Abstract
Coinfection with human immunodeficiency virus type 1 (HIV-1) and opportunistic mycobacteria, especially Mycobacterium tuberculosis, is a cause of high morbidity and mortality worldwide. Both mycobacteria and HIV-1 may infect macrophages, and thus, coinfection may generate conditions that reciprocally influence the intracellular replication of the pathogens. Elucidation of the interaction between HIV-1 and mycobacteria in their common target cell is important for understanding pathogenesis in coinfected individuals. In this study, we investigated the effects of in vitro HIV-1 infection on the growth of M. tuberculosis, M. avium, and M. paratuberculosis in human peripheral blood monocyte-derived macrophages. Interestingly, HIV-1 infection induced a greater bacterial burden in coinfected cell cultures for all of the mycobacterial species tested and specifically induced accelerated growth of M. tuberculosis with a reduced mean generation time. The interaction of HIV-1 and M. tuberculosis was especially detrimental to the host cell, causing a significant synergistic reduction in macrophage viability. Also, in M. tuberculosis/HIV-1-coinfected cultures, increased levels of interleukin-1β (IL-1β), IL-6, IL-8, and granulocyte-macrophage colony-stimulating factor were observed and viral replication was enhanced. Overall, the present data suggest that HIV-1 infection of macrophages may impair their ability to contain mycobacterial growth. Furthermore, coinfection with HIV-1 and M. tuberculosis seems to give rise to synergistic effects at the cellular level that mutually enhance the replication of both pathogens. This may, in part, contribute to the increased morbidity and mortality seen in coinfected individuals.
Human immunodeficiency virus type 1 (HIV-1) has caused a global pandemic, and approximately 33.4 million people were living with HIV infection by the end of 2008 (34). HIV+ individuals have an increased susceptibility to a wide range of opportunistic bacterial infections (reviewed in references 2 and 25). However, infections with certain opportunistic organisms like Mycobacterium tuberculosis are of particular concern, as people in resource-poor countries are often struck disproportionately hard, with increased morbidity and mortality due to the relatively high prevalence of HIV-1 and mycobacterial coinfections and limited access to appropriate treatment.
In the clinical context of HIV-1 infection, M. tuberculosis and M. avium complex (MAC) infections are the most frequent. These mycobacteria display various propensities to cause disease. Notably, M. tuberculosis is the leading cause of death in HIV+ patients and a major public health concern, while MAC infections are typically seen in the later stages of HIV infection, when the adaptive immunity of the host is compromised. By current classical genetic criteria, the MAC consists of mainly two species, M. intracellulare and M. avium, and M. avium is further subdivided into three subsets, M. avium subsp. avium (here referred to as M. avium) M. avium subsp. paratuberculosis (here referred to as M. paratuberculosis), and the nonhuman pathogen M. avium subsp. silvaticum (33). Among the MAC isolates from AIDS patients, M. avium is the most predominant, followed by M. intracellulare, while there is only one case report of M. paratuberculosis infection (27). However, and interestingly, several studies of MAC isolates derived from AIDS patients considered to be M. avium subsp. avium indicate that a considerable proportion (25% according to Roiz et al. [28], 58% according to Naser et al. [23], and 73% according to Hampson et al. [11]) carry the IS900 element or an IS900-like sequence element, which is traditionally considered a genetic hallmark of M. paratuberculosis. Hence, data suggest that clinical M. avium subsp. avium isolates, especially those derived from HIV/AIDS patients, may be even more closely related to M. avium subsp. paratuberculosis than generally acknowledged. Thus, in this study, we included M. paratuberculosis partially to explore the possible interaction with HIV-1 in macrophages which, to our knowledge, has never been examined before and partially as an avirulent control for M. tuberculosis and M. avium. In terms of clinical virulence, these mycobacteria may be viewed as representing different points on a scale where M. tuberculosis and M. paratuberculosis are two opposite polar extremes representing, respectively, highly virulent and almost avirulent mycobacteria while M. avium would be an intermediate. The ability of mycobacteria to grow within host macrophages is a critical determinant of their pathogenicity. The more virulent mycobacteria, such as M. tuberculosis, grow and survive more efficiently in host macrophages than do less virulent mycobacteria like M. paratuberculosis. As the macrophage is a target cell for both HIV-1 and mycobacteria, it is quite likely that coinfection may generate conditions that reciprocally influence the replication of the pathogens. We (14) and others (10, 13, 24) have previously shown that HIV-1 may induce increased mycobacterial growth in macrophages, whereas other studies did not find enhanced growth (6, 21). Currently, it is not known whether HIV-1 may have an effect on bacterial growth in macrophages that is common to mycobacteria irrespective of their virulence or whether such effects may be specific to virulent mycobacterial species like M. tuberculosis. Therefore, the growth of M. tuberculosis in HIV-1-infected macrophage cultures was compared with that of M. avium and M. paratuberculosis in order to investigate whether HIV-1 has an effect on bacterial growth common to various mycobacterial species in macrophages or whether the in vitro growth might mirror the various characteristics of these mycobacteria to cause clinical disease in HIV+ patients.
MATERIALS AND METHODS
Mycobacterial strains.
M. tuberculosis H37Rv (ATCC 27294, a kind gift from Harleen Grewal, The Gade Institute, University of Bergen, Bergen, Norway) was grown in Middlebrook 7H9 broth (Difco/Becton Dickinson) with albumin-dextrose-catalase enrichment (Difco/BD) and 0.05% Tween 80 and subsequently passed five times through a sterile syringe with a 29-gauge needle to disrupt bacterial clumps before aliquots were frozen at −80°C, and CFU counts were determined by plating on oleic acid-albumin-dextrose-catalase (OADC)-enriched Middlebrook 7H10 agar (Difco/BD). In the present study, we used frozen aliquots with predetermined CFU counts of human clinical isolates. M. avium S72/89 (serovar 4) (a kind gift from Sven Hoffner, Department of Bacteriology/TB Section, Swedish Institute for Infectious Disease Control, Solna, Sweden) was obtained from the blood of an AIDS patient, and M. paratuberculosis Linda (kindly provided by Ingrid Olsen, Department of Animal Health, National Veterinary Institute, Oslo, Norway) was from a patient with Crohn's disease.
Isolation of primary monocytes, in vitro cell differentiation, and HIV-1 infection.
Buffy coats and human serum (HS) from anonymous, healthy, HIV-negative blood donors that undergo routine screening in accordance with national guidelines were obtained from the blood bank at Bergen Hospital Trust, Haukeland University Hospital, Bergen, Norway. Peripheral blood mononuclear cells (PBMC) were isolated from a total of 25 donors (5 donors for each of the five subsets of experiments) by centrifugation on a Lymphoprep (Axis-Shield PoC AS, Oslo, Norway) density gradient as previously described (18). Blood monocytes were differentiated into monocyte-derived macrophages (MDM) by culturing total PBMC in 12-well Nunc plates at a density of 4 × 106 cells/ml at 37°C/5% CO2 in RPMI 1640 medium with glutamine and 20% pooled, heat-inactivated HS for 5 days. On day 5 postisolation, nonadherent cells were removed by washing three times with phosphate-buffered saline (PBS) and the adherent monolayers of monocytes were allowed to mature for an additional 2 days by further culturing in RPMI 1640 medium with glutamine and 10% HS. Seven days postisolation, half of the MDM from each donor were infected with HIV-1 strain BaL at a viral titer corresponding to 5 ng/ml p24 in cell culture medium. Cells were incubated overnight with HIV-1 at 37°C in 5% CO2, and the medium was changed. The cells were left undisturbed for another 7 days in order to get the HIV-1 infection established. Prior to mycobacterial infections, on day 14 postisolation, supernatants were collected from the HIV-1-infected MDM to confirm a productive HIV-1 infection.
Mycobacterial strains and infection.
On day 14 postisolation, MDM either with or without HIV-1 infection were incubated overnight (16 h) at 37°C in 5% CO2 with the specified mycobacteria at an estimated multiplicity of infection (MOI) of 5 CFU per macrophage. The CFU count used for infection was calculated on the basis of the predetermined CFU count of each frozen stock of mycobacteria. However, to asses the viability of the inocula used in each experiment, the inocula were also seeded onto 7H10 agar plates. Expressed as a percentage of the predetermined CFU count, the mean CFU count of the inoculums (range) for the respective mycobacteria were as follows: M. tuberculosis, 65% (60 to 75%); M. avium, 94% (79 to 107%); M. paratuberculosis, 67% (60 to 71%). Hence, the retrospective corrected mean estimated MOIs of the mycobacteria were 3.3, 4.7, and 3.4 CFU per macrophage, respectively. On the next day, the cells were washed three times with PBS to remove any extracellular bacteria. Cells infected with M. tuberculosis H37Rv or M. avium were cultured for 1 week, while cells infected with M. paratuberculosis were cultured for 14 days. At specified time points after mycobacterial infection (days 1, 4, and 7 for M. tuberculosis H37Rv and M. avium and days 1, 7, and 14 for M. paratuberculosis), supernatants were harvested from at least three parallel wells for each donor, sterile filtered, and stored at −80°C for later p24 enzyme-linked immunosorbent assay (ELISA) and cytokine analysis. The remaining adherent control and infected MDM were lysed in autoclaved water, and lysate aliquots were stored in cryotubes containing 250 μl of acid-washed glass beads ≤106 μm in size (Sigma-Aldrich, Norway) at −80°C for later DNA extraction and use in quantitative real-time PCR (qPCR). Appropriate dilutions of the same lysates from mycobacterium-infected MDM were also seeded onto OADC-enriched Middlebrook 7H10 agar plates. For M. paratuberculosis growth determination, the agar plates were additionally enriched with Ferric Mycobactin J (Allied Monitor Inc., Fayette, MO). The CFU of M. tuberculosis H37Rv and M. avium were counted after 3 to 4 weeks, while the CFU of M. paratuberculosis were counted after 6 to 8 weeks.
DNA extraction from cultured cells for qPCR.
Mycobacteria were inactivated by boiling the cryotubes containing lysate and glass microbeads in a water bath for 20 min. Subsequently, the mycobacteria were mechanically disrupted by bead beating using a Ribolyser (Hybaid, United Kingdom) at maximum speed (6.5 m/s) for 45 s and the resulting crude extract containing both mycobacterial and human DNAs was used directly in the qPCR assay without further purification.
Quantification of bacilli and viable human macrophages by qPCR for the mycobacterial heat shock protein 65 gene GroEL2 and the human β-globulin gene.
Mycobacterial bacilli and human cells were enumerated by an in-house duplex qPCR for the mycobacterial heat shock protein 65 gene GroEL2 and the human β-globulin gene using the QuantiTect Multiplex PCR kit (catalog no. 204543; Qiagen, West Sussex, United Kingdom) in accordance with the manufacturer's instructions for TaqMan Probes with final concentrations of all primers and probes of 0.4 and 0.2 μM, respectively. A 150-bp-long segment of the human β-globulin gene was amplified using primers BG-F (5′-TGCCTATCAGAAAGTGGTGGCT-3′) and BG-R (5′-GCTCAAGGCCCTTCATAATATCC-3′) and the BG-TAQ probe (5′-TGGCTAATGCCCTGGCCCACAA-3′) as previously described (8). However, due to the need for multiplexing and optimal performance, a TaqMan-MGB probe tagged with 6-carboxyfluorescein (FAM) was used instead of the FAM- and 6-carboxytetramethylrhodamine-tagged TaqMan probe originally described (8). A 103-bp-long segment of the mycobacterial GroEL2 gene was amplified using primers MycoFP1 (5′-CGAGGCGATGGACAAGGT-3′) and TB 12 (5′-CTTGTCGAACCGCATACCCT-3′) with the VIC-tagged TaqMan-MGB probe MycoPr1 (5′-AACGAGGGCGTCATCACCGTCG-3′) as described by us earlier (1). The duplex qPCR was performed with a 7500 Fast-Real-Time System (Applied Biosystems, Foster City, CA) under the following thermal cycling conditions: 95°C for 15 min to activate Hot Start Taq DNA polymerase, followed by 40 cycles of 94°C for 1 min and 60°C for 1 min. Standard curves were included in each qPCR run and were generated from known dilutions of commercially available human male genomic DNA (Applied Biosystems) and M. tuberculosis H37Rv genomic DNA available through the TB Vaccine Testing and Research Materials Contract (Mycobacteria Research Laboratories, Colorado State University, Fort Collins).The amount of DNA in each human or mycobacterial DNA standard was converted to the number of human or mycobacterial targets by simply dividing by the estimated molecular mass of the haploid human genome (3.3 pg) or mycobacterial genome (4.8 fg), as each amplicon is a single-copy element in the respective genome.
These data were used to calculate the number of bacilli (1:1 ratio of bacterial cells per genome) and macrophages (1:2 ratio of human cells per haploid genome) per well. The number of macrophages thus obtained represents the total number of adherent cells in a given well that were not lost during harvesting. This was used as a measure of viable cells since dead macrophages detach, as previously shown by DesJardin et al. (7) and are lost upon washing.
Cytokine detection in MDM culture supernatants.
Due to cost restraints, it was not possible to run cytokine assays on all of the biological replicates collected from a donor for a given time point and treatment group. Therefore, based on the macrophage viability data, the replicate well closest to the cluster mean of each donor from each time point and treatment group was selected as representative and the culture medium was used for cytokine profile analyses. The amounts of secreted granulocyte-macrophage colony-stimulating factor (GM-CSF), gamma interferon (IFN-γ), interleukin-1β (IL-1β), IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, and tumor necrosis factor alpha (TNF-α) were determined using the human cytokine 10-plex antibody bead kit (LHC0001; Invitrogen) according to the instructions of the manufacturer. Data were acquired on a Luminex 100 System.
Detection of HIV-1 p24 protein.
The HIV-1 p24 protein was used as a marker of productive infection and to estimate viral replication. The amount of HIV-1 p24 in supernatants from HIV-1-infected wells was determined by ELISA with the Vironostika HIV-1 Antigen kit (bioMérieux, France). The assay was performed according to the manufacturer's instructions.
Effect of HIV-1 Tat protein on bacterial growth.
High-performance liquid chromatography purified recombinant HIV-1 His-tagged Tat protein was kindly provided by Dag Helland, Department of Molecular Biology, University of Bergen, Bergen, Norway. To study the effects of the HIV-1 Tat protein on the growth of M. tuberculosis and M. avium, half of the bacterial inocula were incubated with HIV-1 Tat protein at a concentration of 1 μg/ml and incubated for 30 min at room temperature. Subsequently, macrophages were infected overnight as described above before being washed three times with PBS to remove any extracellular bacteria and viral Tat protein. MDM from five different donors were infected on day 7 postisolation, and mycobacterial growth induced by bacterial inocula exposed to Tat protein was compared to growth induced by inocula not exposed to Tat protein. Mycobacterial growth was determined by qPCR on days 1, 4, and 7 after mycobacterial infection.
Statistical analysis.
For statistical analysis, the statistical software packages SPSS version 15.0 (SPSS Inc., Chicago, IL) and R version 2.7.1 (The R Foundation for Statistical Computing, Vienna, Austria) were used. As replicate wells with macrophages from each donor were subjected to four treatments (uninfected control, mycobacterial infection, mycobacterial and HIV-1 coinfection, and HIV-1 infection) and data were collected longitudinally, a linear mixed-effect model (LMM) was fitted to log-transformed data using the Nonlinear and Linear Mixed-Effects (nlme) package for R (26), allowing for clustering at the level of donor and interactions of mycobacterial and HIV-1 infection with each other and with time. A P value of <0.05 was considered significant.
It is possible to perform a priori sample size calculation for LMM by simulations, but then a priori estimates for standard deviations both for repeat measurements within a donor and for measurements between different donors are needed; such information was not available at the planning stage. However, to justify the number of donors used in each subset of experiments, an a priori sample size calculation was based on a paired t test, assuming (i) a 2-fold difference in the mycobacterial CFU count between the singly infected and coinfected groups to be biologically relevant, (ii) pairing correlation set at 0.5, and (iii) the standard deviation of the log CFU count in each group estimated from a previous publication to be 0.3 (30). Under these conditions, the minimum number of donors required in each subset for attaining a power of 80% is four. Thus, a sample size of five donors for each subset of experiments with multiple repeat measurements will most likely have a power of no less than 80% using the LMM procedure, which better utilizes all of the available information in the data than the simpler paired t test does.
RESULTS
M. tuberculosis/HIV-1 coinfection decreases macrophage viability synergistically, while M. avium/HIV-1 coinfection causes an additive reduction and M. paratuberculosis/HIV-1 coinfection has no deleterious effect on macrophage viability.
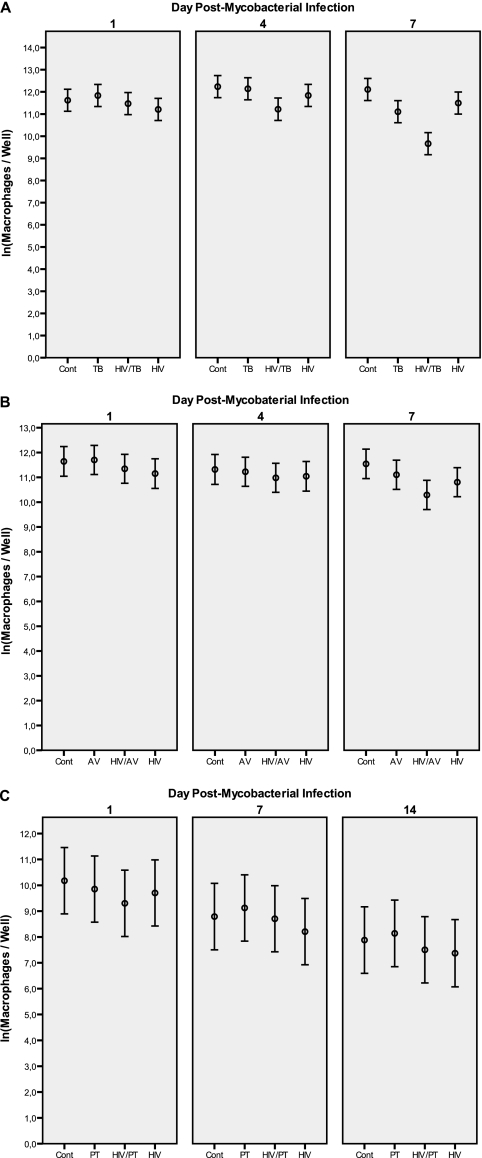
As shown in Fig. 1A, the number of viable macrophages from five separate donors declined gradually in all singly infected or M. tuberculosis/HIV-1 doubly infected cultures during the 7-day period compared to that in uninfected controls. Overall, there was a statistically significant interaction between M. tuberculosis infection and time (P = 0.002), indicating that M. tuberculosis infection by itself caused an accelerated decline in macrophage viability over time, irrespective of HIV-1 infection. However, as summarized in Table 1, on day 7 after M. tuberculosis infection, there was a statistically significant interaction in M. tuberculosis and HIV-1-coinfected cell cultures, showing a considerable synergistic effect of the pathogens which further reduced macrophage viability by a factor of 2.3 (95% confidence interval [CI], 1.1 to 4.6; P = 0.02). Although statistical significance was not reached on day 4 after M. tuberculosis infection, there was a similar synergistic trend of an aggravated decline in macrophage viability by a factor of 1.7 (95% CI, 0.8 to 3.4; P = 0.14).
FIG. 1.
Macrophage viability over time in experiments with M. tuberculosis (A), M. avium (B), and M. paratuberculosis (C). The reduction in the number of viable macrophages is clearly more evident in M. tuberculosis/HIV-1-coinfected cultures. Cont, uninfected control macrophage cultures; HIV, HIV-1-infected cultures; TB, M. tuberculosis-infected cultures, AV, M. avium-infected cultures; PT, M. paratuberculosis-infected cultures.
TABLE 1.
Effects of mycobacterial infection and HIV-1 infection on macrophage viability after 7 days of culture compared to that of uninfected controls
| Treatment or parameter | Relative (n-fold) reduction in no. of viable macrophages vs uninfected controls 7 days after mycobacterial infectiona for data set: |
||
|---|---|---|---|
| Ab (M. tuberculosis) | Bc (M. avium) | Cd (M. paratuberculosis) | |
| Mycobacterial infection only | 2.7 (1.7-4.5; <0.001) | 1.5 (1.03-2.3; 0.037) | 0.7 (0.4-1.3; 0.24) |
| HIV-1 infection onlye | 1.9 (1.1-3.0; 0.015) | 2.1 (1.4-3.1; 0.004) | 1.8 (1.01-3.2; 0.045) |
| Mycobacterial/HIV-1 coinfection | 11.6 (7.0-18.9; <0.0001) | 3.5 (2.3-5.3; <0.0001) | 1.1 (0.6-1.9; 0.77) |
| Synergy factor for mycobacterial/HIV-1 coinfection | 2.3 (1.1-4.6; 0.02) | 1.1 (0.6-1.9; 0.79) | 0.85 (0.4-1.9; 0.68) |
Values are the fitted geometric means (95% CI; P value) derived from the respective data sets.
Data set A consists of a total of 215 observations from replicate wells over time from five different donors, with 17 to 18 observations per group per time point.
Data set B consists of a total of 235 observations from replicate wells over time from five different donors, with 17 to 21 observations per group per time point.
Data set C consists of a total of 166 observations from replicate wells over time from five different donors, with 11 to 15 observations per group per time point.
The data for the HIV-1 infection only group may be pooled for all donors from data sets A, B, and C, totaling 15 donors, as this is common to all donors, irrespective of mycobacterial infection, and give a reduction of 2.0-fold (95% CI, 1.5- to 2.8-fold; P < 0.0001).
Similarly, as illustrated in Fig. 1B, there was a decline in the number of viable cells over the 7-day culture period in M. avium/HIV-coinfected cultures compared to that in uninfected controls. However, the reduction in macrophage viability was additive (Table 1) and not synergistic, as seen for the M. tuberculosis/HIV-1 coinfection.
In contrast to both M. tuberculosis and M. avium, the less virulent species M. paratuberculosis did not affect macrophage viability compared to that of uninfected controls during the 14-day culture period, as shown in Fig. 1C. Also distinct from the other double infections tested, the viability of macrophages remained unaffected in M. paratuberculosis/HIV-1-coinfected cultures at day 7 after M. paratuberculosis infection (Table 1). Although M. paratuberculosis was cultured for a longer time period in macrophages due to its very low replication rate, the overall trend remained similar at day 14.
Greater mycobacterial burden in HIV-1-infected macrophages.
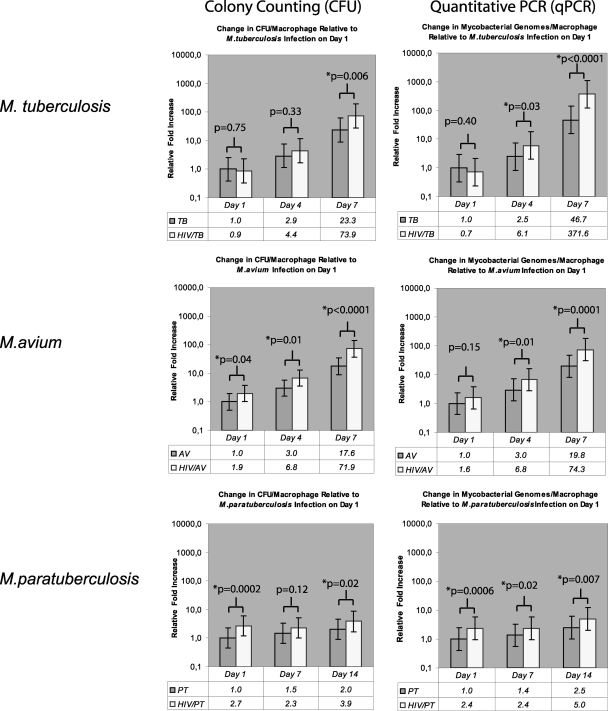
As illustrated in Fig. 2, a greater mycobacterial load, determined by both CFU counting and qPCR, was observed in HIV-1-infected macrophages for all three mycobacterial species tested. At the experimental endpoint, the HIV-1-induced relative increase in the number of mycobacteria was greatest for M. tuberculosis, closely followed by M. avium, but only modestly greater for M. paratuberculosis compared to the respective mycobacterial burdens in non-HIV-1-infected macrophage cultures. For M. tuberculosis, the observed relative increase at the endpoint was 8.0-fold (95% CI, 3.6- to 17.4-fold; P < 0.0001) as measured by qPCR and 3.2-fold (95% CI, 1.4- to 7.1-fold; P = 0.006) as measured by CFU counting. The relative increases determined by qPCR and CFU counting were 3.8-fold (95% CI, 2.0- to 7.2-fold; P < 0.001) and 4.0-fold (95% CI, 2.1- to 7.8-fold; P < 0.0001) for M. avium and 2.0-fold (95% CI, 1.2- to 3.3-fold; P = 0.007) and 1.9-fold (95% CI, 1.1- to 3.2-fold; P = 0.02) for M. paratuberculosis, respectively.
FIG. 2.
Mycobacterial burden, i.e., the number of bacteria at a given time point, in macrophage cultures with HIV-1 coinfection versus HIV-1-negative control cultures as determined by colony counting and qPCR. For ease of comparison of the mycobacterial species, all mycobacterial burden values are presented relative to the respective mycobacterial burden in HIV-1-negative control cultures on day 1 after mycobacterial infection. The P values presented are for the comparison of the mycobacterial burden at each discrete time point in HIV-1-coinfected and HIV-1-negative control cultures. HIV, HIV-1-infected cultures; TB, M. tuberculosis-infected cultures, AV, M. avium-infected cultures; PT, M. paratuberculosis-infected cultures.
HIV-1 infection of macrophages induced an increase in the initial number of cell-associated M. paratuberculosis and M. avium, but not M. tuberculosis, bacilli.
The initial number of cell-associated mycobacteria was determined on day 1 after overnight incubation (16 h) by both CFU counting and qPCR. As all of the mycobacterial species in this study have slow growth kinetics, the contribution of any mycobacterial growth during the overnight incubation was regarded as negligible.
Notably, as shown in Fig. 2, HIV-1 infection of macrophages enhanced the initial number of cell-associated M. paratuberculosis bacilli the most. Measured by CFU counting, a 2.7-fold increase (95% CI, 1.6- to 4.6-fold; P < 0.001) was observed and a similar increase of 2.4-fold (95% CI, 1.5- to 3.8-fold; P < 0.001) was verified by qPCR. For M. avium, HIV-1 infection of macrophages augmented the initial number of cell-associated bacilli 1.9-fold (95% CI, 1.1- to 3.7-fold; P = 0.038) as determined by CFU counting. This trend was confirmed by qPCR with an increase of 1.6-fold (95% CI, 0.8- to 3.0-fold; P = 0.15) without reaching statistical significance. In contrast, no HIV-1-induced initial enhancement of macrophage-associated bacilli was found for M. tuberculosis. Rather, a small and not statistically significant 0.9-fold (95% CI, 0.5- to 2.6-fold; P = 0.75) reduction was observed by CFU counting. Similarly a statistically not significant decrease of 0.7-fold (95% CI, 0.3- to 1.6-fold; P = 0.40) was measured by qPCR.
This may indicate a difference in phagocytosis between the HIV-1-infected and uninfected macrophages for M. paratuberculosis and M. avium, but the long incubation period of 16 h makes the observation uncertain, as differences in the level of intracellular killing of mycobacteria between the singly infected and HIV-1-coinfected cultures may also be a possible explanation.
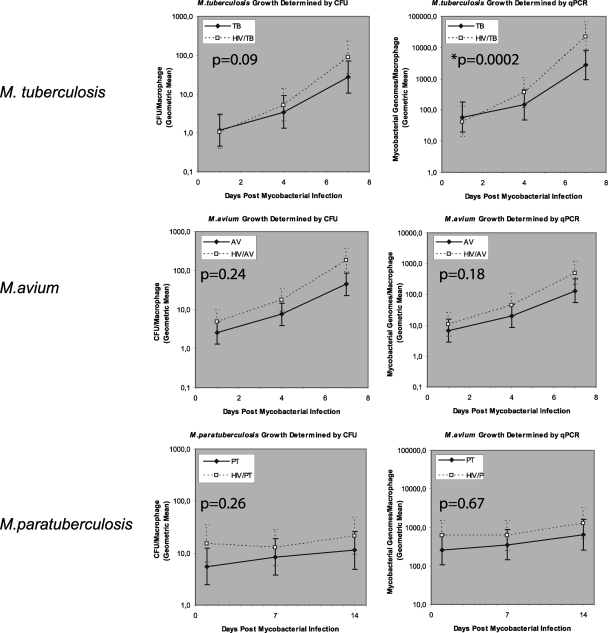
HIV-1 induces accelerated growth of M. tuberculosis but not of M. avium or M. paratuberculosis.
Utilizing the same raw data as in Fig. 2, Fig. 3 depicts the paired growth curves of all three mycobacterial species in macrophage cultures with or without HIV-1 coinfection. However, the statistical analysis in Fig. 3 is distinct from that presented in Fig. 2. While Fig. 2 compares the mycobacterial burdens at each discrete time point, the analysis presented in Fig. 3 evaluates the effect of HIV-1 on the growth rates of the different mycobacteria, i.e., the effect of HIV-1 interacting with time. As shown in Fig. 3, no significant contribution to mycobacterial growth is attributable to the interaction of HIV-1 with time for M. avium or M. paratuberculosis and all P values are above the significance level of 0.05, irrespective of whether growth was determined by qPCR or CFU counting. In contrast, for M. tuberculosis, an interaction of HIV-1 with time does occur which accelerates growth as measured by qPCR (P < 0.001) and is visually reflected in the steeper gradient of the mycobacterial growth curve in doubly infected cultures. The interaction for the entire culture period is marginally insignificant when growth is measured by CFU counting (P = 0.09). To further illustrate the impact of HIV-1 infection on growth kinetics, the mean generation times of M. tuberculosis and M. avium (Table 2) were estimated by assuming a simple first-order bacterial growth model (17). As shown in Table 2 and as expected from the overall P values in Fig. 3, the mean generation time of M. tuberculosis from day 1 to day 7 determined by qPCR is statistically significantly different in HIV-1-coinfected cultures from that in non-HIV-1-infected cultures and the data show nonoverlapping 95% CIs. Notably, the 95% CIs for the corresponding overall generation times determined by CFU counting overlap marginally, as expected.
FIG. 3.
Mycobacterial growth, i.e., the increase in the number of bacteria over time, in infected macrophages without (solid lines) or with HIV-1 coinfection (dashed lines) as determined by colony counting and qPCR. Notably, the raw data presented are the same as in Fig. 2. However, the statistical analysis is distinct, as it evaluates the effect of HIV-1 on the growth rate of the mycobacteria, i.e., the overall effect of HIV-1 coinfection on the gradient of the line graph. The growth curves of M. tuberculosis in HIV-1-negative and HIV-1-coinfected cultures diverge over time, and their gradients are statistically significantly different, as determined by qPCR. HIV, HIV-1-infected cultures; TB, M. tuberculosis-infected cultures, AV, M. avium-infected cultures; PT, M. paratuberculosis-infected cultures.
TABLE 2.
Estimated mean generation times of M. tuberculosis and M. avium in macrophage cultures with HIV-1 coinfection compared to those in non-HIV-1-infected cultures
| Treatment | Mean generation time (h) determined by: |
|||||
|---|---|---|---|---|---|---|
| CFU counting |
qPCR |
|||||
| 1-4 daysa | 4-7 days | 1-7 days | 1-4 days | 4-7 days | 1-7 days | |
| M. tuberculosis | ||||||
| HIV− | 46.4 (26.4-190.7)b | 24.1 (17.3-39.6) | 31.7 (25.2-42.7) | 55.1 (29.6-400.2) | 17.0 (13.4-23.1) | 26.0 (21.6-32.6) |
| HIV+ | 30.9 (20.4-63.3) | 17.7 (13.7-25.0) | 22.5 (19.0-27.6) | 23.3 (17.0-36.9) | 12.1 (10.2-15.0) | 16.0 (14.1-18.2) |
| M. avium | ||||||
| HIV− | 45.7 (29.0-108.3) | 28.1 (20.6-43.8) | 34.8 (28.4-44.7) | 45.6 (28.8-108.7) | 26.4 (19.7-40.1) | 33.5 (27.5-42.7) |
| HIV+ | 39.8 (26.5-80.0) | 21.2 (16.7-29.1) | 27.7 (23.7-33.6) | 34.6 (24.0-61.8) | 20.8 (16.4-28.5) | 26.0 (22.3-31.2) |
Time after mycobacterial infection.
The values in parentheses are 95% CI.
Also of note in Table 2, although HIV-1 seems to induce a reduction in the mean generation time of M. avium, this trend is not statistically significant.
M. tuberculosis, but not M. avium or M. paratuberculosis, increases HIV-1 replication in coinfected cultures.
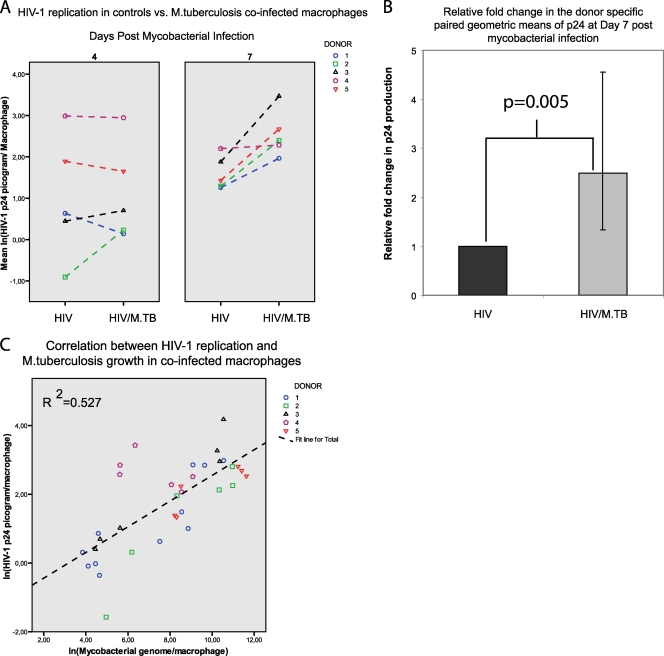
The effect of mycobacterial coinfection on HIV-1 replication was investigated in macrophages from five separate blood donors. As shown in Fig. 4, no effect was observed at day 4 after M. tuberculosis infection. However, 7 days after mycobacterial infection, M. tuberculosis induced an increase in HIV-1 replication of 2.5-fold (95% CI, 1.3- to 4.6-fold; P = 0.005), as determined by p24 production in culture supernatants relative to that of cultures infected with HIV-1 only. A strong correlation (Pearson correlation coefficient = 0.73; P < 0.001) between HIV-1 replication and M. tuberculosis growth as determined by qPCR was observed in coinfected macrophage cultures. The correlation was similar (Pearson correlation coefficient = 0.63; P < 0.001) when M. tuberculosis growth was determined by CFU counting. Coinfection with M. avium and M. paratuberculosis had no effect on HIV-1 replication (data not shown).
FIG. 4.
Donor-specific paired comparison of HIV-1 replication in HIV-1-infected control cultures versus HIV-1/M. tuberculosis-coinfected (HIV/M.TB) cultures (A) shows no difference at day 4 after mycobacterial infection. However, a difference is shown at day 7 and coinfection with M. tuberculosis induces a statistically significant 2.5-fold increase in viral replication, as determined by HIV-1 p24 protein production in culture supernatants (B; error bar, 95% CI). There is a strong correlation (Pearson correlation coefficient = 0.73; P < 0.001) between HIV-1 replication and M. tuberculosis growth in coinfected macrophages (C).
Increased proinflammatory cytokines in HIV-1/M. tuberculosis-coinfected cultures.
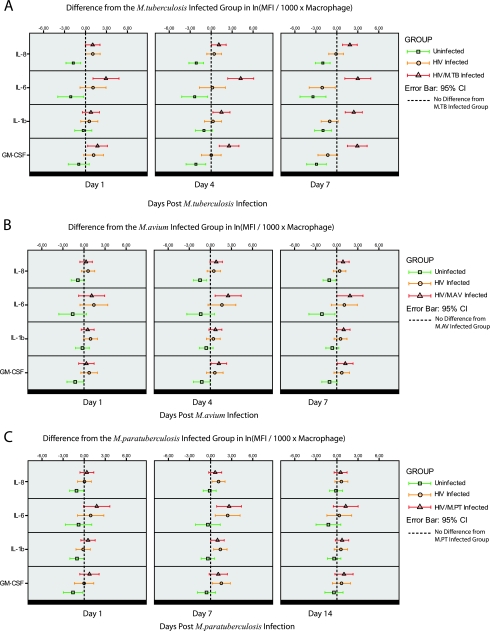
HIV/AIDS patients are known to have a chronically activated immune system, and coinfections with mycobacteria may induce macrophages to produce proinflammatory cytokines (19, 31). Hence, culture supernatants from one representative biological sample for each time point and treatment group from each donor (see Materials and Methods for selection criteria) were analyzed for macrophage-associated proinflammatory cytokines (GM-CSF, IL-1β, IL-6, IL-8, and TNF-α). Additionally, as we employed classical plastic adherence for MDM generation (a method which is prone to some contamination by T cells), we also examined cytokines typical of T-helper 1/T-helper 2 responses (IL-2, IL-4, IL-5, IL-10, and IFN-γ). Since T-cell-derived cytokines have profound effects on the phenotype of macrophages and their ability to control mycobacterial growth, it is important to control for this when interpreting the data and acknowledge the influence of any contaminating T cells. All of the measured T-helper 1/T-helper 2 cytokines were below the level of detection or borderline in our macrophage cultures, which confirmed the light microscopic examination finding that the cultures consisted of macrophages only. Four of the proinflammatory cytokines were detected (IL-6, IL-8, IL-1β, and GM-CSF), and the results are summarized in Fig. 5. Figure 5A shows that HIV-1/M. tuberculosis-coinfected cultures displayed statistically significantly increased levels of IL-6, IL-8, IL-1β, and GM-CSF compared to those in only M. tuberculosis-infected cultures over the 7-day time period. Compared to that in the M. tuberculosis-infected cultures, the mean fluorescence intensity (MFI) per 1,000 macrophages in the coinfected cultures, measured 7 days after bacterial infection, increased 6.5-fold for IL-8 (95% CI, 2.3- to 18.5-fold; P < 0.001), 20.1-fold for IL-6 (95% CI, 3.1- to 129.0-fold; P = 0.002), 11.3-fold for IL-1β (95% CI, 3.3- to 38.5-fold; P < 0.001), and 18.7-fold for GM-CSF (95% CI, 4.6- to 76.4-fold; P < 0.001). A similar tendency of increased cytokine levels was noted in HIV-1/M. avium-coinfected cultures (Fig. 5B), but at day 7, statistical significance was reached only for GM-CSF with a 3.4-fold (95% CI, 1.1- to 10.2-fold; P = 0.03) increase in MFI per 1,000 macrophages. No difference was found between HIV-1/M. paratuberculosis-coinfected cultures and M. paratuberculosis-infected cultures at day 14.
FIG. 5.
Cytokine profiles of different treatment groups compared to those of the respective singly mycobacterium-infected groups. The dashed line represents the line of no difference from the respective mycobacterium-infected group. A statistically significant increase in the MFI per 1,000 macrophages for IL-8, IL-6, IL-1β, and GM-CSF is shown for the HIV-1/M. tuberculosis-coinfected group (A), as the respective CI does not intersect with the dashed line of no difference. HIV, HIV-1-infected cultures; M.TB, M. tuberculosis-infected cultures, M.AV, M. avium-infected cultures; M.PT, M. paratuberculosis-infected cultures.
HIV-1 Tat protein had no effect on macrophage viability or the growth of M. tuberculosis and M. avium.
In a previous study, it was shown that the HIV-1 Tat protein specifically binds to the surface of M. avium and enhances intracellular mycobacterial growth (5). We therefore wanted to examine the effect of purified HIV-1 Tat protein on macrophage viability and the growth of M. tuberculosis and M. avium. Growth was determined by qPCR. We found no significant effect of HIV-1 Tat protein on macrophage viability or the growth of M. tuberculosis or M. avium (data not shown).
DISCUSSION
In the present study, we found that HIV-1 and M. tuberculosis interact in macrophage cultures to synergistically reduce macrophage viability, to reciprocally enhance the ability of the pathogens to replicate, and to induce increased levels of proinflammatory cytokines. Furthermore, in HIV-1-infected macrophage cultures, subsequent coinfection with M. avium or M. paratuberculosis results in a greater mycobacterial burden. The mechanism underlying this increased mycobacterial burden seems to be distinct for the species examined, as we found that HIV-1 induces accelerated growth of M. tuberculosis, whereas the growth kinetics of M. avium and M. paratuberculosis remained unchanged. Also, in contrast to the situation seen for M. tuberculosis, for M. avium and M. paratuberculosis, HIV-1 coinfection induced no or only marginal increases in the levels of proinflammatory cytokines.
Macrophages play an important role in the pathogenesis of both M. tuberculosis infection and HIV-1 infection. Not only are they key innate immune cells responsible for sensing and initiating appropriate immune responses, but they also function as a reservoir for the intracellular replication of both pathogens. There is considerable evidence supporting the notion that HIV-1 infection of monocytes/macrophages impairs innate immune responses to bacterial infections (reviewed in reference 25). Thus, it is tempting to suggest that the basis for the increased susceptibility of HIV+ patients to mycobacterial infections may at least partly be linked to dysfunction of the antimicrobial activities of their macrophages. Our data support such a view, especially for M. tuberculosis. However, there are several conflicting publications related to the effects of HIV-1 on the ability of macrophages to control mycobacterial infection. Some studies indicate that HIV-1 has no effect (6, 15, 21), whereas other reports, in agreement with our findings, show that HIV-1 infection of macrophages enhances mycobacterial growth (4, 10, 13, 14, 24). A direct comparison of these studies is difficult, as the experimental conditions and the sources of the macrophages used differ. Hence, the possibility that the different experimental setups may partly explain the conflicting results cannot be excluded. However, some general considerations as to why there are differing findings in the literature should be taken into account. First, one needs to appreciate that, biologically, macrophages consist of a heterogeneous population of cells with tissue-specific phenotypes coupled with a dynamic activation spectrum depending on the local cytokine/chemokine milieu (reviewed in reference 22). Many studies, including this one, have used monocyte-derived macrophages, which are classically activated by adherence to plastic, while others, like the study by Denis and Ghadirian (6), used alveolar macrophages. Hence, discordant results may arise simply because the macrophage phenotypes differ in vitro. Second, we studied mycobacterial growth over time with measurements at several time points. This is important, as measuring mycobacterial growth at only one time point may give misleading conclusions if the time allowed for growth is too short. Since the mean generation time of mycobacteria is relatively long and in our experience there is also an initial lag phase with especially slow growth when thawed mycobacterial stocks are used for inoculation, appropriate time needs to be allowed for adequate growth to occur in order for differences to be detectable. Thus, had we also done our measurements only at day 4 postinfection, as in the study by Kalsdorf et al. (15), we would have also found no difference between the M. tuberculosis- and M. tuberculosis/HIV-1-infected cultures. Third, some studies, like that of Meylan et al. (21), have concluded that HIV does not enhance mycobacterial growth. However, the statistical power of such a conclusion is questionable since the experiments were conducted with macrophages from only three donors. Nonetheless, clinically it is well established that there is an unfortunate synergism between HIV-1 and M. tuberculosis already at an early time point in coinfected patients prior to the collapse of adaptive immunity and that these patients experience greater morbidity and mortality. Our findings of a reciprocal enhancement of replication of both M. tuberculosis and HIV-1 in coinfected macrophage cultures are in line with this and may, in part, explain the cellular basis for the accelerated pathology seen in coinfected patients.
Subsequent to phagocytosis of pathogens, macrophages typically express membrane markers of activation and secrete proinflammatory cytokines and chemokines. Normally, these cytokines and chemokines act in vivo to control microbial dissemination by recruiting peripheral blood lymphocytes and monocytes to the site of inflammation and thereby help to initiate an appropriate immune response. However, several studies have shown that, in the context of M. tuberculosis/HIV-1 coinfection, these proinflammatory signals may be dysregulated and strongly enhance viral and/or mycobacterial replication (9, 19, 20, 32). Our findings of increased HIV-1 replication and simultaneously elevated levels of IL-6, IL-8, IL-1β, and GM-CSF in M. tuberculosis-coinfected cultures are consistent with these reports and imply a correlation between increased proinflammatory cytokines/chemokines and enhanced transcriptional activation of HIV-1 in macrophages. Interestingly, the long terminal repeats (LTR) of the integrated proviral HIV-1 DNA have binding sites for transcriptional factors like NF-κB, NF-IL-6, and C/EBP, which are also commonly present in the genes of several proinflammatory cytokines and chemokines (reference 12 and references therein). Several studies have linked increased HIV-1 replication in macrophages to the induction of these transcription factors and shown a correlation to simultaneous increases in proinflammatory cytokines and chemokines (reference 12 and references therein). M. tuberculosis has also been shown to be specifically able to activate the LTR of HIV-1 and enhance viral replication (16). Notably, there was no increase in these cytokines/chemokines in cultures coinfected with either M. avium or M. paratuberculosis relative to those in cultures infected with HIV-1 alone. Also, correspondingly, there was no increase in viral replication in these doubly infected cultures.
HIV-1 induced accelerated growth of M. tuberculosis and reduced its mean generation time in coinfected macrophage cultures. To our knowledge, this has not been demonstrated previously, although increased growth of M. tuberculosis in HIV-1-infected macrophage cultures has been shown (13). Previous studies on M. tuberculosis H37Rv growth in macrophages without HIV-1 coinfection have shown mean generation times similar to our findings (3). It has also been shown that treatment of macrophages with glucocorticoid steroids may accelerate the growth of M. tuberculosis (29). Hence, it is biologically conceivable that immunosuppression at the cellular level may accelerate the growth kinetics of M. tuberculosis in macrophages.
In conclusion, this study presents data demonstrating that coinfection of macrophages with HIV-1 and mycobacteria may give rise to an increased cellular bacterial burden. Coinfection with HIV-1 and M. tuberculosis is especially detrimental to the host cell, and the microorganisms mutually enhance each other's replication. The synergism at the cellular level between HIV-1 and M. tuberculosis seems to be specific and not shared by other mycobacterial species like M. avium or M. paratuberculosis. In an attempt to explore the mechanism of this synergism, the effects of the viral Tat protein were investigated but the results of these experiments were inconclusive. Interestingly, our overall findings mirror the ability of these mycobacteria to cause clinical disease in HIV+ patients. Still, additional studies are needed to further elucidate the complex interactions between HIV-1 and mycobacteria.
Acknowledgments
We thank Karl Albert Brokstad for his guidance and technical assistance when running Luminex assays.
This study was supported by the University of Bergen and Regional Health Authorities of Western Norway project 911239 (to S.P.).
There are no conflicts of interest for any of us.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 12 July 2010.
REFERENCES
- 1.Baba, K., S. Pathak, L. Sviland, N. Langeland, A. A. Hoosen, B. Asjo, A. M. Dyrhol-Riise, and T. Mustafa. 2008. Real-time quantitative PCR in the diagnosis of tuberculosis in formalin-fixed paraffin-embedded pleural tissue in patients from a high HIV endemic area. Diagn. Mol. Pathol. 17:112-117. [DOI] [PubMed] [Google Scholar]
- 2.Berger, B. J., F. Hussain, and K. Roistacher. 1994. Bacterial infections in HIV-infected patients. Infect. Dis. Clin. North Am. 8:449-465. [PubMed] [Google Scholar]
- 3.Chanwong, S., N. Maneekarn, L. Makonkawkeyoon, and S. Makonkawkeyoon. 2007. Intracellular growth and drug susceptibility of Mycobacterium tuberculosis in macrophages. Tuberculosis (Edinburgh) 87:130-133. [DOI] [PubMed] [Google Scholar]
- 4.Crowle, A. J., D. L. Cohn, and P. Poche. 1989. Defects in sera from acquired immunodeficiency syndrome (AIDS) patients and from non-AIDS patients with Mycobacterium avium infection which decrease macrophage resistance to M. avium. Infect. Immun. 57:1445-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denis, M. 1994. Tat protein from HIV-1 binds to Mycobacterium avium via a bacterial integrin. Effects on extracellular and intracellular growth. J. Immunol. 153:2072-2081. [PubMed] [Google Scholar]
- 6.Denis, M., and E. Ghadirian. 1994. Interaction between Mycobacterium avium and human immunodeficiency virus type 1 (HIV-1) in bronchoalveolar macrophages of normal and HIV-1-infected subjects. Am. J. Respir. Cell Mol. Biol. 11:487-495. [DOI] [PubMed] [Google Scholar]
- 7.DesJardin, L. E., T. M. Kaufman, B. Potts, B. Kutzbach, H. Yi, and L. S. Schlesinger. 2002. Mycobacterium tuberculosis-infected human macrophages exhibit enhanced cellular adhesion with increased expression of LFA-1 and ICAM-1 and reduced expression and/or function of complement receptors, FcgammaRII and the mannose receptor. Microbiology 148:3161-3171. [DOI] [PubMed] [Google Scholar]
- 8.Eishi, Y., M. Suga, I. Ishige, D. Kobayashi, T. Yamada, T. Takemura, T. Takizawa, M. Koike, S. Kudoh, U. Costabel, J. Guzman, G. Rizzato, M. Gambacorta, R. du Bois, A. G. Nicholson, O. P. Sharma, and M. Ando. 2002. Quantitative analysis of mycobacterial and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J. Clin. Microbiol. 40:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrait, V., J. Cadranel, H. Esvant, I. Herry, P. Morinet, C. Mayaud, and D. Israel-Biet. 1997. Tuberculosis generates a microenvironment enhancing the productive infection of local lymphocytes by HIV. J. Immunol. 159:2824-2830. [PubMed] [Google Scholar]
- 10.Ghassemi, M., B. R. Andersen, V. M. Reddy, P. R. Gangadharam, G. T. Spear, and R. M. Novak. 1995. Human immunodeficiency virus and Mycobacterium avium complex coinfection of monocytoid cells results in reciprocal enhancement of multiplication. J. Infect. Dis. 171:68-73. [DOI] [PubMed] [Google Scholar]
- 11.Hampson, S. J., F. Portaels, J. Thompson, E. P. Green, M. T. Moss, J. Hermon-Taylor, and J. J. McFadden. 1989. DNA probes demonstrate a single highly conserved strain of Mycobacterium avium infecting AIDS patients. Lancet 1:65-68. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, A. J., X. Zou, and K. L. Calame. 1995. C/EBP proteins activate transcription from the human immunodeficiency virus type 1 long terminal repeat in macrophages/monocytes. J. Virol. 69:5337-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imperiali, F. G., A. Zaninoni, L. La Maestra, P. Tarsia, F. Blasi, and W. Barcellini. 2001. Increased Mycobacterium tuberculosis growth in HIV-1-infected human macrophages: role of tumour necrosis factor-alpha. Clin. Exp. Immunol. 123:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Källenius, G., T. Koivula, K. J. Rydgard, S. E. Hoffner, A. Valentin, B. Asjo, C. Ljungh, U. Sharma, and S. B. Svenson. 1992. Human immunodeficiency virus type 1 enhances intracellular growth of Mycobacterium avium in human macrophages. Infect. Immun. 60:2453-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalsdorf, B., T. J. Scriba, K. Wood, C. L. Day, K. Dheda, R. Dawson, W. A. Hanekom, C. Lange, and R. J. Wilkinson. 2009. HIV-1 infection impairs the bronchoalveolar T cell response to mycobacteria. Am. J. Respir. Crit. Care Med. 180:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitaura, H., N. Ohara, N. Nakao, N. Yoshida, and T. Yamada. 2002. Changed activation of HIV-1 LTR in monocytoid cells by mycobacteria with temporal progression of infection. New Microbiol. 25:357-361. [PubMed] [Google Scholar]
- 17.Lambrecht, R. S., J. F. Carriere, and M. T. Collins. 1988. A model for analyzing growth kinetics of a slowly growing Mycobacterium sp. Appl. Environ. Microbiol. 54:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukey, P. T., and E. U. Hooker. 2001. Macrophage virulence assays, p. 271-280. In T. Parish and N. G. Stoker (ed.), Methods in molecular medicine, vol. 54: Mycobacterium tuberculosis protocols. Humana Press, Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 19.Mariani, F., D. Goletti, A. Ciaramella, A. Martino, V. Colizzi, and M. Fraziano. 2001. Macrophage response to Mycobacterium tuberculosis during HIV infection: relationships between macrophage activation and apoptosis. Curr. Mol. Med. 1:209-216. [DOI] [PubMed] [Google Scholar]
- 20.Meddows-Taylor, S., D. J. Martin, and C. T. Tiemessen. 1999. Dysregulated production of interleukin-8 in individuals infected with human immunodeficiency virus type 1 and Mycobacterium tuberculosis. Infect. Immun. 67:1251-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meylan, P. R., J. R. Munis, D. D. Richman, and R. S. Kornbluth. 1992. Concurrent human immunodeficiency virus and mycobacterial infection of macrophages in vitro does not reveal any reciprocal effect. J. Infect. Dis. 165:80-86. [DOI] [PubMed] [Google Scholar]
- 22.Mosser, D. M., and J. P. Edwards. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8:958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naser, S. A., J. Felix, H. Liping, C. Romero, N. Naser, A. Walsh, and W. Safranek. 1999. Occurrence of the IS900 gene in Mycobacterium avium complex derived from HIV patients. Mol. Cell. Probes 13:367-372. [DOI] [PubMed] [Google Scholar]
- 24.Newman, G. W., T. G. Kelley, H. Gan, O. Kandil, M. J. Newman, P. Pinkston, R. M. Rose, and H. G. Remold. 1993. Concurrent infection of human macrophages with HIV-1 and Mycobacterium avium results in decreased cell viability, increased M. avium multiplication and altered cytokine production. J. Immunol. 151:2261-2272. [PubMed] [Google Scholar]
- 25.Noursadeghi, M., D. R. Katz, and R. F. Miller. 2006. HIV-1 infection of mononuclear phagocytic cells: the case for bacterial innate immune deficiency in AIDS. Lancet Infect. Dis. 6:794-804. [DOI] [PubMed] [Google Scholar]
- 26.Pinheiro, J. C., and D. M. Bates. 2000. Mixed-effects models in S and S-PLUS. Springer, New York, NY.
- 27.Richter, E., J. Wessling, N. Lugering, W. Domschke, and S. Rusch-Gerdes. 2002. Mycobacterium avium subsp. paratuberculosis infection in a patient with HIV, Germany. Emerg. Infect. Dis. 8:729-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roiz, M. P., E. Palenque, C. Guerrero, and M. J. Garcia. 1995. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J. Clin. Microbiol. 33:1389-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rook, G. A., J. Steele, M. Ainsworth, and C. Leveton. 1987. A direct effect of glucocorticoid hormones on the ability of human and murine macrophages to control the growth of M. tuberculosis. Eur. J. Respir. Dis. 71:286-291. [PubMed] [Google Scholar]
- 30.Silver, R. F., Q. Li, and J. J. Ellner. 1998. Expression of virulence of Mycobacterium tuberculosis within human monocytes: virulence correlates with intracellular growth and induction of tumor necrosis factor alpha but not with evasion of lymphocyte-dependent monocyte effector functions. Infect. Immun. 66:1190-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toossi, Z. 2003. Virological and immunological impact of tuberculosis on human immunodeficiency virus type 1 disease. J. Infect. Dis. 188:1146-1155. [DOI] [PubMed] [Google Scholar]
- 32.Toossi, Z., J. L. Johnson, R. A. Kanost, M. Wu, H. Luzze, P. Peters, A. Okwera, M. Joloba, P. Mugyenyi, R. D. Mugerwa, H. Aung, J. J. Ellner, and C. S. Hirsch. 2001. Increased replication of HIV-1 at sites of Mycobacterium tuberculosis infection: potential mechanisms of viral activation. J. Acquir. Immune Defic. Syndr. 28:1-8. [DOI] [PubMed] [Google Scholar]
- 33.Turenne, C. Y., R. Wallace, Jr., and M. A. Behr. 2007. Mycobacterium avium in the postgenomic era. Clin. Microbiol. Rev. 20:205-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. 2009. AIDS epidemic update: December 2009. World Health Organization, Geneva, Switzerland. http://data.unaids.org/pub/Report/2009/JC1700_Epi_Update_2009_en.pdf.