity-purified IgG was added to bacteria in 1 ml of antibiotic- and serum-free HaCaT or tonsil culture medium and incubated with gentle agitation for 1 h at room temperature. Bacteria were then recovered by centrifugation (16,000 × g) for 1 min, resuspended in 1 ml of the appropriate antibiotic- and serum-free medium, and used to infect cells as described previously (1).
For protein binding assays, confluent cell monolayers on glass coverslips were incubated at 37°C, 5% CO2, in antibiotic- and serum-free medium for 3 h before incubating for a further 3 h in the same medium supplemented with 100 μg of the appropriate recombinant protein. After removing unbound recombinant protein by washing three times in PBS, cells were fixed, permeabilized, and blocked as described above. To detect bound recombinant protein, cells were incubated for 1 h with 20 μg of the appropriate affinity-purified IgG antibody, followed by washing three times in PBS. Cells were then blocked again in 3% (vol/vol) goat serum, before incubating for 1 h in the dark with 50 μg/ml of rhodamine-labeled phalloidin (Sigma) to stain cells and 5 μg/ml of FITC-labeled goat anti-rabbit IgG (Molecular Probes) to detect IgG bound to recombinant protein. Following three washes in PBS, cells were mounted and analyzed as described above.
RESULTS
Only anti-rSpy0125 inhibits S. pyogenes adhesion.
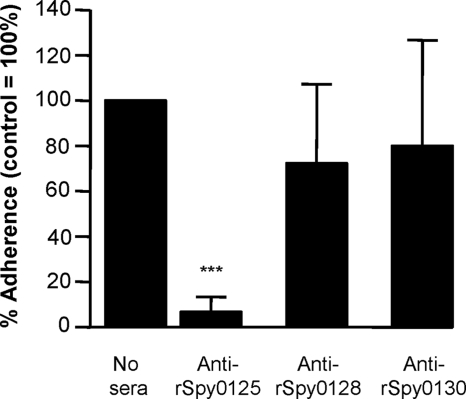
Both minor pilins (Spy0125 and Spy0130) from S. pyogenes strain SF370, but not the major shaft subunit (Spy0128), have been reported to bind to the human pharyngeal Detroit cell line (29). Consistent with this, during the present study purified rSpy0125 and rSpy0130 proteins, but not the rSpy0128 protein, bound to intact human tonsil epithelium and to HaCaT cells (data not shown). However, when the abilities of specific antisubunit sera to inhibit bacterial adhesion were examined, only anti-rSpy0125 serum consistently inhibited adhesion of S. pyogenes strain SF370 to human tonsil epithelium (n = 7 from four separate experiments) and HaCaT cells (n = 6 from three separate experiments) (Fig. 1). Differences in adhesion between the control and S. pyogenes cells preincubated with either anti-rSpy0128 or anti-rSpy0130 serum were not significant.
FIG. 1.
Effects of antisera to recombinant pilus subunits on adhesion of S. pyogenes SF370 to HaCaT cells. Adhesion is expressed relative to S. pyogenes in the absence of antiserum (100%), using duplicate samples from three independent experiments (n = 6). Asterisks indicate significant differences relative to the no-serum control (***, P < 0.0001). Similar results were obtained in assays with tonsil epithelium.
Spy0130 acts as a wall linker.
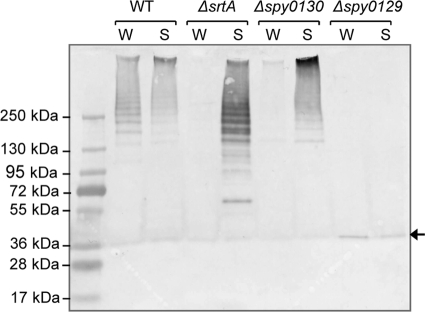
As subunits are joined covalently during pilus assembly in Gram-positive bacteria, pili of various lengths can be detected as high-molecular-weight ladders upon immunoblotting of cell wall preparations following separation by SDS-PAGE. As seen in Fig. 2, immunoblotting with anti-rSpy0128 serum detected polymerized pili in both the cell wall and, to a lesser extent, concentrated culture supernatant fractions of the wild-type S. pyogenes strain SF370. Loss of some pili from the bacterial surface during growth is mirrored by loss of other wall-anchored proteins, such as M protein, which was also detected in both wall and culture supernatant fractions (data not shown). Deletion of spy0129 from the bacterial chromosome abolishes the sortase encoded within the pilus gene cluster. No high-molecular-mass bands were detected in either wall or supernatant fractions of the Δspy0129 mutant, while a band corresponding to the size of the Spy0128 monomer was seen in both. This confirms the previously reported role of Spy0129 as a pilin polymerase (1, 32). A deletion mutant lacking the housekeeping sortase gene (ΔsrtA) retains the ability to polymerize pili but, in contrast to the wild-type parent strain, pili are found predominantly in culture supernatants (Fig. 2). This reveals a role for SrtA in anchoring pili to the cell wall. A mutant lacking the minor pilin subunit Spy0125 displayed the same pattern as the wild-type parent strain (not shown), but in a mutant lacking the minor pilin subunit Spy0130 the pattern was the same as with the ΔsrtA mutant, with pili found predominantly in the culture supernatants rather than wall fractions (Fig. 2). This indicates a role for Spy0130 as a wall linker at the base of pili, enabling them to be anchored to the wall by SrtA.
FIG. 2.
Localization of pili in wild-type (WT) S. pyogenes and various deletion mutants. Proteins in wall (W) and culture supernatant (S) fractions were separated by SDS-PAGE on a 4 to 15% (wt/vol) gel and immunoblotted with anti-rSpy0128. The arrow at the right indicates unpolymerized Spy0128 monomers. The intensity of the band detected in the Δspy0129 mutant suggests that monomers may be degraded if not incorporated into pili.
Localization of Spy0125 at the pilus tip.
Pilus polymerization in S. pyogenes strain SF370 involves cleavage of the major shaft subunit (Spy0128) between Thr311 and Gly312 of its C-terminal CWS motif (EVPTG), followed by covalent linkage of Thr311 to the side chain ɛ-amino group of Lys161 in a second copy of Spy0128. The Thr311 of this second copy is then similarly linked to Lys161 of another secreted Spy0128 monomer. Repeated rounds of transpeptidation occur, incorporating further monomers at the base to extend the pilus (18).
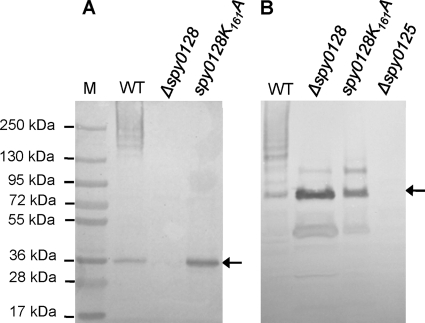
Unlike Spy0130 (which has a canonical LPXTG motif), the Val-Val-Pro-Thr-Gly-Val (VVPTGV) CWS motif of minor subunit Spy0125 is similar to the Glu-Val-Pro-Thr-Gly-Val (EVPTGV) CWS motif in Spy0128, and additional sequence homologies are apparent between the C-terminal regions of these subunits. This raised the possibility that incorporation of Spy0125 during pilus biogenesis also involves the side chain of Lys161 in Spy0128. To test this, site-directed and allelic replacement mutagenesis were combined to replace the Spy0128 Lys161 with Ala, by constructing point mutation spy0128.K161A in the chromosome of S. pyogenes strain SF370 (see Materials and Methods). Expression of the resulting mutant protein (Spy0128.K161A) was confirmed by immunoblotting SDS-PAGE gels of separated cell wall proteins with anti-rSpy0128 serum (Fig. 3A). While a ladder of high-molecular-weight bands indicative of polymerized pili was readily detected in wall fractions from the wild-type parent strain (and Spy0128 was not detectable in the Δspy0128 negative control), only subunit monomers were detected in the case of the spy0128.K161A mutant. Thus, the Spy0128.K161A mutant subunit was expressed but, as expected, could not be polymerized into pili.
FIG. 3.
Lys161 of Spy0128 is required for incorporation of Spy0125 into pili. Proteins in wall fractions of wild-type S. pyogenes (WT) and the indicated mutants were separated as described for Fig. 2 and immunoblotted with either anti-rSpy0128 (A) or anti-rSpy0125 (B) serum. Arrows at the right of each panel indicate unincorporated Spy0128 (A) and Spy0125 (B) monomers.
Figure 3B shows a duplicate of the SDS-PAGE gel from Fig. 3A, where anti-rSpy0125 serum, rather than anti-rSpy0128 serum, was used to detect proteins. High-molecular-weight ladders indicative of polymerized pili from the wild-type parent strain are visible in both panels A and B, confirming that these pili incorporate both Spy0128 and Spy0125. High-molecular-weight ladders were not detected by anti-Spy0125 serum in either the Δspy0128 deletion mutant or the spy0128.K161A mutant. In both these mutants, anti-rSpy0125 serum identified an intense band with an apparent molecular mass of ∼94 kDa, which corresponds to monomeric Spy0125 (as outlined below, this protein runs slowly on SDS-PAGE gels), as well as smaller faint bands corresponding to Spy0125 degradation products (see below). A larger band with an apparent molecular mass of ∼125 kDa was also visible. This was also present in the Δspy0128 mutant, showing that it cannot correspond to a Spy0125-Spy0128 heterodimer. Moreover, this band was not detected by anti-rSpy0128 serum (Fig. 3A). Therefore, like Spy0128, incorporation of Spy0125 into pili also requires Lys161 of Spy0128, and it was predicted that this would involve a sortase-mediated reaction linking Thr723 from the CWS motif of Spy0125 to the ɛ-amino group of Lys161 in Spy0128.
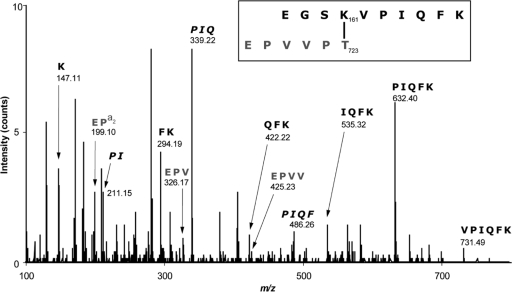
This linkage was confirmed in pili purified from the wild-type strain SF370, based on tryptic digest followed by LC-MS/MS analysis. This analysis identified an m/z 585.73+ species, which is consistent with the noncontinuous dipeptide containing the predicted isopeptide bond (Fig. 4; Table 2). The molecular mass of the cross-linked peptide was 18 Da lower than the sum of the two peptides, consistent with the loss of a water molecule due to isopeptide bond formation between the carboxylate of Thr723 (Spy0125) and the ɛ-amino group of Lys161 (Spy0128). MS/MS sequencing confirmed that this peptide contains components of both the C-terminal sequence of Spy0125 following sortase cleavage (i.e., Glu-Pro-Val-Val-Pro-Thr; residues 718 to 723) and a sequence around the nucleophilic Lys161 of Spy0128 (Glu-Gly-Ser-Lys-Val-Pro-Ile-Gln-Phe-Lys; residues 158 to 167). Although not definitively excluding linkage to the nearby Lys167 at the end of the Spy0128-derived peptide, the data indicate that this is highly unlikely, since Lys167 with a free ɛ-amino group (but not free Lys161) was detected in multiple daughter peptides (Table 2). Moreover, Lys167 was found in only two of nine aligned Spy0128 homologues from other S. pyogenes strains, in contrast to Lys161, which is completely conserved (18). These results suggest strongly that Spy0125 is confined to the tip of the pilus, as also demonstrated recently for the equivalent protein in the serotype M3 S. pyogenes T3 pilus (37).
FIG. 4.
MS/MS spectra of the peptide containing an intermolecular isopeptide bond between Spy0125 and Spy0128. The boxed sequence shows the tryptic peptide containing the Lys161-Thr723 intersubunit linkage at m/z 585.73+. Following loss of a water molecule, due to isopeptide bond formation, this peptide has an expected mass of 1,754.97 Da, which agrees with the observed mass of 1,754.04 Da. Internal fragments are shown in italics, and daughter ions are summarized in Table 2.
TABLE 2.
Daughter ions produced during MS/MS analysis of a peptide at m/z 585.73+ containing the intersubunit isopeptide bond between Spy0128 (Lys161) and Cpa (Thr723)
| Observed m/za | Charge state | Calculated m/zb | Δ(obs−calc)c | Proposed structure |
|---|---|---|---|---|
| 147.11 | +1 | 147.11 | 0 | K |
| 199.10 | +1 | 199.11 | −0.01 | EP (a2 ion) |
| 211.15 | +1 | 211.14 | 0.01 | PId |
| 294.19 | +1 | 294.18 | 0.01 | FK |
| 326.17 | +1 | 326.17 | 0 | EPV |
| 339.22 | +1 | 339.20 | 0.02 | PIQ |
| 422.22 | +1 | 422.24 | −0.02 | QFK |
| 425.23 | +1 | 425.24 | −0.01 | EPVV |
| 486.26 | +1 | 486.27 | −0.01 | PIQF |
| 535.32 | +1 | 535.32 | 0 | IQFK |
| 632.40 | +1 | 632.38 | 0.02 | PIQFK |
| 731.49 | +1 | 731.45 | 0.04 | VPIQFK |
Monoisoptic masses of the observed ions.
Calculated ions. Monoisotopic masses were calculated using the Fragment Ion Calculator (http://db.systemsbiology.net:8080/proteomicsToolkit/FragIonServlet.html).
Difference between observed ion mass and calculated ion mass.
Internal ions are shown in italics.
Spy0125 domains.
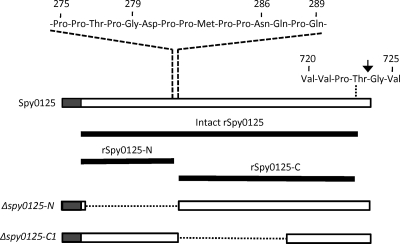
Purified rSpy0125 comprises residues Lys48 to Val725 of the spy0125 gene product (it excludes the N-terminal secretion signal and the 32 C-terminal residues downstream from the CWS motif), with four additional cloning vector-encoded residues (Gly-Ser-His-Met) incorporated at the N terminus (Fig. 5). The amino acid sequence indicates this protein has a molecular mass of 76,722 Da, but it displayed an apparent molecular mass of ∼94 kDa when run on SDS-PAGE gels. Despite purification to apparent homogeneity, storage of the protein for more than a few days at 4°C resulted in degradation, leading eventually to two major products with molecular masses of ∼50 kDa and ∼25 kDa. These were readily separated by size exclusion chromatography. Intact mass spectrometry and Edman sequencing were used to characterize the two purified fragments and determine their origins in Spy0125. Edman sequencing indicated the N terminus of the larger fragment to be Asn-Gln-Pro-Gln-Thr-Thr-, which corresponds to the sequence beginning at Asn286 of Spy0125. Fourier transform ion cyclotron resonance (FT-ICR)-MS determined the mass of this fragment to be 49,769 Da, which corresponds closely, although not exactly, to the theoretical mass of 49,354 Da for the Spy0125 sequence extending from Asn286 to Val725. The 415-Da discrepancy between the determined and theoretical masses might be explained by the determined mass being an average of several unresolved species with slightly varied ends, or a posttranslational modification, but this was not explored further, as the data were sufficient to show that the large fragment was derived from the C-terminal two-thirds of Spy0125. The mass of the smaller (∼25-kDa) fragment was also determined by FT-ICR-MS and precisely describes a fragment comprising Gly52 to Gly279 of intact Spy0125, defining the N-terminal one-third of Spy0125. It is notable that these fragments are separated in intact Spy0125 by a short proline-rich sequence (Fig. 5), which is likely to be an unstructured region, indicating that the fragments may form distinct domains in Spy0125. These regions were designated Spy0125-N and Spy0125-C, respectively.
FIG. 5.
Spy0125 recombinant proteins and deletion mutants. The upper horizontal box represents Spy0125, with shading highlighting the N-terminal secretion sequence. The cell wall sorting motif is shown above the box on the right, with the arrow highlighting the sortase cleavage site between Thr723 and Gly724. The Pro-rich sequence separating the N- and C-terminal domains is shown at the top, with the positions of residues mentioned in the text numbered. The solid lines below the box show the extents of the indicated recombinant proteins, which include a few additional vector-encoded residues at their N-terminal ends (Gly-Ser-His-Met in intact rSpy0125 and Gly-Ser in both rSpy0125-N and rSpy0125-C). The lower horizontal boxes describe the indicated deletion mutants, with the deleted sequences represented by dotted lines.
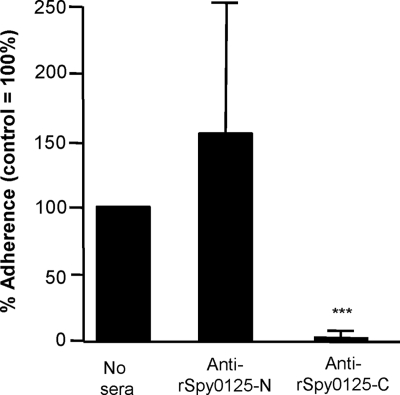
To explore the roles of Spy0125-N and Spy0125-C in cell adhesion, PCR-amplified spy0125 sequences encoding each domain were cloned separately in an expression vector, and the resulting recombinant proteins (Fig. 5) were purified and used to raise specific antisera. The purified rSpy0125-N protein, but not purified rSpy0125-C, bound readily to HaCaT cells (data not shown). It was, therefore, surprising that only the anti-Spy0125-C serum inhibited binding of the parent S. pyogenes strain SF370 to HaCaT cells, while anti-rSpy0125-N serum had little or no effect on bacterial adhesion (Fig. 6).
FIG. 6.
Effects of antisera to the recombinant N- and C-terminal regions of Spy0125 on adhesion of S. pyogenes SF370 to HaCaT cells. Adhesion is expressed relative to S. pyogenes in the absence of antiserum (100%), using duplicate samples from three independent experiments (n = 6). Asterisks indicate significant differences relative to the no-serum control (***, P < 0.0001).
Localization of the adhesin domain within Spy0125.
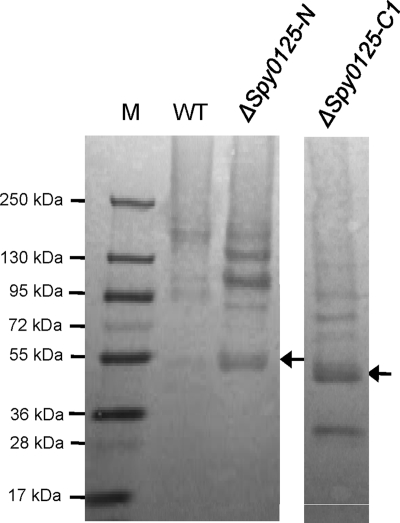
In order to further explore the roles of the different regions of Spy0125, allelic replacement mutagenesis was used to construct two deletion mutants in the S. pyogenes SF370 genome (Fig. 5). Both were designed to retain the Spy0125 secretion signal sequence (residues 1 to 48) and C-terminal sequences that might be required for the secreted (mutant) proteins to be recognized by the pilin polymerase. One deletion, designated Δspy0125-N, encoded a mutant Spy0125 lacking N-terminal residues extending from Leu53 (just downstream of the secretion signal peptide) to Pro285 (at the junction of the two putative domains). The second, designated Δspy0125-C1, encoded a mutant Spy0125 lacking central sequences extending from Asn286 to Pro559. Proteins from cell wall fractions of the parent and mutant strains were separated on SDS-PAGE gels and immunoblotted with anti-rSpy0125 (Fig. 7). High-molecular-weight ladders (indicative of polymerized pili) were detected in all three strains, indicating that the deleted sequences had not prevented incorporation of the mutant Spy0125 derivatives into pili. The lower band apparent in the Δspy0125-C1 lane corresponds in size to Spy0125-N, suggesting that this separates from the remaining C-terminal sequences at the linking Pro-rich sequence.
FIG. 7.
Anti-rSpy0125 immunoblot of wall fractions from the wild-type (WT) S. pyogenes parent strain and spy0125 deletion mutants. The arrows on the right of each panel indicate the positions of the unincorporated mutant Spy0125 monomers. M, molecular mass markers.
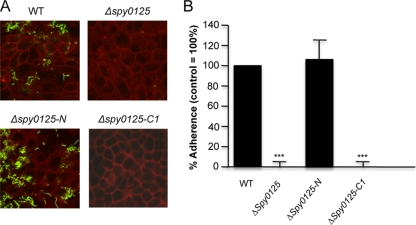
The abilities of the Δspy0125-N and Δspy0125-C1 mutant strains to adhere to HaCaT cells were determined. Despite the previously observed binding of purified rSpy0125-N (above), the Δspy0125-N mutant, which lacks this domain, displayed no significant reduction in its capacity to adhere to HaCaT cells, binding at least as efficiently as the wild-type S. pyogenes SF370 parent strain (Fig. 8). In contrast, the Δspy0125-C1 mutant failed to adhere to HaCaT cells, displaying the same phenotype as the mutant lacking the entire spy0125 gene (Fig. 8). Together with the observation that only anti-rSpy0125-C serum was able to inhibit bacterial binding, these results reveal that Spy0125-mediated adhesion is determined by a region within the center of the molecule.
FIG. 8.
Adhesion of S. pyogenes wild-type (WT) parent and spy0125 mutants to HaCaT cells. (A) Bacteria (green fluorescence) adhering to HaCaT cells (red fluorescence) in the case of the wild-type parent and the Δspy0125-N deletion mutant. Very few adherent bacteria are visible in the case of the Δspy0125 control or the Δspy0125-C1 mutant. (B) Extent of adherence relative to the wild type (100%), based on five independent experiments (n = 15). Asterisks indicate significant differences from the wild-type parent strain (***, P < 0.0001).
DISCUSSION
The contribution of the major shaft subunit (Spy0128) in pili from the M1 S. pyogenes strain SF370 to pilus length and stability is now well established (1, 18, 29, 32), but the roles of the minor pilin subunits (Spy0125 and Spy0130) have not been clearly defined. Previous studies showed that recombinant purified forms of both minor subunits bind to the human pharyngeal Detroit cell line (29), suggesting both may act as pilus adhesins. If this were the case, it might be expected that deletion of either minor subunit alone would result in a partial, but not complete, reduction in S. pyogenes adhesion to target cells. However, as reported previously (1), neither the Δspy0125 nor the Δspy0130 deletion mutant binds to target cells to any significant extent. This suggested that the minor subunits play distinct but equally essential roles. The results presented here reveal that Spy0125 is clearly a pilus adhesin, and they provide evidence that Spy0130 acts as a wall linker protein.
Spy0130 is the only pilin subunit to possess a canonical Leu-Pro-Ser-Thr-Gly-Glu CWS motif that is typically a substrate for class A sortases, such as the housekeeping sortase (SrtA), that attach proteins to lipid II peptidoglycan precursors (8). Experiments reported here show that both Spy0130 and SrtA are required to anchor pili in the S. pyogenes strain SF370 cell wall but are not needed for pilus polymerization. Both other subunits possess very distinct CWS motifs (Val-Val-Pro-Thr-Gly-Val and Glu-Val-Pro-Thr-Gly-Val) that are typical substrates for class B sortases, such as the pilin polymerase encoded by spy0129 (7, 8). Studies on class A and class B sortases from Staphylococcus aureus highlight their marked substrate specificities (8, 30), indicating it is highly unlikely that Spy0129 would recognize the Spy0130 CWS motif as a substrate. Therefore, the only location where Spy0130 could be incorporated into pili would be at the base, where a specific Lys residue (to be identified) in Spy0130 can act as the target that resolves the acyl-enzyme intermediate, including the terminal pilus shaft (Spy0128) subunit. It is worth noting that the pilin polymerases found in other Gram-positive bacteria, such as C. diphtheriae, S. agalactiae, and S. pneumoniae, in which wall linker subunits have been observed by immunogold labeling at intervals along the pilus are, in all cases, class C sortases that display greater functional overlap with class A sortases. Moreover, in the case of S. pneumoniae, earlier reports on the localization of minor subunits were not consistent, and more recent studies have shown that they are limited to either end of the assembled pilus, where one acts as an adhesin and the second as a wall linker (15). Taken together, available data suggest strongly that Spy0130 is limited to the base of assembled pili, where it is very unlikely to contribute directly to adhesion. The previously reported adhesion defect of Δspy0130 mutants (1) is more likely to reflect a failure to anchor pili firmly on the bacterial surface.
In C. diphtheriae, a minor wall linker subunit (SpaB) limits pilus length by acting as the terminal subunit at the base of the pili, and mutants lacking this subunit release longer-than-normal pili into the culture medium (26). The lengths of pili recovered from culture supernatants of the Δspy0130 mutant were not measured during the present study, but it was notable that in immunoblot assays these pili consistently stained more intensely in the higher-molecular-mass region near the tops of gels than pili produced by the wild-type parent (Fig. 2). This suggests that, like SpaB in C. diphtheriae, Spy0130 may limit pilus length in S. pyogenes. Similarly, immunoblot assay results also suggested that a higher proportion of shorter pili are produced by the ΔsrtA mutant than by the parent strain, implying that Spy0130 may be incorporated more readily at the base of pili in the absence of SrtA. The reasons for this are not understood, and further studies will be required to explore interactions between the individual subunits and sortases that may regulate pilus biogenesis.
A common characteristic of pilus-associated adhesins in other bacteria is their localization at the tip of the pilus, where they project away from the bacterial surface in a manner that presumably facilitates interactions with target receptors. Recent studies have demonstrated that the homologue of Spy0125 (called Cpa) in a serotype M3 S. pyogenes strain is localized at the tip of pili (37). There is considerable diversity among the loci (known as FCT regions) encompassing pilus gene clusters in different strains of S. pyogenes, and the FCT-3 region of M3 S. pyogenes differs from the FCT-1 region of the M1 strain SF370 studied here (9, 21). Nevertheless, by using LC-MS/MS analysis and the spy0128K161A mutant, Spy0125 was shown to be joined to the pilus shaft through an intermolecular isopeptide bond linking its C-terminal Thr723 to the ɛ-amino group of Lys161 in Spy0128. Because this same Lys residue is utilized as the target in polymerizing Spy0128 subunits (18), addition of Spy0125 would block further linkages at this site and, consequently, Spy0125 could only be incorporated at the tip of polymerized pili. However, it remains possible that Spy0125 is linked directly to Spy0130 by the pilin polymerase, employing the same target as used to link Spy0128 to Spy0130. Although yet to be confirmed, it is speculated that the ∼125-kDa band detected in wall fractions of the Δspy0128 and Δspy0128K161A mutants may represent a wall-anchored Spy0125-Spy0130 heterodimer. However, previous studies showing that all three pilus subunits are required for adhesion (1) argue strongly that Spy0125 functions only when presented by pili. This differs from S. agalactiae, for which a pilus adhesin anchored to the wall in the absence of the pilus shaft subunit mediated attachment to epithelial cells as effectively as the piliated parent strain in standard adhesion assays, although pilus integrity was essential under certain stress conditions (7, 20).
Very recently, a crystal structure of RrgA, the pilus tip adhesin from S. pneumoniae strain TIGR4, was reported, and the existence of homologues in S. pyogenes was highlighted (17). While a minority of S. pyogenes strains (those possessing an FCT-6 pilus gene cluster) do encode RrgA homologues, the majority of S. pyogenes strains do not. Neither the Spy0125 adhesin described here nor its homologues (sometimes designated Cpa) found in many other S. pyogenes strains share any significant sequence similarity with RrgA. Consequently, Spy0125 represents a very distinct type of pilus tip adhesin.
Following purification, rSpy0125-N degrades into multiple small peptides within a few weeks, suggesting that this region of Spy0125 is unstable, whereas rSpy0125-C remains stable for many months upon storage at 4°C (data not shown). Frequently, immunoblotting of S. pyogenes cell wall fractions with anti-rSpy0125 serum detected a band corresponding in size to the more stable C-terminal breakdown fragment (Fig. 6, WT lane), suggesting that breakdown of Spy0125 can also occur in vivo. Apart from the apparently unstructured nature of the Pro-rich region between residues 275 and 285, it is not clear why this region is prone to spontaneous breakdown. The failure of the Δspy0125-N deletion to have any apparent effect on adhesion indicates that the unstable N-terminal region is entirely dispensable and raises intriguing questions concerning its role in Spy0125. In this context, it is noteworthy that spy0125 gene homologues in S. pyogenes serotypes M5 and M18 lack sequences encoding this N-terminal region (16, 42).
The abilities of the various antisera employed in this study to inhibit adhesion of wild-type bacteria to HaCaT cells were entirely consistent with the effects on adhesion of the Δspy0125, Δspy0125-N, and Δspy0125-C1 deletion mutants (1) (Fig. 6). These findings indicate clearly that Spy0130 and the N-terminal region of Spy0125 make little (if any) direct contribution to bacterial adhesion. The reasons why the purified rSpy0130 and rSpy0125-N proteins can bind to HaCaT cells are not clear but may reflect nonspecific interactions of partially unfolded proteins, as neither rSpy0125-N (above) nor rSpy0130 (43) adopt a stable or well-folded stable structure. A more unexpected observation was that the purified rSpy0125-C failed to bind to HaCaT cells, despite clear evidence that this region encompasses the key bacterial adhesin. It has to be presumed that there is a difference between the recombinant protein expressed and purified from E. coli and the corresponding region of Spy0125 presented by S. pyogenes at the pilus tips, or that the assay conditions were not conducive to recombinant protein binding (e.g., the protein produced in vitro may be more susceptible to tissue culture media components than when presented on pili). This, together with the results described above for rSpy0125-N and rSpy0130, highlights the importance of testing the functions of proteins in their natural in vivo context rather than relying solely on purified recombinant proteins. Further studies to more precisely define the mechanism of Spy0125-mediated adhesion will, therefore, require the construction and analysis of additional mutants in S. pyogenes.
Acknowledgments
This work was supported by the Medical Research Council (MRC) UK, through grant G0400849. J.P. was supported by an MRC studentship (G0600309). M.J.B. is supported by the Royal Society (United Kingdom) through the award of a University Research Fellowship.
We thank J. Gray, Pinnacle Unit, Newcastle University, for assistance with MS analysis of purified Spy0125 fragments.
Footnotes
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Abbot, E. L., W. D. Smith, G. P. Siou, C. Chiriboga, R. J. Smith, J. A. Wilson, B. H. Hirst, and M. A. Kehoe. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 9:1822-1833. [DOI] [PubMed] [Google Scholar]
- 2.Bagnoli, F., M. Moschioni, C. Donati, V. Dimitrovska, I. Ferlenghi, C. Facciotti, A. Muzzi, F. Giusti, C. Emolo, A. Sinisi, M. Hilleringmann, W. Pansegrau, S. Censini, R. Rappuoli, A. Covacci, V. Masignani, and M. A. Barocchi. 2008. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J. Bacteriol. 190:5480-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 103:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budzik, J. M., L. A. Marraffini, and O. Schneewind. 2007. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66:495-510. [DOI] [PubMed] [Google Scholar]
- 5.Budzik, J. M., S. Y. Oh, and O. Schneewind. 2008. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J. Biol. Chem. 283:36676-36686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401-1413. [DOI] [PubMed] [Google Scholar]
- 8.Dramsi, S., S. Magnet, S. Davison, and M. Arthur. 2008. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol. Rev. 32:307-320. [DOI] [PubMed] [Google Scholar]
- 9.Falugi, F., C. Zingaretti, V. Pinto, M. Mariani, L. Amodeo, A. G. Manetti, S. Capo, J. M. Musser, G. Orefici, I. Margarit, J. L. Telford, G. Grandi, and M. Mora. 2008. Sequence variation in group A Streptococcus pili and association of pilus backbone types with Lancefield T serotypes. J. Infect. Dis. 198:1834-1841. [DOI] [PubMed] [Google Scholar]
- 10.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 12.Fontaine, M. C., J. J. Lee, and M. A. Kehoe. 2003. Combined contributions of streptolysin O and streptolysin S to virulence of serotype M5 Streptococcus pyogenes strain Manfredo. Infect. Immun. 71:3857-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaspar, A. H., and H. Ton-That. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 188:1526-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttilla, I. K., A. H. Gaspar, A. Swierczynski, A. Swaminathan, P. Dwivedi, A. Das, and H. Ton-That. 2009. Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in gram-positive bacteria. J. Bacteriol. 191:5603-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilleringmann, M., P. Ringler, S. A. Muller, G. De Angelis, R. Rappuoli, I. Ferlenghi, and A. Engel. 2009. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 28:3921-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holden, M. T., A. Scott, I. Cherevach, T. Chillingworth, C. Churcher, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, K. Mungall, M. A. Quail, C. Price, E. Rabbinowitsch, S. Sharp, J. Skelton, S. Whitehead, B. G. Barrell, M. Kehoe, and J. Parkhill. 2007. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain Manfredo. J. Bacteriol. 189:1473-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izore, T., C. Contreras-Martel, L. El Mortaji, C. Manzano, R. Terrasse, T. Vernet, A. M. Di Guilmi, and A. Dessen. 2010. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 18:106-115. [DOI] [PubMed] [Google Scholar]
- 18.Kang, H. J., F. Coulibaly, F. Clow, T. Proft, and E. N. Baker. 2007. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 318:1625-1628. [DOI] [PubMed] [Google Scholar]
- 19.Kang, H. J., N. G. Paterson, A. H. Gaspar, H. Ton-That, and E. N. Baker. 2009. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. U. S. A. 106:16967-16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konto-Ghiorghi, Y., E. Mairey, A. Mallet, G. Dumenil, E. Caliot, P. Trieu-Cuot, and S. Dramsi. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kratovac, Z., A. Manoharan, F. Luo, S. Lizano, and D. E. Bessen. 2007. Population genetics and linkage analysis of loci within the FCT region of Streptococcus pyogenes. J. Bacteriol. 189:1299-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnan, V., A. H. Gaspar, N. Ye, A. Mandlik, H. Ton-That, and S. V. Narayana. 2007. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure 15:893-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B Streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 24.LeMieux, J., D. L. Hava, A. Basset, and A. Camilli. 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 74:2453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LeMieux, J., S. Woody, and A. Camilli. 2008. Roles of the sortases of Streptococcus pneumoniae in assembly of the RlrA pilus. J. Bacteriol. 190:6002-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandlik, A., A. Das, and H. Ton-That. 2008. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2007. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 64:111-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manetti, A. G., C. Zingaretti, F. Falugi, S. Capo, M. Bombaci, F. Bagnoli, G. Gambellini, G. Bensi, M. Mora, A. M. Edwards, J. M. Musser, E. A. Graviss, J. L. Telford, G. Grandi, and I. Margarit. 2007. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64:968-983. [DOI] [PubMed] [Google Scholar]
- 30.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 32.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U. S. A. 102:15641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson, A. L., J. Ries, F. Bagnoli, S. Dahlberg, S. Falker, S. Rounioja, J. Tschop, E. Morfeldt, I. Ferlenghi, M. Hilleringmann, D. W. Holden, R. Rappuoli, S. Normark, M. A. Barocchi, and B. Henriques-Normark. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66:329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nobbs, A. H., R. Rosini, C. D. Rinaudo, D. Maione, G. Grandi, and J. L. Telford. 2008. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect. Immun. 76:3550-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry, A. M., H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 277:16241-16248. [DOI] [PubMed] [Google Scholar]
- 37.Quigley, B. R., D. Zahner, M. Hatkoff, D. G. Thanassi, and J. R. Scott. 2009. Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Mol. Microbiol. 72:1379-1394. [DOI] [PubMed] [Google Scholar]
- 38.Rosini, R., C. D. Rinaudo, M. Soriani, P. Lauer, M. Mora, D. Maione, A. Taddei, I. Santi, C. Ghezzo, C. Brettoni, S. Buccato, I. Margarit, G. Grandi, and J. L. Telford. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61:126-141. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Scott, J. R., and T. C. Barnett. 2006. Surface proteins of gram-positive bacteria and how they get there. Annu. Rev. Microbiol. 60:397-423. [DOI] [PubMed] [Google Scholar]
- 41.Scott, J. R., and D. Zahner. 2006. Pili with strong attachments: Gram-positive bacteria do it differently. Mol. Microbiol. 62:320-330. [DOI] [PubMed] [Google Scholar]
- 42.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. U. S. A. 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solovyova, A. S., J. A. Pointon, P. R. Race, W. D. Smith, M. A. Kehoe, and M. J. Banfield. 2009. Solution structure of the major (Spy0128) and minor (Spy0125 and Spy0130) pili subunits from Streptococcus pyogenes. Eur. Biophys. J. 39:469-480. [DOI] [PubMed] [Google Scholar]
- 44.Swaminathan, A., A. Mandlik, A. Swierczynski, A. Gaspar, A. Das, and H. Ton-That. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 66:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 46.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53:251-261. [DOI] [PubMed] [Google Scholar]
- 47.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 48.Whatmore, A. M., and M. A. Kehoe. 1994. Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in Vir regulons. Mol. Microbiol. 11:363-374. [DOI] [PubMed] [Google Scholar]