Abstract
Paracoccidioidomycosis (PCM), caused by the dimorphic fungus Paracoccidioides brasiliensis, is a disseminated, systemic disorder that involves the lungs and other organs. The ability of the pathogen to interact with host components, including extracellular matrix (ECM) proteins, is essential to further colonization, invasion, and growth. Previously, enolase (EC 4.2.1.11) was characterized as a fibronectin binding protein in P. brasiliensis. Interaction of surface-bound enolase with plasminogen has been incriminated in tissue invasion for pathogenesis in several pathogens. In this paper, enolase was expressed in Escherichia coli as a recombinant glutathione S-transferase (GST) fusion protein (recombinant P. brasiliensis enolase [rPbEno]). The P. brasiliensis native enolase (PbEno) was detected at the fungus surface and cytoplasm by immunofluorescence with an anti-rPbEno antibody. Immobilized purified rPbEno bound plasminogen in a specific, concentration-dependent fashion. Both native enolase and rPbEno activated conversion of plasminogen to plasmin through tissue plasminogen activator. The association between PbEno and plasminogen was lysine dependent. In competition experiments, purified rPbEno, in its soluble form, inhibited plasminogen binding to fixed P. brasiliensis, suggesting that this interaction required surface-localized PbEno. Plasminogen-coated P. brasiliensis yeast cells were capable of degrading purified fibronectin, providing in vitro evidence for the generation of active plasmin on the fungus surface. Exposure of epithelial cells and phagocytes to enolase was associated with an increased expression of surface sites of adhesion. In fact, the association of P. brasiliensis with epithelial cells and phagocytes was increased in the presence of rPbEno. The expression of PbEno was upregulated in yeast cells derived from mouse-infected tissues. These data indicate that surface-associated PbEno may contribute to the pathogenesis of P. brasiliensis.
Microbial adhesion to host tissues is the initial event of most infectious process (39). Interaction with extracellular matrix (ECM) proteins has been correlated with the invasive abilities of different organisms (28, 40). ECM underlines epithelial and endothelial cells and surrounds connective tissues, and its major components are the collagens, laminin, fibronectin, and proteoglycans (52). After adherence, the next step must be to overcome the barriers imposed by epithelial tissues and ECM. The proteolytic activity achieved by subversion of host proteases by pathogens, such as plasmin, has been shown to be important during the process of many types of infections (47, 51).
Paracoccidioides brasiliensis is the causative agent of paracoccidioidomycosis (PCM), a human systemic mycosis that constitutes a major health problem in South America (44). Clinical manifestations of PCM are related to chronic granulomatous reactions with involvement of the lungs and the reticuloendothelial system, as well as mucocutaneous areas and other organs (22). In the soil, the fungus grows as saprobic mycelium, resulting in the formation of infectious propagules. After penetrating the host, the fungus differentiates into its yeast form, a fundamental step for the successful establishment of the disease (46).
Although P. brasiliensis is not traditionally considered a typical intracellular pathogen, independent studies have demonstrated that P. brasiliensis yeast cells have the capacity to adhere and invade host cells (4, 24, 31). P. brasiliensis may actively penetrate the mucocutaneous surface and parasitize epithelial cells, thus evading the host defenses and reaching deeper tissues.
Fungal ECM-binding adhesins have been characterized in different models, including P. brasiliensis. Vicentini et al. (49) showed specific binding of the protein gp43 to laminin, which is correlated to the adhesiveness of the fungus in vitro as well as to an enhancement of pathogenic potential. We have been systematically searching for new adhesion proteins in P. brasiliensis that have the potential to play roles in the fungal virulence, and proteins such as P. brasiliensis malate synthase (PbMLS) (34), PbDfg5p (defective for filamentous growth protein) (10), triosephosphate isomerase (PbTPI) (41), and glyceraldehyde-3-phosphate dehydrogenase (PbGAPDH) (4) were found to associate with ECM components. In particular, enolase from P. brasiliensis (PbEno) is a fibronectin-binding protein, as characterized by affinity ligand assays (17).
The importance of plasminogen in infectious diseases is supported by the fact that many pathogens manifest the ability to bind plaminogen (47, 13). Plasminogen is a single-chain glycoprotein with a molecular mass of 92 kDa. Its protein structure comprises an N-terminal preactivation peptide, five consecutive disulfide-bonded triple-loop kringle domains, and a serine-protease domain containing the catalytic triad (48). The kringle domains of plasminogen mediate its attachment to cells surfaces by binding proteins with accessible carboxyl-terminal or internal lysine residues. The plasminogen system displays a unique role in the host defense by dissolving fibrin clots and serving as an essential component to maintain homeostasis (43). Activation of the fibrinolytic system is dependent on the conversion of plasminogen to the serine-protease plasmin by the physiological activators urokinase-type plasminogen activator (uPA) and tissue-type plasminogen activator (tPA) (9). Plasmin is involved in fibrinolysis homeostasis and degradation of the extracellular matrix and basement membrane. The mammalian plasminogen-plasmin proteolytic system plays a crucial role in extracellular matrix degradation which is exploited by invasive pathogens, including fungi (25, 47). Microbe-derived plasminogen conversion to plasmin may promote dissemination of the pathogen within the host (1).
Among several proteins, enolase has been found to play a major role in microbial recruitment of plasminogen (32). By serving as a key surface receptor for plasminogen recruitment, enolase has been shown to function as mediator of microbial virulence (6, 15). The potential of P. brasiliensis to recruit human plasminogen for invasion and virulence has not been studied until now. In this study, we demonstrated for the first time that P. brasiliensis is capable of recruiting plasminogen and activating the plasminogen fribrinolytic system in a process, at least in part, mediated by the cell wall-localized enolase. Furthermore, recombinant PbEno (rPbEno) promoted an increase in the adhesion/invasion of P. brasiliensis in in vitro models of infection, a process that seems to be associated with the enolase ability of modifying the surface of host cells. These data suggest that PbEno may play a role in mediating the P. brasiliensis recruitment of plasminogen as well as in attachment and internalization of the fungus to host tissues, potentially playing a role in the establishment of PCM.
MATERIALS AND METHODS
Fungal isolate and growth conditions.
Yeast cells were obtained by growing P. brasiliensis isolate 01 (ATCC MYA-826) in Fava-Netto's medium for 4 days at 36°C as described previously (4).
Cloning cDNA containing the complete enolase coding region into an expression vector.
The enolase cDNA (GenBank accession number EF558735.1), obtained from a library from yeast cells of P. brasiliensis (14), was amplified by PCR using oligonucleotide sense (5′-GTC GAC ATG GCT ATC ACC AAA ATC CAC G-3′; SalI restriction site underlined) and antisense (5′-GCG GCC GCT TAC ATA TTA ATA GCT GCC C-3′; NotI restriction site underlined) primers. The PCR product was cloned in frame with the glutathione S-transferase (GST) coding region of the pGEX-4T-3 vector (GE Healthcare) to yield the pGEX-4T-3-PbEno construct. The Escherichia coli strain BL21(pLys) competent cells were transformed with the expression construct.
Expression and characterization of the recombinant enolase.
Bacteria transformed with the pGEX-4T-3-PbEno construct were grown in LB medium supplemented with ampicillin (100 μg/ml) and glucose (20 mM) at 37°C at 200 rpm. Protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. The GST-PbEno protein was affinity purified using glutathione Sepharose 4B (GE Healthcare), and the GST was cleaved by the addition of thrombin (Sigma Aldrich).
Antibody production.
The purified rPbEno was used to generate specific rabbit polyclonal serum. Rabbit preimmune serum was obtained and stored at −20°C. The purified protein was injected into rabbits with Freund's adjuvant three times at 2-week intervals. The serum, containing monospecific anti-rPbEno polyclonal antibodies (5.3 μg/μl), was stored at −20°C.
Preparation of P. brasiliensis protein fractions.
The P. brasiliensis crude protein extract was obtained by disruption of frozen yeast cells in the presence of protease inhibitors 50 μg/ml N-α-p-tosyl-l-lysine chloromethyl ketone (TLCK), 1 mM 4-chloromercuribenzoic acid (PCMB), 20 mM leupeptin, 20 mM phenylmethylsulfonyl fluoride (PMSF), and 5 mM iodoacetamide in homogenization buffer (20 mM Tris-HCl, pH 8.8, 2 mM CaCl2). The mixture was centrifuged at 12,000 × g at 4°C for 10 min, and the supernatant was used for further analysis of proteins.
Cell wall fractionation was performed basically as described previously (42).
The culture filtrate was processed as described previously (33) with modifications. Yeast cells were harvested from the solid medium and transferred to Fava Netto's liquid medium. After 1 day of growth at 37°C with gentle agitation, the proteins from the supernatant were precipitated with 10% (wt/vol) trichloroacetic acid (TCA) during overnight (o/n) incubation at 4°C. The precipitate was centrifuged for 10 min at 10,000 × g. The pellet was washed twice with acetone and air dried prior to resuspension in electrophoresis dissolving buffer. Protein samples were then subjected to SDS-PAGE. The protein content was quantified using the Bradford assay (8).
Two-dimensional (2D) gel electrophoresis and MALDI-TOF mass spectrometry analysis.
Samples containing 200 μg of P. brasiliensis yeast protein crude extract were separated by isoelectric focusing as described by O'Farrell in 1975 (37). The second dimension was performed as described by Laemmli in 1970 (27). Protein spots were excised from the gel and submitted to reduction, alquilation, and in-gel digestion with trypsin (Promega, Madison, WI). The resulting tryptic peptides were extracted and submitted to mass spectrometry (MS) analysis. The protein tryptic fragments were analyzed using a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Reflex IV; Bruker Daltonics, Karlsruhe, Germany). The peptide mass list obtained for each spectrum was searched against the SwissProt database (http://expasy.org/sprot) by using the MASCOT search engine (Matrix Science).
Western blot and ligand binding analyses.
Proteins fractionated by gel electrophoresis were transferred to nylon membranes. Blots were sequentially incubated with the rabbit polyclonal anti-rPbEno antibodies, anti-rabbit immunoglobulin G (IgG) coupled to alkaline phosphatase (Sigma), and developed with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (BCIP-NBT). Rabbit preimmune serum was used as a negative control.
For ligand blot analysis, the membrane was washed three times with 0.1% (vol/vol) Tween 20 in phosphate-buffered saline (PBS) and incubated o/n with human plasminogen (hPlg; Sigma) (35 μg/ml) in PBS-1% bovine serum albumin (BSA) (wt/vol). The blot was washed and incubated with mouse anti-human plasminogen monoclonal antibody (MAb) (1 μg/ml) (R&D Systems) in PBS-1% BSA (wt/vol) for 1 h at room temperature. The membrane was next incubated with anti-mouse IgG coupled to alkaline phosphatase (Sigma). The blots were developed with BCIP-NBT.
Immunofluorescence detection of PbEno and plasminogen on the surface of P. brasiliensis yeast cells.
An immunofluorescence assay was performed using a modification of a described procedure (33). Briefly, 2 × 107 yeast cells were fixed with 4% (vol/vol) paraformaldehyde in PBS for 10 min. The cells were washed twice in PBS and blocked with BSA for 1 h at room temperature prior to incubation with (i) rabbit anti-rPbEno antibodies or (ii) plasminogen followed by incubation with antiplasminogen monoclonal antibodies. The cells were then washed three times with PBS and treated for 1 h at 37°C with affinity-purified fluorescein isothiocyanate-conjugated goat anti-rabbit IgG or anti-mouse IgG (Sigma) diluted 1:1,000. Finally, yeast cells were washed twice with PBS and visualized using the Olympus BX41 microscope at ×100 magnification.
Plasminogen binding assay.
Cellular assays were performed after coating the wells of multititer plates with fungal cells followed by fixation. Briefly, 1 × 108 yeast cells in PBS were added to the wells and incubated for 1 h, and glutaraldehyde was added to the wells to a final concentration of 1% (vol/vol) for 10 min. After three washes with PBS, the wells were blocked with 1% (wt/vol) BSA in PBS for 1 h. Different amounts of hPlg (0.05 to 1.0 μg) were added to the wells, which were incubated for 1 h. Competition experiments were performed by the addition of increasing concentrations (0.5 μg to 3 μg) of rPbEno for 1 h prior to the addition of 1 μg of hPlg. Binding was determined by incubation with antiplasminogen monoclonal antibody. Plates were washed three times with 0.1% Tween 20 (vol/vol) in PBS. Horseradish peroxidase was added to the wells and incubated for 1 h. The absorbance was measured at A405 by using a microplate reader (Bio Tek Instruments Inc., Winooski, VT).
In another set of experiments, wells of multititer plates were coated with 1 μg of rPbEno diluted in carbonate buffer. After blocking and washing as described above, different amounts of hPlg (1 μg to 4 μg) were added to the plates. Alternatively, the plates were coated with 1 μg of hPlg diluted in carbonate buffer and incubated o/n at 4°C. A range of concentrations (1 μg to 4 μg) of rPbEno diluted in 1% BSA (wt/vol) in PBS were added to the hPlg-coated wells, incubated for 1 h, and washed with 0.1% (vol/vol) Tween 20 in PBS. Protein-protein interactions were determined by incubation with anti-rPbEno polyclonal antibodies. Competition experiments were performed by the addition of increasing concentrations (5 mM to 20 mM) of the lysine analogue ɛ-aminocaproic acid (ɛ-ACA) (Sigma) to the rPbEno-coated wells. The wells were incubated for 1 h followed by the addition of hPlg. Another set of competition experiments included the addition of specific rPbEno rabbit polyclonal antibodies prior to the addition of hPlg. All reactions were carried out at 37°C. Binding was determined by incubation with antiplasminogen monoclonal antibody. Following three washes, the wells were developed as described above. All final volumes for the enzyme-linked immunosorbent assay (ELISA) reactions were 100 μl.
Plasminogen activation assay.
Plasminogen activation was performed by measuring the amidolytic activity of the generated plasmin. Wells of multititer plates were coated with 1 μg of rPbEno or fixed P. brasiliensis and incubated with 1 μg hPlg (Sigma), 3 μg of plasmin substrate (d-valyl-l-lysyl-p-nitroaniline hydrochloride) (Sigma), and 15 ng of tissue plasminogen activator (tPA) (Sigma). Control experiments were performed by measuring the generation of plasmin either in the absence of tPA or in the presence of ɛ-ACA. Plates were incubated at room temperature for 2 h and read at A405.
Degradation of fibrin in jellified matrices.
Fibrinolysis was assayed using previously described methods with minor modifications (25). Briefly, 107 P. brasiliensis cells were preincubated with hPlg (50 μg) for 3 h in the presence or absence of tPA (50 ng) and the serine proteinase inhibitors aprotinin (1 μg) and PMSF (50 mM) in a final volume of 1 ml. Thereafter, the mixtures were washed three times with PBS to remove free plasminogen. The resulting cell pellets were placed in wells of a fibrin substrate matrix gel that contained 1.25% (wt/vol) low-melting-temperature agarose, hPlg (100 μg), and fibrinogen (4 mg; Sigma) in a final volume of 2 ml. Controls consisted of untreated cells (no plasminogen incubation) or incubation systems where no cells were added. The jellified matrix was incubated in a humidified chamber at 37°C for 12 h. Plasmin activity was detected by the observation of clear hydrolysis haloes within the opaque jellified-fibrin-containing matrix.
Influence of enolase on the interaction of P. brasiliensis with host cells.
Human type II alveolar cells (A549 lineage) and murine macrophage-like cells (RAW 264.7 [RAW] lineage) were obtained from the American Type Culture Collection (ATCC). Cultures were maintained and grown to confluence in 25-cm2 culture flasks containing Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) at 37°C with 5% CO2. To evaluate the effects of rPbEno on the interaction of P. brasiliensis with host cells, RAW and A549 lineages were first exposed to the enzyme and then probed with succinylated wheat germ agglutinin (S-WGA). S-WGA has affinity for β1,4-N-acetylglucosamine (GlcNAc) oligomers, which are recognized by the P. brasiliensis adhesin paracoccin (23). Mammalian cells were placed in a 24-well plate (105 cells/well) and treated with various concentrations of rPbEno (1 and 50 μg/ml) for 1 h at 37°C. Controls were exposed to medium alone for the same amount of time. The cells were detached from plastic surfaces, fixed, and blocked as described previously (3) and then incubated for 30 min at 37°C in 100 μl of a 5-μg/ml solution of tetramethyl rhodamine isothiocyanate (TRITC)-labeled S-WGA (EY Laboratories). The cells were washed in PBS and analyzed by flow cytometry as previously described (3). To analyze the effects of rPbEno on the interaction of P. brasiliensis with host cells, the culture medium of RAW or A549 cells was replaced with fresh media for further incubation with P. brasiliensis yeast cells. For flow cytometry experiments, the cell wall of P. brasiliensis was stained with 0.5 mg/ml fluorescein isothiocyanate (FITC; Sigma) (3, 11) in PBS (25°C) for 10 min (11). Fungal suspensions were prepared in DMEM to generate a ratio of 10 yeasts per host cell. Interactions between fungal and host cells occurred at 37°C with 5% CO2 for 18 h. Cells were washed three times with PBS to remove nonadherent yeasts. Fungus-host cell complexes were treated for 10 min at 25°C with trypan blue (200 μg/ml) to discriminate between surface-associated and intracellular yeast cells (3, 11). After removal from the plastic surface with a cell scrapper, the cells were analyzed by flow cytometry as described previously (3). Control preparations were developed as described above by using uninfected cells and nonstained yeast (data not shown). For analysis of the morphological aspects of infected cells, the complexes were fixed with paraformaldehyde and stained with 25 μM calcofluor white (Invitrogen, Life Technologies). Control or infected cells were finally observed with an Axioplan 2 (Zeiss, Germany) fluorescence microscope, following conditions previously described (3).
Infection of mice with P. brasiliensis and RNA extraction.
Mice were infected as described previously (16). Female BALB/c mice were infected intraperitoneally with 1 × 108 yeast cells and intranasally with 5 × 107 yeast cells and killed on the seventh day after infection; livers and spleens were removed from mice infected intraperitoneally, and lungs were removed from mice infected intranasally. One hundred milliliters of this suspension was plated onto brain heart infusion (BHI) agar (Becton-Dickinson, MD) supplemented with 1% (wt/vol) glucose. After 7 days, total RNA was extracted from the yeast cells (1 × 1010). Control cDNA was prepared by removing P. brasiliensis yeast cells from Fava-Netto cultures and plating to BHI agar as described above.
Quantitative analysis of RNA transcripts by qRT-PCR.
Total RNAs were treated with DNase, and cDNA was prepared using Superscript II reverse transcriptase (Invitrogen) and oligo(dT)15 primer. Quantitative real-time reverse transcription-PCR (qRT-PCR) analysis was performed on a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA) in triplicate. Values were averaged from three biological replicates. PCR thermal cycling was performed at 40 cycles of 95°C for 15 s followed by 60°C for 1 min. Ten picomoles of each primer and 40 ng of template cDNA in a total volume of 25 μl SYBR green PCR master mix (Applied Biosystems) were used for each experiment. A melting curve analysis was performed to confirm a single PCR product. The data were normalized with transcript encoding tubulin amplified in each set of qRT-PCR experiments. A nontemplate control was also included. Relative expression levels of the genes of interest were calculated using the standard curve method for relative quantification (7).
Statistical analysis.
Experiments were performed in triplicate with samples in triplicates. Results are presented as means ± standard deviations. Statistical comparisons were performed using Student's t test. Statistical significance was accepted for P values of <0.05.
RESULTS
Expression and purification of the P. brasiliensis enolase and production of polyclonal antibodies.
Previous studies identified PbEno (EC 4.2.1.11) as a fibronectin binding protein. In the present work to further investigate the role of PbEno in fungus-host interaction, we first expressed the protein in order to create antibodies specific for PbEno. The cDNA encoding PbEno (GenBank accession number EF558735.1) was cloned into the expression vector pGEX-4T-3 to obtain the recombinant fusion protein GST-PbEno. The fusion protein was affinity purified, and the 47-kDa rPbEno was obtained by digestion with thrombin (data not shown). The purified rPbEno was used to generate rabbit polyclonal antibodies.
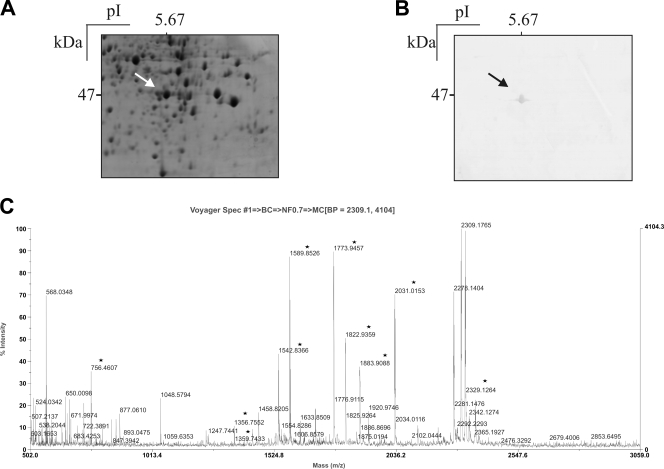
Antibody specificity was evaluated in serological tests using protein extracts from cell lysates resolved by 2D gel electrophoresis. A protein with a pI of 5.67 was recognized by polyclonal antibodies raised to rPbEno (Fig. 1 A and B). The protein was analyzed by mass spectrometry (Fig. 1C). Experimental masses were searched against data from public gene databases by using MASCOT. The peptides obtained (Table1) matched PbEno.
FIG. 1.
Identification of enolase in the P. brasiliensis proteome by two-dimensional gel electrophoresis. (A) Protein staining with Coomassie blue. (B) Reactivity of the P. brasiliensis total extract with rabbit polyclonal antibodies to enolase raised to the recombinant protein. Numbers to the left of panels A and B refer to the molecular mass of the enolase. At the top is the isoelectric point of the protein. Arrows point to enolase. (C) Peptide mass spectrum generated from tryptic digestion of PbEno. The protein reacting with polyclonal antibodies was removed from the gel and submitted to mass spectrometry analysis after trypsin digestion. The black stars indicate molecular mass values, and each peak corresponds to a peptide. Experiments represent three gels from independent protein preparations. BP, base peak.
TABLE 1.
Identification of P. brasiliensis enolase by peptide mass fingerprinta
| Positionb | Identified amino acid sequencec | Mass |
|
|---|---|---|---|
| Exptl (in-gel digestion) | Theoretical (in silico digestion) | ||
| 16-32 | R.GNPTVEVDVVTETGLHR.A | 1,822.9489 | 1,822.9293 |
| 33-50 | R.AIVPSGASTGQHEACELR.D | 1,882.9548 | 1,825.8861 |
| 90-103 | K.VDEFLNKLDGTPNK.S | 1,589.8751 | 1,588.8678 |
| 106-120 | K.LGANAILGVSLAIAK.A | 1,410.9979 | 1,410.8678 |
| 164-184 | R.LAFQEFMIVPTAAPSFSEALR.Q | 2,325.2178 | 2,325.1947 |
| 243-254 | K.IALDIASSEFYK.A | 1,356.7701 | 1,356.7045 |
| 274-285 | K.WLTYEQLADLYK.K | 1,542.8513 | 1,542.7838 |
| 314-331 | K.TCDLQVVADDLTVTNPIR.I | 2,030.0239 | 1,973.0008 |
| 377-393 | R.SGETEDVTIADIVVGLR.A | 1,773.9489 | 1,773.9228 |
| 411-416 | K.LNQILR.I | 756.4464 | 756.4726 |
Protein scores higher than 76 are significant (P < 0.05). Peptide masses matched with PbEno (GenBank accession number EF558735.1), presenting a score of 113 and coverage of 34.25% of the whole deduced sequence.
Position corresponds to the peptide fragment detected by mass spectrometry.
The sequence within the periods represents the amino acids of the detected fragment. The amino acids outside of the periods represent the amino acids which remain within the protein following tryptic digestion.
Detection of PbEno on fungal surface.
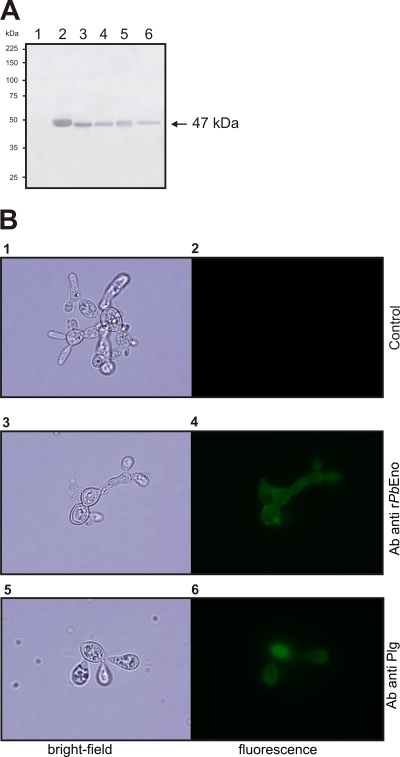
In order to determine the cellular distribution of PbEno, we probed different fractions of fungal cells by Western blot analysis. PbEno was detected in total protein extract (Fig. 2 A, lane 3), cell wall-enriched fraction (Fig. 2A, lane 4), cytoplasmic fraction (Fig. 2A, lane 5), and the culture filtrate (Fig. 2A, lane 6). Bovine serum albumin (Fig. 2A, lane 1) and rPbEno (Fig. 2A, lane 2) were employed as negative and positive controls, respectively. Altogether, these results suggest that PbEno is associated with the cell wall and is secreted to the extracellular space in addition to its expected intracellular distribution in P. brasiliensis.
FIG. 2.
Detection of PbEno and plasminogen binding at the cell surface of P. brasiliensis. Experiments were performed in triplicate. (A) Western blot analysis of bovine serum albumin (BSA; lane 1), rPbEno (lane 2), P. brasiliensis crude protein extract (lane 3), cell wall-enriched fraction proteins (lane 4), the soluble cytoplasmic fraction (lane 5), and secreted proteins (lane 6) blotted onto a nylon membrane and detected with rabbit polyclonal anti-recombinant enolase antibodies. The arrow indicates enolase. (B) Paraformaldehyde-fixed, nonpermeabilized cells were incubated with rPbEno antibodies (panels 3 and 4) or treated with human plasminogen followed by incubation with an antibody raised to this protein (panels 5 and 6). Control systems were obtained with anti-rabbit immunoglobulin G (IgG) coupled to alkaline phosphatase antibody only (panels 1 and 2). Bright-field microscopy is shown in left panels. The same cells are shown under the fluorescence mode in the right panels.
To further validate PbEno's association with the fungal surface, immunofluorescence was performed. As shown in Fig. 2B (panel 4), rabbit polyclonal antibodies reacted with the surface of the organism. Plasminogen antibodies (Fig. 2B, panel 6) also reacted with the surface of plasminogen-treated organisms, suggesting an ability of P. brasiliensis to recognize this molecule. No fluorescence and immunoreactivity were detected when yeast cells were incubated with the secondary antibody alone (Fig. 2B, panel 2).
P. brasiliensis and PbEno bind plasminogen.
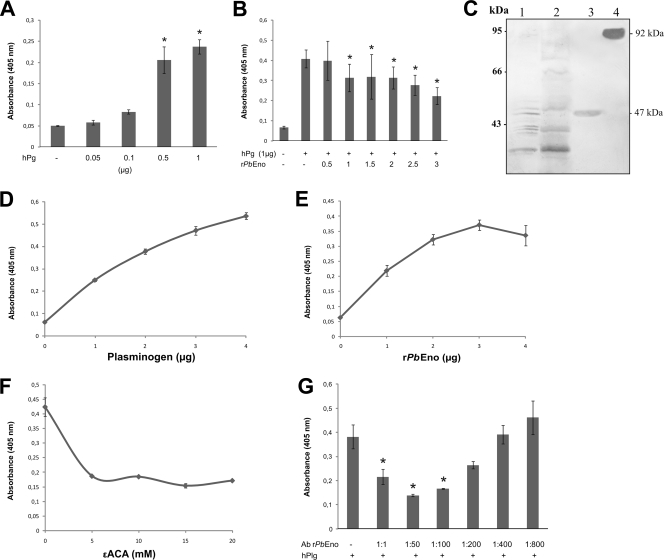
By the immunofluorescence assay, we discovered that P. brasiliensis binds to hPlg. To further characterize this phenomenon, P. brasiliensis yeast cells were fixed to the wells of multititer plates and increasing concentrations of hPlg were added. Figure 3 A shows a dose-dependent pattern of binding of hPlg to fixed fungal cells. The addition of increasing concentrations of rPbEno decreased hPlg binding to P. brasiliensis in a dose-dependent manner (Fig. 3B). These data further confirmed that enolase was involved in P. brasiliensis binding to hPlg. To support this supposition, we performed a ligand blot assay using crude protein extracts, the cell wall-enriched fraction, and rPbEno (Fig. 3C). Different P. brasiliensis proteins, including enolase, interacted with hPlg. The presence of several proteins in the ligand blot assay implicated the existence of other plasminogen binding proteins in P. brasiliensis, as described for other organisms (15). Therefore, the ability of rPbEno to bind hPlg was tested in an ELISA. Increasing concentrations of hPlg bound to immobilized rPbEno in a dose-dependent fashion (Fig. 3D). The same increasing pattern was observed when the wells were coated with hPlg and increasing concentrations of rPbEno were added (Fig. 3E).
FIG. 3.
Plasminogen binding assays. Microtiter plates were coated with fixed P. brasiliensis yeast cells as detailed in Materials and Methods. (A) Plasminogen (0.05 to 1.0 μg) binds to fixed P. brasiliensis in a concentration-dependent manner. (B) In a competition assay, binding of plasminogen is inhibited by increasing amounts of rPbEno (0.5 to 3.0 μg). (C) Binding of P. brasiliensis proteins to plasminogen. P. brasiliensis crude protein extract (lane 1), cell wall-enriched fraction proteins (lane 2), rPbEno (lane 3), and hPlg (lane 4) were sequentially incubated with plasminogen and a mouse monoclonal anti-human plasminogen antibody. The numbers on the left side are molecular size markers. (D) Plasminogen (1 to 4 μg) binds to rPbEno (1 μg) immobilized on microtiter well plates in a concentration-dependent manner. ELISAs were developed at A405 with antiplasminogen antibody. (E) rPbEno (1 to 4 μg) binds to immobilized plasminogen (1 μg) in a similar fashion. The assay was developed at A405 with antibodies to rPbEno. (F) Effects of different ɛ-ACA concentrations (5 to 20 mM) on plasminogen binding. (G) Plasminogen binding to immobilized rPbEno is specifically inhibited by anti-rPbEno. Microtiter plates were coated by overnight incubation with 1 μg of rPbEno. After blocking, the wells were incubated with decreasing concentrations of rabbit polyclonal rPbEno antibodies. Reactions were developed after incubation with hPlg, the antiplasminogen antibody followed by secondary antibodies. Panels A, B, D, E, F, and G show the averages of three independent experiments performed in triplicates. The error bars indicate the standard deviations from three independent experiments performed in triplicate. *, significantly different from control, at a P value of <0.05.
Previous work has shown that enolase binds to hPlg through lysine residues (33, 47). We thus examined if binding of rPbEno was lysine dependent by using competitive antagonism with the lysine analog ɛ-ACA. The results shown in Fig. 3F indicate that lysine residues present on rPbEno may have a role in plasminogen recruitment by P. brasiliensis.
To further validate rPbEno binding to hPlg, competition experiments were also performed by the addition of specific rPbEno rabbit polyclonal antibodies to the experimental system. The presence of enolase-specific antibodies dose-dependently decreased hPlg binding to rPbEno (Fig. 3G). In the presence of preimmune sera, no effects were observed (data not shown). These data confirmed that enolase specifically binds hPlg.
Plasminogen activation and fibrinolysis.
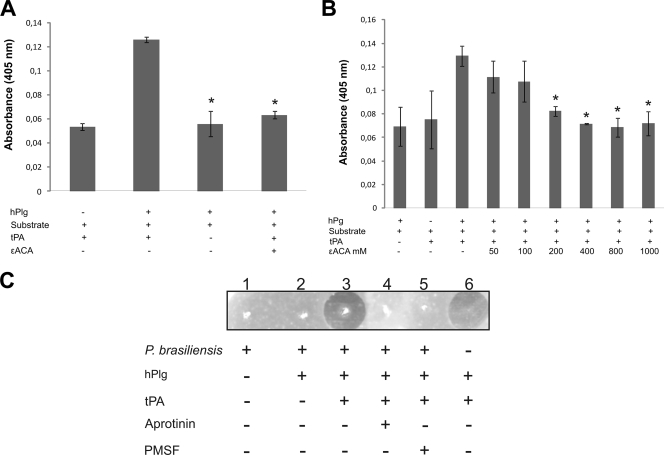
Once yeast cells and rPbEno had been seen to bind hPlg, we had expected that this interaction could also activate hPlg. For this reason, an ELISA was performed to determine the abilities of P. brasiliensis and rPbEno to produce plasmin from hPlg. In the presence of tPA, rPbEno was able to generate plasmin (Fig. 4 A). The addition of ɛ-ACA inhibited plasmin generation. hPlg activation was evaluated in assays using fixed fungal cells in the presence of tPA, confirming that interaction with fixed P. brasiliensis also resulted in hPlg activation (Fig. 4B). The addition of increasing concentrations of ɛ-ACA to the experimental system inhibited plasmin generation in a dose-dependent manner. These results suggest that P. brasiliensis and rPbEno mediate activation of plasminogen to plasmin and that lysine residues are involved in binding and activation of hPlg (Fig. 4B).
FIG. 4.
Plasminogen activation assays. (A) rPbEno (1 μg) generates plasmin from plasminogen in the presence of tPA and in the absence of ɛ-ACA. (B) P. brasiliensis converts plasminogen into plasmin in the presence of tPA. Various concentrations of ɛ-ACA (50 mM to 1,000 mM) were added to wells containing fixed P. brasiliensis, followed by the addition of plasminogen, and an ELISA was performed as described in Materials and Methods. The error bars indicate the standard deviations from three independent experiments performed in triplicate. *, significantly different from control, at a P value of <0.05. (C) Fibrinolytic activity of plasminogen-bound P. brasiliensis. Lane 1, P. brasiliensis cells in the absence of plasminogen; lane 2, P. brasiliensis cells after binding to plasminogen. Lanes 3, 4, and 5 are similar to lane 2, except that they reflect the presence of tPA, tPA plus aprotinin, and tPA plus PMSF, respectively. Lane 6, controls consisting of plasminogen and tPA.
Fibrinogen is one of the major substrates of plasminogen/plasmin in vivo, and jellified matrices containing fibrinogen have been used to examine plasmin activity (1, 25). As demonstrated in Fig. 4C, the association of P. brasiliensis with plasminogen and tPA promoted increased fibrinolysis (Fig. 4C, lane 3). Aprotinin (lane 4) and PMSF (lane 5) inhibited proteolysis, indicating the specificity of the reaction. No proteolysis was observed either when P. brasiliensis yeast cells were used alone or in the presence of plasminogen (lanes 1 and 2, respectively). Figure 4C, lane 6, shows a control consisting of plasminogen and tPA.
Enolase influences the interaction of P. brasiliensis with host cells.
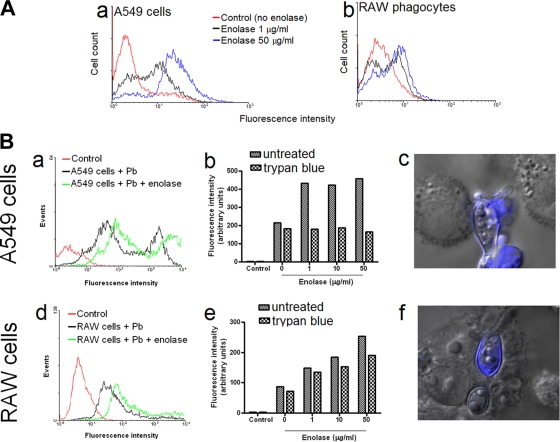
Previous studies had demonstrated that antibodies raised to a 54-kDa enolase from P. brasiliensis isolate Pb18 abolished 80% of adhesion to A549 epithelial cells (17). In this work, the participation of enolase in the infection of host cells by P. brasiliensis was evaluated by flow cytometry and fluorescence microscopy. Exposure of human epithelial cells (Fig. 5 A, panel a) and murine phagocytes (Fig. 5A, panel b) to rPbEno resulted in an expressive increase in their reactivity with WGA, suggesting that the enzyme modifies the surface of host cells to promote an enhanced exposure of GlcNAc residues, which are recognized by a P. brasiliensis adhesin (23). We therefore asked whether exposure to the enzyme would make mammalian cells more susceptible to infection by P. brasiliensis.
FIG. 5.
Exposure of host cells to rPbEno enhances the efficacy of association of P. brasiliensis to host cells. (A) Treatment of A549 epithelial cells (panel a) or RAW phagocytes (panel b) resulted in increased reactivity with WGA, indicating enhanced exposure of GlcNAc residues. (B) Effects of rPbEno on the infection of host cells by P. brasiliensis. Panels a and d demonstrate that P. brasiliensis (Pb) efficiently infects epithelial (A549) and macrophage-like (RAW) cells. Histograms of control cells (noninfected) are shown in red. Exposure of host cells to rPbEno (10 μg/ml, green histogram) results in their increased association with fungi, as determined by the comparison with infection systems prepared in the absence of enolase (black histograms). Exposure of cells infected with FITC-P. brasiliensis to trypan blue (b and e) resulted in an accentuated reduction of fluorescence levels in A549 cells but not macrophages. The suggestive internalization of P. brasiliensis by macrophages, but not by epithelial cells, was supported by fluorescence microscopy (c and f). In this analysis, yeast fluorescence appears in blue.
Incubation of FITC-stained P. brasiliensis yeast cells with epithelial or macrophage-like cells under control conditions resulted in high levels of infection. Approximately 85% of the epithelial cells became fluorescent after interaction with fungi. This index corresponded to almost 90% of infected cells when phagocytes were used. Pretreatment of the cells with rPbEno caused an increase in the percentage of infected cells to approximately 97% in both epithelial and macrophage-like systems. More importantly, the intensity of fluorescence in infected cells clearly increased when they were first exposed to rPbEno (Fig. 5B, panels a and d). Although this characteristic was common to both systems of infections, there was a clearer increase in the dose-response profile of fluorescence in the macrophage system after exposure to the enzyme (Fig. 5B, panels b and e).
To evaluate if P. brasiliensis yeast cells were internalized by epithelial cells, the fungus was treated with trypan blue. Exposure to this dye caused an expressive decrease in the levels of fluorescence of infected A549 cells, suggesting that fungal cells adhered to but were not internalized by alveolar epithelial cells. In contrast, the fluorescence levels of infected macrophages were barely affected by exposure to trypan blue. This indicates that internalization of P. brasiliensis by the phagocytes, and consequent protection against fluorescence quenching, occurred efficiently. Results shown in Fig. 5B are representative of two independent experiments producing the same fluorescence profile in flow cytometry measurements. Statistics were not included because, although the profiles described in Fig. 5B were similar in all experiments, the absolute fluorescence values may differ considerably in different assays, impairing calculation of reliable average values. Data interpretation was confirmed by fluorescence microscopy (Fig. 5B, panels c and f). In either system, the viability of host cells was not affected by the fungal infection (data not shown).
Assessment of PbEno by real-time PCR in models of infection.
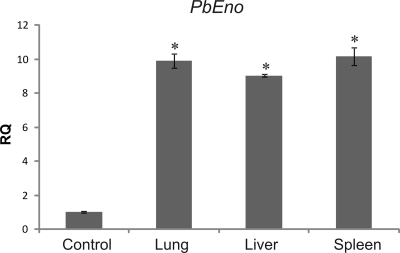
We speculated that if PbEno was required for efficient fungal attachment and invasion of host cells, the upregulation of the gene during infection would be necessary. Relative quantification of gene transcripts was examined by real-time PCR in yeast cells of P. brasiliensis derived from lungs, spleens, and livers of infected mice (Fig. 6). Enolase expression was upregulated in yeast cells derived from tissues at 7 days postinoculation.
FIG. 6.
Analysis of enolase transcripts by quantitative real time RT-PCR. qRT-PCR plot of PbEno expression levels of transcripts from yeast cells of P. brasiliensis derived from lungs, livers, and spleens of mice after 7 days of infection. Control systems consisted of yeast cells from cultures inoculated in BHI agar. The primers were as follows: sense, 5′-GATTTGCAGGTTGTCGCCGA-3′; antisense, 5′-TGGCTGCCTGGATGGATTCA-3′. The expression values were standardized using the expression values for the constitutive gene encoding the protein tubulin. The relative quantification (RQ) of the experiment was performed in triplicate. The error bars indicate the standard deviations from three independent experiments performed in triplicate. *, significantly different from the control, at a P value of <0.05.
DISCUSSION
The present study describes characterization of enolase as a plasminogen binding molecule on the surface of P. brasiliensis. The presence of enolase on the surface of cells is not without precedent. Pitarch et al. (42) analyzed cell wall fractions of Candida albicans and concluded that enolase can be loosely associated with the cell surface, as it was released when the cells were treated with SDS. The enzyme was also found to be tightly entrapped within the glucan-chitin network, which is consistent with the identification of enolase as a glucan-associated integral component of the cell wall of C. albicans (2). The question of how proteins lacking any signal peptide are exported on the cell surface is unresolved. It is clear, however, that fungal cells express many molecules with apparently conflicting functions (35). The nuclear histone-like protein H2B, for instance, is also found at the cell wall of Histoplasma capsulatum, where it functions as a target for protective antibodies (36). In P. brasiliensis, the mitochondrial protein Mdj1p and the cytosolic enzymes GAPDH and TPI were also characterized as cell wall components (4, 5, 41). This multiplicity in cellular distribution and functions is also common to enolase because this protein functions in sugar metabolism but is also present at the cell surface (30) and in secretory vesicles that reach the extracellular space (45). Enolase has been also described as a component in bacterial cell walls (26), where it mediates the interaction of Streptococcus pneumoniae with human plasminogen (13). The dual location in the cytosol and on the cell surface indicated the pivotal roles of enolase in glycolysis and pathogenesis, respectively. However, an important and challenging important issue that needs to be addressed further is the discerning of the mechanism of its export to the cell surface.
PbEno has previously been characterized as a 54-kDa fibronectin binding protein (17). Differences in molecular mass related in the previous work could be related to the potential sites for glycosylation and myristoylation, present in the deduced sequence of the protein (data not shown). We demonstrated that PbEno was not the only adherence protein but it is involved in P. brasiliensis binding to plasminogen. Similarly, enolase is a predominant plasminogen binding and cell wall protein in C. albicans, Aspergillus fumigatus, and Pneumocystis carinii (19, 21, 25, 30). Plasminogen is abundant in the circulation, and its activation by invasive pathogens could increase the organism's potential of tissue invasion. The binding of plasminogen to mammalian and bacterial cells is mediated by its five kringle domains, which have affinity for lysine (43). Lysine-dependent binding is characteristic of the plasminogen-pathogen interaction (50). We observed that the lysine analogue ɛ-ACA inhibited plasminogen binding to both P. brasiliensis and rPbEno while it also inhibited activation to plasmin. These data suggest that plasminogen binding to the surface of P. brasiliensis might involve lysine residues, since the majority of all of the plasminogen receptor proteins identified have carboxy-terminal lysine residues (25, 33, 38). With these data taken together, we hypothesized that P. brasiliensis may take advantage of the plasminogen-clotting system during invasion of host tissues.
In addition to describing the localization and functional characterization of PbEno, we described the fibrinolytic potential of P. brasiliensis mediated by the surface-associated enolase. Our studies with jellified matrices provided more evidence that plasminogen can perform proteolytic activity while bound to P. brasiliensis. Functional studies to address the significance of plasminogen binding in the invasiveness of Cryptococcus neoformans demonstrated that plasmin-coated organisms possess an increased potential to penetrate the ECM in vitro (47). There are remarkable studies showing that host susceptibility to invasive aspergillosis is strongly influenced by the plasminogen system and that plasminogen activation on the surfaces of both A. fumigatus and C. albicans promotes ECM invasion (25, 53). In agreement with this finding, Esgleas et al. (20) showed that enolase was important for the adhesion and invasion of brain microvascular endothelial cells by Streptococcus suis. Although multiple factors contribute to fungal virulence, including the expression of extracellular proteases, morphological switching, and adherence, the ability of fungal pathogens to subvert the host plasminogen system suggests that plasminogen binding may be an additional mechanism used by fungi to promote dissemination and tissue invasion during infection (19, 21, 25, 30, 53). The capture of plasminogen by adhesins such as enolase and its conversion to plasmin has in fact been described for different pathogens (20).
In the current study, exposure of epithelial cells and phagocytes to rPbEno enhanced the efficacy of P. brasiliensis association with host components. Treatment of host cells with enolase caused an increase in exposure of surface N-acetylglucosamine. Although the mechanisms connecting exposure to enolase with changes in surface carbohydrates are unclear, this observation echoes previous findings showing that animal infection with S. pneumoniae, an enolase-producing pathogen (6), results in an increased surface exposure of N-acetylglucosamine residues by host tissues (29). Since P. brasiliensis uses N-acetylglucosamine as a surface site of adhesion in host cells (12, 18, 23), we hypothesized that treatment of host cells with enolase could result in increased infectivity.
Yeast cells preferentially adhered to epithelial cells and were internalized by phagocytes. The mechanisms explaining how an enzyme could alter surface interactions of a fungal pathogen with host cells are still obscure. Although the mechanisms by which enolase interferes with steps of the interaction of pathogens with host cells are unknown, it is evident from the current literature that this enzyme may be involved in adhesion to and microbial infection in host elements. Our analysis demonstrated that enolase is a surface-secreted protein in P. brasiliensis as it is in C. neoformans (45). In this way, release of the enzyme to the extracellular space, as demonstrated here, could somehow increase the availability of adhesion sites in host cells. This putative phenomenon would result in higher efficacy of association of fungi with host cells, as currently described in our manuscript.
The results described in this paper expand current knowledge on the adhesion and invasion processes of P. brasiliensis. The transcript encoding PbEno was upregulated in yeast cells derived from tissues from infected mice. Overall, the present work is the first study, to our knowledge, to demonstrate the plasminogen binding and activation activity of P. brasiliensis enolase and shows that, similar to its role in other microbes, enolase may contribute to the virulence of P. brasiliensis. In summary, we have shown that P. brasiliensis can borrow the plasminogen system from the host in a process mediated by the surface protein enolase.
Acknowledgments
This work at the Universidade Federal de Goiás was supported by grants from Financiadora de Estudos e Projetos (FINEP 0106121200 and 0107055200) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 472947/2007-9 and 558405/2008-8). M.L.R. was supported by grants from the Brazilian agencies FAPERJ and CNPq.
We thank Tereza Cristina Rezende for helpful suggestions.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 6 July 2010.
REFERENCES
- 1.Agarwal, S., P. Kulshreshtha, D. Bambah Mukku, and R. Bhatnagar. 2008. α-Enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim. Biophys. Acta 1784:986-994. [DOI] [PubMed] [Google Scholar]
- 2.Angiolella, L., M. Facchin, A. Stringaro, B. Maras, N. Simonetti, and A. Cassone. 1996. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J. Infect. Dis. 173:684-690. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa, F. M., F. L. Fonseca, C. Holandino, C. S. Alviano, L. Nimrichter, and M. L. Rodrigues. 2006. Glucuronoxylomannan-mediated interaction of Cryptococcus neoformans with human alveolar cells results in fungal internalization and host cell damage. Microbes Infect. 8:493-502. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa, M. S., S. N. Báo, P. F. Andreotti, F. P. Faria, M. S. Felipe, L. S. Feitosa, M. J. S. Mendes-Giannini, and C. M. A. Soares. 2006. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batista, W. L., A. L. Matsuo, L. Ganiko, T. F. Barros, T. R. Veiga, E. Freymuller, and R. Puccia. 2006. The PbMDJ1 gene belongs to a conserved MDJ1/LON locus in thermodimorphic pathogenic fungi and encodes a heat shock protein that localizes to both the mitochondria and cell wall of Paracoccidioides brasiliensis. Eukaryot. Cell 5:379-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 7.Bookout, A. L., C. L. Cumming, D. J. Mangelsdorf, J. M. Pesola, and M. F. Kramer. 2006. High-throughput real-time quantitative reverse transcription PCR, p. 15.8.1-15.8.28. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Hoboken, NJ. [DOI] [PubMed]
- 8.Bradford, M. M. 1976. A dye binding assay for protein. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Castellino, F. J., and V. A. Ploplis. 2005. Structure and function of the plasminogen/plasmin system. Thromb. Haemost. 93:647-654. [DOI] [PubMed] [Google Scholar]
- 10.Castro, N. D. S., M. S. Barbosa, Z. A. Maia, S. N. Bao, M. S. Felipe, J. M. Santana, M. J. S. Mendes-Giannini, M. Pereira, and C. M. A. Soares. 2008. Characterization of Paracoccidioides brasiliensis PbDfg5p, a cell-wall protein implicated in filamentous growth. Yeast 25:141-154. [DOI] [PubMed] [Google Scholar]
- 11.Chaka, W., J. Scharringa, A. F. Verheul, J. Verhoef, A. G. Van Strijp, and I. M. Hoepelman. 1995. Quantitative analysis of phagocytosis and killing of Cryptococcus neoformans by human peripheral blood mononuclear cells by flow cytometry. Clin. Diagn. Lab. Immunol. 2:753-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coltri, K. C., A. S. Casabona-Fortunato, M. L. Gennari-Cardoso, C. F. Pinzan, L. P. Ruas, V. S. Mariano, R. Martinez, J. C. Rosa, A. Panunto-Castelo, and M. C. Roque-Barreira. 2006. Paracoccin, a GlcNAc-binding lectin from Paracoccidioides brasiliensis, binds to laminin and induces TNF-alpha production by macrophages. Microbes Infect. 8:704-713. [DOI] [PubMed] [Google Scholar]
- 13.Cork, A. J., S. Jergic, S. Hammerschmidt, B. Kobe, V. Pancholi, J. L. Benesch, C. V. Robinson, N. E. Dixon, J. A. Aquilina, and M. J. Walker. 2009. Defining the structural basis of human plasminogen binding by streptococcal surface enolase. J. Biol. Chem. 284:17129-17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa, M., C. L. Borges, A. M. Bailão, G. V. Meirelles, Y. A. Mendonça, S. F. I. M. Dantas, F. P. Faria, M. S. S. Felipe, E. E. W. I. Molinari-Madlum, M. J. S. M. Giannini, R. B. Fiúza, W. S. Martins, M. Pereira, and C. M. A. Soares. 2007. Transcriptome profiling of Paracoccidioides brasiliensis yeast-phase cells recovered from infected mice brings new insights into fungal response upon host interaction. Microbiology 153:4194-4207. [DOI] [PubMed] [Google Scholar]
- 15.Crowe, J. D., I. K. Sieywright, G. C. Auld, N. R. Moore, N. A. Gow, and N. A. Booth. 2003. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol. Microbiol. 47:1637-1651. [DOI] [PubMed] [Google Scholar]
- 16.Dantas, S. F., T. C. Vieira de Rezende, A. M. Bailão, C. P. Taborda, R. S. Santos, K. C. Pacheco, and C. M. A. Soares. 2009. Identification and characterization of antigenic proteins potentially expressed during the infectious process of Paracoccidioides brasiliensis. Microbes Infect. 11:895-903. [DOI] [PubMed] [Google Scholar]
- 17.Donofrio, F. C., A. C. Calil, E. T. Miranda, A. M. Almeida, G. Benard, C. P. Soares, S. V. Nogueira, C. M. A. Soares, and M. J. Mendes-Giannini. 2009. Enolase from Paracoccidioides brasiliensis: isolation and identification as a fibronectin-binding protein. J. Med. Microbiol. 58:706-713. [DOI] [PubMed] [Google Scholar]
- 18.dos Reis Almeida, F. B., L. L. de Oliveira, M. Valle de Sousa, M. C. Barreira, and E. S. Hanna. 2010. Paracoccin from Paracoccidioides brasiliensis; purification through affinity with chitin and identification of N-acetyl-beta-D-glucosaminidase activity. Yeast 27:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eroles, P., M. Sentandreu, M. V. Elorza, and R. Sentandreu. 1997. The highly immunogenic enolase and Hsp70p are adventitious Candida albicans cell wall proteins. Microbiology 143:313-320. [DOI] [PubMed] [Google Scholar]
- 20.Esgleas, M., Y. Li, M. A. Handock, J. Harel, J. D. Dubreuil, and M. Gottschalk. 2008. Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiology 154:2668-2679. [DOI] [PubMed] [Google Scholar]
- 21.Fox, D., and A. G. Smulian. 2001. Plasminogen-binding activity of enolase in the opportunistic pathogen Pneumocystis carinii. Med. Mycol. 39:495-507. [DOI] [PubMed] [Google Scholar]
- 22.Franco, M. 1987. Host-parasite relationships in paracoccidioidomycosis. J. Med. Vet. Mycol. 25:5-18. [DOI] [PubMed] [Google Scholar]
- 23.Ganiko, L., R. Puccia, V. S. Mariano, O. A. Sant'Anna, E. Freymuller, M. C. Roque-Barreira, and L. R. Travassos. 2007. Paracoccin, an N-acetyl-glucosamine-binding lectin of Paracoccidioides brasiliensis, is involved in fungal growth. Microbes Infect. 9:695-703. [DOI] [PubMed] [Google Scholar]
- 24.Hanna, S. A., J. L. Monteiro da Silva, and M. J. Mendes-Giannini. 2000. Adherence and intracellular parasitism of Paracoccidioides brasiliensis in Vero cells. Microbes Infect. 2:877-884. [DOI] [PubMed] [Google Scholar]
- 25.Jong, A. Y., S. H. M. Chen, M. F. Stins, K. S. Kim, T. L. Tuan, and S. H. Huang. 2003. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J. Med. Microbiol. 52:615-622. [DOI] [PubMed] [Google Scholar]
- 26.Kesimer, M., N. Kilic, R. Mehrotra, D. J. Thornton, and J. K. Sheehan. 2009. Identification of salivary mucin MUC7 binding proteins from Streptococcus gordonii. BMC Microbiol. 9:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lima, O. C., C. C. Figueiredo, J. O. Previato, L. Mendonça-Previato, V. Morandi, and L. M. L. Bezerra. 2001. Involvement of fungal cell wall components in adhesion of Sporothrix schenckii to human fibronectin. Infect. Immun. 69:6874-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linder, T. E., R. L. Daniels, D. J. Lim, and T. F. DeMaria. 1994. Effect of intranasal inoculation of Streptococcus pneumoniae on the structure of the surface carbohydrates of the chinchilla eustachian tube and middle ear mucosa. Microb. Pathog. 16:435-441. [DOI] [PubMed] [Google Scholar]
- 30.López-Villar, E., L. Monteoliva, M. R. Larsen, E. Sachon, M. Shabaz, M. Pardo, J. Pla, C. Gil, P. Roepstorff, and C. Nombela. 2006. Genetic and proteomic evidences support the localization of yeast enolase in the cell surface. Proteomics 6(Suppl. 1):S107-S118. [DOI] [PubMed] [Google Scholar]
- 31.Mendes-Giannini, M. J. S., S. A. Hanna, J. L. da Silva, P. F. Andretti, L. R. Vicentini, G. Bernard, H. L. Lenzi, and C. P. Soares. 2004. Invasion of epithelial mammalian cells by Paracoccidioides brasiliensis leads to cytoskeletal rearrangement and apoptosis of the host cell. Microbes Infect. 6:882-891. [DOI] [PubMed] [Google Scholar]
- 32.Miles, L. A., C. M. Dahberg, J. Plescia, J. Felez, K. Kato, and E. F. Plow. 1991. Role of cell-surface lysines in plasminogen binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry 30:1682-1691. [DOI] [PubMed] [Google Scholar]
- 33.Mundodi, V., A. S. Kucknoor, and J. F. Alderete. 2008. Immunogenic and plasminogen-binding surface-associated α-enolase of Trichomonas vaginalis. Infect. Immun. 76:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neto, B. R. D. S., J. F. Silva, M. J. Mendes-Giannini, H. L. Lenzi, C. M. A. Soares, and M. Pereira. 2009. The malate synthase of Paracoccidioides brasiliensis is a linked surface protein that behaves as an anchorless adhesin. BMC Microbiol. 9:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nimrichter, L., M. L. Rodrigues, E. G. Rodrigues, and L. R. Travassos. 2005. The multitude of targets for the immune system and drug therapy in the fungal cell wall. Microbes Infect. 7:789-798. [DOI] [PubMed] [Google Scholar]
- 36.Nosanchuk, J. D., J. N. Steenbergen, L. Shi, G. S. Deepe, Jr., and A. Casadevall. 2003. Antibodies to a cell surface histone-like protein protect against Histoplasma capsulatum. J. Clin. Invest. 112:1164-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 38.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 39.Patti, J. L., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 40.Peñalver, M. C., J. E. O'Connor, J. P. Martinez, and M. L. Gil. 1996. Binding of human fibronectin to Aspergillus fumigatus conidia. Infect. Immun. 64:1146-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereira, L. A., S. N. Báo, M. S. Barbosa, J. L. Silva, M. S. Felipe, J. M. Santana, M. J. S. Mendes-Giannini, and C. M. A. Soares. 2007. Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res. 7:1381-1388. [DOI] [PubMed] [Google Scholar]
- 42.Pitarch, A., M. Sánchez, C. Nombela, and C. Gil. 2002. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell Proteomics 1:967-982. [DOI] [PubMed] [Google Scholar]
- 43.Plow, E. F., T. Redlitz, L. A. Miles, and J. L. Hoover-Plow. 1995. The cell biology of the plasminogen system. FASEB J. 9:939-945. [DOI] [PubMed] [Google Scholar]
- 44.Restrepo, A., J. G. McEwen, and E. Castañeda. 2001. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med. Mycol. 39:233-241. [DOI] [PubMed] [Google Scholar]
- 45.Rodrigues, M. L., E. S. Nakayasu, D. L. Oliveira, L. Nimrichter, J. D. Nosanchuk, I. C. Almeida, and A. Casadevall. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 7:58-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.San-Blas, G., G. Nino-Veja, and T. Iturriaga. 2002. Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 40:225-242. [DOI] [PubMed] [Google Scholar]
- 47.Stie, J., G. Bruni, and D. Fox. 2009. Surface-associated plasminogen binding of Cryptococcus neoformans promotes extracellular matrix invasion. PLoS One 3:e5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vassalli, J. D., A. P. Sappino, and D. Belin. 1991. The plasminogen activator/plasmin system. J. Clin. Invest. 88:1067-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vicentini, A. P., J. L. Gesztesi, M. F. Franco, W. Souza, J. Z. Moraes, L. R. Travassos, and J. D. Lopes. 1994. Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect. Immun. 4:1465-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieira, M. L., S. A. Vasconcellos, A. P. Gonçales, Z. M. de Morais, and A. L. Nascimento. 2009. Plasminogen acquisition and activation at the surface of leptospira species lead to fibronectin degradation. Infect. Immun. 77:4092-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker, M. J., J. D. McArthur, F. Mckay, and M. Ranson. 2005. Is plasminogen deployed as a Streptococcus pyogenes virulence factor? Trends Microbiol. 13:308-313. [DOI] [PubMed] [Google Scholar]
- 52.Westerlund, B., and T. K. Korhonen. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 4:687-694. [DOI] [PubMed] [Google Scholar]
- 53.Zaas, A. K., G. Liao, J. W. Chien, C. Weinberg, D. Shore, S. S. Giles, K. A. Marr, J. Usuka, L. H. Burch, L. Pereira, J. R. Perfect, G. Peltz, and D. A. Schwartz. 2008. Plasminogen alleles influence susceptibility to invasive aspergillosis. PLoS Genet. 4:e1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]