Abstract
Pasteurella multocida is the causative agent of a number of diseases in animals, including fowl cholera. P. multocida strains simultaneously express two lipopolysaccharide (LPS) glycoforms (glycoforms A and B) that differ only in their inner core structure. Glycoform A contains a single 3-deoxy-d-manno-octulosonic acid (Kdo) residue that is phosphorylated by the Kdo kinase, KdkA, whereas glycoform B contains two unphosphorylated Kdo residues. We have previously shown that P. multocida mutants lacking the heptosyltransferase, HptA, produce full-length glycoform B LPS and a large amount of truncated glycoform A LPS, as they cannot add heptose to the glycoform A inner core. These hptA mutants were attenuated in chickens because the truncated LPS made them vulnerable to host defense mechanisms, including antimicrobial peptides. However, here we show that birds inoculated with high doses of the hptA mutant developed fowl cholera and the P. multocida isolates recovered from diseased birds no longer expressed truncated LPS. Sequencing analysis revealed that the in vivo-derived isolates had mutations in kdkA, thereby suppressing the production of glycoform A LPS. Interestingly, a number of the spontaneous KdkA mutant strains produced KdkA with a single amino acid substitution (A112V, R123P, H168Y, or D193N). LPS structural analysis showed that complementation of a P. multocida kdkA mutant with wild-type kdkA restored expression of glycoform A to wild-type levels, whereas complementation with any of the mutated kdkA genes did not. We conclude that in P. multocida KdkA, the amino acids A112, R123, H168, and D193 are critical for Kdo kinase function and therefore for glycoform A LPS assembly.
Pasteurella multocida is a Gram-negative pathogen that is the causative agent of a wide range of diseases in animals, including bovine hemorrhagic septicemia, fowl cholera, and porcine atrophic rhinitis (4). The major virulence determinants in P. multocida include the capsule and lipopolysaccharide (LPS), both of which vary in composition and structure between strains (2, 5, 7-8, 11, 21-23). The LPS of P. multocida is composed of an inner and an outer core region, and like the LPS produced by species within the Neisseria and Haemophilus genera, P. multocida LPS lacks the polymeric O antigen that is attached to the distal end of the LPS structure in most other Gram-negative bacteria (6, 9, 17, 20, 23). Structural analysis of the LPS isolated from a number of P. multocida strains revealed that most simultaneously produce two LPS glycoforms that differ only in their inner core structure (23; A. Cox, unpublished observations). The key difference between the two structures is that the glycoform A inner core contains a single phosphorylated 3-deoxy-d-manno-octulosonic acid (Kdo) residue (lipid A-Kdo1-P), with the phosphate group frequently being substituted with phosphoethanolamine (PEtn), whereas the glycoform B inner core contains two Kdo residues (lipid A-Kdo1-Kdo2) (22).
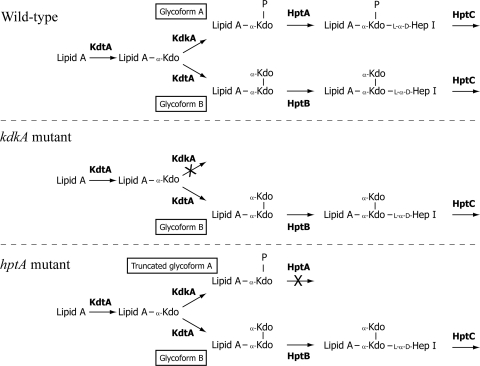
The enzymes required for assembly of the LPS inner core have been identified and include a Kdo transferase, KdtA, that is predicted to add both the first and the second (glycoform B only) Kdo residues; a Kdo kinase, KdkA, that adds the phosphate group to the lipid A-Kdo1 for assembly of glycoform A; and two heptosyltransferases, HptA and HptB, specific for the addition of the first heptose to glycoform A and glycoform B, respectively (Fig. 1) (10). Previous data suggest that the amount of each glycoform produced is primarily dependent upon the phosphorylation activity of the glycoform A-specific Kdo kinase: if the lipid A-Kdo1 acceptor molecule is phosphorylated by KdkA, the molecule is recognized by HptA and glycoform A is assembled, whereas if lipid A-Kdo1 remains unphosphorylated, an additional Kdo is added by the Kdo transferase (KdtA) and glycoform B is assembled (Fig. 1) (10).
FIG. 1.
Schematic representation of LPS inner core assembly in the P. multocida wild-type (VP161), kdkA mutant (AL721), and hptA mutant (AL836). Two LPS inner core structures, glycoforms A and B, are observed. The enzymes required for selected steps in the biosynthesis of each glycoform are shown in boldface. Residues are heptose (Hep), 3-deoxy-d-manno-octulosonate (Kdo), and phosphate (P).
In previous studies with P. multocida strain VP161, we systematically inactivated each of the LPS transferase genes and the Kdo kinase gene (kdkA) and analyzed the effects of these mutations on the LPS structure and virulence in chickens (3, 10). In kdkA mutants, there is no phosphorylation of lipid A-Kdo1, so all lipid A-Kdo1 acceptor molecules are used to produce full-length glycoform B (Fig. 1). These kdkA mutants are fully virulent in chickens infected by the intramuscular (i.m.) route (10). In contrast, P. multocida hptA mutants produce full-length glycoform B but lack the first heptosyltransferase (HptA) required for assembly of glycoform A beyond lipid A-Kdo1-P (Fig. 1) (10). These hptA mutants are fully attenuated in chickens due to the production of a large amount of truncated LPS, rendering them vulnerable to host defense mechanisms, such as antimicrobial peptides (10).
In the present study, we show that under in vivo selective pressure, avirulent P. multocida hptA mutants can spontaneously revert to full virulence by way of secondary kdkA suppressor mutations. These virulent isolates produce full-length glycoform B LPS and no longer produce any truncated LPS. Sequencing analysis of the mutated kdkA genes amplified from each of the recovered in vivo hptA mutants demonstrated that they all contained single nucleotide substitutions or deletions. Importantly, four of the mutated kdkA genes were intact, but each encoded a single amino acid substitution. Further analysis confirmed that each amino acid substitution resulted in the loss of Kdo kinase activity. This is the first report that defines the amino acids essential for bacterial Kdo kinase activity, a critical enzyme in LPS assembly.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown routinely in Luria-Bertani broth. P. multocida strains were grown in brain heart infusion (BHI) broth. Solid media were obtained by the addition of 1.5% agar. When required, the media were supplemented with spectinomycin (100 μg/ml), kanamycin (50 μg/ml), or tetracycline (2.5 μg/ml).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | deoR endA1 gyrA96 hsdR17(rK− mK+) recA1 relA1 supE44 thi-1 (lacZYA-argFV169), φ80dlacZΔM15 F− | Bethesda Research Laboratories |
| P. multocida | ||
| VP161 | Serotype A:1, Vietnamese isolate from chickens | 27 |
| AL435 | VP161 carrying a Tn916 insertion in gene pm1417, fully virulent | 10 |
| AL721 | kdkA mutant of AL435, Specr | 10 |
| AL836 | Nonpolar hptA mutant of AL435, Specr | 10 |
| AL839 | AL721 containing pAL99, Specr Kanr | 10 |
| AL840, AL1808 | AL721 containing pAL462, Specr Kanr | 10 |
| AL1774, AL1775 | Spontaneous A112V kdkA mutants of AL836, Specr | This study |
| AL1777, AL1778 | Spontaneous R123P kdkA mutants of AL836, Specr | This study |
| AL1780, AL1781 | Spontaneous D193N kdkA mutants of AL836, Specr | This study |
| AL1782, AL1783 | Spontaneous H168Y kdkA mutants of AL836, Specr | This study |
| AL1799 | AL721 containing pAL856, Specr Kanr | This study |
| AL1800 | AL721 containing pAL857, Specr Kanr | This study |
| AL1801 | AL721 containing pAL858, Specr Kanr | This study |
| AL1802 | AL721 containing pAL860, Specr Kanr | This study |
| AL1803 | AL721 containing pAL861, Specr Kanr | This study |
| AL1804 | AL721 containing pAL862, Specr Kanr | This study |
| AL1805 | AL721 containing pAL863, Specr Kanr | This study |
| AL1806 | AL721 containing pAL864, Specr Kanr | This study |
| Plasmids | ||
| pAL99 | P. multocida expression plasmid (Kanr), constitutive tpiA promoter upstream of cloning site | 11 |
| pAL462 | Wild-type kdkA, cloned into pAL99 | 10 |
| pAL856, pAL857 | kdkA A112V, cloned into pAL99, independent clonings | This study |
| pAL858, pAL860 | kdkA R123P, cloned into pAL99, independent clonings | This study |
| pAL861, pAL862 | kdkA H168Y, cloned into pAL99, independent clonings | This study |
| pAL863, pAL864 | kdkA D193N, cloned into pAL99, independent clonings | This study |
| Primers | ||
| BAP2782 | GCCCTACACAAATTGGGAGA, pUA826-specific primer | 10 |
| BAP3327 | GCACGCCTGCTGTGCTTTAG, reverse primer in kdkA gene | This study |
| BAP3914 | AAAGGGATCCGTGGCCATTCGCTATCTCTG, forward primer flanking kdkA for expression, contains BamHI site | 10 |
| BAP3915 | TATGTGTCGACCTTATGCTTGATATCCCGC, reverse primer flanking kdkA for expression, contains SalI site | 10 |
| BAP3966 | CACACAGGATCCATTGGACGAGATAATGATGCCG, forward primer flanking hptA, contains BamHI site | 10 |
| BAP3967 | ATAACAGTCGACGGTTCATCATTGAACGTC, reverse primer flanking hptA, contains SalI site | 10 |
DNA manipulations.
Restriction digests and ligations were performed according to the manufacturers' instructions using enzymes obtained from NEB (Ipswich, MA) or Roche Diagnostics (Mannheim, Germany). Plasmid DNA was prepared using Qiagen plasmid minikits (Hamburg, Germany), while genomic DNA was prepared using the cetyltrimethylammonium bromide (CTAB) method (1). PCR amplification of DNA was performed using Taq DNA polymerase or an Expand high-fidelity PCR system (Roche Diagnostics) and purified using a QIAquick PCR purification kit from Qiagen GmbH (Hamburg, Germany). The oligonucleotides used in this study are listed in Table 1. Sequencing reactions were performed using the Applied Biosystems Prism BigDye Terminator mix, version 3.1. Electropherograms were generated on an Applied Biosystems 3730S genetic analyzer and were analyzed using the Vector NTI Advance, version 10, program (Invitrogen, Carlsbad, CA).
In trans complementation of the P. multocida kdkA mutant.
We previously constructed P. multocida kdkA mutant AL721 and showed that complementation of this strain with the wild-type kdkA gene, encoded on plasmid pAL462, restored expression of glycoform A LPS (10). For the complementation experiments with the mutated kdkA genes, two independent plasmid constructs were generated for each mutation, as follows. The kdkA gene was amplified directly from each of the P. multocida hptA kdkA double mutants (Table 1), using oligonucleotides BAP3914 and BAP3915. Each amplified product was then ligated into SalI- and BamHI-digested pAL99 (Table 1), such that transcription of kdkA would be driven by the constitutive P. multocida tpiA promoter. The nucleotide sequence of each of the recombinant plasmids was determined to check the fidelity of the cloned gene and the tpi promoter region, and then each plasmid was introduced into P. multocida kdkA mutant AL721, generating complemented strains AL1799 through AL1806 (Table 1). As a control, the pAL99 vector was introduced separately into the kdkA mutant, generating strain AL839 (Table 1).
Structural analyses of LPS.
LPS was isolated from plate-grown cells as described previously (3, 10). Sugars were identified as their alditol acetate derivatives and linkage analysis determined following methylation analysis by gas-liquid chromatography-mass spectrometry (MS), as described previously (22). Capillary electrophoresis (CE)-electrospray (ES)-MS analysis was performed as described previously (22).
Assessment of virulence.
Virulence trials with chickens were performed as described previously (10). To check the phenotypes of the strains recovered from infected chickens, muscle swab and blood samples were plated onto BHI agar and BHI agar containing spectinomycin. The genotypes of the recovered strains with respect to the hptA gene were assessed using two different PCR assays. To identify the P. multocida hptA mutant, a primer located outside hptA (primer BAP3327) was paired with a primer specific for the integrated plasmid located within hptA (primer BAP2782). The amplification of a 700-bp product indicated that the strain still contained an insertion within hptA. To identify P. multocida wild-type revertant strains, primers flanking the hptA gene (primers BAP3966 and BAP3967) were used. P. multocida wild-type revertant strains were identified by the PCR amplification of a 1,050-bp product, indicating the loss of the plasmid insertion within hptA. To determine the LPS phenotype of the recovered strains, proteinase K-treated whole-cell lysates of the bacteria recovered from each in vivo site were separated by SDS-PAGE and visualized by carbohydrate silver staining, as described previously (25).
Bioinformatic analysis.
Amino acid sequence alignments were generated using COBALT, a constraint-based multiple-alignment tool that utilizes conserved domain and local sequence similarity information (18).
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of the P. multocida VP161 kdkA gene is HM448896.
RESULTS
P. multocida hptA mutants isolated from the blood of chickens with fowl cholera express only full-length LPS.
We previously observed that birds injected i.m. with 70 CFU of the P. multocida hptA single-crossover insertion mutant AL836 displayed no signs of disease (10). To extend these data, a virulence trial was conducted with chickens to determine the 50% infectious dose of the hptA mutant. Three groups of seven birds each were injected i.m. with 70, 7 × 103, or 7 × 105 CFU. As expected (10), there were no signs of disease in the birds injected with the lowest dose of the hptA mutant, but two birds injected with 7 × 103 CFU and all birds injected with 7 × 105 CFU developed acute fowl cholera. As the hptA mutant was known to be attenuated in chickens, we hypothesized that the causative agent of disease was a wild-type revertant strain that had lost the single-crossover insertion from within hptA. Indeed, we have reported in a previous study that under in vivo selective pressure, wild-type revertant strains can arise spontaneously within a population of P. multocida single-crossover mutants and cause disease (3). Virulent, wild-type revertant strains are no longer spectinomycin resistant, as they have lost the single crossover insertion that encodes the spectinomycin resistance gene aadA. Therefore, to determine the phenotype of the P. multocida strain responsible for disease, blood and muscle (from the site of injection) tissue samples were collected at necropsy from six of the diseased birds and cultured on BHI agar and BHI agar with spectinomycin. In all birds, the P. multocida organism recovered from the site of injection was able to grow on BHI agar with spectinomycin, indicating the presence of the hptA mutant in the muscle tissue. Furthermore, spectinomycin-resistant P. multocida was recovered from the blood of four of the six diseased birds, suggesting that the hptA mutant was able to cause systemic disease. To determine if both mutant and wild-type revertant P. multocida isolates were present in the samples, the bacteria on the nonselective plates were patched onto selective (spectinomycin, 50 μg/ml) and nonselective plates (Table 2). Four of the six birds (birds 18, 21, 25, and 53) had the hptA mutant present in the blood and at the intramuscular injection site. In addition, bird 18 also had wild-type revertant P. multocida present at both sites. Although there was evidence that the hptA mutant was present at the injection site in birds 23 and 24, only wild-type revertant P. multocida was detected in the blood and these bacteria were not investigated further (Table 2). The spectinomycin-resistant P. multocida isolates from the blood of four birds (birds 18, 21, 25, and 53) were chosen for further study.
TABLE 2.
Analysis of in vivo-derived P. multocida isolatesa
| Bird | Time of death (h)b | Muscle |
Blood |
||
|---|---|---|---|---|---|
| % Specrc | No. positive for pUA826:: hptA/total no. testedd | % Specr | No. positive for pUA826:: hptA/total no. testedd | ||
| 18 | 28 | 50 | 3/3 | 40 | 10/10 |
| 21 | 37 | 100 | 3/3 | 100 | 5/5 |
| 25 | 27 | 100 | 3/3 | 100 | 3/3 |
| 53 | >48 | 100 | 3/3 | 100 | 3/3 |
| 23 | 37 | 10 | NDe | 0 | ND |
| 24 | 33 | 20 | ND | 0 | ND |
Spectinomycin-resistant colonies isolated from each tissue site were analyzed by PCR for the presence of pUA826 integrated into the hptA gene.
All birds showing terminal signs of fowl cholera were euthanized, in accordance with animal ethics requirements.
Percentage of spectinomycin-resistant P. multocida isolates in tissue sample.
Number of spectinomycin-resistant colonies positive by PCR for pUA826 integrated into hptA gene.
ND, not done.
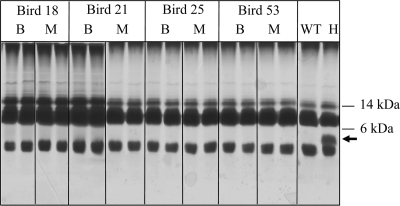
The hptA region of the genome in six spectinomycin-resistant isolates from each bird (three per site) was analyzed by PCR. The results confirmed that the spectinomycin-resistant P. multocida isolates recovered from the blood and muscle tissue of birds 18, 21, 25, and 53 had retained pUA826 integrated into the hptA gene (Table 2). We have previously shown that the heptosyltransferase HptA is essential for addition of heptose to the glycoform A inner core. P. multocida hptA mutants are fully attenuated, as inactivation of hptA results in a large amount of truncated glycoform A (lipid A-Kdo1-P) present on the surface of these mutants (Fig. 1) (10). To determine if the hptA mutants recovered from the infected chickens still produced truncated LPS, we analyzed whole-cell lysates from a number of the in vivo-derived hptA mutants by SDS-PAGE and carbohydrate silver staining. The virulent hptA mutants recovered from both the blood and muscle of the infected birds no longer produced truncated LPS at detectable levels (Fig. 2). This finding was confirmed by mass spectrometry analysis of the LPS isolated from six P. multocida hptA mutants derived from the blood of bird 18 and from two mutants derived from the blood of bird 21, which clearly showed the lack of glycoform A (Table 3). On the basis of previous analysis of the assembly of the LPS inner core in P. multocida, these data indicated that the in vivo-derived hptA mutants each had a compensatory mutation which prevented the phosphorylation of the first Kdo residue by KdkA.
FIG. 2.
Analysis of P. multocida LPS produced by in vivo-derived hptA mutants by PAGE and carbohydrate silver staining of whole-cell lysates. Truncated LPS is indicated by the arrow. WT, wild-type strain (VP161); H, in vitro-grown hptA mutant (AL836); B, blood isolate; M, muscle isolate. The positions of the relevant standard molecular mass markers (kDa) are shown on the right.
TABLE 3.
Negative ion CE-ES-MS data and proposed compositions of O-deacylated LPS (LPS-OH) from the P. multocida parent (AL435), the hptA mutant (AL836), in vivo-derived kdkA and hptA double mutants, the kdkA mutant (AL721), and complemented kdkA mutant strains
| Description, strain | Observed ions (m/z) |
Molecular mass (Da)a |
Relative intensity | Proposed composition | ||
|---|---|---|---|---|---|---|
| (M-2H)2− | (M-3H)3− | Observed | Calculated | |||
| Parent, AL435 | 1,488.6 | 991.9 | 2,978.9 | 2,977.6 | 0.1 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| 1,499.4 | 999.2 | 3,000.7 | 2,999.5 | 0.1 | 2PCho, 4Hex, 4Hep, Kdo-P, lipid A-OH | |
| 1,548.9 | 1,032.9 | 3,100.7 | 3,100.6 | 0.2 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn | |
| 1,560.0 | 1,040.1 | 3,122.6 | 3,122.6 | 0.6 | 2PCho, 4Hex, 4Hep, Kdo-P-PEtn, lipid A-OH | |
| 1,621.5 | 1,081.2 | 3,245.8 | 3,245.6 | 1.0 | 2PCho, 4Hex, 4Hep, Kdo-P-PEtn, lipid A-OH-PEtn | |
| hptA mutant, AL836 | 616.8 | 1,235.6 | 1,252.2 | 0.1 | Kdo-P, lipid A-OH | |
| 678.4 | 451.6 | 1,358.8 | 1,375.2 | 0.2 | Kdo-P-PEtn, lipid A-OH | |
| 739.8 | 492.6 | 1,481.2 | 1,498.3 | 0.4 | Kdo-P-PEtn, lipid A-OH-PEtn | |
| 1,487.4 | 992.1 | 2,978.1 | 2,977.6 | 0.3 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH | |
| 1,549.9 | 1,033.4 | 3,102.6 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn | |
| In vivo-derived kdkA hptA double mutants | ||||||
| A112V KdkA, AL1774 | 1,487.4 | 992.1 | 2,978.1 | 2,977.6 | 0.3 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| (AL1775),b bird 18 | 1,548.9 | 1,032.9 | 3,100.7 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
| R123P KdkA, AL1777 | 1,487.7 | 992.1 | 2,978.3 | 2,977.6 | 0.4 (0.3)c | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| (AL1778), bird 18 | 1,549.5 | 1,032.6 | 3,100.9 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
| D193N KdkA, AL1780 | 1,487.7 | 991.5 | 2,977.5 | 2,977.6 | 0.4 (0.5) | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| (AL1781), bird 18 | 1,549.2 | 1,032.3 | 3,100.1 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
| H168Y KdkA, AL1782 | 1,487.7 | 991.5 | 2,977.5 | 2,977.6 | 0.4 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| (AL1783), bird 21 | 1,549.2 | 1,032.3 | 3,100.1 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
| KdkA complementation data | ||||||
| kdkA mutant, AL721 | 992.1 | 2,979.3 | 2,977.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH | |
| 1,032.9 | 3,101.7 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn | ||
| kdkA mutant + vector, | 1,487.7 | 991.5 | 2,977.5 | 2,977.6 | 0.4 (0.3) | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| AL839 (AL1807) | 1,548.9 | 1,032.9 | 3,100.7 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
| kdkA mutant + wild-type | 991.8 | 2,978.4 | 2,977.6 | 0.1 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH | |
| KdkA, AL840 (AL1808) | 1,032.9 | 3,101.7 | 3,100.6 | 0.3 (0.2) | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn | |
| 1,560.0 | 1,040.1 | 3,122.6 | 3,122.6 | 0.6 (0.5) | 2PCho, 4Hex, 4Hep, Kdo-P-PEtn, lipid A-OH | |
| 1,621.5 | 1,081.2 | 3,245.8 | 3,245.6 | 1.0 | 2PCho, 4Hex, 4Hep, Kdo-P-PEtn, lipid A-OH-PEtn | |
| kdkA mutant + A112V | 1,488.0 | 991.5 | 2,977.7 | 2,977.6 | 0.6 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| KdkA, AL1799 (AL1800) | 1,549.2 | 1,032.6 | 3,100.6 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
| 1,040.1 | 3,123.3 | 3,122.6 | 0.3 (0.2) | 2PCho, 4Hex, 4Hep, Kdo-P-PEtn, lipid A-OH | ||
| 1,621.5 | 1,080.9 | 3,245.4 | 3,245.6 | 0.4 (0.5) | 2PCho, 4Hex, 4Hep, Kdo-P-PEtn, lipid A-OH-PEtn | |
| kdkA mutant+R123P | 1,488.0 | 991.5 | 2,977.7 | 2,977.6 | 0.3 (0.4) | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| KdkA, AL1801 (AL1802) | 1,548.9 | 1,032.6 | 3,100.4 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
| kdkA mutant + H168Y | 1,487.7 | 991.5 | 2,977.5 | 2,977.6 | 0.4 (0.5) | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| KdkA, AL1803 (AL1804) | 1,549.2 | 1,032.6 | 3,100.6 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
| kdkA mutant + D193N | 1,487.7 | 991.2 | 2,977.0 | 2,977.6 | 0.4 (0.3) | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| KdkA, AL1805 (AL1806) | 1,548.9 | 1,032.6 | 3,100.4 | 3,100.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH-PEtn |
Molecular mass was determined from the observed ions or calculated from the proposed composition. Average mass units were used for calculation of molecular weight, based on the proposed composition, as follows: lipid A-OH, 952.00; hexose (Hex), 162.15; heptose (Hep), 192.17; Kdo-P, 300.13; Kdo, 220.18; PEtn, 123.05; phosphocholine (PCho), 165.05.
Duplicate strains were analyzed, but data for only one strain are shown.
Where the relative intensity of each glycoform differed between duplicate strains, the value for the second strain is shown in parentheses.
All virulent hptA mutants isolated from chickens have compensatory mutations in the kdkA gene.
Sequencing of the kdkA gene from 12 independent in vivo-derived hptA mutants lacking truncated glycoform A LPS revealed that 7 isolates contained a point mutation within the kdkA open reading frame which resulted in truncation of the KdkA protein (Table 4). Interestingly, the remaining in vivo-derived P. multocida hptA mutants expressed a complete kdkA gene with a single point mutation, resulting in one of the following amino acid substitutions: A112V, R123P, D193N, or H168Y (Table 4). These data indicate that all these amino acid substitutions result in the loss of KdkA kinase activity.
TABLE 4.
Nucleotide changes observed in the kdkA gene amplified from in vivo-derived P. multocida hptA kdkA double mutants
| Bird | Sample | Nucleotide change | Result | Strain designations |
|---|---|---|---|---|
| 18 | Muscle | T78 deleted | Frame shift after F26 | |
| 18 | Blood | C64T | Stop | |
| 18 | Blood | C335T | A112V | AL1774, AL1775 |
| 18 | Blood | G368C | R123P | AL1777, AL1778 |
| 18 | Blood | C424T | Stop | |
| 18 | Blood | G577A | D193N | AL1780, AL1781 |
| 21 | Muscle | C502T | H168Y | |
| 21 | Blood | C502T | H168Y | AL1782, AL1783 |
| 25 | Muscle | C478T | Stop | |
| 25 | Blood | C538T | Stop | |
| 53 | Muscle | C466T | Stop | |
| 53 | Blood | C466T | Stop |
Complementation of a defined P. multocida kdkA mutant with the R123P, H168Y, or D193N kdkA fails to restore production of glycoform A.
To determine if the KdkA transferases containing the amino acid changes were functional, expression plasmids containing KdkA with one of the amino acid substitutions indicated above were constructed in duplicate. These plasmids, along with a plasmid containing wild-type kdkA (pAL462), were then introduced separately into the P. multocida kdkA mutant, AL721, which can no longer phosphorylate the first Kdo residue and which is therefore unable to produce glycoform A LPS (Fig. 1) (10). CE-ES-MS analyses of isolated LPS showed that wild-type levels of both glycoform A and B LPS were produced when the kdkA mutant was complemented in trans with the wild-type kdkA gene (Table 3). However, complementation in trans with a kdkA gene encoding an R123P, H168Y, or D193N amino acid substitution failed to restore the expression of glycoform A LPS, confirming that amino acids R123, H168, and D193 in KdkA are essential for kinase function (Table 3). Although LPS structural analysis showed that the A112V kdkA mutants AL1774 and AL1775 did not express observable amounts of the glycoform A inner core, complementation experiments showed that providing this gene in trans on the high-copy-number expression plasmid pAL99 allowed some expression of glycoform A LPS (Table 3, AL1799 and AL1800). However, the relative intensity values obtained from the CE-ES-MS data suggest that the amount of glycoform A produced by these strains was significantly less than the amount of glycoform A expressed when wild-type KdkA was provided in trans (approximately 30% and 82% of total LPS, respectively) (Table 3).
P. multocida KdkA is a member of the protein kinase superfamily.
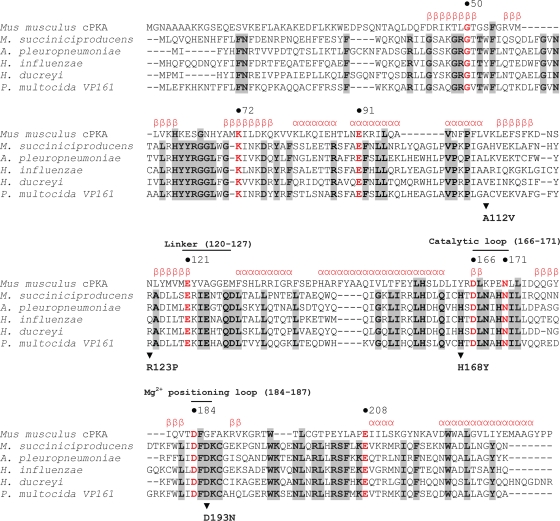
Previous bioinformatic analyses indicated that bacterial LPS kinases are homologs of eukaryotic protein kinases, a large group of regulatory enzymes that are bilobal in structure, consisting of a small N-terminal subdomain and a large C-terminal subdomain separated by a linker or hinge region (18). The amino acids essential for function in the eukaryotic proteins of this superfamily are also highly conserved across the bacterial LPS kinases (15). Using the COBALT multiple-alignment tool (18), we compared the amino acid sequence of P. multocida KdkA with the amino acid sequences of Kdo kinases from a number of different bacterial species and the amino acid sequence of the catalytic subunit of the eukaryotic protein kinase A (cPKA) (Fig. 3). This alignment showed that 23 amino acids were conserved among all of the aligned kinases and revealed that three of the four substitutions (R123P, H168Y, and D193N) identified in the P. multocida kdkA mutants were in regions of the protein that aligned with critical residues within cPKA. The nonconservative substitution R123P (in the position equivalent to N115 in cPKA) is predicted to be positioned at the start of the 5th beta sheet immediately preceding the linker region (12). The relatively conservative H168Y substitution (in the position equivalent to Y164 in cPKA) is positioned in the predicted catalytic loop of the kinase where phosphate transfer from the ATP donor to the acceptor molecule occurs (Fig. 3). The D193N substitution in P. multocida KdkA aligns within the region predicted to form the Mg2+ positioning loop and is situated only 2 amino acids downstream from the essential D184 in cPKA. Interestingly, the fourth mutation, A112V, which resulted in a significant reduction in kinase activity (Table 3), does not appear to be located in a critical region of the kinase. Moreover, the equivalent residue that aligns with A112 in both cPKA and the Haemophilus ducreyi KdkA is also valine.
FIG. 3.
Amino acid sequence alignment of the catalytic subunit of cyclic AMP-dependent protein kinase (cPKA) from Mus musculus (GenBank accession no. AAA39936.1) aligned with the KdkA proteins from Mannheimia succiniciproducens (GenBank accession no. Q65R39.2), Actinobacillus pleuropneumoniae (GenBank accession no. YP_001651918.1), Haemophilus influenzae (GenBank accession no. NB_438429.1), Haemophilus ducreyi (GenBank accession no. NB_873578.1), and Pasteurella multocida strain VP161 (GenBank accession no. HM448896). Numbers above the alignment refer to the amino acid positions relative to the cPKA sequence. Conserved amino acids are shown in boldface and are highlighted in gray. Highly conserved amino acids within the kinase superfamily are red and are shown in boldface, and critical regions of the kinase are marked with a solid line. The known secondary structure of the cPKA subunit is shown in red above the alignment; α, alpha helical, β, beta sheet. The positions of amino acid substitutions A112V, R123P, H168Y, and D193N in P. multocida KdkA are indicated with arrowheads, and the substitutions are shown underneath the P. multocida VP161 sequence.
DISCUSSION
P. multocida simultaneously produces LPS with two different inner core structures, termed glycoforms A and B. Glycoform A has a single phosphorylated Kdo residue attached to the lipid A (lipid A-Kdo1-P), whereas the glycoform B structure contains two unphosphorylated Kdo residues (lipid A-Kdo1-Kdo2) (10). In Gram-negative bacteria, glycoform B is the most commonly occurring LPS core structure, whereas glycoform A is produced by relatively few bacterial species, including Haemophilus influenzae, Vibrio cholerae, and the veterinary pathogens Mannheimia haemolytica and Actinobacillus pleuropneumoniae. The production of both the glycoform A and glycoform B inner core structures simultaneously has been detected only in P. multocida and M. haemolytica, both of which belong to the Pasteurellaceae family (16, 23). H. influenzae, which is also a member of the Pasteurellaceae family, can synthesize only a glycoform A inner core, as the H. influenzae Kdo transferase is monofunctional and therefore unable to add a second Kdo residue (26).
The kinase required for the phosphorylation of the first Kdo residue in P. multocida glycoform A is Kdo kinase (KdkA). Following Kdo phosphorylation, the first heptose residue is added by the glycoform A-specific heptosyltransferase, HptA (10). In a previous study, we showed that P. multocida hptA mutant AL836 produced full-length glycoform B LPS but was rendered avirulent by the large amount of truncated glycoform A LPS present on its surface. In contrast, P. multocida kdkA mutant AL721 was fully virulent; this mutant produced full-length glycoform B but did not express any truncated glycoform A, as it was unable to phosphorylate the first Kdo residue (10). In the present study, we found that introduction of the hptA mutant into chickens at high doses resulted in the natural selection and growth of spontaneous kdkA hptA double mutants that no longer assembled any truncated glycoform A.
Homologs of P. multocida KdkA are present in most species within the Pasteurellaceae family, including members of the genera Pasteurella, Haemophilus, Actinobacillus, and Mannheimia. Outside of the Pasteurellaceae family, KdkA homologs are also expressed by the human gastrointestinal pathogens V. cholerae and Vibrio parahaemolyticus, as well as by a number of species within the order Alteromonadales.
Previous bioinformatic analyses of the P. multocida Kdo kinase showed that KdkA is distantly related to eukaryotic protein kinases, of which cPKA is the best characterized (13-15, 24). In eukaryotic species, cPKA is required to catalyze the transfer of phosphate from MgATP to the target protein substrate. The structure of cPKA has been determined and consists of two lobes, one small and one large, with the catalytic site or ATP binding pocket being located in the active cleft between the two lobes (13-14). Most of the highly conserved amino acids identified in the cPKA structure are located in the active cleft and are required for MgATP binding and catalysis (14). In this study we identified P. multocida mutants that encode a nonfunctional KdkA with an H168Y substitution. In cPKA, the amino acid in the equivalent position (position 164) is also tyrosine, which is located within the active cleft and close to the catalytic aspartate 166 (Fig. 3). The active site in cPKA is dependent upon a number of hydrogen bond networks for correct conformation. To achieve this conformation, the amino acid in position 164 of cPKA needs to be an efficient hydrogen-bonding amino acid, such as tyrosine or histidine (14, 19). There is no structure available for any bacterial Kdo kinase, but it is likely that the fully conserved histidine at position 168 of the P. multocida KdkA kinase is also important for the function of the active cleft. Although the hydrogen-bonding capability of the amino acid in this position has probably been conserved, the total lack of kinase activity by H168Y KdkA suggests that the substitution to tyrosine results in destabilization of the catalytic loop or other conformational changes that disrupt function.
The Kdo kinase expressed by P. multocida spontaneous mutants AL1777 and AL1778 had the amino acid substitution R123P. The amino acid at the equivalent position (position 115) in cPKA is asparagine (Fig. 3), which is located at the start of the highly conserved 5th beta strand, which immediately precedes the linker region (cPKA amino acids 120 to 127) (19). This region is important for the correct interlobe movement, allowing both an open form, for ATP to access the catalytic site, and a closed form, so that key amino acids are brought into the correct conformation for ATP alignment and subsequent catalysis (12). Thus, the nonconservative R123P substitution in KdkA is likely to have a significant impact on the secondary structure of this kinase, and it is probable that the mutation affects the hinge action of the linker region.
The Kdo kinase expressed by AL1780 and AL1781 contained a D193N substitution that led to the complete loss of kinase activity. Analysis of the alignment with cPKA in this region showed that there are two aspartate residues in close proximity to each other in all the bacterial KdkA kinases examined. In the COBALT alignment, D191 aligns directly with the cPKA essential catalytic site amino acid D184, and the KdkA D193, present in all KdkA proteins examined, aligns 2 amino acids downstream. In cPKA, D184 is required to chelate the activating Mg2+ ion, which in turn binds to the β- and γ-phosphates of ATP (28). In the present study, we showed that the D193N substitution leads to a complete loss of KdkA kinase activity. This D193 residue may be critical for kinase function because of its close proximity to the predicted catalytic site amino acid D191, or alternatively, D193 rather than D191 may be the essential catalytic site amino acid in bacterial Kdo kinases.
The Kdo kinase mutants AL1774 and AL1775 expressed no glycoform A LPS as a result of an A112V substitution within KdkA. However, when this mutant kdkA gene was expressed from a high-copy-number expression plasmid, a low level of KdkA activity was observed. On the basis of the COBALT alignment of the bacterial Kdo kinases with the eukaryotic cPKA, the A112V substitution was not in a highly conserved or structurally critical region of the protein, and valine is also in the equivalent position in both cPKA and the H. ducreyi KdkA. Nevertheless, it is clear that in P. multocida KdkA, A112 is required for full enzyme activity.
In conclusion, the use of a fowl cholera natural host infection model has allowed us the unique opportunity to rapidly identify essential amino acids in the P. multocida Kdo kinase, KdkA, required for the addition of phosphate to the Kdo1 of glycoform A LPS. Four of the kdkA mutants identified in this study encoded complete kdkA genes, but each had a single nucleotide change leading to an amino acid substitution. On the basis of analysis of the LPS produced by the kdkA mutants, we have shown that these changes in the KdkA protein either abolish (R123P, H168Y, and D193N) or result in the significant loss (A112V) of kinase activity on the lipid A-Kdo1 acceptor molecule. Furthermore, on the basis of comparisons with the eukaryotic protein kinases, three of the mutations that abolish kinase activity are predicted to be in or near two important regions of the kinase molecule, namely, the linker region hinging the two lobes of the kinase molecule and the active cleft containing the ATP binding pocket. This is the first report of the identification of residues critical for the function of the bacterial Kdo kinase KdkA and will form a solid basis for future structural studies and the identification of inhibitors for this important LPS kinase.
Acknowledgments
This work was funded in part by the Australian Research Council, Canberra, Australia.
We thank Marietta John for excellent technical assistance, Perry Fleming (Core Bacterial Culture Facility) for bacterial growth, Jacek Stupak for recording CE-ES-MS, and Gerald Murray for critical reading of the manuscript.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 21 June 2010.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, NY.
- 2.Boyce, J. D., and B. Adler. 2000. The capsule is a virulence determinant in the pathogenesis of Pasteurella multocida M1404 (B:2). Infect. Immun. 68:3463-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, J. D., M. Harper, F. St. Michael, M. John, A. Aubry, H. Parnas, S. M. Logan, I. W. Wilkie, M. Ford, A. D. Cox, and B. Adler. 2009. Identification of novel glycosyltransferases required for assembly of the Pasteurella multocida A:1 lipopolysaccharide and their involvement in virulence. Infect. Immun. 77:1532-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyce, J. D., M. Harper, I. W. Wilkie, and B. Adler. 2010. Pasteurella. In C. L. Gyles, J. F. Prescott, G. Songer and C. O. Thoen (ed.), Pathogenesis of bacterial infections in animals, 4th ed. Wiley-Blackwell, Ames, IA.
- 5.Chung, J. Y., I. Wilkie, J. D. Boyce, K. M. Townsend, A. J. Frost, M. Ghoddusi, and B. Adler. 2001. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect. Immun. 69:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, A. D., H. Masoud, P. Thibault, J. R. Brisson, M. van der Zwan, M. B. Perry, and J. C. Richards. 2001. Structural analysis of the lipopolysaccharide from the nontypable Haemophilus influenzae strain SB 33. Eur. J. Biochem. 268:5278-5286. [DOI] [PubMed] [Google Scholar]
- 7.DeAngelis, P. L. 1996. Enzymological characterization of the Pasteurella multocida hyaluronic acid synthase. Biochemistry 35:9768-9771. [DOI] [PubMed] [Google Scholar]
- 8.DeAngelis, P. L., N. S. Gunay, T. Toida, W. J. Mao, and R. J. Linhardt. 2002. Identification of the capsular polysaccharides of type D and F Pasteurella multocida as unmodified heparin and chondroitin, respectively. Carbohydr. Res. 337:1547-1552. [DOI] [PubMed] [Google Scholar]
- 9.Gamian, A., M. Beurret, F. Michon, J. R. Brisson, and H. J. Jennings. 1992. Structure of the L2 lipopolysaccharide core oligosaccharides of Neisseria meningitidis. J. Biol. Chem. 267:922-925. [PubMed] [Google Scholar]
- 10.Harper, M., J. D. Boyce, A. D. Cox, F. St. Michael, I. W. Wilkie, P. J. Blackall, and B. Adler. 2007. Pasteurella multocida expresses two lipopolysaccharide glycoforms simultaneously, but only a single form is required for virulence: identification of two acceptor-specific heptosyl I transferases. Infect. Immun. 75:3885-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harper, M., A. D. Cox, F. St. Michael, I. W. Wilkie, J. D. Boyce, and B. Adler. 2004. A heptosyltransferase mutant of Pasteurella multocida produces a truncated lipopolysaccharide structure and is attenuated in virulence. Infect. Immun. 72:3436-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, L. N., M. E. Noble, and D. J. Owen. 1996. Active and inactive protein kinases: structural basis for regulation. Cell 85:149-158. [DOI] [PubMed] [Google Scholar]
- 13.Knighton, D. R., J. H. Zheng, L. F. Ten Eyck, V. A. Ashford, N. H. Xuong, S. S. Taylor, and J. M. Sowadski. 1991. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:407-414. [DOI] [PubMed] [Google Scholar]
- 14.Knighton, D. R., J. H. Zheng, L. F. Ten Eyck, N. H. Xuong, S. S. Taylor, and J. M. Sowadski. 1991. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:414-420. [DOI] [PubMed] [Google Scholar]
- 15.Krupa, A., and N. Srinivasan. 2002. Lipopolysaccharide phosphorylating enzymes encoded in the genomes of Gram-negative bacteria are related to the eukaryotic protein kinases. Protein Sci. 11:1580-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logan, S. M., W. Chen, A. Aubry, M. A. Gidney, S. Lacelle, F. St. Michael, R. Kuolee, M. Higgins, S. Neufeld, and A. D. Cox. 2006. Production of a d-glycero-d-manno-heptosyltransferase mutant of Mannheimia haemolytica displaying a veterinary pathogen specific conserved LPS structure; development and functionality of antibodies to this LPS structure. Vet. Microbiol. 116:175-186. [DOI] [PubMed] [Google Scholar]
- 17.Michon, F., M. Beurret, A. Gamian, J. R. Brisson, and H. J. Jennings. 1990. Structure of the L5 lipopolysaccharide core oligosaccharides of Neisseria meningitidis. J. Biol. Chem. 265:7243-7247. [PubMed] [Google Scholar]
- 18.Papadopoulos, J. S., and R. Agarwala. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23:1073-1079. [DOI] [PubMed] [Google Scholar]
- 19.Scheeff, E. D., and P. E. Bourne. 2005. Structural evolution of the protein kinase-like superfamily. PLoS Comput. Biol. 1:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schweda, E. K., A. C. Sundstrom, L. M. Eriksson, J. A. Jonasson, and A. A. Lindberg. 1994. Structural studies of the cell envelope lipopolysaccharides from Haemophilus ducreyi strains ITM 2665 and ITM 4747. J. Biol. Chem. 269:12040-12048. [PubMed] [Google Scholar]
- 21.St. Michael, F., J. Li, and A. D. Cox. 2005. Structural analysis of the core oligosaccharide from Pasteurella multocida strain X73. Carbohydr. Res. 340:1253-1257. [DOI] [PubMed] [Google Scholar]
- 22.St. Michael, F., J. Li, E. Vinogradov, S. Larocque, M. Harper, and A. D. Cox. 2005. Structural analysis of the lipopolysaccharide of Pasteurella multocida strain VP161: identification of both Kdo-P and Kdo-Kdo species in the lipopolysaccharide. Carbohydr. Res. 340:59-68. [DOI] [PubMed] [Google Scholar]
- 23.St. Michael, F., E. Vinogradov, J. Li, and A. D. Cox. 2005. Structural analysis of the lipopolysaccharide from Pasteurella multocida genome strain Pm70 and identification of the putative lipopolysaccharide glycosyltransferases. Glycobiology 15:323-333. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, S. S., and E. Radzio-Andzelm. 1994. Three protein kinase structures define a common motif. Structure 2:345-355. [DOI] [PubMed] [Google Scholar]
- 25.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 26.White, K. A., I. A. Kaltashov, R. J. Cotter, and C. R. Raetz. 1997. A mono-functional 3-deoxy-d-manno-octulosonic acid (Kdo) transferase and a Kdo kinase in extracts of Haemophilus influenzae. J. Biol. Chem. 272:16555-16563. [DOI] [PubMed] [Google Scholar]
- 27.Wilkie, I. W., S. E. Grimes, D. O'Boyle, and A. J. Frost. 2000. The virulence and protective efficacy for chickens of Pasteurella multocida administered by different routes. Vet. Microbiol. 72:57-68. [DOI] [PubMed] [Google Scholar]
- 28.Zheng, J., D. R. Knighton, L. F. ten Eyck, R. Karlsson, N. Xuong, S. S. Taylor, and J. M. Sowadski. 1993. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry 32:2154-2161. [DOI] [PubMed] [Google Scholar]