Abstract
The Gram-negative anaerobic bacterium Porphyromonas gingivalis is a major pathogen in periodontal disease, one of the biofilm-caused infectious diseases. The bacterium possesses potential virulence factors, including fimbriae, proteinases, hemagglutinin, lipopolysaccharide (LPS), and outer membrane vesicles, and some of these factors are associated with biofilm formation; however, the precise mechanism of biofilm formation is still unknown. Colonial pigmentation of the bacterium on blood agar plates is related to its virulence. In this study, we isolated a nonpigmented mutant that had an insertion mutation within the new gene PGN_1251 (gtfB) by screening a transposon insertion library. The gene shares homology with genes encoding glycosyltransferase 1 of several bacteria. The gtfB mutant was defective in biosynthesis of both LPSs containing O side chain polysaccharide (O-LPS) and anionic polysaccharide (A-LPS). The defect in the gene resulted in a complete loss of surface-associated gingipain proteinases, strong autoaggregation, and a marked increase in biofilm formation, suggesting that polysaccharide portions of LPSs influence attachment of gingipain proteinases to the cell surface, autoaggregation, and biofilm formation of P. gingivalis.
Porphyromonas gingivalis is a major pathogen in severe forms of periodontal disease and refractory periapical perodontitis (28, 39). This Gram-negative anaerobic bacterium possesses several virulence factors, including fimbriae, proteinases, hemagglutinin, lipopolysaccharide (LPS), and outer membrane vesicles (7, 13, 16, 27). P. gingivalis forms black-pigmented colonies on blood agar plates. Colonial pigmentation is caused by accumulation of μ-oxo heme dimer on the cell surface (58). Nonpigmented mutants of P. gingivalis have been isolated and characterized by a number of researchers (5, 17, 51, 56, 62-64). Colonial pigmentation on blood agar plates has been shown to be linked with hemagglutination and activities of major proteinases, Arg-gingipain (Rgp) and Lys-gingipain (Kgp), and other virulence factors, suggesting that colonial pigmentation is associated with the presence of gingipain-adhesin complexes on the cell surface (3, 11, 60).
Pigmentation-related genes that have been characterized are classified into three categories: gene expression, membrane translocation, and surface attachment of gingipain-adhesin complexes (51). Gingipain-adhesin complexes comprise Rgp and Kgp proteinases encoded by rgpA, rgpB, and kgp and adhesins encoded by rgpA, kgp, and hagA. kgp single and rgpA rgpB kgp triple mutants form less- and nonpigmented colonies, respectively, whereas an rgpA rgpB double mutant forms pigmented colonies (42, 55). Smalley et al. (59) found that Rgp activity is crucial for converting oxyhemoglobin into the methemoglobin form, which is rendered more susceptible to Kgp degradation for the eventual release of iron(III) protoporphyrin IX and production of μ-oxo heme dimer.
A defect in membrane translocation of gingipain-adhesin complexes causes nonpigmentation. The three genes porT, sov, and PG0027, mutants of which exhibit nonpigmentation, have been reported to be involved in membrane translocation of gingipain-adhesin complexes (18, 49, 53). High-molecular-weight forms of Rgp, Kgp, and adhesins accumulate in the periplasmic space of those mutants (49, 53). Very recently, we found a novel protein secretion system, the Por secretion system (PorSS), mediating secretion for gingipain-adhesin complexes, and the porK, porL, porM, porN, porP, porQ, porU, porW, porX, and porY genes have been added to this category (52).
The porR, vimA, vimE, vimF, rfa, and ugdA genes, mutants of which lose colonial pigmentation, appear to be involved in the formation of extracellular polysaccharides and glycan additions of gingipain-adhesin complexes, resulting from a lack of immunoreactivity to MAb 1B5, which reacts with anionic surface polysaccharide (APS) (51, 56, 62-64). Also, Chen et al. (5) isolated a nonpigmented mutant having a transposon insertion at a gene homologous to a glycosyl (rhamnosyl) transferase-encoding gene that showed reduced levels of Rgp activity and hemagglutination.
In this study, we isolated a nonpigmented mutant that has a Tn4400′ insertion mutation within an uncharacterized gene, PGN_1251, by screening a transposon insertion library, and we characterized the properties of the PGN_1251 mutant.
MATERIALS AND METHODS
Strains and culture conditions.
All bacterial strains used in this study are shown in Table 1. P. gingivalis cells were grown anaerobically (10% CO2, 10% H2, and 80% N2) in enriched brain heart infusion (BHI) medium and on enriched tryptic soy agar (36). For blood plates, defibrinated laked sheep blood was added to enriched tryptic soy agar at 5%. For selection and maintenance of antibiotic-resistant strains, antibiotics were added to the medium at the following concentrations: ampicillin, 50 μg/ml; erythromycin (Em), 10 μg/ml; and tetracycline (Tc), 0.7 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| E. coli | ||
| S17-1 | RP4-2-Tc::Mu aph::Tn7 recA, Smr | 57 |
| HB101/RK231, pYT646B | Tn4400′ donor strain | 4 |
| P. gingivalis | ||
| 33277 | Wild type | ATCC |
| KDP136 | kgp-2::cat rgpA2::[ermF ermAM] rgpB2::tetQ, Cmr Emr Tcr | 55 |
| KDP354 | porP::ermF, Emr | 52 |
| KDP355 | porK::ermF, Emr | 52 |
| KDP356 | porL::ermF, Emr | 52 |
| KDP357 | porM::ermF, Emr | 52 |
| KDP358 | porN::ermF, Emr | 52 |
| KDP359 | porW::ermF, Emr | 52 |
| KDP390 | gtfB::Tn4400', Tcr | This study |
| KDP400 | ΔgtfB::ermF, Emr | This study |
| KDP401 | KDP400 fimA::[gtfB+ tetQ], Emr Tcr | This study |
| KDP402 | KDP400/pKD906, Emr Tcr | This study |
| KDP403 | KDP400/pKD909, Emr Tcr | This study |
Transposon mutagenesis and gene-directed mutagenesis.
Tn4400′ transposon mutagenesis of P. gingivalis strain 33277 using Escherichia coli HB101 harboring RK231 and pYT646B (4) and gene-directed mutagenesis of P. gingivalis strains with electroporation were done as described previously (4, 36).
Construction of bacterial strains and plasmids.
A P. gingivalis PGN_1251 (gtfB) deletion mutant was constructed as follows. gtfB-upstream and gtfB-downstream DNA regions were PCR amplified from strain 33277 chromosomal DNA with the primer pair GUF and GUR for the gtfB-upstream region and with the primer pair GDF and GDR for the gtfB-downstream region. DNA primers used in this study are listed in Table S1 in the supplemental material. The amplified DNAs were digested with NotI and BamHI for the gtfB-upstream region and with BamHI and KpnI for the gtfB-downstream region and then inserted into the NotI-KpnI region of pBluescript II SK(−) to yield pKD901. The 1.5-kb BamHI ermF DNA cartridge was inserted into the BamHI site of pKD901, resulting in pKD902 (ΔgtfB::ermF). P. gingivalis 33277 was then transformed with the NotI-linearized pKD902 DNA to yield strain KDP400. To construct the gtfB+-complementing strain, a 1.13-kb gtfB region was PCR amplified from 33277 chromosomal DNA with the primer pair C1F and C1R. The amplified DNA fragment was cloned into pGEM-T Easy vector (Promega), resulting in pKD903. The gtfB region DNA obtained by BamHI digestion was inserted into the BamHI site of pKD713 (21) to yield pKD904 (fimA::[gtfB+ tetQ]). The pKD904 plasmid DNA was linearized by BssHII digestion and introduced into strain KDP400 by electroporation, resulting in strain KDP401 (ΔgtfB::ermF fimA::[gtfB+ tetQ]). To construct another gtfB+-complemented strain, the gtfB gene region was PCR amplified from 33277 chromosomal DNA with the primer pair C2F and C2R. The amplified DNA fragment was cloned into the pGEM-T Easy vector, resulting in pKD905. The gtfB region DNA (1.13 kb) obtained by NotI and BamHI digestion was inserted into the NotI-BamHI region of pTCB (32) to yield pKD906 (gtfB+). The pKD906 plasmid DNA was introduced into strain KDP400 by conjugation with E. coli S17-1 (57) harboring pKD906 as a donor strain, resulting in strain KDP402 (ΔgtfB::ermF/gtfB+). For construction of a gtfB′-′myc chimera gene, the gtfB gene region was PCR amplified from 33277 chromosomal DNA with the primer pair GMF and GMR. The amplified DNA fragment was cloned into the pGEM-T Easy vector, resulting in pKD907. The gtfB region of pKD907 obtained by KpnI and HindIII digestion was inserted into the KpnI-HindIII region of pKD858 (51) to yield pKD908. The gtfB′-′myc region of pKD908 obtained by KpnI and NotI digestion was inserted into the KpnI-NotI region of pTCB to yield pKD909. The pKD909 plasmid DNA was introduced into strain KDP400 by conjugation, resulting in strain KDP403 (ΔgtfB::ermF/gtfB′-′myc).
DNA probes and Southern blot hybridization.
A DNA fragment (1.2 kb) comprising the gtfB gene was PCR amplified from 33277 chromosomal DNA with the primer pair GSF and GSR. An ermF DNA fragment (1.1 kb) was PCR amplified from pKD355 (61) with the primer pair EMF and EMR. These fragments were labeled with the AlkPhos Direct system for chemiluminescence (GE Healthcare). Southern blotting was performed by using a nylon membrane and developed with the CDP-star detection reagent.
Hemagglutination.
Overnight cultures of P. gingivalis strains in enriched BHI medium were centrifuged, washed with phosphate-buffered saline (PBS), and suspended in PBS at an optical density at 540 nm of 0.3. The bacterial suspensions were then diluted in a 2-fold series with PBS. A 100-μl aliquot of each suspension was mixed with an equal volume of defibrinated laked sheep erythrocyte suspension (1% in PBS) and incubated in a round-bottom microtiter plate at room temperature for 3 h.
Enzymatic assay.
Kgp and Rgp activities were determined using the synthetic substrates t-butyl-oxycarbonyl-l-valyl-leucyl-l-lysine-4-methyl-7-coumarylamide (Boc-Val-Leu-Lys-MCA) and carbobenzoxy-l-phenyl-l-arginine-4-methyl-7-coumarylamide (Z-Phe-Arg-MCA). The released 7-amino-4-methylcoumarin was measured at 460 nm (excitation at 380 nm).
Subcellular fractionation.
Preparation of vesicle-containing culture supernatants and subcellular fractionation of P. gingivalis cells were performed as described previously (53). Particle-free culture supernatants were prepared as described previously (46).
Antibodies.
Monoclonal antibody (MAb) anti-C Myc was obtained from Sigma. MAb 1B5 (6) was kindly provided by Michael A. Curtis. Preparations of antibodies against Hgp44 and Kgp and antibody against gingipain containing Rgp and Kgp have been described by Sato et al. (53) and Abe et al. (1), respectively.
Real-time quantitative reverse transcription-PCR (real-time qRT-PCR).
Real-time qRT-PCR was performed essentially according to the method of Kondo et al. (23). Primers for real-time qRT-PCR were shown in Table S1 in the supplemental material. All qRT-PCRs were carried out in triplicate.
LPS analysis.
LPS of P. gingivalis cells was purified essentially according to the method of Darveau and Hancock (8) and separated on a 15% SDS-PAGE gel containing 4 M urea.
Hydrophobicity assay.
A surface hydrophobicity assay was performed essentially according to the method of Kawabata and Hamada (20). Briefly, bacterial cells were suspended with 30 mM urea and 0.8 mM MgSO4 in 100 mM sodium phosphate buffer at a final concentration of 0.6 mg/ml and mixed with n-hexadecane. Following vigorous shaking, optical densities of the aqueous phases were measured at 550 nm (A550). Percent hydrophobicity was calculated as follows: [(A550 without n-hexadecane − A550 with n-hexadecane)/A550 without n-hexadecane] × 100.
Transmission electron microscopy.
P. gingivalis cells were washed three times with 0.1 M cacodylate buffer and fixed with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer, followed by embedding in 3% agar. After dehydration, specimens were embedded in LR White (EM Polysciences, Warrington, PA). Thin sections were contrasted with uranyl acetate and lead citrate. Images were obtained with transmission electron microscopy (TEM) (H-7650; Hitachi, Japan).
Biofilm formation.
Mature biofilm of P. gingivalis was prepared with a flow cell system on hydroxyapatite (HA) disks and celluloid disks (Sumilon Celltite C-1; Sumitomo Bakelite Co. Ltd., Tokyo, Japan) using the modified Robbins device (MRD) (10). HA disks and celluloid disks were incubated with human saliva, which had been centrifuged at 2,000 × g for 10 min and filter sterilized, for 24 h at 37°C. The HA disks and celluloid disks were placed face down in the MRD, and culture medium (500 ml) containing P. gingivalis cells (3.0 × 106 CFU/ml) was circulated using a peristaltic pump at a flow rate of 3.3 ml/min (in the lumen) in an anaerobic incubator. The culture medium was changed every 2 days, and the disks were perfused for 14 days to allow biofilm growth.
Examination by a CLSM.
The biofilm on the saliva-coated celluloid disks was stained with a bacterial viability kit (BacLight Live/Dead bacterial viability kit L-7007; Molecular Probes) and imaged with a confocal laser scanning microscope (CLSM) (LSM510, Carl Zeiss, Oberkochem, Germany). The three-dimensional structure of the biofilm was reconstructed from CLSM images with the IMARIS software program (Bitplane AG, Switzerland).
Examination by a SEM.
For scanning electron microscope (SEM) examination, P. gingivalis cells on the saliva-coated HA disks were washed three times with 0.1 M cacodylate buffer and fixed with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 30 min at room temperature and subsequently washed three times in 0.1 M cacodylate buffer. After fixation, all specimens were dehydrated in a graded series of aqueous ethanol, dried, ion coated with platinum, and examined by SEM (JSM-6390LV; Jeol, Japan), as previously described (40).
RESULTS
Identification of a gene disrupted by Tn4400′ transposon insertion.
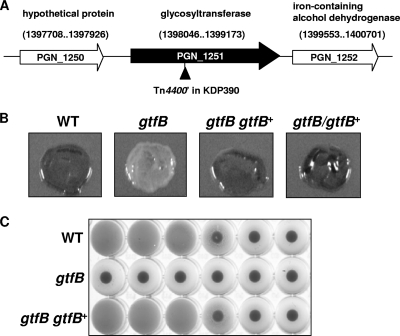
A collection of P. gingivalis 33277 random-transposon mutants was screened on blood agar plates for colonial pigmentation. Chromosomal DNA of nonpigmented mutants was digested with HindIII, self ligated, and cloned by the marker rescue method with the bla gene on Tn4400′ DNA. Sequencing of the cloned DNA fragments revealed that the insertion site of an insertion mutant (KDP390) was located 428 bp downstream of the first nucleotide residues of the initiation codon of PGN_1251 (Fig. 1 A). The PGN_1251 gene encoded a putative group 1 family glycosyltransferase (33) and was designated gtfB.
FIG. 1.
Physical map around the gtfB gene and properties of the gtfB mutant. (A) A physical map around the gtfB gene region. A triangle indicates the Tn4400′ insertion site of strain KDP390. (B) Colonial pigmentation. P. gingivalis cells were anaerobically grown on blood agar plates at 37°C. (C) Hemagglutination. P. gingivalis cells were grown in enriched BHI broth, washed with PBS, and suspended in PBS at an optical density at 540 nm of 0.3. The suspension and its dilutions in a 2-fold series were applied to the wells of a microtiter plate from left to right and mixed with sheep erythrocyte suspension.
Construction of a gtfB mutant by gene-directed mutagenesis.
To determine whether nonpigmentation of KDP390 was attributable to gtfB, we constructed a mutant in which gtfB was replaced by the ermF DNA cartridge (KDP400). Strain KDP400 showed no pigmentation on blood agar plates. To further confirm the relation between gtfB and colonial pigmentation, we constructed a complemented strain (KDP401) in which the wild-type gtfB gene was introduced into the fimA locus of KDP400. In addition, we constructed another complemented strain (KDP402) with the wild-type gtfB gene on the pTCB shuttle plasmid. The complemented strains KDP401 and KDP402 showed the same colonial pigmentation as the wild-type strain (Fig. 1B). KDP400 showed no hemagglutination (Fig. 1C). These results showed that the gtfB gene product is required for colonial pigmentation and hemagglutination of P. gingivalis.
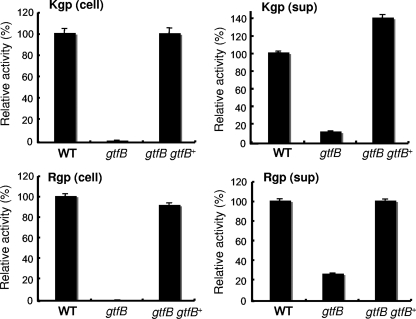
Kgp and Rgp activities of the gtfB mutant.
Several studies have indicated that activities of Kgp and Rgp proteinases on the cell surface were associated with colonial pigmentation on blood agar plates (42, 55). We therefore determined Kgp and Rgp activities in the intact cells and culture supernatants of the gtfB mutant. KDP400 (ΔgtfB::ermF) showed no detectable activities of Kgp or Rgp in intact cells and reduced activities in the vesicle-containing culture supernatants. KDP401 (ΔgtfB::ermF fimA::gtfB+) exhibited almost the same Kgp and Rgp activities as the wild-type strain (Fig. 2). We determined the mRNA levels of the kgp, rgpA, and rgpB genes by real-time qRT-PCR. The kgp, rgpA, and rgpB mRNAs in the gtfB mutant were 3.4-, 12.8-, and 3.2-fold upregulated, respectively, compared to those in the wild-type strain, suggesting that a decrease of Rgp and Kgp activities of the gtfB mutant cannot be explained by expression of the rgpA, rgpB, and kgp genes at the transcriptional level (see Fig. S1 in the supplemental material).
FIG. 2.
Kgp and Rgp activities. P. gingivalis cells were anaerobically grown overnight in enriched BHI medium at 37°C. Kgp and Rgp activities of the cell lysates (cell) and vesicle-containing culture supernatants (sup) of 33277 (wild type), KDP400 (gtfB), and KDP401 (gtfB gtfB+) were measured.
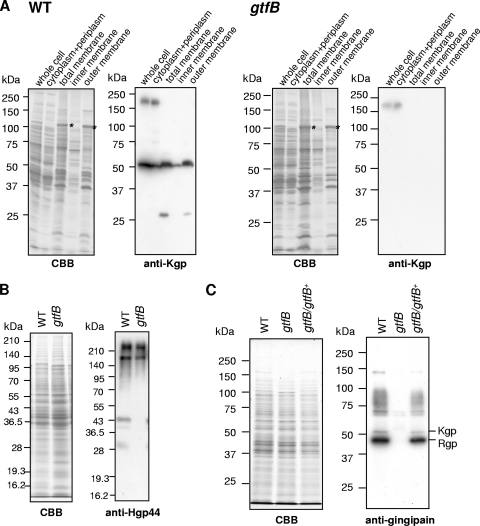
Immunoblot analysis using anti-Kgp, anti-gingipain, and anti-Hgp44 antibodies.
The kgp and rgpA genes, comprising 5,193-bp and 5,118-bp coding sequences (CDSs), respectively, encode polyproteins that consist of four segments: signal peptide, propeptide, proteinase, and adhesin domains (41, 45). The C-terminal adhesin domains comprise four subdomains (Hgp44/A1, Hgp15 [HbR]/A2, Hgp17/A3, and Hgp27/A4) that are involved in hemagglutination and hemoglobin binding (37, 50, 66). The rgpB gene comprises a 2,208-bp CDS and lacks most of the adhesin domain (35). Cultures of the gtfB mutant and the wild-type strain were separated into bacterial cells and vesicle-containing culture supernatants. The bacterial cells were lysed and separated into fractions enriched for cytoplasm/periplasm, inner membrane, and outer membrane, and immunoblot analyses with anti-Kgp and anti-Hgp44 were performed. In the wild-type strain, the 190-kDa proprotein and the 50-kDa processed protein immunoreactive to anti-Kgp antibody were found mainly in the cytoplasm/periplasm and outer membrane fraction, respectively (Fig. 3 A). On the other hand, the 190-kDa Kgp proprotein was found in the cytoplasm/periplasm fraction, whereas the 50-kDa Kgp protein was not detected in any fraction of the gtfB mutant (Fig. 3A). The whole-cell lysate of the gtfB mutant contained anti-Hgp44-immunoreactive proteins with molecular masses of more than 210 kDa and 190 kDa, which appear to be HagA proprotein and RgpA/Kgp proproteins, respectively, as much as those of the wild-type strain, whereas there was no processed/mature Hgp44 (44 kDa), Kgp (50 kDa), or Rgp (43 kDa) in the whole-cell lysate of the gtfB mutant (Fig. 3B and C).
FIG. 3.
Immunoblot analysis with P. gingivalis cells. (A) The whole-cell, cytoplasm/periplasm, total membrane, inner membrane, and outer membrane fractions of 33277 (wild type) and KDP400 (gtfB) were subjected to SDS-PAGE and immunoblot analysis with anti-Kgp antibody. Asterisks indicate a major outer membrane protein, RagA. (B) The whole-cell lysates of 33277 (wild type) and KDP400 (gtfB) were subjected to SDS-PAGE and immunoblot analysis with anti-Hgp44 antibody. (C) The whole-cell lysates of 33277 (wild type), KDP400 (gtfB), and KDP402 (gtfB/gtfB+) were subjected to SDS-PAGE and immunoblot analysis with antigingipain antibody. WT, wild type; CBB, Coomassie brilliant blue.
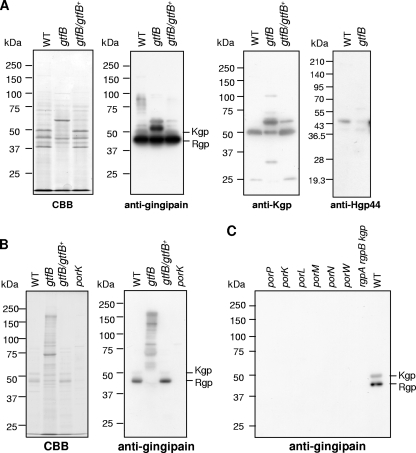
The vesicle-containing culture supernatant of the gtfB mutant contained a larger amount of a 59-kDa intermediate protein immunoreactive to anti-Kgp antibody than the 50-kDa Kgp protein, whereas the 50-kDa Kgp protein was dominant in the wild-type strain (Fig. 4 A). Immunodetection with anti-gingipain antibody, which reacted mainly with Rgp, showed a similar result. The Hgp44 region is translated as a subdomain in the adhesin domain of the rgpA gene and then processed by activities of gingipains in the wild type. The vesicle-containing culture supernatant of the gtfB mutant showed a faint protein band of processed/mature Hgp44 (Fig. 4A).
FIG. 4.
Immunoblot analysis with vesicle-containing and particle-free culture supernatants. (A) Vesicle-containing culture supernatants of P. gingivalis strains were subjected to SDS-PAGE and immunoblot analysis with antigingipain, anti-Kgp, and anti-Hgp44 antibodies. (B and C) Particle-free culture supernatants of P. gingivalis strains were subjected to SDS-PAGE and immunoblot analysis with antigingipain antibody. WT, wild type; CBB, Coomassie brilliant blue.
The particle-free culture supernatant of the gtfB mutant contained various intermediate precursor forms of Rgp and Kgp, whereas processed/mature Rgp (43 kDa) and Kgp (50 kDa) were detected in those of the wild type and the complemented strain (Fig. 4B). PorSS-deficient mutants (porP, porK, porL, porM, porN, and porW) showed no proteins immunoreactive to antigingipain antibody in the particle-free culture supernatants (Fig. 4B and C). We determined gene expression of porP, porK, porL, porM, porN, porT, and sov, which encode proteins involved in PorSS, in the gtfB mutant, the wild-type strain, and the gtfB/gtfB+ complemented strain (see Fig. S1 in the supplemental material). The genes were well expressed in the gtfB mutant. These results suggest that PorSS is functioning in the gtfB mutant.
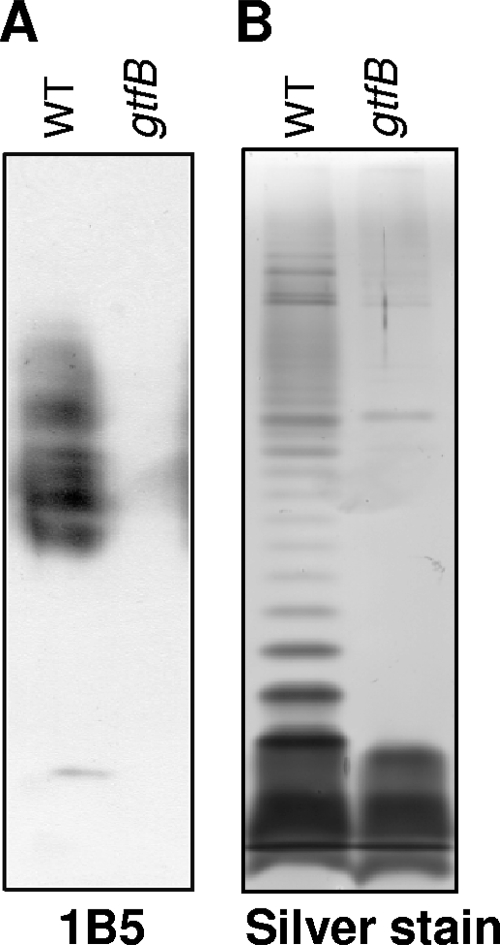
Analysis of APS and LPS of the gtfB mutant.
Since previous studies indicated that several nonpigmented strains showed alteration in anionic polysaccharide (APS) and LPS profiles (51, 56, 62-64), we examined the gtfB mutant for APS and LPS profiles using immunoblot analysis and silver staining. Curtis et al. (6) isolated an MAb 1B5 that reacts with APS. The gtfB mutant showed no MAb 1B5-immunoreactive molecules (Fig. 5 A). LPS of the gtfB mutant showed two bands, a core and a core plus one truncated O repeating unit, which differed from the typical ladder pattern of wild-type LPS (Fig. 5B). The PGN1302 (PG1051; encoding O-antigen ligase), PGN1242 (PG1142; encoding O-antigen polymerase), and rfa (PGN1255 and PG1155; encoding putative ADP heptose-LPS heptosyltransferase) genes are involved in the biosynthesis of A-LPS and O-LPS in P. gingivalis (44, 51). Real-time qRT-PCR analysis revealed that the mRNA levels of the PGN1302, PGN1242, and rfa genes of the gtfB mutant were 3.2-, 1.9-, and 11.4-fold upregulated, respectively, compared to those of the wild-type strain (see Fig. S1 in the supplemental material).
FIG. 5.
APS and LPS of the gtfB mutant. (A) APS profiles of the gtfB mutant. The cell lysates of 33277 (wild type) and KDP400 (gtfB) were separated by SDS-PAGE and analyzed for APS using MAb 1B5. (B) LPS profiles. LPS fractions were extracted from 33277 (wild type) and KDP400 (gtfB) as previously described (7) and subjected to SDS-PAGE followed by silver staining.
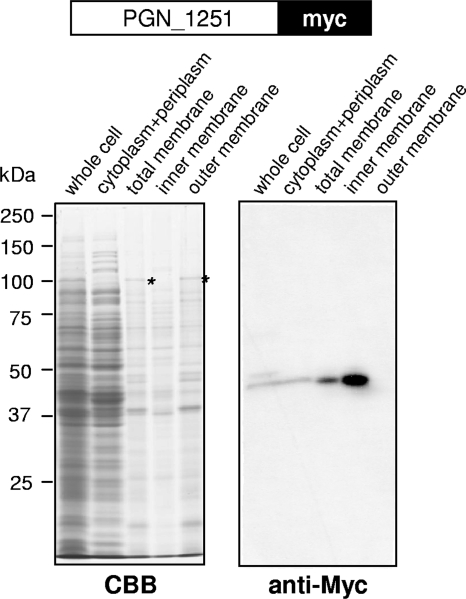
Subcellular localization of GtfB.
To determine the subcellular localization of the GtfB protein, we constructed strain KDP403 (ΔgtfB::ermF/gtfB′-′myc), which expressed a GtfB-Myc chimera protein in the gtfB mutant. Strain KDP403 showed the same phenotype in colonial pigmentation and hemagglutination as the wild type (data not shown). Cell lysates of KDP403 were fractionated into the cytoplasm/periplasm, inner membrane, and outer membrane and then subjected to SDS-PAGE and immunoblot analysis with anti-Myc antiserum (Fig. 6). A 46-kDa anti-Myc-immunoreactive protein was found in the inner membrane fraction. This result suggested that GtfB was an inner membrane-associated protein.
FIG. 6.
Subcellular localization of the GtfB protein. Whole-cell, cytoplasm/periplasm, total membrane, inner membrane, and outer membrane fractions of KDP403 (gtfB′-′myc) were subjected to immunoblot analysis with anti-Myc antiserum. Asterisks indicate a major outer membrane protein, RagA.
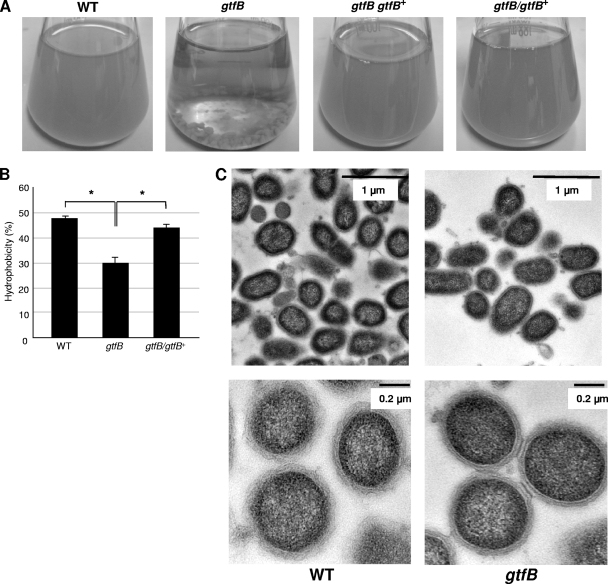
Autoaggregation and TEM analysis.
The gtfB mutant (KDP400) showed very strong autoaggregation compared to that of the wild-type strain (33277) and the complemented strains (KDP401 and KDP402), while the mutant cells did not adhere to glass (Fig. 7 A). The hydrophobicity assay revealed that the gtfB mutant (KDP400) was less hydrophobic than the wild-type strain (33277) and the complemented strain (KDP402) (Fig. 7B). Next, we examined the gtfB mutant and the wild-type parent strain for cell structure by transmission electron microscopy. As shown in Fig. 7C, the gtfB mutant exhibited tight adherence among bacterial cells.
FIG. 7.
Autoaggregation of the gtfB mutant. (A) Autoaggregation. P. gingivalis cells were anaerobically grown in BHI broth at 37°C overnight. The optical densities at 600 nm of bacterial cultures of 33277 (wild type), KDP400 (gtfB), and KDP402 (gtfB/gtfB+) were 1.92 ± 0.05, 0.12 ± 0.00, and 1.89 ± 0.10, respectively. (B) Hydrophobicity. The graph shows the percentages of cells partitioning into the hydrophobic solvent n-hexadecane. **, P < 0.05. (C) Transmission electron microscopic images of 33277 (wild type) and KDP400 (gtfB). The cells were stained with uranium acetate and lead citrate.
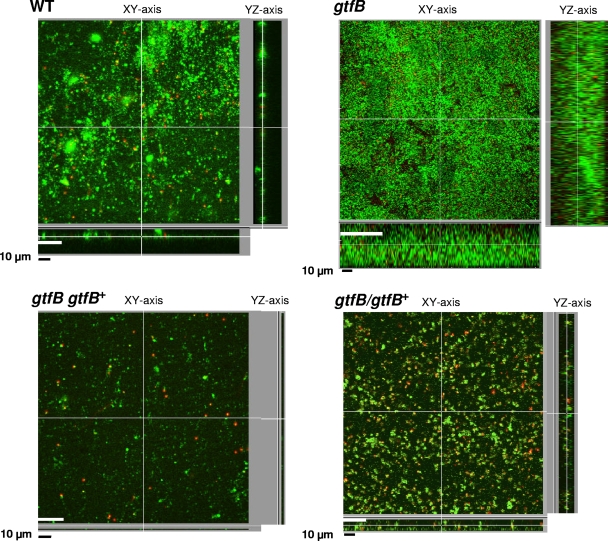
Biofilm formation.
We examined the ability of the gtfB mutant to form a biofilm on the saliva-coated celluloid disks by using CLSM. BacLight Live/Dead-stained biofilms of the wild-type strain (33277) and the gtfB mutant (KDP400) were imaged by using the Imaris software program and are shown in Fig. 8. In the gtfB mutant, biofilm formation on the saliva-coated celluloid disks was markedly increased compared to that of the wild-type strain, whereas the complemented strain (KDP402) exhibited almost the same biofilm formation as the wild-type strain (Fig. 8). The other gtfB+-complemented strain (KDP401), which had a gtfB+ insertion in the fimA gene, showed reduced biofilm formation, which is consistent with results of a previous study (25). Live cells, which have intact cell membranes, are stained with Syto9 and emit green fluorescence when they are stained with the BacLight Live/Dead strains. Cells with damaged membranes are stained with propidium iodide and show red fluorescence. The ratio of live and dead cells in biofilms was not significantly different between the gtfB mutant and the wild-type strain (Fig. 8).
FIG. 8.
Confocal laser scanning microscopic image of biofilms on saliva-coated celluloid disks using the MRD. The bacterial cells were stained with a Molecular Probes Live/Dead BacLight bacterial viability kit.
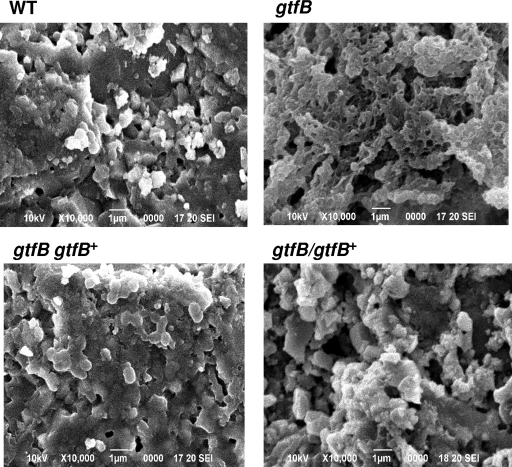
SEM analysis of biofilms on the saliva-coated HA disks revealed that the extracellular matrix-like structure of the gtfB mutant was different from that of the wild-type strain and that the gtfB mutant formed an extracellular matrix-like structure that was rougher than that of the wild-type strain (Fig. 9). The complemented strains exhibited almost the same extracellular matrix-like structure as the wild-type strain (Fig. 9).
FIG. 9.
Scanning electron microscopic image of biofilms on saliva-coated HA disks using the MRD. The biofilms were fixed with 2% paraformaldehyde and 2.5% glutaraldehyde and ion coated with platinum.
DISCUSSION
P. gingivalis has at least three sugar macromolecules on the cell surface: LPS, anionic cell surface polysaccharide (APS), and capsular polysaccharide (CPS). APS is distinct from LPS and CPS (43). MAb 1B5, which was raised against one of the five isoforms of Rgps, immunoreacts with APS (6, 43). LPS purified by a procedure described previously by Darveau and Hancock (8) shows no immunoreactivity to MAb 1B5, indicating that APS and LPS are two different polysaccharides (43). Recently APS was found to be associated with lipid A, and LPS with APS repeating units is designated A-LPS (44, 47). LPS biosynthesis in Gram-negative bacteria involves a large number of enzymes, encoded by more than 40 genes (14, 15, 67). As revealed by O-LPS profile analysis, O-LPS of the gtfB mutant had a single repeating unit of the O side chain with a length shorter than that of the wild-type parent strain, suggesting that GtfB is a glycosyltransferase involved in biosynthesis of a repeating unit of the O side chain. These results taken, together with the results showing that the gtfB mutant had no substance immunoreactive to MAb 1B5, suggest that GtfB is involved in the biosynthesis of polysaccharide portions of both O-LPS and A-LPS.
The kgp and rgpA genes encode polyproteins comprising the signal peptide, propeptide, proteinase, and adhesin domains (41, 45). The rgpB gene encodes a protein comprising the signal peptide, propeptide, and proteinase domains (31, 35). Therefore, Kgp and Rgp are synthesized first in the cytoplasm as preproenzymes, which are translocated across the inner and outer membranes and secreted into the extracellular milieu as mature proteinases or located on the cell surface as complexes noncovalently associated with adhesin domain proteins (66). Results of several studies on attachment of gingipain-adhesin complexes to the cell surface have been reported (56). A-LPS formation and glycosylation of gingipain-adhesin complexes are required for attachment of gingipain-adhesin complexes to the cell surface. The porR, vimA, vimE, and vimF genes appear to be involved in the formation of A-LPS and glycan additions of gingipain-adhesin complexes since mutants of these genes lose colonial pigmentation, have altered distribution of gingipain activities in subcellular fractions, and show no immunoreactivity to MAb 1B5 (56, 62-64). The vimF and porR genes share homology with genes encoding glycosyltransferase and transaminase involved in biosynthesis of the sugar portion of LPSs and aminoglycosides, respectively (56, 62). A C-terminal adhesin domain protein of the rgpA gene product, RgpA27, shows MAb 1B5-immunoreactive diffuse protein bands, suggesting that a glycosylation site(s) is located at the C-terminal domain region of the RgpA polyprotein (6). These findings suggest that attachment of gingipain-adhesin complexes to the cell surface is caused by binding of the polysaccharide portion of A-LPS to the C-terminal domain region. Seers et al. (54) found that a number of P. gingivalis proteins have amino acid sequence similarity in the C-terminal region with RgpA27 and named them C-terminal domain (CTD) proteins. Very recently, we found a novel protein secretion system (PorSS), by which the CTD proteins appear to be secreted (52).
The gtfB mutant showed decreased activities of Kgp and Rgp in the vesicle-containing culture supernatants and no Kgp or Rgp activity in the cell lysates. In addition, the gtfB mutant had no 44-kDa Hgp44 domain protein associated with the cells. The deficiency in colonial pigmentation and hemagglutination of the gtfB mutant can be explained by a lack of cell-associated activity of Rgp or Kgp and of a cell-associated form of processed Hgp44, since the Rgp/Kgp-null mutant (rgpA rgpB kgp) shows a deficiency in colonial pigmentation and hemagglutination and since processed Hgp44 is responsible for hemagglutination (50, 55). Subcellular fractionation analysis revealed that anti-Kgp-immunoreactive proteins were absent in the membrane fraction of gtfB mutant cells whereas 50-kDa Kgp was present mainly in the outer membrane fraction of the wild-type strain. The absence of Kgp proteins on the surface of the gtfB mutant supports the idea that A-LPS contributes to attachment of gingipain-adhesin complexes to the cell surface because of a lack of A-LPS in the mutant. In the vesicle-containing culture supernatants of the wild-type and gtfB mutant strains, there were anti-Kgp-immunoreactive proteins with molecular masses of 50 and 59 kDa. The amount of 50-kDa Kgp (mature form of Kgp) in the culture supernatant of the wild-type strain was much greater than that of the gtfB mutant, whereas the amount of 59-kDa Kgp (intermediate precursor form of Kgp) in the gtfB mutant was greater than that of the wild-type strain, which may explain the decreased Kgp activity in the vesicle-containing culture supernatant of the gtfB mutant. Unprocessed/immature intermediate precursor forms of Kgp and Rgp were detected in the particle-free culture supernatant of the gtfB mutant, whereas there were no proteins immunoreactive to anti-gingipain antibody in those of the PorSS-deficient mutants, suggesting that GtfB is not involved in PorSS. Interestingly, transcription of the rgpA, rgpB, and kgp genes was upregulated in the gtfB mutant, although Rgp and Kgp activities of the mutant markedly decreased, suggesting a regulatory mechanism in the expression of the rgpA, rgpB, and kgp genes. In addition, we found that transcription of the rfa and PGN1302 genes, which are involved in the biosynthesis of polysaccharide portions of A-LPS and O-LPS, was upregulated in the gtfB mutant defective in biosynthesis.
The P. gingivalis gtfB mutant exhibited strong autoaggregation and a marked increase in biofilm formation. This phenotype of the gtfB mutant is clearly different from that of a P. gingivalis gtfA mutant showing reduced autoaggregation (38). It has been reported that fimbriae and gingipains seem to act coordinately to regulate biofilm formation of P. gingivalis (25). On the other hand, little is known about the roles of LPS in biofilm formation of the bacterium, although a P. gingivalis galE mutant, which had an O side chain that was shorter than that of the wild type, was reported to exhibit enhanced biofilm formation (34). Pseudomonas aeruginosa is one of the major causative agents of mortality and morbidity in hospitalized patients due to a multiplicity of virulence factors associated with both chronic and acute infections. LPS of P. aeruginosa consists of lipid A, core oligosaccharide, and two distinct O side chains: the shorter A-band homopolymer is the so-called “common polysaccharide antigen” among the bacterium and consists of repeating d-rhamnose subunits, while the longer B-band heteropolymer is composed of repeating tri- to pentasaccharide subunits that vary among serotypes of P. aeruginosa (29). A B-band-deficient mutant forms luxurious biofilms compared to the wild-type strain (48). Very recently, P. aeruginosa mutants with LPS core variants have been reported to show enhanced cell adhesion and cohesion and altered viscoelasticity in early biofilms (29). In the P. gingivalis gtfB mutant, polysaccharide portions of A-LPS and O-LPS were deficient and bacterial cells were found to adhere tightly to one another by TEM analysis. These results suggest that alteration in the bacterial cell surface structure of the gtfB mutant leads to tight attachment of bacteria and enhancement of autoaggregation. Several studies have indicated that hydrophobicity of bacterial cells is related to autoaggregation (12, 22, 30). However, the gtfB mutant showed a significant decrease in surface hydrophobicity, suggesting that autoaggregation of the gtfB mutant cannot be explained by its surface hydrophobicity.
The P. gingivalis 33277 mutant of the PG0106 (PGN_0223) gene, which shares high sequence similarity to a glycosyltransferase gene and is involved in CPS synthesis, exhibited decreased autoaggregation (9). Strain 33277 and strain 381 are classified in the K− group (2, 10, 19, 24, 26, 65). The K− group is typified by the ability to autoaggregate and adhere to the pocket epithelium and other oral bacteria (2). The PGN_0223 mutants of strains 33277 and 381 show altered phenotypes of autoaggregation and biofilm formation (9), suggesting that even K− strains produce some surface component encoded by the chromosomal region including the PGN_0223 gene. Tiling microarray analysis revealed that the chromosomal region (PGN_0223-PGN_0234) in 33277 was polycistronically transcribed (unpublished data). It is likely that the surface component encoded by the chromosomal region of 33277 also influences autoaggregation and biofilm formation.
In this study, we isolated a nonpigmented mutant that has a Tn4400′ insertion mutation within an uncharacterized gene, PGN_1251, by screening a transposon insertion library. This gene, designated gtfB, shares homology with glycosyltransferase 1 genes from several bacteria. A gtfB mutant of P. gingivalis showed defects in polysaccharide portions of O-LPS and A-LPS and a complete loss of surface-associated gingipain-adhesin complexes. In addition, a lack of the gene resulted in strong autoaggregation and a marked increase in biofilm formation. These results strongly suggest that alteration of the surface structure, especially surface polysaccharides, of the gtfB mutant causes autoaggregation and increased biofilm formation.
Supplementary Material
Acknowledgments
We thank Michael A. Curtis and Michael H. Malamy for the gifts of monoclonal antibody 1B5 and the Tn4400′ transposon mutagenesis system, respectively, and Eiji Oiki for technical assistance for TEM.
This work was supported by Grants-in-Aid (21791793 to K.S., 20249076 to S.E., and 18018032 and 20249073 to K.N.) for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Global COE Program at Nagasaki University to K.N.
Editor: A. Camilli
Footnotes
Published ahead of print on 12 July 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abe, N., T. Kadowaki, K. Okamoto, K. Nakayama, M. Ohishi, and K. Yamamoto. 1998. Biochemical and functional properties of lysine-specific cysteine proteinase (Lys-gingipain) as a virulence factor of Porphyromonas gingivalis in periodontal disease. J. Biochem. 123:305-312. [DOI] [PubMed] [Google Scholar]
- 2.Aduse-Opoku, J., J. M. Slaney, A. Hashim, A. Gallagher, R. P. Gallagher, M. Rangarajan, K. Boutaga, M. L. Laine, A. J. Van Winkelhoff, and M. A. Curtis. 2006. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 74:449-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhogal, P. S., N. Slakeski, and E. C. Reynolds. 1997. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology 143(Pt. 7):2485-2495. [DOI] [PubMed] [Google Scholar]
- 4.Chen, T., H. Dong, Y. P. Tang, M. M. Dallas, M. H. Malamy, and M. J. Duncan. 2000. Identification and cloning of genes from Porphyromonas gingivalis after mutagenesis with a modified Tn4400 transposon from Bacteroides fragilis. Infect. Immun. 68:420-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, T., H. Dong, R. Yong, and M. J. Duncan. 2000. Pleiotropic pigmentation mutants of Porphyromonas gingivalis. Microb. Pathog. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 6.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramonov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutler, C. W., J. R. Kalmar, and C. A. Genco. 1995. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 3:45-51. [DOI] [PubMed] [Google Scholar]
- 8.Darveau, R. P., and R. E. Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey, M. E., and M. J. Duncan. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 188:5510-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dierickx, K., M. Pauwels, M. L. Laine, J. Van Eldere, J. J. Cassiman, A. J. van Winkelhoff, D. van Steenberghe, and M. Quirynen. 2003. Adhesion of Porphyromonas gingivalis serotypes to pocket epithelium. J. Periodontol. 74:844-848. [DOI] [PubMed] [Google Scholar]
- 11.Farquharson, S. I., G. R. Germaine, and G. R. Gray. 2000. Isolation and characterization of the cell-surface polysaccharides of Porphyromonas gingivalis ATCC 53978. Oral Microbiol. Immunol. 15:151-157. [DOI] [PubMed] [Google Scholar]
- 12.Gallant, C. V., M. Sedic, E. A. Chicoine, T. Ruiz, and K. P. Mintz. 2008. Membrane morphology and leukotoxin secretion are associated with a novel membrane protein of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190:5972-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grenier, D., and D. Mayrand. 1987. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect. Immun. 55:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinrichs, D. E., J. A. Yethon, P. A. Amor, and C. Whitfield. 1998. The assembly system for the outer core portion of R1- and R4-type lipopolysaccharides of Escherichia coli. The R1 core-specific beta-glucosyltransferase provides a novel attachment site for O-polysaccharides. J. Biol. Chem. 273:29497-29505. [DOI] [PubMed] [Google Scholar]
- 15.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 16.Holt, S. C., and T. E. Bramanti. 1991. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit. Rev. Oral Biol. Med. 2:177-281. [DOI] [PubMed] [Google Scholar]
- 17.Hoover, C. I., and F. Yoshimura. 1994. Transposon-induced pigment-deficient mutants of Porphyromonas gingivalis. FEMS Microbiol. Lett. 124:43-48. [DOI] [PubMed] [Google Scholar]
- 18.Ishiguro, I., K. Saiki, and K. Konishi. 2009. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol. Lett. 292:261-267. [DOI] [PubMed] [Google Scholar]
- 19.Katz, J., D. C. Ward, and S. M. Michalek. 1996. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:309-318. [DOI] [PubMed] [Google Scholar]
- 20.Kawabata, S., and S. Hamada. 1999. Studying biofilm formation of mutans streptococci. Methods Enzymol. 310:513-523. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, Y., N. Ohara, K. Sato, M. Yoshimura, H. Yukitake, E. Sakai, M. Shoji, M. Naito, and K. Nakayama. 2005. Novel stationary-phase-upregulated protein of Porphyromonas gingivalis influences production of superoxide dismutase, thiol peroxidase and thioredoxin. Microbiology 151:841-853. [DOI] [PubMed] [Google Scholar]
- 22.Kolenbrander, P. E., R. J. Palmer, Jr., A. H. Rickard, N. S. Jakubovics, N. I. Chalmers, and P. I. Diaz. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47-79. [DOI] [PubMed] [Google Scholar]
- 23.Kondo, Y., N. Ohara, K. Sato, M. Yoshimura, H. Yukitake, M. Naito, T. Fujiwara, and K. Nakayama. 2010. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect. Immun. 78:2846-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kremer, B. H., and T. J. van Steenbergen. 2000. Peptostreptococcus micros coaggregates with Fusobacterium nucleatum and non-encapsulated Porphyromonas gingivalis. FEMS. Microbiol. Lett. 182:57-62. [DOI] [PubMed] [Google Scholar]
- 25.Kuboniwa, M., A. Amano, E. Hashino, Y. Yamamoto, H. Inaba, N. Hamada, K. Nakayama, G. D. Tribble, R. J. Lamont, and S. Shizukuishi. 2009. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC. Microbiol. 9:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laine, M. L., B. J. Appelmelk, and A. J. van Winkelhoff. 1996. Novel polysaccharide capsular serotypes in Porphyromonas gingivalis. J. Periodontal Res. 31:278-284. [DOI] [PubMed] [Google Scholar]
- 27.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamont, R. J., and H. F. Jenkinson. 2000. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol. 15:341-349. [DOI] [PubMed] [Google Scholar]
- 29.Lau, P. C., T. Lindhout, T. J. Beveridge, J. R. Dutcher, and J. S. Lam. 2009. Differential lipopolysaccharide core capping leads to quantitative and correlated modifications of mechanical and structural properties in Pseudomonas aeruginosa biofilms. J. Bacteriol. 191:6618-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ljungh, A., S. Hjerten, and T. Wadstrom. 1985. High surface hydrophobicity of autoaggregating Staphylococcus aureus strains isolated from human infections studied with the salt aggregation test. Infect. Immun. 47:522-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikolajczyk-Pawlinska, J., T. Kordula, N. Pavloff, P. A. Pemberton, W. C. Chen, J. Travis, and J. Potempa. 1998. Genetic variation of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol. Chem. 379:205-211. [DOI] [PubMed] [Google Scholar]
- 32.Nagano, K., Y. Murakami, K. Nishikawa, J. Sakakibara, K. Shimozato, and F. Yoshimura. 2007. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J. Med. Microbiol. 56:1536-1548. [DOI] [PubMed] [Google Scholar]
- 33.Naito, M., H. Hirakawa, A. Yamashita, N. Ohara, M. Shoji, H. Yukitake, K. Nakayama, H. Toh, F. Yoshimura, S. Kuhara, M. Hattori, T. Hayashi, and K. Nakayama. 2008. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 15:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakao, R., H. Senpuku, and H. Watanabe. 2006. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect. Immun. 74:6145-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama, K. 1997. Domain-specific rearrangement between the two Arg-gingipain-encoding genes in Porphyromonas gingivalis: possible involvement of nonreciprocal recombination. Microbiol. Immunol. 41:185-196. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama, K., T. Kadowaki, K. Okamoto, and K. Yamamoto. 1995. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J. Biol. Chem. 270:23619-23626. [DOI] [PubMed] [Google Scholar]
- 37.Nakayama, K., D. B. Ratnayake, T. Tsukuba, T. Kadowaki, K. Yamamoto, and S. Fujimura. 1998. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol. Microbiol. 27:51-61. [DOI] [PubMed] [Google Scholar]
- 38.Narimatsu, M., Y. Noiri, S. Itoh, N. Noguchi, T. Kawahara, and S. Ebisu. 2004. Essential role for the gtfA gene encoding a putative glycosyltransferase in the adherence of Porphyromonas gingivalis. Infect. Immun. 72:2698-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noguchi, N., Y. Noiri, M. Narimatsu, and S. Ebisu. 2005. Identification and localization of extraradicular biofilm-forming bacteria associated with refractory endodontic pathogens. Appl. Environ. Microbiol. 71:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noiri, Y., A. Ehara, T. Kawahara, N. Takemura, and S. Ebisu. 2002. Participation of bacterial biofilms in refractory and chronic periapical periodontitis. J. Endod. 28:679-683. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto, K., T. Kadowaki, K. Nakayama, and K. Yamamoto. 1996. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain). J. Biochem. 120:398-406. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto, K., K. Nakayama, T. Kadowaki, N. Abe, D. B. Ratnayake, and K. Yamamoto. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273:21225-21231. [DOI] [PubMed] [Google Scholar]
- 43.Paramonov, N., M. Rangarajan, A. Hashim, A. Gallagher, J. Aduse-Opoku, J. M. Slaney, E. Hounsell, and M. A. Curtis. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 58:847-863. [DOI] [PubMed] [Google Scholar]
- 44.Paramonov, N. A., J. Aduse-Opoku, A. Hashim, M. Rangarajan, and M. A. Curtis. 2009. Structural analysis of the core region of O-lipopolysaccharide of Porphyromonas gingivalis from mutants defective in O-antigen ligase and O-antigen polymerase. J. Bacteriol. 191:5272-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavloff, N., J. Potempa, R. N. Pike, V. Prochazka, M. C. Kiefer, J. Travis, and P. J. Barr. 1995. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J. Biol. Chem. 270:1007-1010. [DOI] [PubMed] [Google Scholar]
- 46.Potempa, J., R. Pike, and J. Travis. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rangarajan, M., J. Aduse-Opoku, N. Paramonov, A. Hashim, N. Bostanci, O. P. Fraser, E. Tarelli, and M. A. Curtis. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 190:2920-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saiki, K., and K. Konishi. 2007. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol. Immunol. 51:483-491. [DOI] [PubMed] [Google Scholar]
- 50.Sakai, E., M. Naito, K. Sato, H. Hotokezaka, T. Kadowaki, A. Kamaguchi, K. Yamamoto, K. Okamoto, and K. Nakayama. 2007. Construction of recombinant hemagglutinin derived from the gingipain-encoding gene of Porphyromonas gingivalis, identification of its target protein on erythrocytes, and inhibition of hemagglutination by an interdomain regional peptide. J. Bacteriol. 189:3977-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato, K., N. Kido, Y. Murakami, C. I. Hoover, K. Nakayama, and F. Yoshimura. 2009. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis. Microbiology 155:1282-1293. [DOI] [PubMed] [Google Scholar]
- 52.Sato, K., M. Naito, H. Yukitake, H. Hirakawa, M. Shoji, M. J. McBride, R. G. Rhodes, and K. Nakayama. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato, K., E. Sakai, P. D. Veith, M. Shoji, Y. Kikuchi, H. Yukitake, N. Ohara, M. Naito, K. Okamoto, E. C. Reynolds, and K. Nakayama. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 280:8668-8677. [DOI] [PubMed] [Google Scholar]
- 54.Seers, C. A., N. Slakeski, P. D. Veith, T. Nikolof, Y. Y. Chen, S. G. Dashper, and E. C. Reynolds. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 188:6376-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 56.Shoji, M., D. B. Ratnayake, Y. Shi, T. Kadowaki, K. Yamamoto, F. Yoshimura, A. Akamine, M. A. Curtis, and K. Nakayama. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183-1191. [DOI] [PubMed] [Google Scholar]
- 57.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 58.Smalley, J. W., A. J. Birss, A. S. McKee, and P. D. Marsh. 1996. Haemin binding as a factor in the virulence of Porphyromonas gingivalis. FEMS Microbiol. Lett. 141:65-70. [DOI] [PubMed] [Google Scholar]
- 59.Smalley, J. W., A. J. Birss, B. Szmigielski, and J. Potempa. 2007. Sequential action of R- and K-specific gingipains of Porphyromonas gingivalis in the generation of the haem-containing pigment from oxyhaemoglobin. Arch. Biochem. Biophys. 465:44-49. [DOI] [PubMed] [Google Scholar]
- 60.Takii, R., T. Kadowaki, A. Baba, T. Tsukuba, and K. Yamamoto. 2005. A functional virulence complex composed of gingipains, adhesins, and lipopolysaccharide shows high affinity to host cells and matrix proteins and escapes recognition by host immune systems. Infect. Immun. 73:883-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ueshima, J., M. Shoji, D. B. Ratnayake, K. Abe, S. Yoshida, K. Yamamoto, and K. Nakayama. 2003. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun. 71:1170-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanterpool, E., F. Roy, and H. M. Fletcher. 2005. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect. Immun. 73:3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanterpool, E., F. Roy, L. Sandberg, and H. M. Fletcher. 2005. Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect. Immun. 73:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanterpool, E., F. Roy, W. Zhan, S. M. Sheets, L. Sangberg, and H. M. Fletcher. 2006. VimA is part of the maturation pathway for the major gingipains of Porphyromonas gingivalis W83. Microbiology 152:3383-3389. [DOI] [PubMed] [Google Scholar]
- 65.van Winkelhoff, A. J., B. J. Appelmelk, N. Kippuw, and J. de Graaff. 1993. K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol. Immunol. 8:259-265. [DOI] [PubMed] [Google Scholar]
- 66.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitfield, C., and M. A. Valvano. 1993. Biosynthesis and expression of cell-surface polysaccharides in gram-negative bacteria. Adv. Microb. Physiol. 35:135-246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.