Abstract
Despite excellent biocompatibility and mechanical properties, the poor in vitro and in vivo degradability of cellulose has limited its biomedical and biomass conversion applications. To address this issue, we report a metabolic engineering-based approach to the rational redesign of cellular metabolites to introduce N-acetylglucosamine (GlcNAc) residues into cellulosic biopolymers during de novo synthesis from Gluconacetobacter xylinus. The cellulose produced from these engineered cells (modified bacterial cellulose [MBC]) was evaluated and compared with cellulose produced from normal cells (bacterial cellulose [BC]). High GlcNAc content and lower crystallinity in MBC compared to BC make this a multifunctional bioengineered polymer susceptible to lysozyme, an enzyme widespread in the human body, and to rapid hydrolysis by cellulase, an enzyme commonly used in biomass conversion. Degradability in vivo was demonstrated in subcutaneous implants in mice, where modified cellulose was completely degraded within 20 days. We provide a new route toward the production of a family of tailorable modified cellulosic biopolymers that overcome the longstanding limitation associated with the poor degradability of cellulose for a wide range of potential applications.
A variety of biopolymers, such as polysaccharides, polyesters, and polyamides, have been produced by bacteria. Due to less complexity, genetic manipulation in these microbes opens up an enormous potential to tailor biopolymers for high-value medical applications and drug delivery systems (32). Cellulose is the most abundant biopolymer on Earth, recognized as the major component of plant biomass but also as a representative of microbial extracellular polymers. Bacterial cellulose (BC), produced by Gluconacetobacter xylinus (formerly Acetobacter xylinum) is extruded as fibrils that accumulate to form microfibrils. These microfibrils then aggregate into ribbon structures, which can be disrupted by incorporation of various compounds during de novo synthesis (34). BC has been demonstrated to be a remarkably versatile biomaterial and can be used in a wide variety of applied endeavors, such as paper products, electronics, acoustics, and biomedical devices (4). Due to its unique nanostructure and properties that closely resemble the structure of native extracellular matrices, BC has been considered for numerous medical and tissue-engineering applications, such as wound healing, skin replacement, blood vessel replacement, cartilage engineering, and guided tissue regeneration (GTR) (4, 5, 7, 17, 33).
Despite the excellent biocompatibility and mechanical properties of BC, the lack of cellulose-hydrolyzing enzymes in the human body and the high crystallinity restrict its utility (10). Therefore, cellulose with low crystallinity and more facile degradability could be an important polymer for biomass conversion and tissue engineering applications. While in biofuel refineries plant cellulose is the main source for cellulosic biomass, this source is still not efficient due to crystallinity, inhibited access to enzymes, and poor purity of plant cellulose (24).
Advances in metabolic engineering over the last 2 decades have led to high-level production of specific metabolites at commercially viable levels. More recently, advancements in genetic engineering combined with those in metabolic engineering have led to the generation of microbes that express heterologous pathways, resulting in the production of natural products beyond the genetic confines of the natural host (29, 39). Examples of this include the production of biofuels (ethanol), adhesives like xanthan gum, and biodegradable plastics, such as poly(3-hydroxybutyric acid) (PHA) (9, 22, 26, 43). In the case of ethanol production, metabolic alterations have been carried out to expand the substrate range utilized by a microbe that naturally ferments sugars into alcohol and thus have improved the overall biomass conversion yield. Another metabolic engineering strategy successfully produced a fast-growing and technically amenable Escherichia coli strain which normally does not produce alcohols at high yields (6).
To address the challenges associated with BC, prior efforts were made to modify the polymer based on chemical modifications or on feeding strategies, with limited success (16, 27). It is our hypothesis that the most likely cause of the limitations to these studies was related to insufficient levels of UDP-charged nonglucose monomers available to the cellulose synthase. While these studies proved a limited ability to incorporate nonglucose monomers, they did demonstrate that the cellulose synthase (glycosyltransferase) of G. xylinus can utilize both UDP-glucose and UDP-N-acetylglucosamine (UDP-GlcNAc) as substrates. Therefore, in the present work, an attempt was made to engineer G. xylinus to rationally redesign the flow of cellular metabolites to incorporate GlcNAc sugar residues into cellulose during de novo synthesis. The goal was to generate modified cellulose with more facile enzyme degradation and improved degradability in vivo. To achieve this goal, an operon containing three genes from Candida albicans (Table 1) for UDP-GlcNAc synthesis was expressed in G. xylinus to produce activated cytoplasmic UDP-GlcNAc monomers accessible to cellulose synthase to produce a chimeric polymer comprising both glucose and GlcNAc (see Fig. S1 in the supplemental material). The presence of GlcNAc not only enables BC to be susceptible to lysozyme but also disrupts the highly ordered cellulose crystalline structure. Moreover, in contrast to plant-derived cellulose, the coupling between biosynthesis and material processing in this bacterium offers an experimentally accessible system (1).
TABLE 1.
Candida albicans genes constructed into a heterologous operon for GlcNac incorporation into cellulose fibrils
| Gene | Enzyme | Function |
|---|---|---|
| NAG5 | GlcNac kinase | Conversion of GlcNac to GlcNac-6-P |
| AGM1 | Phosphoacetyl-glucosamine mutase | Conversion of GlcNac-6-P to GlcNac-1-P |
| UAP1 | UDP-GlcNac pyrophosphorylase | Charges the monomer with UDP for incorporation by the synthase |
MATERIALS AND METHODS
Bacterial strains.
The cellulose-positive G. xylinus strain 10245 was obtained from ATCC (Manassas, VA). Transformants of E. coli Top10 cells (Invitrogen, Carlsbad, CA) were used for cloning purposes. Kanamycin (50 μg/ml), tetracycline (20 μg/ml), and/or ampicillin (50 μg/ml) was included in the culture medium for antibiotic selection. G. xylinus was cultured in Hestrin and Schramm medium (HS medium) with 0.1% (vol/vol) celluclast cellulase (Sigma, St. Louis, MO) and grown at 30°C with or without shaking (11). For cellulose production, HS medium without cellulase was used. For cellulose production, the cell suspension was added to fresh medium to a final A600 of 0.3 to 0.4.
Construction of pBBR-GlcNAc plasmid.
Primers and plasmids used in this study are listed in Table S1 and Table S2 of the supplemental material. For cloning purposes, the multiple cloning site (MCS) of pET-30 was cloned into the pUC19 backbone. The β-lactamase constitutive promoter (bla) region and full-length sequences of AGM1, NAG5, and UAP1 were amplified by PCR and cloned into the pUC-MCS plasmid at SphI-NdeI, NdeI-BamHI, and BamHI-HindIII and HindIII-NotI sites, respectively. The SphI and PsPOMI (compatible to NotI) sites were introduced in the pBBR-Tet plasmid by PCR for the subcloning of the bla-AGM1-NAG5-UAP1 cartridge. For subcloning, the SphI- and NotI-digested bla-AGM1-NAG5-UAP1 cassette was introduced in SphI- and PsPOMI-digested pBBR-Tet, and the resulting plasmid was named pBBR-GlcNAc.
Transformation of G. xylinus and screening.
Transformation of G. xylinus was carried out by electroporation as previously described (40). For screening, five colonies were selected from the selection plates for plasmid isolation. Plasmids were isolated (Qiagen, Valencia, CA), and PCR was carried out using sequence-specific primers to confirm the presence of promoter, AGM1, NAG5, UAP1, and Tetr.
Expression studies: reverse transcription-PCR (RT-PCR).
For expression studies, engineered G. xylinus cells were grown with either glucose, GlcNAc, or both as carbon source for 24 h. Total RNA was isolated using a RiboPure bacteria kit (Ambion, Austin, TX). First-strand cDNA synthesis was conducted using the SuperScript III first-strand synthesis kit (Invitrogen, Carlsbad, CA) followed by PCR with gene-specific primers.
Cell growth, cellulose production, and purification.
For cellulose production in static culture, cells were grown in 90-mm culture dishes containing 20 ml of HS medium at 30°C for 1 week. The cellulose pellicles were purified by treating twice with 4% SDS solution at 70°C for 4 h and 4% NaOH at 70°C for 4 h to remove the entrapped bacteria, followed by several washings with deionized water (16). Purified cellulose mats were dried at 70°C for 30 h and weighed. The mass obtained was normalized based on the culture volume.
Cell morphology.
For cell morphology, cells were immobilized on a mica surface by using 0.01% gelatin and observed by atomic force microscopy (AFM; Dimension V; Veeco Instruments Inc., Plainview, NY) in air (8). A 225-μm-long silicon cantilever with a spring constant of 2.8 N/m was used in tapping mode.
SEM.
The surface morphologies of BC and modified bacterial cellulose (MBC) membranes were studied with scanning electron microscopy (SEM; model 982; LEO-FESEM) at the Center for Nanoscale Systems (Harvard University, Cambridge, MA).
Measurements of intracellular UDP-GlcNAc and UDP-glucose.
Both engineered and control cells were grown to mid-logarithmic phase and isolated by centrifugation. Cells were lysed by sonication using 2 ml sonication buffer (100 mM KCl, 1 mM EDTA, 50 mM KH2PO4; pH 7.5) at 4°C. The samples were deproteinized by addition of 1 volume of 0.1 M perchloric acid and subsequent centrifugation for 30 min at 13,000 rpm at 4°C. The resulting supernatants were diluted with 10 volumes of 10 mM KH2PO (pH 2.5), and the final pH was adjusted to 2.5 for each sample. Intracellular UDP-GlcNAc and UDP-glucose levels were measured by standard procedures (28). Briefly, two Supelco LC-18T columns in series (25 cm by 4.6 mm, with 5-μm particle size; Sigma) were used to separate and quantify UDP-sugar with the mobile phase of 0.1 mol/liter KH2PO4 buffer containing 2 mmol/liter tetrabutylammonium phosphate, pH 6.2, at a flow rate of 1 ml/min.
Acid hydrolysis of cellulose and quantitative analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Both BC and MBC were hydrolyzed with 70% H2SO4 at 4°C for 30 min. Acid was neutralized with Ba(OH)2, and the final pH was adjusted to 5 to 6. Fragment separation was achieved on a Waters Atlantis dC18 column (150 by 2.1 mm, with 5-μm particle size; Waters, Milford, MA) with an Agilent high-performance LC (HPLC) 1200 system (Agilent, Santa Clara, CA). Mass spectrometric detection was performed on an API 3200 triple quadrupole instrument (Applied Biosystems, Bedford, MA) using multiple reaction monitoring (MRM). A TurboSpray interface with negative ionization mode was used. The precursor-to-product ion transitions m/z 220 to 119 for GlcNAc and 179 to 89 for glucose were analyzed with the selected ion chromatogram (SIC) mode. The main MS working parameters are listed in Table S3 of the supplemental material. A linear calibration standard curve for glucose and GlcNAc ranging up to 100 μg/ml was set up for the quantification (see Fig. S2 in the supplemental material).
Lectin staining.
To access the lectin binding to GlcNAc residues, both BC and MBC films were incubated with fluorescent dye-conjugated wheat germ agglutinin (WGA) conjugated to Alexa Fluor 488 (Molecular Probes, Carlsbad, CA) at 50 μg/ml in phosphate-buffered saline (PBS) for 30 min. The samples were subsequently washed several times with PBS and then analyzed using a Zeiss (Thornwood, NY) Axiovert S100 fluorescence microscope at 495-nm excitation.
XRD analysis.
Purified BC and MBC mats were washed with distilled water and dried at 70°C for 30 h. X-ray diffraction (XRD) studies in reflection mode were performed using a Phillips PW1830 generator operated at 40 kV and 45 mA, using an optically encoded diffractometer at room temperature. Fast scans were performed from 5°C to 40°C. The range of interest was then narrowed to 12°C to 28°C, and slow scans were conducted to improve the signal-to-noise ratio. D-spacings were calibrated with silicon powder reference standards.
FT-IR.
Infrared spectroscopy of both BC and MBC mats was carried out using a Fourier transform infrared spectrometry (FT-IR) 6200 apparatus (JASCO, Japan) with MIRacle ATR (Pike Technologies, Madison, MI) and attenuated total reflectance in the wave number range of 1,250 to 3,000 cm−1 at a rate of 32 scans per second and 4 cm−1 resolution (13).
Enzyme hydrolysis and LC-MS/MS analysis.
The enzymatic hydrolysis rates of BC and MBC were determined using either cellulase, a mixture of celluclast (from Trichoderma reesei; Sigma, St. Louis, MO) and cellobiase (from Aspergillus niger; Sigma, St. Louis, MO), or chicken egg white lysozyme (Sigma, St. Louis, MO). For digestion, 0.1% (vol/vol) cellulase or 0.5 mg/ml chicken egg white lysozyme was added to cellulose mats directly. The hydrolysis reaction was carried out at 30°C for cellulase and 37°C for chicken egg white lysozyme for up to 24 h with shaking. The digested slurry was analyzed by LC-MS/MS as described for the acid hydrolysis experiment. The main ions for mono- and oligosaccharides results were as follows: m/z 179 for glucose; m/z 220 for GlcNAc; m/z 382 for the glucose-GlcNAc dimer and m/z 544 for the glucose-GlcNAc-glucose trimer. The main MS working parameters are listed in Table S4 of the supplemental material.
In vivo biodegradation of cellulose constructs.
Engineered and control strains were fed with 2% GlcNAc, purified as described above, and lyophilized. Implants were sterilized by soaking in 70% ethanol prior to implantation subcutaneously on the back of 6- to 8-week-old female BALB/c mice. Implants were excised and examined for gross degradation at 10 and 20 days postimplantation (n = 3 per time point). All animal studies were conducted under approved animal care protocols.
Statistical analysis.
Comparisons between two experimental groups were performed using a one-way analysis of variance (GraphPad; InStat Software, La Jolla, CA). Group means were deemed to be statistically significant when P was <0.001.
RESULTS
Cloning and expression of the UDP-GlcNAc synthesis operon in G. xylinus.
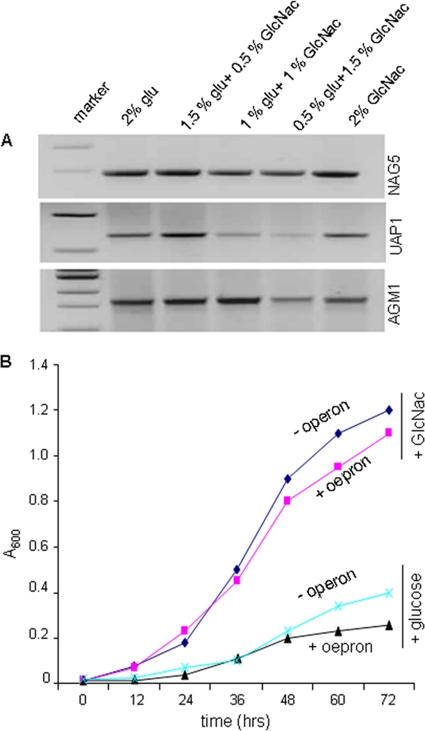
To express the UDP-GlcNAc synthetic operon of C. albicans in G. xylinus, all three genes involved in UDP-GlcNAc synthesis, i.e., AGM1, UAP1, and NAG5, were cloned under the control of the constitutive bla (β-lactamase) promoter in plasmid pBBR-Tet to construct the recombinant pBBR-GlcNAc plasmid (see Fig. S3a in the supplemental material). G. xylinus cells were transformed by electroporation with either pBBR-Tet (without operon) or pBBR-GlcNAc plasmid (with operon) and screened by PCR to confirm successful transformation (see Fig. S3b in the supplemental material). Due to the constitutive nature of the bla promoter, consistent high transcript levels were observed for all three genes regardless of substrate, indicating successful cloning and expression of the operon in G. xylinus (Fig. 1 A). To assess the impacts of the heterologous operon on G. xylinus morphology and growth, AFM was performed and the A600 was measured, respectively. No obvious morphological changes were observed between engineered (+operon) and control (-operon, plasmid-only) cells (see Fig. S3c). In the presence of glucose, both engineered and normal cells exhibited a typical bacterial growth pattern, whereas in the presence of GlcNAc, both cell types grew more slowly (Fig. 1B). Not surprisingly, G. xylinus preferred glucose as the carbon source over GlcNAc (23). Taken together, the growth experiments indicated that expression of the heterologous operon does not impair cell growth or morphology.
FIG. 1.
Expression of the UDP-GlcNAc biosynthesis operon in G. xylinus. (A) RT-PCR expression of NAG5; UAP1 and AGM1 in G. xylinus were grown for 24 h in the presence of different amounts of glucose and GlcNAc. (B) Growth patterns of engineered (+ operon) and normal (- operon) G. xylinus fed either glucose or GlcNAc.
Metabolic flux analysis and polymer composition.
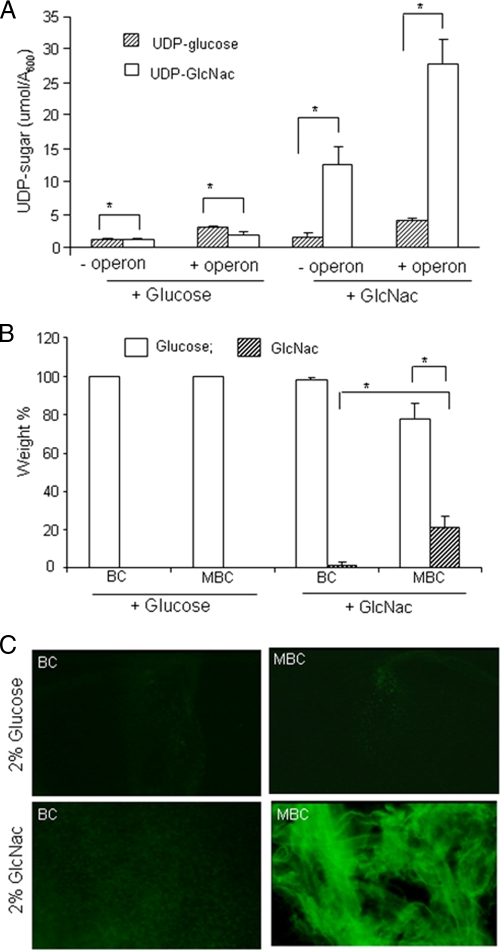
The role of the heterologous operon on UDP-sugar production in G. xylinus was assessed by measuring the cytoplasmic UDP-GlcNAc and UDP-glucose levels using HPLC. Under glucose feed conditions, low levels of both UDP-GlcNAc and UDP-glucose were observed in both cell types, while with GlcNAc-fed cultures, a high level of UDP-GlcNAc was found in the engineered cells compared to normal cells (Fig. 2A). These data revealed that the C. albicans operon is not only expressed at the transcript level but also is translated into functional enzymes that result in a high level of UDP-GlcNAc synthesis. We did not observe any disruptive impacts of the operon on UDP-glucose production. The chemical compositions of both BC and MBC were determined by acid hydrolysis followed by LC-MS/MS. On a weight percent basis, GlcNAc incorporation was maximum in cellulose produced from cells in the GlcNAc-fed medium. Compared to BC (1.1% ± 0.25%, wt/wt), the GlcNAc content in the MBC was more than 18-fold higher (20.6% ± 3.9%, wt/wt) (Fig. 2B). Incorporation of GlcNAc residues into MBC directly correlated with the formation of the activated UDP-GlcNAc monomer by the operon. The presence of GlcNAc in MBC was further confirmed by lectin staining, as fluorescently labeled WGA, which has specificity for GlcNAc residues, failed to bind to either BC or MBC produced with a glucose feed or BC produced with a GlcNAc feed but strongly bound to MBC produced with a GlcNAc feed (Fig. 2C). Mass spectrometry data together with lectin binding confirmed the presence of GlcNAc residues in MBC extruded from the GlcNAc-fed culture.
FIG. 2.
Metabolic flux study and cellulose chemical composition determination. (A) Quantification of cytosolic levels of UDP-glucose and UDP-GlcNAc in both engineered (+ operon) and control (- operon) G. xylinus cells fed either glucose or GlcNAc as the carbon source. Error bars represent standard deviations of three replicates (P < 0.0001). (B) Quantitative LC-MS/MS for measurement and the relative weight percent (wt%) of glucose and GlcNAc in acid-hydrolyzed BC and MBC extruded from either glucose- or GlcNAc-fed cultures. Error bars represent standard deviations of three replicates (*, P < 0.001). (C) Binding of fluorescent WGA to BC and MBC films.
Polymer structure and production.
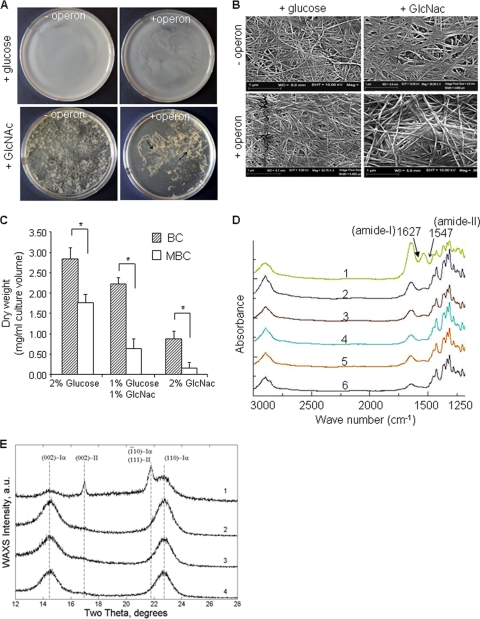
The morphologies of MBC pellicles produced from engineered G. xylinus cells (with operon) under either glucose or GlcNAc feeding conditions were different from BC produced from control cells (without operon) (Fig. 3A). Compared to BC pellicles, MBC pellicles were thin and poorly associated. Interestingly, with the GlcNAc feed, MBC was unable to remain as an intact pellicle and therefore remained submerged in the culture medium (Fig. 3A). Additional characterization of pellicles generated with supplemental GlcNAc by SEM showed the arrangement of fibers in MBC to have a more diffuse fiber structure than BC (Fig. 3B). The presence of GlcNAc in the polymer chain also likely altered the hydrogen bonding between fibrils, as observed by the scattered arrangement of fibers in MBC films. Based on these observations, we concluded that the presence of GlcNAc in MBC decreases the stability of fibril-fibril interactions, resulting in a lower crystallinity of MBC than BC. The impact of the heterologous operon on biomass production was evaluated by comparing the dry weight of the extruded polymer. Partially or fully substituting GlcNAc for glucose as the carbon source significantly lowered total cellulose production in both engineered and wild-type cells (Fig. 3C). With glucose-fed cultures, MBC production (1.8 ± 0.2 mg/ml culture medium) was 35% lower than wild-type BC production (2.8 ± 0.4 mg/ml culture medium). The drop in total production was greater in GlcNAc-fed cultures, with a >70% decrease in MBC production (0.25 ± 0.15 mg/ml culture medium) compared to BC (0.7 ± 0.3 mg/ml culture medium). In absolute terms, insertion of the operon reduced cellulose synthesis efficiency under all feeding conditions.
FIG. 3.
Cellulose production, morphology, and structure. (A) Morphological appearance of cellulose pellicles produced after 1 week by G. xylinus with the operon or without the operon in the presence of either glucose or GlcNAc as the carbon source. (B) Scanning electron micrograph of purified BC and MBC mats produced in glucose- or GlcNAc-fed cultures. (C) Cellulose production efficiency from engineered (+ operon) and normal (- operon) G. xylinus cells supplemented with either glucose, GlcNAc, or both as the carbon source. Error bars represent standard deviations of three replicates (*, P < 0.001). (D) FT-IR spectra of BC (curves 2, 4, and 6) and MBC (curves 1, 3, and 5) produced by G. xylinus grown in the presence of either 2% glucose (curves 5 and 6) or 2% GlcNAc (curves 1 and 2) or 1% each (curves 3 and 4). (E) Wide-angle XRD spectra of both BC (curves 2 and 4) and MBC (curves 1 and 3) prepared from G. xylinus cultured with GlcNAc (curves 1 and 2) or glucose (curves 3 and 4) as the carbon source.
Polymer properties.
The FT-IR spectra of MBC and BC produced in glucose-fed cultures showed no detectable differences. In the case of MBC, increasing the GlcNAc content in the culture medium produced two strong absorption bands at 1,660 cm−1 and 1,550 cm−1, corresponding to amide I and amide II signals and confirming the presence of amine group sugars in MBC (Fig. 3D). Similar absorption bands were also observed in the FT-IR spectra of chitin-cellulose blends (19). Wide-angle XRD studies were performed in reflection mode (λ, 0.1542 nm) for purified MBC and BC membranes (Fig. 3E). Both BC and MBC displayed two broad peaks (Fig. 3E, curves 1 to 4) consistent with triclinic cellulose form Iα, at 2θ = 14.3° and 22.7° (Miller indices of 100 and 110, respectively) (31). The appearance of these peaks is similar to the results obtained by algae-bacteria-type cellulose (38, 42). Interestingly, MBC extruded from GlcNAc-fed cultures gave two additional peaks at 2θ = 17.0° and 21.8° (curve 1). The reflections at 17.0° (002) and 21.8° (111) are consistent with the monoclinic cellulose II (15). The XRD data demonstrate that the presence of GlcNAc transforms MBC from highly crystalline type Iα to a less crystalline form of cellulose type II. To confirm these findings, real-time wide-angle XRD studies were performed in transmission mode to determine the degree of crystallinity (Xc) in both BC and MBC, which was found to be 0.70 ± 0.05 and 0.35 ± 0.05, respectively.
Enzymatic degradation.
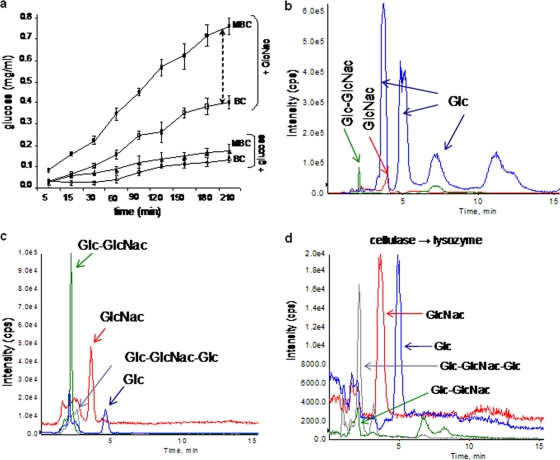
Due to its amorphous nature, MBC produced with GlcNAc feed exhibited faster hydrolysis when treated with cellulases from Trichoderma sp. compared to BC, while no significant differences were observed with a glucose feed (Fig. 4a). The susceptibility of MBC, produced from a 2% GlcNAc-fed culture, to biodegradation was investigated using chicken egg white lysozyme and cellulases from Trichoderma sp. Cellulase treatment produced glucose with a small amount of Glc-GlcNAc (Fig. 4b; see also Fig. S4a and b in the supplemental material). These results were consistent with the specificity of the cellulases, which do not recognize the 1→4 β-glycosidic bond between Glc-GlcNAc (36). With lysozyme, most of the polymer remains undigested, as neither BC nor MBC is a natural substrate for lysozyme. But the LC-MS data indicated fractions comprised of Glc, GlcNAc, Glc-GlcNaGlc, and Glc-GlcNAc (Fig. 4c; see also Fig. S4c to e). Unlike BC, which was completely digested by cellulase, MBC resisted complete hydrolysis, suggesting the presence of large fragments containing Glc-GlcNAc, which will need to be confirmed in more comprehensive biophysical evaluations of MBC. The undigested portion of cellulase-treated MBC was further hydrolyzed with chicken egg white lysozyme and analyzed by LC-MS (Fig. 4d). The mass spectra showed two major peaks for Glc and GlcNAc, together with peaks for Glc-GlcNAc and Glc-GlcNAc-Glc (see Fig S4f and g in the supplemental material). These results indicate that MBC is susceptible to both lysozyme and cellulases.
FIG. 4.
Enzyme degradability of BC and MBC mats. (a) After addition of cellulase, the amount of glucose liberated was measured at different times. The black dotted arrow shows the difference in hydrolysis rates between BC and MBC. (b) LC-MS profile of MBC hydrolyzed with Tricoderma reesei cellulase. (c) LC-MS profile of MBC treated with chicken egg white lysozyme. A mixture of oligomers of up to 950-Da mass was obtained. (d) MBC was hydrolyzed with Tricoderma reesei cellulase, and the remaining undigested portion was treated with chicken egg white lysozyme. The LC-MS profile of lysozyme-treated undigested polymer is shown.
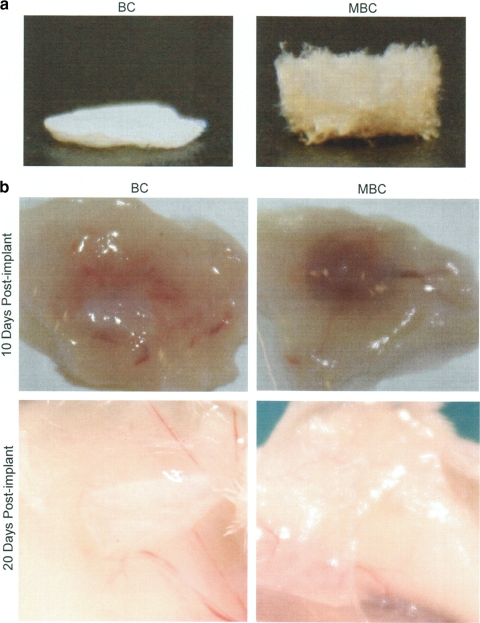
In vivo degradation.
The susceptibility of MBC to in vivo degradation was examined in a subcutaneous implant model in mice. Lyophilized and sterilized cellulose membranes (4-mm diameter) produced from 2% GlcNAc cultures (Fig. 5a) were implanted subcutaneously in the backs of female BALB/c mice (6 to 8 weeks old). Constructs were removed at 10 and 20 days postimplantation. Visual inspection of implant sites indicated little to no degradation of the cellulose produced from the control strain at either time point, while the modified cellulose from the engineered strain was almost entirely degraded at 10 days and was completely undetectable at 20 days (Fig. 5b). Both constructs induced a temporary inflammatory response at the 10-day time point which was completely resolved by 20 days (Fig. 5b).
FIG. 5.
In vivo degradation of cellulose and modified cellulose constructs. (a) Purified modified and control cellulose constructs from cultures fed 2% GlcNAc were lyophilized prior to subcutaneous implantation in a BALB/c mouse. (b) Constructs were examined at 10 and 20 days postimplantation to determine gross degradation. Three animals were used per time point. Images are representative of each replicate.
DISCUSSION
Many examples from metabolic engineering in microbes have proven that the capacity for biochemical synthesis is extensive and highly amenable to modification (18). The current challenge was to expend the substrate utilization efficiency of cellulose synthase of the bacterium G. xylinus through the de novo design of biosynthetic pathways for which natural pathways are intractable. While the UDP-GlcNAc synthesis machinery already exists in this bacterium, it does not produce excess UDP-GlcNAc that would be accessible to the cellulose synthase. As a result, cellulose synthase of G. xylinus does not generally utilize UDP-GlcNAc as a substrate even though it has the potential to recognize this substrate (16, 27).
To enhance the cytoplasmic UDP-GlcNAc level, we engineered G. xylinus through the expression of enzymes responsible for UDP-GlcNAc synthesis from C. albicans. Cellulose produced from engineered cells was evaluated for the presence of GlcNAc. When fed with GlcNAc, a greater-than-18-fold increase in GlcNAc content in the MBC compared to BC was observed, revealing the successful incorporation and expression of this heterologous operon in the engineered strain. The synthesis of high levels of the activated UDP-GlcNAc monomer substrate was clearly accessible to cellulose synthase, resulting in a cellulose-chitin copolymer. The relatively low GlcNAc assimilation from culture medium by engineered cells could be the result of feedback inhibition of elevated cytoplasmic UDP-GlcNAc levels or the result of diversion of intermediates in the heterologous pathway into the normal energy metabolism of the cell. To overcome low MBC productivity, modifications in medium composition or heterologous expression of genes related to higher cellulose production could be considered (2, 35).
As determined by XRD, MBC exhibited diffraction peaks from both crystal types, indicating it is a mixture of cellulose Iα and II, although the bulk is cellulose Iα. The peaks for cellulose II showed much narrower diffraction profiles, indicating larger crystals than those of Iα. Based on these data, we conclude that the addition of GlcNAc contributed to the structural transformation of MBC from cellulose type Iα to type II, which is less crystalline in nature than Iα. Earlier studies involving wide-angle XRD showed that the crystallinity of cellulose-chitin blends change from highly crystalline to a more amorphous state with chitin content (21). Although the exact mechanism is not known, it is believed that the presence of GlcNAc in the cellulose chains may influence the second and third stages of the cellulose synthesis process, thus altering fibril-fibril interactions (12, 41). The data presented here led to similar findings, as MBC exhibited half the crystallinity of BC.
In cellulosic biofuel refineries, crystallinity and purity of cellulose are two major issues related to cost-effective production of biofuel, due to the inefficient enzymatic digestion of cellulosic matter (25). The use of cellulose biomass could provide an alternative route to starch-based biofuel production, avoiding competition with food supplies while still offering a renewable source of biomass (30). In this regard, production of less-crystalline cellulose from metabolically engineered G. xylinum may provide new routes for biofuel production using microbial cellulose. Obviously, these results are preliminary but provide an initial proof-of-concept for the utilization of modified polymer for biofuel or other renewable energy sources.
The concept of a completely in vivo-degradable cellulosic material for tissue engineering applications is attractive. The degradation of bacterial cellulose has not been fully evaluated in vitro or in vivo, although previous studies showed lysozyme susceptibility of bacterial cellulose produced with GlcNAc-fed and P-chitin-fed cultures (20). The most common substrates for lysozyme are chitin and bacterial cell wall derivatives (peptidoglycan). These sugar polymers contain β(1→4)-linked N-acetyl hexosaminyl residues and are structural analogs of cellulose. Lysozyme has a particularly strong affinity for fragments containing consecutive GlcNAc residues (n = 1 to 6) (3). The cleavage of cellobioses is at least six times slower than that of GlcNAc-Glc, while GlcNAc-GlcNAc-GlcNAc-Glc-Glc residues can be hydrolyzed rapidly (14, 37). In our analysis of MBC hydrolysis by lysozyme, we found all possible GlcNAc-Glc derivatives up to 950 Da. These data support the susceptibility of MBC to lysozyme and also suggest that the cellulose synthase incorporates GlcNAc in MBC on a random basis.
While in vitro degradation of a biopolymer is an important first step in characterizing its biological properties, in vivo studies are required for a better understanding of its potential for biotechnology applications, particularly with regard to tissue engineering. The results of the preliminary in vivo study presented here are an exciting development, demonstrating in vivo degradation of a bacterial cellulosic biomaterial within 3 weeks. In addition, the implant was well tolerated, inducing only a transient inflammatory response that was completely resolved by 20 days. The potential for future cellulosic constructs in biomedical applications is extraordinary, and the genetic and metabolic controls inherent in the engineered strains presented here are without precedent in previous work with bacterial cellulose.
The formation of novel cellulose-chitin copolymers provides opportunities that bridge the properties of the respective homopolymers, cellulose and chitin/chitosan. Deacetylation of MBC would generate a material with reactive amine groups on the surface for in vitro biomimetic engineering for various applications, such as scaffolds for tissue engineering, biosensors for small molecule detection, novel biocomposites, drug delivery vehicles, and adjuvants for vaccines, as well as countless other options. With normal cellulose, this phenomenon is achieved only through chemical treatment, resulting in disruption of the cellulose native structure, while with engineered cellulose this can be achieved in a single-step process without destructive alterations.
In summary, the production of lysozyme and in vivo-degradable and multifunctional cellulose-chitin copolymers represents a significant leap forward in cellulosic research. Further control of GlcNAc incorporation as well as genetic and metabolic engineering approaches to increase yield and efficiency of production will enhance the potential of this copolymer to be used in several biomedical and biotechnological applications.
Supplementary Material
Acknowledgments
We thank Keiji Numata and Aneta J. Mieszawska, Department of Biomedical Engineering, Tufts University, for help with AFM and SEM, respectively. SEM imaging was performed at the Center for Nanoscale Systems, Harvard University.
This work was funded by the National Institutes of Health (grant R21 GM0791471).
Footnotes
Published ahead of print on 23 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Cannon, R. E., and S. M. Anderson. 1991. Biogenesis of bacterial cellulose. Crit. Rev. Microbiol. 17:435-447. [DOI] [PubMed] [Google Scholar]
- 2.Chien, L.-J., H.-T. Chen, P.-F. Yang, and C.-K. Lee. 2006. Enhancement of cellulose pellicle production by constitutively expressing Vitreoscilla hemoglobin in Acetobacter xylinum. Biotechnol. Prog. 22:1598-1603. [DOI] [PubMed] [Google Scholar]
- 3.Chipman, D. M., and N. Sharon. 1969. Mechanism of lysozyme action. Science 165:454-465. [DOI] [PubMed] [Google Scholar]
- 4.Czaja, W., A. Krystynowicz, S. Bielecki, and R. M. Brown, Jr. 2006. Microbial cellulose: the natural power to heal wounds. Biomaterials 27:145-151. [DOI] [PubMed] [Google Scholar]
- 5.Czaja, W. K., D. J. Young, M. Kawecki, and R. M. Brown, Jr. 2007. The future prospects of microbial cellulose in biomedical applications. Biomacromolecules 8:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Dien, B. S., M. A. Cotta, and T. W. Jeffries. 2003. Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63:258. [DOI] [PubMed] [Google Scholar]
- 7.Dieter, K., S. Dieter, U. Ulrike, and M. Silvia. 2001. Bacterial synthesized cellulose: artificial blood vessels for microsurgery. Prog. Polym. Sci. 26:1561-1603. [Google Scholar]
- 8.Doktycz, M. J., C. J. Sullivanc, P. R. Hoytc, D. A. Pelletiera, S. Wud, and D. P. Allison. 2003. AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy 97:209-216. [DOI] [PubMed] [Google Scholar]
- 9.Hassler, R. A., and D. H. Doherty. 1990. Genetic engineering of polysaccharide structure: production of variants of xanthan gum in Xanthomonas campestris. Biotechnol. Prog. 6:182-187. [DOI] [PubMed] [Google Scholar]
- 10.Helenius, G., H. Bäckdahl, A. Bodin, U. Nannmark, P. Gatenholm, and B. Risberg. 2006. In vivo biocompatibility of bacterial cellulose. J. Biomed. Mater. Res. 76:431-438. [DOI] [PubMed] [Google Scholar]
- 11.Hestrin, S., and M. Schramm. 1954. Synthesis of cellulose by Acetobacter xylinum: preparation of freeze dried cells capable of polymerizing glucose to cellulose. Biochem. J. 58:345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirai, A., M. Tsuji, and F. Horii. 1997. Culture conditions producing structure entities composed of cellulose I and II in bacterial cellulose. Cellulose 4:239-243. [Google Scholar]
- 13.Kataoka, Y., and Y. Kondo. 1998. FT-IR microscopic analysis of changing cellulose crystalline structure during wood cell wall formation. Macromolecules 31:760-764. [Google Scholar]
- 14.Kobayashi, S., and M. Ohmae. 2006. Enzymatic polymerization to polysaccharides. Adv. Polym. Sci. 194:159-210. [Google Scholar]
- 15.Kolpak, F. J., and J. Blackwell. 1975. The structure of regenerated cellulose. Macromolecules 8:563-564. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J. W., F. Deng, W. G. Yeomans, A. L. Allen, R. A. Gross, and D. L. Kaplan. 2001. Direct incorporation of glucosamine and N-acetylglucosamine into exopolymers by Gluconacetobacter xylinus (Acetobacter xylinum) ATCC 10245: production of chitosan-cellulose and chitin-cellulose exopolymers. Appl. Environ. Microbiol. 67:3970-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, P. 1997. Wound healing: aiming for perfect skin regeneration. Science 276:75-81. [DOI] [PubMed] [Google Scholar]
- 18.Michalodimitrakis, K., and M. Isalan. 2009. Engineering prokaryotic gene circuits. FEMS Microbiol. Rev. 33:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishi, N., A. Ebinaa, S.-I. Nishimuraa, A. Tsutsumi, O. Hasegawaa, and S. Tokura. 1986. Highly phosphorylated derivatives of chitin, partially deacetylated chitin and chitosan as new functional polymers: preparation and characterization. Int. J. Biol. Macromol. 8:311-317. [Google Scholar]
- 20.Ogawa, R., Y. Miura, S. Tokura, and T. Koriyama. 1992. Susceptibilities of bacterial cellulose containing N-acetylglucosamine residues for cellulolytic and chitinolytic enzymes. Int. J. Biol. Macromol. 14:343-347. [DOI] [PubMed] [Google Scholar]
- 21.Pang, F.-J., C.-J. He, and Q.-R. Wang. 2003. Preparation and properties of cellulose/chitin blend fiber. J. Appl. Polym. Sci. 90:3430-3436. [Google Scholar]
- 22.Reinecke, F., and A. Steinbüchel. 2009. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J. Mol. Microbiol. Biotechnol. 16:91-108. [DOI] [PubMed] [Google Scholar]
- 23.Ross, P., R. Mayer, and M. Benziman. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55:35-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubin, E. M. 2008. Genomics of cellulosic biofuels. Nature 454:841-845. [DOI] [PubMed] [Google Scholar]
- 25.Schubert, C. 2006. Can biofuels finally take center stage? Nat. Biotechnol. 24:777-784. [DOI] [PubMed] [Google Scholar]
- 26.Shaw, A. J., K. K. Podkaminer, S. G. Desai, J. S. Bardsley, S. R. Rogers, P. G. Thorne, D. A. Hogsett, and L. R. Lynd. 2008. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc. Natl. Acad. Sci. U. S. A. 105:13769-13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirai, A., M. Takahashi, H. Kaneko, S.-l. Nishimura, M. Ogawa, N. Nishi, and S. Tokura. 1994. Biosynthesis of a novel polysaccharide by Acetobacter xylinum. Int. J. Biol. Macromol. 16:297-300. [DOI] [PubMed] [Google Scholar]
- 28.Span, P. N., M.-J. J. M. Pouwels, A. J. Olthaar, R. R. Bosch, A. R. M. M. Hermus, and C. G. J. Sweep. 2001. Assay for hexosamine pathway intermediates (UDP-GlcNAc) in small samples of human muscle tissue. Clin. Chem. 47:944-946. [PubMed] [Google Scholar]
- 29.Steinbuchel, A., and T. Lutke-Eversloh. 2003. Metabolic engineering and pathway construction for biotechnological production of relevant polyhydroxyalkanoates (PHA) in microorganisms. Biochem. Eng. J. 14:454-459. [Google Scholar]
- 30.Sticklen, M. B. 2008. Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat. Rev. Genet. 9:433-443. [DOI] [PubMed] [Google Scholar]
- 31.Sugiyama, J., R. Vuong, and H. Chanzy. 1991. Electron diffraction study on the two crystalline phases occurring in native cellulose from an algal cell wall. Macromolecules 24:4168-4175. [Google Scholar]
- 32.Suzuki, Y., and Y. Makino. 1999. Mucosal drug delivery using cellulose derivatives as a functional polymer. J. Control. Release 62:101-107. [DOI] [PubMed] [Google Scholar]
- 33.Svensson, A., E. Nicklasson, T. Harrah, B. Panilaitis, D. L. Kaplan, M. Brittberg, and P. Gatenholm. 2005. Bacterial cellulose as a potential scaffold for tissue engineering of cartilage. Biomaterials 26:419-431. [DOI] [PubMed] [Google Scholar]
- 34.Tokoh, C., K. Takabe, M. Fujita, and H. Saiki. 1998. Cellulose synthesized by Acetobacter xylinum in the presence of glucomannan. Cellulose 5:249-261. [Google Scholar]
- 35.Tomonori, N., N. Tonouchi, T. Konishi, Y. Kojima, T. Tsuchida, F. Yoshinaga, F. Sakai, and T. Hayashi. 1999. Enhancement of cellulose production by expression of sucrose synthase in Acetobacter xylinum. Proc. Natl. Acad. Sci. U. S. A. 96:14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Die, I., R. D. Cummings, A. van Tetering, C. H. Hokke, C. A. M. Koeleman, and D. H. van den Eijnden. 2000. Identification of a novel UDP-Glc:GlcNAc β1→4-glucosyltransferase in Lymnaea stagnalis that may be involved in the synthesis of complex-type oligosaccharide chains. Glycobiology 10:263-271. [DOI] [PubMed] [Google Scholar]
- 37.van Eikeren, P., and H. McLaughlin. 1977. Analysis of the lysozyme-catalyzed hydrolysis and transglycosylation of N-acetyl-D-glucosamine oligomers by high-pressure liquid chromatography. Anal. Biochem. 77:513-522. [DOI] [PubMed] [Google Scholar]
- 38.Wada, M., J. Sugiyama, and T. Okano. 1993. Native celluloses on the basis of two crystalline phase (Iα/Iβ) system. J. Appl. Polym. Sci. 49:1491-1496. [Google Scholar]
- 39.Watts, K. T., B. N. Mijts, and C. Schmidt-Dannert. 2003. Current and emerging approaches for natural biosynthesis in microbial cells. Adv. Synth. Catal. 347:927-940. [Google Scholar]
- 40.Wong, H. C., A. L. Fear, R. D. Calhoon, G. H. Eichinger, R. Mayer, D. Amikam, M. Benziman, D. H. Gelfand, J. H. Meade, and A. W. Emerick. 1990. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. U. S. A. 87:8130-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto, H., F. Horii, and A. Hirai. 1996. In situ crystallization of bacterial cellulose. II. Influences of polymeric additives with different molecular weights on the formation of celluloses Iα and Iβ at the early stage of incubation. Cellulose 3:229-242. [Google Scholar]
- 42.Yamamoto, H., F. Horii, and H. Odani. 1989. Structural changes of native cellulose crystals induced by annealing in aqueous alkaline and acidic solutions at high temperatures. Macromolecules 22:4130-4132. [Google Scholar]
- 43.Zhang, M., C. Eddy, K. Deanda, M. Finkelstein, and S. Picataggio. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240-243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.