Abstract
Enzymes catalyzing the conversion of organohalogen compounds are useful in the chemical industry and environmental technology. Here we report the occurrence of a new reduced flavin adenine dinucleotide (FAD) (FADH2)-dependent enzyme that catalyzes the removal of a halogen atom from an unsaturated aliphatic organohalogen compound by the addition of a water molecule to the substrate. A soil bacterium, Pseudomonas sp. strain YL, inducibly produced a protein named Caa67YL when the cells were grown on 2-chloroacrylate (2-CAA). The caa67YL gene encoded a protein of 547 amino acid residues (Mr of 59,301), which shared weak but significant sequence similarity with various flavoenzymes and contained a nucleotide-binding motif. We found that 2-CAA is converted into pyruvate when the reaction was carried out with purified Caa67YL in the presence of FAD and a reducing agent [NAD(P)H or sodium dithionite] under anaerobic conditions. The reducing agent was not stoichiometrically consumed during this reaction, suggesting that FADH2 is conserved by regeneration in the catalytic cycle. When the reaction was carried out in the presence of H218O, [18O]pyruvate was produced. These results indicate that Caa67YL catalyzes the hydration of 2-CAA to form 2-chloro-2-hydroxypropionate, which is chemically unstable and probably spontaneously dechlorinated to form pyruvate. 2-Bromoacrylate, but not other 2-CAA analogs such as acrylate and methacrylate, served as the substrate of Caa67YL. Thus, we named this new enzyme 2-haloacrylate hydratase. The enzyme is very unusual in that it requires the reduced form of FAD for hydration, which involves no net change in the redox state of the coenzyme or substrate.
Dehalogenases catalyze the removal of halogen atoms from organohalogen compounds. These enzymes have been attracting a great deal of attention partly because of their possible applications to the chemical industry and environmental technology. Several dehalogenases have been discovered and characterized (6, 11, 14, 17, 22). Some of them act on unsaturated aliphatic organohalogen compounds in which a halogen atom is bound to an sp2-hybridized carbon atom. Examples include various corrinoid/iron-sulfur cluster-containing reductive dehalogenases (1, 7), cis- and trans-3-chloroacrylic acid dehalogenases (4, 19), and LinF (maleylacetate reductase), which acts on 2-chloromaleylacetate (5).
In order to gain more insight into the enzymatic dehalogenation of unsaturated aliphatic organohalogen compounds, we searched for microorganisms that dissimilate 2-chloroacrylate (2-CAA) as a sole source of carbon and energy (8). 2-CAA is a bacterial metabolite of 2-chloroallyl alcohol, an intermediate or by-product in the industrial synthesis of herbicides (26). Rats treated orally with the herbicides sulfallate, diallate, and triallate excrete urinary 2-CAA (16). Various halogenated acrylic acids are produced by a red alga (27). We obtained three 2-CAA-utilizing bacteria as a result of screening (8). For one of these bacteria, Burkholderia sp. strain WS, we previously discovered a new NADPH-dependent enzyme, 2-haloacrylate reductase (12, 13). Although this enzyme does not directly remove a halogen atom from the substrate, it is supposed to participate in the metabolism of 2-CAA by catalyzing the conversion of 2-CAA into l-2-chloropropionate, which is subsequently dehalogenated by l-2-haloacid dehalogenase.
Another bacterium that we obtained, Pseudomonas sp. strain YL, also dissimilates 2-CAA. However, the metabolic fate of 2-CAA in this bacterium remains unclear. In the present study, we analyzed proteins from 2-CAA- and lactate-grown cells of Pseudomonas sp. YL by two-dimensional polyacrylamide gel electrophoresis (PAGE) and identified a 2-CAA-inducible protein. We found that the protein catalyzes the dehalogenation of 2-CAA by the addition of a water molecule to the substrate, representing a new family of dehalogenases that act on unsaturated aliphatic organohalogen compounds. Remarkably, the enzyme requires reduced flavin adenine dinucleotide (FAD) (FADH2) for its activity, although the reaction does not involve a net change in the redox state of the coenzyme or substrate. Here we describe the occurrence and characteristics of this unusual flavoenzyme.
MATERIALS AND METHODS
Materials.
2-CAA was purchased from Lancaster Synthesis Ltd. (Lancashire, United Kingdom). H218O (99atom%) was obtained from Taiyo Nippon Sanso Corporation (Tokyo, Japan). All other chemicals were of analytical grade.
Microorganism and culture conditions.
Pseudomonas sp. YL, isolated from soil as a 2-CAA-utilizing bacterium (8), was grown at 28°C in a medium containing either 2-CAA or lactate as the sole carbon source, as described previously (12).
Two-dimensional PAGE.
Proteins from 2-CAA- and lactate-grown cells were analyzed by two-dimensional PAGE. First-dimensional isoelectric focusing was performed with IPG ReadyStrip at pH 3 to 10 (Bio-Rad Laboratories, Inc., Hercules, CA), and the gel was subjected to second-dimensional SDS-PAGE.
Determination of amino acid sequences.
The proteins in the two-dimensional PAGE gel were blotted onto an Immobilon-PSQ membrane (Millipore, Bedford, MA) and stained with Coomassie brilliant blue R-250. The spot of Caa67YL was excised, and the N-terminal amino acid sequence was determined with a Shimadzu (Kyoto, Japan) PPSQ-21 protein sequencer. Internal amino acid sequencing was performed by APRO Life Science Institute, Inc. (Naruto, Japan).
Sequencing of the gene coding for Caa67YL.
A part of the caa67YL gene was amplified by degenerate PCR with a sense primer (5′-ATGYTIGAYTTYYTIGTIAC-3′ or 5′-GAYTTYYTIGTIACIGAYGT-3′), an antisense primer (5′-GGIACYTGRTAIGCYTCIAT-3′ or 5′-TTRTCIACIGGIACYTGRTA-3′), the genomic DNA of Pseudomonas sp. YL, and TaKaRa LA Taq DNA polymerase (TaKaRa Bio, Otsu, Japan). PCR products of the predicted size were obtained with any set of the above-described primers and used as sequencing templates. The flanking region of these PCR products was amplified by inverse PCR (20) with self-ligated AatII-digested genomic DNA as a template and the following primers: 5′-CACGAAGGCTTCGATCGTGC-3′ and 5′-GCCGTCTCCGAGCAAGATGA-3′ for the first PCR and 5′-AAGCGATGTCGCGGACCACA-3′ and 5′-ACCCTCCTCGCTCGCAGAAA-3′ for the second nested PCR. The sizes of the DNA obtained were 1.7 kbp for the first PCR and 1.6 kbp for the second PCR. The product was sequenced, and the flanking region of this PCR product was amplified by inverse PCR (20) with self-ligated BamHI-digested genomic DNA as a template and the following primers: 5′-GCAAAGCAGCGCAGCAAG-3′ and 5′-TTCATCGACGAGACGCCT-3′ for the first PCR and 5′-CGATCAAGCTGTCTGACGG-3′ and 5′-TCGCTCGCAGAAAGGGCC-3′ for the second nested PCR. The sizes of the DNA obtained were 1.2 kbp for the first PCR and 0.7 kbp for the second PCR. AatII and BamHI were used for inverse PCR because the recognition sites for these restriction enzymes were not found in the partial sequence of the caa67YL gene available before inverse PCR.
Construction of a plasmid for the overproduction of Caa67YL.
The caa67YL gene was amplified by PCR by using the total genomic DNA of Pseudomonas sp. YL as a template; a forward primer, 5′-GGGAATTCCATATGTTGGATTTTCTTGTAAC-3′ (the NdeI site is underlined); and a reverse primer, 5′-CCGCCGCTCGAGCTAGACCGGGACGTCCTCGA-3′ (the XhoI site is underlined). The PCR product was digested with NdeI and XhoI and inserted into pET-21a(+) (Novagen, Darmstadt, Germany). The plasmid obtained was introduced into Escherichia coli BL21(DE3) cells.
Expression and purification of Caa67YL.
Recombinant E. coli cells were cultivated in 5 liters of Luria-Bertani medium containing 100 μg/ml ampicillin at 18°C until the A600 reached 0.5. After the addition of 50 μM isopropyl-1-thio-β-d-galactopyranoside, the cells were cultured for 24 h at 18°C. The cells were harvested, washed, and lysed by sonication in 70 ml of ice-cold 5 mM potassium phosphate buffer (KPB) (pH 7.1) containing 1 mM dithiothreitol (DTT). The following purification procedures were performed at 4°C. After centrifugation, the crude extract was treated with streptomycin sulfate (1% [wt/vol]), and nucleic acids were removed by centrifugation. The resulting supernatant was applied onto a DEAE-Toyopearl 650 M column (Tosoh, Tokyo, Japan) equilibrated with 5 mM KPB (pH 7.1) containing 1 mM DTT. Unbound proteins were washed out with 5 mM KPB (pH 7.1) containing 1 mM DTT. Chromatography was carried out with a linear gradient of 5 to 60 mM KPB (pH 7.1) containing 1 mM DTT. Proteins in chromatographic fractions were separated by SDS-PAGE and stained with Coomassie brilliant blue G-250. Caa67YL was eluted at about 30 to 60 mM KPB. The fractions containing Caa67YL were collected as the purified enzyme. The purified enzyme was dialyzed against 60 mM KPB (pH 7.1) containing 1 mM DTT, concentrated to 10 mg/ml, and stored at −80°C until use.
Quantification of FAD bound to Caa67YL.
Purified Caa67YL (0.17 mM) was incubated with 2.9 mM FAD in 60 mM KPB (pH 7.1) for 12 h at 4°C. After removing excess FAD not bound to the protein by gel filtration with a Bio-Spin 6 column (Bio-Rad Laboratories, Inc.), the protein was denatured by heating at 100°C for 10 min to release protein-bound FAD. The FAD thus obtained was quantified by measuring the absorbance at 450 nm (ɛ450 = 11,300 M−1 cm−1) (15). The content of FAD in the purified protein not incubated with externally added FAD was also determined by the same method.
Enzyme and protein assays.
Enzyme tests of Caa67YL were carried out in a glove box at oxygen levels of less than 2 ppm. For determinations of the enzyme activity, halide ions released from 2-CAA were measured according to a method described previously by Iwasaki et al. (10). In addition, the enzymatic conversions of 2-CAA and its analogs were monitored by electrospray ionization mass spectrometry (ESI-MS) with a triple-quadrupole Sciex API3000 liquid chromatography (LC)/tandem mass spectrometry (MS/MS) system (Applied Biosystems, Foster City, CA). The standard assay mixture (100 μl) contained 3.5 mM 2-CAA, 3.5 mM NaOH (to neutralize 2-CAA), 60 mM Tris-sulfate buffer (pH 9.0), purified Caa67YL (50 to 100 μg), 0.1 mM FAD, and 10 mM NADH to reduce FAD. Such a high concentration of NADH was added to the assay mixture for the nonenzymatic reduction of FAD to produce FADH2 (23). NADH was replaced by 10 mM NADPH or 10 mM sodium dithionite when the cofactor requirement was examined. For measurements of halide ions, the reaction was carried out at 35°C for 1 to 5 min and terminated by the addition of 11.1 μl of 1.5 M sulfuric acid. FAD in the assay mixture, which interferes with the colorimetric assay of halide ions due to its yellow color, was removed with charcoal powder. One unit of the enzyme activity was defined as the amount of enzyme that catalyzes the dehalogenation of 1 μmol of the substrate per minute. The activity toward other halogenated substrates, 2-bromoacrylate, 2-chloro-1-propene, and 2-chloroacrylonitrile, was assayed by the same method. Activities toward 2-fluoroacrylate, acrylate, methacrylate, fumarate, phosphoenolpyruvate, 2-chloropropionate, and lactate were examined by ESI-MS. The identities of the products obtained from 2-CAA and 2-bromoacrylate were also determined by ESI-MS. For mass spectrometric analysis of the reaction, the standard assay mixture was incubated for 240 min at 35°C. The reaction was terminated by the addition of 200 μl acetonitrile to the mixture, and the mixture was centrifuged, filtered, diluted with acetonitrile-10 mM ammonium acetate (1:1), and then introduced into the mass spectrometer in the negative-ion mode at 5 μl/min. For 18O incorporation experiments, the reaction was carried out in a standard assay mixture containing 50% (vol/vol) H218O.
The protein concentration was determined according to the method of Bradford, with bovine serum albumin as a standard (3).
Effects of pH and temperature on enzyme stability and activity.
To examine the effect of pH on the stability of Caa67YL, the enzyme was incubated for 30 min at 30°C in the following buffers (60 mM): citrate-NaOH (pH 5.5 to 6.5), potassium phosphate (pH 6.5 to 8.0), Tris-sulfate (pH 8.0 to 9.0), and glycine-NaOH (pH 9.0 to 10.5). After incubation, the incubation buffer was replaced with 60 mM KPB (pH 7.1) by use of a Microcon filter device (Millipore), and the remaining activity was measured by a standard assay. To analyze the effect of pH on activity, the initial reaction velocities were measured with the standard assay mixture containing the above-described buffers instead of the standard buffer. The effect of the temperature on the stability of the enzyme was determined by incubating the enzyme at different temperatures from 10°C to 60°C for 30 min prior to the standard assay. The effect of temperature on activity was examined by performing the standard assay at different temperatures from 10°C to 60°C. In all experiments, the reaction was started by the addition of the enzyme after the complete reduction of FAD to eliminate the effect of different temperatures and pH values on the reduction of FAD by NADH.
Molecular weight determination.
The subunit molecular weight of Caa67YL was determined by SDS-PAGE and ESI-MS. The molecular weight of the native enzyme was analyzed by gel filtration with an ÄKTA Explorer 10S system (GE Healthcare UK Ltd., Buckinghamshire, United Kingdom) equipped with a HiLoad 16/60 Superdex 200-pg column (GE Healthcare UK Ltd.). Molecular mass marker proteins (Oriental Yeast Co. Ltd., Tokyo, Japan) consisting of glutamate dehydrogenase (290 kDa), lactate dehydrogenase (142 kDa), enolase (67 kDa), myokinase (32 kDa), and cytochrome c (12.4 kDa) were used as standards.
Nucleotide sequence accession number.
The sequence of the caa67YL gene was registered in GenBank with the accession number AB519652.
RESULTS
Identification of a protein inducibly synthesized in 2-CAA-grown Pseudomonas sp. YL and cloning of its gene.
To identify proteins inducibly synthesized in Pseudomonas sp. YL cells grown on 2-CAA, proteins from 2-CAA-grown cells and lactate-grown cells were compared by two-dimensional PAGE. One major protein (Fig. 1) was found only in the 2-CAA-grown cells, suggesting its involvement in the metabolism of 2-CAA. This protein was named Caa67YL. The N-terminal amino acid sequence was MLDFLVTDVLVVGE, and the following internal amino acid sequence was determined: EMAELIEAYQVPVDK. Degenerate primers were designed based on the partial amino acid sequences of Caa67YL, and the gene coding for the protein was cloned as described in Materials and Methods. The caa67YL gene contained an open reading frame of 1,644 nucleotides coding for 547 amino acid residues (Mr of 59,301). A putative Shine-Dalgarno sequence, AAGGAGG, was found in the upstream region of the initiation codon of the caa67YL gene.
FIG. 1.
Two-dimensional PAGE analysis of the proteins of 2-CAA- and lactate-grown Pseudomonas sp. YL. Soluble proteins from 2-CAA-grown cells (A) and lactate-grown cells (B) were analyzed. The arrowhead indicates the spot of a 2-CAA-inducible protein, which we named Caa67YL.
Structural characteristics of Caa67YL.
A homology search revealed that Caa67YL shares 84.6% sequence identity with a 2-CAA-inducible protein from Burkholderia sp. WS (Caa67WS) (GenBank accession number BAD91550), whose function is unknown (12). Both proteins have a nucleotide-binding motif (VXGXGXXGXXXA) probably involved in binding FAD or NAD(P) in the region from positions 13 to 18. In addition, Caa67YL showed weak but significant sequence similarity to various flavoproteins, such as l-aspartate oxidase, the succinate dehydrogenase flavoprotein subunit, and thiol:fumarate reductase subunit A. The sequence identities to l-aspartate oxidase from Rhodopseudomonas palustris BisA53 (accession number YP_783305), the succinate dehydrogenase flavoprotein subunit from Methanothermobacter thermautotrophicus strain ΔH (accession number AAB85977) (21), and thiol:fumarate reductase subunit A from Methanothermobacter thermautotrophicus strain Marburg (accession number CAA04398) (9) were 23.2%, 23.6%, and 22.9%, respectively. The sequence similarity to various flavoproteins and the occurrence of a nucleotide-binding motif suggested that Caa67YL requires FAD for its function.
Reaction catalyzed by Caa67YL.
We tested whether Caa67YL catalyzes the degradation of 2-CAA in the presence of FAD. Caa67YL was overproduced in recombinant E. coli cells, and the crude extract was incubated with 2-CAA. Under aerobic conditions, 2-CAA was not degraded at all, as judged by ESI-MS analysis. In contrast, when the reaction was carried out under anaerobic conditions in the presence of high concentrations of NADH (10 mM) for the nonenzymatic reduction of FAD (0.1 mM), the peaks of 2-CAA {m/z = 105 (2-[35Cl]CAA) and 107 (2-[37Cl]CAA)} disappeared, and a new peak appeared at m/z 89, which was likely due to the formation of lactate (data not shown). The release of a chloride ion from 2-CAA was also observed by a colorimetric assay under the same conditions. The reaction did not proceed when the cell extract from E. coli harboring pET21a(+) without the caa67YL gene was used.
We purified Caa67YL from recombinant E. coli cells overproducing Caa67YL by monitoring the Caa67YL-dependent release of a chloride ion from 2-CAA (Table 1). The protein was purified 2.2-fold with 43% recovery. The final preparation was shown to be homogeneous by SDS-PAGE (Fig. 2). The specific activity of the purified enzyme was 0.96 units/mg.
TABLE 1.
Purification of Caa67YL from recombinant E. coli cellsa
| Purification step | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Yield (%) | Fold purification |
|---|---|---|---|---|---|
| Crude extract | 51 | 120 | 0.43 | 100 | 1 |
| DEAE-Toyopearl | 22 | 23 | 0.96 | 43 | 2.2 |
The enzyme activities were determined by measuring halide ions released from 2-CAA.
FIG. 2.

SDS-PAGE analysis of Caa67YL. Purified protein (10 μg) was loaded onto the gel.
Purified Caa67YL contained an oxidized form of FAD, as judged by its absorption spectrum. The molar ratio of FAD to the protein was 0.25. The ratio increased to 0.91 after incubation with externally added FAD, as described in Materials and Methods. Despite the presence of FAD in the purified protein, no enzyme activity was detected without a reduction of FAD.
To identify the product of the reaction catalyzed by Caa67YL, the purified protein was incubated with 2-CAA in the presence of 0.1 mM FAD and 10 mM NADH under anaerobic conditions. The reduction of FAD under these conditions was confirmed by the disappearance of the absorption peaks at 370 and 450 nm, characteristic of oxidized FAD. The reaction was monitored by ESI-MS. We found that the peaks of 2-CAA disappeared and a new peak appeared at m/z 87 only in the presence of Caa67YL (Fig. 3 A and B). When lactate dehydrogenase was added to the assay mixture, the peak at m/z 87 disappeared and a new peak appeared at m/z 89 (data not shown). Judging from the molecular mass and the reactivity with lactate dehydrogenase, the product of the Caa67YL-catalyzed reaction was concluded to be pyruvate. When the crude extract from recombinant E. coli cells was used instead of purified Caa67YL in the above-mentioned experiment, pyruvate was probably converted into lactate by endogenous lactate dehydrogenase.
FIG. 3.
Mass spectrometric monitoring of the conversion of 2-CAA with Caa67YL. 2-CAA was incubated with or without purified Caa67YL in the presence of 0.1 mM FAD and 10 mM NADH under anaerobic conditions as described in Materials and Methods. (A and B) The mixture was analyzed after a 4-h reaction without (A) and with (B) the addition of Caa67YL by ESI-MS in the negative-ion mode. (C) To determine the incorporation of an oxygen atom of a water molecule into the substrate, the reaction was carried out in the presence of 50% H218O, and the solution was analyzed after a 4-h reaction. Because chlorine has two isotopes with mass numbers of 35 and 37 at a ratio of 3:1, 2-CAA has two peaks at m/z 105 and 107. The peaks at m/z 87 and 89 are due to pyruvate and 18O-labeled pyruvate, respectively, as described in the text. The peak at m/z 97 is due to hydrogen sulfate in the reaction buffer and dihydrogen phosphate in the enzyme preparation.
Caa67YL did not catalyze the conversion of 2-CAA unless the reaction mixture contained a reduced form of FAD: the reaction proceeded when NADH was replaced by NADPH or sodium dithionite, but the lack of all of these reducing agents completely abolished the reaction. To investigate the dependence on FAD, we prepared the apoenzyme of Caa67YL by dialyzing the solution of purified Caa67YL against 2 M KBr. The UV-visible spectrum of the apoenzyme did not show a peak around 450 nm, a characteristic absorption of FAD, indicating that FAD was removed from Caa67YL. The apoenzyme showed no activity even when NADH, NADPH, or sodium dithionite was added to the reaction mixture. However, the activity was restored to the original level when 0.1 mM FAD was added to the assay mixture together with the reducing agent. When FAD was replaced with flavin mononucleotide (FMN) (0.1 mM), the enzyme did not recover its activity. Thus, Caa67YL catalyzes the conversion of 2-CAA into pyruvate in a reduced FAD-dependent manner.
Substrate specificity of Caa67YL.
The activities of Caa67YL toward various 2-haloacrylates and their analogs were determined. Caa67YL catalyzed the conversion of 2-bromoacrylate into pyruvate. However, 2-fluoroacrylate, methacrylate, acrylate, 2-chloroacrylonitrile, 2-cloro-1-propene, fumarate, and phosphoenolpyruvate were inert as substrates. Also, d- and l-2-chloropropionate and d- and l-lactate did not serve as the substrates. The velocity-versus-substrate plot for 2-CAA and 2-bromoacrylate showed typical Michaelis-Menten kinetics when the substrate concentration was low (2-CAA, <3.5 mM; 2-bromoacrylate, <4.0 mM). The apparent Km and Vmax values for 2-CAA were 0.47 mM and 1.2 units/mg, respectively, and those for 2-bromoacrylate were 1.3 mM and 1.6 units/mg, respectively. The enzyme activity was inhibited by the high concentrations of 2-CAA (>3.5 mM) and 2-bromoacrylate (>4.0 mM).
Molecular weight and subunit structure of Caa67YL.
The molecular weight of purified Caa67YL was estimated to be about 61,000 by SDS-PAGE, which agrees well with the value (molecular weight of 59,301) calculated from the deduced primary structure of the enzyme. Mass spectrometric analysis showed two peaks for Caa67YL: one at a molecular weight of 59,311, corresponding to the apoenzyme, and the other at a molecular weight of 60,099, probably due to the enzyme binding to FAD. The molecular weight determined by gel filtration was 52,000, suggesting that the enzyme is monomeric.
Effects of pH and temperature on Caa67YL.
The effect of pH on the activity of Caa67YL was examined over the pH range from 5.5 to 10.5. The enzyme was relatively stable between pH 8.5 and 9.5 for 30 min at 30°C and showed maximum activity at pH 9.0 (Fig. 4 A and B). The optimum temperature was found to be 35°C, and the enzyme was fairly stable at 20°C or lower for 30 min (Fig. 4C and D).
FIG. 4.
Effects of pH and temperature on Caa67YL. (A and B) Effects of pH on the activity (A) and stability (B) were determined by using the following buffers (60 mM): citrate-NaOH (closed circles) (pH 5.5 to 6.5), potassium phosphate (closed squares) (pH 6.5 to 8.0), Tris-sulfate (closed triangles) (pH 8.0 to 9.0), and glycine-NaOH (closed diamonds) (pH 9.0 to 10.5). The enzyme activity after treatment with Tris-sulfate (pH 9.0) was taken as 100% in A. (C and D) Effects of temperature on the activity (C) and stability (D) were determined as described in Materials and Methods. The enzyme activity at 40°C was taken as 100% in C.
Identification of the substrate providing oxygen in the Caa67YL-catalyzed reaction.
Caa67YL catalyzes the conversion of 2-CAA and 2-bromoacrylate into pyruvate as described above. In this reaction, an oxygen atom is incorporated into the substrate. To determine the source of the oxygen atom, the reaction was carried out in the presence of 50% (vol/vol) H218O. Mass spectrometric analysis of the reaction mixture showed that two peaks appeared after the reaction: one at m/z 87, corresponding to unlabeled pyruvate, and the other at m/z 89, corresponding to pyruvate containing 18O (Fig. 3C). The identity of the product as pyruvate was confirmed by the addition of lactate dehydrogenase to the reaction mixture: two peaks appeared at m/z 89 and 91, corresponding to unlabeled and 18O-labeled lactate, respectively (data not shown). The incorporation of 18O into pyruvate indicates that the oxygen atom of a water molecule is introduced into the substrate in the Caa67YL-catalyzed reaction.
Conservation of the reducing agent in the Caa67YL-catalyzed reaction.
A reducing agent is required for the reduction of FAD for the Caa67YL-catalyzed reaction. To examine whether the reduced form of FAD is consumed during the reaction, the amount of the reducing agent (NADH) in the reaction mixture was monitored: an aliquot of the reaction mixture was diluted, and the absorbance was measured at 340 nm for the quantification of NADH. We found that NADH did not decrease stoichiometrically for the release of chloride ions from 2-CAA (Fig. 5), indicating that the reduced form of FAD is not consumed during the reaction.
FIG. 5.
Amount of NADH consumed for dehalogenation of 2-CAA. 2-CAA was incubated with Caa67YL in the presence of 0.1 mM FAD and 10 mM NADH under anaerobic conditions, and the consumption of NADH and the formation of chloride ions were monitored. The concentrations of NADH and chloride ions in the reaction mixture are indicated by closed triangles and closed circles, respectively.
DISCUSSION
Occurrence of 2-haloacrylate hydratase.
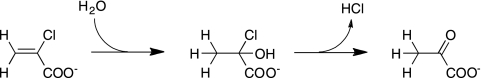
We found a novel flavoenzyme that catalyzes the conversion of 2-CAA into pyruvate. The 18O incorporation experiment showed that an oxygen atom of a water molecule is introduced into the substrate, indicating that Caa67YL catalyzes the hydration of the substrate. 2-Chloro-2-hydroxypropionate, a geminal halohydrin produced by the hydration of 2-CAA, is chemically unstable and probably spontaneously decomposes into pyruvate by the removal of HCl (Fig. 6). We named this novel enzyme 2-haloacrylate hydratase because the enzyme specifically acts on 2-CAA and 2-bromoacrylate.
FIG. 6.
Reaction catalyzed by 2-haloacrylate hydratase.
2-Haloacrylate hydratase has an absolute requirement for the reduced form of FAD for its catalytic reaction, which involves no net change in the redox state of the coenzyme or substrate. This cofactor requirement is notable because most flavoenzymes catalyze the net redox reactions, including oxidations, reductions, oxygenations, and electron transfers. A conceivable mechanism is that 2-CAA is first reduced to form 2-chloropropionate, which is subsequently hydrolyzed to form lactate, and lactate is further converted into pyruvate by oxidation. Although this explains the production of pyruvate from 2-CAA and the incorporation of an oxygen atom of a water molecule into the product, the mechanism is unlikely because 2-chloropropionate and lactate did not serve as the substrate of the enzyme. Another possible explanation for the FADH2 requirement for 2-haloacrylate hydratase is that FADH2 functions as a general acid-base catalyst. A general acid-base role of the flavin was recently shown for type 2 isopentenyl diphosphate isomerase (24, 25). FADH2 may be involved in the activation of a water molecule that attacks the C-2 atom of 2-CAA or the protonation of the C-3 position of 2-CAA. A more plausible mechanism is that FADH2 acts as a radical catalyst. One electron transfer from FADH2 to 2-CAA and protonation at the C-3 position would produce a 2-chloropropionate radical, which may be hydroxylated in the following step to produce 2-chloro-2-hydroxypropionate. Such a free-radical redox role of the reduced flavin has been reported for other flavoenzymes such as chorismate synthase (2, 18). Further studies such as stopped-flow kinetic analysis and crystallographic analysis are required to elucidate the reaction mechanism of this very unusual flavoenzyme.
Metabolism of 2-CAA.
Caa67YL is inducibly produced when the cells are grown on 2-CAA and catalyzes the conversion of 2-CAA into pyruvate, suggesting that 2-CAA is metabolized by Caa67YL in Pseudomonas sp. YL. In contrast, another 2-CAA-utilizing bacterium, Burkholderia sp. WS, inducibly produces two proteins, named Caa43 and Caa67, which we call Caa67WS in this paper to avoid confusion, when the cells are grown on 2-CAA (12). We previously reported that Caa43 catalyzes the reduction of 2-CAA to form l-2-chloropropionate by using NADPH as a cosubstrate and named this enzyme 2-haloacrylate reductase. l-2-Chloropropionate produced by 2-haloacrylate reductase is probably further metabolized to d-lactate by l-2-haloacid dehalogenase occurring in this bacterium. On the other hand, the function of Caa67WS is unknown. Considering its high sequence similarity with Caa67YL, it is very likely that Caa67WS also catalyzes the conversion of 2-CAA into pyruvate. The contribution of Caa43 and Caa67WS to the metabolism of 2-CAA in Burkholderia sp. WS remains to be examined in future studies.
In contrast with Burkholderia sp. WS, Pseudomonas sp. YL does not produce 2-haloacrylate reductase when grown on 2-CAA, as judged by the results of activity measurement and two-dimensional PAGE analysis. The gene coding for Caa43 is located immediately downstream of the gene coding for Caa67WS on the genome of Burkholderia sp. WS (12), whereas the gene coding for a Caa43 homolog was not found in the corresponding region on the genome of Pseudomonas sp. YL (data not shown). Taken together, 2-haloacrylate hydratase, but not 2-haloacrylate reductase, probably plays a principal role in 2-CAA metabolism in Pseudomonas sp. YL.
Comparison with cis- and trans-3-chloroacrylic acid dehalogenases.
cis-3-Chloroacrylic acid dehalogenase (4) and trans-3-chloroacrylic acid dehalogenase (19) catalyze the conversion of 3-chloroacrylate into 3-chloro-3-hydroxypropionic acid, which is decomposed to malonate semialdehyde and a chloride ion (4, 19). They share low but significant sequence similarity with each other and are supposed to evolve from a common ancestor. 2-Haloacrylate hydratase, discovered in the present study, resembles these enzymes in that it catalyzes dehalogenation by the addition of a water molecule to the substrate. However, it is significantly different from 3-chloroacrylic acid dehalogenases not only in its substrate specificity but also in its primary structure and cofactor requirement: 3-chloroacrylic acid dehalogenases require no cofactor for their catalytic activities, whereas 2-haloacrylate hydratase depends on a reduced form of FAD. Thus, 2-haloacrylate hydratase represents a new class of dehalogenase that degrades unsaturated aliphatic organohalogen compounds.
Acknowledgments
This work was supported in part by a Grant-in-Aid for the Global COE Program Integrated Materials Science (B-09) (to N.E. and A.M.M.), a Grant-in-Aid for Scientific Research (B) (grant 17404021) from the JSPS (to T.K.), and a Grant for Research for Promoting Technological Seeds from the JST (to T.K.). N.E. is a recipient of the 2009 Humboldt Research Award.
N.E. gratefully acknowledges Wolfgang Buckel (Max Planck Institute for Terrestrial Microbiology, Marburg, Germany), Rudolf K. Thauer (Max Planck Institute for Terrestrial Microbiology), Roland Lill (Philipps University Marburg, Marburg, Germany), and Antonio J. Pierik (Philipps University Marburg) for their kind suggestions concerning this study. We sincerely thank Hideyuki Hayashi (Osaka Medical College, Takatsuki, Japan) for helpful discussion.
Footnotes
Published ahead of print on 23 July 2010.
REFERENCES
- 1.Banerjee, R., and S. W. Ragsdale. 2003. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 72:209-247. [DOI] [PubMed] [Google Scholar]
- 2.Bornemann, S. 2002. Flavoenzymes that catalyse reactions with no net redox change. Nat. Prod. Rep. 19:761-772. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.de Jong, R. M., P. Bazzacco, G. J. Poelarends, W. H. Johnson, Jr., Y. J. Kim, E. A. Burks, H. Serrano, A. M. Thunnissen, C. P. Whitman, and B. W. Dijkstra. 2007. Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase. Structural basis for substrate specificity and inactivation by (R)-oxirane-2-carboxylate. J. Biol. Chem. 282:2440-2449. [DOI] [PubMed] [Google Scholar]
- 5.Endo, R., M. Kamakura, K. Miyauchi, M. Fukuda, Y. Ohtsubo, M. Tsuda, and Y. Nagata. 2005. Identification and characterization of genes involved in the downstream degradation pathway of γ-hexachlorocyclohexane in Sphingomonas paucimobilis UT26. J. Bacteriol. 187:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa, K. 2006. Oxygenases and dehalogenases: molecular approaches to efficient degradation of chlorinated environmental pollutants. Biosci. Biotechnol. Biochem. 70:2335-2348. [DOI] [PubMed] [Google Scholar]
- 7.Futagami, T., M. Goto, and K. Furukawa. 2008. Biochemical and genetic bases of dehalorespiration. Chem. Rec. 8:1-12. [DOI] [PubMed] [Google Scholar]
- 8.Hasan, A. K. M. Q., H. Takada, H. Koshikawa, J. Q. Liu, T. Kurihara, N. Esaki, and K. Soda. 1994. Two kinds of 2-halo acid dehalogenases from Pseudomonas sp. YL induced by 2-chloroacrylate and 2-chloropropionate. Biosci. Biotechnol. Biochem. 58:1599-1602. [Google Scholar]
- 9.Heim, S., A. Kunkel, R. K. Thauer, and R. Hedderich. 1998. Thiol:fumarate reductase (Tfr) from Methanobacterium thermoautotrophicum—identification of the catalytic sites for fumarate reduction and thiol oxidation. Eur. J. Biochem. 253:292-299. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki, I., S. Utsumi, K. Hagino, and T. Ozawa. 1956. A new spectrophotometric method for the determination of small amounts of chloride using the mercuric thiocyanate method. Bull. Chem. Soc. Jpn. 29:860-864. [Google Scholar]
- 11.Janssen, D. B., I. J. Dinkla, G. J. Poelarends, and P. Terpstra. 2005. Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities. Environ. Microbiol. 7:1868-1882. [DOI] [PubMed] [Google Scholar]
- 12.Kurata, A., T. Kurihara, H. Kamachi, and N. Esaki. 2005. 2-Haloacrylate reductase, a novel enzyme of the medium chain dehydrogenase/reductase superfamily that catalyzes the reduction of a carbon-carbon double bond of unsaturated organohalogen compounds. J. Biol. Chem. 280:20286-20291. [DOI] [PubMed] [Google Scholar]
- 13.Kurata, A., T. Kurihara, H. Kamachi, and N. Esaki. 2004. Asymmetric reduction of 2-chloroacrylic acid to (S)-2-chloropropionic acid by a novel reductase from Burkholderia sp. WS. Tetrahedron Asymmetry 15:2837-2839. [Google Scholar]
- 14.Kurihara, T., and N. Esaki. 2008. Bacterial hydrolytic dehalogenases and related enzymes: occurrences, reaction mechanisms, and applications. Chem. Rec. 8:67-74. [DOI] [PubMed] [Google Scholar]
- 15.Macheroux, P. 1999. UV-visible spectroscopy as a tool to study flavoproteins, p. 1-7. In S. K. Chapman and G. A. Reid (ed.), Flavoprotein protocols, vol. 131. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 16.Marsden, P. J., and J. E. Casida. 1982. 2-Haloacrylic acids as indicators of mutagenic 2-haloacrolein intermediates in mammalian metabolism of selected promutagens and carcinogens. J. Agric. Food Chem. 30:627-631. [DOI] [PubMed] [Google Scholar]
- 17.Nagata, Y., R. Endo, M. Ito, Y. Ohtsubo, and M. Tsuda. 2007. Aerobic degradation of lindane (γ-hexachlorocyclohexane) in bacteria and its biochemical and molecular basis. Appl. Microbiol. Biotechnol. 76:741-752. [DOI] [PubMed] [Google Scholar]
- 18.Osborne, A., R. N. Thorneley, C. Abell, and S. Bornemann. 2000. Studies with substrate and cofactor analogues provide evidence for a radical mechanism in the chorismate synthase reaction. J. Biol. Chem. 275:35825-35830. [DOI] [PubMed] [Google Scholar]
- 19.Poelarends, G. J., and C. P. Whitman. 2004. Evolution of enzymatic activity in the tautomerase superfamily: mechanistic and structural studies of the 1,3-dichloropropene catabolic enzymes. Bioorg. Chem. 32:376-392. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, J. N. Reeve, et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson, P. E. 1999. Dehalogenases applied to industrial-scale biocatalysis. Curr. Opin. Biotechnol. 10:365-369. [DOI] [PubMed] [Google Scholar]
- 23.Taga, M. E., N. A. Larsen, A. R. Howard-Jones, C. T. Walsh, and G. C. Walker. 2007. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446:449-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thibodeaux, C. J., S. O. Mansoorabadi, W. Kittleman, W. C. Chang, and H. W. Liu. 2008. Evidence for the involvement of acid/base chemistry in the reaction catalyzed by the type II isopentenyl diphosphate/dimethylallyl diphosphate isomerase from Staphylococcus aureus. Biochemistry 47:2547-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unno, H., S. Yamashita, Y. Ikeda, S. Y. Sekiguchi, N. Yoshida, T. Yoshimura, M. Kusunoki, T. Nakayama, T. Nishino, and H. Hemmi. 2009. New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase. J. Biol. Chem. 284:9160-9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Waarde, J. J., R. Kok, and D. B. Janssen. 1993. Degradation of 2-chloroallylalcohol by a Pseudomonas sp. Appl. Environ. Microbiol. 59:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woolard, F. X., R. E. Moore, and P. P. Roller. 1979. Halogenated acetic acid and acrylic acids from the red alga Asparagopsis taxiformis. Phytochemistry 18:617-620. [Google Scholar]