Abstract
Histamine, a toxic compound that is formed by the decarboxylation of histidine through the action of microbial decarboxylases, can accumulate in fermented food products. From a total of 69 Streptococcus thermophilus strains screened, two strains, CHCC1524 and CHCC6483, showed the capacity to produce histamine. The hdc clusters of S. thermophilus CHCC1524 and CHCC6483 were sequenced, and the factors that affect histamine biosynthesis and histidine-decarboxylating gene (hdcA) expression were studied. The hdc cluster began with the hdcA gene, was followed by a transporter (hdcP), and ended with the hdcB gene, which is of unknown function. The three genes were orientated in the same direction. The genetic organization of the hdc cluster showed a unique organization among the lactic acid bacterial group and resembled those of Staphylococcus and Clostridium species, thus indicating possible acquisition through a horizontal transfer mechanism. Transcriptional analysis of the hdc cluster revealed the existence of a polycistronic mRNA covering the three genes. The histidine-decarboxylating gene (hdcA) of S. thermophilus demonstrated maximum expression during the stationary growth phase, with high expression levels correlated with high histamine levels. Limited expression was evident during the lag and exponential growth phases. Low-temperature (4°C) incubation of milk inoculated with a histamine-producing strain showed lower levels of histamine than did inoculated milk kept at 42°C. This reduction was attributed to a reduction in the activity of the HdcA enzyme itself rather than a reduction in gene expression or the presence of a lower cell number.
Lactic acid bacteria (LAB) play an essential role in fermented dairy products, with Lactococcus lactis, Streptococcus thermophilus, and Lactobacillus helveticus frequently being used as starter cultures. In particular, S. thermophilus is a very important species, and it is used for the production of fermented milks such as yogurt and for the production of certain cheeses such as mozzarella and Emmental (11). The major role of LAB is to prevent the growth of spoilage organisms and pathogens through rapid acidification and/or production of inhibitory compounds. Additionally, LAB contribute to the organoleptic characteristics of products, and some strains may be probiotic and help to maintain a positive health status in consumers. Nevertheless, the metabolic activity of some LAB strains can produce undesirable or toxic compounds such as biogenic amines (BAs).
BAs are low-molecular-weight nitrogenous compounds present in most living organisms, for which different biological activities have been reported (31). Physiologically, the most important BA is histamine, which is a vasoactive and psychoactive compound (37). Although it has important roles in human metabolism, the intake of food with high concentrations of histamine is associated with a range of toxicological effects (37). These effects can be particularly severe in individuals who, for genetic or pharmacological reasons, are deficient in diamine oxidase, the histamine-degrading enzyme present in the epithelial cells of the intestine (1). In fact, histamine poisoning is the most common food-borne intoxication caused by BAs, due to the fact that it is the most abundant BA found in food products and, in particular, in foods such as fish or fermented dairy products.
At nontoxic doses, the food-borne intake of histamine can cause intolerance symptoms such as diarrhea, hypotension, headache, pruritus, and flushes (24, 38). A level of just 75 mg of histamine, a quantity commonly present in some meals (32), can induce these symptoms in the majority of healthy persons with no history of histamine intolerance (46). Histamine levels are regulated by law for fish and wine; however, for the majority of foods, including fermented dairy products, no regulation exists. Nevertheless, there is a general consensus that high levels of histamine in fermented dairy products are undesirable.
In fermented dairy products, histamine is produced mainly by the decarboxylating activity of certain LAB. These strains may be present in the milk or processing equipment and may thus contaminate dairy products during manufacture (2, 17, 27). The growth of histamine-producing microbiota and the subsequent accumulation of histamine are dependent on environmental (pH, temperature, and availability of substrates, etc.) and technological (pasteurization, proteolysis, and ripening, etc.) factors (8, 13, 28, 43).
The LAB histidine decarboxylases belong to a group of decarboxylases that use a covalently bound pyruvoyl moiety as a prosthetic group. The encoding gene (hdcA) has been identified in several LAB, such as Oenococcus oeni (5), Lactobacillus saerimneri 30a (44), Lactobacillus hilgardii (23), Tetragenococcus halophilus (35), Lactobacillus reuteri JCM1112 and DSM20016 (GenBank accession no. NC010609 and NC_009513, respectively), and Lactobacillus buchneri (25). The hdcA gene in LAB has been shown to be organized into a cluster usually preceded by a transporter (hdcP) and followed by a gene (hdcB) of unknown function (see Fig. S1 in the supplemental material). For lactobacilli, a fourth gene, encoding a histidyl-tRNA synthetase (hisS), has also been described (Fig. S1).
The presence of the hdc gene cluster in bacteria has been shown to improve the growth capacity under conditions of low pH or energy source limitation. In fact, the presence of sugar and organic acids reduces the expression of the hdc gene, while factors such as the availability of histidine, low pH, or ethanol activate it (19). These environmental factors are present in most food fermentations. Therefore, genetic and environmental factors related to histamine biosynthesis in food products and the identification of histamine-producing bacteria merit further study (33).
The present work reports the detection of histamine-producing S. thermophilus strains. Since this species is one of the most important thermophilic starters used for the production of yogurt and many types of cheese, a detailed study has been conducted. The histidine-decarboxylating cluster has been sequenced, and the factors that affect the expression of hdcA and histamine biosynthesis have been investigated.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. thermophilus CHCC1524 and CHCC6483, from the Christian Hansen Culture Collection (Hørsholm, Denmark), were grown at 37°C without aeration in M17 medium (Oxoid, Hampshire, United Kingdom) supplemented with 2 g liter−1 lactose and containing 2 g liter−1 histidine (M17+H).
In order to study the conditions that affect the expression of hdcA, S. thermophilus strain CHCC1524 was cultivated in a Six-Fors bioreactor (Infors AG, Bottmingen, Switzerland) containing 300 ml of chemically defined medium (CDM) (30) supplemented with lactose as a carbon source and with or without histidine at different concentrations. A 1% inoculum was used to inoculate the batch culture. The reactor was maintained at 37°C at 50 rpm and with zero air input. When needed to maintain a fixed pH (6.8 or 5.4), 2 N NaOH was added automatically.
To measure hdcA expression under technological conditions, the strain was grown in skim milk (Oxoid, Hampshire, United Kingdom) at 42°C with or without histidine. The growth in broth was monitored by measuring the optical density at 600 nm (OD600) with a spectrophotometer (Eppendorf, NY), and when indicated, the number of viable cells was calculated by serial dilution on M17 plates.
Screening for histamine-producing S. thermophilus strains.
A 96-well microtiter plate (MTP) screening approach was used in order to screen a total of 69 different S. thermophilus strains. Stock cultures from the Christian Hansen Culture Collection were stored at −80°C in Matrix vials (Thermo Fisher Scientific, NH). For inoculation, the Matrix vials were thawed, and 2 μl was used to inoculate 200 μl of M17 broth (in MTPs) containing 5 g liter−1 glucose and 50 mg liter−1 pyridoxal-5-phosphate, supplemented with 1 g liter−1 histidine, and incubated anaerobically at 37°C for 1 day. In order to ensure the induction of histamine production (if any), the cultures were successively propagated under the same conditions for a total of five times over five consecutive days. On the fifth day, the propagation was scaled up from 200 μl to 1,600 μl (in deep-well MTPs) in order to obtain enough sample volume for subsequent high-performance liquid chromatography (HPLC) analysis. Following the fifth propagation, the cultures were centrifuged at 2,250 × g for 10 min, and 1 ml of supernatant was collected, transferred into alphanumeric Matrix vials, and capped with SepraSeal septa (Thermo Fisher Scientific, NH). Frozen samples were sent to CCFRA Technology Limited (Chipping Campden, Gloucestershire, United Kingdom) for histamine determination by HPLC according to a method previously described (12).
DNA manipulation.
Total DNA of S. thermophilus was isolated according to a method described previously by Hopwood et al. (10). All enzymes for DNA manipulations were used according to the manufacturer's instructions. Restriction enzymes were supplied by Takara (Otsu, Japan). Oligonucleotides used in this work are listed in Table 1 and were synthesized by Sigma Genosys or MWG Biotech (Ebersberg, Germany). PCR amplification of DNA was performed with a DNA thermal cycler (iCycler; Bio-Rad, Hercules, CA). PCR products were purified from an agarose gel by using the QIAquick gel extraction kit (Qiagen, Crawley, United Kingdom). DNA was separated in a 1.5% agarose gel in TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]) and visualized after ethidium bromide staining under UV light.
TABLE 1.
Oligonucleotides used in this study
| Primer | Functiona | Sequence (5′-3′) | Reference |
|---|---|---|---|
| JV16HC | hdcA internal fragment amplification (F) | AGATGGTATTGTTTCTTATG | 20 |
| JV17HC | hdcA internal fragment amplification (R) | AGACCATACACCATAACCTT | 20 |
| Northern blotting | |||
| CHP5 | hdcA probe amplification (F) | CCTGGGAACATCGGGACT | This work |
| CHP1 | hdcA probe amplification (R) | AAATAAGGGCCGACGGTGATG | This work |
| CHP2R | hdcP probe amplification (F) | GAATACGGTTGGCGTTTTGG | This work |
| CHP3R | hdcP probe amplification (R) | CGGTAGTCAAAGCTCGTTTGC | This work |
| CHP4R | hdcB probe amplification (F) | TCATGCTCCTAACGGTGAAGGT | This work |
| CHP6R | hdcB probe amplification (R) | TGGGTGATAATCATCGTATTCCA | This work |
| RT-PCR | |||
| prim1 | hdcA amplification (F) | CCTGGGAACATCGGGACTG | This work |
| prim2 | hdcA amplification (R) | CGCATCTAACCAATCAGGCAG | This work |
| prim3 | hdcP amplification (F) | GAATACGGTTGGGCTTTTGG | This work |
| prim4 | hdcP amplification (R) | CGGTAGTCAAAGCTCGTTTGC | This work |
| prim5 | hdcB amplification (F) | TCATGCTCCTAACGGTGAAGGT | This work |
| prim6 | hdcB amplification (R) | TGGGTGATAATCATCGTATTCCA | This work |
| RT-qPCR | |||
| ST tufF | Normalization | GGTGCGATCCTTGTAGTTGCA | 3 |
| ST tufR | Normalization | ACACCAACCTGACGTGAAAGAA | 3 |
| hdcAQF | hdcA expression analysis (F) | TGCTAACAAAGGTGTGACTGCAT | This work |
| hdcAQR | hdcA expression analysis (R) | TTCACCGGCTTGGAAAGGT | This work |
F, forward; R, reverse.
The primer pair JV16HC and JV17HC (Table 1) was used to PCR amplify a 369-bp fragment within the hdcA gene of S. thermophilus CHCC1524. This primer pair was used previously to amplify an approximately 370-bp fragment within the hdcA gene from histamine-producing L. buchneri, L. saerimneri 30a, Clostridium perfringens, and O. oeni strains (20). Subsequently, the amplified 369-bp fragment within the hdcA gene of S. thermophilus CHCC1524 was sequenced and used as a basis for further primer design for a progressive “gene-walking” sequencing approach. A TOPO Walker kit (Invitrogen A/S, Taastrup, Denmark) was thus used to sequence the entire hdc cluster of S. thermophilus CHCC1524 and the surrounding regions. S. thermophilus CHCC1524 total DNA was digested with EcoRI, BglII, PstI, and AatII and used as a template according to the manufacturer's instructions. PCR rendered suitable bands for further cloning and sequencing. The various PCR fragments generated were sequenced by MWG Biotech (Ebersberg, Germany) and assembled by using the Vector NTI Advance program, version 9.1 (Invitrogen, Taastrup, Denmark). Sequence analysis was performed by using the University of Wisconsin Genetics Computer Group software package (7). The sequence of S. thermophilus CHCC1524 was then used to design new primers in order to amplify and sequence the hdc cluster of the second strain, S. thermophilus CHCC6483. BLAST Suite was used to determine the similarities of the deduced amino acid sequences with those present in the databases. Analysis of transmembrane elements was performed with the TMHMM tool (Danish Technical University, Denmark [http://www.cbs.dtu.dk/services/TMHMM/]).
RNA extraction.
Total RNA was extracted by using Tri reagent according to the manufacturer's instructions (Sigma, Dorset, United Kingdom). S. thermophilus cells were grown in M17+H medium or in CDM for 24 h, except if other time points were indicated, until the stationary phase of growth was reached (OD600 of 4.5) and were harvested by centrifugation. The cells were disrupted with 50-μm-diameter glass beads (Sigma, Dorset, United Kingdom) shaken three times for 1 min at high speed in a bead beater (FastPrep-24 system; MP Biomedicals, Illkirch, France). Samples were kept on ice for 1 min between the shaking intervals. Disrupted samples were then treated according to the manufacturer's instructions (Sigma, Dorset, United Kingdom). Total RNA was obtained from the milk matrix according to a protocol described previously by Calles-Enríquez et al. (3).
Purified RNA was resuspended in RNase-free water (Sigma, Dorset, United Kingdom). The RNA samples were checked for yield and quality by measuring the OD ratio at 260 and 280 nm by using a BioPhotometer (Eppendorf, NY).
Northern blotting.
Total RNA was prepared from S. thermophilus CHCC1524 cells that were grown in M17+H medium and subjected to electrophoresis through a 1.5% agarose gel containing 5% formaldehyde and 1× 3-N-morpholino-propanesulfonic acid (MOPS) buffer (20 mM MOPS, 1 mM EDTA, 5 mM sodium acetate [pH 7.0]).
Twenty micrograms of each sample was mixed with loading buffer (50% deionized formamide, 10% MOPS buffer, 17.5% formaldehyde, 10% glycerol, 0.5% bromophenol blue), denatured at 55°C for 15 min, and finally loaded into the gel together with 0.2 volumes of ethidium bromide (1 mg ml−1).
Transfers and hybridizations were performed according to standard protocols (34). DNA probes were radiolabeled by nick translation, incorporating the [α-32P]dATP nucleotide (Perkin-Elmer, Covina, CA), and the signal was obtained with a Typhoon Trio instrument (GE Healthcare, Piscataway, NJ) after exposure to a storage phosphorscreen.
RT-PCR.
RNA samples (2 μg of total RNA) were treated with 2 U of DNase I (Fermentas, Vilnius, Lithuania) to eliminate any DNA contamination from the sample. cDNA was then synthesized from total RNA by using the Omniscript reverse transcription (RT) kit (Qiagen, Crawley, United Kingdom). Reactions were performed by using 2 μl of the cDNA suspension and 0.4 μM each primer (Table 1). After the reverse transcription reaction, amplifications were performed for 35 cycles (94°C for 30 s, 55°C for 45 s, and 72°C for 1 min), and samples were analyzed on a 1.5% agarose gel in TAE buffer. The absence of contaminating DNA was checked with non-reverse-transcribed PCR, which was performed under the same conditions as those described above but without the addition of reverse transcriptase to the reaction mixture.
Gene expression quantification by RT-qPCR.
Gene expression analysis was carried out by reverse transcription-quantitative real-time PCR (RT-qPCR), as described previously (6), by using SYBR green PCR master mix (Applied Biosystems, Carlsbad, CA), which included ROX (6-carboxy-X-rhodamine) as a passive reference dye, on a 7500 Fast real-time PCR system (Applied Biosystems, Carlsbad, CA).
After the dilution of cDNA (as described above), 5 μl was added to 20 μl of a PCR mixture (12.5 μl of SYBR green supermix, 1 μl of each primer at 7 μM, 5.5 μl of RNase-free water). Four different dilutions of cDNA were included. Specific cDNAs were amplified by real-time PCR with specific primers (Table 1) designed to produce amplicons of equivalent lengths (approximately 100 bp) and selected by using Primer Express software (Applied Biosystems, Carlsbad, CA) using the tuf gene as an internal control to normalize the RNA concentration. Calibration curves were prepared for each of the pairs of primers by using 10-fold serial dilutions of total S. thermophilus CHCC1524 DNA. The standard curves showed a linear relationship of log input DNA versus the threshold cycle (CT). The slopes of the curves and the regression coefficients (R2) were very acceptable and similar for both reactions (hdcA and tuf).
For each run, a negative control with all the reaction components except cDNA was included. A positive control with total S. thermophilus CHCC1524 DNA was also included, and the resulting melting curve was compared with that of PCR products. The amplification was performed by using the default cycling settings established by Applied Biosystems (Carlsbad, CA).
The abundance of mRNA species was calculated as previously described (22). For each experiment, the condition with the lower level of expression was selected as the calibrator. RT-qPCR analysis was performed on RNA purified from three independently grown cultures in each environment.
Histamine quantification.
Quantitative analysis of histamine production was undertaken by reverse-phase HPLC (RP-HPLC) by using a Waters liquid chromatograph controlled by Millennium 32 software (Waters, Milford, MA). The strain was grown in M17 medium or skim milk supplemented with histidine (12.8 mM) for 8 h or 24 h. The cultures were centrifuged at 8,000 × g for 10 min, and the resulting supernatants were filtered through a 0.2-μm Supor membrane (Pall, NY). The resulting samples were derivatized by using dabsyl chloride. Separations were performed by using a Waters Nova-Pack C18 column. The gradient and detection conditions were similar to those described previously by Krause et al. (15).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper are available in the GenBank database (http://www.ncbi.nlm.nih.gov) under accession numbers FN686789 and FN686790 for strains CHCC1524 and CHCC6483, respectively.
RESULTS
Screening for histamine-producing S. thermophilus strains.
From a total of 69 S. thermophilus strains screened, just two strains, CHCC1524 and CHCC6483, were capable of producing histamine from the decarboxylation of histidine. Levels of histamine produced were 6 and 3 mM for CHCC1524 and CHCC6483, respectively. This is the first report of strains of S. thermophilus being able to produce histamine, and this characteristic of S. thermophilus, as in the case for other species of LAB, appears to be highly strain specific.
Sequence analysis and organization of the hdc cluster from S. thermophilus.
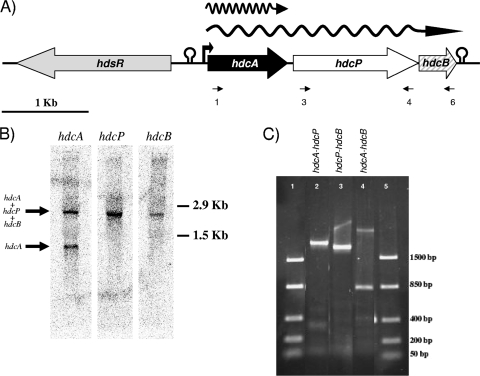
PCR using the oligonucleotides JV16HC and JV17HC (20) was performed by using genomic DNA from S. thermophilus CHCC1524 as a template. A fragment of the expected size was obtained and subsequently sequenced. The 369-bp sequence obtained revealed homology with other hdcA genes from Gram-positive bacteria mentioned previously (data not shown). This PCR fragment was used to design primers for the sequencing of the flanking DNA region. A 7,053-bp DNA fragment was sequenced (GenBank accession no. FN686789) and revealed the presence of four open reading frames (ORFs) (Fig. 1A).
FIG. 1.
Genetic organization and transcriptional analysis of the hdc operon. (A) Organization of the hdc cluster of S. thermophilus CHCC1524. Possible promoters and termination regions are indicated. RT-PCR primers are indicated. (B) Northern blot analysis of the hdc cluster. Internal fragments of hdcA, hdcP, and hdcB were used as probes. (C) RT-PCR amplification with primers designed to amplify the intergenic regions of the genes (hdcA-hdcP, prim1 plus prim4 [lane 2]; hdcP-hdcB, prim3 plus prim6 [lane 3]; hdcA-hdcB, prim1 plus prim6 [lane 4]). Lanes 1 and 5, molecular weight markers (FastRuler low-range DNA ladder; Fermentas, Germany).
(i) hdsR.
The hdsR gene had homology with the R subunit of a type I restriction-modification enzyme that was previously identified in the chromosome of S. thermophilus 4134. The gene is closely associated with a transposase and a cadmium resistance cluster, indicating that this gene is not involved in the decarboxylation of histidine (36).
(ii) hdcA.
Analysis and comparison of the deduced amino acid sequence of hdcA showed that the protein belonged to the group of bacterial histidine decarboxylases that use a covalently bound pyruvoyl residue as a prosthetic group. It showed the highest identity (67%) with the histidine decarboxylase of L. saerimneri 30a (Swiss-Prot code DCHS_LACS3; EMBL accession no. J02613).
(iii) hdcP.
The hdcP gene had homology with several amino acid/biogenic amine antiporter sequences from GenBank. Analyses of HdcP using the TMHMM tool predicted that HdcP could be a transmembrane protein with 13 transmembrane elements.
(iv) hdcB.
A comparison of the deduced amino acid sequence of hdcB with those present in GenBank revealed similarities with HdcB proteins of other LAB (see Fig. S1 in the supplemental material). Although some authors associated this protein with the regulation of the operon or the transport of histidine and/or histamine (20, 45), its exact function remains unknown.
The sequence (GenBank accession no. FN686790) from the second strain, CHCC6483, had 99.2% identity at the DNA level to that of CHCC1524. More than half of the differences were located in the intergenic region upstream of the hdc cluster. Within the coding region, point changes were distributed along the sequence (three in hdcA, eight in hdcP, and two in hdcB), with only five of the differences resulting in a change in the amino acid sequence of the encoded proteins (four in HdcP and one in HdcB).
The organization of the S. thermophilus hdc cluster (Fig. 1A) is similar to those observed for Staphylococcus epidermidis, Staphylococcus capitis, and C. perfringens, although in these clusters the hdcB gene is absent and differs from the hdc clusters described for LAB strains such as L. reuteri, L. buchneri, L. hilgardii, L. saerimneri, and Tetragenococcus muriaticus (see Fig. S1 in the supplemental material). The histamine-producing cluster was chromosomally located, since no plasmids were visualized after DNA extraction (total or plasmidic protocols), and a hybridization band using an internal hdcA probe was evident in a pulsed-field gel electrophoresis (PGFE) gel of undigested DNA at the location of the chromosome in the gel (data not shown).
Transcriptional analysis of the hdc cluster.
RNA isolated from S. thermophilus CHCC1524 cells grown in M17 medium supplemented with 12.8 mM histidine was used for Northern blot analysis (Fig. 1B). Internal fragments of hdcA, hdcP, and hdcB were used independently as DNA probes. A hybridization band with the size of hdcA plus hdcP plus hdcB was obtained with the three probes. An additional band, the size of which corresponds to hdcA, was observed only in the case of the hdcA probe (Fig. 1A and B). To confirm these results, total RNA was used in RT-PCR experiments with three sets of primers designed to amplify the intergenic regions (Fig. 1C).
Factors affecting histamine production in S. thermophilus.
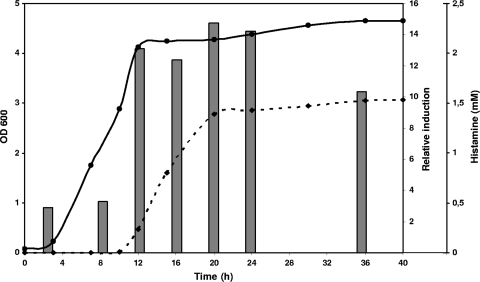
In order to study the conditions that affect histamine production in S. thermophilus CHCC1524 during growth, histidine decarboxylase gene (hdcA) expression and histamine accumulation were measured by RT-qPCR and RP-HPLC, respectively (Fig. 2). Initially, the influence of the growth phase was evaluated. The expression of hdcA was observed at low levels in the lag and exponential growth phases, but no histamine was detected in the medium. There was an increase in the histamine levels detected in the medium at the beginning of the stationary phase that correlated with higher relative levels of induction of the hdcA gene at the beginning of this phase (the value at 3 h was chosen as a calibrator). The expression levels remained elevated, and the concentration of histamine was constant until a maximum concentration of 1.5 mM was reached.
FIG. 2.
Effect of growth phase on the relative induction of hdcA expression and histamine quantity produced by S. thermophilus CHCC1524. Gene expression was measured by RT-qPCR (gray bars), the histamine concentration was measured by RP-HPLC (discontinuous line), and growth was measured by determining the OD600 (continuous line). The culture was grown in M17 broth with 12.8 mM histidine.
Based on these results, an incubation time of 24 h was selected for the analysis of hdcA expression under different experimental conditions.
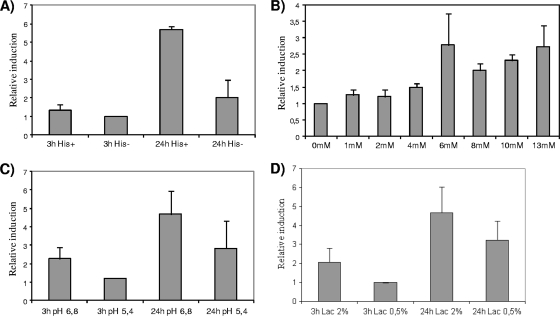
The amino acid substrate of the decarboxylation reaction was previously reported to enhance the expression of decarboxylating genes in several bacteria (4, 18, 21, 26). To test the influence of histidine on the expression of hdcA, S. thermophilus CHCC1524 was grown in CDM with (12.8 mM) or without histidine (Fig. 3A). An increase in the hdcA expression level was observed only in the stationary phase in the presence of histidine. In exponential-phase cultures, hdcA induction was not observed in either the absence or presence of histidine, indicating that under both conditions, the growth phase and the presence of the substrate are needed for its expression.
FIG. 3.
Effect of different factors on the expression level of hdcA from S. thermophilus CHCC1524 cells grown in M17 medium under different conditions measured by RT-qPCR. (A) Presence (12.8 mM) or absence of histidine at two different times, 3 and 24 h after inoculation. (B) Influence of histidine concentrations (0, 1, 2, 4, 6, 8, 10, and 13 mM) on hdcA expression at 24 h after inoculation on CDM. (C) Influence of acidic pH (6.8 versus 5.4) at two different times, 3 and 24 h after inoculation. (D) Influence of carbon source reduction on hdcA expression. Lactose was added at 20 g liter−1 (2%) or 5 g liter−1 (0.5%) to M17 broth, and samples were taken at two different times, 3 and 24 h after inoculation.
In order to quantify the strength of this induction, different histidine concentrations (0, 1, 2, 4, 6, 8, 10, 13, and 15 mM) were added to the medium, and hdcA expression was quantified after 24 h of incubation by RT-qPCR. As shown in Fig. 3B, high histidine concentrations enhanced hdcA expression, and a relatively high increase was evident at a concentration of 6 mM histidine.
Conditions in the stationary phase of LAB cultures often result in a depletion of the carbon source and low pHs. Moreover, acidic pHs are one of the key inductors in the activation of decarboxylase genes of other LAB (21). To test the influence of pH on the expression of hdcA in S. thermophilus, cells were grown in CDM at two different pH values (pH 6.8 and 5.4). There were statistically no differences in expression levels at either pH tested, with only a slightly higher level of expression at a nonacidic pH (Fig. 3C). The influence of the depletion of the carbon source on hdcA expression was tested by growing the cells at two concentrations of lactose (20 and 5 g liter−1) (Fig. 3D). Again, there were no major differences in expression, with the relative induction at the low concentration of the carbon source being only slightly lower than that observed at the high concentration.
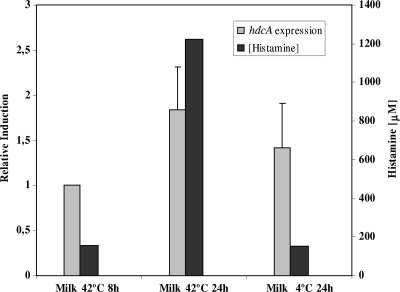
hdcA expression and histamine production under technological conditions.
The accumulation of BAs in dairy products occurs at later stages of the process, when the substrate amino acids are released from casein (9). In the case of yogurt, where S. thermophilus is one of the two starter species normally used, the process consists of a rapid acidification that is performed at 42°C, followed by refrigeration (4°C) until consumption. Low temperatures are one of the technological variables that have been shown to have an important effect in reducing the BA content of dairy products (41). hdcA gene expression was analyzed together with the quantification of the amount of histamine produced under these conditions. S. thermophilus CHCC1524 was inoculated in milk medium supplemented with histidine (12.8 mM) and incubated at 42°C for 8 h. Half of the culture was kept at 42°C for 24 h, while the rest was kept at 4°C for the same time period. The hdcA expression under both conditions was analyzed (Fig. 4). No major differences were observed, with only a slightly lower expression level at 4°C than at 42°C (Fig. 4). There were no differences in cell numbers either: 108 CFU ml−1 in both cases. However, a significant increase in the histamine concentration was observed at 42°C after 24 h. At this temperature the histamine concentration was 10 times higher than that in the culture kept at low temperatures (Fig. 4), thus confirming the importance of low-temperature storage in order to reduce BA accumulation in fermented dairy products such as yogurt.
FIG. 4.
Effect of technological conditions on the expression level of hdcA from S. thermophilus CHCC1524 grown in milk at 42°C for 8 h before a temperature shift (4°C) was performed. Sixteen hours later, the hdcA expression levels and the concentration of histamine were measured.
DISCUSSION
BA accumulation in food requires the presence of decarboxylating microorganisms. In fermented products, these BA-producing strains may be present in raw milk or processing equipment and may develop as a secondary flora during the fermentation and ripening processes (27). In this paper, the first reported histamine-producing strains of S. thermophilus, one of the most important starter culture species, have been characterized, and a detailed analysis of the hdc cluster was conducted.
hdc clusters have been reported to be located either on plasmids or on the chromosome. For L. hilgardii and T. halophilus strains, the hdc cluster is located on plasmids, whereas the hdc cluster for strains belonging to the genera Lactobacillus and Staphylococcus are located on the chromosome. In the S. thermophilus strains reported here, the cluster was located on the chromosome. Interestingly, the DNA sequence upstream of the hdc cluster in these S. thermophilus strains (CHCC1524 and CHCC6483) was homologous to that of a plasmid (pER35) from S. thermophilus 4134 involved in phage resistance (40). The same restriction-modification system (DNA identity over 99%) has also been described for some L. lactis strains, for which it has been reported to be either located on plasmids or incorporated into the chromosome itself. The hdsR gene was associated with a cadmium resistance gene cluster and flanked by transposase elements that have been associated with horizontal gene transfer of genetic material between S. thermophilus strains (29, 36). This indicates that the histidine decarboxylase cluster could be acquired by the same mechanism for S. thermophilus CCHC1524 and CHCC6483.
The genetic organization of the hdc cluster of S. thermophilus more closely resembled those of the genera Clostridium and Staphylococcus than those found for other LAB (see Fig. S1 in the supplemental material). However, in S. thermophilus, there is an hdcB gene that has been found only in histidine decarboxylation clusters of LAB. This chimeric organization also points to a horizontal transfer mechanism as the origin of the histidine decarboxylation cluster in S. thermophilus.
The transcriptional studies demonstrated that hdcA, hdcP, and hdcB constitute an operon transcribed as a polycistronic mRNA. However, an hdcA Northern probe evidenced an additional band that corresponds to a monocistronic transcript. Since the sequence analysis of the hdcA-hdcP intergenic region did not show transcriptional termination structures, it could be due to the posttranscriptional processing of the polycistronic mRNA. This result could reflect the need for higher levels of the histidine decarboxylase enzyme than for the other proteins involved in the production of histamine. A polycistronic mRNA was also found in the L. buchneri histamine operon, but it does not include the antiporter gene, which is transcribed as a monocistronic RNA. In that case, transcriptional termination structures are present in the intergenic regions (25).
The study of the factors affecting the expression of the histidine decarboxylation genes is of great importance in order to establish technological parameters for the reduction of the synthesis and accumulation of histamine in food products. One of the factors that would be of great importance in the case of S. thermophilus is the growth phase. In the strain analyzed (CHCC1524), the expression of hdcA and, consequently, the synthesis and accumulation of histamine were associated with the stationary growth phase. The influence of the growth phase on the expression of hdcA has also been studied for other LAB (18), where for histamine-producing strains isolated from wine, the maximum levels of expression were found at the end of the exponential growth phase (32 and 48 h), and levels decreased during the stationary growth phase. Histamine production in wine is associated with bacteria that develop after alcoholic fermentation and, thus, that grow very slowly. However, for S. thermophilus, which is used as a primary starter culture for yogurt production, the metabolism is adapted to a rapid consumption of sugar moieties and, thus, a fast acidification of the milk.
The increase in the level of hdcA expression at the stationary growth phase could be induced by the environmental conditions of the medium (depletion of nutrients and acidic pH) or by an internal signal more related to survival under these conditions. Low pH values were described previously to be a factor necessary for decarboxylase cluster induction (14, 21). However, in the hdcA expression analysis reported here, no significant differences in expression levels were found, thus indicating that other factors could be implicated in the induction of the hdc cluster. Therefore, for S. thermophilus, this finding may indicate that the physiological function of histidine decarboxylase is more associated with energy generation than with pH control. The lack of a significant induction at a low pH supports this view.
The presence of the substrate amino acid has been shown to influence the expression of the decarboxylase genes of Escherichia coli and Enterococcus durans positively (21, 39). Furthermore, the positive effect on the expression of hdcA genes was also observed for histamine-producing bacteria (14, 18). For a more accurate determination of the role of histidine concentration in hdcA expression, the effect of increasing concentrations was examined in this work. It is noteworthy that concentrations of less than 6 mM were not enough to induce the maximum expression level of hdcA. A similar concentration (1 g liter−1 = 6.4 mM) was reported previously to be the minimum threshold in a study of the HdcA enzymatic activities of other histamine-producing LAB (18). The existence of a minimum substrate amino acid concentration needed to obtain high levels of decarboxylating activity could be explained by the need for a minimum concentration of histidine necessary as a substrate for protein synthesis, while if an excess is present, the bacteria divert it to the histamine-producing pathway. This threshold value, less than 6 mM, was also described for the decarboxylation of tyrosine and lysine by BA-producing strains of E. durans (21) and E. coli (42), respectively.
The technological conditions used in the production of dairy products may favor the synthesis and accumulation of BA. A reduction of the storage and ripening temperatures was proposed to be an effective technological measure to reduce the content of BAs in dairy products (41). In our study the amount of histamine detected at 42°C was significantly higher than that detected in the same culture kept at 4°C. This could be due to a reduction in the gene expression level, the cell growth, or the enzyme activity. However, cell numbers were the same under both conditions, so an increased rate of growth at 42°C compared with that at 4°C does not explain the higher levels of histamine at high temperatures. In fact, cell replication was already stable before the temperature shift (data not shown). The relatively small differences in the expression levels of hdcA in S. thermophilus CCHC1524 seen at 42°C compared to those at 4°C do not sufficiently explain the large difference in the histamine concentrations observed at 42°C. Most probably, the low temperature affects the enzymatic activity of HdcA, indicating that refrigeration could help to reduce the final BA concentration in products that contain histamine-producing S. thermophilus strains. Moreover, the histamine concentration is not very high in milk, and yogurt does not have a ripening period with the subsequent proteolysis and release of amino acids. Therefore, the presence of histamine producers in yogurt should not be a problem. However, the presence of such strains could be a risk in certain situations such as, for example, a breakage in the cold chain or in other dairy products such as cheeses. Nevertheless, the absence of histamine-producing organisms from food is the most secure way to avoid histamine production.
In summary, this work describes the first reported histamine-producing strains of S. thermophilus, one of the most important thermophilic starters used for the production of yogurt and certain cheese varieties (11). The presence of strains with the capacity to decarboxylate histidine could result in products containing histamine produced during manufacture or during storage before consumption. This underlies the importance of using only histamine-negative strains in the manufacture of fermented dairy products. The present results contribute to a better understanding of the regulation affecting histamine production by S. thermophilus as well as to the design of rational strategies for the production of safe and high-quality dairy foods free of toxic compounds such as histamine.
Supplementary Material
Acknowledgments
This research was performed with financial support from the Ministry of Education and Science, Spain (grant AGL2010-01024), and the European Community's Seventh Framework Programme (grant KBBE-CT-2007-211441). V.L. is the beneficiary of an I3P-CSIC contract financed by the European Social Fund, and M.C.-E. is the recipient of a fellowship from the Spanish Ministry of Education and Science.
We thank Sussi Hansen for her excellent technical assistance and Kim Ib Sørensen for his valuable advice on various scientific aspects of the work.
Footnotes
Published ahead of print on 23 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bodmer, S., C. Imark, and M. Kneubühl. 1999. Biogenic amines in foods: histamine and food processing. Inflamm. Res. 48:296-300. [DOI] [PubMed] [Google Scholar]
- 2.Burdychova, R., and T. Komprda. 2007. Biogenic amine-forming microbial communities in cheese. FEMS Microbiol. Lett. 276:149-155. [DOI] [PubMed] [Google Scholar]
- 3.Calles-Enríquez, M., V. Ladero, M. Fernández, M. C. Martín, and M. A. Alvarez. 2010. Extraction of RNA from fermented milk products for in situ gene expression analysis. Anal. Biochem. 400:307-309. [DOI] [PubMed] [Google Scholar]
- 4.Copeland, W. C., J. D. Domena, and J. D. Robertus. 1989. The molecular cloning, sequence and expression of the hdcB gene from Lactobacillus 30a. Gene 85:259-265. [DOI] [PubMed] [Google Scholar]
- 5.Coton, E., G. C. Rollan, and A. Lonvaud-Funel. 1998. Histidine decarboxylase of Leuconostoc oenos 9204: purification, kinetic properties, cloning and nucleotide sequence of the hdc gene. J. Appl. Microbiol. 84:143-151. [DOI] [PubMed] [Google Scholar]
- 6.Desroche, N., C. Beltramo, and J. Guzzo. 2005. Determination of an internal control to apply reverse transcription quantitative PCR to study stress response in the lactic acid bacterium Oenococcus oeni. J. Microbiol. Methods 60:325-333. [DOI] [PubMed] [Google Scholar]
- 7.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández, M., D. M. Linares, B. del Río, V. Ladero, and M. A. Alvarez. 2007. HPLC quantification of biogenic amines in cheeses: correlation with PCR-detection of tyramine-producing microorganisms. J. Dairy Res. 74:276-282. [DOI] [PubMed] [Google Scholar]
- 9.Fernández-García, E., J. Tomillo, and M. Nuñez. 2000. Formation of biogenic amines in raw milk Hispanico cheese manufactured with proteinases and different levels of starter culture. J. Food Prot. 63:1551-1555. [DOI] [PubMed] [Google Scholar]
- 10.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces. A laboratory manual, John Innes Institute. The John Innes Foundation, Norwich, United Kingdom.
- 11.Iyer, R., S. K. Tomar, T. U. Maheswari, and S. Rameshwar. 2010. Streptococcus thermophilus strains: multifunctional lactic acid bacteria. Int. Dairy J. 20:133-141. [Google Scholar]
- 12.Izquierdo-Pulido, M., M. C. Vidal-Carou, and A. Mariné-Font. 1993. Determination of biogenic amines in beers and their raw materials by ion-pair liquid chromatography with post-column derivatization. J. AOAC Int. 76:1027-1032. [Google Scholar]
- 13.Joosten, H. M. L. J., and M. D. Northolt. 1987. Conditions allowing the formation of biogenic amines in cheese. 2. Decarboxylative properties of some non starter bacteria. Neth. Milk Dairy J. 41:259-280. [Google Scholar]
- 14.Kimura, B., H. Takahashi, S. Hokimoto, Y. Tanaka, and T. Fuji. 2009. Induction of the histidine decarboxylase genes of Photobacterium damselae subsp. damselae (formally P. histaminum) at low pH. J. Appl. Microbiol. 107:485-497. [DOI] [PubMed] [Google Scholar]
- 15.Krause, I., A. Bockhardt, H. Neckermann, T. Henleand, and H. Klostermeyer. 1995. Simultaneous determination of amino acids and biogenic amines by reversed phase high-performance liquid chromatography of the dabsyl derivatives. J. Chromatogr. A 715:67-79. [Google Scholar]
- 16.Reference deleted.
- 17.Ladero, V., M. Fernández, and M. A. Alvarez. 2009. Effect of post-ripening processing on the histamine and histamine-producing bacteria contents of different cheeses. Int. Dairy J. 19:759-762. [Google Scholar]
- 18.Landete, J. M., I. Pardo, and S. Ferrer. 2006. Histamine, histidine, and growth phase mediated regulation of the histidine decarboxylase gene in lactic acid bacteria isolated from wine. FEMS Microbiol. Lett. 260:84-90. [DOI] [PubMed] [Google Scholar]
- 19.Landete, J. M., B. de las Rivas, A. Marcobal, and R. Muñoz. 2008. Updated molecular knowledge about histamine biosynthesis by bacteria. Crit. Rev. Food Sci. Nutr. 48:697-714. [DOI] [PubMed] [Google Scholar]
- 20.Le Jeune, C., A. Lonvaud-Funel, B. ten Brink, H. Hofstra, and J. M. B. M. van der Vossen. 1995. Development of a detection system for histidine decarboxylating lactic acid bacteria based on DNA probes, PCR and activity test. J. Appl. Bacteriol. 78:316-326. [DOI] [PubMed] [Google Scholar]
- 21.Linares, D. M., M. Fernández, M. C. Martín, and M. A. Álvarez. 2009. Tyramine biosynthesis is transcriptionally regulated by pH and tyrosine concentration in Enterococcus durans IPLA655. Microb. Biotech. 2:625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 23.Lucas, P. M., W. A. Wolken, O. Claisse, J. S. Lolkema, and A. Lonvaud-Funel. 2005. Histamine-producing pathway encoded on an unstable plasmid in Lactobacillus hilgardii 0006. Appl. Environ. Microbiol. 7:1417-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maintz, L., and N. Novak. 2007. Histamine and histamine intolerance. Am. J. Clin. Nutr. 85:1185-1196. [DOI] [PubMed] [Google Scholar]
- 25.Martín, M. C., M. Fernández, D. M. Linares, and M. A. Alvarez. 2005. Sequencing, characterization and transcriptional analysis of the histidine decarboxylase operon of Lactobacillus buchneri. Microbiology 151:1219-1228. [DOI] [PubMed] [Google Scholar]
- 26.Neely, M. N., and E. R. Olson. 1996. Kinetics of expression of the Escherichia coli cad operon as a function of pH and lysine. J. Bacteriol. 178:5522-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novella-Rodríguez, S., M. T. Veciana-Nogués, A. Roig-Sagués, A. Trujillo-Mesa, and M. C. Vidal-Carou. 2002. Influence of starter and non starter bacteria on the formation of biogenic amines in goat cheese during ripening. J. Dairy Sci. 85:2471-2478. [DOI] [PubMed] [Google Scholar]
- 28.Novella-Rodríguez, S., M. T. Veciana-Nogués, A. J. Trujillo-Mesa, and M. C. Vidal-Carou. 2003. Profile of biogenic amines in goat cheese made from pasteurized and pressurized milks. J. Food Sci. 67:2940-2944. [Google Scholar]
- 29.Pavlovic, G., V. Burrus, B. Gintz, B. Decaris, and G. Guedon. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759-774. [DOI] [PubMed] [Google Scholar]
- 30.Poolman, B., and W. N. Konings. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Premont, R. T., R. R. Gainetdinov, and M. G. Caron. 2001. Following the trace of elusive amines. Proc. Natl. Acad. Sci. U. S. A. 98:9474-9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauscher-Gabernig, E., R. Grossgut, F. Bauer, and P. Paulsen. 2009. Assessment of alimentary histamine exposure of consumers in Austria and development of tolerable levels in typical foods. Food Control 20:423-429. [Google Scholar]
- 33.Recsei, P. A., and E. E. Snell. 1972. Histidine decarboxylaseless mutants of Lactobacillus 30a: isolation and growth properties. J. Bacteriol. 112:624-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Satomi, M., M. Furushita, H. Oikawa, M. Yoshikawa-Takahashi, and Y. Yano. 2008. Analysis of a 30 kbp plasmid encoding histidine decarboxylase gene in Tetragenococcus halophilus isolated from fish sauce. Int. J. Food Microbiol. 126:202-209. [DOI] [PubMed] [Google Scholar]
- 36.Schirawski, J., W. Hagens, G. F. Fitzgerald, and D. van Sinderen. 2002. Molecular characterization of cadmium resistance in Streptococcus thermophilus strain 4134: an example of lateral gene transfer. Appl. Environ. Microbiol. 68:5508-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shalaby, A. R. 1996. Significance of biogenic amines in food safety and human health. Food Res. Int. 29:675-690. [Google Scholar]
- 38.Silla-Santos, M. H. 1996. Biogenic amines: their importance in foods. Int. J. Food Microbiol. 29:213-231. [DOI] [PubMed] [Google Scholar]
- 39.Soksawatmaekhin, W., A. Kuraishi, K. Sakata, K. Kashiwagi, and K. Igarashi. 2004. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol. Microbiol. 51:1401-1412. [DOI] [PubMed] [Google Scholar]
- 40.Solow, B. T., and G. A. Somkuti. 2001. Molecular properties of Streptococcus thermophilus plasmid pER35 encoding a restriction modification system. Curr. Microbiol. 42:122-128. [DOI] [PubMed] [Google Scholar]
- 41.Stratton, J. E., R. W. Hutkins, and S. L. Taylor. 1991. Biogenic amines in cheese and other fermented foods: a review. J. Food Prot. 54:460-470. [DOI] [PubMed] [Google Scholar]
- 42.Tetsch, L., C. Koller, I. Haneburger, and K. Jung. 2008. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol. Microbiol. 67:570-583. [DOI] [PubMed] [Google Scholar]
- 43.Valsamaki, K., A. Michaelidou, and A. Polychroniadou. 2000. Biogenic amine production in Feta cheese. Food Chem. 71:259-266. [Google Scholar]
- 44.Vanderslice, P., W. C. Copeland, and J. D. Robertus. 1986. Cloning and nucleotide sequence of wild type and a mutant histidine decarboxylase from Lactobacillus 30a. J. Biol. Chem. 261:15186-15191. [PubMed] [Google Scholar]
- 45.Van Poelje, P. D., and E. E. Snell. 1990. Cloning, sequencing, expression, and site-directed mutagenesis of the gene from Clostridium perfringens encoding pyruvoyl-dependent histidine decarboxylase. Biochemistry 29:132-139. [DOI] [PubMed] [Google Scholar]
- 46.Wöhrl, S., W. Hemmer, M. Focke, K. Rappersberger, and R. Jarisch. 2004. Histamine intolerance-like symptoms in healthy volunteers after oral provocation with liquid histamine. Allergy Asthma Proc. 25:305-311. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.