Abstract
Group A streptococci (GAS) can cause a wide variety of human infections ranging from asymptomatic colonization to life-threatening invasive diseases. Although antibiotic treatment is very effective, when left untreated, Streptococcus pyogenes infections can lead to poststreptococcal sequelae and severe disease causing significant morbidity and mortality worldwide. To aid the development of a non-M protein-based prophylactic vaccine for the prevention of group A streptococcal infections, we identified novel immunogenic proteins using genomic surface display libraries and human serum antibodies from donors exposed to or infected by S. pyogenes. Vaccine candidate antigens were further selected based on animal protection in murine lethal-sepsis models with intranasal or intravenous challenge with two different M serotype strains. The nine protective antigens identified are highly conserved; eight of them show more than 97% sequence identity in 13 published genomes as well as in approximately 50 clinical isolates tested. Since the functions of the selected vaccine candidates are largely unknown, we generated deletion mutants for three of the protective antigens and observed that deletion of the gene encoding Spy1536 drastically reduced binding of GAS cells to host extracellular matrix proteins, due to reduced surface expression of GAS proteins such as Spy0269 and M protein. The protective, highly conserved antigens identified in this study are promising candidates for the development of an M-type-independent, protein-based vaccine to prevent infection by S. pyogenes.
Streptococcus pyogenes is a Gram-positive pathogenic bacterium belonging to group A streptococci (GAS) that causes uncomplicated infections like pharyngitis and impetigo. However, S. pyogenes can also trigger severe infections, such as streptococcal toxic shock syndrome, sepsis, or poststreptococcal sequelae resulting in rheumatic heart disease, arthritis, and glomerulonephritis (11). Although the global burden of GAS disease is unknown, it was estimated that S. pyogenes causes well over 500 million cases of pharyngitis and more than 100 million cases of pyoderma per year (9). Severe GAS diseases, including rheumatic heart disease and invasive infections, cause at least an estimated 500,000 deaths each year. The fatality rate of invasive disease ranges from 15 to 30% but can exceed 50% in cases of streptococcal toxic shock syndrome (9, 45). Prevention of severe diseases relies on the diagnosis and fast treatment with penicillin. Although S. pyogenes so far remains susceptible to penicillin, resistance to different antibiotics has been reported with an increasing frequency (1, 32, 41, 65). Most significantly, approximately 20% of antibiotic prescriptions for acute respiratory illnesses in the Unites States are attributed to GAS pharyngitis (24). Therefore, vaccination clearly constitutes an attractive alternative strategy to control GAS infections not only to significantly reduce the burden of invasive and noninvasive disease but also to reduce antibiotic use and thus development of resistance in group A streptococci and other important human pathogens.
The initial step during the infection process by GAS is the adherence of the bacterium to pharyngeal or dermal epithelial cells via surface proteins, the hyaluronic acid capsule or fibronectin-binding proteins, which is followed by colonization and invasion and finally the spread throughout other tissues of the host (5). The involved surface molecules are good targets for protective humoral immune responses to prevent infection and disease. The best-studied protein mediating protection against GAS infection is the surface M protein. Its variable N-terminal as well as its conserved carboxy-terminal region has been studied as a possible vaccine candidate (2, 4, 12). However, the existence of more than 100 M protein serotypes of S. pyogenes and the link between M protein-induced humoral and cellular immune responses and autoimmune poststreptococcal sequelae hinder M protein-based vaccine development (13, 18, 42). Several other group A streptococcal surface proteins were also shown to induce protective immune responses in animals and are therefore considered vaccine candidates; among them are the extracellular pyrogenic exotoxins, streptococcal superantigens, C5a peptidase, and the streptococcal fibronectin-binding protein SfbI (5, 10, 25, 36, 56). Since protein candidates such as SfbI and other fibronectin-binding proteins either are not present in the majority of GAS strains or show large variability in their amino acid sequences or in their levels of surface expression among different GAS isolates, they have not been considered single-vaccine candidates. Although candidates such as C5A peptidase are highly conserved among GAS strains, due to the heterogeneity of GAS evidenced by the existence of more than 150 emm types, with the highest diversity observed in developing countries, and the frequent emergence of new emm types, a broadly protective vaccine will most likely require a combination of antigens.
Several approaches were recently applied to identify novel vaccine candidates from GAS based on proteomic methodologies or on reverse vaccinology; advantage was taken of the availability of several genomic GAS sequences (39, 51, 57, 58). These studies have provided evidence for the surface localization of numerous group A streptococcal proteins, some of them without predictable signatures for surface localization. In spite of these efforts, so far only one of the identified surface proteins, Spy0416 (ScpC), was shown to mediate protection against S. pyogenes infection (51).
We have applied the Antigenome technology, which successfully identified protective vaccine candidates from Staphylococcus aureus (16, 35), Streptococcus pneumoniae (23), and several additional bacterial pathogens (unpublished data), to S. pyogenes for the comprehensive identification of novel conserved and protective antigens suitable for vaccine development to prevent GAS infections. For immune selection, we used human serum antibodies obtained from patients who recovered from common S. pyogenes infections and healthy, noncolonized parents of small children. These studies led to the discovery of eight novel antigens in addition to Spy0416/ScpC, all of which are highly conserved among GAS clinical isolates and provide significant protection in murine challenge models. Gene deletion studies have furthermore provided evidence for an important role for one of the protective antigens, Spy1536, in modulating the surface expression of GAS proteins and the interaction of streptococcal cells with host proteins.
MATERIALS AND METHODS
Bacterial strains.
The S. pyogenes strain SF370 was obtained from the American Type Culture Collection. S. pyogenes M49591 was provided by Bernd Kreikemeyer, University of Rostock, Germany. Clinical GAS isolates were obtained from Franz-Josef Schmitz (Klinikum Minden, Germany) and Rodger Novak (Medical University of Vienna, Austria). The strain A20-MA is a spontaneous streptomycin-resistant derivative of S. pyogenes DSM 2071 (M type 23; German Culture Collection). The virulence of the S. pyogenes NS192 strain in mice was increased by several passages in mice, resulting in the mouse-adapted strain A147-MA (M type 106). The original S. pyogenes strain NS192 is a blood isolate of a patient from the Australian Northern Territory. The S. pyogenes AP1 strain was characterized as serotype M1 and obtained from L. Björck, University of Lund, Sweden.
Human sera.
Patient serum samples were collected by Zsófia Pusztai, Betheseda Children's Hospital, Budapest, Hungary, Rodger Novak, Department of Microbiology and Genetics, University of Vienna, Austria, and John Goodacre, University of Central Lancashire, Preston, United Kingdom. All sera were collected according to the general national ethical guidelines and upon consent from individual subjects. Research with human sera was carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association.
Preparation of streptococcal extracts.
Total bacterial lysate was prepared from S. pyogenes SF370 grown overnight in Todd-Hewitt broth and lysed by repeated freeze-thaw cycles (incubation on a dry ice-ethanol mixture until frozen [1 min] and then thawing at 37°C [5 min]; repeated 3 times). This was followed by sonication and collection of the supernatant by centrifugation at 2,600 × g for 15 min at 4°C. Culture supernatants were generated by removal of bacteria from overnight-grown bacterial cultures via centrifugation, following precipitation of the supernatant with ice-cold ethanol by mixing 1 part supernatant with 3 parts absolute ethanol and incubation overnight at −20°C. Precipitates were collected by centrifugation at 2,600 × g for 15 min. Dry pellets were dissolved either in phosphate-buffered saline (PBS) for enzyme-linked immunosorbent assay (ELISA) or in urea and SDS sample buffer for SDS-PAGE and immunoblotting. The protein concentrations of samples were determined by Bradford assay.
Detection of antigen-specific antibodies by ELISA.
Antigen-specific antibody titers in sera were determined by ELISA using 96-well Nunc-Immuno MaxiSorp assay plates (Nunc, Germany) coated with 0.5 to 1 μg/well of the corresponding antigen in coating buffer (bicarbonate, pH 9.4). After overnight incubation at 4°C, plates were blocked with 1% bovine serum albumin (BSA) in PBS (pH 7.4) for 1 h at 37°C. Appropriate dilutions of mouse serum in PBS with 1% BSA were added (100 μl/well), and plates were incubated for 2 h at 37°C. After four washes, secondary biotinylated antibodies were added, followed by 1 h of incubation at 37°C. After six washes, 50 μl/well of peroxidase-conjugated streptavidin diluted 1:1,000 (Pharmingen, United States) was added, and plates were further incubated for 45 min at room temperature. Alternatively to the last two steps, horseradish peroxidase (HRPO)-conjugated anti-human IgG or anti-human IgA were used for human sera. After six final washes, the substrate ABTS [2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)] in 0.1 M citrate-phosphate buffer containing 0.1% H2O2 was added, and plates were incubated for 30 to 60 min at room temperature. Antigen-antibody complexes were quantified by measuring the conversion of ABTS to a colored product at an optical density at 405 nm (OD405).
Preparation of antibodies from human sera.
For purification of immunoglobulins (IgGs and IgAs), human serum pools were heat inactivated at 56°C for 30 min and centrifuged to remove precipitated proteins. The supernatant was passed through a 0.2-μm filter. Antibodies against Escherichia coli proteins were removed by incubating the heat-inactivated sera with whole E. coli cell suspensions (DH5alpha, transformed with pHIE11 [17, 23]). For IgG purification, samples were diluted 1:3 with binding buffer and applied to equilibrated UltraLink-immobilized-protein G columns (Pierce, United States). The flowthrough was used for IgA purification. After the column was washed with a 20× bed volume of binding buffer, bound IgGs were recovered by elution. The eluted fractions were adjusted immediately to physiologic pH by adding 1/10 volume of neutralization buffer (1 M Tris, pH 8 to 9). Samples with the highest absorbance were pooled and dialyzed against PBS. For IgA purification, the flowthrough of the immobilized-protein G column was loaded onto a HiTrap streptavidin column coupled with biotinylated anti-human IgA (Southern Biotech, United States). Wash, elution, and neutralization steps were performed in the same way as for the IgG purification. Pooled fractions were dialyzed against PBS. The efficiency of depletion and purification was checked by SDS-PAGE, Western blotting, ELISA, and protein concentration measurements. Pooled IgG and IgA fractions were biotinylated according to the manufacturer's instructions (EZ-Link Sulfo-NHS-LC-Biotin; Pierce, United States).
Library construction and MACS.
Streptococcal genomic DNA, DNA inserts, and genomic libraries were generated and magnetic activated cell sorting (MACS) was performed as described previously (16, 17, 29). In brief, genomic DNA fragments were mechanically sheared using sonication or mild DNase I treatment. Fragments were blunt ended twice using T4 DNA polymerase followed by ligation into the vector pMAL4.1 or pMAL4.31 for frame selection. Genomic DNA fragments were then transferred into plasmid pMAL9.1 (LamB), pMAL10.1 (BtuB), or pHIE11 (FhuA) for bacterial surface display screens. For MACS, approximately 2 × 107 cells from a given library were incubated with 10 μg of biotinylated human serum at 4°C. Ten microliters of MACS microbeads coupled with streptavidin (Miltenyi Biotech, Germany) was added, and the MACS microbead cell suspension was loaded onto a mass spectrometry (MS) column (Miltenyi Biotech, Germany). The column was washed, and selected cells were eluted thereafter. The MACS selection was repeated a second time prior to cells being plated onto LB agar plates supplemented with 50 μg/ml kanamycin.
Western blot analysis.
For serum characterization, 10 to 25 μg/lane total protein of bacterial lysate or culture supernatant samples from in vitro-grown S. pyogenes SF370 cells was subjected to SDS-PAGE, and proteins were transferred to a nitrocellulose membrane (GE Healthcare, England). After being blocked overnight in 5% milk in PBS, human sera were added at a 2,000× dilution and HRPO-labeled anti-human IgG was used for detection.
In order to confirm the immunoreactivities of E. coli clones by Western blot analysis, approximately 10 to 20 μg of total cellular protein was separated by 10% SDS-PAGE and blotted onto a Hybond C membrane (GE Healthcare, England). The LamB, BtuB, and FhuA fusion proteins were detected using human serum as the primary antibody at a dilution of 1:5,000 and anti-human IgG antibodies coupled to HRPO at a dilution of 1:5,000 as secondary antibodies. Detection was performed using the ECL detection kit (GE Healthcare, England). Alternatively, rabbit anti-FhuA, rabbit anti-BtuB, or mouse anti-LamB antibodies were used as primary antibodies in combination with the respective secondary antibodies coupled to HRPO for the detection of the fusion proteins.
For the detection of M protein, proteins were separated by SDS-PAGE using 4 to 20% Tris-glycine gradient gels (PAGEr Duramide precast gels; Cambrex Bio Science, United States). Subsequently, proteins were transferred onto nitrocellulose membranes for Western blot analysis. M1-specific polyclonal mouse antibodies and secondary conjugated Affinipure F(ab′)2 fragment goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, United States) were diluted 1:1,000 and 1:5,000, respectively, in 5% (wt/vol) milk powder in PBS containing 0.1% Tween 20. M protein was visualized using the ECL detection kit (GE Healthcare, England).
Gene distribution of S. pyogenes antigens by PCR.
PCR was performed using S. pyogenes genomic DNA with primers specific for the gene of interest. S. pyogenes isolates, covering selected serotypes most frequently present in patients, were serotyped by sequencing the 5′ end of the gene encoding M protein. Oligonucleotide sequences as primers were designed for all identified open reading frames (ORFs) using the public program Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_slow.cgi). The PCR was performed in a reaction volume of 25 μl using Taq polymerase (1 U), 200 nM deoxynucleoside triphosphates (dNTPs), 10 pmol of each oligonucleotide, and the kit according to the manufacturer's instructions (Invitrogen, Netherlands). As a standard, 30 cycles (1 cycle of 5 min at 95°C, 30 cycles of 30 s at 95°C, 30 s at 56°C, and 30 s at 72°C, and 1 cycle of 4 min at 72°C) were performed, unless conditions had to be adapted for individual primer pairs. The DNA samples were subsequently visualized by electrophoresis on a 1.6% agarose gel and staining with ethidium bromide.
Expression and purification of recombinant S. pyogenes proteins.
The gene fragment of interest was amplified from genomic DNA of S. pyogenes SF370 by PCR using gene-specific primers and cloned into the pET28b(+) vector (Novagen, United States). Upon transformation of the recombinant plasmid into E. coli BL21 star cells (Invitrogen, Netherlands), a single colony was inoculated in 5 ml LB supplemented with kanamycin, incubated overnight, and subsequently expanded to a volume of 500 ml or up to 5 liters. Cells were induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at an OD600 of 0.8, and growth continued for 3 h. The cells were harvested by centrifugation, and the pellet was resuspended (∼1/20 of the culture volume) in 20 mM Tris, 0.9% NaCl buffer containing DNase and EDTA-free protease inhibitors (Roche, Germany). Lysis was performed by a combination of the freeze-thaw method and treatment with BugBuster (Novagen, United States). The lysate was separated by centrifugation at 12,000 × g for 30 min into soluble (supernatant) and insoluble (pellet) fractions.
For the proteins that were localized in the soluble fraction, purification of the protein was performed by binding the supernatant to Ni-agarose beads (Ni-nitrilotriacetic acid [NTA]-agarose; Qiagen, Germany) as recommended by the manufacturer. For proteins in the insoluble fraction, the pellet was washed 3 times with 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5% Triton X-100 and then solubilized in a suitable buffer containing 8 M urea. The purification was performed under denaturing conditions in buffer containing 8 M urea. The eluate was concentrated and dialyzed to remove all urea in a gradual and stepwise manner.
Animal challenge experiments. (i) Systemic immunization followed by intravenous or intranasal challenge of mice.
Female NMRI or CD-1 mice (6 to 8 weeks of age; Harlan Winkelman GmbH, Germany) were immunized three times at 2-week intervals (days 0, 14, and 28) subcutaneously (flank) with 50 μg M1 or M23 protein (positive controls), 50 μg PBS (negative control), or 50 μg of the respective recombinant antigens with complete Freund's adjuvant-incomplete Freund's adjuvant (CFA-IFA) or aluminum hydroxide. One week after the last booster immunization (day 35), hyperimmune sera were taken to determine antigen-specific total IgG levels. At days 37 to 42, mice were challenged either with S. pyogenes AP-1 intravenously (100 μl challenge volume) or with S. pyogenes A20-MA intranasally (20-μl challenge volume). For intranasal application, mice were anesthetized with isoflurane (inhalation anesthesia). Survival was monitored for 14 days and is expressed as a percentage of the total number of animals included in the experiment. All systemic immunization experiments were performed according to Austrian law (37a).
(ii) Intranasal immunization followed by intranasal challenge of mice.
Female BALB/c mice (4 weeks of age, Harlan Winkelman GmbH, Germany), were immunized by the intranasal route (10 μl/nostril) with 40 μg of antigen coadministered with IC31 (10 nmol KLK, 0.4 nmol ODN1a; 1:1, vol/vol) (55) or 0.5 μg of a synthetic derivative of the TLR2/6 agonist MALP-2 as mucosal adjuvants on days 0, 7, 14, and 28. Control animals received PBS alone. In order to exclude severe toxic effects of the administered vaccine formulations, the motility and the weight development of the immunized mice were recorded daily. Serum samples were collected on day 37 and stored at −20°C prior to determination of antigen-specific antibodies. The pulmonary challenge was performed 10 days after the last boost. Vaccinated animals were anesthetized using Isofluran (CuraMed Pharma GmbH, Germany) and intranasally challenged with the heterologous virulent S. pyogenes strain A147-MA or A20-MA in 40 μl of PBS (20 μl/nostril). Mortality was recorded daily up to 14 days after the challenge.
Statistical significance was calculated with the Kaplan-Meier log rank (Mantel-Cox) test using GraphPad Prism5 software. A P value of <0.05 was considered statistically significant.
Generation of gene deletion constructs for spy0895 and spy1536.
A 3-step PCR strategy was employed to amplify the N- and C-terminal regions of the gene of interest from strain SF370. For spy0895, a 662-bp fragment was amplified using primers 5412 (5′-ATATAGGTACCTGCCTTCGATCTCTATCTATGCTCG-3′, with KpnI underlined) and 5413 (5′-TGGCATTCATGTCATCAAAGTTAACATCTCTAGTAAATAGAGGGCAG-3′). The second fragment was amplified using primers 5414 (5′-CTGCCCTCTATTTACTAGAGATGTTAACTTTGATGACATGAATGCCA-3′) and 5415 (5′-TATATGGATCCTCTCTTTATCAACGACTATAACCGAGAT-3′, with BamHI underlined). In a third PCR, the two fragments were ligated using primers 5412 and 5415, yielding a 1,273-bp fragment. For the spy1536 gene, a 589-bp fragment was amplified using primers 5416 (5′-ATATAGGTACCCGCTGTCAAGTTAGATATGTTTTATGTA-3′, with KpnI underlined) and 5417 (5′-AAATGCCTGGAGGCGCTTACTGGGATTGGCATTGCCTTGAC) and a 284-bp fragment was amplified using primers 5418 (5′-GTCAAGGCAATGCCAATCCCAGTAAGCGCCTCCAGGCATTT-3′) and 5419 (5′-TATATGGATCCTTGAAACCGTCTATTTGATATCAAG-3′, with BamHI underlined). The two fragments were then ligated using primers 5416 and 5419, resulting in the 832-bp large spy1536 knockout construct. The two deletion mutant constructs were ligated into the Topo vector using Invitrogen's Topo TA cloning kit and transformed into TOP10 chemically competent E. coli cells (Invitrogen, Netherlands). The spy0895 and spy1536 knockout constructs were finally transferred via digestion with BamHI and KpnI into the respectively digested pGhost5 vector, transformed into electrocompetent DH5α cells (Invitrogen, Netherlands), and selected on LB agar containing 500 μg/ml erythromycin.
Generation of the gene deletion mutants in the S. pyogenes strain M49591 using the temperature-sensitive shuttle vector pGhost5 was adapted from Biswas and colleagues (7). In brief, 150 μl of competent S. pyogenes cells and 1 μg of the pGhost5 vector, carrying the gene deletion construct, were used for each transformation. Electroporation was carried out at 2 kV using Bio-Rad's (United States) E. coli Pulser. The transformed cells were kept on ice for 5 min and incubated for 5 h in prewarmed THY medium (Todd-Hewitt broth containing 5% yeast extract) at 28°C, at which temperature the shuttle vector pGhost5 is maintained extrachromosomally. Cells were plated on THY agar plates containing 5 μg/ml erythromycin and incubated at 28°C with 5% CO2. For the integration of the construct into the chromosome of S. pyogenes, cells were grown in THY medium containing 5 μg/ml erythromycin overnight at 28°C with 5% CO2 and diluted 1:100 in THY medium without erythromycin until they reached an optical density (600 nm) of between 0.3 and 0.5. A temperature shift from 28°C to 37°C for 150 min induced integration of the vector into the GAS chromosome, and serial dilutions of cells were plated on THY medium containing 5 μg/ml erythromycin and incubated overnight at 37°C with 5% CO2. Successful integration of the vector into the chromosome was confirmed via PCR using primers amplifying fragments across the borders of the M49591 genome and the integrated pGhost5 vector sequences. For the spy0895 gene, primers 6005 (5′-ACATGGCGTTAGTTCCCTAAATTTCAGTC-3′) and 6007 (5′-GAGCGGATAACAATTTCACACAGG-3′) were used, giving either a 1,863-bp or a 2,467-bp fragment, depending on the direction of pGhost5 integration. Primers 6006 (5′-AGGGTTTTCCCAGTCACGACGTT-3′) and 6008 (5′-TGTACTTATGAGAATTCAACAACTGCATTA-3′) amplified 1,767 and 2,374 bp, respectively. For the spy1536 genome region, primers 6009 (5′-TCCTTTTTGAGAAATATACCTGGAGACTGTT-3′) and 6007 amplified 1,294 bp and 1,794 bp, respectively. Primers 6006 and 6010 (5′-CAAACGGTCATCTCGATATTGTTAAAC-3′) amplified 1,198 bp and 1,698 bp, respectively. After verification of pGhost5 integration into the chromosome, another temperature shift back to 28°C induced excision of pGhost5. Cells were plated on agar plates with and without erythromycin for the selection of cells with the desired recombination event. Positive cells, growing only on THY medium plates containing no erythromycin, were identified by PCR. For the spy0895 gene deletion, mutant primers 6005 and 6008 were used; for spy1536 gene deletion, primers 6009 and 6010 were used. Gene deletion was further verified by sequencing and Southern blot analysis.
Binding of S. pyogenes to extracellular host proteins.
ELISA plates were coated with human plasma and extracellular matrix proteins (10 μg in 100 μl; fibronectin, fibrinogen, laminin, type I and IV collagen, plasminogen, and haptoglobin; Sigma-Aldrich, Germany) and incubated overnight at 4°C. The plates were washed once with PBS and blocked with 10% BSA in PBS for 2 h at 37°C. For labeling with fluorescein isothiocyanate (FITC) isomer I (Sigma-Aldrich, Germany), overnight cultures of wild-type S. pyogenes M49591 and ΔSpy0895 and ΔSpy1536 cells were washed once with PBS and incubated with 1 ml FITC isomer I [1 mg/ml Na(CO3)2] for 20 min at room temperature in the dark. After incubation, the culture was washed with PBS until the supernatant appeared transparent again and 6 × 107 CFU/100 μl was incubated on the ELISA plates previously coated with human proteins for 1 h at 37°C. Finally, the plates were washed 4 times with 0.1% Tween 20 in PBS and growth was measured with the BioTek (United States) Synergy2 ELISA reader using the following settings: excitation at 485/20 nm, emission at 560/60 nm, and gains of 72, 80, 75, and 77.
Immunofluorescent microscopy.
Immunostaining was adapted from Harry and colleagues (28). In brief, bacterial cells from an overnight culture of the M49591 wild type and ΔSpy0895 and ΔSpy1536 strains were harvested by centrifugation at 1,000 × g for 10 min and washed once with PBS. After the cells were fixed for 15 min at room temperature and 45 min on ice using 5 ml 3% (vol/vol) paraformaldehyde, 30 mM sodium phosphate (pH 7.5), they were washed three times with 100 mM glycine in PBS and once with PBS only. For the detection of streptococcal proteins on the bacterial surface, cells were resuspended in 50 mM glucose, 20 mM Tris-HCl (pH 7.5), 10 mM EDTA, and ∼5 × 107 CFU/100 μl was transferred immediately onto poly l-lysine-coated Poly-Prep slides (Sigma-Aldrich, Germany) and allowed to settle for 15 min. The slides were washed twice with PBS and dipped in 0.1% Triton X-100 in PBS for 5 min. After another wash with PBS, the slides were blocked overnight at 4°C with 2% BSA in PBS. For the 1-h incubation with the primary antibody at room temperature, M23-, M1-, and Spy0269-specific hyperimmune mouse sera generated in CD-1 mice were diluted 1:25 in 100 μl 1% BSA in PBS. Slides were washed three times for 5 min with 0.1% Tween 20 in PBS, dried, and incubated with Texas Red dye-conjugated AffiniPure goat anti-mouse antibody (Jackson Immunoresearch Laboratories, United States) diluted 1:100 with 1% BSA in PBS for 1 h in the dark at room temperature. Slides were washed three times with 0.1% Tween 20 in PBS for 5 min, followed by another 3 washes with PBS for 5 min. Control cells were stained with 4′,6-diamidino-2′-phenylindole (DAPI)-dihydrochloride by mounting the slides with ProLong gold antifade reagent containing DAPI (Invitrogen, Netherlands). Polymerization was allowed for 24 h at room temperature in the dark. Immunostaining was visualized with a Zeiss Axiovert 200M microscope equipped with a Zeiss LSM 510 META confocal laser-scanning unit and a Plan-Fluor 100×/1.45 oil (aperture, 0.11 mm) or a Plan-Apochromat 63×/1.40 oil DIC MC27 (aperture, 0.19 mm) objective (Zeiss, Germany).
Preparation of subcellular fractions from S. pyogenes.
Bacteria were grown overnight in Todd-Hewitt broth, and cell pellets were harvested by centrifugation at 1,800 × g and washed once with PBS. After being frozen overnight at −80°C, the bacterial cell pellets were resuspended in 10 mM Tris-HCl, pH 7.6, and lysed by sonication. Lysates were centrifuged, and the supernatants containing cytoplasmic proteins were concentrated using the Nanosep centrifugal device with an Omega membrane (molecular weight cutoff of 10,000; Pall Life Sciences, United States). The pellet from the lysate was washed twice with 10 mM Tris-HCl, pH 7.6, resuspended in 50 mM Tris, 5 mM EDTA, 10 mM NaCl, pH 8.1, containing 250 μg lysozyme and 50 U mutanolysin, and incubated at 37°C for 4 h. After centrifugation, cell wall proteins contained in the supernatant were concentrated in the same way as the cytoplasmic fractions. The protein concentrations of lysates and cytoplasmic and cell wall fractions were measured using the BCA protein assay kit (Thermo Scientific, United States).
RESULTS
Genome-wide selection of GAS antigens by human sera.
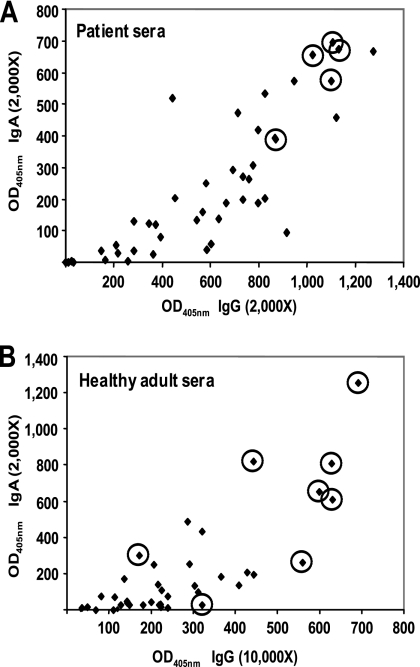
We have employed the Antigenome technology in order to identify novel group A streptococcal antigens for vaccine development. Three genomic libraries were constructed with the LamB, BtuB, and FhuA platforms, representing 5 × 105, 1.5 × 105, and 3.5 × 105 clones with average sizes of 40, 100, and 350 bp, respectively, covering the S. pyogenes SF370 genome more than 50 times (29). Human sera were collected from patients with acute S. pyogenes infections, such as pharyngitis, wound infections, and bacteremia (based on medical microbiological tests), who recovered from these GAS infections. As another source for human sera, we chose healthy adults without nasopharyngeal colonization at the time of sampling. The presence of GAS-specific antibodies in the latter donor group is not surprising, since group A streptococcal infections are common and antibodies are present as a consequence of natural immunization from previous encounters. Interestingly, we found that most of the high-titer healthy adult donors were parents of small children. The 310 and 110 serum samples from patients and healthy individuals, respectively, were characterized for anti-S. pyogenes antibodies by ELISA (Fig. 1) and Western blot analysis (data not shown). This led to the compilation of 7 antibody pools (4 IgG and 3 IgA pools), each with five individual sera from pharyngitis patients or healthy individuals that were used to screen the three genomic surface display libraries. As we aimed to use sera from individuals who would not develop poststreptococcal complications, we selected sera from pharyngitis patients, as poststreptococcal complications are rare events among these patients.
FIG. 1.
Characterization of human sera for anti-S. pyogenes antibodies as measured by ELISA. Total anti-S. pyogenes IgG and IgA antibody levels were measured by standard ELISA using total bacterial lysates or culture supernatant fractions as coating antigens prepared from S. pyogenes SF370. Serum samples from patients with a positive anti-streptolysin O titer and sera from healthy adults were analyzed at two different serum dilutions. Results of representative ELISAs are shown. (A) Patient sera with total bacterial lysate proteins; (B) sera from healthy adults with culture supernatant proteins. Data are expressed as ELISA units calculated from absorbance at 405 nm at a serum dilution in the linear range of detection (2,000× or 10,000×). Individual sera used for antigen screening are circled.
All together, 15 bacterial surface display screens were carried out: 4 IgG and 2 IgA screens each with the LamB and FhuA libraries and 3 IgG screens with the BtuB library. More than 10,000 bacterial clones were selected, and their streptococcal DNA insert was sequenced. The immunogenicity of the epitopes was confirmed by Western blot analysis of almost 1,500 clones by using the human serum Ig pools. The screens identified 95 antigen candidates annotated in the S. pyogenes SF370 genome and 55 peptides that could not be assigned to an annotated open reading frame (Tables 1 and A1). Approximately 75% of the most frequently selected antigens were identified by more than one platform protein (Table 1). Antigens selected only by the smaller-insert libraries in LamB and BtuB (e.g., Spy0433, Spy0488, and Spy1054) may not be properly expressed as a larger fragment in FhuA and were thus absent from this library. On the other hand, antigens selected only or mainly from the large-insert-containing FhuA library (e.g., Spy0019, Spy0166, and Spy1813) may form structural epitopes and be disrupted by generation of shorter fragments of the respective genes. The detailed analysis of the screen data revealed that the BtuB library contributed only two unique antigens (Spy0171, Spy0292) to the GAS antigenome, whereas the vast majority of the antigens that were identified by FhuA or LamB displayed GAS libraries indicating that almost full coverage of the genome was achieved by the LamB and FhuA platforms. We further noticed that very few proteins were selected by IgA antibodies only, and none of these belonged to the most frequently selected antigens (Table 1). Among the most frequently identified antigens, we detected most of the previously published antigens and protective proteins, such as M1 protein (Spy2018 [37]), C5A peptidase (Spy2010 [10]), streptolysin O (Spy0167 [19]), exotoxin B (Spy2036 [36]), Spy0843 (48), and ScpC (Spy0416 [51]). The last observation clearly confirmed that our approach selected valuable vaccine candidates.
TABLE 1.
Most frequently selected immunogenic Streptococcus pyogenes proteins
| ORFa | Common name | No. of clonesb | No. of screensc | Gene distributiond | Reference; patente |
|---|---|---|---|---|---|
| Spy0167 | Streptolysin O | 1,465 | 14 | 50/50 | 19; pat |
| Spy1054 | Putative collagen-like protein (SclC) | 1,295 | 9 | 26/50 | |
| Spy0843 | Cell surface protein | 1,079 | 14 | 50/50 | 48; pat |
| Spy2018 | M1 protein | 783 | 12 | Not determined | 37; pat |
| Spy0416 | Putative cell envelope serine proteinase | 462 | 13 | 50/50 | 51; pat |
| Spy0433 | Hypothetical protein | 295 | 9 | 21/50 | pat |
| Spy2016 | Inhibitor of complement (Sic) | 185 | 10 | 17/50 | pat |
| Spy2010 | C5A peptidase precursor | 172 | 11 | Not determined | 10; pat |
| Spy1983 | Collagen-like surface protein (SclD) | 169 | 6 | 50/50 | |
| Spy2025 | Immunogenic secreted protein precursor | 155 | 7 | 50/50 | |
| Spy0469 | Putative 42-kDa protein | 145 | 7 | 50/50 | pat |
| Spy0437 | Hypothetical protein | 99 | 5 | 19/50 | |
| Spy0747 | Extracellular nuclease | 98 | 4 | 50/50 | pat |
| Spy2006 | Hypothetical protein | 95 | 9 | 50/50 | pat |
| Spy0019 | Putative secreted protein | 83 | 5 | 50/50 | pat |
| Spy0166 | Hypothetical protein | 82 | 5 | 50/50 | |
| Spy0488 | Hypothetical protein | 78 | 7 | 50/50 | |
| Spy0872 | Putative secreted 5′-nucleotidase | 75 | 9 | 50/50 | pat |
| Spy1798 | Hypothetical protein | 69 | 6 | 50/50 | |
| Spy1361 | Putative internalin A precursor | 62 | 5 | 50/50 | pat |
| Spy0230 | Putative ABC transporter (ATP binding) | 46 | 1 | 50/50 | |
| Spy1357 | Protein GRAB (protein G-related alpha 2 M-binding protein) | 41 | 4 | 49/50 | |
| Spy0269 | Putative surface exclusion protein | 39 | 6 | 50/50 | |
| Spy1494 | Hypothetical protein | 39 | 7 | 50/50 | |
| Spy0430 | Hypothetical protein | 35 | 3 | 13/50 | |
| Spy1813 | Hypothetical protein | 34 | 3 | 46/50 | |
| Spy1228 | Putative lipoprotein | 33 | 1 | 49/50 | pat |
| Spy2039 | Pyrogenic exotoxin B | 30 | 3 | Not determined | 36; pat |
| Spy0115 | Putative glutamyl-aminopeptidase | 29 | 2 | 50/50 | |
| Spy2191 | Hypothetical protein | 28 | 3 | 50/50 | |
| Spy1801 | Immunogenic secreted protein precursor | 27 | 4 | 50/50 | pat |
| Spy0727 | Putative DNA gyrase, subunit B | 26 | 1 | Not determined | |
| Spy1536 | Conserved hypothetical protein | 22 | 2 | 50/50 | |
| Spy1972 | Pullulanase | 22 | 4 | 50/50 | pat |
| Spy0012 | Hypothetical protein | 19 | 3 | 50/50 | |
| Spy0737 | Putative extracellular matrix-binding protein | 19 | 3 | 29/50 | |
| Spy1979 | Streptokinase A precursor | 19 | 3 | 50/50 | pat |
| Spy2009 | Hypothetical protein | 18 | 4 | 39/50 | |
| Spy1371 | Putative NADP-dependent GAPDH | 17 | 2 | 50/50 |
Antigens selected by more than 15 individual clones.
Number of selected clones for an antigen.
Number of screens in which the antigen was selected.
Number of strains in which the gene was detected by PCR/number of tested strains.
References are shown for antigens published as protective. pat, claimed in a published patent.
TABLE A1.
Immunogenic proteins of Streptococcus pyogenes identified by the Antigenome technology
| ORF | Common name | No. of clonesa | No. of screensb | Library:no. of clones per ORFc | Location of immunogenic region (aa)d |
|---|---|---|---|---|---|
| Spy0012 | Hypothetical protein | 19 | 3 | A:12, I:5, N:2 | 1-114 |
| Spy0019 | Putative secreted protein (cell division and antibiotic tolerance) | 83 | 5 | F:2, I:16, K:24, N:29, P:12 | 29-226 |
| Spy0025 | Putative phosphor-ribosylformylglycinamidine synthase II | 3 | 1 | D:3 | 919-929 |
| Spy0031 | Putative choline-binding protein | 9 | 3 | I:3, K:3, N:3 | 145-305 |
| Spy0103 | Putative competence protein | 8 | 1 | A:8 | 71-81 |
| Spy0112 | Putative pyrroline carboxylate reductase | 4 | 1 | B:4 | 173-186 |
| Spy0115 | Putative glutamyl-aminopeptidase | 29 | 2 | A:3, C:26 | 316-331 |
| Spy0166 | Hypothetical protein | 82 | 5 | I:22, K:7, N:17, O:31, P:5 | 21-99 |
| Spy0167 | Streptolysin O | 1,465 | 14 | A:118, B:14, C:18, D:37, F:141, G:79, H:92, I:97, K:123, L:5, M:21, N:225, O:230, P:265 | 9-264 |
| Spy0168 | Hypothetical protein | 11 | 2 | K:4, N:7 | 1-112 |
| Spy0171 | Hypothetical protein | 2 | 1 | H:2 | 21-56 |
| Spy0183 | Putative glycine betaine/proline ABC transporter | 6 | 1 | C:6 | 23-39 |
| Spy0230 | Putative ABC transporter (ATP-binding protein) | 46 | 1 | C:46 | 474-489 |
| Spy0269 | Putative surface exclusion protein | 39 | 6 | A:2, B:12, D:3, F:11, H:5, N:6 | 37-241 |
| 409-534 | |||||
| 582-604 | |||||
| 743-804 | |||||
| Spy0287 | Conserved hypothetical protein | 1 | 1 | K:1 | 202-337 |
| Spy0292 | Penicillin-binding protein | 2 | 1 | F:2 | 1-48 |
| Spy0295 | Oligopeptide permease | 3 | 1 | A:3 | 203-217 |
| Spy0348 | Putative aminodeoxychorismate lyase | 14 | 4 | D:5, I:3, M:3, P:3 | 261-273 |
| Spy0416 | Putative cell envelope serine proteinase | 462 | 13 | A:3, B:4, C:30, D:13, F:134, G:120, H:93, I:9, K:13, M:2, N: 14, O:8, P:19 | 1-414 |
| 443-614 | |||||
| 997-1392 | |||||
| Spy0430 | Hypothetical protein | 35 | 3 | B:7, I:10, P:18 | 1-164 |
| Spy0433 | Hypothetical protein | 295 | 9 | A:138, B:8, C:67, D:11, E:13, F:35, G:10, H:5, M:8 | 126-207 |
| Spy0437 | Hypothetical protein | 99 | 5 | A:29, B:10, C:21, D:24, E:15 | 180-204 |
| Spy0469 | Putative 42-kDa protein | 145 | 7 | B:5, F:77, I:8, K:15, M:3, N:17, O:20 | 11-197 |
| 204-219 | |||||
| 258-372 | |||||
| Spy0488 | Hypothetical protein | 72 | 7 | A:17, B:11, C:23, D:11, G:4, H:6 | 195-289 |
| Spy0515 | Putative sugar transferase | 8 | 2 | B:5, I:3 | 12-130 |
| Spy0580 | Conserved hypothetical protein | 5 | 1 | C:5 | 434-444 |
| Spy0621 | Conserved hypothetical protein | 3 | 1 | C:3 | 360-375 |
| Spy0630 | Putative PTSe-dependent N-acetyl-galactosamine IIC | 2 | 1 | C:2 | 254-260 |
| Spy0681 | Hypothetical protein, phage associated | 8 | 1 | A:8 | 369-382 |
| Spy0683 | Putative minor capsid protein, phage associated | 15 | 2 | B:11, D:4 | 270-312 |
| Spy0702 | Hypothetical protein | 2 | 1 | L:2 | 486-598 |
| Spy0710 | Conserved hypothetical protein, phage associated | 10 | 1 | B:10 | 378-396 |
| Spy0711 | Pyrogenic exotoxin C precursor, phage associated (speC) | 2 | 1 | K:2 | 75-235 |
| Spy0720 | Conserved hypothetical protein | 2 | 1 | D:2 | 30-51 |
| Spy0727 | Putative DNA gyrase, subunit B | 26 | 1 | M:26 | 208-219 |
| Spy0737 | Putative extracellular matrix-binding protein | 19 | 3 | B:5, E:3, K:11 | 396-533 |
| 1342-1502 | |||||
| 1672-1920 | |||||
| Spy0747 | Extracellular nuclease | 98 | 4 | A:72, B:17, H:6, O:3 | 1-113 |
| 210-232 | |||||
| 250-423 | |||||
| 536-564 | |||||
| Spy0777 | Putative ATP-dependent exonuclease, subunit A | 6 | 2 | C:4, E:2 | 617-635 |
| Spy0789 | Putative ABC-transporter (permease protein) | 3 | 1 | A:3 | 190-203 |
| Spy0839 | Putative glycerophosphodiester phosphodieste | 9 | 2 | A:7, D:2 | 385-398 |
| Spy0843 | Cell surface protein | 1,079 | 14 | A:11, B:3, C:5, D:4, F:50, H:19, G:49, I:112, K:102, L:10, M:3, N:213, O:188, P:310 | 12-190 |
| 276-283 | |||||
| 666-806 | |||||
| Spy0872 | Putative secreted 5′-nucleotidase | 75 | 9 | A:6, D:2, F:5, H:14, I:9, K:10, L:1, N:16, O:12 | 30-80 |
| 89-105 | |||||
| 111-151 | |||||
| Spy0895 | Histidine protein kinase | 11 | 1 | C:11 | 195-203 |
| Spy0972 | Putative terminase, large subunit, phage | 2 | 1 | B:2 | 32-50 |
| Spy0981 | Hypothetical protein, phage associated | 9 | 2 | A:7, B:2 | 75-90 |
| Spy1008 | Streptococcal exotoxin H precursor (speH) | 11 | 1 | C:11 | 69-88 |
| Spy1032 | Extracellular hyaluronate lyase | 11 | 3 | B:3, K:3, M:5 | 96-230 |
| 361-491 | |||||
| 572-585 | |||||
| Spy1054 | Putative collagen-like protein (SclC) | 1,295 | 9 | A:71, B:13, C:233, D:41, E:163, F:200, G:442, H:129, N:3 | 102-210 |
| Spy1063 | Putative periplasmic-iron-binding protein | 4 | 1 | A:4 | 240-248 |
| Spy1162 | Putative RNase HII | 8 | 2 | B:3, C:5 | 182-198 |
| Spy1206 | Putative ABC transporter | 2 | 1 | A:2 | 41-56 |
| Spy1228 | Putative lipoprotein | 33 | 1 | M:33 | 202-217 |
| Spy1245 | Putative phosphate ABC transporter | 6 | 2 | I:3, K:3 | 1-127 |
| Spy1315 | Hypothetical protein | 4 | 1 | B:4 | 297-458 |
| Spy1357 | Protein GRAB (protein G-related alpha 2 M-binding protein) | 41 | 4 | G:27, H:8, K:2, N:4 | 24-135 |
| Spy1361 | Putative internalin A precursor | 62 | 5 | F:21, G:26, H:6, K:4, N:5 | 176-330 |
| Spy1371 | Putative NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | 17 | 2 | D:14, H:3 | 46-62 |
| 296-341 | |||||
| Spy1375 | Putative ribonucleotide reductase alpha-c | 2 | 1 | A:2 | 667-684 |
| Spy1389 | Putative alanyl-tRNA synthetase | 5 | 2 | B:2, P:3 | 258-416 |
| Spy1390 | Putative protease maturation protein | 8 | 3 | A:3, B:2, D:3 | 278-295 |
| Spy1422 | Putative recombination protein | 2 | 1 | C:2 | 183-195 |
| Spy1436 | Putative DNase | 1 | 1 | K:1 | 63-238 |
| Spy1494 | Hypothetical protein | 39 | 7 | G:3, I:5, K:6, M:5, N:10, O:6, P:4 | 1-141 |
| Spy1523 | Cell division protein | 2 | 1 | I:2 | 231-368 |
| Spy1536 | Conserved hypothetical protein | 22 | 2 | A:19, C:3 | 247-260 |
| Spy1564 | Conserved hypothetical protein | 4 | 1 | C:4 | 64-72 |
| Spy1604 | Conserved hypothetical protein | 4 | 2 | B:2, K:2 | 222-362 |
| 756-896 | |||||
| Spy1607 | Conserved hypothetical protein | 5 | 1 | D:5 | 153-170 |
| Spy1615 | Putative late competence protein | 4 | 1 | C:4 | 56-73 |
| Spy1666 | Conserved hypothetical protein | 2 | 1 | D:2 | 298-312 |
| Spy1727 | Conserved hypothetical protein | 6 | 1 | B:5 | 141-157 |
| Spy1785 | Putative ATP-dependent DNA helicase | 3 | 1 | D:3 | 433-440 |
| 572-593 | |||||
| Spy1798 | Hypothetical protein | 69 | 6 | A:12, I:12, K:7, N:17, O:13, P:8 | 17-319 |
| 417-563 | |||||
| Spy1801 | Immunogenic secreted protein precursor homolog | 27 | 4 | H:2, I:8, K:6, N:11 | 46-187 |
| Spy1813 | Hypothetical protein | 34 | 3 | I:16, K:12, N:6 | 21-244 |
| 381-499 | |||||
| 818-959 | |||||
| Spy1821 | Putative translation elongation factor EF-P | 6 | 1 | C:6 | 118-136 |
| Spy1916 | Putative phospho-beta-d-galactosidase | 8 | 1 | C:8 | 147-155 |
| Spy1972 | Pullulanase | 22 | 4 | A:6, I:2, K:5, N:9 | 74-438 |
| Spy1979 | Streptokinase A precursor | 19 | 3 | I:6, M:3, N:10 | 156-420 |
| Spy1983 | Collagen-like surface protein (SclD) | 169 | 6 | A:81, B:24, F:19, G:41, I:2, K:2 | 79-348 |
| Spy1991 | Anthranilate synthase component II | 2 | 1 | D:2 | 53-70 |
| Spy2000 | Surface lipoprotein | 5 | 2 | B:3, N:2 | 183-341 |
| Spy2006 | Hypothetical protein | 95 | 9 | A:15, B:9, C:5, D:3, F:18, G:25, H:5, M:10, N:5 | 92-231 |
| 618-757 | |||||
| Spy2009 | Hypothetical protein | 18 | 4 | B:2, I:7, K:7, P:2 | 41-170 |
| Spy2010 | C5A peptidase precursor | 172 | 11 | A:47, B:10, D:3, F:48, G:20, H:4, I:6, K:13, M:5, N:10, P:6 | 20-487 |
| 757-1153 | |||||
| Spy2016 | Inhibitor of complement (Sic) | 185 | 10 | A:11, B:38, C:16, F:56, G:27, H:13, K:5, N:2, O:3, P:14 | 26-74 |
| 91-100 | |||||
| 105-303 | |||||
| Spy2018 | M1 protein | 783 | 12 | A:316, B:26, C:107, D:12, E:49, F:88, G:118, H:6, I:7, K:2, M:48, N:4 | 10-223 |
| 231-251 | |||||
| 264-297 | |||||
| 312-336 | |||||
| Spy2025 | Immunogenic secreted protein precursor | 155 | 7 | F:7, G:16, H:7, K:63, L:2, N:18, O:42 | 22-344 |
| Spy2039 | Pyrogenic exotoxin B | 30 | 3 | I:15, K:3, N:12 | 1-151 |
| Spy2043 | Mitogenic factor MF1 (speF) | 1 | 1 | K:1 | 91-263 |
| Spy2059 | Penicillin-binding protein 2a | 4 | 2 | D:2, E:2 | 261-272 |
| Spy2110 | Putative anaerobic ribonucleoside-triphosphate reductase | 7 | 1 | E:7 | 541-551 |
| Spy2127 | Hypothetical protein | 8 | 2 | I:6, P:2 | 84-254 |
| Spy2191 | Hypothetical protein | 28 | 3 | C:20, E:3, M:5 | 61-78 |
| Spy2211 | Transmembrane protein | 3 | 1 | A:3 | 568-580 |
Number of hits for a particular antigen.
Number of screens in which the antigen was selected.
A, LSPy-70 library in lamB with IC3-IgG (number of clones successfully sequenced, 1,588); B, LSPy-70 library in lamB with IC3-IgA (1,539); C, LSPy-70 library in lamB with IC6-IgG (1,173); D, LSPy-70 library in lamB with P4-IgG (1,138); E, LSPy-70 library in lamB with P4-IgA (981); F, LSPy-150 library in btuB with IC3-IgG (991); G, LSPy-150 library in btuB with IC6-IgG (1,036); H, LSPy-150 library in btuB with P4-IgG (681); I, LSPy-400 library in fhuA with IC3-IgG (559); K, LSPy-400 library in fhuA with IC6-IgG (543); L, LSPy-400 library in fhuA with P4-IgG (20); M, LSPy-70 library in lamB with P13-IgG (796); N, LSPy-400 library in fhuA with P13-IgG (762); O, LSPy-400 library in fhuA with P13-IgA (652); P, LSPy-400 library in fhuA with IC3-IgA (741).
Amino acid (aa) positions of the ORF borne by selected clones.
PTS, phosphotransferase system.
Selection of candidate antigens from the Antigenome by in vitro assays.
In order to select the most promising candidate antigens for further evaluation in animal models of group A streptococcal disease, we performed peptide ELISA with human sera and gene conservation analysis. In addition, we analyzed all antigens by bioinformatics for the lack of homology with human proteins and for novelty with regard to intellectual property.
Selected epitopes corresponding to 84 annotated ORFs and 23 nonannotated sequences in the S. pyogenes SF370 genome were evaluated with individual human sera used for the antigen screens. Streptavidin-coated ELISA plates were coated with synthetic peptides of approximately 25 to 30 amino acids in length, with an N-terminal biotin tag representing the selected immunogenic epitopes. Immune reactivities of individual synthetic peptides were assessed with the following individual human sera: P227, P256, P268, P269, P273, P274, P275, P279, P290, P884, P911, IC24, IC35.A, IC37A, IC38, IC39, IC44A, IC48, IC54A, IC58A, IC62A, and N33 (Table 2). The peptides showed a wide range of reactivity, from highly and widely reactive, e.g., Spy0416, Spy0872, Spy1063, Spy2010, Spy2016, and Spy2018, to weakly positive or negative (Table 2). The peptide ELISA data correlated largely also with the results from the Western blot analyses for the selected antigen epitopes, as the most immunogenic peptides were often also selected most frequently in the screens, e.g., Spy0843, Spy1983, Spy2010, and Spy2016 (Table 1). In fewer cases, highly immunogenic epitopes were observed by ELISA from less frequently selected proteins (e.g., Spy0720, Spy1063, and Spy1666), but only in rare cases did overlapping synthetic peptides from frequently selected long epitopes not display high antibody levels in ELISAs, most probably due to the presence of conformational, discontinuous epitopes (e.g., Spy1054 or Spy0437). When we analyzed the 24 most immunogenic peptides from the above-described ELISAs with a set of five sera, each obtained from patients with acute rheumatic fever and glomerulonephritis, 13 peptides showed reactivity comparable with that of sera from individuals without poststreptococcal sequelae, whereas 5 showed a strongly reduced reactivity and 6 an intermediate reduction (unpublished data). This indicates that the immune response in individuals developing poststreptococcal sequelae may differ from that in healthy individuals not contracting or recovering from initial GAS disease. Further studies will be necessary to validate these observations.
TABLE 2.
ELISA data for the 50 most immunogenic peptides from GAS
ELISA units are coded as follows: white. <200 ELISA U; light gray, 200 to 350 ELISA U; dark gray, 350 to 500 ELISA U; black, >500 ELISA U. ELISA units are calculated from OD405 readings and a serum dilution of 1:200 after correction for background.
Scores were calculated as average values of all single ELISA units obtained for the individual peptides.
As a second selection criterion, we determined the presence of the genes encoding the identified antigens in a set of 50 GAS isolates encompassing 15 different emm types (1, 1T1, 3.1, 3.2, 12, 22.2, 28, 49, 81, 83, 85, 89, 89.3, 94, T25) collected from hospitals in Europe. PCR analyses for 90 annotated genes of the Antigenome revealed that 70 genes were present in all or >90% of tested clinical isolates (gene-specific amplification was confirmed by sequencing the PCR product from at least one strain for each gene) (Table 3). Twenty genes were detected in less than 90% of the clinical isolates tested and therefore excluded from further evaluation.
TABLE 3.
S. pyogenes vaccine candidates selected based on in vitro assays
| ORF | Common name | Motif(s)a | Amino acids | Serology score(s)b | Gene distributionc | Reference(s)d |
|---|---|---|---|---|---|---|
| Spy0012 | Hypothetical protein | SP | 428 | 342, 336 | 50/50 | |
| Spy0019 | SibA (secreted immunoglobulin-binding protein) | SP | 398 | 252 | 50/50 | 39 |
| Spy0031 | Putative choline-binding protein | SP | 374 | 59, 108, 3, 30 | 50/50 | |
| Spy0103 | Putative competence protein | SP | 108 | 130 | 50/50 | |
| Spy0269 | Putative surface exclusion protein | SP | 873 | 104, 493 | 50/50 | |
| Spy0292 | Penicillin-binding protein | SP | 410 | 64 | 50/50 | |
| Spy0348 | Putative aminodeoxychorismate lyase | None | 524 | 309 | 50/50 | |
| Spy0416 | Putative cell envelope proteinase | SP, LPXTG | 1,647 | 376, 492, 362 | 50/50 | 39 |
| Spy0488 | Hypothetical protein | None/SP | 331 | 0, 238, 181, 200, 23 | 50/50 | |
| Spy0720 | Conserved hypothetical protein | None | 313 | 382 | 50/50 | |
| Spy0872 | Putative secreted 5′-nucleotidase | SP, LPXTG | 670 | 547, 199 | 50/50 | 39, 47 |
| Spy0895 | Histidine protein kinase | None | 262 | 239 | 50/50 | |
| Spy1063 | Putative periplasmic-iron-binding protein | None | 323 | 614 | 49/50 | |
| Spy1245 | Putative phosphate ABC transporter | SP | 288 | 11, 5, 3, 38 | 50/50 | 38 |
| Spy1315 | Hypothetical protein | SP | 724 | 2 | 50/50 | |
| Spy1357 | GRAB protein | SP, LPXTG | 217 | 73 | 49/50 | 63 |
| Spy1361 | Putative internalin A precursor | SP | 792 | 35, 79, 108 | 50/50 | 47 |
| Spy1390 | Putative protease maturation protein | SP | 351 | 312 | 50/50 | 38 |
| Spy1494 | Hypothetical protein | SP, LPXTG | 313 | 12, 40, 164, 99, 263 | 50/50 | |
| Spy1536 | Conserved hypothetical protein | SP | 345 | 274 | 50/50 | |
| Spy1607 | Conserved hypothetical protein | None | 258 | 319 | 50/50 | |
| Spy1666 | Conserved hypothetical protein | None | 337 | 491 | 50/50 | |
| Spy1727 | Conserved hypothetical protein | None | 263 | 273 | 50/50 | |
| Spy1798 | Hypothetical protein | SP | 1,275 | 34, 120, 105, 116, 0, 59 | 50/50 | |
| Spy1813 | Hypothetical protein | SP | 995 | 149 | 46/50 | |
| Spy1972 | Putative pullulanase | SP, LPXTG | 1,165 | 376, 228, 94 | 50/50 | 39, 47 |
| Spy1979 | Streptokinase A precursor | SP | 440 | 75, 104 | 50/50 | 39 |
| Spy2000 | Surface lipoprotein | None | 542 | 107, 0, 67, 150, 33, 43, 0, 200 | 50/50 | |
| Spy2025 | Immunogenic secreted protein precursor | SP | 541 | 304, 99, 110, 82 | 50/50 | |
| Spy2191 | Hypothetical protein | SP | 204 | 449 | 50/50 | |
| Spy2211 | Conserved hypothetical protein | SP, LPXTG | 858 | 108 | 50/50 | |
| Spy2018 | M1 protein | SP, LPXTG | 484 | 124, 210, 519, 26 | Not determined | 37 |
SP, signal peptide; LPXTG, cell wall-anchoring motif.
Scores for individual peptides were calculated as average values of all single ELISA units obtained for individual peptides.
Number of PCR-positive strains of 50 analyzed GAS strains.
References are shown for ORFs previously described as an antigen.
Based on the ELISA data, gene conservation, bioinformatic analysis, and novelty, we selected 31 antigens for further evaluation in animal models (Table 3). These vaccine candidates were cloned into the pET28b vector for expression of recombinant proteins. Whenever it was possible, full-length proteins were expressed; however, ancillary sequences encoding, e.g., signal peptides and cell wall anchor sequences clipped off by sortases were removed. Some of the genes were cloned in several smaller fragments due to their large size, which was predicted or shown to hinder efficient expression of the encoded full-length protein. The M protein (derived from strain AP1 or A20-MA) was used for all animal experiments as a positive control.
Nine Antigenome-derived candidate GAS antigens provide protection against group A streptococcal infection in mouse sepsis models.
In order to select protective antigens from the 31 candidates, we established several different animal models using three S. pyogenes challenge strains, AP1 (serotype M1), A147-MA (M106), and A20-MA (M23), two different immunization (subcutaneous or intranasal) and challenge (intravenous or intranasal) routes, and three different mouse strains. Protection was evaluated based on survival rates on day 14 postchallenge.
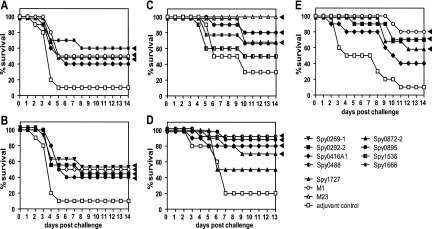
We adapted the A20-MA and A147-MA strains by serially passaging them in mice so that they would be suitable for inducing invasive GAS infection upon intranasal challenge of CD-1 mice. Animals were immunized subcutaneously with the recombinant antigens adjuvanted with CFA-IFA, aluminum hydroxide (AlOH3), or no adjuvant (up to four individual experiments; 10 mice/group). These studies showed that six GAS proteins, namely, Spy0269, Spy0292, Spy0416, Spy0872, Spy0895, and Spy1666, were capable of inducing significant protection against intranasal A20-MA GAS infection (Fig. 2 A and B). The protection levels ranged from 30% above the control level to a maximum of 50% survival in individual experiments.
FIG. 2.
Protection by candidate antigens in intravenous and intranasal challenge models. CD-1 (A and B), BALB/c (C and D), and NMRI (E) mice (10 mice/group) were immunized subcutaneously (A, B, E) or intranasally (C, D) either with 50 μg M1 or M23 protein (positive control) or with 50 μg candidate antigens with 1% aluminum hydroxide (A, B), IC31 (10 nmol KLK, 0.4 nmol ODN1a) (C), 0.5 μg of MALP-2 (D), or CFA-IFA (E). For the negative control, PBS with adjuvant was used. About 2 weeks after the last booster immunization, mice were challenged intranasally with 1 × 108 CFU S. pyogenes A20-MA (M23) (A, B), intranasally with 7 × 105 CFU S. pyogenes A20-MA (C), intranasally with 7.6 × 107 CFU S. pyogenes A147-MA (M106) (D), or intravenously with 7.5 × 107 CFU S. pyogenes AP-1 (M1) (E). Numbers of surviving mice are plotted as a percentage of total mice. Statistically significant differences based on the Kaplan-Meier log rank (Mantel-Cox) test are indicated by arrowheads.
Some of the same antigens, Spy0416, Spy0872, Spy0895, and Spy1666, but also three additional ones, Spy0488, Spy1536, and Spy1727, protected mice against the A147-MA or A20-MA challenge upon intranasal immunization with two different adjuvants, IC31 (55) and MALP-2 (49) (two independent experiments) (Fig. 2C and D). Levels of protection for the seven antigens ranged from 40 to 70% above the adjuvant control level.
In addition, we also evaluated these antigens in a further animal model using a mouse-adapted M1 serotype GAS strain (AP1) upon intravenous bacterial challenge of NMRI mice after subcutaneous immunization with CFA-IFA (up to four independent experiments). In this stringent model, three antigens, Spy0292, Spy0416, and Spy0872, provided protection, with an average of 44 to 50% survival (Fig. 2E).
Based on all the animal protection data, we selected nine group A streptococcal proteins, Spy0269, Spy0292, Spy0416, Spy0488, Spy0872, Spy0895, Spy1536, Spy1666, and Spy1727, as the most promising candidates for further vaccination studies. Experiments are ongoing to further characterize the protective efficacies of the candidates individually and in combination.
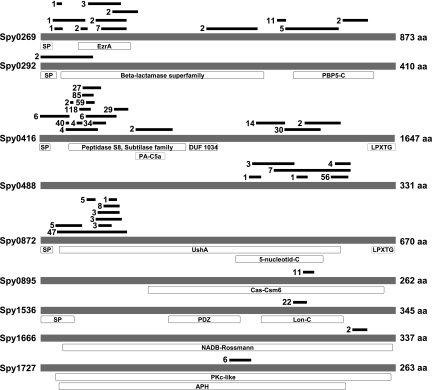
The nine selected vaccine candidates have not been characterized previously in more detail, with the exception of Spy0416, which was recently shown to possess interleukin 8 (IL-8)-degrading activity (15, 21, 30). The other remaining eight proteins have no or only putative functions assigned (Fig. 3). Spy0488, Spy1536, Spy1666, and Spy1727 were described as hypothetical or conserved hypothetical proteins and show only very limited similarity to proteins with known functions. Spy0269 was described as a putative surface exclusion protein and possesses a short 130-amino-acid region showing similarity with the EzrA protein, which interacts with the cell division protein FtsZ (40). Spy0292 was annotated as penicillin-binding protein, but no beta-lactamase activity has yet been reported for this protein. The Spy0872 protein shows similarity to secreted 5′-nucleotidases, enzymes located at the cell surface, such as UshA of E. coli (8), with an important function in nucleotide salvage or growth on nucleosides as a carbon source. Spy0895 displays weak similarity with several proteins and was thus annotated as a putative histidine kinase, the transcription regulator LytR, or a hypothetical protein, but no function has so far been experimentally determined.
FIG. 3.
Epitope map and structural features of the 9 protective antigens. The schematic drawings are based on the protein sequences encoded by the SF370 genome. aa, amino acids; SP, signal peptide; EzrA, septation ring formation regulator EzrA; PBP5-C, penicillin-binding protein 5, C-terminal domain; PA-C5a, protease-associated domain of C5a-like proteins; DUF 1034, domain of unknown function; LPXTG, cell wall anchor motif; UshA, 5′-nucleotidase-2′,3′-cyclic phosphodiesterase and related esterases; 5-nucleotid-C, C-terminal domain of 5′-nucleotidases; Cas-Csm6, CRISPR (clustered regularly interspaced short palindromic repeats)-associated protein; PDZ, PDZ (postsynaptic density disc-large zo-1′) domain, also called DHR (discs-large homologous region); Lon-C, Lon protease (S16) C-terminal proteolytic domain; NADB-Rossmann, Rossmann fold of NAD(P)+-binding proteins; PKc-like, catalytic domain of protein kinase C superfamily; APH, aminoglycoside-2"-phosphotransferase enzyme family. Black bars represent epitope regions identified in the surface display screens with human immunoglobulins, with the numbers representing the numbers of individual clones selected for the depicted regions.
Genomic conservation of GAS vaccine candidates.
The goal of the present study was the selection of highly conserved protective vaccine candidates. We thus extended the gene distribution study beyond the detection of the encoding genes via PCR and evaluated the sequence conservation of the nine protective proteins in a selected, representative set of clinical isolates of S. pyogenes. While this work was in progress, 12 genome sequences of S. pyogenes became available in addition to SF370, allowing a comparison of the selected nine candidates in 13 GAS strains of 10 different M serotypes (31, 43). Table 4 shows that Spy0269, Spy0416, Spy0872, Spy1536, Spy1666, and Spy1727 display at least 98% amino acid sequence identity in all published genomes, with only a few genomes encoding a protein with a potentially different length, most likely due to a sequencing error or aberrant annotation. For Spy0292, Spy0488, and Spy0895, the level of sequence identity is larger than 96 to 98% in all genomes, but some genes are predicted to encode distinctively smaller proteins (Table 4). In addition to including the published genomes in our analyses (derived from strains originating from Canada, the United States, New Zealand, and Japan), we sequenced the nine candidate genes in up to 51 different GAS strains collected from hospitals in Europe, including the AP1 strain isolated in Australia (emm types 1, 3, 3.1, 4, 5, 6, 9, 11, 12, 19, 22, 22.2, 25, 28, 44, 49, 66, 76, 78, 81, 83, 85, 89, 90, 94, 118). The obtained data confirmed the high level of sequence conservation of all 9 proteins (Spy0269, >98.7%; Spy0292, >97.3%; Spy0416, 98.1%; Spy0488, >85.4%; Spy0872, >98.2%; Spy0895, >98.9%; Spy1536, >99.1%; Spy1666, >98.2%; Spy1727, 100%).
TABLE 4.
Gene sequence conservation of the nine vaccine candidates
| ORFa | Amino acid length (SF370-M1) | % amino acid sequence identityb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Manfredo (M5) | MGAS 10270 (M2) | MGAS 10394 (M6) | MGAS 10750 (M4) | MGAS 2096 (M12) | MGAS 315 (M3) | MGAS 5005 (M1) | MGAS 6180 (M28) | MGAS 8232 (M18) | MGAS 9429 (M12) | NZ131 (M49) | SSI-1 (M3) | ||
| Spy0269 | 873 | 99.4 | 99.2 | 99.5 | 99.5 | 99.4 | 99.1 | 100 | 99.4 | 99.7 | 99.4 | 99.8 | 99.1 |
| Spy0292 | 410 | 99.5 | 99.5 | 99.3 | 99.3 | 100 (271)c | 99.8 | 99.8 | 99.3 | 99.5 | 99.8 | 97.2 (359)c | 99.8 |
| Spy0416 | 1,647 | 98.5 | 98.1 | 98.8 | 97.6 (911)c | 98.7 | 98.3 (1,622)d | 99.9 | 98.5 | 98.6 | 98.7 | 98.3 (1,621)d | 98.3 (1,622)c,d |
| Spy0488 | 331 | 98.4 (315) | 96.8 (315) | 98.4 (315) | 97.8 (315) | 98.4 (315) | 98.1 (315) | 100 | 96.5 (315) | 96.2 (315) | 98.4 (315) | 96.8 (315) | 98.1 (315) |
| Spy0872 | 670 | 99.1 | 99.6 | 99.3 | 98.2 (661)d | 98.5 | 99.6 | 100 | 98.5 | 99.1 | 98.5 | 99.3 | 99.6 |
| Spy0895 | 262 | 19.4 (129)c | 99.6 | 99.2 | 98.9 | 98.9 | 99.5 (217)d | 100 | 98.9 | 99.6 | 98.9 | 100 | 100 |
| Spy1536 | 345 | 100 | 100 | 100 | 99.4 | 99.7 | 100 | 100 | 99.7 | 100 | 99.7 | 99.4 | 100 |
| Spy1666 | 337 | 100 (316)d | 100 | 99.4 | 98.2 | 100 | 100 | 100 | 100 | 100 | 100 | 99.7 (316)d | 100 |
| Spy1727 | 263 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.6 (254)d |
Sequences for alignment were derived from the S. pyogenes strain SF370.
Pair-wise comparison of sequences from SF370 and homologous sequences from 12 published genomes. Values in parentheses show lengths of incomplete alignments (in amino acids).
The sequence annotated in the genome was shorter due to a frameshift possibly caused by a sequencing error.
The sequence was shorter due to a differently annotated start codon.
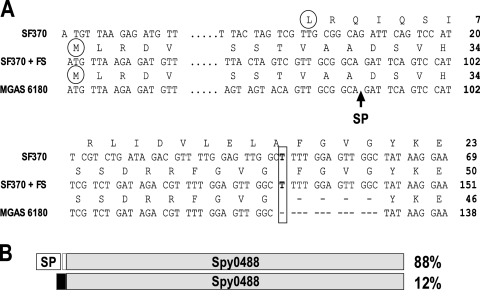
In agreement with the published genomes, the sequencing and comparison of the spy0488 gene indicated that certain genomes (SF370, MGAS5005, and 6 clinical isolates [7 strains of serotype M1 and 1 of serotype M4]) may possess a frameshift within the signal peptide containing the sequence of the gene. Such programmed frameshifts have been observed previously, e.g., for Escherichia coli prfB (RF2) (53). In the above-listed eight strains, the spy0488 gene could be expressed only with a signal peptide, under the assumption that a programmed frameshift occurs at bp position 48 of the currently annotated genes (if not, all 8 sequences contain the same sequencing artifact), where the sequence displays a duplication of 13 bp relative to the other genomes (Fig. 4). All other genomes predict a signal peptide sequence of 30 or 32 amino acids for the spy0488 gene. Preliminary immunoblot analysis of total lysates prepared from different GAS strains, including the ones with an aberrant spy0488 genomic sequence, revealed that Spy0488-specific rabbit hyperimmune sera detected a protein of an apparently identical size, providing the first evidence for the postulated frameshift (data not shown).
FIG. 4.
Multiple gene sequences predict a frameshift (FS) in the spy0488 gene. (A) Alignment of the spy0488 gene and protein sequences from the SF370 strain and the clinical isolate MGAS6180. The postulated start codon is circled, and the predicted signal peptide (SP) cleavage site is indicated by the arrow. Numbers refer to positions in the gene or full-length protein. The observed 13-bp duplication in the spy0488 gene from SF370 requires the predicted frameshift to restore the signal peptide-encoding sequence as annotated for the MGAS6180 gene. (B) Schematic presentation of the predicted frameshift in the spy0488 gene. The percentage of strains showing the SF370 or MGAS6180 spy0488 gene type is listed (in total, 66 gene sequences were analyzed).
Deletion of the spy1536 gene reduces in vitro binding of S. pyogenes to human plasma and extracellular matrix proteins.
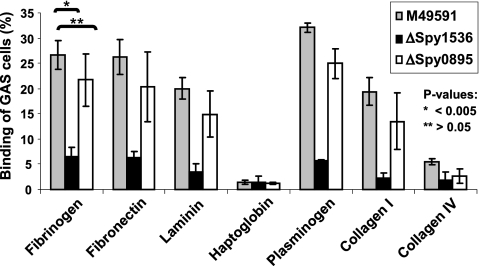
In order to understand the role of the selected vaccine candidates in the pathogenesis and virulence of S. pyogenes, we generated spy0416, spy0895, and spy1536 deletion mutants with the M49591 GAS strain. While our studies confirmed the recently published reduced virulence of the ΔSpy0416 mutant (30), we could not observe reduced virulence or an altered in vitro growth phenotype for the ΔSpy0895 and ΔSpy1536 mutants. As both recombinant proteins mediated protection in animal studies, we initiated a more thorough analysis of both mutants by assessing their possible role as interaction partners with host proteins. Binding of the FITC-labeled M49591 wild type and ΔSpy0895 and ΔSpy1536 cells to selected human plasma and extracellular matrix proteins was therefore analyzed in vitro by an ELISA-based assay. ΔSpy1536 cells showed significantly (P value < 0.005, 2 clones tested in 3 independent experiments) reduced binding to human fibronectin, fibrinogen, laminin, plasminogen, and type I collagen compared to that of the wild type and spy0895 gene deletion mutant cells (Fig. 5). The specificity of the interaction of GAS with these human proteins was supported by our observation that no interaction was detected with haptoglobin or type IV collagen (Fig. 5). The reduced binding of ΔSpy1536 cells was accompanied by a 2- to 3-fold increase in fluorescent light emission of FITC-labeled ΔSpy1536 cells measured at 560 nm in comparison to that of wild-type M49591 and ΔSpy0895 cells, indicating significant changes in the composition of the cell surface of the mutant ΔSpy1536 GAS cells.
FIG. 5.
In vitro binding of wild-type, ΔSpy1536, and ΔSpy0895 GAS cells to human plasma and extracellular matrix proteins. Comparison of levels of binding of FITC-labeled M49591 wild-type and spy0895 and spy1536 gene deletion mutant cells (6 × 107 CFU) to fibrinogen, fibronectin, laminin, haptoglobin, plasminogen, and type I and IV collagen on ELISA plates (10 μg protein in 100 μl per well). Results for 3 independent experiments testing 2 individual clones of each strain were obtained in triplicates. P values were calculated using Student's t test, showing that the reduced binding of ΔSpy1536 cells is highly significant for all proteins except haptoglobin and collagen IV.
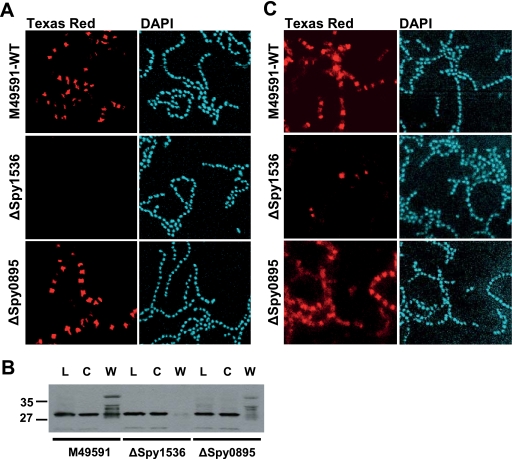
In the absence of spy1536, surface expression of M protein and Spy0269 is drastically reduced on S. pyogenes cells.
The reduced binding of ΔSpy1536 cells to selected human extracellular proteins prompted us to investigate whether this was due to changes of the exposure of group A streptococcal proteins on the bacterial surface. We therefore assessed the surface localization of the major virulence factor of S. pyogenes, M protein, and of two candidate antigens, Spy0269 and Spy1666, by immunofluorescence. Wild-type M49591 and ΔSpy0895 cells showed similar localizations of M protein at the cell surface as visualized by M protein-specific immunofluorescent staining (Fig. 6 A). In contrast, ΔSpy1536 cells displayed no M protein-specific staining under the same experimental conditions (Fig. 6A). Western blot analysis of bacterial lysates and cytoplasmic fractions showed that M protein was expressed at comparable levels in both wild-type and mutant cells (Fig. 6B), indicating that the lack of surface expression was not due to the transcriptional or translational modulation of M protein/gene expression. The cell wall fraction, in contrast, showed a strongly reduced amount of M protein for ΔSpy1536 cells, in agreement with the immunofluorescence data. Similarly to what we observed for M protein, we observed a strong reduction in the surface localization of the Spy0269 protein on ΔSpy1536 cells (Fig. 6C), whereas that of the Spy1666 protein was not affected by the deletion of the spy1536 gene (unpublished data).
FIG. 6.
Localization of M protein and Spy0269 on the surfaces of wild-type (WT), ΔSpy1536, and ΔSpy0895 GAS cells. (A) S. pyogenes cells of an overnight culture of M49591 and ΔSpy1536 and ΔSpy0895 gene deletion mutants were subjected to immunofluorescence analysis with M23-specific polyclonal mouse antibodies, Texas Red dye-conjugated goat anti-mouse antibodies, and DAPI, following microscopic visualization. (B) Western blot analysis of cell fractions of wild-type M49591 and ΔSpy1536 and ΔSpy0895 cells to detect M protein, total lysate (lanes L), the cytoplasmic fraction (lanes C), and the cell wall fraction (lanes W) with M1 protein-specific polyclonal mouse antibodies. For all fractions, an amount of ∼30 μg total protein as measured by the BCA assay was loaded. Numbers at the left indicate molecular weights (in thousands). (C) Immunofluorescence analysis of WT M49591 and ΔSpy1536 and ΔSpy0895 gene deletion mutants with Spy0269-specific polyclonal mouse antibodies, Texas Red dye-conjugated goat anti-mouse antibodies, and DAPI.
DISCUSSION
Development of vaccines to prevent group A streptococcal diseases, although highly warranted, is hindered by the observation that poststreptococcal sequelae are possibly caused by molecular mimicry between GAS antigens and human tissue proteins. The most commonly recognized antigenic structure to induce both humoral and cellular immunological cross-reactivity, especially with heart and joint tissues, is the M protein (11, 13, 18, 42). Besides this link of autoimmunity to poststreptococcal sequelae, the existence of more than 100 different M types hinders M protein-based vaccine development.
While individual non-M protein protective antigens, such as C5A peptidase and SfbI, have been identified by traditional technologies, large-scale proteomic approaches building on the available genomic sequence information have identified novel surface proteins from S. pyogenes on a broader scale, but protection data have been reported for only a few novel antigens (39, 51, 57). Therefore, we embarked on a genome-wide selection of antigens, employing sera from patients with streptococcal pharyngitis, as well as from healthy individuals exposed to, but not colonized by, S. pyogenes. In order to avoid any potential shortcomings by the lack of in vitro expression or proper bioinformatic annotation of protein function, we applied the Antigenome technology (16, 23, 44) to select novel protective GAS vaccine candidates on a genomic scale. As a result, we selected 95 antigenic proteins from S. pyogenes, including most of the already known protective antigens. Among the eight most frequently selected antigens in our screens were five published protective proteins (Table 1), confirming the validity of the technology to identify relevant candidates. Applying several in vitro assays and three different murine infection models, we identified nine novel proteins capable of providing protection against GAS infection (Fig. 2, 3).
One of the most frequently selected antigens, Spy0416 (ScpC), was also identified recently by a proteomic approach and shown to provide protection against an M23 S. pyogenes strain in mice (51). In addition, Spy0416 was shown to mediate cleavage of IL-8, thereby impairing clearance from infected tissues and promoting resistance to neutrophil killing (30, 66). We provided further evidence that this activity was also exhibited by the recombinant form of the Spy0416 protein and required the concerted actions of the enzymatic and ancillary domains of the large ScpC protein (21). These studies further strengthen the selection of Spy0416 as a candidate for a GAS vaccine.
In addition to Spy0416, two proteins, Spy0292 and Spy0872, were found to be efficacious in all three murine models that we applied in this study, including the model using intravenous challenge with the AP1 strain. Two antigens, Spy0895 and Spy1666, protected a significant number of animals in the two lethal sepsis models induced by intranasal challenge with the A20-MA GAS strain. An additional four GAS proteins were selected as vaccine candidates based on protection in one of the models using the A20-MA GAS strain following either subcutaneous (Spy0269 and Spy1536) or intranasal (Spy0488 and Spy1727) immunization of NMRI or BALB/c mice, respectively.
To our knowledge, none of these antigens (except Spy0416) have been shown to protect against GAS infection or have been characterized with regard to their biochemical function and role in streptococcal virulence. We thus initiated the generation of gene deletion mutants for the novel candidates and report here the first results for Spy0895 and Spy1536, annotated in the SF370 genome as a putative histidine protein kinase and conserved hypothetical protein, respectively (Fig. 3). Since we could not observe obvious in vitro growth phenotypes for either of the two deletion mutants or a reduced virulence in mice compared to that of the wild-type M49591 strain, we characterized these mutants in functional assays. The interaction with and adherence to epithelial cells of S. pyogenes are generally accepted as crucial mechanisms for colonization and invasion of the human host and consequently cause diseases. Many different surface molecules mediate the attachment of GAS to human extracellular matrix components, including fibronectin, fibrinogen, laminin, and collagen, and the interaction with human plasma proteins (33, 62). Among these streptococcal surface proteins, M protein binds to fibrinogen and plasminogen (50, 54), the adhesin protein F binds to fibronectin (26, 46), Lbp has been described to bind laminin (60), PAM binds to plasminogen (3, 22), and Cpa binds to type I collagen (34, 64). Based on in vitro binding assays, we observed that deletion of the spy1536 gene resulted in a drastically reduced interaction of S. pyogenes with all human extracellular matrix proteins tested. In contrast to this broad effect of Spy1536 deletion, insertional inactivation of the prtF gene, encoding the major fibronectin-binding protein of GAS, resulted in the selective loss of fibronectin binding and a consequently lower capacity to adhere to respiratory epithelial cells (26, 27). The broad effect on binding to several extracellular matrix proteins (fibronectin, fibrinogen, laminin, plasminogen, and type I collagen) indicated that Spy1536 could be involved in a more general process relevant for the proper surface expression of streptococcal proteins. The lack of M protein and Spy0269 on the bacterial surfaces of ΔSpy1536 cells is consistent with this assumption. It was shown previously that mutation of the sagA gene, encoding streptolysin S (6), did not affect transcription of the emm gene but resulted in the C-terminal truncation of M protein, thus preventing peptidoglycan anchoring and surface localization. The detection of M protein at similar levels in the cytoplasm of wild-type and ΔSpy1536 cells (Fig. 6), but not in the supernatant, indicates that Spy1536 also does not influence the transcription of the M protein gene but might be involved in the process of transporting M protein to the bacterial cell surface or localizing it in the cell wall. Similarly, Spy1536 may affect the surface localization of other proteins, such as Spy0269, using the same or yet another mechanism. Although Spy1536 is annotated in the SF370 genome as a conserved hypothetical protein, it shows similarity to Lon proteases, including a Lon protease PDZ domain, for which reason it was annotated in subsequently published GAS genomes as ATP-dependent endopeptidase Lon. Lon proteases are multidomain enzymes (14, 20, 52) found in all living organisms and are involved in E. coli in the degradation of naturally unstable and misfolded proteins (52, 61). So far, we could not detect any proteolytic activity with the recombinant Spy1536 protein used in these studies, but our data clearly show that the protein is crucial for the surface expression of streptococcal surface proteins and the binding of streptococci to human extracellular matrix proteins. Further, preliminary data from gene array studies confirmed that expression of the Spy0269 and emm genes was unchanged in ΔSpy1536 cells, whereas several surface proteins were downregulated at the transcriptional level, indicating that deletion of the spy1536 gene may also influence transcriptional regulation of surface protein expression.
All nine GAS vaccine candidates described in this study are highly conserved not only in the 13 published S. pyogenes genomes but also in up to 51 further clinical isolates analyzed. Thus, it can be assumed that an immune response against any of these candidates will be able to recognize the pathogen if the respective protein is surface accessible during infection. The observed high level of sequence conservation further argues that these proteins provide an important function to the pathogen. Despite the present lack of understanding regarding their precise role in GAS virulence, our studies clearly showed that all 9 candidates can individually induce protection in mice upon active immunization. Since mice do not develop acute pharyngitis mimicking human disease, all three animal models used in this study relied on mortality as a readout for protection. The only animal model of acute GAS pharyngitis reported to date uses nonhuman primates, which are not suitable for antigen screening (59). In order to avoid elimination of an antigen based on negative data in one model and due to the observed variability of the sepsis models, we applied models using different routes of challenge, adjuvants, and two different GAS challenge strains. As S. pyogenes has evolved many mechanisms of evading the immune system of its human host, including more than one protein in a vaccine will increase the likelihood of inducing broadly protective immune responses and prevention of disease.
The nine identified protective antigens from S. pyogenes are highly conserved in a representative selection of GAS serotypes, and several of the identified candidates were shown or predicted to be involved in the pathogenesis of GAS. This makes them suitable candidates for a group A streptococcal vaccine, and we therefore currently evaluate and select combinations of GAS proteins to achieve a high level of protection against the numerous S. pyogenes serotypes for the development of a vaccine to lower the burden of group A streptococcal diseases worldwide.
Acknowledgments
We thank Christine Triska, Simon Rittmann, Ulrike Stierschneider, Michael Schunn, Karina Watzke, and Barbara Maierhofer for technical help; Ulrike Schirmer for critical reading of the manuscript; Wolfgang Schüler for bioinformatic support; Franz-Josef Schmitz (Klinikum Minden, Germany) and Rodger Novak (Department of Microbiology and Genetic, Vienna) for providing GAS clinical isolates; Zsófia Pusztai (Bethesda Children's Hospital, Hungary) and Rodger Novak for providing human sera; Lars Björck for the S. pyogenes AP-1 strain; Bernd Kreikemeyer for the M49591 strain; Dieter Reinscheid for plasmid pGhost5; and Josef Gotzmann for technical help with confocal microscopy.
The authors affiliated with Intercell AG, a biotechnology company, declare a potential conflict of financial interest.
APPENDIX
Table A1 lists all bacterial clones which were selected by the antigen screens for a particular ORF of S. pyogenes.
Editor: A. Camilli
Footnotes
Published ahead of print on 12 July 2010.
REFERENCES
- 1.Albertí, S., C. García-Rey, M. A. Domínguez, L. Aguilar, E. Cercenado, M. Gobernado, and A. García-Perea. 2003. Survey of emm gene sequences from pharyngeal Streptococcus pyogenes isolates collected in Spain and their relationship with erythromycin susceptibility. J. Clin. Microbiol. 41:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beachey, E. H., J. M. Seyer, J. B. Dale, W. A. Simpson, and A. H. Kang. 1981. Type-specific protective immunity evoked by synthetic peptide of Streptococcus pyogenes M protein. Nature 292:457-459. [DOI] [PubMed] [Google Scholar]
- 3.Berge, A., and U. Sjobring. 1993. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem. 268:25417-25424. [PubMed] [Google Scholar]
- 4.Bessen, D., and V. A. Fischetti. 1990. Synthetic peptide vaccine against mucosal colonization by group A streptococci. I. Protection against a heterologous M serotype with shared C repeat region epitopes. J. Immunol. 145:1251-1256. [PubMed] [Google Scholar]
- 5.Bisno, A. L., M. O. Brito, and C. M. Collins. 2003. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 3:191-200. [DOI] [PubMed] [Google Scholar]
- 6.Biswas, I., P. Germon, K. McDade, and J. R. Scott. 2001. Generation and surface localization of intact M protein in Streptococcus pyogenes are dependent on sagA. Infect. Immun. 69:7029-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns, D. M., and I. R. Beacham. 1986. Nucleotide sequence and transcriptional analysis of the E. coli ushA gene, encoding periplasmic UDP-sugar hydrolase (5′-nucleotidase): regulation of the ushA gene, and the signal sequence of its encoded protein product. Nucleic Acids Res. 14:4325-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carapetis, J. R., A. C. Steer, E. K. Mulholland, and M. Weber. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685-694. [DOI] [PubMed] [Google Scholar]
- 10.Cleary, P. P., Y. V. Matsuka, T. Huynh, H. Lam, and S. B. Olmsted. 2004. Immunization with C5a peptidase from either group A or B streptococci enhances clearance of group A streptococci from intranasally infected mice. Vaccine 22:4332-4341. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale, J. B. 2008. Current status of group A streptococcal vaccine development. Adv. Exp. Med. Biol. 609:53-63. [DOI] [PubMed] [Google Scholar]
- 13.Dinkla, K., S. R. Talay, M. Morgelin, R. M. Graham, M. Rohde, D. P. Nitsche-Schmitz, and G. S. Chhatwal. 2009. Crucial role of the CB3-region of collagen IV in PARF-induced acute rheumatic fever. PloS One 4:e4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebel, W., M. M. Skinner, K. P. Dierksen, J. M. Scott, and J. E. Trempy. 1999. A conserved domain in Escherichia coli Lon protease is involved in substrate discriminator activity. J. Bacteriol. 181:2236-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, R. J., G. W. Taylor, M. Ferguson, S. Murray, N. Rendell, A. Wrigley, Z. Bai, J. Boyle, S. J. Finney, A. Jones, H. H. Russell, C. Turner, J. Cohen, L. Faulkner, and S. Sriskandan. 2005. Specific C-terminal cleavage and inactivation of interleukin-8 by invasive disease isolates of Streptococcus pyogenes. J. Infect. Dis. 192:783-790. [DOI] [PubMed] [Google Scholar]
- 16.Etz, H., D. B. Minh, T. Henics, A. Dryla, B. Winkler, C. Triska, A. P. Boyd, J. Sollner, W. Schmidt, U. von Ahsen, M. Buschle, S. R. Gill, J. Kolonay, H. Khalak, C. M. Fraser, A. von Gabain, E. Nagy, and A. Meinke. 2002. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 99:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etz, H., D. B. Minh, C. Schellack, E. Nagy, and A. Meinke. 2001. Bacterial phage receptors, versatile tools for display of polypeptides on the cell surface. J. Bacteriol. 183:6924-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fae, K. C., D. D. da Silva, S. E. Oshiro, A. C. Tanaka, P. M. Pomerantzeff, C. Douay, D. Charron, A. Toubert, M. W. Cunningham, J. Kalil, and L. Guilherme. 2006. Mimicry in recognition of cardiac myosin peptides by heart-intralesional T cell clones from rheumatic heart disease. J. Immunol. 176:5662-5670. [DOI] [PubMed] [Google Scholar]
- 19.Falconer, A. E., R. Carson, R. Johnstone, P. Bird, M. Kehoe, and J. E. Calvert. 1993. Distinct IgG1 and IgG3 subclass responses to two streptococcal protein antigens in man: analysis of antibodies to streptolysin O and M protein using standardized subclass-specific enzyme-linked immunosorbent assays. Immunology 79:89-94. [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer, H., and R. Glockshuber. 1994. A point mutation within the ATP-binding site inactivates both catalytic functions of the ATP-dependent protease La (Lon) from Escherichia coli. FEBS Lett. 356:101-103. [DOI] [PubMed] [Google Scholar]
- 21.Fritzer, A., B. Noiges, D. Schweiger, A. Rek, A. J. Kungl, A. von Gabain, E. Nagy, and A. L. Meinke. 2009. Chemokine degradation by the Group A streptococcal serine proteinase ScpC can be reconstituted in vitro and requires two separate domains. Biochem. J. 422:533-542. [DOI] [PubMed] [Google Scholar]
- 22.Fu, Q., M. Figuera-Losada, V. A. Ploplis, S. Cnudde, J. H. Geiger, M. Prorok, and F. J. Castellino. 2008. The lack of binding of VEK-30, an internal peptide from the group A streptococcal M-like protein, PAM, to murine plasminogen is due to two amino acid replacements in the plasminogen kringle-2 domain. J. Biol. Chem. 283:1580-1587. [DOI] [PubMed] [Google Scholar]
- 23.Giefing, C., A. L. Meinke, M. Hanner, T. Henics, M. D. Bui, D. Gelbmann, U. Lundberg, B. M. Senn, M. Schunn, A. Habel, B. Henriques-Normark, A. Ortqvist, M. Kalin, A. von Gabain, and E. Nagy. 2008. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205:117-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzales, R., D. C. Malone, J. H. Maselli, and M. A. Sande. 2001. Excessive antibiotic use for acute respiratory infections in the United States. Clin. Infect. Dis. 33:757-762. [DOI] [PubMed] [Google Scholar]
- 25.Guzman, C. A., S. R. Talay, G. Molinari, E. Medina, and G. S. Chhatwal. 1999. Protective immune response against Streptococcus pyogenes in mice after intranasal vaccination with the fibronectin-binding protein SfbI. J. Infect. Dis. 179:901-906. [DOI] [PubMed] [Google Scholar]
- 26.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanski, E., P. A. Horwitz, and M. G. Caparon. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60:5119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harry, E. J., K. Pogliano, and R. Losick. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henics, T., B. Winkler, U. Pfeifer, S. R. Gill, M. Buschle, A. von Gabain, and A. L. Meinke. 2003. Small-fragment genomic libraries for the display of putative epitopes from clinically significant pathogens. Biotechniques 35:196-206. [DOI] [PubMed] [Google Scholar]
- 30.Hidalgo-Grass, C., I. Mishalian, M. Dan-Goor, I. Belotserkovsky, Y. Eran, V. Nizet, A. Peled, and E. Hanski. 2006. A streptococcal protease that degrades CXC chemokines and impairs bacterial clearance from infected tissues. EMBO J. 25:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holden, M. T., A. Scott, I. Cherevach, T. Chillingworth, C. Churcher, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, K. Mungall, M. A. Quail, C. Price, E. Rabbinowitsch, S. Sharp, J. Skelton, S. Whitehead, B. G. Barrell, M. Kehoe, and J. Parkhill. 2007. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain manfredo. J. Bacteriol. 189:1473-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikebe, T., K. Hirasawa, R. Suzuki, J. Isobe, D. Tanaka, C. Katsukawa, R. Kawahara, M. Tomita, K. Ogata, M. Endoh, R. Okuno, and H. Watanabe. 2005. Antimicrobial susceptibility survey of Streptococcus pyogenes isolated in Japan from patients with severe invasive group A streptococcal infections. Antimicrob. Agents Chemother. 49:788-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreikemeyer, B., M. Klenk, and A. Podbielski. 2004. The intracellular status of Streptococcus pyogenes: role of extracellular matrix-binding proteins and their regulation. Int. J. Med. Microbiol. 294:177-188. [DOI] [PubMed] [Google Scholar]
- 34.Kreikemeyer, B., M. Nakata, S. Oehmcke, C. Gschwendtner, J. Normann, and A. Podbielski. 2005. Streptococcus pyogenes collagen type I-binding Cpa surface protein. Expression profile, binding characteristics, biological functions, and potential clinical impact. J. Biol. Chem. 280:33228-33239. [DOI] [PubMed] [Google Scholar]
- 35.Kuklin, N. A., D. J. Clark, S. Secore, J. Cook, L. D. Cope, T. McNeely, L. Noble, M. J. Brown, J. K. Zorman, X. M. Wang, G. Pancari, H. Fan, K. Isett, B. Burgess, J. Bryan, M. Brownlow, H. George, M. Meinz, M. E. Liddell, R. Kelly, L. Schultz, D. Montgomery, J. Onishi, M. Losada, M. Martin, T. Ebert, C. Y. Tan, T. L. Schofield, E. Nagy, A. Meineke, J. G. Joyce, M. B. Kurtz, M. J. Caulfield, K. U. Jansen, W. McClements, and A. S. Anderson. 2006. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect. Immun. 74:2215-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo, C. F., J. J. Wu, K. Y. Lin, P. J. Tsai, S. C. Lee, Y. T. Jin, H. Y. Lei, and Y. S. Lin. 1998. Role of streptococcal pyrogenic exotoxin B in the mouse model of group A streptococcal infection. Infect. Immun. 66:3931-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lancefield, R. C. 1962. Current knowledge of type-specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 37a.Landeskultur und Wasserrecht (Magistratsabteilung 58), Republik Österreich. Tierversuchsgesetz. Bundesgesetzblatt für die Republik Osterreich no. 501/1989. Landeskultur und Wasserrecht, Republik Österreich, Vienna, Austria.
- 38.Lei, B., M. Liu, G. L. Chesney, and J. M. Musser. 2004. Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J. Infect. Dis. 189:79-89. [DOI] [PubMed] [Google Scholar]
- 39.Lei, B., S. Mackie, S. Lukomski, and J. M. Musser. 2000. Identification and immunogenicity of group A streptococcus culture supernatant proteins. Infect. Immun. 68:6807-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin, J. M., M. Green, K. A. Barbadora, and E. R. Wald. 2002. Erythromycin-resistant group A streptococci in schoolchildren in Pittsburgh. N. Engl. J. Med. 346:1200-1206. [DOI] [PubMed] [Google Scholar]
- 42.Martins, T. B., J. L. Hoffman, N. H. Augustine, A. R. Phansalkar, V. A. Fischetti, J. B. Zabriskie, P. P. Cleary, J. M. Musser, L. G. Veasy, and H. R. Hill. 2008. Comprehensive analysis of antibody responses to streptococcal and tissue antigens in patients with acute rheumatic fever. Int. Immunol. 20:445-452. [DOI] [PubMed] [Google Scholar]
- 43.McShan, W. M., J. J. Ferretti, T. Karasawa, A. N. Suvorov, S. Lin, B. Qin, H. Jia, S. Kenton, F. Najar, H. Wu, J. Scott, B. A. Roe, and D. J. Savic. 2008. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J. Bacteriol. 190:7773-7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meinke, A., M. Storm, T. Henics, D. Gelbmann, S. Prustomersky, Z. Kovacs, D. B. Minh, B. Noiges, U. Stierschneider, M. Berger, A. von Gabain, L. Engstrand, and E. Nagy. 2009. Composition of the ANTIGENome of Helicobacter pylori defined by human serum antibodies. Vaccine 27:3251-3259. [DOI] [PubMed] [Google Scholar]
- 45.O'Loughlin, R. E., A. Roberson, P. R. Cieslak, R. Lynfield, K. Gershman, A. Craig, B. A. Albanese, M. M. Farley, N. L. Barrett, N. L. Spina, B. Beall, L. H. Harrison, A. Reingold, and C. Van Beneden. 2007. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000-2004. Clin. Infect. Dis. 45:853-862. [DOI] [PubMed] [Google Scholar]
- 46.Ozeri, V., A. Tovi, I. Burstein, S. Natanson-Yaron, M. G. Caparon, K. M. Yamada, S. K. Akiyama, I. Vlodavsky, and E. Hanski. 1996. A two-domain mechanism for group A streptococcal adherence through protein F to the extracellular matrix. EMBO J. 15:989-998. [PMC free article] [PubMed] [Google Scholar]
- 47.Reid, S. D., N. M. Green, J. K. Buss, B. Lei, and J. M. Musser. 2001. Multilocus analysis of extracellular putative virulence proteins made by group A streptococcus: population genetics, human serologic response, and gene transcription. Proc. Natl. Acad. Sci. U. S. A. 98:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid, S. D., N. M. Green, G. L. Sylva, J. M. Voyich, E. T. Stenseth, F. R. DeLeo, T. Palzkill, D. E. Low, H. R. Hill, and J. M. Musser. 2002. Postgenomic analysis of four novel antigens of group A streptococcus: growth phase-dependent gene transcription and human serologic response. J. Bacteriol. 184:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rharbaoui, F., B. Drabner, S. Borsutzky, U. Winckler, M. Morr, B. Ensoli, P. F. Muhlradt, and C. A. Guzman. 2002. The Mycoplasma-derived lipopeptide MALP-2 is a potent mucosal adjuvant. Eur. J. Immunol. 32:2857-2865. [DOI] [PubMed] [Google Scholar]
- 50.Ringdahl, U., and U. Sjobring. 2000. Analysis of plasminogen-binding M proteins of Streptococcus pyogenes. Methods 21:143-150. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Ortega, M. J., N. Norais, G. Bensi, S. Liberatori, S. Capo, M. Mora, M. Scarselli, F. Doro, G. Ferrari, I. Garaguso, T. Maggi, A. Neumann, A. Covre, J. L. Telford, and G. Grandi. 2006. Characterization and identification of vaccine candidate proteins through analysis of the group A streptococcus surface proteome. Nat. Biotechnol. 24:191-197. [DOI] [PubMed] [Google Scholar]
- 52.Rotanova, T. V., I. Botos, E. E. Melnikov, F. Rasulova, A. Gustchina, M. R. Maurizi, and A. Wlodawer. 2006. Slicing a protease: structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains. Protein Sci. 15:1815-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanders, C. L., and J. F. Curran. 2007. Genetic analysis of the E site during RF2 programmed frameshifting. RNA 13:1483-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanderson-Smith, M. L., K. Dinkla, J. N. Cole, A. J. Cork, P. G. Maamary, J. D. McArthur, G. S. Chhatwal, and M. J. Walker. 2008. M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. FASEB J. 22:2715-2722. [DOI] [PubMed] [Google Scholar]
- 55.Schellack, C., K. Prinz, A. Egyed, J. H. Fritz, B. Wittmann, M. Ginzler, G. Swatosch, W. Zauner, C. Kast, S. Akira, A. von Gabain, M. Buschle, and K. Lingnau. 2006. IC31, a novel adjuvant signaling via TLR9, induces potent cellular and humoral immune responses. Vaccine 24:5461-5472. [DOI] [PubMed] [Google Scholar]
- 56.Schulze, K., E. Medina, G. S. Chhatwal, and C. A. Guzman. 2003. Stimulation of long-lasting protection against Streptococcus pyogenes after intranasal vaccination with non adjuvanted fibronectin-binding domain of the SfbI protein. Vaccine 21:1958-1964. [DOI] [PubMed] [Google Scholar]
- 57.Severin, A., E. Nickbarg, J. Wooters, S. A. Quazi, Y. V. Matsuka, E. Murphy, I. K. Moutsatsos, R. J. Zagursky, and S. B. Olmsted. 2007. Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J. Bacteriol. 189:1514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steer, A. C., M. R. Batzloff, K. Mulholland, and J. R. Carapetis. 2009. Group A streptococcal vaccines: facts versus fantasy. Curr. Opin. Infect. Dis. 22:544-552. [DOI] [PubMed] [Google Scholar]
- 59.Sumby, P., A. H. Tart, and J. M. Musser. 2008. A non-human primate model of acute group A Streptococcus pharyngitis. Methods Mol. Biol. 431:255-267. [DOI] [PubMed] [Google Scholar]
- 60.Terao, Y., S. Kawabata, E. Kunitomo, I. Nakagawa, and S. Hamada. 2002. Novel laminin-binding protein of Streptococcus pyogenes, Lbp, is involved in adhesion to epithelial cells. Infect. Immun. 70:993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsilibaris, V., G. Maenhaut-Michel, and L. Van Melderen. 2006. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 157:701-713. [DOI] [PubMed] [Google Scholar]
- 62.Vercellotti, G. M., J. B. McCarthy, P. Lindholm, P. K. Peterson, H. S. Jacob, and L. T. Furcht. 1985. Extracellular matrix proteins (fibronectin, laminin, and type IV collagen) bind and aggregate bacteria. J. Pathol. 120:13-21. [PMC free article] [PubMed] [Google Scholar]
- 63.Virtaneva, K., M. R. Graham, S. F. Porcella, N. P. Hoe, H. Su, E. A. Graviss, T. J. Gardner, J. E. Allison, W. J. Lemon, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2003. Group A streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect. Immun. 71:2199-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visai, L., S. Bozzini, G. Raucci, A. Toniolo, and P. Speziale. 1995. Isolation and characterization of a novel collagen-binding protein from Streptococcus pyogenes strain 6414. J. Biol. Chem. 270:347-353. [DOI] [PubMed] [Google Scholar]
- 65.Yan, S. S., M. L. Fox, S. M. Holland, F. Stock, V. J. Gill, and D. P. Fedorko. 2000. Resistance to multiple fluoroquinolones in a clinical isolate of Streptococcus pyogenes: identification of gyrA and parC and specification of point mutations associated with resistance. Antimicrob. Agents Chemother. 44:3196-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zinkernagel, A. S., A. M. Timmer, M. A. Pence, J. B. Locke, J. T. Buchanan, C. E. Turner, I. Mishalian, S. Sriskandan, E. Hanski, and V. Nizet. 2008. The IL-8 protease SpyCEP/ScpC of group A streptococcus promotes resistance to neutrophil killing. Cell Host Microbe 4:170-178. [DOI] [PMC free article] [PubMed] [Google Scholar]