Abstract
A point mutation (E115K) resulting in slower growth of Escherichia coli DH5α and XL1-Blue in minimal media was identified in the purB gene, coding for adenylosuccinate lyase (ASL), through complementation with an E. coli K-12 genomic library and serial subcultures. Chromosomal modification reversing the mutation to the wild type restored growth phenotypes in minimal media.
The Escherichia coli DH5α strain possesses many beneficial genotypes (recA, deoR, gyrA, and endA1) and has been widely used for many purposes, such as gene cloning and protein production (5). However, E. coli DH5α also exhibits inferior growth phenotypes, especially in minimal media, compared to other E. coli strains. As such, the utilization of this bacterium has been limited to the laboratory despite its numerous advantages. We can assume that these inferior growth phenotypes have resulted from unknown accumulated mutations during the strain development process (5). Some of those mutations, which might impact growth in minimal media, have been characterized, including the phenotypes for thiamine requirement and relaxed amino acid synthesis (5). Still, there may be other uncharacterized mutations whose interactions hamper the growth of E. coli DH5α in minimal media.
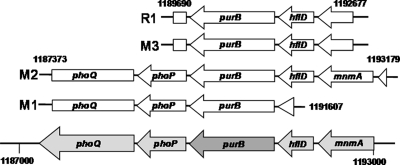
Based on successful identifications (6, 7) of gene targets for metabolic engineering (3), we performed serial subcultures of E. coli DH5α transformants with an E. coli K-12 genomic library based on a multicopy plasmid (9) to isolate genes that improve growth phenotypes in minimal media. The M9 minimal medium and R medium (11) were chosen for enrichment experiments because of their popular use in metabolic engineering (1, 2, 7) and in high-cell-density fermentation (8, 10, 11). After 11 serial transfers of the transformants in the M9 medium, and 27 transfers in the R medium, cultured cells were diluted and plated onto LB agar for single-colony isolation. Although more than 10 colonies were picked, only three distinctive plasmids, containing different inserts, were isolated from the transformants enriched in M9 medium. In the case of R medium enrichment, all isolated plasmids were identical. Sequencing of the isolated plasmids revealed the exact genome coordinates of each insert. A diagram of the inserts in the context of the E. coli genome sequence is shown in Fig. 1. Interestingly, all of the isolated plasmids contained similar regions of genomic DNA. mnmA (tRNA 5-methylaminomethyl-2-thiouridylate-methyltransferase), purB (adenylosuccinate lyase), and hflD (lysogenization regulator) were the annotated genes in the overlapping region among distinctive isolated fragments. However, since the N-terminal portions of mnmA and hflD were truncated in some of the inserts, we selected only the M3 and R1 plasmids for further experimentation. These two plasmids were retransformed into E. coli DH5α for confirmation of their beneficial effects on growth of E. coli in minimal media. The newly transformed strains showed growth phenotypes almost identical to those of the previously isolated transformants. When cultured in flasks, the specific growth rate of E. coli DH5α with the R1 plasmid was 1.5-fold higher (0.53 versus 0.36 h−1) than the rate of cells transformed with a control plasmid (pZE). The R1 transformant reached the stationary phase much earlier, arriving at an optical density at 600 nm (OD600) of 10 within 16 h, whereas the control transformant reached this cell density after 24 h. However, the final cell densities were almost equivalent. Acetate accumulation, as well as glucose consumption, by the R1 transformant was much higher than that of the control transformant (2.2 versus 0.3 g acetate/liter). The increased accumulation of acetate could be the result of increased cell density. These findings confirm that the enhanced growth phenotypes of the isolated transformants were conferred not by accumulated spontaneous mutations in the genome during enrichment but by the introduced plasmids.
FIG. 1.
Diagram of open reading frames in the identified genomic DNA fragments. M1, M2, and M3 were isolated from the serial subculture using M9 medium. R1 was isolated from the serial subculture using R medium.
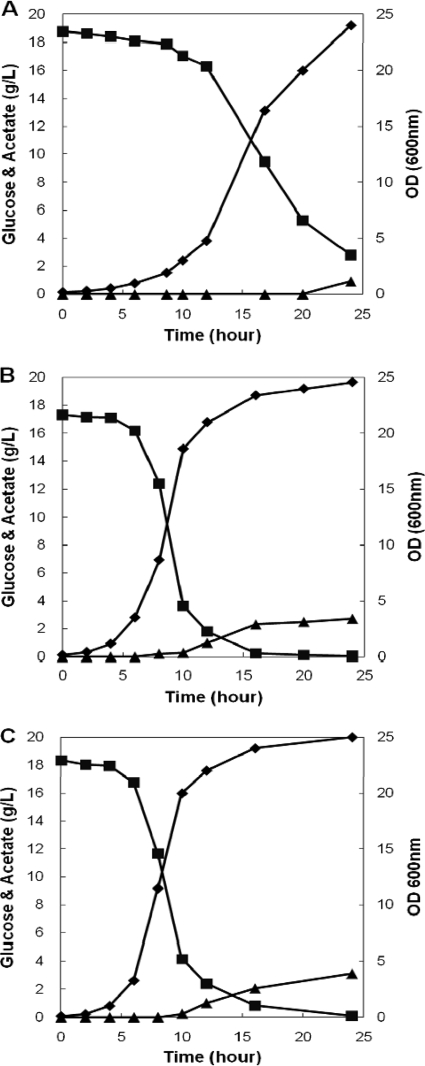
The open reading frame (ORF) of purB was amplified and cloned into a multicopy plasmid under the control of a strong promoter (rrnB). Transformation of the resulting plasmid (pZE-purB) into E. coli DH5α resulted in a growth phenotype almost identical to that of the R1 transformant. This result suggested that overexpression of purB is a specific genetic perturbation improving growth phenotypes of E. coli DH5α in minimal media. We also performed 1-liter batch fermentation experiments with three DH5α transformants: one containing the control plasmid (pZE), one with the isolated plasmid (R1), and a third with the purB overexpression plasmid (pZE-purB). Growth phenotypes of these strains were very similar to results obtained from shaker flask experiments (Fig. 2). Next, we tested whether the overexpression of purB is beneficial to the growth of other E. coli strains by introducing the R1 and pZE-purB plasmids into various other strains (K-12, BL21, and XL1-Blue) that are commonly used in biotechnological research. Among the four strains tested in our various experiments, the positive effects of purB overexpression on growth phenotypes were observed only in DH5α and XL1-Blue, both of which have been favored in molecular cloning. These results suggest that an uncharacterized mutation might have been introduced into both strains during strain development. This unknown mutation might cause growth inhibition, which can be suppressed by the overexpression of purB. Therefore, we concluded that expression of an exogenous, K-12-derived copy of the purB gene under a constitutive promoter can enhance growth phenotypes of E. coli DH5α and XL1-Blue strains in minimal media.
FIG. 2.
Comparison of levels of cell growth (♦) (OD600), glucose consumption (▪) (g/liter), and acetate production (▴) (g/liter) by E. coli DH5α transformants with a control plasmid (A), the isolated R1 plasmid (B), and the pZE-purB plasmid (C) in R medium with glucose in a bioreactor.
However, it is plausible that a mutation is located in the purB locus of DH5α and XL1-Blue that decreases the activity of the encoded enzyme. In order to identify a putative mutation in purB, we sequenced the chromosomal purB gene of DH5α and XL1-Blue. A point mutation resulting in the transition of nucleotide 343 of purB from guanine (G) to adenine (A) was identified in the genomes of both strains. This mutation causes a change of the 115th residue of adenylosuccinate lyase from glutamate to lysine (E115K). This finding explains why the expression of exogenous, K-12-derived purB in DH5α and XL1-Blue strains enhances growth phenotypes in minimal media. The E115K mutation of purB was named purB20 for simple notation.
Chromosomal modification of the mutant allele in E. coli DH5α or XL1-Blue might be desirable for practical applications. To this end, the purB20 mutant allele was replaced by purB amplified from E. coli K-12 through recombination based on phage lambda Red recombinase (4). The resulting strain (SC1) showed growth phenotypes similar to those of E. coli DH5α strains harboring the pZE-purB or R1 plasmid. The specific growth rate of SC1 in M9 medium was 40% higher than that of DH5α (0.50 versus 0.36 h−1). These results show that what we had originally interpreted as overexpression of the purB gene was actually complementation of the mutant purB20 allele with wild-type purB. We also tested whether the modification from purB20 to wild-type purB elicits a change in the transformation efficiency. Chemically induced competent SC1 cells exhibited approximately 2.5-fold lower transformation efficiency than E. coli DH5α cells did when induced under identical conditions (1.8 ± 0.1 × 106 versus 4.6 ± 0.3 × 106 CFU/μg pUC19 DNA). Still, the transformation efficiency of the SC1 strain was of the same order of magnitude as that of E. coli DH5α, suggesting that the SC1 strain would be useful for many biotechnological applications, such as the mass production of DNA vectors and recombinant proteins.
Acknowledgments
This work was supported by the MIT Energy Initiative and NSF grant CBET-0730238, as well as a Korea Research Foundation grant funded by the Korean Government (KRF-2006-331-F00057).
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Alper, H., Y. S. Jin, J. F. Moxley, and G. Stephanopoulos. 2005. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 7:155-164. [DOI] [PubMed] [Google Scholar]
- 2.Alper, H., K. Miyaoku, and G. Stephanopoulos. 2006. Characterization of lycopene-overproducing E. coli strains in high cell density fermentations. Appl. Microbiol. Biotechnol. 72:968-974. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, J. E. 1991. Toward a science of metabolic engineering. Science 252:1668-1675. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid tranformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 6.Jin, Y. S., H. Alper, Y. T. Yang, and G. Stephanopoulos. 2005. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl. Environ. Microbiol. 71:8249-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin, Y. S., and G. Stephanopoulos. 2007. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab. Eng. 9:337-347. [DOI] [PubMed] [Google Scholar]
- 8.Lee, S. Y. 1996. High cell-density culture of Escherichia coli. Trends Biotechnol. 14:98-105. [DOI] [PubMed] [Google Scholar]
- 9.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I-1-I-2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riesenberg, D. 1991. High-cell density cultivation of Escherichia coli. Curr. Opin. Biotechnol. 2:380-384. [DOI] [PubMed] [Google Scholar]
- 11.Riesenberg, D., K. Menzel, V. Schulz, K. Schumann, G. Veith, G. Zuber, and W. A. Knorre. 1990. High cell-density fermentation of recombinant Escherichia coli expressing human interferon alpha-1. Appl. Microbiol. Biotechnol. 34:77-82. [DOI] [PubMed] [Google Scholar]