Abstract
Two almost identical gene clusters, tphRICIA2IA3IBIA1I and tphRIICIIA2IIA3IIBIIA1II, are responsible for the conversion of terephthalate (TPA) to protocatechuate in Comamonas sp. strain E6. In the present study, we investigated the transcriptional regulation of the tphRIICIIA2IIA3IIBIIA1II gene cluster. Reverse transcription-PCR analysis suggested that the tphRIICIIA2IIA3IIBIIA1II genes form two transcriptional units, the tphCIIA2IIA3IIBIIA1II catabolism operon and tphRII, with the latter encoding an IclR-type transcriptional regulator (ITTR). The transcription start site of the tphII catabolism operon was mapped at 21 nucleotides upstream of the initiation codon of tphCII. The lacZ transcriptional fusion experiments showed that tphRII encodes a transcriptional activator of the tphII catabolism operon and that TPA acts as an inducer. On the other hand, TphRII appeared to repress its own transcription regardless of the presence of TPA. The analysis of mutant derivatives of E6 indicated that tphRII is essential for the transcriptional activation of the tphII catabolism operon and the growth on TPA of a tphI-deficient derivative of E6. Purified His-tagged TphRII bound specifically to the tphRII-tphCII intergenic region containing a 21-bp inverted repeat sequence. Alignment of the inverted repeat sequences in the binding sites for TphRII and other members of ITTRs revealed highly conserved nucleotides. The substitution of conserved nucleotides resulted in significantly reduced TPA-dependent transcriptional activation from the tphCII promoter and reduced binding to His-tagged TphRII. These results clearly indicate that the conserved nucleotides are required for the inducible expression of the tphII catabolism operon regulated by TphRII.
The phthalate isomers terephthalate (TPA), isophthalate, and o-phthalate are widely used as plasticizers and are degraded by microbial catabolic pathways via protocatechuate (PCA) (8, 9, 26, 30). The degradation of PCA is catalyzed by one of the following three aromatic ring cleavage pathways: the PCA 2,3-cleavage (17), the PCA 3,4-cleavage (14), or the PCA 4,5-cleavage (19, 20) pathway. 2-Pyrone-4,6-dicarboxylate (PDC), which appears as an intermediate metabolite in the PCA 4,5-cleavage pathway (19, 20), has been considered to be a useful chemical building block for the synthesis of biodegradable and high-function polymers (15, 22, 23). Due to the potentiality of phthalate isomers to act as starting materials for the production of PDC, it has become increasingly more important to elucidate the microbial catabolism genes for phthalates and their regulation.
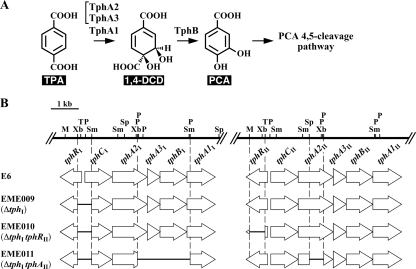
Previously, our research group isolated a TPA degrader, Comamonas sp. strain E6, which is able to grow on TPA, isophthalate, and o-phthalate as the sole source of carbon and energy; and these compounds are degraded via the PCA 4,5-cleavage pathway (9, 30). We reported on the characterization of two almost identical TPA catabolism gene clusters (>94% identity) of E6, tphRICIA2IA3IBIA1I (GenBank accession no. AB238678) and tphRIICIIA2IIA3IIBIIA1II (GenBank accession no. AB238679) (30). The catabolism of TPA is initiated by the transformation of TPA into 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate (1,4-DCD) by TPA 1,2-dioxygenase (oxygenase component, TphA2 and TphA3; reductase component, TphA1), and 1,4-DCD is converted to PCA by 1,4-DCD dehydrogenase (TphB) (Fig. 1 A). The resulting PCA is further degraded via the PCA 4,5-cleavage pathway. On the basis of the amino acid sequence similarity and the findings of expression studies, tphC and tphR were suggested to code for a receptor and a regulator involved in TPA catabolism: a periplasmic TPA binding receptor and an IclR-type transcriptional regulator (ITTR), respectively.
FIG. 1.
TPA catabolic pathway of Comamonas sp. strain E6 (A) and schematic diagrams of the gene disruptions in E6 (B). (A) TphA2, oxygenase large subunit of TPA 1,2-dioxygenase; TphA3, oxygenase small subunit of TPA 1,2-dioxygenase; TphA1, reductase component of TPA 1,2-dioxygenase; TphB, 1,4-DCD dehydrogenase. (B) The horizontal bars in the thick arrows indicate the regions deleted in EME009, EME010, and EME011. Abbreviations for restriction enzymes: M, MunI; P, PstI; Sm, SmaI; Sp, SphI; T, Tth111I; Xb, XbaI.
The TPA catabolic pathway has also been reported in Comamonas testosteroni T-2 (32), C. testosteroni YZW-D (37), Delftia tsuruhatensis T7 (33), Rhodococcus sp. strain DK17 (4), and Rhodococcus jostii RHA1 (13). The TPA catabolism gene clusters were identified in YZW-D, DK17, and RHA1; and all of these gene clusters include a gene encoding a putative ITTR, which appeared to be involved in TPA catabolism. Several ITTRs involved in the degradation of aromatic compounds have been described. This family includes, among others, PcaU and PcaR, activators of β-ketoadipate pathway genes in Acinetobacter baylyi ADP1 (5, 10) and Pseudomonas putida PRS2000 (29), respectively; PobR, an activator of the p-hydroxybenzoate 3-hydroxylase gene, pobA, from A. baylyi ADP1 (6); as well as CatR, a repressor of the catechol ortho-cleavage pathway genes in Rhodococcus erythropolis CCM2595 (36). However, the transcriptional regulation of TPA catabolism genes has not yet been reported.
In this report, we describe the transcriptional control of one of the tph catabolism gene clusters, tphCIIA2IIA3IIBIIA1II, regulated by the tphRII gene product. This is the first report on the characterization of the regulation of TPA catabolism genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Comamonas sp. strain E6 was grown in Luria-Bertani (LB) medium or in W minimal salt medium (27) containing 10 mM TPA or 10 mM succinate at 30°C. Escherichia coli strains were grown in LB medium at 37°C. For cultures of cells carrying antibiotic resistance markers, the media for E. coli transformants were supplemented with 100 mg of ampicillin (Ap)/liter or 25 mg of kanamycin (Km)/liter, and the media for E6 transformants were supplemented with 50 mg of Km/liter or 30 mg of chloramphenicol/liter.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Comamonas sp. | ||
| E6 | Wild type, TPA+ | 30 |
| EME009 | E6 derivative, ΔtphRItphCI | This study |
| EME010 | EME009 derivative, ΔtphRII | This study |
| EME011 | EME009 derivative, ΔtphA2IA3IBIA1ItphA2II | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 12 |
| XL2-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr) Amy Cmr] | Stratagene |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3), T7 RNA polymerase gene under control of the lacUV promoter | 35 |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector, Apr | 34 |
| pT7Blue | Cloning vector, Apr T7 promoter | Novagen |
| pK19mobsacB | oriT sacB Kmr | 31 |
| pKT230MC | IncQ broad-host-range cloning vector, Kmr | 25 |
| pJB866 | RK2 broad-host-range expression vector, Tcr PmxylS | 2 |
| pET16b | Expression vector, N-terminal His10 tag, Apr T7 promoter | Novagen |
| pPR9TZ | Broad-host-range promoter probe vector, RK2 ori Apr Cmr | 17 |
| pSS92R | pBluescript II SK(+) with a 4.0-kb SalI fragment carrying tphRI and tphCI | This study |
| pK19RCI | pK19mobsacB with a 2.5-kb EcoRV-HindIII fragment of pSS92R | This study |
| pDTRCI | pK19RCI with a deletion of a 0.5-kb XbaI-SmaI fragment carrying the tphRI-tphCI intergenic region | This study |
| pEJ89 | pJB866 with a 8.9-kb EcoRI fragment carrying tphRIICIIA2IIA3IIBIIA1II | 30 |
| pEJ84 | pEJ89 with a deletion of a 0.6-kb MunI-Tth111I fragment carrying part of tphRII | 30 |
| pDTRII | pK19mobsacB with a 2.3-kb EcoRI-HindIII fragment of pEJ84 | This study |
| pK19A2II | pK19mobsacB with a 2.0-kb HindIII-XhoI fragment carrying tphA2II | This study |
| pDTA2II | pK19A2II with a deletion of a 0.5-kb SphI-PstI fragment carrying part of tphA2II | This study |
| pK19ABIF | pK19mobsacB with a 4.3-kb HindIII fragment carrying tphA2IA3IBIA1I | This study |
| pDTABI | pK19ABIF with a deletion of a 1.9-kb PstI fragment carrying tphA2IA3IBIA1I | This study |
| pKTRII | pKT230MC with a 1.8-kb KpnI fragment carrying tphRII | This study |
| pKTtphII | pKT230MC with a 8.9-kb EcoRI fragment carrying tphRIICIIA2IIA3IIBIIA1II | This study |
| pTRII | pT7Blue with a 779-bp PCR fragment generated by the TphRII-F and TphRII-R primer pair | This study |
| pETtprII | pET16b with a 772-bp NdeI-XhoI fragment of pTRII | This study |
| pTTC | pT7Blue with a 393-bp PCR fragment carrying the tphRII-tphCII intergenic region | This study |
| pZTC | pPR9TZ with a 381-bp HindIII-KpnI fragment of pTTC | This study |
| pTTR | pT7Blue with a 500-bp PCR fragment carrying the tphRII-tphCII intergenic region | This study |
| pZTR | pPR9TZ with a 379-bp HindIII-KpnI fragment of pTTR | This study |
| pTTCT5C | Site-directed mutant of pTTC generated by the SMT5C-F and SMT5C-R primer pair | This study |
| pTTCG6A | Site-directed mutant of pTTC generated by the RM315-F and RM315-R primer pair | This study |
| pTTCC7T | Site-directed mutant of pTTC generated by the SMC7T-F and SMC7T-R primer pair | This study |
| pTTCG8A | Site-directed mutant of pTTC generated by the SMG8A-F and SMG8A-R primer pair | This study |
| pT5C | pPR9TZ with a 381-bp HindIII-KpnI fragment of pTTCT5C | This study |
| pG6A | pPR9TZ with a 381-bp HindIII-KpnI fragment of pTTCG6A | This study |
| pC7T | pPR9TZ with a 381-bp HindIII-KpnI fragment of pTTCC7T | This study |
| pG8A | pPR9TZ with a 381-bp HindIII-KpnI fragment of pTTCG8A | This study |
Abbreviations: TPA+, able to grow on TPA as the sole carbon source; Apr, Kmr, Tcr, and Cmr, resistance to ampicillin, kanamycin, tetracycline, and chloramphenicol, respectively.
DNA manipulations and nucleotide sequencing.
DNA manipulations, including total DNA isolation, construction of deletion derivatives, and nucleotide sequencing, were performed as described previously (18). Analysis of nucleotide sequences was carried out as described previously (1).
Generation of mutants.
The 2.5-kb EcoRV-HindIII fragment carrying tphRI and tphCI of pSS92R was inserted into pK19mobsacB to generate pK19RCI. The 0.5-kb XbaI-SmaI fragment including the intergenic region between tphRI and tphCI of pK19RCI was deleted. The resulting plasmid, pDTRCI, was introduced into E6 cells by electroporation, and candidates for the deletion mutant of the tphRI-tphCI intergenic region (strain EME009) were isolated (Fig. 1B) as described previously (21). In order to disrupt the tphRII gene in EME009, the 0.6-kb MunI-Tth111I fragment of the tphRII internal region in pEJ89 was deleted, and then the 2.3-kb EcoRI-HindIII fragment carrying part of tphRII was cloned into pK19mobsacB. The resulting plasmid, pDTRII, was further introduced into EME009 cells, and candidates for the mutant (strain EME010) were isolated (Fig. 1B). To obtain a mutant which lacked the ability to transform TPA, the tphA2II disruption plasmid, pDTA2II, was introduced into EME009 cells. Furthermore, the tphA2IA3IBIA1I disruption plasmid, pDTABI, was introduced into the resulting mutant cells to obtain EME011 (Fig. 1B).
Disruption of the genes was examined by Southern hybridization analysis. To confirm the disruptions of the tphRI-tphCI intergenic region, tphRII, and tphA2II, total DNA of candidates for mutants were digested with HincII-HindIII. To confirm the tphA2IA3IBIA1I deletion, total DNA of candidates for the mutant was digested with HindIII. The 0.8-kb XbaI-PvuII fragment carrying the tphRI-tphCI intergenic region, the 1.3-kb PstI fragment carrying tphRII, and the 0.9-kb SmaI-NcoI fragment carrying tphA2II were labeled with a digoxigenin (DIG) system (Roche, Mannheim, Germany) and used as probes.
Reverse transcription (RT)-PCR.
EME009 cells were grown in W minimal salt medium supplemented with 10 mM TPA or 10 mM succinate at 30°C. When the absorbance at 600 nm (A600) of the culture was 1.0, the cells were harvested by centrifugation at 5,000 × g at 4°C for 10 min. Total RNA was isolated with the Isogen reagent (Nippon Gene, Toyama, Japan), according to the manufacturer's instructions. Purified RNA was then treated with RNase-free DNase I (Takara Bio Inc., Otsu, Japan) to remove contaminating DNA. Single-stranded cDNA was synthesized from 5.0 μg of total RNA utilizing PrimeScript reverse transcriptase (Takara Bio Inc.) with random primers in a 20-μl reaction mixture. PCR amplification was performed using 2.0 μl of the cDNA mixture, specific primers (Table 2), and Ex Taq DNA polymerase (Takara Bio Inc.) under the following conditions: 95°C for 30 s and 35 cycles of 95°C for 60 s, 62°C for 60 s, and 72°C for 60 s. A control without reverse transcriptase was used for each reaction to verify the absence of genomic DNA contamination. Samples from the PCR were electrophoresed on a 1.8% agarose gel and visualized with ethidium bromide.
TABLE 2.
Primer sequences used in this study
| Primer use and primer | Sequence (5′-3′)a |
|---|---|
| RT-PCR analysis | |
| insxRF | CCTAACAGCAATGCTCATGG |
| insRR | ACTACTGGATGCACTGTCGG |
| intRCF | ACAGTGCATCCAGTAGTGGC |
| intRCR | AGGCTAGCACAGTCATTCCG |
| insCF | ACGGAATGACTGTGCTAGCC |
| insCR | GGATGAACTGAGCAGTCTTGG |
| intCA2F | CGAGAAGTGGAAGAAGGTGC |
| intCA2R | TTGTGAGAGGCGATTCAGC |
| intA2A3F | GCTGCCTAACAACTGGAAGC |
| intA2A3R | GCAATAGTGGCAATCCTTGG |
| intA3BF | TGTCGACAACCATGATGAGG |
| intA3BR | ACGATGTCTTGAAGAAGCGG |
| intBA1F | ACGACCTGTATGTAGCCATGC |
| intBA1R | ATAGTGCTGAAGCGACCACC |
| insA1F | GATCCATATCCACGACTCCG |
| insA1R | ATGGCGCTAGGAAGATACAGG |
| Primer extension | |
| PEtphC110 | GGCACGACGATCTTGAGAGG |
| PEtphR160 | GCTGTACCAGTGTGCTGAGC |
| Construction of promoter-lacZ fusion plasmids | |
| HRC-F | AAGCTTGAGCGAACGTCTGG (HindIII) |
| HRC-R | GGAACACTAGATCGGCACC |
| KRCH-F | GGTACCGATAGCCAAGCTGTACC (KpnI) |
| KRCH-R | AAGCTTCGACGATCTTGAGAGG (HindIII) |
| Construction of tphRII expression plasmids | |
| TphRII-F | CATATGCAGGACAAGAACTTTGTGG (NdeI) |
| TphRII-R | CTCGAGCACTACAACCCCTGCG (XhoI) |
| EMSAs | |
| RMF01 | GTTCTTGTCCTGCATAGCG |
| RMF11 | GACATCCTCATACTGCAGTTCC |
| RMR01 | GTCTGCGAATAGATTCGTTGC |
| RMR11 | AACTGCAGTATGAGGATGTCG |
| RM31-F | CAACATTTTTGCGCATAGCGCAAAAACAGGT |
| RM31-R | ACCTGTTTTTGCGCTATGCGCAAAAATGTTG |
| RM315-F | CAACATTTTTaCGCATAGCGCAAAAACAGGT |
| RM315-R | ACCTGTTTTTGCGCTATGCGtAAAAATGTTG |
| Site-directed mutagenesis | |
| SMT5C-F | GGTGTTTTCAACATTTTcGCGCATAGCGCAAAAACAGG |
| SMT5C-R | CCTGTTTTTGCGCTATGCGCgAAAATGTTGAAAACACC |
| SMC7T-F | GGTGTTTTCAACATTTTTGtGCATAGCGCAAAAACAGG |
| SMC7T-R | CCTGTTTTTGCGCTATGCaCAAAAATGTTGAAAACACC |
| SMG8A-F | GGTGTTTTCAACATTTTTGCaCATAGCGCAAAAACAGG |
| SMG8A-R | CCTGTTTTTGCGCTATGtGCAAAAATGTTGAAAACACC |
Engineered restriction sites are underlined, and the corresponding restriction enzymes are shown in parentheses. Mutated nucleotides are in lowercase.
Primer extension.
E6 cells harboring pKTtphII were grown in W minimal salt medium containing 10 mM succinate either with or without 10 mM TPA at 30°C. After 4 h of incubation, total RNA was isolated as described above. The primers used to detect the start sites of tphCII and tphRII mRNAs were PEtphC110 and PEtphR160 (Table 2), respectively, which are complementary to the nucleotide sequences between positions +90 and +110 and between positions +138 and +157 relative to the start codons of tphCII and tphRII, respectively. The primers were labeled at their 5′ ends with dye D4 (Beckman Coulter Inc., Fullerton, CA). A labeled primer (2 pmol) was hybridized to 5.0 μg of the total RNA preparation. A primer extension reaction was performed by incubating the annealing mixture with 200 U of PrimeScript reverse transcriptase at 42°C for 45 min. After the extension, the enzyme was inactivated by being exposed to a temperature of 95°C for a period of 10 min and ethanol precipitated. The products were then analyzed by a CEQ2000XL genetic analysis system (Beckman Coulter Inc.).
LacZ transcriptional fusions and β-galactosidase activity assays.
To determine the promoter activities of tphCII and tphRII, pZTC and pZTR, which expressed the tphCII promoter-lacZ fusion and the tphRII promoter-lacZ fusion, respectively, were introduced into cells of E6 and its mutant derivatives by electroporation. The resulting transformants were grown in W minimal salt medium containing 10 mM succinate either with or without 10 mM TPA, phthalate, isophthalate, or PCA at 30°C. After 4 h of incubation, cells were harvested and resuspended in 20 mM Tris-HCl buffer (pH 8.0), broken by an ultrasonic disintegrator (UD-201; Tomy Seiko Co., Tokyo, Japan), and centrifuged at 15,000 × g for 15 min. The resulting supernatants were used as crude enzymes. The protein concentration was determined by the Bradford method (3).
β-Galactosidase activity assays with 4-methylumbelliferyl-β-d-galactopyranoside (4MUG) as the substrate were performed as follows. A 300-μl assay mixture contained 20 mM Tris-HCl (pH 8.0), crude extract (50 μg or 1 mg of protein), and 300 μM 4MUG. After incubation of said mixture for 15 min, 200 μl of the reaction mixture was transferred to 1.8 ml of 100 mM glycine-NaOH buffer (pH 10.0), and then the production of 4-methylumbelliferone (4MU) was detected with an RF-1500 spectrofluorometer (Shimadzu, Kyoto, Japan). One unit of enzyme activity was defined as the amount of enzyme that resulted in the production of 1 μmol of 4MU per min. Specific activity was expressed in units per milligram of protein.
Expression of His tag-fused tphRII in E. coli.
The coding region of tphRII was amplified by PCR using Ex Taq DNA polymerase together with pKTtphII as the template as well as the TphRII-F and TphRII-R primer pair (Table 2). The 779-bp PCR product was cloned in pT7Blue and sequenced. The 772-bp NdeI-XhoI fragment of the resulting plasmid was inserted into pET16b to generate pETtprII, which fused the codons for 10 histidines (His tag) at the 5′ end of tphRII. E. coli BL21(DE3) cells harboring pETtprII were grown in 1 liter of LB medium containing 100 mg of Ap/liter at 30°C. When the A600 of the culture reached 0.5, the expression of His tag-fused tphRII was induced for 2 h by adding 1 mM isopropyl-β-d-thiogalactopyranoside. After the incubation, cells were harvested and resuspended in 50 mM Tris-HCl buffer (pH 7.5), broken by three passages through a French pressure cell (Aminco, Urbana, IL), and centrifuged at 26,000 × g for 20 min. The resulting supernatants were used as crude enzymes.
Purification of His-TphRII.
To remove nucleic acids, streptomycin sulfate was added to the crude enzyme to a final concentration of 1%. The lysate was kept on ice for 10 min and centrifuged at 15,000 × g for 15 min. The supernatant was applied to an Ni Sepharose 6 Fast Flow column (GE Healthcare, Buckinghamshire, United Kingdom) previously equilibrated with buffer A, consisting of 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 40 mM imidazole. Proteins were allowed to bind for 1 min at 4°C while they were rotated, followed by washing five times in 5 ml of buffer B, consisting of 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 100 mM imidazole. His-tagged TphRII (His-TphRII) was eluted with 5 ml of buffer C, consisting of 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 500 mM imidazole; and 500-μl fractions were pooled and concentrated by centrifugal filtration with a Microcon YM-30 apparatus (Millipore, Billerica, MA). The purity of the enzyme preparation was examined by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE).
Electrophoretic mobility shift assays (EMSAs).
DNA fragments containing the tphRII-tphCII intergenic region were synthesized by PCR with the specific primer pairs (Table 2). To prepare the RM31 and G6A probes, the RM31-F and RM31-R and the RM315-F and RM315-R oligonucleotide pairs, respectively, were annealed to each other. To prepare probes T5C, C7T, and G8A, the DNA fragments were amplified from pTTCT5C, pTTCC7T, and pTTCG8A, respectively, using the RMF01 and RMR01 primer pair. The 3′ ends of the fragments of probes were labeled with DIG-11-ddUTP utilizing the 2nd-generation DIG gel shift kit (Roche), according to the manufacturer's instructions. Binding reactions were performed in 10-μl reaction mixtures containing 20 ng of purified His-TphRII (65 nM monomer), 0.1 or 1 nM DIG-labeled probes, 1 μg of poly[d(I-C)], 0.1 μg of poly-l-lysine, and 1× reaction buffer (20 mM Tris-HCl [pH 8.0], 100 mM KCl, 10% glycerol, 5 mM EDTA). The mixtures were then incubated at 20°C for 20 min, loaded onto 10% polyacrylamide gels in 0.5× Tris-borate-EDTA buffer, and then separated by gel electrophoresis at 80 V for 120 to 150 min. Subsequently, the DIG-labeled DNA was transferred to a nylon membrane (Hybond-N; GE Healthcare) at 200 mA for 1 h with a Trans Blot SD apparatus (Bio-Rad, Hercules, CA) and then visualized by an enzyme immunoassay using a DIG gel shift kit, following the manufacturer's instructions. For detection of the labeled DNA, X-ray film was used. In competitive assays, a 1,000-fold molar excess of unlabeled probe was added to the preformed protein-DNA complexes.
Site-directed mutagenesis of the tphRII-tphCII intergenic region.
Site-directed substitutions in the inverted repeat sequence occurring in the tphRII-tphCII intergenic region were introduced using a QuikChange site-directed mutagenesis kit (Stratagene, San Diego, CA) and plasmid pTTC as the template, according to the manufacturer's instructions. The oligonucleotides used for site-directed mutagenesis are listed in Table 2. All mutational changes were verified by DNA sequencing.
RESULTS AND DISCUSSION
RT-PCR analysis of tphII genes.
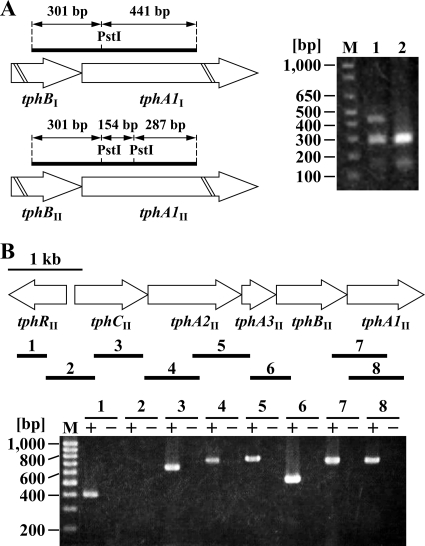
Because tphI and tphII gene clusters are organized as tphR and tphCA2A3BA1, which are divergently transcribed from one another, the promoter regions for tphR and tphC were expected to be present in the tphR-tphC intergenic regions. Due to the fact that the nucleotide sequences of the tphRI-tphCI and tphRII-tphCII intergenic regions (109 bp) are identical and the amino acid sequence identity between TphRI and TphRII is 99%, the transcriptional regulation of both operons was thought to be the same. For all of these reasons, we focused on the regulation of the tphII gene cluster. In order to characterize the transcription of the tphII genes, we constructed a mutant of Comamonas sp. strain E6, EME009, in which the tphRI-tphCI intergenic region was deleted (Fig. 1B). To verify the effect of the deletion of the tphRI-tphCI intergenic region on E6 cells, RT-PCR was performed using total RNAs extracted from the cells of E6 and EME009 and the intBA1F and intBA1R primer pair, which was designed to amplify a 742-bp region of the tphI and tphII gene clusters (Fig. 2 A). The resulting RT-PCR products were digested with PstI and then analyzed by agarose gel electrophoresis. In the case of the wild-type strain, fragments of 301 bp and 441 bp derived from tphI and fragments of 301 bp, 287 bp, and 154 bp derived from tphII were observed. However, the 441-bp fragment was missing in the digested fragments obtained from EME009 cells, indicating that the tphI genes were no longer transcribed in EME009 cells (Fig. 2A). The growth rate of EME009 cells on TPA was almost identical to that of the wild-type strain (data not shown). This result indicates that the tphII gene cluster is sufficient to support the normal growth of E6 on TPA.
FIG. 2.
RT-PCR analysis of the tph gene clusters. (A) Agarose gel electrophoresis of the RT-PCR products digested with PstI. RT-PCR was performed using total RNAs extracted from the cells of E6 and EME009 grown on TPA and the intBA1F and intBA1R primer pair. Lane M, molecular size markers; lanes 1 and 2, PstI digests of RT-PCR products obtained from E6 and EME009 cells, respectively. (B) Total RNA used for cDNA synthesis was isolated from EME009 cells grown on TPA. The results of agarose gel electrophoresis of the products of RT-PCR assays with primers targeting tphRII (lane 1; expected size, 403 bp), tphRII-tphCII (lane 2; expected size, 666 bp), tphCII (lane 3; expected size, 657 bp), tphCII-tphA2II (lane 4; expected size, 745 bp), tphA2II-tphA3II (lane 5; expected size, 771 bp), tphA3II-tphBII (lane 6; expected size, 533 bp), tphBII-tphA1II (lane 7; expected size, 742 bp), and tphA1II (lane 8; expected size, 738 bp) are shown. Lane M, molecular size markers; lanes + and −, RT-PCR with and without reverse transcriptase, respectively.
RT-PCR experiments were performed with total RNA extracted from EME009 cells grown on TPA and primers complementary to neighboring tphII genes, as shown in Fig. 2B. RT-PCR products of the expected sizes were detected for the internal regions of tphRII, tphCII, and tphA1II and the intergenic regions of tphCII-tphA2II, tphA2II-tphA3II, tphA3II-tphBII, and tphBII-tphA1II (Fig. 2B). However, no product was observed when a primer pair that spans tphRII-tphCII was used. These results suggest that all the tphII catabolism genes except tphRII are organized in the same transcriptional unit, with tphRII being transcribed divergently from tphCII.
Determination of transcription start site of tphII catabolism operon.
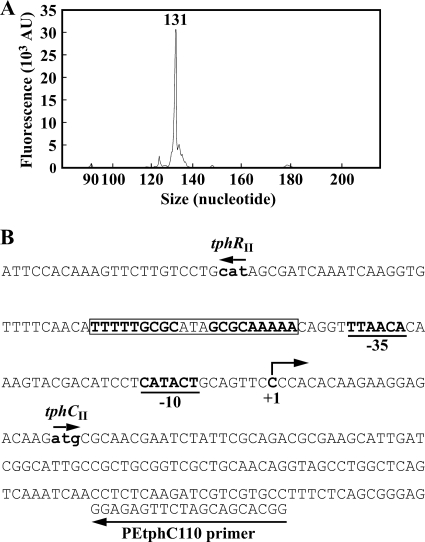
The transcription start site of the tphII catabolism operon was determined by primer extension analysis using a primer, PEtphC110, that anneals to tphCII sequences. Due to the fact that no significant extension product was observed when total RNA from E6 cells was used as the template, total RNAs were prepared from E6 cells harboring plasmid pKTtphII, which carries tphRIICIIA2IIA3IIBIIA1II. In this analysis, a single major product was observed using RNA isolated from the cells grown in W minimal salt medium containing succinate and TPA (Fig. 3 A), while no product was obtained with RNA isolated from the cells grown on succinate (data not shown). The transcription start site of the tphII catabolism operon was mapped at the C residue located at 21 nucleotides upstream from the ATG start codon of tphCII (Fig. 3B). Putative −35 (TTAACA) and −10 (CATACT) sequences were found upstream of the transcription start site. We also carried out a primer extension analysis to determine the transcription start site of tphRII; however, no extension product was obtained under our experimental conditions.
FIG. 3.
Determination of the transcription start site of the tphCII operon. (A) Fluorescent primer extension was performed with D4-labeled primer PEtphC110 and 5 μg of total RNA isolated from E6 cells harboring pKTtphII grown in W minimal salt medium containing 10 mM succinate with 10 mM TPA. The transcription start site was determined by comparing the retention time of the D4-labeled primer extension product with the retention times of DNA size standards. AU, arbitrary fluorescence units. (B) Nucleotide sequence of the tphRII-tphCII intergenic region. The initiation codons for tphRII and tphCII are indicated by boldface lowercase letters. The transcription start site (position +1), putative −10 and −35 regions of the tphCII promoter, and sequence of PEtphC110 are indicated. The inverted repeat sequence is boxed.
Promoter activities of tphII catabolism operon and tphRII.
The tphRII-tphCII intergenic region amplified by PCR was cloned into the promoter-probe vector pPR9TZ to generate a transcriptional fusion to the promoterless lacZ reporter gene. The resulting tphCII promoter-lacZ fusion plasmid, pZTC, included a DNA fragment which spans from positions −254 to +127 in relation to the transcription start site of tphCII. The levels of expression of the tphCII promoter were examined in E6 and EME009 cells harboring pZTC. β-Galactosidase activity of E6 cells harboring pZTC was detected only in the cells grown in the presence of TPA (Table 3); therefore, the cis-acting region necessary for the transcriptional activation of the tphII catabolism operon appeared to be in the tphRII-tphCII intergenic region. tphCII promoter activity in response to TPA was also detected in EME009 cells harboring pZTC (Table 3), suggesting that the presence of tphRII is sufficient to control the inducible expression of the tphII catabolism operon. In the cells of EME011 (Fig. 1B) harboring pZTC, which lacks the ability to transform TPA, tphCII promoter activity was observed by supplementation with TPA (Table 3). This result demonstrates that TPA itself acts as an inducer for the tphII catabolism operon. In addition, the tphCII promoter activities observed in the presence of isophthalate, o-phthalate, or PCA were negligible; and these were also found in the absence of either compound (data not shown). The tphRII promoter activity was constitutively expressed at a low level in E6 and EME009 cells, both harboring the tphRII promoter-lacZ fusion plasmid pZTR (Table 3).
TABLE 3.
Promoter activities of tphCII and tphRII
| Promoter | Strain | β-Galactosidase activity (mU/mg) |
|
|---|---|---|---|
| With TPA | Without TPA | ||
| tphCII (pZTC) | E6 | 2.03 ± 0.14 | NDa |
| EME009 | 2.02 ± 0.12 | ND | |
| EME011 | 2.53 ± 0.18 | ND | |
| EME010 | ND | ND | |
| tphRII (pZTR) | E6 | 0.04 ± 0.01 | 0.04 ± 0.01 |
| EME009 | 0.05 ± 0.01 | 0.06 ± 0.01 | |
| EME010 | 0.11 ± 0.02 | 0.09 ± 0.003 | |
ND, not detected.
Disruption of tphRII.
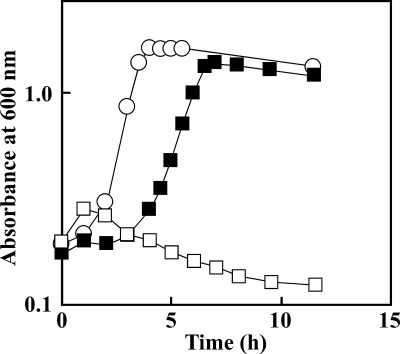
The tphRII gene in EME009 was inactivated by the gene replacement technique with the tphRII disruption plasmid pDTRII so as to evaluate the role of tphRII in TPA catabolism. The ability of the resulting mutant, EME010 (Fig. 1B), to grow on TPA was completely lost (Fig. 4). To determine if this growth deficiency was caused by the disruption of tphRII, pKTRII, which contains the 1.8-kb KpnI fragment carrying tphRII in pKT230MC, was introduced into EME010 cells. Complementation of EME010 with tphRII restored the growth of the mutant on TPA (Fig. 4). These results indicate that tphRII is essential for the TPA catabolism by EME009 cells.
FIG. 4.
Growth of E6 and EME010 cells on TPA. E6 (open circles), EME010 (open squares), and EME010 cells harboring pKTRII carrying tphRII (closed squares) were grown in W minimal salt medium containing 10 mM TPA. The results are the means of the representatives of three independent experiments. Error bars are hidden behind the symbols.
In order to confirm whether the tphRII gene product plays a role in the transcriptional regulation of the tphII catabolism operon, the tphCII promoter activity in EME010 cells harboring pZTC was measured. When the cells were grown in the presence or absence of TPA, no activity was observed (Table 3), suggesting that tphRII encodes a transcriptional activator of the tphII catabolism operon. On the other hand, the tphRII promoter activity of EME010 cells harboring pZTR was about 2.5-fold greater than that of cells of E6 and EME009 harboring the same plasmid (Table 3). TphRII appeared to negatively autoregulate the transcription from its own promoter, regardless of the presence of TPA, similar to that which was previously reported for ITTRs (24).
Binding of His-TphRII to the tphRII-tphCII intergenic region.
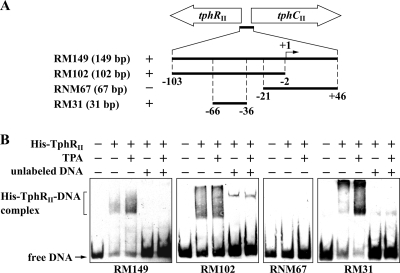
The His tag-fused tphRII gene was expressed in E. coli BL21(DE3) cells harboring pETtprII. Production of a 30-kDa protein in E. coli cells was observed by SDS-PAGE (data not shown). The size of this product was close to the predicted molecular mass of His-TphRII (Mr, 30,696). EMSAs were carried out using purified His-TphRII and the DIG-labeled RM149, RM102, RNM67, and RM31 probes covering nucleotide positions −103 to +46, −103 to −2, −21 to +46, and −66 to −36, respectively (position +1 is the tphCII transcription start site) (Fig. 5 A). The binding experiments showed that the mobilities of the RM149, RM102, and RM31 fragments were retarded upon the addition of TphRII, whereas no retardation was observed when the RNM67 probe was used (Fig. 5B). In these analyses, multiple complexes were observed. Because His-TphRII tended to aggregate during desalting, formation of multiple complexes might be related to the nature of His-TphRII. The retardations were almost completely abolished when a 1,000-fold excess of the same unlabeled probe was added to the reaction mixtures. This confirmed that His-TphRII directly binds to the region from positions −66 to −36, which contains an inverted repeat sequence, 5′-TTTTTGCGCATAGCGCAAAAA-3′, centered at position −51 from the tphCII transcription start site. In the case of the binding sites for several ITTRs, including PcaU, PcaR, and PobR, an external direct sequence repetition of the half site of the inverted repeat sequences was observed (7, 11, 16, 28); and this direct sequence repetition was required for the binding of PcaU (28). No such direct repeat sequence was found in the tphRII-tphCII intergenic region. In the case of other ITTRs, effector molecules had no effect on the affinity of the protein-DNA interactions (24). TPA also had no obvious effect on the binding of His-TphRII to the tphRII-tphCII intergenic region.
FIG. 5.
Binding of His-TphRII to the tphRII-tphCII intergenic region. (A) Schematic diagrams of the DNA fragments used in EMSAs. The sizes of the DNA probes are indicated in parentheses. The binding (+) and nonbinding (−) of the respective fragments are shown. The transcription start site (position +1) of tphCII is indicated. (B) EMSAs of the binding of purified His-TphRII to the tphRII-tphCII intergenic region. The positions of the unbound probe (free DNA) and His-TphRII-DNA complex are shown. His-TphRII (65 nM monomer), 100 μM TPA, and a 1,000-fold molar excess of the same unlabeled DNA fragment were added as indicated.
Site-directed mutagenesis of the inverted repeat sequence.
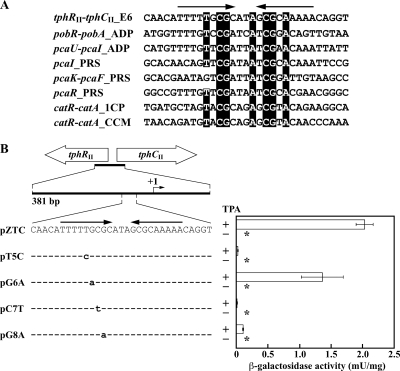
The existence of an inverted repeat sequence in the binding sites for PobR, PcaU, PcaR, and CatR has been reported (7, 10, 11, 36). Alignment of the inverted repeat sequences in the binding sites for TphRII and these ITTRs revealed that T, C, G, A, C, G, and A at positions −57, −55, −54, −50, −48, −47, and −45 from the tphCII transcriptional start site, respectively, are highly conserved (Fig. 6 A). To investigate the involvement of these conserved nucleotides in the transcriptional activation from the tphCII promoter, conserved nucleotides T, C, and G, at positions −57, −55, and −54, respectively, were substituted by site-directed mutagenesis (Fig. 6B). The tphCII promoter activities of E6 cells harboring mutated tphCII promoter-lacZ fusion plasmid pT5C (T to C at position −57), pCT7 (C to T at position −55), or pG8A (G to A at position −54) and grown in the presence of TPA were only 0.9, 0.4, and 4.7%, respectively, of the activity obtained by E6 cells harboring pZTC grown under the same conditions. On the other hand, a single change from G at position −56, which is not conserved, to A (pG6A) resulted in only a slightly reduced β-galactosidase level (Fig. 6B). These results indicate that the T at residue −57, the C at residue −55, and the G at residue −54, which are conserved in the inverted repeat sequences, are important for TPA-dependent transcriptional activation from the tphCII promoter.
FIG. 6.
β-Galactosidase activities from the wild-type and mutated tphCII promoters. (A) Alignment of the inverted repeat sequence of the tphRII-tphCII intergenic region with the sequences involved in the binding of other ITTRs. Identical nucleotides are indicated by dark shading. The arrows indicate the inverted repeat sequence. Abbreviations (GenBank accession numbers): tphRII-tphCII_E6, the tphRII-tphCII intergenic region of E6 (AB238679); pobR-pobA_ADP, the pobR-pobA intergenic region of Acinetobacter baylyi ADP1 (L05770); pcaU-pcaI_ADP, the pcaU-pcaI intergenic region of ADP1 (L05770); pcaI_PRS, the pcaI promoter region of Pseudomonas putida PRS2000 (M88763); pcaK-pcaF_PRS, the pcaK-pcaF intergenic region of PRS2000 (U10895); pcaR_PRS, the pcaR promoter region of PRS2000 (L33795); catR-catA_1CP, the catR-catA intergenic region of Rhodococcus opacus 1CP (X99622); catR-catA_CCM, the catR-catA intergenic region of Rhodococcus erythropolis CCM2595 (AJ605581). (B) Effect of mutations in the inverted repeat sequence on tphCII promoter activity. Mutated nucleotides are in lowercase. The arrows indicate the inverted repeat sequence. The transcription start site (position +1) of the tphCII promoter is indicated. E6 cells harboring each plasmid were grown in W minimal salt medium containing 10 mM succinate either with or without 10 mM TPA. Each value is the average ± standard deviation (error bars) based on at least three independent experiments. Asterisks, activities lower than the limit of detection.
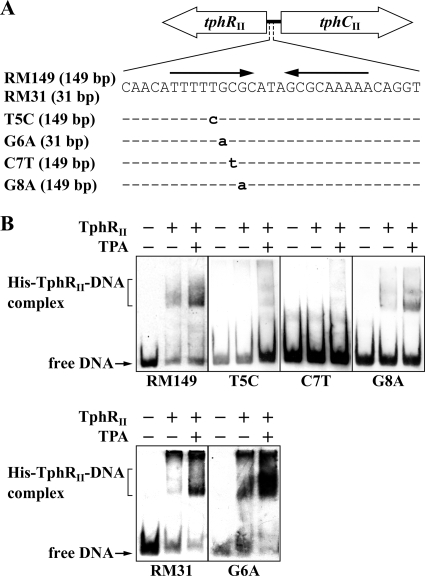
To investigate whether these nucleotides play a role in the formation of the His-TphRII-DNA complex, EMSAs were performed with the mutated tphCII promoter probes (Fig. 7 A). As shown in Fig. 7B, the C7T (C to T at position −55) probe was not bound by His-TphRII, and significantly weak bindings were observed for the T5C (T to C at position −57) and G8A (G to A at position −54) probes. On the other hand, His-TphRII formed high-affinity complexes with the G6A (G to A at position −56) probe, similar to that which was found for the wild-type probe (Fig. 7B). These binding profiles correlated with the tphCII promoter activities. On the basis of these results, it is concluded that the conserved nucleotides are crucial for the ability to form the His-TphRII-DNA complex. Furthermore, the binding of TphRII to the inverted repeat sequence is required for the inducible expression of the tphII catabolism operon in E6. The promoter region of the TPA catabolism gene cluster of C. testosteroni YZW-D (GenBank accession no. AY923836) showed 100% identity with the promoter regions of the tphI/II gene clusters, and the deduced amino acid sequences between E6 tphRII and YZW-D tphR showed 97% identity. These observations suggest that a common regulatory mechanism for the TPA catabolism genes exists in strains E6 and YZW-D.
FIG. 7.
EMSAs of the binding of His-TphRII to the tphCII promoter regions containing mutated inverted repeat sequences. (A) Schematic diagrams of the DNA fragments used in EMSAs. Mutated nucleotides are in lowercase. The sizes of the DNA probes are indicated in parentheses. The arrows indicate the inverted repeat sequence. (B) EMSAs of the binding of purified His-TphRII to the RM149 and RM31 probes and their mutant derivatives. The positions of the unbound probe (free DNA) and His-TphRII-DNA complex are indicated. His-TphRII (65 nM monomer) and 100 μM TPA were added as indicated.
Conclusions.
The TPA-induced expression of the tphCIIA2IIA3IIBIIA1II operon of Comamonas sp. strain E6 is controlled by TphRII, which belongs to the ITTR family. His-TphRII specifically binds to the inverted repeat sequence within the tphCII-tphRII intergenic region. The nucleotides in the inverted repeat sequences highly conserved among the promoter regions, which interact with PcaU, PcaR, PobR, and CatR, are essential for the TPA-dependent transcriptional activation from the tphCII promoter and the formation of the His-TphRII-DNA complex.
Footnotes
Published ahead of print on 23 July 2010.
REFERENCES
- 1.Abe, T., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2005. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, P. Karunakaran, and S. Valla. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in Gram-negative bacteria. Plasmid 38:35-51. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Choi, K. Y., D. Kim, W. J. Sul, J. C. Chae, G. J. Zylstra, Y. M. Kim, and E. Kim. 2005. Molecular and biochemical analysis of phthalate and terephthalate degradation by Rhodococcus sp. strain DK17. FEMS Microbiol. Lett. 252:207-213. [DOI] [PubMed] [Google Scholar]
- 5.Dal, S., G. Trautwein, and U. Gerischer. 2005. Transcriptional organization of genes for protocatechuate and quinate degradation from Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 71:1025-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiMarco, A. A., B. Averhoff, and L. N. Ornston. 1993. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J. Bacteriol. 175:4499-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMarco, A. A., and L. N. Ornston. 1994. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J. Bacteriol. 176:4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton, R. W. 2001. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183:3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuhara, Y., K. Inakazu, N. Kodama, N. Kamimura, D. Kasai, Y. Katayama, M. Fukuda, and E. Masai. 2010. Characterization of the isophthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 76:519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerischer, U., A. Segura, and L. N. Ornston. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 180:1512-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, Z., and J. E. Houghton. 1999. PcaR-mediated activation and repression of pca genes from Pseudomonas putida are propagated by its binding to both the −35 and the −10 promoter elements. Mol. Microbiol. 32:253-263. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Hara, H., L. D. Eltis, J. E. Davies, and W. W. Mohn. 2007. Transcriptomic analysis reveals a bifurcated terephthalate degradation pathway in Rhodococcus sp. strain RHA1. J. Bacteriol. 189:1641-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 15.Hishida, M., K. Shikinaka, Y. Katayama, S. Kajlta, E. Masai, M. Nakamura, Y. Otsuka, S. Ohara, and K. Shigehara. 2009. Polyesters of 2-pyrone-4,6-dicarboxylic acid (PDC) as bio-based plastics exhibiting strong adhering properties. Polym. J. 41:297-302. [Google Scholar]
- 16.Jerg, B., and U. Gerischer. 2008. Relevance of nucleotides of the PcaU binding site from Acinetobacter baylyi. Microbiology 154:756-766. [DOI] [PubMed] [Google Scholar]
- 17.Kasai, D., T. Fujinami, T. Abe, K. Mase, Y. Katayama, M. Fukuda, and E. Masai. 2009. Uncovering the protocatechuate 2,3-cleavage pathway genes. J. Bacteriol. 191:6758-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasai, D., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2005. Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187:5067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kersten, P. J., S. Dagley, J. W. Whittaker, D. M. Arciero, and J. D. Lipscomb. 1982. 2-Pyrone-4,6-dicarboxylic acid, a catabolite of gallic acids in Pseudomonas species. J. Bacteriol. 152:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masai, E., Y. Katayama, and M. Fukuda. 2007. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71:1-15. [DOI] [PubMed] [Google Scholar]
- 21.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michinobu, T., M. Bito, M. Tanimura, Y. Katayama, E. Masai, M. Nakamura, Y. Otsuka, S. Ohara, and K. Shigehara. 2009. Mechanical properties of poly(l-lactide) films controlled by blending with polyesters of lignin-derived stable metabolic intermediate, 2-pyrone-4,6-dicarboxylic acid (PDC). Polym. J. 41:843-848. [Google Scholar]
- 23.Michinobu, T., M. Hishida, M. Sato, Y. Katayama, E. Masai, M. Nakamura, Y. Otsuka, S. Ohara, and K. Shigehara. 2008. Polyesters of 2-pyrone-4,6-dicarboxylic acid (PDC) obtained from a metabolic intermediate of lignin. Polym. J. 40:68-75. [Google Scholar]
- 24.Molina-Henares, A. J., T. Krell, M. Eugenia Guazzaroni, A. Segura, and J. L. Ramos. 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 30:157-186. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa, S., T. Sonoki, T. Kasahara, T. Obi, S. Kubota, S. Kawai, N. Morohoshi, and Y. Katayama. 1998. Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl. Environ. Microbiol. 64:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patrauchan, M. A., C. Florizone, M. Dosanjh, W. W. Mohn, J. Davies, and L. D. Eltis. 2005. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. J. Bacteriol. 187:4050-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popp, R., T. Kohl, P. Patz, G. Trautwein, and U. Gerischer. 2002. Differential DNA binding of transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J. Bacteriol. 184:1988-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 176:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasoh, M., E. Masai, S. Ishibashi, H. Hara, N. Kamimura, K. Miyauchi, and M. Fukuda. 2006. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 72:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schäfer, A., A. Tauch, W. J. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 32.Schläfli, H. R., M. A. Weiss, T. Leisinger, and A. M. Cook. 1994. Terephthalate 1,2-dioxygenase system from Comamonas testosteroni T-2: purification and some properties of the oxygenase component. J. Bacteriol. 176:6644-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shigematsu, T., K. Yumihara, Y. Ueda, S. Morimura, and K. Kida. 2003. Purification and gene cloning of the oxygenase component of the terephthalate 1,2-dioxygenase system from Delftia tsuruhatensis strain T7. FEMS Microbiol. Lett. 220:255-260. [DOI] [PubMed] [Google Scholar]
- 34.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. λ ZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 36.Veselý, M., M. Knoppová, J. Nešvera, and M. Pátek. 2007. Analysis of catRABC operon for catechol degradation from phenol-degrading Rhodococcus erythropolis. Appl. Microbiol. Biotechnol. 76:159-168. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Y. Z., Y. Zhou, and G. J. Zylstra. 1995. Molecular analysis of isophthalate and terephthalate degradation by Comamonas testosteroni YZW-D. Environ. Health Perspect. 103(Suppl. 5):9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]