Abstract
Clostridium perfringens is a medically important clostridial pathogen and an etiological agent causing several diseases in humans and animals. C. perfringens and its toxins have been listed as potential biological and toxin warfare (BTW) agents; thus, efforts to develop strategies for detection and protection are warranted. Forty-eight extracellular proteins of C. perfringens type A and type C strains have been identified here using a 2-dimensional gel electrophoresis-mass spectrometry (2-DE-MS) technique. The SagA protein, the DnaK-type molecular chaperone hsp70, endo-beta-N-acetylglucosaminidase, and hypothetical protein CPF_0656 were among the most abundant proteins secreted by C. perfringens ATCC 13124. The antigenic component of the exoproteome of this strain has also been identified. Most of the extracellular proteins were predicted to be involved in carbohydrate transport and metabolism (16%) or cell envelope biogenesis or to be outer surface protein constituents (13%). More than 50% of the proteins were predictably secreted by either classical or nonclassical pathways. LipoP and TMHMM indicated that nine proteins were extracytoplasmic but cell associated. Immunization with recombinant ornithine carbamoyltransferase (cOTC) clearly resulted in protection against a direct challenge with C. perfringens organisms. A significant rise in IgG titers in response to recombinant cOTC was observed in mice, and IgG2a titers predominated over IgG1 titers (IgG2a/IgG1 ratio, 2). The proliferation of spleen lymphocytes in cOTC-immunized animals suggested a cellular immune response. There were significant increases in the levels of gamma interferon (IFN-γ) and interleukin 2 (IL-2), suggesting a Th1 type immune response.
Clostridium perfringens is a medically important clostridial pathogen and an etiological agent causing several diseases in humans and animals; the former include gas gangrene, food poisoning, necrotizing enterocolitis of infants, and enteritis necroticans (28, 37, 45). C. perfringens is an obligately anaerobic rod-shaped bacterium commonly found in the gastrointestinal tracts of both animals and humans and widely distributed in soil and sewage. The ability of C. perfringens to cause disease is associated with the production of a variety of extracellular toxins (13 different toxins have been reported so far). On the basis of differential production of toxins, the strains of C. perfringens can be divided into five types, A through E (35), of which type A and type C strains are implicated in human diseases while other types are of veterinary importance. Type A strains cause gas gangrene, the most destructive of all clostridial diseases, which is characterized by rapid destruction of tissue with production of gas (4, 42). The incidence of disease ranged from 1% or less of wounded personnel during World War II to 10% of wounded personnel during World War I (27). Hundreds of thousands of soldiers died of gas gangrene as a result of battlefield injuries, and C. perfringens was widely recognized as the most important causal organism of the disease. Besides gas gangrene, type A strains also cause gastrointestinal diseases in humans (food poisoning, antibiotic-associated diarrhea, sporadic diarrhea, sudden infant death syndrome) and animals (diarrhea in foals and pigs, etc.). C. perfringens type C strains cause necrotic enteritis in humans and animals, in addition to enterotoxemia in sheep. Moreover, C. perfringens and its toxins have been listed as potential biological and toxin warfare (BTW) agents; therefore, efforts to develop strategies for detection and protection are warranted.
Interest in a vaccine against gas gangrene has been intermittent; most efforts were made during World Wars I and II and were devoted to the therapeutic use of antisera. Such antisera, raised against toxoids of all five species of clostridia associated with gas gangrene, were shown to have benefits if they were given soon after trauma (20). Active immunization against the disease has received little attention until a few years ago (32, 43, 44). A number of clinical studies of other pathogenic bacteria, including Clostridium difficile, have highlighted the importance of nontoxin protein antigens in disease expression, especially in colonization by the bacterium (9, 12, 16, 26).
Despite a sudden spurt of activity in the proteomic characterization of bacterial pathogens, for reasons unknown, clostridia have been largely ignored. Clostridium difficile is the only clostridial species whose proteome has been analyzed to some extent (34). Proteomic methodology has been used to elucidate proteins regulated by the VirR/VirS system in Clostridium perfringens (40).
To invade, multiply in, and colonize host tissues, a pathogen must be able to evade the host immune system and obtain nutrients essential for growth. The factors involved in these complex processes are largely unknown and of crucial importance for the understanding of microbial pathogenesis. The exoproteins of Gram-positive bacteria are likely to contain some of these key factors. The term “secretome” refers to and takes into account both the protein secretion systems and the secreted proteins; in monoderm bacteria (Gram-positive cell envelope architecture), these proteins can also be found in the membrane and/or cell wall. The proteins found in the extracellular milieu of Gram-positive bacteria are hence extracellular proteins, or exoproteins, which form the exoproteome; these exoproteins are not necessarily secreted by known secretion systems (15). This terminology is used for the subproteome described in this study.
Further, the correct identification of C. perfringens pathovars is critical for epidemiological studies and for the development of effective preventive measures, including vaccination. It is not well understood why C. perfringens produces illnesses that differ so greatly in severity. Specific illnesses may reflect the particular type of tissue that high numbers of the organism are able to invade. Clearly, the presence of certain toxins is one explanation, but it falls short in many instances. For example, strains that cause food poisoning may differ from those that cause gas gangrene only by the presence of an enterotoxin gene in the former, yet food-poisoning strains have never been found to cause gas gangrene. Elucidation of the exoproteins of this bacterium is likely to reveal factors responsible for the host specificity of the different C. perfringens pathovars.
The present investigation was carried out with the following objectives: (i) identification of dominant exoproteins from the C. perfringens type A strain ATCC 13124 (a gas gangrene isolate and the species type strain) and comparison with those of type C strains, (ii) elucidation of immunogenic components of the exoproteome, (iii) in silico analysis of the identified proteins with respect to localization, predicted function, and likely potential as surface markers for detection and as vaccine candidates, and (iv) validation of selected candidates with respect to vaccine potential in an experimental mouse model of gas gangrene. To the best of our knowledge, ours is the first proteomic elucidation of the C. perfringens exoproteome. The identification of extracellular proteins of C. perfringens type A and type C strains in the present investigation advances our understanding of the pathogenesis of the disease and opens significant opportunities for the identification of diagnostic markers and the development of vaccines against gas gangrene in humans.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Clostridium perfringens ATCC 13124 was obtained from Becton Dickinson India Pvt. Ltd., India. C. perfringens type A (AG03-4, AG04-3, AL05-13, AL08-13, AL08-16) and type C (AG02-3, AG07-3, AL05-1, H2-3, AL06-1) strains were isolated from environmental samples obtained from diverse ecological niches from the plains of northern India (see Table S1 in the supplemental material). For isolation and purification, cells were grown on SPS agar (Difco, France) at 30°C. The SPS agar was composed of tryptose (15 g), yeast extract (10 g), ferric citrate (0.5 g), sodium sulfite (0.5 g), sodium thioglycolate (0.1 g), polysorbate 80 (0.05 g), sulfadiazine (0.12 g), polymyxin B sulfate (0.01 g), agar (15 g), and distilled water (1,000 ml). The inoculation was carried out in an anaerobic workstation (Don Whitley Scientific Ltd., Shipley, England) operating at 30°C as described previously (2). After incubation, black colonies were anaerobically transferred to 3 ml of thioglycolate broth (TGB) containing 5 g of yeast extract, 15 g of tryptone, 5.5 g of glucose, 0.5 g of sodium thioglycolate, 2.5 g of sodium chloride, 0.5 g of l-cystine, 0.001 g of resazurin, 0.75 g of agar, and 1,000 ml of distilled water. The cultures were maintained at 4°C in cooked meat medium (CMM) containing 454 g of beef heart infusion, 20 g of proteose peptone, 2 g of dextrose, 5 g of sodium chloride, and 1,000 ml of distilled water. All the media were procured either from Oxoid Ltd., England, or from Difco Laboratories, France, and were prepared anaerobically by standard methods using a gassing manifold and serum vials. For preparation of the exoproteome, C. perfringens cells were grown in RPMI 1640 broth medium at 37°C under anaerobic conditions as described previously (40).
Strains were identified as C. perfringens on the basis of diagnostic morphological and biochemical tests and sequencing of 16S rRNA and alpha-toxin genes. Toxinotyping was carried out by PCR using specific primer sets for alpha-toxin (plc), beta-toxin (cpb and cpb2), epsilon-toxin (etx), iota-toxin (iA), and enterotoxin (cpe) according to the method of Schoepe et al. (38).
Immunization and raising of hyperimmune sera.
Animal experiments were approved by the institutional Animal Ethical Committee at the Defence Research & Development Establishment (DRDE), Gwalior, India. All the polyclonal antibodies were raised in 4-week-old female BALB/c mice by active immunization on a 4-week immunization schedule. For the anti-culture (anti-WC) serum, cells were grown in RPMI 1640 broth medium. After growth at 37°C to exponential phase (optical density at 600 nm [OD600], 0.8 to 1.0), the number of bacteria in the culture was determined by plating 10-fold serial dilutions onto SPS agar (Difco) plates. Heat inactivation was accomplished in a water bath at 60°C for 30 min. No live bacteria were detected after this suspension was plated onto agar plates. Cells were injected intraperitoneally (i.p.) using Freund's complete adjuvant (Sigma-Aldrich, India) for the first immunization and Freund's incomplete adjuvant for booster immunizations. On days 1 and 7, 103 CFU (100-μl cell suspension in phosphate-buffered saline [PBS] and 100 μl adjuvant) was injected in each mouse; on days 14 and 27, the dose was increased to 105 CFU. One week after the administration of the last booster, 10 animals were anesthetized by halothane inhalation, and a blood specimen (500 μl) was obtained from each by means of retro-orbital puncture. Sera from these animals were pooled and used for Western blot analysis of surface proteins. Sham-immunized animals received an equal volume of adjuvant alone in parallel on the same immunization schedule, and serum was collected after 5 weeks. Another serum (anti-gangrene) was obtained from mice surviving gas gangrene infection as follows. C. perfringens ATCC 13124 cells were grown in TPYG broth (30 g of pancreatic digest of casein, 5 g of yeast extract, 20 g of glucose, 1 g of sodium thioglycollate, 250 mg of cycloserine, 76 mg of sulfamethoxazole, 4 mg of trimethoprim, and 1,000 ml of distilled water) at 37°C and were harvested in exponential phase. Four-week-old female BALB/c mice in groups of 6 each were given intramuscular injections of 105, 106, and 107 CFU of washed C. perfringens cells, in a volume of 0.1 ml anaerobically prepared saline, into the right hindquarter with a 26-gauge needle (41). Mice infected with C. perfringens cells developed swollen hemorrhagic thighs, and 3 of those receiving 106 cells survived the infection. Serum was collected after 4 weeks of inoculation from all the mice that developed gas gangrene and survived.
Polyclonal sera were raised against SagA protein (anti-SagA) using spots separated by 2-dimensional gel electrophoresis (2-DE) as described by Amero et al. (3). Serum was also raised in mice against recombinant ornithine carbamoyltransferase (cOTC) (anti-ornith) by using 100 μg nickel-nitrilotriacetic acid (Ni-NTA)-purified proteins per mouse on a 4-week immunization schedule as described above.
Expression and purification of recombinant ornithine carbamoyltransferase.
Total DNA was extracted from C. perfringens ATCC 13124 according to the procedure of Marmur (29), and the full-length ornithine carbamoyltransferase gene was PCR amplified using primers Ornith4 for (ATGGCAGTTAACTTAAAAGGAAG) and Ornith4 rev (TTATCTTCCAGCAGTTGCTACC). The 996-bp product, corresponding to the full-length mature protein, was purified by gel extraction using a commercial kit (Qiagen, Germany) and was cloned into the pQE30-UA vector (Qiagen, Germany) according to the manufacturer's instructions. The ligated products were transformed into Escherichia coli M15 cells. Plasmids were isolated from 10 randomly selected clones and were tested for the presence of the insert by size determination on an agarose gel (1.5%) and PCR amplification of the target gene. Two clones positive for the insert in the correct orientation were subjected to double-pass sequencing to check for possible mismatches using the commercial services (MWG, India) employing an automated sequencer (ABI Prism, model 3730). The expression of recombinant protein was induced for 4 h at 30°C in 250-ml LB medium cultures containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The overexpressed recombinant protein was purified to near-homogeneity by using Ni-NTA Sepharose resin (Qiagen, Germany) according to the manufacturer's instructions.
Preparation of the extracellular protein fraction.
Extracellular protein was prepared from C. perfringens as described by Shimizu et al. (40) with some modifications. Cells were grown in RPMI 1640 medium at 37°C under anaerobic conditions and were harvested by centrifugation at 10,000 × g for 10 min at 4°C in the mid-exponential-growth phase. The extracellular proteins in the supernatant were precipitated with 10% (wt/vol) trichloroacetic acid overnight at 4°C and were centrifuged at 10,000 × g for 10 min at 4°C. The resulting protein precipitate was washed twice with cold acetone containing 0.07% β-mercaptoethanol, air dried, and stored at −80°C until use. The total-protein concentration was determined according to the method of Bradford (8) by using the Quick Start Bradford protein assay kit (Bio-Rad) according to the manufacturer's instructions. The protein concentration was calculated using bovine serum albumin (BSA) as the standard.
2-DE.
Just before the isoelectric focusing step, the protein pellet was resuspended in sample rehydration buffer {8 M urea, 2% [wt/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 15 mM dithiothreitol [DTT], and 0.5% [vol/vol] IPG buffer [pH 3 to 10]}. Isoelectric focusing was performed using immobilized pH gradient (IPG) strips (Bio-Rad). IPG strips with a pH range from 4 to 7 were used for all the experiments. For the first dimension, 500 μg of protein samples in 150 μl of rehydration solution was used to rehydrate the IPG strip (7 cm [length]; pH 4 to 7). The IPG strips were passively rehydrated overnight, and then the proteins were focused for 10,000 V·h at 20°C under mineral oil. After focusing, the strips were incubated for 10 min in 2 ml of equilibrium buffer I (6 M urea, 30% [wt/vol] glycerol, 2% [wt/vol] sodium dodecyl sulfate [SDS], and 1% [wt/vol] DTT in 50 mM Tris-HCl buffer [pH 8.8]), followed by equilibrium buffer II (6 M urea, 30% [wt/vol] glycerol, 2% [wt/vol] SDS, and 4% [wt/vol] iodoacetamide in 375 mM Tris-HCl buffer [pH 8.8]). After the equilibration steps, the strips were transferred to a 12% SDS-polyacrylamide gel electrophoresis (PAGE) gel for the second dimension by the method of Blackshear (7).
Protein spots were visualized by staining with Coomassie brilliant blue G-250. Gel images were captured by a GS800 densitometer (Bio-Rad). The relative abundances of the spots were determined by PD Quest software (Bio-Rad). Two independent experiments were carried out for the exoproteome study, and replicate gels (n, 4) were generated from each independent experiment.
Immunoblotting.
Extracellular proteins were immunoblotted using a standard method as described previously (2). Sera (anti-WC or anti-gangrene) obtained from mice were used at a 1:1,000 dilution (or as indicated otherwise) in blocking buffer. A horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Dako) was used as a secondary antibody at a 1:30,000 dilution. Bound antibodies were detected by chemiluminescence using an ECL Western blot kit (Millipore, India) and Hyperfilm ECL (Amersham) according to the manufacturer's instructions.
Identification of protein spots by mass spectrometry.
Protein spots were excised by use of thin-walled PCR tubes (200 μl) appropriately cut at the bottom with a fresh surgical scalpel blade. Care was taken not to contaminate the spots with adjoining proteins or with skin keratin. The gel spots were washed with proteomic-grade deionized water, and proteins were identified by mass spectrometry (MS) using a matrix-assisted laser desorption ionization—tandem time-of-flight (MALDI-TOF-TOF) mass spectrometer (Ultraflex III; Bruker Daltonics, Germany). The gel piece containing the protein was destained, reduced/alkylated, and digested with trypsin using the Montage In-Gel digest kit (Millipore) by following the kit's instructions. For MALDI-TOF mass spectrometry, 1 μl of the digest was mixed with 2 μl of the matrix solution (5 mg α-cyano-4-hydroxycinnamic acid in 80% [vol/vol] acetonitrile and 0.1% [wt/vol] trifluoroacetic acid [TFA]), and 1 μl of this mixture was deposited onto the MALDI target. The spectrum was obtained in the mass range of 500 to 4,000 Da and was calibrated using a calibration mixture containing angiotensin I, Substance P, adrenocorticotropin(1-17) [ACTH(1-17)], ACTH(18-39), and somatostatin (28).
Spectra were analyzed with Mascot sequence-matching software from Matrix Science using a nonidentical protein sequence database based on MSDB in the taxonomy group of Bacteria. Search parameters were a maximum of one missed cleavage by trypsin, fixed modification of oxidation, a charged state of +1, a peptide mass tolerance of 50 ppm, and a fragment mass tolerance of ±0.6 Da. Proteins were identified using one or more of the tandem MS (MS-MS) platforms shown in Table S2 in the supplemental material. Peptide mass fingerprinting (PMF) data for trypsin-digested proteins and MS-MS analysis results for one or more selected peptides were used for the database search as described above. In most cases, proteins were identified as C. perfringens strain 13 proteins with significant Mowse scores, and the same protein match was observed for PMF and MS-MS data for both peptides (see Table S2 in the supplemental material).
Bioinformatic analysis.
Homology searches were carried out using the BLAST and PSI-BLAST protein algorithms against the GenBank nonredundant protein database (http://wwww.ncbi.nlm.nih.gov), and close homologs in C. perfringens ATCC 13124 were listed against the corresponding proteins (see Table S2 in the supplemental material). The theoretical molecular weights (MWs) and isoelectric points were determined using the Compute pI/Mw algorithm (http://ca.expasy.org/).
Clusters of orthologous groups (COGs) for the identified proteins were determined using the COGnitor program at http://www.ncbi.nlm.nih.gov/COG/. The identified proteins were also used to search the Pfam database (http://pfam.sanger.ac.uk/), a large collection of protein families (18). Pattern/profile, posttranslational modification, and topology searches were carried out using ExPASy Proteomics tools at http://www.expasy.ch. The presence of a signal peptide was predicted by SignalP, version 3.0, whereas nonclassical protein secretion was predicted by SecretomeP. The number of predicted transmembrane helices in proteins was screened using the TMHMM server, version 2.0, and lipoproteins and signal peptides were predicted using LipoP, version 1.0.
Hypothetical proteins identified in the exoproteome were further analyzed for secondary-structure predictions using the GOR IV secondary-structure prediction method at http://npsa-pbil.ibcp.fr/. The grand average of hydropathicity (GRAVY) was obtained from the online tool at http://www.expasy.ch/tools/protparam.html, and leucine zippers were predicted using an online tool at http://2zip.molgen.mpg.de/index.html. The disulfide bonding state, solvent accessibility, and globularity were predicted using PredictProtein software at http://www.predictprotein.org/. The antigenicity index was determined using the ANTIGENpro tool at http://scratch.proteomics.ics.uci.edu/.
In vivo studies and whole-cell enzyme-linked immunosorbent assays (ELISA).
In vivo expression of SagA protein was studied in rabbits by comparing intraperitoneal fluid obtained from healthy and infected animals. Cells were grown in TGB at 37°C, harvested in late-exponential phase, and washed with anaerobically prepared saline. One milliliter (108 cells) of the cell suspension was injected (i.p.). Soon after the onset of gas gangrene symptoms, characterized by abdominal swelling, change in the skin coloration, and difficulty in movement, peritoneal fluid was collected. Equal volumes (3 ml) of peritoneal fluids from healthy and infected rabbits were desalted and partially purified using Econo-Pac 10DG columns (Bio-Rad, India). The polyacrylamide gel matrix excluded >6-kDa solutes and partially separated high-molecular-weight globular host proteins from the bacterial secretory components. Fractions were subjected to Western blot analysis using an anti-SagA serum (1:1,000) as described above.
Proteins were detected on the bacterial surface by whole-cell ELISA according to the method of Severin et al. (39). Preimmune serum was used at the respective hyperimmune serum dilutions, and the mean values (n, 3) were subtracted from the mean test values (n, 3) to reflect the binding by specific antibodies.
Immunization of mice with recombinant cOTC protein.
A group of 30 mice was immunized intraperitoneally with purified recombinant cOTC in a 1:1 emulsion with Freund's complete adjuvant (Sigma-Aldrich, India) for the first immunization and Freund's incomplete adjuvant for booster immunizations. On days 1 and 7, 20 μg protein was injected into each mouse, while on days 21 and 35, the dose was increased to 50 μg. On day 48, sera were collected from 10 animals as described above. Sera from these specimens were pooled and used to verify specific antibody production by Western blot analysis. The control group received an equal volume of adjuvant with PBS in parallel on the same immunization schedule, and serum was collected on day 48. Five animals from each group were sacrificed 30 days after the last dose for lymphoproliferation and cytokine assays, and the rest of the animals were taken for the challenge studies.
Determination of antibody responses to recombinant cOTC.
An indirect ELISA was used to quantify the antibody response to cOTC protein. Briefly, the microtitration plates (Maxisorp; Nunc) were coated with recombinant ornithine carbamoyltransferase protein antigen (10 μg/ml) in 0.05 M carbonate buffer (pH 9.6) and were incubated at 37°C for 2 h. The plates were blocked with PBS containing 1% (wt/vol) BSA and were washed three times with PBS containing 0.05% (vol/vol) Tween 20 (PBST). The test sera, serially diluted (2-fold) in PBS containing 0.1% BSA starting from 1:100, were appended in triplicate wells (100 μl/well) for1 h at 37°C. The wells were washed five times with PBST. Goat anti-mouse (IgG) antibodies conjugated to horseradish peroxidase and isotype conjugates (IgG1, IgG2a, IgG2b, and IgG3, labeled with HRP or alkaline phosphatase [AP]), obtained from Sigma and Pharmingen, respectively, were diluted as recommended by the manufacturers and were added to the wells. After incubation for 1 h at 37°C and subsequent washes, the plate was developed with ortho-phenylenediamine (0.4 mg/ml) and H2O2 (6%; 0.4 μl/ml) or in diethanolamine buffer (10 mM; pH 9.5) containing 0.5 mM MgCl2 with para-nitrophenyl phosphate (1 mg/ml). The absorbance was measured at 492 or 405 nm in an ELISA reader, and values are presented as endpoint titers. The highest dilution of a test serum with an OD greater than the mean OD of the preimmune serum (1:100 dilution) in the same group was considered the endpoint titer.
Lymphoproliferation and cytokine assays.
For the lymphoproliferation assay, five mice from each group (except the vaccine group) were sacrificed, and their spleens were removed aseptically. Spleen cell proliferation was determined by a method described previously (25). Experiments were carried out in triplicate wells for each mouse, and the results are expressed as the mean specific absorbance by subtracting the absorbance at 600 nm from that at 570 nm, after subtracting the reading for cells without antigen at each wavelength. Supernatants from parallel cultures were harvested after 72 h and stored at −70°C until they were assayed for specific cytokines. The levels of interleukin-2 (IL-2), interleukin-4 (IL-4), interleukin-10 (IL-10), and interferon-γ (IFN-γ) were determined in culture supernatants by using a sandwich ELISA (OptEIA; Pharmingen) according to the manufacturer's instructions.
Active protection assay.
On day 54, the mice were challenged intraperitoneally in groups of 12 with 1.2 × 108 or 1.2 × 107 CFU of freshly grown C. perfringens cells. The cells for challenge were grown in TGB at 37°C, harvested in late-exponential phase, and washed with anaerobically prepared phosphate-buffered saline (pH 7.2) before injection. The number of cells (CFU) was determined by serial dilution plating on SPS agar as described above. Death, as well as gas gangrene symptoms, was recorded intermittently for 7 days. Animals that survived for the entire 7 days were considered survivors.
Statistical analysis.
Statistical analysis to compare the groups was performed using a t test (SigmaStat; Jandel Scientific). The Mann-Whitney rank-sum test was used to determine the P values wherever the normality test failed.
RESULTS
Comparison of the exoproteomes of C. perfringens type A and type C strains.
Proteins secreted into the extracellular milieu by selected C. perfringens type A and type C strains in the exponential phase of growth on RPMI medium were studied using 1-DE and immunoblotting as described above. The protein profiles of the exoproteomes on a Coomassie-stained 1-dimensional (1-D) gel were very different for different strains (data not shown). Western blotting of exoproteins using anti-WC (whole cell plus supernatant) and anti-gangrene sera also indicated qualitative and quantitative differences in immunogenic protein components among the strains (data not shown).
We also compared the 2-DE exoproteome map of C. perfringens ATCC 13124 to those of the two other environmental type A strains (AG04-3 and AL08-16) and to three type C strains (AL06-1, AG02-3, and H2-3) after growing the cells at 37°C on RPMI medium to late-exponential phase. The protein patterns indicated global differences between the exoproteomes of type A and type C strains, though some consensus spots were seen among isolates belonging to the same type (data not shown). We could not perform a specific comparative quantitative analysis of the gels, because the profiles were significantly different and beyond the analytical capacity of PDQuest software. The 2-DE profiles of the type A strain ATCC 13124 (Fig. 1) and pooled type C strains (Fig. 2) reflect the overall differences between the exoproteomes of the two types (A and C).
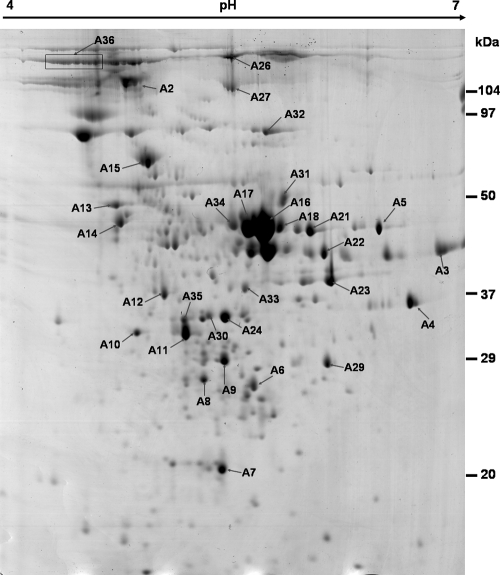
FIG. 1.
Two-dimensional Coomassie-stained gel of proteins in the culture supernatant of C. perfringens ATCC 13124. Proteins were separated in the first dimension by a pH 4 to 7 immobilized pH gradient gel (length, 17 cm) and then in the second dimension by a 12% polyacrylamide gel. Spots were excised, and the corresponding proteins were identified by MALDI-TOF-MS and database searches. The spots are labeled on the gel according to the numbers presented in Table 1 and Table S2 in the supplemental material.
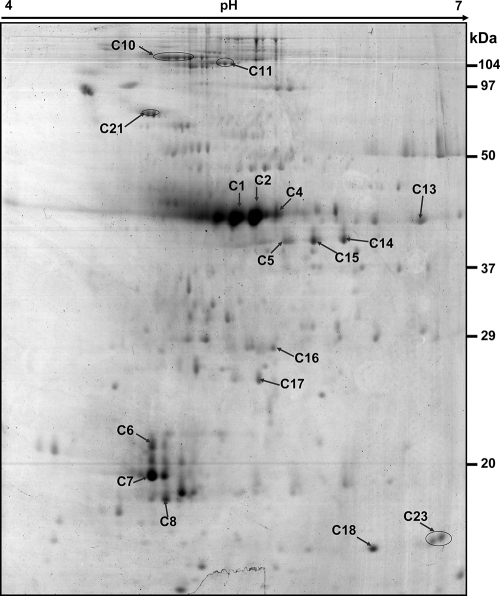
FIG. 2.
Two-dimensional Coomassie-stained gel of proteins in the pooled culture supernatants of C. perfringens type C isolates. Proteins were separated in the first dimension by a pH 4 to 7 immobilized pH gradient gel (length, 17 cm) and then in the second dimension by a 12% polyacrylamide gel. Spots were excised, and the corresponding proteins were identified by MALDI-TOF-MS and database searches. The spots are labeled on the gel according to the numbers presented in Table 1 and Table S2 in the supplemental material.
Identification of proteins.
A total of 31 dominant extracellular proteins of C. perfringens ATCC 13124 and 17 dominant spots from the pooled type C exoproteome (strains AL06-1, AG02-3, and H2-3) were marked and analyzed (Fig. 1 and 2) after in-gel digestion with trypsin using MALDI-TOF-MS as shown in Table 1 and Table S1 in the supplemental material. For most of the proteins, the identification was based on the PMF result for the trypsin-digested protein and MS-MS analysis of 2 or 3 matched peptides; the independent data sets resulted in the same protein matches with significant scores in the Mascot database search (see Table S2 in the supplemental material). Further, our protein identification was based on an ion search at the NCBI nonredundant database in the taxonomic group of Firmicutes (3,239,079 entries), so the chances of false-positive hits are substantially reduced.
TABLE 1.
Extracellular proteins identified from C. perfringens ATCC 13124 and pooled exoproteomes of type C strains
| Spot no. | Protein name (accession no.) |
Mr/pI |
COG (functional category)b | Pfam match | |
|---|---|---|---|---|---|
| Theoreticala | Observed | ||||
| A2 (IM1)c | Endo-beta-N-acetylglucosaminidase (YP_695266) | 126/4.66 | 120/4.8 | No related COG | Uncharacterized protein conserved in bacteria |
| A3 (IM6)c | Acetyl-CoA acetyltransferase (YP_696874) | 40/6.06 | 43/6.6 | Acetyl-CoA acetyltransferases, COG0183 (I) | Thiolase, N-terminal domain |
| A4 | Polysaccharide deacetylase family protein (ABG83723) | 34/6.93 | 37/6.7 | Predicted xylanase/chitin deacetylase, COG0726 (G) | Polysaccharide deacetylase |
| A5 | Putative lipoprotein (YP_696216) | 51/6.08 | 48/6.5 | No related COG | No Pfam match |
| A6 | Hypothetical protein CPF_1441 (YP_695886) | 25/5.21 | 28/5.6 | NifU homologs involved in Fe-S cluster formation, COG0822 (C) | NifU-like N terminal domain |
| A7 | Putative rubrerythrin (YP_697309) | 19/5.43 | 21/5.42 | Rubrerythrin, COG1592 (C) | Rubrerythrin |
| A8 | Triosephosphate isomerase (YP_695952) | 27/5.01 | 27/5.3 | Triosephosphate isomerase, COG0149 (C) | Triosephosphate isomerase |
| A9 | Fructose-bisphosphate aldolase (YP_696000) | 30/5.02 | 29/5.4 | Fructose bis-phosphate aldolase, COG0191 (G) | Fructose-bisphosphate aldolase class II |
| A10 | Hypothetical protein CPF_2918 (YP_697282) | 31/4.88 | 43/4.9 | No related COG | Protein of unknown function |
| A11 | SagA protein (YP_694734) | 46/5.61 | 43/4.2 | Cell wall-associated hydrolases (invasion-associated proteins), COG0791 (M) | NlpC/P60 family |
| A12 | Probable ion uptake ABC transporter (YP_694897) | 38/5.20 | 37/5.08 | ABC-type iron/thiamine transport systems, COG1840 (I) | Bacterial extracellular solute-binding protein |
| A13 (IM16)c | Fructose-bisphosphate aldolase (YP_696000) | 30/5.02 | 49/4.01 | Fructose bis-phosphate aldolase, COG0191 (G) | Fructose-bisphosphate aldolase class II |
| A14 (IM18)c | Extracellular solute-binding protein (YP_696758) | 54/4.7 | 47/4.79 | Sugar-binding periplasmic proteins/domains COG1653 (G) | Bacterial extracellular solute-binding protein |
| A15 | DnaK-type molecular chaperone hsp70 (YP_696713) | 66/4.75 | 80/4.9 | Molecular chaperone, COG0443 (O) | Hsp70 protein |
| A16 (IM13)c | SagA protein (YP_694734) | 46/5.61 | 48/5.75 | Cell wall-associated hydrolases (invasion-associated proteins), COG0791 (M) | NlpC/P60 family |
| A17 (IM12)c | Phospholipase C (YP_694509) | 42/5.24 | 45/5.6 | No related COG | No Pfam match |
| A18 | SagA protein (YP_694734) | 46/5.61 | 48/5.8 | Cell wall-associated hydrolases (invasion-associated proteins), COG0791 (M) | NlpC/P60 family |
| A21 | M24/M37 family peptidase (YP_694655) | 44/5.67 | 48/6.0 | Membrane proteins/ metalloendopeptidase, COG0739 (M) | Peptidase family M23 |
| A22 (IM9)c | Ornithine carbamoyltransferase (YP_694626) | 37/5.4 | 43/6.1 | Ornithine carbamoyltransferase, COG0078 (E) | Aspartate/ornithine carbamoyltransferase |
| A23 | Glyceraldehyde-3-phosphate dehydrogenase (YP_695954) | 35/5.5 | 38/6.15 | Glyceraldehyde-3-phosphate dehydrogenase, COG0057 (G) | Glyceraldehyde 3-phosphate dehydrogenase |
| A24 | Hypothetical protein CPF_0656 (YP_695109) | 37/5.53 | 35/5.49 | No related COG | No Pfam match |
| A26 | F5/8 type C domain-containing protein (YP_695932) | 189/5.15 | 110/5.5 | No related COG | No Pfam match |
| A27 | Hypothetical protein CPF_2364 (YP_696787) | 123/5.18 | 105/5.5 | No related COG | No Pfam match |
| A29 | Purine-nucleoside phosphorylase (YP_697182) | 29/5.63 | 29/6.1 | Purine nucleoside phosphorylase, COG0005 (F) | Phosphorylase superfamily |
| A30 | Hypothetical protein CPF_0656 (YP_695109) | 37/5.53 | 35/5.34 | No related COG | No Pfam match |
| A31 | Cell wall binding repeat-containing protein (YP_695059) | 63/5.37 | 50/5.82 | Predicted carboxypeptidase, COG2866 (E) | Putative cell wall binding repeat |
| A32 | Exo-alpha-sialidase (YP_695173) | 77/5.43 | 95/5.7 | No related COG | No Pfam match |
| A33 (IM14)c | Translation elongation factor (YP_696386) | 33/5.13 | 37/5.6 | Translation elongation factor Ts, COG0264 (J) | Elongation factor TS |
| A34 | S-Adenosylmethionine synthetase (YP_696856) | 37/5.17 | 45/5.5 | S-Adenosylmethionine synthetase, COG0192 (H) | S-Adenosylmethionine synthetase, N-terminal domain |
| A35 | Peptidyl-prolyl cis-isomerase family protein (YP_694723) | 27/4.89 | 34/5.2 | Parvulin-like peptidyl-prolyl isomerase, COG0760 (O) | Rotamase |
| A36 | F5/8 type C domain-containing protein (YP_695932) | 189/5.15 | 110/4.5 | No related COG | No Pfam match |
| C1 | Alpha-clostripain (NP_562197) | 59/5.88 | 40/5.5 | No related COG | No Pfam match |
| C2 | Alpha-clostripain trans (NP_562197) | 59/5.88 | 40/5.65 | No related COG | No Pfam match |
| C4 | Alpha-clostripain (NP_562197) | 59/5.88 | 40/4.8 | No related COG | No Pfam match |
| C5 | Glyceraldehyde-3-phosphate dehydrogenase (YP_695954) | 35/5.5 | 38/5.82 | Glyceraldehyde-3-phosphate dehydrogenase, COG0057 (G) | Glyceraldehyde 3-phosphate dehydrogenase |
| C6 | Alpha-clostripain (NP_562197) | 59/5.88 | 21/4.98 | No related COG | No Pfam match |
| C7 | Alpha-clostripain (NP_562197) | 59/5.88 | 20/4.98 | No related COG | No Pfam match |
| C8 | Alpha-clostripain (NP_562197) | 59/5.88 | 18/5.05 | No related COG | No Pfam match |
| C10 | Collagenase A (NP_561089) | 125/4.93 | 110/5.15 | PKD repeat proteins, COG3291 (R) | Peptidase_M9 |
| C11 | Collagenase A (NP_561089) | 125/4.93 | 105/4.5 | PKD repeat proteins, COG3291 (R) | Peptidase_M9 |
| C13 | Acetyl-CoA acetyltransferase (YP_696874) | 40/6.06 | 39/6.7 | Acetyl-CoA acetyltransferases, COG0183 (I) | Thiolase, N-terminal domain |
| C14 | Glyceraldehyde-3-phosphate dehydrogenase (YP_695954) | 35/5.5 | 38/6.21 | Glyceraldehyde-3-phosphate dehydrogenase, COG0057 (G) | Glyceraldehyde 3-phosphate dehydrogenase |
| C15 | Glyceraldehyde-3-phosphate dehydrogenase (YP_695954) | 35/5.5 | 38/6.01 | Glyceraldehyde-3-phosphate dehydrogenase, COG0057 (G) | Glyceraldehyde 3-phosphate dehydrogenase |
| C16 | Hypothetical protein CPF_2364 (YP_696787) | 123/5.18 | 28/5.79 | No related COG | No Pfam match |
| C17 | Probable heme oxygenase (YP_694668) | 25/5.18 | 27/5.70 | No related COG | No Pfam match |
| C18 | HPr family phosphocarrier protein (YP_696356) | 9/6.5 | 15/6.4 | Phosphotransferase system, HPr-related proteins, COG1925 (G) | PTS-HPr |
| C21 | DnaK-type molecular chaperone hsp70 | 66/4.79 | 95/4.95 | Molecular chaperone, COG0443 (O) | Hsp70 protein |
| C23 | HPr family phosphocarrier protein (YP_696356) | 9/6.59 | 15/6.83 | Phosphotransferase system, HPr-related proteins, COG1925 (G) | PTS-HPr |
Theoretical values were obtained with an online tool at http://expasy.org/sprot/.
COGs were assigned after a COGnitor search. Functional role categories, assigned according to the descriptions on the COG page at http://www.ncbi.nlm.nih.gov/COG, are as follows: E, amino acid transport and metabolism; M, cell envelope biogenesis, outer membrane; G, carbohydrate transport and metabolism; O, posttranslational modification, protein turnover, chaperones; C, energy production and conversion; J, translation, ribosomal structure and biogenesis; H, coenzyme transport and metabolism; I, lipid metabolism; F, nucleotide transport and metabolism; R, general function prediction only.
Immunoreactive to serum raised against C. perfringens whole-cell culture in mice.
The proteins identified exhibited matches with significant scores in the C. perfringens strain 13 proteome, since this is the only strain of this pathogen represented in the MSDB. Homologs of identified proteins had high amino acid sequence identity (94 to 100%), with significant E-values (except for spot A7), with the corresponding proteins from C. perfringens ATCC 13124 (see Table S2 in the supplemental material), as revealed by blastp analysis.
The MW and pI values of the protein spots on the 2-DE gels were determined and compared with the theoretical MW and pI values of corresponding proteins from C. perfringens ATCC 13124. Most of the experimental values matched well with theoretical values, indicating unambiguous identification (Table 1). Significant differences between theoretical and experimental values were observed for seven of the identified proteins; these might have been caused by posttranslational proteolytic processing and modification. The differences between the two pI values might be attributed to the cleavage of alkaline regions and the phosphorylation of multiple residues.
Extracellular proteome of C. perfringens ATCC 13124.
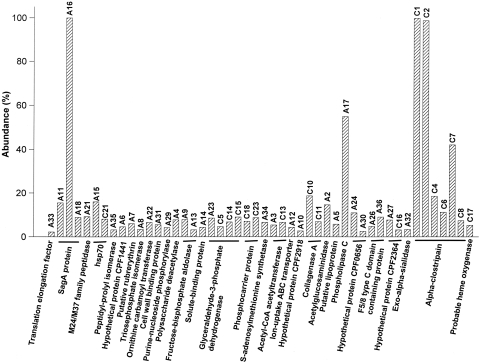
The relative spot density analysis on the 2-DE gel and the identification data for the exoproteins suggest that SagA protein (spots A11, A16, and A18), phospholipase C (A17), DnaK-type molecular chaperone hsp70 (A15), endo-beta-N-acetylglucosaminidase (A2), and hypothetical protein CPF_0656 (A23) were among the most abundant proteins in the extracellular fraction of C. perfringens ATCC 13124 (Table 1; Fig. 1 and 3). The 2-DE gel pattern also indicated that SagA protein (A16 and A18), hypothetical protein CPF_0656 (A24 and A30), and F5/8 type C domain-containing protein (A26 and A36) existed as multiple electropherotypes. Curiously, 10 of the 31 (32%) identified exoproteins were either hypothetical or uncharacterized proteins with putative functions. The C. perfringens ATCC 13124 extracellular proteins were also grouped into major cellular functions (see Table S4 in the supplemental material). Leaving apart proteins for which a cluster of orthologous groups (COG) could not be assigned (26%), most of the exoproteins were predicted to be involved in carbohydrate transport and metabolism (16%) or in cell envelope biogenesis or to be outer surface protein constituents (13%).
FIG. 3.
Relative abundances of identified exoproteins from the C. perfringens ATCC 13124 (spot numbers beginning with A) and pooled type C (spot numbers beginning with C) exoproteomes. The abundance is relative to the level of the most abundant protein on the respective gel. Spot volume was estimated as average OD × area (mm2) in replicate gels by using the spot density determination tool in PDQuest software. The protein numbers refer to Table 1.
Extracellular proteome of C. perfringens type C strains.
The pooled exoproteome of three selected C. perfringens type C strains showed an abundance of charge variants and possible degradation products of alpha-clostripain (C1, C2, C4, C6, C7, and C8) in addition to collagenase A (C10 and C11) (Table 1; Fig. 2 and 3). It is difficult to predict whether the different electropherotypes of the former are strain specific or are secreted in multiple charged forms by one or more of the isolates. Some other abundant proteins in the extracellular fraction of type C strains were glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (C5, C14, and C15), hypothetical protein CPF_2364 (C16), acetyl coenzyme A (acetyl-CoA) acetyltransferase (C13), and the DnaK-type molecular chaperone hsp70 (C21); these were also present in the ATCC 13124 exoproteome. Most of the abundant extracellular proteins (47%) of type C strains could not be assigned a COG, and as in the case of ATCC 13124, most of them (29%) were predicted to be involved in carbohydrate transport and metabolism.
Immunogenic proteins.
Immunogenic protein components in the C. perfringens ATCC 13124 exoproteome were identified by immunoblotting of 2-DE-separated proteins and identification of gel-excised spots from the corresponding Coomassie-stained gel (Table 1). Endo-beta-N-acetylglucosaminidase (A2), acetyl-CoA acetyltransferase (A3), SagA protein (A16), phospholipase C (A17), ornithine carbamoyltransferase (A22), translation elongation factor (A33), fructose-bisphosphate aldolase (A13), and extracellular solute-binding protein (A14) were found to be immunogenic secretory proteins when probed with mouse anti-WC serum. We could not identify a few other immunogenic proteins visible on the Western blot (data not shown).
In silico analysis of protein similarity and features.
Based on the blastp search results, most of the proteins identified in the present investigation appeared to be highly conserved (showing 93 to 100% amino acid identity and 96 to 100% amino acid similarity) among C. perfringens strains and were not strain specific (based on whole-genome sequence data for 9 strains available in the database) (see Table S3 in the supplemental material). In contrast, conservation of protein sequences across related clostridial species, showing close homologs, was observed for only 33% of the proteins identified. Curiously, hypothetical proteins CPF_2918, CPF_0656, and CPF_2364, as well as cell wall binding repeat-containing protein and exo-alpha-sialidase, showed no significant similarity with any other protein in the database by use of blastp.
All the proteins identified were analyzed using various bioinformatics software programs, such as PROSITE, SignalP, SecretomeP, TMHMM, LipoP, and PSORT, for predicting protein secretion and localization (Table S4). A total of 17 (56.6%) proteins were predicted by SignalP to be secreted in the classical Sec pathway, which is characterized by the presence of a signal peptide. However, 4 proteins were shown to possess the cleavage site for signal peptidase II (SpII), while 5 proteins were predicted to have transmembrane helices (TMHMM), indicating an extracytoplasmic but membrane-associated location. In contrast to the predictions of SignalP and SecretomeP, PSORT predicted 1 cell wall-associated protein, 4 extracellular proteins, 15 cytoplasmic proteins, 2 proteins with multiple locations, and 8 proteins of unknown localization.
To summarize, more than 50% of the proteins were predictably secreted by either Sec or nonclassical pathways as indicated by the presence of a signal peptide (SignalP) and/or nonsignal peptide triggered protein secretion (SecretomeP). Nevertheless, several proteins (43.3%) from the C. perfringens exoproteome have metabolic functions and are predicted to be cytoplasmic by all the algorithms tested.
It is noteworthy that for 7 out of 8 hypothetical and uncharacterized proteins; an extracellular localization was predicted by using one or more of the bioinformatic tools (data not shown). Primary- and secondary-structure predictions for these 8 proteins indicated a low (e.g., cell wall binding protein, hypothetical protein CPF_0656) to moderate (e.g., SagA protein, hypothetical protein CPF_1441) content of alpha-helical structures. No beta-bridge, beta turn, or disulfide bonding state was seen in any of the hypothetical proteins, and a leucine zipper was observed in SagA protein alone. High antigenicity indices were observed for SagA protein, cell wall binding protein, hypothetical protein CPF_2918, and F5/8 type C domain-containing protein by use of the ANTIGENpro algorithm.
Expression of SagA protein.
We decided to examine the expression of SagA protein in different strains and in an in vivo mouse model, because this protein (i) was poorly characterized, (ii) was the most abundant of the extracellular proteins, (iii) was assigned the COG of cell wall-associated hydrolases (invasion-associated proteins), and (iv) has been shown previously to be regulated by the VirR/VirS system in Clostridium perfringens (40). Using anti-SagA serum, the expression of the protein in the culture supernatants of nine selected RCM (reinforced clostridial medium)-grown C. perfringens type A and type C strains was studied. The protein was secreted by all the strains tested and was generally abundant. We also examined the secretion of SagA protein in vivo, using rabbits as model hosts, by allowing freshly grown and washed C. perfringens ATCC 13124 cells to colonize and grow in the peritoneal cavity, as described in Materials and Methods. A prominent immune reaction with anti-SagA serum was observed, indicating in vivo secretion of the protein in infected animals (data not shown).
Immunogenicity of recombinant cOTC and protection study.
Recombinant cOTC was cloned and successfully expressed in the E. coli M15 host. The protein was purified to near-homogeneity from the soluble fraction of an induced cell lysate after sonication. On the SDS-PAGE gel, the purified band had the expected size of the recombinant protein (∼34 kDa) as revealed by Coomassie blue staining, and immunoblotting using HRP-conjugated RGS-His (Qiagen, Germany) revealed the presence of the fusion protein (data not shown). The expressed recombinant protein was also confirmed by double-pass sequencing of the construct and peptide mass fingerprinting of the gel-extracted band. The level of expression of recombinant cOTC was estimated to be 25 to 50 μg/ml of cell culture by densitometric analysis of Coomassie-stained gels.
Using recombinant cOTC, the Western blot was developed with anti-gangrene and anti-WC sera, which clearly indicated that the protein elicits an immune response in mice, both in an experimental gas gangrene model and also when a whole-cell bacterial culture was used for immunization. In a whole-cell ELISA performed by coating washed C. perfringens cells and probing with anti-ornith serum, binding was observed even at the 1:16,000 serum dilution, indicating the presence of the antigen on the cell surface as well. A direct ELISA was performed to determine the relative density of the antigen on the cell surface, using different concentrations of recombinant cOTC at an antibody (anti-ornith) dilution of 1:16,000 (data not shown). When ∼105 cells per well were used in a whole-cell ELISA, the OD492 corresponded to approximately 1.2 ng of antigen, indicating a high concentration of protein on the cell surface.
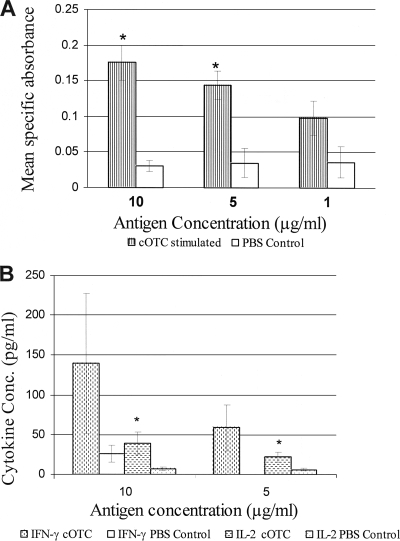
The immune response to recombinant cOTC protein was studied in BALB/c mice. A significant rise in titers of IgG to recombinant cOTC was observed in mice 15 or 30 days after last dose. To further understand the nature of the immune response, isotyping was carried out. The results are summarized in Table 2. Levels of all antibody isotypes (IgG1, IgG2a, IgG2b, and IgG3) were significantly higher in animals immunized with cOTC than in control animals. However, IgG2a titers predominated over IgG1 titers (IgG2a/IgG1 ratio, 2), suggesting a bias toward a Th1 type immune response (Table 2). The proliferation of spleen lymphocytes in cOTC-immunized animals was observed (Fig. 4 A), suggesting a cellular immune response. There were significant increases in the levels of IFN-γ and IL-2 (Fig. 4B), suggesting a Th1 type immune response. There was no rise in IL-4 or IL-10 levels.
TABLE 2.
Antibody response to ornithine carbamoyltransferase protein in cOTC- or PBS-immunized BALB/c mice
| Immunization groupa | Log10 antibody titerb |
IgG2a/IgG1 ratio | ||||
|---|---|---|---|---|---|---|
| IgG | IgG1 | IgG2a | IgG2b | IgG3 | ||
| cOTC antigen + FA | 5.1 ± 0.007, 4.8 ± 0.031 | 3.9 ± 0.068, 3.6 ± 0.046 | 4.2 ± 0.009, 3.9 ± 0.02 | 4.5 ± 0.006, 4.5 ± 0.012 | 3.3 ± 0.007, 3.6 ± 0.0075 | 2, 2 |
| PBS control | <2 | <2 | <2 | <2 | <2 | |
FA, Freund's adjuvant.
Titers were determined by ELISA at days 15 and 30 after the last dose.
FIG. 4.
(A) Lymphocyte proliferation assay. Splenocytes were prepared 1 month after the last immunization and were cultured with cOTC (final concentration, 1 or 10 μg/ml) for 72 h. Splenocyte proliferation was observed by adding Alamar Blue for 15 to 18 h and was calculated as described in Materials and Methods. Results are expressed as mean specific absorbance (at 570 to 600 nm) and represent means ± standard errors. Significant differences between the antigen group (cOTC plus Freund's adjuvant) and the PBS group, determined by the Student t test, are indicated by asterisks (P < 0.05). (B) IFN-γ and IL-2 production in cultured spleen cells of mice injected with cOTC plus Freund's adjuvant or the PBS control. Results are expressed in picograms per milliliter and are means ± standard errors. Significant differences obtained by comparison with unstimulated cells, as determined by a paired Student t test, are indicated by asterisks (P < 0.05).
The survival of animals immunized with recombinant cOTC upon challenge with viable C. perfringens cells is shown in Table 3. Immunization with cOTC clearly resulted in protection against a direct challenge with C. perfringens organisms. There was complete protection when 1.2 × 107 CFU of the organism was used, whereas mice were partially protected when the challenge dose was increased to 1.2 × 108 CFU: 58% survived, and the time to death was increased for mice that did not survive the higher dose.
TABLE 3.
Results of C. perfringens challenge of mice immunized with ornithine carbamoyltransferase
| Immunization group | Challenge dose (no. of C. perfringens cells given i.p.) | No. of animals challenged | No. (%) of animals that surviveda | Time to death (h) |
|---|---|---|---|---|
| Recombinant ornithine carbamoyltransferase | 1.2 × 108 | 12 | 7 (58.3 ± 8.33)* | 10-12 |
| 1.2 × 107 | 12 | 12 (100 ± 0.0)* | ||
| Adjuvant control | 1.2 × 108 | 12 | 0 (0 ± 0) | 4-6 |
| 1.2 × 107 | 12 | 0 (0 ± 0) | 4-6 |
Each challenge dose was given to 3 groups of 4 animals each. Percentages of animals surviving are means ± standard errors. An asterisk indicates that the result was significantly different from that for the control by Student's t test(P ≤ 0.001).
DISCUSSION
The extracellular proteomes of several Gram-positive (monoderm) pathogenic bacteria have been the subject of recent proteomic studies (17, 19, 30, 36, 46, 47, 48). It is postulated that several extracellular proteins may potentially be involved in the adherence of the bacteria to host cells, while some may be required for the suppression of the host's defense mechanisms. In the present study, the use of mass spectrometry led to the identification of 48 proteins altogether and the determination of their relative abundances during the exponential-growth phase of the bacterium.
Analysis of the C. perfringens extracellular proteins identified here revealed many new exoproteins beyond the findings of previous proteomic studies on monoderm bacteria. Some of the exoproteins reported here are also shown in the proteomic elucidation of secretory fractions of other Gram-positive and Gram-negative pathogenic bacteria, often as virulence and/or antigenic determinants (see Table S3 in the supplemental material). As shown in Fig. 1 and 3, SagA and phospholipase C were the most abundant proteins secreted by the gas gangrene clinical isolate C. perfringens ATCC 13124. Both these predominant secretory proteins have been shown previously to be regulated by one of the two-component systems of C. perfringens, VirR/VirS, which globally controls the production of several virulence factors (6, 40). In their proteomic study on extracellular proteins of C. perfringens strain 13 regulated by the VirR/VirS two component system, Shimizu et al. (40) identified hypothetical protein CPE1529 (putative lipoprotein) and fructose-bisphosphate aldolase, which have also been identified in our study as predominant exoproteins of C. perfringens ATCC 13124.
Alpha-clostripain and collagenase A were the most abundant proteins identified in type C strains. Homologs of these two proteins have also been shown to be positively regulated in wild-type C. perfringens strain 13 compared with a VirR mutant (40). There are global qualitative and quantitative differences between the exoproteomes of C. perfringens type A and type C strains, which are likely to be correlated with discrete disease presentations by the two types of strains. The difference between the exoproteomes of C. perfringens type A and type C strains seems to arise from the differential regulation and/or secretion of proteins, since many of the proteins identified in type C strains were also identified in the type A strain (though with different abundances) or had homologs in all 9 genomes of C. perfringens, which include representative strains of all the types (A through E) of the bacterium (see Table S3 in the supplemental material).
Some of the C. perfringens extracellular proteins identified here showed metabolic functions that would typically place them in the cytoplasm. There is growing evidence that such “housekeeping” enzymes have a role in the pathogenesis of, or immunity to, other infections (5, 10, 39, 40, 48). Besides, some cytoplasmic proteins have been shown to actually moonlight on the bacterial cell surface or to have more than one role in an organism (24). For example, GAPDH and enolase are moonlighting proteins involved in plasmin binding when present on the cell surfaces of some Gram-positive bacteria (31, 33). The results shown in the present investigation are not sufficient to rule out the possibility of cell lysis. However, few such predictably cytoplasmic proteins reported in this study have also been detected on the bacterial surface or in extracellular fractions in previous proteomic analyses (see Table S3 in the supplemental material), and in some of the studies, cell lysis was excluded as a reason for this observation by strong experimental evidence. For example, acetyl-CoA acetyltransferase was identified in the extracellular fractions of Mycobacterium tuberculosis and Burkholderia cenocepacia, and triosephosphate isomerase was found in the extracellular proteomes of Bacillus anthracis, M. tuberculosis, and Streptococcus suis, while glyceraldehyde-3-phosphate dehydrogenase was shown to be secreted by Streptococcus pneumoniae, Staphylococcus aureus, B. anthracis, and S. suis (see references in Table S3 in the supplemental material). In their study on the B. anthracis extracellular proteome, Walz et al. (48) observed several predictably cytoplasmic proteins in the extracellular culture supernatant of the bacterium, and based on their experimental evidence, cell lysis was excluded as the major contributor to the protein accumulation in the exoproteome. In spite of a growing list of cytoplasmic proteins identified on the bacterial surface or secreted out, the mechanism of their export to the cell exterior remains unclear. Internal signal sequences, posttranslational acylation, or an association with an extracellular protein are hypothesized as possible means (33).
Four proteins are shown here to possess the cleavage site for signal peptidase II (SpII) and are predicted to be bound at the surface of the cytoplasmic membrane. Surface-localized lipoproteins are deemed important vaccine candidates, and during the development of a Neisseria meningitidis vaccine, a search of the genomic sequence revealed more than 25 novel antigens with vaccine potential, the majority of which were lipoproteins (1).
The presence of several hypothetical proteins in the extracellular fraction of C. perfringens is of particular significance. Their exact roles in virulence, niche adaptation, and host specificity need to be explored. Our data demonstrate for the first time that hypothetical proteins CPF_1441, CPF_2918, CPF_0656, and CPF_2364 are genuine proteins of C. perfringens. These proteins should be characterized in detail with respect to their roles in pathogenesis and their prophylactic potentials. Hsu et al. (21) have recently identified a hypothetical protein (NMB1468) as a novel lipoprotein ubiquitous in Neisseria meningitidis with vaccine potential. Eight secretory proteins with putative and unknown functions, identified in this study, were subjected to bioinformatic analysis in order to assess secondary structure and general features. Five out of eight such proteins were not assigned any known function either by a COGnitor search or by a Pfam search. Curiously, the cell wall binding protein has been predicted to be a carboxypeptidase and contains conserved aromatic residues and multiple tandem copies of glycines in a number of homologs. These repeats are implicated in the specific recognition of choline-containing cell walls. Whether this protein plays any role in virulence and aids in membrane-damaging activity needs to be tested.
The invasion-associated protein SagA, shown here as the most abundant exoprotein of C. perfringens ATCC 13124, is immunogenic and is expressed in vivo during infection in a rabbit model (Table 1; Fig. 1 and 3). Curiously, SagA is also a virulence determinant of C. perfringens regulated by the two component VirR/VirS regulon and has been described as an invasion or invasion-associated protein in other bacteria (COG functional category in Table 1). An argument for the vaccine potential of SagA needs to be substantiated by more data.
Currently there is no subunit vaccine for human or animal use to protect against gas gangrene infection caused by C. perfringens. In some countries, serum raised against a crude culture supernatant (toxoid) is used for passive immunization of trauma patients suffering from deep and crushing injuries (45). Subunit vaccines based on selected antigens and/or their respective genes are recognized as the safest type of antibacterial vaccine. The bottleneck in the development of effective subunit vaccines is the choice of the antigens. A proper candidate for immunization must possess a wide range of different properties, such as extracytoplasmic localization, abundant presence in the cell, immunogenicity (i.e., ability to stimulate the immune system), and conservation among different pathogen serotypes/genotypes. Further, a candidate vaccine antigen has to be expressed in vivo during infection, when the pathogen is present in the host organism. In the present study, ornithine carbamoyltransferase (cOTC) was an abundant immunogenic protein secreted into the extracellular milieu by the bacterium (Fig. 1; Table 1) and was arguably also surface localized at a high concentration, as revealed by whole-cell ELISA. The recombinant protein elicited a predominantly Th1 type immune response, as revealed by the high titers of anti-cOTC IgG2a antibodies and secretion of IFN-γ and IL-2 in the spleen cells of cOTC-immunized animals. Further, the immunized mice were found to be protected from developing gas gangrene disease after intraperitoneal inoculation with a lethal dose of C. perfringens ATCC 13124, suggesting that cOTC may be a promising vaccine candidate for the prevention of gangrenous infection. In our earlier study, cOTC was identified as an immunogenic surface protein of this bacterium and was upregulated in CMM-grown cells (2). In another study, cOTC was isolated as a putative adhesin from a surface molecule preparation of Staphylococcus epidermidis and was shown to be protective in active immunization challenge experiments for the pathogen (11, 23). The homolog of this protein has also been identified as an immunogenic protein in outer surface protein preparations of Streptococcus agalactiae and Streptococcus pyogenes (10, 22). Taken together, these findings make cOTC a putative vaccine candidate against C. perfringens infection.
Conclusion.
Several proteins identified here in the exoproteomes of C. perfringens type A and type C strains have not been reported in the exoproteome of any other Gram-positive bacterium. The nature of the C. perfringens exoproteome suggests that this organism is capable of thriving in both carbohydrate- and protein-rich environments, since several proteins were assigned the function of carbohydrate transport and metabolism, while peptidases and enzymes required for the metabolism of amino acids were also identified. The putative lipoprotein, SagA protein, the M24/M37 family peptidase, and the hypothetical protein CPF_0656 can be excellent protein markers for specific detection of C. perfringens in food samples, since they share very low amino acid sequence identity with their nearest homologs (<50%) and are conserved among C. perfringens strains (see Table S3 in the supplemental material). In future studies, an exhaustive genomic analysis of the protein secretion systems in C. perfringens would cover the secretome of this species, as has been done for Clostridium acetobutylicum (13) and Listeria monocytogenes (14). This should provide indications of the systems involved in the secretion of exoproteins, which is particularly relevant information considering that a significant proportion of exoproteins could only be predicted to be transported by nonclassical secretion pathways and that the Sec pathway is the only protein secretion route uncovered in C. perfringens.
Some of these exoproteins have features characteristic of virulence determinants, suggesting that in addition to the “classic” extracellular toxins, a large number of proteins may be essential for C. perfringens virulence. Further empirical studies are required to test their potential as vaccine candidates. The presence in high abundance of several hypothetical proteins described here in the exoproteome of this bacterium is worth noting and warrants investigation not only with respect to their roles in virulence and pathogenesis but also with respect to their potential as candidates for therapeutics and vaccines. The present investigation surely demonstrates the strength of proteomics in the rapid elucidation of vaccine candidates that can be further studied for their potential in the prevention of disease. Ornithine carbamoyltransferase has been shown here to be surface associated and protective in mice. This knowledge will ultimately lead to the development of a specific, safer, and highly efficacious vaccine against C. perfringens. In addition, the identification of high-abundance extracellular proteins with sequences unique to this species may aid in the development of specific detection methods and diagnostic kits.
Supplementary Material
Acknowledgments
We thank R. Vijayaraghavan, Director, DRDE, Gwalior, India, for providing all the facilities and support required for this study.
This work was funded by the Defence Research and Development Organization, Government of India.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 6 July 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Adu-Bobie, J., B. Capecchi, D. Serruto, R. Rappuoli, and M. Pizza. 2003. Two years into reverse vaccinology. Vaccine 21:605-610. [DOI] [PubMed] [Google Scholar]
- 2.Alam, S. I., S. Bansod, R. B. Kumar, N. Sengupta, and L. Singh. 2009. Differential proteomic analysis of Clostridium perfringens ATCC 13124; identification of dominant, surface and structure associated proteins. BMC Microbiol. 9:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amero, S. A., T. C. James, and S. C. R. Elgin. 1988. Production of antibodies using proteins in gel bands. Methods Mol. Biol. 3:355-362. [DOI] [PubMed] [Google Scholar]
- 4.Awad, M. M., A. E. Bryant, D. L. Stevens, and J. I. Rood. 1995. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15:191-202. [DOI] [PubMed] [Google Scholar]
- 5.Banu, S., K. Ohtani, H. Yaguchi, T. Swe, S. T. Cole, H. Hayashi, and T. Shimizu. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 35:854-864. [DOI] [PubMed] [Google Scholar]
- 6.Ba-Thein, W., M. Lyristis, K. Ohtani, I. T. Nisbet, H. Hayashi, J. I. Rood, and T. Shimizu. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 178:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackshear, P. J. 1984. Systems for polyacrylamide gel electrophoresis. Methods Enzymol. 104:237-255. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Calabi, E., and N. Fairweather. 2002. Patterns of sequence conservation in the S-layer proteins and related sequences in Clostridium difficile. J. Bacteriol. 184:3886-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, J. N., R. D. Ramirez, B. J. Currie, S. J. Cordwell, S. P. Djordjevic, and M. J. Walker. 2005. Surface analyses and immune reactivities of major cell wall-associated proteins of group A Streptococcus. Infect. Immun. 73:3137-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, J. N., A. Henningham, C. M. Gillen, V. Ramachandran, and M. J. Walker. 2008. Human pathogenic streptococcal proteomics and vaccine development. Proteomics Clin. Appl. 2:387-410. [DOI] [PubMed] [Google Scholar]
- 12.DelVecchio, V. G., J. P. Connolly, T. G. Alefantis, A. Walz, M. A. Quan, G. Patra, J. M. Ashton, J. T. Whittington, R. D. Chafin, X. Liang, T. Grewal, A. S. Khan, and C. V. Mujer. 2006. Proteomic profiling and identification of immunodominant spore antigens of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Appl. Environ. Microbiol. 72:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desvaux, M., A. Khan, A. Scott-Tucker, R. R. Chaudhuri, M. J. Pallen, and I. R. Henderson. 2005. Genomic analysis of the protein secretion systems in Clostridium acetobutylicum ATCC 824. Biochim. Biophys. Acta 1745:223-253. [DOI] [PubMed] [Google Scholar]
- 14.Desvaux, M., and M. Hébraud. 2006. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol. Rev. 30:774-805. [DOI] [PubMed] [Google Scholar]
- 15.Desvaux, M., M. Hébraud, R. Talon, and I. R. Henderson. 2009. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17:139-145. [DOI] [PubMed] [Google Scholar]
- 16.Drudy, D., E. Calabi, L. Kyne, S. Sougioultzis, E. Kelly, N. Fairweather, and C. P. Kelly. 2004. Human antibody response to surface layer proteins in Clostridium difficile infection. FEMS Immunol. Med. Microbiol. 41:237-242. [DOI] [PubMed] [Google Scholar]
- 17.Dumas, E., M. Desvaux, C. Chambon, and M. Hébraud. 2009. Insight into the core and variant exoproteomes of Listeria monocytogenes species by comparative sub-proteomic analysis. Proteomics 9:3136-3155. [DOI] [PubMed] [Google Scholar]
- 18.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, A. Heger, J. E. Pollington, O. L. Gavin, P. Gunasekaran, G. Ceric, K. Forslund, L. Holm, E. L. L. Sonnhammer, S. R. Eddy, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gohar, M., N. Gilois, R. Graveline, C. Garreau, V. Sanchis, and D. Lereclus. 2005. A comparative study of Bacillus cereus, Bacillus thuringiensis and Bacillus anthracis extracellular proteomes. Proteomics 5:3696-3711. [DOI] [PubMed] [Google Scholar]
- 20.Hall, I. C. 1945. An experimental evaluation of American commercial bivalent and pentavalent gas gangrene anti-toxins. Surg. Gynecol. Obstet. 81:487-499. [Google Scholar]
- 21.Hsu, C. A., W. R. Lin, J. C. Li, Y. T. Tseng, C. M. Chang, Y. S. Lee, and C. Y. Yang. 2008. Immunoproteomic identification of the hypothetical protein NMB1468 as a novel lipoprotein ubiquitous in Neisseria meningitidis with vaccine potential. Proteomics 8:2115-2125. [DOI] [PubMed] [Google Scholar]
- 22.Hughes, M. J. G., J. C. Moore, J. D. Lane, R. Wilson, P. K. Pribul, Z. N. Younes, R. J. Dovson, P. Everest, A. J. Reason, J. M. Redfern, F. M. Greer, T. Paxton, M. Panico, H. R. Morris, R. J. Fieldman, and J. D. Santangelo. 2002. Identification of major outer surface proteins of Streptococcus agalactiae. Infect. Immun. 70:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain, M., G. Peters, G. S. Chhatwal, and M. Herrmann. 1999. A lithium chloride-extracted, broad-spectrum-adhesive 42-kilodalton protein of Staphylococcus epidermidis is ornithine carbamoyltransferase. Infect. Immun. 67:6688-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeffery, C. J. 2003. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19:415-417. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Balakrishna, G. S. Agarwal, S. Merwyn, G. P. Rai, H. V. Batra, A. A. Serdesai, and J. Gowrishankar. 2009. Th1-type immune response to infection by pYV-cured phoP-pohQ null mutant of Yersinia pseudotuberculosis is defective in mouse model. Antonie Van Leeuwenhoek 95:91-100. [DOI] [PubMed] [Google Scholar]
- 26.Kyne, L., M. Warny, A. Qamar, and C. P. Kelly. 2001. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhea. Lancet 357:189-193. [DOI] [PubMed] [Google Scholar]
- 27.MacLennan, J. D. 1943. Anaerobic infections of war wounds in the Middle East. Lancet ii:123-126. [Google Scholar]
- 28.MacLennan, J. D. 1962. The histotoxic clostridial infections of man. Bacteriol. Rev. 26:177-276. [PMC free article] [PubMed] [Google Scholar]
- 29.Marmur, J. 1961. Procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 30.Mattow, J., U. E. Schaible, F. Schmidt, and K. Hagens. 2003. Comparative proteome analysis of culture supernatant proteins from virulent Mycobacterium tuberculosis H37Rv and attenuated M. bovis BCG Copenhagen. Electrophoresis 24:3405-3420. [DOI] [PubMed] [Google Scholar]
- 31.Modun, B., and P. Williams. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neeson, B. N., G. C. Clark, H. S. Atkins, B. Lingard, and R. W. Titball. 2007. Analysis of protection afforded by a Clostridium perfringens alpha-toxoid against heterologous clostridial phospholipases C. Microb. Pathog. 43:161-165. [DOI] [PubMed] [Google Scholar]
- 33.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 34.Péchiné, S., C. Janoir, and A. Collignon. 2005. Variability of Clostridium difficile surface proteins and specific serum antibody response in patients with Clostridium difficile-associated disease. J. Clin. Microbiol. 43:5018-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petit, L., M. Gibert, and M. R. Popoff. 1999. Clostridium perfringens toxinotype and genotype. Trends Microbiol. 7:104-110. [DOI] [PubMed] [Google Scholar]
- 36.Pocsfalvi, G., G. Cacace, M. Cuccurullo, G. Serluca, A. Sorrentino, G. Schlosser, G. Blaiotta, and A. Malorni. 2008. Proteomic analysis of exoproteins expressed by enterotoxigenic Staphylococcus aureus strains. Proteomics 8:2462-2476. [DOI] [PubMed] [Google Scholar]
- 37.Rood, I. R., and S. T. Cole. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55:621-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoepe, H., C. Pache, A. Neubauer, H. Potschka, T. Schlapp, L. H. Wieler, and G. Baljer. 2001. Naturally occurring Clostridium perfringens nontoxic alpha-toxin variant as a potential vaccine candidate against alpha-toxin-associated diseases. Infect. Immun. 69:7194-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Severin, A., E. Nickbarg, J. Wooters, S. A. Quazi, Y. V. Matsuka, E. Murphy, I. K. Moutsatsos, R. J. Zagursky, and S. B. Olmsted. 2007. Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J. Bacteriol. 189:1514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu, T., K. Shima, K. Yoshino, K. Yonezawa, T. Shimizu, and H. Hayashi. 2002. Proteome and transcriptome analysis of the virulence genes regulated by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 184:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens, D. L., K. A. Maier, B. M. Laine, and J. E. Mitten. 1987. Comparison of clindamycin and rifampicin, tetracycline, metronidazoles and penicillin for efficacy in prevention of experimental gas gangrene due to Clostridium perfringens. J. Infect. Dis. 155:220-228. [DOI] [PubMed] [Google Scholar]
- 42.Stevens, D. L., R. K. Tweten, M. M. Awad, J. I. Rood, and A. E. Bryant. 1997. Clostridial gas gangrene: evidence that alpha and theta toxins differentially modulate the immune response and induce acute tissue necrosis. J. Infect. Dis. 176:189-195. [DOI] [PubMed] [Google Scholar]
- 43.Stevens, D. L., R. W. Titball, M. Jepson, C. R. Bayer, S. M. Hayes-Schroer, and A. E. Bryant. 2004. Immunization with C-domain of α-toxin prevents lethal infection, localizes tissue injury, and promotes host response to challenge with Clostridium perfringens. J. Infect. Dis. 190:767-773. [DOI] [PubMed] [Google Scholar]
- 44.Titball, R. W., C. E. Naylor, D. Moss, and E. D. Williamson. 1998. Mechanism of protection against disease caused by Clostridium perfringens. Immunology 95:34. [Google Scholar]
- 45.Titball, R. W., and J. I. Rood. 2002. Clostridium perfringens: wound infections, p. 1875-1903. In M. Sussman (ed.), Molecular medical microbiology. Academic Press, London, United Kingdom.
- 46.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tjalsma, H. 2007. Feature-based reappraisal of the Bacillus subtilis exoproteome. Proteomics 7:73-81. [DOI] [PubMed] [Google Scholar]
- 48.Walz, A., C. V. Mujer, J. P. Connolly, T. Alefantis, R. Chafin, C. Dake, J. Whittington, S. P. Kumar, A. S. Khan, and V. G. DelVecchio. 2007. Bacillus anthracis secretome time course under host-simulated conditions and identification of immunogenic proteins. Proteome Sci. 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.