Abstract
Yersinia pestis, the causative agent of plague, is a facultative intracellular pathogen. Previous studies have indicated that the ability of Y. pestis to survive inside macrophages may be critical during the early stages of plague pathogenesis. To gain insights into the biology of intracellular Y. pestis and its environment following phagocytosis, we determined the genome-wide transcriptional profile of Y. pestis KIM5 replicating inside J774.1 macrophage-like cells using DNA microarrays. At 1.5, 4, and 8 h postinfection, a total of 801, 464, and 416 Y. pestis genes were differentially regulated, respectively, compared to the level of gene expression of control bacteria grown in tissue culture medium. A number of stress-response genes, including those involved in detoxification of reactive oxygen species, as well as several metabolic genes involved in macromolecule synthesis, were significantly induced in intracellular Y. pestis, consistent with the presence of oxidative stress and nutrient starvation inside Yersinia-containing vacuoles. A putative stress-induced operon consisting of y2313, y2315, and y2316 (y2313-y2316), and a previously unidentified open reading frame, orfX, was studied further on the basis of its high level of intracellular expression. Mutant strains harboring either deletion, Δy2313-y2316 or ΔorfX, exhibited diverse phenotypes, including reduced effector secretion by the type III secretion system, increased intracellular replication, and filamentous morphology of the bacteria growing inside macrophages. The results suggest a possible role for these genes in regulating cell envelope characteristics in the intracellular environment.
Yersinia pestis is a Gram-negative bacterium and the causative agent of plague. In humans, bubonic plague can occur following the bite of an infected flea, while pneumonic plague can be spread through the inhalation of infectious droplets and is highly fatal (44). Although natural outbreaks of plague have declined in recent years, plague is still endemic in regions of North and South America, Africa, and Asia (44). In addition, the potential use of plague as a biological weapon has become a serious concern (30). Therefore, it is important to better understand plague pathogenesis and to develop new preventative measures and treatments for the disease.
A number of virulence determinants of Y. pestis have been identified, and many such factors have been found to promote extracellular growth of Y. pestis. Examples include those involved in the acquisition of iron from the body fluids and a plasmid-encoded type III secretion system (T3SS) (44). The T3SS injects effectors known as Yops into macrophages, which inhibit phagocytosis and modulate proinflammatory responses (63). Y. pestis also produces a capsule composed of F1 protein, which inhibits phagocytosis (12). However, these antiphagocytic factors are highly expressed only after cultivation at 37°C and not at an ambient temperature (e.g., 28°C) found in the flea vector, and therefore, the bacteria are likely to be taken up by phagocytes when they first enter the host (49). Thus, it has been suggested that some Y. pestis bacteria initially survive within macrophages before being released and replicate extracellularly in the host (49). In fact, it has long been established that Y. pestis is a facultative intracellular pathogen capable of surviving and replicating inside macrophages both in vivo and in vitro (5, 16, 60, 61). In a recent study, Lukaszewski et al. reported that a large number of Y. pestis bacteria are found inside macrophages in the spleens of mice during the first several days of infection (38). Furthermore, inactivation of phoP, the transcriptional regulator required for survival of Y. pestis inside macrophages, results in the attenuation of Y. pestis in both a subcutaneous and an intranasal mouse infection model (42; J. P. Grabenstein et al., unpublished data). These results indicate that intracellular survival of Y. pestis may be critical for plague disease progression. In addition to phoP, the rip operon, located within the pigmentation (pgm) locus of the Y. pestis chromosome, has been shown to promote the survival of Y. pestis inside activated macrophages (50). However, except for the rip operon and several PhoP-regulated genes (22), the bacterial factors required for intracellular replication of Y. pestis remain largely unknown.
When a bacterium is phagocytosed by a macrophage, the bacterium-containing phagosome quickly matures into a phagolysosome through a series of interactions with components of the endocytic pathway (17). During maturation, phagosomes acidify and acquire many hydrolytic enzymes (e.g., cathepsin D) and reactive oxygen species (ROSs) that degrade bacteria (17). To survive in phagosomes, pathogenic bacteria have evolved diverse strategies to counteract host bactericidal mechanisms (17). For example, Salmonella enterica serovar Typhimurium segregates its phagosomes from the late endocytic pathway to prevent acquisition of lysosomal hydrolytic enzymes, creating vacuoles that are less hostile (4). In the case of Y. pestis, less is known about its phagosome maturation processes or the strategies it employs to survive inside macrophages. Y. pestis-containing vacuoles (YCVs) likely interact with components of the endocytic pathway to some extent, since previous studies showed that YCVs fuse with thorium dioxide-labeled lysosomes (61) and that YCVs colocalize with cathepsin D and lysosomal-associated membrane protein 1 (LAMP1), the markers of late endosomes and lysosomes (22), respectively. Some YCVs also colocalize with an autophagosome marker, LC3 (51). However, YCVs fail to acidify below a pH of 7 (51), suggesting that they probably do not acquire all the typical characteristics of phagolysosomes.
DNA microarray techniques have proven to be a powerful tool for assessing the global transcriptional responses of intracellular bacteria to their host environment. Genome-wide transcriptional profiles have been obtained and studied for many intracellular pathogens, including S. Typhimurium, Salmonella enterica serovar Typhi, Shigella flexneri, Mycobacterium tuberculosis, Bacillus anthracis, and Francisella tularensis (2, 13, 15, 37, 57, 64). By observing the global changes in gene expression that occur as the bacterium enters the host cells, one can identify metabolic pathways and regulatory circuits that are critical for bacterial adaptation to the intracellular environment and draw inferences about the composition of the phagocytic vacuoles containing the pathogens (7). In some cases, the genes highly expressed in the intraphagosomal environment were also found to play an important role in the intracellular survival and/or the virulence of the pathogens (2, 64). In Y. pestis, microarray studies have been applied to various in vitro growth conditions as well as in vivo animal infection models, but no study has been done on the bacteria growing inside the mammalian host cells (32, 40, 52, 59, 66, 67).
To better understand the genetic basis for Y. pestis survival inside the macrophages and to gain insights into the environment that Y. pestis encounters during macrophage infection, genome-wide transcriptional profiling was carried out using DNA microarrays. This study provides the first global transcriptional analysis of any Yersinia species growing inside host cells. Differential regulation of a large number of metabolic and stress-response genes was observed, illustrating the unique nutritional requirements and stresses posed by the intracellular environment. Furthermore, we have identified novel factors that are induced in intracellular Y. pestis and analyzed their roles in the growth of Y. pestis inside macrophages.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Y. pestis strains used in this study (Table 1) are derived from strain KIM (molecular group 2.MED [1]). Y. pestis strains were grown at 28°C on either heart infusion (HI; Difco) or Luria-Bertani (LB) agar plates. Y. pestis cultures were grown in HI broth with aeration at the indicated temperature. Escherichia coli strains S17-1 and NovaBlue (Novagen) were grown on LB agar or LB broth at 37°C. The bacterial growth media were supplemented with kanamycin (Kan; 25 μg/ml), tetracycline (20 μg/ml), ampicillin (25 μg/ml for Y. pestis, 100 μg/ml for E. coli), and chloramphenicol (10 μg/ml for Y. pestis, 30 μg/ml for E. coli), when necessary.
TABLE 1.
Yersinia strains used in this study
| Strain name | Relevant characteristicsa | Reference or source |
|---|---|---|
| KIM6+ | Pgm+ pCD1− pMT1+ pPCP1+ | 21 |
| KIM6+ Δy2313-y2316 | Deletion of y2313 nt 70-y2316 nt 903 | This work |
| KIM6+ ΔorfX | Deletion of nt 2550560-2550725 in Y. pestis KIM chromosomeb | This work |
| KIM6+ ΔfadAB | Deletion of fadA (y0463) nt 49-fadB (y0464) nt 1098 | This work |
| KIM6+ Δy655 | Deletion of y655 nt 37-504 | This work |
| KIM6+ Δy3553-y3555 | Deletion of y3553 nt 55-y3555 nt 368 | This work |
| KIM6 ΔpsaABC::aph pGFP | Δpgm ΔpsaABC::aph pGFP Kanr Apr | 35 |
| KIM6+ Δy2313-y2316/pComp | Δy2313-y2316 pMMB207 y2313-y2316 Camr | This work |
| KIM5 | Δpgm pMT1+ pPCP1+ pCD1Ap+ Apr | 33 |
| KIM5/pGFP | Δpgm pGFP Kanr Apr | 33 |
| KIM5 Δy2313-y2316 | Δy2313-y2316 Apr | This work |
| KIM5 ΔorfX | ΔorfX Apr | This work |
| KIM5 mJ | yopJ C172A (Cys172 to Ala172 codon change) Apr | 33 |
| KIM5 yopB | yopB18 (in-frame deletion of yopB nt 496-774) Apr | 33 |
| KIM5 Δy2313-y2316/pComp | Δy2313-y2316 pMMB207 y2313-y2316 Camr Apr | This work |
| KIM5 Δy2313-y2316/pVec | Δy2313-y2316 pMMB207 Camr Apr | This work |
| KIM5 mJ/yopJ-bla | yopJC172A pMMB207 yopJC172A-M45-bla Camr Apr | This work |
| KIM5 mJ Δy2313-y2316/yopJ-bla | Δy2313-y2316 yopJC172A pMMB207 yopJC172A-M45-bla Camr Apr | This work |
+, positive for indicated element; −, negative for the indicated element; nt, nucleotides.
The 165-bp deletion includes most of the putative orfX gene, starting immediately after the stop codon of the upstream flanking gene (y2316) and ending at nucleotide 120 of orfX.
Construction of Y. pestis mutants.
The primers used to create deletion strains are listed in Table S2 in the supplemental material. Two DNA fragments of approximately 500 bp, each flanking KIM6+ open reading frames (ORFs) (Table 1), were amplified by PCR using the whole KIM6+ extract as a DNA template. The two PCR products were joined by splicing by overlap extension (SOEing) (28) to create an ∼1-kb deletion DNA construct, which was inserted into the suicide plasmid pSB890 (43). The KIM6+ deletion mutants were generated through allelic exchange, as described previously (23), and the deletions were verified by PCR. For creation of KIM6+ ΔorfX, approximately 500-bp-long DNA regions flanking orfX were amplified by PCR from the genomic DNA of Y. pseudotuberculosis IP2666 (48) and subcloned into the pUC18 plasmid. The Kan resistance gene sequence flanked by FLP recombinase recognition sequences was amplified by PCR from plasmid pKD4 (10) and inserted between the cloned orfX homology sequences. The resulting orfX-Kan cassette was then transferred into the suicide vector pSB890. KIM6+ orfX-Kan was created by allelic exchange, and the sequence was confirmed by PCR and sequencing. The Kan region was then removed from the chromosome of the knockout strain by expression of FLP recombinase from plasmid pFLP2 (27), and subsequently, the latter plasmid was cured. KIM5 with the y2313, y2315, and y2316 deletion (Δy2313-y2316) was generated from KIM5 through allelic exchange. KIM5 ΔorfX was generated from KIM6+ ΔorfX by first isolating its Δpgm derivative, KIM6 ΔorfX, and then transforming it with plasmid pCD1Ap, as described previously (22).
Construction of complemented strains.
For y2313-y2316 complementation analysis, y2313-y2316 ORFs were first amplified by PCR using primers y2313_5 and y2313_6 (see Table S2 in the supplemental material) and cloned between the EcoRI and PstI sites of pMMB207. The resulting plasmid, pMMB207 y2313-y2316, was introduced into Y. pestis Δy2313-y2316 strains by electroporation. Expression of y2313-y2316 ORFs was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). For orfX complementation experiments, the orfX gene was amplified using ORFX-start-EcoRI and ORFX-stop-BamHI (see Table S2 in the supplemental material) and cloned between the EcoRI and BamHI sites of pMMB207. The resulting vector, pMMB207orfX, was introduced into orfX deletion strains of Yersinia. The pMMB207orfX vector was also subjected to single-base insertion mutagenesis to introduce a stop codon at the third-amino-acid-encoding position of the orfX gene using primers orfXmutS and orfXmutAS (see Table S2 in the supplemental material), and the resulting vector was named pMMB207orfXmut.
Macrophage infection and analysis of bacterial survival.
Murine macrophage-like cell line J774A.1 and murine bone marrow-derived macrophages (BMMs) were cultured and infected with Y. pestis, as described previously, unless noted otherwise (23, 48). The CFU assays, as well as the detection of intracellular bacteria by fluorescent microscopy, were also performed as described previously (48), with the following modifications. BMMs were seeded in 24-well tissue culture plates at a concentration of 1.5 × 105 cells/well and infected with Y. pestis at multiplicities of infection (MOIs) of 5 to 10. Infection was performed with Y. pestis grown overnight at 28°C in HI broth (default) or with Y. pestis that had been subcultured for 2 h at 37°C in the presence of 2.5 mM CaCl2. For the CFU assays, the results were log transformed prior to statistical analysis.
Determination of extracellular versus intracellular bacteria (inside-outside staining).
J774A.1 cells were seeded on glass coverslips at 2 × 105 cells/ml medium in 24-well cell culture plates and incubated overnight. Macrophages were infected at an MOI of 50 with KIM5/pGFP, as described previously (48). Green fluorescent protein (GFP) expression was induced 1 h prior to fixation. At various time points postinfection, the wells were washed with phosphate-buffered saline (PBS) twice and fixed with 2.5% paraformaldehyde in PBS for 20 min. The fixed but nonpermeabilized cells were treated with anti-Yersinia antiserum SB349, as described previously (48), in order to specifically label extracellular yersiniae. The slides were examined by epifluorescence microscopy using a Zeiss Axioplan2 microscope to identify the macrophage-associated intracellular bacteria (green due to GFP only) and extracellular bacteria (red and green overlay).
RNA isolation.
J774A.1 cells were seeded on 10-cm tissue culture plates at 4 × 106/plate and grown overnight. Approximately 2 × 107 J774A.1 cells (five tissue culture plates) were infected at an MOI of 50 with Y. pestis KIM5 that had been grown overnight at 28°C in HI broth supplemented with ampicillin. After 20 min of infection, 8 μg/ml of gentamicin was added to inhibit extracellular growth. The gentamicin concentration was reduced to 2 μg/ml at 1.5 h postinfection. At 1.5, 4, and 8 h postinfection, the infected macrophages were lysed and the RNA was stabilized by incubation in 0.1% SDS-0.1% acidic phenol-19% ethanol on ice, as described previously (13). Bacteria were collected by centrifugation at 3,800 × g and 4°C for ∼20 min. The bacterial cells were lysed with 3 to 5 mg/ml lysozyme (Boehringer Mannheim) in Tris-EDTA for 5 min at room temperature, and total RNA was isolated using a Qiagen RNeasy minikit. On-column digestion of genomic DNA was performed using an RNase-free DNase set (Qiagen). Control bacteria were grown to logarithmic phase (4 h) in tissue culture medium under the same conditions but without macrophages or gentamicin, and RNA was isolated using the same stabilization and purification procedures. The integrity and purity of the isolated RNA were determined using an Agilent 2100 bioanalyzer. Four independent biological replicates were collected at each time point.
Generation of fluorescent cDNA probes and microarray hybridization.
The 70-mer oligonucleotide microarrays representing all ORFs from Y. pestis (Y. pestis microarray, version 2) were obtained from Pathogen Functional Genomics Resource Center/J. Craig Venter Institute. Fluorescent cDNA probes were prepared by an indirect aminoallyl labeling method, as described previously (22), except that 2 to 5 μg of total RNA was used for cDNA synthesis. Microarray slide preparation, hybridization, and processing were also carried out as described previously (22), except that 250 pmol each of Cy3- or Cy5-labeled cDNA was used for hybridization. For each time point, two of the control and intracellular RNA samples were labeled with Cy3 and Cy5, respectively, and the reciprocal labeling was done with the remaining two pairs of samples to correct for dye bias.
Analyses of the microarray data.
Statistical analyses of the microarray data were performed as described previously (22). A heat map diagram depicting gene expression levels was created for those genes that had a P value of less than 0.05 for the entire experiment and had at least one time point where there was a 2-fold difference in intensity values compared to the level of expression of the control. The heat map was generated using the heatmap.2 function of the gplots library of R. The expression values for each gene at each time point were used as input for the hierarchical clustering used in heatmap.2. Genes with similar expression patterns are grouped together on the basis of the clustering, and the dendrogram presented in Fig. 2 shows those relationships. The biological terms overrepresented in the lists of differentially regulated genes were identified using the functional annotation clustering program of DAVID (http://david.abcc.ncifcrf.gov) (11, 29). The analysis was performed by inputting Entrez gene identifiers for genes identified as being up- or downregulated by the transcriptional profiling and comparing the genes on that list to all Y. pestis genes in the genome. Gene lists were analyzed under the default setting using default gene sets, which include gene ontology, Biocarta, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Interpro and Protein Information Resource (PIR) superfamily names. Fisher's exact tests were used to calculate P values, and the enrichment score was computed as the negative log of the geometric mean of the P values for each clustered group. The clustered groups with enrichment scores of ≥1.3 (mean P value < 0.05) are listed in Tables 4 and 5. Since DAVID does not give a name for each clustered group, a representative annotation from each group is shown in Table 4 and 5.
FIG. 2.
Isolation of Y. pestis RNA. Representative chromatograms of total RNA isolated from uninfected J774A.1 murine macrophage-like cells (lane 2), Y. pestis KIM5 grown in Dulbecco modified Eagle tissue culture medium (lane 3), or KIM5 recovered from inside J774A.1 cells at 4 h postinfection (lane 4). Lane 1, molecular size standards.
TABLE 4.
Results of annotation enrichment analysis for the lists of differentially regulated genes at 1.5 h postinfection
| Overrepresented functional family or pathwaya | No. of genes | No. of genes in group/no. of genes in genome (%) | P valueb |
|---|---|---|---|
| Upregulated inside macrophages at 1.5 h | |||
| Citrate cycle (TCA cycle) | 16 | 3.3 | 2.4E−10 |
| ABC transporters, general | 54 | 11.1 | 1.1E−08 |
| Carboxylic acid metabolic process | 39 | 8.0 | 9.6E−07 |
| Cellular respiration/coenzyme catabolic process | 8 | 1.7 | 6.1E−06 |
| Branched-chain-family amino acid metabolic process | 9 | 1.9 | 1.4E−05 |
| Oxidoreductase | 25 | 5.2 | 2.5E−05 |
| Amino acid biosynthetic process | 20 | 4.1 | 4.2E−05 |
| Flavoprotein | 11 | 2.3 | 3.7E−04 |
| Fatty acid metabolism/valine, leucine and isoleucine degradation | 7 | 1.4 | 1.1E−03 |
| Glutamine family amino acid biosynthetic process | 7 | 1.4 | 2.4E−03 |
| Glyoxylate and dicarboxylate metabolism | 7 | 1.4 | 3.8E−03 |
| Hydrolyase | 7 | 1.4 | 3.9E−03 |
| Anion transport | 7 | 1.4 | 7.4E−03 |
| Carbohydrate metabolic process | 23 | 4.7 | 1.5E−02 |
| Glyoxylate metabolic process | 3 | 0.6 | 2.0E−02 |
| Vitamin binding/thiamine pyrophosphate | 6 | 1.2 | 2.4E−02 |
| Downregulated inside macrophages at 1.5 h | |||
| Virulence (ysc and lcr genes) | 10 | 3.9 | 7.0E−07 |
| Iron ion transport | 9 | 3.5 | 1.3E−05 |
| Protein biosynthesis | 10 | 3.9 | 7.3E−05 |
| Purine metabolism | 13 | 5.1 | 3.5E−03 |
| tRNA metabolic process | 10 | 3.9 | 7.0E−03 |
| Translation | 14 | 5.5 | 9.7E−03 |
| Pyrimidine metabolism | 9 | 3.5 | 1.1E−02 |
| TonB-dependent receptor, plug | 4 | 1.6 | 1.2E−02 |
| Ribonucleoside metabolic process | 4 | 1.6 | 1.6E−02 |
Underlined categories are overrepresented only at 1.5 h postinfection and not at 4 or 8 h postinfection.
P values were calculated using Fisher's exact test (see Materials and Methods for details).
TABLE 5.
Results of annotation enrichment analysis for the lists of differentially regulated genes at 4 h postinfection
| Overrepresented functional family or pathwaya | No. of genes | No. of genes in group/no. of genes in genome (%) | P valueb |
|---|---|---|---|
| Upregulated inside macrophages at 4 h | |||
| Glutamine family amino acid biosynthetic process | 11 | 3.5 | 5.0E−09 |
| ABC transporters, general | 45 | 14.2 | 1.2E−08 |
| Flavoprotein | 10 | 3.2 | 7.8E−05 |
| Fatty acid metabolism | 7 | 2.2 | 2.3E−04 |
| Citrate cycle (TCA cycle)/oxidative phosphorylation | 8 | 2.5 | 1.4E−03 |
| Aspartate family amino acid biosynthetic process | 6 | 1.9 | 3.7E−03 |
| Anion transport | 6 | 1.9 | 5.8E−03 |
| Glyoxylate and dicarboxylate metabolism | 6 | 1.9 | 6.0E−03 |
| Lysine-arginine-ornithine-binding periplasmic protein | 3 | 1.0 | 8.0E−03 |
| Glyoxylate cycle | 3 | 1.0 | 8.8E−03 |
| Branched-chain-family amino acid metabolic process | 5 | 1.6 | 9.6E−03 |
| Tryptophan metabolism | 5 | 1.6 | 1.1E−02 |
| Thiamine pyrophosphate enzyme, C-terminal TPP binding | 3 | 1.0 | 1.5E−02 |
| Nitrogen metabolism | 6 | 1.9 | 1.7E−02 |
| Benzoate degradation via hydroxylation | 4 | 1.3 | 2.0E−02 |
| Downregulated inside macrophages at 4 h | |||
| Glycolysis/gluconeogenesis | 8 | 6.8 | 2.3E−05 |
| Iron ion transport | 6 | 5.1 | 8.4E−05 |
| Transport | 20 | 17.1 | 2.0E−04 |
| TonB-dependent receptor, plug | 4 | 3.4 | 8.9E−04 |
| Fructose and mannose metabolism | 6 | 5.1 | 2.2E−03 |
Underlined categories are overrepresented at 4 and 8 h postinfection but not at 1.5 h postinfection.
P values were calculated using Fisher's exact test (see Materials and Methods for details).
qRT-PCR.
The relative levels of transcription of Y0052 (yopB), y0464 (fadB), y0509, y0655, y0850, y1895 (yfeC), y2316, y3120, and y3553 were determined by quantitative reverse transcription-PCR (qRT-PCR). The primers used for qRT-PCR were designed using the Primer3 Plus program (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) (56) and are listed in Table S2 in the supplemental material. Each primer set was designed to have a melting temperature difference of 2°C or less and product sizes of between 250 and 350 bp. cDNA synthesis was performed with 0.6 μg RNA and SuperScript II reverse transcriptase, according to the manufacturer's instruction, except for the following changes. Three micrograms of random primers (Invitrogen) was used, and the reaction was carried out overnight at 42°C. Each of the 25-μl qPCR mixtures contained 2.0 μl of a 1:20 dilution of cDNA, 0.3 μM gene-specific primers, and 12.5 μl of 2× QuantiTect SYBR green (Qiagen). The reaction was performed according to the manufacturer's instruction and monitored using an ABI Prism Applied Biosystem 7500 SDS real-time PCR machine. The results were normalized to those for Y. pestis 16S RNA, and the relative fold change for each gene was calculated using the Pfaffl method (46), as follows: gene expression ratio = [(Etarget gene)ΔCT (control − experimental)]/[(E16S RNA)ΔCT (control − experimental)], where E represents the primer efficiency of the primer set specific for the gene, and CT is the threshold cycle. Serial dilutions of cDNA were used to determine the amplification efficiency of each primer set. For each target gene, qPCR measurements were made in duplicate with three independent RNA samples.
LDH release assay.
Supernatants from infected macrophages were collected at 24 h postinfection and analyzed for lactate dehydrogenase (LDH) release using the CytoTox-96 nonradioactive cytotoxicity assay (Promega), as described previously (33), except for the following change. The percentage of LDH released per infection condition was calculated as [(infected-cell LDH release − medium blank)/(total LDH release − medium blank)] × 100.
β-Lactamase (bla) assay.
BMMs were seeded in black 96-well tissue culture plates at a concentration of 0.3 × 105 cells/well and infected with Y. pestis strains at an MOI of 10. The strains used for the assay carried the mutant yopJ gene on pCD1, encoding the catalytically inactive form of YopJ (C172A), to prevent YopJ-induced macrophage cell death, and plasmid pMMB207-yopJ (C172A)-M45-bla. Construction of the plasmids (our unpublished data) will be described elsewhere. Infection was performed with Y. pestis strains that had been grown at 37°C for 2 h in the presence of 2.5 mM CaCl2, and 0.5 mM IPTG was present throughout the infection. At ∼4 h postinfection, CCF2-AM dye (Invitrogen) was prepared according to the manufacturer's instructions and added directly to live cells to reach a 1× final concentration. The plate was incubated at room temperature in the dark for an additional 45 to 60 min. The live cells were examined by epifluorescence microscopy using a Zeiss Axiovert 100 microscope equipped with a ×32 objective and a 4′,6-diamidino-2-phenylindole filter for excitation. The images were captured using a Spot camera (Diagnostic Instruments, Inc.) and processed using Adobe Photoshop CS2 software. The percentage of blue macrophages was calculated by counting three independent fields (a minimum of 180 macrophages) in each experiment.
Infection of mice.
All experiments were carried out with the approval of the Stony Brook. University IACUC. Eight-week-old female BALB/c mice (Taconic) were used. To prepare bacterial cultures for mouse infection, Y. pestis strains from frozen stocks were plated on HI medium plates containing ampicillin and allowed to form single colonies at 28°C for 2 days. Isolated colonies were used to inoculate HI broth containing ampicillin, and the cultures were incubated at 28°C overnight with aeration. Twenty-four hours later, the cultures were diluted in HI broth supplemented with ampicillin to an optical density at 600 nm of ∼0.02 and incubated overnight at 28°C with aeration. On the morning of infection, the cultures were centrifuged and the bacterial pellets were suspended and serially diluted in PBS. One hundred microliters of the dilution containing 104 CFU/ml bacteria was injected into the tail vein of each mouse, and the health of the mice was monitored daily for 3 weeks. The actual numbers of input CFU were determined by plating 1:10 dilutions of the inputs and counting the colonies.
Statistical analyses of the data.
When appropriate, statistical tests were performed using Student's t test or one-way analysis of variance (ANOVA) with Tukey's multiple comparison test (Prism software, version 4.0c, for Macintosh; GraphPad Software, San Diego, CA).
Microarray data accession number.
All microarray data described in this study have been submitted to Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE22168.
RESULTS AND DISCUSSION
Isolation of RNA from intracellular Y. pestis.
During the first few hours following phagocytosis, Y. pestis does not replicate but remains viable (48). Vacuoles containing Y. pestis are thought to undergo some interaction with the components of the endocytic pathway and acquire several markers of late endosomes or lysosomes, but they fail to acidify (22, 51, 61). By 3 to 5 h postinfection, intracellular phoP mutant Y. pestis begins to die (22, 42), suggesting that at least some of the genes required for the intracellular survival of Y. pestis should be expressed by this time point. Subsequently, intracellular Y. pestis begins replication between 5 and 8 h postinfection (48). To capture the transcriptional changes that take place in response to these events inside YCVs, global gene expression profiling was performed with RNA samples isolated from intracellular Y. pestis after infection of macrophages.
J774A.1 macrophage-like cells were infected with Y. pestis KIM5, an attenuated strain lacking the pgm locus (Table 1), that had been grown to stationary phase at 28°C in HI broth. An MOI of 50 was used in order to gain maximal bacterial yields while minimizing the toxic effects of the bacteria on macrophages. After the initial engulfment of bacteria, low concentrations of gentamicin were added to the tissue culture medium to prevent extracellular Y. pestis replication. A large number of bacteria were taken up by J774A.1 cells under these infection conditions; and the vast majority of them remained intracellular at 1.5, 4, and 8 h postinfection, although small numbers of Y. pestis organisms were found on the extracellular surface of the macrophages at the 8-h time point (Fig. 1). At 1.5, 4, and 8 h postinfection, the macrophages were lysed and the bacterial RNA was quickly stabilized by adding a mixture of SDS, phenol, and ethanol, as previously described by Eriksson et al. (13). Bacterial cells were then collected by centrifugation, and RNA was isolated. This differential lysis approach allows enrichment of bacterial RNA via physical separation of the bacteria from the host cells and has successfully been used for the gene expression profiling of intracellular S. Typhimurium and S. flexneri (13, 37). Control bacteria were grown to logarithmic phase (4 h) in tissue culture medium under the same conditions but without macrophages or gentamicin, and RNA was isolated using the same stabilization and purification procedures. The analyses of the isolated RNA showed that the intracellular Y. pestis RNA was highly enriched with a minimum amount of degradation or host RNA contamination (Fig. 2), indicating that the isolated RNA was suitable for use in microarray analyses.
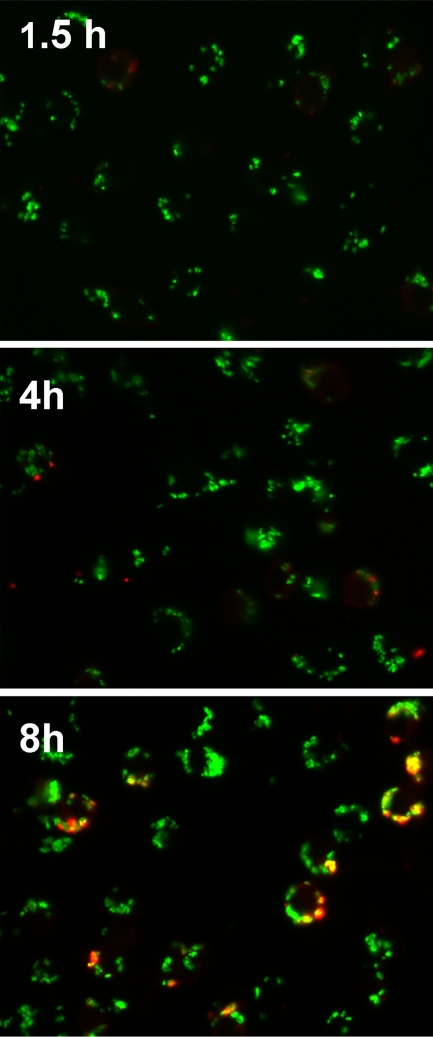
FIG. 1.
Determination of extracellular versus intracellular Y. pestis associated with macrophages. J774A.1 cells were infected with KIM5/pGFP at an MOI of 50 and fixed with 2.5% paraformaldehyde at the indicated time points. GFP expression was induced 1 h prior to fixation. The fixed but nonpermeabilized cells were treated with anti-Yersinia antiserum SB349 (48) to specifically label extracellular yersiniae. The slides were examined by epifluorescence microscopy to identify the macrophage-associated intracellular bacteria (green due to GFP only) and extracellular bacteria (red and green overlay). Three independent experiments were performed, and representative pictures are shown.
Global expression profiles.
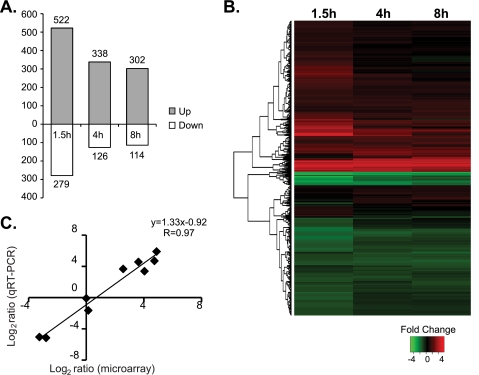
The transcriptional profiles of intracellular Y. pestis at various times postinfection were compared to those of control Y. pestis using the 70-mer oligonucleotide microarrays representing all ORFs from Y. pestis, which were obtained from Pathogen Functional Genomics Resource Center/J. Craig Venter Institute. Differential gene expression of more than 2-fold and a P value of <0.05 were considered significant. At 1.5, 4, and 8 h postinfection, a total of 801, 464, and 416 genes of intracellular Y. pestis showed significant up- or downregulation, respectively (Fig. 3 A; see Table S1 in the supplemental material). These account for approximately 9 to 18% of the Y. pestis genome. The relative changes in gene expression levels between the intracellular bacteria and the control bacteria ranged from −25.3 to +44.6 fold. A large number of differentially regulated genes underscored the drastic difference between the intracellular and extracellular environments. Substantially fewer genes exhibited significant changes in expression levels between different time points. About 200 genes altered their expression levels between 1.5 and 4 h, and 56 genes altered their expression levels between 4 and 8 h, with the fold changes ranging from −6.5 to +9 (see Table S1 in the supplemental material). Hierarchical clustering analysis of the microarray data also illustrated that the strongest induction/repression of many genes occurred at 1.5 h postinfection and that the transcriptional profiles of intracellular Y. pestis at 4 and 8 h are more similar to each other than they are at 1.5 h (Fig. 3B). This might be because more genes are required during the initial adaptation to the intracellular environment than at the later time points, or it might reflect the different replicative states of intracellular Y. pestis between these time points (48). Alternatively, since the bacteria infecting macrophages were pregrown at 28°C and were compared to the control bacteria grown at 37°C for 4 h, some of the genes that appear to be differentially regulated at 1.5 h could be the thermoregulated genes that had not been acclimated to the host cell temperature of 37°C. (Out of 164 genes that showed lower expression levels at 1.5 h but that later had expression levels comparable to the level for the control, 35 genes were induced at 37°C in tissue culture media but not at 28°C in HI medium [data not shown], suggesting that these genes may fall into such a category.) Overall, however, the majority of the genes were consistently up- or downregulated over the course of 8 h, and only a small number of genes changed the direction of the fold change. These results indicate that although the biggest transcriptional changes occur during the initial stage of macrophage infection, the transcriptional profiles of Y. pestis are grossly similar over the 8-h period.
FIG. 3.
Transcriptional profiling of intracellular Y. pestis KIM5. (A) Numbers of Y. pestis genes significantly up- or downregulated inside macrophages at various times postinfection compared to the level of expression by Y. pestis cells grown in Dulbecco modified Eagle tissue culture medium. (B) Hierarchical clustering of differentially regulated Y. pestis genes. Each horizontal line indicates one gene. The fold change in the mRNA expression level of each gene is represented according to the color key shown below the heat map. Genes with similar expression patterns are grouped together on the basis of the clustering, and the dendrogram on the left shows those relationships. (C) Validation of microarray results by qRT-PCR. qRT-PCR was performed with nine genes that encompassed a range of expression levels in the microarray experiments. The results were normalized to the level of expression of 16S rRNA. Eight out of nine genes showed the differential regulation in the same direction. The only exception was Y0052 (yopB), which had +1.1-fold change by microarray analysis but a −1.8-fold change by qRT-PCR. The results of the linear regression analysis are shown above the graph.
Transcriptome analyses have previously been carried out with Y. pestis cells that were isolated from an infected rat bubo (59) or mouse lung (32). The comparison between the lists of differentially regulated genes in these studies and ours showed no strong overlap. Out of 464 Y. pestis genes that were either up- or downregulated at 4 h postinfection in our experiments, only 10 and 28 genes were also up- or downregulated in the same direction in rat bubo and in mouse lung, respectively (see Table S1 in the supplemental material). In these animal infection studies, bacteria are found mainly in the extracellular environments (32, 59). The lack of overlap likely reflects the distinct challenges that the pathogen faces in the intracellular and extracellular environments.
Microarray results were confirmed by performing qRT-PCR analyses of nine genes that encompassed a range of expression levels in the microarray experiments. The linear regression analysis of the microarray and qRT-PCR measurements resulted in a correlation coefficient (R value) of 0.97, showing that the two sets of data correlated positively and closely (Fig. 3C).
Virulence-related genes.
A number of known virulence-related genes were differentially regulated inside macrophages (Table 2). Among the most highly upregulated genes were the members of the psaABC operon, which encodes proteins necessary for the assembly of pH 6 antigen pili. The role of pH 6 pili inside macrophages is unclear, but our microarray data are in agreement with the findings presented in a previous report that showed the induction of pH 6 antigen expression inside macrophages (34). The insecticidal toxin-like genes tcaA1, tcbA, and tcaC1 were also upregulated inside macrophages. These genes were previously shown to be induced at 26°C in vitro and in the flea midgut and were implicated in the antiphagocytic activity of Y. pestis during transmission from fleas to the mammalian host (40, 62). It has also been reported that the protein products of these genes can be translocated into mammalian cells via the T3SS (20). Our data raise the possibility that, like Yops, these insecticidal toxin-like genes could have a role during infection of the mammalian host in addition to during transmission. In contrast, the caf1AMR operon, which encodes F1 capsule assembly, was downregulated at all time points. Downregulation of the caf1AMR operon was unexpected, as this operon is usually induced at 37°C. Although the results of anti-F1 antibody staining and the comparison of our expression data with those for bacteria grown at 28°C suggest that the caf1AMR operon is still expressed at some levels inside macrophages (data not shown), it is plausible that, in addition to temperature, factors specific to the intraphagosomal environment may be contributing to the regulation of this operon.
TABLE 2.
Differential expression of the selected virulence-related Y. pestis genes during macrophage infection
| Gene location and locus tag | Predicted function | Symbol | Fold change in expression levela |
||
|---|---|---|---|---|---|
| 1.5 h | 4 h | 8 h | |||
| pCD1 plasmid | |||||
| Y0009 | Secreted protein kinase | ypkA | 1.1 | 1.0 | 1.2 |
| Y0010 | Targeted effector protein | yopJ | −3.1 | −2.5 | −1.1 |
| Y0013 | Secreted effector protein | yopH | 1.4 | 1.2 | 2.0 |
| Y0050 | Secreted effector protein | lcrV | −2.7 | −1.1 | 1.0 |
| Y0052 | Secreted effector protein | yopB | −1.0 | 1.1 | 1.2 |
| Y0053 | Secreted effector protein | yopD | −1.3 | −1.0 | −1.0 |
| Y0059 | Yop targeted effector protein | yopM | −7.9 | −3.6 | −1.5 |
| Y0065 | Yop targeted effector | yopT | −1.3 | −1.3 | 1.0 |
| Y0066 | Yop targeting protein | yopK | −1.8 | −1.2 | 1.4 |
| Y0077 | Secreted effector | yopE | 1.0 | 1.1 | 1.4 |
| pPCP1 plasmid, YPKp07 | Plasminogen activator protease precursor | pla | 2.8 | 1.3 | 2.8 |
| pMT1 plasmid | |||||
| YPKMT065 | F1 capsule antigen | caf1 | −4.2 | −1.5 | −1.1 |
| YPKMT066 | F1 capsule protein | caf1A | −19.9 | −6.8 | −2.4 |
| YPKMT067 | F1 capsule protein | caf1M | −13.5 | −7.8 | −5.0 |
| Y1097 | F1 capsule positive regulator | caf1R | −3.2 | −4.3 | −2.1 |
| YPKMT083 | Murine toxin | tox | −1.8 | −1.6 | −2.2 |
| Chromosomal | |||||
| y2880 | Outer membrane usher protein | psaC | 19.1 | 18.0 | 9.6 |
| y2881 | pH 6 antigen chaperone protein | psaB | 20.5 | 11.7 | 5.1 |
| y2882 | pH 6 antigen fimbrial subunit | psaA | 22.1 | 15.4 | 40.3 |
| y2883 | Hypothetical protein | psaF | 1.7 | 1.7 | 1.5 |
| y2884 | Putative regulator | psaE | 1.6 | 2.0 | 1.5 |
| y0183 | Putative toxin subunit | tcaA1 | 7.6 | 6.6 | 2.4 |
| y0184 | Putative toxin subunit | tcbA | 3.9 | 5.6 | 2.3 |
| y0185 | Putative toxin subunit | tcaC1 | 1.7 | 1.8 | 1.3 |
The fold changes with P values of <0.05 are indicated in boldface.
The T3SS and Yop effectors, encoded by the ysc and yop genes on virulence plasmid pCD1, are some of the most critical virulence factors in Y. pestis. Our microarray data showed that the expression levels of many ysc/yop genes in intracellular Y. pestis were slightly lower than those of the control bacteria at 1.5 h postinfection but were comparable to those of the control bacteria at later time points (Table 2). Our unpublished analysis showed that the transcription of the ysc and yop genes is highly induced under our control condition (37°C tissue culture medium) compared to the level of induction of in vitro growth at 28°C (data not shown). Therefore, the ysc and yop genes should also be induced at similar levels in intracellular Y. pestis at these time points. This is interesting, since the previous studies, using either J774A.1 cells or HeLa cells, have indicated that intracellular Y. pestis does not produce Yop effectors in large amounts or effectively deliver Yops into the host cells (8, 55). Consistent with the lack of Yop activities originating from intracellular Y. pestis, Das et al. investigated the host cell response to intracellular Y. pestis using DNA microarray analysis and found many proinflammatory cytokine genes to be induced in macrophages containing Y. pestis (9). Our study indicates that although Yop proteins may not be produced in large amounts, transcription of yop genes is induced in intracellular Y. pestis. It is possible that induction of the ysc and yop genes inside macrophages primes the T3SS so that it can exert maximal activity when the bacteria escape from macrophages and enter the extracellular phase of growth.
It should be noted that the strain used in this study, KIM5, lacks another virulence determinant, the pgm locus; and thus, we were unable to determine the transcriptional regulation of genes within this locus. The pgm locus contains the rip operon, which promotes bacterial survival in activated macrophages, hms genes that are required for biofilm formation inside the flea vector, and several other genes predicted to be involved in bacterial metabolism. Analysis of intracellular expression of these genes, as well as the effects caused by the absence of the pgm locus on the transcription of other Y. pestis genes, requires further study using a fully virulent strain.
Stress-response genes.
The Y. pestis genes that were upregulated inside macrophages included many stress-response genes (Table 3). Most notably, genes involved in the detoxification of ROSs, including superoxide dismutases (sodB and sodC) and catalase (katG), were highly upregulated throughout the time course. This suggested that the YCVs may contain elevated levels of oxidative stress. Consistent with the presence of oxidative stress, Das et al. have observed strong induction of an antioxidant gene, that for thioredoxin peoxidase 2, in macrophages containing Y. pestis (9). Interestingly, however, the levels of the superoxide-responsive transcriptional regulator oxyR and SOS response genes (sulA, recA, etc.) that repair the DNA damage inflicted by ROSs were not significantly induced. The reason for this is unclear, but it is possible that the concentration of the superoxide and other ROSs is not sufficient to activate transcription of oxyR or the SOS response. In contrast, the level of transcription of genes associated with detoxification of nitric oxide (NO), such as hmp, was unchanged in our study. This may be because the increase in the NO level requires activation of macrophages by gamma interferon. Several general stress-response genes encoding acid, heat, and cold shock proteins were moderately induced. Some of these genes showed increased expression levels early, while the expression levels of others increased at later time points, suggesting that intracellular Y. pestis may be subjected to different kinds of stress over time.
TABLE 3.
Differential expression of the selected stress-response Y. pestis genes during macrophage infection
| Gene function and locus tag | Predicted function | Symbol | Fold change in expression levela |
||
|---|---|---|---|---|---|
| 1.5 h | 4 h | 8 h | |||
| Stress response | |||||
| y0870 | Catalase, hydroperoxidase HPI(I) | katG | 7.0 | 8.7 | 10.7 |
| y1951 | Superoxide dismutase, iron | sodB | 20.2 | 8.7 | 3.6 |
| y0815 | Superoxide dismutase precursor (Cu-Zn) | sodC | 9.4 | 3.2 | 6.1 |
| y2014 | Acid shock protein | asr | 4.2 | 1.3 | 1.2 |
| y1226 | Acid shock protein precursor | asr | 6.3 | −1.5 | −1.3 |
| y2774 | Cold shock protein | cspE | −1.0 | 3.3 | 3.8 |
| y0224 | Cold shock-like protein | cspI | 1.1 | 2.4 | 3.2 |
| y2076 | Carbon starvation protein | cstA | 2.8 | 3.6 | 3.9 |
| y3859 | Universal stress protein | uspA | 4.1 | 1.8 | 1.5 |
| y3860 | Universal stress protein UspB | uspB | 3.4 | 2.2 | 2.0 |
| y1677 | DNA protection under starvation conditions | dps | 2.9 | 1.6 | 1.3 |
| y4101 | Heat shock protein | ibpB | 2.6 | 3.1 | 4.4 |
| y4102 | Heat shock protein | ibpA | 3.8 | 4.1 | 4.6 |
| y2966 | Outer membrane protein 1b (Ib;c) | ompC | 10.4 | 7.8 | 2.4 |
| y1820 | Putative transport ATPase | mgtC | 3.8 | 2.5 | 3.4 |
| y1821 | Mg2+ transport ATPase | mgtA | 6.7 | 11.8 | 7.9 |
| Transcriptional regulators | |||||
| y1794 | Transcriptional regulatory protein | phoP | 2.5 | 2.6 | 2.0 |
| y1793 | Sensor protein PhoQ | phoQ | 2.5 | 2.4 | 2.1 |
| y3916 | Osmolarity response regulator | ompR | 1.4 | 1.3 | 1.3 |
| y3917 | Osmolarity sensor protein | envZ | −1.0 | 1.3 | 1.3 |
| y0834 | RNA polymerase sigma factor | rpoS | 4.9 | 2.0 | 3.2 |
| Iron uptake | |||||
| y1208 | Ferric uptake regulator | fur | 1.7 | 1.3 | 1.5 |
| y3910 | Ferrous iron transport protein B | feoC | 2.5 | 1.7 | −1.2 |
| y3911 | Ferrous iron transport protein B | feoB | 2.0 | 1.5 | −1.1 |
| y3912 | Ferrous iron transport protein A | feoA | 7.2 | 1.8 | −1.4 |
| y0539 | ATP-binding protein of ABC transporter | hmuV | −4.5 | −2.4 | −1.7 |
| y0540 | ABC-type permease | hmuU | −2.5 | −1.8 | −1.6 |
| y0541 | Periplasmic binding protein | hmuT | −1.7 | −1.3 | −1.2 |
| y0543 | TonB-dependent outer membrane receptor | hmuR | −2.6 | −2.0 | −1.5 |
| y0544 | Hemin uptake system component | hmuP | −5.5 | −2.8 | −1.8 |
| y1891 | Putative regulator of yfeABCD | yfeE | 1.6 | 1.4 | 1.3 |
| y1894 | Permease for iron and manganese ABC transporter | yfeD | −12.0 | −9.2 | −7.8 |
| y1895 | Permease for iron and manganese ABC transporter | yfeC | −6.3 | −6.9 | −5.3 |
| y1896 | ATP-binding protein for iron/manganese ABC transporter | yfeB | −9.8 | −7.2 | −4.3 |
| y1897 | Periplasmic-binding protein for iron/manganese transporter | yfeA | −9.0 | −4.5 | −5.1 |
| y1524 | ATP-binding protein of iron ABC transporter | yfuC | −1.0 | 1.5 | 1.3 |
| y1525 | Inner membrane permease of iron ABC transporter | yfuB | −1.2 | 1.2 | 1.2 |
| y1526 | Solute-binding periplasmic protein for iron ABC transporter | yfuA | 1.7 | 1.4 | 1.2 |
| y2872 | Putative outer membrane iron/siderophore receptor | yiuR | −25.3 | −21.5 | −10.4 |
| y2873 | ATP-binding protein of iron/siderophore ABC transporter | yiuC | −6.0 | −5.3 | −3.6 |
| y2874 | Inner membrane permease of iron/siderophore transporter | yiuB | −4.8 | −4.4 | −3.6 |
| y2875 | Solute-binding protein of iron/siderophore transporter | yiuA | −11.4 | −6.0 | −4.6 |
The fold changes with P values of <0.05 are indicated in boldface.
A number of global transcriptional regulators are known to play crucial roles in response to environmental stresses. For example, the PhoPQ two-component system promotes growth of Y. pestis and/or Y. pseudotuberculosis under low-Mg2+-concentration conditions and resistance to antimicrobial peptides and is essential for survival inside macrophages (23, 26, 42). The level of expression of phoP and phoQ, as well as the majority (56%) of the phoP-regulated genes identified in our previous in vitro microarray study (22), was mildly increased inside macrophages (Table 3). The upregulated PhoP targets included mgtC, whose gene product is involved in Mg2+ transport. The OmpR/EnvZ two-component system and the alternative sigma factor RpoS are also involved in the bacterial response to stresses such as low pH and hydrogen peroxide. The ompR and envZ genes were not upregulated in our study, but their transcriptional target, ompC, was highly induced (Table 3), suggesting that ompR and envZ may be regulated at a posttranscriptional level. The expression of rpoS was moderately increased in intracellular Y. pestis. The results are consistent with the important roles that these global regulators may be playing in allowing Y. pestis to adapt to the intracellular environment.
Pathway analyses.
To gain further insights into the physiology of Y. pestis growing inside macrophages and the characteristics of YCVs, we subjected the lists of up- or downregulated genes identified in our microarray experiments to an online annotation enrichment analysis program, DAVID (http://david.abcc.ncifcrf.gov) (11, 29). The program searches for biological pathways or functional gene families that are overrepresented within a given gene list on the basis of several different annotation categories, including gene ontology terms.
Several biological themes were identified to be significantly overrepresented (P < 0.05 by Fisher's exact test) in our lists (Table 4 and 5). The list of enriched themes at 8 h was similar to that at 4 h (data not shown), while the list contained several unique pathways at 1.5 h postinfection. For example, it was found that the pathways involved in protein biosynthesis and purine/pyrimidine metabolism were downregulated at 1.5 h postinfection compared to the level of expression by control bacteria that were logarithmically growing in the tissue culture medium (Table 4). At 4 and 8 h postinfection, these pathways were no longer overrepresented in the downregulated gene list (Table 5). This might reflect the fact that intracellular Y. pestis cells do not start replicating until 3 to 5 h postinfection under our experimental conditions (48) or could be due to the other factors discussed in earlier sections. Also as discussed earlier, components of T3SS were also downregulated only at the 1.5-h time point.
The genes involved in iron transport (hmu, yfe, and yiu) were highly overrepresented in the downregulated gene lists at all time points (Tables 3 to 5). The finding was unexpected because the intracellular environment is predicted to be limited in iron and upregulation of iron-uptake genes has been observed in many intracellular bacteria (2, 57, 64). A Y. pestis mutant lacking Ybt, Feo, and Yfe iron-uptake systems fails to grow inside J774A.1 cells, also suggesting the importance of iron-uptake systems in intracellular survival (45). However, similar downregulation of iron-uptake genes inside macrophages has been reported in S. Typhimurium replicating inside J774A.1 cells (13) and in S. Typhi replicating inside human monocyte cell line THP-1 (15). In Y. pestis, iron-limiting conditions are known to cause upregulation of the hmu, yfe, and yiu operons (67), so downregulation of these genes might indicate that Y. pestis inside J774A.1 cells is not starved for iron. Alternatively, the iron-uptake systems other than hmu, yfe, and yiu may be responsible for the acquisition of iron inside YCVs. The major iron-acquisition system, Ybt, is missing from the KIM5 Δpgm strain, but feo genes are mildly upregulated inside macrophages (Table 3). Therefore, it is possible that the expression of the Feo system is sufficient for intracellular Y. pestis to acquire necessary amounts of iron.
The glycolysis pathway and fructose mannose metabolism are downregulated at 4 and 8 h, consistent with the idea that glucose may be limited in the intraphagosomal environment (Table 5). In contrast, amino acid biosynthesis pathways and the tricarboxylic acid (TCA) cycle are upregulated in intracellular Y. pestis. The TCA cycle not only allows the cells to utilize various carbon sources, but it also provides precursors for many macromolecules (41). Upregulation of the TCA cycle and amino acid biosynthetic pathways may therefore indicate that Y. pestis inside YCVs has limited access to macromolecules such as amino acids and needs to synthesize them de novo. The fatty acid β-oxidation pathway and the glyoxylate pathway, which together allow conversion of fatty acids into the TCA cycle intermediates, are both highly upregulated at all time points (Table 4 and 5). This suggests that fatty acids could perhaps be used as alternative carbon sources for the intracellular Y. pestis. This is an intriguing possibility, given that isocitrate lyase, the critical enzyme in the glyoxylate pathway, is required for the persistence of M. tuberculosis inside activated macrophages (39) and for chronic infection of mice by S. Typhimurium (14). However, it has been shown that aceA, a gene encoding the Y. pestis homolog of isocitrate lyase, is not required for the virulence of Y. pestis (58). In addition, deletion of the β-oxidation pathway components fadA and fadB did not affect the ability of Y. pestis to survive inside macrophages (Table 6). The role of β-oxidation/glyoxylate pathways in Y. pestis pathogenesis therefore remains unclear and warrants further study. Nonetheless, the observed upregulation of the TCA cycle and the catabolic pathways for alternative carbon sources share trends similar to the changes observed by Motin et al. in their microarray studies of Y. pestis during a temperature shift from 26°C to 37°C (40). As proposed by Motin et al. (40) as well as by Brubaker (3), the bacteria inside the mammalian host may utilize a variety of the carbon sources present in host tissues, and these metabolic adaptations may perhaps play a critical part in plague pathogenesis.
TABLE 6.
Forty Y. pestis genes that are the most highly upregulated inside macrophages at 4 h postinfection
| Symbol | Locus taga | Predicted functionb | Fold change in expression level | Survival of deletion mutant in BMMsc |
|---|---|---|---|---|
| y1048 | 50S ribosomal protein L31 | 31.5 | ND | |
| fadB | y0464 | Four-enzyme protein involved in fatty acid oxidation | 29.4 | + |
| y2316 | Hypothetical protein | 26.6 | ++ | |
| mglB | y2662 | Periplasmic d-galactose-binding ABC transport protein | 24.1 | ND |
| ilvN | y2127 | Acetolactate synthase small subunit | 23.7 | ND |
| y2315 | Hypothetical protein | 21.0 | ++ | |
| ilvB | y2126 | Acetolactate synthase large subunit | 20.5 | ND |
| mglB | YPO1507 | Galactose-binding protein | 19.2 | ND |
| y1430 | Putative solute-binding periplasmic protein of ABC transporter | 19.1 | ND | |
| acs | y0510 | Acetyl coenzyme A synthetase | 18.8 | ND |
| y2313 | Hypothetical protein | 18.6 | ++ | |
| psaC | y2880 | Outer membrane usher protein | 18.0 | + |
| YPO3135 | 50S ribosomal protein L36 | 17.5 | ND | |
| argI | y0740 | Ornithine carbamoyltransferase | 17.4 | ND |
| ilvN | y2127 | Acetolactate synthase small subunit | 17.3 | ND |
| y1048 | 50S ribosomal protein L31 | 17.3 | ND | |
| fadE | y0946 | Acyl coenzyme A dehydrogenase | 17.2 | ND |
| cspD | YPO1366 | Cold shock-like protein | 17.1 | ND |
| hisJ | y1607 | Histidine-binding protein of histidine transport system | 16.6 | ND |
| y0655 | Hypothetical protein | 16.4 | + | |
| psaA | y2882 | pH 6 antigen fimbrial subunit | 15.4 | + |
| gltI | y1189 | Solute-binding protein of glutamate/aspartate ABC transporter | 14.8 | ND |
| glnK | y1041 | Nitrogen regulatory protein P-II 2 | 14.8 | ND |
| argC | y0310 | N-Acetyl-gamma-glutamyl-phosphate reductase | 14.0 | ND |
| hisJ | y1607 | Histidine-binding protein of histidine transport system | 13.6 | ND |
| rmf | YPO1423a | Putative ribosome modulation factor | 13.4 | ND |
| y3555 | Aspartate aminotransferase | 12.6 | + | |
| y0509 | Hypothetical protein | 12.3 | ND | |
| aceA | y0016 | Isocitrate lyase | 12.1 | ND |
| fadA | y0463 | Acetyl coenzyme A acetyltransferase | 12.1 | + |
| y0655 | Hypothetical protein | 11.8 | + | |
| mgtA | y1821 | Mg2+ transport ATPase | 11.8 | ND |
| psaB | y2881 | pH 6 antigen chaperone protein | 11.7 | + |
| ivbL | y2125 | ilvB operon leader peptide | 11.5 | ND |
| glnA | y3804 | Glutamine synthetase | 11.4 | ND |
| fadD | y2236 | Acyl coenzyme A synthase | 11.1 | ND |
| aceB | y0015 | Malate synthase | 10.7 | ND |
| YPO3681 | Putative insecticidal toxin | 10.6 | ND | |
| dctA | y3836 | C4-dicarboxylate transport protein | 10.5 | ND |
| y2591 | Hypothetical protein | 9.7 | ND |
Locus tags of Y. pestis genes from which the oligonucleotide probes are designed. Genes that appear twice on the list have two probes corresponding to them on the microarray. YPO designations are used for the probes that are based on CO92-specific sequences.
Predicted function is based on annotation by Pathogen Functional Genomics Resource Center/J. Craig Venter Institute.
+, bacterial replication in BMMs similar to that of wild-type bacteria; ++, increased intracellular replication; ND, not determined.
In summary, our data highlighted the hostile aspects of the YCVs, which are replete in oxidative and other stresses but limited in nutrient availability. The transcriptional response of intracellular Y. pestis in YCVs shares some common characteristics with that of other pathogens in their respective phagosomes. For example, high levels of induction of oxidative stress-response genes were found in intracellular F. tularensis (64), and the glyoxylate pathway is upregulated in many intracellular pathogens, including S. Typhi, M. tuberculosis, and the fungal pathogen Candida albicans (15, 36, 57). However, we also found a large number of hypothetical genes or genes of unknown function to be upregulated in intracellular Y. pestis, suggesting that they may have novel roles in intracellular replication.
Deletion of Y. pestis KIM6+ y2313-y2316-orfX putative operon results in increased intracellular proliferation and filamentous morphology.
As shown in Table 6, the list of the most highly upregulated genes in intracellular Y. pestis included many genes of unknown function, as well as known genes whose roles in the intracellular environment are undefined. To assess the roles of these genes in intracellular survival, deletions were created in selected genes from the list, and the resulting mutants were tested for viability and growth inside murine BMMs. The deletions were created in KIM6+, which does not carry the virulence plasmid pCD1 (Table 1) and which replicates well in macrophages. The deletions were introduced into KIM6+ because this strain can be reconstituted to full virulence (by reintroduction of pCD1), thereby facilitating future studies of pathogenesis in mouse infection models. Deletion of fadAB, psaABC, y0655, and y3553-y3555 in Y. pestis KIM6+ caused no obvious defect in intracellular growth compared to that of the parental strain (Table 6). However, deletion of the putative operon y2313-y2316 resulted in an unusual intracellular growth phenotype, as described below.
y2313, y2315, and y2316 are among the most highly upregulated genes in intracellular Y. pestis at all time points postinfection. They are located next to each other on the Y. pestis chromosome (Fig. 4 A) and most likely form an operon, on the basis of the findings of previous studies by Han et al., which showed that expression of these genes is coordinately regulated under different environmental conditions and that they are transcribed as a single transcript, determined on the basis of RT-PCR analysis (24). In addition to y2313-y2316, we identified a novel putative short ORF of 47 amino acids, which we called orfX, located 46 bp downstream of the stop codon of y2316 on the same DNA strand. (This sequence is listed as a predicted protein-encoding gene in a number of Y. pestis and Y. pseudotuberculosis genomic databases.) In a separate whole-genome microarray-based transcriptome analysis of Y. pseudotuberculosis, we found that the chromosomal region of this organism equivalent to y2313-y2316-orfX is highly transcribed under T3SS-inducing conditions (see Fig. S1A and B in the supplemental material; data not shown). The apparent coordinated expression and the physical proximity of orfX to y2316 suggest that orfX may also be part of the y2313-y2316 operon. Interestingly, this putative operon is highly conserved in all Y. pestis and Y. pseudotuberculosis strains but is not found in other Yersinia species.
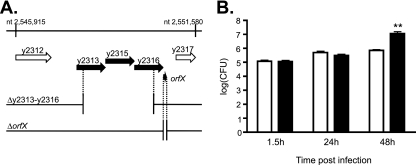
FIG. 4.
Characterization of the putative y2313-y2315-y2316-orfX operon. (A) ORF map of region from nucleotides (nt) 2545915 to 2551580 of the Y. pestis KIM chromosome (GenBank accession number AE009952.1). ORFs are shown as arrows. The coordinates of each gene are as follows: y2313, nucleotides 2547779 to 2548684; y2315, nucleotides 2548690 to 2549622; y2316, nucleotides 2549609 to 2550559; and orfX, nucleotides 2550605 to 2550559. The predicted 117-bp y2314 ORF (nucleotides 2548874 to 2548990) is on the opposite strand. The deletion strains used in this study are shown below the map, with black dotted lines indicating the endpoints of the deletions. (B) CFU assay. BMMs prepared from female C57BL/6 mice were infected with KIM6+ (white bar) or KIM6+ Δy2313-y2316 (black bar) at an MOI of 5 for 20 min and incubated in the presence of gentamicin as described in Materials and Methods. The infected cells were lysed at the indicated times, and serial dilutions of the lysates were plated to determine the number of intracellular bacteria by the CFU assay. The data represent the averages of three independent experiments. The asterisks indicate a significant difference compared to the result for KIM6+ (P < 0.01), as determined by Student's t test. The error bars indicate the standard errors of the mean (SEMs).
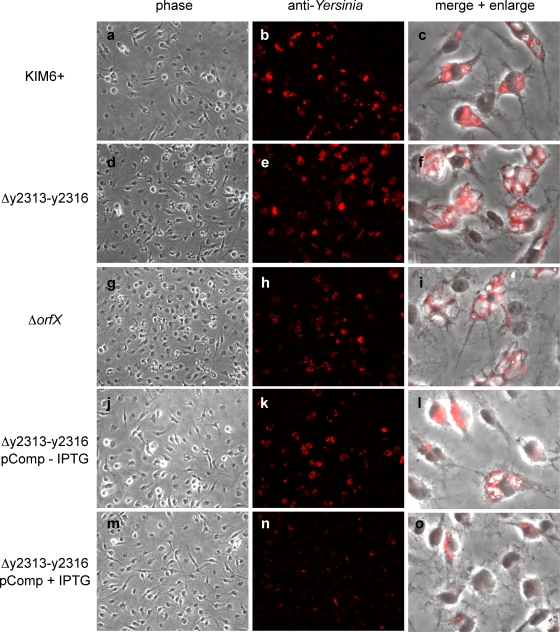
A deletion mutant lacking most of the y2313-y2316 predicted ORFs, as well as a mutant lacking most of the orfX ORF, were created in Y. pestis KIM6+ (Fig. 4A), and the intracellular replication of these mutants in BMMs was followed by a CFU-counting assay and fluorescent microscopy. We hypothesized that deletion of the genes that are normally induced under the intracellular conditions may result in decreased intracellular viability. Surprisingly, the deletion of both y2313-y2316 and orfX resulted in increased intracellular accumulation of the bacteria, with their numbers of CFU being increased by more than 10-fold compared to the numbers of CFU of parental strain KIM6+ at 48 h postinfection (Fig. 4B; see Fig. S2A in the supplemental material). In addition, the mutant bacteria exhibited an unusual filamentous morphology inside macrophages, suggesting a possible defect in intracellular bacterial cell division or septation (Fig. 5 a to i). The phagosomes containing the filamentous bacteria were enlarged into spacious vacuoles. The increased bacterial replication was less apparent by the CFU-counting assay at 24 h postinfection; however, stronger fluorescence was observed in the mutant-infected BMMs at this time point in immunofluorescent microscopic analysis (Fig. 5a to i). It is possible that the CFU assay underestimates the number of bacteria in the mutant-infected BMMs at earlier time points because individual bacteria are not completely separated. Both the Δy2313-y2316 and the ΔorfX mutants exhibited growth rates and morphologies similar to those of the parental wild-type strain during growth in various liquid culture media (data not shown); thus, the increased proliferation and filamentation phenotypes were specific to the intracellular environment.
FIG. 5.
Intracellular replication phenotype of KIM6+ Δy2313-y2316 and KIM6+ ΔorfX mutants. BMMs were infected with KIM6+ or the indicated KIM6+-derived strains (Table 1) at an MOI of 5 in the presence or absence of IPTG. At 24 h postinfection, the samples were fixed, stained with anti-Yersinia antibody, and examined by phase-contrast (a, d, g, j, m) or fluorescence (b, e, h, k, n) microscopy. The overlays of the phase-contrast and fluorescence images are magnified 4× (c, f, i, l, o).
To determine whether the observed phenotypes of the Δy2313-y2316 mutants are due to the loss of the y2313-y2316 region itself or the polar effect on expression of orfX and/or other downstream genes, a plasmid carrying all the y2313-y2316 ORFs under the control of a tac promoter was constructed. The introduction of the complementing plasmid into the mutant strain abolished the filamentous morphology and resulted in decreased replication, as determined by microscopy, while the introduction of the empty vector did not, indicating that loss of the y2313-y2316 region itself was the cause of these phenotypes (Fig. 5 and data not shown). The complementation was observed in both the absence and the presence of the IPTG-mediated induction, possibly due to the leakiness of the tac promoter (Fig. 5j to o). In the presence of 0.5 mM IPTG, the intracellular replication was severely inhibited. However, this amount of IPTG induction also slowed the growth of the complemented strain in HI broth (data not shown), suggesting that overexpression of these genes may be generally toxic to the bacteria. Similar complementation experiments conducted with orfX deletion mutants of both Y. pestis and Y. pseudotuberculosis (see Fig. S2A and B in the supplemental material) indicate that this novel short ORF plays a similar role as y2313-y2316 during intracellular growth of the bacteria. In addition, when a nonsense mutation was generated in the copy of the orfX gene carried on the plasmid used in the complementation assay, the resulting vector failed to complement the intracellular growth phenotypes of the orfX deletion, confirming the protein-coding nature of the predicted orfX gene (see Fig. S2B in the supplemental material).
Taken together these data indicate that Y. pestis genes y2313-y2316 and orfX, while they are not essential for intracellular survival, may play a role in intracellular growth, division, or exit of the pathogen from the macrophage cells.
Deletion of y2313-y2316-orfX causes decreased YopJ translocation.
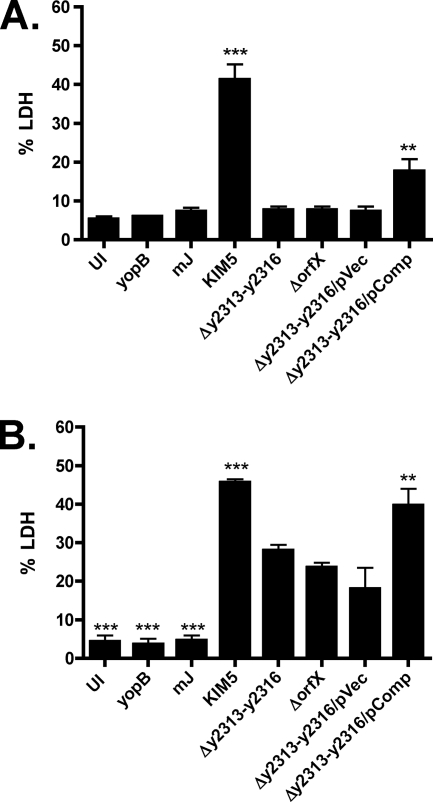
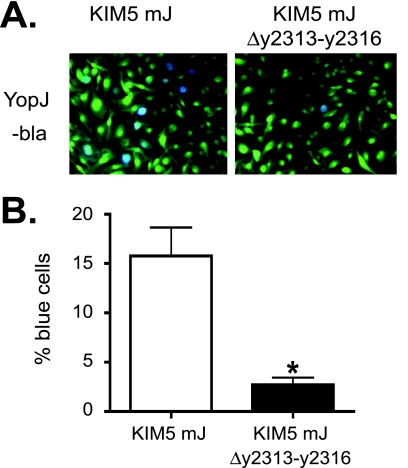
In Y. pestis, the T3SS encoded by the genes of virulence plasmid pCD1 allows the pathogen to inject the Yop effector proteins and disrupt cellular signaling of host cells, such as macrophages (63). In particular, YopJ blocks mitogen-activated protein kinases and NF-κB signaling pathways, leading to cell death (65). To assess if y2313-y2316 and orfX are required for the proper functioning of the T3SS, deletions in these genes were created in KIM5 (Table 1), and the ability of the mutants to induce cell death in BMMs was determined by the LDH release assay. BMMs infected with KIM5 Δy2313-y2316 or KIM5 ΔorfX mutants showed markedly decreased levels of YopJ-dependent cell death compared to the levels for those infected with the parental strain KIM5 when the macrophages were infected with bacteria grown at 28°C (Fig. 6 A). The defect was less severe when the bacteria were grown at 37°C in the presence of 2.5 mM calcium to prime T3SS expression prior to macrophage infection. However, even under this condition, YopJ-dependent cytotoxity was reduced significantly in both mutants (Fig. 6B). The ability to induce cell death was partially restored in the Δy2313-y2316 mutant by the introduction of the complementing plasmid (Fig. 6A and B). To test if the Δy2313-y2316 mutant is defective in translocating Yops into BMMs, we expressed YopJ β-lactamase (YopJ-bla) fusion proteins in either KIM5 mJ, which carries a mutant yopJ gene encoding a catalytically inactive YopJ, or KIM5 mJ Δy2313-y2316 and compared the efficiency of Yop injection by staining BMMs with the β-lactamase substrate CCF2-AM. Upon diffusion into mammalian cells, CCF2-AM gets cleaved into CCF and emits green fluorescence, but when Yop-bla fusion proteins are injected into these cells, CCF can be cleaved further, changing the fluorescent emission to blue (6). As expected, the percentage of blue cells in the BMMs infected with the Δy2313-y2316 mutant expressing YopJ-bla was significantly less than that of BMMs infected with the parental strain, indicating that the mutant bacteria translocated YopJ less efficiently (Fig. 7 A and B). Interestingly, Yops were secreted at similar levels in the mutant and the parental strains in an in vitro secretion assay performed under low-calcium condition (data not shown). These results suggest that loss of y2313-y2316 causes a specific defect in the translocation of Yops into macrophages.
FIG. 6.
KIM5 Δy2313-y2316 and KIM5 ΔorfX mutants show decreased YopJ-dependent cytotoxicity. BMMs were infected with Y. pestis grown overnight at 28°C (A) or subcultured for 2 h at 37°C in the presence of 2.5 mM calcium (B) at an MOI of 5. BMMs were left uninfected (UI) or were infected with KIM5 or the indicated KIM5-derived strains (Table 1). Supernatants collected after 24 h of infection were used to measure cell death by LDH release assay. The data represent the averages of three independent experiments. Error bars represent standard errors of the mean (SEMs). The asterisks indicate a significant difference compared to the results for KIM5 Δy2313-y2316 (**, P < 0.01; ***, P < 0.001), as determined by one-way ANOVA with Tukey's multiple comparison test.
FIG. 7.
Determination of Yop translocation using bla assay. BMMs were infected with Y. pestis strains (KIM5 mJ or KIM5 mJ Δy2313-y2316) expressing YopJ-bla (Table 1) at an MOI of 10. The CCF2-AM dye was added at 4 h postinfection. The cells were incubated for an additional hour, and images were taken using fluorescent microscopy. Bacteria were grown at 37°C for 2 h in the presence of 2.5 mM calcium prior to macrophage infection. (A) Representative pictures of the infected BMMs. Blue indicates the translocation of the Yop-bla fusion protein into the BMMs. (B) Percent blue cells. The results are the averages of three independent experiments, in which cells were counted in three independent fields. Error bars represent standard errors of the mean (SEMs). The asterisk indicates a significant difference compared to the result for the wild type (P < 0.05), as determined by Student's t test.
To assess the role of y2313-y2316 in the virulence of Y. pestis, we compared the virulence of KIM5 Δy2313-y2316 to that of KIM5 in the mouse intravenous infection model. There was a slight increase in the median survival time (wild type = 7 days, Δy2313-y2316 = 8 days) and in the number of surviving mice, but the increase was not statistically significant (data not shown). Therefore, the effect of deletion of these genes is minimal, if any, under this experimental condition. It remains to be seen whether deletion of these genes in the fully virulent strain has any effect in the subcutaneous or intranasal routes of infection. Interestingly, a transposon insertion mutant of y2316 was found previously in a signature-tagged mutagenesis (STM) screen for mutants that are defective in spleen colonization in the mouse subcutaneous infection model, using a fully virulent Y. pestis strain, Kimberley53 (18). This mutant was not attenuated in virulence when it was tested singly (18), but it is possible that the insertion caused only a partial loss of function of the operon and that the complete loss of function could result in the subtle defect in virulence, dissemination, or tissue colonization.
From the results combined, we conclude that the loss of function in y2313, y2315, y2316, and orfX results in subtle yet diverse changes in the behavior of Y. pestis, affecting both the intracellular replication and the translocation of some important virulence factors into its host cells.
The molecular mechanisms by which this putative operon controls such diverse cellular functions in Y. pestis are unknown at this moment. Each of its members, y2313, y2315, y2316, and orfX, is predicted to encode a protein of unknown function; and their predicted amino acid sequences reveal little information about their biochemical activity. y2313 is predicted to encode an armadillo (ARM)-like repeat-containing protein that bears no similarity to other known proteins. The predicted amino acid sequences of y2315 and y2316 are similar to each other (37% identity and 57% similarity), but the only other protein that shares significant similarity to y2315 and y2316 is a putative ISXo8 transposase from Xanthomonas oryzae (43% identity and 55% similarity over 117 amino acids) and other bacteria. orfX also encodes a novel protein. The y2315 protein product was designated an integral or peripheral membrane protein on the basis of fractionation analysis of Y. pestis proteins (47), but the subcellular localization of either y2313, y2316, or OrfX is unknown. More biochemical studies are needed to elucidate the function of each of the proteins encoded by this operon.
Filamentous growth of bacteria occurs when cell growth continues in the absence of septation. Filamentation has been observed in many bacteria, including Salmonella, Mycobacterium, as well as uropathogenic E. coli (UPEC), and can be due to a wide range of conditions, such as DNA damage followed by the SOS response, metabolic changes, and exposure to subinhibitory concentrations of antibiotics (31). In the case of Salmonella, filamentous growth can occur inside activated macrophages in response to the phagocyte oxidase activity and antimicrobial peptides (53, 54). Therefore, it is possible that the filamentation of the y2313-y2316 mutant is triggered by similar stress factors produced by the host cells. The y2313-y2316 operon is strongly induced in response to stressful conditions, including nutrient starvation and chloramphenicol treatment, while it is repressed in the ompR mutant (24, 52), consistent with the idea that it may play a role in dealing with stressful environments. Other studies of S. Typhimurium and E. coli showed that instability of the outer membrane could have an effect on FtsZ septation ring assembly and on cell division (19, 25). As stated above, the protein product of y2315 has been found in a membrane fraction in a proteomic analysis of Y. pestis proteins (47), raising the possibility that the proteins encoded by the y2313-y2316 operon could have a role in the localization and/or stability of other membrane proteins, such as FtsZ, and T3SS. This hypothesis is consistent with our observation that the loss of the operon also affects the translocation efficiency of YopJ. By fine-tuning the cell envelope characteristics, the y2313-y2316 operon products may be able to regulate, or perhaps coordinate, intracellular growth and Yop translocation, both of which are energy-costing processes. Of course, this hypothesis is highly speculative at this moment, and there are numerous other possibilities for the function of this operon. Additional studies of the subcellular localization of each gene product of the operon may help decipher the basis for the pleiotropic effects caused by its inactivation.
In conclusion, our study provides a comprehensive analysis of the transcriptome of Y. pestis during the intracellular growth stage of infection and uncovers a number of known pathways as well as novel factors that are important for various aspects of intracellular growth. Further analysis of these pathways and factors will bring us closer to having a full understanding of mechanisms of intracellular survival and infection of Yersinia pestis.
Supplementary Material
Acknowledgments
We thank Dieter Schifferli for providing the KIM6 psaABCΔ::aph mutant strain; Yue Zhang for creating Yop-bla fusion constructs; Adam Rosebrock, Bruce Futcher, Isabelle Hautefort, and Jay Hinton for technical advice on the microarray experiments; Hong Wang for carrying out cDNA synthesis and microarray hybridizations; Galina Romanov for excellent technical assistance in isolating BMMs; Patricio Mena and Maya Ivanov for carrying out mouse infection experiments; Sheila Firoosan for carrying out qRT-PCR experiments; Sireesha Teegala for the help in creating deletion mutants; and Raedeen Russell for performing in vitro Yop secretion assays. We are grateful to Yue Zhang and Joe McPhee for constructive comments on the manuscript and the members of the Bliska and Karzai laboratories for helpful suggestions on our project.
This research was supported by grant P01 AI055621 to A.W.K. and J.B.B. from the NIH. H.S.F. was supported in part by funding from the Northeast Biodefense Center (U54 AI057158-Lipkin).
Editor: S. M. Payne
Footnotes
Published ahead of print on 21 June 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergman, N. H., E. C. Anderson, E. E. Swenson, B. K. Janes, N. Fisher, M. M. Niemeyer, A. D. Miyoshi, and P. C. Hanna. 2007. Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect. Immun. 75:3434-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brubaker, R. R. 2005. Influence of Na(+), dicarboxylic amino acids, and pH in modulating the low-calcium response of Yersinia pestis. Infect. Immun. 73:4743-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumell, J. H., and S. Grinstein. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7:78-84. [DOI] [PubMed] [Google Scholar]
- 5.Cavanaugh, D. C., and R. Randall. 1959. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of flea-borne plague. J. Immunol. 83:348-363. [PubMed] [Google Scholar]
- 6.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 8.Cowan, C., A. V. Philipovskiy, C. R. Wulff-Strobel, Z. Ye, and S. C. Straley. 2005. Anti-LcrV antibody inhibits delivery of Yops by Yersinia pestis KIM5 by directly promoting phagocytosis. Infect. Immun. 73:6127-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, R., A. Dhokalia, X. Z. Huang, R. Hammamieh, N. Chakraborty, L. E. Lindler, and M. Jett. 2007. Study of proinflammatory responses induced by Yersinia pestis in human monocytes using cDNA arrays. Genes Immun. 8:308-319. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis, G., Jr., B. T. Sherman, D. A. Hosack, J. Yang, W. Gao, H. C. Lane, and R. A. Lempicki. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:P3. [PubMed] [Google Scholar]
- 12.Du, Y., R. Rosqvist, and A. Forsberg. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 70:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 14.Fang, F. C., S. J. Libby, M. E. Castor, and A. M. Fung. 2005. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect. Immun. 73:2547-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faucher, S. P., S. Porwollik, C. M. Dozois, M. McClelland, and F. Daigle. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. U. S. A. 103:1906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finegold, M. J. 1969. Pneumonic plague in monkeys. An electron microscopic study. Am. J. Pathol. 54:167-185. [PMC free article] [PubMed] [Google Scholar]
- 17.Flannagan, R. S., G. Cosio, and S. Grinstein. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7:355-366. [DOI] [PubMed] [Google Scholar]
- 18.Flashner, Y., E. Mamroud, A. Tidhar, R. Ber, M. Aftalion, D. Gur, S. Lazar, A. Zvi, T. Bino, N. Ariel, B. Velan, A. Shafferman, and S. Cohen. 2004. Generation of Yersinia pestis attenuated strains by signature-tagged mutagenesis in search of novel vaccine candidates. Infect. Immun. 72:908-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujishima, H., A. Nishimura, M. Wachi, H. Takagi, T. Hirasawa, H. Teraoka, K. Nishimori, T. Kawabata, K. Nishikawa, and K. Nagai. 2002. kdsA mutations affect FtsZ-ring formation in Escherichia coli K-12. Microbiology 148:103-112. [DOI] [PubMed] [Google Scholar]
- 20.Gendlina, I., K. G. Held, S. S. Bartra, B. M. Gallis, C. E. Doneanu, D. R. Goodlett, G. V. Plano, and C. M. Collins. 2007. Identification and type III-dependent secretion of the Yersinia pestis insecticidal-like proteins. Mol. Microbiol. 64:1214-1227. [DOI] [PubMed] [Google Scholar]
- 21.Gong, S., S. W. Bearden, V. A. Geoffroy, J. D. Fetherston, and R. D. Perry. 2001. Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect. Immun. 69:2829-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabenstein, J. P., H. S. Fukuto, L. E. Palmer, and J. B. Bliska. 2006. Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect. Immun. 74:3727-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabenstein, J. P., M. Marceau, C. Pujol, M. Simonet, and J. B. Bliska. 2004. The response regulator PhoP of Yersinia pseudotuberculosis is important for replication in macrophages and for virulence. Infect. Immun. 72:4973-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, Y., J. Qiu, Z. Guo, H. Gao, Y. Song, D. Zhou, and R. Yang. 2007. Comparative transcriptomics in Yersinia pestis: a global view of environmental modulation of gene expression. BMC Microbiol. 7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry, T., F. Garcia-Del Portillo, and J. P. Gorvel. 2005. Identification of Salmonella functions critical for bacterial cell division within eukaryotic cells. Mol. Microbiol. 56:252-267. [DOI] [PubMed] [Google Scholar]
- 26.Hitchen, P. G., J. L. Prior, P. C. Oyston, M. Panico, B. W. Wren, R. W. Titball, H. R. Morris, and A. Dell. 2002. Structural characterization of lipo-oligosaccharide (LOS) from Yersinia pestis: regulation of LOS structure by the PhoPQ system. Mol. Microbiol. 44:1637-1650. [DOI] [PubMed] [Google Scholar]
- 27.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 28.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 29.Huang, D. W., B. T. Sherman, and R. A. Lempicki. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44-57. [DOI] [PubMed] [Google Scholar]
- 30.Inglesby, T. V., D. T. Dennis, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, J. F. Koerner, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, M. Schoch-Spana, and K. Tonat. 2000. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA 283:2281-2290. [DOI] [PubMed] [Google Scholar]
- 31.Justice, S. S., D. A. Hunstad, L. Cegelski, and S. J. Hultgren. 2008. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6:162-168. [DOI] [PubMed] [Google Scholar]
- 32.Lathem, W. W., S. D. Crosby, V. L. Miller, and W. E. Goldman. 2005. Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 102:17786-17791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lilo, S., Y. Zheng, and J. B. Bliska. 2008. Caspase-1 activation in macrophages infected with Yersinia pestis KIM requires the type III secretion system effector YopJ. Infect. Immun. 76:3911-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindler, L. E., and B. D. Tall. 1993. Yersinia pestis pH 6 antigen forms fimbriae and is induced by intracellular association with macrophages. Mol. Microbiol. 8:311-324. [DOI] [PubMed] [Google Scholar]
- 35.Liu, F., H. Chen, E. M. Galvan, M. A. Lasaro, and D. M. Schifferli. 2006. Effects of Psa and F1 on the adhesive and invasive interactions of Yersinia pestis with human respiratory tract epithelial cells. Infect. Immun. 74:5636-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83-86. [DOI] [PubMed] [Google Scholar]
- 37.Lucchini, S., H. Liu, Q. Jin, J. C. Hinton, and J. Yu. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 73:88-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukaszewski, R. A., D. J. Kenny, R. Taylor, D. G. Rees, M. G. Hartley, and P. C. Oyston. 2005. Pathogenesis of Yersinia pestis infection in BALB/c mice: effects on host macrophages and neutrophils. Infect. Immun. 73:7142-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 40.Motin, V. L., A. M. Georgescu, J. P. Fitch, P. P. Gu, D. O. Nelson, S. L. Mabery, J. B. Garnham, B. A. Sokhansanj, L. L. Ott, M. A. Coleman, J. M. Elliott, L. M. Kegelmeyer, A. J. Wyrobek, T. R. Slezak, R. R. Brubaker, and E. Garcia. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munoz-Elias, E. J., and J. D. McKinney. 2006. Carbon metabolism of intracellular bacteria. Cell. Microbiol. 8:10-22. [DOI] [PubMed] [Google Scholar]
- 42.Oyston, P. C., N. Dorrell, K. Williams, S. R. Li, M. Green, R. W. Titball, and B. W. Wren. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer, L. E., S. Hobbie, J. E. Galan, and J. B. Bliska. 1998. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol. Microbiol. 27:953-965. [DOI] [PubMed] [Google Scholar]
- 44.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry, R. D., I. Mier, Jr., and J. D. Fetherston. 2007. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. Biometals 20:699-703. [DOI] [PubMed] [Google Scholar]
- 46.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pieper, R., S. T. Huang, D. J. Clark, J. M. Robinson, H. Alami, P. P. Parmar, M. J. Suh, S. Kuntumalla, C. L. Bunai, R. D. Perry, R. D. Fleischmann, and S. N. Peterson. 2009. Integral and peripheral association of proteins and protein complexes with Yersinia pestis inner and outer membranes. Proteome Sci. 7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pujol, C., and J. B. Bliska. 2003. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect. Immun. 71:5892-5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pujol, C., and J. B. Bliska. 2005. Turning Yersinia pathogenesis outside in: subversion of macrophage function by intracellular yersiniae. Clin. Immunol. 114:216-226. [DOI] [PubMed] [Google Scholar]
- 50.Pujol, C., J. P. Grabenstein, R. D. Perry, and J. B. Bliska. 2005. Replication of Yersinia pestis in interferon gamma-activated macrophages requires ripA, a gene encoded in the pigmentation locus. Proc. Natl. Acad. Sci. U. S. A. 102:12909-12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pujol, C., K. A. Klein, G. A. Romanov, L. E. Palmer, C. Cirota, Z. Zhao, and J. B. Bliska. 2009. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect. Immun. 77:2251-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu, J., D. Zhou, L. Qin, Y. Han, X. Wang, Z. Du, Y. Song, and R. Yang. 2006. Microarray expression profiling of Yersinia pestis in response to chloramphenicol. FEMS Microbiol. Lett. 263:26-31. [DOI] [PubMed] [Google Scholar]
- 53.Rosenberger, C. M., and B. B. Finlay. 2002. Macrophages inhibit Salmonella typhimurium replication through MEK/ERK kinase and phagocyte NADPH oxidase activities. J. Biol. Chem. 277:18753-18762. [DOI] [PubMed] [Google Scholar]
- 54.Rosenberger, C. M., R. L. Gallo, and B. B. Finlay. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc. Natl. Acad. Sci. U. S. A. 101:2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosqvist, R., A. Forsberg, M. Rimpilainen, T. Bergman, and H. Wolf-Watz. 1990. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol. Microbiol. 4:657-667. [DOI] [PubMed] [Google Scholar]
- 56.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 57.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sebbane, F., C. O. Jarrett, J. R. Linkenhoker, and B. J. Hinnebusch. 2004. Evaluation of the role of constitutive isocitrate lyase activity in Yersinia pestis infection of the flea vector and mammalian host. Infect. Immun. 72:7334-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sebbane, F., N. Lemaitre, D. E. Sturdevant, R. Rebeil, K. Virtaneva, S. F. Porcella, and B. J. Hinnebusch. 2006. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. U. S. A. 103:11766-11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Straley, S. C., and P. A. Harmon. 1984. Growth in mouse peritoneal macrophages of Yersinia pestis lacking established virulence determinants. Infect. Immun. 45:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Straley, S. C., and P. A. Harmon. 1984. Yersinia pestis grows within phagolysosomes in mouse peritoneal macrophages. Infect. Immun. 45:655-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vadyvaloo, V., C. Jarrett, D. E. Sturdevant, F. Sebbane, and B. J. Hinnebusch. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog. 6:e1000783. [DOI] [PMC free article] [PubMed]
- 63.Viboud, G. I., and J. B. Bliska. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59:69-89. [DOI] [PubMed] [Google Scholar]
- 64.Wehrly, T. D., A. Chong, K. Virtaneva, D. E. Sturdevant, R. Child, J. A. Edwards, D. Brouwer, V. Nair, E. R. Fischer, L. Wicke, A. J. Curda, J. J. Kupko III, C. Martens, D. D. Crane, C. M. Bosio, S. F. Porcella, and J. Celli. 2009. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11:1128-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., and J. B. Bliska. 2005. Role of macrophage apoptosis in the pathogenesis of Yersinia. Curr. Top. Microbiol. Immunol. 289:151-173. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, D., Y. Han, J. Qiu, L. Qin, Z. Guo, X. Wang, Y. Song, Y. Tan, Z. Du, and R. Yang. 2006. Genome-wide transcriptional response of Yersinia pestis to stressful conditions simulating phagolysosomal environments. Microbes Infect. 8:2669-2678. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, D., L. Qin, Y. Han, J. Qiu, Z. Chen, B. Li, Y. Song, J. Wang, Z. Guo, J. Zhai, Z. Du, X. Wang, and R. Yang. 2006. Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol. Lett. 258:9-17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.