Abstract
Neisseria meningitidis is a leading cause of septicemia and meningitis worldwide. N. meningitidis capsular polysaccharides have been classified into 13 distinct serogroups which are defined by antibody reactivity and structural analysis, and the capsule plays an important role in virulence. Serogroups A, B, C, W135, and Y have been reported to be clinically important. Several newly identified serogroup C isolates belonging to the unique sequence type 7 (ST-7) were identified in China. Since most ST-7 isolates from China belonged to serogroup A, the newly identified ST-7 serogroup C strains were proposed to have arisen from those belonging to ST-7 serogroup A. In this study, six ST-7 serogroup C and three ST-7 serogroup A isolates were analyzed by pulsed-field gel electrophoresis to confirm their sequence type. In order to clarify the genetic basis of capsular switching between ST-7 serogroup A and C strains, the whole capsular gene clusters and surrounding genes of the two representative ST-7 strains belonging to serogroups A and C, respectively, were sequenced and compared. Potential recombination sites were analyzed using the RDP3 beta software, and recombination-related regions in two other ST-7 serogroup A and five ST-7 serogroup C strains were also sequenced and compared to the representative ST-7 serogroup A and C strain sequences.
Neisseria meningitidis is a leading cause of septicemia and meningitis (13), and the extracellular polysaccharide capsule is a prerequisite for meningococcal virulence (19). As a consequence of capsular polysaccharide structural differences, 13 N. meningitidis serogroups have been identified, and the isolates most frequently associated with human disease belong to serogroups A, B, C, W135, and Y (9, 14). Serogroups B and C are responsible for most human infections in Europe and America, and serogroup A and C infections are more common in Africa and Asia (11). Antigenic diversity between N. meningitidis strains resulting from DNA transformation and/or subsequent recombination of capsular genes has been described (2, 15). Although the genetic mechanisms involved in the switching of capsular genes from B to C followed by switching from C to W135 have been described, the recombination sites associated with these events have not been clearly defined (2, 15).
In China, the predominant genotype of serogroup C N. meningitidis (menC) is sequence type 4821 (ST-4821), accounting for about 75% of menC found in 12 provinces (10). Previously, we identified several isolates in China belonging to the unique ST-7 and serogroup C by using multilocus sequence typing (MLST), porA typing, and comparative genomic hybridization (CGH) analyses (10). Since the majority of ST-7 isolates in China belong to serogroup A, we hypothesized that the newly identified ST-7 serogroup C arose from ST-7 serogroup A strains.
In this report, we analyze several ST-7 serogroup A and C isolates by comparing their respective whole capsular gene clusters and their surrounding regions. This analysis describes the genetic basis of the capsular switching events that resulted in the establishment of ST-7 serogroup C strains and identifies the recombination sites involved in the genetic exchange which gave rise to this novel serogroup.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All N. meningitidis strains used in this study were obtained from the National Institute for Communicable Disease Control and Prevention, Beijing, China, and are listed in Table 1. N. meningitidis strains were streaked on 5% sheep blood agar plates and incubated overnight in 5% CO2 at 37°C prior to DNA extractions that were performed using a QIAamp DNA blood mini kit as described by the manufacturer (Qiagen, Germany).
TABLE 1.
N. meningitidis isolates used in this study
| Isolate | Yr of isolation | Origin | Sitea | Clinical source | Serogroup | ST | Lineage name |
|---|---|---|---|---|---|---|---|
| 510602 | 2006 | Sichuan | TS | Close contact | C | 7 | ST-5 complex/subgroup III |
| 510603 | 2006 | Sichuan | TS | Close contact | C | 7 | ST-5 complex/subgroup III |
| 510604 | 2006 | Sichuan | TS | Close contact | C | 7 | ST-5 complex/subgroup III |
| 510605 | 2006 | Sichuan | TS | Close contact | C | 7 | ST-5 complex/subgroup III |
| 510606 | 2006 | Sichuan | TS | Close contact | C | 7 | ST-5 complex/subgroup III |
| 510609 | 2006 | Sichuan | TS | Close contact | C | 7 | ST-5 complex/subgroup III |
| 630501 | 2005 | Qinhai | CSF | Patient | A | 7 | ST-5 complex/subgroup III |
| 530514 | 2005 | Yunnan | TS | Close contact | A | 7 | ST-5 complex/subgroup III |
| 620523 | 2005 | Gansu | TS | Close contact | A | 7 | ST-5 complex/subgroup III |
TS, throat swab; CSF, cerebrospinal fluid.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed according to the method described by Shao et al. (12). Briefly, chromosomal DNA was digested with NheI and separated on 1.0% SeaKem gold agarose in 0.5× Tris-borate-EDTA buffer using the CHEF-DR III system from Bio-Rad (Hercules, CA) with the following parameters: initial pulse, 1 s; final pulse, 25 s with linear ramping; voltage, 6 V/cm; time of electrophoresis, 16 h; and temperature, 14°C. Clustering was performed using the Dice coefficient with a 1.2% optimization setting, and the dendrogram was generated by the unweighted pair group method using arithmetic averages (UPGMA).
Capsular gene sequencing and genetic analysis.
Capsular gene cluster sequencing of the 630501 and 510602 capsular gene clusters was carried out using 41 and 51 PCR walking primers based on the genome sequences of strains Z2491 and FAM18, respectively (Table 2). For strains 530514 and 620523, PCR walking primers wl_11591, wl_11593, wl_12006, wl_13151, wl_13152, and wl_13422 were used to identify upstream recombination sequences and primers wl_12014, wl_12016, wl_12018, wl_13426, wl_13427, wl_13428, wl_13429, and wl_13430 were used to identify downstream recombination sequences. For strains 510603, 510604, 510605, 510606, and 510609, PCR walking primers wl_11591, wl_11593, wl_13151, and wl_13152 were used for sequences upstream of the recombination region and primers wl_11967, wl_11969, wl_11971, wl_11973, wl_13160, wl_13161, wl_13162, wl_13232, and wl_13233 were used to identify sequences downstream of the recombination region. The PCR conditions were as follows: denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min for 30 cycles. Sequencing was carried out using an ABI 3730 automated DNA sequencer (Applied Biosystems, Foster City, CA), and sequencing data were analyzed as described previously (4).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Templatea |
|---|---|---|
| wl_11942 | ATGAGTAATCAAGATTTTTATGCG | A, B |
| wl_11943 | CCCCATACCCCATTGAAA | A, B |
| wl_11945 | GATAAAGTGGTAAAACGACACG | A |
| wl_11949 | TATGCCGTGCAACCCAGT | A, B |
| wl_11951 | ATGTCGACCGTTCTATCGG | A, B |
| wl_11952 | GCTGCTTCAAGCAGATTGA | B |
| wl_11953 | CGCATTGATTCGGTGATT | A, B |
| wl_11954 | ATAATATTTCATCGGCTCGG | A |
| wl_11955 | CATTTCTTGGAAGGCTTGC | B |
| wl_11957 | GTATGCAACGGGAGTAACAAT | B |
| wl_11959 | GGGCAAATCGTGATTGAT | B |
| wl_11963 | ATAATTGTTTTTTTATCGCCC | B |
| wl_11964 | CTGCCTGTTGTAAATCAAGC | B |
| wl_11965 | CTCCCATCCTTAGAAGAAGTC | B |
| wl_11966 | TTCTGTAGTCACGGGGTGA | B |
| wl_11967 | CGGCCTTTAATAATTTCCTG | B |
| wl_11968 | GGTACGGTTTCTGTGCCG | B |
| wl_11969 | AGCCTCTTCCTCGCCAC | A, B |
| wl_11970 | CGGTGCGTGAGTTTTATGA | A, B |
| wl_11971 | GCCAAGCCATCAGCATATA | A, B |
| wl_11972 | TTGTGATTATGGCGGTATTG | B |
| wl_11973 | CGGTAGCGGGCGATAAA | B |
| wl_11976 | GTCGATTTATTCCGCGTC | B |
| wl_11977 | GTAATTGATGATGGGGGAA | B |
| wl_11978 | CGGATTCCAACAATTGCT | B |
| wl_11979 | GTTGTTGGTATTCATCGTGA | B |
| wl_11980 | CGCAATGATTTTTTCAGG | B |
| wl_11981 | CAAGCAGATTGAATGTGCC | B |
| wl_12006 | TGGCGGGCATGATAATG | A |
| wl_12007 | CTTACGCATTTTATATATTTTGG | A |
| wl_12008 | CGCAATGCGAACACTCTC | A |
| wl_12010 | GGCGTAAACTTAAAAGAGACC | A |
| wl_12011 | CTTTGCAAATTTTTTAGCAG | A |
| wl_12012 | AAAAACGTGATAAGCTCCTAA | A |
| wl_12013 | GACGGTTAAGACTTTCATTGG | A |
| wl_12014 | CGCTGCATAGAAAATAGCG | A |
| wl_12015 | GGTTATGGCAGTGAGGCA | A |
| wl_12016 | GTTTGTTCGGCTGGGAA | A |
| wl_12017 | GATGACTTCTTTGGAAAGTGC | A |
| wl_12018 | ATCGTGCAAGACTGTCCG | A |
| wl_13148 | GCCATATTGTTGTCGAAACG | A, B |
| wl_13150 | TCGCGTTGAATCTGCAAA | A, B |
| wl_13151 | GGTTTTTCTTCGATGGAAAC | A |
| wl_13152 | GCGGTTTCGGTAAACAAAT | A, B |
| wl_13153 | GGATATCCGTCAAAATGCG | B |
| wl_13154 | TGCTTCGAAAGGTTGTCATG | B |
| wl_13155 | AGCAAGATACTGCGCATATT | B |
| wl_13157 | TCGGCATTTTCATATTTTGT | B |
| wl_13158 | GGAATGACTTGTTTGGCCT | B |
| wl_13160 | GTTCAAATTCCTGAAGTGGAG | A, B |
| wl_13161 | ATTTCGTTGGTGTATTTCGG | A, B |
| wl_13162 | TTTGCGATTACTGGCTATCC | B |
| wl_13165 | GGCGCAAAACATAAAATG | B |
| wl_13166 | ACCGCCTTTATTGGCAAA | B |
| wl_13228 | CGTTTGCGATGAAGCTGT | A, B |
| wl_13229 | AAGACCTTGCCGGCAAA | A, B |
| wl_13230 | ACTGGCCAGTTGCCAATA | B |
| wl_13231 | GATAATCGGCTGACTTTTCAGT | B |
| wl_13232 | GTGCCAGTGTCATTATTAGGG | B |
| wl_13233 | TTAACCCTATATTCCAACGAA | B |
| wl_13234 | TTCGCTGCCTTAAAGCGA | A, B |
| wl_13235 | GGATAGGCCTCCTGCTCC | B |
| wl_13236 | GACGATTCCGTATGAAGTCA | B |
| wl_13355 | AGTCGCAGGACGTGGAAG | A, B |
| wl_13356 | GCGATTTGGCGAGCTGG | B |
| wl_13357 | CACCCAAGATTCCGTCGT | B |
| wl_13422 | CGATGTGCCGTAAGGGC | A |
| wl_13423 | GGAAGTTACTGTTGTCTGCAA | A |
| wl_13424 | TAGAGATAGCCCGTTACTGC | A |
| wl_13425 | CCGATTTTGTTTTAAGGTTG | A |
| wl_13426 | GCATTTCACGATGCTGTG | A |
| wl_13427 | GCTTTCTGAAGCCATTGG | A |
| wl_13428 | GAAATCTCGCAACAAATGA | A |
| wl_13429 | CGACAGCGTCACGACTTAC | A |
| wl_13430 | CATCAGAATCGCACGCG | A |
A, 630501; B, 510602.
Recombination analysis.
The sequences were compared using the Cluster X program as described previously (16), and recombination events were analyzed using the RDP3 beta recombination detection program, which includes the methods RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, PhylPro, LARD, and 3Seq, according to the instruction manual (8).
Nucleotide sequence accession numbers.
The DNA sequences corresponding to the capsular genes from strains 510602 and 630501 were deposited in GenBank under accession numbers HM037270 and HM037271, respectively.
RESULTS AND DISCUSSION
PFGE analysis of ST-7 serogroup A and C isolates.
For genotype confirmation, nine isolates, including 6 isolates belonging to serogroup C and 3 belonging to serogroup A, were analyzed by PFGE (Table 1 and Fig. 1). This analysis confirmed that all the strains used in this study had almost identical PFGE patterns (Fig. 1), supporting our previous MLST analysis which demonstrated that these isolates possessed the same sequence type, ST-7 (10).
FIG. 1.
PFGE analysis of N. meningitidis strains used in this study. Chromosomal DNA from the respective strains was digested with NheI and subjected to electrophoresis. Clustering was performed using the Dice coefficient and a 1.2% optimization setting. The dendrogram was generated using the unweighted pair group method using arithmetic averages (UPGMA). The numbers on the scale represent the Dice coefficient, which reflects the similarity of electronic bands.
Sequencing of the capsular gene clusters for two N. meningitidis ST-7 serogroup A and C strains.
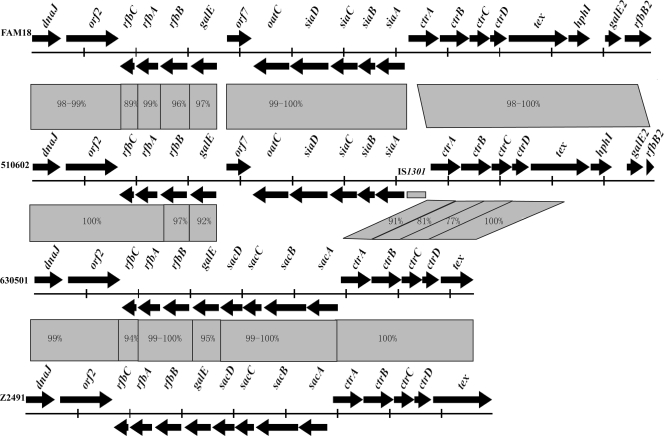
ST-7 strains 630501 and 510602 belong to serogroups A and C, respectively. Since most ST-7 strains belong to serogroup A, we hypothesized that the ST-7 serogroup C strains were newly emerged by capsular switching from serogroup A, probably as a consequence of capsular gene recombination events. The capsular gene clusters (and their corresponding surrounding regions) from these two strains were sequenced using the primers described herein (Table 1). Sixteen primer sequences were shared between 510602 and 630501. A 16,900-base-pair (bp) sequence containing 15 open reading frames (ORFs), including dnaJ, rfbC, rfbA, rfbB, galE, sacD, sacC, sacB, sacA, ctrA, ctrB, ctrC, ctrD, tex, and orf2 (encoding a putative oligopeptide transporter), was obtained for strain 630501 (Fig. 2). A 23,930-bp sequence containing 20 ORFs, including dnaJ, rfbC, rfbA, rfbB, galE, oatC, siaD, siaC, siaB, siaA, ctrA, ctrB, ctrC, ctrD, tex, hphI (truncated), galE2, rfbB2, orf2 (encoding a putative oligopeptide transporter), and orf7 (function unknown), was obtained for strain 510602 (Fig. 2). The ctrA gene shares 100% identity with those of different isolates from serogroups C, B, Y, and W135 but not serogroup A. An insertion of IS1301 was found between the siaA and ctrA genes, which was reported to lead to an increase in the amount of serogroup C N. meningitidis capsular polysaccharide in a previous study (18) (Fig. 2). It is noted that the sequence, including 147 bp of the siaA 5′ end, 139 bp of the ctrA 5′ end, and a 1,004-bp region between siaA and ctrA in strain 510602, shared 99% identity with the sequence from N. meningitidis strain 95-325, except that a 32-bp sequence from bp 170 to 201 in this region of 510602 was absent in strain 95-325. Strain 95-325 belongs to serogroup C:ET15, and the serotype of the isolate is 2a:P1.2 (17). Twelve and 16 DNA uptake sequences (DUS), which promote the initial stages of DNA uptake and therefore facilitate DNA transformation and recombination events (1), were found in the capsular gene clusters of 510602 and 630501, respectively. DUS were frequently found to be present in the N. meningitidis genome, especially in poorly conserved genes (5), and they are the molecular basis of preferential uptake of “self” DNA over foreign DNA in N. meningitidis (3, 7).
FIG. 2.
Comparison of capsular genes of N. meningitidis strains 510602, 630501, FAM18, and Z2491. The sequences were aligned using the Cluster X program. DNA identities of different genes are shown between them. The gray rectangle in the directional arrows of the gene cluster of 510602 is IS1301.
Analysis of capsular gene and recombination site sequences.
The capsular gene cluster and flanking DNA sequences of ST-7 strains 630501 and 510602 (proposed to have undergone capsular switching) were fully sequenced and compared to the homologous gene clusters of the published genome sequences (NC_003116 and NC_008767) from strains Z2491 (ST-4 serogroup A) and FAM18 (ST-11 serogroup C), respectively. Except for different capsular biosynthesis genes, the identities for the rfbB (97%), galE (92%), ctrA (91%), ctrB (81%), and ctrC (77%) gene sequences of 630501 and 510602 were lower than those found for other flanking genes. These five gene sequences shared higher identities with sequences from strains of the same serogroups (95 to 100%), and the rfbB upstream sequences shared lower identities with those from the same serogroups (89 to 99%) (Fig. 2). Based on these data, the serogroup C-associated recombination event appeared to include the rfbB, galE, and siaABCD sequences and at least the ctrABC sequences from the ctr gene operon (which are responsible for transporting polysaccharide with phospholipid substitutions across the inner and outer membranes) (6). A putative upstream recombination site seemed to be located in the rfbB gene; however, downstream recombination sites seemed unlikely since the downstream gene sequences of ctrC shared 100% identity in all strains examined.
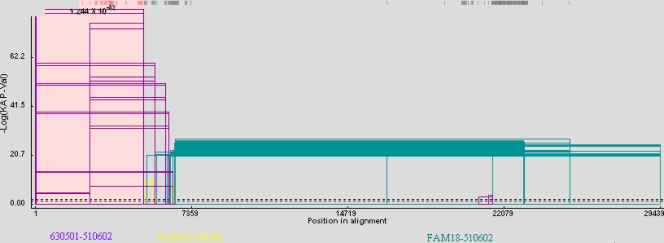
We examined the possibility that 510602 (ST-7 serogroup C) evolved from 630501 (ST-7 serogroup A) as a consequence of gene cluster recombination by analyzing the recombination events resulting in strain 510602. A recombination graph was generated, and a potential upstream breakpoint was predicted at position 5055 in strain 510602 and at position 5072 in strain 630501 located 3′ from rfbB (Fig. 3). This breakpoint was also confirmed using the GENECONV method and other methods in RDP3 beta (Table 3). The nucleotide sequences before the breakpoint in strain 510602 were almost identical to the 630501 sequences (Fig. 4). A predicted downstream breakpoint region was not identified using the techniques described.
FIG. 3.
Analysis of the recombination event in N. meningitidis strain 510602. Sequences of N. meningitidis strains 510602, 630501, and FAM18 were compared and analyzed using the GENECONV method (the sequence of 630501 was proposed as the acceptor). The x axis shows genome length (nucleotide numbers of the consensus sequence), and the y axis shows the negative log of the probability value (P-Val). The beginning and end of the fragment that underwent recombination are connected by a line to form an open rectangle, and the height of the open rectangle is proportional to the negative log of the P value. The purple open rectangles represent the recombination events between strains 630501 and 510602, the yellow ones represent the events between strains 630501 and FAM18, and the green ones represent the events between strains 510602 and FAM18. The dashed lines represent the P value cutoffs.
TABLE 3.
Probability of recombination events in strain 510602 analyzed by all the methods in RDP3 beta software
| Method | No. of recombination events | Breakpoint | P value |
|---|---|---|---|
| RDP | 1 | 5055 | 2.900 × 10−84 |
| GENECONV | 1 | 5055 | 1.243 × 10−82 |
| BootScan | 1 | 5055 | 1.677 × 10−84 |
| MaxChi | 1 | 6537 | 2.506 × 10−27 |
| Chimaera | 1 | 5056 | 1.789 × 10−27 |
| SiScan | 1 | 4006 | 1.608 × 10−33 |
| PhylPro | |||
| LARD | |||
| 3Seq | 1 | 5055 | 9.993 × 10−08 |
FIG. 4.
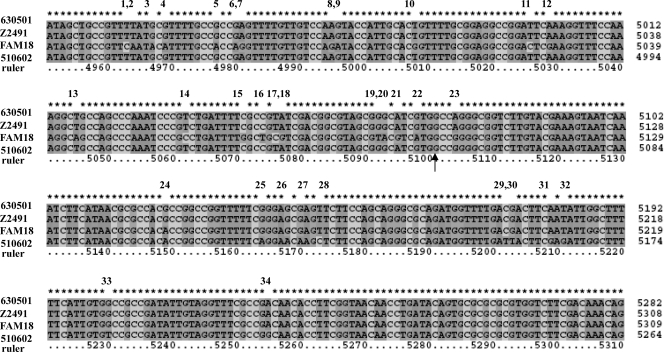
Nucleotide comparison between N. meningitidis strains 510602, 630501, FAM18, and Z249 near the recombination sites. The respective sequences were aligned, and the arrow indicates the breakpoint. The positions of each of the four sequences are on the right of the sequence alignments, and “ruler” means nucleotide positions in the consensus sequence. The numbers 1 to 34 above the sequence alignments present the nucleotide polymorphisms in this region. From numbers 1 to 22, which are before the breakpoint, the nucleotides of 510602 and 630501 are the same, and from 23 to 34, after the breakpoint, the nucleotides of 510602 and 630501 are different.
Confirmation of the identified recombination sites.
We sequenced potential up- and downstream recombination regions from two additional ST-7 serogroup A strains and from five ST-7 serogroup C strains to confirm the recombination events described herein. DNA sequences of 3,645 bp (upstream recombination site) and 4,591 bp (downstream recombination site) were identified from ST-7 serogroup A strains 530514 and 620523, respectively. DNA sequences of 2,452 bp corresponding to the upstream recombination site and of 5,776 bp corresponding to the downstream recombination site were identified for the ST-7 serogroup C strains 510603, 510604, 510605, 510606, and 510609.
The sequences from strains 530514 and 620523 were 100% identical to the corresponding sequences of 630501 (at positions 4389 to 8033 and 11501 to 16091). The sequences from strains 510603, 510604, 510605, 510606, and 510609 shared 100% identity to the corresponding 510602 sequences (at positions 4371 to 6822 and 13468 to 19243) (data not shown). These data confirmed that the recombination sites were conserved following the ST-7 strain A-to-C capsular change.
The data presented above suggested that the ST-7 serogroup C strains, including 510602, 510603, 510604, 510605, 510606, and 510609, arose from ST-7 serogroup A strains such as 630501, 530514, and 620523. The region between siaA and ctrA, including IS1301 in 510602, shares 99% identity with that of strain 95-325, indicating that strain 95-325 may be the putative capsular gene cluster donor.
The data presented in this report suggest that capsular switching from serogroup A to C strains is the result of capsular gene rearrangement. In this study, the genetic basis of capsular switching from A to C was clarified and the recombination sites were described.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (grants 30900255, 30788001, and 30870070), the National 863 Program (grants 2007AA02Z106 and 2007AA021303), the National Key Programs for Infectious Diseases of China (grants 2008ZX10004-002, 2008ZX10004-009, 2009ZX10004-108, 2008ZX10003, and 2008ZX10001-004), and Tianjin Research Program of Application Foundation and Advanced Technology (grant 10JCYBJC10100).
Editor: J. N. Weiser
Footnotes
Published ahead of print on 12 July 2010.
REFERENCES
- 1.Ambur, O. H., S. A. Frye, and T. Tonjum. 2007. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J. Bacteriol. 189:2077-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beddek, A. J., M. S. Li, J. S. Kroll, T. W. Jordan, and D. R. Martin. 2009. Evidence for capsule switching between carried and disease-causing Neisseria meningitidis strains. Infect. Immun. 77:2989-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danner, D. B., R. A. Deich, K. L. Sisco, and H. O. Smith. 1980. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11:311-318. [DOI] [PubMed] [Google Scholar]
- 4.Feng, L., S. N. Senchenkova, J. Yang, A. S. Shashkov, J. Tao, H. Guo, G. Zhao, Y. A. Knirel, P. Reeves, and L. Wang. 2004. Structural and genetic characterization of the Shigella boydii type 13 O antigen. J. Bacteriol. 182:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Findlay, W. A., and R. J. Redfield. 2009. Coevolution of DNA uptake sequences and bacterial proteomes. Genome Biol. Evol. 2009:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frosch, M., U. Edwards, K. Bousset, B. Krausse, and C. Weisgerber. 1991. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol. Microbiol. 5:1251-1263. [DOI] [PubMed] [Google Scholar]
- 7.Goodman, S. D., and J. J. Scocca. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 85:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heath, L., E. van der Walt, A. Varsani, and D. P. Martin. 2006. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 80:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peltola, H. 1983. Meningococcal disease: still with us. Rev. Infect. Dis. 5:71-91. [DOI] [PubMed] [Google Scholar]
- 10.Peng, J., X. Zhang, Z. Shao, L. Yang, and Q. Jin. 2008. Characterization of a new Neisseria meningitidis serogroup C clone from China. Scand. J. Infect. Dis. 40:63-66. [DOI] [PubMed] [Google Scholar]
- 11.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, T. Popovic, and J. M. Hughes. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378-1388. [DOI] [PubMed] [Google Scholar]
- 12.Shao, Z., W. Li, J. Ren, X. Liang, L. Xu, B. Diao, M. Li, M. Lu, H. Ren, Z. Cui, B. Zhu, Z. Dai, L. Zhang, X. Chen, B. Kan, and J. Xu. 2006. Identification of a new Neisseria meningitidis serogroup C clone from Anhui province, China. Lancet 367:419-423. [DOI] [PubMed] [Google Scholar]
- 13.Stephens, D. S. 1999. Uncloaking the meningococcus: dynamics of carriage and disease. Lancet 353:941-942. [DOI] [PubMed] [Google Scholar]
- 14.Swartley, J. S., L. J. Liu, Y. K. Miller, L. E. Martin, S. Edupuganti, and D. S. Stephens. 1998. Characterization of the gene cassette required for biosynthesis of the (alpha1→6)-linked N-acetyl-d-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J. Bacteriol. 180:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swartley, J. S., A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. U. S. A. 94:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyler, S., and R. Tsang. 2004. Genetic analysis of Canadian isolates of C:2a:P1.2,5 and B:2a:P1.2,5 Neisseria meningitidis strains belonging to the hypervirulent clone of ET-15. Can. J. Microbiol. 50:433-443. [DOI] [PubMed] [Google Scholar]
- 18.Uria, M. J., Q. Zhang, Y. Li, A. Chan, R. M. Exley, B. Gollan, H. Chan, I. Feavers, A. Yarwood, R. Abad, R. Borrow, R. A. Fleck, B. Mulloy, J. A. Vazquez, and C. M. Tang. 2008. A generic mechanism in Neisseria meningitidis for enhanced resistance against bactericidal antibodies. J. Exp. Med. 205:1423-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel, U., S. Hammerschmidt, and M. Frosch. 1996. Sialic acids of both the capsule and the sialylated lipooligosaccharide of Neisseria meningitis serogroup B are prerequisites for virulence of meningococci in the infant rat. Med. Microbiol. Immunol. 185:81-87. [DOI] [PubMed] [Google Scholar]