Abstract
Summary: Platforms for pathogen discovery have improved since the days of Koch and Pasteur; nonetheless, the challenges of proving causation are at least as daunting as they were in the late 1800s. Although we will almost certainly continue to accumulate low-hanging fruit, where simple relationships will be found between the presence of a cultivatable agent and a disease, these successes will be increasingly infrequent. The future of the field rests instead in our ability to follow footprints of infectious agents that cannot be characterized using classical microbiological techniques and to develop the laboratory and computational infrastructure required to dissect complex host-microbe interactions. I have tried to refine the criteria used by Koch and successors to prove linkage to disease. These refinements are working constructs that will continue to evolve in light of new technologies, new models, and new insights. What will endure is the excitement of the chase. Happy hunting!
INTRODUCTION
In 1926, Paul de Kruif published Microbe Hunters (40a), a book that captured the imagination of generations of budding microbiologists. Subsequent contributions, including Microbe Hunters—Then and Now, by Koprowski and Oldstone (68a), have summarized advances and disease overviews possible to that time. However, recent increases in the rate of speed of pathogen discoveries call for us to take a deep breath and see where we are. This paper summarizes and compares methods now in use and suggests possible directions in which we might travel over the next few years, providing rationales and examples of discoveries. It is weighted more toward viruses than other microbes because this reflects the experience of the author; nonetheless, I have tried to emphasize issues generic to the field.
In 1967, U.S. Surgeon General William Stewart was reported to have said that given advances in antibiotics and vaccines, it was time to “close the book” on infectious diseases so as to free resources to focus on chronic illness (22). There is no formal annotation of the remark, and Stewart himself disavowed any knowledge of making it. Nonetheless, the fact that this apocryphal quote survives suggests that at least some held similar views at the time. The fallacy of such misplaced optimism was amply illustrated in the years that followed, as a host of new plagues were discovered, for example, HIV/AIDS and bovine spongiform encephalopathy, as well as old ones in new clothing, such as antibiotic-resistant staphylococci, various mycobacteria, and Escherichia coli O157:H7. Perhaps more intriguing than these discoveries has been the recognition that the notion of infectious diseases as always being acute is too narrow. Microbes have been linked to a wide range of chronic illnesses, including ulcers, cancer, cardiovascular disease, and mental illness. Thus, rather than closing the book on infectious diseases, new chapters are being written.
DIVERSITY OF THE MICROBIAL UNIVERSE
Can we actually “know” the universe? My God, it's hard enough finding your way around in Chinatown.
—Woody Allen
The introduction of cultivation-independent methods for microbial discovery and surveillance has dramatically altered our view of the breadth and diversity of the microbial world. Not only can we now find and characterize disease agents for which we have no culture system, we can also more rapidly survey ourselves and the larger biosphere. These advances have enabled extraordinary revelations. One example is the extent to which humans represent microbial vessels. Whereas the number of cells of the human body has been estimated to be 1014, our bacterial passengers on internal and external surfaces number at least 1015 (115). Furthermore, up to 10% of our genomes comprise retroviral sequences (52). 16S rRNA gene analyses of the oropharynx (1), esophagus (105), stomach (11), intestine and colon (43), vagina (94), and skin (49) indicate differences in human bacterial microflora by anatomical location, individual, and area of residence.
These analyses have also revealed that bacterial composition is dynamic. It varies over time and can be modified as a function of diet (including the use of probiotics), antibiotics (58), hygiene, and, in the instance of intestinal microflora, surgical interventions such as bypass procedures (145). The mouth alone has been shown to harbor more than 600 species of bacteria (102). Recent improvements in throughput, reductions in costs, and investments in metagenomic sequencing will almost certainly drive this figure much higher. Environmental sampling has revealed bacteria and fungi that thrive in extreme temperatures and in the presence of radioactivity, organic compounds, and heavy metals not tolerated by higher organisms (40, 91, 111).
Unlike bacteria, viruses do not comprise regions of sequence conservation that enable surveillance and discovery by a method analogous to 16S rRNA gene PCR. Thus, with a few notable exceptions in which agents have been shown to be present because investigators invested in more complex analyses (e.g., subtractive cloning [76] or consensus PCR [cPCR] using sequences of related agents) based on clues from immunohistochemistry (90), studies of viral diversity have come into their own only more recently with the introduction of high-throughput sequencing. Even here, however, we are limited by our capacity to recognize similarities between what we observe for a sample and what is present in a database. We have only begun to scratch the surface of virus discovery. Figure 1 illustrates this point by tracking the annual growth since 1982 of the viral sequence database vis-à-vis selected seminal discoveries and improvements in sequencing technology. The number of vertebrate species is estimated to be >50,000 (P. Daszak, personal communication). If each one is associated with only 20 endemic viruses, the vertebrate virome would exceed 1 million. Viruses are present in concentrations as high as 1010 particles per 100 ml in superficial coastal waters and are estimated to comprise 1 to 10% of the total prokaryotic biomass (approximately 200 megatons of carbon). The extent to which these viruses pose threats to human health remains to be determined. Most are phages; nonetheless, the sheer mass and diversity are staggering. In addition to serving as potential reservoirs for the emergence of novel pathogens, these viruses may serve a critical function in global as well as marine ecosystems by controlling prokaryote multiplication (124).
FIG. 1.
Growth of the viral sequence database mapped to seminal discoveries and improvements in sequencing technology. EM, electron microscopy. (Courtesy of Omar Jabado, reproduced with permission.)
PROOF OF CAUSATION
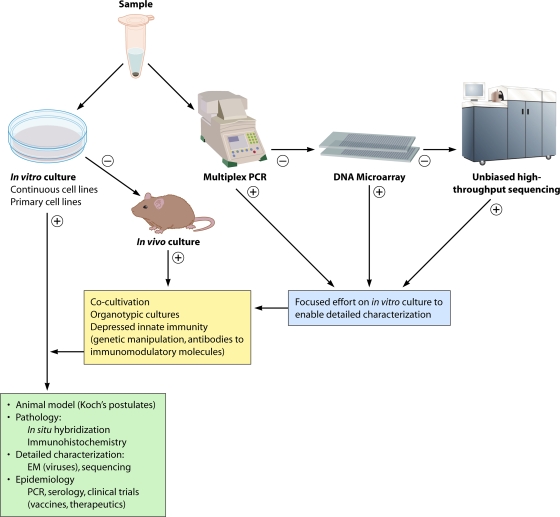
Finding an organism in association with disease is only the first step in establishing a causal relationship (Fig. 2). Pasteur, Koch, and Loeffler proposed precise criteria that define a causative relationship between agent and disease. In what are now known as Koch's postulates, these criteria require that an agent be present in every case of the disease, be specific for that disease, and be propagated in culture and proven capable of replicating the original disease upon inoculation into a naïve host (68). Rivers modified these criteria for specific application to viruses (110), focusing on the presence of neutralizing antibodies as evidence of specificity. Fredericks and Relman adapted them to reflect the introduction of PCR, requiring the presence of microbial sequences rather than an intact, infectious microorganism (47) (Table 1). Although the original Koch's postulates remain the most compelling criteria for proving causation, there are many examples, such as HIV/AIDS, where there is a clear consensus of a causal relationship despite a failure to fulfill them due to ethical or biological constraints. For a detailed history of proof of causation that remains timely despite its publication more than 3 decades ago, the reader is referred to an excellent review by Evans (45).
FIG. 2.
Staged strategy for pathogen discovery and proof of causation. In the molecular era of pathogen discovery, culture and molecular methods are pursued in parallel until an agent is detected, isolated, and characterized. +, positive result; −, negative result. ssRNA, single-stranded RNA; dsRNA, double-stranded RNA.
TABLE 1.
Criteria for proof of causation
| Criterion reported by: | ||
|---|---|---|
| Koch (1890) | Rivers (1936) | Fredericks and Relman (1996) |
| A microbe must be present in every case of a disease | A virus must be associated with a disease with a degree of regularity | Candidate sequences should be present in most cases of disease and at sites of disease pathology |
| It must not be found in association with any other disease | The association cannot be incidental | Few or no sequences should be present in host or tissue without disease |
| After isolation and propagation in pure culture, the microbe must be capable of inducing the same disease | Rivers predicted that methods for proving causal relationships would evolve with improvements in technology; at the time of his writing, he invoked seroconversion and experimental inoculations but acknowledged the limitations of both | Sequences should diminish in frequency with resolution of disease and increase with relapse |
| Sequences should be present prior to the onset of disease | ||
Signs and symptoms are rarely pathognomonic of infection with a specific agent. One cannot, for example, differentiate among the many potential causes of encephalitis using clinical criteria alone. Additionally, the manifestations of infection may vary with genetic susceptibility (e.g., herpes simplex virus encephalitis in human UNC-93B deficiency [29]), age (West Nile virus [WNV] encephalitis [89]), nutrition, previous exposure of the host to similar agents that confer partial protection or exacerbate disease (e.g., dengue hemorrhagic fever [53]), or coinfections that lead to increased pathogenicity (e.g., measles or HIV and opportunistic infections [78]). Infectious agents may also have effects that are difficult to appreciate because of a temporal delay (birth defects [23, 32] or neoplasia [147]), action at a distance from the site of infection mediated by toxins (botulism [119] or tetanus [95]), or autoimmune responses (rheumatologic disorders or Guillain-Barré syndrome [2]).
As an ideal, implication of an infectious agent can be viewed as a four-step process.
- Step 1. Detect an agent or its footprints in association with disease
- Isolate and propagate the agent in culture or in animal models
- Image the agent or its gene products in situ by using immunological (immunohistochemistry) or molecular (in situ hybridization) methods
- Demonstrate an agent-specific adaptive immune response (e.g., IgM or an increase in IgG titer, cell-mediated immunity)
- Step 2. Provide a plausible mechanism for an explanation of disease
- Demonstrate the presence of the agent, a gene product of the agent (e.g., a toxin), or a host product attributable to infection (e.g., a cross-reactive antibody or cytotoxic T cell) at site(s) of pathology
- Demonstrate that the same or a similar agent can induce disease in a model system
Step 3. Demonstrate that modulation of the agent concentration, or of a factor that can be attributed to the presence of the agent (e.g., an antibody), influences the presence or severity of disease
Step 4. Demonstrate that preventing infection prevents disease
However, as it is unlikely that many candidates would complete this gauntlet in a time frame relevant to clinical medicine or public health, I prefer to think in terms of levels of confidence in establishing a link between candidate and disease wherein evidence is scored as possible, probable, or definitive. Early in the course of a pathogen discovery project, a candidate will almost certainly be scored as possibly causal. As evidence accumulates and confidence increases, legitimate candidates will shift categories through probable to definitive.
Possible Causal Relationship
In a possible causal relationship, a disease is statistically associated with the presence of an agent or its molecular or serological footprints and with signs and symptoms reminiscent of disease caused by a related agent in the same host or in a model organism. Many important discoveries begin with a statistically significant observation. However, confidence that an observation represents more than a simple marker for disease is greatly enhanced if there is precedent to indicate biological plausibility for a causal relationship.
Probable Causal Relationship
These are instances where one can actively test the hypothesis that a particular disease is due to infection with an agent through use of strategies designed to ameliorate or prevent infection.
Interventional evidence.
Interventional evidence shows that the implementation of a drug is specific for an agent associated with disease and aborts the disease process.
(i) Challenges.
Some interventions may affect the replication of multiple agents or modify host responses, resulting in clinical improvements that could be misleading. Ribavirin, for example, has activity against a wide range of RNA viruses, including arenaviruses, bunyaviruses, orthomyxoviruses, paramyxoviruses, picornaviruses, and reoviruses (36, 44, 104, 122, 126, 142). Reports that amantadine, an agent with well-documented activity against orthomyxoviruses (39), inhibited Borna disease virus (BDV) replication while improving clinical status were used as evidence that BDV caused unipolar depression (13). However, the effects of amantadine on BDV replication could not be replicated (37), and the antidepressant properties of amantadine likely reflected its promotion of dopamine release (51). Thus, with the exception of interventions that target specific nucleic acid sequences or epitopes (e.g., RNA interference [RNAi] or monoclonal antibodies), it may be difficult to ensure the specificity of an intervention.
Prophylactic evidence.
Prophylactic evidence shows that the application of a vaccine or a drug that is specific for an agent associated with disease prevents the disease. Sequences of human papillomaviruses (HPVs) can be identified in close to 100% of cervical cancers (147). However, many women may be infected with HPV and never develop cervical cancer. Thus, while the epidemiological evidence supports a statistically significant association, the presence of HPV sequences alone cannot predict cervical cancer. One explanation for this phenomenon is that some viral strains may have a higher propensity to cause disease than do others. In fact, some strains are more highly correlated with cancer; however, even where one considers only those strains that are more prevalent in cervical cancer, HPV is not the sole determinant of disease. The most compelling evidence of causation came with international longitudinal studies of more than 17,000 women without previous infection with HPV-16 or -18 who were immunized and protected from high-grade cervical neoplasia (14). Another example is HIV/AIDS. The strong statistical association between HIV and AIDS and the development of simian models for disease using a related lentivirus provided compelling evidence of a causal relationship. The argument was further buttressed by the observation that specific antiretroviral therapy prevents both HIV infection and AIDS after occupational or gestational exposure.
(i) Challenges.
Diseases that have low incidence rates or are postulated to reflect late sequelae of infection may require large multisite studies and several years to attain statistical significance.
Definitive Causal Relationship
These are instances wherein one can fulfill Koch's postulates or variants thereof: (i) an agent isolated from a host with a disease and propagated in culture causes the same disease when introduced into a similar host, and (ii) an agent found to be associated with a disease using molecular, serological, or other methods and, through genetic engineering, recreated as an infectious entity causes the same or a similar disease when introduced into a similar host.
(i) Challenges.
In the most straightforward examples, the agent or its footprints will be found at a high concentration in the affected tissue at the time when disease is manifest, and the host range will be sufficiently broad to facilitate testing in a tractable animal model within the same subphylum (e.g., Vertebrata) or class (e.g., Mammalia). However, more stringent phylogenetic restrictions may apply, such as order (e.g., primates) or family (e.g., Hominidae). Modifying factors, such as age, genetic background, immune status, or coinfection, may also confound modeling efforts, particularly for those regarding viruses causing persistent infections.
ISOLATION OR VISUALIZATION OF INFECTIOUS AGENTS
Microbe hunters employ a wide range of media and tissue culture systems, including complex organotypic cultures (16, 84, 123, 146), to isolate and grow prokaryotic and eukaryotic organisms. When these efforts fail, alternative strategies may include inoculation of immature or genetically modified higher organisms that possess innate immune responses that are inefficient or disabled (e.g., newborn [15] and knockout [50] mice) or transgenes that are introduced to express products essential to the entry or replication of viruses (80, 108) or prions (118). The choice of an in vitro versus an in vivo strategy for the isolation of infectious agents can have a profound impact on what one can find. For example, whereas the surveillance of human stool for enteroviruses by the inoculation of suckling mice favors detection of human enterovirus type A, tissue culture favors the detection of human enterovirus type B (140). If the sequence of the pathogen candidate is known, genomic reconstruction can circumvent the need to isolate it as a viable organism (56). This approach has enabled a new field of archaeovirology wherein infectious retroviruses have been built from endogenous retroviral sequences (77), and the 1918 pandemic influenza strain was rebuilt and analyzed for pathogenetic properties (127). When an agent cannot be propagated by any of these methods, one may nonetheless find evidence of its presence by imaging it morphologically via light or electron microscopy or imaging its proteins or nucleic acids through immunohistochemistry or in situ hybridization, respectively. In some instances a candidate agent is sufficiently similar to known ones such that available antibodies to the latter are cross-reactive with the former. Indeed, immunohistochemistry has been used not only to confirm the presence of an agent or determine its anatomic distribution but also as a clue to its identity. Prominent examples include Sin Nombre virus (35), Nipah virus (103), and West Nile virus (19), for which the screening of tissues from victims of unrecognized infectious diseases with broadly reactive sera led investigators to focus on candidate viral families by consensus PCR.
COMPLEXITIES OF PATHOGENETIC MECHANISMS MAY CONFOUND DISCOVERY AND IMPLICATION OF MICROBES IN DISEASE
In the most straightforward pathogen discovery expeditions, an agent is present in high concentrations at a site where pathology is readily apparent and organ dysfunction is dramatic. Classical examples include infections with polioviruses and motor neuron disease, an influenza virus or Streptococcus pneumoniae and acute respiratory disease, and a rotavirus or Shigella sp. and diarrhea. Viruses may kill cells directly through intracellular replication and lysis, the induction of apoptosis, or autophagy. They may also do so indirectly, by presenting antigens that are recognized by cytotoxic T lymphocytes or that become bound to antibodies and trigger the activation of the classical complement cascade. Causal links may be more difficult to establish when damage is indirect, particularly when effects are manifest at sites other than the replication site. Clostridium botulinum and Clostridium tetani bacteria, for example, grow in the skin or the gastrointestinal tract and release zinc metalloproteases that have distal effects on motor function by modulating neurotransmitter release (botulism [119] and tetanus [24]).
Another example is Sin Nombre virus, a hantavirus that induces the expression of cytokines that in turn promote pulmonary capillary leakage, culminating in an acute respiratory distress syndrome (86). Microbes can elicit immune responses that break tolerance to self, resulting in autoimmune disease. A well-known example is group A beta-hemolytic streptococcus (GABHS). GABHS infection of the oropharynx may cause local inflammation or be asymptomatic. In either case, infection in susceptible individuals elicits a humoral immune response that can cause both cardiac valvular damage (rheumatic heart disease) and abnormalities in movement and behavior (Sydenham's chorea) (106).
Another example is Campylobacter jejuni and Guillain-Barré syndrome (GBS). GBS is an acute demyelinating neuropathy treated by plasmapheresis or the administration of intravenous immunoglobulin (26). More than 25% of individuals with GBS are infected with C. jejuni. Of those, up to 70% report a history of diarrheal illness consistent with C. jejuni infection (2). C. jejuni elicits an immune response that cross-reacts with the ganglioside GM1 in host neural tissue (144).
Infection with one organism may increase vulnerability to others. HIV/AIDS is an extreme form of this phenomenon, in which immunosuppression sets the stage for opportunistic infection with Toxoplasma gondii, Pneumocystis jirovecii, human herpesvirus 8, or Cryptococcus neoformans. HIV is not unique (or even the first example) in this respect. Indeed, in 1908 Von Pirquet reported that measles was associated with a loss of delayed-type hypersensitivity to tuberculin antigen and suggested that impaired immunity might explain the dissemination of tuberculosis in individuals with measles (132).
Infection may also facilitate local invasion. S. pneumoniae invasion in influenza, for example, is linked to damage to respiratory tract epithelium and is correlated with the sialidase activity of influenza virus neuraminidase (81). Infections may have long-term as well as acute effects. Viruses can express gene products (oncoproteins) that impair cell cycle regulation (139) or integrate into the host genome (147) to promote neoplasia. Inflammation associated with persistent bacterial, parasitic, or viral infection has also been implicated in cancer (79).
During vulnerable periods of embryogenesis, any of a variety of agents may cause similar types of structural damage to the central nervous or cardiovascular system, damage that continues long after the infection has cleared. In TORCH syndrome, for example, neurological effects of prenatal infection with T. gondii, rubella virus, cytomegalovirus, or herpes simplex virus cannot be distinguished by clinical criteria. In some animal models of autism, schizophrenia, and attention deficit hyperactivity disorder, the neurological effects of prenatal infection with RNA viruses and Gram-negative bacteria can be recreated by using the double-stranded RNA virus mimic, polyinosine/cytosine (41), and lipopolysaccharide (27), respectively. It is unknown whether sequelae in these examples are mediated by the loss of somatic or stem cells (or both), by altered signaling that impedes the trafficking of cells to their appropriate destinations, or by another mechanism. Therefore, it may be that any heuristic is perhaps too stringent if it requires an exclusive relationship between a pathogen and a specific outcome.
MICROBIAL PROSPECTING
Microbial prospecting takes several forms. Research centers such as ours frequently receive requests to investigate outbreaks of acute diseases in human or animal populations for which a cause is not immediately apparent. These projects may be technically challenging; however, achieving possible proof of causation can be straightforward. In such instances the agent itself or associated nucleic acids or proteins are frequently present in several affected individuals, providing the statistical power needed to establish an association between an agent and a disease. An additional advantage is that epidemiologists deployed to areas of outbreaks of infectious diseases are experts in methods for investigating causality; thus, even before laboratories focused on discovery become engaged, it is likely that biologically plausible models for pathogenesis are already in place, appropriate samples for culture and molecular characterization have been collected, and efforts have been initiated to collect acute-phase and convalescent-phase serum samples needed to test for adaptive immune responses to candidate agents.
We are more circumspect with respect to requests to address single cases of acute disease. In many instances we find a known agent that was simply missed because the appropriate conventional assays were not employed. This can be a source of embarrassment for others and difficult to report. In other instances when we find a new agent, it can be difficult to rule out a chance association. Notable exceptions include the detection of agents related to those classically associated with the syndrome observed, for example, a new filovirus in an individual with hemorrhagic fever or meningitis in which the presence of any agent in the cerebrospinal fluid (CSF) is cause for concern. Nonetheless, unless discovery efforts have the potential to alter clinical management, single-case investigations can be disappointing, expensive, and time-consuming investments. Chronic diseases can also be difficult but for different reasons. Many have already been extensively investigated; thus, agents present at sites of pathology are likely to have been found. Additionally, pathogenetic mechanisms are typically more complex than those in acute diseases. Hence, triggers of disease may no longer be present in affected tissues, or the effects may reflect infection at sites other than those where manifestations of disease are observed.
PATHOGEN DE-DISCOVERY
Although finding a causal relationship between an agent and disease is more gratifying than disproving it, the latter is no less critical to the process of pathogen discovery. Pathogen de-discovery is particularly challenging because the failure to detect a relationship may reflect differences in the site(s) or timing of sample collection, methods for sample processing or storage, assay design, or technical competence. There is also the issue of political sensitivity associated with the discounting of the work of a colleague and of the judgment of a funding agency or an advocacy group seeking solutions to a problem of medical, professional, or personal interest. It is not surprising, therefore, that journals are frequently reluctant to publish such work. In my own experience with pathogens linked to affective disorders such as schizophrenia (75), chronic fatigue syndrome (46), amyotrophic lateral sclerosis (137), and autism (61), the process of bringing a refutation study to publication takes 1 to 2 years, with much of that time being devoted to finding an editor who can be persuaded of the value of the enterprise. An additional wrinkle is that unlike pathogen discoveries, which open new avenues for basic and translational research, de-discoveries foreclose opportunities. They rarely culminate in more than one publication or in support for additional exploratory work; thus, it is difficult to persuade investigators to undertake them. Nonetheless, doing so is a community service given that results can influence the allocation of resources, preserve the reputation of the field, and, in some instances, prevent the inappropriate or unnecessary use of drugs that are toxic or in short supply.
There are three objectives in pursuing de-discovery projects. The first is to test, rigorously and objectively, a candidate hypothesis; the second is to persuade one's peers that the first objective has been achieved; the third is to persuade an educated lay audience that you and your scientific peers have considered the hypothesis in a fair and balanced fashion. Many scientists fail in impact because they consider only themselves and their peers in the study design. Over the course of many validation/invalidation projects, we have developed the following guidelines for study design.
At the outset, engage other scientists, including those responsible for the original findings, as well as representatives of professional, funding, and advocacy organizations. Ensure that key partners agree on study design as well as plans for monitoring of progress, release of results, and conflict resolution.
Collect samples (under code at a location not affiliated with the laboratory sites) from subjects who meet strict case definitions accepted by key partners. When appropriate to obviate the risk of geographic or temporal bias, use samples collected from different regions and in different seasons and different populations.
Establish concordance across laboratories for sensitivity and specificity using positive and negative controls. For PCR assays, we insert non-wild-type sequences into positive-control template constructs to differentiate bona fide versus control products.
Prepare several aliquots for each sample using a minimum of two codes. This enables blinded duplicate assays and facilitates blinded reassessments in the event that results of assays are discordant within or between laboratories.
Distribute duplicate, differently coded aliquots of positive and negative controls and samples. To facilitate interlaboratory comparisons, we typically send replicates of multiwell plates wherein each control or sample is placed into two nonadjacent wells.
Report results to a central biostatistics core for an assessment of intra- and interlaboratory consistency. As needed to achieve intralaboratory consistency, assays should be done with coded specimens.
Release results simultaneously to key partners.
Challenges.
Some key partners may suggest that it is easier to independently test samples previously found to be positive than to invest in a new sample collection. This path should be discouraged because it does not address issues of diagnostic stability, geographic or temporal bias, or sample contamination. Some key partners may differ with respect to assay design. Where resources are not limiting, it is best to employ assay formats proposed by all key investigators. However, in the event that this is not feasible, it is important to appreciate that the critical laboratory is the one in which the finding originated. This laboratory may have the most sensitive assay. It also typically has the confidence of the advocacy community. In my experience, spurious results most commonly arise not from failure in technical expertise but rather from the selection of inappropriate cases and controls or from sample contamination. At the end of the day, the success of such endeavors depends not only on the quality of the science but also on the extent to which participants can be engaged and encouraged to maintain open minds with respect to processes and outcomes.
MOLECULAR METHODS FOR PATHOGEN DISCOVERY
Singleplex Assays
The most common singleplex assays employed in clinical microbiology and microbial surveillance are PCR assays, wherein DNA strand replication results in either the cleavage or release of a fluorescence-labeled oligonucleotide probe bound to a sequence between the forward and reverse primers. Equipment needs are simple (thermal cycler, fluorescent reader, and laptop computer), and rugged instruments have been implemented for field use with battery power. Loop-mediated isothermal amplification (LAMP) does not require programmable thermal cyclers (54, 93, 120). In laboratory settings, LAMP products are detected in ethidium bromide-stained agarose gels. However, in the field, changes in the turbidity of the amplification solution may be sufficient; assays in which the accumulation of product can be detected by eye have been described (63). The most sensitive assays are those wherein primers and/or probes perfectly match a single genetic target. Fluorescence reporter-based TaqMan or molecular beacon singleplex PCR assays, for example, typically have detection thresholds of <10 RNA molecules. Although ideal for detecting the presence of a specific agent and for quantitating burden (57, 128), these assays may nonetheless fail with RNA viruses characterized by high mutation rates and genetic variability. Consensus PCR assays are less likely to be confounded by sequence divergence but are less sensitive than specific PCR assays. Furthermore, given that many potential pathogens can overlap in clinical presentation, unless one has the sample mass, resources, and time to invest in many singleplex assays for different agents, there is the risk that a spurious candidate or candidates will be selected. 16S rRNA gene assays have been particularly powerful tools in bacteriology with such seminal contributions as the discovery of Tropheryma whippeli (107); they have become more powerful yet, with the introduction of sequencing technologies that enable the description of microbial communities.
Nested PCR, in which two amplification reactions are pursued sequentially with either one (heminested) or two (fully nested) primers located 3′ with respect to the original primer set, may be more sensitive than fluorescent reporter dye singleplex assays. However, because the original reaction vessels must be opened to add reagents for the second, nested reaction (28, 125), the risk for contamination is high, even in laboratories with scrupulous experimental hygiene.
Multiplex Assays
Signs and symptoms of disease are rarely pathognomonic of a single agent, particularly early in the course of an illness. Multiplex assays may be helpful in such situations because they may be used to entertain many hypotheses simultaneously. The number of candidates considered ranges from 10 to 100 with multiplex PCR, to thousands with microarrays, to the entire tree of life with unbiased high-throughput sequencing. In multiplex assays many genetic targets compete for assay components (e.g., nucleotides, polymerases, and dyes), in some instances with variable efficiencies. Thus, multiplex assays tend to be less sensitive than singleplex assays.
Multiplex PCR Assays
Multiplex PCR assays are more difficult to establish because primer sets may differ in optimal reaction conditions (e.g., annealing temperature and magnesium concentration). Furthermore, complex primer mixtures are more likely to result in primer-primer interactions that reduce the assay sensitivity and/or specificity. To enable multiplex primer design, we developed Greene SCPrimer, a software program that automates consensus primer design over a multiple-sequence alignment and allows users to specify the primer length, melting temperature, and degree of degeneracy (62).
Gel-based multiplex PCR assays, wherein products are distinguished by mass, can detect as many as 10 distinct targets (28, 125). Fluorescence reporter-based multiplex assays are more sensitive but are limited by the number of fluorescent emission peaks that can be unequivocally separated. At present, up to four fluorescent reporter dyes are detected simultaneously. “Sloppy molecular beacons” can address this limitation in part by binding to related targets at different melting temperatures (114); however, they cannot detect targets that differ by more than a few nucleotides, and thus, their applications are limited.
Two platforms that combine PCR and mass spectroscopy (MS) for the sensitive, simultaneous detection of several targets have been established. The Ibis T5000 biosensor system uses matrix-assisted laser desorption-ionization (MALDI) MS to directly measure the molecular weights of PCR products obtained in an experimental sample and to compare them with a database of known or predicted product weights (59, 113, 131). MassTag PCR uses atmospheric pressure chemical ionization (APCI) MS to detect molecular weight reporter tags attached to PCR primers (20, 97). Whereas the Ibis system is confined to specialized laboratories, MassTag PCR can be performed by using smaller, less expensive instruments and does not require sophisticated operators. The Ibis system has an advantage in that it can detect novel variants of known organisms via a divergent product weight; nonetheless, like MassTag PCR, it too requires sequencing for a detailed characterization. Syndrome-specific MassTag PCR panels have been established for the detection of viruses, bacteria, fungi, and parasites associated with acute respiratory diseases, diarrheas, encephalitides/meningitides, and hemorrhagic fevers (20, 69, 97).
The Bio-Plex (also known as Luminex) platform employs flow cytometry to detect PCR amplification products bound to matching oligonucleotides on fluorescent beads (25, 55, 73). Assay panels that allow the detection of up to 50 genetic targets simultaneously have been developed.
Although multiplex PCR methods are designed to detect known agents, they can nonetheless facilitate pathogen discovery. The use of MassTag PCR to investigate influenza-like illness in New York State revealed the presence of a novel rhinovirus clade. This discovery enabled follow-up studies across the globe wherein this novel genetic clade was implicated not only in influenza-like illnesses but also in asthma, pediatric pneumonia, and otitis media (3, 12, 21, 42, 65, 66, 71, 72, 82, 83, 85, 109, 116, 143).
Microarrays
Microarray technology runs the gamut from assays that comprise hundreds to those comprising millions of probes. Probes can be designed to discriminate differences in sequence that allow virus speciation or to detect thousands of agents across the tree of life. An example of the former application is respiratory virus resequencing arrays, where specific genetic targets are amplified by multiplex consensus PCR, and the resultant products are hybridized to oligonucleotide probes less than 25 nucleotides (nt) in length (33, 34, 74, 141). These arrays are easily implemented when one considers only a limited number of known agents. However, because the signal is dependent on precise complementarity between probes and their genetic targets, these arrays are not ideal for pathogen discovery. In contrast, arrays comprising longer probes (e.g., >60 nt) are more tolerant of sequence mismatches and may detect agents that have only modest similarity to those already known.
Two longer probe array platforms are in common use: the GreeneChip and the Virochip (101, 138). Although they differ in design, both employ random amplification strategies to allow an unbiased detection of microbial targets. This is critical to exploiting the broad probe repertoire of these arrays; nonetheless, because host and microbe sequences are amplified with similar efficiencies, the sensitivity for microbial detection in tissues is lower than that for multiplex consensus PCR methods employed with resequencing arrays. Host DNA can be eliminated by enzymatic digestion; however, host rRNA remains a major confounder. Thus, these platforms have been most successful with acellular template sources, such as virus cell culture supernatant, serum, plasma, cerebrospinal fluid, or urine. Methods for depleting host rRNA prior to amplification through subtraction or the use of random primers selected for the lack of complementarity to rRNA have been described (4). Whether these interventions will sufficiently enhance sensitivity to enable pathogen discovery in tissues remains to be determined. At present, hybridization to probes representing pathogen targets is detected by binding of fluorescent label; however, platforms that will detect hybridization as changes in electrical conductance are in development. These platforms may enhance both ease of use and sensitivity. During a Marburg virus outbreak, the GreeneChip array, a panmicrobial array, implicated Plasmodium falciparum in a fatal case of hemorrhagic fever that was not resolved by using standard diagnostic methods (101); a variant of the GreeneChip array recently facilitated the discovery of Ebola virus Reston in a porcine respiratory illness outbreak in the Philippines (9). The Virochip was employed in the characterization of the severe acute respiratory syndrome (SARS) coronavirus in 2003 (138).
Unbiased High-Throughput Sequencing
The power of unbiased high-throughput sequencing has enabled unique advances in microbial surveillance and discovery. Applications include metagenomic characterization of environmental and clinical samples, rapid and comprehensive sequence analysis of microbial strains and isolates, and pathogen discovery. Unlike cPCR or array methods, whereby investigators are limited by known sequence information and must choose the pathogens to be considered in an experiment, high-throughput sequencing can be unbiased and allow an opportunity to inventory the entire tree of life. We have chiefly used the 454 Life Sciences pyrosequencing system; however, applications and principles for sample preparation and data analysis are similar across platforms. Although we have employed primers designed to amplify phyla (e.g., for 16S rRNA gene analyses of gastrointestinal flora) or specific viruses (e.g., for characterizations of influenza or dengue virus isolates), we more commonly focus on pathogen discovery. As in microarray applications based on unbiased PCR amplification strategies, host nucleic acid can be a critical impediment to sensitivity. The same caveats and potential solutions also apply. After amplification and sequencing, raw sequence reads are clustered into nonredundant sequence sets. Unique sequence reads are assembled into contiguous sequences, which are then compared to databases using programs that examine homology at the nucleotide and amino acid levels using all six potential reading frames (98). A truly novel pathogen might elude this level of analysis. Thus, we and others are exploring ways in which insights into the identity of agents may be determined by features such as nucleotide composition or predicted secondary or tertiary structures.
VIGNETTES IN PATHOGEN DISCOVERY
Molecular Characterization of Borna Disease Virus, a Novel Neurotropic Virus
In 1985, Rott and colleagues reported serological evidence that patients with bipolar disorder were infected with Borna disease virus (BDV) (112). At that time, BDV was simply an unclassified, filterable agent. Named after the town of Borna in Saxony (Eastern Germany), where a prominent outbreak of encephalitis in horses crippled the Prussian cavalry (75), BDV has been shown to induce behavioral disturbances vaguely reminiscent of bipolar disorder in a Lewis rat model (18). Intrigued by both the concept that an infectious process might be implicated in a neuropsychiatric disease and the fact that established methods for virus isolation had failed, we began to pursue characterization using molecular tools. Through the use of the phenol emulsion reassociation technique, BDV nucleic acids were isolated by subtractive hybridization. This effort, the first successful application of purely genetic methods in pathogen discovery (76), relied upon cDNA cloning with home-brew kits, as it preceded the advent of PCR and ready access to DNA sequencing technologies.
The relationship between candidate nucleic acids and disease was achieved by demonstrating that (i) candidate cDNAs competed with RNA extracted from brains of infected rats for the transcription and translation of a protein present in brains of rats and horses with Borna disease (hybrid arrest experiments), (ii) the distribution of candidate nucleic acids correlated with pathology in brains of experimentally infected rats and naturally infected horses (in situ hybridization), and (iii) no signal was obtained in Southern hybridization experiments wherein normal brain was probed with candidate clones (76). Based on Northern hybridization experiments with strand-specific probes, the genome was variously reported as an 8.5-kb negative-polarity RNA (76) or an 11-kb positive-polarity RNA (130). Over the next 5 years, the genome was cloned, and the virus was visualized and classified as the prototype of a new family of nonsegmented, negative-strand RNA viruses with unusual properties: nuclear replication/transcription, posttranscriptional modification of selected mRNA species by splicing, low-level productivity, broad host range, neurotropism, and capacity for persistence (117).
It was widely held that the introduction of specific reagents for BDV, such as recombinant proteins, antibodies, primers, and probes, would allow the rapid assessment of the role of BDV in human disease. However, in a classical example of the pitfalls of PCR diagnostics, particularly using nesting methods, BDV was implicated in a wide variety of disorders that included major depressive disorder, bipolar disorder, schizophrenia, chronic fatigue syndrome (CFS), AIDS encephalopathy, multiple sclerosis, motor neuron disease, and brain tumors (glioblastoma multiforme) (75). Following a report of BDV RNA in the blood of more than 50% of CFS subjects, which raised concerns for blood product safety (87, 88), we were recruited to pursue a replication study. We found no BDV nucleic acid or specific immunoreactivity in a blinded analysis of well-characterized CFS subjects. At the time of this writing, there is no conclusive evidence that BDV infects humans. However, new highly divergent strains of BDV were recently identified in birds by microarray (67) and pyrosequencing (60), strains that would not have been detected using PCR assays designed to detect viruses previously described for ungulates. Thus, it is conceivable that there are human strains yet to be found. It is worth noting that the 2 years of subtractive cloning required to identify BDV in the late 1980s was collapsed into 2 weeks with the pyrosequencing of avian isolates. However, all strains of BDV found in mammals and birds to date are so divergent from other viruses that they would not be identified by cPCR, microarray, or pyrosequencing without prior knowledge of the genus Bornavirus.
West Nile Virus in North America
In the summer of 1999, health officials in New York City reported an outbreak of encephalitis accompanied by profound motor weakness (http://www.nyc.gov/html/doh/html/cd/cdsten2.shtml). This outbreak was not detected because of an increase in the frequency of encephalitis in the greater metropolitan area but because Deborah Asnis, an infectious diseases physician at Flushing Hospital Medical Center, and Marcelle Layton, Assistant Commissioner, Communicable Disease Program, New York City Department of Health, appreciated the appearance of a distinctive clinical syndrome. Their critical role in the recognition of the outbreak underscores the unheralded importance of clinicians and public health practitioners in the process of pathogen discovery.
Investigation by serology led to an announcement of a St. Louis encephalitis virus (SLEV) outbreak (8). Investigation of the outbreak epicenter revealed sites of active mosquito breeding, and early victims of the outbreak had histories consistent with mosquito exposure. Concurrently, large numbers of crows began dying in the greater metropolitan area. There were also several deaths of exotic birds in the Bronx Zoo. Tracy McNamara, a veterinary pathologist at the Wildlife Conservation Society (located at the Bronx Zoo), performed a histological analysis of crows and flamingos and found meningoencephalitis, gross hemorrhage of the brain, splenomegaly, and myocarditis (121). When McNamara was unable to persuade her colleagues in human infectious disease surveillance to review materials, she forwarded tissue samples from diseased birds to the U.S. Department of Agriculture (USDA) National Veterinary Service Laboratory in Ames, IA, where the virus was cultured, and electron micrographs showed consistence with the presence of either a togavirus or a flavivirus. Thereafter, the avian virus was forwarded from the USDA to the CDC, Fort Collins, for molecular analysis (70).
On 13 to 15 September 1999, the CDC Encephalitis Project (comprised of centers in California, New York, and Tennessee) held its annual meeting in Albany, NY. Data emerging from both California and New York over an 18-month survey period indicated that an etiological agent was never identified for 70% of all cases of encephalitis despite culture, serology, and molecular analyses. In this context, our group was invited to discuss methods for the identification of unknown pathogens and to consider the application of a new method for amplifying viral nucleic acids, domain-specific differential display (DSDD), to project samples. Sherif Zaki of the CDC in Atlanta, GA, had demonstrated the presence of flavivirus protein in brains of human victims of the New York City outbreak; however, efforts to amplify SLEV or other flaviviral sequences by conventional reverse transcription (RT)-PCR had been unsuccessful. Employing several degenerate primer sets designed to target highly conserved domains in the NS3, NS5, and 3′-untranslated regions of flaviviruses by DSDD, we obtained positive results for four of the five New York patients in only a few hours (http://www.health.state.ny.us/press/releases/1999/nile.htm). Sequence analysis confirmed the presence of a lineage 1 WNV (18, 19). Concurrently, our colleagues at the CDC in Fort Collins reported WNV-like sequences in cell lines infected with homogenates from New York birds (70). In concert, these findings implicated WNV as the cause of the outbreak.
Thereafter, we built real-time PCR assays for the sensitive, high-throughput detection of the virus in clinical materials and mosquito pools. Although analysis of blood samples from infected humans revealed the presence of WNV sequences in late 1999 (17), the significance of human-to-human transmission was not appreciated until 2002, when evidence of transmission through organ transplants and blood transfusions led to the adoption of blood screening (30, 31). An important sequela of the 1999 WNV outbreak was a new impetus to promote the One Medicine/One Health Initiative (http://www.onehealthinitiative.com/index.php). The notion that the health of humans and the health of other animals are linked (i.e., “zoonotic”) dates back to antiquity; nonetheless, the belated recognition in the summer of 1999 of the link between disease in humans, wildlife, and domestic animals led to a new emphasis on enhancing communication between the human and veterinary comparative medicine communities.
Novel Enterovirus in Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS) is a disorder characterized by the progressive loss of motor neurons and muscle atrophy. Although inherited forms have been described, the majority of cases are spontaneous and idiopathic. Treatment is confined to supportive care, and victims typically have a relentless course that culminates in death from respiratory failure within 5 years of diagnosis. In 2000, Berger and colleagues reported the presence of novel echovirus sequences in the spinal cord of 15 of 17 French patients with ALS, versus only 1 of 29 patients with other neurological diseases (10). The work had practical implications. If confirmed, individuals with ALS might be candidates for treatment with pleconaril, a drug that had recently been shown to have activity against picornaviruses. We undertook a replication study with colleagues at Columbia University and the University of Pittsburgh under the auspices of the NIH. Our experience with BDV, in which problems with PCR hygiene led to spurious links to disease, was invaluable in directing experimental design. Whereas the Berger group had used an RNA template extracted from sections cut on cryostats and analyzed by nested PCR in the same laboratory, we collected frozen tissues from two tissue banks, extracted RNA in an independent laboratory, and performed blinded real-time PCR analyses in yet another laboratory. Analysis of spinal cord and motor cortex from 20 subjects with ALS and 14 controls revealed no echovirus sequences (137). The performance of this work led to accusations that we were trying to discredit the investigators rather than test a clinically significant finding; its publication required 1 year, chiefly because we had difficulty in finding a journal interested in reporting a “negative study.” Our results were subsequently confirmed by Nix et al. (92).
Dandenong Virus, a New Lymphocytic Choriomeningitis Virus
In December 2006, three patients in Melbourne, Australia, were transplanted with solid organs from a single donor on the same day. The donor was reported to be in good health until he died of a hemorrhagic stroke approximately 10 days after returning to Australia following a 3-month trip through the former territory of Yugoslavia. All three organ recipients died 3 to 4 weeks after transplantation following a clinical course characterized by fever and encephalopathy. Extensive laboratory analyses with bacterial and viral cultures and PCR for a wide range of bacterial and viral pathogens were uninformative. When MassTag PCR and GreeneChip assays of RNA from recipient organs, plasma, and CSF yielded no evidence of infection, the same RNA was subjected to unbiased pyrosequencing on the 454 platform. A total of more than 100,000 nucleotide sequences were obtained. Using bioinformatic algorithms, human sequences were subtracted, and nucleotide and deduced amino acid sequences were compared with genetic databases to identify related microbial sequences (98). Whereas the S-segment sequence was recognizable at the nucleotide level, footprints of the L segment were detected only at the amino acid level, consistent with a novel lymphocytic choriomeningitis virus. Specific PCR analyses confirmed the presence of the same virus in all recipients. Tissue homogenates from organs with the highest viral RNA titers were used to inoculate cell cultures. Infected cells were used to develop an indirect immunofluorescence assay for serology and to obtain electron micrographs of the agent. In this example of pathogen discovery using unbiased sequencing technology, only 14 of more than 100,000 sequences obtained represented the pathogen; the vast majority represented rRNA.
Measles-Mumps-Rubella Vaccine and Autism
In 1998, Wakefield and colleagues reported intestinal abnormalities, including reactive lymphoid hyperplasia in ileum and chronic inflammation in colon, in children with autism and other developmental disturbances (5-7, 48, 134-136). These findings, combined with parent-reported associations of the onset of behavioral abnormalities to the timing of measles, mumps, and rubella (MMR) vaccine administration, led some to conclude that the MMR vaccine contributed to the pathogenesis of autism (136). The model proposed was that by replicating in the intestines, measles virus altered the permeability of the intestinal tract, enabling the trafficking of neuroactive molecules from the lumen to the circulation and, ultimately, to the brain. Although over 25 epidemiological studies found no relationship between the MMR vaccine and autism spectrum disorder (ASD) (64, 96, 129, 133), the role of the MMR vaccine in ASD pathogenesis remains controversial, continues to influence public perceptions of vaccine safety, and has contributed to vaccine-preventable measles outbreaks in the United States and Europe. In an effort to address the relationship between the MMR vaccine and autism, we tried to replicate the original work with the explicit oversight of an advisory committee comprising representatives from academia, public health institutions, professional medical societies, and autism advocacy groups that crafted and approved the study design. Ileal and cecal samples were obtained from children with autism and gastrointestinal disturbances and from children with gastrointestinal disturbances but without developmental disabilities. All diagnoses were confirmed by using standard diagnostic and research instruments as well as clinical evaluation. Historical data regarding immunization, medication, and child and family health were acquired; additionally, histopathological ratings were obtained. Coded specimens of RNA extracted from bowel were tested in three laboratories (the original laboratory reporting the association, an independent academic laboratory, and a government laboratory with specific expertise in measles diagnostics). Real-time PCR assays were designed to detect RNA corresponding to two regions of the viral genome. One primer set included primers used in the original study reporting the association; the other was newly designed. Sequence analysis was pursued to ensure that products obtained from clinical samples did not represent positive-control transcripts. Discordant results across the three laboratories were resolved through repeat testing using newly coded samples. The first issue that we addressed was the temporal relationship between vaccination, gastrointestinal disease, and autism. Vaccination preceded autism or gastrointestinal disease in approximately 50% of cases. In only 20% of cases did vaccination precede both gastrointestinal disease and autism. Viral RNA was detected in ileal biopsy specimens from only two children: one case and one control. Levels of viral sequence were orders of magnitude lower than those reported by the original study. These results were released simultaneously to all clinical and laboratory investigators and members of the advisory board by an independent biostatistician. Several journals rejected the paper with critiques to the effect that although the science was sound, the notion that the MMR vaccine could cause autism had already been discounted. One journal rejected the paper because it was in litigation relating to the publication of the original findings. It was published coincident with an outbreak of measles in the United States in children not vaccinated due to concerns about vaccine safety.
Streptococci and Novel H1N1 Pathogenicity
When the novel influenza virus strain H1N1pdm (pdm for “pandemic”) first appeared in the spring of 2009, the case fatality rate (CFR) was estimated to be 0.6%, similar to that of seasonal influenza. Within a few weeks, however, Argentina reported 3,056 cases with 137 deaths, representing a CFR of 4.5%. Potential explanations for the increased CFR included virus reassortment, genetic drift, or infection of a more vulnerable population. Virus genomic sequencing of samples representing both severe disease and mild disease indicated no evidence of reassortment, mutations associated with resistance to antiviral drugs, or genetic drift that might contribute to virulence. We examined nasopharyngeal swab (NPS) samples from 199 cases of H1N1pdm infection from Argentina with MassTag PCR, testing for 33 additional microbial agents. At least one additional agent of potential pathogenic importance was identified in 152 samples (76%), including Streptococcus pneumoniae (n = 62), Haemophilus influenzae (n = 104), human respiratory syncytial viruses A (n = 11) and B (n = 1), human rhinovirus A (n = 1) and B (n = 4), human coronaviruses 229E (n = 1) and OC43 (n = 2), Klebsiella pneumoniae (n = 2), Acinetobacter baumannii (n = 2), Serratia marcescens (n = 1), Staphylococcus aureus (n = 35), and methicillin-resistant S. aureus (MRSA) (n = 6) (99). The presence of S. pneumoniae was strongly correlated with severe disease. S. pneumoniae was present in 56.4% of severe cases, versus 25% of mild cases; more than one-third of H1N1pdm NPS samples with S. pneumoniae were obtained from subjects with severe disease (22 of 62 S. pneumoniae-positive NPS samples; P = 0.0004). For subjects 6 to 55 years of age, the adjusted odds ratio (OR) of severe disease in the presence of S. pneumoniae was 125.5 (95% confidence interval [CI], 16.95, 928.72; P < 0.0001). Although the association of S. pneumoniae with morbidity and mortality was established by current and previous influenza pandemics, these findings were the first to demonstrate the prognostic significance of a noninvasive antemortem diagnosis of S. pneumoniae infection and may provide insights into clinical management.
FUTURE PERSPECTIVES
Molecular platforms are rapidly evolving, with enhancements in sensitivity and throughput at a lower cost. Such improvements are facilitating the decentralization of technology such that studies now restricted to a few specialized laboratories will soon be feasible on a global scale. This technology transfer will, in turn, circumvent logistical and political issues relating to specimen transfer that can delay informed responses to outbreaks of acute disease. Multiplex PCR is relatively mature; thus, advances are likely to be incremental. In contrast, microarray technology is less advanced. Predictable near-term improvements include higher-density arrays, automation, microfluidic sample processing, and alternatives to imaging of results, such as the direct measurement of conductance changes associated with hybridization. Data management and bioinformatics will become increasingly important as each of these platforms becomes more complex. This article has not addressed the emerging fields of proteomics and host response profiling, nor has it discussed new platforms for serology. It is conceivable that biomarkers will be found that are specific for classes of infectious agents and/or provide insights that can guide clinical management. Although less advanced, there are efforts to develop high-density serological arrays that offer the promise of a historical perspective of microbial exposures to a wide range of pathogens. There is an increasing appreciation for the fact that individuals can respond differently to infectious agents based on genetic and epigenetic factors, nutritional status, age, exposure history, and simultaneous infections with other microbes. This is particularly true for chronic diseases. Thus, it is anticipated that many substantive advances may come not from technical improvements but from investments in prospective serial sample collections and an appreciation that many diseases reflect intersections of genes and the environment in a temporal context.
Acknowledgments
I am grateful to my friend Charlie Calisher for inspiration and editorial comments (helpful and otherwise), to Katrina Ciraldo for ensuring that the manuscript was intelligible, to Omar Jabado for the gift of Fig. 1, to current and former members of the Center for Infection and Immunity and our collaborators for their contributions and scientific fellowship, and to the members of study sections and program officers at the National Institutes of Health.
I thank the National Institutes of Health (grants AI051292, AI57158 [Northeast Biodefense Center—Lipkin], and R24-EY017404-03), the Bill and Melinda Gates Foundation, the Department of Defense, and Google.org for their support of our fishing expeditions.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allos, B. 1997. Association between Campylobacter infection and Guillain-Barré syndrome. J. Infect. Dis. 176:125-128. [DOI] [PubMed] [Google Scholar]
- 3.Arden, K. E., P. McErlean, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2006. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 78:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armour, C., J. Castle, R. Chen, T. Babak, P. Loerch, S. Jackson, J. Shah, J. Dey, C. Rohl, and J. Johnson. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat. Methods 6:647-649. [DOI] [PubMed] [Google Scholar]
- 5.Ashwood, P., A. Anthony, A. Pellicer, F. Torrente, J. Walker-Smith, and A. Wakefield. 2003. Intestinal lymphocyte populations in children with regressive autism: evidence for extensive mucosal immunopathology. J. Clin. Immunol. 23:504-517. [DOI] [PubMed] [Google Scholar]
- 6.Ashwood, P., A. Anthony, F. Torrente, and A. Wakefield. 2004. Spontaneous mucosal lymphocyte cytokine profiles in children with autism and gastrointestinal symptoms: mucosal immune activation and reduced counter regulatory interleukin-10. J. Clin. Immunol. 24:664-673. [DOI] [PubMed] [Google Scholar]
- 7.Ashwood, P., and A. Wakefield. 2006. Immune activation of peripheral blood and mucosal CD3+ lymphocyte cytokine profiles in children with autism and gastrointestinal symptoms. J. Neuroimmunol. 173:126-134. [DOI] [PubMed] [Google Scholar]
- 8.Asnis, D. S., R. Conetta, A. A. Teixeira, G. Waldman, and B. A. Sampson. 2000. The West Nile virus outbreak of 1999 in New York: the Flushing Hospital experience. Clin. Infect. Dis. 30:413-418. [DOI] [PubMed] [Google Scholar]
- 9.Barrette, R. W., S. A. Metwally, J. M. Rowland, L. Xu, S. R. Zaki, S. T. Nichol, P. E. Rollin, J. S. Towner, W. J. Shieh, B. Batten, T. K. Sealy, C. Carrillo, K. E. Moran, A. J. Bracht, G. A. Mayr, M. Sirios-Cruz, D. P. Catbagan, E. A. Lautner, T. G. Ksiazek, W. R. White, and M. T. McIntosh. 2009. Discovery of swine as a host for the Reston ebolavirus. Science 325:204-206. [DOI] [PubMed] [Google Scholar]
- 10.Berger, M. M., N. Kopp, C. Vital, B. Redl, M. Aymard, and B. Lina. 2000. Detection and cellular localization of enterovirus RNA sequences in spinal cord of patients with ALS. Neurology 54:20-25. [DOI] [PubMed] [Google Scholar]
- 11.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U. S. A. 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blomqvist, S., C. Savolainen-Kopra, A. Paananen, T. Hovi, and M. Roivainen. 2009. Molecular characterization of human rhinovirus field strains isolated during surveillance of enteroviruses. J. Gen. Virol. 90:1371-1381. [DOI] [PubMed] [Google Scholar]
- 13.Bode, L., D. Dietrich, R. Stoyloff, H. Emrich, and H. Ludwig. 1997. Amantadine and human Borna disease virus in vitro and in vivo in an infected patient with bipolar depression. Lancet 349:178-179. [DOI] [PubMed] [Google Scholar]
- 14.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, and K. V. Shah. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 15.Bowen, E., G. Lloyd, W. Harris, G. Platt, A. Baskerville, and E. Vella. 1977. Viral hemorrhagic fever in southern Sudan and northern Zaire: preliminary studies on the aetiological agent. Lancet 309:571-573. [DOI] [PubMed] [Google Scholar]
- 16.Braun, E., T. Zimmerman, T. B. Hur, E. Reinhartz, Y. Fellig, A. Panet, and I. Steiner. 2006. Neurotropism of herpes simplex virus type 1 in brain organ cultures. J. Gen. Virol. 87:2827-2837. [DOI] [PubMed] [Google Scholar]
- 17.Briese, T., W. G. Glass, and W. I. Lipkin. 2000. Detection of West Nile virus sequences in cerebrospinal fluid. Lancet 355:1614-1615. [DOI] [PubMed] [Google Scholar]
- 18.Briese, T., M. Hornig, and W. Lipkin. 1999. Bornavirus immunopathogenesis in rodents: models for human neurological diseases. J. Neurovirol. 5:604-612. [DOI] [PubMed] [Google Scholar]
- 19.Briese, T., X. Y. Jia, C. Huang, L. J. Grady, and W. I. Lipkin. 1999. Identification of a Kunjin/West Nile-like flavivirus in brains of patients with New York encephalitis. Lancet 354:1261-1262. [DOI] [PubMed] [Google Scholar]
- 20.Briese, T., G. Palacios, M. Kokoris, O. Jabado, Z. Liu, N. Renwick, V. Kapoor, I. Casas, F. Pozo, R. Limberger, P. Perez-Brena, J. Ju, and W. I. Lipkin. 2005. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg. Infect. Dis. 11:310-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briese, T., N. Renwick, M. Venter, R. Jarman, D. Ghosh, S. Kn̂dgen, S. Shrestha, A. Hoegh, I. Casas, E. Adjogoua, et al. 2008. Global distribution of novel rhinovirus genotype. Emerg. Infect. Dis. 14:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bristol, N. 2008. William H Stewart. Lancet 372:110. [Google Scholar]
- 23.Brown, A. S., M. D. Begg, S. Gravenstein, C. A. Schaefer, R. J. Wyatt, M. Bresnahan, V. P. Babulas, and E. S. Susser. 2004. Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch. Gen. Psychiatry 61:774-780. [DOI] [PubMed] [Google Scholar]
- 24.Brüggemann, H., S. Bäumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. MartÌnez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. U. S. A. 100:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunstein, J., and E. Thomas. 2006. Direct screening of clinical specimens for multiple respiratory pathogens using the Genaco Respiratory Panels 1 and 2. Diagn. Mol. Pathol. 15:169-173. [DOI] [PubMed] [Google Scholar]
- 26.Buchwald, B., R. Ahangari, A. Weishaupt, and K. Toyka. 2002. Intravenous immunoglobulins neutralize blocking antibodies in Guillain-Barre syndrome. Ann. Neurol. 51:673-680. [DOI] [PubMed] [Google Scholar]
- 27.Cai, Z., Z. L. Pan, Y. Pang, O. B. Evans, and P. G. Rhodes. 2000. Cytokine induction in fetal rat brains and brain injury in neonatal rats after maternal lipopolysaccharide administration. Pediatr. Res. 47:64-72. [DOI] [PubMed] [Google Scholar]
- 28.Casas, I., A. Tenorio, J. Echevarria, P. Klapper, and G. Cleator. 1997. Detection of enteroviral RNA and specific DNA of herpesviruses by multiplex genome amplification. J. Virol. Methods 66:39-50. [DOI] [PubMed] [Google Scholar]
- 29.Casrouge, A., S. Zhang, C. Eidenschenk, E. Jouanguy, A. Puel, K. Yang, A. Alcais, C. Picard, N. Mahfoufi, N. Nicolas, et al. 2006. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314:308-312. [DOI] [PubMed] [Google Scholar]
- 30.CDC. 2003. Detection of West Nile virus in blood donations—United States, 2003. MMWR Morb. Mortal. Wkly. Rep. 52:769-772. [PubMed] [Google Scholar]
- 31.CDC. 2004. Update: West Nile virus screening of blood donations and transfusion-associated transmission—United States, 2003. MMWR Morb. Mortal. Wkly. Rep. 53:281-284. [PubMed] [Google Scholar]
- 32.Chess, S., P. Fernandez, and S. Korn. 1978. Behavioral consequences of congenital rubella. J. Pediatr. 93:699-703. [DOI] [PubMed] [Google Scholar]
- 33.Chiu, C. Y., A. A. Alizadeh, S. Rouskin, J. D. Merker, E. Yeh, S. Yagi, D. Schnurr, B. K. Patterson, D. Ganem, and J. L. DeRisi. 2007. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J. Clin. Microbiol. 45:2340-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiu, C. Y., S. Rouskin, A. Koshy, A. Urisman, K. Fischer, S. Yagi, D. Schnurr, P. B. Eckburg, L. S. Tompkins, B. G. Blackburn, J. D. Merker, B. K. Patterson, D. Ganem, and J. L. DeRisi. 2006. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin. Infect. Dis. 43:e71-e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chizhikov, V., C. Spiropoulou, S. Morzunov, M. Monroe, C. Peters, and S. Nichol. 1995. Complete genetic characterization and analysis of isolation of Sin Nombre virus. J. Virol. 69:8132-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crotty, S., D. Maag, J. Arnold, W. Zhong, J. Lau, Z. Hong, R. Andino, and C. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 37.Cubitt, B., and J. De la Torre. 1997. Amantadine does not have antiviral activity against Borna disease virus. Arch. Virol. 142:2035-2042. [DOI] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Davies, W. L., R. R. Grunert, R. F. Haff, J. W. McGahen, E. M. Neumayer, M. Paulshock, J. C. Watts, T. R. Wood, E. C. Hermann, and C. E. Hoffmann. 1964. Antiviral activity of 1-adamantanamine (amantadine). Science 144:862-863. [DOI] [PubMed] [Google Scholar]
- 40.Degryse, E., N. Glansdorff, and A. Pièrard. 1978. A comparative analysis of extreme thermophilic bacteria belonging to the genus Thermus. Arch. Microbiol. 117:189-196. [DOI] [PubMed] [Google Scholar]
- 40a.de Kruif, P. 1926. Microbe hunters. Harcourt, Brace Co., New York, NY.
- 41.De Miranda, J., K. Yaddanapudi, M. Hornig, and W. I. Lipkin. 2009. Astrocytes recognize intracellular polyinosinic-polycytidylic acid via MDA-5. FASEB J. 23:1064-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dominguez, S., T. Briese, G. Palacios, J. Hui, J. Villari, V. Kapoor, R. Tokarz, M. Glodé, M. Anderson, and C. Robinson. 2008. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J. Clin. Virol. 43:219-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eriksson, B., E. Helgstrand, N. G. Johansson, A. Larsson, A. Misiorny, J. O. Noren, L. Philipson, K. Stenberg, G. Stening, S. Stridh, and B. Öberg. 1977. Inhibition of influenza virus ribonucleic acid polymerase by ribavirin triphosphate. Antimicrob. Agents Chemother. 11:946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans, A. S. 1976. Causation and disease: the Henle-Koch postulates revisited. Yale J. Biol. Med. 49:175-195. [PMC free article] [PubMed] [Google Scholar]
- 46.Evengard, B., T. Briese, G. Lindh, S. Lee, and W. Lipkin. 1999. Absence of evidence of Borna disease virus infection in Swedish patients with chronic fatigue syndrome. J. Neurovirol. 5:495-499. [DOI] [PubMed] [Google Scholar]
- 47.Fredericks, D. N., and D. A. Relman. 1996. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin. Microbiol. Rev. 9:18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furlano, R., A. Anthony, R. Day, A. Brown, L. McGarvey, M. Thomson, S. Davies, M. Berelowitz, A. Forbes, and A. Wakefield. 2001. Colonic CD8 and gammadelta T-cell infiltration with epithelial damage in children with autism. J. Pediatr. 138:366-372. [DOI] [PubMed] [Google Scholar]
- 49.Gao, Z., C. H. Tseng, Z. Pei, and M. J. Blaser. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. U. S. A. 104:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glaser, L., G. Conenello, J. Paulson, and P. Palese. 2007. Effective replication of human influenza viruses in mice lacking a major 2,6 sialyltransferase. Virus Res. 126:9-18. [DOI] [PubMed] [Google Scholar]
- 51.Grelak, R. P., R. Clark, J. M. Stump, and V. G. Vernier. 1970. Amantadine-dopamine interaction: possible mode of action in Parkinsonism. Science (New York) 169:203-204. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths, D. 2001. Endogenous retroviruses in the human genome sequence. Genome Biol. 2:1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagiwara, M., H. Sasaki, K. Matsuo, M. Honda, M. Kawase, and H. Nakagawa. 2007. Loop-mediated isothermal amplification method for detection of human papillomavirus type 6, 11, 16, and 18. J. Med. Virol. 79:605-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han, J., D. C. Swan, S. J. Smith, S. H. Lum, S. E. Sefers, E. R. Unger, and Y. W. Tang. 2006. Simultaneous amplification and identification of 25 human papillomavirus types with Templex technology. J. Clin. Microbiol. 44:4157-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 58.Hoban, D. 2003. Antibiotics and collateral damage. Clin. Cornerstone 5:12-20. [DOI] [PubMed] [Google Scholar]
- 59.Hofstadler, S. A., K. A. Sannes-Lowery, and J. C. Hannis. 2005. Analysis of nucleic acids by FTICR MS. Mass Spectrom. Rev. 24:265-285. [DOI] [PubMed] [Google Scholar]
- 60.Honkavuori, K. S., H. L. Shivaprasad, B. L. Williams, P. L. Quan, M. Hornig, C. Street, G. Palacios, S. K. Hutchison, M. Franca, M. Egholm, T. Briese, and W. I. Lipkin. 2008. Novel Borna virus in psittacine birds with proventricular dilatation disease. Emerg. Infect. Dis. 14:1883-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hornig, M., T. Briese, T. Buie, M. Bauman, G. Lauwers, U. Siemetzki, K. Hummel, P. Rota, W. Bellini, and J. O'Leary. 2008. Lack of association between measles virus vaccine and autism with enteropathy: a case-control study. PLoS One 3:e3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jabado, O. J., G. Palacios, V. Kapoor, J. Hui, N. Renwick, J. Zhai, T. Briese, and W. I. Lipkin. 2006. Greene SCPrimer: a rapid comprehensive tool for designing degenerate primers from multiple sequence alignments. Nucleic Acids Res. 34:6605-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jayawardena, S., C. Y. Cheung, I. Barr, K. H. Chan, H. Chen, Y. Guan, J. S. Peiris, and L. L. Poon. 2007. Loop-mediated isothermal amplification for influenza A (H5N1) virus. Emerg. Infect. Dis. 13:899-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawashima, H., T. Mori, Y. Kashiwagi, K. Takekuma, A. Hoshika, and A. Wakefield. 2000. Detection and sequencing of measles virus from peripheral mononuclear cells from patients with inflammatory bowel disease and autism. Dig. Dis. Sci. 45:723-729. [DOI] [PubMed] [Google Scholar]
- 65.Khetsuriani, N., X. Lu, W. G. Teague, N. Kazerouni, L. J. Anderson, and D. D. Erdman. 2008. Novel human rhinoviruses and exacerbation of asthma in children. Emerg. Infect. Dis. 14:1793-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kistler, A., P. C. Avila, S. Rouskin, D. Wang, T. Ward, S. Yagi, D. Schnurr, D. Ganem, J. L. DeRisi, and H. A. Boushey. 2007. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J. Infect. Dis. 196:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kistler, A. L., A. Gancz, S. Clubb, P. Skewes-Cox, K. Fischer, K. Sorber, C. Y. Chiu, A. Lublin, S. Mechani, Y. Farnoushi, et al. 2008. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol. J. 5:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koch, R. 1891. Ueber bakteriologische Forschung, Verhandl. des X. Interna. Med. Congr., Berlin 1890. August Hirschwald, Berlin, Germany.
- 68a.Koprowski, H., and M. B. Oldstone. 1996. Microbe hunters—then and now. Medi-Ed Press, Bloomington, IL.
- 69.Lamson, D., N. Renwick, V. Kapoor, Z. Liu, G. Palacios, J. Ju, A. Dean, K. St. George, T. Briese, and W. I. Lipkin. 2006. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J. Infect. Dis. 194:1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 71.Lau, S. K., C. C. Yip, H. W. Tsoi, R. A. Lee, L. Y. So, Y. L. Lau, K. H. Chan, P. C. Woo, and K. Y. Yuen. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 45:3655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee, W., C. Kiesner, T. Pappas, I. Lee, K. Grindle, T. Jartti, B. Jakiela, R. Lemanske, Jr., P. Shult, and J. Gern. 2007. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS One 2:e966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li, H., M. A. McCormac, R. W. Estes, S. E. Sefers, R. K. Dare, J. D. Chappell, D. D. Erdman, P. F. Wright, and Y. W. Tang. 2007. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J. Clin. Microbiol. 45:2105-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin, B., K. M. Blaney, A. P. Malanoski, A. G. Ligler, J. M. Schnur, D. Metzgar, K. L. Russell, and D. A. Stenger. 2007. Using a resequencing microarray as a multiple respiratory pathogen detection assay. J. Clin. Microbiol. 45:443-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lipkin, W. I., and T. Briese. 2007. Bornaviridae, p. 1829-1851. In D. M. Knipe and R. M. Howley (ed.), Virology, 5th ed. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 76.Lipkin, W. I., G. H. Travis, K. M. Carbone, and M. C. Wilson. 1990. Isolation and characterization of Borna disease agent cDNA clones. Proc. Natl. Acad. Sci. U. S. A. 87:4184-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Löwer, R., J. Löwer, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lucas, S. B., C. S. Peacock, A. Hounnou, K. Brattegaard, K. Koffi, M. Honde, J. Andoh, J. Bell, and K. M. De Cock. 1996. Disease in children infected with HIV in Abidjan, Cote d'Ivoire. BMJ 312:335-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mantovani, A., P. Allavena, A. Sica, and F. Balkwill. 2008. Cancer-related inflammation. Nature 454:436-444. [DOI] [PubMed] [Google Scholar]
- 80.Martina, Y., K. T. Marcucci, S. Cherqui, A. Szabo, T. Drysdale, U. Srinivisan, C. A. Wilson, C. Patience, and D. R. Salomon. 2006. Mice transgenic for a human porcine endogenous retrovirus receptor are susceptible to productive viral infection. J. Virol. 80:3135-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCullers, J., and K. Bartmess. 2003. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 187:1000-1009. [DOI] [PubMed] [Google Scholar]
- 82.McErlean, P., L. Shackelton, E. Andrews, D. Webster, S. Lambert, M. Nissen, T. Sloots, and I. Mackay. 2008. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C). PLoS One 3:e1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McErlean, P., L. A. Shackelton, S. B. Lambert, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2007. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J. Clin. Virol. 39:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meyers, C. 1996. Organotypic (raft) epithelial tissue culture system for the differentiation-dependent replication of papillomavirus. Methods Cell Sci. 18:201-210. [Google Scholar]
- 85.Miller, E., K. Edwards, G. Weinberg, M. Iwane, M. Griffin, C. Hall, Y. Zhu, P. Szilagyi, L. Morin, and L. Heil. 2009. A novel group of rhinoviruses is associated with asthma hospitalizations. J. Allergy Clin. Immunol. 123:98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mori, M., A. Rothman, I. Kurane, J. Montoya, K. Nolte, J. Norman, D. Waite, F. Koster, and F. Ennis. 1999. High levels of cytokine-producing cells in the lung tissues of patients with fatal hantavirus pulmonary syndrome. J. Infect. Dis. 179:295-302. [DOI] [PubMed] [Google Scholar]
- 87.Nakaya, T., H. Takahashi, Y. Nakamura, S. Asahi, M. Tobiume, H. Kuratsune, T. Kitani, K. Yamanishi, and K. Ikuta. 1996. Demonstration of Borna disease virus RNA in peripheral blood mononuclear cells derived from Japanese patients with chronic fatigue syndrome. FEBS Lett. 378:145-149. [DOI] [PubMed] [Google Scholar]
- 88.Nakaya, T., H. Takahashi, Y. Nakamura, H. Kuratsune, T. Kitani, T. Machii, K. Yamanishi, and K. Ikuta. 1999. Borna disease virus infection in two family clusters of patients with chronic fatigue syndrome. Microbiol. Immunol. 43:679-689. [DOI] [PubMed] [Google Scholar]
- 89.Nash, D., F. Mostashari, A. Fine, J. Miller, D. O'Leary, K. Murray, A. Huang, A. Rosenberg, A. Greenberg, M. Sherman, et al. 2001. The outbreak of West Nile virus infection in the New York City area in 1999. N. Engl. J. Med. 344:1807-1814. [DOI] [PubMed] [Google Scholar]
- 90.Nichol, S. T., C. F. Spiropoulou, S. Morzunov, P. E. Rollin, T. G. Ksiazek, H. Feldmann, A. Sanchez, J. Childs, S. Zaki, and C. J. Peters. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914-917. [DOI] [PubMed] [Google Scholar]
- 91.Nicholson, W., N. Munakata, G. Horneck, H. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nix, W. A., M. M. Berger, M. S. Oberste, B. R. Brooks, D. M. McKenna-Yasek, R. H. Brown, Jr., R. P. Roos, and M. A. Pallansch. 2004. Failure to detect enterovirus in the spinal cord of ALS patients using a sensitive RT-PCR method. Neurology 62:1372-1377. [DOI] [PubMed] [Google Scholar]
- 93.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oakley, B., T. Fiedler, J. Marrazzo, and D. Fredricks. 2008. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74:4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oguma, K., Y. Fujinaga, and K. Inoue. 1995. Structure and function of Clostridium botulinum toxins. Microbiol. Immunol. 39:161-168. [DOI] [PubMed] [Google Scholar]
- 96.O'Leary, J., V. Uhlmann, and A. Wakefield. 2000. Measles virus and autism. Lancet 356:772. [DOI] [PubMed] [Google Scholar]
- 97.Palacios, G., T. Briese, V. Kapoor, O. Jabado, Z. Liu, M. Venter, J. Zhai, N. Renwick, A. Grolla, T. W. Geisbert, C. Drosten, J. Towner, J. Ju, J. Paweska, S. T. Nichol, R. Swanepoel, H. Feldmann, P. B. Jahrling, and W. I. Lipkin. 2006. MassTag polymerase chain reaction for differential diagnosis of viral hemorrhagic fever. Emerg. Infect. Dis. 12:692-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palacios, G., J. Druce, L. Du, T. Tran, C. Birch, T. Briese, S. Conlan, P. Quan, J. Hui, J. Marshall, J. Simons, M. Egholm, C. Paddock, W. Shieh, C. Goldsmith, S. Zaki, M. Catton, and W. Lipkin. 2008. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 358:991-998. [DOI] [PubMed] [Google Scholar]
- 99.Palacios, G., M. Hornig, D. Cisterna, N. Savji, A. Bussetti, V. Kapoor, J. Hui, R. Tokarz, T. Briese, E. Baumeister, and W. I. Lipkin. 2009. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS One 4:e8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reference deleted.
- 101.Palacios, G., P. L. Quan, O. J. Jabado, S. Conlan, D. L. Hirschberg, Y. Liu, J. Zhai, N. Renwick, J. Hui, H. Hegyi, A. Grolla, J. E. Strong, J. S. Towner, T. W. Geisbert, P. B. Jahrling, C. Buchen-Osmond, H. Ellerbrok, M. P. Sanchez-Seco, Y. Lussier, P. Formenty, M. S. Nichol, H. Feldmann, T. Briese, and W. I. Lipkin. 2007. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 13:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paton, N., Y. Leo, S. Zaki, A. Auchus, K. Lee, A. Ling, S. Chew, B. Ang, P. Rollin, T. Umapathi, I. Sng, C. C. Lee, E. Lim, and T. G. Ksiazek. 1999. Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet 354:1253-1256. [DOI] [PubMed] [Google Scholar]
- 104.Patterson, J., and R. Fernandez-Larsson. 1990. Molecular mechanisms of action of ribavirin. Rev. Infect. Dis. 12:1139-1146. [DOI] [PubMed] [Google Scholar]
- 105.Pei, Z., E. J. Bini, L. Yang, M. Zhou, F. Francois, and M. J. Blaser. 2004. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. U. S. A. 101:4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pichichero, M. E. 1998. Group A beta-hemolytic streptococcal infections. Pediatr. Rev. 19:291-302. [DOI] [PubMed] [Google Scholar]
- 107.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 108.Ren, R. B., F. Costantini, E. J. Gorgacz, J. J. Lee, and V. R. Racaniello. 1990. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell 63:353-362. [DOI] [PubMed] [Google Scholar]
- 109.Renwick, N., B. Schweiger, V. Kapoor, Z. Liu, J. Villari, R. Bullmann, R. Miething, T. Briese, and W. Lipkin. 2007. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J. Infect. Dis. 196:1754-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rivers, T. 1937. Viruses and Koch's postulates. J. Bacteriol. 33:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rothschild, L., and R. Mancinelli. 2001. Life in extreme environments. Nature 409:1092-1101. [DOI] [PubMed] [Google Scholar]
- 112.Rott, R., S. Herzog, B. Fleischer, A. Winokur, J. Amsterdam, W. Dyson, and H. Koprowski. 1985. Detection of serum antibodies to Borna disease virus in patients with psychiatric disorders. Science 228:755-756. [DOI] [PubMed] [Google Scholar]
- 113.Sampath, R., T. A. Hall, C. Massire, F. Li, L. B. Blyn, M. W. Eshoo, S. A. Hofstadler, and D. J. Ecker. 2007. Rapid identification of emerging infectious agents using PCR and electrospray ionization mass spectrometry. Ann. N. Y. Acad. Sci. 1102:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saunders, N., and A. Jeffries. 2000. The growing utility of microbial genome sequences. Genome Biol. 1:reports 410.1-reports 410.3.
- 115.Savage, D. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 116.Savolainen-Kopra, C., S. Blomqvist, T. Kilpi, M. Roivainen, and T. Hovi. 2009. Novel species of human rhinoviruses in acute otitis media. Pediatr. Infect. Dis. J. 28:59-61. [DOI] [PubMed] [Google Scholar]
- 117.Schneemann, A., P. Schneider, R. Lamb, and W. Lipkin. 1995. The remarkable coding strategy of Borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology 210:1-8. [DOI] [PubMed] [Google Scholar]
- 118.Scott, M., D. Foster, C. Mirenda, D. Serban, F. Coufal, M. Wälchli, M. Torchia, D. Groth, et al. 1989. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59:847-857. [DOI] [PubMed] [Google Scholar]
- 119.Segelke, B., M. Knapp, S. Kadkhodayan, R. Balhorn, and B. Rupp. 2004. Crystal structure of Clostridium botulinum neurotoxin protease in a product-bound state: evidence for noncanonical zinc protease activity. Proc. Natl. Acad. Sci. U. S. A. 101:6888-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shirato, K., H. Nishimura, M. Saijo, M. Okamoto, M. Noda, M. Tashiro, and F. Taguchi. 2007. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. J. Virol. Methods 139:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Steele, K. E., M. J. Linn, R. J. Schoepp, N. Komar, T. W. Geisbert, R. M. Manduca, P. P. Calle, B. L. Raphael, T. L. Clippinger, T. Larsen, J. Smith, R. S. Lanciotti, N. A. Panella, and T. S. McNamara. 2000. Pathology of fatal West Nile virus infections in native and exotic birds during the 1999 outbreak in New York City, New York. Vet. Pathol. 37:208-224. [DOI] [PubMed] [Google Scholar]
- 122.Stephen, E. L., D. E. Jones, C. J. Peters, G. A. Eddy, P. S. Loizeaux, and P. B. Jarling. 1980. Ribavirin treatment of toga-, arena- and bunyavirus infections in subhuman primates and other laboratory animal species, p. 169-183. In R. A. Smith and R. A. Kirkpatrick (ed.), Ribavirin: a broad spectrum antiviral agent. Academic Press Inc., New York, NY.
- 123.Straub, T. M., K. H. zu Bentrup, P. Orosz-Coghlan, A. Dohnalkova, B. K. Mayer, R. A. Bartholomew, C. O. Valdez, C. J. Bruckner-Lea, C. P. Gerba, M. Abbaszadegan, and C. A. Nickerson. 2007. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect. Dis. 13:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Suttle, C. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 125.Templeton, K. E., S. A. Scheltinga, M. F. Beersma, A. C. Kroes, and E. C. Claas. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 42:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Toltzis, P., K. O'Connell, and J. Patterson. 1988. Effect of phosphorylated ribavirin on vesicular stomatitis virus transcription. Antimicrob. Agents Chemother. 32:492-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tumpey, T., C. Basler, P. Aguilar, H. Zeng, A. Solorzano, D. Swayne, N. Cox, J. Katz, J. Taubenberger, and P. Palese. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80. [DOI] [PubMed] [Google Scholar]
- 128.Tyagi, S., and F. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 129.Uhlmann, V., C. M. Martin, O. Sheils, L. Pilkington, I. Silva, A. Killalea, S. B. Murch, J. Walker-Smith, M. Thomson, A. J. Wakefield, and J. J. O'Leary. 2002. Potential viral pathogenic mechanism for new variant inflammatory bowel disease. Mol. Pathol. 55:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.VandeWoude, S., J. A. Richt, M. C. Zink, R. Rott, O. Narayan, and J. E. Clements. 1990. A Borna virus cDNA encoding a protein recognized by antibodies in humans with behavioral diseases. Science 250:1278-1281. [DOI] [PubMed] [Google Scholar]
- 131.Van Ert, M., S. Hofstadler, Y. Jiang, J. Busch, D. Wagner, J. Drader, D. Ecker, J. Hannis, L. Huynh, and J. Schupp. 2004. Mass spectrometry provides accurate characterization of two genetic marker types in Bacillus anthracis. Biotechniques 37:642-644, 646, 648. [DOI] [PubMed] [Google Scholar]
- 132.Von Pirquet, Ñ. 1908. Das Verhalten der kutanen Tuberkulin-Reaktion warend der Masern. Dtsch. Med. Wochenschr. 34:1297-1300. [Google Scholar]
- 133.Wakefield, A. 2002. Enterocolitis, autism and measles virus. Mol. Psychiatr. 7:44-46. [DOI] [PubMed] [Google Scholar]
- 134.Wakefield, A., A. Anthony, S. Murch, M. Thomson, S. Montgomery, S. Davies, J. O'Leary, M. Berelowitz, and J. Walker-Smith. 2000. Enterocolitis in children with developmental disorders. Am. J. Gastroenterol. 95:2285-2295. [DOI] [PubMed] [Google Scholar]
- 135.Wakefield, A., P. Ashwood, K. Limb, and A. Anthony. 2005. The significance of ileo-colonic lymphoid nodular hyperplasia in children with autistic spectrum disorder. Eur. J. Gastroenterol. Hepatol. 17:827-836. [DOI] [PubMed] [Google Scholar]
- 136.Wakefield, A., S. Murch, A. Anthony, J. Linnell, D. Casson, M. Malik, M. Berelowitz, A. Dhillon, M. Thomson, and P. Harvey. 1998. Ileal-lymphoid-nodular hyperplasia, non-specific colitis, and pervasive developmental disorder in children. Lancet 351:637-641. [DOI] [PubMed] [Google Scholar]
- 137.Walker, M., R. Schlaberg, A. Hays, R. Bowser, and W. Lipkin. 2001. Absence of echovirus sequences in brain and spinal cord of amyotrophic lateral sclerosis patients. Ann. Neurol. 49:249-253. [PubMed] [Google Scholar]
- 138.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. U. S. A. 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weinberg, R. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 140.Witsø, E., G. Palacios, K. Rønningen, O. Cinek, D. Janowitz, M. Rewers, B. Grinde, and W. Lipkin. 2007. Asymptomatic circulation of HEV71 in Norway. Virus Res. 123:19-29. [DOI] [PubMed] [Google Scholar]
- 141.Wong, C. W., T. J. Albert, V. B. Vega, J. E. Norton, D. J. Cutler, T. A. Richmond, L. W. Stanton, E. T. Liu, and L. D. Miller. 2004. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 14:398-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wray, S., B. Gilbert, and V. Knight. 1985. Effect of ribavirin triphosphate on primer generation and elongation during influenza virus transcription in vitro. Antiviral Res. 5:39-48. [DOI] [PubMed] [Google Scholar]
- 143.Xiang, Z., R. Gonzalez, Z. Xie, Y. Xiao, L. Chen, Y. Li, C. Liu, Y. Hu, Y. Yao, and S. Qian. 2008. Human rhinovirus group C infection in children with lower respiratory tract infection. Emerg. Infect. Dis. 14:1665-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yuki, N., K. Susuki, M. Koga, Y. Nishimoto, M. Odaka, K. Hirata, K. Taguchi, T. Miyatake, K. Furukawa, and T. Kobata. 2004. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barré syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:11404-11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang, H., J. DiBaise, A. Zuccolo, D. Kudrna, M. Braidotti, Y. Yu, P. Parameswaran, M. Crowell, R. Wing, and B. Rittmann. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.zu Bentrup, H. 2006. Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microbes Infect. 8:1813-1825. [DOI] [PubMed] [Google Scholar]
- 147.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]