Abstract
Shewanella oneidensis is able to conserve energy for growth by reducing a wide variety of terminal electron acceptors during anaerobic respiration, including several environmentally hazardous pollutants. This bacterium employs various electron transfer mechanisms for anaerobic respiration, including cell-bound reductases and secreted redox mediators. The aim of this study was to develop rapid tools for profiling the key metabolic changes associated with these different growth regimes and physiological responses. Initial experiments focused on comparing cells grown under aerobic and anaerobic conditions. Fourier transform infrared (FT-IR) spectroscopy with cluster analysis showed that there were significant changes in the metabolic fingerprints of the cells grown under these two culture conditions. FT-IR spectroscopy clearly differentiated cells of S. oneidensis MR-1 cultured at various growth points and cells grown using different electron acceptors, resulting in different phenotypic trajectories in the cluster analysis. This growth-related trajectory analysis is applied successfully for the first time, here with FT-IR spectroscopy, to investigate the phenotypic changes in contrasting S. oneidensis cells. High-performance liquid chromatography (HPLC) was also used to quantify the concentrations of flavin compounds, which have been identified recently as extracellular redox mediators released by a range of Shewanella species. The partial least-squares regression (PLSR) multivariate statistical technique was combined with FT-IR spectroscopy to predict the concentrations of the flavins secreted by cells of S. oneidensis MR-1, suggesting that this combination could be used as a rapid alternative to conventional chromatographic methods for analysis of flavins in cell cultures. Furthermore, coupling of the FT-IR spectroscopy and HPLC techniques appears to offer a potentially useful tool for rapid characterization of the Shewanella cell metabolome in various process environments.
Shewanella oneidensis, a Gram-negative dissimilatory metal-reducing bacterium, is able to conserve energy for growth by reducing a variety of terminal electron acceptors during aerobic and anaerobic respiration (24), including several environmentally hazardous pollutants (1). The terminal electron acceptors used during anaerobic respiration vary, ranging from nitrate, fumarate, trimethylamine N-oxide (TMAO), dimethyl sulfoxide (DMSO), and sulfur compounds to fuel cell anodes and various metals and metalloids, including insoluble metal oxides (5, 33, 34).
There has been intense interest in the versatile metabolism of this bacterium and its potential to respire and bioremediate toxic environmental chemicals and metals, such as U(VI) and Cr(VI) (25, 43), under anoxic conditions. Riboflavin (RF), flavin mononucleotide (FMN), and flavin-adenine dinucleotide (FAD) were identified as the dominant electron shuttles secreted by a diversity of Shewanella cells and shown to mediate extracellular reduction of insoluble Fe(III) minerals and organic molecules, including azo dyes (23, 40). Field and Brady tested riboflavin and found that its presence during anaerobic reduction of azo dyes improves the overall kinetics of the reduction process (9). von Canstein et al. recently found that Shewanella cells are able to secrete flavins, FAD, FMN, and riboflavin as extracellular redox mediators and quantified these using high-performance liquid chromatography-mass spectrometry (HPLC-MS) (44). The experiments showed that the production profiles of these three chemicals were different under anaerobic and aerobic regimes, and this can be used to distinguish the two metabolic pathways with oxygen or fumarate as electron acceptors.
Although we are gaining a deeper understanding of the genetic and biochemical basis of the diverse respiration pathways of this organism, supported by the recent availability of the complete genome sequence (14), complementary metabolomic approaches have not been used to identify or quantify the metabolic changes expected with major physiological shifts in this organism under contrasting growth regimes. Fourier transform infrared (FT-IR) spectroscopy was chosen for this study, as this method offers the advantage of minimal sample preparation, as well as being rapid, nondestructive, readily automatable, relatively inexpensive, and quantitative, compared with other metabolic profiling techniques (8, 12, 15, 35). The infrared absorbance spectra generated by FT-IR spectroscopy have been used to identify specific biochemical features and also provide a global biochemical “fingerprint” for mixed, complex samples (11, 27) in many research areas (10, 11). For instance, FT-IR spectroscopy has proved sensitive enough for analysis of the chemical composition of a single strain of Escherichia coli after exposure to ionic liquids (6), for functional genomics screening (22), for measuring abiotic perturbations in algae (39), and for characterization of microbial degradation pathways (16). We believe that this technique offers considerable potential for rapid differentiation of metabolic changes of bacteria responding to contrasting growth regimes in natural and engineered environments, although this remains to be demonstrated.
The FT-IR spectra generated from microorganisms have very complex profiles that can be related back to biochemical components which one would expect to be detected in the samples. While some subtle quantitative differences can be observed between the spectra, there are few, if any, qualitative differences to be seen, and so it is almost impossible to interpret these data with the naked eye (19, 20, 45). Therefore, multivariate statistical techniques are needed to model the relationship between the phenotypic changes occurring in this organism during growth with the observations recorded by FT-IR spectroscopy.
In the present study, we investigated the ability of FT-IR spectroscopy to analyze the S. oneidensis MR-1 cell metabolome and distinguish significant metabolic changes associated with anaerobic and aerobic growth conditions. Analyses were conducted directly on the cells for the endometabolome in assessing the metabolic fingerprint. Both unsupervised and supervised learning methods (viz., principal component analysis [PCA] [21] and discriminant function analysis [DFA] [26]) were used to identify any differences between the FT-IR spectra from cells grown under the two regimes, and the contributions of a range of biomolecules were elucidated. In addition to metabolic fingerprinting of whole cells by FT-IR, key components of the extracellular metabolome (the so-called exometabolome, or metabolic footprint) were also quantified by HPLC analysis. Here, riboflavin, FMN, and FAD in culture supernatants were quantified, and partial least-squares regression (PLSR) was used to identify correlations between the cell's FT-IR spectra and flavin concentrations assessed using HPLC data. These studies show for the first time that FT-IR analyses can be used for rapid identification of metabolic shifts in both the intracellular and the extracellular metabolomes of S. oneidensis MR-1 cultures.
MATERIALS AND METHODS
Bacterial strains and medium.
S. oneidensis strain MR-1 was obtained from the Manchester University Geomicrobiology group culture collection and stored at −80°C in 50% glycerol prior to use. Cells were incubated at 30°C in LB agar and then kept at room temperature. Starter cultures were grown in tryptic soy broth (TSB; 30 g/liter and 50 ml in a 250-ml Erlenmeyer conical flask). The TSB aerobic culture was incubated overnight at 30°C while being shaken at 150 rpm (C24KC refrigerated incubator shaker; New Brunswick Scientific). All the experiments were conducted using a fully defined minimal medium, based on the method of Myers and Nealson (34). Amino acid solution (20 mg/liter l-arginine hydrochloride, 20 mg/liter l-glutamate, 20 mg/liter l-serine) was added. Sodium dl-lactate (100 mM) was used as the carbon source and the electron donor, and fumarate (20 mM) or oxygen was used as the electron acceptor. The pH of the medium was adjusted to 7.6 before sterilization. All chemicals were purchased from Sigma unless noted otherwise.
Aerobic bath culture studies.
Inocula were grown overnight in TSB liquid medium to late log phase at 30°C. A 1-ml suspension was transferred into 250-ml flasks containing 50 ml of sterile minimal medium and grown overnight at 25°C with shaking at 130 rpm. Cultures (three biological replicates) were incubated at 20°C with shaking at 130 rpm until the biomass density (optical density at 600 nm [OD600]) indicated that the cultures had entered late exponential phase (∼18 h). For growth measurements, new cultures were started by transferring 5 ml of late-exponential-phase culture to a 250-ml Erlenmeyer flask containing 50 ml of minimal medium and incubated at 130 rpm at 25°C. Subsamples were periodically removed and analyzed for turbidity (OD600). In addition, 1-ml samples were centrifuged, and cell pellets were stored at −20°C for subsequent analysis of total cellular protein, while the supernatant was also frozen for HPLC analysis of extracellular flavins. Biomass samples for FT-IR and HPLC analysis of intracellular flavins were also prepared by centrifugation and washed with sterile 0.9% NaCl prior to storage at −80°C.
Anaerobic batch culture studies.
Experiments were conducted in a Coy anaerobic chamber under an atmosphere of 5% hydrogen-95% nitrogen (vol/vol), unless otherwise noted. Anaerobic cultures were grown in 80 ml of defined minimal medium in 100-ml serum bottles (flushed with N2 for 10 min and sealed with a butyl rubber stopper before use; Bello Glass, Inc.). Inocula for these experiments were grown overnight anaerobically at 30°C to early exponential phase. A 10-ml aliquot of the “starter” culture was used to inoculate three biological replicate “experimental” cultures, which were incubated without agitation at 30°C. Samples were withdrawn regularly for OD600 analysis. In addition, samples (1 ml) were centrifuged at 13,000 × g in a microcentrifuge and cell pellets stored at −20°C for subsequent total cellular protein analysis. The samples for FT-IR and HPLC analysis were prepared as described for the aerobic cultures.
Cell metabolism analysis by FT-IR spectroscopy.
The metabolic profiles of cells grown aerobically and anaerobically were recorded by FT-IR spectroscopy periodically. Samples (5 ml) were collected and centrifuged at 4°C at 12,000 × g for 10 min, the supernatant was removed, and the cell pellet was washed twice with sterile 0.9% NaCl solution and centrifuged again prior to storage at −80°C. After all the samples had been collected, they were defrosted and resuspended in sterile 0.9% NaCl solution to 0.8 g/liter biomass (dry weight).
All FT-IR spectroscopy analysis was conducted using an Equinox 55 infrared spectrometer equipped with a high-throughput motorized microplate module, HTS-XT (Bruker Optics, Coventry, United Kingdom). The motorized module of this instrument introduces the plate into the airtight optics of the instrument, in which tubes of desiccant are contained to provide a moisture-free environment (13). A deuterated triglycine sulfate (DTGS) detector was employed for transmission measurements of the samples to be acquired. A 96-well Si sample plate was washed thoroughly with 2-propanol and deionized water and allowed to dry at room temperature prior to use. Twenty microliters (0.016 g biomass) of each bacterial sample was evenly applied in triplicate onto the plate (so-called technical replicates). The plate was consequently dried at 60°C in an oven for 10 min and loaded into the motorized module for FT-IR spectroscopy. Spectra were collected over the wavelength range of 4,000 to 600 cm−1 under the control of a computer programmed with Opus 4, operated under Microsoft Windows 2000. Spectra were acquired at a resolution of 4 cm−1, and 64 spectra were coadded and averaged to improve the signal-to-noise ratio. The collection time for each spectrum was approximately 1 min.
Quantification of flavins by HPLC.
Cultures for the analyses of extracellular flavins were grown in minimal medium as described above. Reverse-phase high-performance liquid chromatography (HPLC) was used to separate and quantify the flavins. A Gemini 5μ C18 110A column (dimensions, 250 by 10.0 mm; Phenomenex, United Kingdom) was used, fitted to a GP50 gradient pump and a UVD170U UV-visible-light (UV-vis) detector, both from Dionex, United Kingdom. Samples (1 ml) were harvested by centrifugation at 13,000 × g for 20 min and the supernatants analyzed for extracellular flavins. To analyze intracellular flavins, the cell pellets were washed in HEPES buffer (100 mM, pH 7.4) twice and split into aliquots of 1.0 ml, pelleted again at 12,000 × g for 3 min, resuspended in 490 μl NaOH (0.1 M), mixed thoroughly for 10 s, and then acidified by the addition of 510 μl HCl (0.1 M) to stabilize the flavins. All the samples were analyzed by HPLC using a nonlinear gradient of methanol versus an aqueous solution of 20 mM NH4 acetate, pH 6.4. An initial isocratic step at 5% methanol for 6 min was followed by an increase to 34.5% methanol (by 12 min) and then to 37% methanol (at 28 min) and 95% methanol (maintained between 33 and 38 min), followed by a rapid drop to 5% methanol (by 39 min), which was maintained for a further 16 min. Flavins were detected at 280 nm. The column was calibrated using standard FAD, FMN, and riboflavin, and the retention times were found to be 13.7, 20.7, and 30.6 min, respectively.
FAD, FMN, and riboflavin (Sigma, United Kingdom) standard solutions of 0.01, 0.1, 1.0, 10.0, and 100 μM were separated using HPLC, and the peak area was calculated using the software program Chromeleon (version 6.50; Dionex). The concentrations of unknown samples were calculated by comparison with a graph prepared from the standards.
Data processing.
The ASCII data were imported into Matlab version 6 (The MathWorks). The FT-IR spectra were normalized such that the lowest recorded absorbance was set to 0 and the highest was set to 1 for each spectrum. Baseline correction, extended multiplicative scatter correction (EMSC) (29), and row normalization preprocessing steps were applied before exportation of the data to PyChem v3.0.3 for further analysis (18).
Discriminant analysis.
An unsupervised data reduction method, principal component analysis (PCA), was employed to reduce the dimensionality of the FT-IR data from 1,764 absorbance measurements at different wavelengths to 10 principal components (PCs), representing 99% of the total variance in the FT-IR data. These PCs were used as inputs for discriminant function analysis (DFA), which is a supervised technique that discriminates groups by use of a priori knowledge of class membership. The algorithm works to maximize between-group variance and minimize within-group variance. In this study, knowledge of the sampling time was passed to the algorithm, and all models were cross-validated by 3-fold cross-validation, using each biological repeat measurement for model testing in each fold (37). The a priori class structure used by the DFA algorithm provided information on the biological sample membership (that is to say, 17 groups), and this allowed any similarities between the different biological samples to be modeled. Thus, in this process, as we conducted the experiment in triplicate (3 biological replicates, each analyzed 3 times) during calibration (or training) of the chemometric models, two of these biological replicates (with all technical replicates, i.e., 6 spectra in total) were used for training and the third biological replicate (containing all 3 of its technical replicates) was used to test whether the model was valid. On the basis of this, we rotated the test data 3 times, and this allowed any similarities between the different biological samples to be modeled.
Calculation of peak areas.
A range of peak areas from identifiable FT-IR bands were integrated in MATLAB version 6, using a function written in-house, whereby the extent of each peak was identified manually, a linear trend subtracted from the base of the peak to remove baseline variation, and the total intensity within this region summed. Quantitative information on cellular proteins was computed from amide I and II peak integrals from absorbances at 1,630 to 1,654 cm−1 and 1,530 to 1,550 cm−1, respectively. Lipid content was estimated using the integrals of absorbance from 1,370 to 1,400 cm−1 (COO− stretch and CH3 symmetric bending in protein and lipids), the area of the CH2 asymmetric/CH3 asymmetric stretch (2,885 to 2,945 cm−1), and aliphatic CH vibrations between 2,948 and 2,984 cm−1. Phosphate-related vibrations, including asymmetric PO2− (1,270 to 1,200 cm−1) and symmetric PO2− (1,135 to 1,000 cm−1), were also calculated. Finally, information on the levels of RNA and DNA in the cells were estimated from the areas between 1,710 cm−1 to 1,716 cm−1. The ratios of these values were calculated and plotted.
Partial least-squares regression.
The supervised multivariate linear regression approach of partial least-squares regression (PLSR), as detailed in references 3, 17, and 28, was used to model the relationship between the concentrations of flavins on the basis of HPLC analysis and FT-IR spectra. PLSR was calibrated with FT-IR data from the bacterial fingerprints to predict the known flavin concentrations measured by HPLC. As with PC-DFA, models were cross-validated by 3-fold cross validation. For each model, the optimal number of PLSR factors was determined with reference to the minimum of the root mean squared (RMS) error for the cross-validation samples. The RMS errors for calibration and cross-validation predictions, as well as the equations for the least-squares method, show strong linear correlations. The root mean squared error of prediction (RMSEP) was also used to compare the performances of different models.
RESULTS AND DISCUSSION
Profiles of growth under aerobic and anaerobic conditions.
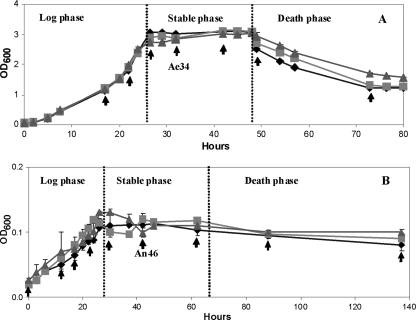
Prior to metabolomic analyses, the growth kinetics and biomass yields of S. oneidensis MR-1 under aerobic and anaerobic conditions were compared (Fig. 1). A fully defined minimal medium containing lactate and fumarate was used, with fumarate as the sole electron acceptor under anaerobic conditions. The lag phases of the two contrasting growth regimes were markedly different, lasting 4 h in the aerobic cultures and 8 h in the anaerobic cultures, while the doubling times were approximately 5 and 7.5 h, respectively. The final biomass yields were also very informative of the growth condition; an OD600 of 3.12 was reached at 26 h in the aerobic culture and 0.15 at 50 h in the anaerobic culture. This organism has fairly low growth rates in the above-defined regimes, which makes studying the phenotypic variation of the cells via their metabolic fingerprints readily achievable.
FIG. 1.
Profile of S. oneidensis MR-1 growth under aerobic (A) and anaerobic (B) conditions. Each point shows the biological repeats from 3 measurements, and the error bars indicate standard deviations. The arrows represent samples collected for metabolic analysis. The coding is such that Ae34 is from cells grown aerobically for 34 h and An46 from cells grown anaerobically for 46 h.
FT-IR metabolic fingerprinting.
To make broad comparisons of the metabolic states of S. oneidensis MR-1 grown under aerobic and anaerobic conditions, 17 samples (with 3 biological repeats for each) were collected at various points in the growth cycle, including the log, exponential, stationary, and death phases, and analyzed by FT-IR spectroscopy. In Fig. 2, examples of the FT-IR absorbance spectra are shown, and these were typical of bacterial FT-IR spectra reported previously (7, 36). The biological characteristic bands, which included the acyl vibration bands from fatty acids and lipids (C=O and N—H from amides I and II as well as the P=O nucleic acid vibration and phospholipids), were readily observed in these spectra (Fig. 2) and demonstrate that from the cells a whole organism fingerprint has been generated (11).
FIG. 2.
FT-IR spectra of cells of S. oneidensis MR-1 collected at different time intervals from aerobic and anaerobic cultures. The bars at the top of the figure relate to the following biochemical regions: FA, fatty acids; A1, amide I; A2, amide II; A3, amide III; MP, carboxylic groups of proteins and fatty acids (phosphate rich); PS, polysaccharides; L, RNA and DNA. The six spectra (from bottom to top) are as follows: Ae12, aerobic conditions in exponential phase (12 h); Ae34, aerobic conditions in stationary phase (34 h); Ae50, aerobic conditions in death phase (50 h); An17, anaerobic conditions in exponential phase (17 h); An48, anaerobic conditions in stationary phase (48 h); An72, anaerobic conditions in death phase (72 h). These spectra are offset so that the spectral features can be clearly seen.
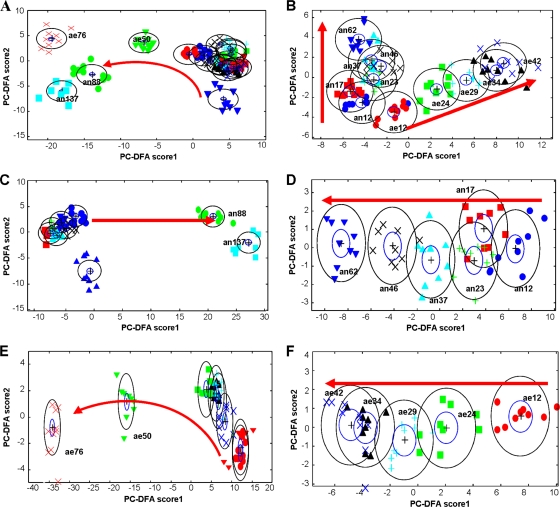
Visible inspection of these FT-IR data is difficult, as all spectra are qualitatively very similar. Therefore, PC-DFA models based upon the FT-IR data were used to observe the overall relationships between S. oneidensis MR-1 samples. The cross-validation data were all correctly classified with the training samples, falling within the 95% confidence intervals (data not shown). Plotting the first two PC-DFA scores recovered from all of the samples (Fig. 3A) showed clear separation of the growing cells from those in the death phase, rather than revealing the effect of the electron acceptors introduced to the growth medium. This is hardly surprising, given that the cells' fingerprints are totally different when the bacteria are dying, and this is irrespective of the medium. Thus, these “outlying” spectra need to be removed from the cluster analysis.
FIG. 3.
PC-DFA plots for FT-IR spectra. (A) Combined aerobic and anaerobic samples; (B) early-exponential- and stationary-phase samples grown aerobically and anaerobically; (C) combined anaerobic samples; (D) early-exponential- and stationary-phase samples grown anaerobically; (E) combined aerobic samples; (F) early-exponential- and stationary-phase samples grown aerobically. The red arrows represent an increase in incubation time. The number after the label stands for the incubation time; labels beginning with “ae” are for aerobic samples, and those beginning with “an” are for anaerobic samples. For example, an12 represents cells grown in anaerobic medium for 12 h, and ae24 represents cells grown in aerobic medium for 24 h. Dark blue ▴, an0; dark blue •, an12; red ▪, an17; green +, an23; light blue ▴, an37; black ×, an46; dark blue ▾, an62; green •, an88; light blue ▪, an137; red ▾, ae0; red •, ae12; green ▪, ae24; blue +, ae29; black ▴, ae34; dark blue ×, ae42; green ▾, ae50; red ×, ae76. PCs 1 to 20 (accounting for 99.5% of the total explained variance) were used by the DFA algorithm with a priori knowledge of machine replicates (i.e., 1 class per sample point, giving 17 classes in total). The different symbols represent the different sample points. The circles represent the 95% χ2 confidence interval about the mean.
After removing these “death phase” samples, the remaining aerobic and anaerobic samples were reanalyzed by PC-DFA (Fig. 3B). It is clear from this plot that the first PC-DF score represents the difference between the anaerobic and aerobic cultures, while PC-DF2 accounts for growth time. This proved that the highest variation among the samples is dependent on the electron acceptors and that the second-highest variation is caused by growth time. The plots showed clearly that the anaerobically and aerobically grown cells fall into two groups. All the anaerobic samples were located in the negative area of PC-DF1, while the aerobic samples were clustered in the positive area. This indicates that FT-IR spectroscopy is able to detect the metabolic changes in bacterial cells associated with the delivery of different electron acceptors (oxygen in aerobic medium and fumarate in anaerobic medium).
To reveal the metabolic changes of the Shewanella cells as a function of growth time, the FT-IR spectra of anaerobic (Fig. 3C and D) and aerobic (Fig. 3E and F) samples were analyzed separately. Coincident with the above observation, the largest variation was on the lag and dying phases for both cultures (Fig. 3C and E). Therefore, reanalysis of the cells harvested during early log phase and stationary phase was conducted (Fig. 3D and F). Figure 3D shows the PC-DF1 scores versus the PC-DF2 scores calculated from the FT-IR spectra of MR-1 cells cultivated after growing for 12, 17, 23, 37, 46, and 62 h under anaerobic conditions with fumarate as the electron acceptor. These samples were located in the early exponential and stationary phases during anaerobic growth. The cluster plot shows that there was a clear trend in the plots among the samples that correlates with growth time (as indicated by the arrow). The same trend was also seen for aerobically grown MR-1 cells (Fig. 3F). Thus, it would seem that FT-IR is sensitive enough to detect the metabolic changes in whole cells associated with growth time.
The observed growth-related trends (or trajectories) can be interpreted in terms of the underlying chemical changes occurring under these different conditions, which in turn can be linked to specific biomolecules, including proteins, lipids, carbohydrates, fatty acids, phospholipids, and nucleic acids (4). Changes in these specific compounds will reflect important biochemical shifts in S. oneidensis MR-1 in response to changes in metabolism (38). To reveal this biochemical information, the PC-DFA loading matrices need to be investigated further.
Interpretation of the FT-IR spectra.
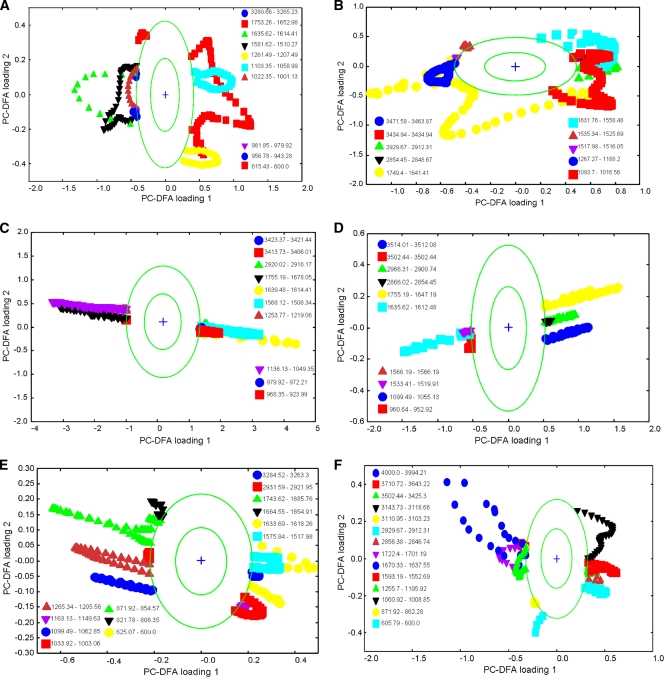
Decomposition of the spectral data during the application of PC-DFA yields a matrix of spectral loadings, which gives an indication of the relative importance of the spectral variables for derivation of each latent variable (26). The spectral loadings for the first two PC-DFA variables, which account for the majority of the variance in the data, are plotted against one another in Fig. 4. The bands recovered by PC-DFA loading were assigned to specific molecular vibrations, and these results are detailed in Table 1, along with their biological relevance.
FIG. 4.
PC-DFA plots for FT-IR spectra of S. oneidensis MR-1 cells. PCs 1 to 20 (accounting for 99.5% of the total explained variance) were used by the DFA algorithm with a priori knowledge of machine replicates (i.e., 1 class per sample point, giving 17 classes in total). The different symbols represent the different sample points. The circles represent a boundary of 2 standard deviations from the loading centroid, and this is used to give a nonstatistical approximation of a 95% confidence interval (this aid to visualization highlights bands which are changing the most). The order of these plots is the same as that in Fig. 3: (A) combined aerobic and anaerobic samples; (B) early-exponential- and stationary-phase samples grown aerobically and anaerobically; (C) combined anaerobic samples; (D) early-exponential- and stationary-phase samples grown anaerobically; (E) combined aerobic samples; (F) early-exponential- and stationary-phase samples grown aerobically.
TABLE 1.
Tentative assignment of biochemical bands identified by FT-IR spectroscopy
| Biomolecule group | Wavelength (cm−1) | Assignment(s)a |
|---|---|---|
| Protein | 3,200 | Amide A ν(NH) |
| 3,129 | Amide B ν(NH) | |
| 3,070 | Amide B CNH | |
| 1,667 | Amide I ν(CO) coupled to δ(NH2) in-plane | |
| 1,655 | Amide I (α helix) | |
| 1,637 | Amide I (β structure) | |
| 1,550 | Amide II α helix | |
| 1,530 | Amide II, β structure | |
| 1,330 | Amide III | |
| 1,267 | Amide III (α helix) | |
| 1,250 | Amide III (β sheet) | |
| Phospholipids and lipid proteins | 2,960 | ν(CH3) membrane phospholipids |
| 2,920 | ν(CH2) membrane phospholipids | |
| 2,875 | νs(CH3) | |
| 2,850 | νs(CH2) membrane phospholipids | |
| 1,740 | ν(CO) esters | |
| 1,467 | δ(CH2) lipids and proteins | |
| 1,455 | δ(CH3) and δ(CH2) scissoring lipids and proteins | |
| 1,420 | νa(CH3) lipids and aromatics | |
| 1,397 | δ(CH3) methyl bond | |
| 1,370-1,400 | δ(COO−) and δ(CH3) lipids and proteins | |
| Phosphate and carbohydrate | 1,235 | νa(PO32−) |
| 1,170 | νa(COOC) phospholipids and/or cholesterol esters | |
| 1,150 | ν(CO) saccharides | |
| 1,054 | νs(COOC) | |
| 970 | ν(CC), ν(CN), and ν(PO32−) | |
| 935 | CC residue α helix | |
| 906 | CC stretch proline | |
| RNA and DNA | 1,710 | ν(CO) RNA (esters) |
| 1,690 | ν(CO), RNA, and DNA | |
| 1,106 | νs(PO2−) nucleic acids | |
| 1,063 | ν(CO), deoxyribose/ribose DNA, and RNA | |
| Finger print | 600-900 |
The key bands contributing to PC-DFA separation in Fig. 3A were identified by examining the PC-DFA loading plots (Fig. 4A). The first PC-DF loadings showed that the separation between living and dying bacteria is characterized mainly by the regions comprising 1,639 to 1,612 cm−1 and 1,581 to 1,508 cm−1, which represent proteinaceous contributions from the amide I and II vibrations, respectively. In addition, the region comprising 950 to 941 cm−1, which corresponds to carboxylic acids, was also changed during the death phase. This indicates in particular that the protein content of cells was reduced during the death phase.
The PC-DFA loadings from Fig. 4B (corresponding to the PC-DF score plot in Fig. 3B) indicate that the largest differences among anaerobic and aerobic early-exponential- and stationary-phase samples corresponded to changes in the cellular proteins and some membrane phospholipids. Anaerobic samples were characterized mainly by the region comprising 1,683 to 1,651 cm−1 (corresponding to amide I vibrations) and those comprising 1,537 to 1498 cm−1 and 1,265 to 1,188 cm−1 (corresponding to amide II and amide III, respectively). Nucleic acid-related vibrations, ranging between 983 and 866 cm−1, also contributed to the anaerobic sample cluster. The aerobic samples, which were grouped in the positive area of the PC-DF2 axis, were characterized mainly by similarities across the region comprising 1,095 to 1,014 cm−1 of the spectra, which corresponds to the PO2− symmetric stretch [ν(PO2−)] from nucleic acids, ν(CO), and a deoxyribose/ribose DNA absorption region. Differences in the amide II region (1,631 to 1,558 cm−1) also separated the aerobic samples from the anaerobic samples. Smaller contributions based on differences between the regions comprising 2,931 to 2,914 cm−1 and 1,747 to 1,732 cm−1 were also observed, corresponding to the aliphatic CH3 and CH2 stretches from membrane phospholipid and CO lipid vibrations.
As shown in Fig. 3C to F, PC-DFA also showed biochemical similarities in the trajectories observed for aerobic and anaerobic conditions during cell growth in the early exponential and stationary phases. For anaerobic culture, the early-exponential-phase samples were characterized mainly by the CH2 (2,966 to 2,900 cm−1) and CH3 (2,866 to 2,854 cm−1) vibrations from lipids, the PO stretch from nucleic acids (1,099 to 1,055 cm−1), and the CO stretch of esters (1,755 to 1,647 cm−1), carbonic acid, nucleic acids, and amide I absorption regions. After growth, the FT-IR spectra of the bacteria were differentiated by the amide I (1,635 to 1,612 cm−1) and II (1,533 to 1,519 cm−1) regions, with some contribution from aromatic groups at 1,568 to 1,560 cm−1. For aerobic cultures, bands arising from the nucleic acids (1,722 to 1,701 and 872 to 862 cm−1) and amide I (1,670 to 1,637 cm−1) were stronger in the early log phase, but after further growth, lipid (2,929 to 2,912 cm−1) and phospholipid (2,856 to 2,846 cm−1) signals were also elevated. Inspection of both loading plots showed that there were many changes throughout the whole cellular metabolism, including changes within spectral regions relating to protein, fatty acids, nucleic acids, and phospholipids.
The interpretations of these FT-IR spectra signify that specific biochemical information can be gained from this metabolic fingerprinting approach. Therefore, the next stage was to see if it was possible to identify the specific functional changes in the FT-IR spectra as a function of growth time.
Growth-related phenotypes estimated from the FT-IR spectra.
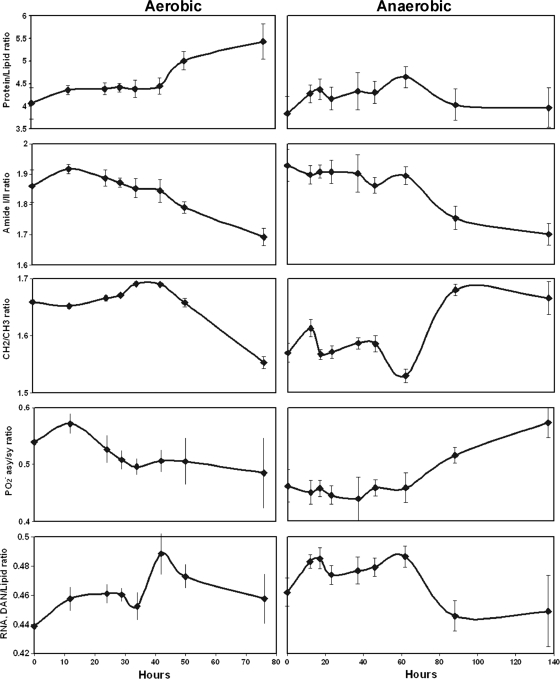
To elucidate further the biochemical differences between samples on the basis of incubation time and culture conditions, specific FT-IR peak areas (highlighted above) were plotted against the growth time (Fig. 5).
FIG. 5.
Quantitative analysis of the absorbance band areas of FT-IR spectra. Protein/lipid ratio, ratio of amide I and II area to lipid area; amide I/II ratio, ratio of band areas of amide I to amide II regions; CH2/CH3 ratio, ratio of band area of CH2 asymmetric/CH3 asymmetric; PO2− asy/sy ratio, ratio of band area of PO2− antisymmetric stretch to PO2− symmetric stretch; RNA, DNA/lipid content ratio, ratio of band area of RNA and DNA to band area of lipids. Points show the means of results from 3 measurements, and the standard deviation error bars are plotted.
The amount of protein and lipid present in the cell membrane plays an important role in maintaining membrane structure and dynamics. Our analysis has shown that the protein-to-lipid ratios in aerobically cultured cells were higher than those observed during anaerobic growth. These ratios were shown to increase subtly with the growth time, and this is in agreement with a previous study (41). In addition, Mourant et al. (32) also found that the ratio of protein to lipid content increases with increasing cellular proliferation. After the cells moved toward the “death phase,” there was a large increase in these peak area ratios for dying aerobic samples, but a small decrease was noted for the anaerobic cells (Fig. 5). In both instances, there were marked decreases in total protein over time.
The CH2/CH3 ratio can be related either to changes in the acyl chain length or to the overall lipid content (31). This information corresponds with the PC-DFA loading (Fig. 4), which showed the intense absorptions arising mainly from lipids at 1,736 cm−1 (ester C=O stretching modes) and/or from acyl chains at 1,469 cm−1 (CH2 bending vibration). During the stationary phase and into the death phase, the CH2/CH3 ratio decreased under aerobic conditions but increased under anaerobic conditions (Fig. 5). The ratio was greater in aerobic samples than in anaerobic cells, which further confirms the high lipid content in the MR-1 cells growing under aerobic conditions.
The nucleic acid content of the cells can also be analyzed by FT-IR spectroscopy. The ratio of the band area of the PO2− antisymmetric stretch to the PO2− symmetric stretch (PO2− asy/sy ratio) showed very different responses in anaerobic and aerobic samples. The starting point for the anaerobic samples was slightly higher than that for the aerobic samples, and with increasing incubation time, this ratio remained constant for anaerobic cells in the exponential and stationary phases and then increased in the death phase. In contrast, for aerobic samples, these ratios decreased during the exponential phase and then remained constant following the stable death phases. These changes showed that the ratios of PO2− asy/sy for the MR-1 cells were affected not only by the different electron acceptors but also by the growth conditions.
The ratio of RNA to lipid content should coincide with the ratio of protein to lipid (32), but this trend was not observed in this study, and we have not been able to ascertain quantitative information indicative of RNA synthesis. This is most likely due to very large numbers of overlapping bands in this region, which could account for the large error bars seen for the RNA-to-lipid spectral area ratios (30).
In summary, S. oneidensis is capable of respiring a range of electron acceptors, including oxygen and fumarate, and previous studies have confirmed that changes in the dominant respiratory process result in well-defined alterations in gene expression (2). We therefore sought to study the impact of these changes on cellular metabolism and phenotype. The ratio of protein to lipid, the CH2/CH3 ratio, and the PO2− asy/sy ratio in the FT-IR spectra all showed very different responses between anaerobic and aerobic samples as a function of growth time, confirming large metabolic differences between these cells.
Quantification of secreted extracellular flavins by HPLC.
In addition to metabolic fingerprinting using FT-IR spectroscopy, experiments were conducted to quantify the concentrations of the flavin compounds riboflavin, flavin-adenine dinucleotide (FAD), and flavin mononucleotide (FMN) secreted by S. oneidensis MR-1, as these have been shown to act as extracellular redox shuttles (44). The concentrations of the three flavins secreted by S. oneidensis MR-1 under anaerobic and aerobic conditions in culture supernatants were analyzed by HPLC (Table 2).
TABLE 2.
FMN and RF concentrations secreted by S. oneidensis MR-1 cells as determined by the reference methods (HPLC)a
| Condition | Sample name | Mean concn (μM/g protein) (SD) |
||
|---|---|---|---|---|
| FMN | RF | Total | ||
| Aerobic | Ae12(Ex) | 0.02 (0.00) | 0.66 (0.01) | 1.18 (0.01) |
| Ae24(Ex) | 0.12 (0.01) | 0.96 (0.02) | 1.56 (0.02) | |
| Ae29(St) | 0.15 (0.01) | 0.95 (0.02) | 1.63 (0.02) | |
| Ae34(St) | 0.17 (0.00) | 0.94 (0.01) | 1.72 (0.02) | |
| Ae42(St) | 0.21 (0.01) | 0.94 (0.02) | 1.83 (0.02) | |
| Anaerobic | An12(Ex) | 1.27 (0.06) | 0.21 (0.01) | 1.47 (0.06) |
| An17(Ex) | 1.41 (0.03) | 0.23 (0.01) | 1.63 (0.04) | |
| An23(Ex) | 1.71 (0.04) | 0.21 (0.01) | 1.91 (0.04) | |
| An37(St) | 1.68 (0.03) | 0.34 (0.01) | 2.03 (0.04) | |
| An46(St) | 1.72 (0.03) | 0.51 (0.01) | 2.23 (0.03) | |
| An62(St) | 1.93 (0.01) | 0.72 (0.03) | 2.65 (0.01) | |
Ex, samples harvested at the exponential phase; St, samples harvested at the stationary phase. The number after the label stands for the incubation time; labels beginning with “Ae” are for aerobic samples, and those with “An” are for anaerobic samples. For example, An12 represents cells grown in anaerobic medium for 12 h, and Ae24 represents cells grown in aerobic medium for 24 h. Numbers in brackets indicate standard deviations of measurements from 3 biological repeats.
The predominant flavin secreted by S. oneidensis MR-1 under aerobic and anaerobic conditions was FMN, followed by riboflavin. FAD occurred only at trace concentrations, typically as cells entered the death phase, presumably due to release of cellular FAD by lysed cells (data not shown). The extracellular concentrations of flavins were 3 to 5 times higher in aerobic cultures than in anaerobic cultures (Table 2). These results are in accordance with those from previous studies from our laboratory (44).
In this study, we measured the concentrations of the flavin components throughout the aerobic and anaerobic cultures to generate a series of values against which to calibrate the FT-IR spectra for rapid and accurate screening (Table 2). Under aerobic conditions, the extracellular concentrations of both FMN and riboflavin increased during the exponential phase. However, the riboflavin concentrations leveled off as the cells entered the stationary phase, while FMN concentrations continued to increase. Nevertheless, riboflavin was present at higher concentrations throughout, e.g., riboflavin was present at 0.96 μM/g protein, versus FMN at 0.21 μM/g protein, after 42 h (corresponding to the stationary phase). In anaerobic cultures, FMN dominated throughout, with both FMN and riboflavin increasing through the exponential and stationary phases and reaching 1.93 and 0.72 μM/g protein, respectively (at 62 h, in the stationary phase).
Quantification of flavins by use of PLSR analysis of the FT-IR spectra.
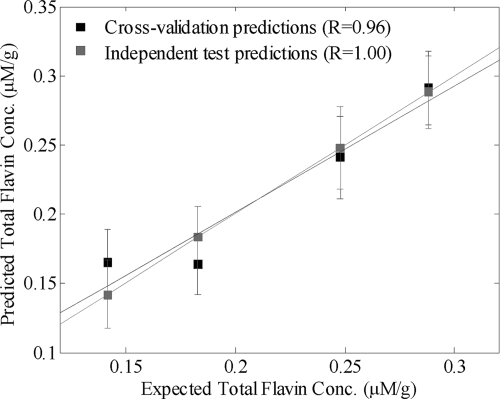
The correlation between the FT-IR spectra of S. oneidensis MR-1 and the extracellular flavin concentrations measured by HPLC were quantified using the supervised learning method of PLSR (26). The FT-IR spectra were used as the input to the PLSR models (X-variables), and the individual concentrations of (i) FMN, (ii) riboflavin, and (iii) total flavin content were used as the target output (y variable) in three different PLSR models. A plot of predicted total flavin content versus measured flavin content for cells grown under anaerobic conditions is shown in Fig. 6, where it can be clearly seen that the model's results lie on the expected y = x line and so graphically show excellent prediction; the same was also seen for predictions of the FMN and riboflavin concentrations (data not shown).
FIG. 6.
Actual total flavin concentrations calculated from HPLC analyses plotted against the predicted concentration values using a calibration model based on FT-IR spectra analyzed by PLSR. In this PLSR model, 8 latent variables are employed (Table 3).
The statistics for the three PLSR calibration models developed for extracellular FMN, riboflavin, and total flavin concentrations under both aerobic and anaerobic conditions are summarized in Table 3. The number of PLSR factors (latent variables) used, the percentage of the RMSEP, the calibration error, the cross-validation error, and the intercepts and slopes were calculated from self-prediction (calibration) and cross-validation. The root mean square error of prediction (RMSEP) is the most unbiased parameter and so was used to compare the performances of different models.
TABLE 3.
Partial least-squares regression results for FT-IR spectra versus flavin concentrationsa
| Condition | No. of PLSR factors | RMSEP | RMSE |
LSR value (error) |
||||
|---|---|---|---|---|---|---|---|---|
| Calibration | Validation | Train intercept | Validation intercept | Train slope | Validation slope | |||
| FMN in An | 8 | 7.00 | 0.01 | 0.05 | 0.00 (0.01) | 0.08 (0.08) | 1.00 (0.01) | 0.95 (0.05) |
| RF in An | 8 | 15.00 | 0.01 | 0.01 | 0.00 (<0.001) | 0.01 (0.01) | 1.00 (0.01) | 0.97 (0.02) |
| Total in An | 8 | 10.42 | 0.02 | 0.05 | 0.00 (0.02) | 0.06 (0.07) | 1.00 (0.01) | 0.96 (0.04) |
| FMN in Ae | 10 | 3.61 | 0 | 0 | 0.00 (0.01) | −0.03 (0.08) | 1.00 (0.01) | 1.03 (0.09) |
| RF in Ae | 6 | 7.14 | 0.02 | 0.02 | 0.04 (0.03) | 0.04 (0.04) | 0.92 (0.05) | 0.92 (0.08) |
| Total in Ae | 6 | 11.89 | 0.03 | 0.03 | 0.09 (0.07) | 0.05 (0.13) | 0.94 (0.04) | 0.97 (0.08) |
An, anaerobic medium; Ae, aerobic medium; RMSEP, percentage of the root mean square error of prediction; RMSE, root mean squared error; LSR: least-squares regression.
As shown in Table 3, all of the PLSR models produced excellent predictive results, underlining the correlation between the spectra and the concentrations of the flavins, and this clearly showed that FT-IR could provide an accurate means of determining the relative concentrations of the flavins secreted by S. oneidensis MR-1 without recourse to time-consuming chromatographic separations and could play a role in the high-throughput screening of microbial cultures for the flavins.
Conclusions.
This study demonstrates that the use of FT-IR spectroscopy in combination with multivariate statistical methods is a powerful approach for distinguishing significant metabolic changes associated with anaerobic and aerobic growth conditions for S. oneidensis MR-1. This is the first time that FT-IR spectroscopy has been applied in analyzing the S. oneidensis MR-1 metabolic fingerprinting profiles under these two regimes. It is also clear that FT-IR spectroscopy can be used to distinguish cells collected at different growth phases due to their significantly metabolic fingerprint variations. Changes in phenotype that were highlighted include altered protein and lipid profiles in these cells. Furthermore, using PLSR, secreted flavins could be quantified accurately without the need to use complicated and time-consuming chromatographic separations. This study reveals that metabolic fingerprinting using FT-IR spectroscopy has excellent potential for future use in rapid detection of the Shewanella cell metabolome in a range of engineered and natural environments.
Acknowledgments
This research was supported by BBSRC grant BBS/B/03718 and a Manchester-CSC Ph.D. scholarship to H. Wang.
Harald von Canstein is acknowledged for his assistance in HPLC analysis.
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Beliaev, A. S., D. M. Klingeman, J. A. Klappenbach, L. Wu, M. F. Romine, J. M. Tiedje, K. H. Nealson, J. K. Fredrickson, and J. Zhou. 2005. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J. Bacteriol. 187:7138-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beliaev, A. S., D. K. Thompson, T. Khare, H. Lim, C. C. Brandt, G. Li, A. E. Murray, J. F. Heidelberg, C. S. Giometti, J. Yates, K. H. Nealson, J. M. Tiedje, and J. Zhou. 2002. Gene and protein expression profiles of Shewanella oneidensis during anaerobic growth with different electron acceptors. OMICS 6(1):39-60. [DOI] [PubMed] [Google Scholar]
- 3.Brereton, R. G. 2000. Introduction to multivariate calibration in analytical chemistry. Analyst 125:2125-2154. [Google Scholar]
- 4.Choo-Smith, L. P., K. Maquelin, T. van Vreeswijk, H. A. Bruining, G. J. Puppels, N. A. N. Thi, C. Kirschner, D. Naumann, D. Ami, A. M. Villa, F. Orsini, S. M. Doglia, H. Lamfarraj, G. D. Sockalingum, M. Manfait, P. Allouch, and H. P. Endtz. 2001. Investigating microbial (micro)colony heterogeneity by vibrational spectroscopy. Appl. Environ. Microbiol. 67:1461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coates, J. D., and L. A. Achenbach. 2002. The biogeochemistry of aquifer systems: manual of environmental microbiology. ASM Press, Washington, DC.
- 6.Cornmell, R. J., C. L. Winder, G. J. T. Tiddy, R. Goodacre, and G. Stephens. 2008. Accumulation of ionic liquids in Escherichia coli cells. Green Chem. 10:836-841. [Google Scholar]
- 7.Delille, A., F. Quiles, and F. Humbert. 2007. In situ monitoring of the nascent pseudomonas fluorescens biofilm response to variations in the dissolved organic carbon level in low-nutrient water by attenuated total reflectance-Fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 73:5782-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiehn, O. 2001. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genomics 2:155-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field, J. A., and J. Brady. 2003. Riboflavin as a redox mediator accelerating the reduction of the azo dye Mordant Yellow 10 by anaerobic granular sludge. Water Sci. Technol. 48:187-193. [PubMed] [Google Scholar]
- 10.Goodacre, R., L. Roberts, D. I. Ellis, D. Thorogood, S. M. Reader, H. Ougham, and I. King. 2007. From phenotype to genotype: whole tissue profiling for plant breeding. Metabolomics 3:489-501. [Google Scholar]
- 11.Goodacre, R., E. M. Timmins, R. Burton, N. Kaderbhai, A. M. Woodward, D. B. Kell, and P. J. Rooney. 1998. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology 144:1157-1170. [DOI] [PubMed] [Google Scholar]
- 12.Goodacre, R., S. Vaidyanathan, W. B. Dunn, G. G. Harrigan, and D. B. Kell. 2004. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends. Biotechnol. 22:245-252. [DOI] [PubMed] [Google Scholar]
- 13.Harrigan, G. G., R. H. LaPlante, G. N. Cosma, G. Cockerell, R. Goodacre, J. F. Maddox, J. P. Luyendyk, P. E. Ganey, and R. A. Roth. 2004. Application of high-throughput Fourier-transform infrared spectroscopy in toxicology studies: contribution to a study on the development of an animal model for idiosyncratic toxicity. Toxicol. Lett. 146:197-205. [DOI] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. DeBoy, T. R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 15.Hollywood, K., D. R. Brison, and R. Goodacre. 2006. Metabolomics: current technologies and future trends. Proteomics 6:4716-4723. [DOI] [PubMed] [Google Scholar]
- 16.Huang, W. E., D. Hopper, R. Goodacre, M. Beckmann, A. Singer, and J. Draper. 2006. Rapid characterization of microbial biodegradation pathways by FT-IR spectroscopy. J. Microbiol. Methods 67:273-280. [DOI] [PubMed] [Google Scholar]
- 17.Jarvis, R. M., E. W. Blanch, A. P. Golovanov, J. Screen, and R. Goodacre. 2007. Quantification of casein phosphorylation with conformational interpretation using Raman spectroscopy. Analyst 132:1053-1060. [DOI] [PubMed] [Google Scholar]
- 18.Jarvis, R. M., D. Broadhurst, H. Johnson, N. M. O'Boyle, and R. Goodacre. 2006. PYCHEM: a multivariate analysis package for Python. Bioinformatics 22:2565-2566. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis, R. M., and R. Goodacre. 2004. Ultra-violet resonance Raman spectroscopy for the rapid discrimination of urinary tract infection bacteria. FEMS Microbiol. Lett. 232:127-132. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis, R. M., and R. Goodacre. 2004. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem. 76:40-47. [DOI] [PubMed] [Google Scholar]
- 21.Jolliffe, I. T. 1986. Principal component analysis. Springer, New York, NY.
- 22.Kaderbhai, N. N., D. I. Broadhurst, D. I. Ellis, R. Goodacre, and D. B. Kell. 2003. Functional genomics via metabolic footprinting: monitoring metabolite secretion by Escherichia coli tryptophan metabolism mutants using FT-IR and direct injection electrospray mass spectrometry. Comp. Funct. Genomics 4:376-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lloyd, J. R., and D. R. Lovley. 2001. Microbial detoxification of metals and radionuclides. Curr. Opin. Biotechnol. 12:248-253. [DOI] [PubMed] [Google Scholar]
- 24.Lovley, D. R. 2006. Bug juice: harvesting electricity with microorganisms. Nat. Rev. Microbiol. 4:497-508. [DOI] [PubMed] [Google Scholar]
- 25.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 26.Manly, B. F. J. 1994. Multivariate statistical methods: a primer. Chapman and Hall, London, United Kingdom.
- 27.Maquelin, K., C. Kirschner, L. P. Choo-Smith, N. van den Braak, H. P. Endtz, D. Naumann, and G. J. Puppels. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255-271. [DOI] [PubMed] [Google Scholar]
- 28.Martens, H., and T. Naes. 1989. Multivariate calibration. John Wiley & Sons, Chichester, United Kingdom.
- 29.Martens, H., and E. Stark. 1991. Extended multiplicative signal correction and spectral interference subtraction-new preprocessing methods for near infrared spectroscopy. J. Pharm. Biomed. Anal. 9:625-635. [DOI] [PubMed] [Google Scholar]
- 30.Meade, A., F. Lyng, P. Knief, and H. Byrne. 2007. Growth substrate induced functional changes elucidated by FTIR and Raman spectroscopy in in-vitro cultured human keratinocytes. Anal. Bioanal. Chem. 387:1717-1728. [DOI] [PubMed] [Google Scholar]
- 31.Melin, A. M., A. Allery, A. Perromat, C. Bébéar, G. Déléris, and B. de Barbeyrac. 2004. Fourier transform infrared spectroscopy as a new tool for characterization of mollicutes. J. Microbiol. Methods 56:73-82. [DOI] [PubMed] [Google Scholar]
- 32.Mourant, J. R., Y. R. Yamada, S. Carpenter, L. R. Dominique, and J. P. Freyer. 2003. FTIR spectroscopy demonstrates biochemical differences in mammalian cell cultures at different growth stages. Biophys. J. 85:1938-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers, C. R., and J. M. Myers. 1993. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 108:15-21. [Google Scholar]
- 34.Myers, C. R., and K. H. Nealson. 1990. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J. Bacteriol. 172:6232-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterisation by FTIR spectroscopy. Nat. Biothechnol. 351:81-82. [DOI] [PubMed] [Google Scholar]
- 36.Neal, A. L., S. N. Dublin, J. Taylor, D. J. Bates, J. L. Burns, R. Apkarian, and T. J. DiChristina. 2007. Terminal electron acceptors influence the quantity and chemical composition of capsular exopolymers produced by anaerobically growing Shewanella spp. Biomacromolecules 8:166-174. [DOI] [PubMed] [Google Scholar]
- 37.Nicolaou, N., and R. Goodacre. 2008. Rapid and quantitative detection of the microbial spoilage in milk using Fourier transform infrared spectroscopy and chemometrics. Analyst 133:1424-1431. [DOI] [PubMed] [Google Scholar]
- 38.Oust, A., T. Moretro, K. Naterstad, G. D. Sockalingum, I. Adt, M. Manfait, and A. Kohler. 2006. Fourier transform infrared and Raman spectroscopy for characterization of Listeria monocytogenes strains. Appl. Environ. Microbiol. 72:228-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, S. A., F. Currie, N. Thakker, and R. Goodacre. 2008. Spatial metabolic fingerprinting using FT-IR spectroscopy: investigating abiotic stresses on Micrasterias hardyi. Analyst 133:1707-1713. [DOI] [PubMed] [Google Scholar]
- 40.Pearce, C. I., R. Christie, C. Boothman, H. von Canstein, J. T. Guthrie, and J. R. Lloyd. 2006. Reactive azo dye reduction by Shewanella strain J18 143. Biotechnol. Bioeng. 95:692-703. [DOI] [PubMed] [Google Scholar]
- 41.Szalontai, B., Y. Nishiyama, Z. Gombos, and N. Murata. 2000. Membrane dynamics as seen by Fourier transform infrared spectroscopy in a cyanobacterium, Synechocystis PCC 6803: the effects of lipid unsaturation and the protein-to-lipid ratio. Biochim. Biophys. Acta 1509:409-419. [DOI] [PubMed] [Google Scholar]
- 42.Reference deleted.
- 43.Tiedje, J. 2002. Shewanella—the environmentally versatile genome. Nat. Biotechnol. 20:1093-1094. [DOI] [PubMed] [Google Scholar]
- 44.von Canstein, H., J. Ogawa, S. Shimizu, and J. R. Lloyd. 2008. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74:615-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weckwerth, W., and K. Morgenthal. 2005. Metabolomics: from pattern recognition to biological interpretation. Drug Discov. Today 10:1551-1558. [DOI] [PubMed] [Google Scholar]