Abstract
Xylella fastidiosa is an insect-borne bacterium that colonizes xylem vessels of a large number of host plants, including several crops of economic importance. Chitin is a polysaccharide present in the cuticle of leafhopper vectors of X. fastidiosa and may serve as a carbon source for this bacterium. Biological assays showed that X. fastidiosa reached larger populations in the presence of chitin. Additionally, chitin induced phenotypic changes in this bacterium, notably increasing adhesiveness. Quantitative PCR assays indicated transcriptional changes in the presence of chitin, and an enzymatic assay demonstrated chitinolytic activity by X. fastidiosa. An ortholog of the chitinase A gene (chiA) was identified in the X. fastidiosa genome. The in silico analysis revealed that the open reading frame of chiA encodes a protein of 351 amino acids with an estimated molecular mass of 40 kDa. chiA is in a locus that consists of genes implicated in polysaccharide degradation. Moreover, this locus was also found in the genomes of closely related bacteria in the genus Xanthomonas, which are plant but not insect associated. X. fastidiosa degraded chitin when grown on a solid chitin-yeast extract-agar medium and grew in liquid medium with chitin as the sole carbon source; ChiA was also determined to be secreted. The gene encoding ChiA was cloned into Escherichia coli, and endochitinase activity was detected in the transformant, showing that the gene is functional and involved in chitin degradation. The results suggest that X. fastidiosa may use its vectors' foregut surface as a carbon source. In addition, chitin may trigger X. fastidiosa's gene regulation and biofilm formation within vectors. Further work is necessary to characterize the role of chitin and its utilization in X. fastidiosa.
The vector-borne plant-pathogenic bacterium Xylella fastidiosa colonizes insect and plant hosts (6). As a xylem-limited organism, X. fastidiosa interacts with plant structural polysaccharides and has proteins implicated in cell adhesion to carbohydrate-coated surfaces and enzymes involved in the degradation of pectin, glucan, and cellulose (35). Studies on X. fastidiosa interactions with plants have demonstrated that this organism is capable of degrading and potentially exploiting host plant polysaccharides as carbon sources (32). Similar to the xylem vessels in plants, the surface of the vectors' foregut also contains structural polysaccharides. These polysaccharides may be enzymatically degraded by X. fastidiosa and used as nutrient sources. In addition, they may affect the bacterium's biology: pectin for example, a plant structural polysaccharide, induces a regulon that controls vector transmission of X. fastidiosa (22). However, our understanding of how this pathogen interacts with its insect vectors lags significantly behind other aspects of its biology.
Xylem-sap-feeding leafhoppers and spittlebugs are vectors of X. fastidiosa to plants (1). After acquisition from an infected plant, the bacterium colonizes a narrow canal in the chitinous mouthparts (foregut) of vectors (2). The foregut's chitinous lining is shed during each molt, together with the rest of the insect's exoskeleton (30). The exoskeleton of insects has a complex chemical composition, but it is primarily a matrix of chitin and proteins covered by a thin epicuticle that includes mucopolysaccharides (3). Microscopy studies have indicated that X. fastidiosa initially attaches to the foregut of vectors laterally. Moreover, in older colonies, cells are always polarly attached (2). Afimbrial surface adhesins have been demonstrated to be important for early colonization of vectors but had no observable role in biofilm maturation (23).
Chitin is one of the most abundant polysaccharides in nature, second to cellulose (12); it is present in a wide range of organisms, including fungi, nematodes, and arthropods. Chitin is composed of N-acetylglucosamine units that form long chains through 1,4-β-linked glycosidic bonds. As a plentiful biopolymer in terrestrial and aquatic environments, it serves as a major organic carbon and nitrogen source for microbes (12). The chitinous surface of arthropods may also be colonized by bacteria, facilitating microbial persistence in the environment (20) or transmission between hosts in the case of insect vectors (23). Degradation of chitin is performed by glycoside hydrolases (GHs) (chitinases; EC 3.2.1.14), which break down polymers to smaller units or monomers. Chitinases are classified into two glycoside hydrolase families (GH18 and GH19) based on sequence similarity and protein structure (14, 15). GH19 has been found primarily in plants; plant chitinases have been shown to impact fungal colonization (5, 9). GH18 includes chitinases present in bacteria, fungi, animals, and plants (19, 28). Proteins in the GH18 family more often than not consist of a catalytic and a chitin-binding domain, which are often connected by linkers (19). The chitin-binding domain is not involved in catalyzing the hydrolysis of the glycosidic bonds but has been shown to increase enzyme kinetics (25).
In this study, evidence is presented supporting X. fastidiosa degradation and use of chitin as a carbon source. Additionally, X. fastidiosa's phenotype and gene expression profile were modified in the presence of chitin. A functional GH18 homolog in the genome of X. fastidiosa was identified in silico; furthermore, homologs of it were found in other members of the family Xanthomonadaceae. The significance of these findings is discussed, as is the possible role of chitin utilization in X. fastidiosa.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Luria broth (LB; Difco Laboratories) and solid Luria medium (1.5% [wt/vol] agar) were used to grow Escherichia coli JM109 cells at 37°C. The sequenced X. fastidiosa grape strain Temecula (35) was used and cultured on a modified version of the Periwinkle wilt solid medium (PWG) (16) and the liquid medium XFM (22). Cultures were incubated at 28°C. PWG was supplemented with 30 μg/ml of kanamycin in the case of rpfF mutant KLN61 (26), green fluorescent protein (GFP)-labeled X. fastidiosa KLN59.3 (27), and GFP-labeled rpfF mutant KLN121 (7). For media with chitin, XFM and LB were supplemented with 0.1 g/liter of colloidal chitin, which was prepared as described by Ramirez et al. (31). In certain experiments, liquid XFM medium was used without its carbon sources (disodium succinate and sodium citrate; XFM Δ), which was also supplemented with colloidal chitin as a single carbon source (XFM Δ-chitin).
Growth curve and adhesion assays.
Determination of X. fastidiosa's growth curve in XFM, XFM-chitin, XFM Δ, and XFM Δ-chitin media was performed as previously described (22). The total number of planktonic and attached cells in each flask was estimated as described by Chatterjee et al. (8). Analysis of variance (ANOVA) was used to compare planktonic and attached cell numbers (log transformed) in the presence or absence of chitin. A post hoc test (Tukey's test) was performed to conduct pairwise comparisons of growth types. Log-transformed cell numbers were normally distributed (Shapiro-Wilk test; P > 0.05). Cell adhesion to glass at the air-medium interface of flasks kept in a shaker was used as another indicator of X. fastidiosa adhesion to surfaces as previously described (22). The medium XFM-chitin without cells was included as a control for the adhesion of colloidal chitin to flasks; minimal staining with crystal violet was observed when compared to the treatments tested with cells added, XFM, and XFM-chitin (figure not shown). To determine if there were any transcriptional changes between X. fastidiosa grown in XFM and XFM-chitin, quantitative PCR was used to determine differences in the transcription level of selected genes (see the supplemental material for details).
Detection of chitinolytic activity.
The PD1826 gene was identified in silico as a chitinase gene in X. fastidiosa's genome. Methodological details of the in silico analyses are described in the supplemental material. PD1826 was transformed into E. coli using an expression vector following standard procedures (see the supplemental material for details). To detect chitinolytic activity by X. fastidiosa and transformed E. coli, cells were harvested from liquid media and resuspended in a protease inhibitor extraction buffer (1 mM ethylene diamine tetraacetic acid [EDTA], 10 mM Tris-HCl, pH 7.4, 1 μM phenylmethylsulfonyl fluoride [PMSF]). The suspension was then subjected to 5 cycles of freezing at −20°C and thawing at room temperature. The total protein concentration was determined according to Bradford (4). Proteins were separated using native polyacrylamide gel electrophoresis following standard protocols (13). To determine if PD1826 was secreted, liquid medium suspensions were filtrated through 0.22-μm filters. The filtrate was centrifuged at 14,000 rpm for 20 min at 4°C, and the pellet was suspended in native electrophoresis sample buffer. Detection of chitinolytic activity was performed by overlaying the gel with 2 ml of 50°C-heated 1% agarose in 100 mM sodium acetate (pH 4.8), containing 300 μg of the substrate 4-methylumbelliferyl N-acetyl-β-d-N,N′,N"-triacetylchitotriose [4-MU(GlcNAc)3] (Sigma-Aldrich), and incubated in 37°C for 10 min. Upon cleavage by endochitinases, the fluorescent product methylumbelliferyl (4-MU) is released into the gel from the trimeric substrate 4-MU(GlcNAc)3 and visualized under UV light (13). At large concentrations, however, enzymes with less affinity for this substrate may also hydrolyze it (17). A chitin-yeast extract-agar medium (10) was also used to visually detect chitonolytic activity on agar plates.
Biofilm formation on leafhopper wings.
A previously developed cell adhesion assay that uses the chitinous hindwings of leafhopper vectors as a proxy for the small region of the foregut colonized by X. fastidiosa was used to study cell attachment and biofilm formation (23). X. fastidiosa vector Homalodisca vitripennis individuals (collected from Bakersfield, CA, and reared in a greenhouse) had their hindwings removed and washed with distilled water. Wings were attached onto the concavities of agglutination slides using ParaplastX-TRA (Fisher Scientific). GFP-labeled X. fastidiosa and GFP-labeled rpfF mutant cells grown for 10 days in PWG were suspended in XFM Δ liquid medium (no carbon source); suspensions were adjusted to an optical density at 600 nm (OD600) of 0.075, and 2 μl of the suspensions was loaded onto each wing. Slides were stored in box and incubated at 28C° for 10 days. Sets of 20 independent wings from each treatment were investigated after 2 days, 5 days, and 10 days. Drops were removed and wings were washed with distilled water at 10 days, and the presence of GFP-labeled cells in drops (prior to wash) and biofilm formation (after wash) was detected with a UV light and appropriate GFP filters. Images were acquired with an epifluorescence stereomicroscope in the Biological Imaging Facility at The University of California—Berkeley.
RESULTS
Effect of chitin on X. fastidiosa growth and phenotype.
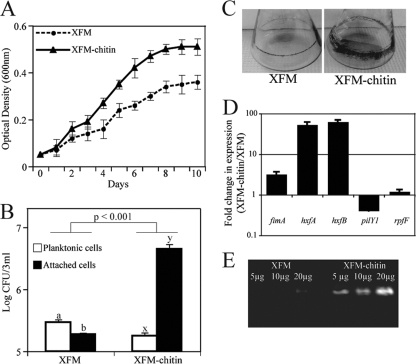
X. fastidiosa grew to larger populations in XFM-chitin than XFM, suggesting it might use chitin as a carbon source (F1, 44 = 133.1; P < 0.0001) (Fig. 1 A). Cell adhesion assays showed that X. fastidiosa cells grown in XFM-chitin attached to glass significantly more than those in XFM (Fig. 1B and C). Furthermore, planktonic cells occurred at larger numbers than attached ones in XFM (P < 0.042), while the opposite was observed for XFM-chitin (P < 0.001). The transcription level of the genes coding for fimbrial and afimbrial adhesins, fimA, hxfA, and hxfB, was increased in XFM-chitin compared to XFM (Fig. 1D). The transcription level of pilY1 was decreased, suggesting lower cell motility (6) under this condition (Fig. 1D). These results partly explain the change of phenotype, similarly to what was previously observed with the addition of pectin to XFM (22). On the other hand, the expression level of rpfF was not affected by the presence of chitin (Fig. 1D). RpfF synthesizes the cell-cell signaling molecule DSF (diffusible signaling factor) (26). For assays on chitinase activity using 4-MU(GlcNAc)3 an increase in fluorescence proportional to increments in protein concentration was observed for cells grown in XFM-chitin (Fig. 1E). A faint fluorescent band was detected for cells from XFM at the highest protein concentration used, indicating a basal level of chitinase production.
FIG. 1.
Effect of chitin on Xylella fastidiosa. (A) Growth curve of X. fastidiosa in XFM in presence or absence of colloidal chitin. (B) Log-transformed cell numbers present as planktonic or attached as a biofilm on glass in liquid XFM. Different letters on bars indicate statistically different treatments. (C) Biofilm formation of X. fastidiosa in a shaking culture. The biofilm is stained at the broth-air interface. (D) Chitin-induced transcriptional changes in X. fastidiosa; (E) in-gel chitinase activity ([4-MU(GlcNAc)3] cleavage) of X. fastidiosa whole-cell culture extracts.
In silico analyses of a chitinase and related genes in X. fastidiosa.
PD1826 was initially annotated as a hypothetical protein in the strain used in this study (Temecula 1), but it is present in other X. fastidiosa genomes and was annotated as an enzyme in the GH18 family. Based on sequence similarity, chiA was used to refer to PD1826 in this article. A database search revealed that the deduced ChiA protein was 74% identical to the Xanthomonas campestris ortholog, while identity to GH18 of other bacteria was lower, ranging from 28% with GH18 of Halotheromothrix orenii H186 to 49% with GH18 of Clostridium acetobutylicum ATCC 824. A phylogenetic analysis (maximum parsimony) showed monophyly of chiA orthologs in X. fastidiosa and Xanthomonas spp.; these genera also formed a monophyletic clade when compared to other chiA orthologs, indicating vertical descent of the gene in these genera (data not shown).
The predicted molecular mass of ChiA was ∼40 kDa (351 amino acids), with an isoelectric point of 8.36. ChiA had a signal peptide of 19 amino acids. In addition, the hydropathy plot suggested that the protein is secreted (data not show). The alignment with other bacterial GH18s showed similarity in the conserved catalytic regions with other GH18s, including the essential Glu residue (36), which acts as a proton donor (see Fig. S1A in the supplemental material). Interestingly, ChiA had no significant homology to chitin-binding domains identified in other bacterial chitinases (37). The predicted secondary structure with potential catalytic domains is shown in Fig. S1B in the supplemental material, while Fig. S1C represents the surface structure of the enzyme. These results indicate that the inferred catalytic site of X. fastidiosa's ChiA is conserved and functional.
chiA is physically located within an ∼10-kb locus in X. fastidiosa's chromosome (see Fig. S1D in the supplemental material). This carbohydrate-processing locus contains 4 open reading frames (ORFs) with sequence orthology to different glycoside hydrolase families (see Table S1 in the supplemental material). In that locus, chiA is followed by nahA, bmnA, and bgl2. nahA encodes a β-N-acetylhexosaminidase (EC 3.2.1.52), which has a β-N-acetylhexosaminidase glycoside hydrolase family 20 conserved domain. bmnA encodes a β-mannosidase (EC 3.2.1.25) with a glycoside hydrolase family 2 conserved domain, while bgl2 encodes a glucan 1,4-β-glucosidase (EC 3.2.1.74) with a glycoside hydrolase family 3 domain. Enzymatic activity of ChiA followed by NahA would result in degradation of chitin polymers to N-acetylglucosamine monomers. The ∼10-kb locus was found in both X. fastidiosa and different Xanthomonas species. However, in other sequenced Xanthomonadaceae members, such as Stenotrophomonas spp., the locus is absent, but homologs to chiA and the other three genes in the locus are still present elsewhere in the genome. Other genes hypothesized to be involved in chitin utilization by X. fastidiosa are also present in other members of the Xanthomonadaceae (Table S1; see Fig. 4A).
X. fastidiosa degrades chitin and uses it as a carbon source.
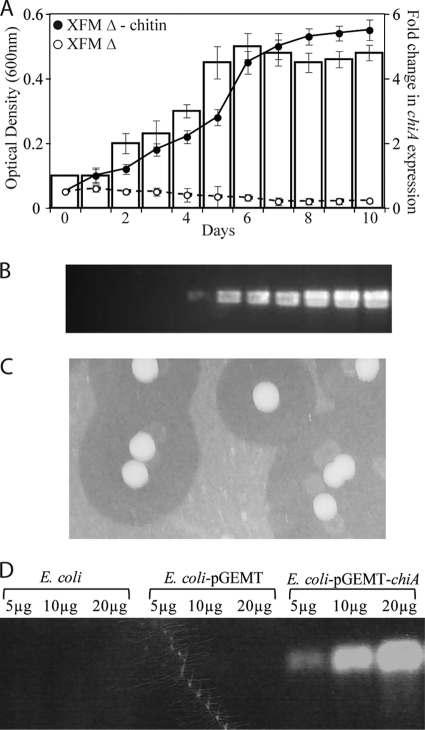
X. fastidiosa cells only grew in XFM Δ when supplemented with colloidal chitin as a single carbon source (XFM Δ-chitin) (Fig. 2 A). For X. fastidiosa growing in XFM Δ-chitin, samples were collected daily and chiA expression increased over time (Fig. 2A). The cell-free suspension was used to detect chitinolytic activity using the fluorescent-labeled substrate 4-MU(GlcNAc)3; an increasing gradient of enzyme activity was detected in the cell-free suspension, indicating the secretion of ChiA (Fig. 2B). Additionally, clearing zones around X. fastidiosa colonies grown on chitin-yeast extract-agar medium plates were observed, indicating chitin hydrolysis by a secreted enzyme (Fig. 2C).
FIG. 2.
Chitin utilization by Xylella fastidiosa. (A) Bacterial growth curve in XFM medium without carbon sources (XFM Δ) or with chitin added as the only carbon source (XFM Δ-chitin). Bars (second y axis) show the progression of chiA expression over time. (B) In-gel chitinase activity [4-MU(GlcNAc)3 cleavage] over time for X. fastidiosa culture filtrates in liquid XFM Δ-chitin, demonstrating ChiA secretion; (C) X. fastidiosa colonies grown on chitin-agar medium. The clear zones around colonies indicate chitin degradation. (D) In-gel chitinase activity (4-MU(GlcNAc)3 cleavage) for Escherichia coli strains grown in LB medium.
Chitinolytic activity of E. coli expressing chiA.
In order to confirm that the predicted ChiA is an active chitinase, E. coli was transformed with chiA. The E. coli transformants carrying recombinant chiA grew in LB supplied with colloidal chitin and were active in the hydrolysis of 4-MU(GlcNAc)3, while E. coli JM109 and the transformant carrying the vector alone did not hydrolyze the substrate (Fig. 2D).
X. fastidiosa forms a biofilm on chitinous surfaces: chiA is regulated by DSF.
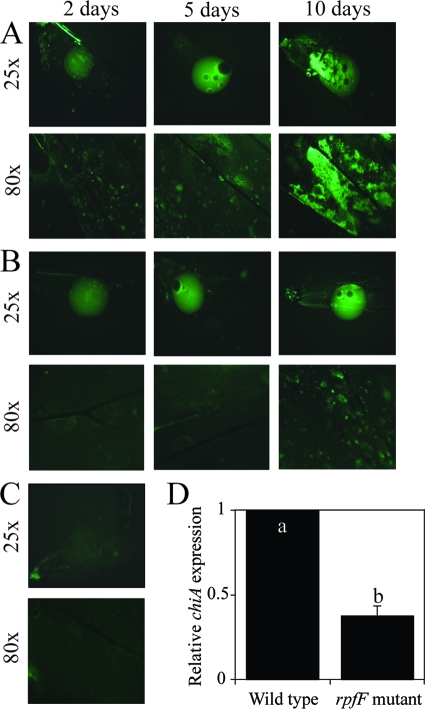
GFP-labeled cells were suspended in XFM Δ, and drops of suspension were loaded onto wings: cell growth was inferred based on the presence of fluorescence on the wings (Fig. 3). Growth was observed in both the wild-type (Fig. 3A) and rfpF mutant (Fig. 3B) treatments, while only minor background fluorescence was observed in the no-cell control (Fig. 3C). These results corroborate other observations indicating that X. fastidiosa cells multiply in and colonize a chitin-dominated environment. However, the rpfF mutant was affected in colonization (drop; magnification, ×25) and biofilm formation (after wing wash; magnification, ×80). This mutant was deficient in adhesion to surfaces and vectors (23, 26), which may have impaired its utilization of this substrate. In addition, expression of chiA in the rpfF mutant was only 40% of that of the wild type (Fig. 3D). The decrease was significant, considering the role of chitin in X. fastidiosa's adhesion and gene expression and the fact that the presence of chitin in the environment did not affect rpfF transcription (1.2-fold in relation to control) (Fig. 1D).
FIG. 3.
Xylella fastidiosa growth and biofilm formation on hindwings of leafhopper vectors. Cells were suspended in XFM Δ, and drops were placed on wings. Wild-type (A) and cell-cell signaling rpfF mutant (B) cells were incubated up to 10 days on wings. (C) Control medium XFM Δ without cells. The upper pictures were taken at ×25 magnification and are suspension droplets; the lower pictures were taken at ×80 magnification after removing the drop of medium and rinsing the wings with water. (D) chiA expression in the cell-cell signaling mutant rpfF compared to the wild type. Different letters on bars represent statistically different treatments.
DISCUSSION
Although X. fastidiosa has been shown to degrade plant polysaccharides such as pectin and glucan (32, 38), there is no direct evidence that those carbohydrates are used as carbon sources by this bacterium. A recent study showed that the addition of pectin to a simple defined medium did not increase X. fastidiosa's population size (22), but the medium used in that study included other carbon sources. On the other hand, within its vectors, it has been assumed that X. fastidiosa obtains nutrients from xylem sap flowing through the insect's mouthparts; the possibility of this bacterium utilizing chitin as a source of nutrients has not been discussed in the literature, despite the presence of glycoside hydrolases in its genome.
We found that X. fastidiosa grew to larger populations when chitin was added to the defined medium XFM. This finding indicates that chitin is being used as a nutrient source even though other organic acids were available, which was not the case when pectin and glucan were added to the same medium (22). However, similar to the effect of pectin on X. fastidiosa's phenotype, the presence of chitin in the environment resulted in more biofilm formation, suggesting that this polysaccharide induces changes in gene regulation in X. fastidiosa. Although the expression of rpfF was not affected by chitin, fimbrial and afimbrial adhesins were upregulated, while pilY1, which is associated with cell motility, was downregulated. This transcriptional pattern is associated with the cell adhesion phenotype observed and is similar to that induced by pectin and glucan (22). Chitinolytic activity increased in the presence of chitin in the environment. These results suggest that chitin or its by-products have a regulatory function in X. fastidiosa independent of the cell-cell signaling cascade controlled by DSF, although it is evident that the phenotypic and transcriptional changes observed here also match those under DSF control (6). Pectin had a similar effect on X. fastidiosa (22); thus, cell-cell signaling is likely an overarching regulatory system that regulates cell chitinolytic and pectinolytic activity, although both chitin and pectin seem to also have an important regulatory role in this pathogen.
A chitin-binding domain was not identified in chiA of X. fastidiosa. However, it has been shown that the removal of the chitin-binding domain from Clostridium paraputrificum's ChiB did not significantly affect hydrolytic activity when colloidal chitin was used as a substrate (25). Furthermore, a noncatalytic chitin-binding protein, CBP21, in Serratia marcescens was found to be essential for chitin degradation by ChiA and ChiC, and it was essential for the full degradation by ChiB (34). In fact, several fungal and bacterial chitinases have been shown to lack a binding domain (11, 24). Thus, it is possible that X. fastidiosa's ChiA does not possess a chitin-binding domain to facilitate hydrolytic activity. However, X. fastidiosa has noncatalytic chitin-binding proteins that may serve this role. Because ChiA is secreted by cells, afimbrial adhesins HxfA and HxfB (hemagglutinin-like proteins), which are important for chitin binding and cell adhesion to insect vectors (23), may have a fundamental role in chitin degradation by this pathogen. Moreover, a search for chitin-binding proteins in the cell membrane revealed the presence of several chitin-binding proteins in X. fastidiosa (unpublished data). It is also possible that ChiA has a chitin-binding domain with no sequence homology to other known proteins with that function. Further research is necessary to explore the role of chitin-binding proteins in the chitinolytic activity of X. fastidiosa.
In medium with chitin as the sole carbon source, XFM Δ-chitin, X. fastidiosa grew to similar optical density as in XFM-chitin, which has two other organic acids as carbon sources. Together with other evidence presented, chitin exploitation as a carbon source while colonizing insect vectors seems likely in X. fastidiosa. The media used included glutamine and asparagine, so that even if X. fastidiosa could incorporate NH3 into amino acids, thus using chitin's by-products as a nitrogen source, as predicted based on its genome, the experiments conducted here did not test that hypothesis. Because cells are directly attached to the insect's cuticle in a biofilm while colonizing vectors, it is unlikely that ChiA would be lost to the environment despite the fact it lacks a chitin-binding domain. Evidence for chitinolytic activity of X. fastidiosa in insects is limited (Fig. 4 B); one explanation is that studies looking at X. fastidiosa's colonization of vectors have focused on insects already fully colonized. In the former case, cells are in close contact with the cuticle of vectors and any evidence of chitinolytic activity would be covered by a biofilm of adhered cells (2).
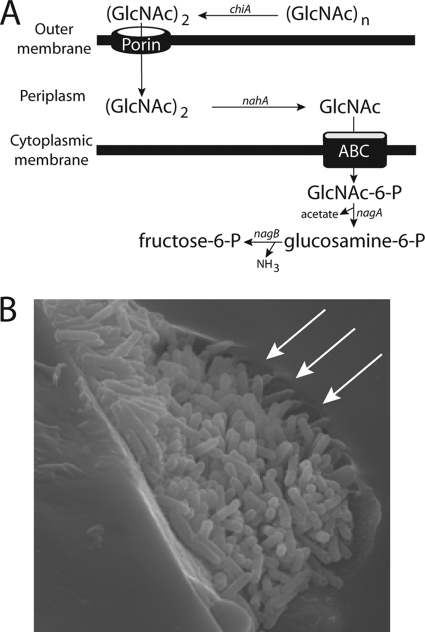
FIG. 4.
(A) Hypothetical model for chitin utilization in X. fastidiosa. Chitin is hydrolyzed to chitobiose outside the cell by chiA and passively transported into the periplasmic space as a dimer. nahA converts the substrate into N-acetylglucosamine, which is phosphorylated and transported into the cytoplasm via an ABC transporter. (B) Scanning electron microscopy micrograph of X. fastidiosa cells colonizing the mouthparts of a leafhopper vector. The arrows indicate potential degradation of the chitinous surface at the fringe of the microcolony. The picture is an unpublished image obtained in a previous study (2).
Chitin utilization is well studied in marine bacteria, and it includes several extracellular chitinases, noncatalytic chitin-binding proteins, chitodextrinases, and periplasmic and cytoplasmic N-acetylglucosaminidases and N-acetylhexosaminidases (20, 21, 33). Using these systems as models, it seems that X. fastidiosa also has a complete chitin-processing machinery consisting of several enzymes, in addition to noncatalytic chitin-binding proteins. In X. fastidiosa, chitin is hypothesized to be degraded into chitobiose extracellularly and transported into the periplasm via non-specific porins (Fig. 4A). In the periplasm, chitobiose would be hydrolyzed into N-acetylglucosamine, which would be phosphorylated and transported into the cytoplasm by sugar ABC transporters. Deacetylation and deamination of the substrate within cells result in fructose-6-phosphate, acetate, and NH3, which are linked to different metabolic pathways, as shown for other chitinolytic bacteria such as Vibrio spp. (18, 20).
Although the existence of a chitin-processing machinery in X. fastidiosa is not biologically surprising, given that it colonizes insects, the finding that it also present in the much-better-studied plant pathogen Xanthomonas was unexpected. Xanthomonas spp. are epiphytic bacteria that also colonize apoplastic space within leaf tissue and xylem vessels (6), but they are not known to require arthropod vectors for dispersal or to be associated with insects. Because the chiA locus is conserved in all Xanthomonas spp. and X. fastidiosa (single species in genus), and these two genera are monophyletic (29), this locus must have been shared by the common ancestor of these bacteria. However, X. fastidiosa seems to have exploited this metabolic pathway to diverge and colonize insects. Thus, it is possible that X. fastidiosa split from Xanthomonas after it started colonizing insects. We speculate that Xanthomonas uses ChiA to inhibit the growth of epiphytic fungi competing with cells for resources on the surface of leaves, although a knockout mutant is needed to understand its biological role.
In regard to X. fastidiosa transmission, N-acetylglucosamine and other sugars may act as competitors for cell binding sites within vectors, as shown by the lower X. fastidiosa attachment to insect wings in suspensions supplemented with chitin (23). It is possible that free N-acetylglucosamine released from chitin degradation acts as a molecular competitor to partially detach laterally bound cells from the cuticle of vectors (early stages of biofilm formation), resulting in polar attachment of cells, as observed in mature biofilms within leafhoppers (2). In these late stages, fimbrial proteins would contribute to cell adhesion to cuticle. The role of ChiA and chitin in X. fastidiosa's biology should be investigated in detail given the potential relevance to the pathogen's colonization of insect vectors. In vitro and in vivo studies using chiA and other mutants will also be necessary to dissect its role in this system.
ADDENDUM IN PROOF
While this manuscript was under revision, work by Boulanger et al. (J. Bacteriol. 192:1487-1497, 2010) describing the GlcNAc utilization pathway in Xanthomonas campestris pv. campestris was published. Their research suggests that transportation of GlcNAc through the inner membrane in Xylella fastidiosa may not occur via ABC transporters but rather through an MFS transporter.
Supplementary Material
Acknowledgments
We thank Steve Lindow, Sandy Purcell, and our lab colleagues for helpful discussions and suggestions. We acknowledge and thank Johan Leveau for providing us with the chitin-yeast extract-agar plates used here.
This work was supported with funding from the California Department of Food and Agriculture (PDRP).
Footnotes
Published ahead of print on 23 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Almeida, R. P. P., M. J. Blua, J. R. S. Lopes, and A. H. Purcell. 2005. Vector transmission of Xylella fastidiosa: applying fundamental knowledge to generate disease management strategies. Ann. Entomol. Soc. Am. 98:775-786. [Google Scholar]
- 2.Almeida, R. P. P., and A. H. Purcell. 2006. Patterns of Xylella fastidiosa colonization on the precibarium of sharpshooter vectors relative to transmission to plants. Ann. Entomol. Soc. Am. 99:884-890. [Google Scholar]
- 3.Anderson, S. O. 1979. Biochemistry of insect cuticle. Annu. Rev. Entomol. 24:29-59. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Broekaert, W. F., J. V. Parijs, A. K. Allen, and W. J. Peumans. 1988. Comparison of some molecular, enzymatic and antifungal properties of chitinase from thorn-apple, tobacco and wheat. Physiol. Mol. Plant Pathol. 33:319-331. [Google Scholar]
- 6.Chatterjee, S., R. P. P. Almeida, and S. E. Lindow. 2008. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 46:243-271. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, S., K. L. Newman, and S. E. Lindow. 2008. Cell-to-cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol. Plant Microbe Interact. 21:1309-1315. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, S., C. Wistrom, and S. E. Lindow. 2008. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc. Natl. Acad. Sci. U. S. A. 105:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinge, D. B., K. M. Kragh, J. D. Mikkelsen, K. K. Nielsen, U. Rasmussen, and K. Vad. 1993. Plant chitinases. Plant J. 3:31-40. [DOI] [PubMed] [Google Scholar]
- 10.de Boer, W., J. H. J. Leveau, G. A. Kowalchuk, P. J. A. K. Gunnewiek, E. C. A. Abeln, M. J. Figge, K. Sjollema, J. D. Janse, and J. A. van Veen. 2004. Collimonas fungivorans gen. nov., sp. nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int. J. Syst. Evol. Microbiol. 54:857-864. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, I., J. M. Lora, J. De la Cruz, T. Benitez, A. Llobell, and J. A. Pintor-Toro. 1994. Cloning and characterization of a chitinase (CHIT42) cDNA from the mycoparasitic fungus Trichoderma harzianum. Curr. Genet. 27:83-89. [DOI] [PubMed] [Google Scholar]
- 12.Gooday, G. W. 1990. The ecology of chitin degradation. Adv. Microb. Ecol. 11:387-430. [Google Scholar]
- 13.Haran, S., H. Schickler, A. Oppenheim, and I. Chet. 1995. New components of the chitinolytic system of Trichoderma harzianum. Mycol. Res. 99:441-446. [Google Scholar]
- 14.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill, B. L., and A. H. Purcell. 1995. Multiplication and movement of Xylella fastidiosa within grapevine and four other plants. Phytopathology 85:1368-1372. [Google Scholar]
- 17.Hoell, I. A., S. S. Klemsdal, G. Vaaje-Kolstad, S. J. Horn, and V. G. Eijsink. 2005. Overexpression and characterization of a novel chitinase from Trichoderma atroviride strain P1. Biochim. Biophys. Acta 1748:180-190. [DOI] [PubMed] [Google Scholar]
- 18.Hunt, D. E., D. Gevers, N. M. Vahora, and M. F. Polz. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 74:44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson, M., and J. Stenlid. 2009. Evolution of family 18 glycoside hydrolases: diversity, domain structures and phylogenetic relationships. J. Mol. Microbiol. Biotechnol. 16:208-223. [DOI] [PubMed] [Google Scholar]
- 20.Keyhani, N. O., and S. Roseman. 1999. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473:1008-1022. [DOI] [PubMed] [Google Scholar]
- 21.Keyhani, N. O., and S. Roseman. 1996. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation, and characterization of a periplasmic beta-N-acetylglucosaminidase. J. Biol. Chem. 271:33425-33432. [DOI] [PubMed] [Google Scholar]
- 22.Killiny, N., and R. P. P. Almeida. 2009. Host structural carbohydrate induces vector transmission of a bacterial plant pathogen. Proc. Natl. Acad. Sci. U. S. A. 106:22416-22420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killiny, N., and R. P. P. Almeida. 2009. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl. Environ. Microbiol. 75:521-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Limon, M. C., J. A. Pintor-Toro, and T. Benitez. 1999. Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology 89:254-261. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto, K., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 1997. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J. Bacteriol. 179:7306-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman, K. L., R. P. P. Almeida, A. H. Purcell, and S. E. Lindow. 2004. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. U. S. A. 101:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman, K. L., R. P. P. Almeida, A. H. Purcell, and S. E. Lindow. 2003. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl. Environ. Microbiol. 69:7319-7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohno, T., S. Armand, T. Hata, N. Nikaidou, B. Henrissat, M. Mitsutomi, and T. Watanabe. 1996. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J. Bacteriol. 178:5065-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pieretti, I., M. Royer, V. Barbe, S. Carrere, R. Koebnik, S. Cociancich, A. Couloux, A. Darrasse, J. Gouzy, M. A. Jacques, E. Lauber, C. Manceau, S. Mangenot, S. Poussier, B. Segurens, B. Szurek, V. Verdier, M. Arlat, and P. Rott. 2009. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell, A. H., and A. Finlay. 1979. Evidence for non-circulative transmission of Pierce's disease bacterium by sharpshooter leafhoppers. Phytopathology 69:393-395. [Google Scholar]
- 31.Ramírez, M. G., L. I. R. Avelizapa, N. G. R. Avelizapa, and R. C. Camarillo. 2004. Colloidal chitin stained with Remazol Brilliant Blue R, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J. Microbiol. Methods 56:213-219. [DOI] [PubMed] [Google Scholar]
- 32.Roper, M. C., L. C. Greve, J. G. Warren, J. M. Labavitch, and B. C. Kirkpatrick. 2007. Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol. Plant Microbe Interact. 20:411-419. [DOI] [PubMed] [Google Scholar]
- 33.Techkarnjanaruk, S., and A. E. Goodman. 1999. Multiple genes involved in chitin degradation from the marine bacterium Pseudoalteromonas sp. strain S91. Microbiology 145:925-934. [DOI] [PubMed] [Google Scholar]
- 34.Vaaje-Kolstad, G., S. J. Horn, D. M. van Aalten, B. Synstad, and V. G. Eijsink. 2005. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280:28492-28497. [DOI] [PubMed] [Google Scholar]
- 35.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, L. E. A. Camargo, A. C. R. da Silva, D. H. Moon, M. A. Takita, E. G. M. Lemos, M. A. Machado, M. I. T. Ferro, F. R. da Silva, M. H. S. Goldman, G. H. Goldman, M. V. F. Lemos, H. El-Dorry, S. M. Tsai, H. Carrer, D. M. Carraro, R. C. de Oliveira, L. R. Nunes, W. J. Siqueira, L. L. Coutinho, E. T. Kimura, E. S. Ferro, R. Harakava, E. E. Kuramae, C. L. Marino, E. Giglioti, I. L. Abreu, L. M. C. Alves, A. M. do Amaral, G. S. Baia, S. R. Blanco, M. S. Brito, F. S. Cannavan, A. V. Celestino, A. F. da Cunha, R. C. Fenille, J. A. Ferro, E. F. Formighieri, L. T. Kishi, S. G. Leoni, A. R. Oliveira, V. E. Rosa, F. T. Sassaki, J. A. D. Sena, A. A. de Souza, D. Truffi, F. Tsukumo, G. M. Yanai, L. G. Zaros, E. L. Civerolo, A. J. G. Simpson, N. F. Almeida, J. C. Setubal, and J. P. Kitajima. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe, T., K. Kobori, K. Miyashita, T. Fujii, H. Sakai, M. Uchida, and H. Tanaka. 1993. Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J. Biol. Chem. 268:18567-18572. [PubMed] [Google Scholar]
- 37.Watanabe, T., W. Oyanagi, K. Suzuki, K. Ohnishi, and H. Tanaka. 1992. Structure of the gene encoding chitinase D of Bacillus circulans WL-12 and possible homology of the enzyme to other prokaryotic chitinases and class III plant chitinases. J. Bacteriol. 174:408-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wulff, N. A., H. Carrer, and S. F. Pascholati. 2006. Expression and purification of cellulase Xf818 from Xylella fastidiosa in Escherichia coli. Curr. Microbiol. 53:198-203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.