Abstract
Both in mice and humans, two major SPO11 isoforms are generated by alternative splicing: SPO11α (exon 2 skipped) and SPO11β. Thus, the alternative splicing event must have emerged before the mouse and human lineages diverged and was maintained during 90 million years of evolution, arguing for an essential role for both isoforms. Here we demonstrate that developmental regulation of alternative splicing at the Spo11 locus governs the sequential expression of SPO11 isoforms in male meiotic prophase. Protein quantification in juvenile mice and in prophase mutants indicates that early spermatocytes synthesize primarily SPO11β. Estimation of the number of SPO11 dimers (ββ/αβ/αα) in mutants in which spermatocytes undergo a normal number of double strand breaks but arrest in midprophase due to inefficient repair argues for a role for SPO11β-containing dimers in introducing the breaks in leptonema. Expression kinetics in males suggested a role for SPO11α in pachytene/diplotene spermatocytes. Nevertheless, we found that both alternative transcripts can be detected in oocytes throughout prophase I, arguing against a male-specific function for this isoform. Altogether, our data support a role for SPO11α in mid- to late prophase, presumably acting as a topoisomerase, that would be conserved in male and female meiocytes.
Meiotic recombination is initiated at the onset of prophase I by the introduction of SPO11-dependent double strand breaks (DSBs) throughout the genomic DNA (20). These breaks are repaired by homologous recombination, with the eukaryotic recombinases RAD51 and DMC1 catalyzing the invasion and strand exchange reaction between nonsister chromatids on homologous chromosomes, the earliest steps toward the generation of crossovers. Disruption of Spo11 in mice results in male and female infertility (8, 31). SPO11-deficient spermatocytes are unable to generate DSBs, their homologous chromosomes fail to recombine and synapse, and they undergo massive apoptosis in mid-prophase I. Spermatocytes carrying mutations in several genes required for early processing of meiotic DSBs, such as Dmc1, Hop2, and Atm, are also arrested in midprophase (2, 4, 27, 28), while those deficient in processing late recombination intermediates, such as an Mlh1 mutant, complete prophase but are arrested in metaphase I (1, 14).
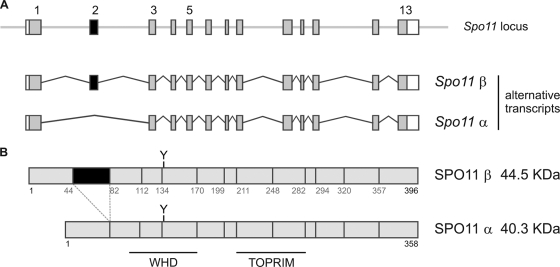
In Saccharomyces cerevisiae, where SPO11 has been studied most extensively, the Spo11 gene is intronless (as are 95% of all loci in this organism), whereas in the mouse it spans 13 exons and can generate two major SPO11 isoforms by alternative splicing (30, 31) (Fig. 1). The longer transcript, including all 13 exons, is translated into the SPO11β isoform (44.5 kDa), while skipping of exon 2 results in a 12-exon transcript that is translated into the smaller isoform, SPO11α (40.3 kDa). Both alternative mRNAs include exon 5, which codes for the catalytic tyrosine essential for DSB formation (10; M. Bellani and R. D. Camerini-Otero, unpublished data). Hence, both isoforms might be capable of introducing breaks.
FIG. 1.
Scheme of the mouse Spo11 locus and the alternative transcripts/polypeptides for SPO11α and -β isoforms. (A) Mouse Spo11 locus and alternative transcripts for Spo11α and -β. The Spo11β transcript includes all 13 exons, whereas the Spo11α transcript excludes exon 2. (B) SPO11α and -β polypeptides. The region encoded by exon 2 is in black, and the catalytic tyrosine encoded by exon 5 is indicated by Y. The regions encompassing the WHD and TOPRIM domains are also shown.
The alternative splicing giving rise to SPO11α and -β isoforms in the mouse is conserved in the orthologous gene in humans (30) (Ensembl; corroborated by quantitative PCR [qPCR] in this work). This indicates that the alternative form emerged before the mouse and human lineages diverged, 90 million years ago, and that the alternative exon remained fixed during evolution. This conservation strongly suggests that there are functional roles for both isoforms (21), yet to date, hardly any progress has been made toward understanding the function and relevance of each isoform.
There are two published reports that have addressed to some extent the existence of alternatively spliced SPO11 isoforms in the mouse. Our lab showed that some meiotic mutants (Dmc1, Morc, and Mei1 mutants) contained mainly Spo11β transcripts, yet it must be noted that this was a nonquantitative reverse transcription-PCR (RT-PCR) assessment (31). Later, Neale et al. reported detection of exclusively the SPO11β polypeptide in Dmc1−/− testis extracts (24). A minor fraction of SPO11β isoforms were covalently attached to a DNA oligonucleotide, suggesting that the SPO11β isoform could introduce DSBs in the absence of SPO11α. Nevertheless, there is no information on the kinetics of expression of each isoform during wild-type meiosis, and no attempt has been made to clarify the need for a second isoform, if indeed SPO11β is the one introducing the breaks.
In plants, where there is limited alternative splicing, most species exhibit extensive whole or partial genome duplication events. These gene duplications can presumably satisfy the requirement for diversification in the absence of widespread alternative splicing (21). Arabidopsis thaliana carries three Spo11 genes, two of which, Spo11-1 and Spo11-2, are expressed during meiosis. They are both essential for meiotic recombination to occur (18), suggesting that in higher eukaryotes two different SPO11 proteins might be required to proceed through meiosis I. In mammals, the alternatively spliced isoforms α and β might fulfill the role these two gene products play in plants.
Based on the structure of its archaeal homolog, topoisomerase VIA, SPO11 is predicted to form a dimer, containing two bipartite active sites that can catalyze the concerted scission of the two DNA strands to generate a DSB (25). The region encoded by exon 2 does not include any of the residues involved in the monomer-monomer interphase of the dimer (25); hence, in principle the SPO11α and -β isoforms could form either homodimers (α/α and β/β) or heterodimers (α/β). One possibility is that one of the isoforms is the active endonuclease, while the other plays a regulatory role, sequestering the active isoform in an inactive or incorrectly localized heterodimer. On the other hand, the structure of the full-length topoisomerase VI (A2B2 tetramer) suggests that differences in the N termini of SPO11α and -β could influence the ability of SPO11 dimers to interact with other proteins (13), so alternative partners could dictate specific localization or activities for these complexes. In order to evaluate these possibilities and shed some light on the function and relevance of SPO11 isoforms in mammalian meiosis, we have characterized the kinetics of expression of SPO11.
We report distinct sequential expression kinetics for the SPO11β and -α isoforms in mouse spermatocytes undergoing prophase. SPO11β is the major isoform expressed in early spermatocytes, consistent with SPO11β introducing the breaks that initiate meiotic recombination at the onset of prophase I. SPO11α becomes prevalent after spermatocytes progress past midprophase, when meiotic DSBs have already been repaired, suggesting an alternative role for this isoform in pachytene/diplotene spermatocytes. The fact that both splicing variants can be detected in oocytes undergoing prophase argues against a male-specific role for SPO11α. We propose a topoisomerase function for SPO11α in mid- to late prophase, which would be conserved in male and female meiocytes.
MATERIALS AND METHODS
Mice.
Mice used in this study were as follows: Spo11 knockout (KO) (31), Dmc1 KO (28), Hop2 KO (27), Atm KO (5), and Mlh1 KO (1).
Quantification of Spo11 transcripts by qPCR.
Testes were homogenized in Trizol reagent (Invitrogen) using an Omni International power homogenizer, flash frozen, and stored at −70°C. Total RNA was isolated using Trizol as indicated by the manufacturer and was further purified using a total RNA isolation kit (Agilent Technologies, Inc.). Total RNA was quantified by measuring absorbance at 260 nm, and RNA quality was assessed on an Agilent 2100 bioanalyzer. Prior to cDNA synthesis, samples were treated with amplification grade DNase (Invitrogen). Reverse transcription was done using the Superscript II first-strand synthesis system for RT-PCR (Invitrogen). The resulting cDNA was used as a template for qPCRs using TaqMan probes targeting specific exon boundaries: exon 11-12 boundary (total Spo11), exon 1-3 boundary (Spo11α), or exon 2-3 boundary (Spo11β). Real-time PCRs were done in quadruplicate in an Applied Biosystems 7500. The comparative threshold cycle (CT) method of relative quantification (ΔΔCT method) was used to compare the amounts of transcripts in a certain sample and another sample used as a calibrator. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an endogenous control. Efficiencies of the three TaqMan assays were assessed by generating calibration curves (CT versus quantity of cDNA) spanning 7 logs (10 ng to 10 fg) of template (serially diluted Spo11α or -β cDNA). The efficiencies for the TaqMan assays for Spo11α, Spo11β, total Spo11, and GAPDH are 92%, 93%, 88.5%, and 98%, respectively. The sequences of the primers/probes used in the different TaqMan assays can be provided upon request. Regarding the use of GAPDH as the endogenous control, it displayed fairly constant CT values (18.2 to 19.2) in testes from mice at 1 to 18 days postpartum (dpp), whereas CT values for total Spo11, Spo11α, and Spo11β spanned 6, 8, and 6 CTs, respectively. This rules out the possibility that the increase in Spo11 transcripts observed might be an artifact due to decreasing GAPDH transcripts as prophase I progresses. In fact, the bar graphs with and without normalization to GAPDH are comparable.
In the case of enriched cell populations, cells were spun down and resuspended in the lysis buffer of the RNeasy Plus microkit (Qiagen). RNA was isolated by following the manufacturer's instructions. For reverse transcription we used the Omniscript RT kit (Qiagen). Fetal ovaries were lysed and processed with the RNeasy Plus microkit (Qiagen) by following the manufacturer's instructions. For reverse transcription we used the Omniscript RT kit (Qiagen).
Ho33342/FACS to generate cell populations enriched in spermatocytes undergoing certain stages of prophase I.
Isolation of spermatogonia (Sg) and spermatocytes was done according to reference 7. Fluorescence-activated cell sorting (FACS) was performed on a FACSVantage SE (Becton Dickinson) (351-nm UV laser, dichroic mirror [610SP filter], 630/22 filter). Spermatogonia were identified based on their characteristic side population (SP) phenotype (6, 7). Spermatocytes were sorted into four populations. An aliquot of each cell population (10,000 cells) was used to prepare spreads that were immunostained with antibodies against SCP3 and γH2AX, and the remaining cells were resuspended in lysis buffer (RNeasy Plus microkit; Qiagen), flash frozen, and stored at −70°C until processed for RNA isolation. Between 150 and 250 cells in spreads from each cell population were scored and classified as preleptotene, leptotene, zygotene, early or late pachytene, or diplotene spermatocytes (according to axial/later element characteristics and γH2AX staining) in order to estimate the percentages of each type of spermatocytes in the different cell populations. Spermatocyte spreads were prepared as described previously (9). Immunostaining for spermatocyte stage classification was done as described previously (9).
Isolation of fetal ovaries carrying oocytes in the successive stages of prophase I.
We isolated ovaries from embryos (embryonic day 15.5 [E15.5], E16, E17, E18, E19, and E20) and newborn female mice, flash froze them in liquid nitrogen, and stored them at −70. Between 6 and 12 ovaries were pooled for each embryonic time point. One ovary from each time point was used to prepare oocyte spreads (26). Staining with antibodies against SCP3 and γH2AX permitted classification of oocytes into the different stages of prophase, which in turn was used to stage each pool of ovaries as specified in Fig. 6. Spo11α and -β mRNA kinetics were confirmed by analyzing a second independent set of embryos.
FIG. 6.
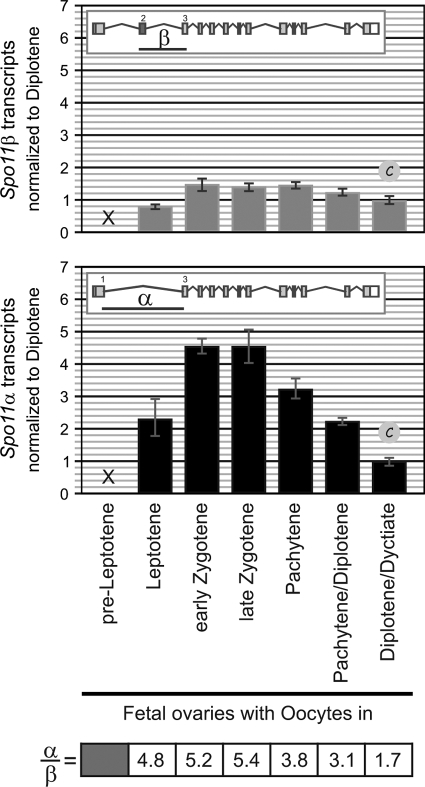
Oocytes synthesize Spo11α and -β transcripts during prophase I. TaqMan assays targeting isoform-specific exon junctions were used to quantify Spo11α and -β transcripts by RT-qPCR in fetal ovaries (E14.5 to E20/newborn females) carrying oocytes in the successive stages of prophase I. Results were normalized to those for the diplotene/dictyate oocyte sample. The table shows the relative ratios of mRNA levels (α/β).
Isolation of GV and metaphase II (MII) oocytes from hormonally stimulated females.
Ten sexually mature C57BL/6 females were injected with 5 IU pregnant mare's serum gonadotropin (PMSG), and 48 h later, ovarian follicles were punctured to release oocytes. Around 150 oocytes were collected in M2 media, pooled together, and flash frozen. Differential interference contrast (DIC) microscopy, as well as fixing and staining with propidium iodide (DNA) and antitubulin antibodies, confirmed they were germinal vesicle (GV) oocytes. GV oocytes are committed to resume meiosis and have grown in size 70-fold during follicle maturation.
Ten sexually mature C57BL/6 females were injected with 5 IU PMSG and 48 h later were injected with 5 IU human chorionic gonadotropin (hCG) to induce ovulation. Twelve hours later, a total of 150 metaphase II ovulated oocytes were collected from the oviducts onto M2 media, pooled together, and flash frozen. A subset was fixed and stained with propidium iodide and antitubulin antibodies, and observation of the extruded first polar body under DIC, plus the metaphase II spindle by immunofluorescence, confirmed they were metaphase II oocytes.
Oocytes were lysed and processed with an RNeasy Plus microkit (Qiagen), and total RNA was isolated by following the manufacturer's instructions. For reverse transcription we used the Omniscript RT kit (Qiagen). Upon qPCR analysis, GAPDH transcript levels in GV and MII oocytes were comparable to those of prophase I oocytes, yet Spo11α and -β transcripts were undetectable.
Cot1-RNA FISH combined with immunostaining.
Cot-1 RNA fluorescent in situ hybridization (FISH) labels all nascent RNA transcripts, and the intensity of the signal is correlated with overall transcription rate. Cot-1 RNA FISH was carried out according to reference 35 with few modifications. Briefly, after removal of the tunica albuginea, the seminiferous tubules were minced in 1× phosphate-buffered saline (PBS) containing a cocktail of protease inhibitors (Complete EDTA free; Roche). Large tissue pieces in the cell suspension were allowed to settle, and the supernatant was transferred to a new tube and spun down. The pelleted cells were resuspended in a minimal volume of hypotonic solution (30 mM Tris-HCl, 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, pH 8.2) for 10 min at 37°C. Cells were then resuspended in an equal volume of 100 mM sucrose, pH 8.2. Approximately 40 μl of this suspension was dropped on boiled slides and allowed to attach to the slides for 10 min at 4°C. Slides were covered in cold CSK buffer [100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES {piperazine-N,N′-bis(2-ethanesulfonic acid)}, 0.5% Triton X-100, 1 mM EGTA, and 2 mM vanadyl ribonucleoside (pH 6.8)] for 10 min, and then in cold 4% paraformaldehyde (pH 7 to7.4) for 10 min. Slides were then rinsed in 1× PBS, dehydrated through an ethanol series (twice at 70%, 80%, 96%, and 100%), and air dried. Biotinylated Cot-1 probes were prepared using the biotin nick translation kit (Roche). Hybridization reaction mixtures consisted of 0.4 mg of labeled Cot-1 and 10 mg of salmon sperm DNA in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10% dextran sulfate, 50% formamide, 1 mg/ml bovine serum albumin, and 2 mM vanadyl ribonucleoside, and reactions were carried out in humid chambers for 3 h at 37°C. We then subjected slides to stringency washes at 42°C (three washes with 2× SSC and 50% formamide and three washes with 2× SSC) and transferred them to 4× SSC and 0.1% Tween 20. Slides were blocked in 4× SSC, 4 mg/ml bovine serum albumin, and 0.001% Tween 20, for 30 min at 37°C. Probe detection was carried out using streptavidin-AF488 (1:100), followed by amplification using biotinylated antibody to streptavidin (1:150) and then one further round of streptavidin-AF488 (1:100). Each of these steps involved incubation for 30 min at 37°C and three washes for 2 min each in 4× SSC and 0.1% Tween 20.
Generation of a rabbit monoclonal antibody against SPO11 (MB36-4).
Insoluble His-tagged SPO11β, purified from inclusion bodies in guanidine hydrochloride using a nickel column, was used to immunize two prescreened rabbits (Covance). Protein extracts from HEK293T cells transiently transfected with two plasmids expressing hemagglutinin (HA)-murine SPO11α (mSPO11α) and HA-mSPO11β were used for all the screenings. First, we identified a rabbit antiserum capable of immunoprecipitating overexpressed HA-SPO11α and -β from transiently transfected HEK293T cells. After the last bleed was collected, the spleen from the selected rabbit was sent to Epitomics (Burlingame, CA) to generate rabbit hybridomas. We subjected hybridoma supernatants to two screening rounds. Once we identified a hybridoma producing a monoclonal antibody (MB36-4) that could immunoprecipitate overexpressed HA-SPO11α and -β from cell extracts, it was subcloned, expanded, and subsequently cultured to produce the monoclonal antibody, which was purified on a protein A affinity column.
Immunoprecipitation of SPO11 from testis extracts.
Testes from one to four adult males (≥7 weeks old) were used for each immunoprecipitation. Testes were decapsulated, flash frozen in liquid nitrogen, and stored at −70°C until processed. Each testis sample (consisting of 2 wild-type or 4 to 8 mutant testes) was homogenized in 350 μl of lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], Complete protease inhibitor cocktail), sonicated in a Bioruptor (Diagenode, Belgium) twice for 30 s on high, and centrifuged at 10,000 × g for 5 min at 4°C. The supernatant was centrifuged a second time, and the resulting supernatant was incubated with anti-SPO11 antibody (MB36-4; 50 μg/sample) immobilized on M280 tosyl-activated magnetic beads (Invitrogen) at 4°C for 1 h. Beads were washed twice with lysis buffer and three times with wash buffer (1× PBS, 0.1% Triton X-100, 2 mM EDTA, Complete protease inhibitor cocktail). Beads were boiled in 30 μl of 1× NuPAGE loading buffer and fractionated on a 10% Bis-Tris NuPAGE gel. The protein was transferred to a polyvinylidene difluoride (PVDF) membrane in 10 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid; pH 11] and 10% methanol, probed with anti-SPO11 antibody (MB36-4, diluted 1:250), and detected with AF680 goat anti-rabbit IgG secondary antibody (diluted 1:10,000). The membranes were scanned and quantified using the Odyssey scanner (LI-COR) and associated software. The intensity levels were determined for the α and β bands in each lane, and the background fluorescence in the Spo11−/− lane was subtracted. Protein concentration in the extracts was determined by the Bradford method (Quickstart Bradford dye reagent; Bio-Rad).
Mass spectrometry analysis.
A wild-type testis extract was immunoprecipitated with an anti-SPO11 rabbit monoclonal antibody or nonspecific rabbit IgG and loaded onto two halves of a gel. Half of the gel was transferred to a membrane and analyzed by Western blotting with the antibody against SPO11. The bands on the Western blot were used to triangulate onto the silver-stained half of the gel so as to cut the corresponding regions in the lanes pertaining to the SPO11 pulldown and the IgG control and subject them to mass spectrometry analysis.
Estimation of the number of molecules of SPO11α and -β polypeptides per spermatocyte.
We estimated the amounts of SPO11α and -β polypeptides per mouse by referring the intensities of α and β bands in an immunoprecipitate/Western blot to that of a standard curve run in parallel consisting of serial dilutions of purified His-mSPO11β (1, 10, and 100 fmol). We calculated the number of molecules of each isoform per mouse, considering that 1 fmol of polypeptide contains 3.022 × 108 molecules. Finally, we divided the number of molecules of polypeptide per mouse by the number of primary spermatocytes in the testes of the corresponding mouse strain. In order to estimate the number of ββ, αβ, and αα dimers per spermatocyte, the proportion of each type of dimer was calculated as described in Fig. S2 in the supplemental material and multiplied by the total number of dimers ([no. of β molecules + no. of α molecules]/2).
To determine the number of primary spermatocytes per mouse, we prepared a testicular cell suspension, determined the total number of cells, fixed and stained the cell suspension with propidium iodide, and measured the proportions of 4N, 2N, and 1N cells by flow cytometry. The number of primary spermatocytes per mouse was calculated as the proportion of 4N cells multiplied by the total number of cells (see Fig. S3A in the supplemental material).
RESULTS
Total Spo11 transcripts peak in late prophase I, past the stage in which meiotic DSBs are introduced.
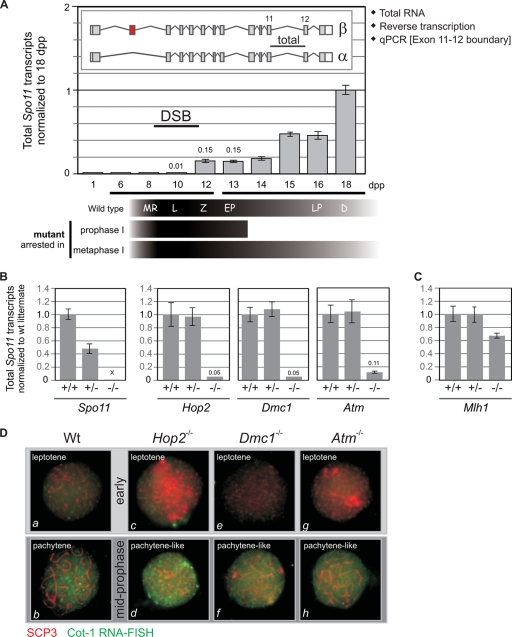
By studying prepuberal mice (6 to 18 days old), it is possible to analyze fairly synchronized spermatocytes as they progress through the first meiotic division (15). Hence, in order to define the kinetics of transcription of Spo11 during prophase I, we quantified Spo11 transcripts in testes of juvenile mice undergoing the first wave of spermatogenesis. Immunostaining of murine spermatocytes with antibodies against γH2AX (marker for DSBs) and RAD51 and DMC1 (markers for early meiotic recombination intermediates) has shown that SPO11-dependent DSBs are generated very early in prophase (leptonema) and that most of them are repaired before entering pachynema (22, 23). Therefore, Spo11 transcription would be expected to peak before or around leptonema, corresponding to 8 to 10 days postpartum (dpp) in juvenile mice. Yet the largest amount of Spo11 transcripts is detected at day 18, when the majority of germ cells are in late prophase, past the stage in which DSBs are introduced (Fig. 2 A). This result is unexpected in view of the only function currently accepted for SPO11, the introduction of DSBs that initiate meiotic recombination, as this would call for a peak of expression in early spermatocytes.
FIG. 2.
In wild-type mouse spermatocytes Spo11 transcripts peak past the stage in prophase in which meiotic DSBs are introduced. (A) Total Spo11 transcripts quantified in juvenile mice peak in late prophase I. Total Spo11 transcripts were quantified in testes of juvenile mice at 1 to 18 dpp. Total RNA was reverse transcribed and subjected to quantitative PCR (qPCR) using a TaqMan assay targeting the exon 11-12 boundary. Data were normalized to the 18-dpp sample. The scheme below the x axis indicates the predominant cell type corresponding to each age in wild-type juvenile mice, according to reference 15. Most prophase I mutants are arrested at stage IV (corresponding to early pachytene spermatocytes); therefore, their cell type composition would be comparable to that of a 13-dpp wild-type pup, whereas mutants arrested in metaphase I contain spermatocytes in all stages of prophase I. MR, meiotic replication; L, leptonema; Z, zygonema; EP, early pachynema; LP, late pachynema; D, diplonema. (B) Mutants arrested in prophase I display a 10-fold reduction in total Spo11 transcripts. Total Spo11 transcripts were quantified by reverse transcription-qPCR (RT-qPCR) in testes of several mutants arrested in prophase I. The ΔSpo11 strain was used as a negative control. Transcript levels for each homozygous mutant were normalized to those of its wild-type (wt) littermate. (C) Mutants arrested in metaphase I do not display the drastic reduction in total Spo11 transcript levels observed for prophase I mutants. Total Spo11 transcript levels in the testes of Mlh1−/− mutant mice were compared to those of a wild-type littermate. (D) The low levels of total Spo11 transcripts detected in mutants arrested in prophase I are not a reflection of lower overall transcription levels due to the arrest. Cot-1 DNA consists of repetitive elements ubiquitously represented throughout the genome that can be used to detect nascent RNA transcripts in the nucleus by RNA FISH. The intensity of the nuclear Cot-1 signal correlates to the rate of transcription. Combined Cot-1 RNA FISH and SCP3 immunostaining were performed on wild-type and mutant spermatocyte spreads to assess overall transcription levels in leptotene and more advanced spermatocytes. Spermatocytes were classified as early (a, c, e, and g) or pachytene-like (b, d, f, and h) according to the SCP3 staining pattern.
If late spermatocytes contain the highest levels of Spo11 transcripts in the testis, mutants arrested in midprophase and therefore lacking late spermatocytes would be expected to have greatly reduced total Spo11 transcripts. Indeed, we found that testes from several meiotic mutants arrested in midprophase contain 5 to 11% of the total Spo11 transcripts detected in wild-type littermates (Fig. 2B). On the other hand, mutants arrested in metaphase I, and thereby containing spermatocytes in all stages of prophase I, should have normal levels of Spo11 transcripts. Consistent with this prediction, an Mlh1−/− mutant, which is arrested in metaphase I (14), contains over 60% of the total Spo11 transcripts detected in a wild-type littermate (Fig. 2C).
Overall transcript levels are low at the beginning of prophase and significantly increase when spermatocytes enter pachynema (35) (Fig. 2D, a and b). To assess the possibility that mutants arrested in prophase might have an overall reduction in transcription, we compared nascent transcript levels in mutant and wild-type spermatocytes by Cot1-RNA fluorescent in situ hybridization (FISH). We found that mutant spermatocytes display low transcript levels in leptonema (Fig. 2D, c, e, and g) but reach high overall transcript levels, comparable to those of wild-type pachytene spermatocytes, in more-advanced stages (Fig. 2D, d, f, and h). Thus, we ruled out the possibility that the low total Spo11 transcript levels observed in prophase mutants reflect a shutting down of overall transcription as a consequence of the arrest.
Data presented in Fig. 2 demonstrate that, in wild-type testes, most of the Spo11 transcripts are generated in late prophase, past the stage in which meiotic DSBs are introduced, and that meiotic mutants arrested in prophase display greatly reduced Spo11 transcripts because they lack the late spermatocytes that transcribe Spo11 to high levels in wild-type testes.
Isoform-specific transcripts have distinct kinetics: Spo11β transcripts are prevalent in early spermatocytes, whereas Spo11α transcripts increase in pachytene spermatocytes and peak in late prophase.
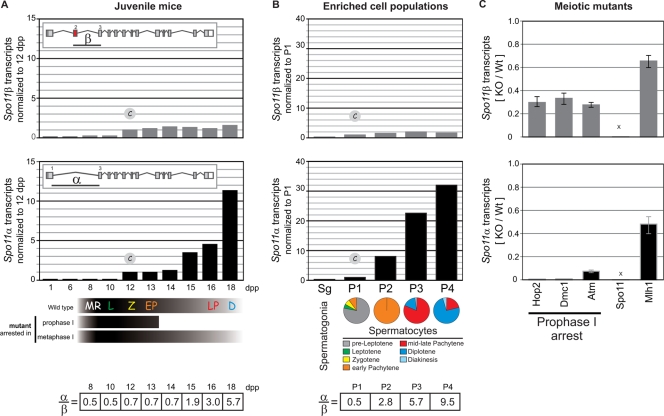
Previous reports have detected two alternative SPO11 isoforms in mouse testes (24, 30, 31). In order to examine Spo11β and -α transcript kinetics, we designed isoform-specific TaqMan assays (see Fig. S1A in the supplemental material). In juvenile mice, we found that Spo11β transcripts reach maximum levels in early stages and remain fairly constant throughout prophase I, whereas Spo11α transcripts peak in late prophase (Fig. 3 A). The α/β transcript ratio is 0.5 in early prophase, increases to 0.7 in midprophase, and rises to 5.7 in late prophase. These results suggest that progression past midprophase triggers exon 2 skipping and results in a shift toward an increase in the steady-state levels of Spo11α transcripts in late prophase.
FIG. 3.
Alternative splicing determines a switch from Spo11β to Spo11α transcripts in mid-prophase I. (A) Differential isoform-specific kinetics of transcription in juvenile mice. TaqMan assays targeting isoform-specific exon junctions were used to quantify Spo11α and -β transcripts by RT-qPCR in the testes of juvenile mice. Results were normalized to those for the 12-dpp sample. The table shows the relative ratios of mRNA levels (α/β). (B) Quantification of isoform-specific transcripts in isolated spermatocytes corroborates that Spo11α transcripts peak in late spermatocytes. Isoform-specific transcript levels were assessed by RT-qPCR on enriched cell populations isolated by FACS. Pie graphs show the percentages of spermatocytes undergoing the different stages of prophase in each cell population. Bar graphs show the relative levels of Spo11α and -β transcripts in each cell population normalized to those in P1. The table shows the relative ratios of mRNA levels (α/β). (C) Mutants arrested in prophase I contain only Spo11β transcripts, indicating that early spermatocytes synthesize solely Spo11β transcripts. Spo11α and -β transcripts were quantified by RT-qPCR in testes of different mutants arrested in prophase I or metaphase I. Bars represent the ratios of Spo11α or -β transcripts in homozygous mutants versus a wild-type littermate (KO/Wt).
We went one step further by measuring isoform-specific transcript levels in enriched cell populations isolated from wild-type mouse testes (Fig. 3B). We used fluorescence-activated cell sorting, as described previously (7), to isolate five cell populations enriched in spermatogonia (Sg) or spermatocytes in preleptonema (P1), early pachynema (P2), mid- to late pachynema (P3), and diplonema (P4). We analyzed Spo11α and -β transcript levels in each cell population and normalized them to those observed in P1. It was not possible to isolate spermatocytes undergoing leptonema devoid of contaminating preleptotene/pachytene spermatocytes to confirm a peak of Spo11β transcripts in leptonema. Nevertheless, we verified that Spo11β transcripts do not increase significantly at late stages, such as mid- to late pachynema and diplonema. On the other hand, we corroborated that Spo11α transcripts become prevalent in pachytene spermatocytes and peak in diplotene spermatocytes (30-fold difference between P1 and P4).
The kinetics observed for Spo11α transcripts in juvenile mice (Fig. 3A) resembles that of total Spo11 transcripts (Fig. 2A), suggesting that Spo11α transcripts are more abundant than Spo11β transcripts. This would explain why mutants arrested in prophase display such a low level of total Spo11 transcripts (5 to 11% compared to their wild-type littermates) (Fig. 2B). Wild-type adult testes carry spermatocytes in all stages, including the late spermatocytes, which contain high levels of Spo11α transcripts, whereas mutant testes contain mostly Spo11β transcripts at roughly 10-fold-lower levels than those of late spermatocytes.
Indeed, when we analyzed several meiotic mutants arrested in prophase I, we found that Spo11α transcripts were almost undetectable in the homozygous mutants (1 to 6% of the wild-type levels). On the other hand, Spo11β transcripts were detected in all prophase-arrested mutants, albeit at reduced levels (28 to 33% of the level observed in wild-type littermates). Finally, transcripts for both isoforms were detected in the Mlh1−/− mutant, arrested in metaphase I (Fig. 3C).
These results, together with the transcription kinetics shown for each isoform in juvenile mouse testes and enriched cell populations, indicate that early spermatocytes synthesize primarily Spo11β transcripts but that, past the arrest point of prophase mutants, Spo11α transcripts become prevalent throughout pachynema and still continue to increase in late prophase.
Protein levels mirror transcript kinetics: SPO11β is expressed in early spermatocytes, whereas the SPO11α polypeptide is hardly synthesized until past early pachynema.
In order to determine whether protein levels mirrored the kinetics of transcription described above for Spo11α and -β, we generated a monoclonal antibody against SPO11 to examine protein levels in testis extracts. Using this monoclonal antibody to immunoprecipitate and immunodetect SPO11 in wild-type testis extracts (Fig. 4 A), we detected two major bands with estimated molecular masses of 38 and 42.2 kDa, which are in the range expected for the SPO11α and SPO11β splicing isoforms (40.3 and 44.5 kDa, respectively) (Fig. 4C). We could also detect two minor bands with intermediate mobilities (Fig. 4A), which seem to be minor splicing isoforms of SPO11α (see Fig. S1 in the supplemental material). All four bands were absent in a mock immunoprecipitation (wild-type extract immunoprecipitated with nonspecific IgG) and in Spo11−/− testis extracts (Fig. 4A).
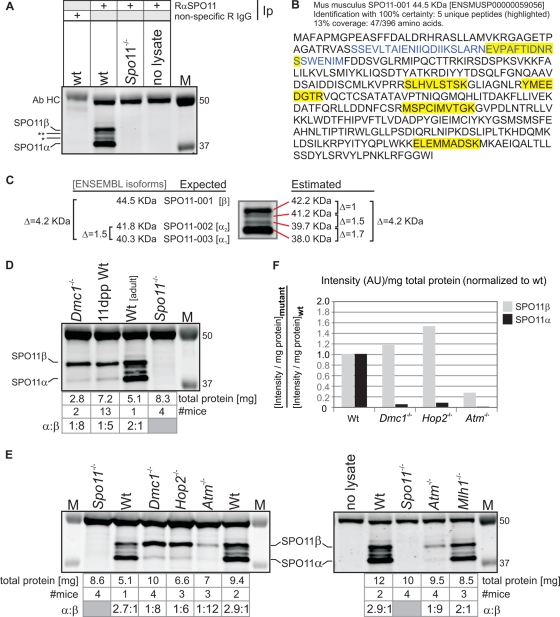
FIG. 4.
SPO11β polypeptide is expressed in early prophase I whereas SPO11α is only expressed once spermatocytes advance past early pachynema. (A) Generation of a monoclonal antibody (Ab) that can immunoprecipitate (Ip)/immunodetect SPO11 in mouse testis extracts. Total testis extracts (6 to 10 mg of total protein) from wild-type (wt) and Spo11−/− mice were immunoprecipitated with anti-SPO11 (or nonspecific rabbit [R] IgG) and subsequently analyzed by Western blotting with the same antibody, anti-SPO11. M, molecular weight marker. (B) Presence of SPO11 in anti-SPO11 immunoprecipitates confirmed by mass spectrometry. Five peptides mapping to different areas of the protein (including exon 2 [blue]) identified SPO11 with 100% certainty in a wild-type extract immunoprecipitated with anti-SPO11 antibody (see Materials and Methods). The relatively low coverage (13%) did not allow identification of specific isoforms, given that a coverage above 95% would be required to identify SPO11α through the absence of peptides mapping to exon 2. (C) The estimated molecular masses of the proteins detected by the SPO11 antibody are in the range expected for SPO11 isoforms. Estimated molecular masses of the four bands detected by the antibody and expected molecular masses for the SPO11 isoforms annotated in Ensembl. (D) Testes from wild-type juvenile mice containing spermatocytes in early stages of prophase I contain primarily the SPO11β polypeptide. Testis extracts from juvenile mice at 11 dpp (early prophase I) and Dmc1−/− and wild-type adult mice were immunoprecipitated/blotted with anti-SPO11 antibody. (E) Mutants arrested in midprophase express primarily the SPO11β polypeptide, whereas a mutant arrested in metaphase I expresses both isoforms. Total testis extracts from wild-type, Spo11−/−, Dmc1−/−, Hop2−/−, Atm−/−, and Mlh1−/− mice (5 to 10 mg of total protein) were precipitated/blotted with anti-SPO11 antibody. Mutant testes are 0.2 to 0.5 the size of wild-type testes, so 3 or 4 mutant mice were used in order to immunoprecipitate extracts containing comparable amounts of total protein. The table specifies the number of mice used for each sample, the milligrams of total protein used for the Ip, and the α/β ratios. (F) Amount of SPO11 isoform precipitated from each extract normalized to total protein and to the wild-type extract. Shown is a comparison of the amounts of SPO11 isoform precipitated from mutant and wild-type extracts, taking into consideration the difference in total protein between extracts. For each isoform, the band intensity was divided by the amount of total protein in the extract and this ratio was divided by the corresponding ratio obtained for the wt sample. AU, arbitrary units.
In order to verify the identities of these bands, we separated anti-SPO11 immunoprecipitates from wild-type testis extracts on an SDS-PAGE gel and confirmed by mass spectrometry the presence of peptides diagnostic of SPO11 among the bands with estimated molecular masses ranging between 38 and 45 kDa (Fig. 4B). SPO11 was not detected in the negative control, consisting of a wild-type extract immunoprecipitated with nonspecific IgG.
Using our antibody against SPO11, we corroborated an earlier report (24) that Dmc1−/− testes contain primarily the SPO11β polypeptide (Fig. 4D, lane 1), suggesting that this is the major isoform expressed in early spermatocytes. To determine if this was also true in a wild-type background, we looked at wild-type juvenile mice. We found that testes from 11-dpp pups, containing spermatocytes in early stages of prophase, express primarily the SPO11β polypeptide (Fig. 4D, lane 2). We detect five times more SPO11β than SPO11α (α/β ratio of 1:5). Assuming there is no bias for homodimer versus heterodimer formation, this α/β ratio would result in an excess of ββ homodimers (ββ, 70%; αβ, 30%; αα, 3%) (see Fig. S2 in the supplemental material).
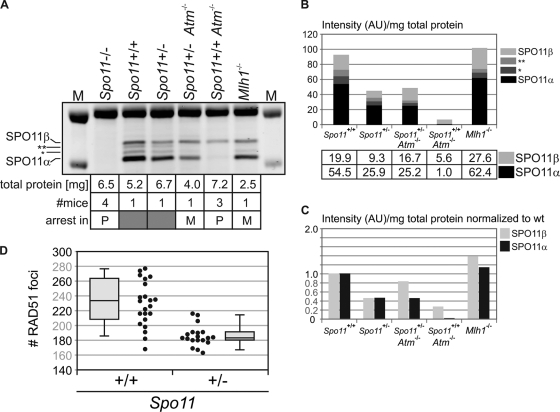
When we compared wild-type and mutant testis extracts (Fig. 4E), we found that extracts from the three mutants arrested in prophase I, the Dmc1−/−, Hop2−/−, and Atm−/− mutants, contain primarily SPO11β. In fact, SPO11β is in a 6- to 12-fold excess over SPO11α (α/β ratio, 1:6 to 1:12), which is barely detectable (the comparatively lower level of SPO11β observed in the Atm−/− mutant is addressed below). This confirms that early spermatocytes express an excess of the SPO11β isoform. Conversely, in the Mlh1−/− mutant, arrested in metaphase, both isoforms are detected (α/β ratio, 2:1) and in approximately the same relative amounts as in wild-type extracts, in which SPO11α is consistently in a 2- to 3-fold excess over SPO11β (α/β ratio, ∼2.6:1) (Fig. 4E). Both in Mlh1−/− and wild-type adult testes about 70% of spermatocytes are in pachynema (due to the longer duration of pachynema than of the other stages of prophase), so the α/β ratios observed for these mice most likely reflect an excess of the α isoform over β in pachytene spermatocytes.
Overall, SPO11 protein levels are consistent with the transcription data presented above. Quantification of SPO11α and -β polypeptides in mouse testis extracts revealed that SPO11β is the major isoform expressed early in prophase, both in wild-type juvenile mice and in several mutants arrested in prophase I, suggesting that it is the isoform introducing the DSBs that initiate meiotic recombination. On the other hand, SPO11α is hardly expressed until past early pachynema (point of arrest of prophase mutants), raising the question of what might be the role of a SPO11 isoform expressed so late in prophase when meiotic DSBs have already been repaired.
Regardless of an ATM deficiency, SPO11α and -β levels are reduced by only one-half in Spo11+/− testes.
Even though Spo11β transcript levels are comparable in Dmc1−/−, Hop2−/−, and Atm−/− mutants (Fig. 3C), the Atm−/− protein extract contained less than 20% of the amount of SPO11β polypeptide observed in the other two prophase mutants (Fig. 4E and F). This was confirmed in three independent experiments. Interestingly, we and others have previously reported a genetic interaction between Atm and Spo11 (3, 9), namely, Spo11 heterozygosity can partially rescue the meiotic arrest of Atm−/− spermatocytes. At the time, we speculated that reduced expression from the single Spo11 allele might result in a delayed or decreased number of DSBs, thus allowing the repair-deficient Atm−/− spermatocytes to proceed through prophase I. Yet, there is no published evidence for reduced expression of Spo11 due to hemizygosity, and it is plausible that increased transcription or translation from the functional allele might compensate for the null allele. Therefore, we decided to examine Spo11 transcript and protein levels in a Spo11+/− background. We found that both total Spo11 transcript levels (Fig. 2B) and SPO11β and -α polypeptide levels (Fig. 5 A, lanes 3 and 4, B, and C) are reduced by 50% in Spo11+/− testes compared to those in a wild-type littermate. To determine if this reduction in the amount of SPO11β polypeptide translated into an equivalent decrease in the number of DSBs, we counted RAD51 foci on spermatocyte spreads. Even though we found a significant difference in the number of RAD51 foci between the wild-type and Spo11+/− backgrounds among leptotene spermatocytes (12-dpp pups) (Fig. 5D), the reduction did not amount to 50%. We observed 18% fewer RAD51 foci in Spo11+/− leptotene spermatocytes (186 ± 14 foci in Spo11+/− cells [19 cells] versus 227 ± 25 foci in Spo11+/+ cells [22 cells]; mean ± standard deviation [SD]; P < 0.0001, t test).
FIG. 5.
SPO11β is expressed in excess such that a reduction by 50% in its level of expression does not translate into a comparable decline in the number of meiotic DSBs. (A) SPO11α and -β polypeptides are reduced by one-half in Spo11+/− testes, irrespective of ATM deficiency. Total testis extracts from Spo11+/+ and Spo11+/− littermates as well as Atm−/− Spo11+/−, Atm−/− Spo11+/+, and Mlh1−/− mice were immunoprecipitated/blotted with anti-SPO11 antibody. * and ** (also for panel B), minor splicing isoforms of SPO11α. (B) Amounts of SPO11 isoform precipitated from mutant and wild-type extracts, taking into consideration the difference in total protein between extracts. For each isoform, the band intensity (in arbitrary units) was divided by the amount of total protein in the extract. The normalized values for the α and β isoforms are documented in the table. (C) Amount of SPO11 isoform precipitated from each extract normalized to the wild-type (wt) extract. For each isoform, the band intensity (in arbitrary units) was divided by the amount of total protein in the extract and this ratio was divided by the corresponding ratio obtained for the wt sample: (intensity/mg total protein)mutant/(intensity/mg total protein)wt. (D) A 50% reduction in SPO11β polypeptide does not translate into a comparable decline in the number of DSBs. Spermatocyte spreads from Spo11+/+ and Spo11+/− (12-dpp) pups were stained for SCP3 and RAD51. The graph reports RAD51 focus numbers from 22 and 19 nuclei, respectively.
The major reduction (≥80%) in the amount of SPO11β polypeptide observed in Atm−/− testes (Fig. 4C) suggested that, in the absence of functional ATM, either SPO11 translation or protein turnover might be affected, significantly reducing steady-state levels of SPO11β. We wondered whether the same effect would be evident in Atm−/− Spo11+/− testes. As mentioned above, Spo11 heterozygosity allows Atm−/− spermatocytes to proceed through prophase I, but they are arrested in metaphase I. Given the metaphase arrest, they would be expected to carry both SPO11β and -α polypeptides, like the Mlh1−/− mutant. Yet, due to the ATM deficiency, SPO11 protein levels might be significantly below those expected for a metaphase mutant. Hence, we compared SPO11 protein levels in Atm−/− Spo11+/−, Atm−/− Spo11+/+, Mlh1−/−, and Spo11+/− mice (Fig. 5A). We found that the Atm−/− Spo11+/− mice did not display the dramatic reduction in SPO11 protein levels observed in the Atm−/− single mutant (Fig. 5A, lanes 5 and 6, B, and C). Instead, levels resembled those in the Mlh1−/− metaphase mutant, in that both isoforms were present yet their levels were reduced by 50% compared to those in the Mlh1−/− extract, which is consistent with Spo11 hemizygosity (note that, since metaphase mutants do not contain postmeiotic cells, a higher proportion of total protein comes from meiotic cells; hence, normalization to total protein yields amounts of α and β polypeptides apparently higher than those for a wild-type background). The fact that in Atm−/− Spo11+/− testes SPO11 protein levels are not reduced beyond what is expected given the Spo11 heterozygosity rules out the possibility that the ATM deficiency per se might cause a decline in the steady-state levels of the SPO11 polypeptides.
Overall, these results confirm that metaphase I mutants express both SPO11 isoforms and indicate that SPO11α and -β polypeptides are reduced by 50% in Spo11+/− testes, irrespective of ATM. Halving the complement of SPO11β polypeptide does not result in an equivalent decline in the number of DSBs, suggesting that wild-type spermatocytes express SPO11β in excess of what is required to ensure an adequate number of DSBs to guarantee the formation of at least one crossover between every homolog pair.
In early spermatocytes SPO11β-containing dimers are present in excess of what would be necessary to introduce meiotic DSBs.
To test our prediction that primary spermatocytes express an excess of SPO11, we estimated the number of SPO11 dimers present in primary spermatocytes in early to midprophase. In wild-type testes, most spermatocytes are past the stage in which the breaks are introduced, so we quantified the amount of SPO11 polypeptides and the number of primary spermatocytes present in Hop2−/− testes (see Materials and Methods) and estimated the number of SPO11α and -β molecules per primary spermatocyte (Table 1). Given that Hop2−/− testes contain exclusively early and mid-prophase I spermatocytes, these numbers represent a snapshot of SPO11 polypeptide levels around the time of DSB formation and can be used to estimate the numbers of ββ, αβ, and αα dimers that primary spermatocytes contain in early prophase I (Table 1).
TABLE 1.
Quantification of SPO11 polypeptides in Hop2−/− mouse testesa
| Isoform or dimer | No. of SPO11 moleculesb or dimers per primary spermatocyte |
|---|---|
| β | 43,200 |
| α | 6,800 |
| ββ | 18,300 (0.73) |
| βα | 6,200 (0.25) |
| αα | 500 (0.02) |
We estimated the amount of each SPO11 isoform in Hop2−/− immunoprecipitates by contrasting the intensities of the bands in a Western blot to those of a standard curve generated with purified His-mSPO11β. The estimated moles of SPO11α and -β per mouse were used to calculate the number of molecules per mouse, and normalization to the number of primary spermatocytes revealed the number of molecules of each SPO11 isoform per primary spermatocyte. Given the estimated proportions of ββ, αβ, and αα dimers (in parentheses), we calculated the expected number of each type of dimer per primary spermatocyte.
β/α ratio, 6.3:1.
Only 26% of Hop2−/− primary spermatocytes are in leptonema (see Fig. S3C in the supplemental material), and we have established that SPO11α synthesis begins around midprophase (Fig. 4D and E); therefore, the number of α molecules is likely overestimated. Hence, in Hop2−/− testes, early spermatocytes would contain barely enough αα dimers to generate the reported number of meiotic DSBs, which is around 350 (23). Furthermore, in Atm−/− testes, where SPO11 polypeptide levels are reduced by more than 80% compared to those in Hop2−/− testes (and the α/β ratio indicates an even greater excess of SPO11β to -α than in Hop2−/− testes) (Fig. 4E), the number of αα dimers per spermatocyte is expected to drop below 100. Nevertheless, we observe no significant decline in the number of RAD51 foci (data not shown). This observation strengthens the notion that, in spermatocytes, SPO11 αα dimers are not essential to achieve the normal number of DSBs at the beginning of prophase I.
On the other hand, Hop2−/− primary spermatocytes contain SPO11 ββ and αβ dimers in excess (50- and 15-fold, respectively) of what would be required to introduce 350 DSBs in leptonema (and so do Atm−/− spermatocytes). This excess of SPO11 dimers is consistent with our observation that reducing the SPO11 complement by 2- to 5-fold (Spo11+/− and Atm−/− spermatocytes, respectively) does not result in an equivalent decline in the number of DSBs (Fig. 5A, C, and D). Our estimates suggest that, based on the level of expression, either SPO11 ββ or αβ dimers could account for the observed number of DSBs. Yet, given the kinetics of expression described for SPO11 in spermatocytes undergoing prophase (Fig. 3 and 4D and E) and the evident excess of ββ dimers over αβ dimers in early spermatocytes (Table 1; see Fig. S2 in the supplemental material), we favor the notion that SPO11 ββ dimers are responsible for introducing meiotic DSBs in spermatocytes.
The fact that SPO11 is expressed in such an excess hints at additional layers of control of SPO11 activity to limit the number of breaks. These most likely involve interaction with one or more proteins that would affect its subcellular localization (cytoplasmic/nuclear translocation) or its ability to load onto the chromatin.
Sexual dimorphism in Spo11α transcript kinetics in prophase I.
In order to determine whether one of the isoforms had a gender-specific role during meiosis, we analyzed Spo11 transcript kinetics in females. We quantified Spo11α and -β transcripts in fetal ovaries containing oocytes in the different stages of prophase I (Fig. 6). We found that both alternative transcripts can be detected in oocytes during prophase I. Spo11α and -β transcripts can first be detected in fetal ovaries containing leptotene oocytes and increase 2-fold in early zygonema. Spo11β mRNA levels are maintained throughout late zygonema/pachynema and decrease slightly in diplotene and dictyate oocytes (30% reduction), a profile very similar to that observed in spermatocytes (Fig. 3A). On the other hand, Spo11α transcripts are maintained through zygonema but steadily decline as oocytes progress into mid- to late prophase I (80% reduction by dictyate). Hence, in contrast to the increase in Spo11α transcripts observed in spermatocytes in late prophase, we found that in oocytes α transcripts decrease in late prophase. Nevertheless, it should be noted that there is an excess of Spo11α transcripts throughout prophase in oocytes (addressed in Discussion).
The peak of α transcripts observed in spermatocytes could indicate a role in mid- to late prophase I or in the imminent cell divisions. Given that oocytes can remain arrested in dictyate for months until being hormonally activated to reinitiate meiosis, we quantified Spo11 transcripts in oocytes that have been induced to resume meiosis (GV) and in metaphase II-arrested oocytes (see Materials and Methods) to determine whether Spo11 mRNAs could be detected past prophase I. Neither Spo11α nor -β transcripts could be detected in these oocytes (data not shown), indicating that in females, Spo11 transcription is restricted to oocytes undergoing prophase I in fetal ovaries. This is in agreement with publicly available microarray data reporting absence of Spo11 transcripts in GV-MII oocytes and 1- to 8-cell embryos (Geo datasets: GDS1265, GDS1677, and GDS813).
The fact that both male and female meiocytes generate Spo11 alternative transcripts during prophase I argues against a gender-specific function for SPO11 isoforms. The sexual dimorphism in terms of the kinetics of Spo11α transcripts and the α/β transcript ratio could be related to differences in timing and duration of prophase I between spermatocytes and oocytes (see Discussion).
DISCUSSION
Developmental regulation of alternative splicing at the Spo11 locus governs the sequential expression of SPO11 isoforms in male meiotic prophase I.
In wild-type mouse testes, total Spo11 transcripts peak in spermatocytes undergoing mid- to late prophase (Fig. 2A). Interestingly, when we quantified Spo11 isoform-specific transcripts, we found that they have a distinct temporal pattern. Spo11β transcripts reach maximum levels in early spermatocytes and remain fairly constant throughout prophase, whereas Spo11α transcripts become prevalent around midprophase and peak in late spermatocytes (Fig. 3A and B). Consequently, mutants arrested in metaphase contain transcripts for both isoforms, whereas mutants arrested in midprophase contain almost exclusively Spo11β transcripts (Fig. 3C). The fact that Spo11α transcripts are barely detectable in Dmc1−/−, Hop2−/−, and ATM−/− mutants indicates that Spo11α transcripts are not synthesized until past the arrest point of these mutants.
Overall, protein levels follow the kinetics determined by the alternative splicing pattern throughout prophase: evidence gathered from juvenile mice and several prophase mutants (Fig. 4D and E) indicates that early spermatocytes synthesize primarily the SPO11β polypeptide. On the other hand, judging by results obtained for metaphase mutants and wild-type adult mice carrying spermatocytes in all stages of prophase, a major fraction of which correspond to pachytene spermatocytes, pachytene/diplotene spermatocytes express more SPO11α than -β. The implications and questions raised by this sequential expression pattern are discussed below.
Sexual dimorphism in Spo11α transcript kinetics and relative α/β mRNA levels during prophase I.
When we quantified Spo11 transcripts in females, we found that β transcript kinetics were similar in oocytes and spermatocytes but that Spo11α mRNA steady-state levels had distinct profiles (Fig. 3A and B and 6). In males, Spo11α transcripts become prevalent around midprophase in pachytene spermatocytes and peak in diplonema, over a period of 9 days, whereas in females, Spo11α transcripts peak in zygonema and decline thereafter as prophase I progresses over a period of 3 days. Nevertheless, in oocytes, in contrast to spermatocytes, they constitute the major splicing product throughout prophase I (with the α/β ratio decreasing from 5 to 1.7 as α transcripts decline toward the end of prophase I).
The sensitivity of our SPO11 immunoprecipitation/Western blot procedure does not allow us to detect SPO11 polypeptides in fetal ovaries to determine whether protein levels mirror transcript kinetics in oocytes. Assuming this is the case (as it is in males), SPO11α expression in oocytes argues against a male-specific role for this isoform (elaborated below). The fact that Spo11α transcripts predominate from the beginning of prophase in females but do not become prevalent until midprophase in males could be related to the difference in the lengths of prophase I in male and female meiocytes. Spermatocytes take 12 days to complete prophase, 8 of which are spent in pachynema. On the other hand, in females, oocytes undergo prophase in 5 days (E15.5 to E20/newborns), spending roughly a day per prophase substage and another 24 h developing the arrested diplotene stage denominated dictyate. If SPO11α has a role in mid- to late prophase, expression in males could be postponed until midprophase, because spermatocytes take over a week to complete pachynema, but it might need to be expressed earlier in females, because oocytes reach and complete pachynema in 2 to 3 days.
Evidence for SPO11 ββ homodimers introducing the breaks at the onset of prophase I.
Immunostaining for γH2AX and RAD51/DMC1 on wild-type spermatocytes has demonstrated that the breaks that initiate meiotic recombination are introduced in leptonema (22, 23). We report here that, in testes from 11-dpp wild-type pups carrying leptotene and zygotene spermatocytes, we detect primarily the SPO11β isoform. The α/β ratio (1:5) suggests that almost 70% of SPO11 expressed at this stage of prophase would form ββ homodimers (see Fig. S2A and B in the supplemental material). Furthermore, in Dmc1−/−, Hop2−/−, and Atm−/− mutants, in which meiotic DSBs are introduced but cannot be efficiently processed and repaired (5, 27, 28), testis extracts contain an excess of the β isoform, with α/β ratios (1:6 to 1:12) implying that 70 to 85% of the SPO11s expressed in these mutants are ββ homodimers (see Fig. S2A and B in the supplemental material). This suggests that SPO11 ββ dimers would likely be responsible for introducing meiotic DSBs.
We found additional support for this notion when we quantified the numbers of SPO11α and -β molecules per primary spermatocyte (Table 1). The fact that, in Hop2−/− (G. Petukhova, personal communication) and Atm−/− (data not shown) mice, two mutants in which a normal number of breaks are introduced, primary spermatocytes are estimated to contain barely or not enough SPO11 αα dimers to account for 350 DSBs argues against a role for αα dimers in the generation of DSBs in early prophase. On the other hand, the 15- and 50-fold excesses of αβ and ββ dimers with respect to breaks suggest that either of these two dimers could be introducing them. Although we cannot rule out αβ dimers, given the kinetics of expression and the 3-fold excess of ββ over αβ dimers in early spermatocytes, we favor the notion that SPO11 ββ dimers are responsible for introducing meiotic DSBs in leptonema.
A role for SPO11α in mid- to late prophase I in male and female meiocytes.
In wild-type adult testes, containing a majority of pachytene spermatocytes (∼10% leptotene, ∼10% zygotene, ∼70% pachytene, and ∼10% diplotene) there are two to three α polypeptides per β monomer, suggesting that in late spermatocytes (pachytene/diplotene) there is an excess of the SPO11α isoform, which is predicted to result in ∼40% αβ heterodimers and ∼50% αα homodimers (see Fig. S2A and B in the supplemental material). Equivalent results were obtained for the Mlh1−/− mutant. Altogether, these data suggest that SPO11α levels increase past midprophase and that SPO11α must have a role in pachytene/diplotene spermatocytes.
Pachynema, the stage in prophase in which a fraction of recombination intermediates are matured into crossovers, is extended for 8 days in spermatocytes but is completed in 1 day in oocytes. The difference in duration is presumably related to the challenge of recombining, repairing, and silencing the mostly heterologous XY sexual chromosomes in males (16, 34). Given that SPO11α expression increases in midprophase spermatocytes, as they enter pachynema, we weighed the possibility that it might somehow be involved in dealing with the XY pair. We reasoned that, if this was the case, SPO11α would not be expected to be highly expressed in oocytes, where the completely homologous XX pair behaves just like any autosomal bivalent. Yet, when we characterized Spo11 transcription in oocytes undergoing prophase I in fetal ovaries, we found that an excess of Spo11α transcripts are synthesized in oocytes since early prophase I, which argues against a male-specific role for SPO11α.
What could be the role for SPO11α in male and female meiocytes? There are two alternative functions SPO11α could have in mid- to late prophase. The first would be titration of the β isoform in an inactive or incorrectly localized heterodimer, so as to prevent formation of unwanted breaks in mid- to late prophase. We do not favor this hypothesis because we have shown that SPO11β is expressed in excess in early spermatocytes, which suggests that an interacting protein(s) would limit the number of DSBs by restricting its access to the nucleus/chromatin. Hence, there would not be a need for a titrator subunit to restrict SPO11β DSB-generating activity. Also, the fact that Spo11α transcripts are prevalent from the beginning of prophase in oocytes is not consistent with a titrator role for SPO11α. A second possibility requires assuming that interaction of a SPO11 dimer with another protein might turn it into a topoisomerase (much like the topoisomerase VI A2B2 tetrameric topoisomerase in Archaea). This complex could condense the chromosomes in late prophase or even during diakinesis, in preparation for metaphase. Alternatively, it could resolve tangles generated during the maturation of recombination intermediates that takes place during pachynema. Both the mitotic At-SPO11-3 in Arabidopsis and the archaeal homolog topoisomerase VI have been reported to act as decatenases (11, 17, 33).
Consistent with a “topoisomerase” activity, we and others have previously reported localization of SPO11 (29, 31, 32) to the chromosome cores in pachytene spermatocytes. Yet, the monoclonal antibody against SPO11 developed for this work does not stain the synaptonemal complex in either spread or squashed spermatocyte preparations (data not shown). On testis sections, this monoclonal antibody stains only the cytoplasm of pachytene and diplotene spermatocytes (data not shown). Nevertheless, the fact that we do not detect it in the nuclei of late spermatocytes does not rule out the possibility that it might localize to the nucleus transiently or in small quantities. In fact, even though we know that SPO11 is required to generate meiotic DSBs in early spermatocytes (31), we do not detect the protein in the nuclei of early spermatocytes either. In contrast to what has been described for Sordaria SPO11 (32), mouse SPO11 appears to lack a canonical nuclear localization signal, as several servers for subcellular localization prediction failed to identify any localization motif in either SPO11 isoform (PSORT II [19], LOCSVMPS I [36], and ESLpred [12]). Thus, the cytoplasmic signal we observe might reflect localization of the bulk of SPO11, while a small proportion of the protein (relying on interaction with a partner protein) transiently relocates to the nucleus.
Our analysis of SPO11 expression in mouse meiocytes demonstrates a role for SPO11β in generating the breaks that initiate meiotic recombination in early spermatocytes and indicates that SPO11α has an additional role in mid- to late prophase that is conserved in oocytes. The generation of knock-in mice carrying a modified Spo11 locus, such that it expresses either the SPO11α or -β isoform, will enable us to dissect the roles of these isoforms in prophase I.
Supplementary Material
Acknowledgments
The Intramural Research Program of the NIDDK, NIH, supported this research.
We thank Bruce Raaka for his help with the FACS of testicular cells, Eric Anderson for his help with the mass spectrometry analysis, and Huiyan Lu for her help with oocyte isolation. We are grateful to Michael Lichten for a critical reading of our manuscript. Our thanks go to John Schimenti for providing us with Dmc1−/− mice, to Florencia Pratto for countless insightful discussions, and to Linda Robinson for her assistance.
Footnotes
Published ahead of print on 20 July 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris, X. Yao, D. M. Christie, C. Monell, N. Arnheim, A. Bradley, T. Ashley, and R. M. Liskay. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13:336-342. [DOI] [PubMed] [Google Scholar]
- 2.Barchi, M., S. Mahadevaiah, M. Di Giacomo, F. Baudat, D. G. de Rooij, P. S. Burgoyne, M. Jasin, and S. Keeney. 2005. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell. Biol. 25:7203-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barchi, M., I. Roig, M. Di Giacomo, D. G. de Rooij, S. Keeney, and M. Jasin. 2008. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 4:e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlow, C., S. Hirotsune, R. Paylor, M. Liyanage, M. Eckhaus, F. Collins, Y. Shiloh, J. N. Crawley, T. Ried, D. Tagle, and A. Wynshaw-Boris. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86:159-171. [DOI] [PubMed] [Google Scholar]
- 5.Barlow, C., M. Liyanage, P. B. Moens, M. Tarsounas, K. Nagashima, K. Brown, S. Rottinghaus, S. P. Jackson, D. Tagle, T. Ried, and A. Wynshaw-Boris. 1998. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 125:4007-4017. [DOI] [PubMed] [Google Scholar]
- 6.Barroca, V., B. Lassalle, M. Coureuil, J. P. Louis, F. Le Page, J. Testart, I. Allemand, L. Riou, and P. Fouchet. 2009. Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nat. Cell Biol. 11:190-196. [DOI] [PubMed] [Google Scholar]
- 7.Bastos, H., B. Lassalle, A. Chicheportiche, L. Riou, J. Testart, I. Allemand, and P. Fouchet. 2005. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65:40-49. [DOI] [PubMed] [Google Scholar]
- 8.Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. [DOI] [PubMed] [Google Scholar]
- 9.Bellani, M. A., P. J. Romanienko, D. A. Cairatti, and R. D. Camerini-Otero. 2005. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J. Cell Sci. 118:3233-3245. [DOI] [PubMed] [Google Scholar]
- 10.Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386:414-417. [DOI] [PubMed] [Google Scholar]
- 11.Bergerat, A., D. Gadelle, and P. Forterre. 1994. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J. Biol. Chem. 269:27663-27669. [PubMed] [Google Scholar]
- 12.Bhasin, M., and G. P. Raghava. 2004. ESLpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucleic Acids Res. 32:W414-W419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett, K. D., P. Benedetti, and J. M. Berger. 2007. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat. Struct. Mol. Biol. 14:611-619. [DOI] [PubMed] [Google Scholar]
- 14.Eaker, S., J. Cobb, A. Pyle, and M. A. Handel. 2002. Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: insights into regulation of spermatogenic progress. Dev. Biol. 249:85-95. [DOI] [PubMed] [Google Scholar]
- 15.Goetz, P., A. C. Chandley, and R. M. Speed. 1984. Morphological and temporal sequence of meiotic prophase development at puberty in the male mouse. J. Cell Sci. 65:249-263. [DOI] [PubMed] [Google Scholar]
- 16.Handel, M. A. 2004. The XY body: a specialized meiotic chromatin domain. Exp. Cell Res. 296:57-63. [DOI] [PubMed] [Google Scholar]
- 17.Hartung, F., K. J. Angelis, A. Meister, I. Schubert, M. Melzer, and H. Puchta. 2002. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 12:1787-1791. [DOI] [PubMed] [Google Scholar]
- 18.Hartung, F., R. Wurz-Wildersinn, J. Fuchs, I. Schubert, S. Suer, and H. Puchta. 2007. The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19:3090-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton, P., and K. Nakai. 1997. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5:147-152. [PubMed] [Google Scholar]
- 20.Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1-53. [DOI] [PubMed] [Google Scholar]
- 21.Kim, E., A. Goren, and G. Ast. 2008. Alternative splicing: current perspectives. Bioessays 30:38-47. [DOI] [PubMed] [Google Scholar]
- 22.Mahadevaiah, S. K., J. M. Turner, F. Baudat, E. P. Rogakou, P. de Boer, J. Blanco-Rodriguez, M. Jasin, S. Keeney, W. M. Bonner, and P. S. Burgoyne. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27:271-276. [DOI] [PubMed] [Google Scholar]
- 23.Moens, P. B., D. J. Chen, Z. Shen, N. Kolas, M. Tarsounas, H. H. Heng, and B. Spyropoulos. 1997. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106:207-215. [DOI] [PubMed] [Google Scholar]
- 24.Neale, M. J., J. Pan, and S. Keeney. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols, M. D., K. DeAngelis, J. L. Keck, and J. M. Berger. 1999. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 18:6177-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters, A. H., A. W. Plug, M. J. van Vugt, and P. de Boer. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5:66-68. [DOI] [PubMed] [Google Scholar]
- 27.Petukhova, G. V., P. J. Romanienko, and R. D. Camerini-Otero. 2003. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 5:927-936. [DOI] [PubMed] [Google Scholar]
- 28.Pittman, D. L., J. Cobb, K. J. Schimenti, L. A. Wilson, D. M. Cooper, E. Brignull, M. A. Handel, and J. C. Schimenti. 1998. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1:697-705. [DOI] [PubMed] [Google Scholar]
- 29.Prieler, S., A. Penkner, V. Borde, and F. Klein. 2005. The control of Spo11's interaction with meiotic recombination hotspots. Genes Dev. 19:255-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanienko, P. J., and R. D. Camerini-Otero. 1999. Cloning, characterization, and localization of mouse and human SPO11. Genomics 61:156-169. [DOI] [PubMed] [Google Scholar]
- 31.Romanienko, P. J., and R. D. Camerini-Otero. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6:975-987. [DOI] [PubMed] [Google Scholar]
- 32.Storlazzi, A., S. Tesse, S. Gargano, F. James, N. Kleckner, and D. Zickler. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17:2675-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugimoto-Shirasu, K., N. J. Stacey, J. Corsar, K. Roberts, and M. C. McCann. 2002. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 12:1782-1786. [DOI] [PubMed] [Google Scholar]
- 34.Turner, J. M. 2007. Meiotic sex chromosome inactivation. Development 134:1823-1831. [DOI] [PubMed] [Google Scholar]
- 35.Turner, J. M., S. K. Mahadevaiah, O. Fernandez-Capetillo, A. Nussenzweig, X. Xu, C. X. Deng, and P. S. Burgoyne. 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37:41-47. [DOI] [PubMed] [Google Scholar]
- 36.Xie, D., A. Li, M. Wang, Z. Fan, and H. Feng. 2005. LOCSVMPSI: a web server for subcellular localization of eukaryotic proteins using SVM and profile of PSI-BLAST. Nucleic Acids Res. 33:W105-W110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.