Abstract
Chlamydia trachomatis is the leading cause of infectious blindness worldwide and is the most commonly reported pathogen causing sexually transmitted infections. Tarp (translocated actin recruiting phosphoprotein), a type III secreted effector that mediates actin nucleation, is central to C. trachomatis infection. The phylogenetic analysis of tarP from reference strains as well as ocular, genital, and lymphogranuloma venereum (LGV) clinical isolates demonstrated an evolutionary relationship with disease phenotype, with LGV and ocular isolates branched into clades that were separate from the urogenital isolates. The sequence analysis of Tarp indicated a high degree of variability and identified trends within clinical groupings. Tarps from LGV strains contained the highest number of tyrosine-rich repeat regions (up to nine) and the fewest (two) predicted actin binding domains. The converse was noted for Tarp proteins from ocular isolates that contained up to four actin binding domains and as few as one tyrosine-rich repeat region. The results suggest that Tarp is among the few known genes to play a role in C. trachomatis adaptations to specific niches within the host.
Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections (STIs) and preventable blindness worldwide (52). The species C. trachomatis consists of more than 15 serologically defined variants, or serovars, associated with different disease states and anatomical sites of infection (31, 52, 69, 70). Despite a high degree of synteny among the genomes of C. trachomatis strains, the different serovars exhibit a remarkable degree of tissue tropism (55). Distinct diseases caused by C. trachomatis include trachoma (serotypes A to C), sexually transmitted diseases (serotypes D to K), and lymphogranuloma venereum (LGV; L1 to L3). Infections by the ocular and urogenital strains, collectively referred to as the trachoma biovar, are restricted to mucosal epithelial cells of the conjunctivae and genital tracts, respectively. The LGV biovar is more invasive and disseminates via the infection of macrophages to regional lymph nodes, where they establish a chronic granulomatis disease (52).
The serological typing of C. trachomatis strains is based on the major outer membrane protein (MOMP) (11, 71-73). Despite MOMP being the immunodominant surface antigen, the phylogenetic categorization of MOMP is not concordant with pathobiotypes or tissue tropism (1, 6, 8, 24, 44, 50, 58). Numerous typing techniques have been applied to C. trachomatis to better understand the epidemiology and pathogenesis of disease. In addition to serological typing, the sequencing of ompA (which encodes MOMP) can detect numerous trachoma and serovar genotypes. Restriction fragment length polymorphism (RFLP) (26), pulsed-field gel electrophoresis (PFGE) (48), and multilocus sequence typing (MLST) (39) analysis of ompA have been used to discriminate C. trachomatis strains within serotypes but correlate poorly with disease phenotype.
Although the diseases associated with C. trachomatis serovars are unique, the completed genome sequence of three serovars representing ocular, urogenital, and LGV strains exhibit more than 99% identity (14, 56, 66) with a high degree of synteny in gene order and content. The observed differences in tissue tropism and pathobiology of C. trachomatis serovars therefore likely are due to a relatively small number of loci. One 50-kb region of the chlamydial genome, termed the plasticity zone, exhibits a much higher degree of divergence than the remainder of the genome (47). In C. trachomatis serovar D, the plasticity zone encompasses the region between ycfV (CT152) and dbsB (CT177) (47). Within this plasticity zone are at least two loci that have been correlated with tissue tropism and disease. One of these loci, first identified in Chlamydia muridarum (47), includes three copies of a putative cytotoxin with homology to the large clostridial toxins. In C. trachomatis serovar D, only a single, disrupted partial cytotoxin-like gene is expressed (3, 13). This locus is highly polymorphic, in that all urogenital serovars express both the UDP-glucose binding and glucosyltransferase domains of the toxin, while the ocular serovars, with the exception of serovar B, encode only the UDP-glucose binding domain, and the LGV strains have both domains entirely deleted (13). Also within the plasticity zone is the trpAB operon, which encodes tryptophan synthase and a repressor. The trpAB operon distinguishes genital and LGV strains from ocular strains, as only genital and LGV strains produce a functional synthase (12, 22, 53). The phylogenetic analysis of the C. trachomatis polymorphic membrane protein (pmp) genes, which are dispersed throughout the genome, has revealed that six of the nine pmp genes (pmpB, pmpC, pmpF, pmpG, pmpH, and pmpI) from 18 serovars correlate with disease groups (29, 30, 59).
C. trachomatis, like many Gram-negative bacterial pathogens, utilizes a type III secretion system to deliver an arsenal of bacterial gene-encoded effector proteins directly into the cytosol of the host cell (4, 16, 17, 23). Tarp (translocated actin recruiting protein [CT456]), a type III secreted effector, directly induces actin polymerization at the site of chlamydial invasion (16, 37). Tarp has been found in all C. trachomatis serovars to date in addition to orthologs present in Chlamydophila pneumoniae, Chlamydophila caviae, and C. muridarum (15). The biochemical and sequence analyses of Tarp have revealed three functionally distinct domains consisting of an N-terminal tyrosine-rich repeat region, a proline-rich domain, and C-terminal Wasp homology 2 (WH2) actin binding domains (15, 37, 38).
Overall, there are variations in the number of tyrosine-rich repeats and actin binding domains of Tarp between different C. trachomatis serovars examined to date (15, 16, 37). This suggests that Tarp is adapting to the selective pressures in the host and therefore is a good candidate protein to examine for tissue tropism. Here, we present a phylogenetic analysis of tarP from reference strains as well as ocular, urogenital, and LGV clinical isolates and demonstrate a correlation between tarP and clinical phenotype. We demonstrate that LGV isolates contain the greatest number of tyrosine-rich repeat regions and the fewest actin binding domains, which is in contrast to the ocular isolates that contain up to four predicted actin binding domains and the fewest tyrosine-rich repeats. Taken together, these findings identify Tarp as another C. trachomatis gene that varies in relation to disease and tissue tropism.
MATERIALS AND METHODS
Clinical samples.
The clinical isolates used in this study are listed in Table 1. Ocular, genital, urethral, and anorectal swabs from male and female patients were obtained as previously described (7, 9, 18, 21, 28, 34, 54, 62, 67) and used for the PCR amplification of tarP. Sequences analyzed in this study were derived from specimens obtained at clinical sites in the North Bank, Lower River, Central River, and Upper River Regions, The Gambia; Nairobi, Kenya; Rombo District, Tanzania; Winnipeg, Manitoba, Canada; Ottawa, Ontario, Canada; Indianapolis, IN; San Francisco, CA; Seattle, WA; and Northern Territory, Australia. Collection sites included sexually transmitted disease clinics, family planning clinics, emergency rooms, community clinics, obstetrics and gynecology clinics, and adolescent clinics. All personal identifiers were removed from the sample before analysis.
TABLE 1.
Strains and Tarp characteristics
| Strain | Serovar | Geographical source | Clinical phenotype | Tyrosine repeat region(s)a | No. of actin binding domainsb | Tarp accession no. |
|---|---|---|---|---|---|---|
| A/HAR-13 | A | Ocular | 3(4, 4, 4) | 4 | AAX50730.1 | |
| B/TW-5/OT | B | Ocular | 3(4, 4, 4) | 4 | HM567159 | |
| Ba/AP-2/OT | Ba | Ocular | 3(4, 4, 4) | 4 | HM567160 | |
| C/TW-3/OT | C | Ocular | 3(4, 4, 4) | 3 | EU121609 | |
| D/UW-3/CX | D | Genital | 3(4, 4, 4) | 2 | AAC68056 | |
| E/BOUR | E | Genital | 2(4, 4) | 3 | HM567161 | |
| F/IC Cal-3 | F | Genital | 3(4, 4, 4) | 3 | HM567162 | |
| G/UW-524/CX | G | Genital | 3(4, 4, 4) | 3 | HM567163 | |
| H/UW-4/CX | H | Genital | 3(4, 4, 4) | 2 | HM567164 | |
| I/UW-12/UR | I | Genital | 3(4, 4, 4) | 3 | HM567165 | |
| J/UW-36/CX | J | Genital | 3(4, 4, 4) | 3 | HM567166 | |
| K/UW-31/CX | K | Genital | 3(4, 4, 4) | 3 | HM567167 | |
| L1/LGV440 | L1 | LGV | 9(4*, 4, 4, 5, 4, 4, 4, 4, 4) | 2 | HM567168 | |
| L2/LGV434 | L2 | LGV | 6(4*, 4, 5, 4, 4, 5) | 2 | AY623902 | |
| L3/LGV404 | L3 | LGV | 9(4*, 5, 4, 4, 4, 4, 4, 5, 5) | 2 | HM567169 | |
| 1 | B | The Gambia | Ocular | 2(4, 4) | 4 | HM543594 |
| 2 | B | The Gambia | Ocular | 2(4, 4) | 4 | HM543595 |
| 3 | B | The Gambia | Ocular | 2(4, 4) | 4 | HM543596 |
| 4 | A | The Gambia | Ocular | 2(4, 4) | 4 | HM543597 |
| 5 | A | The Gambia | Ocular | 2(4, 4) | 4 | HM543598 |
| 6 | A | The Gambia | Ocular | 2(4, 4) | 4 | HM543599 |
| 7 | A | The Gambia | Ocular | 2(4, 4) | 4 | HM543600 |
| 8 | B | The Gambia | Ocular | 3(4, 4, 4) | 4 | HM543601 |
| 9 | B | The Gambia | Ocular | 3(4, 4, 4) | 4 | HM543602 |
| 10 | B | The Gambia | Ocular | 3(4, 4, 4) | 4 | HM543603 |
| 11 | B | The Gambia | Ocular | 2(4, 4) | 4 | HM543676 |
| 12 | A | The Gambia | Ocular | 2(4, 4) | 4 | HM543677 |
| 13 | A | The Gambia | Ocular | 2(4, 4) | 4 | HM543678 |
| 14 | A | The Gambia | Ocular | 3(4, 4, 4) | 4 | HM543679 |
| 15 | B | The Gambia | Ocular | 3(4, 4, 4) | 4 | HM543680 |
| 16 | A | Tanzania | Ocular | 1(4) | 4 | HM543681 |
| 17 | A | Tanzania | Ocular | 1(4) | 4 | HM543682 |
| 18 | A | Tanzania | Ocular | 3(4, 4, 4) | 4 | HM543683 |
| 19 | A | Tanzania | Ocular | 3(4, 4, 4) | 4 | HM543684 |
| 20 | A | Tanzania | Ocular | 3(4, 4, 4) | 4 | HM543685 |
| 21 | Ba | Australia | Ocular | 2(4, 4) | 2 | HM543686 |
| 22 | Ba | Australia | Ocular | 2(4, 4) | 2 | HM543687 |
| 23 | C | Australia | Ocular | 2(4, 4) | 2 | HM543688 |
| 24 | C | Australia | Ocular | 2(4, 4) | 2 | HM543689 |
| 25 | C | Australia | Ocular | 2(4, 4) | 2 | HM543690 |
| 26 | C | Australia | Ocular | 2(4, 4) | 2 | HM543691 |
| 27 | C | Australia | Ocular | 2(4, 4) | 2 | HM543692 |
| 28 | C | Australia | Ocular | 2(4, 4) | 2 | HM543693 |
| 29 | C | Australia | Ocular | 2(4, 4) | 2 | HM543694 |
| 30 | C | Australia | Ocular | 2(4, 4) | 2 | HM543695 |
| 31 | C | Australia | Ocular | 2(4, 4) | 2 | HM543666 |
| 32 | C | Australia | Ocular | 2(4, 4) | 2 | HM543667 |
| 33 | C | Australia | Ocular | 2(4, 4) | 2 | HM543668 |
| 34 | C | Australia | Ocular | 2(4, 4) | 2 | HM543669 |
| 35 | C | Australia | Ocular | 2(4, 4) | 2 | HM543670 |
| 36 | C | Australia | Ocular | 2(4, 4) | 2 | HM543671 |
| 37 | E | Winnipeg | Genital | 1(4) | 3 | HM543672 |
| 38 | E | Winnipeg | Genital | 2(4, 4) | 3 | HM543673 |
| 39 | E | Winnipeg | Genital | 2(4, 4) | 3 | HM543674 |
| 40 | D | Winnipeg | Genital | 3(4, 4, 4) | 3 | HM543675 |
| 41 | D | Winnipeg | Genital | 3(4, 4, 4) | 3 | HM543604 |
| 42 | F | Winnipeg | Genital | 3(4, 4, 4) | 3 | HM543605 |
| 43 | F | Winnipeg | Genital | 2(4, 4) | 3 | HM543606 |
| 44 | I | Winnipeg | Genital | 2(4, 4) | 3 | HM543607 |
| 45 | Ia | Winnipeg | Genital | 2(4, 4) | 3 | HM543608 |
| 46 | E | Kenya | Urethral | 2(4, 4) | 3 | HM543609 |
| 47 | E | Kenya | Urethral | 2(4, 4) | 3 | HM543610 |
| 48 | E | Kenya | Urethral | 2(4, 4) | 3 | HM543611 |
| 49 | E | Kenya | Urethral | 2(4, 4) | 3 | HM543612 |
| 50 | E | Kenya | Urethral | 2(4, 4) | 3 | HM543613 |
| 51 | E | Kenya | Urethral | 2(4, 4) | 3 | HM543614 |
| 52 | E | Kenya | Urethral | 2(4, 4) | 3 | HM543615 |
| 53 | E | Kenya | Urethral | 2(4, 4) | 3 | HM543616 |
| 54 | F | Kenya | Urethral | 2(4, 4) | 3 | HM543617 |
| 55 | Ia | Kenya | Urethral | 2(4, 4) | 3 | HM543618 |
| 56 | B | Seattle | Genital | 2(4, 4) | 3 | HM543619 |
| 57 | B | Seattle | Genital | 2(4, 4) | 3 | HM543620 |
| 58 | B | Seattle | Genital | 2(4, 4) | 3 | HM543621 |
| 59 | F | Seattle | Genital | 3(4, 4, 4) | 3 | HM543622 |
| 60 | K | Seattle | Genital | 3(4, 4, 4) | 3 | HM543623 |
| 61 | J | Seattle | Genital | 3(4, 4, 4) | 3 | HM543624 |
| 62 | J | Seattle | Genital | 3(4, 4, 4) | 3 | HM543625 |
| 63 | J | Seattle | Genital | 3(4, 4, 4) | 3 | HM543626 |
| 64 | J | Seattle | Genital | 3(4, 4, 4) | 3 | HM543627 |
| 65 | J | Seattle | Genital | 3(4, 4, 4) | 3 | HM543628 |
| 66 | J | Seattle | Genital | 3(4, 4, 4) | 3 | HM543629 |
| 67 | J | Seattle | Genital | 3(4, 4, 4) | 3 | HM543630 |
| 68 | H | Seattle | Genital | 3(4, 4, 4) | 3 | HM543631 |
| 69 | H | Seattle | Genital | 3(4, 4, 4) | 3 | HM543632 |
| 70 | H | Seattle | Genital | 3(4, 4, 4) | 3 | HM543633 |
| 71 | H | Seattle | Genital | 3(4, 4, 4) | 3 | HM543634 |
| 72 | Ja | Seattle | Genital | 2(4, 4) | 3 | HM543635 |
| 73 | Ja | Seattle | Genital | 2(4, 4) | 3 | HM543636 |
| 74 | Ja | Seattle | Genital | 2(4, 4) | 3 | HM543637 |
| 75 | D | Indianapolis | Genital | 3(4, 4, 4) | 3 | HM543638 |
| 76 | D | Indianapolis | Genital | 3(4, 4, 4) | 3 | HM543639 |
| 77 | D | Indianapolis | Genital | 2(4, 4) | 2 | HM543640 |
| 78 | E | Indianapolis | Genital | 2(4, 4) | 3 | HM543641 |
| 79 | E | Indianapolis | Genital | 1(4) | 3 | HM543642 |
| 80 | E | Indianapolis | Genital | 2(4, 4) | 3 | HM543643 |
| 81 | F | Indianapolis | Genital | 3(4, 4, 4) | 3 | HM543644 |
| 82 | F | Indianapolis | Genital | 4(4, 4, 4, 4) | 2 | HM543645 |
| 83 | H | Indianapolis | Genital | 2(4, 4) | 3 | HM543646 |
| 84 | H | Indianapolis | Genital | 3(4, 4, 4) | 3 | HM543647 |
| 85 | H | Indianapolis | Genital | 4(4, 4, 4, 3) | 3 | HM543648 |
| 86 | H | Indianapolis | Genital | 3(4, 4, 4) | 3 | HM543649 |
| 87 | I | Indianapolis | Genital | 2(4, 4) | 3 | HM543650 |
| 88 | I | Indianapolis | Genital | 2(4, 4) | 2 | HM543651 |
| 89 | K | Indianapolis | Genital | 3(4, 4, 4) | 3 | HM543652 |
| 90 | K | Indianapolis | Genital | 3(4, 4, 4) | 3 | HM543653 |
| 91 | D | Ottawa | Genital | 3(4, 4, 4) | 2 | HM543654 |
| 92 | G | Ottawa | Genital | 3(4, 4, 4) | 3 | HM543655 |
| 93 | J | Ottawa | Genital | 2(4, 4) | 3 | HM543656 |
| 94 | I | Ottawa | Genital | 3(4, 4, 4) | 2 | HM543657 |
| 95 | D | Ottawa | Rectal | 3(4, 4, 4) | 3 | HM543658 |
| 96 | G | Ottawa | Rectal | 3(4, 4, 4) | 3 | HM543659 |
| 97 | J | Ottawa | Rectal | 3(4, 4, 4) | 3 | HM543660 |
| 98 | L2 | Ottawa | Rectal | 5(4*, 4, 4, 4, 5) | 2 | HM543661 |
| 99 | L2b | Amsterdam | Rectal | 6(4*, 4, 5, 4, 4, 5) | 2 | HM543662 |
| 100 | L2b | San Francisco | Rectal | 6(4*, 4, 5, 4, 4, 5) | 2 | HM543663 |
| 101 | L2b | San Francisco | Rectal | 6(4*, 4, 4, 4, 4, 5) | 2 | HM543664 |
| 102 | L2b | San Francisco | Rectal | 5(4*, 4, 4, 4, 5) | 2 | HM543665 |
The number of tyrosine repeat regions followed by the number of tyrosines in each repeat. An asterisk indicates a partial tyrosine-rich repeat region in which the first 8 amino acids are missing.
The predicted number of actin binding domains predicted in silico as described by Jewett et al. (38).
Sequencing of tarP from clinical isolates.
The template for PCR was prepared using 100 μl of culture resuspended in lysis buffer containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.022% gelatin, 1.5% Nonidet P-40, and 0.5% Tween 20. The mixture was digested with proteinase K (100 μg/ml) for 2 h at 55°C and then boiled for 10 min. The nested PCR amplification of tarP was performed using primers listed in Table 2. The entire tarP gene was amplified using primers CT456-nested-F and CT456-nested-R in the first round of PCR followed by CT456-F and CT456-R for the second round of PCR. The nested PCR products were purified and sequenced using the forward sequencing primers CT456F-Seq1, CT456F-Seq2, CT456F-Seq3, and CT456F-Seq1A and reverse primers CT456R-Seq1, CT456R-Seq2, CT456R-Seq3, and CT456R-Seq1A. For PCR amplifications, approximately 50 ng of DNA was amplified in 50-μl reaction mixtures containing 1× PCR buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1 μM primers, and 2.6 U of expand high-fidelity Taq (Roche). For both PCRs an initial denaturation of 94°C for 2 min was followed by 25 cycles of 94°C for 30 s, 66°C (first round of PCR) or 69°C (second round of PCR) for 30 s, 72°C for 2 min (with a 5-s cycle elongation for each successive cycle), and a final elongation at 72°C for 10 min. Purified nested PCR products were sequenced using an ABI PRISM 377 DNA sequencer at the DNA Core Facility (Health Canada, Winnipeg, Manitoba, Canada).
TABLE 2.
Primer sequences
| Primer | Sequence (5′-3′) |
|---|---|
| CT456-F | GCT CCT GAC ACG CGC ACA GAC C |
| CT456-R | GCG CCT TGT CGA TTG TGA TGA GG |
| CT456nested-F | CAC TTG CGC TTG CTG ATC CC |
| CT456nested-R | GAT TGA CTG TGG AGG ACA GG |
| CT456F-Seq1 | GAT ACC AGG CAT TGC AGT TCC |
| CT456F-Seq2 | GGC CTA GTA GCG AAG ACG ATG GC |
| CT456F-Seq3 | CAG CAG GAG GAA GTG GTA GCG TAC |
| CT456R-Seq1 | GAA CAG ACT TGG TCC CAA TTT CCG |
| CT456R-Seq2 | CCA TTG ACT CCA CCA GCT CCG |
| CT456R-Seq3 | GGC TTA GGC ATT CAC GAG CAG |
| CT456F-Seq1A | CAT ATC CCT AGC GAT TAC G |
| CT456R-Seq1A | CCT CCT GAA CCA GTT TCT TGG CGG |
Alignment and phylogenetic analysis.
Nucleic acid sequence alignments for ompA and tarP were generated using ClustalW (65) and analyzed using the Molecular Evolutionary Genetics Analysis software package, MEGA4 (63). The analysis of ompA was done using the reference strains A/HAR13 (CP000051.1), B/TWR/OT (DQ064281.1), Ba/Apache-2 (DQ064282.1), C/TW-3/OT (DQ064283.1), D/UW-3/CX (AE001273.1), E/BOUR (DQ064286.1), F/IC-CAL3 (DQ064287.1), G392 (DQ064288), H/UW-4 (AF304857.1), I/UW-12 (DQ064290), J/UW-36/CX (DQ064292.1), K/UW31 (DQ064293), L1/LGV440 (DQ064294), L2/LGV434 (AM884176.1), L3/LGV404 (DQ064296.1), Chlamydophila abortus (NC_004552.2), C. muridarum (AE002160.2), C. caviae (AF269282.1), Chlamydophila felis (AP006861), and C. pneumoniae (AE001363.1). Phylogenetic trees were constructed using the neighbor-joining method (49). Evolutionary distances are in the units of the number of base pair substitutions per site and were computed using the maximum composite likelihood method (64). Branching pattern confidence levels were estimated by the bootstrap resampling of data based on 1,000 random replicates. Dot matrix analysis on Tarp protein sequences was performed with the AA coding comparison in DNAMAN (Lynnon Corporation, Quebec, Canada). Multiple protein alignments were generated by ClustalW and visualized with Geneious Pro 3.0.6 (A. J. Drummond, B. Ashton, M. Cheung, J. Heled, M. Kearse, R. Moir, S. Stones-Havas, T. Thierer, and A. Wilson, 2007). Sequences obtained in this study were deposited into GenBank (Table 1).
RESULTS
Phylogenetic analysis of tarP.
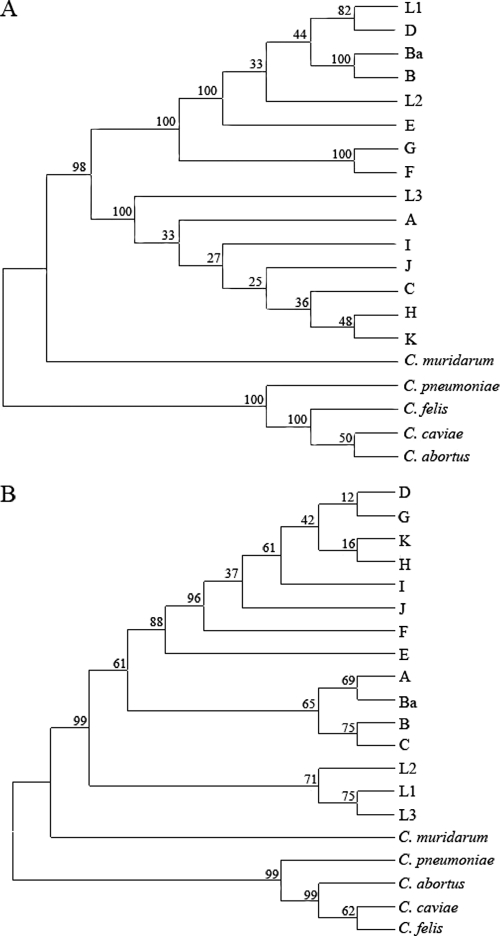
Phylogenetic analysis was performed on ompA and tarP from reference strains of C. trachomatis as well as C. muridarum, C. felis, C. pneumoniae, C. abortus, and C. caviae using the neighbor-joining method in MEGA4 (Fig. 1). The analysis of tarP and ompA produced very different phylograms. In both clusters, the non-C. trachomatis reference strains appeared to group separately in an outgroup distinct from the C. trachomatis reference strains, with C. muridarum being more closely related than the others. As has been demonstrated previously (1, 6, 8, 24, 44, 50, 58), no association between genotype and clinical phenotype could be found with ompA, as each of the two major clades contained ocular, genital, and LGV reference strains (Fig. 1A). The analysis did separate the chlamydial strains into known serologic groupings, with the top clade consisting of B complex and intermediate strains and the middle clade consisting of C complex strains. The phylogenetic analysis of tarP, however, produced three distinct clades, which were grouped according to clinical biotype (Fig. 1B). The clusters grouped according to disease with one clade containing strains from genital reference serovars D to K, another clade containing the ocular reference strains A to C, and a third clade containing the LGV strains L1 to L3 (Fig. 1B).
FIG. 1.
Phylogenetic analysis of ompA and tarP from reference strains of C. trachomatis as well as C. muridarum, C. felis, C. pneumoniae, C. abortus, and C. caviae. The neighbor-joining method was used in MEGA4 to generate unrooted phylogenetic trees for ompA (A) and tarP (B) from DNA sequences. Relevant bootstrap values (as a percentage of 1,000 replicates) are provided. Initial nucleotide alignments were generated using ClustalW.
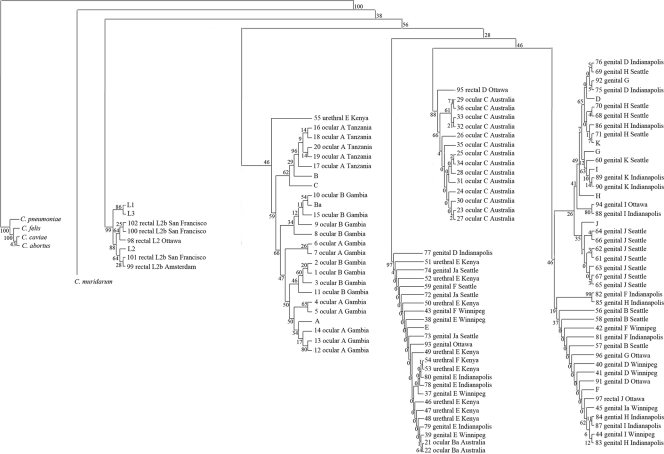
To further investigate the relationship of the phylogenics of tarP, tarP was amplified and sequenced from 36 ocular isolates (5 from Tanzania, 16 from Australia, and 15 from The Gambia), 57 genital isolates (10 from Kenya, 9 from Winnipeg, 16 from Indianapolis, 18 from Seattle, and 4 from elsewhere), 3 rectal isolates, and 5 LGV isolates (Table 1). Phylogenetic analysis was performed using the neighbor-joining method in MEGA4 on all clinical isolates and reference strains (Fig. 2). The ocular and LGV clinical isolates and reference strains clustered separately (Fig. 2), similarly to what was observed for the reference strains alone (Fig. 1B). The LGV isolates from San Francisco, Ottawa, and Amsterdam formed a separate clade with the LGV reference strains (L1 to L3). The LGV clade appears to be distinct from the other isolates (ocular, rectal, and genital), as they are the first C. trachomatis strains to diverge on the phylogram (Fig. 2). The ocular isolates from The Gambia and Tanzania were the next to diverge and formed a number of smaller clades, which branch with the Gambian and Tanzanian ocular isolates, forming separate clusters that grouped according to serotype. Interestingly, tarP genes from the reference strains belonging to serovars A, B, Ba, and C clustered with the Gambian and Tanzanian ocular isolates but not the Australian ocular isolates. The ocular serovar C isolates from Australia formed a distinct clade with the exception of 21 and 22 (serovar Ba), which fell in a clade consisting of a mixture of genital isolates from Kenya, Indianapolis, Winnipeg, and Seattle. The genital isolates formed two separate clades, one of which contained predominantly clinical isolates from serovars E and F as well as the reference serovar E. The second genital clade contained a greater diversity of serologically defined isolates as well as the remaining reference strains from serovars D to K.
FIG. 2.
Phylogenetic analysis of tarP from C. trachomatis clinical isolates and reference strains. The neighbor-joining method was used by MEGA4 to generate an unrooted phylogenetic tree for tarP based on DNA sequences. For illustration purposes, the initial branches of the phylogenetic tree were redrawn from the original version but the clades and bootstrap values remain untouched. Relevant bootstrap values (as a percentage of 1,000 replicates) are provided. Initial nucleotide alignments were generated using ClustalW.
Variability of Tarp across C. trachomatis serovars.
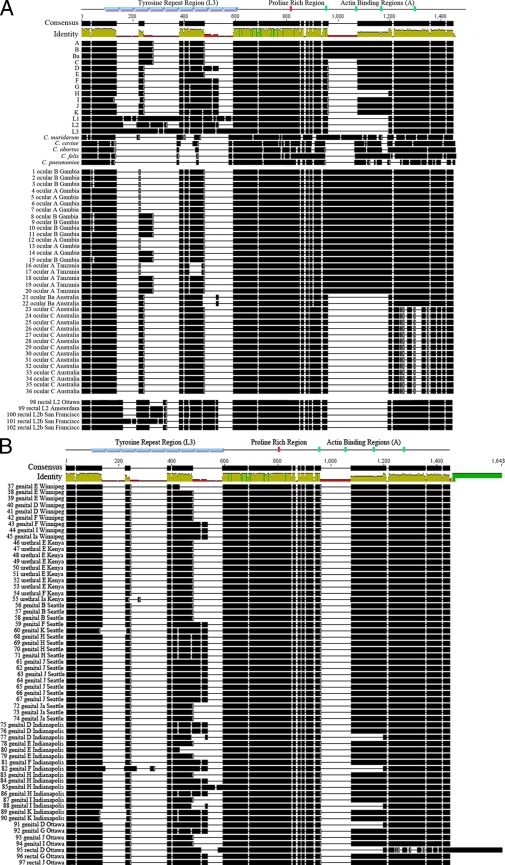
The greatest variations in C. trachomatis Tarp sequences are in the numbers of tyrosine-rich repeat regions and actin binding domains (15, 37). In this study, the number of tyrosine-rich repeat regions varied from as many as nine (L1 and L3) to as few as one (genital isolates 37 and 79 and ocular isolates 16 and 17) (Table 1, Fig. 3). The number of tyrosines within individual repeat units also varied from three (isolate 85) to five (LGV strains). The ocular and genital reference strains each contained three tyrosine-rich repeat regions with the exception of serovar E, which contained two. There appeared to be a greater diversity among Tarp proteins from the ocular and genital clinical isolates, as they ranged in the number of tyrosine-rich repeats from one to three and one to four, respectively. Overall, there was a trend toward a reduction in the number of tyrosine-rich repeats in Tarp from ocular and genital clinical isolates compared to those from the reference strains as a whole or within serovars (Table 1).
FIG. 3.
Variability of Tarp protein sequences. Tarp protein sequences were aligned with ClustalW and visualized with Geneious 3.0.6. Black regions represent the presence of amino acids, and blank regions represent gaps in the protein sequence. At the top, regions corresponding to the tyrosine-rich repeats are shown in blue (based on nine tyrosine repeat regions of L3), the proline-rich region in red, and the actin binding domains in green (based on laboratory strain serovar D). All regions are shown where they appear in the consensus sequence obtained in the ClustalW alignment. Regions of consensus and percent identity are indicated at the top and also are based on the aligned protein sequences. Reference and non-C. trachomatis strains and ocular and LGV clinical isolates are shown in panel A; genital and rectal isolates are shown in panel B.
Each isolate was further examined for the presence of the proline-rich region and actin binding domains as defined by Jewett et al. (37). All isolates contained a single proline-rich region, which was conserved among all reference strains and clinical isolates. A trend emerged as most of the ocular isolates (reference and clinical) contained four predicted actin binding domains (Table 1, Fig. 4A), with the exception of the Australian ocular isolates, which contained two (Table 1). Ocular isolates from Australia also exhibited the greatest sequence divergence from the reference strains in the C-terminal portion containing the actin binding domains, which was not seen in any of the other isolates except for rectal isolate 95 (Fig. 3B). The reference serovars D through K and the D to K genital and rectal clinical isolates contained either two or three predicted actin binding domains in the C terminus of Tarp (Table 1, Fig. 4A). The LGV reference strains (L1 to L3) and LGV clinical isolates all contained two putative actin binding domains (Table 1).
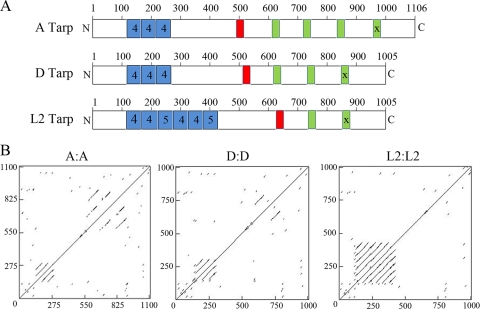
FIG. 4.
Schematic representation and dot matrix comparison of Tarp sequences. (A) The number and location of tyrosine-rich repeats (blue), the proline rich region (red), and actin binding domains (green) identified for an ocular strain (reference strain serovar A), a genital strain (reference strain serovar D), and an LGV strain (reference strain L2) are shown. The “X” on the last actin binding domain of each Tarp illustrates biochemical inactivity (38). (B) Dot matrix comparison of reference strain serovars A to A, D to D, and L2 to L2 showing regions of repetition along the length of Tarp as generated by DNAMAN.
Dot matrix comparisons were performed on representative reference strains from serovars A, D, and L2 to further investigate the changes in the number of tyrosine-rich repeat regions and actin binding domains (Fig. 4B). Dot matrix analysis aligns protein (or nucleotide) sequences against themselves such that similar sequences appear as a parallel lines above or below a diagonal representing identity. The tyrosine-rich repeat regions can be seen in the bottom left corner of each matrix, with the WH2 actin binding domains appearing near the top right corner (Fig. 4B). The trend for increased numbers of repeat units within the tyrosine-rich regions can be seen in the Tarp proteins from the ocular strain (A), the genital strain (D), and the LGV strain (L2), which contains the greatest number of repeats. The converse is seen with duplications of the C-terminal actin binding WH2 domains, in which serovar A contains four putative WH2 domains, D contains three, and serovar L2 shows the fewest number of WH2 domains, with two.
The analysis of the tarP sequences from all isolates and reference strains identified single-nucleotide polymorphisms (SNPs) that correlate with groupings identified in the phylogenetic analysis (see Fig. S1 in the supplemental material). A total of 18 SNPs were identified that were specific to the LGV isolates. The majority of these SNPs were nonsynonymous (16/18), and eight of these SNPs were found to occur in the tyrosine-rich repeat regions. Furthermore, six additional SNPs were identified in the LGV isolates, which also were present in all ocular isolates from The Gambia, Tanzania, and the ocular reference strains. These six SNPs also were present in urogenital isolate 55 but were absent from all other genital isolates as well as the Australian ocular isolates. The sharing of these six SNPs is consistent with a closer evolutionary link between the LGV and ocular tarP sequences, as was suggested in the phylogram in which the clade containing the ocular isolates diverged from the tree after the LGV isolates. Thirteen ocular-specific SNPs (for the Gambian, Tanzanian, and reference strains) were identified that were absent from all other strains, including the Australian ocular isolates. Of these 13, 12 were nonsynonymous. Urogenital isolate 55, however, contained 7 of these 13 SNPs, which may account for the close phylogenetic grouping of this isolate with the ocular isolates. The Tanzanian ocular isolates also contained three additional SNPs that were absent from the other ocular isolates and that may explain the separation of the Tanzanian and Gambian isolates in their clade. There were no SNPs that were shared by all genital isolates.
DISCUSSION
Chlamydial Tarp is a type III secreted effector protein that is translocated at the time of the initial contact of elementary bodies (EBs) with the host cell and is implicated in the mobilization of actin that is essential for chlamydial internalization (15, 16, 37). C. trachomatis Tarp is tyrosine phosphorylated over a region of tyrosine-rich repeat units by host cell src family kinases, but this feature is not conserved in Tarp orthologs from C. caviae, C. muridarum, or C. pneumoniae (15, 16, 37). C. trachomatis L2 Tarp contains a single Wasp homology domain-2 (WH2) actin binding domain that acts in conjunction with an upstream proline-rich oligomerization domain to nucleate actin filament formation (37). All chlamydial Tarp orthologs contain one or more predicted WH2 domains and the proline-rich domain (38). C. trachomatis serovars representative of different diseases and tissue tropisms have shown variation in the number of tyrosine-rich repeat units as well as in the number of predicted WH2 domains. We compared Tarp structures from reference strains and 102 clinical isolates, which were representative of 16 serovars of C. trachomatis taken from various geographical locations and anatomical sites. Tarp showed a remarkable degree of heterogeneity. The phylogenetic analysis of C. trachomatis reference strains segregated ocular, urogenital, and LGV disease groups. With few exceptions, the clinical isolates similarly clustered in concordance with tissue tropism and disease. For the most part, the greatest variation was in the number of tyrosine-rich repeat units and in the number of putative WH2 domains. A trend emerged with the trachoma biovars tending toward fewer tyrosine-rich repeats and more WH2 domains, while the LGV biovar displayed a greater number of tyrosine-rich repeats and fewer WH2 domains. The genital strains displayed intermediate numbers of tyrosine-rich repeats and WH2 domains.
C. trachomatis displays remarkable genome sequence similarity and synteny considering the different diseases and tissue tropisms exhibited by the different serovars (55). Comparative genomics has identified a relatively small number of loci that vary according to biovar or tissue tropism and disease manifestation. Microarray-based genomic comparisons have identified only 31 genes that varied according to tissue tropism (5). Interestingly, tarP was not among these. The failure to detect tarP in microarray analysis likely was due to the tarP sequences differing primarily in the duplication of sequence rather than the presence of unique sequences that might be more readily detected in microarrays.
MOMP is the predominant surface antigen of chlamydiae and a target for neutralizing antibody (10). Its proposed roles include structural stability (32, 33, 45), porin activity (2), and attachment to host cells (60, 61). It is also one of the most polymorphic chlamydial genes known (24). It has been proposed that this variability is due to the antigenicity of MOMP and selective pressure of the immune system (6). As a result, the phylogeny of MOMP is not in agreement with tissue tropism or disease. Although there have been multiple attempts to correlate MOMP genotype with disease, the associations have been somewhat limited and inconsistent (1, 19, 27, 40, 44). Tarp also exhibits a high degree of variability, although the selective pressure to induce this is unclear. Tarp is antigenic and recognized by antisera from humans infected with chlamydiae (68) but does not appear to be surface exposed or a target for neutralizing antibody (16). Polyclonal antisera to intact Tarp show reactivity with multiple regions of the protein yet do not react with intact EBs unless the EBs are first allowed to attach to host cells at 37°C to trigger Tarp secretion and exposure. Furthermore, the surface proteolysis of viable EBs cleaves surface-exposed domains of MOMP, but Tarp is not degraded (16). Thus, in contrast to MOMP, the evolutionary pressure favoring Tarp variation may involve tissue tropism rather than the evasion of immune surveillance.
The selective pressures for Tarp selection that might favor specific tissue tropisms are unclear. At least one of the WH2 domains is functional in actin binding for all serovars tested, and all nucleate actin filament formation (38). Where multiple WH2 domains were predicted in C. trachomatis Tarp proteins, in all cases tested (serovars L2, D, and A), the most C-terminal of these did not sequester actin. The presence of multiple WH2 domains on the same polypeptide of some serovars suggests alternative mechanisms or at least the potential for the enhancement of actin nucleation by sequestering multiple actin monomers in immediate juxtaposition to favor nucleation (38). Unfortunately, rates of entry cannot be determined with sufficient resolution to determine effects on internalization, and isogenic variants differing only in the number of WH2 domains cannot yet be engineered in chlamydiae. Regardless of the potential for hybrid mechanisms of actin nucleation, all reference and clinical isolates retain a single copy of the proline-rich domain required for Tarp oligomerization and actin nucleation.
The function of the tyrosine-rich repeat region also is unknown. Models have been proposed that implicate this domain in host Arp2/3 complex recruitment to promote entry (20, 41). Tarp phosphorylation is not required for the entry of strains other than C. trachomatis (15). Furthermore, the inhibition of C. trachomatis Tarp tyrosine phosphorylation had no effect on entry (36, 42). We hypothesized that the requirement for Tarp tyrosine phosphorylation was for events and cell signaling pathways that are active postentry. Indeed, multiple host proteins bind ectopically expressed tyrosine-phosphorylated Tarp, presumably via SH2 domains on Tarp (19a). The retention of Tarp on the nascent inclusion has led to suggestions that it plays a role in some early event that does not require de novo chlamydial protein synthesis, such as the avoidance of lysosomal fusion (16). How multiple copies of this domain on individual Tarp monomers might enhance any signaling pathway is unknown. It is tempting to speculate, however, that the amplification of an as-yet unidentified host signaling pathway accounts for the increased survival of LGV strains in monocytes and macrophages.
The phylogenetic analysis performed on the clinical isolates produced similar trends, in that tarP genes from ocular and LGV isolates formed clades distinct from those of the urogenital strains. The LGV tarP sequences diverged from the phylogenic tree first with a separate clade, as was observed for the reference strains. It is interesting that even though the ocular isolates from The Gambia, Tanzania, and Australia formed separate clades, they did not group together within a single branch on the phylogram. The Australian ocular isolates were notable in that they formed a distinct clade apparently more closely related to the urogenital strains. This suggests that even though ocular strains contain enough differences from most of the urogenital and LGV strains to cluster separately, there also are some sequence variations among ocular tarP genes.
Tissue tropism for C. trachomatis serovars is not exclusive. Genital strains have been isolated from ocular sites, although these have not been implicated as progressing to trachoma (52). Conversely, ocular strains are rarely isolated from the genital tract (51). In this study, three of the clinical isolates of serovar B (56, 57, and 58) were recovered from genital sites. The B, Ba, and C serovars previously have been isolated from genital infections and classified as variants based upon the failure to conform to the prototype B strain in microimmunofluorescence with a panel of B antibodies (46) or by the genotyping of MOMP (25, 35, 43, 58). Intriguingly, tarP from the three Seattle B strains described here resembled the structure of urogenital isolates and phylogenetically clustered with urogenital Tarps sequences. These particular B isolates were previously characterized by Caldwell et al. as possessing intact tryptophan synthase genes identical to those found in the genital serovars. The sequence analysis of ompA of these strains revealed a single-nucleotide difference from the D and H genotypes, but they were identical to the ompA sequence of serovars B and Ba over the remainder of the gene. The conclusion was that these appeared to be true genital strains that had acquired an ocular MOMP sequence (9). The tarP sequence is consistent with this assumption. Conversely, two urogenital strains, 55 (a urethral isolate from Kenya) and 95 (a rectal isolate from Ottawa), clustered with ocular strains from The Gambia and Australia, respectively, and two Australian ocular Ba isolates clustered (21 and 22) with genital/urethral isolates. It also is possible that with the high rates of C. trachomatis transmission, chlamydiae can be transferred from one tissue to another and that the site of isolation is the original niche of the isolate.
tarP genes from ocular isolates from The Gambia, Tanzania, and Australia appeared to be relatively similar within their phylogenetic groupings but distinct from each other. This may be due to the isolates being obtained from geographical regions where the majority of the populations are relatively restricted to local travel. tarP genes from Australian ocular isolates were phylogenetically distinct from the Gambian and Tanzanian ocular isolates as well as from the ocular reference strains (A, B, Ba, and C), suggesting that a distinct lineage was causing these ocular infections. A similar observation for serovar C conjunctival strains from Australia evolving from a unique lineage was found in a study characterizing ompA genotypes from remote Australian communities (57). Despite this high degree of relatedness among localized ocular outbreaks, there was a remarkable degree of sequence dissimilarity within these groups (see Fig. S1 and S2 in the supplemental material), suggesting that tarP exhibits a high rate of evolution. tarP from urogenital isolates did not show a similar phylogenetic clustering. For example, the Kenyan urethral isolates fell into a separate clade; however, that clade also contained genital isolates from Winnipeg, Seattle, and Indianapolis and two ocular isolates from Australia, suggesting that factors other than geographical isolation were impacting tarP relatedness. Behavioral or sociological factors may play a role in the wider dissemination of sexually transmitted infections and obfuscate geographic variations in tarP.
Tissue tropism and infections caused by C. trachomatis appear to be controlled by a relatively small number of loci (5). Although it is unlikely that any one gene product confers tissue tropism, tarP shows clear phylogenetic grouping with tissue tropism compared among reference strains. The picture becomes more complex when clinical isolates are compared, yet clear trends are established. It is probable that multiple loci function together to favor specific tissue tropism, and the whole-genome analysis of clinical isolates may be required to decipher complex interactions defining chlamydial disease. Tarp appears to be among those chlamydial factors worthy of consideration in efforts to correlate disease and genotype and also may prove useful in epidemiological studies.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the NIAID/NIH. This work also was supported in part by research grants from the Canadian Institutes of Health Research (GR-13301 to G. McClarty) and the Public Health Agency of Canada Office of the Chief Scientist Fund (to G. McClarty).
We thank Harlan Caldwell for the laboratory reference strains and S. Tabrizi, B. Van Der Pol, R. Jones, J. Wylie, S. Moses, I. Maclean, D. Mabey, R. Bailey, N. Faal, M. Ragonnet, M. Burton, A. Solomon, and J. Schachter for clinical specimens. We also thank H. Caldwell and J. Carlson for the critical review of the manuscript.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 6 July 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Andreasen, A. A., M. J. Burton, M. J. Holland, S. Polley, N. Faal, D. C. Mabey, and R. L. Bailey. 2008. Chlamydia trachomatis ompA variants in trachoma: what do they tell us? PLoS Negl. Trop. Dis. 2:e306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bavoil, P., A. Ohlin, and J. Schachter. 1984. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 44:479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U. S. A. 98:13984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts, H. J., K. Wolf, and K. A. Fields. 2009. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr. Opin. Microbiol. 12:81-87. [DOI] [PubMed] [Google Scholar]
- 5.Brunelle, B. W., T. L. Nicholson, and R. S. Stephens. 2004. Microarray-based genomic surveying of gene polymorphisms in Chlamydia trachomatis. Genome Biol. 5:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunelle, B. W., and G. F. Sensabaugh. 2006. The ompA gene in Chlamydia trachomatis differs in phylogeny and rate of evolution from other regions of the genome. Infect. Immun. 74:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunham, R. C., J. Kimani, J. Bwayo, G. Maitha, I. Maclean, C. Yang, C. Shen, S. Roman, N. J. Nagelkerke, M. Cheang, and F. A. Plummer. 1996. The epidemiology of Chlamydia trachomatis within a sexually transmitted diseases core group. J. Infect. Dis. 173:950-956. [DOI] [PubMed] [Google Scholar]
- 8.Brunham, R. C., and J. Rey-Ladino. 2005. Immunology of chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nature 5:149-161. [DOI] [PubMed] [Google Scholar]
- 9.Burton, M. J., M. J. Holland, P. Makalo, E. A. Aryee, N. D. Alexander, A. Sillah, H. Faal, S. K. West, A. Foster, G. J. Johnson, D. C. Mabey, and R. L. Bailey. 2005. Re-emergence of Chlamydia trachomatis infection after mass antibiotic treatment of a trachoma-endemic Gambian community: a longitudinal study. Lancet 365:1321-1328. [DOI] [PubMed] [Google Scholar]
- 10.Caldwell, H. D., and L. J. Perry. 1982. Neutralization of Chlamydia trachomatis infectivity with antibodies to the major outer membrane protein. Infect. Immun. 38:745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldwell, H. D., and J. Schachter. 1982. Antigenic analysis of the major outer membrane protein of Chlamydia spp. Infect. Immun. 35:1024-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Investig. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson, J. H., S. Hughes, D. Hogan, G. Cieplak, D. Sturdevant, G. McClarty, H. D. Caldwell, and R. J. Belland. 2004. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 72:7063-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson, J. H., S. F. Porcella, G. McClarty, and H. D. Caldwell. 2005. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 73:6407-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clifton, D. R., C. A. Dooley, S. S. Grieshaber, R. A. Carabeo, K. A. Fields, and T. Hackstadt. 2005. Tyrosine phosphorylation of chlamydial Tarp is species specific and not required for the recruitment of actin. Infect. Immun. 73:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clifton, D. R., K. A. Fields, S. Grieshaber, C. A. Dooley, E. Fischer, D. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cocchiaro, J. L., and R. H. Valdivia. 2009. New insights into Chlamydia intracellular survival mechanisms. Cell Microbiol. 11:1571-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conway, D. J., M. J. Holland, R. L. Bailey, A. E. Campbell, O. S. Mahdi, R. Jennings, E. Mbena, and D. C. Mabey. 1997. Scarring trachoma is associated with polymorphism in the tumor necrosis factor alpha (TNF-alpha) gene promoter and with elevated TNF-alpha levels in tear fluid. Infect. Immun. 65:1003-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean, D., E. Oudens, G. Bolan, N. Padian, and J. Schachter. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J. Infect. Dis. 172:1013-1022. [DOI] [PubMed] [Google Scholar]
- 19a.Dooley, C. A., D. R. Clifton, R. A. Carabeo, and T. Hackstadt. 2004. Abstr. 104th Gen. Meet. Am. Soc. Microbiol., abstr. D-228.
- 20.Elwell, C. A., A. Ceesay, J. H. Kim, D. Kalman, and J. N. Engel. 2008. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 4:e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faal, N., R. L. Bailey, D. Jeffries, H. Joof, I. Sarr, M. Laye, D. C. Mabey, and M. J. Holland. 2006. Conjunctival FOXP3 expression in trachoma: do regulatory T cells have a role in human ocular Chlamydia trachomatis infection? PLoS Med. 3:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 23.Fields, K. A., D. Mead, C. A. Dooley, and T. Hackstadt. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671-683. [DOI] [PubMed] [Google Scholar]
- 24.Fitch, W. M., E. M. Peterson, and L. M. de la Maza. 1993. Phylogenetic analysis of the outer-membrane-protein genes of Chlamydiae, and its implication for vaccine development. Mol. Biol. Evol. 10:892-913. [DOI] [PubMed] [Google Scholar]
- 25.Frost, E. H., S. Deslandes, D. Gendron, D. Bourgaux-Ramoisy, and P. Bourgaux. 1995. Variation outside variable segments of the major outer membrane protein distinguishes trachoma from urogenital isolates of the same serovar of Chlamydia trachomatis. Genitourin Med. 71:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frost, E. H., S. Deslandes, S. Veilleux, and D. Bourgaux-Ramoisy. 1991. Typing Chlamydia trachomatis by detection of restriction fragment length polymorphism in the gene encoding the major outer membrane protein. J. Infect. Dis. 163:1103-1107. [DOI] [PubMed] [Google Scholar]
- 27.Geisler, W. M., R. J. Suchland, W. L. Whittington, and W. E. Stamm. 2003. The relationship of serovar to clinical manifestations of urogenital Chlamydia trachomatis infection. Sex. Transm. Dis. 30:160-165. [DOI] [PubMed] [Google Scholar]
- 28.Geisler, W. M., W. L. Whittington, R. J. Suchland, and W. E. Stamm. 2002. Epidemiology of anorectal chlamydial and gonococcal infections among men having sex with men in Seattle: utilizing serovar and auxotype strain typing. Sex. Transm. Dis. 29:189-195. [DOI] [PubMed] [Google Scholar]
- 29.Gomes, J. P., W. J. Bruno, M. J. Borrego, and D. Dean. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 186:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomes, J. P., A. Nunes, W. J. Bruno, M. J. Borrego, C. Florindo, and D. Dean. 2006. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 199:275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grayston, J. T., and S. Wang. 1975. New knowledge of chlamydiae and the diseases they cause. J. Infect. Dis. 132:87-105. [DOI] [PubMed] [Google Scholar]
- 32.Hackstadt, T., W. J. Todd, and H. D. Caldwell. 1985. Disulfide-mediated interactions of the chlamydial major outer membrane protein: role in the differentiation of chlamydiae? J. Bacteriol. 161:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatch, T. P., I. Allan, and J. H. Pearce. 1984. Structural and polypeptide differences between envelopes of infective and reproductive life cycle forms of Chlamydia spp. J. Bacteriol. 157:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh, Y. H., L. D. Bobo, T. C. Quinn, and S. K. West. 2001. Determinants of trachoma endemicity using Chlamydia trachomatis ompA DNA sequencing. Microbes Infect. 3:447-458. [DOI] [PubMed] [Google Scholar]
- 35.Ikehata, M., K. Numazaki, and S. Chiba. 2000. Analysis of Chlamydia trachomatis serovars in endocervical specimens derived from pregnant Japanese women. FEMS Immunol. Med. Microbiol. 27:35-41. [DOI] [PubMed] [Google Scholar]
- 36.Jewett, T. J., C. A. Dooley, D. J. Mead, and T. Hackstadt. 2008. Chlamydia trachomatis tarp is phosphorylated by src family tyrosine kinases. Biochem. Biophys. Res. Commun. 371:339-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jewett, T. J., E. R. Fischer, D. J. Mead, and T. Hackstadt. 2006. Chlamydial TARP is a bacterial nucleator of actin. Proc. Natl. Acad. Sci. U. S. A. 103:15599-15604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jewett, T. J., N. J. Miller, C. A. Dooley, and T. Hackstadt. The conserved Tarp actin binding domain is important for chlamydial invasion. PLoS Pathog., in press. [DOI] [PMC free article] [PubMed]
- 39.Klint, M., H. H. Fuxelius, R. R. Goldkuhl, H. Skarin, C. Rutemark, S. G. Andersson, K. Persson, and B. Herrman. 2007. High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J. Clin. Micriobiol. 45:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lampe, M. F., K. G. Wong, and W. E. Stamm. 1995. Sequence conservation in the major outer membrane protein gene among Chlamydia trachomatis strains isolated from the upper and lower urogenital tract. J. Infect. Dis. 172:589-592. [DOI] [PubMed] [Google Scholar]
- 41.Lane, B. J., C. Mutchier, S. Al Khodor, S. S. Grieshaber, and R. A. Carabeo. 2008. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 4:e1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehlitz, A., S. Banhart, S. Hess, M. Selbach, and T. F. Meyer. 2008. Complex kinase requirements for Chlamydia trachomatis Tarp phosphorylation. FEMS Microbiol. Lett. 289:233-240. [DOI] [PubMed] [Google Scholar]
- 43.Millman, K., C. M. Black, R. E. Johnson, W. E. Stamm, R. B. Jones, E. W. Hook, D. H. Martin, G. Bolan, S. Tavare, and D. Dean. 2004. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 186:2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millman, K., C. M. Black, W. E. Stamm, R. B. Jones, E. W. R. Hook, D. H. Martin, G. Bolan, S. Tavare, and D. Dean. 2006. Population-based genetic epidemiologic analysis of Chlamydia trachomatis serotypes and lack of association between ompA polymorphisms and clinical phenotypes. Microb. Infect. 8:604-611. [DOI] [PubMed] [Google Scholar]
- 45.Newhall, W. J., and R. B. Jones. 1983. Disulfide-linked oligomers of the major outer membrane protein of chlamydiae. J. Bacteriol. 154:998-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persson, K. 1990. Epidemiology of serovars of Chlamydia trachomatis, p. 559-562. In W. R. Bowie, H. D. Caldwell, R. B. Jones, P.-H. Mardh, G. L. Ridgway, J. Schachter, W. E. Stamm, and M. E. Ward (ed.), Proceedings of the Seventh International Symposium on Human Chlamydial Infections. Cambridge University Press, Cambridge, United Kingdom.
- 47.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez, P., A. Allardet-Servent, B. de Barbeyrac, M. Ramuz, and C. Bebear. 1994. Genetic variability among Chlamydia trachomatis reference and clinical strains analyzed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 32:2921-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 50.Schachter, J. 1978. Chlamydial infections (first of three parts). N. Engl. J. Med. 298:428-434. [DOI] [PubMed] [Google Scholar]
- 51.Schachter, J. 1978. Chlamydial infections (second of three parts). N. Engl. J. Med. 298:490-495. [DOI] [PubMed] [Google Scholar]
- 52.Schachter, J. 1999. Infection and disease epidemiology, p. 139-169. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC.
- 53.Shaw, A. C., G. Christiansen, P. Roepstorff, and S. Birkelund. 2000. Genetic differences in the Chlamydia trachomatis tryptophan synthase alpha-subunit can explain variations in serovar pathogenesis. Microbes Infect. 2:581-592. [DOI] [PubMed] [Google Scholar]
- 54.Solomon, A. W., M. J. Holland, N. D. Alexander, P. A. Massae, A. Aguirre, A. Natividad-Sancho, S. Molina, S. Safari, J. F. Shao, P. Courtright, R. W. Peeling, S. K. West, R. L. Bailey, A. Foster, and D. C. Mabey. 2004. Mass treatment with single-dose azithromycin for trachoma. N. Engl. J. Med. 351:1962-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephens, R. S. 1999. Genomic autobiographies of chlamydiae, p. 9-27. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC.
- 56.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 57.Stevens, M. P., S. N. Tabrizi, R. Muller, V. Krause, and S. M. Garland. 2004. Characterization of Chlamydia trachomatis omp1 genotypes detected in eye swab samples from remote Australian communities. J. Clin. Microbiol. 42:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stothard, D. R., G. Boguslawski, and R. B. Jones. 1998. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 66:3618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stothard, D. R., G. A. Toth, and B. E. Batteiger. 2003. Polymorphic membrane protein H has evolved in parallel with the three disease-causing groups of Chlamydia trachomatis. Infect. Immun. 71:1200-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su, H., L. Raymond, D. D. Rockey, E. Fischer, T. Hackstadt, and H. D. Caldwell. 1996. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 93:11143-11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su, H., N. G. Watkins, Y. X. Zhang, and H. D. Caldwell. 1990. Chlamydia trachomatis-host cell interactions: role of the chlamydial major outer membrane protein as an adhesin. Infect. Immun. 58:1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suchland, R. J., B. M. Jeffrey, M. Xia, A. Bhatia, H. G. Chu, D. D. Rockey, and W. E. Stamm. 2008. Identification of concomitant infection with Chlamydia trachomatis IncA-negative mutant and wild-type strains by genomic, transcriptional, and biological characterizations. Infect. Immun. 76:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura, K., M. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 64.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomson, N. R., M. T. Holden, C. Carder, N. Lennard, S. J. Lockey, P. Marsh, P. Skipp, C. D. O'Conner, I. Goodhead, H. Norbertzake, B. Harris, D. Ormond, R. Rance, M. A. Quail, J. Parkhill, R. S. Stephens, and I. N. Clarke. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Der Pol, B., D. V. Ferrero, L. Buck-Barrington, E. Hook III, C. Lenderman, T. Quinn, C. A. Gaydos, J. Lovchik, J. Schachter, J. Moncada, G. Hall, M. J. Tuohy, and R. B. Jones. 2001. Multicenter evaluation of the BDProbeTec ET system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J. Clin. Microbiol. 39:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, J., L. Chen, F. Chen, X. Zhang, Y. Zhang, J. Baseman, S. Perdue, I. T. Yeh, R. Shain, M. Holland, R. Bailey, D. Mabey, P. Yu, and G. Zhong. 2009. A chlamydial type III-secreted effector protein (Tarp) is predominantly recognized by antibodies from humans infected with Chlamydia trachomatis and induces protective immunity against upper genital tract pathologies in mice. Vaccine 27:2967-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, S.-P., and J. T. Grayston. 1963. Classification of trachoma virus strains by protection of mice from toxic death. J. Immunol. 90:849-856. [PubMed] [Google Scholar]
- 70.Wang, S.-P., and J. T. Grayston. 1971. Classification of TRIC and related strains with micro immunofluorescence, p. 305-321. In R. L. Nichols (ed.), Trachoma and related disorders caused by chlamydial agents. Excerpta Medica, Amsterdam, The Netherlands.
- 71.Wang, S.-P., and J. T. Grayston. 1991. Three new serovars of Chlamydia trachomatis: Da, Ia, and L2a. J. Infect. Dis. 163:403-405. [DOI] [PubMed] [Google Scholar]
- 72.Wang, S.-P., C.-C. Kuo, and J. T. Grayston. 1973. A simplified method for immunological typing of trachoma-inclusion conjunctivitis-lymphogranuloma venereum organisms. Infect. Immun. 7:356-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, S. P., C. C. Kuo, R. C. Barnes, R. S. Stephens, and J. T. Grayston. 1985. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J. Infect. Dis. 152:791-800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.