Abstract
Short repetitive sequences are common in the human genome, and many fall within transcription units. We have previously shown that transcription through CAG repeat tracts destabilizes them in a way that depends on transcription-coupled nucleotide excision repair and mismatch repair. Recent observations that antisense transcription accompanies sense transcription in many human genes led us to test the effects of antisense transcription on triplet repeat instability in human cells. Here, we report that simultaneous sense and antisense transcription (convergent transcription) initiated from two inducible promoters flanking a CAG95 tract in a nonessential gene enhances repeat instability synergistically, arrests the cell cycle, and causes massive cell death via apoptosis. Using chemical inhibitors and small interfering RNA (siRNA) knockdowns, we identified the ATR (ataxia-telangiectasia mutated [ATM] and Rad3 related) signaling pathway as a key mediator of this cellular response. RNA polymerase II, replication protein A (RPA), and components of the ATR signaling pathway accumulate at convergently transcribed repeat tracts, accompanied by phosphorylation of ATR, CHK1, and p53. Cell death depends on simultaneous sense and antisense transcription and is proportional to their relative levels, it requires the presence of the repeat tract, and it occurs in both proliferating and nonproliferating cells. Convergent transcription through a CAG repeat represents a novel mechanism for triggering a cellular stress response, one that is initiated by events at a single locus in the genome and resembles the response to DNA damage.
Unstable repetitive sequences litter the human genome, mostly without consequence (2), but at 18 genomic loci expansions of trinucleotide repeats (TNRs) trigger severe neurological disease (36, 51). Longer-than-normal TNRs are unstable during intergenerational transmission, giving rise to progeny that carry altered repeat tracts, either with additional units (expansions) or with fewer units (contractions). Expansions, which tend to predominate over contractions in most diseases, lead to more severe disease in the offspring (17, 51). A similar, ongoing expansion-biased instability occurs in aging neurons (38), where it likely exacerbates disease symptoms (64), which often include the compromised function and ultimate death of specific neurons. The mechanisms of repeat instability and cell death are not well defined, and they could differ between the germ line and somatic tissues and vary from tissue to tissue (51, 53, 58).
Extensive studies in bacteria, yeast (Saccharomyces cerevisiae), flies, mammalian cells, and mice have identified several potential contributors to repeat instability in human patients, including DNA replication, DNA repair, recombination, and transcription (8, 38). A common feature of these processes is that they expose DNA single strands, which in repeat tracts can form secondary structures such as hairpins and slipped-strand DNA duplexes (16, 54). These repeat-driven secondary structures in some way interfere with the cell's ability to convert the DNA back to a normal-length duplex. Given the variety of DNA transactions that expose single strands, the different types of secondary structure that repeats can form, and the various ways of resolving such structures, it seems likely that multiple pathways will be found to play a role in the repeat instability observed in human patients—and this does not include additional identified contributors to instability, such as epigenetic modifications, chromatin structure, and local sequence effects (8).
Using a selectable system for repeat contractions in human cells, we have demonstrated that transcription destabilizes CAG repeats in a way that does not depend on DNA replication (39, 42). We have used small interfering RNA (siRNA) knockdowns and chemical inhibition to show that transcription-induced repeat instability requires elements of mismatch repair (MMR) (MSH2 and MSH3) and the transcription-coupled nucleotide excision repair (TC-NER) pathway (CSB, XPA, ERCC1, and XPG), in addition to proteins that may be required to deal with stalled RNA polymerase (RNAP) complexes (TFIIS, the E3 ligase BRCA1/BARD1, and the proteasome machinery) (39, 42). We have also shown that R-loops form between the nascent transcript and the template strand at transcribed CAG repeat tracts in human cells and that their formation promotes repeat instability (37). Recently, transcription has also been shown to destabilize GAA repeats, which cause Friedreich ataxia (13). Collectively, these studies suggest that transcription-induced DNA repair processes serve as key mediators of repeat instability.
Because our assay in human cells models repeat contractions, questions naturally arise as to its relevance to human patients, where repeat expansions are the clinically important disease phenomenon. It is unresolved, and a matter of some controversy, as to whether expansions and contractions arise as alternate outcomes of the same basic mechanism (or mechanisms) or from entirely different mechanisms (46, 53). Nevertheless, our analyses of factors that influence repeat contractions in human cells are largely consistent with observations in model organisms, supporting the general relevance of our studies to human patients (40, 41). For example, knockdown of the MMR components MSH2 or MSH3 in human cells reduced contractions (39), while knockout of MSH2 or MSH3 in mice reduced CAG repeat expansions in germ line and somatic cells (33, 47, 60, 68). Similarly, knockdown of the NER components XPA and XPG in human cells decreased transcription-induced CAG contractions (39, 42), while knockout of XPA in mice reduced expansions of the spinocerebellar ataxia type 1 (SCA1) CAG repeat in the striatum (L. Hubert, Jr., Y. Lin, and J. H. Wilson, unpublished data), and mutation of the Mus201 gene in Drosophila, the fly homolog of XPG, sharply reduced transcription-induced CAG expansions in the fly germ line (29). In addition, knockdown of Dnmt1, the major maintenance DNA methyltransferase, increased CAG contractions in human cells (11, 12, 19), while mice with half the normal amount of Dnmt1 had increased CAG expansions in their germ line (11). Knockdowns and knockouts that had no effect in human cells or mice include the MMR component MSH6 (39, 68); the NER component XPC, which is specific for global genomic NER (15, 39); and the flap endonuclease FEN1 (39, 63, 67). To date, there is a single discordance between our system and mouse studies. Mutation of OGG1, which encodes a glycosylase involved in base excision repair (BER), decreased somatic CAG repeat expansion in a mouse model (32), whereas siRNA knockdowns of OGG1 (or of APEX1, which controls the next step in the BER pathway) did not alter transcription-induced repeat stability in our human cell assay (42). Overall, these comparisons suggest cautious optimism that observations in our selective system in human cells will be useful predictors of processes that influence repeat instability in model organisms and, by extension, in human patients.
In this study, we use our selective assay for contractions in human cells to assess the role of antisense transcription in CAG repeat instability. Recent studies have revealed that surprisingly large numbers of human genes are associated with antisense transcripts (31). Moreover, antisense transcription has been reported in five TNR disease genes (5, 6, 9, 35, 49), and at least 10 other TNR disease genes are represented in the antisense transcriptomes of several human cell lines (27). These observations suggest that simultaneous sense and antisense transcription—which we define as convergent transcription—occur in an unexpectedly high fraction of the human genome, including genes associated with TNR diseases. To determine whether convergent transcription might play a role in repeat instability, we tested a construct in which sense and antisense transcription through a CAG repeat could be independently induced. Here, we show that convergent transcription through a repeat tract not only enhances repeat instability but also causes cell death via apoptosis.
MATERIALS AND METHODS
Cell lines and cell culture.
Most cell lines were derived from the human fibrosarcoma HT1080 cell line. Hypoxanthine phosphoribosyltransferase mutant (HPRT−) HT1080 cells were selected by growth in 6-thioguanine (20 μM) and screened for a nonreverting HPRT mutation. One surviving clone (HT1080/57) arose by spontaneous deletion in the single copy of the endogenous HPRT gene and was used for further construction of cell lines. Five million HT1080/57 cells were electroporated with 1 μg of linearized plasmid pTet-ON (Clontech), which contains the gene for the rtTA protein, a fusion of the herpes simplex virus (HSV) VP16 activation domain and the reverse tetracycline repressor protein (20). Clones resistant to G418 (600 μg/ml) were picked for analysis of rtTA expression using a luciferase reporter assay kit (BD Biosciences). One clone, HT1080/57-6, which gave about a 25-fold increase in luciferase activity upon addition of doxycycline (Dox; 2 μg/ml), was used for construction of the DIT and FLAH cell lines. HT1080/57-6 cells were used to construct FLAH cells as described previously (39).
To construct DIT cells, 5 million HT1080/57-6 cells were electroporated with 1 μg of linearized plasmid pNEBR-R1 (New England Biolabs), which contains the genes for RheoReceptor-1 and RheoActivator (30). Clones resistant to zeocin (300 μg/ml) were picked and analyzed for expression of these regulators using a luciferase reporter assay kit (BD Biosciences). One clone, RS11, which gave a 15-fold increase in luciferase activity upon addition of RSL1 (0.5 μM), was used for construction of the DIT cell lines. Five million RS11 cells were electroporated with 1 μg of linearized pDIT1 and then selected for colonies resistant to puromycin (2.5 μg/ml). Plasmid pDIT1, which has a pBluescript backbone with a selectable puromycin N-acetyltransferase gene, contains an HPRT minigene with a CAG95 repeat inserted in the middle of the intron between fused exons 1 and 2 and fused exons 3 to 9. The HPRT minigene was modified so that the cytomegalovirus (CMV) promoter pTRE-CMVmini, which responds to the inducer doxycycline, controls its sense expression, and the pNEBRX1 promoter, which responds to the inducer RSL1, controls its antisense expression. Individual colonies were analyzed by Southern blotting to identify clones with a single integrated copy of the HPRT minigene that was responsive to both inducers. Two cell lines, DIT7 and DIT3, were used in this study. Many of the colonies that were tested were responsive to doxycycline, but not to RSL1, even though they seemed to have an intact pNEBR-X1 promoter. Two such cell lines, DIT-NAT10 and DIT-NAT26, were used in this study.
The DH7 and DH10 cell lines were constructed by electroporating 1 μg of linearized pAH1 into 5 million FLHA25 cells and then selecting for colonies resistant to zeocin. In addition to a zeocin resistance gene, plasmid pAH1 contains an HPRT minigene that lacks a sense promoter but uses the constitutive pCMV promoter to drive antisense transcription.
The DIT-NR4 and DIT-NR5 cell lines were constructed by electroporating 1 μg of linearized pDIT-NR into 5 million RS11 cells, selecting for individual clones that were resistant to puromycin, and analyzing them by Southern blotting to identify clones with a single integrated copy of the plasmid. The pDIT-NR plasmid was derived from pDIT1 by excising the CAG repeat tract by NotI digestion, which removes the CAG tract along with about 130 bp of flanking sequences.
The DIT7-R cell lines were derived from DIT7 cells by selection for HPRT+ colonies, using hypoxanthine-aminopterin-thymidine (HAT) selection. HPRT+ colonies were screened for length of their contracted CAG repeats by gel electrophoresis of PCR products. The repeat tracts in DIT7-R15, DIT7-R20, and DIT7-R25 were determined by DNA sequencing.
The DITS-H5 and DITS-H8 cell lines were constructed by randomly integrating plasmid pDIT2 into HEK293 F-T cells (Invitrogen), which were modified from HEK293 cells, so that they express the rtTA regulator required for expression of the Tet-ON-inducible system. Plasmid pDIT2 contains a pBluescript backbone with a selectable puromycin N-acetyltransferase gene, along with one copy of the HPRT minigene with a CAG95 tract in its intron and two copies of the pTRE-CMVmini promoter, which flank the minigene and allow both sense and antisense transcription to be controlled by the inducer doxycycline.
In all cases, clones containing a single copy of the randomly integrated construct, as confirmed by Southern blotting, were used in this study (39). All cell lines were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium-F-12 medium (DMEM/F-12) supplemented with 10% fetal bovine serum and 1% MEM nonessential amino acids.
Induction of transcription.
Cells were initially plated in the absence of transcription inducers. The time at which inducers were added to the cultures was defined as day 0. To induce sense transcription in HT1080-derived cell lines, we added doxycycline to final concentrations of 200 ng/ml for DIT and DIT-NR cells and 2,000 ng/ml for DIT-NAT, FLAH, and DH cells. Because the half-life of doxycycline in medium is about 24 h, we added half the starting amount of doxycycline directly into the medium each day until the treatment was completed. To induce antisense transcription fully in cells driven by the Rheo-Switch inducible promoter, RSL1 was added to the medium to a final concentration of 500 nM. No additional RSL1 was required for treatments up to 5 days. In the HEK293-derived DITS-H cells, we induced both sense and antisense transcription simultaneously by addition of 2,000 ng/ml doxycycline.
Viable cell measurements.
Adherent cells (attached to the plate) are operationally defined as viable, and nonadherent cells (present in the medium) are operationally defined as dead. Adherent cells typically contain fewer than 4% of cells that stain with propidium iodide (PI), indicating that >96% of adherent cells are viable. The number of dead cells was determined by counting the nonadherent cells in the medium. The number of viable cells was counted after detachment of adherent cells from the dish by trypsin treatment. The percentage of viable cells was calculated as the number of adherent cells divided by the total number of cells, which is the sum of adherent and nonadherent cells. The data in various figures have not been corrected for the small percentage (<4%) of dead cells in the adherent population.
Real-time RT-PCR.
Total RNA was extracted from about 1 million cells using RNeasy minikits (Qiagen). Induction of HPRT sense or antisense transcription was measured on RNA extracted 1 day after addition of doxycycline or RSL1 to the medium. The levels of sense and antisense transcripts were measured separately using strand-specific real-time reverse transcription-PCR (RT-PCR), as illustrated in Fig. 1 H. To measure the level of sense transcript, we first synthesized cDNA from the sense transcript, using reverse transcriptase and the SSP-H8 primer, which links the HPRT sense-strand-specific H8 sequence to the SSP universal primer, whose sequence is not present in the genome. Reverse transcriptase was then inactivated by incubation at 95°C for 10 min. The cDNA was then amplified using the HPRT-specific H7 primer and the SSP universal primer and quantified by real-time PCR. For measurement of antisense transcripts, we synthesized cDNA from the antisense transcript, using reverse transcriptase and the SSP-H7 primer, in which the universal SSP primer is linked to the antisense-strand-specific HPRT primer H7. The “antisense” cDNA was then amplified using a mixture of the H7 and SSP primers and quantified by real-time PCR. Sequences of the primers are listed in Tables 1 and 2. For strand-specific real-time RT-PCR, total RNA (50 ng per reaction) was reverse transcribed at 50°C for 30 min and assayed using the SYBR green PCR kit (Qiagen). Results were normalized to the concentration of β-actin RNA, which was determined the same way. HPRT sense or antisense transcript levels were expressed relative to the RNA level in untreated DIT7 cells, which was arbitrarily defined as 1 (55). The conditions for real-time PCR were 95°C for 15 min, followed by 45 cycles of 94°C for 15 s, 50°C for 30 s, and 72°C for 30 s.
FIG. 1.
Cell lines for assessing effects of sense and antisense transcription. In all cells, the CAG tract is centered in the 2.1-kb intron in the single, randomly integrated HPRT minigene and the repeat is 1.6 kb downstream of the sense promoter and 2.5 kb upstream of the antisense promoter. (A) DIT cells. Sense and antisense transcription are driven by the inducible promoters pCMVmini, which responds to doxycycline (Dox), and pNEBR-X1, which responds to RSL1. (B) FLAH cells. Sense transcription is driven by pCMVmini; no antisense promoter is present. (C) DIT-NAT cells. Sense transcription is driven by pCMVmini; the antisense promoter pNEBR-X1 is silenced. (D) DH cells. In one HPRT minigene, sense transcription is driven by pCMVmini; no antisense promoter is present. In the other, antisense transcription is constitutively expressed from the pCMV promoter; no sense promoter is present. (E) DIT-NR cells. Sense and antisense transcription are driven by pCMVmini and pNEBR-X1, respectively, but the entire CAG tract plus 120-bp flanking sequence is deleted. (F) DIT7-R cells. These cells were derived by contraction of the repeat in DIT7 cells. (G) DITS-H cells. These HEK293-derived cells have an HPRT minigene in which both sense and antisense transcription are driven by pCMVmini and inducible with doxycycline. (H) Strategy for strand-specific real-time RT-PCR to quantify sense and antisense transcripts from the HPRT minigene. The final PCR product in both cases is 225 bp.
TABLE 1.
Sequences of real-time RT-PCR primers and siRNAs used in this study
| Gene product | Real-time RT-PCR primer sequence |
siRNA sequence | |
|---|---|---|---|
| Forward | Reverse | ||
| β-Actin | AGAGAGGCATCCTCACCCTG | CATGAGGTAGTCAGTCAGGT | |
| ATM | GCAGCATGGAGGAATATGCAG | CTGCGCTTACACATCTCTTCC | GCAACAUUUGCCUAUAUCAGCAAUU |
| CACAAGGTCAGGAGTTCGAG | TCACCTCCCAGGTTCAAGAG | GCGCAGUGUAGCUACUUCUUCUAUU | |
| ATR | TGCTGTTGTCTTACAACTGACTG | CAGTGCCAGTTTCTGAACAG | CCACCUGAGGGUAAGAACAUGUUAA |
| CAGGGCTATTCAGCTAGAACC | CTGCTTCCACTCTGTACGTG | GGGAAAUACUAGAACCUCAUCUAAA | |
| ATRIP | CATCAGACGGAATCCGTTCTAG | CTTTGTCCTTAATTCATTCATCTCTGC | UGACCAGCAGGAACUGCCCAGAGAA |
| GGTTCACAGGCTTAGTGATGG | ACTGGCTTGAAGGTGACC | ||
| CHK1 | AGCCAGACATAGGCATGC | CGAAATACTGTTGCCAAGCC | CCCAGCCCACAUGUCCUGAUCAUAU |
| GTACTCCAGTTCTCAGCCAG | GGTTCTGTGAGGATCCTGG | GGCUUGGCAACAGUAUUUCGGUAUA | |
| CHK2 | CTCCTCTACCAGCACGATG | TCCATCCTGAAGGGCC | GAACCUGAGGAGCCUACCCUU |
| AAGAACCUGAGGACCAAGAACUU | |||
| p53 | AGATGAAGCTCCCAGAATGC | AAACCGTAGCTGCCCTG | GCUUCGAGAUGUUCCGAGAGCUGAA |
| GGAGGCCCATCCTCAC | GCGGAGATTCTCTTCCTCTG | CCGGACGAUAUUGAACAAUGGUUCA | |
| TOPBP1 | GGCTGTTGGAGACTGCTAG | CGTGTGCCAGGATGCTC | GCCUUUACAUGAUUCAGAAAUUGCU |
| AGGCAGTCTCGTCAGACAG | GCCACTGCAAATTGCTGG | ||
| Vimentin | GAAUGGUACAAAUCCAAGU | ||
TABLE 2.
Sequences of strand-specific real-time RT-PCR primers for HPRT
| Primer | Sequence |
|---|---|
| SSP-H7 | CGATGCTTGGACAGCCTGACCAGTCAACAGGGGAC |
| SSP-H8 | CGATGCTTGGACAGCCTGCGTGGGGTCCTTTTCACC |
| SSP | CGATGCTTGGACAGCCTG |
| H7 | ACCAGTCAACAGGGGAC |
| H8 | CGTGGGGTCCTTTTCACC |
Chemical and siRNA treatments.
For chemical treatments, 250,000 DIT7 cells were plated in each well of a 6-well plate on day −1. On day 0, we treated the cells with chemicals at concentrations shown in Table 4. When necessary, we induced convergent transcription by adding doxycycline and RSL1 along with the chemicals on day 0. Because many of the chemicals were dissolved in dimethyl sulfoxide (DMSO), a small amount was carried over into the treated cell cultures. We showed that DMSO at those concentrations had no effect on cell viability, except for serum-starved cells, where DMSO caused some toxicity (see Fig. 6). The siRNAs (Dharmacon or Invitrogen) used in this study are listed in Table 1. For siRNA treatments, we used the procedure described previously (42). Briefly, about 100,000 DIT7 cells were plated in each well of a 6-well plate on day −3. On day −2, we transfected cells with siRNAs at a final concentration of 200 nM, using Oligofectamine (Invitrogen). Treatments with 200 nM vimentin siRNA served as controls. On day 0, the cells were again transfected with siRNA, and cultures were then grown in the presence or absence of doxycycline plus RSL1. We evaluated knockdown of target gene expression and sense and antisense HPRT transcription by analyzing total RNA isolated on day 1.
TABLE 4.
Effects of chemical inhibitors of ATR and ATM pathway components on DIT7 cell viability after induction of convergent transcription
| Chemical (concn) | Target | Transcription (%)a |
% of viable cells |
Significance (P value)e | ||
|---|---|---|---|---|---|---|
| Sense | Antisense | −Dox and RSL1b | +Dox and RSL1 (normalized)c | |||
| DMSO | 100 | 100 | 98 ± 1.4 | 50 ± 3.0 | ||
| Wortmannin (30 μM) | ATM/ATR | 132 | 127 | 91 ± 4.9 | 29 ± 4.8 | <0.01 |
| Caffeine (500 μM) | ATM/ATR | 129 | 95 | 96 ± 1.2 | 24 ± 2.0 | <0.0001 |
| CGK733 (5 μM) | ATM/ATR | 90 | 169 | 76 ± 7.2 | 25 ± 2.1 | <0.0001 |
| UCN-01 (0.1 μM) | CHK1 | 137 | 123 | 67 ± 3.8 | 35 ± 1.1 | <0.0001 |
| CHK2 inhibitor II (20 μM) | CHK2 | 143 | 85 | 78 ± 5.2 | 55 ± 5.0 | NSd |
Sense and antisense transcription through the HPRT minigene in the presence of Dox plus RSL1 was measured by real-time RT-PCR at day 1 and expressed as a percentage of the induced transcript levels measured in the presence of DMSO. Results were averaged from three determinations.
Viable cells were counted 4 days after treatment with chemicals in the absence of doxycycline (Dox) and RSL1. Chemicals were added at day 0. Viable cells after chemical treatment are expressed as a percentage of the count of untreated cells, which was defined as 100%. Mean percentages ± SD were determined from at least four independent measurements.
Cell counts were normalized to (divided by) the cell counts in the absence of Dox and RSL1 and expressed as a percentage of the values for DMSO. Viable cells in the sixth column = (measured viable cells [not shown]) × (viable cells in the noninduced control [DMSO] in the fifth column)/(viable cells in noninduced treatment [specific drug] in the fifth column). Mean percentages ± SD were determined from at least six independent measurements.
NS, not significant.
FIG. 6.
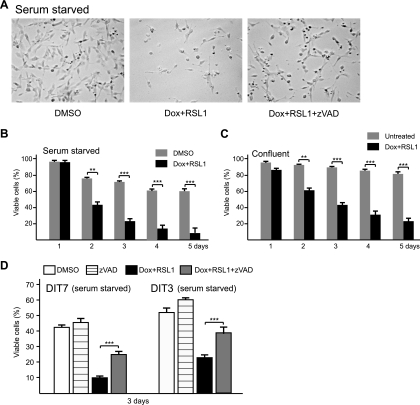
Apoptosis in nonproliferating cells. (A) Convergent transcription and cell death in serum-starved DIT7 cells in the presence and absence of zVAD. Serum-starved DIT7 cells were treated with DMSO, doxycycline plus RSL1 (Dox + RSL1), or doxycycline plus RSL1 and zVAD (Dox + RSL1 + zVAD) and photographed 3 days later. (B) Quantification of cell killing in serum-starved DIT7 cells that were treated with doxycycline plus RSL1 or with DMSO alone. (C) Quantification of cell killing in confluent DIT7 cells that were untreated or treated with doxycycline plus RSL1. (D) Quantification of the effect of zVAD on cell killing in serum-starved DIT7 and DIT3 cells. Serum-starved DIT cells were treated for 3 days with DMSO, zVAD, doxycycline plus RSL1, or doxycycline plus RSL1 and zVAD. Percentages of viable cells were calculated as described in the legend to Fig. 3 and are averaged from at least six independent measurements with error bars indicating standard deviations. Statistical significance is indicated: **, P < 0.001; and ***, P < 0.0001.
Contraction assay.
One million DIT cells were plated in each well of a 6-well plate on day −3. On days −2, −1, and 0, cells were refed with fresh medium daily until they reached confluence. On days 1, 3, and 5, doxycycline (200 ng/ml), RSL1 (500 nM), or both were added. Medium containing the inducers was removed on days 2, 4, 6, 7, and 8 and replaced with fresh medium lacking the inducers. For contraction assays in the presence of 20 μM zVAD [benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethyl ketone], zVAD was present from day 0, when inducers were added, until day 6, when the inducers were removed. On day 9, HPRT+ cells were selected by plating 500,000 DIT cells on 100-mm dishes in HAT medium (0.1 mM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymine) for 2 weeks. Contraction frequencies were calculated as the number of HPRT+ colonies divided by the number of viable cells; they are the average of at least eight experiments. A few dozen individual HPRT+ colonies were grown up, and the lengths of their repeat tracts were assayed by gel electrophoresis and DNA sequencing; all contained shortened CAG repeats, ranging from CAG7 to CAG35.
Apoptosis assays.
In the fluorescein isothioncyanate (FITC)-annexin V-propidium iodide (PI) assay (BD Biosciences), different densities of cells were plated in each well of 6-well plate on day −1. On days 0, 1, 2, 3, and 4, the inducers doxycyline and RSL1 were added for 5-, 4-, 3-, 2-, and 1-day treatments, respectively. On day 5, adherent cells were trypsinized, collected together with nonadherent cells by brief centrifugation (3,000 rpm for 3 min), and resuspended in binding buffer (BD Biosciences) at a concentration of 1 × 106 cells/ml. About 100,000 cells were treated with FITC, annexin V, and PI according to the manufacturer's instructions. Annexin V is a 35- to 36-kDa Ca2+ dependent phospholipid-binding protein that has a high affinity for phosphatidylserine, which is translocated from the inner to the outer leaflet of the plasma membrane in early apoptotic cells. Viable cells have intact membranes and thus exclude PI, while dead cells are permeable to it. Viable cells are both annexin V and PI negative, early apoptotic cells are annexin V positive but PI negative, and dead cells are both annexin V and PI positive. Cells were incubated at room temperature for 15 min before analysis using a FACSCantoII fluorescence-activated cell sorter (BD Biosciences). Frequencies of apoptosis were calculated as the number of apoptotic cells divided by the total number of cells, which is the sum of viable cells, apoptotic cells, and dead cells.
In the Vybrant apoptosis assay (Invitrogen), different densities of cells were plated in each well of 6-well plate on day −1. On days 0, 1, 2, 3, and 4, the inducers doxycyline and RSL1 were added for 5-, 4-, 3-, 2-, and 1-day treatments, respectively. On day 5, the medium was removed and the cells were washed once with 1 ml of phosphate-buffered saline (PBS). Hoechst 33342 and PI were then added according to the manufacturer's instructions. Cells were incubated on ice for 30 min before observation by fluorescence microscopy. Hoechst 33342 stains the condensed chromatin of apoptotic cells more brightly than the chromatin of normal cells. Thus, dead cells are red and bright blue cells, apoptotic cells are bright blue but not red cells, and viable cells are light blue. Frequencies of apoptosis were calculated as the number of apoptotic cells divided by the total number of cells, which is the sum of viable, apoptotic, and dead cells.
Cell cycle assays.
To analyze the distribution of cells in the cell cycle, 250,000 DIT7 cells were plated in each well of a 6-well plate on day −1. On day 0, cells were refed with medium and transcription inducers were added. On day 2, the distribution of cells in the cell cycle was determined using two methods. For the flow cytometry assay (BrdU [bromodeoxyuridine] flow kit; BD Biosciences), BrdU was added to the culture medium at a final concentration of 3 mg/ml 1 h before cells were collected. The cells were then treated according to the manufacturer's instructions. Briefly, cell pellets were washed once with 1 ml of staining buffer. Cells were fixed by resuspension in 100 μl of Cytofix/Cytoperm buffer and incubation at room temperature for 20 min. Cells were washed once with 1 ml of BD Perm/Wash buffer, resuspended in 100 μl of Cytofix/Cytoperm buffer, incubated for 5 min at room temperature, and washed again. To expose BrdU for antibody binding, cells were resuspended in 100 μl of diluted DNase (300 μg/ml), incubated at 37°C for 1 h, and then washed once. Cells were then resuspended in 50 μl of BD Perm/Washer buffer containing diluted (1:50) fluorescent anti-BrdU antibody and incubated at room temperature for 20 min. Cells were washed once with Perm/Wash buffer and resuspended in 1 ml binding buffer containing 20 μl of 7-amino-actinomycin D, which stains DNA. The suspensions were stored at 4°C overnight before analysis by flow cytometry using the FACSCantoII (BD Biosciences). As per the manufacturer's recommendation, cells with values of 110 or higher for BrdU incorporation in the cytometry plot (y axis) were defined as S-phase cells, cells with values higher than about 100 on the x axis, which represents the relative cellular DNA level, were defined as G2/M-phase cells, and cells with values lower than about 100 on the x axis were defined as G0/G1-phase cells.
For the cell cycle staining assay (BD Biosciences cell cycle kit), BrdU was added to the culture medium at a final concentration of 4 mg/ml and incubated at 37°C for 1 h. To fix cells, 250 μl of fixation buffer was added to 3 ml of the medium and incubated at room temperature for 15 min. Cells were washed twice with PBS, followed by incubation with 400 μl of cold Perm buffer III at room temperature for 10 min. Cells were washed twice with PBS and incubated with 400 μl of stain buffer at room temperature for 30 min. To expose BrdU for antibody binding, cells were incubated in 500 μl of PBS containing 0.3 mg/ml DNase at 37°C for 1 h, followed by one wash with PBS. Cells were then stained at room temperature for 15 min with Alexa Fluor 488 mouse anti-BrdU antibody (1:50) for BrdU, Alexa Fluor 647 rabbit anti-histone H3 (S28P) antibody (1:50) for histone H3, and Hoechst 33342 (0.02 mg/ml) for DNA. The conjugated antibody against BrdU generates green fluorescence; thus, cells at S phase are green. The conjugated antibody against histone H3 (S28P), which is specifically phosphorylated at M phase, generates red fluorescence; thus, M-phase cells are red. G-phase cells, which are not bound by antibody against BrdU or antibody against histone H3 (S28P), are blue due to Hoechst staining.
Immunofluorescence microscopy.
For immunofluorescence microscopy, we grew DIT7 cells on poly-l-lysine-coated coverslips, so that they were at about 50% confluence at the time of analysis. After treatment, we fixed cells in methanol for 20 min at −20°C and then rehydrated and washed them with PBS. Fixed cells were incubated with the specific primary antibody in PBS containing 2% goat serum for 16 h at 4°C and then incubated with the corresponding secondary antibody in PBS with 2% goat serum for 1 h at room temperature. Finally, cells were mounted with ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes). The cells were observed by fluorescence microscopy (Nikon Eclipse TE2000-U). In this study, we used rabbit polyclonal antibodies against cleaved caspase 3 (Asp175) (1:50), ATR (ataxia-telangiectasia mutated [ATM] and Rad3 related)-S428P (1:2,000), and CHK1-S345P (1:1,000) and mouse monoclonal antibodies against p53-S15P (1:100) and ATM-S1981P (1:5,000) (Cell Signaling Technologies). Secondary antibodies for immunostaining included Alexa Fluor 488 goat anti-rabbit IgG (1:1,000) or Alexa Fluor 569 goat anti-mouse IgG (1:1,000) (Molecular Probes).
Cells were counted in randomly selected fields on two to three coverslips for each of three different experimentally treated cell populations. For each coverslip, at least 1,000 total cells were evaluated and the results for multiple coverslips were summed. Percentages of positive cells were averaged for the three cell populations and displayed along with their standard deviations (SDs).
ChIP.
DIT7 cells were treated for 48 h in the absence or presence of doxycycline plus RSL1 and then chemically cross-linked by the addition of fresh formaldehyde solution to a final concentration of 1%, followed by gentle shaking at room temperature for 10 min. Cross-linking was stopped by addition of 2 M glycine after rinsing cells twice with cold PBS. Cells were collected using a silicon scraper and then lysed and sonicated to solubilize and shear cross-linked DNA using a Virsonic 600 sonicator. Sonication was carried out at 4°C at power setting 4 for 10 10-s pulses with 30-s pauses between pulses. The whole-cell extract was then precleared by addition of 50 μl protein A beads, 10 μl IgG, 10 μl 5% bovine serum albumin (BSA), and 5 g of sheared salmon sperm DNA and centrifuged at 13,000 rpm for 5 min at 4°C in a benchtop centrifuge. After centrifugation, the supernatant was incubated overnight at 4°C with 30 μl of protein A agarose beads, 3 μg of individual antibodies, 1 μl 5% BSA, and 25 μg of salmon sperm DNA. Beads were washed four times with chromatin immunoprecipitation (ChIP) buffer and once with Tris-EDTA (TE) containing 1 mM dithiothreitol. Bound complexes were eluted from the beads using elution buffer (0.5% SDS, 0.1% NaHCO3), and cross-linking was reversed by overnight incubation at 65°C. Immunoprecipitated DNA and whole-cell extract DNA were then purified using a Qiagen PCR purification kit (Qiagen). The antibodies used for the ChIP assay were directed against RNAP II (Abcam), ATR (Abcam), ATRIP (ATR-interacting protein) (Abcam), TOPBP1 (topoisomerase II binding protein 1) (Abcam), RPA70 (Cell Signaling Technologies), and nonspecific IgG antibodies (Sigma). Immunoprecipitated DNA was quantified by real-time PCR at two sites: ChIP-1, which is adjacent to the CAG repeat tract; and ChIP-2, which is about 1 kb away. The PCR product at ChIP-1 is 82 bp, and the product at ChIP-2 is 106 bp. Enrichment of ChIP-1 DNA in the specific antibody pulldown relative to the IgG pulldown under induced (doxycycline plus RSL1) versus uninduced conditions was calculated from the PCR results as follows: ChIP-1 = [(antibody induced/antibody uninduced)/(IgG induced/IgG uninduced)]. Enrichment of ChIP-2 DNA is calculated as ChIP-2 = [(antibody induced/antibody uninduced)/(IgG induced/IgG uninduced)].
Statistics.
Statistical analyses of significance were conducted using Student's t test to compare the means and standard deviations derived from multiple experiments.
RESULTS
Convergent transcription destabilizes CAG repeats.
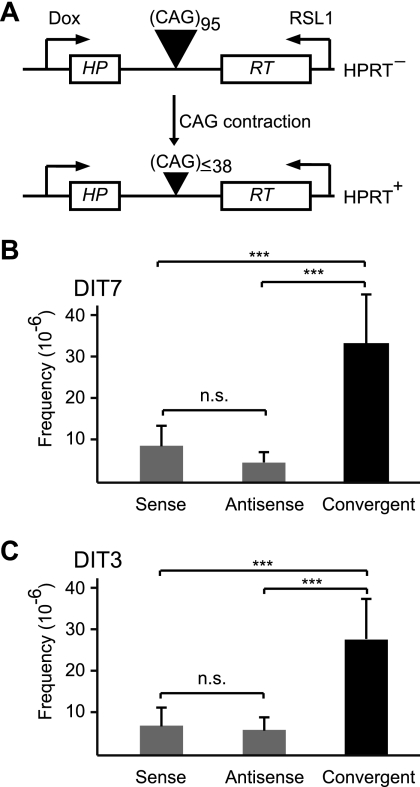
To test the effects of convergent transcription on CAG repeat instability, we constructed two human cell lines—DIT7 and DIT3—in which transcription could be independently induced from either end of a randomly integrated HPRT minigene that harbors a CAG95 repeat tract in its intron (Fig. 1A). Because the CAG95 repeat forces aberrant splicing of the HPRT mRNA, rendering the protein nonfunctional, we can select for contraction to fewer than 39 units, the length compatible with correct splicing and normal HPRT function (Fig. 2 A) (18, 39). In these cells, the doxycycline-inducible Tet-ON promoter controls sense transcription and the RSL1-inducible Rheo-Switch promoter regulates antisense transcription. Strand-specific real-time RT-PCR (Fig. 1H) indicated that doxycycline induced sense transcription 22-fold in DIT7 cells and 15-fold in DIT3 cells and that RSL1 induced antisense transcription 16-fold in DIT7 cells and 10-fold in DIT3 cells (Table 3).
FIG. 2.
Convergent transcription and repeat instability. (A) Selection assay for repeat contraction; (B) CAG contraction in DIT7 cells. Sense transcription was induced by doxycycline (200 ng/ml), antisense transcription by RSL1 (500 nM), and convergent transcription by doxycycline plus RSL1. Frequencies of HPRT+ colonies minus the background in the absence of inducers ([1.8 ± 0.9] × 10−6) are sense ([9.2 ± 5.1] × 10−6), antisense ([4.8 ± 2.4] × 10−6), and convergent ([33 ± 12] × 10−6). Frequencies are mean values of eight independent measurements, with error bars indicating standard deviation. (C) CAG contraction in DIT3 cells. Sense transcription was induced by doxycycline (200 ng/ml), antisense transcription by RSL1 (500 nM), and convergent transcription by doxycycline plus RSL1. Contraction frequencies minus the background in the absence of inducers ([2.1 ± 1.6] × 10−6) were sense ([6.9 ± 3.5] × 10−6), antisense ([5.9 ± 2.3] × 10−6), and convergent ([26 ± 9.2] × 10−6). Frequencies were averaged from six independent experiments, with error bars indicating standard deviation. Statistical significance is indicated: n.s., not significant; and ***, P < 0.0001.
TABLE 3.
Relative sense and antisense transcript levels of HPRT minigene
| Cell line | Relative transcript level in cellsa: |
|||
|---|---|---|---|---|
| Untreated |
Treated with doxycycline + RSL1 |
|||
| Sense | Antisense | Sense | Antisense | |
| DIT7 | 1.0 ± 0.1 | 1.0 ± 0.2 | 22 ± 3.1 | 16 ± 3.3 |
| DIT3 | 0.7 ± 0.1 | 2.2 ± 0.3 | 15 ± 0.5 | 9.7 ± 3.0 |
| FLAH9 | 0.4 ± 0.3 | 16 ± 4.2 | ||
| FLAH25 | 0.6 ± 0.2 | 14 ± 2.7 | ||
| DIT-NAT10 | 0.3 ± 0.1 | 0.2 ± 0.1 | 19 ± 3.6 | 0.5 ± 0.2 |
| DIT-NAT26 | 0.8 ± 0.2 | 0.2 ± 0.1 | 25 ± 5.3 | 0.3 ± 0.1 |
| DIT-NR4 | 0.2 ± 0.1 | 1.2 ± 0.3 | 10 ± 0.5 | 11 ± 0.7 |
| DIT-NR5 | 0.6 ± 0.1 | 1.3 ± 0.5 | 19 ± 2.3 | 14 ± 3.2 |
| DH7 | 1.1 ± 0.1 | 38 ± 8.3 | 11 ± 1.1 | 36 ± 7.7 |
| DH10 | 0.2 ± 0.1 | 24 ± 4.0 | 12 ± 2.3 | 26 ± 1.2 |
| DIT7-R15 | 25 ± 5.9 | 14 ± 2.5 | ||
| DIT7-R20 | 24 ± 4.2 | 16 ± 4.2 | ||
| DIT7-R25 | 27 ± 4.8 | 17 ± 2.8 | ||
| DIT7 (starved) | 2.5 ± 0.4 | 1.9 ± 0.5 | 17 ± 1.3 | 7.2 ± 0.5 |
| DIT3 (starved) | 2.8 ± 0.3 | 1.8 ± 0.2 | 12 ± 1.4 | 9.3 ± 0.9 |
| DIT7 (confluent) | 1.6 ± 0.4 | 1.3 ± 0.3 | 24 ± 4.7 | 20 ± 3.5 |
| DIT7 (zVAD) | 21 ± 3.8 | 16 ± 2.9 | ||
The levels of sense and antisense transcription in DIT7 cells before induction were arbitrarily defined as 1 and used as the basis for comparison. Shown here are the relative levels of sense and antisense transcripts in various cell lines, which were determined using strand-specific RT-PCR as described in the legend to Fig. 1H.
To test the effects of sense, antisense, and convergent transcription on CAG repeat instability, we treated DIT cells with doxycycline, RSL1, or both inducers simultaneously for 24 h every other day for a total of 3 days prior to selection for HPRT+ colonies. DIT7 and DIT3 cells yielded similar results, with the frequencies of HPRT+ colonies induced by antisense transcription (4.8 × 10−6 and 5.9 × 10−6, respectively) somewhat lower than those induced by sense transcription (9.2 × 10−6 and 6.9 × 10−6, respectively) (Fig. 2B and C). In contrast, convergent transcription gave substantially higher frequencies of HPRT+ colonies in both DIT7 cells (33 × 10−6) and DIT3 cells (26 × 10−6). The frequencies induced by convergent transcription were 2.4-fold higher than the sum of those induced by sense and antisense transcription alone for DIT7 cells and 2.1-fold higher for DIT3 cells. In all cases, the HPRT+ colonies isolated after induction of sense, antisense, or convergent transcription were shown to contain contractions of the CAG repeat tract to lengths below 39 units (data not shown). These results suggest a synergistic effect of convergent transcription on repeat instability. This effect is most evident at high levels of sense and antisense transcription (data not shown).
Because there is some cell death in these assays (about 10% for DIT3 and 30% for DIT7) and because cells with shorter repeat tracts survive better than cells with long repeat tracts (see Fig. 4B), we tested the possibility that convergent transcription-induced contractions are overestimated due to their growth advantage relative to the parental cells. When the caspase inhibitor zVAD [benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethyl ketone] (20 μM) was added together with the inducers to prevent apoptosis (see Fig. 5C), the contraction frequency was not significantly different from that observed in the absence of zVAD (data not shown), arguing against any substantial overestimate of the contraction frequency induced by convergent transcription.
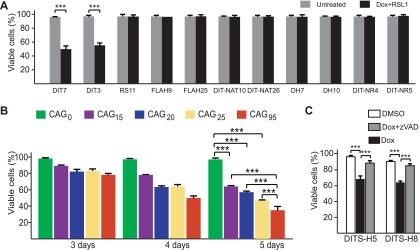
FIG. 4.
Effects of convergent transcription and length of CAG repeats on cell death. (A) Convergent transcription and cell death in various cell lines. Cell lines were treated for 4 days with or without Dox plus RSL1. (B) Correlation between CAG tract length and cell death induced by convergent transcription; (C) convergent transcription and apoptosis in DITS-H cells. DITS-H5 and DITS-H8 cells were treated with DMSO, doxycycline plus DMSO, or doxycycline plus zVAD for 7 days. Percentages of viable cells were calculated as described in the legend to Fig. 3 and are averaged from at least six independent measurements. Error bars show standard deviations. Statistical significance is indicated: ***, P < 0.0001.
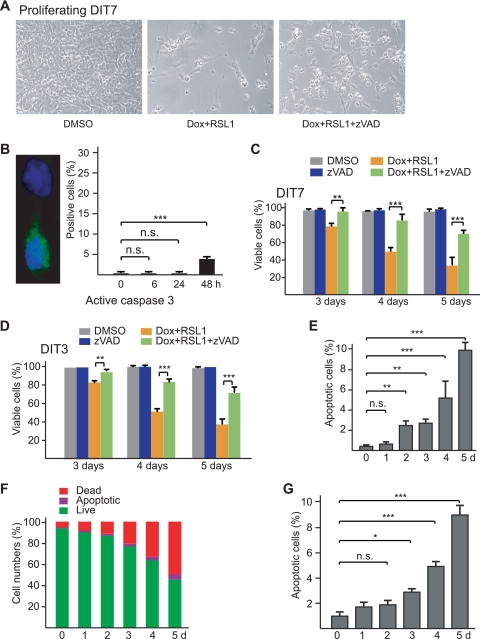
FIG. 5.
Convergent transcription-induced apoptosis. (A) Effect of the caspase inhibitor zVAD (20 μM) on death of proliferating DIT7 cells. Cells were plated at the same initial density, treated with DMSO alone, Dox plus RSL1, or Dox plus RSL1 plus zVAD, and photographed 4 days later. (B) Increased level of active caspase 3 after induction of convergent transcription. The micrograph shows an example of an immunostaining, with a cell negative for active caspase 3 above and a cell positive for active caspase 3 below. The graph shows the mean number ± standard deviation (SD) of cells that were positive for active caspase 3 from 0 to 48 h after induction of convergent transcription. (C) Suppression of convergent-transcription-induced cell killing by zVAD (20 μM) in proliferating DIT7 cells; (D) suppression of convergent-transcription-induced cell killing by zVAD (20 μM) in proliferating DIT3 cells. Percentages of viable cells were calculated as described in the legend to Fig. 3 and are averaged from at least six independent measurements. Error bars show standard deviations. (E) Frequencies of apoptotic cells measured by the PI-Hoechst 33342 staining method. Cells were plated at the same density in six-well plates. Doxycycline (200 ng/ml) and RSL1 (500 nM) were added to different wells at 5 days, 4 days, 3 days, 2 days, and 1 day before staining. For each time point in each experiment, random fields of adherent cells were examined by fluorescence microscopy and a total of at least 1,000 cells were counted. Most dead cells were washed away prior to staining; however, dead cells among the adherent cells increased from 0.3% at day 0 to 3.2% at day 5. Frequencies of apoptotic cells, which are the means of three measurements for each time point, are expressed as a percentage of total adherent cells. Error bars indicate standard deviations. (F) Percentages of live, apoptotic, and dead cells determined by flow cytometry after staining with FTTC, annexin, and PI. The means of three assays are shown. For each time point in each experiment, 10,000 cells were counted by flow cytometry. (G) Frequencies of apoptotic cells measured by flow cytometry after staining with FITC, annexin V, and PI. Frequencies of apoptotic cells increased from 0.3% at day 0 to 8.9% at day 5. Frequencies of apoptotic cells are expressed here as a percentage of live plus apoptotic cells for ease of comparison with the data in panel E. Frequencies are the means of three experiments for each time point, with error bars indicating the standard deviation. For each time point in each experiment, 10,000 cells were counted by flow cytometry. Statistical significance is indicated: n.s., not significant; *, P < 0.05; **, P < 0.001; and ***, P < 0.0001.
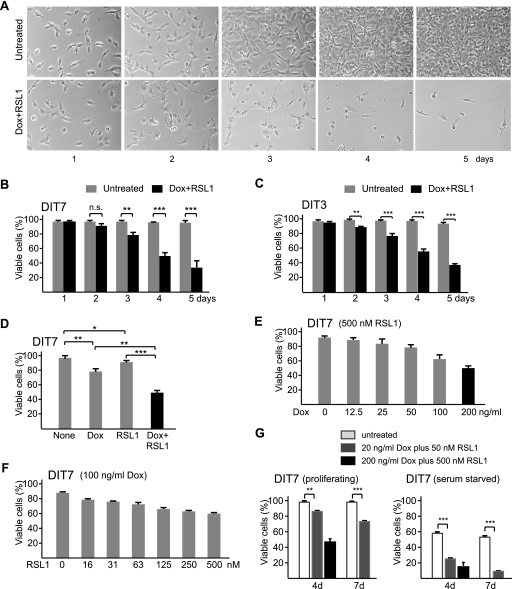
Convergent transcription triggers substantial cell death.
To characterize the cell death induced by convergent transcription, we measured the numbers of adherent (viable) and nonadherent (dead) cells over a 5-day time course in the presence or absence of doxycycline plus RSL1. In the absence of inducers, DIT7 cells proliferated at normal rates with normal cell morphology (Fig. 3 A) and maintained about 97% viability through 5 days (Fig. 3B). When convergent transcription was induced, however, the proportion of viable cells decreased to about 35% over 5 days (Fig. 3B), with abnormal morphological features appearing after 2 days and detectable cell death after 3 days. DIT3 cells gave similar results (Fig. 3C).
FIG. 3.
Convergent transcription and cell death. (A) Convergent transcription in proliferating DIT7 cells. Cells were plated at the same initial density and photographed on different days, as shown. (B) Cell killing with and without convergent transcription in DIT7 cells; (C) cell killing with and without convergent transcription in DIT3 cells; (D) transcription-induced cell death in DIT7 cells. Viable cells were counted after 4 days with no inducer (None), Dox (200 ng/ml), RSL1 (500 nM), or Dox plus RSL1. (E) Cell death as a function of increasing sense transcription with fully induced antisense transcription. (F) Cell death as a function of increasing antisense transcription with highly induced sense transcription. (G) Comparison of cell killing under full or modest induction of convergent transcription in proliferating and serum-starved DIT7 cells. For full induction, 200 ng/ml doxycycline and 500 nM RSL1 were added; for modest induction, 20 ng/ml doxycycline and 50 nM RSL1 were added. For all experiments, percentages of viable cells were calculated as the number of adherent cells divided by the sum of adherent and nonadherent cells and are averaged from at least six independent measurements. Error bars show standard deviations. Statistical significance is indicated: n.s., not significant; *, P < 0.05; **, P < 0.001; and ***, P < 0.0001.
Induction of either sense or antisense transcription alone also caused some cell death, but at lower rates than convergent transcription (Fig. 3D). Because no cell death occurred when sense transcription was fully induced in FLAH cells (Fig. 4 A), which lack an antisense promoter (Fig. 1B), we suspected that cell killing in singly induced DIT cells arose from the combination of fully induced transcription from one direction and basal transcription from the other (Table 3). Consistent with this idea, when DIT7 cells were fully induced for antisense transcription (500 nM RSL1), the amount of cell killing correlated with the induced level of sense transcription, which was controlled using different concentrations of doxycycline (Fig. 3E). Similarly, when DIT7 cells were highly induced for sense transcription (100 ng/ml doxycycline), the amount of cell killing correlated with the induced level of antisense transcription, which was controlled using different concentrations of RSL1 (Fig. 3F). Moreover, when both sense and antisense transcription were modestly induced, DIT7 cells displayed intermediate levels of cell killing in both proliferating and nonproliferating cells (Fig. 3G). These experiments indicate that the level of convergent transcription determines the efficiency of cell killing.
Cell death requires convergent transcription and CAG repeats.
To establish that convergent transcription through a CAG repeat tract caused cell death, we assessed the effects of doxycycline plus RSL1 on cell survival in cell lines that were designed to test particular alternative explanations. To test the possibility that cell killing was unrelated to HPRT transcription, we exposed RS11 cells to doxycycline plus RSL1. RS11 cells were used to construct DIT cells; they contain all of the regulatory components required to induce sense and antisense transcription in DIT cells but lack the HPRT minigene, its CAG repeat, and flanking promoters. RS11 cells grew normally in the presence of doxycycline and RSL1 (Fig. 4A), indicating that one or more components of the HPRT minigene construct are essential for cell killing.
To test whether antisense transcription was required for cell killing, we examined two different types of cell. FLAH9 and FLAH25 cells (39) contain neither the antisense promoter nor the regulatory components necessary to drive antisense transcription (Fig. 1B). DIT-NAT10 and DIT-NAT26 cells, which were isolated from the transfection that generated DIT7 and DIT3 cells, ostensibly contain all of the same elements, but the antisense promoter is impaired for reasons that are not clear (Fig. 1C). DIT-NAT cells have very low basal levels of antisense transcription that do not respond to RSL1 (Table 3). Both FLAH cells and DIT-NAT cells grow normally in the presence of doxycycline and RSL1 (Fig. 4A). Thus, antisense transcription is required for cell death.
To test the possibility that cell killing might be an indirect effect of sense and antisense transcription (for example, via a double-stranded RNA), we constructed DH7 and DH10 cells. These cells, which were derived from FLAH25 cells, contain two copies of the HPRT minigene—each with a CAG95 tract (Fig. 1D). The doxycycline-inducible promoter drives sense transcription from the copy that was present in the parental FLAH25 cells. The constitutive CMV promoter drives antisense transcription from the second copy, which lacks a promoter for sense transcription. In the presence of doxycycline plus RSL1, the levels of sense and antisense transcripts are comparable to those in DIT cells (Table 3); however, DH7 and DH10 cells are not killed (Fig. 4A). These results indicate that sense and antisense transcription (i.e., convergent transcription) through the same HPRT minigene is required for cell killing, and they argue against the possibility that cell death in DIT cells occurs via a double-stranded RNA intermediate. As a more direct test of the possible role of RNA interference, we showed that knockdown of Argonaute 2, Dicer, or Drosha—components involved in generation of cellular siRNAs and microRNAs (miRNAs) (4)—did not decrease cell death (data not shown), as would have been expected if cell killing were mediated by small RNAs processed from a double-stranded RNA intermediate. Finally, the lack of cell killing by high levels of antisense transcription in DH cells (Fig. 4A) rules out the possibility that antisense transcription of the HPRT minigene, by itself, might be toxic.
To examine the requirement for the CAG repeat in cell killing, we constructed DIT-NR4 and DIT-NR5 cells, which carry an HPRT minigene that lacks the CAG95 repeat (Fig. 1E). Doxycycline plus RSL1 efficiently induced convergent transcription in these cells (Table 3) but did not trigger cell death (Fig. 4A). Thus, cell killing requires the presence of a CAG tract. To determine the relationship between tract length and cell killing, we tested three DIT7-derived HPRT+ colonies, DIT7-R15, DIT7-R20, and DIT7-R25, which had repeat tracts of 15, 20, and 25 CAG units, respectively. Sense and antisense transcription in DIT7-R clones could be fully induced (Table 3). Because these clones were derived from DIT7 cells, they differ only in the length of the repeat tract, eliminating possible effects of genomic context. Cell killing induced by convergent transcription in DIT7-R cells was compared with that in DIT7 cells and DIT-NR4 cells (Fig. 4B). After 5 days, for example, the percentages of viable cells for DIT7-R15 (64%), DIT7-R20 (57%), and DIT7-R25 (47%) were significantly higher than that for DIT7 cells (35%; P < 0.0001 for each) and significantly lower than that for DIT-NR4 cells (96%; P < 0.0001 for each). These results show that the CAG repeat tract is required for cell killing and that its length influences the cell's sensitivity to convergent transcription.
To test the possibility that this phenomenon might be peculiar to HT1080 cells, we generated a similar system in HEK293-derived HEK293 F-T cells, which express rtTA, the regulator required for expression of the Tet-ON-inducible system. We tested two cell lines, DITS-H5 and DITS-H8, in which the HPRT minigene with a CAG95 repeat was equipped with doxycycline-responsive promoters to drive both sense and antisense transcription (Fig. 1G). When convergent transcription was induced by addition of doxycycline, these cells, like their HT1080 counterparts, also displayed a cell death phenotype (Fig. 4C). As with DIT3 and DIT7 HT1080-derived cells (Fig. 5 C and D), these HEK-derived cells also survived better in the presence of the caspase inhibitor zVAD (Fig. 4C). Thus, convergent-transcription-induced cell death is not a unique feature of a single human cell line.
Collectively, the studies in this section and the previous one demonstrate that sense and antisense transcription through the same CAG repeat tract are required for cell killing and that the extent of killing depends on both the length of the repeat tract and the level of convergent transcription.
Convergent transcription induces apoptosis.
In our initial experiments, we observed that convergent transcription in DIT7 cells was accompanied by chromatin condensation (data not shown), suggesting that the cells might be dying by apoptosis. To address this possibility, we carried out immunofluorescent staining for active caspase 3, a common product of the main apoptotic pathways (65). As shown in Fig. 5B, active caspase 3 increased in DIT7 cells from 0.2% at 0 h to 3.7% at 48 h after induction of convergent transcription, suggesting that cell death occurs by apoptosis. As a test for caspase-mediated cell death, we induced convergent transcription in DIT cells in the presence and absence of 20 μM zVAD—a general inhibitor of caspases (23). Treatment with zVAD significantly increased the percentage of viable DIT7 cells (Fig. 5A and C) and DIT3 cells (Fig. 5D). Thus, cell death induced by convergent transcription can be partially suppressed (or postponed) by zVAD. As a control, we showed that zVAD does not alter the levels of sense or antisense transcription (Table 3), ruling out the trivial possibility that zVAD blocks cell death indirectly by inhibiting convergent transcription. Taken together, chromatin condensation, activation of caspase 3, and inhibition of cell death by zVAD suggest that convergent transcription induces apoptosis in DIT cells.
To measure the induction of apoptosis and its time course more directly, we assayed for apoptotic cells in two ways. Using the PI-Hoechst 33342 staining assay for apoptotic cells, we showed that convergent transcription increased the proportion of apoptotic DIT7 cells from a background of 0.4% to 2.7% after 2 days, 5.2% after 4 days, and 9.8% after 5 days (Fig. 5E). We confirmed these results using a flow cytometry assay (Fig. 5F and G). These studies show that convergent transcription induces apoptosis in DIT7 and DIT3 cells.
Convergent transcription disrupts the cell cycle.
As is apparent in Fig. 3A, convergent transcription seems to stop cell proliferation. When the data in Fig. 3B are replotted in terms of total and viable cells, it is clear that doxycycline plus RSL1 slows cell proliferation by day 2 but does not cause significant apoptosis until day 3 (data not shown). To analyze the effect of convergent transcription on cell-cycle progression, we determined the distributions of cells in the phases of the cell cycle after 0, 6, 24, and 48 h in the presence of doxycycline plus RSL1. Using two methods, we showed by 48 h that there were small, but significant, decreases in the numbers of cells in S phase (P < 0.001) and M phase (P < 0.05), with corresponding increases in the G phases (data not shown). Thus, convergent transcription causes a measurable disruption in cell cycle progression prior to cell death.
Convergent transcription induces apoptosis in nonproliferating cells.
To test whether cell proliferation is required for convergent transcription-induced cell death, we measured cell killing in DIT cells whose proliferation had been arrested. We used either serum starvation or confluent growth to arrest cell growth (data not shown). As with proliferating cells, induction of convergent transcription also caused cell death in nonproliferating cells arrested by serum starvation (Fig. 6 A and B) or by confluent growth (Fig. 6C). Cell death in these nonproliferating cells also apparently occurs by apoptosis, since zVAD inhibited death in serum-starved DIT7 and DIT3 cells (Fig. 6D). Notably, induction of convergent transcription killed nonproliferating cells more rapidly (compare Fig. 3B with Fig. 6B), with significant cell death already apparent 2 days after addition of doxycycline plus RSL1. This speedier killing occurred even though the induced levels of sense and antisense transcription were comparable to those in proliferating cells (Table 3). These results indicate that convergent transcription can induce apoptosis in both proliferating and nonproliferating cells.
Interference with the ATR pathway increases cell death.
Cell cycle arrest and apoptosis suggest that convergent transcription elicits a stress response in DIT cells. Because the precipitating event occurs on DNA, we tested the two main signaling pathways, ATR-CHK1 and ATM-CHK2, that respond to DNA damage (26). We treated DIT7 cells with chemical inhibitors for ATM/ATR, CHK1, and CHK2 in the presence and absence of doxycycline plus RSL1. Although some of these drugs were slightly toxic to DIT7 cells in the absence of inducers (Table 4, fifth column), when the treated cells were normalized for the drugs' nonspecific toxicity, it was apparent that treatment with each of the three ATM/ATR inhibitors or the CHK1 inhibitor significantly increased cell death (Table 4, sixth column). In contrast, the CHK2 inhibitor did not affect cell death. As shown in Table 4, none of these chemicals substantially altered the induced levels of sense (third column) or antisense (fourth column) transcription though the HPRT minigene, arguing against the trivial possibility that they increased cell death indirectly by stimulating convergent transcription.
To extend these studies and to distinguish between the activities of ATM and ATR, we treated DIT7 cells with gene-specific siRNAs (Table 1) and measured their effects on cell viability. In each case, siRNA treatment knocked down gene expression more than 70%, without substantially affecting the induced levels of sense (third column) and antisense (fourth column) transcription (Table 5). After normalizing for the nonspecific cellular toxicity of some of these siRNAs (Table 5, fifth column), it was clear that knockdown of components of the ATR pathway (ATR, CHK1, ATRIP [ATR-interacting protein], and TOPBP1 [topoisomerase II binding protein 1]) (34, 70) significantly increased cell killing when convergent transcription was induced (Table 5, sixth column). In contrast, knockdown of CHK2 had no effect, while knockdown of ATM or p53 significantly decreased cell killing (Table 5, sixth column).
TABLE 5.
Effects of siRNA knockdown of ATR and ATM pathway components on DIT7 cell viability after induction of convergent transcription
| Target | siRNA | Transcription (%)a |
% of viable cells |
Significance (P value) | ||
|---|---|---|---|---|---|---|
| Sense | Antisense | −Dox and RSL1b | +Dox and RSL1 (normalized)c | |||
| Vimentin | siRNA | 100 | 100 | 97 ± 1.7 | 53 ± 2.5 | |
| ATR | siRNA1 | 108 | 113 | 94 ± 3.2 | 38 ± 3.2 | <0.01 |
| siRNA2 | 87 | 72 | 69 ± 4.3 | 35 ± 2.9 | <0.001 | |
| CHK1 | siRNA1 | 93 | 70 | 59 ± 2.8 | 33 ± 3.3 | <0.0001 |
| siRNA2 | 84 | 54 | 71 ± 3.4 | 30 ± 1.1 | <0.0001 | |
| ATRIP | siRNA1 | 106 | 70 | 98 ± 1.3 | 26 ± 1.5 | <0.0001 |
| TOPBP1 | siRNA1 | 93 | 112 | 95 ± 2.5 | 40 ± 1.2 | <0.0001 |
| ATM | siRNA1 | 89 | 87 | 89 ± 3.6 | 67 ± 2.6 | <0.001 |
| siRNA2 | 94 | 91 | 93 ± 2.1 | 74 ± 3.9 | <0.001 | |
| CHK2 | siRNA1 | 91 | 86 | 89 ± 1.1 | 51 ± 1.2 | NSd |
| siRNA2 | 108 | 94 | 96 ± 1.8 | 56 ± 1.0 | NS | |
| p53 | siRNA1 | 122 | 110 | 83 ± 2.5 | 79 ± 3.5 | <0.001 |
| siRNA2 | 83 | 89 | 96 ± 1.8 | 76 ± 2.6 | <0.0001 | |
Sense and antisense transcription through the HPRT minigene in the presence of Dox plus RSL1 was measured by real-time RT-PCR at day 1 after the second treatment with siRNAs at day 0 and expressed as a percentage of the induced transcript levels measured in the presence of vimentin siRNA. Results were averaged from three determinations.
Viable cells were counted 4 days after treatment with siRNAs (200 nM) in the absence of doxycyline (Dox) plus RSL1. siRNAs were transfected twice: at day −2 and day 0. Viable cells after siRNA treatment are expressed as a percentage of the count of untreated cells, which was defined as 100%. Mean percentages ± SD were determined from at least six independent measurements.
Cell counts were normalized to (divided by) the cell counts in the absence of doxycycline and RSL1 and expressed as a percentage of the values for vimentin siRNA. Viable cells in the sixth column = (measured viable cells [not shown]) × (viable cells in the noninduced control [vimentin siRNA] in the fifth column)/(viable cells in noninduced treatment [specific siRNA] in the fifth column). Mean percentages ± SD were determined from at least six independent measurements.
NS, not significant.
Collectively, these experiments show that the ATR pathway normally acts to minimize death in DIT7 cells experiencing convergent transcription through a CAG repeat tract, which is similar to the effects of ATR pathway inhibitors on the cell death caused by irradiation, hypoxia, and DNA replication stress (24, 50, 62). They also suggest that ATM and p53 may normally act to promote convergent transcription-induced cell death, consistent with the roles ATM and p53 play in promoting apoptosis in other systems (28, 56, 69).
Convergent transcription activates the ATR pathway.
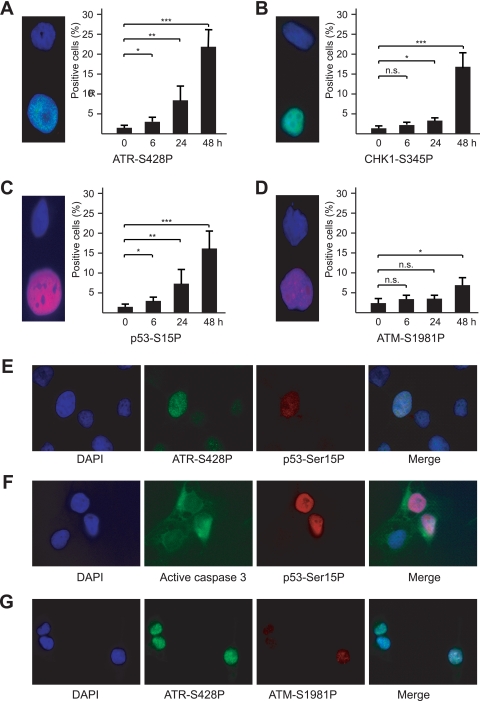
As a direct test of the involvement of the stress response pathway, we induced convergent transcription in DIT7 cells and assayed for the appearance of the active, phosphorylated forms of ATR, CHK1, ATM, and p53 (59), using phosphorylation-specific antibodies and immunofluorescence microscopy. For 2 days after induction of convergent transcription, which is before massive cell death occurs, we followed phosphorylation at serine 428 in ATR (ATR-S428P), serine 345 in CHK1 (CHK1-S345P), serine 1981 in ATM (ATM-S1981P), and serine 15 in p53 (p53-S15P). As shown in Fig. 7, ATR-S428P, CHK1-S345P, and p53-S15P displayed similar kinetics of appearance, becoming visible in some cells by 6 h with increasing numbers of positive cells up to 48 h. In contrast, ATM-S1981P appeared only after about 48 h (Fig. 7D), matching the slower kinetics of appearance observed for cleaved (active) caspase 3—a molecular marker for a major apoptosis pathway in mammalian cells (65) (Fig. 5B). These kinetic differences suggest that ATR plays an earlier role than ATM in the cellular response to convergent transcription. Notably, we observed p53-S15P and ATM-S1981P only in cells positive for ATR-S428P and active caspase 3 only in cells with p53-S15P (Fig. 7E to G). Although these experiments were all conducted in proliferating cells, we observed the same pattern of activation in nonproliferating DIT7 cells (data not shown).
FIG. 7.
ATR response after induction of convergent transcription in DIT7 cells. (A) ATR-S428P; (B) CHK1-S345P; (C) p53-S15P; (D) ATM-S1981P. Individual proteins were assessed at 0, 6, 24, and 48 h by immunostaining. Micrographs show examples of cells that are negative (above) and positive (below). Results were averaged from three determinations, and error bars represent standard deviations. Statistical significance is indicated: n.s., not significant; *, P < 0.05; **, P < 0.001; and ***, P < 0.0001. (E) p53-S15P fluorescence in ATR-S428P-positive cells; (F) ATM-S1981P fluorescence in ATR-S428P-positive cells; (G) active caspase 3 fluorescence in p53-S15P-positive cells.
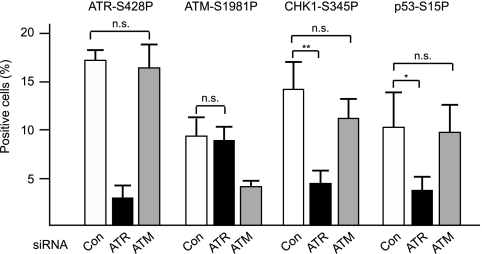
To determine the roles of the kinases, we knocked down ATR or ATM and counted cells positive for phosphorylated forms of ATR, ATM, CHK1, and p53. As shown in Fig. 8, knockdown of ATR significantly reduced ATR-S428P-positive cells, as expected, as well as CHK1-S345P- and p53-S15P-positive cells, but did not alter the number of ATM-S1981P-positive cells. In contrast, knockdown of ATM reduced ATM-S1981P-positive cells, as expected, but did not significantly affect the levels of ATR-S428P-, CHK1-S345P-, or p53-S15P-positive cells. These results suggest that ATR is primarily responsible for phosphorylation of CHK1 and p53 and that ATR and ATM are activated independently of one another in the response to convergent transcription.
FIG. 8.
Roles of ATR and ATM in downstream phosphoryation. DIT7 cells were treated with ATR or ATM siRNAs (or with vimentin siRNA as a control) prior to and during induction of convergent transcription. Cells positive for phosphorylated proteins were counted 2 days after convergent transcription was induced. Results were averaged from three determinations, and error bars represent standard deviations. Statistical significance is indicated: n.s., not significant; *, P < 0.05; and **, P < 0.001.
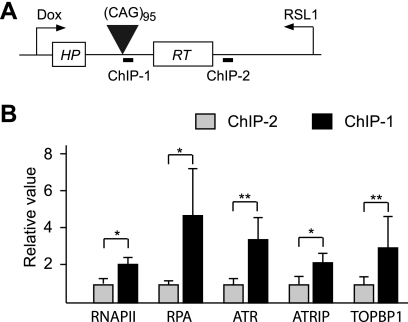
ATR components and RNAP II are recruited to CAG tract.
To link the components of the ATR response pathway to the CAG repeat tract itself, we induced convergent transcription for 2 days and then carried out chromatin immunoprecipitation (ChIP) assays, using antibodies against individual proteins. The amount of DNA pulled down by individual antibodies was analyzed using real-time PCR at a site near the repeat tract (ChIP-1) and another site (ChIP-2) about 1 kb away, but within the same transcription unit (Fig. 9 A). As shown in Fig. 9B, the amount of ChIP-1 DNA pulled down by three ATR pathway factors—ATR, ATRIP, and TOPBP1—was significantly larger than the amount of ChIP-2 DNA, indicating that these proteins were enriched at the repeat region during convergent transcription. Similarly, the antibodies against RNAP II and RPA (a single-stranded DNA binding protein) pulled down significantly larger amounts of ChIP-1 DNA than ChIP-2 DNA. These results suggest that during convergent transcription, ATR components are recruited to CAG repeat tracts that contain stalled RNAP II complexes and RPA-coated single-stranded DNA.
FIG. 9.
Chromatin immunoprecipitation assay. Enrichment of proteins at two sites, ChIP-1 and ChIP-2, was assayed 48 h after induction of convergent transcription. Relative values are the values determined for ChIP-1 and ChIP-2 (as calculated in Materials and Methods) normalized to values at ChIP-2. Results are averaged from three determinations, and error bars show standard deviations. Statistical significance is indicated: *, P < 0.05; and **, P < 0.001.
DISCUSSION
Antisense transcription occurs in a high fraction of the genes in the human genome (31), including most of the genes in which expansions of repeat sequences cause disease (5, 6, 27, 35, 49). These studies led us to test the effects of antisense transcription on triplet repeats in human cells. Here, we show that antisense transcription through a CAG repeat has a similar destabilizing effect to sense transcription. However, when sense and antisense transcription occur simultaneously to produce convergent transcription, the effects of transcription are dramatically enhanced. Convergent transcription through a CAG repeat tract stimulates repeat instability synergistically, arrests the cell cycle, and causes massive cell death via apoptosis. In addition, the effects on repeat instability and cell death do not require cell proliferation, suggesting that they are unlikely to be dependent on DNA replication.
A key question is why convergent transcription through a CAG repeat is so much more detrimental to the cell than sense or antisense transcription alone or than convergent transcription in the absence of a repeat tract. Because a stalled RNAP II can trigger TC-NER (25), we proposed in our previous studies of sense transcription-induced repeat instability (39, 41, 42) that RNAP II stalled at MSH2/MSH3-stabilized CTG hairpins in the template strand, which then served as a signal to elicit TC-NER of the perceived damage, leading to frequent changes in repeat tract length (40). Formation and stabilization of CTG hairpins may be increased by R-loops, which persist in transcribed repeat tracts (37). An analogous scenario triggered by the stalling of RNAP II at stabilized CAG hairpins could plausibly account for the similar level of TNR instability induced by antisense transcription through the repeat tract. We speculate that convergent transcription is much more harmful to the cell than either sense or antisense transcription because it generates stalled RNAP II complexes at repeat-induced hairpins on both template strands. The presence of hairpin-stalled RNAP II on each of the unpaired strands in a repeat tract, perhaps aided by R-loop formation, could create an abnormal bubble in the DNA, a unique structure that could not be produced in the absence of sense transcription, antisense transcription, or a repeat tract. It is possible that such an abnormal DNA structure cannot be repaired by the cell or is repaired inefficiently, ultimately leading to a high frequency of cell death. TC-NER, which we proposed to be involved in the resolution of an RNAP II stalled at a CTG or CAG hairpin (40, 42), would not be expected to operate effectively on the separated stands of such a bubble structure, since NER is dependent on duplex structure in the neighborhood of the target lesion. In the absence of NER, other repair processes that enhance repeat instability may become involved in resolving the bubble. For example, nicks in the two strands of the bubble would generate a double-strand break, whose repair we have previously shown is accompanied by a substantial increase in CAG repeat contractions (48).
The persistence of this or some other abnormal structure is consistent with the idea that convergent transcription stresses the cell in a way that can lead to cell death. The disruption of the cell cycle and eventual apoptosis that accompany convergent transcription through a CAG repeat are consistent with the well-documented transcription-induced stress response. At sites of DNA damage, stalled RNAP II complexes can remain associated with the DNA template for many hours (61), and failure to remove them—as occurs in TC-NER-deficient cells—leads to a strong apoptotic signal (43). This transcriptional stress response apparently does not depend on actual DNA damage since it can be induced by injection of antibodies to RNAP II (10). In addition, the transcriptional stress response is independent of DNA replication (10), consistent with our observation that similar responses occur in both proliferating and nonproliferating cells.
Our studies of convergent transcription through a CAG repeat tract do not define the exact nature of the abnormal DNA structure that triggers the cells' stress response and causes apoptosis. What we do know is that convergent transcription leads to enrichment of RNAP II, ATR, ATRIP, and TOPBP1 at the CAG repeat tract. In addition, ATR is phosphorylated along with its downstream targets CHK1 and p53, suggesting that the structure may be capable of directly activating the ATR response, as has been suggested for the transcriptional stress response (44). Alternatively, ATR activation might require the generation of single-strand breaks, as might plausibly occur via abortive repair by TC-NER, a pathway that we have previously shown to be required for transcription-induced repeat instability (42). The ATR pathway typically acts early to preserve cell viability by affecting cell cycle progression and promoting repair of the detrimental DNA structure; however, if repair is unsuccessful or the “stress” is persistent, the pathway will induce apoptosis (7, 50).
It is remarkable that convergent transcription through a CAG tract present once in the genome is sufficient to trigger a robust apoptotic response. Previous studies have demonstrated that genomewide interference with transcription by UV light, actinomycin D, psoralen, or antibodies against the RNAP II elongation complex can stimulate apoptosis (1, 10, 44, 45). Here, we show that a single site of convergent transcription—a single locus of presumptive RNAP II blockage—is apparently sufficient to induce apoptosis. The critical feature of this locus appears to be the ability of the TNR repeat to form structures capable of blocking transcription on both template strands. By this criterion, convergent transcription through other types of repeats might also be capable of activating the apoptotic response, if the secondary structures on each strand were sufficient to stall RNAP II. Indeed, in vitro studies have shown that transcription stalls at the repeat tract of various types or DNA sequences that can form secondary structures (3, 14, 21, 52, 66), indicating that noncanonical DNA structures can cause problems for RNAP.
Convergent transcription through a CAG tract represents a previously undescribed mechanism for triggering apoptosis. It may be particularly relevant to the pathogenesis associated with the 14 TNR disease genes affected by CAG·CTG tracts. Convergent transcription-induced apoptosis would be expected to occur in all these disease genes at a level dependent on the amount of sense and antisense transcription. Because this mechanism induces apoptosis equally well in proliferating and nonproliferating cells, it could contribute to the loss of differentiated cells—neurons or muscle cells—that is observed in most of these TNR diseases. A recent classification of the pathogenic mechanisms for these diseases lists nine as protein gain of function, two as RNA gain of function, and three as unknown (36). Pathogenesis in these diseases is complicated. Some have been intensely studied, and the proposed link to cell death is convincing. For Huntington's disease (HD), for example, cell toxicity is almost certainly due to the polyglutamine-containing protein encoded by the disease gene, since mutating a specific caspase-6 cleavage site renders the protein nontoxic (22). For myotonic dystrophy type 1 (DM1), an RNA containing an expanded CUG repeat clearly interferes with the function of the splicing factors, CUG-BP1 and MBNL, and disrupts normal splicing patterns (57). The link between this aberrant RNA and cell death is less certain. For the three diseases whose pathogenic mechanism is classified as unknown—SCA8, SCA12, and HD-like 2 (HDL2)—the corresponding genes have been shown to express antisense transcripts (27, 49). For SCA12 and HDL2, the levels of antisense transcripts were equal to or greater than those of sense transcripts in several cell lines (27). Additional studies are needed to test the hypothesis that convergent transcription through repeat tracts contributes to the cell death observed in repeat-associated diseases.
In summary, we have shown that convergent transcription through a CAG repeat in human cells can destabilize the repeat and trigger a cellular stress response that activates the ATR pathway, leading to phosphorylation of ATR, CHK1, and p53. Ultimately, convergent transcription induces apoptosis in a high fraction of proliferating and nonproliferating cells, in a way that depends on the length of repeat tract, the transcription level, and the intactness of ATR pathway. These consequences are triggered by events at a single locus—a single toxic site—in the genome. This novel mechanism of “DNA toxicity” may be potentially relevant to the neuronal death caused by TNR expansion in humans.
Acknowledgments
We thank J. M. Shohet and Z. Chen for helping with real-time RT-PCR. We thank members of the Wilson lab for helpful discussions.
This work was supported by a grant from the NIH (GM38219) to J.H.W.
Footnotes
Published ahead of print on 20 July 2010.
REFERENCES
- 1.Arima, Y., M. Nitta, S. Kuninaka, D. Zhang, T. Fujiwara, Y. Taya, M. Nakao, and H. Saya. 2005. Transcriptional blockade induces p53-dependent apoptosis associated with translocation of p53 to mitochondria. J. Biol. Chem. 280:19166-19176. [DOI] [PubMed] [Google Scholar]
- 2.Bacolla, A., J. E. Larson, J. R. Collins, J. Li, A. Milosavljevic, P. D. Stenson, D. N. Cooper, and R. D. Wells. 2008. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 18:1545-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belotserkovskii, B. P., E. De Silva, S. Tornaletti, G. Wang, K. M. Vasquez, and P. C. Hanawalt. 2007. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 282:32433-32441. [DOI] [PubMed] [Google Scholar]
- 4.Carthew, R. W., and E. J. Sontheimer. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, W. L., J. W. Lin, H. J. Huang, S. M. Wang, M. T. Su, G. J. Lee-Chen, C. M. Chen, and H. M. Hsieh-Li. 2008. SCA8 mRNA expression suggests an antisense regulation of KLHL1 and correlates to SCA8 pathology. Brain Res. 1233:176-184. [DOI] [PubMed] [Google Scholar]
- 6.Cho, D. H., C. P. Thienes, S. E. Mahoney, E. Analau, G. N. Filippova, and S. J. Tapscott. 2005. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell 20:483-489. [DOI] [PubMed] [Google Scholar]
- 7.Cimprich, K. A., and D. Cortez. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleary, J. D., and C. E. Pearson. 2003. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 100:25-55. [DOI] [PubMed] [Google Scholar]
- 9.De Biase, I., Y. K. Chutake, P. M. Rindler, and S. I. Bidichandani. 2009. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One 4:e7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derheimer, F. A., H. M. O'Hagan, H. M. Krueger, S. Hanasoge, M. T. Paulsen, and M. Ljungman. 2007. RPA and ATR link transcriptional stress to p53. Proc. Natl. Acad. Sci. U. S. A. 104:12778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dion, V., Y. Lin, L. Hubert, Jr., R. A. Waterland, and J. H. Wilson. 2008. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum. Mol. Genet. 17:1306-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dion, V., Y. Lin, B. A. Price, S. L. Fyffe, A. Seluanov, V. Gorbunova, and J. H. Wilson. 2008. Genome-wide demethylation promotes triplet repeat instability independently of homologous recombination. DNA Repair (Amst.). 7:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ditch, S., M. C. Sammarco, A. Banerjee, and E. Grabczyk. 2009. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 5:e1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditlevson, J. V., S. Tornaletti, B. P. Belotserkovskii, V. Teijeiro, G. Wang, K. M. Vasquez, and P. C. Hanawalt. 2008. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 36:3163-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragileva, E., A. Hendricks, A. Teed, T. Gillis, E. T. Lopez, E. C. Friedberg, R. Kucherlapati, W. Edelmann, K. L. Lunetta, M. E. Macdonald, and V. C. Wheeler. 2009. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 33:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gacy, A. M., G. Goellner, N. Juranic, S. Macura, and C. T. McMurray. 1995. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81:533-540. [DOI] [PubMed] [Google Scholar]
- 17.Gatchel, J. R., and H. Y. Zoghbi. 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6:743-755. [DOI] [PubMed] [Google Scholar]
- 18.Gorbunova, V., A. Seluanov, V. Dion, Z. Sandor, J. L. Meservy, and J. H. Wilson. 2003. Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells. Mol. Cell. Biol. 23:4485-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbunova, V., A. Seluanov, D. Mittelman, and J. H. Wilson. 2004. Genome-wide demethylation destabilizes CTG·CAG trinucleotide repeats in mammalian cells. Hum. Mol. Genet. 13:2979-2989. [DOI] [PubMed] [Google Scholar]
- 20.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U. S. A. 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabczyk, E., and K. Usdin. 2000. The GAA·TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 28:2815-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham, R. K., Y. Deng, E. J. Slow, B. Haigh, N. Bissada, G. Lu, J. Pearson, J. Shehadeh, L. Bertram, Z. Murphy, S. C. Warby, C. N. Doty, S. Roy, C. L. Wellington, B. R. Leavitt, L. A. Raymond, D. W. Nicholson, and M. R. Hayden. 2006. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125:1179-1191. [DOI] [PubMed] [Google Scholar]
- 23.Green, D. R., and J. C. Reed. 1998. Mitochondria and apoptosis. Science 281:1309-1312. [DOI] [PubMed] [Google Scholar]
- 24.Hammond, E. M., M. J. Dorie, and A. J. Giaccia. 2004. Inhibition of ATR leads to increased sensitivity to hypoxia/reoxygenation. Cancer Res. 64:6556-6562. [DOI] [PubMed] [Google Scholar]
- 25.Hanawalt, P. C., and G. Spivak. 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9:958-970. [DOI] [PubMed] [Google Scholar]
- 26.Harper, J. W., and S. J. Elledge. 2007. The DNA damage response: ten years after. Mol. Cell 28:739-745. [DOI] [PubMed] [Google Scholar]
- 27.He, Y., B. Vogelstein, V. E. Velculescu, N. Papadopoulos, and K. W. Kinzler. 2008. The antisense transcriptomes of human cells. Science 322:1855-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzog, K. H., M. J. Chong, M. Kapsetaki, J. I. Morgan, and P. J. McKinnon. 1998. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science 280:1089-1091. [DOI] [PubMed] [Google Scholar]
- 29.Jung, J., and N. Bonini. 2007. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science 315:1857-1859. [DOI] [PubMed] [Google Scholar]
- 30.Karzenowski, D., D. W. Potter, and M. Padidam. 2005. Inducible control of transgene expression with ecdysone receptor: gene switches with high sensitivity, robust expression, and reduced size. Biotechniques 39:191-192, 194, 196. [DOI] [PubMed] [Google Scholar]
- 31.Katayama, S., Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C. C. Yap, M. Suzuki, J. Kawai, H. Suzuki, P. Carninci, Y. Hayashizaki, C. Wells, M. Frith, T. Ravasi, K. C. Pang, J. Hallinan, J. Mattick, D. A. Hume, L. Lipovich, S. Batalov, P. G. Engstrom, Y. Mizuno, M. A. Faghihi, A. Sandelin, A. M. Chalk, S. Mottagui-Tabar, Z. Liang, B. Lenhard, and C. Wahlestedt. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564-1566. [DOI] [PubMed] [Google Scholar]
- 32.Kovtun, I. V., Y. Liu, M. Bjoras, A. Klungland, S. H. Wilson, and C. T. McMurray. 2007. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovtun, I. V., and C. T. McMurray. 2001. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 27:407-411. [DOI] [PubMed] [Google Scholar]
- 34.Kumagai, A., J. Lee, H. Y. Yoo, and W. G. Dunphy. 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124:943-955. [DOI] [PubMed] [Google Scholar]
- 35.Ladd, P. D., L. E. Smith, N. A. Rabaia, J. M. Moore, S. A. Georges, R. S. Hansen, R. J. Hagerman, F. Tassone, S. J. Tapscott, and G. N. Filippova. 2007. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 16:3174-3187. [DOI] [PubMed] [Google Scholar]
- 36.La Spada, A. R., and J. P. Taylor. 2010. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 11:247-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, Y., S. Y. Dent, J. H. Wilson, R. D. Wells, and M. Napierala. 2010. R loops stimulate genetic instability of CTG·CAG repeats. Proc. Natl. Acad. Sci. U. S. A. 107:692-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, Y., V. Dion, and J. H. Wilson. 2006. Transcription and triplet repeat instability, p. 691-704. In R. Wells and T. Ashizawa (ed.), Genetic instability and neurological diseases. Elsevier, Amsterdam, Netherlands.
- 39.Lin, Y., V. Dion, and J. H. Wilson. 2006. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 13:179-180. [DOI] [PubMed] [Google Scholar]
- 40.Lin, Y., L. Hubert, Jr., and J. H. Wilson. 2009. Transcription destabilizes triplet repeats. Mol. Carcinog. 48:350-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, Y., and J. H. Wilson. 2009. Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair (Amst.) 8:878-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, Y., and J. H. Wilson. 2007. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 27:6209-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ljungman, M. 2005. Activation of DNA damage signaling. Mutat. Res. 577:203-216. [DOI] [PubMed] [Google Scholar]
- 44.Ljungman, M. 2007. The transcription stress response. Cell Cycle 6:2252-2257. [DOI] [PubMed] [Google Scholar]
- 45.Ljungman, M., and D. P. Lane. 2004. Transcription—guarding the genome by sensing DNA damage. Nat. Rev. Cancer 4:727-737. [DOI] [PubMed] [Google Scholar]
- 46.Lopez Castel, A., J. D. Cleary, and C. E. Pearson. 2010. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11:165-170. [DOI] [PubMed] [Google Scholar]
- 47.Manley, K., T. L. Shirley, L. Flaherty, and A. Messer. 1999. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 23:471-473. [DOI] [PubMed] [Google Scholar]
- 48.Mittelman, D., C. Moye, J. Morton, K. Sykoudis, Y. Lin, D. Carroll, and J. H. Wilson. 2009. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc. Natl. Acad. Sci. U. S. A. 106:9607-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moseley, M. L., T. Zu, Y. Ikeda, W. Gao, A. K. Mosemiller, R. S. Daughters, G. Chen, M. R. Weatherspoon, H. B. Clark, T. J. Ebner, J. W. Day, and L. P. Ranum. 2006. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 38:758-769. [DOI] [PubMed] [Google Scholar]
- 50.Myers, K., M. E. Gagou, P. Zuazua-Villar, R. Rodriguez, and M. Meuth. 2009. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 5:e1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orr, H. T., and H. Y. Zoghbi. 2007. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30:575-621. [DOI] [PubMed] [Google Scholar]
- 52.Parsons, M. A., R. R. Sinden, and M. G. Izban. 1998. Transcriptional properties of RNA polymerase II within triplet repeat-containing DNA from the human myotonic dystrophy and fragile X loci. J. Biol. Chem. 273:26998-27008. [DOI] [PubMed] [Google Scholar]
- 53.Pearson, C. E., K. N. Edamura, and J. D. Cleary. 2005. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6:729-742. [DOI] [PubMed] [Google Scholar]
- 54.Pearson, C. E., and R. R. Sinden. 1996. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry 35:5041-5053. [DOI] [PubMed] [Google Scholar]
- 55.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pusapati, R. V., R. J. Rounbehler, S. Hong, J. T. Powers, M. Yan, K. Kiguchi, M. J. McArthur, P. K. Wong, and D. G. Johnson. 2006. ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc. Natl. Acad. Sci. U. S. A. 103:1446-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ranum, L. P., and T. A. Cooper. 2006. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 29:259-277. [DOI] [PubMed] [Google Scholar]
- 58.Riley, B. E., and H. T. Orr. 2006. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 20:2183-2192. [DOI] [PubMed] [Google Scholar]
- 59.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 60.Savouret, C., C. Garcia-Cordier, J. Megret, H. te Riele, C. Junien, and G. Gourdon. 2004. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 24:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selby, C. P., R. Drapkin, D. Reinberg, and A. Sancar. 1997. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 25:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sidi, S., T. Sanda, R. D. Kennedy, A. T. Hagen, C. A. Jette, R. Hoffmans, J. Pascual, S. Imamura, S. Kishi, J. F. Amatruda, J. P. Kanki, D. R. Green, A. A. D'Andrea, and A. T. Look. 2008. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell 133:864-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spiro, C., and C. T. McMurray. 2003. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol. Cell. Biol. 23:6063-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swami, M., A. E. Hendricks, T. Gillis, T. Massood, J. Mysore, R. H. Myers, and V. C. Wheeler. 2009. Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 18:3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor, R. C., S. P. Cullen, and S. J. Martin. 2008. Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9:231-241. [DOI] [PubMed] [Google Scholar]
- 66.Tornaletti, S., S. Park-Snyder, and P. C. Hanawalt. 2008. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 283:12756-12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Broek, W. J., M. R. Nelen, G. W. van der Heijden, D. G. Wansink, and B. Wieringa. 2006. Fen1 does not control somatic hypermutability of the (CTG) (n)·(CAG) (n) repeat in a knock-in mouse model for DM1. FEBS Lett. 580:5208-5214. [DOI] [PubMed] [Google Scholar]
- 68.van den Broek, W. J., M. R. Nelen, D. G. Wansink, M. M. Coerwinkel, H. te Riele, P. J. Groenen, and B. Wieringa. 2002. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11:191-198. [DOI] [PubMed] [Google Scholar]
- 69.Westphal, C. H., S. Rowan, C. Schmaltz, A. Elson, D. E. Fisher, and P. Leder. 1997. atm and p53 cooperate in apoptosis and suppression of tumorigenesis, but not in resistance to acute radiation toxicity. Nat. Genet. 16:397-401. [DOI] [PubMed] [Google Scholar]
- 70.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]