Abstract
Nitrogen is often a limiting nutrient in natural habitats. Therefore, cyanobacteria have developed multiple responses, which are controlled by transcription factor NtcA and the PII-signaling protein, to adapt to nitrogen deficiency. Transcriptional analyses of Synechocystis sp. strain PCC 6803 under nitrogen-deficient conditions revealed a highly induced gene (sll0783) which is annotated as encoding a conserved protein with an unknown function. This gene is part of a cluster of seven genes and has potential NtcA-binding sites in the upstream region. Homologues of this cluster occur in some unicellular, nondiazotrophic cyanobacteria and in several Alpha, Beta-, and Gammaproteobacteria, as well as in some Gram-positive bacteria. Most of the heterotrophic bacteria harboring this gene cluster are able to fix nitrogen and to produce polyhydroxybutyrate (PHB), whereas of the cyanobacteria, only Synechocystis sp. strain PCC 6803 can accumulate PHB. In this work, a Synechocystis sp. strain PCC 6803 sll0783 gene knockout mutant is characterized. This mutant is unable to accumulate PHB, a carbon and energy storage compound. In contrast, the levels of the carbon storage compound glycogen and the PHB precursor acetyl coenzyme A were similar to those of the wild type, indicating that the PHB-deficient phenotype does not likely result from a global deficiency in carbon metabolism. A specific deficiency in PHB synthesis was implied by the fact that the mutant exhibits impaired PHB synthase activity during prolonged nitrogen starvation. However, the expression of PHB synthase-encoding genes was not strongly affected in the mutant, suggesting that the impaired PHB synthase activity observed depends on a posttranscriptional process in which the product of sll0783 is involved.
Cyanobacteria are oxygenic photoautotrophs (32) adapted to a wide range of environments (7, 30). In natural habitats, combined nitrogen is frequently a limiting nutrient, and therefore, cyanobacteria have developed multiple strategies to adapt to nitrogen deprivation. Synechocystis sp. strain PCC 6803, a nondiazotrophic cyanobacterium, responds to the lack of combined-nitrogen sources by bleaching, a process known as chlorosis (3). Concomitantly with bleaching, the cells accumulate excess carbon fixation products in the form of intracellular polymer deposits composed of glycogen or polyhydroxyalkanoates, the latter reported from over 50 cyanobacterial strains from the major phylogenetic subsections (37).
In Synechocystis strain PCC 6803, the polyhydroxyalkanoate PHB (polyhydroxybutyrate) was reported to accumulate under conditions of nitrogen or phosphate deprivation, upon the addition of acetate, or in response to a high photon flux density (38). The pathway of PHB synthesis and its corresponding genes have been elucidated in Synechocystis strain PCC 6803. The β-ketothiolase (slr1993) forms acetoacetyl coenzyme A (acetoacetyl-CoA) from two molecules of acetyl-CoA. Acetoacetyl-CoA reductase (slr1994) then reduces the acetoacetyl-CoA to d-3-hydroxybutyryl-CoA (33), which is subsequently polymerized to PHB. The polymerization reaction is catalyzed by PHB synthase, an enzyme which consists of a heterodimer of PhaC (slr1829) and PhaE (slr1830) (13). The genes for the two subunits of PHB synthase are organized into one operon, as well as the genes of the first two enzymes of the PHB synthesis pathway, β-ketothiolase and acetoacetyl-CoA reductase (33). The regulation of PHB synthesis in cyanobacteria has been poorly investigated. One of the few studies done revealed that PHB synthase activity is stimulated by the addition of acetyl phosphate to extracts of Synechococcus strain MA19 (22).
The response of cyanobacteria to nitrogen starvation largely depends on the NtcA (nitrogen control protein A) transcriptional regulator. NtcA is a transcriptional activator and in some cases a repressor of a large number of genes mainly involved in nitrogen metabolism (15). NtcA belongs to the cyclic AMP receptor protein family of transcriptional regulators (36) and binds to the conserved palindromic sequence GTA-N8-TAC (15). In an attempt to identify novel genes that respond to nitrogen starvation, a proteome analysis of combined-nitrogen-deprived Synechococcus elongatus PCC 7942 cells has been performed. This study revealed several proteins which were highly induced upon starvation (1). The N terminus of one of these proteins, termed Nsi5 (nitrogen starvation-induced 5), is highly similar to the product of the sll0783 gene of Synechocystis strain PCC 6803. Transcriptomic analyses of Synechocystis strain PCC 6803 under nitrogen starvation indicated, that sll0783 is the gene with the highest induction after 4 h of nitrogen starvation (23). Database analyses predict a protein of unknown function with a conserved DsrE domain for the sll0783 product, and close homologues are present in various other unicellular cyanobacteria. The DsrE family members are small soluble proteins involved in intracellular sulfur redox reactions (20).
To reveal the function of sll0783, we analyzed the expression of this gene and constructed an sll0783 knockout mutant of Synechocystis strain PCC 6803. The present report describes the first step in elucidating the function of the product of sll0783. The mutant shows impaired accumulation of PHB following nitrogen starvation, due to impaired PHB synthase activity.
MATERIALS AND METHODS
Organisms and culture conditions.
Wild-type Synechocystis strain PCC 6803 and the corresponding sll0783 mutant were grown photoautotrophically in BG11 medium (27) supplemented with 5 mM NaHCO3 in flasks shaken at 150 rpm at a continuous photon flux density of 50 μmol photons m−2 s−1 at 28°C. For initiation of nitrogen deprivation, 50 ml of exponentially growing cells was harvested by centrifugation (8 min, 4,000 rpm) and the pellet was resuspended in NaNO3-free BG11 medium (BG11-N) and centrifuged again. Finally, the washed cells were resuspended again in BG11-N to an optical density at 750 nm (OD750) of 0.4 and incubated as described above. To examine recovery after 72 h of nitrogen deprivation, the starved cells were diluted in BG11-N to an OD750 of 0.4 and NaNO3 was added to a final concentration of 5 mM.
Bioinformatic data analyses.
The predicted amino acid sequence encoded by sll0783 was used to perform a PSI-BLAST search (4) against the nonredundant protein sequences database of the NCBI homepage (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Conserved domains were automatically detected by CD-Search (21), and protein parameters were obtained from the ExPASy platform (10). The phylogenetic relationships were calculated using the neighbor-joining method (28), which is integrated in the ClustalX v2.0.11 software package (34). The results were visualized as a rectangular tree with the phylogenetic-tree-plotting program TreeView (24).
Growth and pigmentation.
Growth of the cells was monitored by measuring the OD750 of the culture with an LKB Ultrospec III spectrophotometer (Pharmacia). Differences in pigmentation were analyzed by difference spectra as follows. For each sample, a spectrum in the range of 350 to 750 nm was recorded with a Specord 205 (Analytik Jena, Jena, Germany). Thereafter, the WinAspect 2.2.1.0 (Analytik Jena) software was used to correct the wavelength-dependent light scattering by baseline correction using 750 and 400 nm as reference points. Then the corrected spectrum at time point zero was subtracted from the corrected spectra at subsequent time points. From the difference spectra, the peaks at 630 and 680 nm were determined with respect to the baseline at 750 nm. These values quantify the changes in the amounts of phycobilisomes and chlorophyll a.
Construction of the sll0783 knockout mutant.
Inactivation of the sll0783 gene was done by insertion of a kanamycin/bleomycin resistance cassette into the HindIII site of the gene. Plasmid pSLL0783-Kan was constructed by amplification of the sll0783 gene via PCR with primers sll0783-1 (5′-GCGGTACCGCTTTACATAGACAGACCTG-3′) and sll0783-2 (5′-GCGAGCTCTAGGTGACTTTTCCCCATCA-3′), restriction of the purified PCR product with KpnI and SacI, and cloning of the fragment into pBluescript II (Fermentas, St. Leon-Rot, Germany). The kanamycin resistance cassette was inserted into the HindIII restriction site of the sll0783 gene. Wild-type Synechocystis strain PCC 6803 was transformed with the resulting vector according to the method of Golden et al. (11). Segregation was confirmed by Southern blot analyses (19).
Analysis of the 5′ end of the sll0783 transcript.
For determination of the 5′ end of the sll0783 transcript, the 5′/3′RACE (rapid amplification of cDNA ends) kit, second generation (Roche Applied Science, Mannheim, Germany) was used according to the manufacturer's instructions. Wild-type Synechocystis strain PCC 6803 cells in the mid-exponential phase of growth were shifted to nitrogen-free medium, and after 6 h of starvation, 50 ml of culture (OD750 of 0.8) was harvested on ice by centrifugation (4,000 rpm, 10 min, 4°C) and used for RNA extraction. For first-strand cDNA synthesis, 0.5 μg of RNA and an antisense gene-specific primer (sll0783-SP1 [5′-GTAACACCGGGACCATAGAG-3′]) located 205 bp downstream of the ATG start codon were used, and for the nested PCR, the PCR anchor primer (Roche) and an antisense gene-specific primer (sll0783-SP2 [5′-CCTTCAAACGCCACTGTA-3′]) located 115 bp downstream of the ATG start codon were used. Amplification products were checked by agarose gel electrophoresis. The final amplification products were purified with a gel purification kit (Qiagen, Germany) and subcloned into the pGMT-EASY vector using the pGMT-EASY cloning kit (Fermentas). Following transformation of Escherichia coli, insert-containing plasmids were selected and analyzed by sequencing. The sequences were aligned against the upstream region of sll0783 (sequence available at Cyanobase [http://bacteria.kazusa.or.jp/cyano/]).
RNA isolation and cDNA synthesis.
Thirty-five-milliliter volumes of cell cultures grown under different conditions were rapidly cooled by addition of ice and harvested by centrifugation (4,000 rpm, 10 min, 4°C), and RNA was isolated by hot acid-phenol-chloroform extraction and ethanol precipitation combined with the High Pure RNA isolation kit (Roche) according to the manufacturer's instructions (8). cDNA synthesis was performed by using the iScript Select cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions, with 1 μg of total RNA.
qRT-PCR.
Transcript levels were quantified by quantitative reverse transcription-PCR (qRT-PCR) using the iQ5 qRT-PCR detection system (Bio-Rad). Amplifications were performed using IQ SYBR green supermix (Bio-Rad) and gene-specific primers (see Supplement 4 in the supplemental material). Two-step cycling was performed by amplification with an initial preheating step of 10 min at 95°C, 30 cycles at 95°C for 15 s and 60°C for 30 s, and a final 10-min elongation step at 72°C. The level of rnpB was used as a loading control (25). The relative mRNA level (normalized to the level of rnpB) of each specific transcript was determined with the Bio-Rad software according to the 2−ΔΔCT method (18). The transcript level of wild-type cells at time point zero was set to 1. For the correct calculation of transcript abundance, the PCR efficiency was determined by dilution series with genomic DNA. Analyses were performed using triplicate technical replicates from duplicate biological cultures. To determine melting temperatures for the amplification products of the specific primers, the temperature was raised after qRT-PCR from 65 to 95°C, and fluorescence was detected continuously.
Microscopy of PHB granules.
PHB granules in Synechocystis strain PCC 6803 cells were visualized by staining with the fluorescent dye Nile red (31). To 20 μl of cell culture, 6.6 μl of Nile red solution (1 μg ml−1in ethanol) was added. Of this mixture, 15 μl was dropped onto glass slides which had been covered with 1 ml of 2% agarose in H2O and dried. The cells were analyzed by fluorescence microscopy using a Leica DM5500B microscope with a 100×/1.30 oil immersion objective lens (Leica Microsystems, Wetzlar, Germany) and a filter cube with 535/50-nm excitation and 610/75-nm suppression wavelengths. Pictures were taken with a Leica DFC420 camera.
Fluorescence quantification.
For the quantification of Nile red fluorescence, 180 μl of cells was stained with 20 μl of Nile red solution (1 μg ml−1 in ethanol) and fluorescence was measured by a fluorescence microplate reader (Infinite 200; Tecan) with an excitation wavelength of 530 nm and an emission wavelength of 570 nm. Growth medium without cells containing Nile red was used as a blank.
Acetyl-CoA determination.
Acetyl-CoA was determined by a coupled assay in which the formation of oxaloacetate and NADH from malate and NAD was coupled to the formation of citrate from oxaloacetate and acetyl-CoA (6). To perform the assay, 50 ml of cell culture was harvested by centrifugation (4,000 rpm, 10 min, 4°C) and resuspended in 5 ml of 4 M perchloric acid. After 5 min of incubation on ice, the sample was neutralized with KOH and adjusted to a pH value between 6.3 and 6.7 with KHCO3. After centrifugation (4,000 rpm, 5 min, 4°C), the supernatant was freeze-dried and the residue was used for acetyl-CoA determination. The samples were dissolved in 0.5 ml of H2O and mixed with Tris-HCl (pH 8.1; final concentration, 200 mM), l-malate (final concentration, 2.5 mM), and NAD (final concentration, 1.5 mM) to give a final volume of 1 ml. Then 5 μl of malate dehydrogenase was added (final concentration, 0.9 U/ml). When the A339 was constant, the value was recorded (A1). After that, citrate synthase was added (final concentration, 75 mU/ml) and the A339 (A2) was determined when the absorbance was constant. The acetyl-CoA concentration was calculated according to the equation c = 0.325 × (A2 − A1)/v, where c is the concentration of acetyl-CoA and v is the sample volume in the assay mixture.
Glycogen determination.
Two milliliters of a bacterial culture with an OD750 of 0.5 was harvested by centrifugation (2 min, 13,000 × g) and resuspended in 0.5 ml of BG11-N. Ten microliters of 54% H2SO4 was added to 200 μl of this suspension, and the mixture was incubated for 20 min at 100°C. The reaction was stopped by neutralization with 20 μl of 5 M NaOH. For the determination of glucose (derived from acid hydrolysis of glycogen), 1 ml of o-toluidine reagent (Sigma-Aldrich) (17) was added. After 3 min of incubation on ice, the A635 was measured and glucose was quantified with a glucose standard curve.
Enzyme assays.
All steps for preparing cell extracts were performed at 4°C or on ice. Cells of Synechocystis strain PCC 6803 and the sll0783 mutant were harvested from nitrogen-limited or nitrogen-sufficient cultures by centrifugation for 8 min at 4,000 × g. Then they were resuspended in lysis buffer (25 mM Tris/HCl, pH 7.4, 50 mM KCl, 5 mM MgCl2, 0.5 mM EDTA) and disrupted in a Fast-Prep24 apparatus (20 s, intensity setting of 6.5 M/s). Cell debris was removed by centrifugation at 1,000 × g for 1 min. The supernatant was centrifuged again at 20,000 × g for 30 min. The sediment was resuspended in 0.5 ml of Tris/HCl, pH 7.4, and the protein concentration was quantified according to Bradford (5).
Assay of PHB synthase activity was carried out as described previously (35). The assay mixtures (200 μl) contained 20 μg of protein, 100 μM dl-3-hydroxybutyryl-CoA, and 1 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) in 25 mM Tris/HCl, pH 7.4, buffer with 20 mM MgCl2. The reaction mixtures were transferred to microplate wells, and the reaction was started by addition of the substrate dl-3-hydroxybutyryl-CoA. The reaction was recorded in an EL808 microplate reader (BioTek) at a temperature setting of 30°C and the time course of the change in A409 (due to the reaction of the released CoA with DTNB) was monitored for 5 min.
PHB content.
Quantitative analysis of PHB content was done by high-performance liquid chromatography (HPLC) analysis as described previously (33). At different time points, 50- to 100-ml culture volumes were collected by centrifugation (8 min, 4,000 × g, 4°C). The resulting cell pellets were dried overnight at 85°C and then weighed with an analytical balance (BP2110; Sartorius). The dry pellets were boiled in 1 ml of concentrated H2SO4 for 60 min, diluted with 1 ml of 0.014 M H2SO4, and filtered through a polyvinylidene difluoride membrane. Samples were then diluted 1:10 with 0.014 M H2SO4 and analyzed by HPLC with a ReproSil-Pur octyldecyl silane 3.5-μm C18 column. Commercially available PHB, processed in parallel with the samples, was used for quantification, and crotonic acid was used as the standard for HPLC analysis.
RESULTS
Bioinformatic analyses of the sll0783 gene.
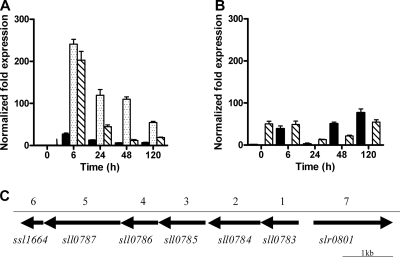
Bioinformatic analyses of the sll0783 gene predict a product of 160 amino acids with a mass of 17.64 kDa and a conserved DsrE domain (pfam02635). It is a putative cytoplasmic soluble protein without any signal sequences. A PSI-BLAST search showed that homologous sequences occur in several unicellular, nondiazotrophic cyanobacteria and in a variety of Alpha-, Beta-, and Gammaproteobacteria, as well as in some Gram-positive bacteria (see Supplement 1 in the supplemental material). To gain further insights into the phylogeny of the sll0783 gene, a phylogenetic tree of the cyanobacterial sll0783 homologues was compared with the 16S phylogeny of these organisms (see Supplement 2 in the supplemental material). This comparison reveals a remarkably similar phylogeny, suggesting early acquisition of this gene during cyanobacterial evolution. Analysis of the surrounding gene region revealed that this gene appears as the first gene of a cluster which had been previously described in the context of the following gene, sll0784 (26). The sll0784 gene encodes a nitrilase which has already been characterized biochemically (14). The genes in the cluster occur in the following order: (i) a hypothetical protein (sll0783), (ii) a nitrilase (sll0784), (iii) a radical S-adenosylmethionine superfamily member (sll0785), (iv) a potential acetyltransferase (sll0786), (v) an AIR synthase-related protein (sll0787), (vi) a hypothetical protein (ssl1464), and (vii) a flavoprotein predicted to be involved in K+ transport (slr0801) (Fig. 1C). In cyanobacteria, the latter gene is placed in front of the cluster on the opposite strand. Interestingly, all heterotrophic bacteria containing this gene cluster are able to produce PHB and/or can fix nitrogen. In contrast, Synechocystis strain PCC 6803 is the only cyanobacterium containing this cluster which harbors PHB synthase genes. Furthermore, all of the cyanobacteria containing this cluster are unicellular and nondiazotrophic. The DNA sequences upstream of the cyanobacterial sll0783 homologues contain perfect NtcA recognition sites (GTA-N8-TAC), indicating that their expression might be NtcA dependent (see Supplement 3A in the supplemental material). Only in Synechocystis strain PCC 6803 does the upstream region of sll0783 display an imperfect NtcA recognition site (see Supplement 3 in the supplemental material) (2, 15).
FIG. 1.
Expression of slr0801 (black bars), sll0783 (dotted bars), and sll0784 (dashed bars) in the Synechocystis strain PCC 6803 wild type (A) and sll0783 mutant (B) under nitrogen-deficient conditions. Levels of mRNA were determined by qRT-PCR and normalized to the rnpB mRNA level. The value at time point zero (immediately before nitrogen step down) was set to 1. For details, see Materials and Methods. (C) In-scale schematic representation of the sll0783 gene cluster.
Expression of sll0783 and surrounding genes.
Beyond the uncertain NtcA recognition site in front of sll0783, in Synechocystis strain PCC 6803, the intergenic region between sll0783 and the potential flavoprotein-encoding gene slr0801 has a size of 465 bp, which is substantially larger than the corresponding intergenic region in other cyanobacteria containing this gene cluster. Therefore, it was necessary to determine experimentally the location of the transcriptional start point of sll0783 to reveal whether it matches the potential NtcA binding site. The 5′ end of the sll0783 transcript was determined by the 5′ RACE method (see Materials and Methods). The putative transcriptional start point, as deduced from this experiment, is located 48 bp upstream of the predicted ATG start codon (see Supplement 3B in the supplemental material). According to this result, the promoter seems to be a noncanonical class I promoter with a −35 site and a putative NtcA binding site 107 bp upstream of the transcriptional start point (16).
The expression of sll0783 and its surrounding genes sll0784 and slr0801 was determined by qRT-PCR in Synechocystis strain PCC 6803 wild-type and sll0783 knockout mutant cells (see Materials and Methods) grown under nitrate-replete conditions and shifted to nitrate-depleted conditions. Under nitrate-replete conditions, the sll0783, sll0784, and slr0801 genes were only slightly expressed, whereas under nitrogen starvation conditions, all three genes were highly induced after 6 h. Transcripts of sll0783 and sll0784 increased about 250-fold and 200-fold, respectively, whereas slr0801 transcripts increased 30-fold. During the course of nitrogen starvation, the expression of these genes regressed again (Fig. 1A).
In the sll0783 mutant, expression of slr0801 was not impaired in comparison to that in the wild type. The sll0784 gene, downstream of sll0783, was constitutively expressed (Fig. 1B) in the mutant as a result of the insertion of a terminator-free, promoter-carrying kanamycin resistance cassette in sll0783, driving transcription in the direction of the downstream (sll0784) genes.
Recovery after nitrogen starvation.
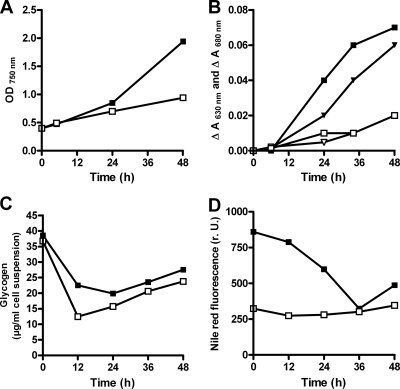
Under standard culture conditions in nitrogen-replete BG11medium, the mutant showed no impairment of growth. Under nitrogen starvation, pigmentation and photosynthetic activity decayed as fast as in the wild type (data not shown). However, after the addition of 5 mM sodium nitrate to cells which had been starved for combined nitrogen for 3 days, the sll0783 mutant was unable to recover as fast as the wild type (Fig. 2A). Following the onset of recovery, the mutant showed a prolonged lag phase compared to the wild-type strain. The impairment of recovery was clearly visible in the time course of chlorophyll a and phycobilisome recovery (Fig. 2B). To further reveal the impaired recovery of the mutant from nitrogen starvation, cells were examined microscopically. Whereas intracellular granules were visible in nitrogen-starved wild-type cells, these structures were absent in the mutant. Staining of the cells with the lipophilic dye Nile red indicated that these were PHB granules (Fig. 3D).
FIG. 2.
Recovery of the Synechocystis strain PCC 6803 wild type (filled squares) and sll0783 mutant (open squares) from previous nitrogen starvation. Sodium nitrate (5 mM) was added to cells nitrogen starved for 72 h. Over time, various parameters were determined. (A) Growth of the wild type and the sll0783 mutant was followed by measuring the OD750 of the cultures. (B) Changes in in vivo chlorophyll a and phycobilisome quantities, as determined by difference spectrum analysis. The change in A680 corresponds to chlorophyll a (triangles), and the change in A630 corresponds to phycobiliproteins (squares). Also shown are (C) glycogen content and (D) quantification of Nile red fluorescence, indicating the PHB level.
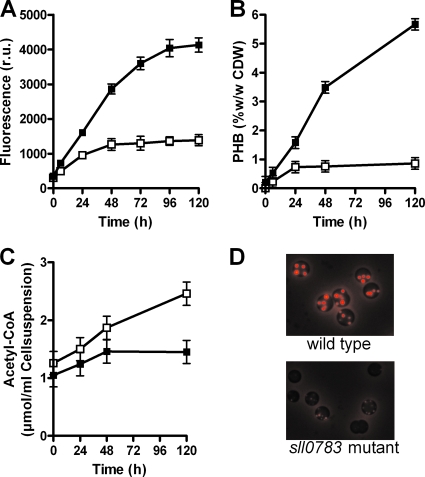
FIG. 3.
Quantification of PHB content and the level of PHB precursor molecules during nitrogen starvation. (A) Spectroscopic quantification of Nile red fluorescence in the wild type (filled squares) and the sll0783 mutant (open squares). (B) HPLC analysis of the PHB content of the wild type (filled squares) and the sll0783 mutant (open squares). (C) Acetyl-CoA quantification in the wild type (filled squares) and the sll0783 mutant (open squares). (D) Fluorescence microscopy of cells starved for nitrogen for 72 h and then stained with Nile red.
Impairment of the sll0783 mutant in PHB accumulation.
During normal cultivation with nitrogen-replete BG11 medium, neither the wild type nor the sll0783 mutant accumulated PHB granules (data not shown). After transfer to combined-nitrogen-free medium, wild-type cells started to accumulate PHB granules, as revealed by fluorescence microscopy of Nile red-stained cells. Over time, the granule size and amount increased. The sll0783 mutant initially started with the production of PHB granules; however, the granules did not grow in size and amount (Fig. 3D).
To quantify the microscopically visible difference in PHB accumulation, the whole PHB content of the cells was determined by HPLC analysis (Fig. 3B). After 120 h of N starvation, the wild type reached a PHB content of 5.7% (wt/wt, cell dry weight) PHB, whereas the amount of PHB in the mutant was about 10 times lower. Furthermore, the Nile red fluorescence of the PHB granules was measured with a fluorescence spectrophotometer. The amount of fluorescence after 120 h of nitrogen starvation was about five times lower in the sll0783 mutant than in the wild type (Fig. 3A) (the smaller difference from the wild type compared to that shown by HPLC analysis may be accounted for by the background Nile red fluorescence of the membranes). To make sure that the reduced accumulation of PHB in the sll0783 mutant was not an indirect effect of reduced CO2 fixation or precursor limitation, the glycogen and acetyl-CoA contents of the cells were determined. For glycogen, there was no difference between the wild type and the sll0783 mutant (data not shown). The level of acetyl-CoA in the wild type only slightly increased during N starvation, whereas in the mutant, the level almost doubled (Fig. 3C). Therefore, the impaired PHB accumulation seen is not likely to be a consequence of limitation of the precursor molecule acetyl-CoA. This was further confirmed by the addition of acetate to nitrate-starved cells, which did not restore PHB accumulation in the sll0783 mutant (data not shown).
To gain more insight into the impaired recovery of the mutant from nitrogen starvation, the levels of PHB and glycogen were analyzed during a recovery experiment. As shown in Fig. 2C, glycogen turnover was apparently not impaired in the sll0783 mutant. Both the wild type and the sll0783 mutant rapidly utilized glycogen in the first 12 h after the addition of nitrogen and thereafter the glycogen level slightly increased again (Fig. 2C). During the same period of time, the wild type degraded the previously accumulated PHB reserves whereas the mutant did not alter its low PHB content (Fig. 2D).
Expression of PHB genes.
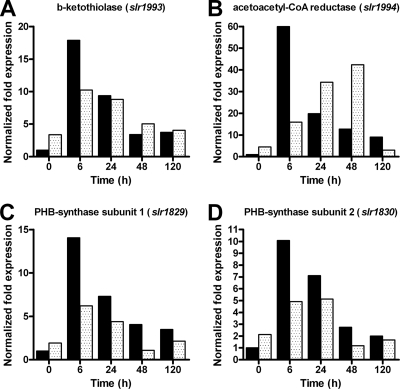
To evaluate the transcript abundance of the four PHB synthesis genes, qRT-PCR with gene-specific primers was performed (Fig. 4). In the wild type, all four genes involved in PHB synthesis were expressed at low levels under nitrogen-replete conditions. After 6 h of nitrogen depletion, the transcript amounts of all four genes were elevated. The highest induction was measured for acetoacetyl-CoA reductase with 60-fold induction, whereas the genes for PHB synthase showed 10-fold induction. In time, the transcript abundance of these four genes decreased but remained above the initial value.
FIG. 4.
Transcript abundance of the PHB synthesis genes in the wild type (black bars) and the sll0783 mutant (spotted bars). Transcript levels were determined by qRT-PCR with gene-specific primers. The transcript levels of the wild type at time point zero were set to 1. All values were normalized to the level of the reference gene rnpB. The following four genes of the PHB synthesis pathway were examined: (A) PHB synthase subunit one (slr1829), (B) PHB synthase subunit two (slr1830), (C) β-ketothiolase (slr1993), and (D) acetoacetyl-CoA reductase (slr1994).
In the sll0783 mutant, the expression of these genes was slightly different from that in the wild type. The mutant showed a higher transcript abundance for all four PHB synthesis genes under nitrogen-replete conditions. Following induction of nitrogen starvation, the transcript levels of all four genes were transiently lower than in the wild-type strain, but during prolonged starvation, the transcript level of the acetoacetyl-CoA reductase gene (slr1994) increased and surpassed the level in the wild type (Fig. 4B).
PHB synthase activity.
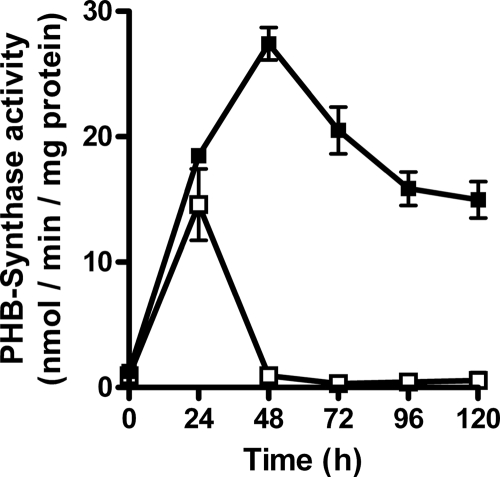
Critical evaluation of the data shown above indicated that neither precursor deficiency nor deficient expression of the PHB synthesis genes could explain the impaired PHB accumulation seen. Therefore, the activity of PHB synthase was investigated. Wild-type cells grown in nitrate-supplemented medium showed only very weak PHB synthase activity. After the cells were shifted to nitrogen-depleted medium, PHB synthase activity rose and reached a maximum after 48 h. Over time, the activity decreased to 50% of the maximal activity. Under nitrogen-sufficient conditions, the sll0783 mutant, like the wild type, showed very low PHB synthase activity. After the cells were shifted to nitrogen-depleted conditions, the activity transiently increased after 24 h, but already after 48 h of N starvation, the activity ceased and was thereafter no longer detectable. This failure to maintain PHB synthase activity matches the PHB-deficient phenotype of the sll0783 mutant (Fig. 5).
FIG. 5.
Activation of PHB synthase following nitrogen step down. Cells were transferred from nitrate-supplemented BG11 medium to combined-nitrogen-free BG11-N medium, and PHB synthase activity was measured in crude membrane fractions from cell lysates of the Synechocystis strain PCC 6803 wild type (filled squares) and sll0783 mutant (open squares) during the course of nitrogen starvation.
DISCUSSION
In this report, we describe a protein of unknown function which is part of a cluster of seven genes. This cluster is scattered throughout bacteria belonging to Proteobacteria, Actinobacteria, and Cyanobacteria. The similarity of the phylogenetic relationships of cyanobacterial sll0783 homologues to the corresponding phylogenetic relationships of the 16S rRNA genes suggests that this gene cluster was probably acquired early in cyanobacterial evolution but was maintained only in those species that could take advantage of its functions. Strikingly, almost all of the heterotrophic bacteria containing this gene cluster are PHB producers and many of them are able to fix nitrogen. This is in marked contrast to cyanobacteria, which all are unicellular, nondiazotrophic organisms. However, there is no clear common property of the habitats from which these strains were isolated. The fact that Prochlorococcus marinus MIT 9301 contains this gene cluster is remarkable: This strain harbors the smallest genome of all known cyanobacteria due to reductive evolution imposed by a static and extremely nutrient-poor environment, causing the loss of dispensable genes (29). Therefore, the sll0783 cluster must be of considerable selective advantage for this organism and is not likely to be a luxury addition. The fortunate fact that the sll0783 mutant of Synechocystis strain PCC6803 shows a robust phenotype allows us to gain our first insights into the function of this gene cluster.
Microarray analyses of Synechocystis strain PCC 6803, which were performed in the context of carbon dioxide utilization and photorespiration, revealed that under CO2-limiting conditions, the genes of this cluster are coherently repressed (9, 12). In addition, microarray analyses of nitrogen-limited Synechocystis strain PCC6803 cells showed that the entire gene cluster is highly induced under nitrogen starvation. The appropriate assumption that sll0783 and the downstream genes may form an operon is corroborated by the transcript analysis of sll0783 and sll0784, which were shown here to be coexpressed. Furthermore, their nitrogen starvation-induced transcription and the identification of a transcriptional start site exhibiting an imperfect NtcA site indicate that these genes may be regulated by NtcA. This assumption is also in accord with the identification of an Sll0783 homologous protein in Synechococcus strain PCC 7942 which accumulates under conditions of nitrogen starvation in an NtcA-dependent manner (1). Furthermore, the conservation of NtcA recognition motifs in all of the intergenic regions between the sll0783 and slr0801 homologues in cyanobacteria indicates that the expression of this gene cluster is controlled by NtcA.
In agreement with its nitrogen starvation-induced expression, the physiological function of sll0783 appears to be related to nitrogen starvation acclimation, as suggested by the slower recovery of the sll0783 mutant from nitrogen depletion (revealed by pigment and cell density analysis). This mutant phenotype may be related to the reduced amount of PHB. Whereas the wild type can use PHB as a source of carbon and reductant for de novo synthesis of amino acids and pigment molecules upon nitrate repletion, the mutant has to derive these building blocks mostly from new CO2 fixation. Apparently, the efficient mobilization of glycogen in the mutant does not compensate for the lack of PHB. This indicates that in Synechocystis strain PCC 6803, PHB degradation yielding acetyl-CoA may be more efficient in providing building blocks for anabolic reactions than is glycogen degradation.
The absence of the sll0783 product results in impaired PHB accumulation in the course of nitrogen starvation, apparently not because of a shortage of the PHB precursor molecule acetyl-CoA but due to impaired PHB synthase activity. The expression of the four genes required for PHB synthesis from acetyl-CoA was far less impaired than the activity of the PHB synthase enzyme, suggesting that regulation of the activity of PHB synthase involves—directly or indirectly—the sll0783 product. A kanamycin resistance cassette was integrated in the sll0783 gene in such a manner that the expression of the downstream genes was not knocked out in the mutant, as confirmed by transcript analysis. Therefore, the pure absence of the sll0783 product is likely responsible for the observed phenotype. Since PHB synthase in the sll0783 mutant was active a short time after the onset of nitrogen starvation but ceased upon prolonged nitrogen starvation, the sll0783 product appears to be required to maintain PHB synthase in an active state. However, knowledge of the regulation of PHB synthase on the activity level is very limited, and therefore, it is unclear how, precisely, the sll0783 gene product is involved in this process. Further studies are required to gain deeper insights into the regulation of the Synechocystis strain PCC 6803 PHB synthase, at the posttranscriptional level and in a wider perspective, to shed light on the function of the sll0783 gene cluster in cyanobacteria, as well as in other bacterial groups.
Supplementary Material
Acknowledgments
We thank D. Jendrossek (Institut für Mikrobiologie, University of Stuttgart) for his help and advice on PHB analysis and M. Valdebenito for help with the HPLC analyses. Furthermore, we thank F. Götz (Interfakultäres Institut für Mikrobiologie und Infektionsmedizin Tübingen) for provision of the fluorescent microplate reader.
This work was supported by DFG grant Fo195/7-1.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aldehni, F., J. Sauer, C. Spielhaupter, R. Schmid, and K. Forchhammer. 2003. Signal transduction protein PII is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942. J. Bacteriol. 185:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldehni, M. F., and K. Forchhammer. 2006. Analysis of a non-canonical NtcA-dependent promoter in Synechococcus elongatus and its regulation by NtcA and PII. Arch. Microbiol. 184:378-386. [DOI] [PubMed] [Google Scholar]
- 3.Allen, M. M., and A. J. Smith. 1969. Nitrogen chlorosis in blue-green algae. Arch. Mikrobiol. 69:114-120. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Decker, K. 1974. Acetyl coenzyme A, 3rd English ed. Academic Press, Inc., New York, NY.
- 7.de Marsac, N. T., and J. Houmard. 1993. Adaptation of cyanobacteria to environmental stimuli—new steps towards molecular mechanisms. FEMS Microbiol. Rev. 104:119-189. [Google Scholar]
- 8.Eisenhut, M., H. Bauwe, and M. Hagemann. 2007. Glycine accumulation is toxic for the cyanobacterium Synechocystis sp. strain PCC 6803, but can be compensated by supplementation with magnesium ions. FEMS Microbiol. Lett. 277:232-237. [DOI] [PubMed] [Google Scholar]
- 9.Eisenhut, M., E. A. von Wobeser, L. Jonas, H. Schubert, B. W. Ibelings, H. Bauwe, H. C. Matthijs, and M. Hagemann. 2007. Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 144:1946-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golden, S. S., J. Brusslan, and R. Haselkorn. 1987. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 153:215-231. [DOI] [PubMed] [Google Scholar]
- 12.Hackenberg, C., A. Engelhardt, H. C. Matthijs, F. Wittink, H. Bauwe, A. Kaplan, and M. Hagemann. 2009. Photorespiratory 2-phosphoglycolate metabolism and photoreduction of O2 cooperate in high-light acclimation of Synechocystis sp. strain PCC 6803. Planta 230:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hein, S., H. Tran, and A. Steinbuchel. 1998. Synechocystis sp. PCC6803 possesses a two-component polyhydroxyalkanoic acid synthase similar to that of anoxygenic purple sulfur bacteria. Arch. Microbiol. 170:162-170. [DOI] [PubMed] [Google Scholar]
- 14.Heinemann, U., D. Engels, S. Burger, C. Kiziak, R. Mattes, and A. Stolz. 2003. Cloning of a nitrilase gene from the cyanobacterium Synechocystis sp. strain PCC6803 and heterologous expression and characterization of the encoded protein. Appl. Environ. Microbiol. 69:4359-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 17.Hultman, E. 1959. Rapid specific method for determination of aldosaccharides in body fluids. Nature 183:108-109. [DOI] [PubMed] [Google Scholar]
- 18.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 20.Marchler-Bauer, A., J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, A. Tasneem, N. Thanki, R. A. Yamashita, D. Zhang, N. Zhang, and S. H. Bryant. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205-D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyake, M., K. Kataoka, M. Shirai, and Y. Asada. 1997. Control of poly-beta-hydroxybutyrate synthase mediated by acetyl phosphate in cyanobacteria. J. Bacteriol. 179:5009-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osanai, T., M. Azuma, and K. Tanaka. 2007. Sugar catabolism regulated by light- and nitrogen-status in the cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 6:508-514. [DOI] [PubMed] [Google Scholar]
- 24.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 25.Paz-Yepes, J., E. Flores, and A. Herrero. 2003. Transcriptional effects of the signal transduction protein P(II) (glnB gene product) on NtcA-dependent genes in Synechococcus sp. PCC 7942. FEBS Lett. 543:42-46. [DOI] [PubMed] [Google Scholar]
- 26.Podar, M., J. R. Eads, and T. H. Richardson. 2005. Evolution of a microbial nitrilase gene family: a comparative and environmental genomics study. BMC Evol. Biol. 5:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rippka, R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol. 167:3-27. [DOI] [PubMed] [Google Scholar]
- 28.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 29.Scanlan, D. J., M. Ostrowski, S. Mazard, A. Dufresne, L. Garczarek, W. R. Hess, A. F. Post, M. Hagemann, I. Paulsen, and F. Partensky. 2009. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73:249-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarz, R., and K. Forchhammer. 2005. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology 151:2503-2514. [DOI] [PubMed] [Google Scholar]
- 31.Spiekermann, P., B. H. Rehm, R. Kalscheuer, D. Baumeister, and A. Steinbuchel. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171:73-80. [DOI] [PubMed] [Google Scholar]
- 32.Stanier, R. Y., and G. Cohen-Bazire. 1977. Phototrophic prokaryotes: the cyanobacteria. Annu. Rev. Microbiol. 31:225-274. [DOI] [PubMed] [Google Scholar]
- 33.Taroncher-Oldenburg, G., K. Nishina, and G. Stephanopoulos. 2000. Identification and analysis of the polyhydroxyalkanoate-specific beta-ketothiolase and acetoacetyl coenzyme A reductase genes in the cyanobacterium Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 66:4440-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valentin, H., and A. Steinbüchel. 1994. Application of enzymatically synthesized short-chain-length hydroxy fatty acid coenzyme A thioesters for assay of polyhydroxyalkanoic acid synthases. Appl. Microbiol. Biotechnol. 40:699-709. [Google Scholar]
- 36.Vega-Palas, M. A., E. Flores, and A. Herrero. 1992. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol. Microbiol. 6:1853-1859. [DOI] [PubMed] [Google Scholar]
- 37.Vincenzini, M., and R. De Philippis. 1999. Polyhydroxyalkanoates, p. 292-312. In R. Z. Cohen (ed.), Chemicals from microalgae. Taylor and Francis, London, United Kingdom.
- 38.Wu, G. F., Q. Y. Wu, and Z. Y. Shen. 2001. Accumulation of poly-beta-hydroxybutyrate in cyanobacterium Synechocystis sp. PCC6803. Bioresour Technol. 76:85-90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.