Abstract
Caenorhabditis elegans is a validated model to study bacterial pathogenicity. We report that Yersinia enterocolitica strains W22703 (biovar 2, serovar O:9) and WA314 (biovar 1B, serovar O:8) kill C. elegans when feeding on the pathogens for at least 15 min before transfer to the feeding strain Escherichia coli OP50. The killing by Yersinia enterocolitica requires viable bacteria and, in contrast to that by Yersinia pestis and Yersinia pseudotuberculosis strains, is biofilm independent. The deletion of tcaA encoding an insecticidal toxin resulted in an OP50-like life span of C. elegans, indicating an essential role of TcaA in the nematocidal activity of Y. enterocolitica. TcaA alone is not sufficient for nematocidal activity because E. coli DH5α overexpressing TcaA did not result in a reduced C. elegans life span. Spatial-temporal analysis of C. elegans infected with green fluorescent protein-labeled Y. enterocolitica strains showed that Y. enterocolitica colonizes the nematode intestine, leading to an extreme expansion of the intestinal lumen. By low-dose infection with W22703 or DH5α followed by transfer to E. coli OP50, proliferation of Y. enterocolitica, but not E. coli, in the intestinal lumen of the nematode was observed. The titer of W22703 cells within the worm increased to over 106 per worm 4 days after infection while a significantly lower number of a tcaA knockout mutant was recovered. A strong expression of tcaA was observed during the first 5 days of infection. Y. enterocolitica WA314 (biovar 1B, serovar O:8) mutant strains lacking the yadA, inv, yopE, and irp1 genes known to be important for virulence in mammals were not attenuated or only slightly attenuated in their toxicity toward the nematode, suggesting that these factors do not play a significant role in the colonization and persistence of this pathogen in nematodes. In summary, this study supports the hypothesis that C. elegans is a natural host and nutrient source of Y. enterocolitica.
Yersinia enterocolitica belongs to the family of Enterobacteriaceae and is a psychrotolerant human pathogen that causes gastrointestinal syndromes ranging from acute enteritis to mesenteric lymphadenitis (5). It infects a number of mammals, and swine was identified as a major source for human infection (6). A multiphasic life cycle, which comprises a free-living phase and several host-associated phases, including cold-blooded and warm-blooded hosts, appears to be characteristic for biovars 1B and 2 to 5 of Y. enterocolitica (7, 24).
Nonmammalian host organisms including Dictyostelium discoideum, Drosophila melanogaster, or Caenorhabditis elegans are increasingly used to study host-pathogen interactions (16, 26). Due to the obvious parallels between the mammalian and invertebrate defense mechanisms, it has been suggested that the bacteria-invertebrate interaction has shaped the evolution of microbial pathogenicity (53). Several human pathogens including Gram-positive and Gram-negative bacteria infect and kill the soil nematode C. elegans when they are supplied as a nutrient source (42). For example, Streptococcus pneumoniae (4), Listeria monocytogenes (50), extraintestinal Escherichia coli (15), and Staphylococcus aureus (43) but not Bacillus subtilis have been shown to kill the nematode. Upon infection of C. elegans with Enterococcus faecalis, Gram-positive virulence-related factors as well as putative antimicrobials have been identified (20, 35). The extensive conservation in virulence mechanisms directed against invertebrates as well as mammals was demonstrated using a screen with Pseudomonas aeruginosa (30). In this study, 10 of 13 genes whose knockout attenuated the nematode killing were also required for full virulence in a mouse model, confirming the suitability of the C. elegans model to study bacterial pathogenicity. C. elegans is also colonized by Salmonella enterica serovar Typhimurium (S. Typhimurium). This process requires Salmonella virulence factors and was used to study the innate immune response of the nematode (1, 2, 49).
The effect of pathogenic Yersinia spp. on C. elegans has also been investigated. It could be demonstrated that both Yersinia pestis and Yersinia pseudotuberculosis block food intake by creating a biofilm around the worm's mouth (13, 27). This biofilm formation requires the hemin storage locus (hms) and has been suggested to be responsible for the blockage of the digestive tract following uptake by fleas, thus acting as a bacterial defense against predation by invertebrates. In a study with 40 Y. pseudotuberculosis strains, one-quarter of them caused an infection of C. elegans by biofilm formation on the worm head (27). In contrast, a similar effect was not observed following nematode infection with 15 Y. enterocolitica strains. Using a Y. pestis strain lacking the hms genes, it could be demonstrated that this mutant can infect and kill the nematode by a biofilm-independent mechanism that includes the accumulation of Y. pestis in the intestine of the worm (47). This pathogenesis model was applied to show that putative virulence factors such as YapH, OmpT, or a metalloprotease, Y3857, but not the virulence plasmids pCD1 and pPCP1, are required for Y. pestis virulence in C. elegans. Six yet unknown genes required for full virulence in C. elegans were also identified, and one of them appeared to be a virulence factor in the mouse infection model.
C. elegans has not been used to study the pathogenicity properties of Y. enterocolitica, mainly due to the fact that many of its virulence factors are upregulated at 37°C in comparison to growth at lower temperatures while C. elegans cannot be cultivated at temperatures above 25°C. In this study, we examined for the first time the infection of C. elegans by Y. enterocolitica strains, demonstrating that this pathogen colonizes and kills C. elegans and that the insecticidal toxin TcaA, which is expressed only at ambient temperature, is required for full nematocidal activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Cultures were grown in Luria-Bertani (LB) broth (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter) or on LB agar (LB broth supplemented with 1.5% [wt/vol] Bacto agar). E. coli OP50 was grown at 37°C, and Y. enterocolitica strains were cultivated at 15°C or 30°C. If appropriate, the media were supplemented with the following antibiotics: 50 μg ml−1 kanamycin, 18 μg ml−1 tetracycline, and 20 μg ml−1 nalidixic acid.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 22 |
| OP50 | Nematode feeding strain | Caenorhabditis Genetics Center, University of Minnesota, MN |
| Y. enterocolitica | ||
| W22703 | Biovar 2, serovar O:9, Nalr Res− Mod+; lacking the pYV virulence plasmid | 10 |
| W22703 ΔtcaA | Mutant of W22703 with a non-polar deletion of tcaA | 7 |
| W22703 tcaA134::Tn5lux | Mutant of W22703 with tcaA knockout upon Tn5lux insertion | 7 |
| W22703 YE2848::Tn5lux | Mutant of W22703 with YE2848 knockout upon Tn5lux insertion | 7 |
| WA314/pYV | Biovar 1B, serovar O:8; Nalr; harbors virulence plasmid pYV | 40 |
| WA314/pYVΔyopE | Mutant lacking pYV-encoded effector protein YopE | 51 |
| WA314/pYVΔyadA | Mutant lacking pYV-encoded adhesin YadA | 38 |
| WA314 Δirp1 | Mutant lacking the chromosomally encoded yersiniabactin synthase HMWP1 | 36 |
| WA314 Δinv | Mutant lacking the chromosomally encoded invasin Inv | 40 |
| Y. pseudotuberculosis | ||
| STM 537 | Strain PB1, serovar O1a | 45 |
| STM 593 | Strain H-Y60/86, serovar O:2c | 3 |
| STM 542 | Strain YpIII(plB1), serovar O:3 | 54 |
| STM 595 | Strain H-Y52/86, serovar O:4 | 3 |
| S. enterica | ||
| Serovar Typhimurium | Wild-type strain ATCC 14028 | DSMZ, Braunschweig, Germany |
| Serovar Dublin | Wild-type strain 98-07710 | Robert Koch-Institut, Wernigerode, Germany |
| C. elegans | ||
| N2 (var. Bristol) | Wild-type strain | Caenorhabditis Genetics Center, University of Minnesota, MN |
| SS104 glp-4(bn2) | Temperature sensitive (sterile at 25°C) mutant | Caenorhabditis Genetics Center, University of Minnesota, MN |
| Plasmids | ||
| pNT-PYE2848::gfp | Promoter probe vector pNT with gfp fused to the low-temperature-induced promoter of YE2848; Kanr | 8 |
| pACYC184/F37.R43(s) | pACYC184 (9) with an EcoRI fragment containing tcaA and its promoter region; Tetr Cams | 7 |
| pBAD/HisA(tet) | pBAD/HisA (Invitrogen, Karlsruhe, Germany); Ampr exchanged for Tetr of pACYC184 | This study |
| pBAD/HisA(tet)-tcaA | pBAD/His(tet) harboring tcaA under the control of the arabinose promoter; Tetr | This study |
| pGreenTIR | Vector with gfp cloning cassette | 33 |
Overexpression of TcaA.
pBAD/HisA was cut with BspHI and ligated to a NcoI (Fermentas, St. Leon-Rot, Germany)-restricted fragment harboring the tetracycline resistance cassette (tet) of pACYC184, resulting in pBAD/HisA(tet). A PCR-amplified tcaA fragment was then ligated to this plasmid via SacI to generate pBAD/HisA(tet)-tcaA. Oligonucleotides used here are shown in Table S1 in the supplemental material. TcaA expression was induced by adding arabinose to a final concentration of 0.2% and confirmed by Western blot analysis performed according to standard procedures with monoclonal His6-tagged antibodies (dianova, Hamburg, Germany) diluted 1:1,000 and alkaline phosphatase-conjugated anti-mouse antibodies (dianova) diluted 1:15,000.
Nematode cultivation.
Maintenance of C. elegans wild-type N2 (var. Bristol) including feeding, transfer, and synchronization was performed according to standard procedures (44). Briefly, E. coli OP50 overnight culture was seeded on nematode growth medium (NGM) agar plates containing 3.0 g of NaCl, 2.5 g of peptone, 1.0 ml of 1 M CaCl2, 1.0 ml of cholesterol (5 mg/ml stock prepared in 95% ethanol), 25.0 ml of 1 M KPO4 buffer (108.3 g/liter KH2PO4 and 35.6 g/liter K2HPO4), pH 6.0, 1.0 ml of 1 M MgSO4, and 17.0 g of high-strength Bacto agar per liter (29). Plates with E. coli OP50 were incubated overnight at room temperature and stored at 4°C. N2 worms were cultivated at 22°C and transferred every 2 to 3 days to new plates. For synchronization, nematodes that were predominantly in the gravid stage were washed off from the plates using sterile M9 buffer (21.5 mM KH2PO4, 46 mM Na2PO4·2H2O, 85.5 mM NaCl, and 1 mM MgSO4 added after autoclaving), followed by frequent washing in M9 buffer to reduce bacterial contamination. The nematodes were then subjected to an alkaline hypochlorite treatment (44) to isolate the eggs, which were transferred to new NGM agar plates with E. coli OP50 and incubated at 22°C for 2 to 3 days, allowing all of the eggs to hatch and grow to larval stage 4 (L4 stage).
Nematode infection and toxicity assay.
Fifty microliters of an overnight culture of bacterial strains was spread on NGM agar plates of 8.5-cm diameter that were then incubated overnight at the appropriate temperature. Plates were equilibrated to room temperature (22°C) before use. C. elegans L4 larvae were individually transferred onto the bacterial lawn. The infection assay was performed at 22°C for 4 h or for the time period indicated in Fig. 1 After infection, nematodes were transferred back to NGM with an OP50 lawn, and then every second day they were transferred to a fresh NGM agar plate with the feeding strain until no more progeny were evident. The number of viable and killed nematodes was determined daily until all worms were dead. Worms were considered dead if they failed to respond to touch. The total number of worms was corrected by subtracting the number of worms mechanically killed during transfer. The time for 50% of the nematodes to die (50% time to death [TD50]) was calculated using the dose-response curve (drc) package of the R software (http://www.r-project.org/). The raw data were analyzed using the GraphPad Prism (version 4.0) computer program and plotted by the Kaplan-Meier method. The curves were compared using a log rank test, which generates a P value testing the null hypothesis that the survival curves are identical. P values of 0.05 or less were considered significantly different from the null hypothesis.
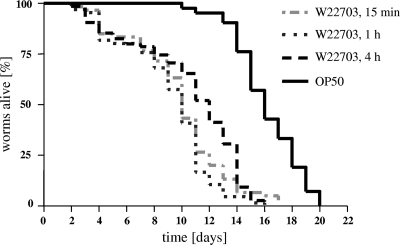
FIG. 1.
Survival of C. elegans fed on Y. enterocolitica W22703 and E. coli OP50. Nematodes were cultivated on W22703 for 15 min, 1 h, and 4 h and then transferred to E. coli OP50. Nematodes fed on only E. coli OP50 served as controls. With respect to the shortest infection time, no significant differences in the TD50 values after 1 h or 4 h of infection were found (P = 0.1505 and P = 0.1003, respectively). The TD50s of the worms infected with yersiniae were calculated in three independent experiments as 9.8, 9.2, and 10.6 days. The Kaplan-Meier plot is based on one experiment with three technical replicates (n ≥60 animals per curve).
Quantification of intestinal bacterial cells.
Nematodes were transferred into 600 μl of ice-cold lysis buffer (M9 buffer with 0.1% Triton X-100) and shaken for 10 min at 400 rpm to release bacteria from the cuticula. Nematodes were sedimented by centrifugation for 2 min at 430 × g, washed twice with M9 buffer, and resuspended in 1 ml of M9 buffer. After 500 μl of 1.0-mm Zirconia silica beads (BioSpec, Bartlesville) was added, nematodes were disrupted in a FastPrep-24 (MP Biomedicals, Solon) for 20 s at maximal speed, and the suspension was placed on ice. This step was repeated twice. Dilutions were made in LB medium, and aliquots of the suspensions were then plated on LB plates containing the appropriate antibiotic to quantify the accumulation of bacteria within the nematodes.
Heat killing and sonication of bacteria.
Y. enterocolitica and E. coli strains were cultivated overnight at 15°C or 37°C in a volume of 10 ml of LB medium until cells reached stationary phase. The cell suspensions were incubated for 90 min in a 60°C water bath, and aliquots of 200 μl were plated on LB agar plates to confirm the absence of viable bacteria. Dead bacteria were concentrated 5-fold by centrifugation for 10 min at 4,000 × g and resuspension in LB medium. A 1:1 (vol/vol) mixture of the debris and viable E. coli OP50 cells was then plated on NGM agar plates. Nematodes were immediately exposed to the lawn, and their viability was monitored until all worms were dead. If appropriate, the worms were transferred to freshly prepared plates of identical composition. The assay was evaluated as described above. Cell-free lysates were prepared by sonication as described recently (7).
In vivo quantification of tcaA expression.
C. elegans SS104 glp-4(bn2) strain was cultivated at 15°C for 4 days. Following egg preparation, the stage 1 larvae (L1) were cocultivated with OP50 at 25°C, leading to sterile adults. When the nematodes had developed to L4 larvae, the plates were overlaid with M9 buffer, and resuspended worms were washed twice in M9 medium to eliminate bacterial contamination. Nematodes were sedimented by centrifugation for 2 min at 430 × g. The nematodes were then exposed for 4 h to lawns of Yersinia strains carrying the luciferase reporter cassette luxCDABE behind the promoter of TcaA or YE2848 to allow infection. Worms were collected in M9 medium and washed twice as described above, and appropriate aliquots were dropped on NGM plates containing OP50. The plates were then incubated at 25°C. To monitor the expression of tcaA, the worms of one plate per strain were collected and washed in M9 medium and distributed in 6 to 12 wells of a microtiter plate per strain. The bioluminescence levels measured as relative light units (RLU) were recorded with an IVIS Lumina (Xenogen, Caliper Life Sciences, Mainz, Germany), and nematodes were enumerated.
Microscopy.
Worms were observed under an M8 binocular microscope (Wild, Heerbrugg, Germany) up to ×40 magnification. For fluorescence microscopy, nematodes were washed three times with M9 buffer, transferred to M9 buffer containing 5 mM levamisole, and studied with a BX51 fluorescence microscope (Olympus, Hamburg, Germany) up to ×65 magnification.
RESULTS
Y. enterocolitica strains W22703 and WA314 kill C. elegans.
We first examined the ability of strain W22703 (biovar 2, serovar O:9) to kill C. elegans. To avoid new progeny interference with the enumeration of live nematodes, L4 larvae were selected for the infection assays. In preliminary experiments, C. elegans fed on W22703 for 4 h, 8 h, or for the whole infection assay lasting at least 16 days. The worms died with similar kinetics independent of the exposure time. Time to death of 50% (TD50) of the worms infected with yersiniae was calculated as 8.2, 8.7, and 7.7 days, respectively. A total of 74 (permanent infection with Y. enterocolitica W22703), 17 (4 h of infection), and 25 (8 h of infection) nematodes were used in the experiments. Thus, the W22703-exposed worms lived only half as long in comparison to worms permanently cultured on feeding strain E. coli OP50 (TD50 of 15.6 days; 42 nematodes). The nematode survival rate following temporary infection for 4 or 8 h did not significantly differ from that upon permanent infection with W22703 (P > 0.0001). In a further experiment, we tried to determine the minimal infection time required to induce slow killing of C. elegans by this human pathogen. Feeding for 15 min revealed to be sufficient for reducing the life span of the nematode, and no significant differences to the TD50 values after 1 h or 4 h of infection were found (Fig. 1 and Table 2). In all subsequent experiments, feeding on Y. enterocolitica strains was limited to 4 h. During all infection assays with this strain, we observed a progressively slower locomotion of the worms. Tested under the conditions established above, strain WA314 (biovar 1B, serovar O:8) also showed nematocidal activity, with a TD50 of 7.4 (Table 2). Taken together, these data demonstrate that Y. enterocolitica strains W22703 and WA314 are toxic toward the nematode and decrease the life span of C. elegans following oral uptake.
TABLE 2.
Statistical analysis of the experiments performed in this study
| Parameter, condition(s), and/or strain | No. of nematodes tested | TD50 (days)a | P value |
|---|---|---|---|
| Nematode survival | |||
| Killing by W22703 infection | |||
| 15 min | 60 | 9.8 | <0.0001 |
| 1 h | 66 | 9.2 | <0.0001 |
| 4 h | 75 | 10.6 | <0.0001 |
| Permanent culture on OP50 feeding strain | 81 | 15.9 | Standard |
| Effect of tcaA deletion on toxicity | |||
| W22703 | 17 | 8.2 | Standard |
| W22703 ΔtcaA | 65 | 13.4 | <0.0001 |
| W22703 ΔtcaA/pACYC/F37.R43(s) | 85 | 7.1 | 0.13 |
| W22703/pACYC184 | 66 | 6.9 | 0.04 |
| OP50 | 81 | 13.5 | <0.0001 |
| Effect of feeding on heat-killed/sonified bacteriab | |||
| DH5α/pBAD; 37°C + OP50 | 91 | 8.6 | Standard |
| DH5α/pBAD/HisA(tet)-tcaA; 37°C + arabinose + OP50 | 94 | 8.9 | 0.54 |
| DH5α/pBAD/HisA(tet)-tcaA; 37°C + arabinose + OP50 (sonified) | 93 | 9.0 | 0.017 |
| W22703/pBAD/HisA(tet)-tcaA; 37°C + OP50 | 96 | 8.2 | 0.33 |
| W22703/pBAD/HisA(tet)-tcaA; 37°C + arabinose + OP50 | 96 | 9.5 | 0.06 |
| W22703/pBAD/HisA(tet)-tcaA; 15°C + OP50 | 186 | 8.5 | 0.37 |
| W22703/pBAD/HisA(tet)-tcaA; 15°C + arabinose + OP50 | 88 | 8.0 | 0.20 |
| W22703; 15°C + OP50 (sonified) | 86 | 12.5 | <0.0001 |
| W22703 ΔtcaA; 15°C + OP50 (sonified) | 93 | 11.3 | 0.0002 |
| W22703 (live); 15°C + OP50 | 90 | 5.8 | <0.0001 |
| Nematocidal activity of WA314 strains | |||
| WA314/pYV | 94 | 7.4 | Standard |
| WA314/pYVΔyadA | 92 | 8.4 | <0.01 |
| WA314/pYVΔyopE | 93 | 6.8 | 0.08 |
| WA314 Δirp1 | 90 | 7.6 | 0.64 |
| WA314 Δinv | 67 | 8.3 | <0.01 |
TD50 values were calculated with the drc package of the R software. The raw data were analyzed using the GraphPad Prism program. P values of <0.01 shown in bold letters are considered significantly different from the survival curve used as the standard.
Overnight cultures for experiments with heat-killed (except as noted) bacteria and untreated OP50 cells were incubated at 37°C or 15°C as indicated. If appropriate, the medium contained arabinose. All experiments were performed in triplicates.
Biofilm formation is not responsible for the nematocidal activity.
It is known that Y. pestis and Y. pseudotuberculosis kill C. elegans by a biofilm-dependent mechanism that also allows Y. pestis to block food intake when it colonizes the flea vector (13). C. elegans was exposed to four Y. pseudotuberculosis strains (Table 1). Only strain STM 542 created an extracellular biofilm around the worm's head (data not shown). The biofilm became visible after several hours of incubation and increased in size with continuing exposure, finally covering the nematode's mouth region completely. In addition to this biofilm-like colonization of the worm, its locomotion was characterized by more narrow windings in comparison to results of infection with the other Y. pseudotuberculosis strains. In contrast, no biofilm formation was observed in any of the numerous independent experiments performed with Y. enterocolitica strains W22703 and WA314 or with their mutants. This observation excludes biofilm formation as the mechanism responsible for the toxic effect of Y. enterocolitica toward C. elegans.
A mutant lacking tcaA is nontoxic toward C. elegans.
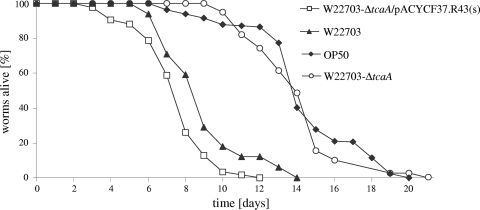
To further investigate the biological role of the insecticidal protein TcaA in the interaction of Y. enterocolitica with C. elegans, an infection assay with a W22703 ΔtcaA strain was performed. Surprisingly, a TD50 of 13.4, nearly identical to that of the control experiment with E. coli OP50 (TD50 of 13.5), was observed (Fig. 2). Similar results were obtained when C. elegans was exposed to a tcaA knockout mutant, W22703 tcaA134::Tn5lux (data not shown). To confirm that the killing phenotype of W22703 is indeed the result of TcaA activity, we fed C. elegans with the W22703 ΔtcaA strain harboring the plasmid pACYC184/F37.R43(s). This construct carries tcaA and has been shown to complement the nontoxic phenotype of a tcaA deletion mutant in the insect Manduca sexta (7). Following infection and recultivation of the nematodes on E. coli OP50 with W22703 Δtca/pACYC184/F37.R43(s), a TD50 of 7.1 was determined, which does not significantly differ from that of strain W22703 (Fig. 2 and Table 2), demonstrating that the in trans complementation of tcaA in the W22703 ΔtcaA strain restores the toxic phenotype of the wild-type strain. Mutant W22703 ΔtcaA transformed with pACYC184 served as a negative control and did not modify the atoxic phenotype of the deletion mutant. Taken together, these results show that TcaA is necessary for toxicity of Y. enterocolitica toward the nematode.
FIG. 2.
TcaA is required for the nematocidal activity of W22730. Feeding of C. elegans on a tcaA deletion mutant (W22730 ΔtcaA) resulted in an OP50-like survival curve (TD50 of 13.4; number of nematodes, 65) that significantly (P < 0.0001) differs from that of strain W22730. The wild-type-like phenotype could be restored using plasmid pACYC184/F37.R43(s) harboring tcaA. Average data of three independent experiments are shown (Table 2). No significant difference (P = 0.1277) between the survival curves following exposure to W22730 or W22730 ΔtcaA/pACYC184/F37.R43(s) was observed. The survival curves for nematodes feeding on W22703 and E. coli OP50 are identical to those in Fig. 1 and 5.
Spatial-temporal analysis of C. elegans infection by Y. enterocolitica.
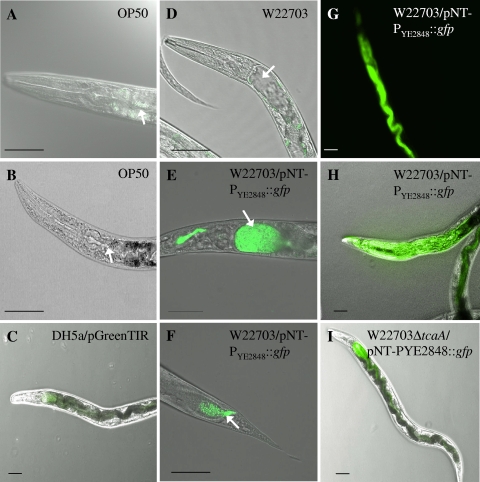
To investigate possible proliferation of Y. enterocolitica after ingestion by the nematode, the W22703 and W22703 ΔtcaA strains were transformed with plasmid pNT-PYE2848::gfp harboring a kanamycin resistance cassette and gfp under the control of a low-temperature-induced promoter that drives the expression of YE2848, a methyl-accepting chemotaxis protein. As a control, DH5α/pGreenTIR constitutively expressing green fluorescent protein (GFP) was used. Overnight cultures of these strains were plated on NGM as described above, and nematodes were exposed to these lawns for 4 h and then transferred to NGM plates with E. coli OP50. Nematodes were monitored each day by fluorescence microscopy (Fig. 3). The absence of fluorescent bacteria from the surfaces of the nematodes was confirmed by fluorescence microscopy, demonstrating that C. elegans movement on NGM plates after infection cures the worms' surfaces of the bacteria to which the nematodes had temporarily been attached (44). From day 2 of permanent infection, a distension of the luminal space was observed, and intact W22703 cells had accumulated throughout the intestine of the worm (Fig. 3D to F). Four days after infection, the highest concentration of yersiniae could be seen in the anterior part of the gut lumen right behind the pharynx and in the posterior part near the anus. In these regions, the luminal distension was maximal 4 to 5 days after infection until the whole nematode was colonized by Y. enterocolitica (Fig. 3H). This phenotype was also obtained with the tcaA negative mutant W22703 ΔtcaA, but it occurred later and was less distinct (Fig. 3I). In repeated experiments, infection with DH5α/pGreenTIR did not result in gut distension. Nematodes were also fed with W22703/pNT-PYE2848::gfp and DH5α/pNT-PYE2848::gfp, each diluted 1:1,000 with unlabeled DH5α. Upon this low-dose infection, Y. enterocolitica also colonized the gut, while E. coli cells could neither be observed under the fluorescence microscope nor recovered from disrupted worms (see below).
FIG. 3.
Y. enterocolitica strain W22703 colonizes the worm intestine. The anterior part of a nematode feeding on OP50 is depicted in panels A and B. Background fluorescence emitted by C. elegans is visible in panel A. (C) Infection with DH5α/pGreenTIR served as a control; no fluorescent bacteria were observed. (D) Distension of the intestinal lumen was observed when worms were fed unlabeled W22703. Four days after infection, an accumulation of W22703/pNT-PYE2848::gfp was detected in the anterior (E) and posterior (F) intestinal regions proximal to the pharynx and the anus of the nematode, the regions exhibiting maximal luminal distension. Approximately from day 4 after infection with Y. enterocolitica, the whole intestine (G) and finally the nematode body (H) are colonized by bacterial cells. Uptake of the W22703 ΔtcaA/pNT-PYE2848::gfp strain results in a less distinct phenotype 7 days after infection (I). Bright-field and fluorescence photographs were not overlaid in panels B and G. Representative photographs are shown. Arrows indicate the intestinal lumen of C. elegans. Scale bar, 50 μm.
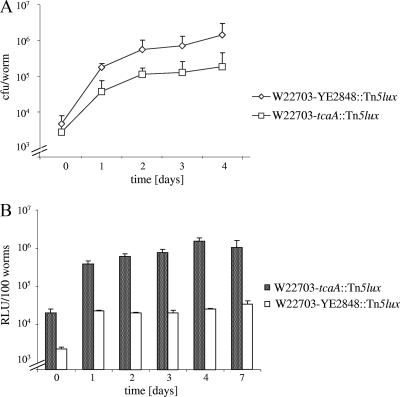
Quantification of bacterial cells within glp-4(bn2) nematodes was then performed using the W22703, W22703 tcaA134::Tn5lux, and DH5α/pNT-PYE2848::gfp strains. After 0, 1, 2, 3, and 4 days, the nematodes were treated with lysis buffer, and no bacteria could be detected when aliquots of the supernatant were plated. Nematodes were disrupted with silica beads to determine the number of viable bacterial cells remaining in the worm. In preliminary tests, no effect of this procedure on the viability of bacterial cells could be observed (data not shown). After 4 days, the number of Yersinia cells had increased from approximately 4 × 103 to more than 106 cells per worm (Fig. 4A). From day 2 onward, the number of tcaA mutant cells was approximately 4-fold lower than the number of wild-type cells. No viable DH5α/pNT-PYE2848::gfp cell was detected after the nematodes had been shifted to an OP50 lawn (data not shown). These data demonstrate that in contrast to E. coli DH5α, Y. enterocolitica is able to survive, persist, and proliferate within the C. elegans intestine after oral uptake and that this phenotype is partially TcaA independent.
FIG. 4.
Proliferation and tcaA expression of W22703 in C. elegans. (A) C. elegans worms were fed for 4 h with W22703 or W22703 tcaA134::Tn5lux and then transferred to plates with E. coli OP50 cells. A total of 20 to 100 nematodes were lysed per time point of each experiment, and the bacterial cell numbers per worm are shown. The number of W22703 cells within the nematode increased to 1.15 × 106 cells per worm in 4 days, and that of mutant W22703 tcaA134::Tn5lux increased to approximately 2.58 × 105 cells per worm. The standard deviation of five independent experiments is indicated. (B) In vivo expression of tcaA was monitored in parallel using mutant W22703 tcaA134::Tn5lux. Bioluminescence of strain W22703 YE2848::Tn5lux served as a control. Average data of five independent experiments are shown.
In vivo expression of tcaA.
To follow the expression of TcaA within C. elegans, glp-4(bn2) nematodes were infected with W22703 tcaA134::Tn5lux and W22703 YE2848::Tn5lux strains and then transferred to NGM plates with the feeding strain OP50. The nematodes of one plate per strain were harvested every day, and bioluminescence was detected. Over a time course of 7 days, TcaA expression increased from approximately 2.0 × 104 RLU/100 worms immediately after infection to a maximum of approximately 1.1 × 106 RLU per 100 worms after 4 days (Fig. 4B). The maximal bioluminescence of the control strain W22703 YE2848::Tn5lux was approximately 3.5 × 104 RLU per 100 worms. Comparison with Fig. 4A indicates that elevated tcaA transcription after the first day of infection correlates with the number of bacteria isolated from the nematode.
Viable yersiniae are a prerequisite for nematocidal activity, and TcaA alone is not sufficient to kill nematodes.
To investigate the role of viable cells and of TcaA in the nematocidal activity of Y. enterocolitica, worms were fed with a lawn of heat-killed bacteria or whole-cell extracts. In a pilot experiment, nematodes were exposed to heat-killed Y. enterocolitica W22703 or E. coli OP50 cells. However, the TD50 value obtained in each experiment was significantly lower than that of the control experiment using viable OP50 cells, suggesting that the worms cannot appropriately feed on cell lysates. In the following experiment, nematodes were therefore exposed to a mixture of heat-killed or sonified bacteria and untreated OP50 cells as described above. When nematodes fed on a mixture of heat-killed DH5α with OP50, the result was a short TD50 of 8.6 days, probably due to a lower uptake rate of viable cells or unknown side effects of lysate uptake. However, a TD50 of 5.8 days was observed when C. elegans was fed a mixture of viable Y. enterocolitica cells with OP50, confirming that these mixtures are appropriate standards for the following experiments. Nematodes were fed heat-killed DH5α overexpressing TcaA and heat-killed W22703/pBAD/HisA(tet)-tcaA. This strain was cultivated at 37°C and 15°C in the absence and presence of the inductor arabinose for maximal TcaA expression. TcaA overexpression was confirmed by Western blot analysis (data not shown). As TcaA might be sensitive to heat treatment, cell extracts of TcaA overexpressing DH5α/pBAD/HisA(tet)-tcaA and of W22703 cultivated at 15°C were also used. Statistical analysis of TD50 values did not reveal a significant difference in any of these experiments between the survival curves following exposure to heat-killed Y. enterocolitica or cell extracts and heat-killed E. coli strains or sonified W22703 ΔtcaA (Table 2), strongly suggesting that Y. enterocolitica-mediated killing requires the direct interaction of viable cells with C. elegans and that TcaA alone is not sufficient for nematode killing by Y. enterocolitica.
YadA, InvA, YopE, and Irp1 have no effect or only a weak effect on Y. enterocolitica toxicity toward C. elegans.
The mouse-virulent Y. enterocolitica strain WA314 (serovar O:8) also showed nematocidal activity (Table 2). This was surprising because none of six biovar 1B strains tested recently harbors tcaA (18), which is essential at least for C. elegans killing by strain W22703. However, six PCRs resulting in fragments amplified from tcaR1, tcaR2, tcaA, and tcaC, as well as two PCRs generating fragments overlapping tcaA-tcaB and tcaB-tcaC, indicated the presence of the toxin complex (TC) genes, and probably of the TC genes of the pathogenicity island of Y. enterocolitica (TC-PAIYe) in strain WA314 (see Table S1 for oligonucleotides).
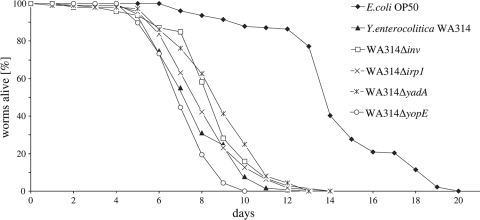
To determine whether the nematode model is useful for the identification and investigation of Y. enterocolitica virulence factors relevant to human disease, we performed a series of C. elegans infection assays with WA314 mutants. The strains tested here lack Inv (WA314 Δinv) and YadA (WA314/pYVΔyadA) involved in invasion and adhesion, respectively, and the effector protein YopE (WA314/pYVΔyopE), all of which have been shown to be involved in mouse virulence (14, 25, 37-39, 51). A further strain (WA314 Δirp1) is unable to synthesize the yersiniabactin required for iron uptake (36). The worms fed on pure WA314 cultures for 4 h and were then transferred to E. coli OP50. The virulence of the strains in the C. elegans model was evaluated by measuring the survival of the nematodes. Mutants WA314 Δinv and WA314/pYVΔyadA exhibited a weakly but significantly diminished toxic activity (P < 0.05) toward C. elegans in comparison to the wild-type strain WA314 (Fig. 5). The prolongation of the nematode's life span by approximately 1 day, however, corresponds to that of L. monocytogenes and S. Typhimurium virulence mutants (28, 50). Mutants WA314 Δirp1, and WA314/pYVΔyopE were nearly as virulent (P > 0.05) as WA314 (Table 2). Taken together, these data suggest that C. elegans is not a feasible model for Y. enterocolitica pathogenicity toward humans.
FIG. 5.
Pathogenicity phenotypes of inv, yadA, irp1, and yopE mutants. C. elegans L4 larvae were exposed to Y. enterocolitica strains WA314, WA314 Δinv (P = 0.0066), WA314/pYVΔyadA (P = 0.0091), WA314 Δirp1 (P = 0.6437), WA314/pYVΔyopE (P = 0.0799), and E. coli OP50 (P < 0.0001). Survival curves are considered significantly different from the survival curve of WA314 when P values are <0.05. Average data of three independent experiments are shown. At least 67 nematodes were used in each experiment (Table 2).
DISCUSSION
Here, we report that Y. enterocolitica establishes an infection in C. elegans that results in a shortened life span of the worm. This infection obviously requires resistance to antimicrobials that are produced by C. elegans (55). Comparison with TD50 values obtained by infection with S. enterica (2, 28) indicates that the oral uptake of Y. enterocolitica and S. enterica results in similar nematode death kinetics. Interestingly, 15 min is sufficient for a W22703 infection while colonization of the nematode by the Y. pestis hms-negative mutant requires feeding for 24 h (47).
It has been reported that certain P. aeruginosa strains can produce toxins of low molecular weight that kill C. elegans within hours, a process that is called fast killing (48). The mechanisms underlying the slow killing of C. elegans by Y. enterocolitica observed here have not been investigated so far. According to our study, accumulation of Y. enterocolitica cells in the nematode digestive tract occurs within a few days after infection. On the hypothesis that such a colonization phenotype includes the activity of adhesin, such a function can be excluded for TcaA because a tcaA mutant also proliferates in the nematode (Fig. 4A). Toxicity of Y. enterocolitica toward larvae of the insect M. sexta upon oral uptake of cell extracts has recently been demonstrated, and TcaA, a subunit of the insecticidal toxin complex (TC) proteins, was shown to be required for this phenotype (7, 18). TcaA transcription in vitro is repressed at 37°C and maximally induced between 10°C and 20°C (7). Interestingly, TcaA is also essential for full nematocidal activity of strain W22703. This is in line with recent data suggesting that TcaA interacts with the gut epithelial cells of invertebrates (18, 52). TcaA expression parallels the increase in the number of bacterial cells within the first 4 days of infection, e.g., approximately 4 days before half of the infected nematodes are dead, and there is no evidence that TcaA expression directly correlates with nematocidal activity. Thus, colonization as well as TcaA expression is only a prerequisite for nematocidal activity possibly caused by a progressive infection and/or the activity of yet unknown toxins. It might also be speculated that the symptom of gut distension disturbs the integrity of the epithelial cell layer, resulting in fluid efflux into the gut lumen and finally in the host's death. The fact that cell extracts of Y. enterocolitica are lethal for M. sexta but not C. elegans might be due to distinct feeding mechanisms rather than host-specific TcaA activity.
In the nematode-associated bacterium Photorhabdus luminescens, TcaA was assumed not to play an important role in nematode pathogenicity because strain TT01 with incomplete tcaA is one of the most pathogenic P. luminescens strains. A P. luminescens strain lacking another insecticidal toxin complex gene belonging to the same homology group, tcdA4, did not decrease worm fitness in comparison to other strains tested. However, this phenotype could not unequivocally be attributed to the tcdA4 deletion (41). In contrast, the insecticidal TcaA protein of Y. enterocolitica is an example not only of a bacterial toxin that is necessary to confer lethality toward C. elegans but also of one whose deletion results in an OP50-like phenotype of infected nematodes. Similar effects have been demonstrated for phenazines, the hydrogen cyanide synthase (HcnC), and other factors of P. aeruginosa (19, 31). The finding of the study presented here is in line with the hypothesis that frameshifts in TC genes of Y. pestis might have allowed its adaptation to insect hosts (53). On the other hand, cells expressing the Y. pseudotuberculosis TC proteins were active against cultured human gut cells (23). The molecular mechanisms underlying the activity of the insecticidal and nematocidal TC proteins of Y. enterocolitica, however, remain to be investigated.
It has been suggested that the interaction of bacteria with invertebrates contributed to the development of virulence factors that have later been adapted to combat defense mechanisms of vertebrate hosts (24, 53). For example, rfaL and ompR mutants of Salmonella are less nematocidal, probably due to reduced resistance to molecules with antibacterial activity (49). Other virulence factors seem to play a role in mammalian hosts only. A listerial actA mutant is not attenuated, probably because intracellular survival and spread are not important for killing C. elegans by L. monocytogenes (50). The absence of a measurable effect of the irp1 deletion on the capacity of WA314 to kill C. elegans suggests that ferric iron acquisition systems are probably not involved in the nematocidal activity of gut-colonizing Y. enterocolitica, thus confirming a similar result in extraintestinal pathogenic E. coli (ExPEC) virulence studies (15). invH and hilA mutants of Salmonella affect toxicity toward the nematode, indicating that pathogenicity island 1 of Salmonella (SPI-1) contributes to C. elegans infection (49). An attenuated sptP mutant found in the same study suggests that the molecular targets of at least some type III secretion system (T3SS) effectors have been conserved (42). However, no such data were reported from P. aeruginosa infection assays (12, 48), and a yopE deletion mutant of Y. enterocolitica WA314 did not show a significant effect on the C. elegans life span (Fig. 5).
Despite several attempts, the causal involvement of tcaA in the nematocidal activity of strain WA314 could not be confirmed. First, the luciferase cassette was inserted behind the putative tcaA promoter, but significant bioluminescence was not observed either in vitro or in vivo. Second, an insertional knockout mutant was preliminarily tested in the C. elegans model but did not show a significant attenuation. A reason for this outcome might be the presence of further insecticidal genes or the involvement of TcaB and related toxins in the toxicity of WA314 toward nematodes.
Temperature is a key environmental clue for the expression of yersiniae genes (32, 46). Several genetic determinants of Y. enterocolitica and Y. pestis are repressed at 37°C but induced at a low temperature, suggesting a role in invertebrates (8, 21, 24, 34). In contrast, the pYV-encoded Yop proteins and YadA are produced only at 37°C. Their transcription is regulated by VirF (LcrF) which is also thermoinduced, and modulated by the histone-like protein YmoA that exerts its down-regulatory activity both at ambient temperature and body temperature (11). YmoA also plays a role in inv expression, which is maximal at 25°C (17). The temperature dependence of Y. enterocolitica calls into question the feasibility of the nematode as an infection model for this pathogen. This is in line with our finding that Inv, YopE, and YadA do not remarkably contribute to C. elegans killing by Y. enterocolitica.
In summary, C. elegans has been demonstrated as a versatile model for identifying novel factors of Y. enterocolitica required for the interaction with invertebrates in the environment. Given that Y. enterocolitica is widely distributed in nature and able to survive for long periods in terrestrial and aquatic environments, it is also tempting to speculate that the nematode may serve as a temporary reservoir of Y. enterocolitica. On the other hand, the data provided here do not support the feasibility of the C. elegans infection model to identify virulence factors involved in Y. enterocolitica pathogenicity toward mammals.
Supplementary Material
Acknowledgments
We thank Jürgen Heesemann for providing WA314 mutants, Geraldine Bresolin for helpful discussions, and Katrin Lasch, Patrick Schiwek, and Manuel Söldenwagner for technical assistance.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (FU 375/4-1).
Footnotes
Published ahead of print on 16 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aballay, A., E. Drenkard, L. R. Hilbun, and F. M. Ausubel. 2003. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr. Biol. 13:47-52. [DOI] [PubMed] [Google Scholar]
- 2.Aballay, A., P. Yorgey, and F. M. Ausubel. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539-1542. [DOI] [PubMed] [Google Scholar]
- 3.Aleksic, S., J. Bockemuhl, and H. H. Wuthe. 1995. Epidemiology of Y. pseudotuberculosis in Germany, 1983-1993. Contrib. Microbiol. Immunol. 13:55-58. [PubMed] [Google Scholar]
- 4.Bolm, M., W. T. Jansen, R. Schnabel, and G. S. Chhatwal. 2004. Hydrogen peroxide-mediated killing of Caenorhabditis elegans: a common feature of different streptococcal species. Infect. Immun. 72:1192-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottone, E. J. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1:323-333. [DOI] [PubMed] [Google Scholar]
- 6.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bresolin, G., J. A. Morgan, D. Ilgen, S. Scherer, and T. M. Fuchs. 2006. Low temperature-induced insecticidal activity of Yersinia enterocolitica. Mol. Microbiol. 59:503-512. [DOI] [PubMed] [Google Scholar]
- 8.Bresolin, G., K. Neuhaus, S. Scherer, and T. M. Fuchs. 2006. Transcriptional analysis of long-term adaptation of Yersinia enterocolitica to low-temperature growth. J. Bacteriol. 188:2945-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis, G., and C. Colson. 1975. Restriction of DNA in Yersinia enterocolitica detected by recipient ability for a derepressed R factor from Escherichia coli. J. Gen. Microbiol. 87:285-291. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis, G. R., C. Sluiters, I. Delor, D. Geib, K. Kaniga, C. Lambert de Rouvroit, M. P. Sory, J. C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. [DOI] [PubMed] [Google Scholar]
- 12.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 14.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 15.Diard, M., S. Baeriswyl, O. Clermont, S. Gouriou, B. Picard, F. Taddei, E. Denamur, and I. Matic. 2007. Caenorhabditis elegans as a simple model to study phenotypic and genetic virulence determinants of extraintestinal pathogenic Escherichia coli. Microbes Infect. 9:214-223. [DOI] [PubMed] [Google Scholar]
- 16.Dorer, M. S., and R. R. Isberg. 2006. Non-vertebrate hosts in the analysis of host-pathogen interactions. Microbes Infect. 8:1637-1646. [DOI] [PubMed] [Google Scholar]
- 17.Ellison, D. W., B. Young, K. Nelson, and V. L. Miller. 2003. YmoA negatively regulates expression of invasin from Yersinia enterocolitica. J. Bacteriol. 185:7153-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, T. M., G. Bresolin, L. Marcinowski, J. Schachtner, and S. Scherer. 2008. Insecticidal genes of Yersinia spp.: taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiol. 8:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U. S. A. 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, Y., D. Zhou, X. Pang, Y. Song, L. Zhang, J. Bao, Z. Tong, J. Wang, Z. Guo, J. Zhai, Z. Du, X. Wang, X. Zhang, J. Wang, P. Huang, and R. Yang. 2004. Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol. Immunol. 48:791-805. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 23.Hares, M. C., S. J. Hinchliffe, P. C. Strong, I. Eleftherianos, A. J. Dowling, R. H. ffrench-Constant, and N. Waterfield. 2008. The Yersinia pseudotuberculosis and Yersinia pestis toxin complex is active against cultured mammalian cells. Microbiology 154:3503-3517. [DOI] [PubMed] [Google Scholar]
- 24.Heermann, R., and T. M. Fuchs. 2008. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heesemann, J., A. Sing, and K. Trülzsch. 2006. Yersinia's stratagem: targeting innate and adaptive immune defense. Curr. Opin. Microbiol. 9:55-61. [DOI] [PubMed] [Google Scholar]
- 26.Hilbi, H., S. S. Weber, C. Ragaz, Y. Nyfeler, and S. Urwyler. 2007. Environmental predators as models for bacterial pathogenesis. Environ. Microbiol. 9:563-575. [DOI] [PubMed] [Google Scholar]
- 27.Joshua, G. W., A. V. Karlyshev, M. P. Smith, K. E. Isherwood, R. W. Titball, and B. W. Wren. 2003. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149:3221-3229. [DOI] [PubMed] [Google Scholar]
- 28.Labrousse, A., S. Chauvet, C. Couillault, C. L. Kurz, and J. J. Ewbank. 2000. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 10:1543-1545. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, J. A., and J. T. Fleming. 1995. Basic culture methods, p. 3-29. In H. F. Epstein and D. C. Shakes (ed.), Caenorhabditis elegans: modern biological analysis of an organism. Academic Press, New York, NY.
- 30.Mahajan-Miklos, S., L. G. Rahme, and F. M. Ausubel. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 37:981-988. [DOI] [PubMed] [Google Scholar]
- 31.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 32.Marceau, M. 2005. Transcriptional regulation in Yersinia: an update. Curr. Issues Mol. Biol. 7:151-177. [PubMed] [Google Scholar]
- 33.Miller, W. G., and S. E. Lindow. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149-153. [DOI] [PubMed] [Google Scholar]
- 34.Motin, V. L., A. M. Georgescu, J. P. Fitch, P. P. Gu, D. O. Nelson, S. L. Mabery, J. B. Garnham, B. A. Sokhansanj, L. L. Ott, M. A. Coleman, J. M. Elliott, L. M. Kegelmeyer, A. J. Wyrobek, T. R. Slezak, R. R. Brubaker, and E. Garcia. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moy, T. I., A. R. Ball, Z. Anklesaria, G. Casadei, K. Lewis, and F. M. Ausubel. 2006. Identification of novel antimicrobials using a live-animal infection model. Proc. Natl. Acad. Sci. U. S. A. 103:10414-10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelludat, C., A. Rakin, C. A. Jacobi, S. Schubert, and J. Heesemann. 1998. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 90:6473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roggenkamp, A., H. R. Neuberger, A. Flügel, T. Schmoll, and J. Heesemann. 1995. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol. Microbiol. 16:1207-1219. [DOI] [PubMed] [Google Scholar]
- 39.Rosqvist, R., A. Forsberg, M. Rimpilainen, T. Bergman, and H. Wolf-Watz. 1990. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol. Microbiol. 4:657-667. [DOI] [PubMed] [Google Scholar]
- 40.Ruckdeschel, K., A. Roggenkamp, S. Schubert, and J. Heesemann. 1996. Differential contribution of Yersinia enterocolitica virulence factors to evasion of microbicidal action of neutrophils. Infect. Immun. 64:724-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sicard, M., S. Hering, R. Schulte, S. Gaudriault, and H. Schulenburg. 2007. The effect of Photorhabdus luminescens (Enterobacteriaceae) on the survival, development, reproduction and behaviour of Caenorhabditis elegans (Nematoda: Rhabditidae). Environ. Microbiol. 9:12-25. [DOI] [PubMed] [Google Scholar]
- 42.Sifri, C. D., J. Begun, and F. M. Ausubel. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13:119-127. [DOI] [PubMed] [Google Scholar]
- 43.Sifri, C. D., J. Begun, F. M. Ausubel, and S. B. Calderwood. 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71:2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stiernagle, T. 11 February 2006, posting date. Maintenance of C. elegans. In WormBook. doi 10.1895/wormbook.1.101.1. The C. elegans Research Community. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 45.Straley, S. C., and R. R. Brubaker. 1981. Cytoplasmic and membrane proteins of yersiniae cultivated under conditions simulating mammalian intracellular environment. Proc. Natl. Acad. Sci. U. S. A. 78:1224-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straley, S. C., and R. D. Perry. 1995. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 3:310-317. [DOI] [PubMed] [Google Scholar]
- 47.Styer, K. L., G. W. Hopkins, S. S. Bartra, G. V. Plano, R. Frothingham, and A. Aballay. 2005. Yersinia pestis kills Caenorhabditis elegans by a biofilm-independent process that involves novel virulence factors. EMBO Rep. 6:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenor, J. L., B. A. McCormick, F. M. Ausubel, and A. Aballay. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 14:1018-1024. [DOI] [PubMed] [Google Scholar]
- 50.Thomsen, L. E., S. S. Slutz, M. W. Tan, and H. Ingmer. 2006. Caenorhabditis elegans is a model host for Listeria monocytogenes. Appl. Environ. Microbiol. 72:1700-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trülzsch, K., T. Sporleder, E. I. Igwe, H. Russmann, and J. Heesemann. 2004. Contribution of the major secreted Yops of Yersinia enterocolitica O:8 to pathogenicity in the mouse infection model. Infect. Immun. 72:5227-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterfield, N., M. Hares, S. Hinchliffe, B. Wren, and R. ffrench-Constant. 2007. The insect toxin complex of Yersinia. Adv. Exp. Med. Biol. 603:247-257. [DOI] [PubMed] [Google Scholar]
- 53.Waterfield, N. R., B. W. Wren, and R. H. ffrench-Constant. 2004. Invertebrates as a source of emerging human pathogens. Nat. Rev. Microbiol. 2:833-841. [DOI] [PubMed] [Google Scholar]
- 54.Wolf-Watz, H., D. A. Portnoy, I. Bolin, and S. Falkow. 1985. Transfer of the virulence plasmid of Yersinia pestis to Yersinia pseudotuberculosis. Infect. Immun. 48:241-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong, D., D. Bazopoulou, N. Pujol, N. Tavernarakis, and J. J. Ewbank. 2007. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 8:R194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.