Abstract
Myxococcus xanthus has to cope with changes in its environment during growth and development. Among these factors, the concentration of copper is crucial due to the essential toxic effect of this metal, which forces the cells to maintain a tight homeostasis. The M. xanthus copper response is more complex than that in other bacteria, which is reflected by the different copper sensitivities of growing and developing cells. In the present study, the participation in copper homeostasis of six heavy metal efflux systems encoded in the M. xanthus genome has been examined. Three of these pumps exhibit the signature sequences in transmembrane domain 4 of the Cus systems (Cus1, Cus2, and Cus3), while the other three exhibit the motifs of the Czc systems (Czc1, Czc2, and Czc3). The Cus2 and Cus3 systems are inducible by copper and monovalent metals, functioning as the main copper efflux pumps, while the Cus1 system is implicated in Zn2+ homeostasis. The Czc systems are also differentially regulated either by divalent metals but not by copper (Czc1), by copper and divalent metals (Czc2), or by starvation (Czc3). The differential regulation of these six efflux systems ensures the proper completion of the M. xanthus life cycle in an environment with fluctuating concentrations of copper and other metals.
Myxococcus xanthus is a rod-shaped deltaproteobacterium considered a model to study social behavior in prokaryotes. M. xanthus cells are found in the topsoil, where they cooperatively feed on decaying organic matter and prey on other microorganisms. However, upon starvation, a developmental process initiates, consisting of the formation of multicellular mound-shaped structures called fruiting bodies, inside which cells differentiate into dormant spherical myxospores. The spores in a fruiting body can germinate and resume growth when nutrients become available (35).
In their habitats, cells have to deal with fluctuations in levels of several metal ions (19). Copper is a crucial trace transition metal required for a number of enzymes involved in energy metabolism and cell detoxification (24) but can cause serious damage through the formation of reactive oxygen species at high concentrations (4, 34). Thus, copper homeostasis has to be tightly regulated to prevent toxic effects. One of the mechanisms that bacteria use to protect themselves from the toxic effect of copper is the active efflux of metals. Two main detoxification mechanisms are known to function in Gram-negative bacteria: (i) P1B-type ATPases, which use ATP energy to pump Cu+ out of the cytoplasm to the periplasm (1), where multicopper oxidases (MCOs) will oxidize this Cu+ to the less toxic form Cu2+ (30), and (ii) CBA transporters (consisting of subunits C, B, and A), which extrude metals from the cytoplasm or the periplasm to outside the outer membrane (5, 9, 22).
The central pump protein of the CBA transporters belongs to the resistance-nodulation-cell division (RND) family, which functions as a transporter energized by proton-substrate antiport (6, 20, 27, 32). The other two components are an outer membrane factor (OMF), which enters into the periplasm and produces an exit duct, and a membrane fusion protein (MFP), which acts as an adaptor to stabilize the contact between the RND and the OMF components. Bacterial CBA efflux systems span the complete cell wall of a Gram-negative bacterium from the cytoplasm to the exterior. Accordingly, they must be able to catalyze transenvelope efflux (from the cytoplasm directly to the exterior), outer membrane efflux (from the periplasm to the outside), or both processes. CBA systems are involved in the export of metal ions, xenobiotics, and drugs, depending on the characteristics of the individual RND protein. The members of this superfamily involved in the transport of hydrophobic and amphiphilic compounds are designated HAE-RND, and those that efflux heavy metals are designated HME-RND (32). The latter family includes pumps that export monovalent metals, known as Cus systems (for Cu sensing), and those that export divalent metals, named Czc systems (for cobalt, zinc, and cadmium). Transmembrane segment 4 (TM4) of monovalent metal RND transporters distinctively differs in sequence from those transporting divalent metal cations or organic compounds (26).
The M. xanthus copper response seems to differ from those described for other bacteria on the basis of several observations. First, developmental cells are more sensitive to copper than are growing cells, since concentrations higher than 900 μM and 60 μM are lethal for growing and developing cells, respectively (30). However, copper-preadapted cells are as resistant to this metal during development as they are during growth, suggesting the induction of several mechanisms that confer metal resistance. Second, copper induces the production of carotenoids in cultures grown in the dark (16). Finally, in silico studies of the complete genome of M. xanthus have revealed the presence of a large number of gene products with sequence similarities to proteins previously reported for other bacteria to be involved in copper detoxification, all of which are redundant. We have identified three MCOs (30), three P1B-type ATPases (15), and six CBA transporters of the HME-RND group. The great abundance of paralogous genes involved in copper resistance would seem to be contradictory with the copper sensitivity of M. xanthus; that is, several bacteria (such as Escherichia coli) that encode only one of each of these elements are more resistant to copper than M. xanthus (26, 30). This observation raises the question of why M. xanthus holds so many paralogous genes to confer copper resistance.
We started the analysis of the M. xanthus global copper response by exploring the family of paralogs of the MCOs, which includes CuoA, CuoB, and CuoC (30). We continue the study by investigating the six HME-RND pumps, three of which exhibit the consensus sequence in TM4 of the Cus systems (designated Cus1, Cus2, and Cus3) and the other three of which exhibit the signature of the Czc systems (named Czc1, Czc2, and Czc3). We have found that these six systems are differentially regulated by copper, other metals, and/or starvation. Some of them are essential for cell viability in the presence of high copper concentrations, and others are essential for the completion of development, even in the absence of external copper.
MATERIALS AND METHODS
Identification of CBA systems in the M. xanthus genome.
Genes encoding CBA transporters in the M. xanthus genome were identified by BLASTP analysis using the sequences of the E. coli proteins CusA (GenBank accession number NP_415107) (RND), CusB (accession number NP_415106.1) (MFP), and CusC (accession number NP_415104.1) (OMF). All sequences obtained with an E value of <3 were back-searched against the NCBI database (http://blast.ncbi.nlm.nih.gov/) to confirm similarities to the corresponding subunits of the CBA systems and against the Pfam database (3) to verify that they matched Pfam accession numbers PF00873 (AcrB/AcrD/AcrF family), in the case of the RND proteins, and PF02321 (outer membrane efflux protein), in the case of the OMF proteins. In addition, genes located adjacent to all the genes identified as belonging to CBA systems were also analyzed. Prediction of transmembrane domains and subcellular localization of the proteins were carried out with the TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/) servers. The consensus sequences in TM4 of the RND proteins were manually analyzed.
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. All M. xanthus strains were grown at 30°C with vigorous shaking (300 rpm) in CTT broth (10). CTT agar plates contained 1.5% Bacto agar (Difco). When necessary, kanamycin (80 μg ml−1), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (100 μg ml−1), or galactose (10 mg ml−1) was added. When required, CTT medium was supplemented with metals at the concentrations indicated in the figure legends and tables. Luria-Bertani broth (28), supplemented with kanamycin (25 μg ml−1) or X-Gal (25 μg ml−1) when needed, was used to grow E. coli strains at 37°C. Standard protocols were followed for nucleic acid manipulations (28).
Construction of in-frame deletion mutants.
The in-frame deletion strains were generated by allelic exchange using the GalK selection method (33). Briefly, sequences upstream and downstream of the genes constituting HME-RND systems were amplified by PCR with PrimeSTARHS high-fidelity polymerase (Takara), using wild-type (WT) chromosomal DNA as a template and the primers listed in Table S2 in the supplemental material. The amplified products were then cloned into KpnI- and HindIII-digested pBJ113 (11) to obtain plasmids harboring deletions of the corresponding CBA transporter (Table S1). The resulting nonreplicating plasmids carrying the deletions were introduced into M. xanthus cells by electroporation (12). Chromosomal integration was selected by plating cells onto CTT plates containing 80 μg ml−1 kanamycin (positive selection). Several randomly chosen kanamycin-resistant (Kmr) merodiploids were analyzed by Southern blot hybridization for the proper recombination event. One positive strain was then grown in the absence of kanamycin and plated onto CTT plates containing 1% galactose for negative selection. Southern blot analysis was used to screen kanamycin-sensitive (Kms) and galactose-resistant (Galr) colonies, searching for the loss of the WT allele.
Development.
The starvation medium CF (8) was used to induce development, which was supplemented with metals at the concentrations indicated in the figure legends and tables when needed. Cells were grown in CTT broth to approximately 3.0 × 108 cells ml−1 (optical density at 600 nm [OD600] of 1) and resuspended to 4.5 × 109 cells ml−1 (OD600 of 15) in TM buffer (10 mM Tris-HCl [pH 7.6], 1 mM MgSO4). Ten microliters of each suspension was spotted onto CF agar plates and incubated at 30°C, with observation on a Wild Heerbrugg dissecting microscope.
Construction and assay of strains harboring lacZ fusions.
The lacZ fusion plasmids were constructed by using vector pKY481 (2). By use of PrimeSTARHS polymerase (Takara), fragments encompassing the upstream CBA transporter regions were amplified from WT chromosomal DNA by the use of the oligonucleotide pairs indicated in Table S2 in the supplemental material. The BamHI site in the primers was introduced at the start codon of the first cus/czc system gene and in frame with the BamHI site existing in the lacZ gene of plasmid pKY481. PCR products were digested with KpnI and BamHI and ligated into vector pKY481 digested with the same enzymes. The resulting plasmids were introduced into M. xanthus cells by electroporation, and the Kmr colonies were analyzed by Southern blotting.
Strains containing lacZ fusions were incubated at 30°C on CTT and CF agar plates containing different metal concentrations. For qualitative analyses of β-galactosidase activity, plates containing 100 μg ml−1 of X-Gal were used. For quantitative analyses, cell extracts were obtained at different times by sonication and assayed for activity as previously reported (16). The amount of protein in the supernatants was determined by using the Bio-Rad protein assay (Bio-Rad, Inc.) with bovine serum albumin as a standard. β-Galactosidase activity was determined as described previously by Kroos et al. (13). Specific activity is expressed as nmol of o-nitrophenol produced per min and mg of protein. The results are the averages of data from three different experiments.
RESULTS
CBA systems in the M. xanthus genome.
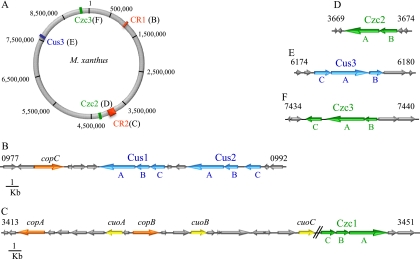
A search for CBA transporters in the M. xanthus genome has revealed the presence of 14 genes that encode proteins with similarities to the RND component, 17 with similarities to MFP, and 14 with similarities to OMF (see Fig. S1 in the supplemental material). By analyzing the sequence motifs in TM4 of all of the RND-like proteins, it was possible to classify six systems into the HME-RND family. Three of these RND proteins have the conserved motif AX5DX6E, which is present in the monovalent metal cation transporters (26). These systems have been designated Cus1, Cus2, and Cus3. The three subunits of each system (RND, MFP, and OMF) are encoded by adjacent genes (Fig. 1B and E). The other three RND proteins contain the consensus motif DX5DX6E, typical of those systems involved in the trafficking of divalent metal cations (26). For that reason, these three systems have been named Czc1, Czc2, and Czc3 (Fig. 1C, D, and F). Two of them, Czc1 and Czc3, are composed of the three above-mentioned subunits encoded by contiguous genes (Fig. 1C and F). In contrast, only the subunits RND and MFP of the Czc2 system are encoded together in the chromosome (Fig. 1D). Nevertheless, as several OMF-like proteins are found as orphans in the genome (Fig. S1), it is plausible to speculate that they might function with the subunits CzcA2 and CzcB2 to extrude metals to the exterior. The rest of the RND proteins contain the sequence signature defined for those CBA systems involved in organic compound detoxification, or they could not be classified into any group because they do not contain any conserved motif (26). These systems have been excluded from this study.
FIG. 1.
Schematic depiction of the location of the six CBA efflux heavy metal systems in the M. xanthus genome. (A) M. xanthus genome map with the positions of copper region 1 (CR1), copper region 2 (CR2), and the dispersed czc2, cus3, and czc3 systems. (B) Genes located in copper region 1. (C) Genes located in copper region 2. (D) Genetic environment of the czc2 system. (E) Genetic environment of the cus3 system. (F) Genetic environment of the czc3 system. The numbers indicated at the top of the first and last genes in each panel correspond to their identifiers in the M. xanthus genome. Paralogous genes are represented with the same color. Blue, cus systems; green, czc systems; orange, P1B-ATPases; yellow, MCOs. The rest of the genes are depicted as gray arrows.
The Cus1 and Cus2 systems are encoded very close in the chromosome, in a region that has been designated copper region 1, where the copper-dependent P1B-ATPase CopC (15) is also found (Fig. 1A and B). Conversely, Czc1 is located in copper region 2 (Fig. 1C), along with the genes for the three MCOs (30) and the P1B-type ATPases CopA and CopB (15). The Cus3, Czc2, and Czc3 systems are dispersed throughout the genome (Fig. 1D to F).
Expression of the HME-RND systems.
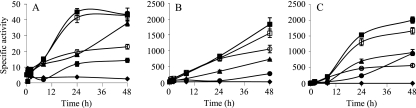
The expression profiles of the cus and czc systems were examined during growth and development in M. xanthus strains harboring in-frame fusions between the first gene of each cluster and the lacZ gene from E. coli. These analyses revealed that the three cus systems exhibited undetectable or very low levels of β-galactosidase activity during growth in the absence of metals (Fig. 2). However, their expression levels increased in a time-dependent manner by the addition of external copper, although the levels reached by cus2 and cus3 were much higher than those reached by cus1. The three Cus systems responded more efficiently to low copper concentrations than to high copper concentrations (Fig. 2).
FIG. 2.
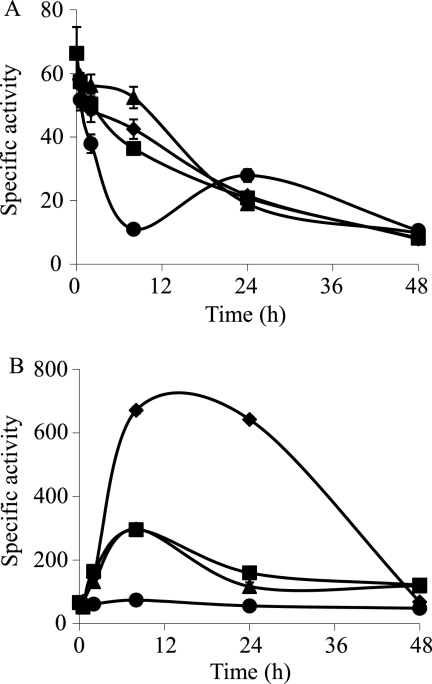
Copper upregulation of cus1 (A), cus2 (B), and cus3 (C) during growth. Strains were incubated on CTT agar plates containing 0 μM (♦), 50 μM (○), 150 μM (□), 300 μM (▪), 600 μM (▴), or 800 μM (•) copper. The specific β-galactosidase activity in cell extracts was determined as described in Materials and Methods. The results are the averages of data from three different experiments. The error bars indicate standard deviations. Note the differences in the scales.
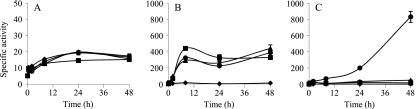
During development, the copper responses of the cus systems were analyzed by using a 10-fold-lower concentration of the metal to prevent cells from dying (30). During this stage, the expression of cus1 was almost undetectable in the absence and in the presence of copper (Fig. 3A). In contrast, cus2 and cus3 responded to the copper supplementation in a manner different from that observed during growth (compare Fig. 2B and C with 3B and C, respectively). cus2 increased its expression level in a time-dependent manner, reaching a plateau at 8 h of starvation (Fig. 3B). In addition, the expression profiles did not exhibit a marked metal concentration dependency. The expression of cus3 increased considerably only in response to copper concentrations of 60 μM and after a 24-h incubation (Fig. 3C).
FIG. 3.
Copper upregulation of cus1 (A), cus2 (B), and cus3 (C) during development. Strains were incubated on CF agar plates containing 0 μM (♦), 20 μM (▪), 40 μM (▴), or 60 μM (•) copper. The specific β-galactosidase activity in the cell extracts was determined as described in Materials and Methods. The results are the averages of data from three different experiments. The error bars indicate standard deviations. Note the differences in the scales.
Of the two redox states of copper, Cu+ is the form that dominates in the reducing cell environment, while Cu2+ predominates outside the cytoplasm (4, 34). As these two redox states can be mimicked by monovalent and divalent metals, respectively, the expression of the cus systems in the presence of Au+, Ag+, Fe2+, Mn2+, Ni2+, Zn2+, Co2+, and Cd2+ was also analyzed. The strains harboring fusions were first tested on agar plates containing X-Gal and several concentrations of each metal, and those concentrations that gave the most intense blue color were chosen to quantify β-galactosidase activity. The results obtained showed that cus1 expression levels were higher with Zn2+ than with copper (Table 1), indicating that this system could be involved in Zn2+ homeostasis. The cus2 promoter responded mainly to copper and gold, suggesting that Cu+ might be the inducer. Conversely, cus3 was strongly induced by copper and Cd2+ (Table 1), pointing to an induction by Cu2+. The other metals tested exerted scant or no effect on the expression of the three M. xanthus cus systems. It should be noted that the responses were not always identical during growth and development (Table 1). Altogether, the obtained results indicate that the three cus systems are differentially regulated by copper and other monovalent and/or divalent metals. Furthermore, a different regulation by metals can be observed during growth and development.
TABLE 1.
Responses of cus promoters to heavy metals
| CBA system | Inducer | Growth |
Development |
||||
|---|---|---|---|---|---|---|---|
| Metal concn (mM) | Avg β-galactosidase activity ± SDb | Fold inductiona | Metal concn (mM) | Avg β-galactosidase activity ± SDb | Fold inductiona | ||
| cus1 | None | 4 ± 0.1 | 19 ± 0.1 | ||||
| CuSO4 | 0.3 | 45 ± 2 | 11 | 0.060 | 20 ± 0.2 | 1 | |
| Ni(NO3)2 | 1.0 | 12 ± 0.5 | 3 | 2.0 | 46 ± 1 | 2 | |
| Zn(NO3)2 | 0.25 | 207 ± 7 | 52 | 0.1 | 203 ± 8 | 11 | |
| Co(NO3)2 | 0.5 | 7 ± 0.1 | 2 | 1.0 | 66 ± 2 | 4 | |
| Cd(NO3)2 | 0.1 | 4 ± 0.3 | 1 | 0.025 | 9 ± 0.3 | 0.5 | |
| FeSO4 | 6.0 | 7 ± 1 | 2 | 6.0 | 118 ± 6 | 6.2 | |
| MnSO4 | 0.6 | ND | 0.8 | ND | |||
| AgNO3 | 0.1 | 3 ± 1 | 1 | 0.01 | 12 ± 1 | 0.6 | |
| AuCl3 | 1.0 | 7 ± 0.5 | 2 | 0.25 | 26 ± 1 | 1 | |
| cus2 | None | 7 ± 1 | 2 ± 0.1 | ||||
| CuSO4 | 0.3 | 641 ± 5 | 92 | 0.020 | 326 ± 33 | 163 | |
| Ni(NO3)2 | 1.0 | ND | 2.0 | 21 ± 2 | 11 | ||
| Zn(NO3)2 | 0.25 | ND | 0.1 | 6 ± 0.1 | 3 | ||
| Co(NO3)2 | 0.5 | ND | 1.0 | 20 ± 1 | 10 | ||
| Cd(NO3)2 | 0.1 | 37 ± 0.5 | 5 | 0.025 | 125 ± 2 | 63 | |
| FeSO4 | 6.0 | 4 ± 0.1 | 1 | 6.0 | ND | ||
| MnSO4 | 0.6 | ND | 0.8 | ND | |||
| AgNO3 | 0.1 | 20 ± 1 | 3 | 0.01 | 49 ± 1 | 25 | |
| AuCl3 | 1.0 | 637 ± 9 | 91 | 0.25 | 1,250 ± 59 | 625 | |
| cus3 | None | 1 ± 0.3 | 0.4 ± 0.2 | ||||
| CuSO4 | 0.3 | 1,533 ± 12 | 1,533 | 0.060 | 200 ± 3 | 500 | |
| Ni(NO3)2 | 1.0 | ND | 2.0 | ND | |||
| Zn(NO3)2 | 0.25 | ND | 0.1 | ND | |||
| Co(NO3)2 | 0.5 | ND | 1.0 | ND | |||
| Cd(NO3)2 | 0.1 | 1,142 ± 8 | 1,142 | 0.025 | 878 ± 56 | 2,195 | |
| FeSO4 | 6.0 | ND | 6.0 | ND | |||
| MnSO4 | 0.6 | ND | 0.8 | ND | |||
| AgNO3 | 0.1 | 1 ± 0.3 | 1 | 0.01 | 76 ± 2 | 190 | |
| AuCl3 | 1.0 | 105 ± 3 | 105 | 0.25 | 187 ± 8 | 468 | |
The fold induction was calculated by dividing the means of the respective β-galactosidase activities by the average value for the uninduced cells (none).
ND, not detectable. β-Galactosidase activity was not quantified when blue color was not detected during the qualitative assay.
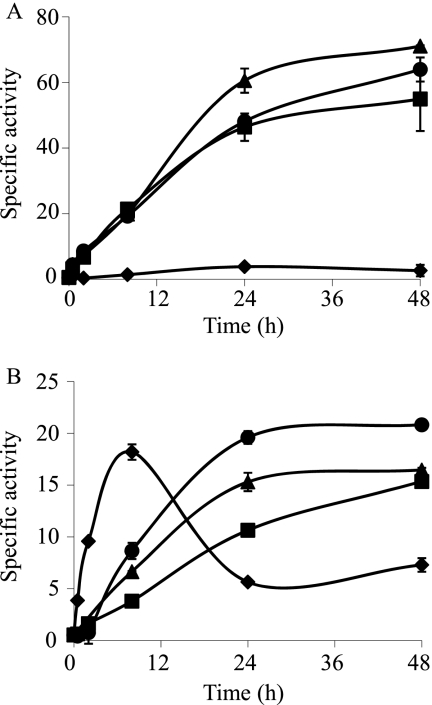
The results obtained for the metal response of the Czc systems are shown in Table 2. czc1 was induced by several divalent metals but not by copper, exhibiting a metal response typical of those of other bacterial Czc systems (21). czc2 showed a small response to copper, Zn2+, and Cd2+. The upregulation of this czc system by copper was clearly detected only during growth but not during development (Fig. 4). Interestingly, czc3 was constitutively expressed during growth, upregulated by nutrient depletion, and repressed by metals (Fig. 5 and Table 2).
TABLE 2.
Responses of czc promoters to heavy metals
| CBA system | Inducer | Growth |
Development |
||||
|---|---|---|---|---|---|---|---|
| Metal concn (mM) | Avg β-galactosidase activity ± SDb | Fold inductiona | Metal concn (mM) | Avg β-galactosidase activity ± SDb | Fold inductiona | ||
| czc1 | None | 1 ± 0.1 | 0.7 ± 0.1 | ||||
| CuSO4 | 0.3 | ND | 0.060 | ND | |||
| Ni(NO3)2 | 1.0 | 483 ± 48 | 483 | 2.0 | ND | ||
| Zn(NO3)2 | 0.25 | ND | 0.1 | ND | |||
| Co(NO3)2 | 0.5 | 618 ± 42 | 618 | 1.0 | 624 ± 37 | 891 | |
| Cd(NO3)2 | 0.1 | 133 ± 3 | 133 | 0.025 | 221 ± 5 | 316 | |
| FeSO4 | 4.0 | 576 ± 36 | 576 | 6.0 | 38 ± 0.4 | 54 | |
| MnSO4 | 0.6 | 458 ± 43 | 458 | 0.8 | 565 ± 19 | 807 | |
| AgNO3 | 0.1 | ND | 0.01 | ND | |||
| AuCl3 | 1.0 | ND | 0.25 | ND | |||
| czc2 | None | 4 ± 0.3 | 18 ± 0.3c | ||||
| CuSO4 | 0.3 | 46 ± 4 | 12 | 0.060 | 20 ± 1 | 1 | |
| Ni(NO3)2 | 1.0 | ND | 2.0 | ND | |||
| Zn(NO3)2 | 0.25 | 19 ± 1 | 5 | 0.1 | ND | ||
| Co(NO3)2 | 0.5 | ND | 1.0 | ND | |||
| Cd(NO3)2 | 0.1 | 27 ± 0.4 | 7 | 0.025 | 55 ± 2 | 3 | |
| FeSO4 | 6.0 | ND | 6.0 | ND | |||
| MnSO4 | 0.6 | ND | 0.8 | ND | |||
| AgNO3 | 0.1 | ND | 0.01 | ND | |||
| AuCl3 | 1.0 | ND | 0.25 | ND | |||
| czc3 | None | 27 ± 1 | 642 ± 9 | ||||
| CuSO4 | 0.3 | 21 ± 1 | 1 | 0.060 | 56 ± 3 | 0.1 | |
| Ni(NO3)2 | 1.0 | 5 ± 1 | 0.2 | 2.0 | 97 ± 1 | 0.2 | |
| Zn(NO3)2 | 0.25 | 9 ± 1 | 0.3 | 0.1 | 48 ± 0.1 | 0.1 | |
| Co(NO3)2 | 0.5 | 5 ± 1 | 0.2 | 1.0 | 36 ± 1 | 0.1 | |
| Cd(NO3)2 | 0.1 | 12 ± 1 | 0.4 | 0.025 | 74 ± 5 | 0.1 | |
| FeSO4 | 6.0 | 11 ± 1 | 0.4 | 6.0 | 70.1 ± 0.3 | 0.1 | |
| MnSO4 | 0.6 | 5 ± 1 | 0.2 | 0.8 | 138 ± 6 | 0.2 | |
| AgNO3 | 0.1 | 8 ± 3 | 0.3 | 0.01 | 56 ± 1 | 0.1 | |
| AuCl3 | 1.0 | 11 ± 0.5 | 0.4 | 0.25 | 127 ± 2 | 0.2 | |
Fold induction was calculated by dividing the means of the respective β-galactosidase activities by the average values for the uninduced cells (none).
ND, not detectable. β-Galactosidase activity was not quantified when blue color was not detected during the qualitative assay.
The maximum level of expression of czc2 in the absence of copper is reached at 8 h. The presence of copper delayed the maximum up to 24 h (Fig. 4). For comparison, we used samples obtained at 8 h as controls for the assay in the absence of copper and at 24 h when copper was added.
FIG. 4.
Copper upregulation of czc2 during growth (A) and development (B). The strain was grown on CTT agar plates containing 0 μM (♦), 300 μM (▪), 600 μM (▴), or 800 μM (•) copper. The copper concentrations added to CF medium during development were 0 μM (♦), 20 μM (▪), 40 μM (▴), or 60 μM (•). The specific β-galactosidase activity in the cell extracts was determined as described in Materials and Methods. The results are the averages of data from three different experiments. The error bars indicate standard deviations. Note the differences in the scales.
FIG. 5.
Copper upregulation of czc3 during growth (A) and development (B). The strain was grown on CTT agar plates containing 0 μM (♦), 300 μM (▪), 600 μM (▴), or 800 μM (•) copper. The copper concentrations added to CF medium during development were 0 μM (♦), 20 μM (▪), 40 μM (▴), or 60 μM (•). The specific β-galactosidase activity in cell extracts was determined as described in Materials and Methods. The results are the averages of data from three different experiments. The error bars indicate standard deviations. Note the differences in the scales.
Role of the Cus systems in metal resistance.
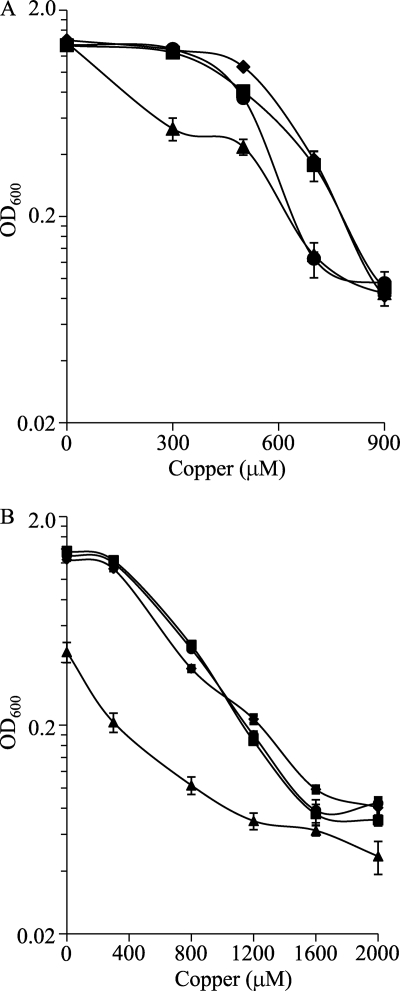
To analyze whether the Cus systems are involved in metal tolerance, three in-frame deletion mutants were constructed (see Table S1 in the supplemental material). When the three mutants and the WT strain were incubated in liquid CTT medium containing several copper concentrations, it was observed that the copper tolerance of the Δcus1 mutant was very similar to that of the WT strain. However, the Δcus3 mutant exhibited a reduced viability at elevated copper concentrations, and the Δcus2 mutant was markedly more sensitive than the other three strains to even low metal concentrations (Fig. 6A). These results are in good agreement with the copper induction exhibited by these systems.
FIG. 6.
Copper sensitivities of the M. xanthus WT strain (▪) and the Δcus1 (♦), Δcus2 (▴), and Δcus3 (•) mutants during growth. (A) Copper tolerance of cells not preadapted to the metal. Cells grown in the absence of copper were diluted to an OD600 of 0.05 in fresh CTT liquid medium containing the indicated copper concentrations. The data shown correspond to the OD600 monitored at 24 h of incubation. (B) Copper tolerance of cells adapted to copper. Strains were grown in the presence of 300 μM copper prior to dilution into fresh CTT liquid medium containing different copper concentrations. The data are expressed as mentioned above for A and are the averages of data from three experiments. The error bars indicate standard deviations.
Since M. xanthus copper adaptation mechanisms are upregulated in cells preincubated with copper (30), mutant cells grown in the presence of this metal should be more sensitive. To test this hypothesis, cells were grown in the presence of 300 μM copper for 24 h. After this preadaptation period, cells were diluted into fresh CTT liquid medium supplemented with higher copper concentrations and incubated for 24 h, when the OD600 of the cultures was monitored. As shown in Fig. 6B, only the Δcus2 mutant was much more sensitive to copper than the WT strain under these conditions.
In order to elucidate the role of the Cus systems during development, the phenotypes of the mutants and the WT strain plated on CF starvation medium were analyzed. The results obtained with both preadapted and nonpreadapted cells were very similar to those described above during growth, as only the Δcus2 mutant turned out to be more sensitive to copper. Details are provided in Fig. S2 in the supplemental material.
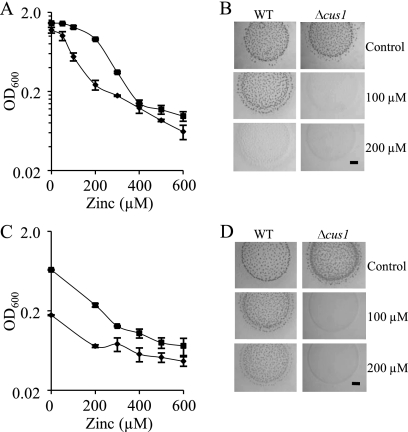
As the Δcus1 and Δcus3 mutants did not exhibit a clear sensitivity to copper, we tested whether they could be implicated in conferring resistance to the other metals that are able to induce their expression. When the growth of the Δcus1 mutant in liquid CTT medium with Zn2+ was monitored, it was observed that these cells tolerated lower metal concentrations than did the WT strain (Fig. 7A). The viability of this mutant was also analyzed with cultures preadapted to the presence of this metal (by preincubation with 0.2 mM Zn2+ for 24 h). Under these conditions, the Δcus1 mutant exhibited a dramatic reduction in viability compared to that of the WT strain (Fig. 7C). Similar results were observed when the phenotype of the Δcus1 mutant grown on starvation medium supplemented with Zn2+ was analyzed (Fig. 7B and D).
FIG. 7.
Zinc sensitivities of the M. xanthus WT strain and the Δcus1 mutant. (A and B) Zinc tolerance of cells not preadapted to the metal during growth (A) and development (B). (A) Cells grown in the absence of metal were diluted to an OD600 of 0.05 in fresh CTT liquid medium containing the indicated zinc concentrations. The data shown correspond to the OD600 monitored at 24 h of incubation of WT (▪) and Δcus1 (♦) strains. (B) Cells not preadapted to zinc were concentrated to an OD600 of 15 in TM buffer and spotted onto CF medium containing the indicated zinc concentrations. (C and D) Zinc tolerance of cells adapted to zinc during growth (C) and development (D). (C) The strains were grown in the presence of 200 μM zinc prior to dilution into fresh CTT liquid medium containing different zinc concentrations. The symbols are the same as those indicated above for A. (D) Preadapted cells were concentrated to an OD600 of 15 in TM buffer and spotted onto CF medium. The data are the averages of data from three experiments, and the error bars indicate standard deviations (A and C). The pictures were taken after a 96-h incubation, and bars represent 1 mm (B and D).
In contrast, although Cd2+ greatly induced the expression of the cus3 system, no differences between the Δcus3 mutant and the WT strain were observed either during growth or during development (data not shown).
Role of the Czc3 system in M. xanthus development.
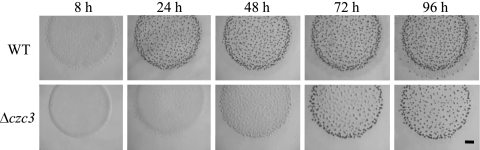
Although the same strategy for the construction of the cus mutants was used to obtain czc deletion mutants, we were unable to obtain the Δczc1 and Δczc2 strains even after analyzing 96 and 128 Kms-Galr colonies, respectively. All these colonies had reverted to the WT genotype, although 50% were expected to evolve to the corresponding deletion mutant. This negative result suggested that both mutations were lethal. In contrast, the Δczc3 mutant could be constructed and phenotypically analyzed. As the expression of this system was repressed by metals (Fig. 5 and Table 2), phenotypic analyses were carried out with no supplementation of metals. Under these conditions, clear differences between the mutant and the WT strain were observed during both growth and development. This mutant grew slower than the WT strain (see Fig. S3 in the supplemental material), exhibiting a generation time of 9 h, while the WT strain duplicated every 6 h. This result is in good agreement with the constitutive expression of czc3 (Fig. 5A). During development, the mutant delayed aggregation more than 48 h, and the fruiting bodies that originated even after a 96-h incubation were immature (Fig. 8). This result also fits well with the induction of this system on CF starvation medium (Fig. 5B).
FIG. 8.
Development of the WT strain and the Δczc3 deletion mutant on CF medium. Pictures were taken at the indicated times. The bar represents 1 mm.
DISCUSSION
In the present study the role of the six M. xanthus HME-RND systems in copper homeostasis has been examined. These efflux pumps have been described to confer resistance to several metals in some Gram-negative bacteria (5, 7, 14, 23, 25, 31), with the CusCFBA system from E. coli (17) being the most investigated. This Cus system is involved in copper homeostasis under conditions of extreme copper stress and is particularly important in anaerobic environments (23). The number of CBA transporters encoded in a genome is highly variable, although most bacteria hold no more than three HME-RND pumps (18). A remarkable exception is Cupriavidus metallidurans, a metal-resistant proteobacterium that has 12 HME-RND systems (21). Nevertheless, it seems that when highly related genes are present in a bacterium, usually one is inducible by heavy metal while the rest are silent or constitutively expressed. The fact that M. xanthus encodes six CBA transporters to extrude metals, all of which are active under specific circumstances, and that cells are not especially resistant to copper and other metals raises the question of why so many paralogous genes have been maintained in this bacterial genome to participate in metal tolerance.
The results obtained for this report, along with results from other studies of M. xanthus MCOs (30) and P1B-type ATPases (15), shed some light on this intriguing question. The variety of expression profiles exhibited by all of the metal-regulated genes of this bacterium indicates that M. xanthus cells must be subjected to a fine-tuned control of the metals allocated not only to the cytoplasm but also to the periplasm. Thus, five different expression patterns can be clearly distinguished for the HME-RND transporters, none of which resembles those of the MCOs or the P1B-type ATPases (15, 30). The reason for this fine-adjustment of metal concentrations in the periplasm may be related to the special structure of the M. xanthus peptidoglycan, which is required to allow the conversion of the long vegetative rods into coccoid myxospores during development (35). It was shown previously that the accumulation of Cu+ in the M. xanthus periplasm originated from the deletion of some MCOs provoked growing cells to change their bacillary morphology and become spherical forms (30), an observation that has not been reported for any other rod-shaped bacteria. The results obtained for M. xanthus homeostasis suggest that this bacterium copes with changing metal concentrations by using several copies of genes that are differentially regulated by environmental fluctuations. As those paralogs that perform the same function under different conditions have been called ecoparalogs (29), this term can be applied to all M. xanthus genes involved in metal homeostasis.
An interesting expression profile (because it cannot be observed for bacteria that do not undergo development) is that exhibited by czc3, which is expressed constitutively, induced by starvation, and repressed by the addition of metals. Although the M. xanthus MCO cuoC is also expressed constitutively and induced by starvation (30), the difference from the czc3 expression profile lies in the fact that cuoC is activated by copper, while czc3 is repressed. The induction of genes encoding proteins involved in copper homeostasis (and homeostasis of other metals) by starvation suggests that M. xanthus cells must also adjust the metal concentration of the dormant myxospores, which would explain why developing cells are much more sensitive to copper than growing cells (30).
Another conclusion that can be deduced from the results obtained for the M. xanthus Cus and Czc systems is that not all of them fit into the classification proposed previously by Rensing and Grass (26), based on the sequence motif of TM4 of the RND subunit. Thus, Cus1 resembles a Czc rather than a Cus system. Therefore, it cannot be ruled out that some of the other CBA systems encoded in the M. xanthus genome that have not been included in this study could also participate in metal homeostasis.
The differential regulation of HME-RND systems, P1B-ATPases, and MCOs indicates that several transcriptional regulators will participate in the adaptation of M. xanthus to changes in the concentrations of copper and other metals. The identification of these factors and the determination of how they sense the fluctuations in metals and nutrients will help to elucidate how this bacterium completes its complex life cycle in soil.
Supplementary Material
Acknowledgments
This work has been supported by grants from the Junta de Andalucía (grant CVI-1377) and the Ministerio de Ciencia y Tecnología (grant BFU2006-00972/BMC), Spain, 70% funded by FEDER, and funded by a grant from the Ministerio de Ciencia e Innovación, Spain, program Consolider-Ingenio 2010 (grant CSD2009-00006). A.M.-M. has been funded by a postdoctoral fellowship from Plan Propio de la Universidad de Granada.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Argüello, J. M., E. Eren, and M. Gonzalez-Guerrero. 2007. The structure and function of heavy metal transport P1B-ATPases. Biometals 20:233-248. [DOI] [PubMed] [Google Scholar]
- 2.Cho, K., and D. R. Zusman. 1999. AsgD, a new two-component regulator required for A-signalling and nutrient sensing during early development of Myxococcus xanthus. Mol. Microbiol. 34:268-281. [DOI] [PubMed] [Google Scholar]
- 3.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finney, L. A., and T. V. O'Halloran. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931-936. [DOI] [PubMed] [Google Scholar]
- 5.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg, M., T. Pribyl, S. Juhnke, and D. H. Nies. 1999. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 274:26065-26070. [DOI] [PubMed] [Google Scholar]
- 7.Gupta, A., K. Matsui, J. F. Lo, and S. Silver. 1999. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 5:183-188. [DOI] [PubMed] [Google Scholar]
- 8.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 9.Hassan, M. T., D. van der Lelie, D. Springael, U. Romling, N. Ahmed, and M. Mergeay. 1999. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene 238:417-425. [DOI] [PubMed] [Google Scholar]
- 10.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U. S. A. 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 13.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 14.Leedjarv, A., A. Ivask, and M. Virta. 2008. Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J. Bacteriol. 190:2680-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moraleda-Muñoz, A., J. Pérez, A. L. Extremera, and J. Muñoz-Dorado. 2010. Expression and physiological role of three Myxococcus xanthus copper-dependent P1B-type ATPases during bacterial growth and development. Appl. Environ. Microbiol. 76:6077-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moraleda-Muñoz, A., J. Perez, M. Fontes, F. J. Murillo, and J. Muñoz-Dorado. 2005. Copper induction of carotenoid synthesis in the bacterium Myxococcus xanthus. Mol. Microbiol. 56:1159-1168. [DOI] [PubMed] [Google Scholar]
- 17.Nies, D. H. 2007. Bacterial transition metal homeostasis, p. 118-142. In D. H. Nies and S. Silver (ed.), Molecular microbiology of heavy metals. Springer-Verlag, Berlin, Germany.
- 18.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 19.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 20.Nies, D. H., A. Nies, L. Chu, and S. Silver. 1989. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc. Natl. Acad. Sci. U. S. A. 86:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nies, D. H., G. Rehbein, T. Hoffmann, C. Baumann, and C. Grosse. 2006. Paralogs of genes encoding metal resistance proteins in Cupriavidus metallidurans strain CH34. J. Mol. Microbiol. Biotechnol. 11:82-93. [DOI] [PubMed] [Google Scholar]
- 22.Nies, D. H., and S. Silver. 1989. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J. Bacteriol. 171:896-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 24.Pena, M. M., J. Lee, and D. J. Thiele. 1999. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 129:1251-1260. [DOI] [PubMed] [Google Scholar]
- 25.Pontel, L. B., M. E. Audero, M. Espariz, S. K. Checa, and F. C. Soncini. 2007. GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol. Microbiol. 66:814-825. [DOI] [PubMed] [Google Scholar]
- 26.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 27.Rensing, C., T. Pribyl, and D. H. Nies. 1997. New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 179:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Sanchez-Perez, G., A. Mira, G. Nyiro, L. Pasic, and F. Rodriguez-Valera. 2008. Adapting to environmental changes using specialized paralogs. Trends Genet. 24:154-158. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez-Sutil, M. C., N. Gomez-Santos, A. Moraleda-Muñoz, L. O. Martins, J. Perez, and J. Muñoz-Dorado. 2007. Differential expression of the three multicopper oxidases from Myxococcus xanthus. J. Bacteriol. 189:4887-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt, T., and H. G. Schlegel. 1994. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J. Bacteriol. 176:7045-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 33.Ueki, T., S. Inouye, and M. Inouye. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153-157. [DOI] [PubMed] [Google Scholar]
- 34.Waldron, K. J., J. C. Rutherford, D. Ford, and N. J. Robinson. 2009. Metalloproteins and metal sensing. Nature 460:823-830. [DOI] [PubMed] [Google Scholar]
- 35.Whitworth, D. E (ed.). 2008. Myxobacteria. Multicellularity and differentiation. ASM Press, Washington, DC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.