Abstract
Summary: In the past several years, we have witnessed an increased interest in understanding the structure and function of the indigenous microbiota that inhabits the human body. It is hoped that this will yield novel insight into the role of these complex microbial communities in human health and disease. What is less appreciated is that this recent activity owes a great deal to the pioneering efforts of microbial ecologists who have been studying communities in non-host-associated environments. Interactions between environmental microbiologists and human microbiota researchers have already contributed to advances in our understanding of the human microbiome. We review the work that has led to these recent advances and illustrate some of the possible future directions for continued collaboration between these groups of researchers. We discuss how the application of ecological theory to the human-associated microbiota can lead us past descriptions of community structure and toward an understanding of the functions of the human microbiota. Such an approach may lead to a shift in the prevention and treatment of human diseases that involves conservation or restoration of the normal community structure and function of the host-associated microbiota.
INTRODUCTION
It has long been known that the human body is host to a wide variety of microbial life (167, 202, 251, 289). These microbes, known generally as the “human-associated microbiota,” outnumber our own cells and are intimately associated with our tissues and organs (see Table 1 for definitions of microbial ecology terms used in this review). They have long been suspected of conferring important functions to the human body, including playing a major role in our nutrition and susceptibility to disease (289). In recent years, there has been a virtual explosion of activity in the study of the human-associated microbiota and microbiome. Through high-profile projects such as the NIH-initiated Human Microbiome Project (HMP) and the international metaHIT initiatives, an awareness of this effort has become widespread among the biomedical research community. With this increased activity has come a much more detailed understanding of the human-associated microbiota and its relationship to health and disease. This recent success is owed in large part to the application of approaches from environmental microbiology to the study of the human body, most notably the use of methods of microbial community characterization that do not require laboratory culture.
TABLE 1.
Microbial ecology definitions
| Term | Definition |
|---|---|
| Biogeography | The study of biodiversity in space and time |
| Diversity | A measure of how much variety is present in a community, irrespective of the identities of the organisms present; consists of richness and evenness |
| Evenness | The distribution of individuals across types |
| Function | An activity or “behavior” associated with a community, e.g., nitrogen fixation or resistance to invasion |
| Invasion | An ecological event characterized by the establishment of a foreign organism in a new community and the persistence and spread of this organism |
| Metagenomics | A culture-independent method used for functional and sequence-based analysis of total environmental (community) DNA (note that this is not the same as amplifying, cloning, and sequencing the 16S rRNA-encoding gene, although metagenomic sequences [e.g., generated via modern sequencing methods] can be probed for 16S rRNA-encoding genes or other phylogenetic markers) |
| Microbiome | The gene complement of a community |
| Microbiota/community | A collection of microorganisms existing in the same place at the same time |
| Resilience | The rate at which a community recovers to its native structure following a perturbation |
| Resistance | The ability of a community to resist change to its structure after an ecological challenge |
| Richness | Number of types (e.g., species) in a community |
| Similarity | A measure that determines the similarity of two or more communities, typically based on shared members, total richness, and sometimes the abundance of members |
| Structure | The composition of the community and the abundance of individual members |
| Temporal stability | The ability of a community to maintain its native structure over time |
The purpose of this review is severalfold. Our primary goal is to summarize the current “state of the art” of the study of the human-associated microbiota, emphasizing examples where work in environmental microbiology has provided an essential foundation for studies of the human-associated microbiota. Next, we identify critical needs and new opportunities for the study of the human-associated microbiota. We then describe what we feel represents the next wave in human-associated microbiota research—the application of ecological theory, especially as it relates to structure and function, to communities associated with the human body. We conclude with a discussion of how knowledge of the human-associated microbiota may impact the future of medical practice, focusing on how management and restoration of the indigenous microbiota may represent a paradigm shift in medical treatment and prevention practices.
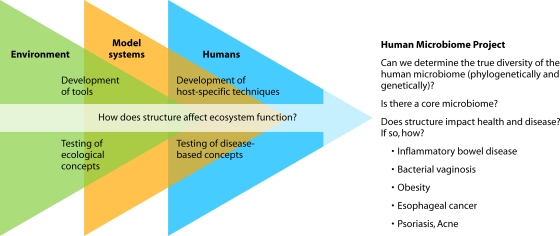
It is our contention that studies of the human microbiota and microbiome do not necessarily represent a revolution in science but rather are part of the continued evolution of a line of investigation initiated by environmental microbiologists (Fig. 1; Table 2). The cross talk between environmental microbiologists and human microbiota researchers has already yielded significant progress. Continued and increased interaction among investigators in these fields will accelerate progress even further.
FIG. 1.
Studies of the human microbiome have benefitted from the work pioneered by environmental microbial ecologists and the knowledge gained from studies conducted in model systems. Despite the many differences among these communities, the questions asked are often very similar conceptually.
TABLE 2.
Ecological concepts shared between human (or model system)-associated communities and environmental communities
| Concept | Community or environmental factor (references) |
|
|---|---|---|
| Human-associated/mouse model | Environmental/other | |
| Temporal stability | Gut (247, 379) | Aquatic communities (41, 49, 160) |
| Oral communities (214, 260, 269) | Soil (114) | |
| Chickens (168) | ||
| Leeches (164) | ||
| Resistance/resilience—structural | Diet (65, 132, 179, 191, 206) | Soil with antibiotics (64, 173, 355, 378) |
| Antibiotics (15, 68) | Soil with minerals/fertilizers (113, 114, 129) | |
| Aquatic communities in dissolved organic carbon/matter (57, 73, 158) | ||
| Aquatic communities in nutrient runoff (i.e., nitrogen and phosphorus) (95, 127, 347) | ||
| Livestock growth-promoting antibiotics (74, 84, 87, 168) | ||
| Livestock growth-promoting antibiotic alternatives (135, 235) | ||
| Resistance/resilience—functional | Aquatic microcosm (217) | |
| Microbial mat (370) | ||
| Soil (114, 278) | ||
| Methanogenic reactor (90, 91) | ||
| Invasion | Potential pathogens/known pathogens (180, 346) | Aquatic microcosm (217) |
| Probiotics (16, 58, 178, 249, 271, 336, 359, 373) | Insect guts (71, 274) | |
| Biocontrol—alfafa protected against oomycete diseases (123, 305) | ||
| Biocontrol—tomatoes protected against foot and root rot (159) | ||
| Plant growth-promoting bacteria (19, 122) | ||
| Growth-promoting probiotics (231) | ||
| Probiotics for infection prevention (5, 299, 360, 376) | ||
MICROBIAL ECOLOGY: A BRIEF HISTORY
One of the first microbial communities to be observed was a host-associated community. When Antonie van Leeuwenhoek scraped his teeth and observed what he termed “animalcules,” he unknowingly launched one of the most interesting fields of study, which still challenges us more than 330 years later—microbiology. More specifically, we are still struggling with the question of how many different kinds of bacteria are associated with the human body as well as with other animal hosts. Understanding the extent of diversity in microbial communities in any environment can be challenging. The techniques utilized in the pursuit of understanding microbial communities of all sorts are shared among microbial ecologists, as are our scientific roots.
Two hundred years after van Leeuwenhoek's discovery, the Golden Era of microbiology was ushered in, most famously, by Louis Pasteur, Robert Koch, and Ferdinand Cohn. The concept of isolating bacteria in pure culture was central to Koch's postulates, which outlined how to determine the etiology of infectious diseases. A few years later, culture of bacteria in isolation and enrichment for specific types of bacteria would be key in the research of Beijerink and Winogradsky, whose work led to the first understandings of chemoautotrophy. Into the mid- to late 20th century, culture of bacteria and microscopic examination were still the primary tools of observation and discovery for microbiologists and, of particular interest for this review, for microbial ecologists, who studied the identities of bacterial communities in specific environments and sought to understand their activities there (including microbe-microbe interactions and microbes' interactions with their environments). Identifying these bacteria was accomplished by direct microscopic evaluation, the use of biochemical tests, and enrichment culture techniques. Interestingly, a number of the bacteria observed microscopically could not be accounted for on culture media. “The great plate anomaly,” as this phenomenon is known, was observed in studies of several environments (312). Eventually, it would be estimated that more than 99% of bacteria have not been cultured (7). Although advances in culture methods increased the diversity of bacteria that were culturable, a far more exciting and fruitful approach to accessing those “unculturable” bacteria was just on the horizon.
By 1980, Carl Woese and colleagues had provided an overview of bacterial phylogeny that was based on 16S rRNA sequences and independent of the morphological and biochemical characteristics that had previously been used to classify bacteria (98, 365). Soon after, researchers began using the sequences of rRNAs to identify bacteria in mixed communities without cultivation (310, 311). As PCR was developed and sequencing became easier, the cloning and sequencing of 16S rRNA-encoding genes (instead of the laborious task of directly extracting and sequencing rRNAs) allowed for more in-depth analyses of microbial communities (112). Although other genes have been used as phylogenetic markers and may provide better resolution at the species and subspecies levels, the cloning and sequencing of 16S rRNA-encoding genes have been considered the gold standard for the characterization of microbial communities and have been used to describe the composition of a variety of communities, including those in insect guts, the human gastrointestinal tract, and microbial mats (33, 61, 128, 194).
Other culture-independent techniques were also developed. Various forms of in situ hybridization that rely on group-specific 16S rRNA probes, such as fluorescent in situ hybridization (FISH), emerged (6, 7, 63, 111). Community fingerprinting techniques such as terminal restriction fragment length polymorphism (T-RFLP) analyses and denaturing gradient gel electrophoresis (DGGE) physically separate fragments of the 16S rRNA-encoding gene and detect variations in the sequence among members of communities (199, 232).
Metagenomics, a culture-independent method of community analysis that is not dependent on the 16S rRNA-encoding gene, is the functional and sequence-based analysis of total environmental DNA and traditionally involves direct cloning of DNAs extracted from environmental samples (125, 273). This direct cloning of environmental DNA was first suggested by Olsen and colleagues in the 1980s (243). The initial implementation of this idea was carried out by Schmidt et al., who cloned DNA from a marine picoplankton community into the bacteriophage λ and screened for 16S rRNA-encoding genes (296). Metagenomics as we know it today was first reported by Stein et al., who identified a 40-kb archaeal genome fragment in a metagenomic library, subcloned and sequenced it, and then revealed the presence of two genes that were not previously known to exist in archaea (316). Since then, metagenomics has been used to explore the genetic capacity and activities of microbes in the human gut, marine planktonic communities, insect guts, and Alaskan soil, among other environments (2, 23, 108, 119, 174, 207).
Recently developed sequencing technologies include those commercialized by 454 Life Sciences/Roche Applied Sciences (454), Illumina Incorporated (Solexa), Applied Biosciences (SOLiD), Dover Systems (Polonator), and Helicos BioSciences Corporation (see references 208 and 302 for reviews of these techniques). These techniques increase the depth of sequencing by orders of magnitude compared to traditional Sanger sequencing. Pyrosequencing, as implemented on the 454 platform, is the technique used by most members of the HMP. This is likely because the 454 chemistry allows for longer reads (∼500 bp) than those of other platforms, such as Solexa (∼100 bp), although Solexa pyrosequencing produces more sequence data, as measured by the number of bases generated per run. Since its development in the late 1990s, pyrosequencing has been used extensively in several fields, including chromatin research and plant biology (228, 246). In microbiology, pyrosequencing has allowed for rapid sequencing of whole genomes, comparisons of multiple strains of bacteria, and quick detection of point mutations responsible for antibiotic resistance, among other advances (59, 121, 227, 322, 333).
In 2006, pyrosequencing and microbial ecology crossed paths for the first time. First, Forest Rohwer and colleagues published a metagenomic analysis of the microbial community in water from an iron mine in Minnesota, and then, a few months later, Mitchell Sogin and colleagues published a study that introduced the ability to access rare members of several microbial communities simultaneously by amplifying and sequencing the V6 hypervariable region of the 16S rRNA gene by use of bar-coded pyrosequencing primers (79, 307). Bar codes (also known as nucleotide keys) are short runs of nucleotides (typically 3 to 8) incorporated directly 5′ of the primer sequence and are used to differentiate samples within a pyrosequencing run. Since 2006, pyrosequencing has been used to explore a variety of communities, including those in hot springs, hamster feces, coral reefs, agricultural soils, the bovine rumen, the mouse gut, and the human gut (14, 15, 35, 72, 212, 224, 275, 331).
Along with the ability to generate community data came the need to analyze such data. Ecological measurements of richness, diversity, and similarity are used to analyze these data. Programs such as EstimateS (http://viceroy.eeb.uconn.edu/estimates) and Canoco (http://www.pri.wur.nl/uk/products/canoco/) were designed to analyze classical ecological data and determine the ecological characteristics of microbial communities as well (323). Borrowing from ecologists, who had long faced the issue of analyzing community data, microbial ecologists modified existing estimators of richness, diversity, and similarity to suit their data sets. Programs like EstimateS and mothur (http://www.mothur.org) allow microbial ecologists to analyze their data using a number of ecologically important estimators, such as those developed by Chao (i.e., the ACE and Chao1 richness estimators) (45-47, 295). Additionally, mothur allows users to compare communities (e.g., to determine whether two communities have the same membership or structure), as does the program QIIME (http://qiime.sourceforge.net/) (40, 295). The application of pyrosequencing to microbial ecology has resulted in massive data sets that require several gigabytes of memory to store and necessitate the use of multiple processors to complete analysis in convenient time frames. Several computer programs and websites have been developed or modified in order to deal with the huge quantity of data generated by pyrosequencing runs. The Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu/), which for many years has been used for classification of 16S rRNA-encoding genes, recently integrated a “pyrosequencing pipeline,” which processes sequences and clusters them based on similarity to sequences in the RDP database (a taxonomy-based approach) (50). Additionally, SILVA (http://www.arb-silva.de/) and greengenes (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) provide comprehensive rRNA gene databases to which 16S rRNA gene data generated by pyrosequencing can be aligned (66, 263). Mitchell Sogin's group at The Josephine Bay Paul Center for Comparative Molecular Biology and Evolution at the Marine Biological Laboratory in Woods Hole, MA, has developed the Visualization and Analysis of Microbial Population Structure (VAMPS) (http://vamps.mbl.edu/index.php) project, which utilizes the Global Alignment for Sequence Taxonomy (GAST) process (147). This Web-based project uses both operational taxonomic unit (OTU; defined by sequence-based phylogenetic distance)- and taxonomy-based approaches to analyze pyrosequencing data generated from the V6 region of the 16S rRNA-encoding gene. VAMPS also allows for easy visualization and presentation of the data and analyses. The aforementioned software packages mothur and QIIME are able to trim, denoise, screen, align, and analyze these sequences as well as to generate visual displays of the analyses (40, 294, 295). These packages allow for complete analysis of an experiment, from the raw data generated by the sequencer all the way to the generation of figures for publication.
Despite advances in microbial ecologists' ability to generate sequence data, there are still challenges and limitations associated with describing communities based on 16S rRNA-encoding gene sequences. For example, the controversy surrounding microbiologists' inability to define a species now extends to sequence data. Multiple similarity cutoffs have been used to define a species-level OTU. Some groups use a phylogenetic distance of 0.03, which is equivalent to a similarity of 97%, to bin sequences into species-level OTUs. Others have used multiple distances, seemingly because it is not clear which distance is appropriate. The distance of 0.03 is appropriate for near-full-length 16S rRNA gene sequences (approximately 1,500 bp), but data generated by pyrosequencing on the 454 platform, which currently produces the longest pyrosequencing read lengths, are at most about 500 bp. Therefore, it is currently possible to pyrosequence only portions of the 16S rRNA gene, and these are typically specific variable regions within the gene. These regions have various amounts of variability, which can be quite different from the amount of variation observed across the full length of the gene. Schloss revealed that phylogenetic distances calculated using individual variable regions rarely correlated well with those calculated using full-length sequences (293). The correlation between the V6 region and full-length sequences was particularly poor. Specifically, distances calculated using the V6 region were approximately 3-fold higher than those calculated using full-length sequences, which suggests that a distance cutoff of 0.10 may be closer to the species level than a cutoff of 0.03 for this region. Schloss further revealed that longer sequences (i.e., multiple regions sequenced as a single read, such as V1-3, V3-5, and V6-9) were better than shorter sequences at relating variations in portions of the gene to the full-length sequence (293). Others reported that analyses of the V1-V2 region and the V6 region overestimated species richness and that analyses of the V3, V7, and V7-V8 regions underestimated richness, while analyses of the V4, V5-V6, and V6-V7 regions resulted in species richness estimates that were comparable to those generated during analysis of full-length sequences (372). In addition, alignment of regions, gap treatment, and application of masks during analysis all have profound effects on downstream diversity analyses (293). These studies show that correct analysis of data is highly dependent on the regions chosen for pyrosequencing. They also highlight the difficulties associated with interpreting sequence data. These issues, however, will become less important as the length of sequencing reads increases.
Another limitation encountered with analyses based on the sequences of the 16S rRNA-encoding gene is that the gene does not contain information that is completely reflective of the remaining genome. Although organisms with similar 16S rRNA- encoding genes typically share similar genomes, they can in fact have vastly different functional capabilities. An example of this can be seen in Escherichia coli. Several strains of this species have nearly identical 16S rRNA-encoding genes but have very different functional capabilities. For instance, strain EDL933 is enterohemorrhagic, CFT073 is a uropathogenic strain, and MG1655 is a substrain of the classic lab strain K-12. These E. coli strains all contain 16S rRNA-encoding genes that are 99% identical, but genomewide, they share only 39% of their proteins (352).
Another common example of function or activity not being represented by 16S rRNA gene similarity is the occurrence of various antibiotic resistance profiles of members of the same species. These sorts of differences are often explained by occurrences of lateral gene transfer, which would have enormous implications for ecosystem organization and function. Even though it is difficult, if not impossible, to draw conclusions about ecosystem functions based solely on analysis of the 16S rRNA-encoding gene, the presence, absence, or fluctuations of specific bacterial groups can be used to ascertain the effects of external and internal forces on the community as a whole. As we describe later, experimentation complementary to sequencing can determine whether bacteria in a community are active and can examine their functions and their interactions with other bacteria and their environments.
WHAT DO WE KNOW ABOUT HOST-ASSOCIATED MICROBIAL COMMUNITIES?
Host-associated communities are essential for a number of functions. Host development and health are dependent on the presence of an intact microbial community. Bacteria play important roles in everything from the development of light organs in the squid Euprymna scolopes to production of vitamins and essential amino acids in humans and the development of lymphatic tissues in mice (22, 107, 175). Additionally, they impact fat and glucose utilization as well as the lymphocyte response to intestinal injury (152, 220, 331). Host-associated microbes also contribute to host susceptibility to inflammatory bowel disease and infection-mediated diseases (103, 288). Furthermore, these bacteria may contribute to host susceptibility to type 1 diabetes, allergies, and cancer (304, 354, 368). Determining how much the human microbiota impacts its host's health and whether there is a core microbiome shared among humans are two of the initiatives put forward by the HMP (330). Using the techniques described above, scientists have revealed that the human body is associated with an estimated 1 × 1014 microbial cells, representing approximately 90% of the cells in the human body (289). This microbial community, the human microbiota, is comprised of all known forms of microorganisms (i.e., bacteria, archaea, protists, and viruses).
In this review, we focus on the bacterial communities that colonize three major body sites: the oral cavity, the vagina, and the gastrointestinal tract. These sites represent three of the five sites currently explored by HMP researchers (the skin and the nasopharyngeal tract are not reviewed here, but see references 56, 99, 118, and 367 for recent studies of these communities). Additionally, communities associated with several nonhuman hosts, which are important for their simplicity (in some cases), reproducibility, and manipulability, are reviewed.
Furthermore, the temporal stability of many of these communities, which is the ability to resist changes to structure and/or function over time, is examined. Many microbial communities associated with hosts change as the host ages and develops. These shifts are likely mediated by host immunity, maternally derived and perhaps otherwise (70, 155). The shifts observed early in the development of the host and community are the result of multiple succession and assembly events. Additionally, the susceptibility of well-established communities to ecological events, such as invasion, periodic shifts in membership, or shifts in member abundance, can also result in temporal variation. Here we present examples of temporal variation associated with host development as well as variation associated simply with some length of time.
Determining the ecological characteristics, such as structure and function, of communities and monitoring change within communities are two examples of how microbial ecologists of all types have learned from each other. This section is representative of the potential benefits of cross talk between microbial ecologists, because the studies described here are direct beneficiaries of earlier studies that sought to characterize environmental samples as described in the previous section.
Oral Community
The human oral microbiota provided some of the first observed bacteria, as stated previously. These bacteria inhabit several locations in the oral cavity, including saliva, the tongue, tooth surfaces, and supra- and subgingival plaque. A recent study investigated the communities in 120 people from 12 locations around the world and revealed that the salivary microbial community from healthy individuals is composed of 6 to 30 species (234). The community is predominated by several members of the genus Streptococcus. Other bacteria present include members of the genera Eikenella, Lautropia, Syngeristes, Bacteroides, Haemophilus, Actinobacillus, Gemella, Neisseria, Prevotella, Megasphaera, Stomatococcus, and Veillonella (234, 279). Interestingly, the communities varied by individual within a location, and this variation was similar to the variation observed among individuals from different locations (234). Most oral bacteria exist in biofilms known as dental plaque (171, 210). Dental plaque forms naturally on the surfaces of teeth and in addition to bacteria is composed of water, polysaccharides, salivary proteins, and glycoproteins (210). One of the first studies using traditional molecular methods indicated that over 450 species of bacteria inhabit subgingival plaque (251). Recently, pyrosequencing data generated from saliva and supragingival plaque samples reported that up to an estimated 19,000 species are present in the collective human oral cavity, but the study did not address how many or which of these species would be expected in an individual microbiota or how many of these might be shared among all individuals in the study (161). Another pyrosequencing-based study examined the tooth surface, cheek, tongue, hard palate, and saliva and revealed that the microbiota in individuals contained over 500 species and that 75% of species observed were present in two of three individuals sampled (374). Additionally, 94% of the pyrosequencing reads from this study were grouped into OTUs shared by all three volunteers (374). These data are consistent with others' findings that oral communities from individuals tend to be more similar to each other than communities from other body sites (56). These findings are strong evidence for the presence of a core oral microbiome.
Furthermore, a study by Zaura et al. determined that the salivary community is more similar to communities on mucosal surfaces than to those on tooth surfaces (374). This is consistent with other findings that the bacterial communities in saliva and plaque are different in structure and in richness (161, 255). Many genera of the oral cavity are found in multiple communities (i.e., salivary, mucosal, and dental communities). For example, Streptococcus is a predominant member in all communities. Other shared members include several of those listed above as well as Rothia, Capnocytophaga, Corynebacterium, Actinomyces, Fusobacterium, Porphyromonas, Campylobacter, and members of the TM7 phylum (161, 374).
Ecological studies of dental plaque have revealed that the architectural organization of bacteria in dental plaque results from complex succession processes. Similar to the case in other microbial communities, bacteria in the oral cavity exhibit both mutualism, which spans the spectrum of obligation, and antagonism (see reference 198 for a review of bacterial interactions in communities, reference 181 for a review of interactions in oral communities, and reference 93 for a review of the microbial ecology of oral biofilms). Nutrient exchange and metabolic cooperation are often drivers for mutualistic interactions (198). For example, under aerobic conditions, Actinomyces naeslundii and Streptococcus oralis, both early colonizers of tooth surfaces, are unable to grow as monocultures in unamended sterile saliva but are able to grow as a coculture (248). Another dental plaque community member, Fusobacterium nucleatum, also requires the presence of A. naeslundii, but not S. oralis, for growth in unamended saliva. Interestingly, although F. nucleatum was not dependent on S. oralis for growth, its biovolume increased significantly when the three bacteria were grown together (257). Furthermore, another plaque community member and periodontopathogen, Porphyromonas gingivalis, could not grow alone or with S. oralis but, when paired with F. nucleatum, grew well in a saliva-fed flow well. Additionally, this pathogen grew even better in the presence of two additional members, Veillonella and Aggregatibacter actinomycetemcomitans, or F. nucleatum. P. gingivalis also exhibits mutualistic interactions with various other bacteria shown to colonize dental plaque at various stages throughout microbial succession in plaque (258).
The temporal dynamics of the oral community during host development have been studied intensely. The first several months of life are characterized by an edentulous oral cavity, and the resident bacteria are associated with mucosal surfaces. The initial bacteria, mostly streptococci, are acquired from the mother and environment, including food. These pioneer bacteria are later joined by a subset of bacteria associated with the adult community. After tooth eruption, others of these adult-associated bacteria begin to be established (209). Early culture-based studies found that the oral community exhibits short-term variation, but this was likely due to technical reasons, ranging from changes in oral hygiene to subjects receiving antibiotic treatments (260). One of the rare studies tracking the microbiota of healthy adult subjects found that the oral community was relatively stable during a period of 7 years (269). In healthy communities not undergoing external perturbations, community composition tends to remain constant and the abundance of members commonly shifts (214, 260, 269).
Vaginal Community
Another well-studied community is that of the vagina. This community plays a large part in protecting the reproductive tract from pathogens and the potentially harmful external environment to which it is exposed (for a review, see reference 364). The healthy vaginal community is comprised of at least 25 members, mostly from the Firmicutes phylum (241, 377). Most vaginal communities in healthy women are dominated by Lactobacillus species, although this is not always the case (149, 165, 309, 364, 377). Lactobacilli are thought to protect the vaginal tract by competitively excluding pathogens from binding mucus and epithelial receptors and by producing various antimicrobial substances (27, 309). Other bacteria that sometimes predominate the healthy community include Bifidobacterium, Gardnerella, Prevotella, Pseudomonas, Streptococcus, Peptostreptococcus, Atopobium, and an uncultured member of the order Clostridiales (133, 149, 270, 377). Vaginal communities typically display one of several structures, and there are significant differences between the community structures in women of different ethnic backgrounds (270, 377). These findings suggest that a core microbiota may not exist for the vaginal community (270). Additionally, the composition of the community varies by location within the vaginal canal (165).
Several studies suggest that the composition of the healthy vaginal microbiota exhibits little temporal variation over short periods ranging from a few days to 3 months. Indeed, others have suggested that vaginal communities likely exist in a state of dynamic equilibrium (270). Interestingly, bacterial vaginosis is characterized, in part, by shifts in the structure of the microbiota (e.g., reduced population of lactobacilli as well as the presence of Gardnerella vaginalis and an assortment of other bacteria), and retrospective studies indicate that approximately one-third of women will experience bacterial vaginosis (4, 11, 176, 240). This suggests that temporal shifts in structure may be more common than is realized. One recent study tracked vaginal community members (G. vaginalis, Atopobium vaginae, and four Lactobacillus spp.) in 12 patients over time and monitored shifts in bacterial vaginosis status (75). Generally, the communities maintained their structure over time, except when patients shifted from being healthy to positive for bacterial vaginosis.
These ecological findings have important implications for medical microbiology and the practice of medicine itself. Because we now know that the “healthy” vaginal community exists in several states, some of which are predominated by bacteria other than species of Lactobacillus and/or contain vaginosis-associated bacteria, and we have access to other ecologically important information (i.e., increased community diversity in patients with obvious symptomatic bacterial vaginosis), methods for diagnosing bacterial vaginosis are being reconsidered (34, 165, 241, 309).
Gastrointestinal Community
Initially, microbiologists thought that the stomach did not contain resident bacteria and that any bacteria detected were simply transients. Despite its extremely low pH, the stomach contains a rich microbiota composed of a diverse bacterial community in addition to several fungal species (24, 289). Although it is not clear which portion of the detected communities are transient or resident, studies using clone libraries and pyrosequencing suggest that the bacterial community may contain up to a few hundred members (14, 24). Members of the community are affiliated with the Firmicutes, Actinobacteria, Bacteroidetes, Proteobacteria, and Fusobacteria phyla (14, 24, 244, 289). Less abundant members belong to the TM7 phylum, Deferribacteres, Deinococcus/Thermus, and others (14, 24). The membership of the gastric community is consistent and does not change with anatomic location (i.e., the corpus and antrum) (24). Interestingly, this community is similar in composition to that in the esophagus, which contains species observed to be components of the oral cavity (14, 253). The presence of bacteria found in both the oral cavity and the esophagus, such as Streptococcus, Actinomyces, Prevotella, and Gemella, indicates that a portion of the stomach community may be a result of translocation of upstream community members (14). However, there are several members of the stomach community that are specifically associated with it (14). Further downstream, the small intestine also contains bacteria similar to those in the stomach (128). However, there is a shift in community structure, as facultative anaerobes are replaced by obligate anaerobes as the predominant members as one moves distally in the gastrointestinal tract from the ileum to the colon (128, 349). The microbial community in the lower gastrointestinal tract has the highest density of organisms (an estimated 1012 bacteria/g) associated with the human body, and recent pyrosequencing data suggest that this community may be composed of more than 5,000 bacterial taxa (defined as reference sequence-based OTUs) (68). Although the human gut community has many members, it is comprised of only a small portion of known bacteria and is predominated by two bacterial phyla—Bacteroidetes and Firmicutes (78). It is thought that the gut environment selects for a community of low diversity at higher phylogenetic levels (∼7 to 9 divisions) and extremely high diversity at lower phylogenetic levels (thousands of strains) (195). Additionally, there is a high level of variation among individuals at lower phylogenetic levels, but not surprisingly, at higher taxonomic levels there is a high level of conservation (24, 78, 142, 366). Despite compositional variation, microbial function in healthy individuals is maintained. This suggests that although there is not a core microbiota in the gut, there may well be a core microbiome.
The gut microbiota of human neonates and infants exhibits little temporal stability; however, within the first 2 years of life, the infant community becomes similar to adult communities (204, 247). Newborns acquire their microbiota from a number of sources. Members of the initial colon community include Escherichia coli, other enterobacteria, enterococci, streptococci, staphylococci, Bacteroides, bifidobacteria, and clostridia (89, 204, 247, 250, 254). It is thought that aerobes and facultative anaerobes reduce the redox potential of neonatal guts such that obligate anaerobes are later able to become established and to grow (314). The composition of the microbiota is in part dependent on the mode of birth (cesarean or vaginal) and feeding (breast or bottle) and on environmental conditions (204). The first days of life are often characterized by the presence of communities that are dominated by one or two taxonomic groups and then become more even as time progresses (89, 247, 250). Interestingly, the communities in infants 11 months after birth contain strains of Bacteroides and Bifidobacterium spp., the dominant gut bacteria, that are different from those in their mothers (332). From infancy to adulthood, the microbiota remains relatively stable (142). Likewise, throughout young to mid-adulthood, the gut microbiota is thought to exhibit very little temporal variation (289, 379). During adulthood, the community exhibits limited variation over extended periods, despite fluctuations of minor members such as Lactobacillus spp. (214, 266, 290, 339). Additionally, the transition to late adulthood is marked by a decrease in the bifidobacterial population (141, 142, 225). (See reference 245 for an in-depth review of community development in the human gastrointestinal tract.)
The complexity of the human gut microbiota makes it difficult to dissect the interactions among its members and with its host. This complexity has led to the gleaning of information from ecological studies of nonhuman host-associated communities. For example, much of what we know about the importance of the gut microbiota to host health has been determined using animal models such as the gnotobiotic mouse. The use of various mouse models has revealed that the community protects against intestinal epithelial injury, influences healing following mucosal injury, impacts energy extraction and storage, and contributes to the development of normal immune function (18, 140, 152, 267, 313, 319). An additional advantage to the mouse model is that within colonies and, in particular, within cages, the communities within individuals are very similar (15). In contrast, humans contain communities that exhibit high levels of interpersonal variation (15, 24, 78, 142, 366).
The classic, culture-based study of temporal changes in the native murine gut community by Schaedler et al. revealed that lactobacilli dominate the gastrointestinal tract within days of birth. In addition to these lactobacilli, there is a population of Flavobacteria that is of equal abundance. By about 1 week of age, however, flavobacterial populations are significantly decreased (291). Additionally, Schaedler et al. found that there was a high degree of temporal variation of enterococci in the colon, although more recent results indicate that this is not always the case (70, 291). The ileum is also a site of temporal variation (70, 291). The Bacteroides population in particular exhibits low temporal stability compared to other ileal community members (70). Not surprisingly, there is also a shift in community structure during weaning of mouse pups (70). Following weaning, the cecal community goes through what appears to be a stage of transition before reaching a period of little temporal variation at about 5 weeks of age, which lasts until at least 8 weeks of age. At 10 weeks, however, there is yet another shift in structure (163). Another study, however, suggests that the period of little temporal variation may range from 6 to 15 weeks of age (17).
Other Host-Associated Communities
In addition to the mouse, several other nonhuman hosts have also been studied for decades. Most of these are important to humans because of their use as food, their economic impact, and the level of interaction they have with humans. The practice of amending feed with antibiotics for growth promotion and prophylactic purposes has been questioned in light of the escalating issue of antibiotic resistance in both animal and human bacterial isolates (9, 12, 37, 281). Although it is not clear how this practice has impacted the reservoir of antibiotic resistance genes or their transfer among bacteria, the issue has led to in-depth investigations of the native microbial communities. Furthermore, as stated previously, these hosts are important components of our ability to stringently test vast hypotheses concerning the structure and function of host-associated communities.
The communities in broiler chickens have been studied since the 1960s (20). These communities are particularly rich in obligate anaerobes and contain members of several phylogenetic groups, including the Bacteroidales order, the Lactobacillaceae family, the Enterobacteriaceae family, and members of the genera Enterococcus, Campylobacter, Atopobium, and Veillonella (363). Temporal stability in broiler chicken communities mirrors that in human communities. Newly hatched chicks obtain their gut microbiota from their mothers and the environment. Within hours, anaerobic bacteria are detected, followed several days later by streptococci and enterobacteria. Predominating members of the cecum within 1 week of hatching include Salmonella, E. coli, Lactobacillus, Clostridium, Atopobium, Enterococcus, and members of the Bacteroides (10, 363). As the birds age, the bifidobacterial population size increases, the Salmonella population decreases, and the community becomes more even (i.e., the abundances of individual members become more similar) (10, 363). The ileum is predominated by lactobacilli from as early as 4 days posthatching (10). This large ileal lactobacillus population remains throughout the life of the chicken (10, 168, 201, 363). During development, the structure of this population shifts and the species of Lactobacillus change (168).
Bovine rumen communities are of interest for the reasons stated above as well as for their potential for harboring enzymes necessary for plant cell wall degradation, which could be useful in biofuel production (35, 308). These communities contain members of the Cytophaga-Flavobacterium-Bacteroides (CFB) and Protebacteria phyla, as well as members of the Firmicutes phylum (35, 321, 356). A recent pyrosequencing study estimated a species richness of about 200 members in the bovine rumen (35). In that study, three rumens were examined for their community structure and metabolic potential. Similar to the variation seen in humans, despite being fed the same diet, the three steers contained communities that were different in structure and metabolic potential. Although there has been limited work investigating the temporal dynamics of the bovine rumen community, there are indications that these communities exhibit variability to some extent during feeding cycles and over a period of several weeks (222, 353).
Microbial communities of invertebrates are also of interest. There are literally millions of insect species, and most of these are likely to be associated with at least one bacterial symbiont (182, 215, 318). Scientists have been studying these symbioses since 1882 and estimate that certain intracellular associations are up to 270 million years old (229, 230, 317). Similar to the microbial community in the human gut, the insect gut community plays a role in host health and is important for nutrient acquisition, detoxification, and colonization resistance (71). Some of the best-studied insect communities are those in termites. Like that of the bovine rumen, the termite gut microbial community has garnered attention due to its enzymatic profile (35, 308). These communities contain archaea, fungi, protists, and dozens to hundreds of bacterial taxa, several of which are unique to the termite gut, and likely contain more novel species (31, 137, 138, 185, 242, 297, 298, 369). Bacterial members of these communities affiliate with more than 15 phyla, including the Spirochaetes, Fibrobacteres, Proteobacteria, CFB, Firmicutes, and Actinobacteria (137, 242, 297, 298, 369). Furthermore, several bacterial members are endo- and ectosymbionts of the protist members (150, 239). The interactions of bacteria and protists in this community have also been studied in detail (see reference 242 for a review). These communities have likely coevolved with their hosts and are essential for termite longevity (86, 138). Additionally, it has been shown that specific groups within the community carry out specific tasks in the gut. For example, hindgut community members from the genus Treponema catalyze the synthesis of acetate, which is a significant energy source for termites (32, 186). Recently, a metagenomic analysis of the hindgut community revealed that it contains several genes necessary for cellulose and xylan hydrolysis, in addition to those for acetogenesis and several other functions related to carbohydrate degradation and nitrogen fixation. Furthermore, metagenomic fragments containing these genes were linked phylogenetically to members of the Spirochaetes and Fibrobacteres (350).
The microbial communities of other insects, including several beetle and cockroach species, have been studied intensely and have informed our understanding of the complex interactions among host-associated community members and with their hosts (82, 104-106, 342). The communities of other insects, such as those from the family Lepidoptera, have gained attention for their relative simplicity (tens of species versus the hundreds observed in termites, beetles, and cockroaches) and ease of manipulation (33, 124, 274). Recently, bacterial signaling in a multispecies community was demonstrated in the midgut of the cabbage white butterfly, Pieris rapae (Lepidoptera: Pieridae), when a community member, Pantoea CWB304, was shown to produce and detect quorum-sensing molecules in vivo (28).
Next Steps
Now that we are beginning to grasp the diversity of host-associated communities and are able to identify members of these communities, our next goals must include linking function with identity, as has been done in the insect system. Although metagenomics allows us to access the functional potential of a community and to identify its members, we have arrived at a point where it is desirable and feasible to assess community functionality and to link that to individual members or groups of members. There are several techniques in widespread use in environmental ecology that combine the identification of bacteria within a community with identification and assignment of function. One of these is the use of stable-isotope probing (SIP), which allows for the identification of community members capable of utilizing a substrate of interest by detecting stable isotopes that have been incorporated into cellular components such as nucleic acids. This technique involves introduction of a stable isotope-labeled substrate, usually containing 13C, to a community, followed by FISH, nucleic acid extraction, and sequence analysis, T-RFLP analysis, or some other method of determining community composition (76, 131, 144, 226, 265). In one example, Pseudomonas fluorescens, Pseudomonas putida, and an uncultured Acidovorax sp. were shown to be capable of naphthalene biodegradation in a groundwater microbial community by stable-isotope probing of mRNA and rRNA coupled with Raman-FISH, which allowed for analysis of community composition, gene expression, and single-cell physiology (144). In vitro models of the human intestine and computer modeling have recently shown the potential for SIP to elucidate function in host-associated communities (61a, 81, 177). The application of SIP to animal and human communities has many challenges, such as potential inhibition of substrate delivery, but with collaboration between environmental microbial ecologists and those that study host-associated communities, several of these should be surmountable (see reference 80 for a review).
Another area of research that is likely to be important includes the study of intracommunity interactions, similar to studies that have revealed symbioses among termite gut inhabitants. These studies are being done in oral cavity communities and would likely prove fruitful if replicated for other body sites. Studies of oral cavity community members go beyond identifying “healthy” and “unhealthy” communities and also investigate interactions among “normal” members and pathogens (e.g., see reference 258). Furthermore, they delve into the complicated interaction web of the “normal” community. Studies of the flagellate and bacterial symbioses in the termite gut further demonstrate our ability to dissect interactions between community members. It seems reasonable that experiments should be conducted to determine whether these interactions also occur in other environments, including the human gut. Understanding not only the composition of communities but also the functions of community members and the factors that shape assembly will be key to a number of applications, including the development of medical treatments such as drugs that affect community structure and “designer” communities.
In the future, understanding the temporal stability of communities will continue to be important. Studies of temporal stability in host-associated communities are often buried in other studies. This is likely because studies of temporal variation may be construed as tedious “fishing expeditions.” However, these studies are necessary in order to understand the dynamics of communities that are essential for the health of their hosts. We are still in the infancy of understanding the nature of these communities. We must ask basic questions about host-associated communities, and journals and funding agencies must support scientists in our quest to understand these fundamental aspects of community dynamics. There is a noticeable paucity of recent, arguably more accurate, molecular biology-based studies of temporal changes in native murine gut communities. This is interesting since so much of what we know about host-associated communities comes from studies of the mouse community (e.g., the links between the microbiota and cancer and the neurological system, as well as the well-established interactions between the microbiota and host development and physiology [8, 18, 205, 313]). Most of what has been published recently on the temporal variation of the murine community seems to be necessary controls for other studies. It seems likely that labs that work on the microbial communities of mice have the data or biological materials necessary to assemble a substantial work investigating the temporal stability of native, untreated communities in mouse colonies, but for some reason they have not published this information. Every aspect of every community cannot be studied, but for the animal that has been so important in our understanding of host-microbe interactions, we argue that an exception must be made and that we are obligated to study this community as thoroughly as we can, just as other microbiologists have studied another workhorse organism, E. coli.
BEYOND DESCRIPTION: THE APPLICATION OF ECOLOGICAL THEORY TO THE HUMAN-ASSOCIATED MICROBIOTA
Researchers participating in the HMP are actively involved in describing the overall diversity of the human-associated microbiota. One goal of the HMP, as stated previously, is to determine whether there is a core microbiome (330). This can be determined by comparing the communities in a large number of individuals and looking for genes with a high prevalence. However, the most exciting questions (What is the association between community structure and disease? How do microbial communities assemble in the human body? How do they respond to disturbance?, etc.) require the ability to detect and understand variation in the microbiota across individuals, rather than common properties. This is a much more difficult task and requires the use of ecological theory. Understanding the principles that govern the formation and persistence of ecological communities is a central goal of the general science of ecology (262). Much progress has been made toward this goal, both in our understanding of the fundamental principles that underlie community structure and in the development of tools, approaches, and theory for the study of communities (116, 143, 198). Here we explore theories that attempt to explain various aspects of community structure and concepts that provide the framework for our understanding of community responses to various ecological challenges, such as perturbation.
Community Structure
Diversity and structure.
Understanding the principles that govern ecological communities requires rigorous metrics for describing community properties. As described previously, communities can be described using the concept of diversity or that of community structure (25, 146, 198). These two concepts are obviously related. One way that they are explicitly linked is through the concept of “components” of diversity. Whittaker first proposed that diversity could be thought of as occurring simultaneously on three scales (357). Local diversity (which he called alpha diversity) is the diversity measured at a particular locality (e.g., the diversity of the microbial community within a single host). Regional or landscape diversity (gamma diversity) is the diversity of a set of these localities (e.g., the diversity of the microbial community found in a population of hosts). Alpha and gamma diversities are linked through beta diversity, the change in community composition across local communities (e.g., the difference in community structure from host to host). For example, a high gamma diversity could be present if local diversity is high and the difference in structure among local communities is low or if local diversity is low and the difference among communities is high. Studying the variation in beta diversity has emerged as a particularly powerful tool for understanding the principles governing ecological communities (116). General ecology has developed a deep theoretical literature regarding the principles that govern beta diversity. This literature suggests a number of hypotheses regarding the processes most likely to determine community structure.
Hypotheses regarding the determinants of community structure.
The community structure of the host-associated microbiota could be uniform across hosts (zero beta diversity), with observations of interindividual variation arising simply through measurement (i.e., sampling) error, or it could vary substantially (nonzero beta diversity). There is growing evidence of nonzero beta diversity among host-associated microbiota populations (56, 69, 100, 192, 329). Nonzero beta diversity can be classified into two types: random and nonrandom. Random differences among communities arise from the effects of random sampling of the “species pool” (those species available for colonization) by local communities (187). For example, each host in a population could be randomly sampling microbial taxa from a common environmental inoculum, and beta diversity in the host-associated microbiota could arise solely from this (187). Alternatively, beta diversity can be nonrandom. Nonrandom beta diversity could arise from two potential sources: dispersal limitation and ecological interactions. Models of dispersal limitation (often referred to as “neutral” models) assume that the primary driver of community differences is heterogeneity in dispersal (145). For example, certain microbial taxa might be more likely to disperse to hosts (and colonize them) than others, perhaps because they are closer to hosts spatially or because they have traits (such as motility) that increase the rate or extent of dispersal relative to those of other microbial taxa. Community patterns consistent with dispersal limitation have been reported for some host-associated communities (e.g., see reference 306). Nonrandom patterns could also result from ecological interactions (e.g., see references 48, 328, and 358). Such interactions could consist of “environmental filtering” by hosts (often referred to as “host selection”), in which the within-host environment allows only certain microbial taxa to colonize or persist. Alternatively, it could be due to interactions among microbial taxa. These interactions could include negative interactions (such as competition for resources) or positive interactions (such as facilitation). Such interactions could result in “priority effects,” where the order in which specific taxa colonize the host determines community structure (280).
Identifying the determinants of community structure.
How does one distinguish among the different hypotheses regarding the causes of beta diversity in host-associated communities? Ecologists have devised a number of approaches (reviewed in reference 187). To determine whether beta diversity is random or nonrandom in nature, there are two basic approaches. Starting with knowledge of the structure of the inoculum source community, simulation can be used to model the random sampling of this inoculum, and the resulting distribution can be compared with the distribution of taxa across individual hosts (e.g., see reference 143). Alternatively, one can use information regarding the source community and statistical theory to predict analytically the distribution resulting from random sampling and then compare this to the actual observed distribution (117). These approaches require knowledge of the source community, and this is not yet understood for most host-associated communities. However, given a large and diverse set of hosts, one can approximate the source community as the sum of all taxa found across hosts.
If one has established that beta diversity is nonrandom, there are several ways to determine the relative importance of different processes (e.g., dispersal limitation, different forms of host selection, etc.) in determining this pattern. One popular approach is variance partitioning (26). This approach uses data regarding the taxa found at each site (e.g., host) and other properties at each site (e.g., location of host, genotype of host, diet of host, etc.) to partition the total variance in community structure across sites into the respective contribution of each site property and their covariations. This approach is very popular among general ecologists, with over 1,500 published uses (256), and it is starting to be used by microbial ecologists as well (148, 162, 169, 183, 268). Two methods have traditionally been used to partition the variation of community composition data: canonical partitioning and distance partitioning (187, 268).
Variation partitioning can be used to estimate the relative contributions of measurable properties of a host to host-associated community structure. But what about the contribution of interactions among microbes (which are often not directly measurable, at least in complex communities)? It is possible to infer the relative importance of some interactions (e.g., competition) from the phylogenetic structure of communities (e.g., see reference 143; also, for an in-depth review of the intersection of community ecology and phylogenetic biology, see reference 44). Competition tends to select for community members that are more distantly related to each other than predicted by random sampling of the inoculum community (a pattern termed phylogenetic overdispersion [130]). Conversely, a community structured primarily by host selection tends to have members that are more closely related than predicted by random sampling (termed phylogenetic clustering [130]). The degree of phylogenetic overdispersion or clustering can be calculated from molecular community structure data (36, 143). Microbial ecologists have investigated the phylogenetic ecology of several environmental systems, including wastewater treatment plants, lakes, and cactus yeast communities (13, 237, 259). Although this topic has not been investigated or addressed explicitly with the human microbiota, the concepts (e.g., the host selecting for a highly related community) have been pondered for several years in host-associated microbial ecology (e.g., see reference 195). It seems that the most direct initial step for the host-associated scientific community will likely be retrospective studies that mine existing sequence data for patterns of phylogenetic overdispersion or clustering.
Biogeographic Patterns
The environmental forces that determine community structure can impose patterns on community structure over space and time. Change in beta diversity is one such pattern. But there are other patterns that may result as well (213). These patterns can be used as tools to better understand how microbial community structure is determined. Collectively, the study of patterns in community structure over space and time is known as biogeography. A number of biogeographical patterns have been documented for microbial communities, including distance-decay, taxon-area, and taxon-time patterns. Since these patterns are also common in plant and animal communities, there is a wealth of general ecological theory that links them to processes of community assembly. However, there has been little attempt to look for such patterns among host-associated microbial communities.
This is likely to change soon. Understanding the biogeography of the human-associated microbiota is an explicit goal of the HMP. A recent example of such work is that of Costello et al. (56). This work utilized 454 pyrosequencing to identify the members of oral, skin (up to 18 locations), fecal, nostril, hair, and ear communities of nine healthy individuals on two consecutive days at two time points 3 months apart. Communities were found to cluster by body site, as opposed to sex, individual, or date of sampling. Within a habitat, there was high variability among individuals. The oral community, however, was more consistent, within and among individuals, than the other body sites. An interesting aspect of this paper was the attempt to determine the impact of current environmental conditions and historical factors, which can confound our ability to understand the effect of spatial variation, on community assembly (56, 213). In this case, the historical factor tested was the presence of certain bacteria. The communities from tongues were applied to disinfected forearms or foreheads. The resultant communities on forearms were more similar to tongue communities, and the resultant communities on foreheads were more similar to forehead communities (56). These results indicate that similar to environmental communities, the spatial variation observed in host-associated communities is due to both environmental (e.g., excretion of sebum, the oily compound secreted by sebaceous glands in skin) and historical factors.
Future work.
How environmental and historical factors impact the communities in sites such as the gastrointestinal tract, the vaginal canal, and the oral cavity, which contain several different communities, remains to be seen. We also do not know whether the influence of these factors changes from site to site, although the work of Costello and colleagues seems to suggest that this is the case. Furthermore, one of the conceptual challenges of applying biogeography to the human microbiota is linking the findings to something less abstract than the theory itself, such as human health. Here we present questions whose answers should inform not only biogeography but also our understanding of health and disease in the context of the microbiota. Healthy individuals are associated with communities that group based on body site, but what about unhealthy individuals? Do the same rules apply to their communities? How do we utilize biogeography patterns to aid in community-based (at least in part) diagnoses? For example, if “healthy” environmental factors strongly suggest that the presence of acid-tolerant bacteria is normal, then the presence of acid-susceptible bacteria would be indicative of a potentially “unhealthy” state. In this case, unexpected biogeographical patterns along with shifts in structure would be the “red flag,” as opposed to just the shift in structure. This sort of additional information could be useful in a number of situations, including diagnoses of bacterial vaginosis.
Resistance and Resilience
In addition to affecting community assembly, environmental factors can also play roles in structural changes in established communities. Community stability is a functional property that focuses on community dynamics in response to perturbation (153, 198). There are several definitions and concepts associated with stability, which have been reviewed elsewhere (e.g. see references 153 and 216). Briefly, stability can be defined as the ability to return to an equilibrium state following perturbation, the ability to resist change (also known as resistance or resistance stability), the rate of return to an equilibrium following perturbation (also known as resilience or resilience stability), or overall system variability (3, 126, 153, 203, 216, 261). Furthermore, stability can be determined by monitoring changes in community structure and function (e.g., see reference 114). Additionally, ecologists have long studied the influence of diversity on stability (e.g., see references 188, 203, 218, and 261). Both empirical and theoretical studies indicate that diversity enhances stability; however, aspects of stability can be affected differently, and these relationships play out in multiple ways in different ecosystems (e.g., see references 101, 139, 156, 292, 325-327, 334, 340, 351, and 375). For example, it has been shown that while diversity increases total community stability, it decreases the stability of populations within a community (156, 188, 340). These ecological concepts have been studied in a wide range of communities (Table 2). This section focuses on what is known about the resistance stability and, to a lesser extent, the resilience stability of host-associated communities in response to diet and antibiotics.
Diet.
The community in the human gastrointestinal tract is influenced by a number of factors, including diet (67, 289). Early studies of the microbial communities in humans examined dietary practices. For example, comparisons of fecal communities from subjects whose nonvegetarian lifestyles were influenced by religion revealed significant differences from those of other nonvegetarian subjects who consumed a standard Western diet (94). As expected, some species within the microbiota are more sensitive to dietary changes than others. A gluten-free diet has a significant impact on certain groups of bacteria, including Bifidobacterium populations, within the gut microbiota and can partially restore the altered microbiota of celiac patients to a microbiota similar to those of healthy individuals (51, 65).
Diet-induced obesity has been linked to the structure of the gut microbiota. In obese subjects, Bacteroidetes populations are smaller and populations of Firmicutes are larger than those in nonobese subjects. This trend of large Firmicutes populations and small Bacteroidetes populations reverses as obese subjects lose weight by switching to lower-fat diets (191). Similarly, mice fed high-fat diets also contain communities with small Bacteroidetes populations and with elevated Firmicutes and Proteobacteria populations (132). This study also revealed that the high-fat diet, not the obese state of the mouse, caused the shift in community structure by using one strain of mouse that did not gain weight as well as one that did (132). Interestingly, although germfree mice conventionalized with the microbiota of mice reared under standard conditions consumed less food than control mice, they gained weight because the microbiota induces lipogenesis (18). These studies of community resistance to dietary fat and community function have revealed how fundamentally important host-microbe interactions are and how relevant community structure can be to host health.
Other dietary components, such as certain carbohydrates, have been studied for their efficacy as prebiotics, defined as nondigestible food ingredients that simulate the growth or activity of beneficial members of the human gut microbiota (172). Inulin and fructo-oligosaccharides, for example, significantly increase Bifidobacterium populations within the gut microbiota (typically determined from fecal samples) of test subjects compared to control subjects or to samples taken prior to administration of the treatment (29, 166, 184, 197, 221). This increased detection of bifidobacteria in response to certain oligosaccharides is also seen in the microbiota of rats and mice and in the microbiota of gnotobiotic rats colonized with human fecal bacteria (38, 284, 335). Additionally, these oligosaccharides can increase populations of lactobacilli and total anaerobes while decreasing total aerobes and other groups of bacteria, such as the sulfite-reducing bacteria (38, 184, 284). Although resistance to inulin and other prebiotics is low, it appears that the gut microbiota exhibits resilience following the conclusion of treatment. Kruse et al. showed that 34 days after a 64-day treatment period, Bifidobacterium population sizes in 8 human subjects began to return to the levels observed before treatment (179). Studies of the mouse gut microbiota also suggest that prebiotic effects are short-lived. Within 1 week of completing a 6-month period of treatment with prebiotics, members of the gut microbiota of mice reverted to population levels similar to basal levels (284).
Fiber, which is a generalized term for indigestible plant components and includes inulin and other oligosaccharides, has been investigated for its anti-colon cancer properties for several years. Not surprisingly, high-fiber diets result in communities that are different from those formed in the presence of low-fiber diets. For example, a high-fiber (from freeze-dried fruit and vegetable extract) and olive oil diet was found to significantly reduce the incidence of intestinal adenomas in mice as well as to alter the microbial community, as determined using DGGE analysis (205). Fiber is also of interest for livestock health, since fiber degradation can contribute a significant portion of energy requirements. For example, in an effort to increase populations of bacteria that degrade fiber, Varel et al. fed pigs a diet high in alfalfa fiber. This resulted in a community that contained more xylanolytic and cellulolytic bacteria within 3 days of beginning the diet treatment than those in pigs fed a control diet (341). The mechanism for fiber-induced shifts in structure is likely linked to the fact that several eukaryotic, archaeal, and bacterial microorganisms are important in the degradation of fiber in the gut and interact trophically (108).
Grain types have also been studied for their influence on microbiota structure in livestock. Pigs fed a rice-based diet have different community structures than pigs fed a wheat- and barley-based control diet (189). Likewise, feeding pigs corn-, wheat-, or barley-based diets also resulted in different communities (134). However, in turkeys, the microbial communities were resistant to changes in grain type (corn versus wheat) (286). Turkey communities did, however, increase in diversity upon the addition of two enzyme preparations thought to promote intestinal health (286). Chicken communities were resistant to changes in diversity and richness when fed either corn or another grain, triticale. Interestingly, changes in diversity and richness were seen when the triticale was fed as whole grains compared to finely ground grain (285).
Microbial communities in invertebrate hosts also exhibit various resistances to changes in diet and dietary components. We revealed that the midguts of cabbage white butterfly larvae fed Brussels sprouts contained communities that were significantly different from those in larvae fed a sterile artificial diet (274). Additionally, sinigrin, a phytochemical found in Brussels sprouts and other cruciferous vegetables, alters the microbiota such that it is more similar in structure to the microbiota observed when larvae are fed Brussels sprouts as opposed to an unamended sterile artificial diet. Similarly, the gypsy moth community varied when larvae were reared on a sterile artificial diet, aspen, larch, oak, or willow (33). Likewise, the structure of the Proteobacteria population within the microbial community of the cricket hindgut was susceptible to changes in diet (283). These results indicate that these communities, or at least certain populations within the communities, have low resistance to phytochemicals.
Antibiotics.
Mass production of antibiotics has resulted in the revolutionary treatment of infectious diseases. However, since the wide-scale introduction of these drugs, it has been noted that treatment often comes with the side effect of diarrhea (i.e., antibiotic-associated diarrhea), and sometimes there are worse effects (e.g., Clostridium difficile infection and colitis) (21, 348). These side effects are indicative of disruptions of the intestinal microbiota. This susceptibility of the microbiota to antibiotics has been well documented (for a thorough review of the effects of antibiotics on various microbial communities associated with the human body, see reference 320). Studies of the effect of antibiotics on the human gut community have covered fluoroquinolones, lincosamides, beta-lactams, oxazolidinones, and nitroimidazoles, among others (154, 196). Typically, these studies show decreases in total bacterial counts and shifts in relative proportions of certain populations. This is not unexpected, since the activities of antibiotics against various bacteria vary due to a number of factors. In general, human-associated microbial communities exhibit low resistance to antibiotics. However, the resilience of these communities seems to be varied. Previously, we reported that a patient was given amoxicillin-clavulanic acid for 10 days, and by day 4 of the treatment, the patient's community exhibited decreases in Bacteroides fragilis, clostridial clusters IV and XIVa, and bifidobacterial populations, while there was an increase in Bacteroides distasonis and Enterobacteriaceae populations (371). Two weeks following the treatment, the community had started to return to its initial structure, with an increase in B. fragilis and clostridia and a decrease in B. distasonis and Enterobacteriaceae. Interestingly, the bifidobacterial population did not return during the experimental period. Similarly, de la Cochetiere et al. also demonstrated high resilience in gut communities (62). Following a 5-day treatment with amoxicillin, the microbial communities in the fecal samples of subjects were altered from pretreatment structures, as determined by DGGE. However, within 30 days posttreatment, the communities had begun to resemble pretreatment structures, with a similarity average of 88%. Interestingly, a 7-day treatment with clindamycin had vastly different results. Jernberg et al. recently reported that the clindamycin-induced shift in community structure persisted for up to 2 years posttreatment (154). Using pyrosequencing, Dethlefsen et al. investigated the reaction of the human microbiota to ciprofloxacin. They found that a 5-day ciprofloxacin treatment typically reduced richness, diversity, and evenness of the community and affected 30% of the OTUs observed. At the end of a 4-week recovery period, the communities had returned to structures that were typically similar to structures observed before treatment began. Interestingly, the same taxa within different individuals responded differently to the antibiotic treatment and exhibited temporal variation in population size before treatment (68). This study was particularly interesting because it also addressed the impact of temporal variability on the response to antibiotics.
Similar to this study is one that examined the ability of the murine gut microbial community to recover following antibiotic treatment, also by use of pyrosequencing. Antonopoulos et al. showed that mice reared in the same cage have remarkably similar communities, and therefore individual-to-individual variation would not be a confounding factor in the analyses of these communities, as it can be in human studies (15). Two antibiotic treatments were examined for their impact on structure: a cocktail of amoxicillin, bismuth, and metronidazole (AMB), formulated into food pellets; and cefoperazone, administered in drinking water. Treatment with the antibiotic cocktail resulted in a shift from a Firmicutes-dominated community to one dominated by the Proteobacteria. A 2-week recovery period resulted in a community that was similar to the pretreatment community. The cefoperazone treatment resulted in a community that could not be detected using bacterium-specific primers and PCR. However, the communities in mice that were allowed to recover in isolation contained fewer members and were dissimilar to the communities in control animals, once again highlighting the point that although there are trends in how antibiotics affect communities (e.g., decreased richness), the effect on specific members varies (15).
In addition to treating infections of individual pathogens, broad-spectrum antibiotics are used to alter entire communities, as in the case of inflammatory bowel disease (276). Additionally, women diagnosed with bacterial vaginosis are commonly given a prescription for the antibiotic metronidazole (16). Although it is not clear that the clinical symptoms commonly attributed to mild bacterial vaginosis are indicative of a disease that should be treated with antibiotics, physicians monitor the effectiveness of antibiotic treatment by a number of criteria, including the composition of the vaginal microbial community. In one study, treatment with a topical metronidazole gel resulted in vaginal communities that were typically dominated by Lactobacillus iners (92). Four of six subjects were considered “cured”; however, in three patients whose pretreatment communities contained Atopobium vaginae, responses to metronidazole were mixed. The treatment of two of these patients “failed,” one because of high vaginal pH and the other because of the presence of clue cells and discharge. Despite the patient not responding clinically to treatment, the community in the first patient seemed to have responded to metronidazole treatment and was dominated by L. iners (92). This result seems to indicate that community structure alone is not determinant of bacterial vaginosis status and supports the idea that bacterial vaginosis is not entirely dependent on an altered community structure but may be dependent on particular structures, including those that may be dominated by health-associated bacteria, such as those belonging to the Lactobacillus genus.
Like human communities, those associated with livestock are commonly exposed to antibiotics. This is because antibiotics are often administered as growth promoters for a variety of animals. The mechanisms by which antibiotics increase livestock productivity are not known, and some speculate that shifts in the microbial community may be key. Chickens and pigs, for example, are routinely given antibiotics as a dietary supplement. In one study, it was found that oxytetracycline had no effect on the richness, diversity, composition, or structure of the intestinal community in chickens, as determined by T-RFLP analysis (87). In contrast, other studies have shown that virginiamycin and a cocktail of bacitracin and salinomycin both alter community structure (74, 84). Yet another study found that the structure of the ileal community shifted with the age of chickens and that the communities in younger chickens exhibited a greater response to antibiotics than did the communities in older chickens (168). Like the communities in chickens, pig communities also shift in response to antibiotic supplements (53). A recent study showed that chlorotetracycline alters porcine community structure in the ileal mucosa and lumen. The antibiotic-fed pigs contained species that were not detected in control animals and exhibited shifts in the population sizes of two Lactobacillus species as well as a Turibacter species (272). In addition to the antibiotics mentioned above, several others, including penicillin, are used for growth promotion (287).
Future work.
It seems clear that the structure of microbial communities is susceptible to change when challenged with a particular stressor. As with the determination of the extent of diversity of a given host-associated microbiota, it is not enough to simply associate shifts in a community with a particular treatment. We must now be dedicated to finding out the underlying mechanisms that drive the shifts and determining what these shifts mean, not only to the host but also to the community itself. In some cases, these shifts may not matter at all. Once again, significance is likely to lie within the specific function of a microbiota that has a given community structure. We noted earlier that there is high interpersonal variation in the human microbiota, yet the communities seem to maintain function in healthy individuals. Fernandez and colleagues addressed the link between function and structure by using methanogenic reactors and found that despite compositional shifts, these communities were consistently able to produce methane and to maintain reactor pH and levels of effluent chemical oxygen demand (90, 91). Furthermore, they later showed that communities with similar functions but different levels of diversity responded differently to perturbation. Reactor communities with more diversity displayed more resistance to perturbation but less functional stability than reactor communities with less diversity. There are important questions provoked by these observations. Although communities with multiple structures may carry out similar functions, are they equivalent with regard to other considerations? For example, are the efficiencies with which they carry out these functions similar? Will host-associated communities with different structures yet similar functions be equivalent in stability following stressors or with regard to interaction with the host immune system?
Although, by design, the research being performed for the HMP involves human subjects, there remains an important role for studying microbial communities in nonhuman systems. The work performed on the human microbiota populations from different individuals clearly illustrates person-to-person variation that likely reflects differing life histories. While such variation is important in approaching questions such as finding a core microbiome, this variation makes it difficult to determine the basic principles that govern the stability of communities. In this case, nonhuman systems that allow reproducible ecological experiments to be conducted can be quite useful. The mouse model allows us to design experiments that ask specific questions about the ecology of microbial communities in several mammalian hosts, including humans. We recently showed that the murine gut microbiota, despite some differences from the human counterpart, can be used to demonstrate reproducible perturbations following an ecological stressor (15). Furthermore, recent work suggests that, at least at a functional level, the microbiota populations from multiple mammals are actually quite similar in many respects (192, 211). The tremendous body of literature and experimental reagents that are available for the murine model system suggest that it would be unwise to completely forgo further investigation of the indigenous mouse microbiota simply because it is not the human microbiota.
Susceptibility to Invasion
Susceptibility to invasion is yet another functional community property that has stimulated much theoretical discussion. Invasion is defined as an ecological process by which an exogenous population establishes itself and persists in a new location, i.e., within an already established community (301, 303). It can be divided into several stages: arrival, establishment, growth, and spread (301). The goals of invasion theory include identification of likely invaders, prediction of community susceptibility to invasion, and predictions of invader efficacy, spread, and impact on the native community (303). Although the invasion literature covers many topics, including dynamics involving invader seed banks (see reference 157 for an example of the importance of dormant bacterial populations within seed banks as invaders and drivers of microbial community diversity), invader speed (determined by population growth and dispersal [e.g., see reference 236]), propagule pressure (the population size of a potential invader exposed to a new location [e.g., see references 85 and 136]), and other invader-centric concepts, we once again take a mostly community-centric approach.
Much of what is known about the nature of invasions was determined at the macro scale and dates back to the mid-20th century (83). One of the major hypotheses concerning invasions presented by Elton, who is a founder of invasion theory, is that more-diverse communities are less susceptible to invasion than less-diverse communities (83). The work of several others also supports this hypothesis (e.g., see reference 203). Some of the best known studies are those that have evaluated the role of diversity in susceptibility to invasion of grassland communities (88, 170, 233). Diversity, however, is not alone in determining the ability of a natural (not experimentally designed) community to resist invasion and sometimes is not a factor (190). Using an aquatic microbial community containing bacteria, protists, and metazoans, McGrady-Steed et al. showed that the abundance of certain community members can be as important as species richness in community resistance to invasion (217). Other factors include disturbance and composition (55, 85). Because the process of invasion in experimental settings can be well controlled and resistance to invasion is a consistent function of a community, it can be used to monitor changes in a community. There are multiple examples of testing of concepts of invasion in both environmental and host-associated communities (Table 2). In the following sections, we discuss invasion in the context of colonization resistance and probiotics.
Colonization resistance.
Invasions in host-associated communities are typically associated with disease. In fact, one of the major functions of the human gastrointestinal and vaginal microbiota is colonization resistance, which is the ability of the community to resist invasion by exogenous and often pathogenic organisms (120, 345). One strategy for studying colonization resistance is to disrupt the community by use of antibiotics and then challenge it with an invader. Cotrimoxazole, for instance, was used to disrupt the human microbiota. This resulted in colonization of the colon by an experimentally introduced invader in all subjects, as well as a Candida albicans infection of the mouth in two of the volunteers (346). Because of the ethical issues faced when infecting humans with known pathogens, animal models are often used to investigate community resistance to pathogen invasion. van der Waaij and colleagues conducted one of the earliest studies of colonization resistance. In this study, they showed that a reduction of the mouse cecal microbiota with antibiotics resulted in a reduction of colonization resistance to three experimentally introduced invaders. They also revealed that as the total numbers of bacteria in the community returned, so did the colonization resistance (338). More recently, it was shown that high doses of ciprofloxacin and levofloxacin, as well as lower doses of gatifloxacin and moxifloxacin, all increased the colonization of mouse ceca with the pathogen Clostridium difficile compared to the saline-treated controls (1). Additionally, using a cynomolgus monkey model, Winberg et al. demonstrated that the colonization resistance of the vaginal community to a pathogenic strain of E. coli was reduced upon community perturbation with antibiotics (361, 362).
Insect gut communities also exhibit colonization resistance. A study of the diversity-stability debate using a gnotobiotic locust system revealed that a more-diverse community was more resistant to invasion by the pathogen Serratia marcescens than a less-diverse community (71). Likewise, S. marcescens was more successful at becoming established in communities of termites that had been fed antibiotics than in the communities of untreated termites (343). Additionally, one study showed that changes in community structure resulting from either a different diet or administration of antibiotics correlated, in some cases, with increased colonization by nonpathogenic invaders (274). Interestingly, this study also showed that antibiotics increased the diversity of the community, as did the diet treatment that increased susceptibility to colonization. This study is consistent with other studies, such as those discussed above, which suggest that diversity is not a lone determinant of a community's susceptibility to invasion (55, 60, 77, 219).
Probiotics and invasive pathogens.
A probiotic is defined as a preparation of living, defined organisms that alters the community associated with a host and confers a health benefit to the host (300). In addition, probiotic strains should be nonpathogenic, nontoxic, viable throughout production and storage of the preparation, and resistant to the host environment (i.e., vaginal pH, gastric acids, etc.) (102, 252). Probiotic organisms are diverse and strain specific and reflect the host environment on which they are expected to act. Lactobacillus strains have been investigated for their probiotic activity in gut and vaginal environments. Treatment with Lactobacillus rhamnosus GG, for instance, results in improved recovery from and reduces the incidence of diarrhea compared to placebo (249, 271, 336). Additionally, strains of L. rhamnosus and Lactobacillus reuteri improved treatment rates of bacterial vaginosis in women who were given metronidazole treatment compared to women who received metronidazole and a placebo (16). Furthermore, a strain of Lactobacillus fermentum reduced the length and severity of respiratory illness in patients and had an immunomodulatory effect (58). Likewise, three Streptococcus strains have shown potential in a probiotic mouthwash that reduces the presence of pathogens in plaque (373), and a strain of Bifidobacterium infantis ameliorated several symptoms associated with irritable bowel syndrome (359). Additionally, a commercially available combination of 8 probiotic species, VSL#3, has been shown to decrease development and reoccurrence of pouchitis as well as pediatric ulcerative colitis (109, 110, 223).
Mechanisms by which probiotics may provide benefits to hosts include modulation of the host immune system and other cellular functions. Additionally, they may prevent pathogen invasion via competition for binding sites or nutrients as well as by production of toxic compounds (see reference 337 for a review that focuses on probiotic mechanisms in inflammatory bowel diseases [which are likely applicable to the role of probiotics elsewhere]) (96, 337). In this way, it is interesting to think of probiotics as altering community susceptibility to invasion without necessarily becoming permanent residents of the community. Whether these beneficial bacteria themselves can be considered invaders depends on the definition of invader used. Here we assert, as have others, that an invader must not only establish but also persist (301). The beneficial effects of probiotics are often short-lived, remaining only a short time after cessation of probiotic administration, which suggests that the bacteria are transient members of the community (264). However, it seems possible that probiotics can, in some cases, successfully establish in a community (temporarily) because of the ability of certain strains to bind to host cells. Several strains adhere to cells and mucus temporarily and, as stated before, competitively exclude pathogens. In vitro studies have shown that several probiotic strains are able to adhere to Caco-2 epithelial colorectal cells and to displace or outcompete pathogens, such as Salmonella enterica serovar Typhimurium and E. coli, or otherwise inhibit their adherence (39, 151). Certain probiotic strains also prevent pathogen binding to intestinal mucus and displace pathogens following adherence (52, 344). Additionally, L. rhamnosus GG, one of these pathogen-displacing probiotic strains, was recently found to have pili that contain human-mucus-binding proteins, which may explain its ability to persist in the gut longer than other L. rhamnosus strains and its ability to outcompete pathogens.
Probiotics are considered a more desirable treatment option than antibiotics because they do not come with the negative effects that can sometimes accompany antibiotics (i.e., disruption of nontarget microbial communities). As stated previously, antibiotic use in livestock husbandry is under fire for its potential to promote antibiotic resistance in human pathogens. Commercially available alternatives to antibiotic treatment for infections in poultry are competitive exclusion (CE) products, which are used to prevent colonization by pathogens. These products are typically comprised of cecal bacteria from domestic fowl (299). The presence of Salmonella and Campylobacter jejuni, for example, was significantly decreased in chickens treated with a CE product and other probiotics (5, 360). In cattle, a mix of various E. coli strains as well as a Proteus mirabilis strain was found to reduce the length of time that E. coli O157:H7 was detected in the rumen and the length of the fecal shedding period (376). Similarly, swine infection with Salmonella enterica serovar Typhimurium was ameliorated with a probiotic mixture containing several Lactobacillus strains and Pediococcus pentosaceus (42). In addition to providing examples of how microbial ecology studies can have a direct impact on human health (e.g., reductions of the human pathogens Campylobacter, Salmonella, and E. coli O157:H7), these animal studies also provided clear, reproducible evidence in support of the administration of probiotics, which was missing for a long time from human-only studies.
Future work.
Recent work revealed that community structure likely plays a role in susceptibility to invasion. One of the key questions that remains to be answered is whether there are certain community structures that are more susceptible to invasion by certain kinds of bacteria. For example, is it more likely that an Enterobacter sp. will successfully invade a community that does not contain another Enterobacter sp. than a community with one or several other Enterobacter spp. under the same conditions? Another way of approaching this question would be to determine whether certain species protect against invasion. Can increasing the population size of a particular community member, for example, Lactobacillus sp. in the mouse cecum, reduce susceptibility to invasion by a particular pathogen? Will that answer hold true for other pathogens? There are several ways of addressing these questions, but none seems likely to provide a conclusive answer. One of these would be to construct communities with the desired structure in previously germfree mice and then to infect them. This sounds simple but only highlights the limits of our knowledge about host-associated communities. The experimental design proposed is predicated on the assumption that the structure would not be altered severely during the assembly period in the host or that we would be able to compensate for these changes. One study that evaluated the ability of the Charles River altered Schaedler flora to establish in a mouse colony found that members of the defined community were detected throughout 7 generations and 4 years; however, the structure of the community was not explored (315). Whether assembly patterns are consistent and reproducible has not yet been determined. An additional challenge is that immune systems in germfree mice are underdeveloped, as are certain other systems. How would this impact our ability to interpret results in the context of a conventional host?
Another potential model would be to alter communities with a targeted approach. For example, one could use vancomycin or clindamycin to reduce the population size of Firmicutes bacteria in a community. The problem with this approach is that these antibiotics will likely have other effects on the community, and perhaps the pathogen. Can we approach these community questions like we approach molecular biology questions, as proposed by Handelsman et al. (124)? Is it possible to “knock out” specific bacteria with bacteriophages or bacteriocins, which tend to be much more specific than antibiotics? Could we “overexpress” a particular member by “preinvading” a community with the desired species and then infecting it with a pathogen? Once again, another question is raised: can we increase the population size of a specific member simply by administering more of the member in pure culture?
A more complicated view of colonization resistance arises when we take into consideration the complex interactions between the microbiota and the host immune system. It is clear that the immune system has an undeniable impact on the microbiota, serving in effect to shape the community structure of the resident microbes. This is expected given the key role of the immune system in recognizing and dealing with microbes (238, 277, 282). However, it has now been recognized that the microbiota can in turn regulate host immune responses. For example, modulation of the resident murine intestinal microbiota by the administration of an antibiotic cocktail containing metronidazole, neomycin, and vancomycin downregulates host expression of the bactericidal lectin RegIIIγ, which specifically targets Gram-positive organisms (43). In another study, it was shown that vancomycin-resistant enterococci (VRE) were able to survive at higher concentrations in mice treated with the same antibiotics (30). This loss of colonization resistance against VRE was not simply due to loss of competition with other members of the microbiota, since administration of lipopolysaccharide (LPS) along with the antibiotic cocktail prevented VRE colonization. The administration of LPS, a product of Gram-negative bacteria, was shown to prevent the downregulation of RegIIIγ. This suggests a mechanism by which the native gut microbiota can influence colonization resistance through modulation of host immune responses. Although we are just beginning to understand the intricate interactions between the resident microbiota and the immune system, it is likely to be important that we consider the ecological implications of these interactions. Because microbe-microbe and host-microbe interactions can serve as drivers of community diversity and function, more complete models of host-associated communities will need to account for both types of interactions.
Next Steps
Although we have begun to explore structure, biogeography, stability, and invasion in the context of the host-associated microbiota, there are a number of other areas with great potential for increasing our understanding of these communities. Coevolutionary theory seeks to understand and predict the reciprocal evolution of interacting organisms. There is some evidence for coevolution between hosts and their microbiota (192). The geographic mosaic theory of coevolution (324) may be particularly relevant to the study of host-associated microbial ecology because it seeks to understand the role in coevolution of dispersal among a mosaic of habitat “patches.” Individual hosts could be thought of as such patches, with transmission among hosts as a source of dispersal. This theory has been applied to microbes in the laboratory (e.g., see reference 97) but not to the host-associated microbiota. Another area of potential application is diversity-ecosystem function theory (200). This area of theory seeks to understand how the diversity and structure of communities result in collective properties such as nutrient cycling, decomposition, and biomass production. Human health could be thought of as a collective property of the human-associated microbiota. This area of theory could be used to guide experiments to rigorously test the linkage between the microbiota and health.
WAR NO MORE: HUMAN MEDICINE IN THE AGE OF ECOLOGY
To this point, the discussion has focused on how principles and approaches from general and microbial ecology can be applied to studies of the indigenous human microbiota. In this final section, we propose how consideration of the ecology of the human microbiota can help to inform future approaches to the prevention and management of human disease.
Understanding the role of microorganisms in human disease represented a revolution in medicine. The application of Koch's postulates gave rise to clinical microbiology and the practice of infectious diseases. However, this also led to the paradigm that views infectious disease therapy based on a war metaphor. Microbial pathogens are viewed as the enemy that needs to be eliminated in order to restore health. Increasingly powerful weapons in the form of antibiotics with increased activity and spectrum were felt to be necessary in order to win this war. However, as has been learned through warfare throughout human history, collateral damage to innocent bystanders increases the cost of success on the battlefield. In terms of the human microbiota, the rise of antibiotic resistance, the appearance of opportunistic organisms such as Clostridium difficile and VRE, and an increase in allergic diseases (via the hygiene hypothesis) and autoimmune diseases, such as inflammatory bowel disease, are all thought to be a reflection of such collateral damage.
As we gain an increased understanding of how complex, host-associated microbial communities assemble and maintain structure and function, it is likely that a new paradigm for the prevention and treatment of microbe-associated diseases will arise. In certain cases, the war metaphor will be replaced by a paradigm where management is the key concept. With this new paradigm, the human body can be considered akin to a national park. The management of the ecology of the human microbiota becomes the focus of both prevention and therapeutics. For example, a focus on maintaining beneficial bacteria during targeted antimicrobial therapy of clinical infections could prevent infection with C. difficile or VRE. Conversely, once the host-associated microbiota has been altered to a deleterious community structure (dysbiosis), a therapeutic approach of restoration ecology could be initiated. Approaches using probiotics and prebiotics, rationally designed based on knowledge of the ecology of the community, could fit under this rubric of restoring community balance.
The Human Genome Project was conducted with the hope that it would usher in the era of personalized medicine. Examples of the dawn of this new era are now appearing. The deciphering of cancer genomes is meant to customize therapy to a specific tumor. Analysis of the host genome is meant to customize therapy based on an individual's potential sensitivity and metabolism of therapeutic agents. It is possible that along with these approaches, customized therapies based on an understanding of an individual's microbiome will be developed in the future. Such an integrated approach would lead to the full realization of (meta)genomic medicine.
Acknowledgments
We acknowledge Nabeetha Nagalingam and Christine Bassis for critical reviews and helpful discussions and Judith Opp for helpful discussions.
This work was supported by NIH grants R01DK070875 (C.J.R. and V.B.Y.) and R01DK070875-03S1 (C.J.R.).
REFERENCES
- 1.Adams, D. A., M. M. Riggs, and C. J. Donskey. 2007. Effect of fluoroquinolone treatment on growth of and toxin production by epidemic and nonepidemic Clostridium difficile strains in the cecal contents of mice. Antimicrob. Agents Chemother. 51:2674-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, H. K., L. A. Moe, J. Rodbumrer, A. Gaarder, and J. Handelsman. 2008. Functional metagenomics reveals diverse [beta]-lactamases in a remote Alaskan soil. ISME J. 3:243-251. [DOI] [PubMed] [Google Scholar]
- 3.Allison, S. D., and J. B. H. Martiny. 2008. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105:11512-11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allsworth, J. E., and J. F. Peipert. 2007. Prevalence of bacterial vaginosis: 2001-2004 National Health and Nutrition Examination Survey data. Obstet. Gynecol. 109:114-120. [DOI] [PubMed] [Google Scholar]
- 5.Al-Zenki, S. F., A. Y. Al-Nasser, A. E. Al-Saffar, F. K. Abdullah, M. E. Al-Bahouh, A. S. Al-Haddad, H. Alomirah, and M. Mashaly. 2009. Effects of using a chicken-origin competitive exclusion culture and probiotic cultures on reducing Salmonella in broilers. J. Appl. Poult. Res. 18:23-29. [Google Scholar]
- 6.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaral, F. A., D. Sachs, V. V. Costa, C. T. Fagundes, D. Cisalpino, T. M. Cunha, S. H. Ferreira, F. Q. Cunha, T. A. Silva, J. R. Nicoli, L. Q. Vieira, D. G. Souza, and M. M. Teixeira. 2008. Commensal microbiota is fundamental for the development of inflammatory pain. Proc. Natl. Acad. Sci. U. S. A. 105:2193-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aminov, R. I., and R. I. Mackie. 2007. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271:147-161. [DOI] [PubMed] [Google Scholar]
- 10.Amit-Romach, E., D. Sklan, and Z. Uni. 2004. Microflora ecology of the chicken intestine using 16S ribosomal DNA primers. Poult. Sci. 83:1093-1098. [DOI] [PubMed] [Google Scholar]
- 11.Amsel, R., P. A. Totten, C. A. Spiegel, K. C. S. Chen, D. Eschenbach, and K. K. Holmes. 1983. Nonspecific vaginitis: diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 74:14-22. [DOI] [PubMed] [Google Scholar]
- 12.Anadon, A. 2006. The EU ban of antibiotics as feed additives (2006): alternatives and consumer safety. J. Vet. Pharmacol. Ther. 29:41-44.16420301 [Google Scholar]
- 13.Anderson, T. M., M. Ä. Lachance, and W. T. Starmer. 2004. The relationship of phylogeny to community structure: the cactus yeast community. Am. Nat. 164:709-721. [DOI] [PubMed] [Google Scholar]
- 14.Andersson, A., M. Lindberg, H. Jakobsson, F. Backhed, P. Nyren, and L. Engstrand. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonopoulos, D. A., S. M. Huse, H. G. Morrison, T. M. Schmidt, M. L. Sogin, and V. B. Young. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anukam, K., E. Osazuwa, I. Ahonkhai, M. Ngwu, G. Osemene, A. W. Bruce, and G. Reid. 2006. Augmentation of antimicrobial metronidazole therapy of bacterial vaginosis with oral probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14: randomized, double-blind, placebo controlled trial. Microbes Infect. 8:1450-1454. [DOI] [PubMed] [Google Scholar]
- 17.Apajalahti, J. H. A., H. Kettunen, A. Kettunen, W. E. Holben, P. H. Nurminen, N. Rautonen, and M. Mutanen. 2002. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl. Environ. Microbiol. 68:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backhed, F., H. Ding, T. Wang, L. V. Hooper, G. Y. Koh, A. Nagy, C. F. Semenkovich, and J. I. Gordon. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 101:15718-15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai, Y., X. Zhou, and D. L. Smith. 2003. Enhanced soybean plant growth resulting from coinoculation of Bacillus strains with Bradyrhizobium japonicum. Crop Sci. 43:1774-1781. [Google Scholar]
- 20.Barnes, E. M. 1972. The avian intestinal flora with particular reference to the possible ecological significance of the cecal anaerobic bacteria. Am. J. Clin. Nutr. 25:1475-1479. [DOI] [PubMed] [Google Scholar]
- 21.Bartlett, J. 1992. Antibiotic-associated diarrhea. Clin. Infect. Dis. 15:573-579. [DOI] [PubMed] [Google Scholar]
- 22.Bauer, H., R. E. Horowitz, H. Popper, and S. M. Levenson. 1963. Response of lymphatic tissue to microbial flora—studies on germfree mice. Am. J. Pathol. 42:471-483. [PMC free article] [PubMed] [Google Scholar]
- 23.Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 24.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U. S. A. 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohannan, B. J. M., and J. Hughes. 2003. New approaches to analyzing microbial biodiversity data. Curr. Opin. Microbiol. 6:282-287. [DOI] [PubMed] [Google Scholar]
- 26.Borcard, D., P. Legendre, and P. Drapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73:1045-1055. [Google Scholar]
- 27.Boris, S., and C. Barbes. 2000. Role played by lactobacilli in controlling the population of vaginal pathogens. Microbes Infect. 2:543-546. [DOI] [PubMed] [Google Scholar]
- 28.Borlee, B. R., G. D. Geske, C. J. Robinson, H. E. Blackwell, and J. Handelsman. 2008. Quorum-sensing signals in the microbial community of the cabbage white butterfly larval midgut. ISME J. 2:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouhnik, Y., L. Raskine, G. Simoneau, E. Vicaut, C. Neut, B. Flourie, F. Brouns, and F. R. Bornet. 2004. The capacity of nondigestible carbohydrates to stimulate fecal bifidobacteria in healthy humans: a double-blind, randomized, placebo-controlled, parallel-group, dose-response relation study. Am. J. Clin. Nutr. 80:1658-1664. [DOI] [PubMed] [Google Scholar]
- 30.Brandl, K., G. Plitas, C. N. Mihu, C. Ubeda, T. Jia, M. Fleisher, B. Schnabl, R. P. DeMatteo, and E. G. Pamer. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breznak, J. A., and A. Brune. 1994. Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 39:453-487. [Google Scholar]
- 32.Breznak, J. A., and J. M. Switzer. 1986. Acetate synthesis from H2 plus CO2 by termite gut microbes. Appl. Environ. Microbiol. 52:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broderick, N. A., K. F. Raffa, R. M. Goodman, and J. Handelsman. 2004. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 70:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brotman, R. M., and J. Ravel. 2008. Editorial commentary: ready or not: the molecular diagnosis of bacterial vaginosis. Clin. Infect. Dis. 47:44-46. [DOI] [PubMed] [Google Scholar]
- 35.Brulc, J. M., D. A. Antonopoulos, M. E. Berg Miller, M. K. Wilson, A. C. Yannarell, E. A. Dinsdale, R. E. Edwards, E. D. Frank, J. B. Emerson, P. Wacklin, P. M. Coutinho, B. Henrissat, K. E. Nelson, and B. A. White. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U. S. A. 106:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryant, J. A., C. Lamanna, H. Morlon, A. J. Kerkhoff, B. J. Enquist, and J. L. Green. 2008. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. U. S. A. 105(Suppl. 1):11505-11511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabello, F. C. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8:1137-1144. [DOI] [PubMed] [Google Scholar]
- 38.Campbell, J. M., G. C. Fahey, Jr., and B. W. Wolf. 1997. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J. Nutr. 127:130-136. [DOI] [PubMed] [Google Scholar]
- 39.Candela, M., F. Perna, P. Carnevali, B. Vitali, R. Ciati, P. Gionchetti, F. Rizzello, M. Campieri, and P. Brigidi. 2008. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 125:286-292. [DOI] [PubMed] [Google Scholar]
- 40.Caporaso, J. G., J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Pena, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld, and R. Knight. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrino-Kyker, S. R., and A. K. Swanson. 2008. Temporal and spatial patterns of eukaryotic and bacterial communities found in vernal pools. Appl. Environ. Microbiol. 74:2554-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casey, P. G., G. E. Gardiner, G. Casey, B. Bradshaw, P. G. Lawlor, P. B. Lynch, F. C. Leonard, C. Stanton, R. P. Ross, G. F. Fitzgerald, and C. Hill. 2007. A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 73:1858-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cash, H. L., C. V. Whitham, C. L. Behrendt, and L. V. Hooper. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavender-Bares, J., K. H. Kozak, P. V. A. Fine, and S. W. Kembel. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12:693-715. [DOI] [PubMed] [Google Scholar]
- 45.Chao, A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 46.Chao, A., and S.-M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 47.Chao, A., M. C. Ma, and M. C. K. Yang. 1993. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 80:193-201. [Google Scholar]
- 48.Chase, J. M., and M. A. Leibold. 2003. Ecological niches. University of Chicago Press, Chicago, IL.
- 49.Chenier, M. R., D. Beaumier, N. Fortin, R. Roy, B. T. Driscoll, J. R. Lawrence, and C. W. Greer. 2006. Influence of nutrient inputs, hexadecane, and temporal variations on denitrification and community composition of river biofilms. Appl. Environ. Microbiol. 72:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole, J., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. Farris, A. Kulam-Syed-Mohideen, D. McGarrell, T. Marsh, and G. Garrity. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collado, M. C., E. Donat, C. Ribes-Koninckx, M. Calabuig, and Y. Sanz. 2009. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J. Clin. Pathol. 62:264-269. [DOI] [PubMed] [Google Scholar]
- 52.Collado, M. C., E. Isolauri, and S. Salminen. 2008. Specific probiotic strains and their combinations counteract adhesion of Enterobacter sakazakii to intestinal mucus. FEMS Microbiol. Lett. 285:58-64. [DOI] [PubMed] [Google Scholar]
- 53.Collier, C. T., M. R. Smiricky-Tjardes, D. M. Albin, J. E. Wubben, V. M. Gabert, B. Deplancke, D. Bane, D. B. Anderson, and H. R. Gaskins. 2003. Molecular ecological analysis of porcine ileal microbiota responses to antimicrobial growth promoters. J. Anim. Sci. 81:3035-3045. [DOI] [PubMed] [Google Scholar]
- 54.Reference deleted.
- 55.Cook, K., J. Garland, A. Layton, H. Dionisi, L. Levine, and G. Sayler. 2006. Effect of microbial species richness on community stability and community function in a model plant-based wastewater processing system. Microb. Ecol. 52:725-737. [DOI] [PubMed] [Google Scholar]
- 56.Costello, E. K., C. L. Lauber, M. Hamady, N. Fierer, J. I. Gordon, and R. Knight. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Covert, J., and M. Moran. 2001. Molecular characterization of estuarine bacterial communities that use high-and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127-139. [Google Scholar]
- 58.Cox, A. J., D. B. Pyne, P. U. Saunders, and P. A. Fricker. 2010. Oral administration of the probiotic Lactobacillus fermentum VRI-003 and mucosal immunity in endurance athletes. Br. J. Sports Med. 44:222-226. [DOI] [PubMed] [Google Scholar]
- 59.Crasta, O. R., O. Folkerts, Z. Fei, S. P. Mane, C. Evans, S. Martino-Catt, B. Bricker, G. Yu, L. Du, and B. W. Sobral. 2008. Genome sequence of Brucella abortus vaccine strain S19 compared to virulent strains yields candidate virulence genes. PLoS One 3:e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crawley, M. J., S. L. Brown, M. S. Heard, and G. R. Edwards. 1999. Invasion-resistance in experimental grassland communities: species richness or species identity? Ecol. Lett. 2:140-148. [Google Scholar]
- 61.Curtis, T. P., I. M. Head, M. Lunn, S. Woodcock, P. D. Schloss, and W. T. Sloan. 2006. What is the extent of prokaryotic diversity? Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2023-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61a.de Graaf, A. A., A. Maathuis, P. de Waard, N. E. P. Deutz, C. Dijkema, W. M. de Vos, and K. Venema. 2010. Profiling human gut bacterial metabolism and its kinetics using [U-13C] glucose and NMR. NMR Biomed. 23:2-12. [DOI] [PubMed] [Google Scholar]
- 62.De La Cochetiere, M. F., T. Durand, P. Lepage, A. Bourreille, J. P. Galmiche, and J. Dore. 2005. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J. Clin. Microbiol. 43:5588-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeLong, E., G. Wickham, and N. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 64.Demoling, L. A., E. Baath, G. Greve, M. Wouterse, and H. Schmitt. 2009. Effects of sulfamethoxazole on soil microbial communities after adding substrate. Soil Biol. Biochem. 41:840-848. [Google Scholar]
- 65.De Palma, G., I. Nadal, M. C. Collado, and Y. Sanz. 2006. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br. J. Nutr. 102:1154-1160. [DOI] [PubMed] [Google Scholar]
- 66.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dethlefsen, L., P. B. Eckburg, E. M. Bik, and D. A. Relman. 2006. Assembly of the human intestinal microbiota. Trends Ecol. Evol. 21:517-523. [DOI] [PubMed] [Google Scholar]
- 68.Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dethlefsen, L., M. McFall-Ngai, and D. A. Relman. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diaz, R. L., L. Hoang, J. Wang, J. L. Vela, S. Jenkins, R. Aranda, and M. G. Martin. 2004. Maternal adaptive immunity influences the intestinal microflora of suckling mice. J. Nutr. 134:2359-2364. [DOI] [PubMed] [Google Scholar]
- 71.Dillon, R. J., C. T. Vennard, A. Buckling, and A. K. Charnley. 2005. Diversity of locust gut bacteria protects against pathogen invasion. Ecol. Lett. 8:1291-1298. [Google Scholar]
- 72.Dinsdale, E., O. Pantos, S. Smriga, R. Edwards, F. Angly, L. Wegley, M. Hatay, D. Hall, E. Brown, and M. Haynes. 2008. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One 3:e1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Docherty, K., K. Young, P. Maurice, and S. Bridgham. 2006. Dissolved organic matter concentration and quality influences upon structure and function of freshwater microbial communities. Microb. Ecol. 52:378-388. [DOI] [PubMed] [Google Scholar]
- 74.Dumonceaux, T., J. Hill, S. Hemmingsen, and A. VanKessel. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dumonceaux, T. J., J. Schellenberg, V. Goleski, J. E. Hill, W. Jaoko, J. Kimani, D. Money, T. B. Ball, F. A. Plummer, and A. Severini. 2009. Multiplex detection of bacteria associated with normal microbiota and with bacterial vaginosis in vaginal swabs by use of oligonucleotide-coupled fluorescent microspheres. J. Clin. Microbiol. 47:4067-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dumont, M. G., and J. C. Murrell. 2005. Stable isotope probing—linking microbial identity to function. Nat. Rev. Microbiol. 3:499-504. [DOI] [PubMed] [Google Scholar]
- 77.Dunstan, P., and C. Johnson. 2004. Invasion rates increase with species richness in a marine epibenthic community by two mechanisms. Oecologia 138:285-292. [DOI] [PubMed] [Google Scholar]
- 78.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edwards, R., B. Rodriguez-Brito, L. Wegley, M. Haynes, M. Breitbart, D. Peterson, M. Saar, S. Alexander, E. Alexander, and F. Rohwer. 2006. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Egert, M., A. A. de Graaf, H. Smidt, W. M. de Vos, and K. Venema. 2006. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 14:86-91. [DOI] [PubMed] [Google Scholar]
- 81.Egert, M., A. A. de Graaf, A. Maathuis, P. de Waard, C. M. Plugge, H. Smidt, N. E. P. Deutz, C. Dijkema, W. M. de Vos, and K. Venema. 2007. Identification of glucose-fermenting bacteria present in an in vitro model of the human intestine by RNA-stable isotope probing. FEMS Microbiol. Ecol. 60:126-135. [DOI] [PubMed] [Google Scholar]
- 82.Egert, M., B. Wagner, T. Lemke, A. Brune, and M. W. Friedrich. 2003. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6659-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Elton, C. S. 1958. The ecology of invasions by animals and plants. Methuen & Co., Ltd., London, United Kingdom.
- 84.Engberg, R., M. Hedemann, T. Leser, and B. Jensen. 2000. Effect of zinc bacitracin and salinomycin on intestinal microflora and performance of broilers. Poult. Sci. 79:1311-1319. [DOI] [PubMed] [Google Scholar]
- 85.Eschtruth, A. K., and J. J. Battles. 2009. Assessing the relative importance of disturbance, herbivory, diversity, and propagule pressure in exotic plant invasion. Ecol. Monogr. 79:265-280. [Google Scholar]
- 86.Eutick, M. L., P. Veivers, R. W. O'Brien, and M. Slaytor. 1978. Dependence of the higher termite, Nasutitermes exitiosus and the lower termite, Coptotermes lacteus on their gut flora. J. Insect Physiol. 24:363-368. [Google Scholar]
- 87.Fairchild, A. S., J. L. Smith, U. Idris, J. Lu, S. Sanchez, L. B. Purvis, C. Hofacre, and M. D. Lee. 2005. Effects of orally administered tetracycline on the intestinal community structure of chickens and on tet determinant carriage by commensal bacteria and Campylobacter jejuni. Appl. Environ. Microbiol. 71:5865-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fargione, J. E., and D. Tilman. 2005. Diversity decreases invasion via both sampling and complementarity effects. Ecol. Lett. 8:604-611. [Google Scholar]
- 89.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fernandez, A. S., S. A. Hashsham, S. L. Dollhopf, L. Raskin, O. Glagoleva, F. B. Dazzo, R. F. Hickey, C. S. Criddle, and J. M. Tiedje. 2000. Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl. Environ. Microbiol. 66:4058-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fernandez, A. S., S. Huang, S. Seston, J. Xing, R. F. Hickey, C. S. Criddle, and J. M. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferris, M. J., J. Norori, M. Zozaya-Hinchliffe, and D. H. Martin. 2007. Cultivation-independent analysis of changes in bacterial vaginosis flora following metronidazole treatment. J. Clin. Microbiol. 45:1016-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Filoche, S., L. Wong, and C. H. Sissons. 2010. Oral biofilms: emerging concepts in microbial ecology. J. Dent. Res. 89:8-18. [DOI] [PubMed] [Google Scholar]
- 94.Finegold, S., V. Sutter, P. Sugihara, H. Elder, S. Lehmann, and R. Phillips. 1977. Fecal microbial flora in Seventh Day Adventist populations and control subjects. Am. J. Clin. Nutr. 30:1781-1792. [DOI] [PubMed] [Google Scholar]
- 95.Fisher, M. M., J. L. Klug, G. Lauster, M. Newton, and E. W. Triplett. 2000. Effects of resources and trophic interactions on freshwater bacterioplankton diversity. Microb. Ecol. 40:125-138. [DOI] [PubMed] [Google Scholar]
- 96.Fooks, L. J., and G. R. Gibson. 2002. Probiotics as modulators of the gut flora. Br. J. Nutr. 88:s39-s49. [DOI] [PubMed] [Google Scholar]
- 97.Forde, S. E., J. N. Thompson, and B. J. M. Bohannan. 2004. Adaptation varies through space and time in a coevolving host-parasitoid interaction. Nature 431:841-844. [DOI] [PubMed] [Google Scholar]
- 98.Fox, G. E., E. Stackebrandt, R. B. Hespell, J. Gibson, J. Maniloff, T. A. Dyer, R. S. Wolfe, W. E. Balch, R. S. Tanner, L. J. Magrum, L. B. Zablen, R. Blakemore, R. Gupta, L. Bonen, B. J. Lewis, D. A. Stahl, K. R. Luehrsen, K. N. Chen, and C. R. Woese. 1980. The phylogeny of prokaryotes. Science 209:457-463. [DOI] [PubMed] [Google Scholar]
- 99.Frank, D. N., L. M. Feazel, M. T. Bessesen, C. S. Price, E. N. Janoff, and N. R. Pace. 2010. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 5:e10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frank, D. N., A. L. St. Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frankow-Lindberg, B. E., C. Brophy, R. P. Collins, and J. Connolly. 2009. Biodiversity effects on yield and unsown species invasion in a temperate forage ecosystem. Ann. Bot. 103:913-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 103.Garner, C. D., D. A. Antonopoulos, B. Wagner, G. E. Duhamel, I. Keresztes, D. A. Ross, V. B. Young, and C. Altier. 2009. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar Typhimurium murine model of infection. Infect. Immun. 77:2691-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Geib, S., M. Jimenez-Gasco, J. Carlson, M. Tien, R. Jabbour, and K. Hoover. 2009. Microbial community profiling to investigate transmission of bacteria between life stages of the wood-boring beetle, Anoplophora glabripennis. Microb. Ecol. 58:199-211. [DOI] [PubMed] [Google Scholar]
- 105.Geib, S. M., M. D. M. Jimenez-Gasco, J. E. Carlson, M. Tien, and K. Hoover. 2009. Effect of host tree species on cellulase activity and bacterial community composition in the gut of larval Asian longhorned beetle. Environ. Entomol. 38:686-699. [DOI] [PubMed] [Google Scholar]
- 106.Gibson, C., and M. Hunter. 2009. Inherited fungal and bacterial endosymbionts of a parasitic wasp and its cockroach host. Microb. Ecol. 57:542-549. [DOI] [PubMed] [Google Scholar]
- 107.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 108.Gill, S. R., M. Pop, R. T. DeBoy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gionchetti, P., F. Rizzello, U. Helwig, A. Venturi, K. M. Lammers, P. Brigidi, B. Vitali, G. Poggioli, M. Miglioli, and M. Campieri. 2003. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology 124:1202-1209. [DOI] [PubMed] [Google Scholar]
- 110.Gionchetti, P., F. Rizzello, A. Venturi, P. Brigidi, D. Matteuzzi, G. Bazzocchi, G. Poggioli, M. Miglioli, and M. Campieri. 2000. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology 119:305-309. [DOI] [PubMed] [Google Scholar]
- 111.Giovannoni, S., E. DeLong, G. Olsen, and N. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 113.Girvan, M. S., J. Bullimore, A. S. Ball, J. N. Pretty, and A. M. Osborn. 2004. Responses of active bacterial and fungal communities in soils under winter wheat to different fertilizer and pesticide regimens. Appl. Environ. Microbiol. 70:2692-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Girvan, M. S., C. D. Campbell, K. Killham, J. I. Prosser, and L. A. Glover. 2005. Bacterial diversity promotes community stability and functional resilience after perturbation. Environ. Microbiol. 7:301-313. [DOI] [PubMed] [Google Scholar]
- 115.Reference deleted.
- 116.Green, J., and B. J. Bohannan. 2006. Spatial scaling of microbial biodiversity. Trends Ecol. Evol. 21:501-507. [DOI] [PubMed] [Google Scholar]
- 117.Green, J. L., and J. B. Plotkin. 2007. A statistical theory for sampling species abundances. Ecol. Lett. 10:1037-1045. [DOI] [PubMed] [Google Scholar]
- 118.Grice, E. A., H. H. Kong, S. Conlan, C. B. Deming, J. Davis, A. C. Young, NISC Comparative Sequencing Program, G. G. Bouffard, R. W. Blakesley, P. R. Murray, E. D. Green, M. L. Turner, and J. A. Segre. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guan, C., J. Ju, B. R. Borlee, L. L. Williamson, B. Shen, K. F. Raffa, and J. Handelsman. 2007. Signal mimics derived from a metagenomic analysis of the gypsy moth gut microbiota. Appl. Environ. Microbiol. 73:3669-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 121.Haanpera, M., P. Huovinen, and J. Jalava. 2005. Detection and quantification of macrolide resistance mutations at positions 2058 and 2059 of the 23S rRNA gene by pyrosequencing. Antimicrob. Agents Chemother. 49:457-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Halverson, L. J., and J. Handelsman. 1991. Enhancement of soybean nodulation by Bacillus cereus UW85 in the field and in a growth chamber. Appl. Environ. Microbiol. 57:2767-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Handelsman, J., S. Raffel, E. H. Mester, L. Wunderlich, and C. R. Grau. 1990. Biological control of damping-off of alfalfa seedlings with Bacillus cereus UW85. Appl. Environ. Microbiol. 56:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Handelsman, J., C. J. Robinson, and K. Raffa. 2005. Microbial communities in lepidopteran guts: from models to metagenomics, p. 143-168. In M. J. McFall-Ngai, B. Henderson, and E. G. Ruby (ed.), Influence of cooperative bacteria on animal host biology. Cambridge University Press, New York, NY.
- 125.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 126.Harrison, G. W. 1979. Stability under environmental stress: resistance, resilience, persistence, and variability. Am. Nat. 113:659-669. [Google Scholar]
- 127.Haukka, K., E. Kolmonen, R. Hyder, J. Hietala, K. Vakkilainen, T. Kairesalo, H. Haario, and K. Sivonen. 2006. Effect of nutrient loading on bacterioplankton community composition in lake mesocosms. Microb. Ecol. 51:137-146. [DOI] [PubMed] [Google Scholar]
- 128.Hayashi, H., R. Takahashi, T. Nishi, M. Sakamoto, and Y. Benno. 2005. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J. Med. Microbiol. 54:1093-1101. [DOI] [PubMed] [Google Scholar]
- 129.He, J.-Z., Y. Zheng, C.-R. Chen, Y.-Q. He, and L.-M. Zhang. 2008. Microbial composition and diversity of an upland red soil under long-term fertilization treatments as revealed by culture-dependent and culture-independent approaches. J. Soils Sediments 8:349-358. [Google Scholar]
- 130.Helmus, M. R., K. Savage, M. W. Diebel, J. T. Maxted, and A. R. Ives. 2007. Separating the determinants of phylogenetic community structure. Ecol. Lett. 10:917-925. [DOI] [PubMed] [Google Scholar]
- 131.Herrmann, S., S. Kleinsteuber, A. Chatzinotas, S. Kuppardt, T. Lueders, H.-H. Richnow, and C. Vogt. 2010. Functional characterization of an anaerobic benzene-degrading enrichment culture by DNA stable isotope probing. Environ. Microbiol. 12:401-411. [DOI] [PubMed] [Google Scholar]
- 132.Hildebrandt, M. A., C. Hoffmann, S. A. Sherrill-Mix, S. A. Keilbaugh, M. Hamady, Y.-Y. Chen, R. Knight, R. S. Ahima, F. Bushman, and G. D. Wu. 2009. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137:1716-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hill, J. E., S. H. Goh, D. M. Money, M. Doyle, A. Li, W. L. Crosby, M. Links, A. Leung, D. Chan, and S. M. Hemmingsen. 2005. Characterization of vaginal microflora of healthy, nonpregnant women by chaperonin-60 sequence-based methods. Am. J. Obstet. Gynecol. 193:682-692. [DOI] [PubMed] [Google Scholar]
- 134.Hill, J. E., S. M. Hemmingsen, B. G. Goldade, T. J. Dumonceaux, J. Klassen, R. T. Zijlstra, S. H. Goh, and A. G. Van Kessel. 2005. Comparison of ileum microflora of pigs fed corn-, wheat-, or barley-based diets by chaperonin-60 sequencing and quantitative PCR. Appl. Environ. Microbiol. 71:867-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hojberg, O., N. Canibe, H. D. Poulsen, M. S. Hedemann, and B. B. Jensen. 2005. Influence of dietary zinc oxide and copper sulfate on the gastrointestinal ecosystem in newly weaned piglets. Appl. Environ. Microbiol. 71:2267-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Holle, B. V., and D. Simberloff. 2005. Ecological resistance to biological invasion overwhelmed by propagule pressure. Ecology 86:3212-3218. [Google Scholar]
- 137.Hongoh, Y., P. Deevong, S. Hattori, T. Inoue, S. Noda, N. Noparatnaraporn, T. Kudo, and M. Ohkuma. 2006. Phylogenetic diversity, localization, and cell morphologies of members of the candidate phylum TG3 and a subphylum in the phylum Fibrobacteres, recently discovered bacterial groups dominant in termite guts. Appl. Environ. Microbiol. 72:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hongoh, Y., P. Deevong, T. Inoue, S. Moriya, S. Trakulnaleamsai, M. Ohkuma, C. Vongkaluang, N. Noparatnaraporn, and T. Kudo. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 71:6590-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hooper, D. U., F. S. Chapin, J. J. Ewel, A. Hector, P. Inchausti, S. Lavorel, J. H. Lawton, D. M. Lodge, M. Loreau, S. Naeem, B. Schmid, H. Setala, A. J. Symstad, J. Vandermeer, and D. A. Wardle. 2005. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75:3-35. [Google Scholar]
- 140.Hooper, L. V. 2004. Bacterial contributions to mammalian gut development. Trends Microbiol. 12:129-134. [DOI] [PubMed] [Google Scholar]
- 141.Hopkins, M. J., and G. T. Macfarlane. 2002. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 51:448-454. [DOI] [PubMed] [Google Scholar]
- 142.Hopkins, M. J., R. Sharp, and G. T. Macfarlane. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Horner-Devine, M. C., and B. J. M. Bohannan. 2006. Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87:100-108. [DOI] [PubMed] [Google Scholar]
- 144.Huang, W. E., A. Ferguson, A. C. Singer, K. Lawson, I. P. Thompson, R. M. Kalin, M. J. Larkin, M. J. Bailey, and A. S. Whiteley. 2009. Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ hybridization. Appl. Environ. Microbiol. 75:234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hubbell, S. P. 2001. The unified neutral theory of biodiversity and biogeography. Princeton University Press, Princeton, NJ.
- 146.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huse, S. M., L. Dethlefsen, J. A. Huber, D. M. Welch, D. A. Relman, and M. L. Sogin. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hwang, C., W. Wu, T. J. Gentry, J. Carley, G. A. Corbin, S. L. Carroll, D. B. Watson, P. M. Jardine, J. Zhou, C. S. Criddle, and M. W. Fields. 2008. Bacterial community succession during in situ uranium bioremediation: spatial similarities along controlled flow paths. ISME J. 3:47-64. [DOI] [PubMed] [Google Scholar]
- 149.Hyman, R. W., M. Fukushima, L. Diamond, J. Kumm, L. C. Giudice, and R. W. Davis. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. U. S. A. 102:7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ikeda-Ohtsubo, W., and A. Brune. 2009. Cospeciation of termite gut flagellates and their bacterial endosymbionts: Trichonympha species and “Candidatus Endomicrobium trichonymphae.” Mol. Ecol. 18:332-342. [DOI] [PubMed] [Google Scholar]
- 151.Ingrassia, I., A. Leplingard, and A. Darfeuille-Michaud. 2005. Lactobacillus casei DN-114 001 inhibits the ability of adherent-invasive Escherichia coli isolated from Crohn's disease patients to adhere to and to invade intestinal epithelial cells. Appl. Environ. Microbiol. 71:2880-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ismail, A. S., C. L. Behrendt, and L. V. Hooper. 2009. Reciprocal interactions between commensal bacteria and γδ intraepithelial lymphocytes during mucosal injury. J. Immunol. 182:3047-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ives, A. R., and S. R. Carpenter. 2007. Stability and diversity of ecosystems. Science 317:58-62. [DOI] [PubMed] [Google Scholar]
- 154.Jernberg, C., S. Lofmark, C. Edlund, and J. K. Jansson. 2007. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1:56-66. [DOI] [PubMed] [Google Scholar]
- 155.Jiang, H.-Q., N. A. Bos, and J. J. Cebra. 2001. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect. Immun. 69:3611-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang, L., and Z. Pu. 2009. Different effects of species diversity on temporal stability in single-trophic and multitrophic communities. Am. Nat. 174:651-659. [DOI] [PubMed] [Google Scholar]
- 157.Jones, S. E., and J. T. Lennon. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 107:5881-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jones, S. E., R. J. Newton, and K. D. McMahon. 2009. Evidence for structuring of bacterial community composition by organic carbon source in temperate lakes. Environ. Microbiol. 11:2463-2472. [DOI] [PubMed] [Google Scholar]
- 159.Kamilova, F., S. Validov, T. Azarova, I. Mulders, and B. Lugtenberg. 2005. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol. 7:1809-1817. [DOI] [PubMed] [Google Scholar]
- 160.Kan, J., M. T. Suzuki, K. Wang, S. E. Evans, and F. Chen. 2007. High temporal but low spatial heterogeneity of bacterioplankton in the Chesapeake Bay. Appl. Environ. Microbiol. 73:6776-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Keijser, B. J. F., E. Zaura, S. M. Huse, J. M. B. M. van der Vossen, F. H. J. Schuren, R. C. Montijn, J. M. ten Cate, and W. Crielaard. 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87:1016-1020. [DOI] [PubMed] [Google Scholar]
- 162.Kent, A. D., A. C. Yannarell, J. A. Rusak, E. W. Triplett, and K. D. McMahon. 2007. Synchrony in aquatic microbial community dynamics. ISME J. 1:38-47. [DOI] [PubMed] [Google Scholar]
- 163.Kibe, R., M. Sakamoto, H. Hayashi, H. Yokota, and Y. Benno. 2004. Maturation of the murine cecal microbiota as revealed by terminal restriction fragment length polymorphism and 16S rRNA gene clone libraries. FEMS Microbiol. Lett. 235:139-146. [DOI] [PubMed] [Google Scholar]
- 164.Kikuchi, Y., and J. Graf. 2007. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl. Environ. Microbiol. 73:1984-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim, T. K., S. M. Thomas, M. Ho, S. Sharma, C. I. Reich, J. A. Frank, K. M. Yeater, D. R. Biggs, N. Nakamura, R. Stumpf, S. R. Leigh, R. I. Tapping, S. R. Blanke, J. M. Slauch, H. R. Gaskins, J. S. Weisbaum, G. J. Olsen, L. L. Hoyer, and B. A. Wilson. 2009. Heterogeneity of vaginal microbial communities within individuals. J. Clin. Microbiol. 47:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kleessen, B., B. Sykura, H. Zunft, and M. Blaut. 1997. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am. J. Clin. Nutr. 65:1397-1402. [DOI] [PubMed] [Google Scholar]
- 167.Kloos, W. E., and M. S. Musselwhite. 1975. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Environ. Microbiol. 30:381-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Knarreborg, A., M. A. Simon, R. M. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Knief, C., A. Ramette, L. Frances, C. Alonso-Blanco, and J. A. Vorholt. 2010. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J. 4:719-728. [DOI] [PubMed] [Google Scholar]
- 170.Knops, J. M. H., D. Tilman, N. M. Haddad, S. Naeem, C. E. Mitchell, J. Haarstad, M. E. Ritchie, K. M. Howe, P. B. Reich, E. Siemann, and J. Groth. 1999. Effects of plant species richness on invasion dynamics, disease outbreaks, insect abundances and diversity. Ecol. Lett. 2:286-293. [DOI] [PubMed] [Google Scholar]
- 171.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kolida, S., and G. R. Gibson. 2007. Prebiotic capacity of inulin-type fructans. J. Nutr. 137:2503S-2506S. [DOI] [PubMed] [Google Scholar]
- 173.Kong, W. D., Y. G. Zhu, B. J. Fu, P. Marschner, and J. Z. He. 2006. The veterinary antibiotic oxytetracycline and Cu influence functional diversity of the soil microbial community. Environ. Pollut. 143:129-137. [DOI] [PubMed] [Google Scholar]
- 174.Konstantinidis, K. T., J. Braff, D. M. Karl, and E. F. DeLong. 2009. Comparative metagenomic analysis of a microbial community from 4000 m at Station ALOHA in the North Pacific Subtropical Gyre. Appl. Environ. Microbiol. 75:5345-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Koropatnick, T. A., J. T. Engle, M. A. Apicella, E. V. Stabb, W. E. Goldman, and M. J. McFall-Ngai. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186-1188. [DOI] [PubMed] [Google Scholar]
- 176.Koumans, E. H., M. R. Sternberg, G. McQuillan, C. Bruce, J. S. Kendrick, M. Y. Sutton, and L. Markowitz. 2006. Prevalence of bacterial vaginosis in the United States, 2001-2002, abstr. 211. Abstr. 2006 Natl. STD Prev. Conf. Centers for Disease Control and Prevention, Atlanta, GA.
- 177.Kovatcheva-Datchary, P., M. Egert, A. Maathuis, M. Rajilic-Stojanovic, A. A. de Graaf, H. Smidt, W. M. de Vos, and K. Venema. 2009. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ. Microbiol. 11:914-926. [DOI] [PubMed] [Google Scholar]
- 178.Kruis, W., P. Fric, J. Pokrotnieks, M. Lukas, B. Fixa, M. Kascak, M. A. Kamm, J. Weismueller, C. Beglinger, M. Stolte, C. Wolff, and J. Schulze. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kruse, H.-P., B. Kleessen, and M. Blaut. 1999. Effects of inulin on faecal bifidobacteria in human subjects. Br. J. Nutr. 82:375-382. [DOI] [PubMed] [Google Scholar]
- 180.Kuehl, C. J., H. D. Wood, T. L. Marsh, T. M. Schmidt, and V. B. Young. 2005. Colonization of the cecal mucosa by Helicobacter hepaticus impacts the diversity of the indigenous microbiota. Infect. Immun. 73:6952-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kuramitsu, H. K., X. He, R. Lux, M. H. Anderson, and W. Shi. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Labandeira, C. C., and J. J. Sepkoski, Jr. 1993. Insect diversity in the fossil record. Science 261:310-315. [DOI] [PubMed] [Google Scholar]
- 183.Lami, R., J. Ghiglione, Y. Desdevises, N. West, and P. Lebaron. 2009. Annual patterns of presence and activity of marine bacteria monitored by 16S rDNA-16S rRNA fingerprints in the coastal NW Mediterranean Sea. Aquat. Microb. Ecol. 54:199-210. [Google Scholar]
- 184.Langlands, S. J., M. J. Hopkins, N. Coleman, and J. H. Cummings. 2004. Prebiotic carbohydrates modify the mucosa associated microflora of the human large bowel. Gut 53:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Leadbetter, J. R., and J. A. Breznak. 1996. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl. Environ. Microbiol. 62:3620-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Leadbetter, J. R., T. M. Schmidt, J. R. Graber, and J. A. Breznak. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686-689. [DOI] [PubMed] [Google Scholar]
- 187.Legendre, P., D. Borcard, and P. R. Peres-Neto. 2005. Analyzing beta diversity: partitioning the spatial variation of community composition data. Ecol. Monogr. 75:435-450. [Google Scholar]
- 188.Lehman, C. L., and D. Tilman. 2000. Biodiversity, stability, and productivity in competitive communities. Am. Nat. 156:534-552. [DOI] [PubMed] [Google Scholar]
- 189.Leser, T. D., R. H. Lindecrona, T. K. Jensen, B. B. Jensen, and K. Moller. 2000. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl. Environ. Microbiol. 66:3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Levine, J. M., and C. M. D'Antonio. 1999. Elton revisited: a review of evidence linking diversity and invasibility. Oikos 87:15-26. [Google Scholar]
- 191.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ley, R. E., M. Hamady, C. Lozupone, P. J. Turnbaugh, R. R. Ramey, J. S. Bircher, M. L. Schlegel, T. A. Tucker, M. D. Schrenzel, R. Knight, and J. I. Gordon. 2008. Evolution of mammals and their gut microbes. Science 320:1647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Reference deleted.
- 194.Ley, R. E., J. K. Harris, J. Wilcox, J. R. Spear, S. R. Miller, B. M. Bebout, J. A. Maresca, D. A. Bryant, M. L. Sogin, and N. R. Pace. 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-848. [DOI] [PubMed] [Google Scholar]
- 196.Lidbeck, A., C. Edlund, J. Å. Gustafsson, L. Kager, and C. E. Nora. 1988. Impact of Lactobacillus acidophilus on the normal intestinal microflora after administration of two antimicrobial agents. Infection 16:329-336. [DOI] [PubMed] [Google Scholar]
- 197.Lindsay, J. O., K. Whelan, A. J. Stagg, P. Gobin, H. O. Al-Hassi, N. Rayment, M. A. Kamm, S. C. Knight, and A. Forbes. 2006. Clinical, microbiological, and immunological effects of fructo-oligosaccharide in patients with Crohn's disease. Gut 55:348-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Little, A. E. F., C. J. Robinson, S. B. Peterson, K. F. Raffa, and J. Handelsman. 2008. Rules of engagement: interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 62:375-401. [DOI] [PubMed] [Google Scholar]
- 199.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Loreau, M., S. Naeem, and P. Inchausti. 2002. Biodiversity and ecosystem functioning: synthesis and perspectives. Oxford University Press, Oxford, United Kingdom.
- 201.Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Luckey, T. D. 1972. Introduction to intestinal microecology. Am. J. Clin. Nutr. 25:1292-1294. [DOI] [PubMed] [Google Scholar]
- 203.MacArthur, R. 1955. Fluctuations of animal populations and a measure of community stability. Ecology 36:533-536. [Google Scholar]
- 204.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035S-1045S. [DOI] [PubMed] [Google Scholar]
- 205.Mai, V., L. H. Colbert, S. N. Perkins, A. Schatzkin, and S. D. Hursting. 2007. Intestinal microbiota: a potential diet-responsive prevention target in ApcMin mice. Mol. Carcinog. 46:42-48. [DOI] [PubMed] [Google Scholar]
- 206.Mai, V., H. A. Katki, H. Harmsen, D. Gallaher, A. Schatzkin, D. J. Baer, and B. Clevidence. 2004. Effects of a controlled diet and black tea drinking on the fecal microflora composition and the fecal bile acid profile of human volunteers in a double-blinded randomized feeding study. J. Nutr. 134:473-478. [DOI] [PubMed] [Google Scholar]
- 207.Manichanh, C., L. Rigottier-Gois, E. Bonnaud, K. Gloux, E. Pelletier, L. Frangeul, R. Nalin, C. Jarrin, P. Chardon, P. Marteau, J. Roca, and J. Dore. 2006. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mardis, E. R. 2008. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9:387-402. [DOI] [PubMed] [Google Scholar]
- 209.Marsh, P. D. 2000. Role of the oral microflora in health. Microb. Ecol. Health Dis. 12:130-137. [Google Scholar]
- 210.Marsh, P. D., and D. J. Bradshaw. 1995. Dental plaque as a biofilm. J. Ind. Microbiol. Biotechnol. 15:169-175. [DOI] [PubMed] [Google Scholar]
- 211.Martin, F. P., M. E. Dumas, Y. Wang, C. Legido-Quigley, I. K. Yap, H. Tang, S. Zirah, G. M. Murphy, O. Cloarec, J. C. Lindon, N. Sprenger, L. B. Fay, S. Kochhar, P. van Bladeren, E. Holmes, and J. K. Nicholson. 2007. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Martinez, I., G. Wallace, C. Zhang, R. Legge, A. K. Benson, T. P. Carr, E. N. Moriyama, and J. Walter. 2009. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl. Environ. Microbiol. 75:4175-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Martiny, J., B. Bohannan, J. Brown, R. Colwell, J. Fuhrman, J. Green, M. Horner-Devine, M. Kane, J. Krumins, C. Kuske, P. Morin, S. Naeem, L. Ovreas, A.-L. Reysenbach, V. Smith, and J. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 214.Maukonen, J., J. Matto, M.-L. Suihko, and M. Saarela. 2008. Intra-individual diversity and similarity of salivary and faecal microbiota. J. Med. Microbiol. 57:1560-1568. [DOI] [PubMed] [Google Scholar]
- 215.Mayhew, P. J. 2007. Why are there so many insect species? Perspectives from fossils and phylogenies. Biol. Rev. 82:425-454. [DOI] [PubMed] [Google Scholar]
- 216.McCann, K. S. 2000. The diversity-stability debate. Nature 405:228-233. [DOI] [PubMed] [Google Scholar]
- 217.McGrady-Steed, J., P. Harris, and P. Morin. 1997. Biodiversity regulates ecosystem predictability. Nature 390:162-165. [Google Scholar]
- 218.McNaughton, S. 1977. Diversity and stability of ecological communities: a comment on the role of empiricism in ecology. Am. Nat. 111:515-525. [Google Scholar]
- 219.Meiners, S. J., M. L. Cadenasso, and S. T. A. Pickett. 2004. Beyond biodiversity: individualistic controls of invasion in a self-assembled community. Ecol. Lett. 7:121-126. [Google Scholar]
- 220.Membrez, M., F. Blancher, M. Jaquet, R. Bibiloni, P. D. Cani, R. G. Burcelin, I. Corthesy, K. Mace, and C. J. Chou. 2008. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 22:2416-2426. [DOI] [PubMed] [Google Scholar]
- 221.Menne, E., and N. Guggenbuhl. 2000. Fn-type chicory inulin hydrolysate has a prebiotic effect in humans. J. Nutr. 130:1197-1199. [DOI] [PubMed] [Google Scholar]
- 222.Michelland, R. J., V. Monteils, A. Zened, S. Combes, L. Cauquil, T. Gidenne, J. Hamelin, and L. Fortun-Lamothe. 2009. Spatial and temporal variations of the bacterial community in the bovine digestive tract. J. Appl. Microbiol. 107:1642-1650. [DOI] [PubMed] [Google Scholar]
- 223.Miele, E., F. Pascarella, E. Giannetti, L. Quaglietta, R. N. Baldassano, and A. Staiano. 2009. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am. J. Gastroenterol. 104:437-443. [DOI] [PubMed] [Google Scholar]
- 224.Miller, S. R., A. L. Strong, K. L. Jones, and M. C. Ungerer. 2009. Bar-coded pyrosequencing reveals shared bacterial community properties along the temperature gradients of two alkaline hot springs in Yellowstone National Park. Appl. Environ. Microbiol. 75:4565-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mitsuoka, T. 1992. Intestinal flora and aging. Nutr. Rev. 50:438-446. [DOI] [PubMed] [Google Scholar]
- 226.Miyatake, T., B. J. MacGregor, and H. T. S. Boschker. 2009. Linking microbial community function to phylogeny of sulfate-reducing deltaproteobacteria in marine sediments by combining stable isotope probing with magnetic-bead capture hybridization of 16S rRNA. Appl. Environ. Microbiol. 75:4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Moder, K.-A., F. Layer, W. Konig, and B. Konig. 2007. Rapid screening of clarithromycin resistance in Helicobacter pylori by pyrosequencing. J. Med. Microbiol. 56:1370-1376. [DOI] [PubMed] [Google Scholar]
- 228.Moore, M., A. Dhingra, P. Soltis, R. Shaw, W. Farmerie, K. Folta, and D. Soltis. 2006. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Moran, N. A., and A. Telang. 1998. Bacteriocyte-associated symbionts of insects. Bioscience 48:295-304. [Google Scholar]
- 230.Moran, N. A., P. Tran, and N. M. Gerardo. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 71:8802-8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Mountzouris, K. C., P. Tsirtsikos, E. Kalamara, S. Nitsch, G. Schatzmayr, and K. Fegeros. 2007. Evaluation of the efficacy of a probiotic containing Lactobacillus, Bifidobacterium, Enterococcus, and Pediococcus strains in promoting broiler performance and modulating cecal microflora composition and metabolic activities. Poult. Sci. 86:309-317. [DOI] [PubMed] [Google Scholar]
- 232.Muyzer, G. 1999. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 233.Naeem, S., J. M. H. Knops, D. Tilman, K. M. Howe, T. Kennedy, and S. Gale. 2000. Plant diversity increases resistance to invasion in the absence of covarying extrinsic factors. Oikos 91:97-108. [Google Scholar]
- 234.Nasidze, I., J. Li, D. Quinque, K. Tang, and M. Stoneking. 2009. Global diversity in the human salivary microbiome. Genome Res. 19:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nava, G. M., M. S. Attene-Ramos, H. R. Gaskins, and J. D. Richards. 2009. Molecular analysis of microbial community structure in the chicken ileum following organic acid supplementation. Vet. Microbiol. 137:345-353. [DOI] [PubMed] [Google Scholar]
- 236.Neubert, M. G., and H. Caswell. 2000. Demography and dispersal: calculation and sensitivity analysis of invasion speed for structured populations. Ecology 81:1613-1628. [Google Scholar]
- 237.Newton, R. J., S. E. Jones, M. R. Helmus, and K. D. McMahon. 2007. Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl. Environ. Microbiol. 73:7169-7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nieuwenhuis, E. E. S., T. Matsumoto, D. Lindenbergh, R. Willemsen, A. Kaser, Y. Simons-Oosterhuis, S. Brugman, K. Yamaguchi, H. Ishikawa, Y. Aiba, Y. Koga, J. N. Samsom, K. Oshima, M. Kikuchi, J. C. Escher, M. Hattori, A. B. Onderdonk, and R. S. Blumberg. 2009. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J. Clin. Invest. 119:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Noda, S., Y. Hongoh, T. Sato, and M. Ohkuma. 2009. Complex coevolutionary history of symbiotic Bacteroidales bacteria of various protists in the gut of termites. BMC Evol. Biol. 9:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Nugent, R. P., M. A. Krohn, and S. L. Hillier. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 29:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Oakley, B. B., T. L. Fiedler, J. M. Marrazzo, and D. N. Fredricks. 2008. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74:4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ohkuma, M. 2008. Symbioses of flagellates and prokaryotes in the gut of lower termites. Trends Microbiol. 16:345-352. [DOI] [PubMed] [Google Scholar]
- 243.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 244.O'May, G. A., N. Reynolds, A. R. Smith, A. Kennedy, and G. T. Macfarlane. 2005. Effect of pH and antibiotics on microbial overgrowth in the stomachs and duodena of patients undergoing percutaneous endoscopic gastrostomy feeding. J. Clin. Microbiol. 43:3059-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.O'Toole, P. W., and M. J. Claesson. 2010. Gut microbiota: changes throughout the lifespan from infancy to elderly. Int. Dairy J. 20:281-291. [Google Scholar]
- 246.Owen-Hughes, T., and M. Engeholm. 2007. Pyrosequencing positions nucleosomes precisely. Genome Biol. 8:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Palmer, C., E. M. Bik, D. B. DiGiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Pant, A. R., S. M. Graham, S. J. Allen, S. Harikul, A. Sabchareon, L. Cuevas, M. T. Med., and C. A. Hart. 1996. Lactobacillus GG and acute diarrhoea in young children in the tropics. J. Trop. Pediatr. 42:162-165. [DOI] [PubMed] [Google Scholar]
- 250.Park, H., S. Shim, S. Kim, J. Park, S. Park, H. Kim, B. Kang, and C. Kim. 2005. Molecular analysis of colonized bacteria in a human newborn infant gut. J. Microbiol. 43:345-353. [PubMed] [Google Scholar]
- 251.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Patterson, J., and K. Burkholder. 2003. Application of prebiotics and probiotics in poultry production. Poult. Sci. 82:627-631. [DOI] [PubMed] [Google Scholar]
- 253.Pei, Z., E. J. Bini, L. Yang, M. Zhou, F. Francois, and M. J. Blaser. 2004. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. U. S. A. 101:4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Penders, J., C. Thijs, C. Vink, F. F. Stelma, B. Snijders, I. Kummeling, P. A. van den Brandt, and E. E. Stobberingh. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511-521. [DOI] [PubMed] [Google Scholar]
- 255.Percival, R. S., S. J. Challacombe, and P. D. Marsh. 1991. Age-related microbiological changes in the salivary and plaque microflora of healthy adults. J. Med. Microbiol. 35:5-11. [DOI] [PubMed] [Google Scholar]
- 256.Peres-Neto, P. R., P. Legendre, S. Dray, and D. Borcard. 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87:2614-2625. [DOI] [PubMed] [Google Scholar]
- 257.Periasamy, S., N. I. Chalmers, L. Du-Thumm, and P. E. Kolenbrander. 2009. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Appl. Environ. Microbiol. 75:3250-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Periasamy, S., and P. E. Kolenbrander. 2009. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J. Bacteriol. 191:6804-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Peterson, S. B., F. Warnecke, J. Madejska, K. D. McMahon, and P. Hugenholtz. 2008. Environmental distribution and population biology of Candidatus Accumulibacter, a primary agent of biological phosphorus removal. Environ. Microbiol. 10:2692-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Petti, S., and G. Tarsitani. 1998. Intra-individual variations of salivary microbial levels in young adults. Eur. J. Oral Sci. 106:616-622. [DOI] [PubMed] [Google Scholar]
- 261.Pimm, S. L. 1984. The complexity and stability of ecosystems. Nature 307:321-326. [Google Scholar]
- 262.Prosser, J. I., B. J. M. Bohannan, T. P. Curtis, R. J. Ellis, M. K. Firestone, R. P. Freckleton, J. L. Green, L. E. Green, K. Killham, and J. J. Lennon. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:384. [DOI] [PubMed] [Google Scholar]
- 263.Pruesse, E., C. Quast, K. Knittel, B. Fuchs, W. Ludwig, J. Peplies, and F. Glockner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Quigley, E. 2008. What is the evidence for the use of probiotics in functional disorders? Curr. Gastroenterol. Rep. 10:379-384. [DOI] [PubMed] [Google Scholar]
- 265.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 266.Rajilic-Stojanovic, M., H. G. H. J. Heilig, D. Molenaar, K. Kajander, A. Surakka, H. Smidt, and W. M. De Vos. 2009. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ. Microbiol. 11:1736-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 268.Ramette, A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Rasiah, I. A., L. Wong, S. A. Anderson, and C. H. Sissons. 2005. Variation in bacterial DGGE patterns from human saliva: over time, between individuals and in corresponding dental plaque microcosms. Arch. Oral Biol. 50:779-787. [DOI] [PubMed] [Google Scholar]
- 270.Ravel, J., P. Gajer, Z. Abdo, G. M. Schneider, S. S. K. Koenig, S. L. McCulle, S. Karlebach, R. Gorle, J. Russell, C. O. Tacket, R. M. Brotman, C. C. Davis, K. Ault, L. Peralta, and L. J. Forney. 3 June 2010, posting date. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed]
- 271.Raza, S., S. M. Graham, S. J. Allen, S. Sultana, L. C. M. Trop, and C. A. Hart. 1995. Lactobacillus GG promotes recovery from acute nonbloody diarrhea in Pakistan. Pediatr. Infect. Dis. J. 14:107-111. [DOI] [PubMed] [Google Scholar]
- 272.Rettedal, E., S. Vilain, S. Lindblom, K. Lehnert, C. Scofield, S. George, S. Clay, R. S. Kaushik, A. J. M. Rosa, D. Francis, and V. S. Brozel. 2009. Alteration of the ileal microbiota of weanling piglets by the growth-promoting antibiotic chlortetracycline. Appl. Environ. Microbiol. 75:5489-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Riesenfeld, C. S., P. D. Schloss, and J. Handelsman. 2004. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38:525-552. [DOI] [PubMed] [Google Scholar]
- 274.Robinson, C. J., P. D. Schloss, Y. Ramos, K. F. Raffa, and J. Handelsman. 2010. Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb. Ecol. 59:199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Roesch, L. F. W., R. R. Fulthorpe, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. H. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Rutgeerts, P., M. Hiele, K. Geboes, M. Peeters, F. Penninckx, R. Aerts, and R. Kerremans. 1995. Controlled trial of metronidazole treatment for prevention of Crohn's recurrence after ileal resection. Gastroenterology 108:1617-1621. [DOI] [PubMed] [Google Scholar]
- 277.Ryu, J.-H., S.-H. Kim, H.-Y. Lee, J. Y. Bai, Y.-D. Nam, J.-W. Bae, D. G. Lee, S. C. Shin, E.-M. Ha, and W.-J. Lee. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319:777-782. [DOI] [PubMed] [Google Scholar]
- 278.Saison, C., V. Degrange, R. Oliver, P. Millard, C. Commeaux, D. Montange, and X. Le Roux. 2006. Alteration and resilience of the soil microbial community following compost amendment: effects of compost level and compost-borne microbial community. Environ. Microbiol. 8:247-257. [DOI] [PubMed] [Google Scholar]
- 279.Sakamoto, M., M. Umeda, I. Ishikawa, and Y. Benno. 2000. Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 44:643. [DOI] [PubMed] [Google Scholar]
- 280.Sale, P. F. 1977. Maintenance of high diversity in coral reef fish communities. Am. Nat. 111:337. [Google Scholar]
- 281.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 282.Salzman, N. H., K. Hung, D. Haribhai, H. Chu, J. Karlsson-Sjoberg, E. Amir, P. Teggatz, M. Barman, M. Hayward, D. Eastwood, M. Stoel, Y. Zhou, E. Sodergren, G. M. Weinstock, C. L. Bevins, C. B. Williams, and N. A. Bos. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11:76-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Santo Domingo, J. W., M. G. Kaufman, M. J. Klug, and J. M. Tiedje. 1998. Characterization of the cricket hindgut microbiota with fluorescently labeled rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:752-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Santos, A., M. San Mauro, and D. M. Dìaz. 2006. Prebiotics and their long-term influence on the microbial populations of the mouse bowel. Food Microbiol. 23:498-503. [DOI] [PubMed] [Google Scholar]
- 285.Santos, F. B. O., B. W. Sheldon, A. A. Santos, Jr., P. R. Ferket, M. D. Lee, A. Petroso, and D. Smith. 2007. Determination of ileum microbial diversity of broilers fed triticale- or corn-based diets and colonized by Salmonella. J. Appl. Poult. Res. 16:563-573. [Google Scholar]
- 286.Santos, A. A., Jr., P. R. Ferket, F. B. O. Santos, N. Nakamura, and C. Collier. 2008. Change in the ileal bacterial population of turkeys fed different diets and after infection with Salmonella as determined with denaturing gradient gel electrophoresis of amplified 16S ribosomal DNA. Poult. Sci. 87:1415-1427. [DOI] [PubMed] [Google Scholar]
- 287.Sarmah, A. K., M. T. Meyer, and A. B. A. Boxall. 2006. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65:725-759. [DOI] [PubMed] [Google Scholar]
- 288.Sartor, R. B. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134:577-594. [DOI] [PubMed] [Google Scholar]
- 289.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 290.Scanlan, P. D., F. Shanahan, C. O'Mahony, and J. R. Marchesi. 2006. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J. Clin. Microbiol. 44:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Schaedler, R. W., R. Dubos, and R. Costello. 1965. The development of the bacterial flora in the gastrointestinal tract of mice. J. Exp. Med. 122:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Schlapfer, F., and B. Schmid. 1999. Ecosystem effects of biodiversity: a classification of hypotheses and exploration of empirical results. Ecol. Appl. 9:893-912. [Google Scholar]
- 293.Schloss, P. D. 2010. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comp. Biol. 6:e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Schloss, P. D. 2009. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One 4:e8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Schloss, P. D., S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn, and C. F. Weber. 2009. Introducing mothur: open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Schmidt, T. M., E. F. DeLong, and N. R. Pace. 1991. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Axial dynamics, stability, and interspecies similarity of bacterial community structure in the highly compartmentalized gut of soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6018-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6007-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Schneitz, C. 2005. Competitive exclusion in poultry—30 years of research. Food Control 16:657-667. [Google Scholar]
- 300.Schrezenmeir, J., and M. de Vrese. 2001. Probiotics, prebiotics, and synbiotics—approaching a definition. Am. J. Clin. Nutr. 73:361S-364S. [DOI] [PubMed] [Google Scholar]
- 301.Shea, K., and P. Chesson. 2002. Community ecology theory as a framework for biological invasions. Trends Ecol. Evol. 17:170-176. [Google Scholar]
- 302.Shendure, J., and H. Ji. 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26:1135-1145. [DOI] [PubMed] [Google Scholar]
- 303.Shigesada, N., and K. Kawasaki. 1997. Biological invasions: theory and practice. Oxford University Press, Oxford, United Kingdom.
- 304.Shreiner, A., G. B. Huffnagle, and M. C. Noverr. 2008. The “microflora hypothesis” of allergic disease, p. 113-134. In GI microbiota and regulation of the immune system. Springer, New York, NY. [DOI] [PubMed]
- 305.Silo-Suh, L. A., B. J. Lethbridge, S. J. Raffel, H. He, J. Clardy, and J. Handelsman. 1994. Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Appl. Environ. Microbiol. 60:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Sloan, W. T., M. Lunn, S. Woodcock, I. M. Head, S. Nee, and T. P. Curtis. 2006. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 8:732-740. [DOI] [PubMed] [Google Scholar]
- 307.Sogin, M., H. Morrison, J. Huber, D. Welch, S. Huse, P. Neal, J. Arrieta, and G. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Somerville, C. 2007. Biofuels. Curr. Biol. 17:R115-R119. [DOI] [PubMed] [Google Scholar]
- 309.Srinivasan, S., and D. Fredricks. 2008. The human vaginal bacterial biota and bacterial vaginosis. Interdiscip. Perspect. Infect. Dis. 2008:1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Stahl, D. A., D. J. Lane, G. J. Olsen, and N. R. Pace. 1984. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science 224:409-411. [DOI] [PubMed] [Google Scholar]
- 311.Stahl, D. A., D. J. Lane, G. J. Olsen, and N. R. Pace. 1985. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl. Environ. Microbiol. 49:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Staley, J., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 313.Stappenbeck, T. S., L. V. Hooper, and J. I. Gordon. 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U. S. A. 99:15451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Stark, P. L., and A. Lee. 1982. The microbial ecology of the large bowel of breastfed and formula-fed infants during the first year of life. J. Med. Microbiol. 15:189-203. [DOI] [PubMed] [Google Scholar]
- 315.Stehr, M., M. C. Greweling, S. Tischer, M. Singh, H. Blocker, D. A. Monner, and W. Muller. 2009. Charles River altered Schaedler flora (CRASF(R)) remained stable for four years in a mouse colony housed in individually ventilated cages. Lab. Anim. 43:362-370. [DOI] [PubMed] [Google Scholar]
- 316.Stein, J., T. Marsh, K. Wu, H. Shizuya, and E. DeLong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Steinhaus, E. A. 1941. A study of the bacteria associated with thirty species of insects. J. Bacteriol. 42:757-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Stork, N. E. 1988. Insect diversity: facts, fiction and speculation. Biol. J. Linn. Soc. Lond. 35:321-337. [Google Scholar]
- 319.Sudo, N., S. Sawamura, K. Tanaka, Y. Aiba, C. Kubo, and Y. Koga. 1997. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J. Immunol. 159:1739-1745. [PubMed] [Google Scholar]
- 320.Sullivan, A., C. Edlund, and C. E. Nord. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1:101-114. [DOI] [PubMed] [Google Scholar]
- 321.Tajima, K., S. Arai, K. Ogata, T. Nagamine, H. Matsui, M. Nakamura, R. I. Aminov, and Y. Benno. 2000. Rumen bacterial community transition during adaptation to high-grain diet. Anaerobe 6:273-284. [Google Scholar]
- 322.Tarnberg, M., T. Jakobsson, J. Jonasson, and U. Forsum. 2002. Identification of randomly selected colonies of lactobacilli from normal vaginal fluid by pyrosequencing of the 16S rDNA variable V1 and V3 regions. APMIS 110:802-810. [DOI] [PubMed] [Google Scholar]
- 323.ter Braak, C. J. F., and P. Šmilauer. 2002. Canoco reference manual and CanoDraw for Windows user's guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, NY.
- 324.Thompson, J. 2005. The geographic mosaic of coevolution. University of Chicago Press, Chicago, IL.
- 325.Tilman, D. 1996. Biodiversity: population versus ecosystem stability. Ecology 77:350-363. [Google Scholar]
- 326.Tilman, D., C. L. Lehman, and C. E. Bristow. 1998. Diversity-stability relationships: statistical inevitability or ecological consequence? Am. Nat. 151:277-282. [DOI] [PubMed] [Google Scholar]
- 327.Tilman, D., P. Reich, and J. Knops. 2006. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441:629-632. [DOI] [PubMed] [Google Scholar]
- 328.Tuomisto, H., K. Ruokolainen, R. Kalliola, A. Linna, W. Danjoy, and Z. Rodriguez. 1995. Dissecting Amazonian biodiversity. Science 269:63. [DOI] [PubMed] [Google Scholar]
- 329.Turnbaugh, P. J., M. Hamady, T. Yatsunenko, B. L. Cantarel, A. Duncan, R. E. Ley, M. L. Sogin, W. J. Jones, B. A. Roe, J. P. Affourtit, M. Egholm, B. Henrissat, A. C. Heath, R. Knight, and J. I. Gordon. 2009. A core gut microbiome in obese and lean twins. Nature 457:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon. 2007. The human microbiome project. Nature 449:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Turnbaugh, P. J., R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis, and J. I. Gordon. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027-1031. [DOI] [PubMed] [Google Scholar]
- 332.Vaishampayan, P. A., J. V. Kuehl, J. L. Froula, J. L. Morgan, H. Ochman, and M. P. Francino. 2010. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol. Evol. 2:53-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Vallenet, D., P. Nordmann, V. Barbe, L. Poirel, S. Mangenot, E. Bataille, C. Dossat, S. Gas, A. Kreimeyer, P. Lenoble, S. Oztas, J. Poulain, B. Segurens, C. Robert, C. Abergel, J.-M. Claverie, D. Raoult, C. Medigue, J. Weissenbach, and S. Cruveiller. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Valone, T. J., and N. A. Barber. 2008. An empirical evaluation of the insurance hypothesis in diversity-stability models. Ecology 89:522-531. [DOI] [PubMed] [Google Scholar]
- 335.Van Craeyveld, V., K. Swennen, E. Dornez, T. Van de Wiele, M. Marzorati, W. Verstraete, Y. Delaedt, O. Onagbesan, E. Decuypere, J. Buyse, B. De Ketelaere, W. F. Broekaert, J. A. Delcour, and C. M. Courtin. 2008. Structurally different wheat-derived arabinoxylooligosaccharides have different prebiotic and fermentation properties in rats. J. Nutr. 138:2348-2355. [DOI] [PubMed] [Google Scholar]
- 336.Vanderhoof, J., D. Whitney, D. Antonson, T. Hanner, J. Lupo, and R. Young. 1999. Lactobacillus GG in the prevention of antibiotic-associated diarrhea in children. J. Pediatr. 135:564. [DOI] [PubMed] [Google Scholar]
- 337.Vanderpool, C., F. Yan, and D. B. Polk. 2008. Mechanisms of probiotic action: implications for therapeutic applications in inflammatory bowel diseases. Inflamm. Bowel Dis. 14:1585-1596. [DOI] [PubMed] [Google Scholar]
- 338.van der Waaij, D., J. M. Berghuis-de Vries, and J. E. C. Lekkerkerk-van der Wees. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. 69:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Vanhoutte, T., G. Huys, E. Brandt, and J. Swings. 2004. Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol. Ecol. 48:437-446. [DOI] [PubMed] [Google Scholar]
- 340.van Ruijven, J., and F. Berendse. 2007. Contrasting effects of diversity on the temporal stability of plant populations. Oikos 116:1323-1330. [Google Scholar]
- 341.Varel, V. H., I. M. Robinson, and H. J. Jung. 1987. Influence of dietary fiber on xylanolytic and cellulolytic bacteria of adult pigs. Appl. Environ. Microbiol. 53:22-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Vasanthakumar, A., J. Handelsman, P. D. Schloss, L. S. Bauer, and K. F. Raffa. 2008. Gut microbiota of an invasive subcortical beetle, Agrilus planipennis Fairmaire, across various life stages. Environ. Entomol. 37:1344-1353. [DOI] [PubMed] [Google Scholar]
- 343.Veivers, P. C., R. W. O'Brien, and M. Slaytor. 1982. Role of bacteria in maintaining the redox potential in the hindgut of termites and preventing entry of foreign bacteria. J. Insect Physiol. 28:947-951. [Google Scholar]
- 344.Vesterlund, S., M. Karp, S. Salminen, and A. C. Ouwehand. 2006. Staphylococcus aureus adheres to human intestinal mucus but can be displaced by certain lactic acid bacteria. Microbiology 152:1819-1826. [DOI] [PubMed] [Google Scholar]
- 345.Vollaard, E. J., and H. A. Clasener. 1994. Colonization resistance. Antimicrob. Agents Chemother. 38:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Vollaard, E. J., H. A. L. Clasener, and A. J. H. M. Janssen. 1992. Co-trimoxazole impairs colonization resistance in healthy volunteers. J. Antimicrob. Chemother. 30:685-691. [DOI] [PubMed] [Google Scholar]
- 347.Waiser, M., and R. Robarts. 1997. Impacts of a herbicide and fertilizers on the microbial community of a saline prairie lake. Can. J. Fish. Aquat. Sci. 54:320-329. [Google Scholar]
- 348.Walk, S., and V. Young. 2008. Emerging insights into antibiotic-associated diarrhea and Clostridium difficile infection through the lens of microbial ecology. Interdiscip. Perspect. Infect. Dis. 2008:125081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Wang, X., S. P. Heazlewood, D. O. Krause, and T. H. J. Florin. 2003. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J. Appl. Microbiol. 95:508-520. [DOI] [PubMed] [Google Scholar]
- 350.Warnecke, F., P. Luginbuhl, N. Ivanova, M. Ghassemian, T. H. Richardson, J. T. Stege, M. Cayouette, A. C. McHardy, G. Djordjevic, N. Aboushadi, R. Sorek, S. G. Tringe, M. Podar, H. G. Martin, V. Kunin, D. Dalevi, J. Madejska, E. Kirton, D. Platt, E. Szeto, A. Salamov, K. Barry, N. Mikhailova, N. C. Kyrpides, E. G. Matson, E. A. Ottesen, X. Zhang, M. Hernandez, C. Murillo, L. G. Acosta, I. Rigoutsos, G. Tamayo, B. D. Green, C. Chang, E. M. Rubin, E. J. Mathur, D. E. Robertson, P. Hugenholtz, and J. R. Leadbetter. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560-565. [DOI] [PubMed] [Google Scholar]
- 351.Weigelt, A., J. Schumacher, C. Roscher, and B. Schmid. 2008. Does biodiversity increase spatial stability in plant community biomass? Ecol. Lett. 11:338-347. [DOI] [PubMed] [Google Scholar]
- 352.Welch, R. A., V. Burland, G. Plunkett, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. T. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Welkie, D. G., D. M. Stevenson, and P. J. Weimer. 2010. ARISA analysis of ruminal bacterial community dynamics in lactating dairy cows during the feeding cycle. Anaerobe 16:94-100. [DOI] [PubMed] [Google Scholar]
- 354.Wen, L., R. E. Ley, P. Y. Volchkov, P. B. Stranges, L. Avanesyan, A. C. Stonebraker, C. Hu, F. S. Wong, G. L. Szot, J. A. Bluestone, J. I. Gordon, and A. V. Chervonsky. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Westergaard, K., A. K. Muller, S. Christensen, J. Bloem, and S. J. Sorensen. 2001. Effects of tylosin as a disturbance on the soil microbial community. Soil Biol. Biochem. 33:2061-2071. [Google Scholar]
- 356.Whitford, M. F., R. J. Forster, C. E. Beard, J. Gong, and R. M. Teather. 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4:153-163. [DOI] [PubMed] [Google Scholar]
- 357.Whittaker, R. H. 1972. Evolution and measurement of species diversity. Taxon 21:213-251. [Google Scholar]
- 358.Whittaker, R. H. 1956. Vegetation of the Great Smoky Mountains. Ecol. Monogr. 26:1-80. [Google Scholar]
- 359.Whorwell, P. J., L. Altringer, J. Morel, Y. Bond, D. Charbonneau, L. O'Mahony, B. Kiely, F. Shanahan, and E. M. M. Quigley. 2006. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am. J. Gastroenterol. 101:1581-1590. [DOI] [PubMed] [Google Scholar]
- 360.Willis, W. L., and L. Reid. 2008. Investigating the effects of dietary probiotic feeding regimens on broiler chicken production and Campylobacter jejuni presence. Poult. Sci. 87:606-611. [DOI] [PubMed] [Google Scholar]
- 361.Winberg, J., L. Gezelius, L. Guldevall, and R. Möllby. 1993. Cephadroxil promotes vaginal colonization with Escherichia coli. Infection 21:201-205. [DOI] [PubMed] [Google Scholar]
- 362.Winberg, J., M. Herthelius-Elman, R. Molby, and C. E. Nord. 1993. Pathogenesis of urinary tract infection—experimental studies of vaginal resistance to colonization. Pediatr. Nephrol. 7:509-514. [DOI] [PubMed] [Google Scholar]
- 363.Wise, M. G., and G. R. Siragusa. 2007. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 102:1138-1149. [DOI] [PubMed] [Google Scholar]
- 364.Witkin, S. S., I. M. Linhares, and P. Giraldo. 2007. Bacterial flora of the female genital tract: function and immune regulation. Best Pract. Res. Clin. Obstet. Gynaecol. 21:347-354. [DOI] [PubMed] [Google Scholar]
- 365.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Woodmansey, E. J., M. E. T. McMurdo, G. T. Macfarlane, and S. Macfarlane. 2004. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl. Environ. Microbiol. 70:6113-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Wos-Oxley, M. L., I. Plumeier, C. von Eiff, S. Taudien, M. Platzer, R. Vilchez-Vargas, K. Becker, and D. H. Pieper. 2010. A poke into the diversity and associations within human anterior nare microbial communities. ISME J. 4:839-851. [DOI] [PubMed] [Google Scholar]
- 368.Wu, S., K.-J. Rhee, E. Albesiano, S. Rabizadeh, X. Wu, H.-R. Yen, D. L. Huso, F. L. Brancati, E. Wick, F. McAllister, F. Housseau, D. M. Pardoll, and C. L. Sears. 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Yang, H., D. Schmitt-Wagner, U. Stingl, and A. Brune. 2005. Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis). Environ. Microbiol. 7:916-932. [DOI] [PubMed] [Google Scholar]
- 370.Yannarell, A. C., T. F. Steppe, and H. W. Paerl. 2007. Disturbance and recovery of microbial community structure and function following Hurricane Frances. Environ. Microbiol. 9:576-583. [DOI] [PubMed] [Google Scholar]
- 371.Young, V. B., and T. M. Schmidt. 2004. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J. Clin. Microbiol. 42:1203-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Youssef, N., C. S. Sheik, L. R. Krumholz, F. Z. Najar, B. A. Roe, and M. S. Elshahed. 2009. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl. Environ. Microbiol. 75:5227-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Zahradnik, R. T., I. Magnusson, C. Walker, E. McDonell, C. H. Hillman, and J. D. Hillman. 2009. Preliminary assessment of safety and effectiveness in humans of ProBiora3tm, a probiotic mouthwash. J. Appl. Microbiol. 107:682-690. [DOI] [PubMed] [Google Scholar]
- 374.Zaura, E., B. Keijser, S. Huse, and W. Crielaard. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Zavaleta, E. S., J. R. Pasari, K. B. Hulvey, and G. D. Tilman. 2010. Sustaining multiple ecosystem functions in grassland communities requires higher biodiversity. Proc. Natl. Acad. Sci. U. S. A. 107:1443-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Zhao, T., M. P. Doyle, B. G. Harmon, C. A. Brown, P. O. E. Mueller, and A. H. Parks. 1998. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J. Clin. Microbiol. 36:641-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Zhou, X., C. J. Brown, Z. Abdo, C. C. Davis, M. A. Hansmann, P. Joyce, J. A. Foster, and L. J. Forney. 2007. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1:121-133. [DOI] [PubMed] [Google Scholar]
- 378.Zielezny, Y., J. Groeneweg, H. Vereecken, and W. Tappe. 2006. Impact of sulfadiazine and chlorotetracycline on soil bacterial community structure and respiratory activity. Soil Biol. Biochem. 38:2372-2380. [Google Scholar]
- 379.Zoetendal, E. G., A. D. L. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]