Abstract
The wood decay basidiomycete Phanerochaete chrysosporium produces a variety of cellobiohydrolases belonging to glycoside hydrolase (GH) families 6 and 7 in the presence of cellulose. However, no inducer of the production of these enzymes has yet been identified. Here, we quantitatively compared the transcript levels of the genes encoding GH family 6 cellobiohydrolase (cel6A) and GH family 7 cellobiohydrolase isozymes (cel7A to cel7F/G) in cultures containing glucose, cellulose, and cellooligosaccharides by real-time quantitative PCR, in order to evaluate the transcription-inducing effect of soluble sugars. Upregulation of transcript levels in the presence of cellulose compared to glucose was observed for cel7B, cel7C, cel7D, cel7F/G, and cel6A at all time points during cultivation. In particular, the transcription of cel7C and cel7D was strongly induced by cellotriose or cellotetraose. The highest level of cel7C transcripts was observed in the presence of cellotetraose, whereas the highest level of cel7D transcripts was found in the presence of cellotriose, amounting to 2.7 × 106 and 1.7 × 106 copies per 105 actin gene transcripts, respectively. These numbers of cel7C and cel7D transcripts were higher than those in the presence of cellulose. In contrast, cellobiose had a weaker transcription-inducing effect than either cellotriose or cellotetraose for cel7C and had little effect in the case of cel7D. These results indicate that cellotriose and cellotetraose, but not cellobiose, are possible natural cellobiohydrolase gene transcription inducers derived from cellulose.
Cellulose existing in the form of microfibrils in plant cell wall is the most abundant renewable carbon source on earth (14). Native cellulose microfibrils are highly crystalline, consisting of parallel-stacked linear chains of β-1,4-linked glucose residues (15). The degradation of cellulose in nature is performed mainly by microorganisms, which produce a set of extracellular cellulose-hydrolyzing enzymes (6, 7, 13, 48). Among these enzymes, cellobiohydrolase (CBH; EC 3.2.1.91) is essential for deconstruction of the crystalline part of cellulose (10, 11, 20), suggesting that it plays an important role in the carbon cycle in nature. Fungal CBHs are mainly classified into glycoside hydrolase (GH) families 7 (Cel7; formally known as CBHI) and 6 (Cel6; formally known as CBHII), respectively, based on amino acid sequence similarities (18-20). Many cellulolytic basidiomycetes and ascomycetes produce one or more GH family 6 and 7 CBHs, as listed in the Carbohydrate-Active enZymes (CAZy) website (http://www.cazy.org/).
The filamentous ascomycete Hypocrea jecorina (anamorph, Trichoderma reesei), which is the best-studied microorganism from the viewpoint of cellulose degradation, has one gene for Cel7A (CBHI) (39, 45) and also one for Cel6A (CBHII) (46). H. jecorina has another gene for GH family 7 hydrolase (Cel7B) (32), but Cel7B was functionally characterized as an endoglucanase (EG; EC 3.2.1.4) (16). CBHs are induced in the presence of cellulose (24), even though cellulose is insoluble and may not be directly recognizable by the fungus. Several studies have indicated that a water-soluble, low-molecular-weight inducer is necessary for efficient production of CBHs (reviewed in reference 42). Sophorose, a β-1,2-diglucoside, is a potent inducer produced from cellulose (26, 40, 41) and may be formed by transglycosylation of cellooligosaccharides catalyzed by a β-glucosidase (BGL) (50), although there is no report of any purified BGL that can convert cellooligosaccharides to sophorose. Antibody competition and antisense RNA experiments have led to the proposal that low basal levels of cellulases (mainly Cel7A and Cel6A) are necessary for further induction (5, 12). Analysis of CBH gene expression by using cellulase deletion mutants strongly suggested that basal expression of Cel6A and GH family 5 EG (Cel5A) is indispensable for the formation of the inducer(s) of CBHs from cellulose (37, 38).
Although basidiomycetes efficiently degrade crystalline cellulose by utilizing CBHs, relatively little is known about the induction of the genes encoding CBHs, compared with the case of ascomycetes. The wood-rotting basidiomycete Phanerochaete chrysosporium has one gene coding for Cel6A (CBHII) (28, 47) and seven genes encoding proteins belonging to GH family 7 (Cel7A to Cel7F/G); the latter are located at seven different loci and include a duplication of the same sequence (cel7F and cel7G) (8, 9, 54). It was found that cel7C and cel7D are coordinately expressed under various conditions (3, 4, 47), and their expression is repressed by glucose (4, 9). The amounts of the cel7 gene transcripts, as estimated by competitive reverse transcription-PCR (RT-PCR) analysis, were different from each other in cellulose medium supplemented with 0.1% cellobiose (9, 52). The cel7D transcripts were the most abundant, while cel7A and cel7B transcripts were expressed at constitutively low levels, and cel7C transcripts were highly expressed in the presence of cellulose. However, no inducer for the expression of cel7 genes has yet been identified.
Since cellobiose and cellooligosaccharides are the major products of cellulose hydrolysis, it is possible that cellulolytic organisms employ them to regulate cellulase production. Indeed, specific transporters of cellobiose and cellotriose have been identified in the cellulolytic bacterium Streptomyces reticuli (35, 36). Moreover, it was reported that Clostridium thermocellum assimilates cellopentaose preferentially during growth on cellulose (57). In addition, the transcription of endoglucanase genes is induced by cellotriose rather than cellobiose in Ruminococcus flavefaciens (56). These findings indicate the presence of selective assimilation and response mechanisms for specific cellooligosaccharides in these bacteria. In H. jecorina, cellobiose was speculated to be a natural inducer (27, 50), although its inductive effect was weaker than that of sophorose. In the case of cellulolytic basidiomycetes, responsiveness to cellooligosaccharides has been reported only in the case of Polyporus arcularius, which showed upregulation of cellobiohydrolase and endoglucanase gene expression in culture medium containing cellopentaose (30, 31).
Here, we performed quantitative transcription analysis of cel7 genes in P. chrysosporium cultured in media containing glucose, cellulose, and cellooligosaccharides and compared the transcription levels of the cel7 genes in the presence of these sugars. In addition, the transcription of cel6A was quantified and compared with that of the cel7 genes.
MATERIALS AND METHODS
Fungal strain and culture conditions.
P. chrysosporium strain K-3 (23) and modified Kremer and Wood medium (25) with the composition described previously (44) were used in this study. For transcription analysis in the presence of cellulose and glucose, 1 × 109 liter−1 spores of the fungus were inoculated in 400 ml of this medium containing 2% cellulose (CF11; Whatman, Fairfield, NJ) or 100 mM glucose (Wako Pure Chemical Industries, Osaka, Japan) as the sole carbon source. The inoculated media were maintained at 37°C and shaken at 150 rpm for 5 days. A 5-ml aliquot of the culture was harvested every 24 h. For cultivation with cellooligosaccharides, the spores were inoculated in 200 ml of the medium and cultivated under the same conditions used for pregrowth. After 3 days of cultivation, the mycelia were harvested, washed three times with 100 ml of the same medium containing no carbon source, and transferred to 200 ml of fresh medium containing 20 mM glycerol (Wako). After 6 h of cultivation, 100 μM glucose (Wako), cellobiose, cellotriose, cellotetraose, or cellopentaose (Seikagaku Corporation, Tokyo, Japan) was added to the medium and cultivation was continued for another 6 h. A 5-ml aliquot of the culture was harvested every hour for the determination of sugar concentration and mRNA extraction.
Measurement of glucose and cellooligosaccharide concentration in the culture supernatant.
The culture supernatant was boiled for 5 min to inactivate enzymes secreted by the fungus. The concentrations of glucose and cellooligosaccharides in the supernatant were measured by high-performance liquid chromatography (HPLC; LC-2000 series; Jasco, Tokyo, Japan), using a corona-charged aerosol detector (ESA Biosciences, Chelmsford, MA) based on our previous report (21). The supernatants were filtered using a MultiScreen HTS 96-well filtration system (Millipore Corporation, Billerica, MA) and then separated on a Shodex Asahipak NH2P-50 4E (Showa Denko K.K., Kanagawa, Japan) with isocratic elution (65% acetonitrile, 35% H2O [vol/vol]). In addition, isocratic elution with 75% acetonitrile-25% H2O was used to separate cellobiose from other disaccharides. The amount of each sugar was quantified by using glucose (BioUltra; Sigma-Aldrich, St. Louis, MO) and cellooligosaccharides with degree of polymerization (DP) values of 2 to 6 (Seikagaku Corporation) as standards.
Real-time RT-PCR analysis of cellulolytic gene transcripts.
Mycelia collected from the culture aliquots were immediately frozen in liquid nitrogen and stored at −80°C to extract RNA. Frozen fungal mycelia were ground to a fine powder using a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan), and then total RNA was extracted with a RNeasy plant mini kit (Qiagen, Valencia, CA) and treated with an RNase-free DNase set (Qiagen), according to the manufacturer's instructions. First-strand cDNA was synthesized using ReverTra Ace (Toyobo, Osaka, Japan), as described previously (44). Real-time RT-PCRs were performed in an Mx3000P real-time QPCR (quantitative PCR) system (Stratagene, La Jolla, CA) as follows. Five microliters of template solution was mixed with 20 μl of solution containing 12.5 μl of Brilliant II Fast SYBR green QPCR master mix (Stratagene), 0.5 μl of 10 μM forward primer, 0.5 μl of 10 μM reverse primer, 0.375 μl of 6-carboxy-X-rhodamine (ROX) reference dye solution, and 6.125 μl of sterile distilled water. The mixtures were initially incubated at 95°C for 2 min, followed by amplification for up to 40 cycles of 95°C for 5 s and 60°C for 20 s. After thermal cycling, reaction mixtures were heated from 60°C to 94°C for 20 min at a constant rate and the fluorescence of SYBR green I was measured continuously during heating for dissociation curve analysis. Fluorescence data were analyzed using MxPro version 4.0 software (Stratagene). The sequences of the oligonucleotide primers used for amplification of the cDNA fragments derived from cel7A, cel7B, cel7C, cel7D, cel7E, cel7F/G, and cel6A genes (GenBank accession no. X54411 for both cel7A and cel7B, Z22528, L22656, Z11727, Z11729, and S76141) were designed based on the corresponding sequences in P. chrysosporium strain K-3 and listed in our previous reports (43, 44). The transcript number of the actin gene was quantified as an internal standard by using the following primers: actin-F (5′-GCATGTGCAAGGCTGGCTTTG-3′) and actin-R (5′-AGGGCGACCAACGATGGATG-3′).
RESULTS
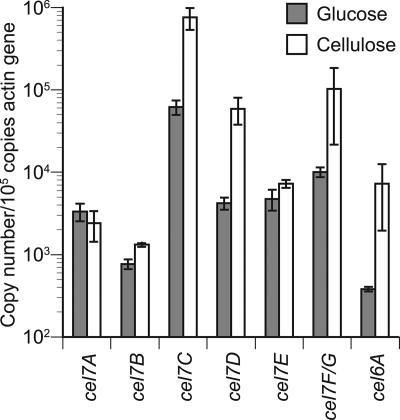
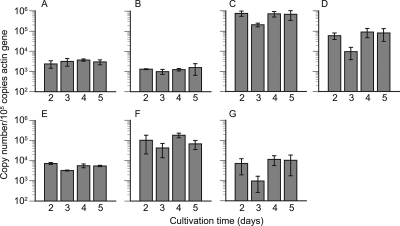
Transcript levels of cel7A to cel7F/G and cel6A were quantified in media containing 2% cellulose and 100 mM glucose by real-time RT-PCR. In 2-day-old cellulose culture, the numbers of gene transcripts of cel7A to -F/G and cel6A were 2.4 × 103, 1.3 × 103, 7.6 × 105, 5.9 × 104, 7.2 × 103, 1.0 × 105, and 7.2 × 103 copies per 105 copies of actin gene transcript, respectively (Fig. 1). The numbers of transcripts were higher in cellulose culture than in glucose culture for most of these genes, although little difference was observed for cel7A and cel7E. Throughout the cultivation, cel7B, cel7C, cel7D, cel7F/G, and cel6A transcript levels were higher in cellulose- versus glucose-grown cultures, while there were no significant differences in the transcript levels of cel7A and cel7E under these conditions. In the case of cellulose culture, the transcript numbers of cel7C, cel7D, cel7E, and cel7F/G were decreased from day 2 to day 3 by 73%, 84%, 55%, and 59%, respectively, and then increased from days 3 to 4 (Fig. 2 C to F). There were no clear alterations during day 4 and day 5, except in the case of cel7F/G, for which the transcript level was decreased again by 63% at this time point. In contrast, no marked alteration of transcript numbers was observed for cel7A and cel7B (Fig. 2A and B). The transcript numbers of cel6A were decreased from day 2 to day 3 by 87% (Fig. 2G). As regards the absolute number of transcripts, cel7C was most abundant, while cel7D and cel7F/G transcripts were at similar levels in cellulose culture.
FIG. 1.
Copy numbers of cel7A to -F/G and cel6A transcripts quantified by real-time PCR in 2-day-old culture with 100 mM glucose or 2% cellulose as a carbon source. The vertical axis indicates the number of transcripts normalized with respect to 105 copies of actin gene transcripts in the same sample. Error bars show the standard deviation in triplicate tests.
FIG. 2.
Copy numbers of cel7A (A), cel7B (B), cel7C (C), cel7D (D), cel7E (E), cel7F/G (F), and cel6A (G) transcripts quantified by real-time PCR during 2 to 5 days of cultivation with 2% cellulose as a carbon source. Template cDNAs were prepared from the culture every 24 h. The vertical axis indicates the number of transcripts normalized with respect to 105 copies of actin gene transcripts in the same sample. Error bars show the standard deviation in triplicate tests.
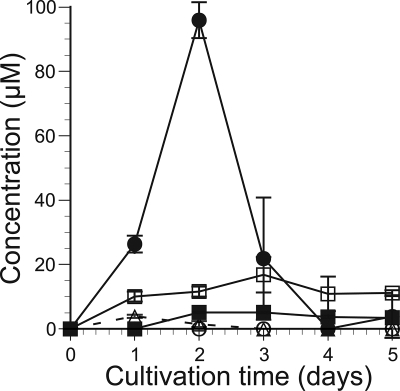
Accumulation of soluble cellooligosaccharides in the supernatant of cellulose-containing culture was quantified by HPLC (Fig. 3). Under the conditions used, approximately 100 μM cellobiose accumulated in 2 days and then was completely lost by day 4. The cellotriose concentration reached about 4 μM in day 1 and then decreased. Production of cellotetraose was observed from day 2 and reached approximately 5 μM during further cultivation. In contrast, no accumulation of cellopentaose or cellohexaose was detected in the culture medium.
FIG. 3.
Concentrations of glucose and cellooligosaccharides in extracellular fluid from culture in 2% cellulose medium. The concentration was quantified by HPLC every 24 h for 5 days. Open squares, glucose; filled circles, cellobiose; open triangles, cellotriose; filled squares, cellotetraose; open circles, cellopentaose. Error bars show the standard deviation in triplicate tests.
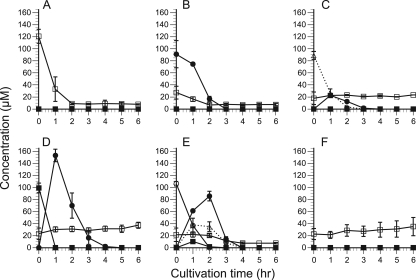
To evaluate the transcript levels of cellulolytic genes in the presence of soluble cellooligosaccharides, P. chrysosporium was cultivated in medium containing 20 mM glycerol as a carbon source supplemented with 100 μM glucose or cellooligosaccharides with a DP value of 2 to 5. The time courses of the concentrations of these sugars were quantified using HPLC during 6 h of cultivation. In the culture with glucose added, the initial concentration of glucose was 120 μM (Fig. 4 A), because 20 μM glucose remained after the precultivation. A similar amount of glucose was present in all media tested (Fig. 4). In the cellobiose culture, cellobiose was completely consumed within 3 h (Fig. 4B). In the culture supplemented with oligosaccharides with DP values of 3 to 5, oligosaccharides with lower DP values appeared during cultivation. As shown in Fig. 4C, cellotriose was totally assimilated or hydrolyzed within 4 h, and 23 μM cellobiose was produced within 1 h in the cellotriose culture. In the culture with cellotetraose, hydrolysis or assimilation of cellotetraose occurred within 1 h and 150 μM cellobiose was produced, although no cellotriose was detected (Fig. 4D). All possible hydrolysis products were detected in the cellopentaose culture (Fig. 4E). In the case of the control cultivation without any additive, only glucose was detected, as shown in Fig. 4F.
FIG. 4.
Time course of concentrations of glucose and cellooligosaccharides in extracellular fluid from 20 mM glycerol culture, supplemented with 100 μM glucose (A), cellobiose (B), cellotriose (C), cellotetraose (D), or cellopentaose (E) or with no addition as a control (F). The concentration was quantified by HPLC for 6 h after the addition of each sugar. Open squares, glucose; filled circles, cellobiose; open triangles, cellotriose; filled squares, cellotetraose; open circles, cellopentaose. Error bars show the standard deviation in triplicate tests.
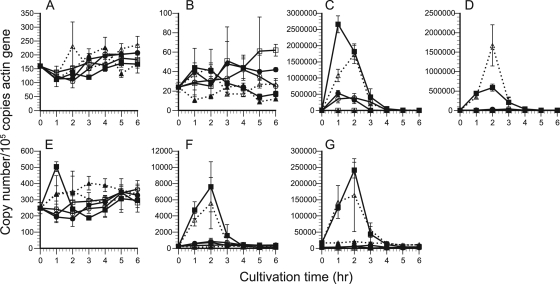
In the culture containing glucose or cellooligosaccharide, the time courses of transcript levels of cellulolytic genes were quantified by real-time RT-PCR. Throughout the cultivation, no apparent alteration of transcript number was detected for any of the genes tested in the control culture containing only glycerol; thus, it was confirmed that glycerol acts as a neutral carbon source for cellulolytic gene expression under the conditions used. The transcript patterns in response to cellooligosaccharides differed among the genes, as shown in Fig. 5. The transcription of cel7C was apparently upregulated by cellotriose and cellotetraose (Fig. 5C), and the maximum amount of cel7C transcripts, corresponding to a 970-fold increase, was obtained after 1 h in cellotetraose culture. Addition of cellobiose caused a 190-fold increase of cel7C transcripts after 1 h. In cellopentaose culture, the number of cel7C transcripts increased to about the same level as in cellobiose culture, although more slowly. In the case of cel7D (Fig. 5D), in contrast, maximum transcript levels (340-fold increase) were seen after 2 h in cellotriose culture rather than in cellotetraose culture (120-fold). Conversely, addition of cellobiose and cellopentaose had little effect on cel7D transcript levels. As for cel7F/G (Fig. 5F), transcription was upregulated in cellotriose and cellotetraose cultures, and the largest number of transcripts was detected after 2 h in cellotetraose culture (30-fold increase), while addition of cellobiose or cellopentaose slightly increased cel7F/G transcripts. The expression of cel6A (Fig. 5G) was upregulated by cellotriose and cellotetraose as a 76-fold increase in cellotetraose culture. However, transcripts of cel7A, cel7B, and cel7E showed little variation among culture conditions throughout the cultivation (Fig. 5A, B and E). In addition, repression of transcript levels in glucose culture was observed only for cel7D and cel6A, for up to 2 h under the culture conditions used.
FIG. 5.
Time courses of the copy numbers of cellulolytic genes during 6 h of cultivation in culture medium supplemented with glucose (open squares), cellobiose (filled circles), cellotriose (open triangles), cellotetraose (filled squares), or cellopentaose (open circles) or with no addition as a control (filled triangles). The transcript numbers of cel7A (A), cel7B (B), cel7C (C), cel7D (D), cel7E (E), cel7F/G (F), and cel6A (G) were quantified by real-time PCR and normalized as described in the legend to Fig. 1. Error bars show the standard deviation in triplicate tests.
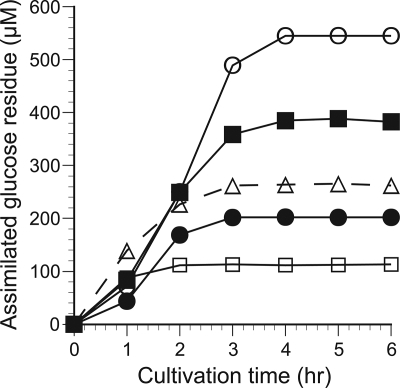
To compare the amounts of substrate metabolized by the fungus in each culture, the cumulative sugar assimilation during cultivation with cellooligosaccharides was calculated as the product of the decrease from the initial concentration and the DP value of each oligosaccharide and expressed in terms of the concentration of glucose residue (Fig. 6). By 2 h, the concentration of assimilated glucose residue was higher in cellotriose or cellotetraose culture than in cellobiose culture, implying that more carbon was metabolized in cellotriose and cellotetraose culture. After 3 h, most of the supplemented sugar was assimilated, so the concentration of assimilated glucose residue was higher in cultures containing cellooligosaccharides with higher DP values.
FIG. 6.
Time course of sugar assimilation during 6 h of cultivation in culture medium supplemented with glucose (open squares), cellobiose (filled circles), cellotriose (open triangles), cellotetraose (filled squares), or cellopentaose (open circles). The vertical axis indicates total assimilated glucose residue from the culture, calculated as the product of the decrease from the initial concentration and the DP value of each oligosaccharide.
DISCUSSION
In cellooligosaccharide-mediated induction of cellulolytic gene expression, two modes were expected: i.e., an extracellular receptor-mediated signaling or uptake and intracellular binding to signaling molecules. There is no information about these signal-transducing molecules in P. chrysosporium at all, as far as we know, and thus the induction mechanism by cellooligosaccharides remains uncertain. Nevertheless, our results indicate that the addition of cellotriose and cellotetraose clearly increasees transcript levels of several cellulolytic genes in P. chrysosporium, although that of cellobiose has little effect. Interestingly, the transcriptional responses of various cel7 genes to these sugars varied. In the case of cel7C, induction in cellobiose culture was clearly observed, but it was 5-fold lower than that in cellotetraose culture (Table 1). Thus, cellobiose, which appeared after 1 h in the cellotetraose culture, probably affected the expression of cel7C in that culture, but its effect was considered to be small compared to that of cellotetraose itself. Moreover, only a small amount of cellobiose was formed in cellotriose culture, so that the potent cel7C-inducing effect is likely to be due to cellotriose itself. Similar results were observed for cel7F/G, although in this case, the inducing effect of cellobiose would be even weaker. Addition of cellobiose had little effect in the cases of cel7D and cel6A, indicating that expression of these genes would not be significantly induced by the formation of cellobiose. In the case of cel7D, cellotriose appeared to be a better inducer than cellotetraose, in contrast to the case of cel7C. The effect of cellopentaose itself was difficult to estimate, because of the complicating effect of shorter oligosaccharides formed during the cultivation. Nevertheless, cellopentaose appeared not to be a potent inducer, because all five genes were only weakly upregulated in cellopentaose culture.
TABLE 1.
Maximum values of gene transcripts during cultivation
| Gene | Maximum no. of copies of transcript in culture with: |
||||
|---|---|---|---|---|---|
| Carbon starvationa | Cellulose | Cellobiose | Cellotriose or cellotetraose | Glycerol | |
| cel7A | 5.22 × 102 | 3.67 × 103 | 2.07 × 102 | 2.36 × 102 | 2.27 × 102 |
| cel7B | 1.30 × 102 | 1.58 × 103 | 4.80 × 101 | 4.40 × 101 | 2.60 × 101 |
| cel7C | 3.78 × 104 | 7.58 × 105 | 5.28 × 105 | 2.66 × 106 | 3.59 × 103 |
| cel7D | 1.39 × 105 | 9.04 × 104 | 2.86 × 104 | 1.67 × 106 | 5.10 × 103 |
| cel7E | 3.28 × 102 | 7.23 × 103 | 3.49 × 102 | 5.04 × 102 | 4.02 × 102 |
| cel7F/G | 2.93 × 103 | 1.83 × 105 | 7.34 × 102 | 7.60 × 103 | 2.81 × 102 |
| cel6A | 1.83 × 105 | 1.15 × 104 | 6.16 × 103 | 2.41 × 105 | 3.18 × 103 |
During the metabolism of cellooligosaccharides by fungi, carbon catabolite repression would be caused by glucose, which is a hydrolysis product of cellooligosaccharides, produced by BGL. In the case of cel7C, for example, expression was upregulated by 2 h of cultivation in the presence of cellobiose, cellotriose, or cellotetraose, and at that time point, larger amounts of glucose residue had been assimilated in cellotriose or cellotetraose culture than in cellobiose culture. In P. chrysosporium, hydrolysis of cellooligosaccharide may be mainly catalyzed by intracellular BGL (BGL1B) rather than extracellular BGL (BGL3A) (49). The catalytic efficiency (kcat/Km) of BGL1B is much greater for cellotriose (21 × 101 s−1 mM−1) and cellotetraose (16 × 101 s−1 mM−1) than for cellobiose (75 s−1 mM−1) (T. Tsukada et al., unpublished data). Thus, the intracellular concentration of glucose is considered to be higher in cellotriose and cellotetraose cultures, which may therefore be subject to higher levels of catabolite repression than that seen in cellobiose culture. Consequently, it appears that the lower inductive activity of cellobiose can not be ascribed to catabolism and consequent repression by glucose.
It was previously shown by transcript analysis that the expression levels of cel7A and cel7E are higher, while that of cel7D is lower, in colonized aspen wood (51) than in submerged cellulose culture (9, 52). Moreover, homology models of the three-dimensional structure of Cel7s have shown several structural differences in the tunnel-forming loops; Cel7A and Cel7B exhibit an endoglucanase-like structure, but four other Cel7s are very similar and might not show marked functional differences (29). Thus, it is expected that Cel7A, Cel7B, and Cel7E have characteristically different roles from other Cel7s in cellulose degradation. The results obtained in the present work strongly support this hypothesis, because the transcription of cel7A, cel7B, and cel7E was not affected by cellooligosaccharides, which apparently induced expression of other cellulolytic genes. Furthermore, it is possible that Cel7A and Cel7B do not participate in cellulose degradation, because their gene expression was only slightly altered during cultivation with cellulose, whereas other genes examined in this work all showed similar changes. However, the relationship between the function and structure of Cel7E requires further investigation.
In addition to the transcript numbers in cellulose- and cellooligosaccharide-supplied culture, we compared the maximum numbers of gene transcripts under a carbon-starved condition (43, 44), which provokes carbon catabolite derepression of cel7C, cel7D, and cel6A (Table 1). In the case of cel7C, the maximum amounts of gene transcripts were higher in cellotriose or cellotetraose culture but lower in carbon-starved culture than in cellulose culture. Therefore, induction by cellotriose and cellotetraose is considered to be a major factor determining cel7C transcription, rather than derepression. The transcripts of cel7D are at a higher level in both cellotriose and carbon-starved culture than in cellulose culture, indicating that the transcription of cel7D is regulated by both induction and derepression. Similarly, the transcript levels of cel6A appear strongly regulated by derepression, as well as by cellotriose- and cellotetraose-mediated induction. In the 2-day-old cellulose culture, transcript levels of cel7C, cel7D, and cel6A were considered to be dominantly affected by cellobiose, because the maximum numbers of transcripts in cellobiose culture were close to those in cellulose culture. Indeed, cellobiose was accumulated, but cellotriose and cellotetraose were produced only in small amounts in cellulose culture medium. In contrast, the expression characteristics of cel7F/G are different from those of cel7C and cel7D. The transcript number of cel7F/G in culture with any cellooligosaccharide or in carbon-starved culture did not reach the levels observed in cellulose cultures. Furthermore, a decrease in the number of transcripts from day 4 to day 5 in cellulose culture was observed only for cel7F/G. These results indicate that, although cel7F/G seems to be related to cellulose degradation, some other soluble compound, not cellooligosaccharides, is involved in the induction of cel7F/G. In addition, the transcript levels of cel7A, cel7B, and cel7E were higher in the presence of cellulose than under other culture conditions. HPLC analysis of the supernatant from the cellulose culture showed several unidentified peaks, in addition to the peaks of cellooligosaccharides. Identification and analysis of the effects of the compounds contained in these peaks will be necessary to understand the modes of expression control of the genes.
Recently, transcriptome and secretome analyses of P. chrysosporium have revealed a complex pattern of production of wood-degrading enzymes (33, 34, 53, 55), but detailed investigation of the regulatory mechanisms involved remains to be undertaken. The results presented here indicate that cellotriose and cellotetraose are candidates for natural inducers generated by cellulose hydrolysis, whereas cellobiose was not effective. In the extracellular cellulolytic system of P. chrysosporium, cellobiose could be oxidized to cellobiono-1,5-lactone if cellobiose dehydrogenase is secreted (1, 2, 17). Cellobiono-1,5-lactone was reported to be an inducer of cellulase production in H. jecorina (22). Accordingly, further investigation of the inductive effect of cellobiono-1,5-lactone on transcription of cel7 and cel6 genes in P. chrysosporium seems worthwhile. It is noteworthy that the cel7 genes of P. chrysosporium were differentially regulated in a complex manner, but not all of the Cel7 enzymes seem to participate in cellulose degradation. Although the machinery of receptors or transporters for cellooligosaccharides remains unknown, the extracellular concentration and/or uptake of cellotriose and cellotetraose could be a signal for cellulolytic enzyme production by P. chrysosporium. However, some of our present results are not consistent with those of a previous study using homokaryotic strain RP-78 or its dikaryotic parent, BKM-F-1767. For example, cel7D was the most abundant transcript among cel7 genes in BKM-F-1767 (52), while cel7C transcripts were most abundant in heterokaryotic strain K-3. Also, cel7E of strain RP-78 is upregulated in cellulose culture compared to glucose culture (53), whereas we found no difference between these carbon sources in strain K-3. Future studies may determine whether these different transcript patterns are due to culture conditions and/or strain variation.
Acknowledgments
This research was supported by a Grant-in-Aid for Scientific Research no. 20380100 to M.S. from the Japanese Ministry of Education, Culture, Sports, and Technology.
Footnotes
Published ahead of print on 23 July 2010.
REFERENCES
- 1.Ayers, A. R., S. B. Ayers, and K. E. Eriksson. 1978. Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum. Eur. J. Biochem. 90:171-181. [DOI] [PubMed] [Google Scholar]
- 2.Bao, W. J., and V. Renganathan. 1993. Cellobiose dehydrogenase of Phanerochaete chrysosporium—purification, characterization, and role in cellulose degradation. Abstr. Pap. Am. Chem. Soc. 205:25-BTEC. [Google Scholar]
- 3.Birch, P. R., P. F. Sims, and P. Broda. 1995. Substrate-dependent differential splicing of introns in the regions encoding the cellulose binding domains of two exocellobiohydrolase I-like genes in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 61:3741-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broda, P., P. R. Birch, P. R. Brooks, and P. F. Sims. 1995. PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: substrate-dependent differential expression within gene families. Appl. Environ. Microbiol. 61:2358-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carle-Urioste, J. C., J. Escobar-Vera, S. El-Gogary, F. Henrique-Silva, E. Torigoi, O. Crivellaro, A. Herrera-Estrella, and H. El-Dorry. 1997. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J. Biol. Chem. 272:10169-10174. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, A. J. 1997. Biodegradation of cellulose: enzymology and biotechnology. Technomic Publishing, Lancaster, PA.
- 7.Coughlan, M. P. 1991. Mechanisms of cellulose degradation by fungi and bacteria. Anim. Feed Sci. Technol. 32:77-100. [Google Scholar]
- 8.Covert, S. F., J. Bolduc, and D. Cullen. 1992. Genomic organization of a cellulase gene family in Phanerochaete chrysosporium. Curr. Genet. 22:407-413. [DOI] [PubMed] [Google Scholar]
- 9.Covert, S. F., A. Vanden Wymelenberg, and D. Cullen. 1992. Structure, organization, and transcription of a cellobiohydrolase gene cluster from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 58:2168-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, G., and B. Henrissat. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:853-859. [DOI] [PubMed] [Google Scholar]
- 11.Divne, C., J. Stahlberg, T. Reinikainen, L. Ruohonen, G. Pettersson, J. K. C. Knowles, T. T. Teeri, and T. A. Jones. 1994. The 3-dimensional crystal structure of the catalytic core of cellobiohydrolase-I from Trichoderma reesei. Science 265:524-528. [DOI] [PubMed] [Google Scholar]
- 12.El-Gogary, S., A. Leite, O. Crivellaro, D. E. Eveleigh, and H. El-Dorry. 1989. Mechanism by which cellulose triggers cellobiohydrolase-I gene expression in Trichoderma reesei. Proc. Natl. Acad. Sci. U. S. A. 86:6138-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriksson, K. E., R. A. Blanchette, and P. Ander. 1990. Microbial and enzymatic degradation of wood and wood components. Springer-Verlag, New York, NY.
- 14.Frey-Wyssling, A. 1954. The fine structure of cellulose microfibrils. Science 119:80-82. [DOI] [PubMed] [Google Scholar]
- 15.Gardener, K. H., and J. Blackwell. 1974. The structure of native cellulose. Biopolymers 13:1975-2001. [Google Scholar]
- 16.Hakansson, U., L. G. Fagerstam, L. G. Pettersson, and L. Andersson. 1979. 1,4-Beta-glucan glucanohydrolase from the cellulolytic fungus Trichoderma viride QM 9414—purification, characterization and preparation of an immunoadsorbent for the enzyme. Biochem. J. 179:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriksson, G., V. Sild, I. J. Szabo, G. Pettersson, and G. Johansson. 1998. Substrate specificity of cellobiose dehydrogenase from Phanerochaete chrysosporium. Biochim. Biophys. Acta 1383:48-54. [DOI] [PubMed] [Google Scholar]
- 18.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrissat, B., and G. Davies. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637-644. [DOI] [PubMed] [Google Scholar]
- 21.Igarashi, K., T. Ishida, C. Hori, and M. Samejima. 2008. Characterization of an endoglucanase belonging to a new subfamily of glycoside hydrolase family 45 of the basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 74:5628-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyayi, C. B., E. E. Bruchmann, and C. P. Kubicek. 1989. Induction of cellulase formation in Trichoderma reesei by cellobiono-1,5-lactone. Arch. Microbiol. 151:326-330. [Google Scholar]
- 23.Johnsrud, S. C., and K. E. Eriksson. 1985. Cross-breeding of selected and mutated homokaryotic strains of Phanerochaete chrysosporium K-3—new cellulase deficient strains with increased ability to degrade lignin. Appl. Microbiol. Biotechnol. 21:320-327. [Google Scholar]
- 24.Kolbe, J., and C. P. Kubicek. 1990. Quantification and identification of the main components of the Trichoderma cellulase complex with monoclonal antibodies using an enzyme-linked immunosorbent assay (ELISA). Appl. Microbiol. Biotechnol. 34:26-30. [DOI] [PubMed] [Google Scholar]
- 25.Kremer, S. M., and P. M. Wood. 1992. Evidence that cellobiose oxidase from Phanerochaete chrysosporium is primarily an Fe(III) reductase. Kinetic comparison with neutrophil NADPH oxidase and yeast flavocytochrome b2. Eur. J. Biochem. 205:133-138. [DOI] [PubMed] [Google Scholar]
- 26.Mandels, M., F. W. Parrish, and E. T. Reese. 1962. Sophorose as an inducer of cellulase in Trichoderma viride. J. Bacteriol. 83:400-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandels, M., and E. T. Reese. 1960. Induction of cellulase in fungi by cellobiose. J. Bacteriol. 79:816-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez, D., L. F. Larrondo, N. Putnam, M. D. Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, P. Ramaiya, J. C. Detter, F. Larimer, P. M. Coutinho, B. Henrissat, R. Berka, D. Cullen, and D. Rokhsar. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695-700. [DOI] [PubMed] [Google Scholar]
- 29.Munoz, I. G., W. Ubhayasekera, H. Henriksson, I. Szabo, G. Pettersson, G. Johansson, S. L. Mowbray, and J. Stahlberg. 2001. Family 7 cellobiohydrolases from Phanerochaete chrysosporium: crystal structure of the catalytic module of Cel7D (CBH58) at 1.32 A resolution and homology models of the isozymes. J. Mol. Biol. 314:1097-1111. [DOI] [PubMed] [Google Scholar]
- 30.Ohnishi, Y., M. Nagase, T. Ichiyanagi, Y. Kitamoto, and T. Aimi. 2007. Transcriptional regulation of two cellobiohydrolase encoding genes (cel1 and cel2) from the wood-degrading basidiomycete Polyporus arcularius. Appl. Microbiol. Biotechnol. 76:1069-1078. [DOI] [PubMed] [Google Scholar]
- 31.Ohnishi, Y., M. Nagase, T. Ichiyanagi, Y. Kitamoto, and T. Aimi. 2007. Transcriptional regulation of two endoglucanase-encoding genes (cel3A and cel4) from the wood-degrading basidiomycete Polyporus arcularius. FEMS Microbiol. Lett. 274:218-225. [DOI] [PubMed] [Google Scholar]
- 32.Penttila, M., P. Lehtovaara, H. Nevalainen, R. Bhikhabhai, and J. Knowles. 1986. Homology between cellulase genes of Trichoderma reesei—complete nucleotide-sequence of the endoglucanase-I gene. Gene 45:253-263. [DOI] [PubMed] [Google Scholar]
- 33.Sato, S., F. A. Feltus, P. Iyer, and M. Tien. 2009. The first genome-level transcriptome of the wood-degrading fungus Phanerochaete chrysosporium grown on red oak. Curr. Genet. 55:273-286. [DOI] [PubMed] [Google Scholar]
- 34.Sato, S., F. Liu, H. Koc, and M. Tien. 2007. Expression analysis of extracellular proteins from Phanerochaete chrysosporium grown on different liquid and solid substrates. Microbiology 153:3023-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlosser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlosser, A., and H. Schrempf. 1996. A lipid-anchored binding protein is a component of an ATP-dependent cellobiose/cellotriose-transport system from the cellulose degrader Streptomyces reticuli. Eur. J. Biochem. 242:332-338. [DOI] [PubMed] [Google Scholar]
- 37.Seiboth, B., S. Hakola, R. L. Mach, P. L. Suominen, and C. P. Kubicek. 1997. Role of four major cellulases in triggering of cellulase gene expression by cellulose in Trichoderma reesei. J. Bacteriol. 179:5318-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seiboth, B., R. Messner, F. Gruber, and C. P. Kubicek. 1992. Disruption of the Trichoderma reesei-Cbh2 gene coding for cellobiohydrolase-II leads to a delay in the triggering of cellulase formation by cellulose. J. Gen. Microbiol. 138:1259-1264. [Google Scholar]
- 39.Shoemaker, S., V. Schweickart, M. Ladner, D. Gelfand, S. Kwok, K. Myambo, and M. Innis. 1983. Molecular cloning of exo-cellobiohydrolase-I derived from Trichoderma reesei strain-L27. Biotechnology 1:691-696. [Google Scholar]
- 40.Sternberg, D., and G. R. Mandels. 1979. Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J. Bacteriol. 139:761-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sternberg, D., and G. R. Mandels. 1980. Regulation of the cellulolytic system in Trichoderma reesei by sophorose: induction of cellulase and repression of β-glucosidase. J. Bacteriol. 144:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suto, M., and F. Tomita. 2001. Induction and catabolite repression mechanisms of cellulase in fungi. J. Biosci. Bioeng. 92:305-311. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, H., K. Igarashi, and M. Samejima. 2009. Quantitative transcriptional analysis of the genes encoding glycoside hydrolase family 7 cellulase isozymes in the basidiomycete Phanerochaete chrysosporium. FEMS Microbiol. Lett. 299:159-165. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, H., K. Igarashi, and M. Samejima. 2008. Real-time quantitative analysis of carbon catabolite derepression of cellulolytic genes expressed in the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 80:99-106. [DOI] [PubMed] [Google Scholar]
- 45.Teeri, T., I. Salovuori, and J. Knowles. 1983. The molecular cloning of the major cellulase gene from Trichoderma reesei. Biotechnology 1:696-699. [Google Scholar]
- 46.Teeri, T. T., P. Lehtovaara, S. Kauppinen, I. Salovuori, and J. Knowles. 1987. Homologous domains in Trichoderma reesei cellulolytic enzymes—gene sequence and expression of cellobiohydrolase-II. Gene 51:43-52. [DOI] [PubMed] [Google Scholar]
- 47.Tempelaars, C. A., P. R. Birch, P. F. Sims, and P. Broda. 1994. Isolation, characterization, and analysis of the expression of the cbhII gene of Phanerochaete chrysosporium. Appl. Environ. Microbiol. 60:4387-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomme, P., R. A. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 49.Tsukada, T., K. Igarashi, M. Yoshida, and M. Samejima. 2006. Molecular cloning and characterization of two intracellular β-glucosidases belonging to glycoside hydrolase family 1 from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 73:807-814. [DOI] [PubMed] [Google Scholar]
- 50.Vaheri, M. P., M. E. O. Vaheri, and V. S. Kauppinen. 1979. Formation and release of cellulolytic enzymes during growth of Trichoderma reesei on cellobiose and glycerol. Eur. J. Appl. Microbiol. Biotechnol. 8:73-80. [Google Scholar]
- 51.Vallim, M. A., B. J. Janse, J. Gaskell, A. A. Pizzirani-Kleiner, and D. Cullen. 1998. Phanerochaete chrysosporium cellobiohydrolase and cellobiose dehydrogenase transcripts in wood. Appl. Environ. Microbiol. 64:1924-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanden Wymelenberg, A., S. Covert, and D. Cullen. 1993. Identification of the gene encoding the major cellobiohydrolase of the white rot fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 59:3492-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanden Wymelenberg, A., J. Gaskell, M. Mozuch, P. Kersten, G. Sabat, D. Martinez, and D. Cullen. 2009. Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl. Environ. Microbiol. 75:4058-4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanden Wymelenberg, A., P. Minges, G. Sabat, D. Martinez, A. Aerts, A. Salamov, I. Grigoriev, H. Shapiro, N. Putnam, P. Belinky, C. Dosoretz, J. Gaskell, P. Kersten, and D. Cullen. 2006. Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveal complex mixtures of secreted proteins. Fungal Genet. Biol. 43:343-356. [DOI] [PubMed] [Google Scholar]
- 55.Vanden Wymelenberg, A., G. Sabat, D. Martinez, A. S. Rajangam, T. T. Teeri, J. Gaskell, P. J. Kersten, and D. Cullen. 2005. The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. J. Biotechnol. 118:17-34. [DOI] [PubMed] [Google Scholar]
- 56.Wang, W., S. J. Reid, and J. A. Thomson. 1993. Transcriptional regulation of an endoglucanase and a cellodextrinase gene in Ruminococcus flavefaciens FD-1. J. Gen. Microbiol. 139:1219-1226. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y. H. P., and L. R. Lynd. 2005. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc. Natl. Acad. Sci. U. S. A. 102:7321-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]