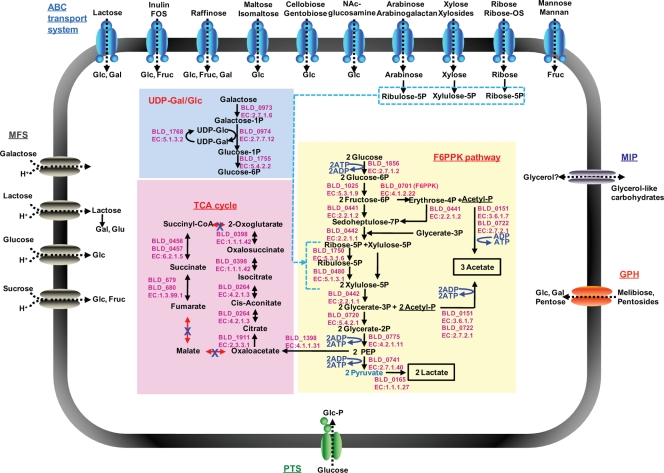
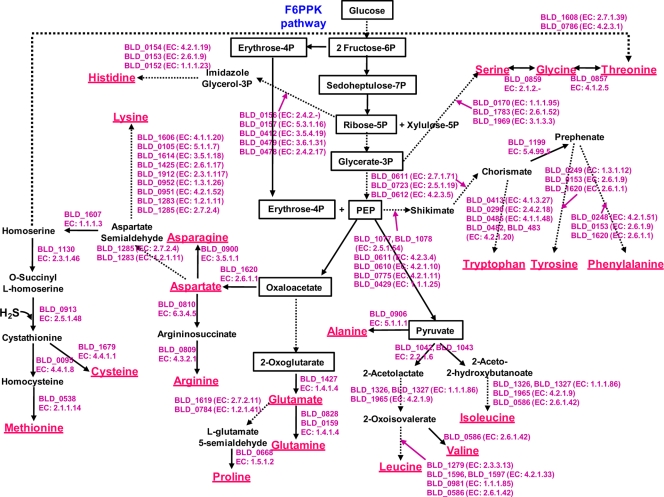
Abstract
Summary: Since the discovery in 1899 of bifidobacteria as numerically dominant microbes in the feces of breast-fed infants, there have been numerous studies addressing their role in modulating gut microflora as well as their other potential health benefits. Because of this, they are frequently incorporated into foods as probiotic cultures. An understanding of their full interactions with intestinal microbes and the host is needed to scientifically validate any health benefits they may afford. Recently, the genome sequences of nine strains representing four species of Bifidobacterium became available. A comparative genome analysis of these genomes reveals a likely efficient capacity to adapt to their habitats, with B. longum subsp. infantis exhibiting more genomic potential to utilize human milk oligosaccharides, consistent with its habitat in the infant gut. Conversely, B. longum subsp. longum exhibits a higher genomic potential for utilization of plant-derived complex carbohydrates and polyols, consistent with its habitat in an adult gut. An intriguing observation is the loss of much of this genome potential when strains are adapted to pure culture environments, as highlighted by the genomes of B. animalis subsp. lactis strains, which exhibit the least potential for a gut habitat and are believed to have evolved from the B. animalis species during adaptation to dairy fermentation environments.
INTRODUCTION
Taxonomy of Bifidobacteria
The phylum Actinobacteria is one of the largest phyla in the domain Bacteria, with 5 subclasses, 6 orders, and 14 suborders, all of which are high-G+C Gram-positive bacteria, except for Tropheryma whipplei, which has a 46.3% G+C content (82, 325). In this phylum, the order Bifidobacteriales contains two families, including Bifidobacteriaceae and Incertae (93). The most recent Taxonomic Outline of Bacteria and Archaea, release 7.7 (TOBA 7.7), suggests that the Bifidobacteriaceae family should be divided into five genera: Bifidobacterium, Aeriscardovia, Gardnerella, Parascardovia, and Scardovia (93). The addition of the genera Aeriscardovia, Parascardovia, and Scardovia was suggested based on their different DNA G+C contents and partial heat shock protein 60 (Hsp60) gene sequences of three Bifidobacterium species, namely, Bifidobacterium aerophilum, B. denticolens, and B. inopinatum, respectively (132, 291). These three species are therefore no longer considered part of the Bifidobacterium genus. The addition of the genus Gardnerella was proposed based on 16S rRNA gene sequence analysis (298). While 16S rRNA gene sequence analysis is the most common approach for calculating microbial phylogeny, a multigene approach can give a more accurate assessment of phylogeny within a genus. This has proven useful for the Bifidobacterium genus, and several genes have been utilized for this purpose, such as recA (154), ldh (259), and tuf (324).
Bifidobacteria were first isolated from the feces of breast-fed infants in 1899, by Henri Tissier, and were designated Bacillus bifidus (314, 315). Even though Orla-Jensen proposed the genus Bifidobacterium in 1924 (213), bifidobacteria were classified into other taxonomic groups, such as Bacillus bifidus (1900), Bacteroides bifidus (1923 to 1934, in the 1st to 4th editions of Bergey's Manual of Systematic Bacteriology [Bergey's Manual]), and Lactobacillus bifidus (1939 to 1957, in the 5th to 7th editions of Bergey's Manual), for several decades. In 1973, Poupard et al. (234), and subsequently the 8th edition of Bergey's Manual (24), reclassified them as a separate taxon and designated the genus Bifidobacterium, consisting of 11 species. Scardovi (269) updated this to 24 species in 1986, and currently there are 31 proposed species that have been isolated from the intestines of humans, animals, and insects, and also from human dental caries and raw milk (Table 1).
TABLE 1.
Currently assigned species of the genus Bifidobacterium
| Species no.a | Nameb | Subspecies | Originc | %G+Cd | Reference |
|---|---|---|---|---|---|
| 1 | B. adolescentis | Intestine of adult | 59.6 ± 0.8 | 248 | |
| 2 | B. angulatum | Human feces | 59.0 ± 0.1 | 270 | |
| 3 | B. animalis | B. animalis subsp. animalis | Animal feces | 60.1 ± 0.3 | 189 |
| B. animalis subsp. lactis | Yogurt | 61.9 | 185 | ||
| 4 | B. asteroides | Intestine of honeybee | 59.0 | 272 | |
| 5 | B. bifidum | Infant feces | 62.3 | 315 | |
| 6 | B. bombi | Intestine of bumblebee | 47.2 | 140 | |
| 7 | B. boum | Rumen of cattle | 60 ± 0.2 | 273 | |
| 8 | B. breve | Intestine of infant | 58.8 ± 0.4 | 248 | |
| 9 | B. catenulatum | Intestine of adult | 54.0 ± 0.2 | 270 | |
| 10 | B. choerinum | Porcine feces | 66.3 ± 0.2 | 273 | |
| 11 | B. coryneforme | Intestine of honeybee | ND | 28 | |
| 12 | B. crudilactis | Raw milk | 56.4 | 68 | |
| 13 | B. cuniculi | Feces of rabbit | 64.1 ± 0.4 | 273 | |
| 14 | B. dentium | Human dental caries | 61.2 ± 0.4 | 270 | |
| 15 | B. gallicum | Human feces | ND | 159 | |
| 16 | B. gallinarum | Chicken cecum | 65.7 ± 1.5 | 343 | |
| 17 | B. indicum | Intestine of honeybee | 60.0 | 272 | |
| 18 | B. longum | B. longum subsp. infantis | Intestine of infant | 60.5 ± 0.3 | 248 |
| B. longum subsp. longum | Intestine of adult | 60.8 ± 0.8 | 248 | ||
| B. longum subsp. suis | Porcine feces | 62.0 | 179 | ||
| 19 | B. magnum | Rabbit feces | 60.0 ± 0.6 | 274 | |
| 20 | B. merycicum | Bovine rumen | ND | 26 | |
| 21 | B. minimum | Sewage | 61.5 | 28 | |
| 22 | B. pseudocatenulatum | Infant feces | 57.5 ± 0.3 | 273 | |
| 23 | B. pseudolongum | B. pseudolongum subsp. globosum | Bovine rumen | 63.8 ± 0.4 | 28 |
| B. pseudolongum subsp. pseudolongum | Porcine feces | 59.5 ± 0.4 | 346 | ||
| 24 | B. pyschraerophilum | Porcine feces | 59.2 | 291 | |
| 25 | B. pullorum | Chicken feces | 67.5 ± 0.4 | 318 | |
| 26 | B. ruminantium | Bovine rumen | ND | 26 | |
| 27 | B. saeculare | Rabbit feces | ND | 27 | |
| 28 | B. scardovii | Human blood | ND | 123 | |
| 29 | B. subtile | Sewage | 61.5 | 28 | |
| 30 | B. thermophilum | Porcine feces | 60.0 | 189 | |
| 31 | B. thermacidophilum | B. thermacidophilum subsp. porcinum | Porcine feces | ND | 354 |
| B. thermacidophilum subsp. thermacidophilum | Sewage | ND | 77 |
There are 31 Bifidobacterium species to date.
B. aerophilum, B. denticolens, and B. inopinatum were reclassified as Aeriscardovia aeriphila, Parascardovia denticolens, and Scardovia inopinata, respectively.
Environment from which the species was initially isolated.
Mean ± SD. ND, not determined.
Relying solely on a single genetic marker, such as the 16S rRNA gene, for bacterial classification does have limitations. Whole-genome approaches can provide much-more-meaningful taxonomic data. Such phylogenetic analyses have been evaluated for many bacterial genomes and have provided evidence for both organism and gene taxonomic families (36, 41). A phylogenomic analysis of the Lactobacillus genus has suggested that GroEL is a better phylogenetic marker for this genus than the 16S rRNA gene (49). Since genome sequences for the Bifidobacterium genus are becoming available, a whole-genome phylogenomic approach will provide insights into the classification of this fast-growing genus.
General Characteristics
Bifidobacteria are nonmotile, non-spore-forming, non-gas-producing, Gram-positive, anaerobic, catalase-negative bacteria with a high G+C content (55 to 67%) (146, 269, 336). Their morphology is generally referred to as bifid or irregular V- or Y-shaped rods resembling branches. The actual reason for the irregular shape of bifidobacteria is not yet clearly understood. However, a few studies have revealed that the absence or low concentrations of N-acetylamino-sugar (100), Ca2+ ions (148-150), or amino acids (alanine, aspartic acid, glutamic acid, and serine) (125) in growth media exclusively induce the bifid shape of bifidobacteria.
Early Studies of Bifidobacteria
After the discovery of bifidobacteria in the feces of breast-fed infants, Tissier (315) suggested that the large number of bifidobacteria in the feces of healthy breast-fed infants was likely the reason for their lower incidence of infantile diarrhea. In his pediatric work, he used bifidobacteria for the treatment of this intestinal diarrhea, and this likely represents the first example of the oral administration of a live microorganism for the treatment of a disease (316). The abundance of bifidobacteria in the feces of breast-fed infants was thought to be due to the Bifidobacterium-stimulating properties of human breast milk (38, 46, 60, 96, 196, 321). Numerous studies have substantiated the higher bifidobacterial counts and lower incidences of gastroenteritis in breast-fed infants than in formula-fed infants (3, 40, 51, 98). Early attempts to make infant formula resemble human breast milk in order to promote bifidobacteria in the intestine were unsuccessful (1, 96). Subsequent studies found the presence of bifidobacterial growth-promoting factors in human breast milk, such as lactulose (151, 231) and N-acetylglucosamine-containing saccharides and other human milk oligosaccharides (HMOs) (108, 109), supporting the association between large bifidobacterial numbers and human breast milk. These HMOs consist of short-chain trisaccharides, such as sialyllactose or fucosyllactose, and complex, high-molecular-weight glycans, such as N-acetyllactosamine polymers (31). The stimulation of bifidobacteria by human breast milk, resulting in their prevalence in the gut, was proposed to be involved in the suppression of undesirable intestinal bacteria (38). This hypothesis was tested by comparing the compositions of intestinal bacteria in the feces of breast-fed and formula-fed infants, and the proposed increase in bifidobacteria and concomitant decrease in undesirable microbes were observed (352). This inhibition effect by bifidobacteria was supported by fecal pH measurements, as the fecal pH of bottle-fed infants was found to be >7.0, and that of breast-fed infants was <6.0, during the first 7 weeks after birth (39). This is most likely due to the production of lactic and acetic acids by bifidobacteria.
Due to the potential health benefits of bifidobacteria, they have been recommended as dietary supplements by many authors throughout the last century (180, 227, 244, 263, 316). Freeze-dried bifidobacterial preparations, sometimes with Lactobacillus acidophilus, have been used for the treatment of gastrointestinal (GI) disorders (235, 246). The addition of lactulose with bifidobacteria in milk was reported to have beneficial effects in the treatment of hepatic encephalopathy (186, 199). Currently, bifidobacteria are added to numerous foods, specifically for their perceived probiotic activities.
Microbial Diversity and the Role of Bifidobacteria in the Large Intestine
The human large intestine is a very complex ecosystem that is still not fully understood, and while its microbial composition consists primarily of just four bacterial phyla, Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria (as well as some Archaea, Eukarya, and viruses), it is highly variable at the genus level between individuals, likely due to factors such as age, health, diet, etc. (25, 99, 127, 244). Therefore, the adaptation capabilities of the intestinal bacteria corresponding to these variable factors likely influence the overall composition of intestinal microflora in the intestine (89).
Culturing and nonculturing analysis.
The composition of the microflora in the human large intestine, as estimated using culturing techniques, is usually dominated by the genera Bacteroides, Eubacterium, and Bifidobacterium, with several other predominant genera, such as Clostridium, Peptostreptococcus, Enterococcus, Lactobacillus, and members of the family Enterobacteriaceae (117, 118). The number of species estimated by culturing techniques is approximately 400. Nonculturing analysis of the gut microflora was greatly facilitated by the direct isolation of DNA from feces and by amplification of the 16S rRNA genes representing the entire microflora. Cloning and sequencing of individual rRNA genes enabled the numerically dominant genera of bacteria to be identified. This molecular analysis of the intestinal microflora in fecal and colonic samples initially suggested that the human large intestine contains more than 500 different bacterial species and that about 75% of them are nonculturable (81, 83). However, a recent extensive metagenomic analysis revealed that this is an overestimation, with most individuals harboring approximately 160 different bacterial species, the majority of which are “known” (239). While the total number of species present in the human gut is not known, recent molecular studies indicate that it is in excess of 1,000 (239, 241), with an upper estimate of 1,150 suggested by an extensive metagenomic analysis of 124 individuals (239).
Unlike the results of culturing studies of the microflora composition in the large intestine, molecular studies utilizing 16S rRNA gene sequencing revealed only a relatively small number of high-G+C Gram-positive Actinobacteria, including bifidobacteria (83, 99). It can be argued that the universal primers used to amplify the 16S rRNA gene may preferentially select certain groups of bacteria, and there may also be biases in DNA isolation due to differences in cell wall characteristics. While some metagenomic studies have found similar microbial distributions to those in 16S rRNA gene analyses (99), others have shown the Actinobacteria to be more prominent (156, 239). These studies substantiate Bacteroides as the major genus of the identifiable gut microbiome in adults and weaned infants, followed by the Firmicutes family, primarily the genera Eubacterium, Ruminococcus, and Clostridium, and the genus Bifidobacterium. In unweaned infants, there was significant individual variation, but Bifidobacterium was generally the most dominant identifiable genus, followed by the members of the Enterobacteriaceae family (156). The recent metagenomic analysis of 124 individuals provided a more comprehensive picture of the composition of the human intestinal microbiota as well as the minimum metagenome shared among people (239). Based on this metagenomic analysis, the human intestinal microbiota consists of two major phyla, Bacteroidetes and Firmicutes, and four other prominent phyla, Actinobacteria, Proteobacteria, Fusobacteria, and Tenericutes, although significant individual variations are present. Interestingly, this analysis showed that the Actinobacteria phylum, including Bifidobacterium, was relatively abundant in all participants, which is quite different from previous reports obtained using 16S rRNA gene sequence analysis (83, 99). This study also revealed an extensive list of bacterial functions, particularly metabolite production, that are important for human health but are encoded only in the metagenome (239). These large-scale studies on the intestinal microflora in different individuals are greatly increasing our knowledge of its diversity and function.
Role of bifidobacteria in the large intestine.
Our understanding of intestinal microbe-host interactions and their symbiosis is growing, but it is still in its infancy. The role of bifidobacteria in the intestinal microbiota is not known, but studies have suggested a likely beneficial role for the host. However, given their significantly higher incidence in the unweaned infant gut than in adults, they may play a more important role in gut microbiota development than in other gut functions. Studies have also revealed bifidobacterial interactions with other gut microbes. For example, cocolonization of gnotobiotic mice with bifidobacteria and Bacteroides revealed that the presence of bifidobacteria expanded the diversity of polysaccharides degraded by Bacteroides, suggesting a synergistic effect of bifidobacteria with other intestinal microbiota on digestion of polysaccharides which are indigestible by the host (295). Another possible role of bifidobacteria in the large intestine is the production of water-soluble vitamins, such as many of the B group of vitamins, as shown previously (67) and also substantiated by the genome analysis discussed below. An important role of bifidobacteria in the large intestine is believed to be modulation of certain bacterial groups that may be detrimental to the host. Numerous studies support their competitive abilities against other intestinal microbes (14, 120, 251, 252). In addition, in vitro studies have shown that bifidobacteria can produce antimicrobial compounds such as organic acids (269), iron-scavenging compounds (214), and bacteriocins (43, 161, 351). An intriguing function of bifidobacteria in the intestine may be in protection against some immune-based disorders, as numerous studies have shown them to stimulate a host innate immune response (13, 116, 209, 295). These inflammatory and immune defense responses are proposed to be triggered by recognition of commensal bifidobacteria via Toll-like receptors (TLRs) of the host innate immune system (242). Furthermore, cell wall constituents and unmethylated CpG DNA motifs of bifidobacteria are also believed to be involved in modulation of innate immune responses (8, 166).
The role of bifidobacteria in controlling the levels of undesirable bacteria, such as clostridia and Escherichia coli, in the intestine is inferred from the reverse correlation of bifidobacterial numbers with these bacteria from clinical feeding studies (6, 45, 176). This correlation is also evident in the intestinal microbial changes that occur as people grow older: specifically, the number of bifidobacteria decreases significantly, while the number of other bacteria, including clostridia and E. coli, increases (127, 190). This correlation has been substantiated by both culturing and nonculturing methods, using fluorescently labeled probes that detect specific microbes directly in feces (121). The levels of bifidobacteria may also correlate with lower levels of putrefactive products, as the levels of ammonia, indole, p-cresol, and phenol, as well as enzymes involved in their production (urease, tryptophanase, and β-glucuronidase), were very low in an infant whose fecal microflora consisted of 96% bifidobacteria compared to those in an adult with a bifidobacterial concentration of 19% (127). This suggests that the balance of the intestinal microflora by supplementation of bifidobacteria is important for maintaining intestinal health (127).
Potential Health Benefits
Numerous studies have suggested that the presence of bifidobacteria in the human large intestine is associated with many human health benefits. However, some of these studies have been criticized for poor design and weak conclusions. This section critically analyzes the scientific credibility of the involvement of bifidobacteria in these possible health benefits.
Prevention of diarrhea.
The prevention and treatment of diarrhea were the first proposed benefits of bifidobacteria (316). Some studies have since examined the role of bifidobacterial supplementation in protection from diarrhea onset and from the most prevalent cause of viral diarrhea, rotavirus (Table 2). The majority of these studies have used animal models, and few show convincing statistically significant data for large groups. The human studies that have been conducted have been either not statistically significant or barely so. While bifidobacteria are likely to protect the intestine from diarrheal diseases, the added protective effect of taking bifidobacterial probiotics is not very convincing based on current studies. It is possible that individuals with high levels of bifidobacteria may not benefit from extra supplementation, and since initial bifidobacterial levels were not considered in any of these studies, this may have resulted in the underwhelming effects.
TABLE 2.
Summary of clinical studies linking bifidobacteria with potential health benefits
| Health benefit | Study summary | Reference |
|---|---|---|
| Prevention of diarrhea | Large murine study (29 to 41 mice per group) showing a statistically significant (P < 0.001) protective effective of B. bifidum and B. longum subsp. infantis supplementation against rotaviral diarrhea | 238 |
| Large murine study (22 to 46 mice per group) with non-statistically significant data, but proposing a protective effect of heat-killed B. breve against rotaviral diarrhea | 347 | |
| Large murine study (52 to 111 mice per group) showing a small, statistically significant (P < 0.05) protective effective of B. bifidum supplementation, with reduced shedding of rotavirus (P < 0.01) | 80 | |
| Small piglet study (8 or 9 piglets per group) showing a moderate, statistically significant (P < 0.01) protective effect of B. animalis subsp. lactis against all forms of diarrhea and a higher titer of antirotaviral antibodies in the feces | 288 | |
| Large human study (26 or 29 infants per group) showing a barely statistically significant (P = 0.035) protective effect of B. bifidum and Streptococcus thermophilus supplementation, with reduced shedding of rotavirus (P = 0.025) | 262 | |
| Large human study (44 or 46 infants per group) showing a non-statistically significant protective effect of B. animalis subsp. lactis | 47 | |
| Large human study (464 or 449 infants per group) with B. breve and S. thermophilus supplementation; did not show a decrease in number of diarrhea episodes over a 5-month period but did show a reduced severity (P < 0.01) | 312 | |
| Small preterm pig study (5 to 13 pigs per group) showing some reduction (P < 0.05) in the incidence of necrotizing enterocolitis from supplementation with B. animalis and 4 species of Lactobacillus | 290 | |
| Establishment of a healthy microflora in premature infants | Small human study (10 preterm infants per group) showing that supplementation with B. breve resulted in establishment of a bifidobacterial flora in the majority of infants during the first week of life, whereas it took the control group several weeks, with only 3 of 9 infants showing bifidobacteria by week 7 | 167 |
| Large human study (33 preterm infants per group) showing that supplementation with B. breve reduced (P < 0.05) fecal butyric acid levels, but only in the subgroup of infants that weighed <2,500 g | 342 | |
| Colon regularity | Small human study (17 subjects with irritable bowel syndrome [IBS] per group) showing some reduction (P < 0.05) in colonic transit times from supplementation with B. animalis subsp. lactis and yogurt cultures | 2 |
| Large human study (132 or 135 subjects with IBS per group) showing no statistically significant reduction in colonic transit times, except in a small subset of 19 subjects who had <3 stools/week initially, from supplementation with B. animalis subsp. lactis and yogurt cultures | 106 | |
| Small human study (15 or 17 women per group) showing some reduction (P < 0.05) in colonic transit times from supplementation with B. animalis subsp. lactis | 172 | |
| Large human study (100 elderly subjects), without a control group, showing a reduction (P < 0.001) in colonic transit after 2 weeks of supplementation with B. animalis subsp. lactis | 184 | |
| Lactose intolerance | Small human study (15 people with lactose intolerance) showing some reduction (P < 0.05) in breath hydrogen from supplementation with B. longum | 133 |
| Small human study (11 Chinese individuals with lactose intolerance) showing some reduction (P < 0.05) in symptom scores from supplementation with B. animalis subsp. lactis and yogurt cultures | 114 | |
| Cholesterol reduction | Small human study (7 subjects per group) showing some reduction (P < 0.05) in serum cholesterol levels following supplementation with B. animalis subsp. lactis and L. acidophilus | 15 |
| Small human study (11 or 18 women per group) showing no reduction in total cholesterol but an increase in high-density lipoprotein (HDL) levels (P = 0.001) | 139 | |
| Medium-sized human study (37 women), without a control group, showing that supplementation with B. longum and L. acidophilus did not affect cholesterol levels | 102 | |
| Immunostimulatory effects | Small mouse study (5 mice per group) showing some reduction (P < 0.05) in CD4+ T cells in the spleen and colon following supplementation with B. bifidum | 142 |
| Small mouse study (10 mice per group) showing some reduction in the proinflammatory cytokines IFN-γ, TNF-α, and IL-12 from supplementation with B. longum subsp. infantis | 182 | |
| Small mouse study (5 mice per group) showing some increase (P < 0.05) in mucosal IgA following supplementation with B. longum | 303 | |
| Large human study (5 groups of 13 to 15 subjects receiving different amounts of B. animalis subsp. lactis and Lactobacillus paracasei) showing no statistically significant changes in cytokine levels | 48 | |
| Small human study (12 or 13 elderly subjects per group) showing some increase (P < 0.05) in the anti-inflammatory cytokine IFN-α and in phagocytic activity following 6 weeks of supplementation with B. animalis subsp. lactis | 13 | |
| Small human study (8 subjects with ulcerative colitis per group) showing no significant change in symptom scores but showing some decrease (P < 0.05) in expression of genes encoding human proinflammatory cytokines from supplementation with B. longum, inulin, and fructooligosaccharides | 90 | |
| Large human study (77 subjects with IBS receiving 1 × 1010 live cells of B. longum subsp. infantis) showing reductions in symptom scores and in the ratio of IL-10 to IL-12 (anti-inflammatory to proinflammatory cytokines), normalized to that of healthy individuals | 210 | |
| Cancer prevention | Medium-sized mouse study (12 mice per group) showing some reduction (P < 0.01) in the incidence of tumors when heat-killed B. infantis cells or cell wall preparations were injected into mice along with tumor cells, with a significant increase (P < 0.001) in the number of mice that were cured of tumors | 279 |
| Large mouse study (4 groups of 15 mice) showing some decrease (P < 0.05) in carcinogen-induced aberrant crypt foci following supplementation with B. longum and a significant decrease (P < 0.001) following cosupplementation with B. longum and inulin | 257 | |
| Large mouse study (30 mice per group) showing a significant reduction (P < 0.001) in carcinogen-induced colonic neoplasms following supplementation with both B. animalis subsp. lactis and resistant starch but no reduction with either supplement individually | 163 | |
| Large human study (4 groups of 18 to 22 colon cancer or polypectomized patients) showing some improvement (P < 0.05) in epithelial barrier function and cell toxicity only in polypectomized patients following supplementation with B. animalis subsp. lactis, L. rhamnosus, and inulin | 240 |
Establishment of a healthy microflora in premature infants.
The rapid establishment of a healthy intestinal microflora in premature infants is believed to be important for mucosal host defense and the prevention of certain intestinal infections (60). Some studies have been conducted to evaluate the effect of early oral supplementation with bifidobacterial probiotics on preterm infants (Table 2). It appears that early supplementation does decrease the time for a bifidobacterial population to develop, but this would be expected when large numbers are ingested into an immature gut. While this may offer some protective effects, supplementation with Bifidobacterium longum and Lactobacillus rhamnosus was not found to reduce the time needed for a feeding tube in a study of 45 infants compared to a 49-infant control group (256).
Colon regularity.
Bifidobacteria may also be useful for the treatment of constipation in the elderly, as their small population of bifidobacteria may be a contributing factor. This was first suggested by the observation of subjects who ingested bifidobacteria and reported a noticeable lubricant effect during fecal passage (216). There have been a number of recent studies which appear to support some effect of bifidobacterial supplementation on reducing colonic transit time in individuals who suffer from long colonic transit times (Table 2). While these studies do suggest a role for bifidobacterial supplementation in colonic transit, they tended to focus on individuals who already had GI problems, not on healthy people. In addition, there was no attempt in any study to understand the mechanism of action. A likely mechanism of action is the production of exopolysaccharides (EPS), which may function as a laxative.
Lactose intolerance.
Although dairy foods are recommended components of a healthy diet, some people have discomfort digesting dairy products due to a shortage of the enzyme lactase (58, 124). Fermented foods appear to be more suitable for lactose-intolerant patients than nonfermented foods because of the decreased lactose concentration. Some studies have suggested that the addition of large numbers of lactose-digesting cultures, such as bifidobacteria, to dairy products may also alleviate the symptoms of lactose intolerance (Table 2). While these studies are quite small and the results are statistically significant only at a P level of <0.05, this effect is generally accepted as scientifically sound, presumably because the mechanism of action is clear.
Cholesterol reduction.
Some studies have been conducted to evaluate if supplementation with bifidobacteria can reduce serum cholesterol levels (Table 2). While some of these studies suggest an effect, it is not generally accepted as significant. Part of the reason is the difficulty in obtaining an accurate baseline cholesterol measurement, as levels are known to fluctuate. This calls the statistical significance of effects into question. A possible mechanism has been proposed, as some bifidobacteria have been shown to assimilate cholesterol into their cell membranes (62).
Immunostimulatory effects.
A number of studies have suggested that the interaction between bifidobacteria and host mucosal cells can result in immunomodulatory effects in the host (Table 2). While the studies are generally small and only just statistically significant, they do suggest some effect, although the one large human study did not show a significant difference. This would be weak evidence, at best, if the effect was mechanistically unclear. However, it is known that the immune system responds to microbes in the gut, and recent molecular evidence obtained using transcription profiling has shown that cecal epithelial cells respond differently to B. longum, which specifically increases gamma interferon (IFN-γ), and Bacteroides thetaiotaomicron, which induces tumor necrosis factor alpha (TNF-α) (295). While both of these cytokines are proinflammatory cytokines, these findings do demonstrate that different bacteria have different immunomodulatory stimulatory effects.
Cancer prevention.
Cancers of the gut are prevalent and are believed to have both genetic and dietary causes. Some studies have examined the potential ability of bifidobacterial supplementation to prevent cancers (Table 2). While these are mainly mouse studies, they suggest that the combination of bifidobacteria and a prebiotic may reduce the probability of carcinogen-induced cancerous cells in mice. However, there are no human trials with convincing evidence showing a protective effect of this type of supplementation.
While numerous studies have found health benefits of ingesting bifidobacteria, some studies have not been able to demonstrate benefits. Differences are believed to be due to the different strains of bifidobacteria used, as many of these effects may be dependent on the strain used as well as on host-specific characteristics. The selection of proper strains and further understanding of the microbial traits necessary for specific health benefits are needed to optimize their potential.
Detection and Identification
Three principal approaches have been developed for isolation, detection, and identification of bifidobacteria from feces: traditional culturing methods using selective media for selection and identification, culture-free molecular methods for detection, and molecular methods for identification and differentiation.
The first selective medium developed for bifidobacteria was lactose-cystine-liver (LCL) agar (30), but it was inhibitory to some bifidobacterial strains. Norris et al. (207) improved Bifidobacterium-selective media by including growth factors (pancreatin, sorbitan monooleate, ascorbic acid, and vitamin B12), and these were inhibitory to other intestinal bacteria, except for enterococci. The identification of specific bifidobacterial growth factors from human milk further enhanced the recovery of bifidobacteria (108, 109). In the last few decades, various selective media have been developed and are listed in Table 3. Some are based on the use of antibiotics, such as neomycin, polymyxin B, nalidixic acid, and/or mupirocin, for selectivity, while others utilize inhibitory agents such as lithium chloride and propionate.
TABLE 3.
Various selective media for isolation and enumeration of bifidobacteria
| Medium | Selectivity | Reference(s) |
|---|---|---|
| Antibiotic-based media | ||
| NPNL | Neomycin, paromomycin, nalidixic acid, LiCl | 311 |
| BIM-25 | Polymyxin B, nalidixic acid, iodoacetic acid, 2,3,5-triphenyltetrazolium chloride | 197 |
| MRS (TPY) + Dic | Dicloxacillin | 297 |
| RMS | Neomycin, paromomycin, LiCl, sodium propionate | 267 |
| AMC | Polymyxin B, nalidixic acid, iodoacetic acid, 2,3,5-triphenyltetrazolium chloride | 11, 12 |
| BL-OG | Oxgall, gentamicin | 168 |
| BSM | Mupirocin | 164 |
| Antibiotic-free media | ||
| mCABa | Propionic acid | 19 |
| LP | LiCl, sodium propionate | 158 |
| TP | Transgalactooligosaccharide, sodium propionate | 131 |
| RB | LiCl, sodium propionate | 112 |
| RAF 5.1 | LiCl, sodium propionate | 258 |
| BFM | LiCl, propionic acid, methylene blue | 204 |
| RCA-aniline blue | Propionic acid, aniline blue | 169 |
Beeren's medium.
The advent of molecular tools has greatly enhanced approaches for detection, differentiation, and identification of bifidobacteria. Many of these are cultivation-free methods allowing in situ analysis, thus removing the limitations of culturing. The development and use of these molecular tools for bifidobacteria are summarized in Table 4.
TABLE 4.
Molecular detection and identification methods used for bifidobacteria
| Methoda | Description | Reference(s) |
|---|---|---|
| DNA fingerprinting methods | ||
| PCR approaches | ||
| AP-PCR | Use of a single arbitrary primer to obtain strain-specific banding patterns; can be subject to reproducibility problems | 341, 344 |
| TAP-PCR | Triplicate AP-PCR incorporating three different annealing temperatures to improve reproducibility of profiles | 59 |
| ERIC-PCR | AP-PCR using ERIC-PCR primers | 289, 333 |
| ARDRA | RFLP analysis of the ldh gene | 259 |
| RFLP analysis of the 16S rRNA gene | 153, 330 | |
| PFGE | Band profile analysis of complete genome by use of rare-cutting enzymes | 292, 338 |
| Molecular identification by sequence analysis of: | ||
| 16S rRNA gene | 136, 174, 338 | |
| groEL/groES | 329 | |
| recA | 154 | |
| grpE/dnaK | 339 | |
| ldh | 259 | |
| tuf | 324 | |
| atpD | 326 | |
| Transaldolase gene | 245 | |
| Molecular quantification by RT-PCR | Real-time PCR quantification of bifidobacteria in feces, using genus- or species-specific primers | 245 |
| FISH | Hybridization of fluorescently labeled genus/species-specific probes to bifidobacteria in feces | 157 |
AP-PCR, arbitrarily primed PCR (also referred to as random amplified polymorphic DNA [RAPD] analysis); ERIC-PCR, enterobacterial repetitive intergenic consensus sequence PCR; ARDRA, amplified rRNA gene restriction analysis; RFLP, restriction fragment length polymorphism; PFGE, pulsed-field gel electrophoresis; FISH, fluorescent in situ hybridization.
PLASMIDS OF BIFIDOBACTERIA
Plasmid Analysis
Plasmids are not commonly detected in bifidobacteria, with approximately 20% of isolated bifidobacteria containing detectable plasmids (281). Currently, plasmids have been detected in 8 of the 31 species of Bifidobacterium, including B. longum subsp. longum, B. pseudolongum subsp. globosum, B. indicum, B. asteroides (282, 283), B. breve (128), B. bifidum (287), B. catenulatum (5), and B. pseudocatenulatum (97). Some plasmids have been sequenced completely, revealing primarily rolling circular plasmids varying in size from 1.8 to 10.2 kb (Table 5).
TABLE 5.
Completely sequenced plasmids from bifidobacteria
| Host species (G+C content [%]a) | Plasmid | Size (bp) | G+C content (%) | Host G+C content (%)b | Reference or GenBank accession no. |
|---|---|---|---|---|---|
| B. longum subsp. longum (60.8 ± 0.8) | pNAC2 | 3,684 | 64.7 | 53 | |
| pTB6 | 3,624 | 65.1 | 309 | ||
| pB44 | 3,624 | 65.1 | 287 | ||
| pKJ36 | 3,625 | 65.1 | 224 | ||
| pMG1 | 3,682 | 65.1 | 222 | ||
| pBLO1 | 3,626 | 64.8 | 60.1 | 275 | |
| P6043B | 3,680 | 65.1 | DQ458911 | ||
| pNAC1 | 3,538 | 58.8 | 53 | ||
| pNAL8L | 3,489 | 59.0 | 104 | ||
| pKJ50 | 4,960 | 61.8 | 225 | ||
| pNAL8 M | 4,910 | 61.9 | 104 | ||
| pBIFA24 | 4,892 | 61.8 | 226 | ||
| p6043A | 4,896 | 61.8 | DQ458910 | ||
| pNAC3 | 10,224 | 62.0 | 53 | ||
| pDOJH10L | 10,073 | 62.2 | 60.2 | 162 | |
| pMB1 | 1,847 | 62.0 | 178 | ||
| pDOJH10S | 3,661 | 66.2 | 60.2 | 162 | |
| pFI2576 | 2,197 | 61.9 | 194 | ||
| B. breve (58.8 ± 0.4) | pCIBb1 | 5,750 | 56.9 | 212 | |
| pNBb1 | 2,297 | 58.7 | E17316 | ||
| pB21a | 5,206 | 56.3 | 287 | ||
| B. pseudolongum subsp. globosum (63.8 ± 0.4) | pASV479 | 4,815 | 59.3 | 268 | |
| B. bifidum (62.3) | pB80 | 4,898 | 61.9 | 287 | |
| pBIF10 | 9,275 | 41.7 | DQ093580 | ||
| B. asteroides (59.0) | pCIBAO89 | 2,111 | 52.3 | 55 | |
| pAP1 | 2,140 | 52.3 | Y11549 | ||
| B. catenulatum (54.0 ± 0.2) | pBC1 | 2,540 | 63.7 | 5 | |
| B. pseudocatenulatum (57.5 ± 0.3) | p4 M | 4,488 | 53.1 | 97 |
Host G+C contents were obtained from Table 1, except for those for pBLO1 (from B. longum subsp. longum NCC2705), pDOJH10L, and pDOJH10S (from B. longum subsp. longum DJO10A).
G+C contents of completely sequenced host chromosomes.
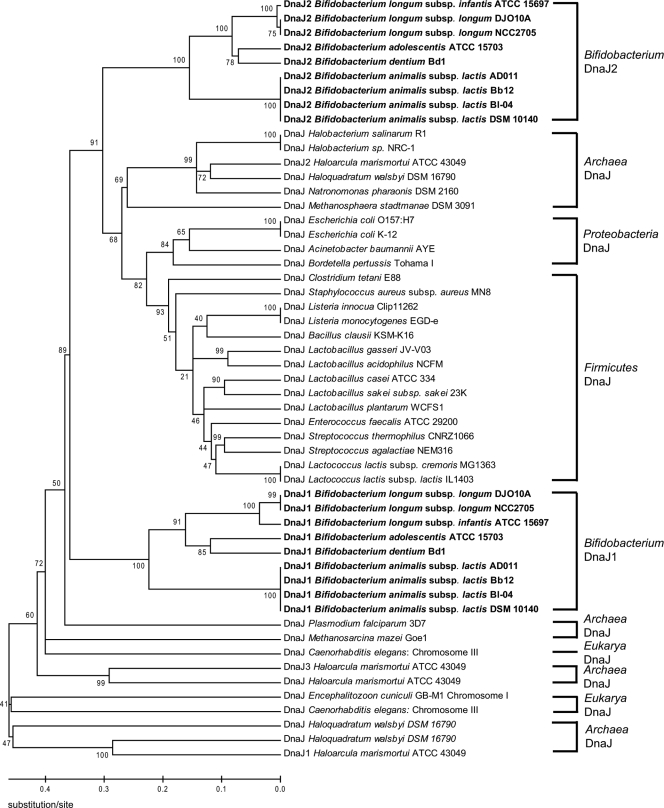
Although the majority of the replication (Rep) proteins identified for plasmids from bifidobacteria are homologous to rolling circle replication (RCR) Rep proteins, a few plasmids encoding proteins that are homologous to theta replication proteins have been reported (5, 53, 162). For a comparative analysis of replication proteins encoded by bifidobacterial plasmids, a phylogenetic analysis of 26 Rep proteins from bifidobacterial plasmids and 15 Rep proteins from closely related plasmids was performed, and it categorized bifidobacterial plasmids into five groups based on their Rep proteins (Fig. 1). This grouping is consistent with previous studies on bifidobacterial plasmids (53, 104). The Rep proteins in group I share the conserved Rep_3 domain from the Pfam database (PF01051), and this is the most commonly detected replicon in bifidobacterial plasmids. This domain is also found in replication proteins from many plasmids of the lactic acid bacteria (LAB), and these plasmids replicate via the RCR mechanism (69). Five of the plasmids in this group, pKJ50, pKJ36, pNAC1, pNAC2, and pNAL8L, were shown experimentally to accumulate single-stranded intermediates in bifidobacteria, consistent with the RCR mode of replication (53, 104, 212, 225). Group II Rep proteins, containing the RepA_C domain (PF04796), are found in a few B. longum and B. asteroides plasmids. One of these plasmids, pDOJH10L, was shown to replicate via RCR, suggesting that the replication mode of plasmids in group II may follow RCR (162). This was further analyzed using a deletion analysis of pCIBAO89 from B. asteroides, defining the minimal replicon and demonstrating binding of the Rep protein by gel mobility shift DNA binding assays (55). The Rep protein of p4M from B. pseudocatenulatum belongs to group III and contains the Viral_Rep domain from the Pfam database (PF02407), which is found in some viral replication proteins. All plasmids from B. breve and B. globosum belong to group IV, whose encoded proteins contain the Rep domain (PF01446) and likely are also involved in RCR, given that single-stranded DNA intermediates were detected for one plasmid from this group (212). Interestingly, the Rep proteins encoded by the group V plasmids pMB1, pDOJH10S, and pBC1, containing the replicase domain (PF03090), are very similar to the theta replicases from Rhodococcus (173), Propionibacterium (138), Corynebacterium (310), and even E. coli (305), suggesting that these plasmids may replicate via theta replication (5, 53, 162). This was subsequently substantiated by demonstrating that pDOJH10S does not accumulate single-stranded DNA intermediates during its replication (162). Based on sequence phylogenetic analysis and G+C content, it was proposed that the B. longum plasmid pDOJH10S may have originated from Rhodococcus rhodochrous (162) (Fig. 1).
FIG. 1.
Phylogenetic analysis of replication proteins expressed by bifidobacterial plasmids. The replication proteins encoded by bifidobacterial plasmids and other homologous replication proteins were compared by ClustalW multiple alignments (313). A phylogenetic tree was generated by the neighbor-joining method, using P distance values (307). The numbers associated with the branches represent the bootstrap values. Bifidobacterial plasmid replicons are indicated in bold. Roman numerals indicate the five different classes of replication proteins expressed from bifidobacterial plasmids.
Mobilization (Mob) protein genes and origins of transfer (oriT), which are involved in plasmid transfer, are frequently found in many bifidobacterial plasmids, suggesting that they may be mobilizable. However, this has not yet been demonstrated. The oriT in bifidobacterial plasmids is highly conserved and consists of a DNA sequence (5′-TAAGTGCGCCCT-3′) and an inverted repeat, consistent with oriT motifs of other plasmids (50, 162). Mob proteins have a highly conserved motif (XPHuHuuuXXu, where “u” represents a hydrophobic amino acid) that was previously implicated in the nicking of oriT (126).
G+C content analysis of the sequenced plasmids revealed that most have quite different G+C contents from those of their host chromosomes, suggesting that they may have originated from other microorganisms in the recent evolutionary past (Table 5). Since only 40% of the plasmids contain a G+C content within 2% of that of the host chromosome, it was suggested that bifidobacteria can readily accept plasmids from other organisms by horizontal gene transfer. Since the plasmid G+C content ranges from 41.7 to 66.2%, the range of potential plasmid donors must be quite large. Given that all of the plasmids contain mob genes but no tra genes, it is possible that they were obtained by utilizing the Tra functions of a helper plasmid either in the donor or in another helper organism. All of the sequenced bifidobacterial plasmids to date are cryptic. Only one possible function connected to bifidobacterial plasmids has been reported, as the 8-kb unsequenced plasmid from B. bifidum NCFB 1454 was proposed to be involved in bifidocin B production (351). The curing of this plasmid corresponded with the loss of bacteriocin production, but bacteriocin immunity was still retained in those strains. While this suggests the involvement of a bifidobacterial plasmid in bacteriocin production, it has yet to be substantiated. Another recent study linked production of a new bacteriocin, bifidin I, to a plasmid in B. longum subsp. infantis BCRC 14602 (43). However, the results of the study do not support the presence of a plasmid in this strain.
Development of Cloning and Expression Vector Systems for Bifidobacteria
A number of shuttle cloning vectors for bifidobacteria with E. coli plasmid replicons have been constructed, and electroporation procedures for bifidobacteria have been developed (4, 162, 177, 178, 188, 222, 225, 254, 255, 268, 287). The high-copy-number ColE1-based replication origin of E. coli was used for the construction of all except one of the E. coli-Bifidobacterium shuttle cloning vectors (4, 177, 178, 225, 255, 287, 309). Interestingly, replication problems in bifidobacteria occurred for two of the vectors, which encoded almost identical Rep proteins (pNAC1 and pNAL8L group I Rep proteins) (Fig. 1), suggesting incompatibility issues with ColE1 (53, 104). Vectors encoding other group I Rep proteins did not exhibit replication issues, suggesting that this incompatibility issue is confined to a small conserved subgroup of group I rep genes.
One vector was constructed using the E. coli p15A ori instead of ColE1. This has a lower copy number in E. coli, which can facilitate cloning of DNA regions that may have stability issues in E. coli. Given that this vector, pDOJHR, also contained a theta replicon from bifidobacteria, it was found to be a stable vector in bifidobacteria, exhibiting high segregational and structural stability without antibiotic pressure (162). Given the low electroporation frequencies for plasmid transfer into bifidobacteria, a number of studies on optimization of electroporation were undertaken, with limited success (9, 253). Recently, methylating plasmids to overcome restriction systems in bifidobacteria were found to greatly improve transformation efficiencies (195, 348).
Several promoter-screening, gene expression, and secretion vectors have been constructed. The reporter gene gusA, encoding the E. coli β-glucuronidase enzyme, has been used to monitor promoter activity in bifidobacteria (147, 268). A strong 16S rRNA gene promoter from bifidobacteria was used for the expression of a cholesterol oxidase gene from Streptomyces coelicolor in B. longum (221). The α-amylase gene from B. adolescentis INT-57 was cloned into B. longum, resulting in extracellular α-amylase production (130, 249). Since the secretion and expression signals from α-amylase were functional, they were used to construct an expression and secretory vector, pBESAF2, which was used to express phytase (223), pediocin (193), and γ-aminobutyric acid (GABA) (220). Currently, no expression vectors with food-grade selection markers have been developed for bifidobacteria.
A vector based on the group V theta-replicating plasmid pBC1 (Fig. 1), containing a luxABCDE operon that originated from Photohapdus luminescens but was optimized for expression in Gram-positive bacteria (237), was modified for expression of lux in Bifidobacterium breve (57). This was used for monitoring of bifidobacteria both in vitro and in vivo in a mouse system. However, cell numbers in vivo were not sufficient to allow real-time monitoring of the ingested bifidobacteria. Future optimization of lux expression in bifidobacteria may increase the sensitivity of this tool.
Another bifidobacterial expression vector system utilized the hup (histone-like protein) gene promoter and was used to express cytosine deaminase in B. longum (202), human interleukin-10 (IL-10) in B. longum (86), the Salmonella flagellin gene in B. animalis (304), and human basic fibroblast growth factor (FGF-2) in B. breve (286). The expression of cytosine deaminase in B. longum was used to treat hypoxic tumors in mice, as previous studies had indicated that bifidobacteria selectively localized only in hypoxic tumor cells, not normal cells, pointing to the tumor-targeting properties of bifidobacteria (56, 165, 349, 350). The function of cytosine deaminase in tumor control is to convert the nontoxic compound 5-fluorocytosine to 5-fluorouracil, which is cytotoxic (202). This vector system was based on a group I replicon from bifidobacteria (pTB6) (Fig. 1) and exhibited stability problems in B. longum, but it was more stable in B. breve (115). A point mutation in the CD gene, at the active site, was found to increase CD activity 10-fold, thus improving the efficacy of this antitumor activity (111). An analogous antitumor strategy was developed for expression of a human endostatin gene in B. longum and B. adolescentis, using the lambda PRPL promoter and a theta-replicating ori from the group V bifidobacterial plasmid pMB1 (Fig. 1). This was found to be effective in the treatment of mouse liver tumors (165, 345).
GENOMES OF BIFIDOBACTERIA
General Characteristics of Bifidobacterial Genomes
Nine complete genome sequences, for B. longum subsp. longum (GenBank accession numbers AE014295 and CP000605), B. adolescentis (AP009256), B. longum subsp. infantis (CP001095), B. dentium (CP001750), and B. animalis subsp. lactis (CP001213, CP001515, CP001606, and CP001853), have been obtained to date and are publically available in the GenBank database (17, 92, 141, 161, 275, 280, 335). In addition, 32 genome sequencing projects with other bifidobacteria are currently ongoing (Table 6).
TABLE 6.
Completed or ongoing bifidobacterial genome projects
| Organism | Strain | Status of project | Sequence availabilitya | Size (Mb)b | Institutionc | Reference or GenBank accession no. |
|---|---|---|---|---|---|---|
| B. adolescentis | ATCC 15703 | Complete | Yes | 2.09 | Gifu University, Japan | AP009256d |
| L2-32 | Ongoing | Yes | 2.39 | Washington University, USA | ||
| B. angulatum | DSM 20098 | Ongoing | Yes | 2.00 | Washington University, USA | |
| JCM7096 | Ongoing | No | 2.00 | University of Tokyo, Japan | ||
| B. animalis subsp. lactis | AD011 | Complete | Yes | 1.93 | KRIBB, South Korea | 141 |
| Bb-12 | Complete | Yes | 1.94 | Integrated Genomics, Inc./Chr. Hansen, USA | 92 | |
| Bl-04 | Complete | Yes | 1.94 | Danisco/Penn State University, USA | 17 | |
| DSM 10140 | Complete | Yes | 1.94 | Danisco/Penn State University, USA | 17 | |
| HN019 | Ongoing | Yes | 1.92 | Fonterra Research Center, New Zealand | ||
| V9 | Ongoing | Yes | 1.94 | Inner Mongolia Agricultural University, China | ||
| B. bifidum | BGN4 | Ongoing | No | KRIBB, South Korea | ||
| DSM 20456 | Ongoing | No | Washington University, USA | |||
| JCM1255 | Ongoing | Yes | 2.00 | University of Tokyo, Japan | ||
| NCIMB 41171 | Ongoing | Yes | 2.19 | Broad Institute, USA | ||
| B. breve | DSM 20213 | Ongoing | Yes | 2.30 | Washington University, USA | |
| JCM1192 | Ongoing | Yes | 2.00 | University of Tokyo, Japan | ||
| UCC2003 | Ongoing | No | 2.42 | University College, Cork, Ireland | ||
| B. catenulatum | DSM 16992 | Ongoing | Yes | 2.06 | Washington University, USA | |
| JCM1194 | Ongoing | Yes | 2.00 | University of Tokyo, Japan | ||
| B. dentium | ATCC 27678 | Ongoing | Yes | 2.62 | Washington University, USA | |
| ATCC 27679 | Ongoing | No | BCM-HGSC, USA | |||
| Bd1 | Complete | Yes | 2.64 | University of Parma, Italy | 335 | |
| JCM1195 | Ongoing | No | University of Tokyo, Japan | |||
| JCVIHMP022 | Ongoing | No | J. Craig Venter Institute, USA | |||
| JCVIHMP023 | Ongoing | No | J. Craig Venter Institute, USA | |||
| B. gallicum | DSM 20093 | Ongoing | Yes | 2.02 | Washington University, USA | |
| B. longum subsp. longum | BORI | Ongoing | No | KRIBB, South Korea | ||
| DJO10A | Complete | Yes | 2.38 | JGI/University of Minnesota, USA | 161 | |
| NCC2705 | Complete | Yes | 2.26 | Nestle/ University of Georgia, USA | 275 | |
| JDM301 | Ongoing | Yes | 2.48 | Shanghai Jiao Tong University, China | ||
| B. longum subsp. infantis | 157F-NC | Ongoing | No | University of Tokyo, Japan | ||
| ATCC 15697 | Complete | Yes | 2.83 | JGI/UC Davis, USA | 280 | |
| ATCC 55813 | Ongoing | Yes | 2.37 | BCM-HGSC, USA | ||
| CCUG 52486 | Ongoing | Yes | 2.45 | Broad Institute, USA | ||
| JCM1217 | Ongoing | No | University of Tokyo, Japan | |||
| JCM1222 | Ongoing | No | University of Tokyo, Japan | |||
| B. pseudocatenulatum | DSM 20438 | Ongoing | Yes | 2.30 | Washington University, USA | |
| B. scardovii | JCM12489 | Ongoing | No | University of Tokyo, Japan | ||
| Bifidobacterium sp. | 12_1_47BFAA | Ongoing | No | Broad Institute, USA | ||
| HM5 | Ongoing | No | University of Tokyo, Japan | |||
| JCM15439 | Ongoing | No | University of Tokyo, Japan |
Complete or draft genome sequences are available in the GenBank database and the Human Metagenome Consortium Japan (HMCJ) database.
Approximate size.
KRIBB, Korea Research Institute of Bioscience & Biotechnology; BCM-HGSC, Baylor College of Medicine-Human Genome Sequencing Center; JGI, Joint Genome Institute, U.S. Department of Energy.
Published in GenBank only.
An overall comparison of the nine completed genome sequences revealed genome sizes ranging from 1.9 to 2.8 Mb (Table 7). Interestingly, the smallest genomes belong to the four strains of B. animalis subsp. lactis. Since strains of this subspecies are believed to have evolved from the B. animalis species in a dairy fermentation environment, this substantiates the tendency of bacteria to undergo genome reduction when exposed to less complex environments (206). This is further substantiated by the case of B. animalis subsp. lactis AD011, which was reported to have only two complete RNA operons, compared to four or five operons in the other genomes. While this may be an anomaly due to possible sequence assembly errors, it may also be the result of adaptation to a simple, constant-nutrient environment, given that extra rRNA operons allow bacteria to adapt more quickly to new nutrient sources (52, 145).
TABLE 7.
General characteristics of bifidobacterial genomes
| Parameter | Value |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| B. adolescentis ATCC 15703 |
B. longum subsp. longum |
B. longum subsp. infantis ATCC 15697 | B. dentium Bd1 |
B. animalis subsp. lactis |
|||||
| DJO10A | NCC2705 | AD011 | Bl-04 | DSM 10140 | Bb-12 | ||||
| Length (bp) | 2,089,645 | 2,375,792 | 2,256,640 | 2,832,748 | 2,636,367 | 1,933,695 | 1,938,709 | 1,938,483 | 1,942,198 |
| Overall G+C content (%) | 59.18 | 60.15 | 60.12 | 59.86 | 58.94 | 60.49 | 60.48 | 60.48 | 60.48 |
| No. of plasmids | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| No. of annotated genes | 1,631 | 1,990 | 1,727 | 2,416 | 2,129 | 1,528 | 1,567 | 1,566 | 1,642 |
| Avg gene length (bp) | 1,109 | 1,031 | 1,115 | 997 | 1,066 | 1,070 | 1,064 | 1,062 | 1,063 |
| Gene density (no. of genes/kb) | 0.780 | 0.838 | 0.765 | 0.852 | 0.807 | 0.790 | 0.808 | 0.807 | 0.845 |
| Gene coding content (%) | 86.5 | 86.4 | 85.3 | 85.1 | 86.1 | 84.6 | 86.0 | 85.8 | 89.9 |
| Gene G+C content (%) | 60.08 | 61.13 | 60.86 | 60.70 | 59.32 | 61.47 | 61.40 | 61.40 | 61.11 |
| No. of rRNA gene operons | 5 | 4 | 4 | 4 | 4 (+ 1 5S rRNA gene) | 2 (+ 1 5S rRNA gene) | 4 | 4 | 4 |
| No. of prophage-like elements | 0 | 1 | 1 (remnant) | 4 | 1 complete + 1 remnant | 1 (remnant) | 1 (remnant) | 1 (remnant) | 1 (remnant) |
| No. of tRNAs | 54 | 58 | 57 | 79 | 55 | 52 | 52 | 52 | 52 |
| No. of tRNA synthetases | 20 | 20 | 21 | 19 | 19 | 21 | 19 | 19 | 19 |
| No. of MICs | 0 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| No. of CRISPRs | 1 (86 repeats) | 1 (43 repeats) | 0 | 0 | 2 (17 and 81 repeats) | 1 (20 repeats) | 1 (22 repeats) | 1 (20 repeats) | 1 (20 repeats) |
| No. of oligonucleotide clustersa | 10 | 11 | 7 | 11 | 10 | 7 | 7 | 7 | 7 |
| No. of polyol clustersb | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
Number of predicted oligosaccharide and other complex carbohydrate utilization gene clusters.
Number of predicted polyol utilization gene clusters.
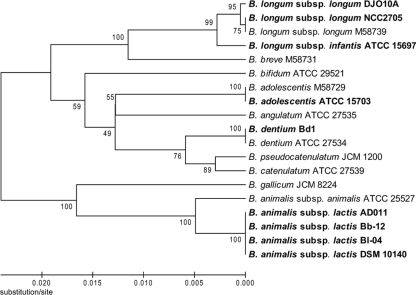
The nine complete bifidobacterial genomes can be divided into three groups based on phylogenetic analysis of their 16S rRNA genes, including the B. longum group (B. longum subsp. longum DJO10A and NCC2705 and B. longum subsp. infantis ATCC 15697), the B. adolescentis group (B. adolescentis ATCC 15703 and B. dentium Bd1), and the B. animalis group (B. animalis subsp. lactis AD011, Bl-04, DSM 10140, and Bb-12) (Fig. 2). A whole-genome alignment of these genomes was consistent with this analysis, with B. animalis subsp. lactis clearly the most distantly related to the other groups. The four strains of B. animalis subsp. lactis exhibited >99% sequence identity over their entire genomes, illustrating a very closely related group (Fig. 3). Interestingly, in the whole-genome comparison of the B. longum group, B. longum subsp. infantis showed a large inverted region (∼1.3 Mb) in the middle of the genome compared to both B. longum subsp. longum strains, suggesting that this possible inversion is a recent evolutionary event within B. longum subsp. infantis, or it may be due to errors in genome assembly (Fig. 3). Large genome inversions were previously observed in a genome comparison of strains of Salmonella enterica, and they were proposed to be due to homologous recombination between identically repeated rRNA operons (70). However, there are no rRNA operons, mobile elements, or repeat regions in the boundaries of the inversion exhibited in the B. longum subsp. infantis genome, suggesting a novel inversion mechanism or sequence assembly issues.
FIG. 2.
Phylogenetic analysis of the 16S rRNA genes of the nine sequenced bifidobacteria, illustrating their separation into three distinct groups. The 16S rRNA gene sequences from complete or draft bifidobacterial genomes were compared by ClustalW multiple sequence alignments as described in the legend to Fig. 1. The completely sequenced bifidobacteria are indicated in bold.
FIG. 3.
Alignments of the complete genome sequences of the B. longum group and the B. animalis subsp. lactis group. Red lines indicate the relative locations of elements that are oriented in the same direction. Blue lines indicate elements orientated in opposite directions.
Mobile Elements
Eight insertion sequence (IS) families were found in the nine bifidobacterial genomes, including the IS3, IS21, IS30, IS110, IS150, IS256, IS607/IS200, and ISL3 families (Table 8). While B. dentium has the smallest set of IS elements, B. longum subsp. infantis contains the most IS elements and the most diversity in IS families, displaying all eight of the families. This is consistent with it containing the largest genome among these nine bifidobacterial strains. Interestingly, the three sequenced strains of B. animalis subsp. lactis contained very few IS elements, and all of these were from a single IS family, ISL3. This family of IS elements was first characterized from the yogurt bacterium Lactobacillus delbrueckii subsp. bulgaricus (95). Since B. animalis subsp. lactis is also used frequently in yogurt manufacturing, the presence of complete ISL3 elements (none of which are present in the other bifidobacterial genomes) in this subspecies may suggest horizontal gene transfer between bacteria in the yogurt fermentation environment.
TABLE 8.
IS elements in bifidobacteria
| IS family | No. of elements (no. of complete elements)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
B. longum subsp. longum |
B. longum subsp. infantis ATCC 15697 | B. adolescentis ATCC 15703 | B. dentium Bd1 |
B. animalis subsp. lactis |
|||||
| DJO10A | NCC2705 | AD011 | Bl-04 | DSM 10140 | Bb-12 | ||||
| IS3 | 13 (1) | 14 (2) | 16 (9) | 8 (3) | 6 (5) | 0 | 0 | 0 | 0 |
| IS21 | 10 (8) | 7 (5) | 7 (1) | 5 (3) | 0 | 0 | 0 | 0 | 0 |
| IS30 | 9 (5) | 5 (3) | 15 (2) | 2 (0) | 0 | 0 | 0 | 0 | 0 |
| IS110 | 0 | 0 | 6 (5) | 0 | 0 | 0 | 0 | 0 | 0 |
| IS256 | 4 (2) | 7 (5) | 7 (2) | 3 (1) | 0 | 0 | 0 | 0 | 0 |
| IS607/IS200 | 1 (1) | 1 (1) | 2 (0) | 0 | 0 | 0 | 0 | 0 | 0 |
| ISL3 | 7 (0) | 12 (0) | 3 (0) | 0 | 0 | 7 (6) | 6 (5) | 6 (5) | 6 (6) |
Numbers in bold indicate species-specific IS elements.
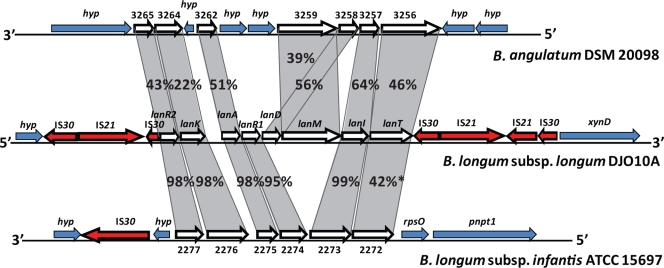
IS elements are frequently involved in genome rearrangement or deletion events (29, 63, 211). During pure culture growth of the intestinal strain B. longum subsp. longum DJO10A, the IS30 element was found to move within the genome, confirming that it was indeed active (161). This hyperactivity of IS30 was also implicated in a genome deletion event in this strain during its adaptation to pure culture growth, as a large gene cluster encoding lantibiotic production, positioned between IS30 elements, was precisely deleted from the genome. This supports the role of IS elements in genome reduction and rapid genome adaptation to new environments.
The genome sequences of both B. longum subsp. longum strains revealed novel mobile integrase cassette (MIC) structures, consisting of three contiguous integrases flanked by an inverted repeat and a palindrome structure sandwiched by two IS3-type IS elements, and their nonlinear positioning within the genomes suggests that they are active (161). Interestingly, these MIC structures are not present in other bifidobacterial genomes, including that of B. longum subsp. infantis, or in other intestinal bacterial genomes, suggesting that this MIC structure is specific to B. longum subsp. longum (161). During pure culture adaptation of B. longum subsp. longum strain DJO10A over 1,000 generations, a MIC element was found to be deleted from the genome along with ∼50 kb of sequence, indicating that these structures can play a role in deletion events (161). This is further substantiated by a predicted deletion from the genome of B. longum subsp. longum strain NCC2705 of a frameshifted tetW gene and bordering MIC element that are present in strain DJO10A (161).
tRNAs and Aminoacyl-tRNA Synthetases
Even though all nine sequenced bifidobacterial genomes encode multiple tRNAs for all 20 amino acids, they do not encode aminoacyl-tRNA synthetases for asparagine and glutamine, suggesting that they may utilize alternative pathways for translation for these amino acids, as has been shown for some other bacteria (187, 294). These alternative pathways utilize GltX (glutamyl-tRNA synthetase) and AspS (aspartyl-tRNA synthetase) for the synthesis of intermediates (Glu-tRNAGln and Asp-tRNAAsn) and GatABC (Glu-tRNAGln/Asp-tRNAAsn amidotransferases) and then convert these intermediates to Gln-tRNAGln and Asn-tRNAAsn for incorporation of these amino acids during translation. Genes encoding GltX, AspS, and GatABC are present in all sequenced bifidobacterial genomes, supporting the utilization of these alternative translation pathways in bifidobacteria (161, 275, 335). Interestingly, the predicted alanyl-tRNA synthetase (AlaS) gene in B. animalis subsp. lactis AD011 is longer than those in the other three B. animalis subsp. lactis genomes because of the presence of two identical 378-bp repeat regions in the gene, which would result in a frameshift, preventing expression of a full alanyl-tRNA synthetase. A perfect sequence match of the alaS gene in strain AD011, after removal of one of the repeat regions, to the alaS genes in the three other B. animalis subsp. lactis genomes suggests that this repeat is probably a genome assembly error.
Prophages
Prophage-like elements were found in eight of the nine complete bifidobacterial genomes (with the B. adolescentis genome being the exception), with the majority being incomplete (Table 7). An incomplete prophage-like element from B. breve UCC2003 has also been reported (332). Recently, a comparative analysis of complete and partial prophage-like elements in three complete bifidobacterial genomes (B. dentium Bd1, B. longum subsp. infantis ATCC 15697, and B. animalis subsp. lactis AD011), as well as 11 elements in six draft bifidobacterial genomes (B. dentium ATCC 27678, B. longum subsp. infantis CCUG 52486, B. bifidum 317B and NCIMB 41171, B. adolescentis L2-32, and B. catenulatum DSM 16992), provided more insights into the taxonomy and evolution of bifidobacterial prophages (334).
It was demonstrated previously that three prophages, in B. longum subsp. longum DJO10A, B. longum subsp. infantis ATCC 15697 (Binf-4), and B. animalis subsp. lactis AD011, are inducible at the transcription level by use of hydrogen peroxide. One prophage in B. dentium Bd1 (Bdent-2) could also be induced at the transcription level by culture in basal medium mucin. None could be demonstrated to complete a full lytic cycle, suggesting that there is a novel signal required for full induction or that they may not be 100% complete (332, 334). It has also been suggested that full induction of the lytic cycle may require a helper phage (334). A microarray analysis of B. dentium Bd1 (Bdent-2) revealed some genes that are known to be required for a lytic lifestyle, but not all were expressed, thus explaining why this prophage DNA sequence cannot produce viable phage particles (334). The prophage in B. longum subsp. longum DJO10A contains 57 genes in a 37.8-kb cluster, and its insertion site has highly conserved direct repeats, representing a likely duplication of its attachment site during its insertion. This attachment site is located in the 3′ region of a tRNAMet gene, resulting in the prophage insertion occurring directly downstream from this gene. Interestingly, an identical insertion site is frequently found in other bifidobacterial prophage-like elements, suggesting that this attachment site is utilized as a general prophage insertion mechanism (332, 334). The insertion of prophages and other DNA elements utilizing certain tRNA genes has been observed in other bacteria, and this attachment site appears to be a favorable insertion position (34). While the prophage-like elements in B. longum subsp. infantis and the three strains of B. animalis subsp. lactis are not inserted bordering this tRNA gene, other DNA elements, possibly of plasmid origin, are inserted in this region, substantiating the hypothesis that certain tRNA genes provide hot spots for insertion of foreign DNA elements.
Comparative sequence analysis of prophage-like elements in bifidobacteria indicated that they can be divided into four groups, with the majority belonging to group II, which is related to the low-GC Firmicutes, such as Lactococcus and Lactobacillus. A number of them also encode products not required for phage functions, such as type I or type II restriction-modification (R-M) systems or metabolic factors such as polyketide biosynthesis enzymes and phosphofructokinase (334). The presence of these extra functions may contribute to the overall cellular advantage of acquiring these elements and also may benefit the ecological fitness of the lysogeny. The prophage present in B. longum subsp. longum DJO10A and one of the prophages (Binf-4) present in B. longum subsp. infantis carry a tRNASer and a tRNATrp gene, respectively (161, 334). The tRNASer gene in the strain DJO10A prophage recognizes the most common Ser codon in the prophage genome, while it is not a common Ser codon in the genome of this strain, suggesting that this was the selective pressure for the prophage acquiring this tRNA gene. This was not the case for the B. longum subsp. infantis prophage, as tRNATrp recognizes only one codon. Since the frequencies of the Trp codon are similar between the prophage and the bacterial genome, this suggests that the acquisition may be of mutual benefit to both the prophage and the bacterium.
Diversity-generating retroelement (DGR) structures have been detected in some bacteriophages, and they are proposed to be involved in a mechanism to rapidly evolve phage tail proteins, which can potentially expand the host range for the phage (79). These structures are composed of genes encoding reverse transcriptase and a tail protein and two direct repeat sequences, one an invariable template repeat (TR) located in the intergenic region between the two genes and the other a variable repeat (VR) located in the 3′ region of the tail protein gene. The TR is proposed to be reverse transcribed, which produces a single-stranded DNA copy with nucleotide substitutions only at adenine residues, and this cDNA switches with the VR region in an event termed “mutagenic homing” (32). Two bifidobacterial prophages (located in B. longum subsp. longum DJO10A and B. adolescentis L2-32) carry DGR structures consisting of putative genes encoding reverse transcriptase and a tail protein and direct repeat sequences consistent with TR and VR regions (332, 334). The only mismatches between the TR and VR sequences in strain DJO10A are located at 13 adenine residues in TR that do not match with VR, consistent with the proposed function of these mutator elements.
CRISPR Structures in Bifidobacteria
CRISPRs are DNA repeats which have been found widely, in more than 40% of bacterial genomes and most archaeal genomes (103, 129, 155). CRISPR structures consist of identically repeated short (24 to 47 bases) DNA sequences interspersed with sequence-variable spacer regions, varying in length from 26 to 72 bases, and with CRISPR-associated cas genes (33, 110, 122, 296). The number of repeat sequences is variable between different bacteria, ranging from as few as 2 to as many as 249 (296). This structure was found in seven of the nine bifidobacterial genomes, with those of B. longum subsp. longum NCC2705 and B. longum subsp. infantis being the exceptions (Table 7). CRISPRs have been demonstrated to be involved in acquiring resistance against infecting bacteriophages by replacing the first spacer sequence with a DNA segment originating from bacteriophage DNA (18, 71). In addition to the completely sequenced bifidobacteria, these structures can also be seen in the draft sequences of B. dentium ATCC 27678, B. adolescentis L2-32, B. animalis subsp. lactis HN019, B. angulatum DSM 20098, and B. catenulatum JCM1194 (Table 9). Interestingly, one of the two CRISPR structures in the B. dentium strain contains a repeat sequence identical to the CRISPR structure in B. longum subsp. longum DJO10A, suggesting a common origin for the CRISPR structures between these two strains. Given their widespread presence in bifidobacteria, these structures may be due to selective pressure to protect these bacteria from bacteriophages in the large intestine. Further substantiation of a functional role for these structures comes from an analysis of bifidobacterial prophages that found 29 regions with >85% sequence identity between 12 prophages and 21 CRISPR spacers (334).
TABLE 9.
Comparison of CRISPR-cas systems in bifidobacteria
| Species | No. of repeats | Groupa | Repeat sequence | Structureb |
|---|---|---|---|---|
| B. longum subsp. longum DJO10A | 43 | I | CAAGCTTATCAAGAAGGGTGAATGCTAATTCCCAGC | c5-c1-c2-repeats |
| B. dentium Bd1 | 17 | I | CAAGTTTATCAAGAAGGGTAGAAGCTAATTCCCAGT | csn1-c1-c2-repeats |
| 81 | II | GTCGCTCTCCTCACGGAGAGCGTGGATTGAAAT | c3-c5-csd1-c1-repeats | |
| B. dentium ATCC 27678 | 15 | I | CAAGTTTATCAAGAAGGGTAGAAGCTAATTCCCAGT | c5-c1-c2-repeats |
| 19 | II | GTCGCTCTCCTCACGGAGAGCGTGGATTGAAAT | c3-c5-csd1-csd2-c4-c1-c2-repeats | |
| B. adolescentis ATCC 15703 | 86 | II | GGTCGCTCTCCTTACGGAGAGCGTGGATTGAAAT | c3-c5-csd1a-csd1b-csd2-c4-c1-c2-repeats |
| B. adolescentis L2-32B | 113 | II | GGTCGCTCTCCTTACGGAGAGCGTGGATTGAAAT | c3-csd2-c4-c1-c2-repeats 2-repeats 1 |
| B. animalis subsp. lactis AD011 | 20 | III | CCCTCAATGAAGCTCCGAAGCCGAGACTTCGGAGAT | c3-c_GSU0053-c1-repeats |
| B. animalis subsp. lactis HN019 | 20 | III | CCCTCAATGAAGCTCCGAAGCCGAGACTTCGGAGAT | c3-hyp-c_GSU0053-c2-c1-repeats |
| B. animalis subsp. lactis Bl-04 | 22 | III | CCCTCAATGAAGCTCCGAAGCCGAGACTTCGGAGAT | csb3-c3-csb2-csb1-c2-c1-repeats |
| B. animalis subsp. lactis DSM 10140 | 20 | III | CCCTCAATGAAGCTCCGAAGCCGAGACTTCGGAGAT | csb3-c3-csb2-csb1-c2-c1-repeats |
| B. animalis subsp. lactis Bb-12 | 20 | III | CCCTCAATGAAGCTCCGAAGCCGAGACTTCGGAGAT | csb3-c3-csb2-csb1-hyp-c1-repeats |
| B. angulatum DSM 20098 | 139? | IV | GTGTTCCCCGCACACGCGGGGATGATCCC | c3-cse1-cse2-cse4-c5-cse3-c1-c2-repeats |
| B. catenulatum JCM1194 | 34 | IV | GTGTTCCCCGCATACGCGGGGATGATCCC | c3-truncated-cse4-c5-cse3-c1-c2-repeats |
CRISPR groups represent elements with identical repeat sequences.
c1 to c5, CRISPR-associated cas1 to cas5 genes; csd1-csd2, CRISPR-associated csd1-csd2 family genes; c_GSU0053, GSU0053 family CRISPR-associated gene; cse1 to cse4, CRISPR-associated cse1 to cse4 family genes; csn1, CRISPR-associated csn1 family gene; hyp, hypothetical protein gene; truncated, incomplete draft sequence.
Since the CRISPR model for protecting against invading bacteriophages involves replacing a spacer region with part of the invading phage DNA, a search for DNA regions identical to spacer regions found several extrachromosomal DNA elements containing these sequences (33, 191). This suggests an expanded role for these structures in protection against all invading DNA elements. Analysis of spacers in bifidobacterial CRISPR structures revealed that there is one 28-bp spacer in B. longum subsp. longum DJO10A (spacer number 5) that has 100% sequence identity with an internal sequence in the putative mob genes carried by the bifidobacterial plasmids p6043B, pNAC2, and pMG1. This further supports an expanded role for CRISPR structures.
Chromosome Origin of Replication
The origin of replication (oriC) in bacteria is generally highly conserved within species, and generally there is more than one oriC cluster per genome (170). Each cluster contains an oriC and multiple DnaA boxes for the binding of DNA polymerase. The origin of replication (oriC) in the B. longum subsp. longum NCC2705 genome was not initially predicted by a total GC skew analysis due to initial errors with genome sequence assembly (105). Geometric analysis for the assembly of whole genome sequences showed that the genome sequence of B. longum NCC2705 was misassembled because of misplacement of several repeated regions. Following a correction of these assembly errors in the revised genome sequence of B. longum NCC2705 (275), the highly conserved oriC region was revealed by bioinformatic predictions. The oriC-associated regions of seven complete genome sequences of bifidobacteria also showed the same organization, with three oriC clusters and seven different kinds of putative DnaA boxes in highly conserved locations, as follows: parB-parA-(oriC cluster 3)-gidB-ssb-oxaA-rnpA-rpmH-(oriC cluster 2)-dnaA-(oriC cluster 1)-dnaN-recF-hyp-gyrB-gyrA. The oriC clusters contain the same number and type of DnaA boxes for each member of a species, but the number and type are species specific (Table 10). The locations of the three oriC clusters in all bifidobacterial genomes are positioned identically within the gene cluster involved in DNA replication. The one apparent exception is B. longum subsp. infantis ATCC 15697, whose annotation does not show an rmpH gene, which encodes the 50S ribosomal protein L34. However, analysis of its sequence shows that the gene is present, indicating that the open reading frame (ORF) was missed during genome annotation.
TABLE 10.
Comparison of DnaA boxes in nine bifidobacterial genomes
| Strain | oriC clustera | No. of DnaA boxesb |
Total no. of DnaA boxes | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | |||
| B. longum subsp. longum DJO10A | 1 | 3 | 1 | 3 | 1 | 0 | 0 | 0 | 8 |
| 2 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 4 | |
| 3 | 6 | 0 | 0 | 0 | 0 | 0 | 1 | 7 | |
| Subtotal | 10 | 2 | 3 | 1 | 1 | 1 | 1 | 19 | |
| B. longum subsp. longum NCC2705 | 1 | 3 | 1 | 3 | 1 | 0 | 0 | 0 | 8 |
| 2 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 4 | |
| 3 | 6 | 0 | 0 | 0 | 0 | 0 | 1 | 7 | |
| Subtotal | 10 | 2 | 3 | 1 | 1 | 1 | 1 | 19 | |
| B. longum subsp. infantis ATCC 15697 | 1 | 3 | 1 | 3 | 1 | 0 | 0 | 0 | 8 |
| 2 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 3 | |
| 3 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | |
| Subtotal | 9 | 1 | 3 | 1 | 1 | 1 | 0 | 16 | |
| B. adolescentis ATCC 15703 | 1 | 5 | 0 | 2 | 0 | 0 | 1 | 0 | 8 |
| 2 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 3 | |
| 3 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 5 | |
| Subtotal | 10 | 0 | 2 | 1 | 2 | 2 | 0 | 17 | |
| B. dentium Bd1 | 1 | 5 | 0 | 1 | 1 | 1 | 0 | 1 | 9 |
| 2 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 3 | |
| 3 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 4 | |
| Subtotal | 8 | 1 | 1 | 1 | 3 | 1 | 1 | 16 | |
| B. animalis subsp. lactis AD011 | 1 | 6 | 0 | 1 | 0 | 1 | 1 | 0 | 9 |
| B. animalis subsp. lactis Bl-04 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| B. animalis subsp. lactis DSM 10140 | 3 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 5 |
| B. animalis subsp. lactis Bb-12 | Subtotal | 10 | 1 | 1 | 1 | 2 | 1 | 0 | 16 |
Locations of oriC clusters in oriC-associated conserved regions are as follows: parB-parA-(oriC cluster 3)-gidB-ssb-oxaA-rnpA-rpmH-(oriC cluster 2)-dnaA-(oriC cluster 1)-dnaN-recF-hyp-gyrB-gyrA.
The DnaA boxes consist of seven types, designated A to G, as follows: A, TTATCCACA; B, TTGTCCACA; C, TTTTCCACA; D, TTACCCACA; E, TTATCCACC; F, TTATTCACA; and G, TTATGCACA.
R-M Systems
R-M systems play an important role in protection of methylated host DNA against invasion of unmethylated foreign DNA, such as plasmids or bacteriophages (236). An analysis of R-M systems in the nine bifidobacterial genomes revealed that only B. longum subsp. longum genomes have type I R-M systems and that all genomes except for that of B. longum subsp. infantis ATCC 15697 have one or two type II R-M systems (Table 11). The lack of any observed restriction-modification system or CRISPR structure in the B. longum subsp. infantis ATCC 15697 genome suggests that it may be more susceptible to phage infection. This is supported by the increased number of prophages in its genome, which is four times that for any of the other genomes (Table 7). The type II R-M systems encoded by the bifidobacterial genomes are analogs of Sau3AI, EcoRII, EcoRI, and KpnI (Table 11). It is intriguing that B. adolescentis carries gene analogs for the two restriction subunits of the multimeric LlaJI restriction enzyme but is missing genes analogous to those for the methylase subunits. This restriction enzyme is encoded by genes for the R1.LlaJI subunit, which recognizes the restriction site; R2.LlaJI, which is required to enable restriction of the DNA; and two methylase subunits (208). The absence of the gene analogs for the methylases suggests that the genes may not be expressed or have developed another function. The positioning of an IS3-type IS element directly upstream from these genes may suggest its role in a deletion event. Since genes comprising a KpnI gene cluster are located directly downstream, this may suggest an evolutionary connection to the formation of unusual multimeric restriction enzymes such as LlaJI. Both of these restriction enzymes are protected by 5 methylcytosines within their recognition sites, and while there are similarities within their recognition sites, they are not identical. Interestingly, the four strains of B. animalis subsp. lactis encode only one R-M system, which is analogous to an EcoRI-type system and is positioned in a predicted integrated plasmid with a significantly (10%) lower G+C content than that of its genome, suggesting that it was acquired from a recent horizontal gene transfer event. Given that bacteriophage infection is very prevalent in dairy fermentation environments (76, 183) and that this subspecies is predicted to have evolved in this environment, these observations suggest that the environment may have provided the selective pressure for acquiring this R-M system.
TABLE 11.
R-M systems in nine complete genomes of bifidobacteria
| R-M system or analog | Locus taga |
|||||
|---|---|---|---|---|---|---|
|
B. longum subsp. longum |
B. longum subsp. infantis ATCC 15697 | B. adolescentis ATCC 15703 | B. dentium Bd1 | B. animalis subsp. lactis AD011, Bl-04, DSM 10140, and Bb-12 | ||
| DJO10A | NCC2705 | |||||
| Type I systems | ||||||
| Mrr | BLD_1958 | BL1781 | ND | ND | BDP_0997 | ND |
| HsdM | BLD_1959 | BL1782 | ND | ND | ND | ND |
| HsdS | BLD_1960 | BL1783 | ND | ND | ND | ND |
| HsdR | BLD_1962 | BL1785 | ND | ND | BDP_1319 | ND |
| Type II analogsb | ||||||
| Sau3AI-R | BLD_1358 | BL0563 | ND | BAD_1232 | ND | ND |
| Sau3AI-M | BLD_1359 | BL0564 | ND | BAD_1233 | ND | ND |
| EcoRII-R | BLD_0355 | BL1472 | ND | ND | ND | ND |
| EcoRII-M | BLD_0356 | BL1473 | ND | ND | ND | ND |
| KpnI-R | ND | ND | ND | BAD_1282 | ND | ND |
| KpnI-M | ND | ND | ND | BAD_1283 | ND | ND |
| EcoRI-R | ND | ND | Blon_1196 (truncated)c | ND | ND | BLA_0644, Balac_1446, Balat_1446, BIF_00872 |
| EcoRI-M | ND | ND | Blon_1197 | ND | ND | BLA_0645, Balac_1447, Balat_1447, BIF_01894 |
| R1.LlaJI | ND | ND | ND | BAD_1280 | ND | ND |
| R2.LlaJI | ND | ND | ND | BAD_1281 | ND | ND |
ND, not detected.
R and M represent restriction enzymes and methyltransferases for type II R-M systems, respectively.
This pseudogene is truncated by insertion of an IS element.
The genes encoding the R-M systems described above are frequently found in bifidobacterial genomes, and they are among the major factors inhibiting the introduction of foreign DNA into these bacteria, resulting in very low electroporation frequencies. To overcome this obstacle, type II methylase genes from B. adolescentis and B. breve were expressed in E. coli to enable the methylation of E. coli-Bifidobacterium shuttle vectors (195, 348). This greatly improved transformation efficiencies, to a level that permitted targeted mutagenesis in bifidobacteria (195).
Biosynthesis of Amino Acids, Vitamins, Purines, and Pyrimidines
Although bifidobacteria have been studied for over 1 century, the lack of genetic information has limited insights into their biosynthetic capabilities. While some biosynthetic capabilities for vitamins and amino acids were elucidated previously (25, 244, 269), the genome analysis of bifidobacteria substantiates these biosynthetic capabilities as well as uncovering all other potential biosynthetic capabilities (275). The bifidobacterial genomes have genes predicted to be involved in the biosynthesis of all 20 amino acids, but since bifidobacteria cannot grow in minimum media, it is likely that not all are functional. It can be predicted from the genome analysis that amino acids can potentially be synthesized from different intermediates. These include ribose-5-phosphate (for histidine), glycerate-3-phosphate (for serine, glycine, and threonine), phosphoenolpyruvate (PEP) (for tyrosine, tryptophan, and phenylalanine), pyruvate (for valine, leucine, isoleucine, and alanine), oxaloacetate (for aspartate, asparagine, arginine, lysine, methionine, and cysteine), and 2-oxoglutarate (for glutamate, glutamine, and proline) (Fig. 4). However, since some core genes involved in sulfur assimilation (ATP sulfurylase, adenosine-5′-phosphosulfate kinase, serine acetyltransferase, and cysteine synthetase) are missing, synthesis of cysteine and methionine could still potentially be achieved in the presence of hydrogen sulfide (H2S), which in the large intestine could be supplied by other intestinal bacteria (301). The H2S produced by the intestinal flora may be assimilated with O-succinyl l-homoserine by cystathionine γ-synthase (EC 2.5.1.48) to produce cystathionine, and then cysteine and methionine biosynthesis could potentially be completed with cystathionine γ-lyase (EC 4.4.1.1), cystathionine β-lyase (EC 4.4.1.8), and methionine synthase (EC 2.1.1.14) in bifidobacteria (Fig. 5).
FIG. 4.
Overview of predicted carbohydrate uptake and metabolism systems in bifidobacteria. The F6PPK pathway, partial TCA cycle, and UDP-glucose/galactose system (UDP-Gal/Glc) are indicated with different background colors (light yellow for F6PPK, light purple for the partial TCA cycle, and sky blue for the UDP-Glc/Gal system). Blue, ABC transport systems; green, PTS; gray, MFS family; dark purple, major intrinsic protein (MIP) family; orange, GPH cation symporter family. Genes encoding predicted metabolic enzymes from B. longum subsp. longum DJO10A are indicated.
FIG. 5.
Predicted amino acid biosynthesis pathways in bifidobacteria. The biosynthesis of amino acids in bifidobacteria utilizes the intermediate products of the F6PPK pathway. Genes predicted to be involved in amino acid biosynthesis in B. longum subsp. longum DJO10A and the EC numbers of their predicted proteins are indicated in purple. Solid arrows indicate single-step reactions, and dotted arrows indicate multistep reactions.
Previous studies of vitamin biosynthesis by bifidobacteria showed that B. bifidum, B. breve, B. adolescentis, B. longum subsp. infantis, and B. longum subsp. longum can produce the vitamins nicotinate, thiamine (B1), pyridoxine (B6), folate (B9), and cobalamin (B12) (67). Analysis of the nine sequenced bifidobacterial genomes showed that they all contain gene analogs associated with the biosynthesis of nicotinate, thiamine (B1), and folate (B9), but they do not carry genes for biosynthesis of cobalamin (B12). This may suggest that production of these vitamins in bifidobacteria occurs by other enzymes, or it may be due to strain differences between the strains used in the previous studies and the sequenced strains. Species and/or strain differences in pyridoxine biosynthesis may also exist, as genes encoding three potential proteins (pyridoxine kinase, PdxT, and PdxS) of the pathway were predicted from the three B. longum genomes. It has also been shown that bifidobacteria have an absolute requirement for riboflavin (B2) (67), pantothenate (B5) (113, 308), and biotin (B7) (107). While seven of the completed genome sequences for bifidobacteria do not contain genes encoding riboflavin biosynthesis enzymes, two of the genomes, B. longum subsp. infantis ATCC 15697 and B. dentium Bd1, carry the genes for the complete riboflavin biosynthesis pathway (280, 335). This may suggest species and strain differences in riboflavin production in bifidobacteria, as five different strains of B. longum subsp. infantis were previously shown to require riboflavin (67).
All nine bifidobacterial genomes carry analogs of all the required genes for biosynthesis of purines and pyrimidines. Surprisingly, a few genes, such as pyrE, pyrF, and pyrD, are duplicated in both sequenced genomes of B. longum subsp. longum, which is not usual for other bacterial genomes (275). These duplications have been suggested to be involved in the evolution of functionally separate genes in bacteria into a multifunctional gene in higher organisms through a process of gene duplication and DNA rearrangement, leading to in-frame gene fusions encoding chimeric proteins, such as dihydroorotate synthase and UMP synthase in mammalian cells (65).
Peptidoglycan Biosynthesis
The cell wall structure of bifidobacteria consists of N-acetyl-d-glucosamine, muramic acid, ornithine, aspartate, glutamate, alanine, and serine, with a ratio of 1:1:1:1:3:2:1 (322). While N-acetyl-d-glucosamine was found to be a growth factor for bifidobacteria (100), they have the capacity to grow without it. In addition, studies have shown that l-rhamnose is a component of bifidobacterial cell walls (200). Eight of the bifidobacterial genomes carry gene analogs for the complete peptidoglycan biosynthesis pathway as well as the l-rhamnose biosynthesis pathway, which produces dTDP-rhamnose for cell wall biosynthesis (Table 12). However, the genome sequence of B. longum subsp. infantis ATCC 15697 does not reveal analogs for two genes involved in the biosynthesis of dTDP-rhamnose, i.e., rfbA and rfbCD. Interestingly, it contains an analog of rfbB, which is arranged in an apparent operon with the other rfb genes in the other eight genomes. This may reflect a deletion event in this strain or, possibly, a sequencing error. In addition, although their DNA sequences were present in the genome sequence of B. animalis subsp. lactis Bb-12, the gene prediction and annotation data did not show murA and murG genes, which are essential genes for peptidoglycan biosynthesis, indicating possible gene prediction and annotation errors in this genome (Table 12).
TABLE 12.
Genes involved in peptidoglycan and rhamnose biosynthesis in bifidobacteria
| Functional category and EC no. | KEGG annotation | Locus tag |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
B. longum subsp. longum |
B. longum subsp. infantis ATCC 15697 | B. adolescentis ATCC 15703 | B. dentium Bd1 |
B. animalis subsp. lactis |
||||||
| DJO10A | NCC2705 | AD011 | Bl-04 | DSM 10140 | Bb-12 | |||||
| Peptidoglycan biosynthesis | ||||||||||
| 2.5.1.7 | murA; UDP-N-acetylglucosamine 1-carboxyvinyltransferase | BLD_1600 | BL1267 | Blon_2321 | BAD_0183 | BDP_0275 | BLA_0233 | Balac_0245 | Balat_0245 | Unannotatedc |
| 1.1.1.158 | murB; UDP-N-acetylmuramate dehydrogenase | BLD_1681 | BL1561 | Blon_2254 | BAD_0294 | BDP_0408 | BLA_0345 | Balac_0364 | Balat_0364 | BIF_00678 |
| 6.3.2.8 | murC; UDP-N-acetylmuramate-alanine ligase | BLD_0182 | BL1324 | Blon_0857 | BAD_1100 | BDP_1541 | BLA_0775 | Balac_1197 | Balat_1197 | BIF_00646 |
| 6.3.2.9 | murD; UDP-N-acetylmuramoylalanine-d-glutamate ligase | BLD_0179 | BL1321 | Blon_0854 | BAD_1103 | BDP_1544 | BLA_0778 | Balac_1200 | Balat_1200 | BIF_00509 |
| 6.3.2.13 | murE; UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase | BLD_0221 | BL1356 | Blon_0893 | BAD_1085 | BDP_1521 | BLA_0746 | Balac_1168 | Balat_1168 | BIF_01077 |
| 6.3.2.4 | ddlA; d-alanine-d-alanine ligase | BLD_1112 | BL0345 | Blon_0320 | BAD_0186 | BDP_0283 | BLA_0236 | Balac_0248 | Balat_0248 | BIF_01368 |
| 6.3.2.10 | murF; UDP-N-acetylmuramoylalanyl-d-glutamyl-2,6-diaminopimelate-d-alanyl-d-alanine ligase | BLD_0177 | BL1319 | Blon_0852 | BAD_1105 | BDP_1546 | BLA_0780 | Balac_1202 | Balat_1202 | BIF_00378 |
| 2.7.8.13 | mraY; phospho-N-acetylmuramoyl-pentapeptide transferase | BLD_0178 | BL1320 | Blon_0853 | BAD_1104 | BDP_1545 | BLA_0779 | Balac_1201 | Balat_1201 | BIF_00379 |
| 2.4.1.227 | murG; UDP-N-acetylglucosamine-N-acetylmuramoyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | BLD_0181 | BL1323 | Blon_0856 | BAD_1101 | BDP_1542 | BLA_0776 | Balac_1198 | Balat_1198 | Unannotatedd |
| 2.4.1.129 | Peptidoglycan glycosyltransferase | BLD_0175 | BL1317 | Blon_0079, Blon_0850 | BAD_0040, BAD_1107 | BDP_1548 | BLA_0077, BLA_0782 | Balac_1204 | Balat_1204 | BIF_00315 |
| 3.6.1.27 | bacA; undecaprenyl-diphosphatase | BLD_0446 | BL0721 | Blon_1151 | BAD_0825 | BDP_1149 | BLA_1353 | Balac_0838 | Balat_0838 | BIF_01744 |
| 3.4.16.4 | dacB; d-alanyl-d-alanine carboxypeptidase | BLD_1845 | BL1679 | Blon_0546 | BAD_0407 | BDP_0546 | BLA_0471 | Balac_0496 | Balat_0496 | BIF_01386 |
| Rhamnose biosynthesis | ||||||||||
| 2.7.7.9 | ugpA; UTP-glucose-1-phosphate uridylyltransferase | BLD_0464 | BL0739 | Blon_1169 | BAD_0816 | BDP_1163 | BLA_1330 | Balac_0808 | Balat_0808 | BIF_00196 |
| 2.7.7.24 | rfbA; glucose-1-phosphate thymidylyltransferase | BLD_1570 | BL0227 | BAD_1509 | BDP_1860 | BLA_0581 | Balac_1376 | Balat_1376 | BIF_01544 | |
| 4.2.1.46 | rfbB; dTDP-glucose 4,6-dehydratase | BLD_1568 | BL0229 | Blon_2390 | BAD_1507 | BDP_1862 | BLA_0583 | Balac_1378 | Balat_1378 | BIF_01541 |
| 5.1.3.13 | rfbC (fuseda); dTDP-4-dehydrorhamnose 3,5-epimerase | BLD_1569 | BL0228 | BAD_1508 | Unannotatedb | BLA_0582 | Balac_1377 | Balat_1377 | BIF_01543 | |
| 1.1.1.133 | rfbD (fuseda); dTDP-4-dehydrorhamnose reductase | |||||||||
rfbC and rfbD are fused in bifidobacteria.
This gene (nucleotides [nt] 2,123,091 to 2,124,545) in B. dentium Bd1 was not annotated in the original annotation data (CP001750).
This gene (nt 928,929 to 930,257) in B. animalis subsp. lactis Bb-12 was not annotated in the original annotation data (CP001853).
This gene (nt 77,792-78,976, complement) in B. animalis subsp. lactis Bb-12 was not annotated in the original annotation data (CP001853).
EPS Production
EPS are produced by numerous bacteria, including LAB and bifidobacteria (265). Bacterial EPS can be either homopolysaccharides containing only one sugar monomer or heteropolysaccharides containing a few or several types of sugar monomers (35). Most LAB produce heteropolysaccharides consisting of glucose, galactose, and rhamnose, generally at a 2:1.3:1 ratio (201, 243, 261, 320). While EPS production in various LAB has been studied extensively, there are very few studies on EPS production in bifidobacteria. Analysis of the EPS produced by B. longum BB-79 revealed that it consisted of galactose and another hexose with lactic acid (250), suggesting that it may be quite different from the EPS produced by the LAB. Interestingly, EPS biosynthesis by a B. animalis subsp. lactis strain was promoted in the presence of bile salt, suggesting that induction of EPS biosynthesis by bile salt may be involved in self-protection against this toxic compound (260). While this may be one function of EPS production by bifidobacteria, another appears to be as a fermentable carbohydrate source for other gut bacteria, including bifidobacteria (264). While some have speculated that bifidobacterial EPS may have immunomodulatory effects on the host, it was not found to stimulate lymphocyte proliferation or cytokine secretion, unlike cell wall extracts (8).
While there is no information on EPS production by the nine bifidobacteria whose genomes have been sequenced, they all contain clusters carrying genes predicted to be involved in EPS biosynthesis. Interestingly, these clusters also contain genes whose annotations suggest involvement in EPS production but which are not present in the EPS gene clusters of the LAB (Fig. 6). This may support a difference in the general EPS structures of bifidobacteria and LAB. The four strains of B. animalis subsp. lactis and B. dentium Bd1 carry EPS gene clusters containing most of the essential genes for EPS biosynthesis, in contrast to the other sequenced bifidobacteria. Compared to the EPS gene cluster of Lactobacillus rhamnosus ATCC 9595, these gene clusters contain all of the genes involved, except for glucose transferase (Fig. 6), as well as some extra genes with annotations suggesting that they may have EPS biosynthesis abilities. The other genomes contain EPS gene clusters but are missing more of the genes than L. rhamnosus ATCC 9595.
FIG. 6.
Comparison of predicted EPS gene clusters in bifidobacterial genomes. The EPS biosynthetic gene cluster in Lactobacillus rhamnosus ATCC 9595 was obtained from the work of Peant et al. (228). All genes were categorized according to their potential functions, as follows: A, chain-length determination; B, Wzx flippase for EPS synthesis; C, glycosyltransferase; D, glucose transferase; E, rhamnosyltransferase; F, polysaccharide polymerase/oligosaccharide repeat unit polymerase; G, polysaccharide pyruvyl transferase; H, dTDP-glucose pyrophosphorylase; I, dTDP-4-dehydrorhamnose-3,5-epimerase; J, dTDP-d-glucose-4,6-dehydratase; K, dTDP-4-keto-l-rhamnose reductase; I/K, fused protein of I and K; and L, priming glycosyltransferase/galactosyltransferase/UDP-galactose phosphotransferase. Genes with red numbers are bifidobacterium-specific genes predicted to be involved in EPS biosynthesis, as follows: 1, extracellular exopolygalacturonase; 2, UDP-glucose/GDP-mannose dehydrogenase; 3, UDP-N-acetylglucosamine/glucuronate/galacturonate-4-epimerase; 4, β-1,4-galactosyltransferase enhancer; and 5, UDP-galactopyranose mutase. Gray arrows indicate genes involved in dTDP-rhamnose precursor biosynthesis. Sky blue arrows indicate hypothetical genes. The red boxes indicate IS mobile elements. MIC, mobile integrase cassette consisting of three contiguous integrase genes (green) sandwiched by two IS3 mobile elements (161). The genome locations of the gene clusters are indicated in kilobases.
Lantibiotic Production
Bacteriocin production is an important characteristic for bacterial competition in natural ecosystems. Bacteriocins are small antimicrobial peptides which inhibit the same or closely related species (144). Lantibiotics are a very broad-spectrum class of bacteriocins, and their composition differs from that of other bacteriocins, as they contain posttranslationally modified amino acids, such as lanthionine and α-methyllanthionine, and cyclic structures (44). Recently, the first bifidobacterial lantibiotic was detected and partially purified from B. longum subsp. longum DJO10A, and it showed a broad antimicrobial spectrum of activity (161). This lantibiotic was encoded by a characteristic lantibiotic operon, with genes encoding production, modification, and regulation functions (161). This operon consists of genes encoding a two-component signal transduction system (lanR2 and lanK), a lantibiotic prepeptide (lanA), a lantibiotic response regulator (lanR1), lantibiotic modification enzymes (lanD and lanM), a lantibiotic immunity protein (lanI), and a lantibiotic transporter with predicted protease activity for removal of the leader peptide during secretion (lanT) (Fig. 7). The comparative genome analysis of bifidobacteria also showed the homologous remnants of this lantibiotic operon in the genomes of B. longum subsp. infantis ATCC 15697 and B. angulatum DSM 20098 (Fig. 7), suggesting the possibility of widespread lantibiotic production in bifidobacteria.
FIG. 7.
Comparison of the lantibiotic operon in B. longum subsp. longum DJO10A with partial operons in B. longum subsp. infantis ATCC 15697 and B. angulatum DSM 20098. Red arrows indicate IS elements, and white arrows indicate the component genes for lantibiotic production, modification, secretion, and immunity. Amino acid sequence identities with B. longum subsp. longum DJO10A are indicated. The asterisk indicates that the lower overall identity is due to an internal gene deletion, with the common gene regions sharing almost 100% identity. Genes: lanR2, response regulator gene; lanK, histidine kinase gene; lanA, lantibiotic prepeptide gene; lanR1, response regulator gene; lanD, prepeptide modification enzyme gene; lanM, lantibiotic modifying enzyme gene; lanI, lantibiotic immunity protein gene; lanT, lantibiotic transporter/protease fusion protein gene. The gene numbers of the gene analogs in B. angulatum DSM 20098 and B. longum subsp. infantis are indicated.
Metabolism of Simple Sugars
The key hexose metabolism pathway in bifidobacteria involves the enzyme fructose-6-phosphate phosphoketolase (F6PPK) and is often called the “bifid shunt,” as bifidobacteria are among the few bacteria to contain this enzyme. Hexoses such as glucose or fructose are metabolized via this pathway to acetate and lactate for energy production (25, 72, 74, 75, 269, 271). The completely sequenced bifidobacterial genomes revealed all the required genes for the F6PPK-mediated bifid shunt, as well as some genes for the trichloroacetic acid (TCA) cycle, except for the genes encoding 2-oxoglutarate dehydrogenase (EC 1.2.4.2), malate dehydrogenase (EC 1.1.1.37), and fumarase (EC 4.2.1.2) (Fig. 4). Since bifidobacteria are among the few intestinal bacteria to have F6PPK, it is the usual diagnostic enzyme used for these bacteria. However, there are some exceptions, such as Gardnerella vaginalis, which has a close phylogenetic relationship with the genus Bifidobacterium (94), and the Gram-negative bacterium Acetobacter xylimum (277). The sequenced bifidobacterial genomes contain genes for the utilization of a wide variety of substrates, such as glucose, fructose, galactose, N-acetylglucosamine, N-acetylgalactosamine, arabinose, xylose, ribose, sucrose, lactose, cellobiose, melibiose, gentobiose, maltose, isomaltose, raffinose, and mannose (Fig. 4). Carbohydrate fermentation studies with B. longum subsp. longum NCC2705 and B. dentium Bd1 experimentally validated many of these substrates (218, 335). Furthermore, a recent molecular study of ribose utilization by B. breve UCC2003 revealed the presence of a functional complete ribose transfer and utilization gene cluster containing rbsACBDK, which is regulated by a LacI-type RbsR transcriptional repressor (232).
While the majority of bacteria preferentially utilize simple sugars such as glucose over more complex forms, many bifidobacteria preferentially utilize di- or oligosaccharides (7, 340). The classical substrate comparison is coculture with glucose and lactose, whereby bacteria generally display catabolite repression of lactose utilization until glucose is exhausted. B. longum does the opposite, whereby glucose utilization experiences catabolite repression by lactose (143, 219). A microarray analysis of B. longum NCC2705 revealed that the only gene downregulated by lactose was a glucose-specific major facilitator superfamily (MFS)-type glucose:H+ symporter gene (glcP), suggesting that only glucose transport experienced catabolite repression by lactose (219). The preference for utilizing di- and oligosaccharides, rather than monosaccharides, is likely an evolutionary adaptation to an environment that is rather devoid of monosaccharides but rich in complex carbohydrates. While lactose is utilized by the host, it is poorly metabolized in many adults and thus frequently reaches the large gut habitat.
Based on complete genome sequence analysis of bifidobacteria and whole-genome microarrays, sugar transportation is predicted to occur via permeases belonging to four families (218). These families are ATP-binding cassette (ABC)-type transport systems for lactose, raffinose, and maltose, a glucose-specific phosphotransferase transport system (PTS) for glucose, MFS systems for lactose, glucose, and sucrose, and the glycoside-pentoside-hexuronide (GPH) cation symporter family for melibiose and pentosides (Fig. 4). Interestingly, the comparative analysis of other complete bifidobacterial genomes also showed that these sugar transport systems are present in all bifidobacterial genomes, except for the four genomes of B. animalis subsp. lactis, which do not show any gene homologs for PTS-type transport systems (Table 13).
TABLE 13.
Comparative analysis of COG carbohydrate metabolism category (category G) in bifidobacterial genomes
| Functional category and COG no. | No. of genes |
COG annotation | ||||||
|---|---|---|---|---|---|---|---|---|
|
B. longum subsp. longum |
B. longum subsp. infantis ATCC 15697 | B. adolescentis ATCC 15703 | B. dentium Bd1 |
B. animalis subsp. lactis |
||||
| DJO10A | NCC2705 | AD011, Bl-04, and DSM 10140 | Bb-12 | |||||
| Non-PTS sugar transport system | ||||||||
| COG0395 | 13 | 10 | 16 | 8 | 19 | 5 | 5 | Sugar permeases |
| COG0477 | 29 | 23 | 41 | 19 | 31 | 15 | 16 | Permeases of the major facilitator superfamily |
| COG0580 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | Glycerol uptake facilitator and related permeases (major intrinsic protein family) |
| COG0697 | 3 | 3 | 4 | 4 | 4 | 1 | 0 | Permeases of the drug/metabolite transporter superfamily |
| COG0738 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | Fucose permease |
| COG1129 | 4 | 4 | 0 | 0 | 0 | 1 | 1 | ABC-type sugar (aldose) transport system, ATPase component |
| COG1134 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | ABC-type polysaccharide/polyol phosphate transport system, ATPase component |
| COG1172 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | Ribose/xylose/arabinose/galactoside ABC-type transport systems, permease components |
| COG1175 | 11 | 8 | 12 | 7 | 16 | 5 | 5 | ABC-type sugar transport systems, permease components |
| COG1653 | 14 | 10 | 20 | 10 | 0 | 4 | 3 | Sugar-binding periplasmic proteins/domains |
| COG1682 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ABC-type polysaccharide/polyol phosphate export systems, permease components |
| COG1879 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | Periplasmic sugar-binding proteins |
| COG2182 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | Maltose-binding periplasmic proteins/domains |
| COG2211 | 1 | 2 | 4 | 2 | 0 | 1 | 2 | Na+/melibiose symporter and related transporters |
| COG2610 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | H+/gluconate symporter and related permeases |
| COG2814 | 2 | 2 | 2 | 1 | 0 | 1 | 1 | Arabinose efflux permease |
| COG3833 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | ABC-type maltose transport systems, permease components |
| COG3839 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | ABC-type sugar transport systems, ATPase components |
| COG4209 | 0 | 0 | 3 | 1 | 3 | 0 | 0 | ABC-type polysaccharide transport systems, permease components |
| COG4213 | 1 | 2 | 1 | 1 | 3 | 1 | 1 | ABC-type xylose transport systems, periplasmic components |
| COG4214 | 1 | 2 | 0 | 0 | 2 | 0 | 0 | ABC-type xylose transport systems, permease components |
| COG4975 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | Putative glucose uptake permease |
| Subtotal | 93 | 78 | 114 | 61 | 91 | 43 | 41 | |
| PTS sugar transport system | ||||||||
| COG1080 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | Phosphoenolpyruvate protein kinase (PTS system EI component in bacteria) |
| COG2190 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | Phosphotransferase system IIA components |
| COG1263 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | Phosphotransferase system IIC components, N-acetylglucosamine specific |
| COG1925 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Phosphotransferase system, HPr-related proteins |
| Subtotal | 3 | 3 | 4 | 2 | 4 | 0 | 0 | |
ABC-type sugar transporters are primary active transporters coupling ATP hydrolysis to efficient uptake of sugars across the cell membrane (64, 134). While ABC transporters are the most popular transport system for sugars in bifidobacteria, there is little functional knowledge of these transporters which is specific to these bacteria. A proteomic analysis of B. longum showed that a fructose-specific ABC sugar-binding protein (BL0033) and an ATPase (BL0034) were highly produced in the presence of fructose compared to those in the presence of glucose, suggesting that this ABC-type transporter is indeed fructose specific and may play an important role in the uptake of fructose (353). In addition to sugar uptake, ABC-type transporters can also function in efflux of cytotoxic compounds out of the cell. A functional study of two genes in B. breve annotated as encoding ABC-type multidrug efflux transporters revealed that coexpression of both genes in Lactococcus lactis resulted in enhanced antibiotic resistance activity, suggesting that this efflux pump is functional for antibiotic resistance in bifidobacteria (171). The existence of these multidrug transporters in gut bacteria is likely an evolutionary response to the use of antimicrobials in medicine and possibly to their use in food production and preservation systems.
While ABC-type transporters are present in all living organisms, the PEP-PTS has been detected only in bacteria (134). The first glucose-specific PTS in B. breve (66) and a potassium-dependent glucose-specific PTS in B. bifidum (152) were demonstrated and characterized biochemically without identification of PTS genes. Transcriptional analysis of a fru operon encoding a fructose-specific PTS in B. breve revealed that it was specifically induced by fructose (181). Subsequent biochemical verification that fructose is phosphorylated at the C-6 position confirmed the functionality of this fructose PTS in B. breve. This differs from the case for B. longum, which does not carry a fru operon and takes up fructose via an ABC transport system, as discussed above. Indeed, B. longum encodes only one PTS, which is glucose specific, suggesting a preference for ABC transporters by this species. Transcriptional analysis of this glucose PTS revealed very little expression, suggesting that glucose may be transported preferentially by the permease system (219). This is substantiated by the catabolite repression of only one gene, the glcP glucose permease gene, during growth on lactose.
Complex Carbohydrate and Polyol Utilization
Successful microbes in any habitat, including the large intestine, must evolve efficient mechanisms for utilizing the available nutrients. Most of the easily digestible simple sugars are generally absorbed or metabolized in the upper gut, and the host-indigestible complex carbohydrates (plant-derived dietary fibers, host-derived glycans, oligosaccharides, resistant starch, cellulose, hemicellulose, xylan, arabinofuran, arabinogalactan, pectins, and gums) and poorly metabolized polyols are utilized by intestinal bacteria in the lower gut (119). While it is generally assumed that competition for these nutrients is a significant competitive factor for microbes in the gut, this has not been demonstrated experimentally. The analysis of the complete genome sequences of bifidobacteria supports this assumption, as they contain numerous gene clusters predicted to be involved in the metabolism of these substrates (146, 161, 275, 280). Previous carbohydrate fermentation studies demonstrated that multiple species of Bifidobacterium have the capacity to utilize a variety of polysaccharides and gums, but in a strain-dependent fashion (54, 266). The capability to utilize host-indigestible complex carbohydrates is believed to be an important competitive feature among bacteria in the large intestine (218).
The sequenced bifidobacterial genomes contain genes predicted to encode many complex carbohydrate utilization enzymes, consistent with the expected biology of these bacteria (Table 14). Many of these genes are located in clusters, and their expression is predicted to be regulated by LacI-type repressors. The nine complete bifidobacterial genomes contain 7 to 11 of these predicted gene clusters, consisting of a LacI-type repressor gene, genes for an ABC-type ATP-dependent transport system, and various types of catabolic genes (Table 7). Interestingly, the B. longum subsp. longum genomes contain a large number of genes predicted to be involved in metabolism of plant-derived dietary fibers, such as xylan, arabinan, and arabinofuran, while the other seven genomes contain very few (Table 14). This suggests that B. longum subsp. longum may be very well adapted to competition in guts with diets rich in plant-derived complex carbohydrates. Conversely, B. longum subsp. infantis carries the most genes predicted to be involved in utilization of HMOs, consistent with it being a dominant member of a breast-fed infant's intestine.
TABLE 14.
Predicted genes for complex carbohydrate and polyol utilization in bifidobacterial genomes
| Enzyme | Substrate | No. of genes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
B. longum subsp. longum |
B. longum subsp. infantis ATCC 15697 | B. adolescentis ATCC 15703 | B. dentium Bd1 |
B. animalis subsp. lactis |
||||||
| DJO10A | NCC 2705 | AD011 | Bl-04 | DSM 10140 | Bb-12 | |||||
| β-Fructofuranosidase/inulinase | Fructofuran | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cyclomaltodextrinase/neopullulanase | Cyclodextrin | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pullulanase/glycogen-debranching enzyme | Pullulan | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 |
| Galactofuranosyltransferase | Galactofuran | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| α-l-Arabinofuranosidase | Arabinofuran | 6 | 4 | 1 | 2 | 3 | 1 | 1 | 1 | 1 |
| Exo-α-l-arabinofuranosidase II | Arabinofuran | 2 | 2 | 0 | 2 | 1 | 0 | 0 | 0 | 0 |
| α-N-Arabinofuranosidase | Arabinofuran | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Arabinogalactan endo-1,4-β-galactosidase | Arabinogalactan | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Arabinan endo-1,5-α-L-arabinosidase | Arabinan | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Glycanase/glycogenase | Glycan | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1,4-α-Glucan branching enzyme | Glucan | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 4-α-Glucanotransferase | Glucan | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Glucan/glycogen phosphorylase | Glucan | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| β-1,4-Endoglucanase/cellulase | Glucan | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| β-1,3-Exoglucanase | Glucan | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 0 |
| Xylan esterase | Xylan | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Endo-1,4-β-xylanase | Xylan | 2 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 1 |
| Extracellular exoxylanase | Xylan | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| β-Glucuronidase | Glycoside | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| α-1,4-Glucosidase | Glycoside | 1 | 1 | 1 | 1 | 6 | 1 | 1 | 1 | 1 |
| Oligo-1,6-glucosidase | Glycoside | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| β-d-glucosidase | Glycoside | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 2 |
| Thermostable β-glucosidase B | Glycoside | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 |
| Endo-β-N-acetylglucosaminidase | Glycoprotein | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| N-Acyl-d-glucosamine 2-epimerase | Glycoprotein | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 |
| Mannitol dehydrogenasea | Mannitol | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| Sorbitol dehydrogenasea | Sorbitol | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| β-N-Acetylhexosaminidaseb | Glycan | 2 | 2 | 2 | 1 | 0 | 1 | 1 | 1 | 1 |
| Lacto-N-biose phosphorylaseb | Glycan | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| α-l-fucosidaseb | Glycan | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 |
| Exo-α-sialidaseb | Glycan | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 38 | 33 | 24 | 21 | 38 | 17 | 17 | 17 | 20 | |
While polyol utilization is predicted to be an important carbon source for microbes in the large intestine, the genomes of B. longum subsp. longum DJO10A, B. adolescentis ATCC 15703, and B. dentium Bd1 are the only ones containing genes predicted to be involved in polyol metabolism (161, 335). The presence of these genes predicts that these bacteria can metabolize mannitol or sorbitol, and microarray analysis and in vitro fermentation studies with B. longum subsp. longum DJO10A and B. dentium Bd1 have demonstrated this phenotype experimentally for these strains (335; J. H. Lee and D. J. O'Sullivan, unpublished data). In addition, an analysis of the draft genome sequences of B. breve DSM 20213, B. catenulatum DSM 16992, B. adolescentis L2-32, and B. dentium ATCC 27678 revealed analogs for the polyol utilization gene cluster, suggesting its prevalence among different species of Bifidobacterium (Table 15).
TABLE 15.
Occurrence of B. longum subsp. longum DJO10A polyol utilization gene analogs in other bifidobacteria
| Genea | Annotation | Locus tag |
|||||
|---|---|---|---|---|---|---|---|
| B. adolescentis ATCC 15703 | B. dentium Bd1 | B. breve DSM 20213 | B. catenulatum DSM 16992 | B. adolescentis L2-32 | B. dentium ATCC 27678 | ||
| BLD_1696 | Mannitol dehydrogenase | BAD_0313 | BDP_0423 | BIFBRE_00261 | BIFDEN_00355 | ||
| BLD_1697 | Mannitol transporter | BAD_0316 | BDP_0424 | BIFBRE_00262 | BIFCAT_01546 | BIFADO_00565 | BIFDEN_00354 |
| BLD_1698 | Lactoylglutathione lyase | BAD_0314 | BIFBRE_00263 | BIFCAT_01548 | BIFADO_00562 | ||
| BLD_1699 | Sorbitol transporter | BAD_0316 | BDP_0424 | BIFBRE_00266 | BIFCAT_01546 | BIFADO_00565 | BIFDEN_00354 |
| BLD_1700 | Sorbitol dehydrogenase | BAD_0317 | BIFBRE_00267 | BIFCAT_01545 | BIFADO_00566 | ||
| BLD_1701 | Transcriptional regulator | BAD_0318 | BDP_0425 | BIFBRE_00268 | BIFCAT_01544 | BIFADO_00567 | |
Polyol utilization genes in B. longum subsp. longum DJO10A.
Secretion Systems and Cell Surface Proteins
Extracellular and cell surface components are predicted to be important for attachment of bacteria to cells and surfaces. The SignalP program (20, 205) detected signal peptides encoded in more than 200 predicted ORFs, including those for 59 surface-associated lipoproteins and 26 solute-binding proteins of ABC transport systems, in B. longum subsp. longum NCC2705 (275). Interestingly, these predicted secretion proteins are analogous to several polymer hydrolysis enzymes, such as xylanase, arabinosidase, arabinogalactan endo-1,4-β-galactosidase, and N-acetylglucosaminidase, which could function to digest plant cell wall material and extracellular polysaccharides to smaller sugars and oligosaccharides.
A functional analysis of all nine bifidobacterial genomes by use of the updated KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database revealed that the protein export systems of bifidobacteria contain Sec-dependent pathways, type I and II signal peptidases, and components of signal recognition particle (SRP)-dependent pathways. These pathways function together for protein secretion, whereby the SRP receptor is proposed to bind to the signal peptide of the prepeptide to facilitate the action of the signal peptidases and the Sec translocase, located in the cell membrane, for secretion of the cleaved peptide (61).
Many surface proteins are covalently linked to the peptidoglycan layer through the action of sortase transpeptidases. The sortase enzymes recognize a conserved LPXTG motif in proteins targeted for cell wall anchoring, cleave proteins between the threonine-glycine bonds, and catalyze transpeptidation to a peptidoglycan cross-linking spacer (146). The LPXTG motif is a signature of one class of cell surface anchoring proteins (317). While the majority of the LPXTG-type proteins encoded by the bifidobacterial genomes are unknown, one is a weakly conserved homolog of FimA and is encoded in all of the genomes except for that of B. longum subsp. infantis. The LPXTG-type proteins are potential cell surface proteins that may be involved in attachment to cells or mucus in the gut. Interestingly, while the bifidobacterial genomes share some analogs encoding putative cell surface proteins with LPXTG motifs, there are also species-specific genes encoding LPXTG-type proteins (Table 16). While both strains of B. longum subsp. longum carry the same gene analogs for LPXTG-type proteins, one of them, in strain NCC2705, is missing the 3′ area of the gene that encodes the LPXTG motif. Given that strain DJO10A was sequenced before extensive culturing outside the gut, unlike strain NCC2705, this observation supports the tendency of bacteria to undergo genome reduction during pure culture growth in regions that are no longer needed for the new environment (161).
TABLE 16.
Genes predicted to be involved in secretion and cell attachment in bifidobacteria
| Functional category and gene annotationa | Locus tagb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
B. longum subsp. longum |
B. longum subsp. infantis ATCC 15697 | B. adolescentis ATCC 15703 | B. dentium Bd1 |
B. animalis subsp. lactis |
|||||
| DJO10A | NCC2705 | AD011 | Bl-04 | DSM 10140 | Bb-12 | ||||
| Sortases | BLD_0779 | BL1028 | Blon_1841 | BDP_0867 | BLA_1023 | Balac_1023 | Balat_1023 | BIF_01167 | |
| srtA1 | BLD_1287 | BL0498 | Blon_0162 | BAD_0087 | BDP_0197 | BLA_0681 | Balac_1486 | Balat_1486 | BIF_01042 |
| BAD_1470 | BDP_2002 | ||||||||
| BDP_0536 | |||||||||
| BDP_0142 | |||||||||
| srtA2 | BLD_1385 | BL0591 | Blon_0075 | BAD_0036 | BDP_0036 | BLA_0073 | Balac_0082 | Balat_0082 | BIF_01566 |
| Blon_1280 | |||||||||
| srtA3 | BLD_1458 | ||||||||
| srtA4 | BLD_1469 | BL0676 | BAD_0122 | BDP_2188 | BLA_0554 | Balac_1349 | Balat_1349 | BIF_00667 | |
| BAD_1551 | BDP_0278 | ||||||||
| LPXTG-type cell surface- | BLD_1189 | BL0387 | Blon_0283 | BIF_01518e | |||||
| anchoring secretion | BLD_1468 | BL0675 | BAD_1467 | BLA_0679 | Balac_1484 | Balat_1484 | BIF_00999 | ||
| proteins | BLD_1637 | BL1539 | BDP_0143 | BLA_0680 | Balac_1485f | Balat_1485f | BIF_00998f | ||
| BLD_1638c | BL1540c | ||||||||
| BLD_1511c | BL0146c,d | ||||||||
| Blon_1005c | |||||||||
| Blon_1227c | |||||||||
| Blon_2163c | |||||||||
| BAD_1451c | |||||||||
| BDP_0144c | |||||||||
| BDP_0200c | |||||||||
| BDP_0276c | |||||||||
| BDP_0277c | |||||||||
| BDP_0535c | |||||||||
| BLA_1540c | Balac_1618c,f | Balat_1618c,f | BIF_01674c,f | ||||||
Based on the B. longum subsp. longum DJO10A annotation.
Genes on the same line are analogs.
Species-specific predicted LPXTG-containing cell surface protein.
LPXTG motif missing because of a predicted ISL3 deletion event.
LPXTG motif is missing.
No predicted secretion ability.
Most Eubacteria and Archaea have a tad (tight adherence) locus involved in pilus construction and believed to be important for adherence to cell surfaces or mucus (135). Interestingly, all of the bifidobacterial genomes contain a few highly conserved sortase genes and gene analogs of tadABC, arranged in an operon, as well as tadV, which may be involved in pilus assembly for cell surface adherence. The tadA gene analog encodes a putative ATPase that is proposed to be involved in the secretion of a FimA prepilin, which is also predicted to be present in bifidobacteria. The tadBC genes are proposed to encode a transport system for the prepilin, and tadV encodes a putative peptidase for maturation of the prepilin. This tad locus is similar in structure to other loci in other actinobacteria, such as Streptomyces coelicolor and Corynebacterium diphtheriae (135). These surface protein export systems and pilus construction capabilities of bifidobacteria may play an important role in their attachment to mucus or cells in the intestine.
Stress Response
Oxygen tolerance.
Even though bifidobacteria are generally described as strict anaerobes, some of them can tolerate some oxygen (269, 293). Their oxygen sensitivities are variable (73, 137, 284), with the fermented milk-adapted organism B. animalis subsp. lactis having the highest oxygen tolerance (185). While anaerobic bacteria differ in their sensitivity to oxygen, some have different degrees of tolerance due to the activities of some enzymes to remove reactive oxygen species (ROS), such as NADH oxidase, NADH peroxidase, and superoxide dismutase (SOD). SOD can eliminate superoxide radicals, and previous studies have reported SOD activity in bifidobacteria (284, 285, 306). However, the nine completed genome sequences of bifidobacteria do not reveal any SOD-related genes. The discrepancy may be explained by the proposal that SOD activity measurements in bifidobacteria were probably false-positive results, based on comparisons with known SOD enzymes (42, 160). This proposal is supported by the genome analysis.
Although previous studies have shown that bifidobacteria utilize NADH oxidase/peroxidase-like systems for the protection of their cells from ROS (284), the complete genome sequences of bifidobacteria revealed a gene analog only for the NADH oxidase gene. The reaction of oxygen and NADH with NADH oxidase produces and results in accumulation of hydrogen peroxide (H2O2), which has many destructive properties, including inhibition of the F6PPK enzyme (73). The absence of catalase activity (or gene analogs for this enzyme) in bifidobacteria indicates that some other mechanism must be present to prevent the accumulation of H2O2 during exposure to oxygen. Previous studies indicated that different Bifidobacterium species did not accumulate H2O2 during exposure to oxygen (284, 285, 306). These studies proposed that this was due to NADH peroxidase activity, which converts H2O2 to water by utilizing NADH. However, none of the genome sequences of bifidobacteria contain gene analogs for NADH peroxidase, suggesting that there may be an alternative mechanism. Interestingly, all of the genome sequences of bifidobacteria, except for that of B. adolescentis ATCC 15703, reveal peroxiredoxin family alkyl hydroperoxide reductase gene analogs, which have been shown to be involved in the reduction of H2O2 in E. coli and Streptococcus mutans (233, 278). An alternative system for H2O2 reduction to H2O is the reduction of thioredoxin by thioredoxin reductase and NADH. Thioredoxin systems are prevalent in nature and are proposed to have multiple functions, including protection from oxidative stress (10, 198). All of the genomes of bifidobacteria carry gene analogs for thioredoxin and thioredoxin reductase, supporting this alternative mechanism for removal of H2O2 from bifidobacteria during exposure to oxygen, and this could explain the limited tolerance of many bifidobacteria to air. In addition, the bifidobacterial genomes contain oxidative damage repair-related genes, including those for nucleoside triphosphate pyrophosphohydrolase (MutT) and DNA-binding ferritin-like protein (Dps), which are involved in removing hydroxyl radicals (91).
Acid tolerance.
The FoF1-type ATPase system is involved in acid tolerance by pumping protons out of the cell. All nine bifidobacterial genomes contain a putative atp operon which carries all genes needed for a complete FoF1-type ATPase system. Previous studies have also demonstrated the activity of the ATPase system in different species of Bifidobacterium in response to acid stress (175, 326). While all nine bifidobacterial genomes contain gene analogs for this system, the acid tolerance of B. animalis subsp. lactis strains is superior to that of all other members that colonize the gut. Interestingly, the genomes of the gut bifidobacteria do not reveal additional stress-related genes, suggesting that these bacteria may have an alternative expression pattern for the atp operon, such that it is active before exposure to more acidic conditions. Alternatively, other novel mechanisms may be present in these “fermentation-friendly” bifidobacteria. Recent genome analysis of B. dentium Bd1 revealed another acid tolerance system, the glutamate-dependent acid resistance system 2, which encodes a glutamate decarboxylase (GadB) and a glutamate/gamma-aminobutyrate antiporter (GadC) (335). This likely gives this oral resident more tolerance to acid than that of the gut bifidobacteria and possibly aids in its ability to induce acid damage to tooth enamel in dental caries.
Temperature tolerance.
While bifidobacteria do not encounter temperature stresses in their normal intestinal environment, they are subjected to temperature stresses when utilized for probiotic applications. All nine complete genome sequences of bifidobacteria carry several temperature stress-related gene analogs, including genes encoding the chaperone families Hsp100 (ClpBCX), Hsp70 (DnaK, GrpE, and DnaJ), and Hsp60 (GroEL/GroES complex) and the SOS response (LexA, RuvA, RecA, and MutY).
The transcription of all genes involved in the Hsp60 and Hsp70 chaperone families of Firmicutes and Proteobacteria is regulated by the HrcA-CIRCE (controlling inverted repeat of chaperone expression) system (328). The Actinobacteria, including all of the sequenced bifidobacteria, utilize the HrcA-CIRCE system for the transcriptional regulation of only some of the Hsp60 chaperone genes (groEL, groES, and dnaJ2). All of the genomes contain a gene encoding a putative HrcA protein, which is a transcriptional repressor of heat shock chaperone genes. The binding site for HrcA is a characteristically conserved inverted repeat (CIRCE) with the consensus sequence TTAGCACTC-N9-GAGTGCTAA (16, 356). CIRCE-like motifs were found in the promoter regions of dnaJ2 (ATTAGCACTC-N9-GAGTGCTAAT) and groES (GTTAGCACTC-N9-GAGTGCTAAC) in bifidobacteria. In addition, a recent computational analysis of the unpublished B. breve UCC2003 genome revealed the presence of conserved CIRCE-like motifs in the promoter regions upstream of the hrcA and groEL genes (355). Interestingly, two CIRCE-like motifs were detected in the promoter region of the groEL gene, and one CIRCE-like motif, as well as a ClgR-binding site, was found in the promoter region of the hrcA gene. The presence of a ClgR-binding site suggested that expression of the hrcA gene may be controlled by the ClgR transcriptional activator. Interestingly, the groEL genes are clustered with cspA (cold shock protein gene) in bifidobacteria, which is unusual but has also been reported for Leifsonia xyli subsp. xyli CTCB07 (192) and Streptomyces coelicolor A3(2) (22). While CIRCE-like elements were found in the promoter regions of dnaJ2 (ATTAGCACTC-N9-GAGTGCTAAT) and groES (GTTAGCACTC-N9-GAGTGCTAAC) in bifidobacteria, the promoter region of the cspA-groEL gene cluster contains two different inverted repeats. These are 10-bp (GCCACCATCA) and 8-bp (CGTTCCCT) inverted repeats, with one 5-bp direct repeat (CGCGA), all of which were previously described for B. breve, which was also shown to cotranscribe cspA and groEL (323, 329). A recent analysis of groEL indicated that it has very different expression levels from those of cspA and also identified an HrcA-regulated promoter (355). In vitro binding of purified HrcA with promoter regions of hrcA, groEL, and groES substantiated this observation. Analysis of B. breve gene expression under different stress conditions (42 to 50°C, ethanol stress, and osmotic stress) by use of a whole-genome microarray showed that the expression of the cspA gene was significantly expressed only during exposure to 50°C, while expression of the groEL gene was high under all stress conditions, supporting the presence of its own promoter (355).
The Hsp70 chaperone family also utilizes another characterized transcription regulatory mechanism, designated HAIR (HspR-associated inverted repeat), with the consensus sequence CTTGAGT-N7-ACTCAAG (37, 101). In Actinobacteria, HspR is an autoregulatory repressor of the dnaK operon (encoding the Hsp70 chaperone), which contains the hspR gene and the clpB gene. Recently, HAIR-like motifs were found in the promoter regions of the dnaK, clpB, and clgR genes of B. breve and were demonstrated experimentally to bind HspR, substantiating the involvement of this regulatory mechanism (355). Interestingly, the promoter regions of the nfo and hrdB genes also contained HAIR-like motifs but did not bind HspR. Since the HAIR-like motifs in the promoter regions of dnaK, clpB, and clgR have 3′-end (T-rich) and 5′-end (A-rich) extensions, unlike the HAIR-like motifs in nfo and hrdB, a role for these extensions in HspR binding was suggested, which was subsequently substantiated by a mutation analysis (355). This proposed a new consensus sequence for HAIR motifs in bifidobacteria, to include these extensions (AAASTTGAGYSW-N5-CTCAASTTTT). While the three HspR-repressed stress genes were found to be upregulated by mild heat stresses, they were significantly upregulated during very high heat stresses (50°C). They were also shown to be induced by ethanol and osmotic stresses (355). Recently, heat shock-tolerant mutants of B. longum subsp. longum were obtained by deliberate exposure to heat stress, and a transcriptome analysis revealed overexpression of the dnaK operon and clpB. Further analysis showed that the HspR repressor contained point mutations which were proposed to prevent its repressor activity, a phenotype that could be reversed by complementation with a wild-type hspR gene (23).
Interestingly, the bifidobacterial genomes carry two different dnaJ gene analogs (dnaJ1 within the dnaK operon and dnaJ2 within the GroEL/GroES gene cluster). The presence of two dnaJ genes, at the same locations, has also been found for the other sequenced genomes from the Actinobacteria phylum (323). However, the other major bacterial phyla within the GI tract, Firmicutes and Proteobacteria, carry only a single dnaJ gene, within the dnaK operon. Multiple dnaJ genes can also be found in the sequenced Archaea genomes and in the Eukarya. A phylogenetic tree of DnaJ protein sequences shows that bifidobacterial DnaJ1 clusters more closely with DnaJ proteins from Archaea and Eukarya, whereas DnaJ2 is not related to these, clustering on a similar branch with Proteobacteria, Firmicutes, and other Archaea DnaJ proteins (Fig. 8). This suggests that the bifidobacterial dnaJ genes have different ancestral origins.
FIG. 8.
Phylogenetic analysis of DnaJ proteins in Proteobacteria, Firmicutes, Archaea, and Eukarya compared with the two DnaJ proteins encoded by the nine complete bifidobacterial genomes (in bold).
A number of gene analogs for the Hsp100 chaperone system are present in the nine bifidobacterial genomes. These include clpC, which is clustered with a uspA analog (encoding universal stress protein), a clpP1-clpP2 operon, and clpX. The expression of this gene system has previously been shown to be regulated at the transcriptional level in bacteria by the ClgR transcription activator, which binds a palindromic sequence motif (CGC-N5-GCG; a ClgR-binding motif) in the promoter regions (84, 337). A clgR gene is also present in the bifidobacterial genomes, as well as a ClgR-binding motif upstream of the clpP1 gene in the apparent clpX-clpP1-clpP2 operon, which has previously been shown to be regulated by ClgR in bifidobacteria (337). Interestingly, this ClgR-binding motif upstream of clpC contains one mismatch (CTC-N5-GCG), which may explain why extracts of clgR overexpressed in E. coli did not bind this promoter region alone but did so when combined with extracts of B. breve, suggesting the requirement for a cofactor or helper protein for ClgR binding (331). Recently, an N-terminally truncated ClgR protein was shown to form complexes without additional cofactors with the upstream regions of the clpC and clpP1 genes, suggesting possible folding differences of this protein in E. coli and confirming that ClgR binds and regulates these promoters (355). A subsequent mutation analysis of the proposed ClgR-binding motif in the promoter region of clpC demonstrated that specific bases are important for interaction with ClgR, suggesting an updated consensus binding motif for bifidobacteria, i.e., TNCGCTNNNGGCGNAA, which is larger than the motif in Streptomyces but similar to the proposed motif in Corynebacterium (355).
Heat stresses are known to induce genes involved in the SOS response (85). For B. breve, a transcriptome analysis indicated that a number of SOS genes were upregulated by heat stress, including recA and recX, which are located directly downstream from the clgR two-gene cluster (355). This suggested the possibility that induction of clgR during heat stress may carry through to the downstream genes, which was substantiated by the similar induction levels for the four genes. This gene organization, clgR-hyp-recA-recX, can be seen in all the bifidobacterial genomes. A LexA SOS repressor is encoded by all the bifidobacterial genomes, and it has been shown to be induced by heat stress in B. breve (355). While a potential LexA-binding motif is present upstream of recA, suggesting a regulatory role, this has not been investigated experimentally.
Small heat shock proteins (sHSPs) are important to prevent irreversible protein denaturation caused by cellular stresses (203). Members of this protein family usually range from 12 to 30 kDa, but larger members have been found. They are not highly conserved, except for conserved α-crystalline domains. While most bacteria encode at least one sHSP, some, such as Helicobacter pylori, do not (203). Computational genome analysis of bifidobacterial genomes revealed that hsp20 gene analogs encoding a family of sHSPs are present in four of the nine completed genomes (all B. longum subspecies and B. adolescentis), and a BLAST analysis also showed analogs in B. breve, B. bifidum, and B. angulatum. In addition, hybridization analysis revealed putative hsp20 analogs in B. catenulatum and B. longum subsp. suis (327). This suggests that hsp20 gene analogs are not widely distributed in bifidobacteria but are relatively conserved in the genus. A phylogenetic analysis of Hsp20 protein sequences in bifidobacteria revealed that they grouped with proteins from the Firmicutes rather than the Actinobacteria, suggesting that these hsp20 gene analogs may have been obtained via horizontal gene transfer from Firmicutes. Expression analysis of the hsp20 analog in B. breve showed significant induction during heat and osmotic stresses (327, 355). However, sequence analysis of its upstream region did not reveal promoter-like sequences or other known motifs, suggesting a novel regulatory mechanism.
GENOME ADAPTATION OF BIFIDOBACTERIA TO THEIR HABITAT
Bifidobacterial Genomes Reveal Features Consistent with a GI Tract Habitat
The microbial population of the human large intestine is limited primarily to host-indigestible nutrients, such as HMOs in infants and plant-derived complex carbohydrates and polyols in adults. In this habitat, intestinal bifidobacteria are efficient utilizers of these types of nutrients, and a significant part of their genomes is dedicated to these capabilities, indicating their adaptation to this habitat (161, 275, 280).
Mammals, including humans, rely primarily on milk for dietary purposes during the infant stage of life. Fecal culturing studies of breast-fed human infants typically show a flora high in bifidobacteria (21, 299). However, the flora is very dynamic during the first year of life, as demonstrated using 16S rRNA gene sequence and PCR-denaturing gradient gel electrophoresis analyses (87). While a nonculturing study of two infants found that both contained high levels of bifidobacteria for up to 6 months during their breast-feeding period, these infants showed variable levels after that. The prominent levels of bifidobacteria in infants were substantiated by analyzing the feces of 1,032 1-month-old breast-feeding infants by quantitative reverse transcription-PCR and finding levels ranging from 106 to 1011 CFU/g feces in 98.6% of subjects (229). Interestingly, a recent study of 14 infants over the first year of life found a low abundance of Actinobacteria (1.28%) by sequence analysis of cloned 16S rRNA genes (217). In addition, this study developed a microarray with the cloned fragments and used it to follow the microbial profile for 1 year, and it did not find very high levels of bifidobacteria. This may reflect low bifidobacterial levels in this small group of infants or possibly some biases in the fecal DNA preparation whereby Actinobacteria were not represented quantitatively. It should be noted that five of these infants were delivered by Caesarian section rather than vaginally, and these infants had the lowest levels of bifidobacteria. A metagenomic study that included four Japanese infants of less than 7 months of age found that two of them contained a very large bifidobacterial population, one had a smaller but still significant population, and one did not show any bifidobacteria (156). This again demonstrates the variability of bifidobacterial populations in infants. In many infants, B. longum subsp. infantis is a very common inhabitant, while B. longum subsp. longum largely replaces this subspecies in adults (174, 247). A comparative genome analysis of both of these subspecies showed significantly more genes predicted to be involved in HMO utilization in B. longum subsp. infantis than in B. longum subsp. longum and, conversely, significantly more genes involved in utilization of plant-derived complex carbohydrates and polyols in B. longum subsp. longum (Table 14). Specifically, B. longum subsp. infantis contains seven genes predicted to be involved in HMO utilization, compared to three genes in B. longum subsp. longum. In addition, the intestinal B. longum subsp. longum strain DJO10A contains nine arabinofuranosidase and three xylanase genes, predicted to be involved in utilization of plant-derived complex carbohydrates, while B. longum subsp. infantis contains just one arabinofuranosidase gene. Furthermore, B. longum subsp. longum carries two polyol utilization genes, while B. longum subsp. infantis does not carry any (Table 14). While ecological conclusions cannot be drawn from only a few genome sequences, these genome characteristics are intriguing, as they suggest that the B. longum subsp. infantis genome may be well adapted for a breast-feeding infant's gut, while the B. longum subsp. longum genome encodes metabolic characteristics consistent with an adult gut.
Another genome insight into the adaptation of B. longum subsp. infantis is that it contains a complete urease gene cluster, which is not present in any of the other bifidobacterial genomes. This was previously demonstrated functionally in a study showing that of more than 100 strains of different species of Bifidobacterium, all 9 strains of B. longum subsp. infantis produced urease and none of the others did, except for 1 strain of B. bifidum among 12 strains tested for this species (302). Given that human milk contains a significant portion of its nitrogen in urea (78), this gene cluster should enable B. longum subsp. infantis to utilize urea for its nitrogen needs. Given the nutritional need of a growing infant for human milk protein, the ability to utilize the available urea may be a competitive advantage.
Genome Adaptation of Intestinal Bifidobacteria to Environments Outside the Gut
Of the nine completed genome sequences from bifidobacteria, only one (B. longum subsp. longum DJO10A) is from an isolate that was purposely minimally cultured outside the gut prior to sequencing (161). It is intriguing that this strain contains the most genes predicted to be involved in the metabolism of complex carbohydrates and polyols (Table 14). Conversely, the complete genome sequences of four strains of B. animalis subsp. lactis revealed that they have the fewest genes predicted to be involved in the utilization of these substrates. Specifically, these strains have limited or no predicted ability to metabolize polymers consisting of arabinofuran, arabinogalactan, arabinan, cyclodextrin, xylan, or sugar alcohols, and they have the only genomes missing a gene encoding β-1,3-exoglucanase, thereby limiting their ability to metabolize glucan-based polymers (Table 14). While B. animalis subsp. lactis was isolated from yogurt, it is not normally associated with traditional fermented milks and likely adapted to this environment following the inclusion of B. animalis subsp. animalis in yogurts for probiotic purposes. Interestingly, it has a high tolerance to oxygen (185), which is unusual for this genus, with the other exception being B. psychroaerophilum, which can even grow on agar media under aerobic conditions (291). While the origin of B. animalis subsp. animalis is animal intestines, the origin of B. animalis subsp. lactis is likely the dairy fermentation environment, and it was first isolated from yogurt. The prolonged exposure of this subspecies to dairy fermentation environments is further supported by the reduced size of the genomes of this subspecies, which is a phenomenon that has been found to occur in organisms that are in less-complex environments where fewer features are needed to be successful, such as the genome reduction of Buchnera aphidicola, which is predicted to be due to environmental changes (230). This may explain the inability of these bacteria to persist in the intestines of human subjects, as they have been observed to disappear rapidly after stoppage of eating foods containing these bacteria (88, 215, 300, 319).
In addition to the differences in complex carbohydrate and polyol utilization, the genome of B. longum subsp. longum DJO10A is the only one of the nine to carry a gene cluster involved in bacteriocin production, specifically that of a broad-spectrum lantibiotic, and gene clusters for arsenic resistance (161). These are features that would be beneficial for an intestinal habitat, as the ability to produce a lantibiotic can give the organism a competitive edge over other intestinal bacteria, and arsenic resistance is common in intestinal bacteria due to low levels of arsenic present in human diets. The predicted superior arsenic resistance of B. longum subsp. longum DJO10A was confirmed experimentally by comparison to strains of B. animalis subsp. lactis that do not contain arsenic resistance gene clusters (161).
Genome adaptation in bifidobacteria via genome reduction was suggested previously by comparative genome analysis of two strains of B. longum subsp. longum, namely, NCC2705, from a culture collection, and DJO10A, cultured minimally outside the gut (161). This analysis revealed that strain NCC2705 has a smaller genome, and it was predicted by a base deviation index analysis to have lost several gene clusters that are predicted to be involved in characteristics important for survival and competition in the gut. This was evaluated experimentally by culturing strain DJO10A in the laboratory, and after 1,000 generations, it was found to have lost two large genome regions, including one that encoded the lantibiotic, in a deletion event very similar to the deletion predicted for strain NCC2705. This deletion mutant of strain DJO10A was shown in a simulated fecal growth environment to have lost a significant proportion of its competitive abilities against other intestinal bacteria (161). This demonstration of genome reduction in bifidobacteria highlights the need to minimize pure culture growth of strains if they are to be utilized as probiotics and expected to function optimally in the GI tract following their ingestion. While commercial production requires large-scale culturing, this limitation of bifidobacteria could be minimized by always starting each culture batch from a frozen stock and developing culturing media that are more representative of the environment of the large gut, such as minimizing exposure to oxygen and acids as well as using a varied complex carbohydrate nutrient profile rather than simple sugars.
CONCLUDING REMARKS
Since bifidobacteria were first discovered in the feces of breast-fed infants more than 100 years ago, there has been a lot of interest in the potential health benefits these bacteria may afford the large intestine. However, surprisingly little is known about which features are important for their survival, colonization, competition, and interactions with other members of the intestinal microbiota. To further this understanding, a genome sequence analysis of bifidobacteria has provided novel molecular insights into their overall characteristics, physiology, and interactions with their habitat. This is exemplified by the presence in bifidobacterial genomes of many genes involved in the uptake and utilization of host-indigestible complex carbohydrates and polyols, supporting their genome adaptation to the human large intestine, where these are the predominant nutrient sources. While the number of genomes is too small to make reliable ecological predictions, the genomes of the two subspecies of B. longum do suggest a possible adaptation, as B. longum subsp. infantis contains a larger number of genes predicted to be involved in human milk oligosaccharide utilization, consistent with it being a dominant member of a breast-fed infant's bifidobacterial flora. Conversely, B. longum subsp. infantis contains fewer genes predicted to be involved in utilization of plant-derived complex carbohydrates and polyols, consistent with B. longum subsp. longum being a prominent member of an adult's bifidobacterial flora. However, since the culturing history of the B. longum subsp. infantis strain prior to sequencing is not known, it is possible that the absence of some of these genes may be due to attenuation. The only bifidobacterial genome sequence that was obtained from a strain that was minimally cultured outside the gut was from B. longum subsp. longum DJO10A, and it carried significantly more genes predicted to be involved in the utilization of complex carbohydrates and polyols, resistance to arsenic, and bacteriocin production, all of which are features predicted to be beneficial for competition in the human GI tract. Growth of this strain in the laboratory demonstrated experimentally that it can lose these features through genome deletion events. This may explain the small genome size of B. animalis subsp. lactis, which contains the fewest genes encoding these features and is predicted to have evolved from B. animalis subsp. animalis from extended exposure to a dairy fermentation environment. This readaptation of the B. animalis subsp. lactis genome may explain the poor colonization abilities that were observed in clinical feeding studies.
The bifidobacterial genome sequences have provided new insights into the biology of these bacteria in the human GI tract. Specifically, they have revealed genomes with extensive features that can be predicted to be important for survival and competition in the large intestine, such as complex carbohydrate utilization and competitive features to protect bifidobacteria from bacteriophages and other bacteria. This is essential to understanding their full role in the gut and how they interact with other members of the microbiota and with the host. Functional investigations of these insights will provide the information needed to effectively utilize this group of bacteria for optimum GI and overall health.
The lack of knowledge regarding the molecular mechanisms of proposed probiotic benefits for bifidobacteria greatly weakens the scientific credibility of health claims. The current explosion in the availability of genome sequences and molecular tools for bifidobacteria provides a means for filling this knowledge void. The immediate future should provide a wealth of information on this important genus and which features are important for both probiotic uses and efficacy. This should enable the identification of strains or groups of strains that can be demonstrated experimentally to carry certain molecular mechanisms relevant to a health claim and also shown clinically to provide health benefits in scientifically valid studies.
Acknowledgments
The University of Minnesota Supercomputing Institute and the University of Minnesota Agricultural Experimental Station are acknowledged for their support of this work.
Biography
 Ju-Hoon Lee received B.S. and M.S. degrees from the Department of Food Science and Technology at Seoul National University, South Korea, and a Ph.D. from the Department of Food Science and Nutrition at the University of Minnesota. Since 2007, he has worked as a Postdoctoral Associate in the Microbial and Plant Genomics Institute at the University of Minnesota. Dr. Lee's major research interest involves microbial genomics and bioinformatics for comparative and functional analysis of bifidobacteria to further understand their evolution, physiology, and ecology as well as their interactions with the host and other intestinal bacteria.
Ju-Hoon Lee received B.S. and M.S. degrees from the Department of Food Science and Technology at Seoul National University, South Korea, and a Ph.D. from the Department of Food Science and Nutrition at the University of Minnesota. Since 2007, he has worked as a Postdoctoral Associate in the Microbial and Plant Genomics Institute at the University of Minnesota. Dr. Lee's major research interest involves microbial genomics and bioinformatics for comparative and functional analysis of bifidobacteria to further understand their evolution, physiology, and ecology as well as their interactions with the host and other intestinal bacteria.
 Daniel J. O'Sullivan is a Professor at the University of Minnesota. He received his Ph.D. in Microbiology from the National University of Ireland, Cork, Ireland, in 1990. He did postdoctoral research work at the National Food Biotechnology Center (Ireland) and at North Carolina State University before joining the faculty at the University of Minnesota in 1994. His research is focused on the molecular analysis of lactic acid bacteria and bifidobacteria, as well as the diversity and host significance of the gut microbiota.
Daniel J. O'Sullivan is a Professor at the University of Minnesota. He received his Ph.D. in Microbiology from the National University of Ireland, Cork, Ireland, in 1990. He did postdoctoral research work at the National Food Biotechnology Center (Ireland) and at North Carolina State University before joining the faculty at the University of Minnesota in 1994. His research is focused on the molecular analysis of lactic acid bacteria and bifidobacteria, as well as the diversity and host significance of the gut microbiota.
REFERENCES
- 1.Adam, A. 1949. Substitute for human milk in infant feedings. A contribution to the establishment of an artificial bifidus flora. Monatsschr. Kinderheilkd. 97:500-507. [Google Scholar]
- 2.Agrawal, A., L. A. Houghton, J. Morris, B. Reilly, D. Guyonnet, N. Goupil Feuillerat, A. Schlumberger, S. Jakob, and P. J. Whorwell. 2008. Clinical trial: the effects of a fermented milk product containing Bifidobacterium lactis DN-173-010 on abdominal distension and gastrointestinal transit in irritable bowel syndrome with constipation. Aliment. Pharmacol. Ther. 29:104-114. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, M. B. 1948. Infantile diarrhoea and vomiting; a review of 456 infants treated in a hospital unit for enteritis. Br. Med. J. 2:973-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Martin, P., A. Belen Florez, A. Margolles, G. del Solar, and B. Mayo. 2008. Improved cloning vectors for bifidobacteria, based on the Bifidobacterium catenulatum pBC1 replicon. Appl. Environ. Microbiol. 74:4656-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez-Martin, P., M. O'Connell-Motherway, D. van Sinderen, and B. Mayo. 2007. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl. Microbiol. Biotechnol. 76:1395-1402. [DOI] [PubMed] [Google Scholar]
- 6.Alvaro, E., C. Andrieux, V. Rochet, L. Rigottier-Gois, P. Lepercq, M. Sutren, P. Galan, Y. Duval, C. Juste, and J. Dore. 2007. Composition and metabolism of the intestinal microbiota in consumers and non-consumers of yogurt. Br. J. Nutr. 97:126-133. [DOI] [PubMed] [Google Scholar]
- 7.Amaretti, A., E. Tamburini, T. Bernardi, A. Pompei, S. Zanoni, G. Vaccari, D. Matteuzzi, and M. Rossi. 2006. Substrate preference of Bifidobacterium adolescentis MB 239: compared growth on single and mixed carbohydrates. Appl. Microbiol. Biotechnol. 73:654-662. [DOI] [PubMed] [Google Scholar]
- 8.Amrouche, T., Y. Boutin, G. Prioult, and I. Fliss. 2006. Effects of bifidobacterial cytoplasm, cell wall and exopolysaccharide on mouse lymphocyte proliferation and cytokine production. Int. Dairy J. 16:70-80. [Google Scholar]
- 9.Argnani, A., R. J. Leer, N. van Luijk, and P. H. Pouwels. 1996. A convenient and reproducible method to genetically transform bacteria of the genus Bifidobacterium. Microbiology 142:109-114. [DOI] [PubMed] [Google Scholar]
- 10.Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. [DOI] [PubMed] [Google Scholar]
- 11.Arroyo, L., L. N. Cotton, and J. H. Martin. 1995. AMC agar—a composite medium for selective enumeration of Bifidobacterium longum. Cult. Dairy Prod. J. 30:12-15. [Google Scholar]
- 12.Arroyo, L., L. N. Cotton, and J. H. Martin. 1994. Evaluation of media for enumeration of Bifidobacterium adolescentis, B. infantis, and B. longum from pure culture. Cult. Dairy Prod. J. 29:20-24. [Google Scholar]
- 13.Arunachalam, K., H. S. Gill, and R. K. Chandra. 2000. Enhancement of natural immune function by dietary consumption of Bifidobacterium lactis (HN019). Eur. J. Clin. Nutr. 54:263-267. [DOI] [PubMed] [Google Scholar]
- 14.Asahara, T., K. Shimizu, K. Nomoto, T. Hamabata, A. Ozawa, and Y. Takeda. 2004. Probiotic bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157:H7. Infect. Immun. 72:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ataie-Jafari, A., B. Larijani, H. Alavi Majd, and F. Tahbaz. 2009. Cholesterol-lowering effect of probiotic yogurt in comparison with ordinary yogurt in mildly to moderately hypercholesterolemic subjects. Ann. Nutr. Metab. 54:22-27. [DOI] [PubMed] [Google Scholar]
- 16.Baldini, R. L., M. Avedissian, and S. L. Gomes. 1998. The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J. Bacteriol. 180:1632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrangou, R., E. P. Briczinski, L. L. Traeger, J. R. Loquasto, M. Richards, P. Horvath, A. C. Coute-Monvoisin, G. Leyer, S. Rendulic, J. L. Steele, J. R. Broadbent, T. Oberg, E. G. Dudley, S. Schuster, D. A. Romero, and R. F. Roberts. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191:4144-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 19.Beerens, H. 1991. Detection of bifidobacteria by using propionic acid as a selective agent. Appl. Environ. Microbiol. 57:2418-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 21.Benno, Y., K. Sawada, and T. Mitsuoka. 1984. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 28:975-986. [DOI] [PubMed] [Google Scholar]
- 22.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 23.Berger, B., D. Moine, R. Mansourian, and F. Arigoni. 2010. HspR mutations are naturally selected in Bifidobacterium longum when successive heat shock treatments are applied. J. Bacteriol. 192:256-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergey, D. H., R. E. Buchanan, and N. E. Gibbons. 1974. Bergey's manual of determinative bacteriology, 8th ed. Williams & Wilkins Co., Baltimore, MD.
- 25.Bezkorovainy, A., and R. Miller-Catchpole. 1989. Biochemistry and physiology of bifidobacteria. CRC Press, Boca Raton, FL.
- 26.Biavati, B., and P. Mattarelli. 1991. Bifidobacterium ruminantium sp. nov. and Bifidobacterium merycicum sp. nov. from the rumens of cattle. Int. J. Syst. Bacteriol. 41:163-168. [DOI] [PubMed] [Google Scholar]
- 27.Biavati, B., P. Mattarelli, and F. Crociani. 1991. Bifidobacterium saeculare--a new species isolated from feces of rabbit. Syst. Appl. Microbiol. 14:389-392. [Google Scholar]
- 28.Biavati, B., V. Scardovi, and W. E. C. Moore. 1982. Electrophoretic patterns of proteins in the genus Bifidobacterium and proposal of 4 new species. Int. J. Syst. Bacteriol. 32:358-373. [Google Scholar]
- 29.Bickhart, D. M., J. P. Gogarten, P. Lapierre, L. S. Tisa, P. Normand, and D. R. Benson. 2009. Insertion sequence content reflects genome plasticity in strains of the root nodule actinobacterium Frankia. BMC Genomics 10:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blaurock, G. 1937. Zur Physiologie der Bifidusbakterien. Monatsschr. Kinderheilkd. 68:304-309. [Google Scholar]
- 31.Bode, L. 2009. Human milk oligosaccharides: prebiotics and beyond. Nutr. Rev. 67:183-191. [DOI] [PubMed] [Google Scholar]
- 32.Boeke, J. D. 2004. Evolution—A is for adaptation. Nature 431:408-409. [DOI] [PubMed] [Google Scholar]
- 33.Bolotin, A., B. Quinquis, A. Sorokin, and S. D. Ehrlich. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551-2561. [DOI] [PubMed] [Google Scholar]
- 34.Boyd, E. F., S. Almagro-Moreno, and M. A. Parent. 2009. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 17:47-53. [DOI] [PubMed] [Google Scholar]
- 35.Broadbent, J. R., D. J. McMahon, D. L. Welker, C. J. Oberg, and S. Moineau. 2003. Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: a review. J. Dairy Sci. 86:407-423. [DOI] [PubMed] [Google Scholar]
- 36.Brown, D., and K. Sjolander. 2006. Functional classification using phylogenomic inference. PLoS Comput. Biol. 2:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bucca, G., A. M. Brassington, H. J. Schonfeld, and C. P. Smith. 2000. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressor. Mol. Microbiol. 38:1093-1103. [DOI] [PubMed] [Google Scholar]
- 38.Bullen, C. L., A. T. Willis, and K. Williams. 1973. The significance of bifidobacteria in the intestinal tract of infants. Soc. Appl. Bacteriol. Symp. Ser. 2:311-325. [PubMed] [Google Scholar]
- 39.Bullen, C. L., and P. V. Tearle. 1976. Bifidobacteria in the intestinal tract of infants: an in-vitro study. J. Med. Microbiol. 9:335-344. [DOI] [PubMed] [Google Scholar]
- 40.Bullen, C. L., and A. T. Willis. 1971. Resistance of the breast-fed infant to gastroenteritis. Br. Med. J. 3:338-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Canback, B., I. Tamas, and S. G. E. Andersson. 2004. A phylogenomic study of endosymbiotic bacteria. Mol. Biol. Evol. 21:1110-1122. [DOI] [PubMed] [Google Scholar]
- 42.Chang, W. S., and J. S. So. 1998. False positive SOD activity of Bifidobacterium spp. grown in MRS medium. J. Microbiol. Biotechnol. 8:305-309. [Google Scholar]
- 43.Cheikhyoussef, A., N. Cheikhyoussef, H. Q. Chen, J. X. Zhao, J. Tang, H. Zhang, and W. Chen. 2010. Bifidin I—a new bacteriocin produced by Bifidobacterium infantis BCRC 14602: purification and partial amino acid sequence. Food Control 21:746-753. [Google Scholar]
- 44.Chen, H., and D. G. Hoover. 2003. Bacteriocins and their food applications. Compr. Rev. Food Sci. Food Saf. 2:82-100. [DOI] [PubMed] [Google Scholar]
- 45.Chen, R. M., J. J. Wu, S. C. Lee, A. H. Huang, and H. M. Wu. 1999. Increase of intestinal Bifidobacterium and suppression of coliform bacteria with short-term yogurt ingestion. J. Dairy Sci. 82:2308-2314. [DOI] [PubMed] [Google Scholar]
- 46.Chierici, R., S. Fanaro, D. Saccomandi, and V. Vigi. 2003. Advances in the modulation of the microbial ecology of the gut in early infancy. Acta Paediatr. 91(Suppl.):56-63. [DOI] [PubMed] [Google Scholar]
- 47.Chouraqui, J. P., L. D. Van Egroo, and M. C. Fichot. 2004. Acidified milk formula supplemented with Bifidobacterium lactis: impact on infant diarrhea in residential care settings. J. Pediatr. Gastroenterol. Nutr. 38:288-292. [DOI] [PubMed] [Google Scholar]
- 48.Christensen, H. R., C. N. Larsen, P. Kaestel, L. B. Rosholm, C. Sternberg, K. F. Michaelsen, and H. Frokiaer. 2006. Immunomodulating potential of supplementation with probiotics: a dose-response study in healthy young adults. FEMS Immunol. Med. Microbiol. 47:380-390. [DOI] [PubMed] [Google Scholar]
- 49.Claesson, M. J., D. van Sinderen, and P. W. O'Toole. 2008. Lactobacillus phylogenomics—towards a reclassification of the genus. Int. J. Syst. Evol. Microbiol. 58:2945-2954. [DOI] [PubMed] [Google Scholar]
- 50.Climo, M. W., V. K. Sharma, and G. L. Archer. 1996. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J. Bacteriol. 178:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins, M. D., and G. R. Gibson. 1999. Probiotics, prebiotics, and synbiotics: approaches for modulating the microbial ecology of the gut. Am. J. Clin. Nutr. 69:1052S-1057S. [DOI] [PubMed] [Google Scholar]
- 52.Condon, C., D. Liveris, C. Squires, I. Schwartz, and C. L. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corneau, N., E. Emond, and G. LaPointe. 2004. Molecular characterization of three plasmids from Bifidobacterium longum. Plasmid 51:87-100. [DOI] [PubMed] [Google Scholar]
- 54.Crociani, F., A. Allesandrini, M. M. B. Mucci, and B. Biavati. 1994. Degradation of complex carbohydrates by Bifidobacterium spp. Int. J. Food Microbiol. 24:199-210. [DOI] [PubMed] [Google Scholar]
- 55.Cronin, M., M. Knobel, M. O'Connell-Motherway, G. F. Fitzgerald, and D. van Sinderen. 2007. Molecular dissection of a bifidobacterial replicon. Appl. Environ. Microbiol. 73:7858-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cronin, M., D. Morrissey, S. Rajendran, S. M. El Mashad, D. van Sinderen, G. C. O'Sullivan, and M. Tangney. 2010. Orally administered bifidobacteria as vehicles for delivery of agents to systemic tumors. Mol. Ther. 18:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cronin, M., R. D. Sleator, C. Hill, G. F. Fitzgerald, and D. van Sinderen. 2008. Development of a luciferase-based reporter system to monitor Bifidobacterium breve UCC2003 persistence in mice. BMC Microbiol. 8:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuatrecasas, P., D. H. Lockwood, and J. R. Caldwell. 1965. Lactase deficiency in the adult. A common occurrence. Lancet i:14-18. [DOI] [PubMed] [Google Scholar]
- 59.Cusick, S. M., and D. J. O'Sullivan. 2000. Use of a single, triplicate arbitrarily primed-PCR procedure for molecular fingerprinting of lactic acid bacteria. Appl. Environ. Microbiol. 66:2227-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai, D., and W. A. Walker. 1999. Protective nutrients and bacterial colonization in the immature human gut. Adv. Pediatr. 46:353-382. [PubMed] [Google Scholar]
- 61.Dalbey, R. E., and M. Y. Chen. 2004. Sec-translocase mediated membrane protein biogenesis. Biochim. Biophys. Acta 1694:37-53. [DOI] [PubMed] [Google Scholar]
- 62.Dambekodi, P. C., and S. E. Gilliland. 1998. Incorporation of cholesterol into the cellular membrane of Bifidobacterium longum. J. Dairy Sci. 81:1818-1824. [DOI] [PubMed] [Google Scholar]
- 63.Darling, A. E., I. Miklos, and M. A. Ragan. 2008. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 4:e1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davidson, A. L., E. Dassa, C. Orelle, and J. Chen. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72:317-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davidson, J. N., K. C. Chen, R. S. Jamison, L. A. Musmanno, and C. B. Kern. 1993. The evolutionary history of the first three enzymes in pyrimidine biosynthesis. Bioessays 15:157-164. [DOI] [PubMed] [Google Scholar]
- 66.Degnan, B. A., and G. T. Macfarlane. 1993. Transport and metabolism of glucose and arabinose in Bifidobacterium breve. Arch. Microbiol. 160:144-151. [DOI] [PubMed] [Google Scholar]
- 67.Deguchi, Y., T. Morishita, and M. Mutai. 1985. Comparative studies on synthesis of water-soluble vitamins among human species of bifidobacteria. Agric. Biol. Chem. 49:13-19. [Google Scholar]
- 68.Delcenserie, V., F. Gavini, H. Beerens, O. Tresse, C. Franssen, and G. Daube. 2007. Description of a new species, Bifidobacterium crudilactis sp. nov., isolated from raw milk and raw milk cheeses. Syst. Appl. Microbiol. 30:381-389. [DOI] [PubMed] [Google Scholar]
- 69.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deveau, H., R. Barrangou, J. E. Garneau, J. Labonte, C. Fremaux, P. Boyaval, D. A. Romero, P. Horvath, and S. Moineau. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Vries, W., S. J. Gerbrandy, and A. H. Stouthamer. 1967. Carbohydrate metabolism in Bifidobacterium bifidum. Biochim. Biophys. Acta 136:415-425. [DOI] [PubMed] [Google Scholar]
- 73.de Vries, W., and A. H. Stouthamer. 1969. Factors determining the degree of anaerobiosis of Bifidobacterium strains. Arch. Mikrobiol. 65:275-287. [DOI] [PubMed] [Google Scholar]
- 74.de Vries, W., and A. H. Stouthamer. 1968. Fermentation of glucose, lactose, galactose, mannitol, and xylose by bifidobacteria. J. Bacteriol. 96:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Vries, W., and A. H. Stouthamer. 1967. Pathway of glucose fermentation in relation to the taxonomy of bifidobacteria. J. Bacteriol. 93:574-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Djordjevic, G. M., and T. R. Klaenhammer. 1997. Genes and gene expression in Lactococcus bacteriophages. Int. Dairy J. 7:489-508. [Google Scholar]
- 77.Dong, X. Z., Y. H. Xin, W. Y. Jian, X. L. Liu, and D. W. Ling. 2000. Bifidobacterium thermacidophilum sp. nov., isolated from an anaerobic digester. Int. J. Syst. Evol. Microbiol. 50:119-125. [DOI] [PubMed] [Google Scholar]
- 78.Donovan, S. M., and B. Lonnerdal. 1989. Non-protein nitrogen and true protein in infant formulas. Acta Paediatr. Scand. 78:497-504. [DOI] [PubMed] [Google Scholar]
- 79.Doulatov, S., A. Hodes, L. Dai, N. Mandhana, M. Liu, R. Deora, R. W. Simons, S. Zimmerly, and J. F. Miller. 2004. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature 431:476-481. [DOI] [PubMed] [Google Scholar]
- 80.Duffy, L. C., M. A. Zielezny, M. Riepenhoff-Talty, D. Dryja, S. Sayahtaheri-Altaie, E. Griffiths, D. Ruffin, H. Barrett, J. Rossman, and P. L. Ogra. 1994. Effectiveness of Bifidobacterium bifidum in mediating the clinical course of murine rotavirus diarrhea. Pediatr. Res. 35:690-695. [DOI] [PubMed] [Google Scholar]
- 81.Duncan, S. H., P. Louis, and H. J. Flint. 2007. Cultivable bacterial diversity from the human colon. Lett. Appl. Microbiol. 44:343-350. [DOI] [PubMed] [Google Scholar]
- 82.Dworkin, M., S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt. 2006. The prokaryotes, vol. 3. Archaea. Bacteria: Firmicutes, Actinomycetes. Springer-Verlag, New York, NY.
- 83.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Engels, S., J. E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor sigma(H). Mol. Microbiol. 52:285-302. [DOI] [PubMed] [Google Scholar]
- 85.Erill, I., S. Campoy, and J. Barbe. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31:637-656. [DOI] [PubMed] [Google Scholar]
- 86.Escogido, M. L. R., A. D. L. Rodriguez, and A. P. B. de la Rosa. 2007. A novel binary expression vector for production of human IL-10 in Escherichia coli and Bifidobacterium longum. Biotechnol. Lett. 29:1249-1253. [DOI] [PubMed] [Google Scholar]
- 87.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fukushima, Y., Y. Kawata, H. Hara, A. Terada, and T. Mitsuoka. 1998. Effect of a probiotic formula on intestinal immunoglobulin A production in healthy children. Int. J. Food Microbiol. 42:39-44. [DOI] [PubMed] [Google Scholar]
- 89.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 90.Furrie, E., S. Macfarlane, A. Kennedy, J. H. Cummings, S. V. Walsh, D. A. O'Neil, and G. T. Macfarlane. 2005. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut 54:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Galperin, M. Y., O. V. Moroz, K. S. Wilson, and A. G. Murzin. 2006. House cleaning, a part of good housekeeping. Mol. Microbiol. 59:5-19. [DOI] [PubMed] [Google Scholar]
- 92.Garrigues, C., E. Johansen, and M. B. Pedersen. 2010. Complete genome sequence of Bifidobacterium animalis subsp. lactis BB-12, a widely consumed probiotic strain. J. Bacteriol. 192:2467-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garrity, G. M., T. G. Lilburn, J. R. Cole, S. H. Harrison, J. Euzeby, and B. J. Tindall. 2007. The Bacteria: phylum Actinobacteria: class “Actinobacteria,” p. 399-541. In Taxonomy outline of the bacteria and archaea, release 7.7. Michigan State University Board of Trustees, East Lansing, MI.
- 94.Gavini, F., M. VanEsbroeck, J. P. Touzel, A. Fourment, and H. Goossens. 1996. Detection of fructose-6-phosphate phosphoketolase (F6PPK), a key enzyme of the bifid-shunt, in Gardnerella vaginalis. Anaerobe 2:191-193. [Google Scholar]
- 95.Germond, J. E., L. Lapierre, M. Delley, and B. Mollet. 1995. A new mobile genetic element in Lactobacillus delbrueckii subsp. bulgaricus. Mol. Gen. Genet. 248:407-416. [DOI] [PubMed] [Google Scholar]
- 96.Gerstley, J. R., K. M. Howell, and B. R. Nagel. 1932. Some factors influencing the fecal flora of infants. Am. J. Dis. Child. 43:555-565. [Google Scholar]
- 97.Gibbs, M. J., V. V. Smeianov, J. L. Steele, P. Upcroft, and B. A. Efimov. 2006. Two families of Rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol. Biol. Evol. 23:1097-1100. [DOI] [PubMed] [Google Scholar]
- 98.Gill, H., and J. Prasad. 2008. Probiotics, immunomodulation, and health benefits. Adv. Exp. Med. Biol. 606:423-454. [DOI] [PubMed] [Google Scholar]
- 99.Gill, S. R., M. Pop, R. T. DeBoy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glick, M. C., T. Sall, F. Zilliken, and S. Mudd. 1960. Morphological changes of Lactobacillus bifidus var. pennsylvanicus produced by a cell-wall precursor. Biochim. Biophys. Acta 37:361-363. [DOI] [PubMed] [Google Scholar]
- 101.Grandvalet, C., V. de Crecy-Lagard, and P. Mazodier. 1999. The clpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol. Microbiol. 31:521-532. [DOI] [PubMed] [Google Scholar]
- 102.Greany, K. A., J. A. Nettleton, K. E. Wangen, W. Thomas, and M. S. Kurzer. 2004. Probiotic consumption does not enhance the cholesterol-lowering effect of soy in postmenopausal women. J. Nutr. 134:3277-3283. [DOI] [PubMed] [Google Scholar]
- 103.Grissa, I., G. Vergnaud, and C. Pourcel. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guglielmetti, S., M. Karp, D. Mora, I. Tamagnini, and C. Parini. 2007. Molecular characterization of Bifidobacterium longum biovar longum NAL8 plasmids and construction of a novel replicon screening system. Appl. Microbiol. Biotechnol. 74:1053-1061. [DOI] [PubMed] [Google Scholar]
- 105.Guy, L., D. Karamata, P. Moreillon, and C. A. H. Roten. 2005. Genometrics as an essential tool for the assembly of whole genome sequences: the example of the chromosome of Bifidobacterium longum NCC2705. BMC Microbiol. 5:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guyonnet, D., O. Chassany, P. Ducrotte, C. Picard, M. Mouret, C. H. Mercier, and C. Matuchansky. 2007. Effect of a fermented milk containing Bifidobacterium animalis DN-173 010 on the health-related quality of life and symptoms in irritable bowel syndrome in adults in primary care: a multicentre, randomized, double-blind, controlled trial. Aliment. Pharmacol. Ther. 26:475-486. [DOI] [PubMed] [Google Scholar]
- 107.Gyllenberg, H., and G. Carlberg. 1958. The nutritional characteristics of the bifid bacteria (Lactobacillus bifidus) in infants. Acta Pathol. Microbiol. Scand. 44:287-292. [DOI] [PubMed] [Google Scholar]
- 108.Gyorgy, P., R. Kuhn, C. S. Rose, and F. Zilliken. 1954. Bifidus factor. II. Its occurrence in milk from different species and in other natural products. Arch. Biochem. Biophys. 48:202-208. [DOI] [PubMed] [Google Scholar]
- 109.Gyorgy, P., R. F. Norris, and C. S. Rose. 1954. Bifidus factor. I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch. Biochem. Biophys. 48:193-201. [DOI] [PubMed] [Google Scholar]
- 110.Haft, D. H., J. Selengut, E. F. Mongodin, and K. E. Nelson. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hamaji, Y., M. Fujimori, T. Sasaki, H. Matsuhashi, K. Matsui-Seki, Y. Shimatani-Shibata, Y. Kano, J. Amano, and S. Taniguchi. 2007. Strong enhancement of recombinant cytosine deaminase activity in Bifidobacterium longum for tumor-targeting enzyme/prodrug therapy. Biosci. Biotechnol. Biochem. 71:874-883. [DOI] [PubMed] [Google Scholar]
- 112.Hartemink, R., B. J. Kok, G. H. Weenk, and F. M. Rombouts. 1996. Raffinose-Bifidobacterium (RB) agar, a new selective medium for bifidobacteria. J. Microbiol. Methods 27:33-43. [Google Scholar]
- 113.Hassinen, J. B., G. T. Durbin, Tomarelli, and F. W. Bernhart. 1951. The minimal nutritional requirements of Lactobacillus bifidus. J. Bacteriol. 62:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.He, T., M. G. Priebe, Y. Zhong, C. Huang, H. J. Harmsen, G. C. Raangs, J. M. Antoine, G. W. Welling, and R. J. Vonk. 2008. Effects of yogurt and bifidobacteria supplementation on the colonic microbiota in lactose-intolerant subjects. J. Appl. Microbiol. 104:595-604. [DOI] [PubMed] [Google Scholar]
- 115.Hidaka, A., Y. Hamaji, T. Sasaki, S. Taniguchi, and M. Fujimori. 2007. Exogeneous cytosine deaminase gene expression in Bifidobacterium breve 1-53-8w for tumor-targeting enzyme/prodrug therapy. Biosci. Biotechnol. Biochem. 71:2921-2926. [DOI] [PubMed] [Google Scholar]
- 116.Hoarau, C., L. Martin, D. Faugaret, C. Baron, A. Dauba, C. Aubert-Jacquin, F. Velge-Roussel, and Y. Lebranchu. 2008. Supernatant from Bifidobacterium differentially modulates transduction signaling pathways for biological functions of human dendritic cells. PLoS One 3:e2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Holzapfel, W. H. 2006. Introduction to prebiotics and probiotics. Taylor & Francis/CRC Press, Boca Raton, FL.
- 118.Holzapfel, W. H., P. Haberer, J. Snel, U. Schillinger, and J. H. Huis in't Veld. 1998. Overview of gut flora and probiotics. Int. J. Food Microbiol. 41:85-101. [DOI] [PubMed] [Google Scholar]
- 119.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 120.Hopkins, M. J., and G. T. Macfarlane. 2003. Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl. Environ. Microbiol. 69:1920-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hopkins, M. J., R. Sharp, and G. T. Macfarlane. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Horvath, P., A. C. Coute-Monvoisin, D. A. Romero, P. Boyaval, C. Fremaux, and R. Barrangou. 2008. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131:62-70. [DOI] [PubMed] [Google Scholar]
- 123.Hoyles, L., E. Inganas, E. Falsen, M. Drancourt, N. Weiss, A. L. McCartney, and M. D. Collins. 2002. Bifidobacterium scardovii sp. nov., from human sources. Int. J. Syst. Evol. Microbiol. 52:995-999. [DOI] [PubMed] [Google Scholar]
- 124.Huang, S. S., and T. M. Bayless. 1968. Milk and lactose intolerance in healthy orientals. Science 160:83-84. [DOI] [PubMed] [Google Scholar]
- 125.Husain, I., J. A. Poupard, and R. F. Norris. 1972. Influence of nutrition on the morphology of a strain of Bifidobacterium bifidum. J. Bacteriol. 111:841-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ishibashi, N., T. Yaeshima, and H. Hayasawa. 1997. Bifidobacteria: their significance in human intestinal health. Mal. J. Nutr. 3:149-159. [Google Scholar]
- 128.Iwata, M., and T. Morishita. 1989. The presence of plasmids in Bifidobacterium breve. Lett. Appl. Microbiol. 9:165-168. [Google Scholar]
- 129.Jansen, R., J. D. Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565-1575. [DOI] [PubMed] [Google Scholar]
- 130.Ji, G. E., H. K. Han, S. W. Yun, and S. L. Rhim. 1992. Isolation of amylolytic Bifidobacterium sp. Int.-57 and characterization of amylase. J. Microbiol. Biotechnol. 2:85-91. [Google Scholar]
- 131.Ji, G. E., S. K. Lee, and I. H. Kim. 1994. Improved selective medium for isolation and enumeration of Bifidobacterium sp. Kor. J. Food Sci. Technol. 26:526-531. [Google Scholar]
- 132.Jian, W., and X. Dong. 2002. Transfer of Bifidobacterium inopinatum and Bifidobacterium denticolens to Scardovia inopinata gen. nov., comb. nov., and Parascardovia denticolens gen. nov., comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 52:809-812. [DOI] [PubMed] [Google Scholar]
- 133.Jiang, T. A., A. Mustapha, and D. A. Savaiano. 1996. Improvement of lactose digestion in humans by ingestion of unfermented milk containing Bifidobacterium longum. J. Dairy Sci. 79:750-757. [DOI] [PubMed] [Google Scholar]
- 134.Jojima, T., C. A. Omumasaba, M. Inui, and H. Yukawa. 2010. Sugar transporters in efficient utilization of mixed sugar substrates: current knowledge and outlook. Appl. Microbiol. Biotechnol. 85:471-480. [DOI] [PubMed] [Google Scholar]
- 135.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9:429-437. [DOI] [PubMed] [Google Scholar]
- 136.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kawasaki, S., T. Mimura, T. Satoh, K. Takeda, and Y. Niimura. 2006. Response of the microaerophilic Bifidobacterium species, B. boum and B. thermophilum, to oxygen. Appl. Environ. Microbiol. 72:6854-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kiatpapan, P., and Y. Murooka. 2002. Genetic manipulation system in propionibacteria. J. Biosci. Bioeng. 93:1-8. [PubMed] [Google Scholar]
- 139.Kiessling, G., J. Schneider, and G. Jahreis. 2002. Long-term consumption of fermented dairy products over 6 months increases HDL cholesterol. Eur. J. Clin. Nutr. 56:843-849. [DOI] [PubMed] [Google Scholar]
- 140.Killer, J., J. Kopecny, J. Mrazek, V. Rada, O. Benada, I. Koppova, J. Havlik, and J. Straka. 2009. Bifidobacterium bombi sp. nov., from the bumblebee digestive tract. Int. J. Syst. Evol. Microbiol. 59:2020-2024. [DOI] [PubMed] [Google Scholar]
- 141.Kim, J. F., H. Jeong, D. S. Yu, S. H. Choi, C. G. Hur, M. S. Park, S. H. Yoon, D. W. Kim, G. E. Ji, H. S. Park, and T. K. Oh. 2009. Genome sequence of the probiotic bacterium Bifidobacterium animalis subsp. lactis AD011. J. Bacteriol. 191:678-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim, N., J. Kunisawa, M. N. Kweon, G. Eog Ji, and H. Kiyono. 2007. Oral feeding of Bifidobacterium bifidum (BGN4) prevents CD4(+) CD45RB(high) T cell-mediated inflammatory bowel disease by inhibition of disordered T cell activation. Clin. Immunol. 123:30-39. [DOI] [PubMed] [Google Scholar]
- 143.Kim, T. B., S. H. Song, S. C. Kang, and D. K. Oh. 2003. Quantitative comparison of lactose and glucose utilization in Bifidobacterium longum cultures. Biotechnol. Prog. 19:672-675. [DOI] [PubMed] [Google Scholar]
- 144.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 145.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Klijn, A., A. Mercenier, and F. Arigoni. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 29:491-509. [DOI] [PubMed] [Google Scholar]
- 147.Klijn, A., D. Moine, M. Delley, A. Mercenier, F. Arigoni, and R. D. Pridmore. 2006. Construction of a reporter vector for the analysis of Bifidobacterium longum promoters. Appl. Environ. Microbiol. 72:7401-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kojima, M., S. Suda, S. Hotta, and K. Hamada. 1968. Induction of pleomorphism in Lactobacillus bifidus. J. Bacteriol. 95:710-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kojima, M., S. Suda, S. Hotta, and K. Hamada. 1970. Induction of pleomorphy and calcium ion deficiency in Lactobacillus bifidus. J. Bacteriol. 102:217-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kojima, M., S. Suda, S. Hotta, K. Hamada, and A. Suganuma. 1970. Necessity of calcium ion for cell division in Lactobacillus bifidus. J. Bacteriol. 104:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kristen, G., and F. Petuely. 1950. Investigations in the bifidus factor; human milk and bifidus bacteria. Osterr. Z. Kinderheilkd. Kinderfuersorge. 4:121-134. [PubMed] [Google Scholar]
- 152.Krzewinski, F., C. Brassart, F. Gavini, and S. Bouquelet. 1997. Glucose and galactose transport in Bifidobacterium bifidum DSM 20082. Curr. Microbiol. 35:175-179. [DOI] [PubMed] [Google Scholar]
- 153.Kullen, M. J., M. M. Amann, M. J. O'Shaughnessy, D. J. O'Sullivan, F. F. Busta, and L. J. Brady. 1997. Differentiation of ingested and endogenous bifidobacteria by DNA fingerprinting demonstrates the survival of an unmodified strain in the gastrointestinal tract of humans. J. Nutr. 127:89-94. [DOI] [PubMed] [Google Scholar]
- 154.Kullen, M. J., L. J. Brady, and D. J. O'Sullivan. 1997. Evaluation of using a short region of the recA gene for rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol. Lett. 154:377-383. [DOI] [PubMed] [Google Scholar]
- 155.Kunin, V., R. Sorek, and P. Hugenholtz. 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kurokawa, K., T. Itoh, T. Kuwahara, K. Oshima, H. Toh, A. Toyoda, H. Takami, H. Morita, V. K. Sharma, T. P. Srivastava, T. D. Taylor, H. Noguchi, H. Mori, Y. Ogura, D. S. Ehrlich, K. Itoh, T. Takagi, Y. Sakaki, T. Hayashi, and M. Hattori. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14:169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lapierre, L., P. Undeland, and L. J. Cox. 1992. Lithium chloride-sodium propionate agar for the enumeration of bifidobacteria in fermented dairy products. J. Dairy Sci. 75:1192-1196. [DOI] [PubMed] [Google Scholar]
- 159.Lauer, E. 1990. Bifidobacterium gallicum sp. nov. isolated from human feces. Int. J. Syst. Bacteriol. 40:100-102. [DOI] [PubMed] [Google Scholar]
- 160.Lee, J., K. T. Hwang, M. Y. Chung, D. H. Cho, and C. S. Park. 2005. Resistance of Lactobacillus casei KCTC 3260 to reactive oxygen species (ROS): role for a metal ion chelating effect. J. Food Sci. 70:388-391. [Google Scholar]
- 161.Lee, J. H., V. N. Karamychev, S. A. Kozyavkin, D. Mills, A. R. Pavlov, N. V. Pavlova, N. N. Polouchine, P. M. Richardson, V. V. Shakhova, A. I. Slesarev, B. Weimer, and D. J. O'Sullivan. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee, J. H., and D. J. O'Sullivan. 2006. Sequence analysis of two cryptic plasmids from Bifidobacterium longum DJO10A and construction of a shuttle cloning vector. Appl. Environ. Microbiol. 72:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Le Leu, R. K., Y. Hu, I. L. Brown, R. J. Woodman, and G. P. Young. 2010. Synbiotic intervention of Bifidobacterium lactis and resistant starch protects against colorectal cancer development in rats. Carcinogenesis 31:246-251. [DOI] [PubMed] [Google Scholar]
- 164.Leuschner, R. G., J. Bew, P. Simpson, P. R. Ross, and C. Stanton. 2003. A collaborative study of a method for the enumeration of probiotic bifidobacteria in animal feed. Int. J. Food Microbiol. 83:161-170. [DOI] [PubMed] [Google Scholar]
- 165.Li, X., G. F. Fu, Y. R. Fan, W. H. Liu, X. J. Liu, J. J. Wang, and G. X. Xu. 2003. Bifidobacterium adolescentis as a delivery system of endostatin for cancer gene therapy: selective inhibitor of angiogenesis and hypoxic tumor growth. Cancer Gene Ther. 10:105-111. [DOI] [PubMed] [Google Scholar]
- 166.Li, Y., X. Qu, H. Yang, L. Kang, Y. Xu, B. Bai, and W. Song. 2005. Bifidobacteria DNA induces murine macrophage activation in vitro. Cell. Mol. Immunol. 2:473-478. [PubMed] [Google Scholar]
- 167.Li, Y., T. Shimizu, A. Hosaka, N. Kaneko, Y. Ohtsuka, and Y. Yamashiro. 2004. Effects of Bifidobacterium breve supplementation on intestinal flora of low birth weight infants. Pediatr. Int. 46:509-515. [DOI] [PubMed] [Google Scholar]
- 168.Lim, K. S., C. S. Huh, Y. J. Baek, and H. U. Kim. 1995. A selective enumeration medium for bifidobacteria in fermented dairy-products. J. Dairy Sci. 78:2108-2112. [DOI] [PubMed] [Google Scholar]
- 169.Locascio, M., A. P. D. Holgado, G. Perdigon, and G. Oliver. 2001. Enteric bifidobacteria: isolation from human infants and challenge studies in mice. Can. J. Microbiol. 47:1048-1052. [DOI] [PubMed] [Google Scholar]
- 170.Mackiewicz, P., J. Zakrzewska-Czerwinska, A. Zawilak, M. R. Dudek, and S. Cebrat. 2004. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 32:3781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Margolles, A., A. B. Florez, J. A. Moreno, D. van Sinderen, and C. G. de los Reyes-Gavilan. 2006. Two membrane proteins from Bifidobacterium breve UCC2003 constitute an ABC-type multidrug transporter. Microbiology 152:3497-3505. [DOI] [PubMed] [Google Scholar]
- 172.Marteau, P., E. Cuillerier, S. Meance, M. F. Gerhardt, A. Myara, M. Bouvier, C. Bouley, F. Tondu, G. Bommelaer, and J. C. Grimaud. 2002. Bifidobacterium animalis strain DN-173 010 shortens the colonic transit time in healthy women: a double-blind, randomized, controlled study. Aliment. Pharmacol. Ther. 16:587-593. [DOI] [PubMed] [Google Scholar]
- 173.Matsui, T., H. Saeki, N. Shinzato, and H. Matsuda. 2006. Characterization of Rhodococcus-E. coli shuttle vector pNC9501 constructed from the cryptic plasmid of a propene-degrading bacterium. Curr. Microbiol. 52:445-448. [DOI] [PubMed] [Google Scholar]
- 174.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Matsumoto, M., H. Ohishi, and Y. Benno. 2004. H+-ATPase activity in Bifidobacterium with special reference to acid tolerance. Int. J. Food Microbiol. 93:109-113. [DOI] [PubMed] [Google Scholar]
- 176.Matsumoto, M., M. Sakamoto, and Y. Benno. 2009. Dynamics of fecal microbiota in hospitalized elderly fed probiotic LKM512 yogurt. Microbiol. Immunol. 53:421-432. [DOI] [PubMed] [Google Scholar]
- 177.Matsumura, H., A. Takeuchi, and Y. Kano. 1997. Construction of Escherichia coli-Bifidobacterium longum shuttle vector transforming B. longum 105-A and 108-A. Biosci. Biotechnol. Biochem. 61:1211-1212. [DOI] [PubMed] [Google Scholar]
- 178.Matteuzzi, D., P. Brigidi, M. Rossi, and D. Digioia. 1990. Characterization and molecular cloning of Bifidobacterium longum cryptic plasmid pMB1. Lett. Appl. Microbiol. 11:220-223. [DOI] [PubMed] [Google Scholar]
- 179.Matteuzzi, D., F. Crociani, G. Zani, and L. D. Trovatelli. 1971. Bifidobacterium suis n. sp.: a new species of the genus Bifidobacterium isolated from pig feces. Z. Allg. Mikrobiol. 11:387-395. [DOI] [PubMed] [Google Scholar]
- 180.Mayer, J. B. 1948. Development of a new infant food with Lactobacillus bifidus. Z. Kinderheilkd. 65:319-345. [Google Scholar]
- 181.Maze, A., M. O'Connell-Motherway, G. F. Fitzgerald, J. Deutscher, and D. van Sinderen. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.McCarthy, J., L. O'Mahony, L. O'Callaghan, B. Sheil, E. E. Vaughan, N. Fitzsimons, J. Fitzgibbon, G. C. O'Sullivan, B. Kiely, J. K. Collins, and F. Shanahan. 2003. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut 52:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2007. Bacteriophages in dairy products: pros and cons. Biotechnol. J. 2:450-455. [DOI] [PubMed] [Google Scholar]
- 184.Meance, S., C. Cayuela, P. Turchet, A. Raimondi, C. Lucas, and J.-M. Antoine. 2001. A fermented milk with a Bifidobacterium probiotic strain DN-173 010 shortened oro-fecal gut transit time in elderly. Microb. Ecol. Health Dis. 13:217-222. [Google Scholar]
- 185.Meile, L., W. Ludwig, U. Rueger, C. Gut, P. Kaufmann, G. Dasen, S. Wenger, and M. Teuber. 1997. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 20:57-64. [Google Scholar]
- 186.Mendez, A., and A. Olano. 1979. Lactulose: a review of some chemical properties and applications in infant nutrition and medicine. Diary Sci. Abstr. 41:531-535. [Google Scholar]
- 187.Min, B., J. T. Pelaschier, D. E. Graham, D. Tumbula-Hansen, and D. Soll. 2002. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc. Natl. Acad. Sci. U. S. A. 99:2678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Missich, R., B. Sgorbati, and D. J. LeBlanc. 1994. Transformation of Bifidobacterium longum with pRM2, a constructed Escherichia coli-B. longum shuttle vector. Plasmid 32:208-211. [DOI] [PubMed] [Google Scholar]
- 189.Mitsuoka, T. 1969. Comparative studies on bifidobacteria isolated from the alimentary tract of man and animals (including descriptions of Bifidobacterium thermophilum nov. spec. and Bifidobacterium pseudolongum nov. spec.). Zentralbl. Bakteriol. Orig. 210:52-64. [PubMed] [Google Scholar]
- 190.Mitsuoka, T., and K. Hayakawa. 1973. The fecal flora in man. I. Composition of the fecal flora of various age groups. Zentralbl. Bakteriol. Orig. A 223:333-342. [PubMed] [Google Scholar]
- 191.Mojica, F. J., C. Diez-Villasenor, J. Garcia-Martinez, and E. Soria. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60:174-182. [DOI] [PubMed] [Google Scholar]
- 192.Monteiro-Vitorello, C. B., L. E. Camargo, M. A. Van Sluys, J. P. Kitajima, D. Truffi, A. M. do Amaral, R. Harakava, J. C. de Oliveira, D. Wood, M. C. de Oliveira, C. Miyaki, M. A. Takita, A. C. da Silva, L. R. Furlan, D. M. Carraro, G. Camarotte, N. F. Almeida, Jr., H. Carrer, L. L. Coutinho, H. A. El-Dorry, M. I. Ferro, P. R. Gagliardi, E. Giglioti, M. H. Goldman, G. H. Goldman, E. T. Kimura, E. S. Ferro, E. E. Kuramae, E. G. Lemos, M. V. Lemos, S. M. Mauro, M. A. Machado, C. L. Marino, C. F. Menck, L. R. Nunes, R. C. Oliveira, G. G. Pereira, W. Siqueira, A. A. de Souza, S. M. Tsai, A. S. Zanca, A. J. Simpson, S. M. Brumbley, and J. C. Setubal. 2004. The genome sequence of the gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol. Plant Microbe Interact. 17:827-836. [DOI] [PubMed] [Google Scholar]
- 193.Moon, G. S., Y. R. Pyun, M. S. Park, G. E. Ji, and W. J. Kim. 2005. Secretion of recombinant pediocin PA-1 by Bifidobacterium longum, using the signal sequence for bifidobacterial alpha-amylase. Appl. Environ. Microbiol. 71:5630-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Moon, G. S., U. Wegmann, A. P. Gunning, M. J. Gasson, and A. Narbad. 2009. Isolation and characterization of a theta-type cryptic plasmid from Bifidobacterium longum FI10564. J. Microbiol. Biotechnol. 19:403-408. [DOI] [PubMed] [Google Scholar]
- 195.Motherway, M. O. C., J. O'Driscoll, G. F. Fitzgerald, and D. van Sinderen. 2009. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2:321-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mountzouris, K. C., A. L. McCartney, and G. R. Gibson. 2002. Intestinal microflora of human infants and current trends for its nutritional modulation. Br. J. Nutr. 87:405-420. [DOI] [PubMed] [Google Scholar]
- 197.Munoa, F. J., and R. Pares. 1988. Selective medium for isolation and enumeration of Bifidobacterium spp. Appl. Environ. Microbiol. 54:1715-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mustacich, D., and G. Powis. 2000. Thioredoxin reductase. Biochem. J. 346:1-8. [PMC free article] [PubMed] [Google Scholar]
- 199.Muting, D. 1977. Clinical picture and therapy of portal-systemic encephalopathy. Leber Magen Darm 7:256-262. [PubMed] [Google Scholar]
- 200.Nagaoka, M., H. Shibata, I. Kimura, S. Hashimoto, K. Kimura, H. Sawada, and T. Yokokura. 1995. Structural studies on a cell wall polysaccharide from Bifidobacterium longum YIT4028. Carbohydr. Res. 274:245-249. [DOI] [PubMed] [Google Scholar]
- 201.Nakajima, H., T. Hirota, T. Toba, T. Itoh, and S. Adachi. 1992. Structure of the extracellular polysaccharide from slime-forming Lactococcus lactis subsp. cremoris SBT 0495. Carbohydr. Res. 224:245-253. [DOI] [PubMed] [Google Scholar]
- 202.Nakamura, T., T. Sasaki, M. Fujimori, K. Yazawa, Y. Kano, J. Amano, and S. Taniguchi. 2002. Cloned cytosine deaminase gene expression of Bifidobacterium longum and application to enzyme/pro-drug therapy of hypoxic solid tumors. Biosci. Biotechnol. Biochem. 66:2362-2366. [DOI] [PubMed] [Google Scholar]
- 203.Narberhaus, F. 2002. Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 66:64-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Nebra, Y., and A. R. Blanch. 1999. A new selective medium for Bifidobacterium spp. Appl. Environ. Microbiol. 65:5173-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Nielsen, H., J. Engelbrecht, S. Brunak, and G. vonHeijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 206.Nilsson, A. I., S. Koskiniemi, S. Eriksson, E. Kugelberg, J. C. Hinton, and D. I. Andersson. 2005. Bacterial genome size reduction by experimental evolution. Proc. Natl. Acad. Sci. U. S. A. 102:12112-12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Norris, R. F., T. Flanders, R. M. Tomarelli, and P. Gyorgy. 1950. The isolation and cultivation of Lactobacillus bifidus; a comparison of branched and unbranched strains. J. Bacteriol. 60:681-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.O'Driscoll, J., D. F. Heiter, G. G. Wilson, G. F. Fitzgerald, R. Roberts, and D. van Sinderen. 2006. A genetic dissection of the LlaJI restriction cassette reveals insights on a novel bacteriophage resistance system. BMC Microbiol. 6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.O'Hara, A. M., P. O'Regan, A. Fanning, C. O'Mahony, J. Macsharry, A. Lyons, J. Bienenstock, L. O'Mahony, and F. Shanahan. 2006. Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology 118:202-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.O'Mahony, L., J. McCarthy, P. Kelly, G. Hurley, F. Luo, K. Chen, G. C. O'Sullivan, B. Kiely, J. K. Collins, F. Shanahan, and E. M. M. Quigley. 2005. Lactobacillus and Bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology 128:541-551. [DOI] [PubMed] [Google Scholar]
- 211.Ooka, T., Y. Ogura, M. Asadulghani, M. Ohnishi, K. Nakayama, J. Terajima, H. Watanabe, and T. Hayashi. 2009. Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res. 19:1809-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.O'Riordan, K., and G. F. Fitzgerald. 1999. Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of mode of replication. FEMS Microbiol. Lett. 174:285-294. [DOI] [PubMed] [Google Scholar]
- 213.Orla-Jensen, S. 1924. La classification des bacteries lactiques. Lait 4:468-474. [Google Scholar]
- 214.O'Sullivan, D. J. 2008. Genomics can advance the potential for probiotic cultures to improve liver and overall health. Curr. Pharm. Des. 14:1376-1381. [DOI] [PubMed] [Google Scholar]
- 215.O'Sullivan, D. J. 2006. Primary sources of probiotic cultures, p. 91-107. In I. Goktepe and V. K. Juneja (ed.), Probiotics in food safety and human health. CRC Press/Taylor & Francis Group, Boca Raton, FL.
- 216.O'Sullivan, D. J. 2001. Screening of intestinal microflora for effective probiotic bacteria. J. Agric. Food Chem. 49:1751-1760. [DOI] [PubMed] [Google Scholar]
- 217.Palmer, C., E. M. Bik, D. B. Digiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Parche, S., J. Amon, I. Jankovic, E. Rezzonico, M. Beleut, H. Barutcu, I. Schendel, M. P. Eddy, A. Burkovski, F. Arigoni, and F. Titgemeyer. 2007. Sugar transport systems of Bifidobacterium longum NCC2705. J. Mol. Microbiol. Biotechnol. 12:9-19. [DOI] [PubMed] [Google Scholar]
- 219.Parche, S., M. Beleut, E. Rezzonico, D. Jacobs, F. Arigoni, F. Titgemeyer, and I. Jankovic. 2006. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. 188:1260-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Park, K. B., G. E. Ji, M. S. Park, and S. H. Oh. 2005. Expression of rice glutamate decarboxylase in Bifidobacterium longum enhances gamma-aminobutyric acid production. Biotechnol. Lett. 27:1681-1684. [DOI] [PubMed] [Google Scholar]
- 221.Park, M. S., B. Kwon, J. J. Shim, C. S. Huh, and G. E. Ji. 2008. Heterologous expression of cholesterol oxidase in Bifidobacterium longum under the control of 16S rRNA gene promoter of bifidobacteria. Biotechnol. Lett. 30:165-172. [DOI] [PubMed] [Google Scholar]
- 222.Park, M. S., H. W. Moon, and G. E. Ji. 2003. Molecular characterization of plasmid from Bifidobacterium longum. J. Microbiol. Biotechnol. 13:457-462. [Google Scholar]
- 223.Park, M. S., J. M. Seo, J. Y. Kim, and G. E. Ji. 2005. Heterologous gene expression and secretion in Bifidobacterium longum. Lait 85:1-8. [Google Scholar]
- 224.Park, M. S., D. W. Shin, K. H. Lee, and G. E. Ji. 2000. Characterization of plasmid pKJ36 from Bifidobacterium longum and construction of an E. coli-Bifidobacterium shuttle vector. J. Microbiol. Biotechnol. 10:312-320. [Google Scholar]
- 225.Park, M. S., D. W. Shin, K. H. Lee, and G. E. Ji. 1999. Sequence analysis of plasmid pKJ50 from Bifidobacterium longum. Microbiology 145:585-592. [DOI] [PubMed] [Google Scholar]
- 226.Park, Y. S., K. H. Kim, J. H. Park, I. K. Oh, and S. S. Yoon. 2008. Isolation and molecular characterization of a cryptic plasmid from Bifidobacterium longum. Biotechnol. Lett. 30:145-151. [DOI] [PubMed] [Google Scholar]
- 227.Parvez, S., K. A. Malik, S. Ah Kang, and H. Y. Kim. 2006. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 100:1171-1185. [DOI] [PubMed] [Google Scholar]
- 228.Peant, B., G. LaPointe, C. Gilbert, D. Atlan, P. Ward, and D. Roy. 2005. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus. Microbiology 151:1839-1851. [DOI] [PubMed] [Google Scholar]
- 229.Penders, J., C. Thijs, C. Vink, F. F. Stelma, B. Snijders, I. Kummeling, P. A. van den Brandt, and E. E. Stobberingh. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511-521. [DOI] [PubMed] [Google Scholar]
- 230.Perez-Brocal, V., R. Gil, S. Ramos, A. Lamelas, M. Postigo, J. M. Michelena, F. J. Silva, A. Moya, and A. Latorre. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314:312-313. [DOI] [PubMed] [Google Scholar]
- 231.Petuely, F., and G. Kristen. 1949. Changing the intestinal flora of infants. Ann. Paediatr. 172:183-184. [PubMed] [Google Scholar]
- 232.Pokusaeva, K., A. R. Neves, A. Zomer, M. O'Connell-Motherway, J. MacSharry, P. Curley, G. F. Fitzgerald, and D. van Sinderen. 2010. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb. Biotechnol. 3:311-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Poole, L. B., M. Higuchi, M. Shimada, M. L. Calzi, and Y. Kamio. 2000. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic. Biol. Med. 28:108-120. [DOI] [PubMed] [Google Scholar]
- 234.Poupard, J. A., I. Husain, and R. F. Norris. 1973. Biology of the bifidobacteria. Bacteriol. Rev. 37:136-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Prevot, A. R. 1971. Les laits modifies par le lactobacille acidophile dans la dietetique hospitaliere. Infect. Diet. 8:19-21. [Google Scholar]
- 236.Price, C., and T. A. Bickle. 1986. A possible role for DNA restriction in bacterial evolution. Microbiol. Sci. 3:296-299. [PubMed] [Google Scholar]
- 237.Qazi, S. N., E. Counil, J. Morrissey, C. E. Rees, A. Cockayne, K. Winzer, W. C. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Qiao, H., L. C. Duffy, E. Griffiths, D. Dryja, A. Leavens, J. Rossman, G. Rich, M. Riepenhoff-Talty, and M. Locniskar. 2002. Immune responses in rhesus rotavirus-challenged BALB/c mice treated with bifidobacteria and prebiotic supplements. Pediatr. Res. 51:750-755. [DOI] [PubMed] [Google Scholar]
- 239.Qin, J., R. Li, J. Raes, M. Arumugam, K. S. Burgdorf, C. Manichanh, T. Nielsen, N. Pons, F. Levenez, T. Yamada, D. R. Mende, J. Li, J. Xu, S. Li, D. Li, J. Cao, B. Wang, H. Liang, H. Zheng, Y. Xie, J. Tap, P. Lepage, M. Bertalan, J. M. Batto, T. Hansen, D. Le Paslier, A. Linneberg, H. B. Nielsen, E. Pelletier, P. Renault, T. Sicheritz-Ponten, K. Turner, H. Zhu, C. Yu, M. Jian, Y. Zhou, Y. Li, X. Zhang, N. Qin, H. Yang, J. Wang, S. Brunak, J. Dore, F. Guarner, K. Kristiansen, O. Pedersen, J. Parkhill, J. Weissenbach, P. Bork, and S. D. Ehrlich. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rafter, J., M. Bennett, G. Caderni, Y. Clune, R. Hughes, P. C. Karlsson, A. Klinder, M. O'Riordan, G. C. O'Sullivan, B. Pool-Zobel, G. Rechkemmer, M. Roller, I. Rowland, M. Salvadori, H. Thijs, J. Van Loo, B. Watzl, and J. K. Collins. 2007. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 85:488-496. [DOI] [PubMed] [Google Scholar]
- 241.Rajilic-Stojanovic, M., H. Smidt, and W. M. de Vos. 2007. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9:2125-2136. [DOI] [PubMed] [Google Scholar]
- 242.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 243.Ramos, A., I. C. Boels, W. M. de Vos, and H. Santos. 2001. Relationship between glycolysis and exopolysaccharide biosynthesis in Lactococcus lactis. Appl. Environ. Microbiol. 67:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Rasic, J. L., and J. A. Kurmann. 1983. Bifidobacteria and their role: microbiological, nutritional-physiological, medical, and technological aspects and bibliography. Birkhäuser Verlag, Basel, Switzerland. [PubMed]
- 245.Requena, T., J. Burton, T. Matsuki, K. Munro, M. A. Simon, R. Tanaka, K. Watanabe, and G. W. Tannock. 2002. Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Reuter, G. 1969. Composition and use of bacterial cultures for therapeutic purposes. Arzneimittelforschung 19:103-109. [PubMed] [Google Scholar]
- 247.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 248.Reuter, G. 1963. Vergleichenden Untersuchung uber die Bifidus-Flora im Sauglings und Erwachsenenstuhl. Zentralbl. Bakteriol. Orig. 191:486-507. [PubMed] [Google Scholar]
- 249.Rhim, S. L., M. S. Park, and G. E. Ji. 2006. Expression and secretion of Bifidobacterium adolescentis amylase by Bifidobacterium longum. Biotechnol. Lett. 28:163-168. [DOI] [PubMed] [Google Scholar]
- 250.Roberts, C. M., W. F. Fett, S. F. Osman, C. Wijey, J. V. Oconnor, and D. G. Hoover. 1995. Exopolysaccharide production by Bifidobacterium longum Bb-79. J. Appl. Bacteriol. 78:463-468. [Google Scholar]
- 251.Romond, M. B., E. Bezirtzoglou, and C. Romond. 1998. Colonization of the murine gut by Bifidobacterium bifidum and Clostridium perfringens during ageing. Microb. Ecol. Health Dis. 10:91-94. [Google Scholar]
- 252.Romond, M. B., Z. Haddou, C. Mielcareck, and C. Romond. 1997. Bifidobacteria and human health: regulatory effect of indigenous bifidobacteria on Escherichia coli intestinal colonization. Anaerobe 3:131-136. [DOI] [PubMed] [Google Scholar]
- 253.Rossi, M., P. Brigidi, and D. Matteuzzi. 1997. An efficient transformation system for Bifidobacterium spp. Lett. Appl. Microbiol. 24:33-36. [Google Scholar]
- 254.Rossi, M., P. Brigidi, and D. Matteuzzi. 1998. Improved cloning vectors for Bifidobacterium spp. Lett. Appl. Microbiol. 26:101-104. [DOI] [PubMed] [Google Scholar]
- 255.Rossi, M., P. Brigidi, A. G. V. Y. Rodriguez, and D. Matteuzzi. 1996. Characterization of the plasmid pMB1 from Bifidobacterium longum and its use for shuttle vector construction. Res. Microbiol. 147:133-143. [DOI] [PubMed] [Google Scholar]
- 256.Rouge, C., H. Piloquet, M. J. Butel, B. Berger, F. Rochat, L. Ferraris, C. Des Robert, A. Legrand, M. F. de la Cochetiere, J. M. N′Guyen, M. Vodovar, M. Voyer, D. Darmaun, and J. C. Roze. 2009. Oral supplementation with probiotics in very-low-birth-weight preterm infants: a randomized, double-blind, placebo-controlled trial. Am. J. Clin. Nutr. 89:1828-1835. [DOI] [PubMed] [Google Scholar]
- 257.Rowland, I. R., C. J. Rumney, J. T. Coutts, and L. C. Lievense. 1998. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 19:281-285. [DOI] [PubMed] [Google Scholar]
- 258.Roy, D., I. Mainville, and F. Mondou. 1997. Selective enumeration and survival of bifidobacteria in fresh cheese. Int. Dairy J. 7:785-793. [Google Scholar]
- 259.Roy, D., and S. Sirois. 2000. Molecular differentiation of Bifidobacterium species with amplified ribosomal DNA restriction analysis and alignment of short regions of the ldh gene. FEMS Microbiol. Lett. 191:17-24. [DOI] [PubMed] [Google Scholar]
- 260.Ruas-Madiedo, P., M. Gueimonde, F. Arigoni, C. G. de los Reyes-Gavilan, and A. Margolles. 2009. Bile affects the synthesis of exopolysaccharides by Bifidobacterium animalis. Appl. Environ. Microbiol. 75:1204-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ruas-Madiedo, P., M. Gueimonde, C. G. de los Reyes-Gavilan, and S. Salminen. 2006. Short communication: effect of exopolysaccharide isolated from “viili” on the adhesion of probiotics and pathogens to intestinal mucus. J. Dairy Sci. 89:2355-2358. [DOI] [PubMed] [Google Scholar]
- 262.Saavedra, J. M., N. A. Bauman, I. Oung, J. A. Perman, and R. H. Yolken. 1994. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in-hospital for prevention of diarrhea and shedding of rotavirus. Lancet 344:1046-1049. [DOI] [PubMed] [Google Scholar]
- 263.Saavedra, J. M., and A. Tschernia. 2002. Human studies with probiotics and prebiotics: clinical implications. Br. J. Nutr. 87:241-246. [DOI] [PubMed] [Google Scholar]
- 264.Salazar, N., M. Gueimonde, A. M. Hernandez-Barranco, P. Ruas-Madiedo, and C. G. de los Reyes-Gavilan. 2008. Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria. Appl. Environ. Microbiol. 74:4737-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Salazar, N., A. Prieto, J. A. Leal, B. Mayo, J. C. Bada-Gancedo, C. G. de los Reyes-Gavilan, and P. Ruas-Madiedo. 2009. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. J. Dairy Sci. 92:4158-4168. [DOI] [PubMed] [Google Scholar]
- 266.Salyers, A. A., S. E. H. West, J. R. Vercellotti, and T. D. Wilkins. 1977. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from human colon. Appl. Environ. Microbiol. 34:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Samona, A., and R. K. Robinson. 1991. Enumeration of bifidobacteria in dairy products. J. Soc. Dairy Technol. 44:64-66. [Google Scholar]
- 268.Sangrador-Vegas, A., C. Stanton, D. van Sinderen, G. F. Fitzgerald, and R. P. Ross. 2007. Characterization of plasmid pASV479 from Bifidobacterium pseudolongum subsp. globosum and its use for expression vector construction. Plasmid 58:140-147. [DOI] [PubMed] [Google Scholar]
- 269.Scardovi, V. 1986. The genus Bifidobacterium Orla-jensen 1924, 472AL, p. 1418-1434. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 270.Scardovi, V., and F. Crociani. 1974. Bifidobacterium catenulatum, Bifidobacterium dentium and Bifidobacterium angulatum: three new species and their deoxyribonucleic acid homology relationships. Int. J. Sys. Bacteriol. 24:6-20. [Google Scholar]
- 271.Scardovi, V., and L. D. Trovatelli. 1965. The fructose-6-phosphate shunt as a peculiar pattern of hexose degradation in the genus Bifidobacterium. Ann. Microbiol. Enzymol. 15:19-29. [Google Scholar]
- 272.Scardovi, V., and L. D. Trovatelli. 1969. New species of bifid bacteria from Apis mellifica L. and Apis indica F. A contribution to the taxonomy and biochemistry of the genus Bifidobacterium. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. 123:64-88. [PubMed] [Google Scholar]
- 273.Scardovi, V., L. D. Trovatelli, B. Biavati, and G. Zani. 1979. Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum--4 new species and their deoxyribonucleic-acid homology relationships. Int. J. Syst. Bacteriol. 29:291-311. [Google Scholar]
- 274.Scardovi, V., and G. Zani. 1974. Bifidobacterium magnum sp. nov., a large, acidophilic bifidobacterium isolated from rabbit feces. Int. J. Syst. Bacteriol. 24:29-34. [Google Scholar]
- 275.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Reference deleted.
- 277.Schramm, M., V. Klybas, and E. Racker. 1958. Phosphorolytic cleavage of fructose-6-phosphate by fructose-6-phosphate phosphoketolase from Acetobacter xylinum. J. Biol. Chem. 233:1283-1288. [PubMed] [Google Scholar]
- 278.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sekine, K., T. Toida, M. Saito, M. Kuboyama, T. Kawashima, and Y. Hashimoto. 1985. A new morphologically characterized cell-wall preparation (whole peptidoglycan) from Bifidobacterium infantis with a higher efficacy on the regression of an established tumor in mice. Cancer Res. 45:1300-1307. [PubMed] [Google Scholar]
- 280.Sela, D. A., J. Chapman, A. Adeuya, J. H. Kim, F. Chen, T. R. Whitehead, A. Lapidus, D. S. Rokhsar, C. B. Lebrilla, J. B. German, N. P. Price, P. M. Richardson, and D. A. Mills. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 105:18964-18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1982. Plasmids in the genus Bifidobacterium. J. Gen. Microbiol. 128:2121-2131. [DOI] [PubMed] [Google Scholar]
- 282.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1986. Related structures in the plasmid profiles of Bifidobacterium asteroides, B. indicum and B. globosum. Microbiologica 9:443-454. [PubMed] [Google Scholar]
- 283.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1986. Related structures in the plasmid profiles of Bifidobacterium longum. Microbiologica 9:415-422. [PubMed] [Google Scholar]
- 284.Shimamura, S., F. Abe, N. Ishibashi, H. Miyakawa, T. Yaeshima, T. Araya, and M. Tomita. 1992. Relationship between oxygen sensitivity and oxygen metabolism of Bifidobacterium species. J. Dairy Sci. 75:3296-3306. [DOI] [PubMed] [Google Scholar]
- 285.Shin, S. Y., and J. H. Park. 1997. Activities of oxidative enzymes related with oxygen tolerance in Bifidobacterium sp. J. Microbiol. Biotechnol. 7:356-359. [Google Scholar]
- 286.Shkoporov, A. N., B. A. Efimov, E. V. Khokhlova, L. I. Kafarskaia, and V. V. Smeianov. 2008. Production of human basic fibroblast growth factor (FGF-2) in Bifidobacterium breve using a series of novel expression/secretion vectors. Biotechnol. Lett. 30:1983-1988. [DOI] [PubMed] [Google Scholar]
- 287.Shkoporov, A. N., B. A. Efimov, E. V. Khokhlova, J. L. Steele, L. I. Kafarskaia, and V. V. Smeianov. 2008. Characterization of plasmids from human infant Bifidobacterium strains: sequence analysis and construction of E. coli-Bifidobacterium shuttle vectors. Plasmid 60:136-148. [DOI] [PubMed] [Google Scholar]
- 288.Shu, Q., F. Qu, and H. S. Gill. 2001. Probiotic treatment using Bifidobacterium lactis HN019 reduces weanling diarrhea associated with rotavirus and Escherichia coli infection in a piglet model. J. Pediatr. Gastroenterol. Nutr. 33:171-177. [DOI] [PubMed] [Google Scholar]
- 289.Shuhaimi, M., A. M. Ali, N. M. Saleh, and A. M. Yazid. 2001. Utilisation of enterobacterial repetitive intergenic consensus (ERIC) sequence-based PCR to fingerprint the genomes of Bifidobacterium isolates and other probiotic bacteria. Biotechnol. Lett. 23:731-736. [Google Scholar]
- 290.Siggers, R. H., J. Siggers, M. Boye, T. Thymann, L. Molbak, T. Leser, B. B. Jensen, and P. T. Sangild. 2008. Early administration of probiotics alters bacterial colonization and limits diet-induced gut dysfunction and severity of necrotizing enterocolitis in preterm pigs. J. Nutr. 138:1437-1444. [DOI] [PubMed] [Google Scholar]
- 291.Simpson, P. J., R. P. Ross, G. F. Fitzgerald, and C. Stanton. 2004. Bifidobacterium psychraerophilum sp. nov. and Aeriscardovia aeriphila gen. nov., sp. nov., isolated from a porcine caecum. Int. J. Syst. Evol. Microbiol. 54:401-406. [DOI] [PubMed] [Google Scholar]
- 292.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2003. Genomic diversity and relatedness of bifidobacteria isolated from a porcine cecum. J. Bacteriol. 185:2571-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2005. Intrinsic tolerance of Bifidobacterium species to heat and oxygen and survival following spray drying and storage. J. Appl. Microbiol. 99:493-501. [DOI] [PubMed] [Google Scholar]
- 294.Skouloubris, S., L. R. de Pouplana, H. de Reuse, and T. L. Hendrickson. 2003. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc. Natl. Acad. Sci. U. S. A. 100:11297-11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sonnenburg, J. L., C. T. Chen, and J. I. Gordon. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4:e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Sorek, R., V. Kunin, and P. Hugenholtz. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181-186. [DOI] [PubMed] [Google Scholar]
- 297.Sozzi, T., P. Brigidi, O. Mignot, and D. Matteuzzi. 1990. Use of dicloxacillin for the isolation and counting of bifidobacteria from dairy-products. Lait 70:357-361. [Google Scholar]
- 298.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 299.Stark, P. L., and A. Lee. 1982. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J. Med. Microbiol. 15:189-203. [DOI] [PubMed] [Google Scholar]
- 300.Su, P., A. Henriksson, J. E. Tandianus, J. H. Park, F. Foong, and N. W. Dunn. 2005. Detection and quantification of Bifidobacterium lactis LAFTI (R) B94 in human faecal samples from a consumption trial. FEMS Microbiol. Lett. 244:99-103. [DOI] [PubMed] [Google Scholar]
- 301.Suarez, F. L., J. Furne, J. Springfield, and M. D. Levitt. 1999. Failure of activated charcoal to reduce the release of gases produced by the colonic flora. Am. J. Gastroenterol. 94:208-212. [DOI] [PubMed] [Google Scholar]
- 302.Suzuki, K., Y. Benno, T. Mitsuoka, S. Takebe, K. Kobashi, and J. Hase. 1979. Urease-producing species of intestinal anaerobes and their activities. Appl. Environ. Microbiol. 37:379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Takahashi, T., E. Nakagawa, T. Nara, T. Yajima, and T. Kuwata. 1998. Effects of orally ingested Bifidobacterium longum on the mucosal IgA response of mice to dietary antigens. Biosci. Biotechnol. Biochem. 62:10-15. [DOI] [PubMed] [Google Scholar]
- 304.Takata, T., T. Shirakawa, Y. Kawasaki, S. Kinoshita, A. Gotoh, Y. Kano, and M. Kawabata. 2006. Genetically engineered Bifidobacterium animalis expressing the Salmonella flagellin gene for the mucosat immunization in a mouse model. J. Gene Med. 8:1341-1346. [DOI] [PubMed] [Google Scholar]
- 305.Takechi, S., and T. Itoh. 1995. Initiation of unidirectional ColE2 DNA replication by a unique priming mechanism. Nucleic Acids Res. 23:4196-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Talwalkar, A., and K. Kailasapathy. 2003. Metabolic and biochemical responses of probiotic bacteria to oxygen. J. Dairy Sci. 86:2537-2546. [DOI] [PubMed] [Google Scholar]
- 307.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 308.Tamura, Z. 1983. Nutriology of bifidobacteria. Bifidobact. Microflora 2:3-16. [Google Scholar]
- 309.Tanaka, K., K. Samura, and Y. Kano. 2005. Structural and functional analysis of pTB6 from Bifidobacterium longum. Biosci. Biotechnol. Biochem. 69:422-425. [DOI] [PubMed] [Google Scholar]
- 310.Tauch, A., A. Puhler, J. Kalinowski, and G. Thierbach. 2003. Plasmids in Corynebacterium glutamicum and their molecular classification by comparative genomics. J. Biotechnol. 104:27-40. [DOI] [PubMed] [Google Scholar]
- 311.Teraguchi, S., M. Uehara, K. Ogasa, and T. Mitsuoka. 1978. Enumeration of bifidobacteria in dairy products. Nippon Saikingaku Zasshi 33:753-761. [PubMed] [Google Scholar]
- 312.Thibault, H., C. Aubert-Jacquin, and O. Goulet. 2004. Effects of long-term consumption of a fermented infant formula (with Bifidobacterium breve c50 and Streptococcus thermophilus 065) on acute diarrhea in healthy infants. J. Pediatr. Gastroenterol. Nutr. 39:147-152. [DOI] [PubMed] [Google Scholar]
- 313.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. ClustalW—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Tissier, H. 1899. Le bacterium coli et la reaction chromophile d'Escherich. Crit. Rev. Soc. Biol. 51:943-945. [Google Scholar]
- 315.Tissier, H. 1900. Recherches sur la flore intestinale des nourrissons (etat normal et pathologique). M.D. thesis. University of Paris, Paris, France.
- 316.Tissier, H. 1906. Traitement des infections intestinales par la methode de la flore bacterienne de l'intestin. Crit. Rev. Soc. Biol. 60:359-361. [Google Scholar]
- 317.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. U. S. A. 96:12424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Trovatelli, L. D., F. Crociani, M. Pedinotti, and V. Scardovi. 1974. Bifidobacterium pullorum sp. nov.: a new species isolated from chicken feces and a related group of bifidobacteria isolated from rabbit feces. Arch. Microbiol. 98:187-198. [DOI] [PubMed] [Google Scholar]
- 319.Turroni, F., E. Foroni, P. Pizzetti, V. Giubellini, A. Ribbera, P. Merusi, P. Cagnasso, B. Bizzarri, G. L. de'Angelis, F. Shanahan, D. van Sinderen, and M. Ventura. 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 75:1534-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.van Casteren, W. H. M., C. Dijkema, H. A. Schols, G. Beldman, and A. G. J. Voragen. 1998. Characterization and modification of exopolysaccharide produced by Lactococcus lactis subsp. cremoris B40. Carbohydr. Polymers 37:123-130. [DOI] [PubMed] [Google Scholar]
- 321.Vandenplas, Y. 2002. Oligosaccharides in infant formula. Br. J. Nutr. 87:S293-S296. [DOI] [PubMed] [Google Scholar]
- 322.Veerkamp, J. H. 1971. The structure of the cell wall peptidoglycan of Bifidobacterium bifidum var. pennsylvanicus. Arch. Biochem. Biophys. 143:204-211. [DOI] [PubMed] [Google Scholar]
- 323.Ventura, M., C. Canchaya, V. Bernini, A. Del Casale, F. Dellaglio, E. Neviani, G. F. Fitzgerald, and D. van Sinderen. 2005. Genetic characterization of the Bifidobacterium breve UCC 2003 hrcA locus. Appl. Environ. Microbiol. 71:8998-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Ventura, M., C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, G. F. Fitzgerald, K. F. Chater, and D. van Sinderen. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Ventura, M., C. Canchaya, D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70:3110-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ventura, M., C. Canchaya, Z. Zhang, G. F. Fitzgerald, and D. van Sinderen. 2007. Molecular characterization of hsp20, encoding a small heat shock protein of Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:4695-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Ventura, M., C. Canchaya, Z. D. Zhang, V. Bernini, G. F. Fitzgerald, and D. van Sinderen. 2006. How high G+C Gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators. FEMS Microbiol. Rev. 30:734-759. [DOI] [PubMed] [Google Scholar]
- 329.Ventura, M., C. Canchaya, R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 70:6197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ventura, M., M. Elli, R. Reniero, and R. Zink. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 331.Ventura, M., G. F. Fitzgerald, and D. van Sinderen. 2005. Genetic and transcriptional organization of the clpC locus in Bifidobacterium breve UCC 2003. Appl. Environ. Microbiol. 71:6282-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Ventura, M., J. H. Lee, C. Canchaya, R. Zink, S. Leahy, J. A. Moreno-Munoz, M. O'Connell-Motherway, D. Higgins, G. F. Fitzgerald, D. J. O'Sullivan, and D. van Sinderen. 2005. Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl. Environ. Microbiol. 71:8692-8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Ventura, M., V. Meylan, and R. Zink. 2003. Identification and tracing of Bifidobacterium species by use of enterobacterial repetitive intergenic consensus sequences. Appl. Environ. Microbiol. 69:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ventura, M., F. Turroni, G. Lima-Mendez, E. Foroni, A. Zomer, S. Duranti, V. Giubellini, F. Bottacini, P. Horvath, R. Barrangou, D. A. Sela, D. A. Mills, and D. van Sinderen. 2009. Comparative analyses of prophage-like elements present in bifidobacterial genomes. Appl. Environ. Microbiol. 75:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Ventura, M., F. Turroni, A. Zomer, E. Foroni, V. Giubellini, F. Bottacini, C. Canchaya, M. J. Claesson, F. He, M. Mantzourani, L. Mulas, A. Ferrarini, B. Gao, M. Delledonne, B. Henrissat, P. Coutinho, M. Oggioni, R. S. Gupta, Z. Zhang, D. Beighton, G. F. Fitzgerald, P. W. O'Toole, and D. van Sinderen. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Van Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 337.Ventura, M., Z. D. Zhang, M. Cronin, C. Canchaya, J. G. Kenny, G. F. Fitzgerald, and D. van Sinderen. 2005. The ClgR protein regulates transcription of the clpP operon in Bifidobacterium breve UCC 2003. J. Bacteriol. 187:8411-8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 68:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ventura, M., R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl. Environ. Microbiol. 71:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Vernazza, C. L., G. R. Gibson, and R. A. Rastall. 2006. Carbohydrate preference, acid tolerance and bile tolerance in five strains of Bifidobacterium. J. Appl. Microbiol. 100:846-853. [DOI] [PubMed] [Google Scholar]
- 341.Vincent, D., D. Roy, F. Mondou, and C. Dery. 1998. Characterization of bifidobacteria by random DNA amplification. Int. J. Food Microbiol. 43:185-193. [DOI] [PubMed] [Google Scholar]
- 342.Wang, C., H. Shoji, H. Sato, S. Nagata, Y. Ohtsuka, T. Shimizu, and Y. Yamashiro. 2007. Effects of oral administration of Bifidobacterium breve on fecal lactic acid and short-chain fatty acids in low birth weight infants. J. Pediatr. Gastroenterol. Nutr. 44:252-257. [DOI] [PubMed] [Google Scholar]
- 343.Watabe, J., Y. Benno, and T. Mitsuoka. 1983. Bifidobacterium gallinarum sp. nov.: a new species isolated from the ceca of chickens. Int. J. Syst. Bacteriol. 33:127-132. [Google Scholar]
- 344.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Xu, Y. F., L. P. Zhu, B. Hu, G. F. Fu, H. Y. Zhang, J. J. Wang, and G. X. Xu. 2007. A new expression plasmid in Bifidobacterium longum as a delivery system of endostatin for cancer gene therapy. Cancer Gene Ther. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 346.Yaeshima, T., T. Fujisawa, and T. Mitsuoka. 1992. Bifidobacterium globosum, subjective synonym of Bifidobacterium pseudolongum, and description of Bifidobacterium pseudolongum subsp. pseudolongum comb. nov. and Bifidobacterium pseudolongum subsp. globosum comb. nov. Syst. Appl. Microbiol. 15:380-385. [Google Scholar]
- 347.Yasui, H., J. Kiyoshima, and H. Ushijima. 1995. Passive protection against rotavirus-induced diarrhea of mouse pups born to and nursed by dams fed Bifidobacterium breve YIT4064. J. Infect. Dis. 172:403-409. [DOI] [PubMed] [Google Scholar]
- 348.Yasui, K., Y. Kano, K. Tanaka, K. Watanabe, M. Shimizu-Kadota, H. Yoshikawa, and T. Suzuki. 2009. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res. 37:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Yazawa, K., M. Fujimori, J. Amano, Y. Kano, and S. Taniguchi. 2000. Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther. 7:269-274. [DOI] [PubMed] [Google Scholar]
- 350.Yazawa, K., M. Fujimori, T. Nakamura, T. Sasaki, J. Amano, Y. Kano, and S. Taniguchi. 2001. Bifidobacterium longum as a delivery system for gene therapy of chemically induced rat mammary tumors. Breast Cancer Res. Treat. 66:165-170. [DOI] [PubMed] [Google Scholar]
- 351.Yildirim, Z., D. K. Winters, and M. G. Johnson. 1999. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J. Appl. Microbiol. 86:45-54. [DOI] [PubMed] [Google Scholar]
- 352.Yoshioka, H., K. Iseki, and K. Fujita. 1983. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics 72:317-321. [PubMed] [Google Scholar]
- 353.Yuan, J., L. Zhu, X. Liu, T. Li, Y. Zhang, T. Ying, B. Wang, J. Wang, H. Dong, E. Feng, Q. Li, H. Wang, K. Wei, X. Zhang, C. Huang, P. Huang, L. Huang, and M. Zeng. 2006. A proteome reference map and proteomic analysis of Bifidobacterium longum NCC2705. Mol. Cell. Proteomics 5:1105-1118. [DOI] [PubMed] [Google Scholar]
- 354.Zhu, L., W. Li, and X. Dong. 2003. Species identification of genus Bifidobacterium based on partial HSP60 gene sequences and proposal of Bifidobacterium thermacidophilum subsp. porcinum subsp. nov. Int. J. Syst. Evol. Microbiol. 53:1619-1623. [DOI] [PubMed] [Google Scholar]
- 355.Zomer, A., M. Fernandez, B. Kearney, G. F. Fitzgerald, M. Ventura, and D. van Sinderen. 2009. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J. Bacteriol. 191:7039-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]