Abstract
Yersinia pestis, the causative agent of plague, has recently diverged from the less virulent enteropathogen Yersinia pseudotuberculosis. Its emergence has been characterized by massive genetic loss and inactivation and limited gene acquisition. The acquired genes include two plasmids, a filamentous phage, and a few chromosomal loci. The aim of this study was to characterize the chromosomal regions acquired by Y. pestis. Following in silico comparative analysis and PCR screening of 98 strains of Y. pseudotuberculosis and Y. pestis, we found that eight chromosomal loci (six regions [R1pe to R6pe] and two coding sequences [CDS1pe and CDS2pe]) specified Y. pestis. Signatures of integration by site specific or homologous recombination were identified for most of them. These acquisitions and the loss of ancestral DNA sequences were concentrated in a chromosomal region opposite to the origin of replication. The specific regions were acquired very early during Y. pestis evolution and were retained during its microevolution, suggesting that they might bring some selective advantages. Only one region (R3pe), predicted to carry a lambdoid prophage, is most likely no longer functional because of mutations. With the exception of R1pe and R2pe, which have the potential to encode a restriction/modification and a sugar transport system, respectively, no functions could be predicted for the other Y. pestis-specific loci. To determine the role of the eight chromosomal loci in the physiology and pathogenicity of the plague bacillus, each of them was individually deleted from the bacterial chromosome. None of the deletants exhibited defects during growth in vitro. Using the Xenopsylla cheopis flea model, all deletants retained the capacity to produce a stable and persistent infection and to block fleas. Similarly, none of the deletants caused any acute flea toxicity. In the mouse model of infection, all deletants were fully virulent upon subcutaneous or aerosol infections. Therefore, our results suggest that acquisition of new chromosomal materials has not been of major importance in the dramatic change of life cycle that has accompanied the emergence of Y. pestis.
Yersinia pestis, the causative agent of plague, is a highly uniform clone that has recently diverged from the enteric pathogen Yersinia pseudotuberculosis (2). Despite their close genetic relationship, the plague bacillus and its recent ancestor differ radically in their pathogenicity and transmission. Y. pseudotuberculosis is transmitted by the fecal-oral route and causes enteritis of moderate intensity in humans (53). In contrast, Y. pestis is transmitted by flea bites or aerosols and causes highly severe infections, i.e., bubonic and pneumonic plague (41).
The transformation of Y. pseudotuberculosis into Y. pestis has been accompanied by the acquisition of two plasmids, pFra and pPla. The 101-kb pFra plasmid is required for colonization of and survival in the flea midgut and appears to increase the efficacy of infection after flea bites (50). However, this plasmid is dispensable for pathogenicity of in vitro-grown Y. pestis upon subcutaneous (s.c.) infection in mice (21) or aerosol infection in African green monkeys (10). The 9.6-kb pPla plasmid, which encodes the plasminogen activator and protease Pla, is critical for the virulence of some Y. pestis strains (63). This plasmid was shown to be required for the translocation of the bacteria from their intradermal site of inoculation to the draining lymph node (54) but not for their direct dissemination to the bloodstream (51, 54, 63). Nonetheless, recent studies have shown that Y. pseudotuberculosis, which is naturally deprived of pPla, can reach the lymph node draining the intradermal site of inoculation as efficiently as Y. pestis (22). Therefore, the stage of the infectious process during which pPla exerts its activity remains unclear. Furthermore, acquisition of pPla is not sufficient to account by itself for the dramatic rise in virulence of Y. pestis, because there are natural isolates of Y. pestis that lack pPla and still are highly virulent to mice (63) or guinea pigs (49) after subcutaneous injection (49, 63), and introduction of pPla in Y. pseudotuberculosis does not increase its virulence in mice (34, 42).
Chromosomal determinants are thus likely to be involved in the modifications in pathogenicity and life cycle that accompanied the emergence of Y. pestis. Surprisingly, it appears that the transformation of the enteropathogen Y. pseudotuberculosis into the plague bacillus has mostly been characterized by extensive loss of genetic material and functions (8). Ten chromosomal loci conserved in the species Y. pseudotuberculosis have been lost by Y. pestis during the course of its differentiation (43, 44), and 208 genes present in the two species have been inactivated during the transformation process (8). The natural inactivation of one of these genes has recently been shown to have participated to the change in Y. pestis mode of transmission (19, 57).
In contrast, only a few new chromosomal genetic materials, designated here as Y. pestis specific, have been acquired during the transformation of an enteropathogen into the hypervirulent plague bacillus. One of these loci encodes a filamentous phage (YpfΦ), which is stably integrated into the chromosome of Y. pestis Orientalis and participates (albeit moderately) to its pathogenicity (13). This phage is mainly extrachromosomal and is lost at high frequencies in the other biovars of Y. pestis (13, 38). Previous comparative genomic analyses of Y. pestis and Y. pseudotuberculosis have identified additional Y. pestis-specific genes (8, 27, 46, 62, 64, 65). Some of these studies involved large arrays of Y. pestis strains (up to 260), but the number of Y. pseudotuberculosis isolates was often much more limited. Similarly, while the number of Y. pestis genome sequences available is increasing rapidly (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?database=386656), only two genomes of Y. pseudotuberculosis have been published until now (8, 17). Nonetheless, since Y. pseudotuberculosis is a much older species, it displays a level of genetic polymorphism much higher than that of the relatively clonal species Y. pestis (2, 62). Therefore, the delineation of Y. pestis-specific chromosomal regions based on the comparison with a few Y. pseudotuberculosis isolates could not be accurate.
In this study, a large panel of Y. pestis and Y. pseudotuberculosis strains was screened to identify chromosomal regions that specify the plague bacillus. The genetic organization, the potential mode of integration into the ancestral bacterial chromosome, the putative function, and the microevolutionary step at which the plague bacillus acquired these regions were studied. Finally, the role of these new genetic materials on Y. pestis growth, the ability to colonize and block fleas, and the pathogenicity in the mouse were evaluated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The natural Y. pestis and Y. pseudotuberculosis isolates used in this study are listed in Tables S1 and S2 in the supplemental material. These isolates were taken from the collection of the Yersinia Research Unit (Institut Pasteur). The various Y. pestis CO92 derivatives constructed here are described in Table S3 in the supplemental material. All bacteria were grown in Luria-Bertani (LB) or M63S broth [KH2PO4, 0.1 M; (NH4)2SO4, 0.2%; MgSO4, 0.02%; FeSO4, 0.00005%] with 0.2% glucose and 0.2% Casamino Acids and on LB agar plates supplemented with 0.002% (wt/vol) hemin at 28°C or 37°C. Chloramphenicol (25 μg/ml), kanamycin (Km; 30 μg/ml), or irgasan (0.1 μg/ml) was added to the medium when necessary. The pigmentation phenotype of Y. pestis was determined after 4 days of growth on Congo red agar plates (58). All experiments involving Y. pestis strains were performed in a biosafety level 3 (BSL3) laboratory.
In silico analysis of Yersinia genomes.
The genome sequences of Y. pestis CO92 (40) and Y. pseudotuberculosis IP32953 (8) were compared pairwise using the Artemis Comparison Tool (ACT) (http://www.sanger.ac.uk/Software/ACT). Reannotation was performed using the AMIGene (annotation of microbial genes) software program (6). DNA and protein sequences were analyzed with BLAST programs (3). The search for syntenies was performed with the tools implemented in the MicroScope platform (61) and available via the MaGe interface (https://www.genoscope.cns.fr/agc/mage/YersiniaScope).
DNA manipulations.
Bacterial genomic DNA was extracted with an Isoquick kit (ORCA Research, Inc.). All primers used for PCR amplification are listed in Table S4 in the supplemental material. PCRs were performed by following a standard procedure, with 1 unit of Taq polymerase (Roche) or 1 unit of a 3:1 mixture of Taq and Pfu (Stratagene) polymerases in the supplier's buffer. DNA sequencing of PCR fragments was done by Millegene.
RNA isolation and transcript analysis.
RNA extraction was performed with a TRI reagent kit (Ambion) on 4 ml of Y. pestis grown to stationary phase in LB at 28°C. DNA was removed from the RNA samples by adding 4 U DNase, as described in the instructions for the DNA-free kit (Ambion). Total RNA was quantified by optical density at 260 nm (OD260), and 200 ng was used for reverse transcription-PCR (RT-PCR) analysis using primer pairs 623/624 (for control of RNA quality and absence of DNA) and 703A/B (for detection of YPO2469 specific mRNA transcript) (see Table S3 in the supplemental material).
Mutagenesis.
Deletion of Y. pestis-specific sequences was done by allelic exchange with a Km (km) resistance cassette, in accordance with the short-flanking-homology procedure (14) for R1pe to R5pe and CDS2pe and with the long-flanking-homology procedure (14) for R6pe and CDS1pe. The Km resistance cassette was obtained by PCR amplification using plasmid pGP704N-km as a template and the primer pairs listed in Table S3 in the supplemental material. The PCR products were introduced by electroporation into Y. pestis CO92(pKOBEG-sacB), as described previously (14). Recombinant colonies were selected on Km agar plates. Deletion of each target region was verified by PCR with primers located (i) on each side of the inserted Km cassette and (ii) within each specific region (see Table S4 in the supplemental material).
Animal infections.
Prior to infection, the presence of the three plasmids was systematically checked by PCR with primer pairs 157A/157B (caf1), 159A/159B (pla), and 160A/160B (yopM). The presence of the high-pathogenicity island (HPI) was determined by plating the Y. pestis clones on Congo red agar plates. The animals used for s.c. and aerosol infections were 5-week-old female OF1 mice (Charles River). The animals were infected s.c. with 10 CFU/ml of bacterial suspensions from cultures grown for 48 h at 28°C on LB agar plates. Aerosol infections were induced by exposing mice for 10 min to an atmosphere containing droplets of a Y. pestis suspension (108 CFU/ml of saline). Droplets (1-μm median size) were generated by a Raindrop nebulizer (Tyco) and were introduced in an all-glass aerosol chamber devised for a nose-only exposure of animals (37). Lethality was recorded daily for 3 weeks. All animal experiments were performed in a BSL3 animal facility.
Flea infections.
Xenopsylla cheopis fleas were infected by allowing them to feed on blood containing ∼5 × 108 CFU/ml of the various Y. pestis CO92 derivatives, using a previously described artificial feeding system (28, 29). A sample of 20 female fleas was collected immediately after the infectious blood meal, placed at −80°C, and subsequently used for CFU plate count determinations of the average infectious dose per flea (28, 29, 33). An additional group of 45 to 113 fleas (approximately equal numbers of males and females) that took an infectious blood meal were maintained at 21°C for 4 weeks, during which time they were allowed to feed on uninfected mice on days 2, 6, 9, 13, 16, 20, 23, and 27. Immediately after each of these feedings, all fleas were individually examined under a dissecting microscope to determine how many had taken a normal blood meal (indicated by the presence of fresh red blood filling the midgut) and how many were blocked (indicated by the presence of fresh red blood only in the esophagus, anterior to the proventriculus) (29). A matched group of uninfected control fleas that fed on blood containing no bacteria was routinely included in each infection experiment. Excess mortality [(percent mortality of infected fleas) − (percent mortality of uninfected control fleas)] was recorded during the 4-week period after the infectious blood meal. After 4 weeks, a final sample of 11 to 20 surviving female fleas was collected to determine the infection rate and average number of CFU per infected flea (28, 29).
RESULTS AND DISCUSSION
Delineation of the Y. pestis-specific chromosomal loci.
A previous in silico comparative analysis of the genomes of Y. pestis (strains CO92 and KIM10+) and Y. pseudotuberculosis (strain IP32953) identified 21 chromosomal clusters specific to the two Y. pestis genomes (8). These loci were either single coding sequences (CDSs) or clusters of two or more adjacent genes (designated Rpe, for Y. pestis-specific regions). When the presence of these 21 clusters was previously tested in a panel of 22 Y. pestis and Y. pseudotuberculosis isolates, only six clusters were found to be present in all Y. pestis and absent from all Y. pseudotuberculosis strains examined (8). Using similar criteria, another study performed on a larger panel of 88 Y. pseudotuberculosis strains identified only two Y. pestis-specific chromosomal clusters (62).
In this study, we wanted to perform an in-depth analysis of chromosomal regions that specify Y. pestis in order to better understand how they might have been acquired and whether they participated to the change in life cycle and pathogenicity of the plague bacillus. These regions could have been horizontally acquired by Y. pestis after its divergence from Y. pseudotuberculosis, but it is also possible that a few of them were first horizontally acquired by the specific Y. pseudotuberculosis strain from which the plague bacillus emerged and then vertically transmitted to Y. pestis because their presence was a prerequisite for the emergence of the plague bacillus. Furthermore, the Y. pestis genome is known to undergo rearrangements that lead to loss of genetic material, as previously shown for the HPI (11) and the filamentous phage YpfΦ (13). Taking all these considerations into account, we considered here as Y. pestis specific a locus present in most Y. pestis strains and absent from almost all Y. pseudotuberculosis isolates studied.
To identify all potentially critical regions, we proceeded in four steps: (i) the in silico comparative analysis of the genomes of CO92 and IP32953 was rerun, (ii) new primers were designed and used to screen the same set of 22 Y. pestis and Y. pseudotuberculosis isolates previously examined (8) to confirm the specificity of the regions studied, (iii) less-stringent criteria were used to select chromosomal loci that might have participated in the emergence of the plague bacillus (i.e., presence in most Y. pestis and absence from almost all Y. pseudotuberculosis strains), and (iv) the specificity and conservation of the selected loci were then tested across a large panel of 76 additional strains of the two species. Since one of these loci corresponds to the Y. pestis filamentous phage YpfΦ, which has already been the subject of an in-depth analysis (13), this genomic region was not included in the specific regions analyzed here.
Our in silico genomic comparison of CO92 and IP32953 allowed us to identify, in addition to the 21 clusters previously found, two additional Y. pestis CDSs, YPO1899 and YPO2469, that were absent from the genome of Y. pseudotuberculosis IP32953. These two CDSs were not retained in the initial comparative analysis (8), probably because their predicted coding potential was low. However, since their functionality is unknown, we decided to keep them for further screening. The first PCR screening was then done to confirm the presence or absence of these 23 loci in the 13 Y. pestis strains belonging to evolutionary branches 1 and 2 (1) and the 9 Y. pseudotuberculosis strains of serotypes I to V previously analyzed, with 51 pairs of primers newly designed and located within one or more open reading frames (ORFs) of each locus (see Table S1 in the supplemental material). As explained above, the absence in a few Y. pestis strains or the presence in a single isolate of Y. pseudotuberculosis was not considered a redhibitory condition for the selection of a given locus. On the basis of these criteria, 10 of the 23 initial loci were not further considered Y. pestis specific (see Table S1). Despite the presence of two nonspecific ORFs (YPO3946 and YPO3947) in locus 21, this region was kept for further analysis because it also contained two specific ORFs (YPO3945 and YPO3948). Altogether, 13 loci were thus retained for the second screening (see Table S1). These included the two (62) or five (8) Y. pestis-specific clusters previously identified as well as eight additional loci, of which two corresponded to the two CDSs newly detected by our in silico comparative analysis.
The specificity of these 13 loci was further investigated using the same set of primers to screen by PCR a panel of 46 additional Y. pestis and 30 additional Y. pseudotuberculosis strains (see Table S2 in the supplemental material). This screen showed that the genomes of some Y. pestis strains (for instance, IP539, IP537, IP540, and IP566) were prone to frequent rearrangements leading to a significant loss of genetic material. These strains were isolated during the 1950s, and their genomic rearrangements most likely occurred upon prolonged storage. It also appeared that some loci (especially YPO2469, YPO2484, and YPO3616) were frequently lost (while others were systematically conserved), suggesting both that they are located in an unstable chromosomal region and that these regions do not carry genes essential for in vitro survival of Y. pestis. Since the number of isolates was large in the second screen, we decided to retain for further physiological analyses the loci for which ≥90% of the 39 Y. pseudotuberculosis strains studied during the first and second screens gave a negative or weak signal. As shown in Table S2 in the supplemental material, eight loci, corresponding to six regions (designated R1pe to R6pe) and two CDSs (CDS1pe and CDS2pe) met this criterion.
After our screening was performed, the genome sequences of three other Y. pseudotuberculosis strains (IP31758 [17], PB1+ [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&cmd=ShowDetailView&TermToSearch=28745], and YPIII [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&cmd=ShowDetailView&TermToSearch=28743]) became available. BLAST analyses between these sequenced genomes were performed and the nucleotide sequences of the eight Y. pestis-specific loci selected. This genome analysis confirmed the results of the PCR screen, except for YPO3945 in R6pe (see Table S2 in the supplemental material). Indeed, a similar sequence (97% nucleotide identity) was found in the genomes of Y. pseudotuberculosis IP31758 and YPIII, although it lacked the start codon present in the Y. pestis sequence. A portion of YPO3945 was also found in the PB1+ genome. Sequence analysis of YPO3945 revealed that one of the primers that we used to amplify this locus was located in a region exhibiting a certain degree of polymorphism, which could explain our negative PCR results. This agrees with the results of the previous study (in which other primer pairs were used), showing the presence of this sequence in several Y. pseudotuberculosis strains (8). Therefore, PCR screening, although sensitive, very convenient, and easy to perform, may overestimate the number of specific regions. YPO3945 should thus not be considered Y. pestis specific, but on the basis of certain peculiarities of R6pe that will be described below, we decided to keep the entire region, and not only YPO3948, for future analyses.
Organization and putative functions of the Y. pestis-specific chromosomal regions.
The results of the BLAST analyses performed on the deduced amino acid sequences of each region and CDS indicated that the majority of the coding sequences are predicted to encode hypothetical proteins or proteins of unknown functions (see Table S5 in the supplemental material). An in-depth in silico analysis of the organization and putative function of each specific chromosomal locus, along with a comparison with the homologous Y. pseudotuberculosis regions in which each Y. pestis-specific locus has inserted itself, was then performed.
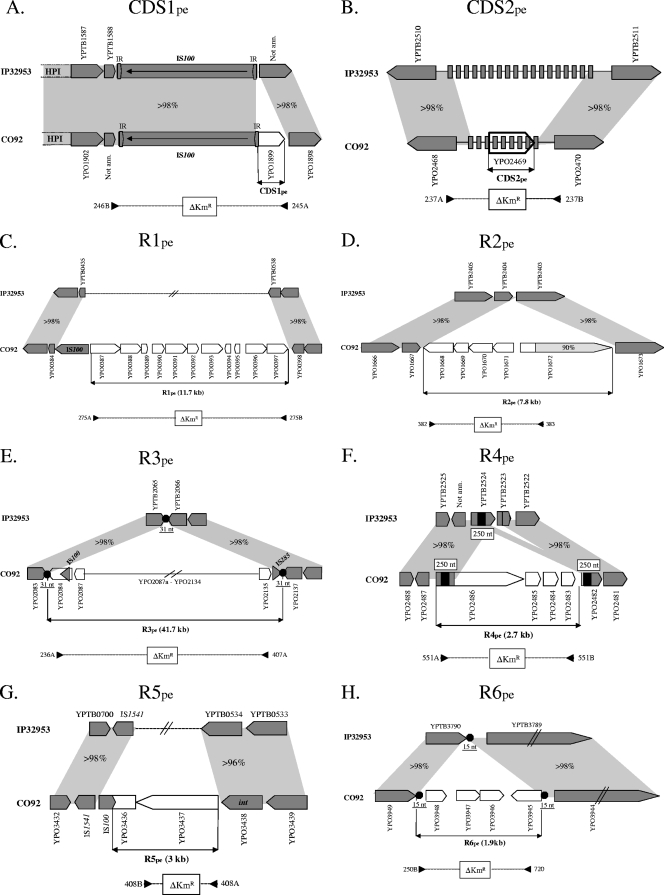
CDS1pe (YPO1899) is located at one extremity of the Y. pestis HPI (7) and, more specifically, at the 5′ end of the IS100 copy (Fig. 1A). This locus most likely corresponds to region A, previously found to be specific for the Y. pestis HPI (24). Its putative start codon is situated within the inverted repeat of IS100. The YPO1899 predicted product shares no significant similarity with proteins in databases, but its DNA sequence contains a 159-bp region similar (70% nucleotide identity) to the iso-IS630 transposase of Salmonella enterica serovar Choleraesuis (GenBank accession no. D10689). To determine whether YPO1899 could be a functional coding sequence, we performed a reverse transcription assay (RT-PCR) on total RNA extracted from Y. pestis CO92 grown to stationary phase in LB at 28°C. No mRNA was detected (data not shown). Our results thus suggest that YPO1899 is not functional, although the possibility that this CDS is transcribed under other conditions cannot be ruled out. It is thus possible that this CDS is a remnant of an insertion sequence (IS) that, like IS100, inserted itself into a hot spot of IS integration. This is reminiscent of the local hopping of IS3 elements found at one extremity of the Yersinia enterocolitica HPI (47).
FIG. 1.
Genetic organization of the eight chromosomal loci acquired by Y. pestis and comparison with the homologous regions in Y. pseudotuberculosis. White arrows indicate CDSs present in Y. pestis CO92 but not in Y. pseudotuberculosis IP32953. Dark gray arrows indicate CDSs harboring >98% nucleotide identity between Y. pestis and Y. pseudotuberculosis. Light gray areas represent the conserved regions on each side of the acquired loci. Dashed lines with arrows below each region indicate the DNA fragments that have been replaced by a kanamycin cassette, and the numbers at each extremity correspond to the primer pairs used to generate the fragment to be deleted. Region not annotated are marked “Not ann.” In panel B, gray rectangles correspond to repeats identical in Y. pseudotuberculosis and Y. pestis. In panel F, the 250-nt-long DNA repeats found at the borders of R4pe are indicated by black rectangles. In panels E and H, black circles represent direct repeated sequences found at the extremities of each region, and their sizes in nucleotides are indicated below. In panel H, dashed arrows indicate ORFs that are conserved in Y. pestis and Y. pseudotuberculosis although they might have been acquired independently (see Fig. S1 in the supplemental material for a more detailed comparison of this region).
CDS2pe (YPO2469) is located within a region containing clustered, regularly interspaced short palindromic repeats (CRISPRs) (Fig. 1B), which consists of 28 bp-long repeated sequences separated by 32- to 33-bp unique spacers (31). Addition of new spacers renders these structures highly plastic and generates a polymorphism which can be used for phylogenetic analyses (45). The fact that several Y. pestis strains were negative during our PCR screens (see Table S2 in the supplemental material) suggests that they had lost (or not acquired) the corresponding spacers. In CO92, the CRISPR region contains nine repeats, whereas in IP32953, there are 18 repeats (Fig. 1B). However, the sequences in the CO92 and IP32953 spacers are different, indicating a different origin. The YPO2469 ORF overlaps several 28-bp-long repeated sequences, and its start and stop codons are located inside specific spacers (Fig. 1B). Because of the peculiar features of YPO2469, we also performed RT-PCR on total RNA extracted from CO92 to see whether this putative ORF is transcribed. A transcript of the expected size was indeed detected (data not shown), thus confirming that, despite its unusual location, CDS2pe is transcribed. A putative function could not be attributed to CDS2pe, because it has no homologs in databases. CRISPRs were shown to provide resistance against phage infections in prokaryotes (5), but such a function has not been demonstrated for Y. pestis.
R1pe is a 11.7-kb-long region which carries 11 ORFs (YPO0387 to -0397) in the two orientations (Fig. 1C). The loci on each side of R1pe are not adjacent on the Y. pseudotuberculosis IP32953 chromosome. It is known that despite the very close genetic relationship between Y. pestis and Y. pseudotuberculosis, their chromosomes are not colinear (8), most probably because of frequent rearrangements between the numerous IS present on the Y. pestis chromosome. The presence of an IS100 copy adjacent to R1pe (or in R1pe) suggests that the integration event was followed by IS-mediated chromosomal rearrangements in Y. pestis. The very low GC content of the central ORFs in R1pe is suggestive of a mosaic structure resulting from successive integration events. This hypothesis is reinforced by the presence of remnants of transposable elements (ISR1 and IS21) in this region (see Table S5 in the supplemental material). Most of the genes on R1pe are predicted to encode products with low or no homology with known proteins. However, three may play a role in DNA restriction/modification. One of them, located in the low-GC-percentage portion of R1pe, would be a methylase (YPO0391), while two adjacent sequences (YPO0387 and -0388) at the left-hand extremity of R1pe have the potential to encode a complete restriction/modification system similar to the McrB system of Escherichia coli (see Table S5). Remarkably, R1 appears to have been exchanged with a Y. pseudotuberculosis-specific region (YPTB0535 to -0537) coding also for a restriction/modification system. This region was shown to play a major role in Y. pseudotuberculosis pathogenicity (44).
R2pe is a 7.8-kb region composed of five CDSs (YPO1668 to -1672) (Fig. 1D). This region has inserted itself between YPTB2403 and YPTB2404 in the genome of the Y. pseudotuberculosis ancestor. No mobile elements adjacent to R2pe could explain its site of integration into the Y. pestis genome. The YPO1672 ORF located at the right-hand extremity of R2pe is predicted to encode a large protein of 1,268 amino acids (aa) which has two paralogs on the CO92 chromosome, YPO0765 (82% identity in 1,206 aa) and YPO0309 (81% identity in 1,199 aa). This identity concerns the last two-thirds of the encoded products, while their NH2 extremities are different. The conserved COOH portion of these proteins exhibits homology with a domain PL2 passenger and an autotransporter beta-domain, suggesting that they encode autotransporter proteins of a type V secretion system. Many proteins secreted by type V secretion systems have been linked to virulence in Gram-negative bacteria (25, 26). Recently, one Y. pestis autotransporter adhesin (YapE, encoded by YPO3984) has been shown to be required for efficient colonization of the host during bubonic plague (36). The other ORFs (YPO1668 to YPO1671) carried by R2pe share similarity (63% to 85% identity) with CDSs of Proteus mirabilis that exhibit the same gene organization and that are putatively forming a sugar transport system (see Table S5 in the supplemental material). Interestingly, one of the Y. pseudotuberculosis-specific regions (R5pst) that was shown to have been lost during the emergence of Y. pestis is also putatively involved in sugar transport (43). However, this Y. pseudotuberculosis-specific region was not found to be important for the physiology or pathogenicity of this species.
R3pe is a very large region (41.7 kb) composed of 52 ORFs (YPO2084 to -2136), which is predicted to be a lambda prophage (Fig. 1E; see also Table S5 in the supplemental material). This phage inserted itself between YPTB2404 and YPTB2405 in the genome of the Y. pseudotuberculosis ancestral strain, probably using its own machinery of insertion via a mechanism of site specific recombination. Indeed, an identical 31-nucleotide (nt) repeat (ATATTGAATAAGCCGAGTTGGGCTAAATAAC) is found at each extremity of the CO92 prophage. This 31-nt sequence is found in two copies flanking the prophage in all Y. pestis sequenced genomes, while this sequence is present in a single copy in the Y. pseudotuberculosis genomes. This suggests that the 31-nt sequence served as an att site for phage integration and, as classically observed during chromosomal insertion of lambda phages, resulted in a duplicated sequence upon integration. This putative prophage also carries at its left-hand extremity two genes predicted to form the phage insertion/excision machinery, i.e., an integrase (YPO2084) and an excisionase (YPO2087) (see Table S5 in the supplemental material). However, the int gene is interrupted by the insertion of an IS100 element which most likely inactivates it, suggesting that this phage is now stabilized in the Y. pestis chromosome. Another IS (IS285) is found at the other extremity of the prophage and has most likely inserted itself secondarily into the phage genome. In addition to int, another prophage-borne gene (YPO2103), predicted to encode a terminase, which is required for cleavage of the cos site and therefore for the encapsidation of the phage DNA, is interrupted by an IS285 insertion. Finally, the YPO2106 CDS (of unknown function) is truncated because of a frameshift mutation in its nucleotide sequence. Upon comparison of the genetic organization of the Y. pestis lambdoid prophage with that of the well-studied E. coli lambda phage, it appears that the regions coding for phage morphogenesis are similar, while the loci involved in lambda replication, regulation, and recombination are absent or divergent in the Y. pestis phage. This, along with the presence of several truncated genes and the fact that the size of the phage is drastically decreased in some Y. pestis strains, such as Antiqua (28.6 kb), Angola (23.6 kb), or Microtus (8.9 kb), suggests that the Y. pestis lambda prophage is nonfunctional and is undergoing a process of reductive evolution. Interestingly, the lambdoid phage closest to that of Y. pestis is found in the genome of Y. enterocolitica Ye8081 (59). Indeed, 25 of the R3pe ORFs have homologs in a region of the Ye8081 genome predicted to be a prophage. Despite a high level of conservation of some genes, some other genes are much less conserved and some regions are different in the two prophages (see Table S5), suggesting that these prophages belong to the same phage family but that they are different or diverged long before independently infecting the Y. enterocolitica and Y. pestis branches.
R4pe is a 2.7-kb region composed of four ORFs (YPO2483 to -2486), all in the same orientation (Fig. 1F). R4pe is flanked by two 98% identical 250-nt-long DNA repeats that are inside the 5′ portions of two coding sequences, one in YPO2486 at the left-hand extremity of R4pe and the other one in YPO2482, bordering the right-hand extremity of R4pe. Remarkably, a segment sharing 97% and 98% nucleotide identity with the left and right 250-nt repeats, respectively, is also found within the YPTB2524 coding sequence on the Y. pseudotuberculosis chromosome (Fig. 1F). This is highly suggestive of the insertion of a circular DNA molecule by homologous recombination between the 250-nt segment in YPTB2524 and the same sequence carried by the incoming replicon. This integration event has probably been accompanied or followed by secondary rearrangements at the site of integration since a single recombination event would not be sufficient to explain the genetic organization of this region in Y. pestis. As could be expected for an integration by a single crossing-over, two fusion ORFs were generated at each extremity, one composed of the entire YPTB2524 sequence, followed by a sequence brought by the replicon, creating YPO2486, and another mosaic ORF (YPO2482) composed of a portion of YPTB2524 fused to a portion of the downstream YPTB2523 CDS (Fig. 1F). Of note, another copy of the 250-nt DNA segment is found within other CDSs in Y. pestis (YPO2490) and in Y. pseudotuberculosis (YPTB2527). Both of these CDSs have the characteristics of being located in the vicinity of the R4pe integration site and of coding for very large proteins predicted to be hemolysins. YPO2486 also shares 38% similarity with a hemolysin/hemagglutinin-like protein HecA precursor from Erwinia chrysanthemi (48). The other three ORFs carried by R4pe (YPO2483 to -2485) are of unknown functions. They all have GC percentages lower than 39 and share little or no homology with proteins in databases, suggesting that they originate from a distantly related donor strain.
R5pe is a 3-kb-long region composed of two ORFs (YPO3436 and -3437). Elements involved in gene mobility are found on each side: YPO3438, at the right-hand extremity encodes a putative integrase, and at the left-hand extremity, YPO3436 is truncated by the insertion of an IS100 element, which is itself followed by an IS1541 copy (Fig. 1G). As already observed for R1pe, the loci on each side of R5pe are not adjacent on the Y. pseudotuberculosis IP32953 genome. Possibly, R5pe was part of a larger mobile element and its integration into the Y. pestis chromosome was associated with intragenomic rearrangements via IS100 recombination. Similarly to the other two CDSs, the putative function of R5pe remains unknown because its amino acid sequence shares no significant similarity with proteins in databases.
R6pe is 1.9-kb long and is composed of four small ORFs (YPO3945 to -3948) in the two orientations (Fig. 1H). This region inserted itself between YPTB3789 and YPTB3790 in the ancestral Y. pseudotuberculosis strain. Interestingly, a perfectly conserved 15-nt-long sequence (GCGAATTCTGCTGTA) is found both between YPTB3789 and YPTB3790 on the Y. pseudotuberculosis IP32953 chromosome and at the two extremities of R6pe on the Y. pestis CO92 chromosome (Fig. 1H). This is consistent with the insertion of a foreign DNA by a mechanism of site-specific recombination. The analysis of the three other Y. pseudotuberculosis sequenced genomes indicates that all three have an element inserted at the same site, flanked by the 15-nt repeat (see Fig. S1 in the supplemental material). This argues for the idea that the 15-nt sequence is a hot spot of integration of foreign DNA. This 15-nt repeat is identical to that of Y. pestis on one side and is slightly divergent (caGtATTCTGCTGTA [lowercase letters represent divergent nucleotides]) on the other side. The imported DNA has a common backbone (represented by strain YPIII), in which different DNA sequences have inserted themselves, either before or after integration at the 15-nt site. Remarkably, in Y. pseudotuberculosis IP31758, the incoming DNA has created a large ORF, either by fusion with the two bordering sequences or, more likely, by allelic replacement between these sequences and a larger ORF having the same extremities. Also remarkable is the observation that in Y. pestis YPO3945, a potential start codon is located just upstream of the 15-nt repeat of Y. pestis, while in Y. pseudotuberculosis, the slight divergence in the repeat introduces a stop codon which leads to a shorter ORF. Therefore, despite the conservation of the region overlapping YPO3945 in Y. pestis and several Y. pseudotuberculosis strains, the coding capacity and function of this DNA sequence might be different. No function could be predicted for the four ORFs carried by R6pe based on BLAST analyses. From the above data, we can thus hypothesize that R6pe corresponds to a mobile element which can insert itself into bacterial chromosomes by site-specific recombination and which carries a conserved core sequence and a variable region in which different CDSs are found inserted. Whether the Y. pestis region was horizontally acquired after its divergence from Y. pseudotuberculosis or whether it was vertically acquired from an ancestor that had an identical region cannot be determined. However, the facts that none of the screened Y. pseudotuberculosis strains harbored YPO3948 and that the 15-nt right-hand repeat is different between the two species argue for the first hypothesis.
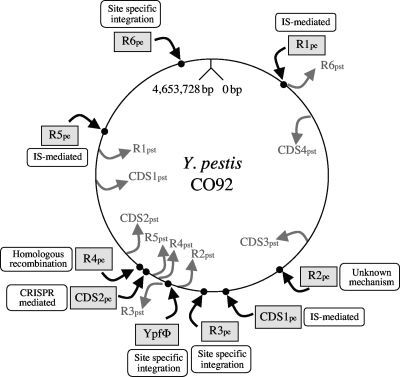
Altogether, it thus appears that, based on the scars left by the insertion of the eight Y. pestis-specific chromosomal loci, a mechanism of acquisition can be predicted for most of them. The position of these loci on the Y. pestis CO92 chromosome indicates that six out of nine loci (when the YpfΦ genome is considered) are located in the region opposite to the origin of replication (Fig. 2). Similarly, most of the regions lost by Y. pestis (i.e., Y. pseudotuberculosis specific) are also located in this part of the chromosome (Fig. 2). The level of genetic polymorphism in bacterial chromosomes is known to be the highest in this portion of the chromosome, which contains the dif site where resolution of chromosomal dimers takes place during bacterial division (56). Indeed, more than one-third of the acquisition or loss of genetic materials (from R4pe to R2pst) (Fig. 2) in Y. pestis are concentrated in a region which represents only 5% of the entire Y. pestis chromosome. In two instances, the acquisition of the Y. pestis-specific loci seems to have occurred by exchange with a Y. pseudotuberculosis-specific region (with R3pst replaced by YpfΦ and with R6pst replaced by R1pe). In terms of functions, R1pe, R2pe, and R3pe putatively encode a restriction/modification system, a sugar transporter, and a lambdoid phage, respectively, but no specific function could be attributed to the other five loci. It is noteworthy that no putative virulence factor was identified on any of the eight Y. pestis-specific chromosomal loci studied.
FIG. 2.
Positions of the regions acquired and lost by Y. pestis on a schematic map of the CO92 chromosome. The positions on the Y. pestis CO92 chromosome of all loci acquired or lost by Y. pestis are shown by black or gray arrows, respectively. The putative mechanism of acquisition is indicated next to each Y. pestis-specific region.
History of the acquisition and modification of the specific regions during Y. pestis microevolution.
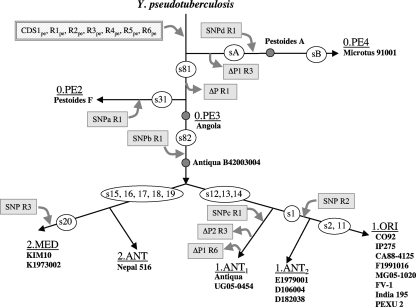
We wondered whether the eight specific regions studied here were acquired early or late after Y. pestis emergence from Y. pseudotuberculosis. A previous molecular phylogenetic study allowed us to design a microevolution tree for Y. pestis and to delineate a set of single-nucleotide polymorphisms (SNPs) that could be used to position a new isolate on this tree (1). Today, the complete genome sequences of 21 Y. pestis strains (Microtus 91001 [55]; Pestoides A, Pestoides F, and Angola [18]; Antiqua B42003004 [16]; Nepal 516 [9]; KIM10 [12]; K1973002 [16]; Antiqua [9]; UG05-0454 and E1979001 [16]; D106004 [52]; D182038 [52]; IP275, CA88-4125, and F1991016 [16]; MG05-1020 and FV-1 [60]; India 195; and PEXU 2 and CO92 [40]) are available (listed at http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?database=386656), but the positions of most of them along the microevolutionary tree are not known. To draw an updated tree, a subset of 16 SNPs defining the main branches and subbranches were selected among those previously identified (1), and their presence/absence in the 21 genomes was checked. This allowed us to position all strains for which no branching was yet assigned in the Y. pestis updated microevolution tree (Fig. 3). The position of Y. pestis Pestoides A was confirmed using a second set of SNPs along the 0.PE4 branch. Furthermore, on the basis of the presence or absence of the s1 SNP, the 1.ANT strains could be differentiated into two subbranches (1.ANT1 and 1.ANT2) (Fig. 3).
FIG. 3.
Y. pestis microevolution tree with the steps at which the specific regions are predicted to have been acquired or to have undergone various mutations. The 21 Y. pestis strains for which a complete genome sequence was available were positioned on the microevolutionary tree based on the presence or absence of a set of 16 SNPs (indicated inside white ovals) shown to characterize the various Y. pestis phylogenetic branches (1). To position strain Pestoides A, additional SNPs defining branch 0.PE4 were examined. Among them, some were present (sA [s95, s96, s97, s99, s102, s104, s105, s114, s116, and s120]), while others were absent (sB [s94, s98, s100, s101, s106, s107, s108, s109, s110, s111, s112, s113, s115, s117, s118, and s119]). The hatched rectangle corresponds to the position in the tree where the Y. pestis-specific regions are predicted to have been acquired. The various mutations in specific regions that characterize phylogenetic branches are indicated by an arrow and a gray rectangle in which the type of mutation (SNP or partial deletion [ΔP]) and the locus involved are indicated. A more detailed description of these mutations is presented in Table S6 in the supplemental material.
CDS2pe is a peculiar case because it is located within a CRISPR, which has the characteristics of being highly plastic and of generating a polymorphism (45). We could nonetheless observe in our PCR screens that this CDS is present in only a portion of Antiqua and Medievalis strains, while it is found in all 23 Orientalis strains analyzed (see Table S2 in the supplemental material). All seven other specific loci were present in the genomes of all 21 sequenced Y. pestis isolates, including Pestoides strains which were not available for our PCR screens (Fig. 3). This indicates that these seven loci were acquired very early during Y. pestis evolution, before the split of the most ancient branch (0.PE4) along branch 0. It also indicates that these regions were stably kept during the microevolution of the plague bacillus.
In a previous analysis of Y. pestis housekeeping genes, we were unable to identify a single polymorphic nucleotide at 21,881 synonymous sites within 36 Y. pestis strains because this species is remarkably monomorphic (2). We wondered whether this was also the case for the seven horizontally acquired regions, or whether peculiar mutations could specify some evolutionary branches. A detailed analysis of these regions at the gene level in the 21 Y. pestis genome sequences indicated a very high degree of nucleotide conservation, with some polymorphisms nonetheless. CDS1pe was completely monomorphic in the 21 genomes analyzed, and R5pe was also identical in 20 genomes except in Angola, which is known to be a very atypical strain (18). Among various reasons that could explain this absence of polymorphism, one could be that these two regions were subjected to a strong selective pressure because they encode functions important for the Y. pestis life cycle. However, another explanation, and a more likely one, is that because of the small sizes of these regions, the chances of acquiring point mutations or deletions/insertions are lower.
For the other regions, a few sequence variations were observed, and some of these variations were informative in terms of Y. pestis microevolution. Indeed several SNPs/deletions in R1pe and R3pe were characteristic of branch 0 and of the 0.PE4 and 0.PE2 branches that split off along this branch early during evolution (Fig. 3; see also Table S6 in the supplemental material). SNPb, one of the three SNPs found in R1pe, occurred before the separation between branches 1 and 2 and thus characterizes all classical Y. pestis strains of branches 1 and 2. Along branch 2, one SNP in R3pe defines the 2.MED subbranch. On branch 1, one SNP in R2pe (see Table S6) specified the 11 strains (3 1.ANT2 and 8 1.ORI strains) as the most recently emerged along branch 1 and confirmed the 1.ANT split evidenced by the s1 SNP (Fig. 3). Curiously, the older 1.ANT1 subbranch was characterized by mutations, including inactivating deletions (see Table S6), in three specific regions (R1pe, R3pe, and R6pe) (Fig. 3).
Overall, R1pe and R3pe were the two specific regions in which the highest rates of genetic polymorphisms were observed. The fact that these regions are the two largest in size could explain at least in part their higher level of polymorphism. R3pe corresponds to the lambdoid prophage genome. The fact that deletions of large regions predicted to encode phage functions required for infection of a bacterial host occurred in different branches (in YPO2095 to YPO2135 in branch 0.PE4 and in YPO2087 to YPO2103 in the 1.ANT1 subbranch) (Fig. 3) suggests a single acquisition event and further argues for a secondary process of reductive evolution. This, along with our above observation that some genes necessary for the phage physiology are inactivated in all strains, supports the hypothesis that a functional phage is not necessary for the Y. pestis life cycle and/or virulence. The level of polymorphism of R1pe was the highest but corresponded mostly to SNPs that occurred in different genes without inactivating them (see Table S6 in the supplemental material). Even the small, 6-nt deletion that arose early along branch 0 (but which could also be a 6-nt insertion in branch 0.PE4) was in-frame and therefore did not disrupt the coding sequence. Except for the presence of a putative DNA restriction/modification system, most of the genes carried by R1pe have no predicted functions. The absence of inactivating mutations in this region, whatever the genes involved and the evolutionary branch, supports the hypothesis that this region may be useful for Y. pestis physiological or pathological processes.
Mutagenesis of the Y. pestis-acquired regions and impact on bacterial physiological and pathogenic properties.
To investigate the role of the eight specific chromosomal loci in the ability of Y. pestis to grow in vitro, to infect fleas, and to kill mice, each locus was individually deleted by allelic exchange with a kanamycin cassette. The ORFs encompassed by the deletions and the primers used to check that the replacements occurred as expected are indicated in Fig. 1 and in Table S4 in the supplemental material. The resulting deletion strains, with their designations, are listed in Table S3 in the supplemental material. To perform the deletions using the lambda red technology, the pKOBEG-sacB plasmid was introduced into Y. pestis strain CO92, yielding CO92p (see Table S3 in the supplemental material). Since the chromosome of Y. pestis is prone to frequent rearrangements (23), and since we found that the presence of pKOBEG-sacB had no significant impact on Y. pestis pathogenicity, we decided not to remove this plasmid in the deletion mutants, in order to limit as much as possible subcultures which would increase the occurrence of genomic reassortments. During our previous analysis of Y. pseudotuberculosis-specific regions, we found that two CDSs (YPTB1495 and YPTB3368) were not deletable, because they are essential for IP32953 survival (44). This was not the case for the Y. pestis-specific loci, as the eight specific regions were successfully deleted.
To determine whether some of these loci play a role in the ability of Y. pestis to multiply in vitro, the CO92p parental strain and the eight deletants were cultured in LB at 28°C (the optimal temperature for Yersinia growth) and 37°C (the in vivo temperature). No growth defects were observed in the various mutants, compared to the level for the CO92p parental strain (data not shown). Similar results were previously obtained with the various Y. pseudotuberculosis strains deleted of each specific region (44). However, when these Y. pseudotuberculosis deletants were grown in the biochemically defined medium M63S, a growth defect could be observed for several mutants. The abilities of the eight Y. pestis deletants and the parental CO92p strain to grow in the M63S medium at 28°C and 37°C were thus tested and compared. The growth curves of all strains were comparable, both at 28°C and at 37°C (data not shown). Therefore, none of the eight Y. pestis-acquired chromosomal loci are required for optimal bacterial growth, at least under the experimental conditions used here.
A major difference during the transformation of Y. pseudotuberculosis into Y. pestis has been a switch in the mode of transmission, from the oral route to flea bites. It could thus be expected that some of the chromosomal regions that specify Y. pestis participated in its adaptation to its new transmission cycle. This has been demonstrated for the two Y. pestis-specific plasmids: pFra is necessary for an efficient colonization of and survival in the flea midgut (30), as well as for an efficient transmission by fleas (50), whereas pPla is important for the dissemination of the bacteria from the intradermal inoculation site to the draining lymph node (51, 54).
The key features of Y. pestis that facilitate flea-borne transmission and that differentiate Y. pestis from Y. pseudotuberculosis are (i) its capacity to colonize the flea gut without causing major damage during the early phase of infection (20) and (ii) its ability to block fleas. To determine whether the acquired regions could play a role in these Y. pestis characteristics in fleas, groups of 45 to 113 Xenopsylla cheopis fleas were artificially fed on blood containing similar inocula (1.5 × 108 to 1 × 109 CFU/ml) of each of the eight deletants or CO92p and were monitored for (i) early mortality 48 h after feeding and (ii) infection and blockage 4 weeks after infection.
Y. pseudotuberculosis exerts acute toxic effects on X. cheopis (20). Soon after a meal on blood artificially infected with Y. pseudotuberculosis, about one-third to one-half of the fleas are moribund. In contrast, Y. pestis has gained the capacity to colonize the flea gut without causing major damage during the early phase of infection. None of the eight deletants or CO92p caused any acute toxicity to the fleas that took an infectious blood meal (data not shown), indicating that none of the Y. pestis-specific chromosomal loci play an essential role in the lower toxicity of Y. pestis for fleas.
The Y. pestis-induced blockage of the flea proventriculus is due to the formation of a biofilm produced by the products of the chromosomal hms genes (32). Although Y. pseudotuberculosis harbors similar hms genes, this species does not block fleas (19), indicating that other genetic determinants participate in this activity. As shown in Table 1, blockage developed in all of the flea groups at a rate expected for the X. cheopis-Y. pestis infection model (29, 30), indicating that none of the Y. pestis-specific loci is required for biofilm development in the flea proventriculus. Because fleas that become blocked can no longer take a blood meal, they rapidly starve to death. Thus, excess mortality can be used as an indirect indicator of blockage. In keeping with their ability to block fleas, all groups of infected fleas had elevated excess mortality rates of 15 to 58%, which is typical for X. cheopis fleas infected with Y. pestis (30). The eight mutants were also able to produce a stable, persistent infection in the flea at rates equivalent to that of CO92p (Table 1). The average bacterial load in infected fleas after 4 weeks was high for all the Y. pestis strains tested, ranging from 1.2 × 104 to 7.4 × 105 CFU/flea (Table 1). These results thus demonstrate that none of the Y. pestis-specific loci are crucial for flea infection or blockage. However, because of the intrinsic high variability observed in flea experiments, the possibility that some of these loci participate in a slightly more efficient capacity of Y. pestis to infect and block fleas cannot be ruled out.
TABLE 1.
Infection and blockage rates in fleas that fed on blood containing wild-type or mutant Y. pestis strains
| Strain | No. of fleas blocked/no. of fleas analyzeda (%) | No. of fleas infected/no. of fleas analyzedb (%) | No. of CFU per infected flea (mean ± SE) |
|---|---|---|---|
| CO92p | 42/100 (42) | 16/20 (80) | (6 ± 1.8) × 104 |
| 11/77 (14) | 14/20 (70) | (1.4 ± 0.4) × 105 | |
| 15/101 (15) | 5/20 (25) | (5 ± 0.7) × 104 | |
| 42/104 (40) | 5/13 (38) | (1.7 ± 0.8) × 105 | |
| CO92ΔR1pe | 25/87 (29) | 8/11 (73) | (1.2 ± 0.6) × 104 |
| CO92ΔR2pe | 37/112 (33) | 19/20 (95) | (7.4 ± 0.7) × 105 |
| CO92ΔR3pe | 44/102 (43) | 5/16 (31) | (1.5 ± 0.6) × 105 |
| CO92ΔR4pe | 18/95 (19) | 11/20 (55) | (8.2 ± 2.2) × 104 |
| CO92ΔR5pe | 36/113 (32) | 11/20 (55) | (5.5 ± 0.8) × 105 |
| CO92ΔR6pe | 15/91 (18) | 9/20 (45) | (6.6 ± 2.8) × 104 |
| CO92ΔCDS1pe | 13/59 (22) | 11/19 (58) | (9 ± 2.6) × 104 |
| CO92ΔCDS2pe | 11/45 (24) | 10/17 (59) | (2.7 ± 1.4) × 105 |
Determined from samples of fleas during a 4-week period following the infectious blood meal.
Determined from samples of surviving fleas collected 4 weeks after the infectious blood meal.
One of the major differences between Y. pestis and its ancestor Y. pseudotuberculosis is the acquisition by the former of an exceptional pathogenicity potential. Although the horizontally acquired pPla plasmid plays a major role in the virulence of most Y. pestis isolates, the facts that it is dispensable in some strains (35, 49, 63) and that its introduction into Y. pseudotuberculosis is not accompanied by an increase in virulence (34, 42) clearly indicate that other factors are required. One of them has been shown to be the YpfΦ filamentous phage (13), but other Y. pestis-acquired chromosomal loci may have also played a role in this gain of virulence. However, when the mouse model of subcutaneous infection was used to mimic bubonic plague, the eight mutants were found to be as virulent as the parental strain CO92p (Table 2). Y. pestis has also acquired the capacity to be transmitted by aerosols and to cause pneumonic plague. The capacity of the eight mutants to produce a lethal infection upon aerosol inoculation was tested by exposing mice to aerosolized droplets of a suspension containing 108 CFU/ml (three times the 50% lethal dose [LD50]). All deletants killed 100% of the infected animals, with a mean time to death similar to that of the CO92p strain (Table 2). Altogether, these results suggest that the acquired regions and CDSs studied here did not participate in the increased pathogenicity of Y. pestis, at least in our experimental mouse models of s.c. and aerosol infections.
TABLE 2.
Virulence for mice of the Y. pestis recombinant strains after subcutaneous injection or aerosol exposure
| Strain | s.c. injection |
Aerosol exposure |
|||
|---|---|---|---|---|---|
| No. of CFU injected | No. of dead animals/total no. of animals infected | No. of suspension-nebulized CFU/ml | No. of dead animals/total no. of animals infected | Time to death (days) | |
| CO92p | 12 | 3/5 | (0.95 ± 0.3) × 108a | 12/12a | 4 ± 1a |
| CO92ΔR1pe | 10 | 3/5 | 1.1 × 108 | 6/6 | 4.5 |
| CO92ΔR2pe | 16 | 3/5 | 2 × 108 | 6/6 | 3 |
| CO92ΔR3pe | 12 | 3/5 | 1.5 × 108 | 6/6 | 3 |
| CO92ΔR4pe | 12 | 3/5 | 1.4 × 108 | 5/5 | 4 |
| CO92ΔR5pe | 9 | 3/5 | 1.4 × 108 | 6/6 | 3 |
| CO92ΔR6pe | 17 | 5/5 | 1.1 × 108 | 6/6 | 3 |
| CO92ΔCDS1pe | 14 | 5/5 | 0.9 × 108 | 6/6 | 4 |
| CO92ΔCDS2pe | 13 | 3/5 | 0.6 × 108 | 6/6 | 3 |
These values are means ± standard deviations for two experiments with six mice each.
Conclusions.
Our study has identified eight chromosomal loci that were acquired soon before or early after the divergence of Y. pestis from Y. pseudotuberculosis. Surprisingly, we were unable to evidence an impact of these regions under in vitro and in vivo conditions. It is possible that acquisition of these regions provided no benefit to the bacterial host and, because Y. pestis is a recently emerged bacterium, that they did not yet have the time to be inactivated and eliminated by gradual mutations and/or deletions. This could be the case for the lambdoid phage (R3pe), in which several genes predicted to encode physiological functions are interrupted, as well as for CDS1pe, which is probably a remnant of an insertion element and which is no longer transcribed. This is also true for many other genes in Y. pestis whose genomes are characterized by a massive gene inactivation (8). However, the fact that the other regions, which were acquired early during Y. pestis microevolution, are conserved and potentially functional in the various evolutionary subbranches suggests that these regions have been subjected to a positive selective pressure for their maintenance and thereby that they should confer advantages at some steps of the Y. pestis biological cycle. If so, our inability to attribute a physiological role to these loci may have several reasons. One possible explanation would be that the functions of two or more acquired regions are redundant. Multiple deletions of the Y. pestis-acquired loci would thus be needed for a defect to be observed. In the flea model, although no effect on toxicity or proventriculus blockage was observed, it is still possible that the deletion of some loci had an impact on the efficiency of flea transmission, as recently observed for the F1 capsule (50). Furthermore, since Y. pestis can be transmitted by many different flea species, some of which do not develop proventricular blockage to the same extent as X. cheopis (15), testing the various mutants in other flea models might also be of interest. The absence of virulence impairment in the various deletants indicates that none of the Y. pestis-specific regions play a major role in Y. pestis pathogenicity. However, because of the extreme pathogenicity of the plague bacillus in the mouse model, minor virulence defects might be difficult to evidence. The use of more-resistant animal models could potentially disable the identification of subtle virulence impediments. Finally, Y. pestis has been shown to be able to survive in naturally contaminated soils for several weeks (4) and in burrows of wild rodents for several years (39). The assumption that the acquired regions could participate to the long-term persistence of the plague bacillus outside an insect vector or a mammalian host may also be considered. Whatever the reasons for the absence of identified roles, our results suggest that, unlike what might have been assumed, the dramatic change of life cycle that has accompanied the emergence of Y. pestis is not attributable to the acquisition of a specific chromosomal locus.
Supplementary Material
Acknowledgments
This study was funded in part by contract DGA 99 01110004709450 from the French Defense Ministry and a fellowship to A.D. from the Pasteur-Weissman Foundation.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 6 July 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayyadurai, S., L. Houhamdi, H. Lepidi, C. Nappez, D. Raoult, and M. Drancourt. 2008. Long-term persistence of virulent Yersinia pestis in soil. Microbiology 154:2865-2871. [DOI] [PubMed] [Google Scholar]
- 5.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 6.Bocs, S., S. Cruveiller, D. Vallenet, G. Nuel, and C. Medigue. 2003. AMIGene: Annotation of MIcrobial Genes. Nucleic Acids Res. 31:3723-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchrieser, C., M. Prentice, and E. Carniel. 1998. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J. Bacteriol. 180:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chain, P. S., P. Hu, S. A. Malfatti, L. Radnedge, F. Larimer, L. M. Vergez, P. Worsham, M. C. Chu, and G. L. Andersen. 2006. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. J. Bacteriol. 188:4453-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, K. J., D. L. Fritz, M. L. Pitt, S. L. Welkos, P. L. Worsham, and A. M. Friedlander. 1996. Pathology of experimental pneumonic plague produced by fraction 1-positive and fraction 1-negative Yersinia pestis in African green monkeys Cercopithecus aethiops. Arch. Pathol. Lab. Med. 120:156-163. [PubMed] [Google Scholar]
- 11.de Almeida, A. M., A. Guiyoule, I. Guilvout, I. Iteman, G. Baranton, and E. Carniel. 1993. Chromosomal irp2 gene in Yersinia: distribution, expression, deletion and impact on virulence. Microb. Pathog. 14:9-21. [DOI] [PubMed] [Google Scholar]
- 12.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derbise, A., V. Chenal-Francisque, F. Pouillot, C. Fayolle, M. C. Prevost, C. Medigue, B. J. Hinnebusch, and E. Carniel. 2007. A horizontally acquired filamentous phage contributes to the pathogenicity of the plague bacillus. Mol. Microbiol. 63:1145-1157. [DOI] [PubMed] [Google Scholar]
- 14.Derbise, A., B. Lesic, D. Dacheux, J. M. Ghigo, and E. Carniel. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38:113-116. [DOI] [PubMed] [Google Scholar]
- 15.Eisen, R. J., S. W. Bearden, A. P. Wilder, J. A. Montenieri, M. F. Antolin, and K. L. Gage. 2006. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc. Natl. Acad. Sci. U. S. A. 103:15380-15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eppinger, M., Z. Guo, Y. Sebastian, Y. Song, L. E. Lindler, R. Yang, and J. Ravel. 2009. Draft genome sequences of Yersinia pestis isolates from natural foci of endemic plague in China. J. Bacteriol. 191:7628-7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eppinger, M., M. J. Rosovitz, W. F. Fricke, D. A. Rasko, G. Kokorina, C. Fayolle, L. E. Lindler, E. Carniel, and J. Ravel. 2007. The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet. 3:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eppinger, M., P. L. Worsham, M. P. Nikolich, D. R. Riley, Y. Sebastian, S. Mou, M. Achtman, L. E. Lindler, and J. Ravel. 2010. Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium. J. Bacteriol. 192:1685-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson, D. L., C. O. Jarrett, B. W. Wren, and B. J. Hinnebusch. 2006. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J. Bacteriol. 188:1113-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erickson, D. L., N. R. Waterfield, V. Vadyvaloo, D. Long, E. R. Fischer, R. Ffrench-Constant, and B. J. Hinnebusch. 2007. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: implications for the evolution of vector-borne transmission of plague. Cell. Microbiol. 9:2658-2666. [DOI] [PubMed] [Google Scholar]
- 21.Friedlander, A. M., S. L. Welkos, P. L. Worsham, G. P. Andrews, D. G. Heath, G. W. Anderson, Jr., M. L. Pitt, J. Estep, and K. Davis. 1995. Relationship between virulence and immunity as revealed in recent studies of the F1 capsule of Yersinia pestis. Clin. Infect. Dis. 21(Suppl. 2):S178-S181. [DOI] [PubMed] [Google Scholar]
- 22.Guinet, F., P. Ave, L. Jones, M. Huerre, and E. Carniel. 2008. Defective innate cell response and lymph node infiltration specify Yersinia pestis infection. PLoS One 3:e1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guiyoule, A., B. Rasoamanana, C. Buchrieser, P. Michel, S. Chanteau, and E. Carniel. 1997. Recent emergence of new variants of Yersinia pestis in Madagascar. J. Clin. Microbiol. 35:2826-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hare, J. M., A. K. Wagner, and K. A. McDonough. 1999. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol. Microbiol. 31:291-303. [DOI] [PubMed] [Google Scholar]
- 25.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinnebusch, B. J., K. L. Gage, and T. G. Schwan. 1998. Estimation of vector infectivity rates for plague by means of a standard curve-based competitive polymerase chain reaction method to quantify Yersinia pestis in fleas. Am. J. Trop. Med. Hyg. 58:562-569. [DOI] [PubMed] [Google Scholar]
- 29.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 30.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 31.Jansen, R., J. D. van Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of a novel family of sequence repeats among prokaryotes. OMICS 6:23-33. [DOI] [PubMed] [Google Scholar]
- 32.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 33.Jarrett, C. O., F. Sebbane, J. J. Adamovicz, G. P. Andrews, and B. J. Hinnebusch. 2004. Flea-borne transmission model to evaluate vaccine efficacy against naturally acquired bubonic plague. Infect. Immun. 72:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kutyrev, V., R. J. Mehigh, V. L. Motin, M. S. Pokrovskaya, G. B. Smirnov, and R. R. Brubaker. 1999. Expression of the plague plasminogen activator in Yersinia pseudotuberculosis and Escherichia coli. Infect. Immun. 67:1359-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kutyrev, V. V., A. A. Filippov, N. Shavina, and O. A. Protsenko. 1989. Genetic analysis and simulation of the virulence of Yersinia pestis. Mol. Gen. Mikrobiol. Virusol. 1989:42-47. (In Russian.) [PubMed] [Google Scholar]
- 36.Lawrenz, M. B., J. D. Lenz, and V. L. Miller. 2009. A novel autotransporter adhesin is required for efficient colonization during bubonic plague. Infect. Immun. 77:317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leif, W. R., and A. P. Krueger. 1950. Studies on the experimental epidemiology of respiratory infections. I. An apparatus for the quantitative study of air-borne respiratory pathogens. J. Infect. Dis. 87:103-116. [DOI] [PubMed] [Google Scholar]
- 38.Li, Y., E. Dai, Y. Cui, M. Li, Y. Zhang, M. Wu, D. Zhou, Z. Guo, X. Dai, B. Cui, Z. Qi, Z. Wang, H. Wang, X. Dong, Z. Song, J. Zhai, Y. Song, and R. Yang. 2008. Different region analysis for genotyping Yersinia pestis isolates from China. PLoS One 3:e2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mollaret, H. H. 1968. Conservation du bacille de la peste durant 28 mois en terrier artificiel: démonstration expérimentale de la conservation interépizootique de la peste dans ses foyers invétérés. C. R. Acad. Sci. Paris 267:972-973. [Google Scholar]
- 40.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 41.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pouillot, F., A. Derbise, M. Kukkonen, J. Foulon, T. K. Korhonen, and E. Carniel. 2005. Evaluation of O-antigen inactivation on Pla activity and virulence of Yersinia pseudotuberculosis harbouring the pPla plasmid. Microbiology 151:3759-3768. [DOI] [PubMed] [Google Scholar]
- 43.Pouillot, F., C. Fayolle, and E. Carniel. 2007. A putative DNA adenine methyltransferase is involved in Yersinia pseudotuberculosis pathogenicity. Microbiology 153:2426-2434. [DOI] [PubMed] [Google Scholar]
- 44.Pouillot, F., C. Fayolle, and E. Carniel. 2008. Characterization of chromosomal regions conserved in Yersinia pseudotuberculosis and lost by Yersinia pestis. Infect. Immun. 76:4592-4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pourcel, C., G. Salvignol, and G. Vergnaud. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653-663. [DOI] [PubMed] [Google Scholar]
- 46.Radnedge, L., P. G. Agron, P. L. Worsham, and G. L. Andersen. 2002. Genome plasticity in Yersinia pestis. Microbiology 148:1687-1698. [DOI] [PubMed] [Google Scholar]
- 47.Rakin, A., S. Schubert, I. Guilvout, E. Carniel, and J. Heesemann. 2000. Local hopping of IS3 elements into the A+T-rich part of the high-pathogenicity island in Yersinia enterocolitica 1B, O:8. FEMS Microbiol. Lett. 182:225-229. [DOI] [PubMed] [Google Scholar]
- 48.Rojas, C. M., J. H. Ham, W. L. Deng, J. J. Doyle, and A. Collmer. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. U. S. A. 99:13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samoilova, S. V., L. V. Samoilova, I. N. Yezhov, I. G. Drozdov, and A. P. Anisimov. 1996. Virulence of pPst+ and pPst− strains of Yersinia pestis for guinea-pigs. J. Med. Microbiol. 45:440-444. [DOI] [PubMed] [Google Scholar]
- 50.Sebbane, F., C. Jarrett, D. Gardner, D. Long, and B. J. Hinnebusch. 2009. The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect. Immun. 77:1222-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sebbane, F., C. O. Jarrett, D. Gardner, D. Long, and B. J. Hinnebusch. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. U. S. A. 103:5526-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen, X., Q. Wang, L. Xia, X. Zhu, Z. Zhang, Y. Liang, H. Cai, E. Zhang, J. Wei, C. Chen, Z. Song, H. Zhang, D. Yu, and R. Hai. 2010. Complete genome sequences of Yersinia pestis from natural foci in China. J. Bacteriol. 192:3551-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smego, R. A., J. Frean, and H. J. Koornhof. 1999. Yersiniosis I: microbiological and clinicoepidemiological aspects of plague and non-plague Yersinia infections. Eur. J. Clin. Microbiol. Infect. Dis. 18:1-15. [DOI] [PubMed] [Google Scholar]
- 54.Sodeinde, O. A., Y. V. Subrahmanyam, K. Stark, T. Quan, Y. Bao, and J. D. Goguen. 1992. A surface protease and the invasive character of plague. Science 258:1004-1007. [DOI] [PubMed] [Google Scholar]
- 55.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Yu, H. Yang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11:179-197. [DOI] [PubMed] [Google Scholar]
- 56.Steiner, W. W., and P. L. Kuempel. 1998. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J. Bacteriol. 180:6269-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun, Y. C., B. J. Hinnebusch, and C. Darby. 2008. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. U. S. A. 105:8097-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Surgalla, M. J., and E. D. Beesley. 1969. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl. Microbiol. 18:834-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Touchman, J. W., D. M. Wagner, J. Hao, S. D. Mastrian, M. K. Shah, A. J. Vogler, C. J. Allender, E. A. Clark, D. S. Benitez, D. J. Youngkin, J. M. Girard, R. K. Auerbach, S. M. Beckstrom-Sternberg, and P. Keim. 2007. A North American Yersinia pestis draft genome sequence: SNPs and phylogenetic analysis. PLoS One 2:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vallenet, D., S. Engelen, D. Mornico, S. Cruveiller, L. Fleury, A. Lajus, Z. Rouy, D. Roche, G. Salvignol, C. Scarpelli, and C. Medigue. 2009. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009:bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, X., Y. Han, Y. Li, Z. Guo, Y. Song, Y. Tan, Z. Du, A. Rakin, D. Zhou, and R. Yang. 2007. Yersinia genome diversity disclosed by Yersinia pestis genome-wide DNA microarray. Can. J. Microbiol. 53:1211-1221. [DOI] [PubMed] [Google Scholar]
- 63.Welkos, S. L., A. M. Friedlander, and K. J. Davis. 1997. Studies on the role of plasminogen activator in systemic infection by virulent Yersinia pestis strain C092. Microb. Pathog. 23:211-223. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, D., Y. Han, E. Dai, D. Pei, Y. Song, J. Zhai, Z. Du, J. Wang, Z. Guo, and R. Yang. 2004. Identification of signature genes for rapid and specific characterization of Yersinia pestis. Microbiol. Immunol. 48:263-269. [DOI] [PubMed] [Google Scholar]
- 65.Zhou, D., Y. Han, Y. Song, Z. Tong, J. Wang, Z. Guo, D. Pei, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, Z. Du, J. Bao, X. Zhang, J. Yu, P. Huang, and R. Yang. 2004. DNA microarray analysis of genome dynamics in Yersinia pestis: insights into bacterial genome microevolution and niche adaptation. J. Bacteriol. 186:5138-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.