Abstract
Herewith we report the expression and screening of microbial enzymes without involving cloning procedures. Computationally predicted putative ω-transaminase (ω-TA) genes were PCR amplified from the bacterial colonies and expressed in a cell-free protein synthesis system for subsequent analysis of their enzymatic activity and substrate specificity. Through the cell-free expression analysis of the putative ω-TA genes, a number of enzyme-substrate pairs were identified in a matter of hours. We expect that the proposed strategy will provide a universal platform for bridging the information gap between nucleotide sequence and protein function to accelerate the discovery of novel enzymes.
Recent advances in genome sequencing technology have accumulated enormous amounts of sequence information (12). Although protein function encoded in nucleotide sequences can be annotated using computational alignment tools, in many cases, significant similarity to proteins with known function is hard to establish (5, 18). To understand the biological function of these unknown proteins, as well as to validate the computer-annotated results, efficient methods that enable rapid translation of genetic information into protein function are in high demand. The availability of high-throughput method for protein generation is also essential for accelerating the discovery and evolution of biocatalysts (3, 4, 6, 14, 22, 23) used in industry. While gene cloning and cultivation of transformed cells have long been used as standard methods for production of recombinant proteins, the vast amount of sequence information from various genome sequencing projects is now demanding a throughput of protein expression that exceeds that of the present in vivo expression techniques.
Compared to cell-based gene expression, cell-free protein synthesis offers substantial advantages in speed and flexibility for the simultaneous expression of multiple proteins (7, 9, 13, 16, 19, 21). As a part of our efforts to extend the application of cell-free protein synthesis into the field of enzyme technology, we report in this paper an integrated methodology for fast expression screening of enzymes using ω-transaminases (ω-TAs) as a model target. Transaminases are pyridoxal-5′-phosphate (PLP)-dependent enzymes that catalyze reversible transfer of amine groups to keto acids, producing diverse proteogenic or nonproteogenic amino acids (1).
In this work, ω-TA genes from microbial colonies were amplified by PCR and directly expressed in a cell-free protein synthesis system. Expressed enzymes were then screened for their activity toward different amine donors by colorimetric measurement of the changes in the concentration of pyruvate, which was used as a common amine acceptor. As a result, analysis of the substrate specificities of the enzymes encoded by 11 ω-TA genes toward 16 amine-donating compounds were completed within a matter of hours, identifying a number of enzyme-substrate matches.
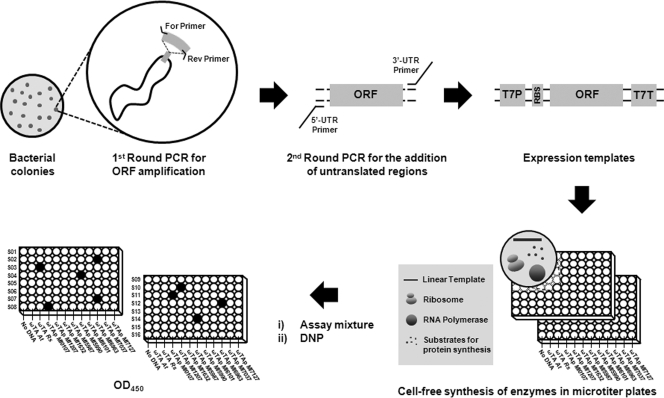
We started by examining whether sufficient amount of proteins could be generated for enzymatic analysis of ω-TAs when the PCR products amplified from the bacterial colonies were used as the template for cell-free synthesis reactions. The efficiency of protein synthesis was compared for reactions programmed with a plasmid-cloned ω-TA gene from Vibrio fluvialis (Vfω-TA) (pIVEX2.3d ω-TA Vf) and reactions programmed with the same gene prepared by two-step PCR from a bacterial colony (Vibrio fluvialis JS17 [20]), as depicted in Fig. 1. The ω-TA genes examined in this study are listed in Table S1 in the supplemental material along with their bacterial sources.
FIG. 1.
Experimental scheme for cloning-independent cell-free expression screening of ω-transaminases. The expression templates for cell-free synthesis were prepared through two-step amplification of the target open reading frame (ORF) from bacterial genomes. PCR products were translated into corresponding enzymes in microtiter plates as described in the text. Upon completion of the synthesis reaction, the reaction mixture was sequentially supplied with the assay mixture and chromogenic compound to determine the residual pyruvate concentration after the amine transfer reaction. Abbreviations: For and Rev Primer, forward and reverse primers, respectively; 5′-UTR, 5′ UTR; T7P, T7 promoter; RBS, ribosome binding site; T7T, T7 terminator; ω-TAp, putative ω-transaminase; ω-TA Rs, ω-transaminase from Rhodobacter sphaeroides; ω-TA At, ω-transaminase from Agrobacterium tumefaciens.
The templates for cell-free synthesis of ω-TA were prepared by colony PCR and subsequent second-round PCR using the MEGA primers flanking the T7 promoter, ribosome binding site, polyhistidine tag, and the T7 terminator. All of the PCRs were carried out using LA Taq DNA polymerase (Takara Bio Inc., Otsu, Japan). PCR products were directly used as the template for protein synthesis without purification. The standard cell-free reaction mixture consisted of the following components in a final volume of 50 μl: 57 mM HEPES-KOH (pH 7.5); 1.2 mM ATP; 0.85 mM (each) CTP, GTP, and UTP; 1.7 mM dithiothreitol; 0.17 mg/ml Escherichia coli total tRNA mixture (from strain MRE600); 90 mM potassium glutamate; 80 mM ammonium acetate; 12 mM magnesium acetate; 34 μg/ml l-5-formyl-5,6,7,8-tetrahydrofolic acid (folinic acid); 1.5 mM (each) 20 amino acids; 2% polyethylene glycol 8000 (PEG 8000); 67 mM creatine phosphate; 3.2 μg/ml creatine kinase; 10 μM l-[U-14C]leucine (11.3 GBq/mmol); 0.5 μg/ml PCR-amplified DNA; and 14 μl of the S12 extract (11). Cell-free synthesized proteins were quantified by measuring trichloroacetic acid (TCA)-insoluble radioactivity (10), and the size and relative solubility of the synthesized protein were determined by Western blot analysis on a 12% Tricine-SDS-polyacrylamide gel (17). Mouse anti-histidine-tagged IgG antibody (Merck KGaA, Darmstadt, Germany) and rabbit anti-mouse IgG conjugated to horseradish peroxidase (HRP) (Sigma, St. Louis, MO) were used as the primary and secondary antibodies, respectively. The PCR products served as translation substrates appropriate for producing as much protein as the corresponding plasmid-cloned gene when expressed in the reaction mixture (see Fig. S1 in the supplemental material).
We next proceeded to amplify 11 ω-TA genes, including 9 putative ones from the colonies of bacterial origins (see Table S2 in the supplemental material for the list of primers used in this study), and express them in the reaction mixture prepared in two microtiter plates. Each of the 11 target genes was added to the plates by column on the plate (columns 2 through 12). Column 1 was used for negative-control reactions without any template DNAs (Fig. 1). After the PCR-amplified template DNAs were added to the plates, the plates were sealed with a plastic film to prevent evaporation and incubated at 37°C. From the measurements of TCA-insoluble radioactivity, it was estimated that 301 (±13) to 501 (±9) μg/ml of the encoded enzymes were produced after 90 min of incubation (see Fig. S2A in the supplemental material). Although most of the cell-free synthesized ω-TAs was highly insoluble in the initial experiments conducted under standard reaction conditions, the relative amounts of the soluble enzymes were markedly improved by using the GroEL/ES-enriched S12 extract (8) (Fig. S2B).
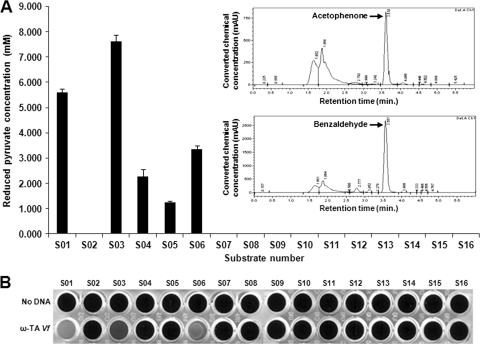
Instead of the conventional high-performance liquid chromatography (HPLC) methods which have limited throughput for handling many reaction samples from different enzyme-substrate combinations, in this study, we used a simple colorimetric method for combinatorial analysis of the cell-free synthesized ω-TAs with different amine-donating substrates. Using Vfω-TA as a model ω-TA, it was first examined whether the progress of amine transfer reaction can be assayed quantitatively by colorimetric measurement of the residual pyruvate concentration. On the basis of our previous finding that Vfω-TA takes amine donors containing aryl groups as effective substrates (20), cell-free synthesized Vfω-TA was incubated with α-methylbenzylamine or benzylamine in the presence of pyruvate, and the residual pyruvate concentration in the assay mixture was determined. In brief, the amine transfer reaction was initiated by adding 50 μl of assay mixture (50 mM Tris-HCl buffer [pH 7.2], 10 mM sodium pyruvate, 10 mM [each] amine donors, and 20 μM pyridoxal-5′-phosphate) to the completed cell-free synthesis reaction mixtures (50 μl) in a 96-well plate. After 3 h of incubation at 37°C, the assay mixture was diluted with an equal volume of distilled water. When 100 μl of the diluted solution was transferred to a fresh plate containing 50 μl of 0.5 mM 2,4-dinitrophenylhydrazine (DNP), a yellow precipitate of pyruvate-dinitrophenylhydrazone (PA-DNPH) derivative was formed instantly. The absorbance at 450 nm was measured in a microplate reader (Bio-Rad, Hercules, CA) and compared with a standard curve to determine the amount of residual pyruvate in each well. Although the sensitivity of the residual pyruvate assay was as low as 0.01 mM, considering the error range, it was determined that a sensitivity of 0.1 mM should be used instead. When the optical density at 450 nm (OD450) was measured after the addition of DNP and referred to a standard curve, the conversion yield based on the amount of residual pyruvate concentration showed good correlation with the results from standard HPLC assay (Table 1 and Fig. 2A, insets) where acetophenone or benzaldehyde generated from the corresponding substrates was separated in the Discovery HS F5 (5-μm particle size; 150- by 4.6-mm inner diameter [i.d.]; Supelco, Bellefonte, PA) column and measured at 254 nm. In addition, the relative amount of pyruvate was also able to be compared visually by adding 100 μl of 4 N NaOH solution, which turned the color of PA-DNPH to dark red (2, 15).
TABLE 1.
Activity comparison of Vibrio fluvialis ω-TA by HPLC and colorimetric method
| Amine donor | Amt (mM) of acetophenone or benzaldehyde converted from the amine donor | Reduced amt (mM) of pyruvatea |
|---|---|---|
| α-Methylbenzylamine | 5.88 ± 0.32b | 5.60 ± 0.46 |
| Benzylamine | 8.30 ± 0.55c | 7.60 ± 0.68 |
Reduced amount of pyruvate after the amine transfer reactions by the colorimetric assay.
Amount of acetophenone converted from α-methylbenzylamine by the HPLC assay.
Amount of benzaldehyde converted from benzylamine by the HPLC assay.
FIG. 2.
(A) Reactivity of 16 amine donors toward Vibrio fluvialis ω-TA. Reduced amounts of pyruvate concentration after the amine transfer reactions are plotted. Substrate number abbreviations: S01, α-methylbenzylamine; S02, α-ethylbenzylamine; S03, benzylamine; S04, 3-phenyl-1-propylamine; S05, phenylbutylamine; S06, 1-aminoindan; S07, ethylamine; S08, propylamine; S09, butylamine; S10, amylamine; S11, isopropylamine; S12, sec-butylamine; S13, β-alanine; S14, 3-amino-n-butyric acid; S15, phenylalanine; S16, 3-amino-3-phenylpropionic acid. The insets show HPLC traces of acetophenone and benzaldehyde after termination of transamination reaction by use of Vibrio fluvialis ω-TA against α-methylbenzylamine and benzylamine. mAU, milliabsorbance units. (B) Photo image of the assay plate after the addition of DNP and NaOH. ω-TA Vf, ω-TA from Vibrio fluvialis.
Next, the substrate specificity of Vfω-TA was examined against 16 amine donors. The colorimetric assay of the reactions using α-methylbenzylamine (S01) and 1-aminoindan (S06) showed that substantial amount of pyruvate is consumed during the assay reaction, indicating that these substrates work as effective amine donors. The relative amount of residual pyruvate after the amine transfer reaction using benzylamine (S03) through phenylbutylamine (S05) showed that the efficiency of amine transfer by Vfω-TA is critically affected by the distance between the amine and aryl groups. Pyruvate consumption during the amine transfer reaction was reduced approximately by half as an additional carbon atom was added between the phenyl and terminal amine groups. Unexpectedly, Vfω-TA showed very little activity toward α-ethylbenzylamine (S02) compared to α-methylbenzylamine (S01), and it was presumed that the length of aliphatic chain at the α-position is also an important factor affecting the substrate binding to the active site of enzyme. Aliphatic amine donors did not cause significant changes in pyruvate concentration (Fig. 2) in accordance with the previous results obtained by HPLC analysis (20).
The colorimetric method described above was applied to the assay of 11 cell-free synthesized ω-TAs against 16 different amine-donating substrates. As a result, a number of compounds were identified to be substrates of the examined enzymes, including the 9 putative enzymes (see Table S1 in the supplemental material). For example, benzylamine, 3-phenyl-1-propylamine, and 4-phenylbutylamine were identified to be the successful amine donating substrates of Agrobacterium tumefaciens ω-TA (Atω-TA). However, similar to the case of Vfω-TA, the presence of an aliphatic chain at the α-position appeared to interfere with the recognition of the substrates by this enzyme, since the assay reactions with the cell-free synthesized α-methylbenzylamine and α-ethylbenzylamine did not show any decrease in pyruvate concentration. Unlike other compounds examined, benzylamine and 1-aminoindan were found to be used as the substrates of various ω-TA enzymes. For example, 1-aminoindan showed reactivity with 7 enzymes, including 5 enzymes from putative genes, and benzylamine served as an amine donor for 5 enzymes, including 3 putative enzymes (Table 2).
TABLE 2.
Substrate specificity of ω-transaminases
| Enzymea | Reduced pyruvate concn (mM) from the following substrateb: |
|||||
|---|---|---|---|---|---|---|
| S03 | S04 | S05 | S06 | S07 | S16 | |
| ω-TAs | ||||||
| At | 1.1 | 0.9 | 2.4 | 0.2 | ||
| Rs | 0.8 | 0.2 | ||||
| ω-TAps | ||||||
| Ml0107 | 0.1 | 0.3 | 0.5 | |||
| Ml1207 | 0.9 | 0.4 | ||||
| Ml1632 | 0.4 | |||||
| Ml5987 | ||||||
| Ml5990 | 0.1 | 0.2 | ||||
| Ml6101 | ||||||
| Ml6963 | 0.1 | |||||
| Ml7037 | ||||||
| Ml7127 | ||||||
ω-Transaminases (ω-TAs) from two species and putative ω-transaminases (ω-TAps) are shown. Ml, Mesorhizobium loti.
10 mM pyruvate and 10 mM of an amine donor were used in the assay mixture, and the reduced amount of pyruvate was measured. Substrate abbreviations: S03, benzylamine; S04, 3-phenyl-1-propylamine; S05, phenylbutylamine; S06, 1-aminoindan; S07, ethylamine; S16, 3-amino-3-phenylpropionic acid.
In this work, we demonstrated the potential of an integrative cell-free protein synthesis strategy as a powerful tool for screening enzymes in a high-throughput manner without involving gene cloning and cell cultivation procedures. From the microbial colonies on agar plates, various ω-transaminases, including computer-predicted tentative enzymes, were selectively expressed and examined for their enzymatic activity toward different substrates. The entire procedures to select enzyme-substrate pairs from a large number of genetic sequences and possible substrates were completed in a matter of hours, successfully identifying a number of potent enzymes for the utilization of an array of amine-donating compounds. Since all of the required steps are amenable to automation, we expect further extension of the versatility of this strategy through the use of automated liquid-handling systems, and the presented strategy can be applied as a universal platform for rapid discovery and engineering of enzymes and other protein species.
Supplementary Material
Acknowledgments
This work was supported by National Research Foundation of Korea (NRF) grants funded by the South Korean government (MEST) (grants 20090082333 and 20090083328).
Footnotes
Published ahead of print on 23 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Christen, P., and D. E. Metzler. 1985. Transaminases. Wiley, New York, NY.
- 2.Crowley, L. V. 1967. The Reitman-Frankel colorimetric transaminase procedure in suspected myocardial infarction. Clin. Chem. 13:482-487. [PubMed] [Google Scholar]
- 3.Entzeroth, M. 2003. Emerging trends in high-throughput screening. Curr. Opin. Pharmacol. 3:522-529. [DOI] [PubMed] [Google Scholar]
- 4.Goddard, J. P., and J. L. Reymond. 2004. Recent advances in enzyme assays. Trends Biotechnol. 22:363-370. [DOI] [PubMed] [Google Scholar]
- 5.Gorbalenya, A. E., E. V. Koonin, and M. M. C. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses—identification of rubivirus and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubivirus, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hertzberg, R. P., and A. J. Pope. 2000. High-throughput screening: new technology for the 21st century. Curr. Opin. Chem. Biol. 4:445-451. [DOI] [PubMed] [Google Scholar]
- 7.Jewett, M. C., K. A. Calhoun, A. Voloshin, J. J. Wuu, and J. R. Swartz. 2008. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang, S. H., D. M. Kim, H. J. Kim, S. Y. Jun, and K. Y. Lee. 2005. Cell-free production of aggregation-prone proteins in soluble and active forms. Biotechnol. Prog. 21:1412-1419. [DOI] [PubMed] [Google Scholar]
- 9.Katzen, F., G. Chang, and W. Kudlicki. 2005. The past, present and future of cell-free protein synthesis. Trends Biotechnol. 23:150-156. [DOI] [PubMed] [Google Scholar]
- 10.Kim, D. M., T. Kigawa, C. Y. Choi, and S. Yokoyama. 1996. A highly efficient cell-free protein synthesis system from Escherichia coli. Eur. J. Biochem. 239:881-886. [DOI] [PubMed] [Google Scholar]
- 11.Kim, T. W., J. W. Keum, I. S. Oh, C. Y. Choi, C. G. Park, and D. M. Kim. 2006. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J. Biotechnol. 126:554-561. [DOI] [PubMed] [Google Scholar]
- 12.Mardis, E. R. 2008. The impact of next-generation sequencing technology on genetics. Trends Genet. 24:133-141. [DOI] [PubMed] [Google Scholar]
- 13.Murthy, T. V., W. Wu, Q. Q. Qiu, Z. Shi, J. LaBaer, and L. Brizuela. 2004. Bacterial cell-free system for high-throughput protein expression and a comparative analysis of Escherichia coli cell-free and whole cell expression systems. Protein Expr. Purif. 36:217-225. [DOI] [PubMed] [Google Scholar]
- 14.Pereira, D. A., and J. A. Williams. 2007. Origin and evolution of high throughput screening. Br. J. Pharmacol. 152:53-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reitman, S., and S. Frankel. 1957. A colorimetric method for the determination of serum glutamic oxalacetic and glutamic pyruvic transaminases. Am. J. Clin. Pathol. 28:56-63. [DOI] [PubMed] [Google Scholar]
- 16.Sawasaki, T., T. Ogasawara, R. Morishita, and Y. Endo. 2002. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. U. S. A. 99:14652-14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu, Y., Y. Kuruma, B. W. Ying, S. Umekage, and T. Ueda. 2006. Cell-free translation systems for protein engineering. FEBS J. 273:4133-4140. [DOI] [PubMed] [Google Scholar]
- 20.Shin, J. S., H. Yun, J. W. Jang, I. Park, and B. G. Kim. 2003. Purification, characterization, and molecular cloning of a novel amine:pyruvate transaminase from Vibrio fluvialis JS17. Appl. Microbiol. Biotechnol. 61:463-471. [DOI] [PubMed] [Google Scholar]
- 21.Spirin, A. S. 2004. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotechnol. 22:538-545. [DOI] [PubMed] [Google Scholar]
- 22.Turner, N. J. 2003. Directed evolution of enzymes for applied biocatalysis. Trends Biotechnol. 21:474-478. [DOI] [PubMed] [Google Scholar]
- 23.Wahler, D., and J. L. Reymond. 2001. High-throughput screening for biocatalysts. Curr. Opin. Biotechnol. 12:535-544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.