Abstract
Three missense SURF1 mutations identified in patients with Leigh syndrome (LS) were evaluated in the yeast homolog Shy1 protein. Introduction of two of the Leigh mutations, F249T and Y344D, in Shy1 failed to significantly attenuate the function of Shy1 in cytochrome c oxidase (CcO) biogenesis as seen with the human mutations. In contrast, a G137E substitution in Shy1 results in a nonfunctional protein conferring a CcO deficiency. The G137E Shy1 mutant phenocopied shy1Δ cells in impaired Cox1 hemylation and low mitochondrial copper. A genetic screen for allele-specific suppressors of the G137E Shy1 mutant revealed Coa2, Cox10, and a novel factor designated Coa4. Coa2 and Cox10 are previously characterized CcO assembly factors. Coa4 is a twin CX9C motif mitochondrial protein localized in the intermembrane space and associated with the inner membrane. Cells lacking Coa4 are depressed in CcO activity but show no impairment in Cox1 maturation or formation of the Shy1-stabilized Cox1 assembly intermediate. To glean insights into the functional role of Coa4 in CcO biogenesis, an unbiased suppressor screen of coa4Δ cells was conducted. Respiratory function of coa4Δ cells was restored by the overexpression of CYC1 encoding cytochrome c. Cyc1 is known to be important at an ill-defined step in the assembly and/or stability of CcO. This new link to Coa4 may begin to further elucidate the role of Cyc1 in CcO biogenesis.
Leigh syndrome (LS) is a highly progressive neurological disorder of infancy characterized by necrotizing lesions in the midbrain and brain stem (32). Humans afflicted with LS have compromised oxidative phosphorylation (OXPHOS) function due to mutations in nuclear or mitochondrial genes encoding respiratory chain components or their assembly factors. Although LS infants are born with a normal appearance, neurological lesions develop within months and dysfunction extends to other organs, resulting in a high mortality rate. LS patients typically have mutations affecting complex I or complex IV (cytochrome c oxidase [CcO]) of the OXPHOS pathway (14). Patients with a specific CcO deficiency most often have mutations in the SURF1 gene that encodes a CcO assembly factor (9, 15, 41).
SURF1 is not absolutely required for CcO biogenesis in humans, since SURF1-deficient patient fibroblasts retain 10 to 15% of residual CcO activity (32). The yeast homolog of SURF1 is Shy1 (SURF1 homolog in yeast) and has a conserved function in CcO biogenesis (24). Yeast lacking Shy1 retain residual CcO activity, but growth of the mutant strain is compromised on respiratory, nonfermentable carbon sources (4).
Insights into the function of SURF1 in human cells have been gleaned through the characterization of stalled CcO assembly intermediates in cells isolated from SURF1 LS patients using blue native (BN) gel electrophoresis. One intermediate, designated S2, which accumulates in SURF1-deficient patient fibroblasts, consists of Cox1 in association with two nuclear CcO subunits, CoxIV and Va (38, 45, 47). A similar stalled assembly intermediate accumulates in CcO-deficient patients with mutations in two other assembly factors, SCO1 and SCO2. These assembly proteins function in the maturation of the mitochondrially encoded Cox2 subunit and the binuclear copper (CuA) site within this subunit. In contrast, studies with patient fibroblasts harboring mutations in the genes encoding Cox10 and Cox15 proteins, which are involved in the biosynthesis of the heme a cofactor used exclusively by CcO (at the heme a and heme a3:CuB sites), show only free Cox1 by BN analysis (1, 2). These data suggest that CcO biogenesis commences with the mitochondrial synthesis and maturation of Cox1, while the other two mitochondrially encoded subunits, Cox2 and Cox3, are added at later stages. The absence of the S2 intermediate in cells with mutations in COX10 or COX15 is consistent with the prediction that the S2 assembly intermediate contains Cox1 with at least the heme a center formed.
The first major clue to the function of SURF1 came from studies with the bacterium Rhodobacter sphaeroides, in which surf1 mutant cells showed impairment in the formation of the heme a3:CuB bimetallic center within Cox1 (33). Specifically, heme a and CuB were observed spectroscopically with surf1 mutant cells, but heme a3 was not present. The CuB site had an altered spectroscopic signature to compensate for the loss of heme a3, as the two cofactors typically coordinate with each other. This study suggests Surf1 is involved in the maturation of the heme a3 site in CcO. In lower eukaryotes, impairment of CcO assembly results in proteolytic degradation of the stalled intermediates (16). Thus, it is not possible to isolate the CcO complex in shy1Δ yeast cells to identify any missing cofactors. However, Shy1 was shown to have a key role in formation of the heterobimetallic CuB:heme a3 center in yeast Cox1 (18). Furthermore, it was recently shown that Surf1 in bacteria is a heme-binding protein (10), although these findings have yet to be confirmed in eukaryotes.
Additional insights into the function of SURF1/Shy1 came from the isolation of genetic suppressors of shy1Δ respiratory deficiency in yeast (3). Respiratory function can be partially restored in shy1Δ cells by enhancing Cox1 translation through the overexpression of MSS51 (6), a dual-function protein that acts as a COX1 translational activator in addition to binding to the newly synthesized Cox1 polypeptide. Suppression of the shy1Δ respiratory defect is also observed with enhanced expression levels of the two CcO subunits Cox5a and Cox6 corresponding to the human S2-containing subunits CoxIV and Va (15). Overexpression of COA2, a recently identified CcO assembly factor shown to interact with Shy1, can also suppress the shy1Δ respiratory defect (30). Finally, overexpression of the COX10 gene that encodes the hydroxyfarnesyl transferase, which generates heme o as the first step in heme a biosynthesis, can partially restore respiratory function in shy1Δ cells. Although overexpression of COX10 has only very weak suppressor activity, a marked synergistic effect was apparent in the overexpression of both MSS51 and COX10 (29).
Shy1 has a secondary function in yeast in the maintenance of the conserved mitochondrial copper storage pool that is used in the copper metallation of Cox1 and Cox2 during CcO biogenesis. Yeast cells lacking Shy1 contain mitochondria with a partially depleted matrix copper storage pool, and the respiratory defect of shy1Δ cells can be partially reversed by growth in the presence of exogenous copper (29). Similarly, liver and muscle samples from patients with SURF1 mutations exhibit a cellular copper deficiency (37). Maintenance of the matrix copper pool is postulated to be linked to active CcO biogenesis in general, as patient tissue with mutations to two other CcO assembly factors, SCO1 and SCO2, result in a cellular copper deficiency as well (22).
Human SURF1 and yeast Shy1 are both mitochondrial proteins tethered to the inner membrane (IM) by two transmembrane (TM) helices with a large central domain projecting into the intermembrane space (IMS). Most LS patients with SURF1 mutations have gene deletions or rearrangements. Missense mutations in SURF1 are quite rare, with only a limited number being reported. These mutations tend to be associated with a mild clinical phenotype, and patient survival is prolonged (28). We selected a subset of known missense mutations, two of which lie within the IMS globular domain and a third that maps to the second TM domain. In an attempt to gain further insights into which functional step of SURF1 was compromised by the missense mutations, we engineered and characterized the corresponding mutations in conserved residues of yeast SHY1. In doing so, we have additionally identified a new member of the CcO assembly factor family, Coa4, that may be linked to the role of cytochrome c in CcO assembly. We show that the respiratory defect of cells lacking Coa4 is specifically suppressed by the overexpression of the IMS electron carrier cytochrome c (CYC1). This is the first time CYC1 has been found as a suppressor of a CcO assembly mutant.
MATERIALS AND METHODS
Yeast strains and vectors.
The Saccharomyces cerevisiae strains used in this study are summarized in Table 1. The SHY1 open reading frame (ORF) was cloned into plasmids pRS413 and pRS423 under the control of its own promoter and terminator (450 base pairs upstream and downstream of the ORF). The G137E, F249T, and Y344D mutations were made in both the high- and low-copy vectors using site-directed mutagenesis. The SHY1 ORF with a 3′-13Myc tag was cloned from genomic DNA of the DY5113 SHY1-13Myc strain into pRS416 and pRS426 vectors under the control of its own promoter and the ADH1 terminator. The G137E, F249T, and Y344D mutations were generated in these vectors by site-directed mutagenesis. The YLR218c/COA4 ORF was cloned into a pRS425 vector under the control of its own promoter and terminator, and the COA4-Myc insert was generated by PCR and cloned into pRS425 under the control of the MET25 promoter and CYC1 terminator. The CYC1 and CYC7 ORFs were amplified by PCR and cloned into pRS425 under the control of their own promoter and terminator. The substitutions of the K78, K79, K92, and K93 residues with Glu in CYC1 were introduced by site-directed mutagenesis. Sequencing was used to confirm all cloning and site-directed mutagenesis products in the vectors created. The COA2 and COX10 vectors used in this study, as well as the construction of the high-copy-number library, have all been described previously (19, 29, 30). Yeast strains were transformed using lithium acetate. Culture conditions for the yeast strains were either rich (YP) medium or synthetic complete (SC) medium lacking the appropriate nutrients for plasmid selection.
TABLE 1.
Yeast strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| W303 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | |
| shy1Δ mutant | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1shy1Δ::URA3 | 24 |
| coa4Δ mutant | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1coa4Δ::KanMX4 | This study |
| coa4Δ shy1Δ mutant | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1coa4Δ::KanMX4 shy1Δ::URA3 | This study |
| cyc1Δ cyc7Δ mutant | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 cyc1Δ::URA3 cyc7Δ:TRP1 | 5 |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| cox11Δ mutant | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cox11Δ::kanMX4 | Invitrogen |
| cox11Δ shy1Δ mutant | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cox11Δ::kanMX4 shy1Δ::URA3 | 30 |
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 lys2Δ0/LYS2 | Invitrogen |
| shy1Δ mutant | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 lys2Δ0/LYS2 shy1Δ::kanMX4/shy1Δ::kanMX4 | Invitrogen |
| coa4Δ mutant | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 lys2Δ0/LYS2 coa4Δ::kanMX4/coa4Δ::kanMX4 | Invitrogen |
| DY5113 | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 | |
| SHY1-13Myc mutant | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 | 30 |
| SHY1-13Myc coa4Δ mutant | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 coa4Δ::KanMX4 | This study |
| COA4-13Myc mutant | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA4-13Myc::KanMX4 | This study |
Suppressor screens.
For screening of the SHY1(G137E)-dependent high-copy-number suppressor, the BY4743 shy1Δ cells containing a single-copy pRS413-SHY1(G137E) were transformed with the coa1Δ genomic DNA library (19, 29, 30). Transformants were replica plated onto the minimal selective plates containing 2% glycerol-lactate as a carbon source and incubated for 6 days at 30°C. To confirm the dependence of the selected respiratory-competent clones on the mutant form of SHY1, the colonies were grown in the medium supplemented with histidine to eliminate pRS413-SHY1(G137E). Plasmids were rescued from 35 transformants, showing strong dependence on SHY1(G137E), and further analyzed by DNA sequencing.
For screening of the high-copy-number suppressor of the coa4Δ allele, a genomic DNA library generated from coa2Δ cells was transformed into BY4743 coa4Δ cells. About 56,000 transformants were obtained, replica plated onto the minimal selective plates containing 2% gylcerol-lactate as a carbon source, and incubated for 4 days at room temperature. The plasmids were shed on the plates containing 5-fluoroorotic acid (5-FOA) to ensure that the revealed suppressors were vector borne. Plasmids were rescued from the cells showing a vector-dependent respiratory growth and subjected to DNA sequencing.
Mitochondrial purification and assays.
Intact mitochondria were isolated from yeast as previously described (13). The standard Bradford assay was used to determine total mitochondrial protein concentration (8). CcO activity was assessed by monitoring the oxidation of reduced cytochrome c (12). The specific activity of CcO was normalized to mitochondrial protein levels and is presented as a percentage of the wild type (WT). Metals were quantified in isolated mitochondria using a Perkin-Elmer Optima inductively coupled plasma optical emission spectrometer (ICP-OES) after nitric acid digestion at 95°C. A standard curve was determined from commercially available standards (Perkin-Elmer), and buffer samples were run in parallel as a control. All values were normalized to milligrams of mitochondrial protein used in the analysis. In vivo mitochondrial translation assays were conducted as described previously (30).
Localization experiments.
Cells were fractionated, and the cytosolic and mitochondrial fractions collected as described previously (13). Purified mitochondria were either diluted in 20 mM HEPES, pH 7.4, or treated with 100 mM sodium carbonate (pH 11.5) and fractionated by high-speed centrifugation (30). The supernatant and pellet were analyzed by immunoblotting for each treatment. The proteinase K assay was performed with isolated mitochondria incubated in isotonic (0.8 M sorbitol, 20 mM HEPES, pH 7.4) and hypotonic buffer (20 mM HEPES, pH 7.4) in the presence or absence of an indicated amount of NaCl for 20 min on ice (13). The treated organelles were collected by centrifugation, resuspended in isotonic buffer, and incubated on ice for 20 min in the presence or absence of proteinase K (0.1 mg/ml). Following centrifugation, the pellets were loaded onto SDS-PAGE, and proteins were detected in the pellet by immunoblotting.
BN-PAGE.
Blue native PAGE (BN-PAGE) was performed essentially as described previously (46), with 1% digitonin or 1% n-dodecyl β-d-maltoside (DDM). After incubation on ice for 15 min and centrifugation (20,000 × g for 15 min at 2°C), supernatants were mixed with sample buffer (5% Coomassie brilliant blue G250, 0.5 M 6-aminocaproic acid, pH 7.0) and then loaded onto a 5 to 13% gradient polyacrylamide gel. Separated complexes were detected by immunoblotting on polyvinylidene difluoride (PVDF) membrane. The high-mass-marker proteins were obtained from GE Healthcare.
Immunoblotting.
Mitochondrial protein samples were separated on 15% polyacrylamide gels and transferred to nitrocellulose. Proteins were visualized using enhanced chemiluminescence (ECL) reagents with horseradish peroxidase-conjugated secondary antibodies. The anti-Myc antibody was purchased from Roche; anti-Por1 and anti-Pgk1 were purchased from Molecular Probes; and anti-Cox1, Cox2, and Cox3 were purchased from Mitosciences. Alex Tzagoloff generously provided antiserum to F1 ATP synthase Atp2 and Cyc1, Carla Koehler kindly provided Cyt1, Cyc1, and Cyb2 antibodies, the Sod2 antibody was a gift from Val Culotta, Bernard Trumpower provided anti-Rip1, and Jerry Kaplan provided anti-Aco1.
Additional assays.
The respiratory competency of strains was determined by growth tests on plates containing 2% glucose or 2% glycerol-2% lactate as a carbon source. Yeast cells were grown overnight in liquid cultures in selective medium containing 2% raffinose-0.2% glucose, adjusted to an optical density at 600 nm (OD600) of 0.5, and serial dilutions were spotted onto the plates and incubated at 30°C or 37°C for 2 days (glucose plates) or 4 to 6 days (glycerol-lactate plates). The oxygen consumption of cells grown to stationary phase was determined using a 5300A biological oxygen monitor (Yellow Springs Instrument Co.). The rate of oxygen consumption (%O2/s/OD600) was calculated from the linear response (17). Sensitivity of yeast strains to hydrogen peroxide was determined as previously described (19). For heme staining, mitochondrial proteins were solubilized in 1× SDS-Laemmli buffer containing 0.1 M dithiothreitol (DTT) and resolved on 12% SDS-PAGE gels. Total protein was visualized with Coomassie brilliant blue G250; peroxidase staining for heme detection of Cyc1 and Cyc7 was performed as described using the 3,3′,5,5′-tetramethylbenzidine (TMBZ)/H2O2 procedure (40).
RESULTS
Characterization of human SURF1 missense mutations in the yeast Shy1 protein.
Missense mutations in SURF1 are very rare, with only a limited number being reported (28). We selected three known missense mutations in SURF1 LS patients that result in G124E, I246T, and Y274D substitutions in SURF1 (31, 39). Gly124 and Tyr274 are conserved residues in SURF1 orthologs, whereas Ile246 is a conserved hydrophobic residue. Gly124 and Ile246 map to the IMS globular domain of SURF1, whereas Tyr274 is predicted to exist within the second TM domain (Fig. 1A and B).
FIG. 1.
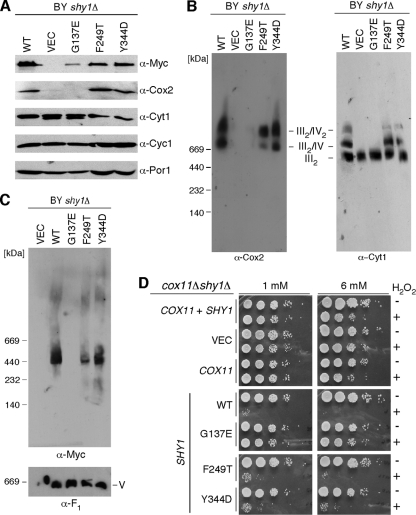
Human SURF1 and yeast Shy1. (A) Alignment of the yeast (Sce) Shy1 and human (Hsp) SURF1 amino acid sequences (ClustalW). Bars indicate the predicted transmembrane domains. The asterisks indicate identically conserved residues; colons and periods represent similarly conserved residues. The arrowheads indicate amino acid residues mutated in Leigh syndrome patients. (B) A schematic view of Shy1. Two predicted transmembrane domains (TM1 and TM2) are indicated along with the IMS (∼250 residues) and the N (∼70-residue)- and C (∼25-residue)- terminal domains. Marked are the substitutions in Shy1 corresponding to pathogenic mutations in SURF1. OM, outer membrane. (C) W303 or BY4743 shy1Δ cells expressing either Myc-tagged wild-type Shy1 or its G137E, F249T, or Y344D mutant derivatives were precultured in selective media containing 2% raffinose-0.2% glucose, serially diluted, and spotted onto the plates containing 2% glucose or 2% glycerol-2% lactate as a carbon source. Pictures were taken after 2 days and 4 days of incubation at 30°C, for glucose and glycerol-lactate, respectively. (D) Mitochondria isolated from the BY4743 transformants mentioned above were assayed (n = 3) for CcO activity (change in the OD550 [ΔOD550]/min/10 mg protein).
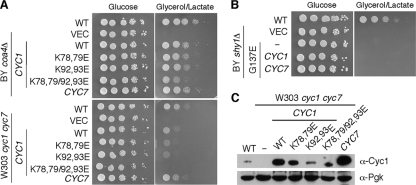
We introduced the corresponding mutations at the conserved residues of the yeast Shy1 protein, yielding G137E, F249T, and Y344D mutations (Fig. 1). Expression of the G137E Shy1 mutant in shy1Δ cells in two distinct genetic backgrounds (BY4743 and W303) resulted in a CcO deficiency that precluded growth on nonfermentable carbon sources (Fig. 1C). In contrast, the respiratory deficiency of shy1Δ cells in the W303 and BY4743 backgrounds was reversed by the introduction of vector-borne SHY1 with F249T and Y344D codon mutations, although BY4743 mutant cells with the Y344D Shy1 mutant were partially impaired in growth on glycerol-lactate medium (Fig. 1C). None of the mutant Shy1 proteins had any dominant negative effects when expressed in wild-type cells (data not shown). The three mutant Shy1 proteins were stably expressed, although the G137E mutant protein was partially attenuated in abundance (Fig. 2A). Cells harboring the F249T and Y344D mutant Shy1 proteins exhibited near-normal oxygen consumption (data not shown) and CcO activity (Fig. 1D). In contrast, cells expressing the G137E Shy1 mutant showed low CcO activity (Fig. 1D) and low oxygen consumption (data not shown), consistent with the lack of growth on glycerol-lactate medium. Furthermore, G137E Shy1-containing cells showed reduced steady-state Cox2 levels (Fig. 2A). In line with these data, BN-PAGE analysis showed that shy1Δ cells expressing the G137E Shy1 mutant as well as the vector control cells lacked the bc1:CcO supercomplex, yet the dimeric bc1 complex was normal (Fig. 2B). Cells expressing either the F249T or Y344D Shy1 mutant showed no major difference in supercomplex formation compared to wild-type cells grown at either 30°C or a more stringent 37°C (Fig. 2B and data not shown).
FIG. 2.
Characterization of the SURF1/Shy1 mutants in yeast. (A) Immunoblot of the mitochondria (30 μg) isolated from the BY4743 (BY) transformants shown in panel C of Fig. 1. Organelles were separated by SDS-PAGE, and steady-state levels of the Cox2, Cyt1, and Cyc1 were assessed with the respective antibodies. Steady-state levels of the WT Shy1 and the three Shy1 mutants were determined using anti-Myc antibodies. The levels of Por1 served as a loading control. (B) Purified mitochondria (100 μg) were subjected to BN-PAGE. The distribution of respiratory complexes was analyzed by Western blotting with antibodies against Cox2 and Cyt1. (C) The Shy1 high-molecular-weight complex was assessed by BN-PAGE as described above for anti-Myc antibodies. The monomeric form of complex V (loading control) was visualized with anti-F1 serum. (D) BY4741 cox11Δ shy1Δ cells, transformed with the episomal vectors expressing wild-type Cox11, Shy1, or the respective mutant forms of the latter were grown to the mid-exponential phase and incubated with or without indicated concentration of H2O2 for 2 h at 30°C. Serial dilutions were spotted onto the plates containing 2% glucose and incubated for 36 to 48 h at 30°C.
As mentioned, Shy1 mediates formation of the heme a3:CuB center in Cox1 (18). This function of Shy1 can be assessed through a hydrogen peroxide assay previously described (19). Cells depleted of Cox11 or Sco1 stall CcO assembly and lead to the accumulation of a pro-oxidant intermediate consisting of Cox1 with reactive heme a3. The hydrogen peroxide sensitivity is reversed by the subsequent depletion of Shy1 (19). cox11Δ shy1Δ double null cells are resistant to hydrogen peroxide treatment because the heme a3:CuB site is not stably formed. The reintroduction of SHY1 restores hydrogen peroxide sensitivity expected of cox11Δ cells (Fig. 2D). Introduction of the F249T and Y344D Shy1 mutants into the cox11Δ shy1Δ null strain restored hydrogen peroxide sensitivity in the cells, consistent with these mutants being nearly wild type in function (Fig. 2D). In contrast, transformation of the double null cells with the G137E Shy1 mutant failed to restore hydrogen peroxide sensitivity, consistent with it being a nonfunctional variant of Shy1.
Shy1-mediated formation of the heme a3:CuB center in Cox1 occurs within a Cox1 assembly intermediate containing Shy1 (18). This complex is visualized on BN-PAGE as an ∼450-kDa band (25, 29, 30) (Fig. 2C). This complex was not detected in cells containing the G137E Shy1 mutant, and cells expressing the F249T or Y344D Shy1-Myc mutant showed a reduction in the abundance of this complex compared to wild-type Shy1 (Fig. 2C).
Cells lacking Shy1 have attenuated levels of mitochondrial copper, and supplementation of the growth medium with exogenous copper yielded a partial reversal of the growth defect on nonfermentable carbon sources (29). Respiratory growth of cells expressing the G137E Shy1 mutant was partially restored by exogenous copper as with the shy1Δ cells (data not shown). Similarly, the G137E Shy1 mutant cells showed low mitochondrial copper levels (55% of WT), and the mitochondrial copper level of shy1Δ mitochondria was determined to be ∼50% of that of the WT. Cells with F249T and Y344D Shy1 mutants contained 80% and 75% of wild-type mitochondrial copper levels, respectively. Thus, the Shy1 role in copper homeostasis is not significantly altered in the F249T and Y344D Shy1 mutant strains but is with the G137E Shy1 mutant, similarly to shy1Δ cells.
Suppressors of the G137E Shy1 respiratory defect.
Gly137 is a highly conserved residue in Shy1 proteins from various species, suggesting that it may be important for Shy1 activity. Since the G137E Shy1 mutant is a nonfunctional variant, we screened for extragenic allele-specific suppressors of the respiratory defect of the G137E Shy1 mutant to identify additional proteins that may be relevant for Shy1 function. Yeast cells harboring the nonfunctional G137E Shy1 mutant were transformed with a high-copy DNA library, and suppressors of the respiratory defect were isolated on SC-glycerol-lactate medium. Respiratory growth was confirmed to be dependent on the library vector by 5-FOA, which resulted in 58 candidates. The vector expressing the G137E Shy1 mutant was shed as well, showing that 23 of the candidates were able to grow on glycerol-lactate without the Shy1 mutant. These candidates were not pursued, since we expected many of these clones to be vector-encoded SHY1 or the well-known MSS51 suppressor (6, 27). The remaining candidates required the presence of the mutant SHY1 (G137E) vector for respiratory growth, and library vectors were isolated and sequenced. Multiple copies of COA2 and COX10 were recovered as suppressors of the respiratory defect (Fig. 3A). Both Coa2 and Cox10 are known CcO assembly factors and have been indicated as weak suppressors of the shy1Δ cell respiratory defect individually on YP-glycerol-lactate (30). The suppressive activity of either COA2 or COX10 in shy1Δ cells is enhanced in the presence of the G137E Shy1 variant when grown on YP- or SC-glycerol-lactate.
FIG. 3.
High-copy-number suppressor for the G137E Shy1 respiratory defect. (A) BY4743 shy1Δ cells transformed with episomal vectors containing COA4, COA2, and COX10 with or without SHY1 (G137E mutant) were precultured, serially diluted, and analyzed as described in the legend to Fig. 1C. (B) Amino acid sequence alignment of Coa4 from different eukaryotic organisms (Kluyveromyces lactis, Candida glabrata, Schizosaccharomyces pombe, Mus musculus, and Homo sapiens). The conserved twin CX9C motif is indicated.
An additional suppressor candidate was determined to contain a library plasmid, encoding the YLR218c ORF that we designated COA4 (cytochrome oxidase assembly 4) (Fig. 3A). Expression of this gene alone does not suppress the respiratory defect of shy1Δ cells; however, in the presence of the G137E Shy1 allele, growth is observed on YP-glycerol-lactate (Fig. 3A). In a recent systematic analysis of yeast proteins with a conserved twin CX9C motif, Coa4 (Cmc3) was identified and shown to be a mitochondrial IMS protein (23) (Fig. 3B). This motif is common to a group of IMS-localized proteins, several of which are involved in CcO assembly (20).
Coa4 is important for CcO assembly.
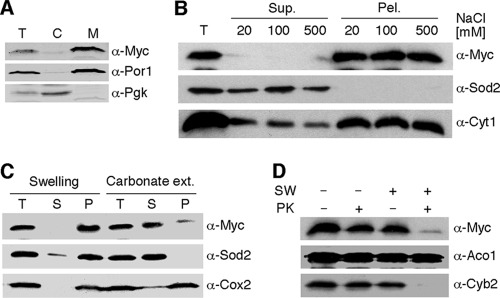
To confirm the IMS localization of Coa4 (23), we tested the mitochondrial localization of a vector-borne Myc-tagged Coa4 allele in coa4Δ cells (Fig. 4). Coa4 was found to be predominantly associated with gradient-purified mitochondria (Fig. 4A) and to be associated with the membrane fraction regardless of high-salt washes (Fig. 4B). The protein was solubilized by sodium carbonate washing and degraded by proteinase K in isolated mitoplasts (Fig. 4C and D). These results confirm the IMS localization of Coa4 and identify an association with the IM. This is consistent with Coa4 lacking any predicted transmembrane helices.
FIG. 4.
Localization and topology of Coa4. (A) BY4743 coa4Δ cells expressing COA4-Myc were lysed and fractionated. Cytosolic and mitochondrial fractions were subjected to SDS-PAGE and analyzed by immunoblotting with anti-Myc, anti-Por1, and anti-Pgk1 antibodies. T, total cell lysate; C, cytosolic fraction; M, mitochondrial fraction. (B) Isolated organelles were resuspended in 20 mM HEPES (pH 7.4) and NaCl of the indicated concentrations and sonicated. The soluble and insoluble fractions were fractionated and analyzed by immunoblotting with anti-Myc, anti-Sod2, and anti-Cyt1 antibodies. (C) Purified mitochondria (50 μg) were incubated on ice for 30 min with 20 mM HEPES (pH 7.4) for swelling or with 0.1 M sodium bicarbonate (pH 11.5) for carbonate extraction (ext.) in the presence of 2 mM PMSF (phenylmethylsulfonyl fluoride). Following incubation, the soluble and insoluble fractions were separated by high-velocity centrifugation and analyzed by immunoblotting. The soluble matrix protein Sod2 and integral membrane protein Cox2 were detected with the respective antibodies. T, total mitochondria; S, supernatant; P, pellet; ext., extraction. (D) Intact (SW−) or osmotically shocked mitochondria (SW+) were incubated with (PK+) or without (PK−) 0.1 mg/ml of proteinase K (PK) for 30 min on ice. Followed centrifugation, the treated organelles were separated by SDS-PAGE and analyzed by immunoblotting with anti-Myc, anti-Cyb2, and anti-Aco1.
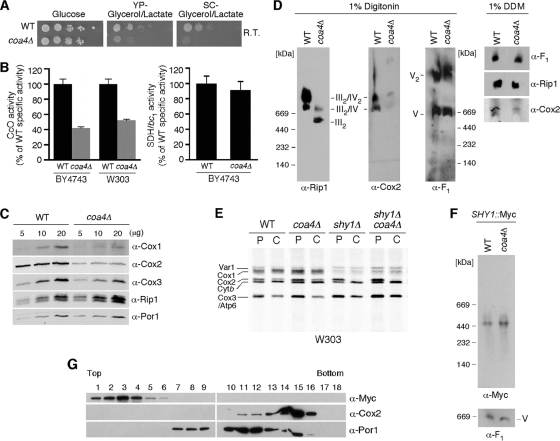
Cells lacking Coa4 in the BY4743 background exhibit a respiratory defect when plated on SC-glycerol-lactate medium at room temperature, but the growth defect was less pronounced at elevated temperatures (Fig. 5A and 6A) (23). No marked growth defect was observed for coa4Δ cells with the W303 background at 23 or 30°C (data not shown), yet CcO activity was depressed to ∼50% of wild-type activity in this background (Fig. 5B). CcO activity in coa4Δ cells in the BY4743 background was reduced to ∼40% of that of the wild type (Fig. 5B). Yeast typically propagate well on glycerol-lactate medium at 30°C when CcO activity is >40% of that of the wild type. The bc1 reductase activity was unchanged in mutant cells of either background (Fig. 5B and data not shown). Consistent with the attenuated CcO activity, the steady-state levels of Cox1, Cox2, and Cox3 were reduced in the coa4 null mutant (Fig. 5C). The abundance of the bc1:CcO supercomplex was also markedly diminished (Fig. 5D) without any change in the ATP synthase monomeric or dimeric complexes. Solubilization of the IM with dodecyl maltoside further confirmed the attenuated levels of CcO (Fig. 5D). Although Coa4 appears to have a role in the assembly or stability of the CcO complex, it is not associated with the CcO supercomplex (Fig. 5G), as Coa4 fractionates in low-mass fractions upon sucrose gradient centrifugation.
FIG. 5.
Cells lacking Coa4 have compromised respiratory functions. (A) Respiratory growth of BY4743 wild-type and coa4Δ cells was analyzed as described in the legend to Fig. 1, except that the plates were incubated at room temperature for 3 days. (B) Mitochondria isolated from the coa4Δ cells cultured at 30°C were used to determine CcO and SDH/bc1 activities. Enzymatic activities are shown as a percentage of wild-type specific activity. The data represent an average of three independent repeats, and the error bars indicate one standard deviation (SD). (C) Mitochondria from WT and coa4Δ BY4743 cells were separated on SDS-PAGE gels, and the steady-state levels of Cox1, Cox2, Cox3, and Rip1 were assessed by immunoblotting. Por1 served as a loading control. (D) Isolated mitochondria (75 μg) were lysed with either 1% digitonin or DDM. Clarified lysates were subjected to BN-PAGE, and separated protein complexes were analyzed by Western blotting. Anti-Cox2 and anti-Rip1 antibodies were used to detect complex IV and III, respectively. Anti-F1 was used for detection of complex V. (E) In vivo labeling of mitochondrial translation products in wild-type, coa4Δ, and shy1Δ cells and coa4Δ shy1Δ double null cells of W303 background. (F) Mitochondria purified from DY5113 SHY1::13Myc wild-type or coa4Δ cells were solubilized with 1% digitonin and analyzed by BN-PAGE as described in the legend to Fig. 2C. V, complex V. (G) Mitochondria from BY4743 coa4Δ cells transformed with an episomal vector expressing Coa4-13Myc were loaded onto a continuous 7 to 30% sucrose gradient and subjected to ultracentrifugation (148,000 × g, 8 h, 2°C). Eighteen fractions were collected, separated on SDS-PAGE, and analyzed by immunoblotting with antisera to Por1, Cox2, and Myc to detect Coa4-Myc.
FIG. 6.
Overexpression of cytochrome c suppresses the respiratory defect in coa4Δ cells. (A) Respiratory growth of the BY4743 wild-type and coa4Δ cells transformed with episomal vectors expressing CYC1 or CYC7 was analyzed as described in the legend to Fig. 5A. RT, room temperature. (B) Mitochondria purified from the aforementioned transformants were used to measure CcO and SDH/bc1 enzymatic activities as described in the legend to Fig. 5B. (C) BN-PAGE analysis of the respiratory complexes as previously described in the legend to Fig. 2. (D) Steady-state levels of Cox1, Cyc1, and Cyt1 analyzed by immunoblotting. (E) Isolated mitochondria (75 μg) were separated by SDS-PAGE and subjected to the in-gel heme staining. A total of 1 μg of horse heart cytochrome c (Cyth) served as a control. Coomassie staining was used to show that the same amount of the total mitochondrial protein is loaded.
Deletions of genes encoding CcO assembly factors often lead to diminished Cox1 translation by trapping Mss51 in Cox1 preassembly complexes (6). In vivo radiolabeling of mitochondrial translation products revealed that normal levels of Cox1 were synthesized during the pulse and short-chase phase of the translation assay (Fig. 5E), suggesting that coa4Δ cells are not subjected to the downregulation of the Cox1 synthesis seen for shy1Δ cells and several CcO assembly mutants (6). Deletion of COA4 in a shy1Δ background retains the attenuated level of Cox1 synthesis seen with shy1Δ cells. Thus, Coa4 is downstream of the Shy1 effect on Cox1 synthesis. To confirm that the Cox1 assembly intermediates, including those stabilized by Mss51 and Shy1 are normal in coa4Δ cells, we performed BN-PAGE on Myc epitope-tagged Shy1 (Fig. 5F). The levels of abundance of the high-mass Shy1-Cox1 complex were similar in wild-type and coa4Δ cells, substantiating the conclusion that Cox1 synthesis and early maturation steps are normal in the mutant cells. Furthermore, no interaction was observed between Coa4 and Shy1 by direct coimmunoprecipitation (data not shown).
Since shy1Δ cells are impaired in mitochondrial copper, we quantified the copper content of mitochondria from coa4Δ cells. Mitochondrial copper was reduced to 50% of wild-type levels in both shy1Δ and coa4Δ cells (data not shown), but supplemental copper failed to enhance the respiratory deficiency of coa4Δ cells.
Genetic suppression of the coa4Δ respiratory growth defect.
As mentioned, the respiratory deficiency of shy1Δ cells can be suppressed by high-copy-number MSS51, COA1, COA2, or COX10 (29). We tested whether overexpression of these genes would enhance the respiratory growth of coa4Δ cells. No marked improvement in respiratory growth was observed with the overexpression of any of these genes (data not shown), including the COX10 mutant encoding the N196K allele (7), all of which are involved in the early assembly stages of Cox1 maturation. This further implies, along with the in vivo translation data, that Coa4 is important in the assembly of CcO after the maturation of Cox1.
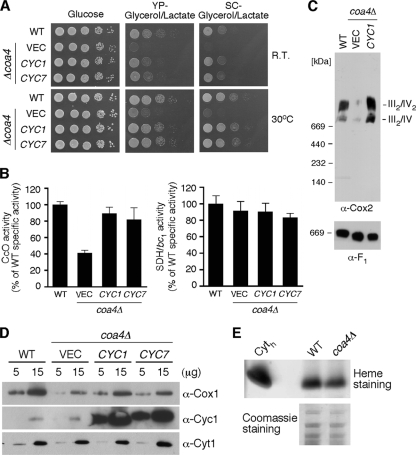
To identify a high-copy-number suppressor that restores respiratory growth of coa4Δ cells, we transformed a high-copy-number DNA library into the null strain and recovered respiratory-competent clones from 56,000 transformants. Twenty clones contained a DNA fragment encompassing the gene CYC1 encoding the soluble electron carrier cytochrome c. CYC1 as well as CYC7 were subcloned into pRS425 and transformed into coa4Δ cells. Cyc1 is the dominant cytochrome c present in aerobic yeast, whereas Cyc7 is an isoform expressed in hypoxic cells (11). Overexpression of either Cyc1 or Cyc7 in coa4Δ cells restored respiratory growth at room temperature and 30°C (Fig. 6A), as well as CcO activity (Fig. 6B). Restoration of the CcO:bc1 supercomplexes was also observed with overexpression of CYC1 or CYC7 in coa4Δ cells (Fig. 6C).
Cytochrome c is known to be important for the assembly and stability of CcO in yeast and mammals (5, 44), although the mechanism remains to be elucidated. To assess whether Cyc1 is deficient in coa4Δ cells, mitochondria isolated from wild-type and coa4Δ cells were subjected to SDS-PAGE for immunoblot analysis and heme staining. Cyc1 protein levels were found to be normal in coa4Δ cells by immunoblotting (Fig. 6D). In addition, heme staining revealed similar levels of hemylated Cyc1 (Fig. 6E) as well as Cyt1, which is a component of the bc1 complex (data not shown).
Cyc1 suppression of coa4Δ cells.
The suppressor activity of Cyc1 in coa4Δ cells may arise from impaired binding to Cox2 within the CcO complex. Near the interface where Cyc1 docks on Cox2 lies the Cox12 subunit that contains a twin CX9C motif analogous to Coa4. Since cox12 mutant cells have impaired CcO activity (21), we questioned whether Cox12 insertion into CcO was impaired in coa4Δ cells. Overexpression of COX12 failed to restore respiratory growth in coa4Δ cells (data not shown).
Cyc1 binding to Cox2 is mediated by electrostatic interactions arising from a constellation of basic residues on Cyc1 and carboxylates on Cox2 (26). In the interaction of Cyc1 and the bc1 complex, oppositely charged residue pairs contribute to long-range coulombic interactions (34). A series of Lys→Glu charge reversal mutations were engineered in Cyc1 to test whether attenuation in this interaction alters the suppressor activity of Cyc1. Two different double mutants carrying K78E K79E and K92E K93E and a quadruple mutant containing the four Lys→Glu substitutions were tested for suppressor activity in coa4Δ cells as well as Cyc1 function in cyc1Δ cyc7Δ double null cells (Fig. 7A). Addition of the quad Cyc1 mutant to the cyc1Δ cyc7Δ double mutant failed to restore respiratory growth, but suppressor activity in coa4Δ cells was only partially impaired (Fig. 7A). The different Cyc1 mutants were all expressed as stable proteins and could be observed by steady-state immunoblotting (Fig. 7C). Expression of a second Cyc1 mutant (the W65S mutant) that impairs Cyc1 function (5, 44) in coa4Δ cells failed to restore respiratory growth (data not shown), although interpretation of this result is complicated by the reported inefficient mitochondrial import of this mutant protein (5, 44). Whereas Cyc1 and Cyc7 were efficient suppressors of the respiratory defect of coa4Δ cells, neither protein alone upon overexpression facilitated respiratory growth of the Shy1 G137E mutant cells (Fig. 7B).
FIG. 7.
Charge reversal mutations in cytochrome c retain residual suppressor activity for coa4Δ cells but not cyc1Δ cyc7Δ mutant cells. (A) Respiratory growth of the BY4743 wild-type and coa4Δ cells transformed with episomal vectors expressing wild-type and mutant forms of CYC1 or wild-type CYC7 was analyzed as described in the legend to Fig. 1. (B) Respiratory growth of the BY4743 wild-type and shy1Δ cells transformed with episomal vectors expressing wild-type CYC1 and CYC7. (C) Expression of the wild-type and mutant forms of Cyc1 in whole-cell extracts assessed by immunoblotting. Whole-cell proteins were precipitated with 20% trichloroacetic acid (TCA) and resolved on 15% SDS-PAGE. Pgk1 was used as a loading control.
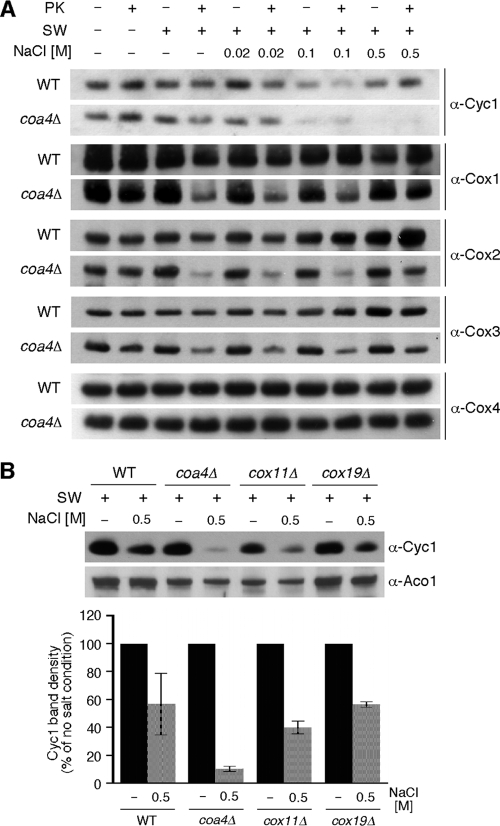
To gain insights into the mechanism by which Cyc1 restores CcO function in coa4Δ cells, we sought to investigate the membrane association of Cyc1 and stability of CcO in WT versus coa4Δ cells. Cyc1 is associated with the inner membrane by forming transient encounter complexes with bc1 and CcO, primarily by electrostatic interactions in addition to hydrophobic contacts with the lipid bilayer that involve cardiolipin (36, 42, 43). First, mitoplasts isolated from each cell were treated with increasing salt concentrations or proteinase K, and membrane association of Cyc1 was assessed by immunoblotting (Fig. 8A). High salt is known to partially dissociate Cyc1 from mitochondrial membranes. Cyc1 membrane association is attenuated in coa4Δ cells relative to WT cells in a salt-dependent fashion (Fig. 8A, top strips). Since the diminution in CcO may contribute to the observed salt-dependent loss of membrane-associated Cyc1 in cells lacking Coa4, we also compared the membrane association of Cyc1 in two other CcO mutants that lack any assembled CcO (Fig. 8B). Cells lacking Cox11 or Cox19 fail to assemble any CcO complex and yet retain appreciable Cyc1 levels in mitoplasts treated with 0.5 M NaCl, unlike coa4Δ cells. Second, mitoplasts were also treated with proteinase K to assess the stability of CcO in WT and coa4Δ cells in the BY4743 background. The addition of proteinase K led to a greater diminution in Cox1 and Cox2 in the mutant cells relative to WT cells. The apparent proteolytic instability of CcO in mutant cells may arise from an effect of Coa4 on complex IV stability or from the destabilization of bc1:CcO supercomplexes. However, the attenuation in membrane association of Cyc1 in coa4Δ cells suggests that Coa4 may have a role in the membrane stabilization of Cyc1, which is likely important for its additional role in CcO assembly.
FIG. 8.
Association of cytochrome c with the inner mitochondrial membrane and CcO stability are decreased in coa4Δ mitochondria. (A) Mitochondria isolated from wild-type and coa4Δ cells were osmotically shocked (SW+) or left intact (SW−). Swollen mitochondria were incubated with NaCl at indicated concentrations for 20 min on ice. Then, all samples were incubated with or without PK as described in the legend to Fig. 4D. Following the incubations, organelles were fractionated, and obtained pellets subjected to SDS-PAGE and analyzed by Western blotting using antisera against Cyc1, Cox1, Cox2, Cox3, and Cox4. Note that the exposure time for the immunoblots with coa4Δ cells was longer due to the reduced steady-state levels of CcO subunits. (B) Mitochondria (50 μg) from BY4743 wild-type, coa4Δ, cox11Δ, and cox19Δ cells were treated as described above and subjected to SDS-PAGE followed by immunoblotting with anti-Cyc1 and anti-Aco1. The bar graph showing the abundances of the membrane-bound Cyc1, retained after the salt treatment of wild-type, coa4Δ, cox11Δ, or cox19Δ mitoplasts. Quantifications were done using Image J software; the error bars indicate SD; n = 2.
DISCUSSION
Studies of three missense SURF1 mutations identified in patients with LS were evaluated with the yeast Shy1 protein. The G137E substitution in Shy1 results in a nonfunctional protein conferring a CcO deficiency. The G137E Shy1 mutation also recapitulates other SHY1 deletion phenotypes, including inefficient heme a insertion and low mitochondrial copper. The inefficient heme a insertion activity likely arises from the destabilization of the ∼450-kDa Shy1/Cox1 assembly intermediate. The lack of a robust CcO deficiency of the F249T and Y344D mutations suggests that the human protein is less tolerant of variations than the yeast protein. Although the F249T Shy1 mutant shows near normal CcO assembly in yeast, the steady-state levels of the Shy1/Cox1 assembly intermediate are partially attenuated. This attenuation of the complex may have a more significant impact on CcO assembly in human LS patients.
The loss of function in the G137E Shy1 mutant created an opportunity for a genetic suppressor screen focusing on allele-specific suppressors that would shed further insights on the Shy1 function in CcO biogenesis and potentially provide information on protein interactions. An abnormal protein can at times be restored to greater functionality by changes in an interacting partner. Presently, COA2, COA4, and COX10 were found to be allelic suppressors of the G137E Shy1 mutant.
Coa4 is a newly identified CcO assembly factor. Its initial characterization was recently reported in a comprehensive study of the 14 twin CX9C motif proteins in yeast (23). As with the other twin CX9C proteins, Coa4 is an IMS protein whose import is dependent on the Mia40 machinery (23). Cells lacking Coa4 in two distinct genetic backgrounds have reduced CcO activity and steady-state protein levels but do not show a reproducible change in the bc1 complex. The respiratory defect is most pronounced at room temperature and minimal at 37°C. Mitochondrial translation of Cox1-Cox3 appears normal in coa4Δ cells. Many CcO assembly mutants are impaired in Cox1 synthesis, but coa4Δ cells do not fall into that group. This is consistent with the lack of significant suppression of the respiratory defect by overexpression of MSS51 or other factors (COX10 and COA1) that stabilize newly synthesized Cox1. In addition, the Shy1/Cox1 assembly intermediate is wild type in coa4Δ cells. Thus, the CcO assembly/stability impairment in coa4Δ cells is not related to a defect in Cox1 maturation. No interaction of Coa4 and Shy1 or the bc1:CcO supercomplex was evident.
Cells lacking Coa4 resemble shy1Δ cells in exhibiting a reduced mitochondrial copper content. The matrix copper content was similarly reduced in these deletion strains as well as coa1Δ cells (∼50% of that of the wild type). Whereas copper supplementation enhanced respiratory growth of coa1Δ and shy1Δ cells, it was less effective in promoting respiratory growth of coa4Δ cells. We postulated previously that Coa1 and Shy1 may, in addition to their roles in Cox1 maturation, modulate Cu(I) import into the matrix or export to the IMS for CcO biogenesis (29). Coa4 may likewise be part of the Cu(I) routing pathway.
A major clue to the function of Coa4 was the isolation of Cyc1 as an extragenic suppressor of the respiratory defect of coa4Δ cells. Cyc1 is important for the assembly and/or stability of CcO (5, 44), and this is the first time that Cyc1 has been found as a suppressor of a CcO mutant. High-copy-number CYC1 is an efficient suppressor of coa4Δ cells but not Shy1 G137E mutant cells. Elevated Cyc1 was effective despite the lack of diminution in chromosomally encoded Cyc1 levels in coa4Δ cells. The suppressor activity of Cyc1 was attenuated but not eliminated by a quadruple Cyc1 mutant with four Lys→Glu substitutions in the docking pocket that affects interactions with Cox2. However, quadruple mutant Cyc1 fails to support respiratory growth in cells lacking WT Cyc1 and Cyc7.
The basis for the suppression of the coa4Δ cell respiratory phenotype by elevated Cyc1 may relate to an observed decrease in membrane association of Cyc1. Membrane association of Cyc1 occurs by electrostatic interactions of encounter complexes with bc1, CcO, and the lipid bilayer as well as hydrophobic effects with the bilayer (36, 42, 43). Cardiolipin contributes to the membrane anchoring of Cyc1 (36). The observed suppressive effect by Cyc1 may indicate that the phospholipid domain encompassing CcO is altered in composition in coa4Δ cells. Alternatively, other membrane proteins have been suggested to mediate the membrane association of Cyc1 with CcO (35). Either membrane lipid composition or a putative protein mediating Cyc1 docking may contribute to the known role of Cyc1 in CcO biogenesis and/or stability. There is no indication that Coa4 contributes directly to binding Cyc1, as no coimmunoprecipitation was observed between these two proteins. The attenuated CcO observed in coa4Δ cells, therefore, may arise from impaired stability of the complex that could have a marked impact on supercomplexes with bc1. Cardiolipin is important for Cyc1 membrane association as well as supercomplex formation, so if Coa4 had a role in the membrane lipid composition, the observed phenotypes of attenuated CcO and Cyc1 membrane association may be explained. Future studies will address whether Coa4 affects the membrane lipid composition of the IM in a way that would be important for CcO assembly.
In addition to COA4, COA2 and COX10 were also found to be allelic suppressors of the G137E Shy1 mutant. COA2 and COX10 were shown previously to be weak suppressors of the shy1Δ respiratory defect (30). Their suppression activity is enhanced by the presence of the G137E Shy1 mutant. Coa2 appears to have a role concurrent with Shy1 in the assembly of the CuB:heme a3 redox center (7). Although Coa2 and Shy1 transiently interact (30), the mechanism of suppression by Coa2 of the G137E Shy1 mutant is unclear. Coa2 is membrane associated on the matrix side of the IM, whereas the Gly137 residue exists within the IMS domain of Shy1. Since the G137E Shy1 mutant fails to stably form the ∼450-kDa Cox1 assembly intermediate visualized by BN-PAGE, high levels of Coa2 may enhance respiratory growth by stabilizing this important assembly intermediate, permitting CuB:heme a3 site formation. Alternatively, the G137E Shy1 mutant may stabilize Coa2, enhancing its Cox1 chaperone activity.
The allelic suppression by COX10 may occur somewhat differently, as no evidence exists for a physical interaction between Cox10 and Shy1. Cox10 functions in the first step in heme a biosynthesis and Shy1 functions in the hemylation of Cox1, so the intersection of Cox10 and Shy1 is rational. High levels of Cox10 may enhance the hemylation of Cox1 that may be compromised in the G137E Shy1 mutant cells. The catalytic function of Cox10 appears linked to its homo-oligomerization (7). We demonstrated that a N196K substitution in Cox10 bypassed the inefficient hemylation of Cox1 in cells lacking Coa2 through enhanced stability of the Cox10 oligomer (7). The G137E Shy1 mutant may likewise contribute in some way to Cox10 oligomerization, a process that is markedly impaired in shy1Δ cells. Another potential mechanism may relate to the recent observation that bacterial Surf1 is a heme-binding protein (10). An H193A Surf1 mutant had reduced heme binding. Mutations of either of the two conserved His residues (H141 and H341) in yeast Shy1 resulted in no defect in Shy1 function, and cells harboring a double His substitution showed only a modest growth impairment on glycerol-lactate (data not shown). Thus, it is unlikely that the G137E Shy1 suppression by Cox10 relates to heme binding to Shy1.
In summary, characterization and allelic suppresion studies of the Shy1 G137E mutation have led to a better understanding of the CcO assembly pathway and the factors involved in this process. A novel assembly factor, Coa4, was identified through these studies and found to be genetically linked to the CcO assembly function of Cyc1.
Acknowledgments
This work was supported by grant from the National Institutes of Health ES03817 to D.R.W. M.B. was supported by predoctoral training grant T32 DK007115 and a University of Utah Graduate Research Fellowship. H.K. is supported by postdoctoral grant T32 DK007115.
We acknowledge the assistance of Pam Smith for ICP measurements of mitochondrial copper.
Footnotes
Published ahead of print on 12 July 2010.
REFERENCES
- 1.Antonicka, H., S. C. Leary, G. H. Guercin, J. N. Agar, R. Horvath, N. G. Kennaway, C. O. Harding, M. Jaksch, and E. A. Shoubridge. 2003. Mutations in COX10 result in a defect in mitochondrial heme a biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 12:2693-2702. [DOI] [PubMed] [Google Scholar]
- 2.Antonicka, H., A. Mattman, C. G. Carlson, D. M. Glerum, K. C. Hoffbuhr, S. C. Leary, N. G. Kennaway, and E. A. Shoubridge. 2003. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 72:101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrientos, A., K. Gouget, D. Horn, I. C. Soto, and F. Fontanesi. 2009. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta 1793:97-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrientos, A., D. Korr, and A. Tzagoloff. 2002. ShyI is necessary for full expression of mitochondrial Cox1 in the yeast model of Leigh's syndrome. EMBO J. 21:43-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrientos, A., D. Pierre, J. Lee, and A. Tzagoloff. 2003. Cytochrome oxidase assembly does not require catalytically active cytochrome c. J. Biol. Chem. 278:8881-8887. [DOI] [PubMed] [Google Scholar]
- 6.Barrientos, A., A. Zambrano, and A. Tzagoloff. 2004. Mss51 and Cox14 jointly regulate mitochondrial Cox1 expression in Saccharomyces cerevisiae. EMBO J. 23:3472-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bestwick, M., O. Khalimonchuk, F. Pierrel, and D. R. Winge. 2010. The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell. Biol. 30:172-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brown, R. M., and G. K. Brown. 1996. Complementation analysis of systemic cytochrome oxidase deficiency presenting as Leigh syndrome. J. Inherit. Metab. Dis. 19:752-760. [DOI] [PubMed] [Google Scholar]
- 10.Bundschuh, F. A., A. Hannappel, O. Anderka, and B. Ludwig. 2009. Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. J. Biol. Chem. 284:25735-25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke, P. V., D. C. Raitt, L. A. Allen, E. A. Kellogg, and P. O. Poyton. 1997. Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J. Biol. Chem. 272:14705-14712. [DOI] [PubMed] [Google Scholar]
- 12.Capaldi, R. A., M. F. Marusich, and J. W. Taanman. 1995. Mammalian cytochrome c oxidase: characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Methods Enzymol. 260:117-132. [DOI] [PubMed] [Google Scholar]
- 13.Diekert, K., A. I. De Kroon, G. Kispal, and R. Lill. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65:37-51. [DOI] [PubMed] [Google Scholar]
- 14.Fernández-Vizarra, E., V. Tiranti, and M. Zeviani. 2009. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta 1793:200-211. [DOI] [PubMed] [Google Scholar]
- 15.Fontanesi, F., C. Jin, A. Tzagoloff, and A. Barrientos. 2008. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum. Mol. Genet. 17:775-788. [DOI] [PubMed] [Google Scholar]
- 16.Fontanesi, F., I. C. Soto, D. Horn, and A. Barrientos. 2006. Assembly of mitochondrial cytochrome c oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 291:C1129-C1147. [DOI] [PubMed] [Google Scholar]
- 17.Horng, Y. C., S. C. Leary, P. A. Cobine, F. B. Young, G. N. George, E. A. Shoubridge, and D. R. Winge. 2005. Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 280:34113-34122. [DOI] [PubMed] [Google Scholar]
- 18.Khalimonchuk, O., M. Bestwick, B. Meunier, T. C. Watts, and D. R. Winge. 2010. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell. Biol. 30:1004-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalimonchuk, O., A. Bird, and D. R. Winge. 2007. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282:17442-17449. [DOI] [PubMed] [Google Scholar]
- 20.Khalimonchuk, O., and D. R. Winge. 2008. Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase. Biochim. Biophys. Acta 1783:618-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaMarche, A. E., M. I. Abate, S. H. Chan, and B. L. Trumpower. 1992. Isolation and characterization of COX12, the nuclear gene for a previously unrecognized subunit of Saccharomyces cerevisiae cytochrome c oxidase. J. Biol. Chem. 267:22473-22480. [PubMed] [Google Scholar]
- 22.Leary, S. C., P. A. Cobine, B. A. Kaufman, G. H. Guercin, A. Mattman, J. Palaty, G. Lockitch, D. R. Winge, P. Rustin, R. Horvath, and E. A. Shoubridge. 2007. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. 5:9-20. [DOI] [PubMed] [Google Scholar]
- 23.Longen, S., M. Bien, K. Bihlmaier, C. Kloeppel, F. Kauff, M. Hammermeister, B. Westermann, J. M. Herrmann, and J. Riemer. 2009. Systematic analysis of the twin Cx(9)C protein family. J. Mol. Biol. 393:356-368. [DOI] [PubMed] [Google Scholar]
- 24.Mashkevich, G., B. Repetto, D. M. Glerum, C. Jin, and A. Tzagoloff. 1997. SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J. Biol. Chem. 272:14356-14364. [DOI] [PubMed] [Google Scholar]
- 25.Mick, D. U., K. Wagner, M. van der Laan, A. E. Frazier, I. Perschil, M. Pawlas, H. E. Meyer, B. Warscheid, and P. Rehling. 2007. ShyI couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26:4347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millett, F., C. de Jong, L. Paulson, and R. A. Capaldi. 1983. Identification of specific carboxylate groups on cytochrome c oxidase that are involved in binding cytochrome c. Biochemistry 22:546-552. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Martinez, X., S. A. Broadley, and T. D. Fox. 2003. Mss51 promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1. EMBO J. 22:5951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piekutowska-Abramczuk, D., M. Magner, E. Popowska, M. Pronicki, E. Karczmarewicz, J. Sykut-Cegielska, T. Kmiec, E. Jurkiewicz, T. Szymanska-Debinska, L. Bielecka, M. Krajewska-Walasek, K. Vesela, J. Zeman, and E. Pronicka. 2009. SURF1 missense mutations promote a mild Leigh phenotype. Clin. Genet. 76:195-204. [DOI] [PubMed] [Google Scholar]
- 29.Pierrel, F., M. L. Bestwick, P. A. Cobine, O. Khalimonchuk, J. A. Cricco, and D. R. Winge. 2007. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26:4335-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierrel, F., O. Khalimonchuk, P. A. Cobine, M. Bestwick, and D. R. Winge. 2008. Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis facilitating the maturation of Cox1. Mol. Cell. Biol. 28:4927-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poyau, A., K. Buchet, M. F. Bouzidi, M. T. Zabot, B. Echenne, J. Yao, E. A. Shoubridge, and C. Godinot. 2000. Missense mutations in SURF1 associated with deficient cytochrome c oxidase assembly in Leigh syndrome patients. Hum. Genet. 106:194-205. [DOI] [PubMed] [Google Scholar]
- 32.Shoubridge, E. A. 2001. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106:46-52. [DOI] [PubMed] [Google Scholar]
- 33.Smith, D., J. Gray, L. Mitchell, W. E. Antholine, and J. P. Hosler. 2005. Assembly of cytochrome c oxidase in the absence of assembly protein Surf1 leads to loss of the active site heme. J. Biol. Chem. 280:17652-17656. [DOI] [PubMed] [Google Scholar]
- 34.Solmaz, S. R., and C. Hunte. 2008. Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J. Biol. Chem. 283:17542-17549. [DOI] [PubMed] [Google Scholar]
- 35.Spaar, A., D. Flock, and V. Helms. 2009. Association of cytochrome c with membrane-bound cytochrome c oxidase proceeds parallel to the membrane rather than in bulk solution. Biophys. J. 96:1721-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speck, S. H., C. A. Neu, M. S. Swanson, and E. Margoliash. 1983. Role of phospholipid in the low affinity reactions between cytochrome c and cytochrome oxidase. FEBS Lett. 164:379-382. [DOI] [PubMed] [Google Scholar]
- 37.Stiburek, L., K. Vesela, H. Hansikova, H. Hulkova, and J. Zeman. 2009. Loss of function of Sco1 and its interaction with cytochrome c oxidase. Am. J. Physiol. Cell Physiol. 296:C1218-C1226. [DOI] [PubMed] [Google Scholar]
- 38.Stiburek, L., K. Vesela, H. Hansikova, P. Pecina, M. Tesarova, L. Cerna, J. Houstek, and J. Zeman. 2005. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 392:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teraoka, M., Y. Yokoyama, S. Ninomiya, C. Inoue, S. Yamashita, and Y. Seino. 1999. Two novel mutations of SURF1 in Leigh syndrome with cytochrome c oxidase deficiency. Hum. Genet. 105:560-563. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 41.Tiranti, V., K. Hoertnagel, R. Carrozzo, C. Galimberti, M. Munaro, M. Granatiero, L. Zelante, P. Gasparini, R. Marzella, M. Rocchi, M. P. Bayona-Bafaluy, J. A. Enriquez, G. Uziel, E. Bertini, C. Dionisi-Vici, B. Franco, T. Meitinger, and M. Zeviani. 1998. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 63:1609-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuominen, E. K., C. J. Wallace, and P. K. Kinnunen. 2002. Phospholipid-cytochrome c interaction: evidence for the extended lipid anchorage. J. Biol. Chem. 277:8822-8826. [DOI] [PubMed] [Google Scholar]
- 43.Tuominen, E. K., K. Zhu, C. J. Wallace, I. Clark-Lewis, D. B. Craig, M. Rytomaa, and P. K. Kinnunen. 2001. ATP induces a conformational change in lipid-bound cytochrome c. J. Biol. Chem. 276:19356-19362. [DOI] [PubMed] [Google Scholar]
- 44.Vempati, U. D., X. Han, and C. T. Moraes. 2009. Lack of cytochrome c in mouse fibroblasts disrupts assembly/stability of respiratory complexes I and IV. J. Biol. Chem. 284:4383-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams, S. L., I. Valnot, P. Rustin, and J. W. Taanman. 2004. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 279:7462-7469. [DOI] [PubMed] [Google Scholar]
- 46.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1:418-428. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, Z., J. Yao, T. Johns, K. Fu, I. De Bie, C. Macmillan, A. P. Cuthbert, R. F. Newbold, J. Wang, M. Chevrette, G. K. Brown, R. M. Brown, and E. A. Shoubridge. 1998. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20:337-343. [DOI] [PubMed] [Google Scholar]