Abstract
The ability to undergo dramatic morphological changes in response to extrinsic cues is conserved in fungi. We have used the model yeast Schizosaccharomyces pombe to determine which intracellular signal regulates the dimorphic switch from the single-cell yeast form to the filamentous invasive growth form. The S. pombe Asp1 protein, a member of the conserved Vip1 1/3 inositol polyphosphate kinase family, is a key regulator of the morphological switch via the cAMP protein kinase A (PKA) pathway. Lack of a functional Asp1 kinase domain abolishes invasive growth which is monopolar, while an increase in Asp1-generated inositol pyrophosphates (PP) increases the cellular response. Remarkably, the Asp1 kinase activity encoded by the N-terminal part of the protein is regulated negatively by the C-terminal domain of Asp1, which has homology to acid histidine phosphatases. Thus, the fine tuning of the cellular response to environmental cues is modulated by the same protein. As the Saccharomyces cerevisiae Asp1 ortholog is also required for the dimorphic switch in this yeast, we propose that Vip1 family members have a general role in regulating fungal dimorphism.
Eucaryotic cells are able to define and maintain a particular cellular organization and thus cellular morphology by executing programs modulated by internal and external signals. For example, signals generated within a cell are required for the selection of the growth zone after cytokinesis in the fission yeast Schizosaccharomyces pombe or the emergence of the bud in Saccharomyces cerevisiae (37, 44, 81). Cellular morphogenesis is also subject to regulation by a wide variety of external signals, such as growth factors, temperature, hormones, nutrient limitation, and cell-cell or cell-substrate contact (13, 34, 66, 75, 81). Both types of signals will lead to the selection of growth zones accompanied by the reorganization of the cytoskeleton.
The ability to alter the growth form in response to environmental conditions is an important virulence-associated trait of pathogenic fungi which helps the pathogen to spread in and survive the host's defense system (7, 32). Alteration of the growth form in response to extrinsic signals is not limited to pathogenic fungi but is also found in the model yeasts S. cerevisiae and S. pombe, in which it appears to represent a foraging response (1, 24).
The regulation of polarized growth and the definition of growth zones have been studied extensively with the fission yeast S. pombe. In this cylindrically shaped organism, cell wall biosynthesis is restricted to one or both cell ends in a cell cycle-regulated manner and to the septum during cytokinesis (38). This mode of growth requires the actin cytoskeleton to direct growth and the microtubule cytoskeleton to define the growth sites (60). In interphase cells, microtubules are organized in antiparallel bundles that are aligned along the long axis of the cell and grow from their plus ends toward the cell tips. Upon contact with the cell end, microtubule growth will first pause and then undergo a catastrophic event and microtubule shrinkage (21). This dynamic behavior of the microtubule plus end is regulated by a disparate, conserved, microtubule plus end group of proteins, called the +TIPs. The +TIP complex containing the EB1 family member Mal3 is required for the delivery of the Tea1-Tea4 complex to the cell tip (6, 11, 27, 45, 77). The latter complex docks at the cell end and recruits proteins required for actin nucleation (46, 76). Thus, the intricate cross talk between the actin and the microtubule cytoskeleton at specific intracellular locations is necessary for cell cycle-dependent polarized growth of the fission yeast cell.
The intense analysis of polarized growth control in single-celled S. pombe makes this yeast an attractive organism for the identification of key regulatory components of the dimorphic switch. S. pombe multicellular invasive growth has been observed for specific strains under specific conditions, such as nitrogen and ammonium limitation and the presence of excess iron (1, 19, 50, 61).
Here, we have identified an evolutionarily conserved key regulator of the S. pombe dimorphic switch, the Asp1 protein. Asp1 belongs to the highly conserved family of Vip1 1/3 inositol polyphosphate kinases, which is one of two families that can generate inositol pyrophosphates (PP) (17, 23, 42, 54). The inositol polyphosphate kinase IP6K family, of which the S. cerevisiae Kcs1 protein is a member, is the “classical” family that can phosphorylate inositol hexakisphosphate (IP6) (70, 71). These enzymes generate a specific PP-IP5 (IP7), which has the pyrophosphate at position 5 of the inositol ring (20, 54). The Vip1 family kinase activity was unmasked in an S. cerevisiae strain with KCS1 and DDP1 deleted (54, 83). The latter gene encodes a nudix hydrolase (14, 68). The mammalian and S. cerevisiae Vip1 proteins phosphorylate the 1/3 position of the inositol ring, generating 1/3 diphosphoinositol pentakisphosphate (42). Both enzyme families collaborate to generate IP8 (17, 23, 42, 54, 57).
Two modes of action have been described for the high-energy moiety containing inositol pyrophosphates. First, these molecules can phosphorylate proteins by a nonenzymatic transfer of a phosphate group to specific prephosphorylated serine residues (2, 8, 69). Second, inositol pyrophosphates can regulate protein function by reversible binding to the S. cerevisiae Pho80-Pho85-Pho81 complex (39, 40). This cyclin-cyclin-dependent kinase complex is inactivated by inositol pyrophosphates generated by Vip1 when cells are starved of inorganic phosphate (39, 41, 42).
Regulation of phosphate metabolism in S. cerevisiae is one of the few roles specifically attributed to a Vip1 kinase. Further information about the cellular function of this family came from the identification of the S. pombe Vip1 family member Asp1 as a regulator of the actin nucleator Arp2/3 complex (22). The 106-kDa Asp1 cytoplasmic protein, which probably exists as a dimer in vivo, acts as a multicopy suppressor of arp3-c1 mutants (22). Loss of Asp1 results in abnormal cell morphology, defects in polarized growth, and aberrant cortical actin cytoskeleton organization (22).
The Vip1 family proteins have a dual domain structure which consists of an N-terminal “rimK”/ATP-grasp superfamily domain found in certain inositol signaling kinases and a C-terminal part with homology to histidine acid phosphatases present in phytase enzymes (28, 53, 54). The N-terminal domain is required and sufficient for Vip1 family kinase activity, and an Asp1 variant with a mutation in a catalytic residue of the kinase domain is unable to suppress mutants of the Arp2/3 complex (17, 23, 54). To date, no function has been described for the C-terminal phosphatase domain, and this domain appears to be catalytically inactive (17, 23, 54).
Here we describe a new and conserved role for Vip1 kinases in regulating the dimorphic switch in yeasts. Asp1 kinase activity is essential for cell-cell and cell-substrate adhesion and the ability of S. pombe cells to grow invasively. Interestingly, Asp1 kinase activity is counteracted by the putative phosphatase domain of this protein, a finding that allows us to describe for the first time a function for the C-terminal part of Vip1 proteins.
MATERIALS AND METHODS
Strains and media.
All strains used are listed in Table 1. New strains were obtained by crossing the appropriate strains and subjecting them to tetrad/random spore analysis or transformation. S. pombe strains were grown in rich media (YE5S) or minimal media (MM) with supplements (52). To repress or derepress, the nmt1+ promoter cells were grown in MM plus 5 μg/ml thiamine or for 18 h at 30°C in thiamine-less MM, respectively. S. cerevisiae strains (Σ1278b background) were grown in YPD or SLAHD (25, 74). Gene deletions were done by PCR-based gene targeting (4) using the kanamycin resistance (Kanr) cassette and plating on 100 mg/ml G418 YE5S, or the his3+ selection marker. 8-Br-cAMP was added to YE5S plates. Unless otherwise indicated, experiments were carried out at 30°C. Sporulation and tetrad analysis of diploid S. cerevisiae strains confirmed the heterozygous deletion of VIP1 or KCS1.
TABLE 1.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| UFY851 | h− mal3-1 ade6-M210 leu1-32 ura4-D6 Ch16[ade6-M216] | U. Fleig |
| UFY1156 | h− asp1Δ::Kanrhis3-D1 ade6-M216 leu1-32 ura4-D18 | This study |
| UFY1511 | h+ asp1D333A/Kanrhis3-D1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1579 | h+ asp1H397A/Kanrhis3-D1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1639 | h+ asp1H397A/Kanrarp3-c1 his3− ade6-M216 leu1-32 ura4− | This study |
| UFY1722 | h− asp1H397A/Kanrsop2-1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1640 | h− asp1H397A/Kanract1-48 leu1-32 ura4-D18 lys1-131 | This study |
| UFY1731 | h+ asp1H397A/Kanrsty1Δ::ura4+ ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1729 | h− aps1Δ::his3+ his3-D1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1725 | h+ asp1H397A/Kanrpmk1Δ::ura4+ leu1− ura4− | This study |
| UFY1580 | h+ asp1D333A-GFP/ura4+ his3-D1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1521 | h− asp1H397A-GFP/ura4+ his3-D1 ade6-M216 leu1-32 ura4-D18 | This study |
| UFY771 | h− asp1-pkGFP/ura4+ his3-D1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1158 | h− mal3Δ::his3+ his3− ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1641 | h− asp1H397A/Kanrmal3Δ::his3+ his3− ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1724 | h− asp1H397A/Kanrtea2Δ::his3+ his3-D1 ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1774 | h− asp1H397A/Kanrtea1Δ::ura4+ ade6-M210 leu1-32 ura4-D18 | This study |
| UFY1723 | h− asp1H397A/Kanrpeg1-1 his3-D1 leu1-32 ura4-D18 | This study |
| UFY1765 | h− asp1H397A/Kanrgpa2Δ::ura4+ his3-D1 ade6-M210 leu1− ura4-D18 | This study |
| UFY1766 | h+ asp1H397A/Kanrcyr1Δ::ura4+ his3-D1 ade6-M216 leu1− ura4-D18 | This study |
| UFY1767 | h+ asp1H397A/Kanrcdc8-110 his3-D1 ade6-M219 leu1-32 ura4-D18 | This study |
| UFY1780 | MATa/MATα ura3-52/ura3-52 vip1Δ::Kanr/VIP1 | This study |
| UFY1781 | MATaura3-52 vip1Δ::Kanr | This study |
| UFY1788 | MATa/MATα ura3-52/ura3-52 kcs1Δ::Kanr/KCS1 | This study |
| KGY425 | h− his3-D1 ade6-M210 leu1-32 ura4-D18 | K. Gould |
| KGY680 | h+ arp3-c1 his3-237 leu1-32 ura4-D18 | K. Gould |
| KGY860 | h− sop2-1 ade6-M210 leu1-32 ura4-D18 | K. Gould |
| KGY1010 | h− act1-48 leu1-32 ura4-D18 lys-131 | K. Gould |
| YUG94 | MATaura3-52 | J. H. Hegemann |
| CGX31 | MATa/MATα ura3-52/ura3-52 | J. H. Hegemann |
| IH1563 | h− peg1-1 leu1-32 ura4-D18 | I. Hagan |
| h− tea2Δ::his3+ his3-D1 ade6-M210 leu1-32 ura4-D18 | J. McIntosh | |
| h+ sty1Δ::ura4+ leu1-32 ura4-D18 | J. Millar | |
| TP319-31A | h− pmk1Δ::ura4+ leu1− ura4− | T. Toda |
| PN1687 | h− tea1Δ::ura4+ ura4-D18 | F. Verde |
| FY7092 | h90 gpa2Δ::ura4+ leu1− ura4-D18 | YGRC |
| FY7461 | h+ cyr1Δ::ura4+ ade6-M216 leu1− ura4-D18 | YGRC |
| FY7728 | h+ spk1Δ::ura4+ leu1− ura4− | YGRC |
| FY12017 | h90 cdc8-110 leu1-32 ura4-D18 | YGRC |
Generation of asp1 mutant strains and plasmids pasp11-364 and pasp1365-920.
A pBSK-based 4,292-bp-long DNA fragment containing wild-type asp1+, asp1D333A, or asp1H397A followed by the Kanr cassette was transformed into strain KG425, thereby replacing the endogenous asp1+ open reading frame (ORF) and generating asp1+/Kanr, asp1D333A/Kanr, and asp1H397A/Kanr strains (asp1D333A and asp1H397A DNA were a gift from John D. York, Duke University). Correct integration was verified by PCR and sequence analysis. To obtain endogenous asp1-gfp fusion variants, the Kanr cassette of the appropriate strain was exchanged with the gfp/ura4+ cassette. For construction of pasp11−364and pasp1365−920, a PCR fragment containing the first 1,092 bp and the last 1,671 bp of the asp1+ ORF, respectively, was cloned into NotI- and XhoI-cut pJR2-3XL (51). Expression of plasmid-encoded asp1 variants was approximately 20-fold higher than that of endogenous asp1+. For expression of the Asp1 kinase domain in vip1Δ/VIP1 cells, asp11−364 was cloned into a CEN plasmid under the control of the MET25 promoter.
Protein analysis.
Protein extracts were prepared as described previously (35). Blots were probed with anti-green fluorescent protein (anti-GFP) antibody (polyclonal rabbit; Invitrogen) followed by alkaline phosphatase (AP)-conjugated secondary antibody (affinity-purified goat anti-rabbit IgG [Fc]; Promega, Madison, WI) and detected with 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NBT) as the substrate.
Invasive growth assays.
A total of 5 μl containing 105 cells or 1 μl containing 106 cells of logarithmically grown overnight (o/n) cultures were spotted on 2% YE5S agar plates or 0.3% and 2% YE5S agar plates, respectively, and incubated. Adhesive and invasive growth assays have been described (61). As the number of invasively growing colonies per strain was dependent on the incubation temperature and the position of the cell spot on the agar plate, we always used an incubation temperature of 30°C and ensured that only cell spots plated on the same position on an agar plate were compared with each other. Quantification of invasive growth was done by determining the number of invasive colonies per strain in at least 3 different experiments.
Enzymatic cell wall degradation assay.
An assay to determine the sensitivity to enzymatic cell wall degradation has been described (58).
Microscopy.
Cells were plated and grown o/n (surface growth) or spotted and grown for 20 days (invasive growth) on YE5S agar plates. Two-millimeter-thick agar blocks were cut out and placed in a temperature-controlled microscope chamber at 30°C (Bioptechs Inc., Butler, PA). Differential interference contrast (DIC) images were obtained with a Zeiss Axiovert200 fluorescence microscope (Carl Zeiss, Jena, Germany) coupled to a charge-coupled-device camera (ORCA-ER; Hamamatsu, Bridgewater, NJ) and Openlab imaging software (Improvision, Coventry, United Kingdom). Photomicrographs of invasively growing or surface colonies were taken with a Zeiss Axioskop.
RESULTS
The Asp1 kinase regulates cell-cell adhesion.
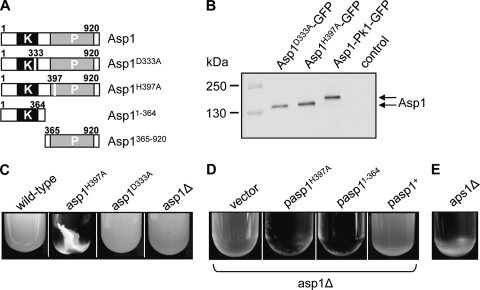
We isolated Asp1 as a multicopy suppressor of the thiabendazole (TBZ) hypersensitivity of a mal3 mutant strain, as overexpression of an Asp1 variant increased the tolerance towards TBZ (Fig. 1). During our experimental analysis of Asp1 variants, we noticed that expression of certain Asp1 mutants led to a severe flocculation phenotype, implying that Asp1 is involved in cell-cell adhesion. We used strains that expressed wild-type Asp1 or endogenous variants with a single point mutation in either the Asp1 kinase domain (Asp1D333A, Asp1 kinase dead) or the Asp1 phosphatase domain (Asp1H397A, Asp1 kinase only) (Fig. 2 A). The D333 amino acid has been identified as a key catalytic residue for Asp1 kinase activity, while H397 was classified as a highly conserved histidine residue of the putative acid phosphatase domain (54). Both Asp1 variants were expressed from the endogenous asp1+ promoter and were generated by replacing the wild-type copy of the asp1+ ORF (see Materials and Methods). As shown by Western blot analysis, the Asp1D333A and Asp1H397A proteins are present in amounts similar to that of the wild-type Asp1 protein (Fig. 2B).
FIG. 1.
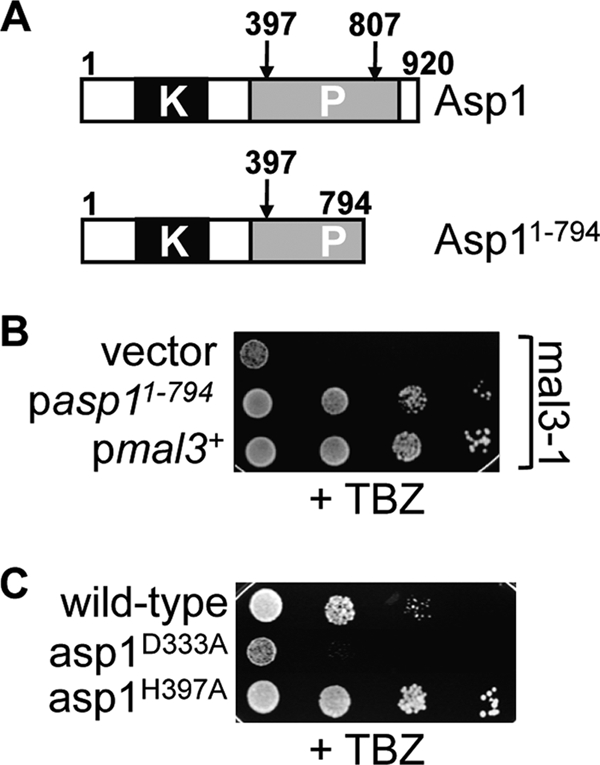
Asp1 kinase activity increases resistance towards TBZ. (A) Diagrammatic representation of wild-type Asp1 (top) and the Asp1 variant that rescues the mal3 mutant TBZ phenotype. Note that the conserved histidine residue H807 is not present in Asp11-794. (B) Serial dilution patch test of a mal3 mutant strain expressing asp11-794 or mal3+. Cells were spotted on plasmid-selective plates containing 8 μg/ml TBZ and incubated for 7 days at 24°C. (C) Wild-type-, asp1D333A-, or asp1H397A-expressing cells were grown on YE5S containing 9 μg/ml TBZ for 5 days at 24°C.
FIG. 2.
Expression of the Asp1 kinase-only variant Asp1H397A leads to flocculation. (A) Diagrammatic representation of Asp1 variants. The Asp1 kinase domain and the putative histidine acid-phosphatase domain are indicated by black and gray boxes, respectively. The changed amino acid in the Asp1 kinase-dead variant (Asp1D333A) and the Asp1 kinase-only variant (Asp1H397A) is indicated. (B) Western blot analysis of strains expressing GFP-tagged endogenous versions of the indicated Asp1 variants. Similar amounts of protein extracts were resolved by SDS-PAGE and probed with an anti-GFP antibody. Asp1-Pk1-GFP runs at 143 kDa (expected, 137 kDa), while the two mutant Asp1-GFP variants run at 137 kDa (expected, 133 kDa). Control, wild-type strain without GFP-tagged asp1+. (C) The indicated strains were grown to stationary phase in rich medium at 30°C. (D) The asp1Δ strain transformed with a vector control or plasmids overexpressing asp1H397A, asp11−364 or asp1+ were grown in plasmid-selective minimal medium at 30°C. (E) The aps1+-deleted strain was grown to stationary phase in rich medium at 30°C.
The strain expressing the Asp1 kinase-only variant Asp1H397A flocculated strongly when the cells were grown in rich medium, and this cell-cell adhesion was dependent on the presence of Ca2+ and galactose residues present in cell wall glycoproteins (Fig. 2C; data not shown). Flocculation was seen most prominently when cells reached stationary phase, implying that it is cell density dependent. The incubation temperature or the growth medium used did not alter the flocculation phenotype (data not shown). Flocculation was observed always for the Asp1H397A-expressing strain and rarely for the isogenic wild-type strain, but not for the Asp1D333A (Asp1 kinase-dead)-expressing strain or a strain in which the entire asp1+ gene had been deleted (asp1Δ strain) (Fig. 2C).
Our results indicated that the presence of a functional Asp1 kinase domain was required for cell-cell adhesion. We therefore tested and found that plasmid-borne overexpression of Asp1H397A induced flocculation in an asp1Δ strain (Fig. 2D). In addition, overexpression of the Asp1 variant Asp11−364, which contains solely the Asp1 kinase domain (Fig. 2A), resulted in flocculating asp1Δ mutant cells (Fig. 2D).
If increased amounts of Asp1-generated inositol pyrophosphates lead to cell-cell adhesion and flocculation, then increasing the intracellular level of these molecules by elimination of a protein that degrades them should give rise to flocculation in wild-type cells. It has been shown that disruption of the S. pombe aps1+ ORF, which encodes a nudix hydrolase, results in a 3-fold increase in the intracellular concentration of IP7 (33). We therefore tested if an aps1Δ strain showed flocculation and found that this was the case (Fig. 2E).
Increased flocculation was observed only when the Asp1 kinase domain was overexpressed or when cells expressed Asp1H397A, in which a highly conserved histidine residue of the putative acid phosphatase domain has been mutated (Fig. 2C and D). This finding implies that (i) the phosphatase domain has a biological function, although no enzymatic activity has been reported, and (ii) the presence of a wild-type phosphatase domain exerts a negative effect on the kinase domain function either directly or indirectly.
The Asp1 kinase activity is required for invasive, pseudohypha-like growth.
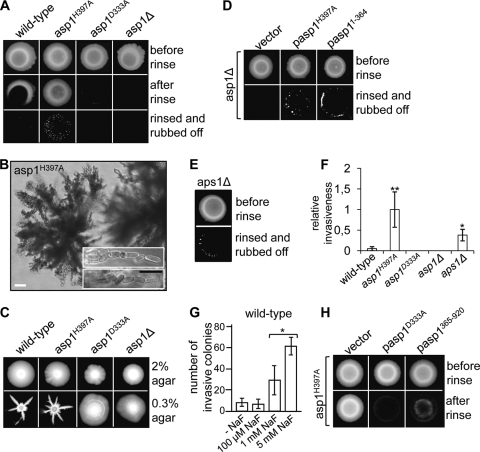
Although the input signals are different for morphogenic events like flocculation, invasive growth, and filamentation, these processes all require adhesion to a substrate (30, 80). We therefore tested if Asp1 also played a role in the adhesion of cells to surfaces such as agar and in invasive, filamentous growth (29). To this end wild-type, asp1H397A, asp1D333A, and asp1Δ cells were spotted on rich media. After 20 days of incubation at 30°C (Fig. 3 A, top), we tested if cells could adhere to the agar by rinsing them with water. We found that the wild-type and asp1H397A strains were able to adhere to the agar, although the Asp1 kinase-only mutant showed significantly increased adhesion compared to the asp1+ strain (Fig. 3A, middle). In contrast, the asp1D333A and the asp1Δ strains showed no adhesion to the agar. These data imply that fission yeast cells are able to adhere to a substrate and that this adhesion requires Asp1 kinase activity.
FIG. 3.
Asp1 kinase activity is needed for pseudohypha-like invasive growth. (A) A total of 105 wild-type, asp1H397A, asp1D333A, and asp1Δ cells were spotted on rich medium and incubated for 20 days at 30°C (top), and the plates were rinsed with water (middle), and the remaining cells were rubbed off (bottom). (B) Photomicrographs of invasively growing colonies show pseudohyphae that grow away from the colony and consist of chains of cells (inset). White bar, 20 μm; black bar, 10 μm. (C) Growth on semisolid 0.3% agar. A total of 106 wild-type, asp1H397A, asp1D333A, or asp1Δ cells were spotted on rich medium with 2% agar (top) or 0.3% agar, pH 7 (bottom), and incubated for 13 days at 30°C. (D) asp1Δ cells overexpressing asp1H397A (with thiamine) or asp11-364 (without thiamine) were grown on plasmid-selective minimal medium for 20 days at 30°C before rinsing and rubbing off the plates. (E) The aps1Δ strain was treated as described in the legend to panel A. (F) Number of invasive colonies formed by the indicated strains were counted and set to 1 for the asp1H397A strain. **, P < 0.01, or *, P < 0.05 for asp1H397A or aps1Δ strains, respectively, compared to wild-type strain as determined using Welch test. (G) Wild-type cells were grown on YE5S with or without NaF for 20 days, and the number of invasive colonies was determined. *, P < 0.05 for 1 mM NaF compared to 5 mM NaF as determined using Welch test. (H) asp1H397A cells overexpressing asp1D333A or asp1365-920 were grown on plasmid-selective minimal medium for 20 days at 30°C and photographed before and after rinsing the plates.
We next tested invasion into the agar by vigorously rubbing cells off the agar (29). Only the asp1H397A strain showed massive invasion into the agar (Fig. 3A, bottom). The wild-type strain was also able to invade a solid substratum, although at a much-reduced frequency compared to the asp1H397A strain. The latter gave rise to approximately 30 times more invasively growing colonies of the type shown in Fig. 3B (Fig. 3F). Invasion into the agar was first observed after 8 days of incubation at 30°C and occurred at the edges of the cell spots possibly because by that time nutrients were available only at the edge of the colony. Microscopic analysis of invasively growing cells revealed that they formed long filament-like structures, which grew away from the colony and consisted of chains of cells separated by septa (Fig. 3B). We named these structures pseudohyphae. Incubating S. cerevisiae cells on low concentrations of agar (0.3%) triggers a form of growth observed in biofilms, namely, the formation of a confluent mat of cells (64). We tested the growth behavior of the S. pombe strains under such conditions and found that wild-type cells did not form mats of cells but instead showed massively invasive growth into the semisolid agar (Fig. 3C, bottom). The asp1H397A strain showed an even stronger invasive growth phenotype than the wild-type strain (Fig. 3C). The center of the colonies consisted of invasively growing cells, while long feather-like structures arose from the edges of the colonies. Again, Asp1 kinase activity was required for invasive growth on 0.3% agar plates, as the asp1D333A and the asp1Δ strains were unable to grow invasively, even under such favorable conditions (Fig. 3C).
Our data suggest that (i) Asp1 kinase activity and therefore, presumably, specific IP7 isomers or IP8 regulate pseudohypha-like, invasive growth and that (ii) increased Asp1 kinase activity or loss of the negative influence of the C-terminal phosphatase domain results in a stronger invasive growth phenotype.
To test if increased Asp1 kinase activity was responsible for increasing the ability of cells to grow in a pseudohypha-like, invasive manner, we (i) overexpressed the Asp11−364 and Asp1H397A variants in the asp1Δ strain and (ii) assayed invasive growth in the aps1Δ strain or in the presence of NaF, which has been shown to inhibit the activity of nudix hydrolases (67).
Overexpression of Asp1H397A or Asp11−364 in the asp1Δ strain strongly increased the number of invasively growing cells compared to the vector control (Fig. 3D). In addition, loss of the nudix hydrolase Aps1 or growth on NaF-containing medium also led to increased invasive growth phenotypes (Fig. 3E, F, and G).
Taken together, these data strongly imply that the inositol pyrophosphate molecules generated by the Asp1 kinase act as regulators/signaling molecules for invasive pseudohyphal growth. The absence of the C-terminal Asp1 domain or mutation of a highly conserved histidine residue in the putative phosphatase domain leads to an increase in cell-cell and cell-substrate adhesion (Fig. 2 and 3). Thus, the putative phosphatase domain appears to have a negative effect on kinase function. Overexpression of the phosphatase-only Asp1 variant Asp1365−920 in the Asp1H397A strain indeed decreased the ability of the strain to adhere to the agar and grow invasively (Fig. 3H and data not shown).
Growth mode of invasively growing cells is predominantly monopolar.
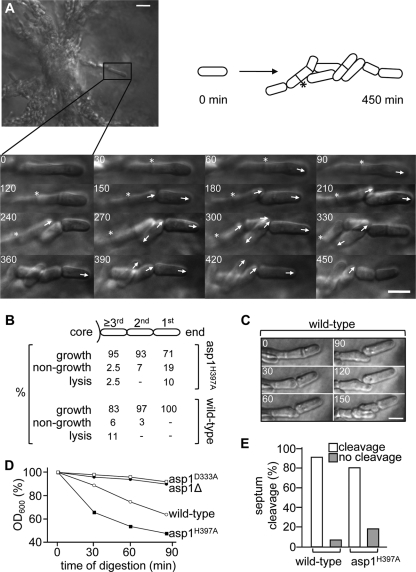
Invasive colonies of the asp1H397A strain consisted of a densely grown center from which long chains of cells irradiated (Fig. 3B). As the growth mode of such S. pombe cells had not been analyzed previously, we imaged invasively growing cells using live cell microscopy (example shown in Fig. 4 A). Wild-type and Asp1H397A-expressing cells were mostly viable and grew at similar rates, with generation times of 2.1 and 2 h, respectively (Fig. 4B). Few major morphological defects, such as round or branched cells, were observed; however, cells adjusted their cell shape to externally present constraints, such as other cells lying in their path of growth (example shown in Fig. 4C).
FIG. 4.
Live cell analysis of invasively growing S. pombe cells. (A) Agar blocks containing invasively growing cells were placed in a temperature-controlled microscope chamber. Growth of cells growing at the end of pseudohyphae was monitored for up to 9 h at 30°C. The asp1H397A cell marked with a box was monitored for 7.5 h. The progeny produced by cells growing away from the colony is shown by photomicrographs and depicted diagrammatically. Numbers indicate minutes, arrows the orientation of growth, and the asterisks a septum that did not resolve during the analysis. Bar, 5 μm. (B) Wild-type or asp1H397A cells were monitored as described in the legend to panel A and growth, nongrowth, or lysis of cells at the very end or within pseudohyphae was counted. (C) Example of morphological alterations in invasively growing wild-type cells. Bar, 5 μm. (D) Cells of the indicated genotypes were treated with zymolyase for 90 min, and decrease of the optical density at 600 nm (OD600) was measured to assess cell lysis. (E) Percentage of cleaved or noncleaved septa of wild-type or asp1H397A cells that were monitored as described in the legend to panel A.
Growth of the pseudohyphae was not restricted to the end-most cells (1st cell, Fig. 4B), and most of the cells analyzed in a pseudohyphal structure were able to divide (Fig. 4B). Wild-type cells that either showed no growth or lysed during the analysis were present mostly within the long chains of cells. In contrast, the vast majority of nongrowing or lysing Asp1H397A cells were the end-most cells (Fig. 4B). The latter phenotype might be caused by changes in the cell wall composition of the Asp1H397A strain, as this strain is hypersensitive to digestion by the beta-1,3-glucan laminaripentaohydrolase containing zymolyase (Fig. 4D). Furthermore, strong overexpression of the Asp1H397A protein leads to cell lysis (54). Absence of Asp1 kinase activity leads to resistance to digestion by zymolyase (Fig. 4D).
Pseudohyphal growth is facilitated by defects in cytokinesis (3). Indeed, we found that 18.5% of Asp1H397A-expressing cells and 8% of wild-type cells formed septa that did not resolve during the course of the experiment (Fig. 4A and E).
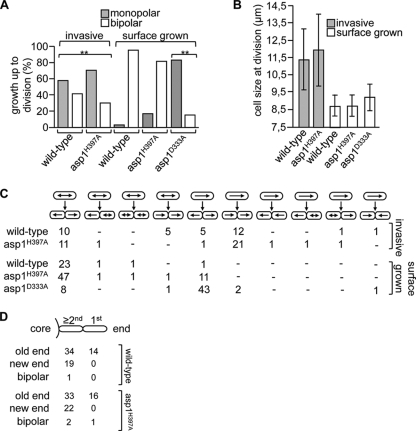
Wild-type cells that grow in liquid or on an agar surface initiate growth after cytokinesis by growing in a monopolar fashion using the old end (the end present before cell division) and then undergo bipolar growth at a transition point named NETO in G2 phase (49). This mode of growth was dramatically altered in invasively growing cells, in which 58% of wild-type and 70.2% of the Asp1H397A cells grew in an exclusively monopolar manner (Fig. 5 A). As only 3.8% of the wild-type and 18% of the Asp1H397A cells showed exclusive monopolar growth when grown on an agar surface, we conclude that invasive growth leads to an alteration of the normal growth pattern (Fig. 5A). Interestingly, 83.6% of surface-grown asp1D333A cells (Asp1 kinase-dead cells) showed monopolar growth, implying that Asp1 kinase activity is required for the switch from mono- to bipolar growth (Fig. 5A and C). As NETO can occur only after cells have attained a minimum size, we measured the cell length at division of invasively and surface-grown cells (Fig. 5B). The invasively growing wild-type and Asp1H397A cells had cell lengths of 11.4 ± 1.7 μm and 11.9 ± 2.1 μm, respectively, cell lengths significantly longer than those of the same strains grown on the agar surface (Fig. 5B). Our data imply that invasive growth leads to an alteration of the normal bipolar pattern and results in predominantly monopolar growth.
FIG. 5.
Analysis of the growth pattern of invasively growing cells. (A) Cells of the indicated strain or genotype were pregrown invasively (YE5S for 20 days at 30°C) or on the agar surface (on YE5S for 16 h at 30°C), and the growth pattern was then determined using time lapse microscopy at 30°C. Invasive growth: wild-type strain, n = 36; asp1H397A strain, n = 57. Surface growth: wild-type strain, n = 26; asp1H397A strain, n = 61; asp1D333A strain, n = 55. **, P < 0.01 for wild-type and asp1H397A strains invasively grown compared to surface grown and for asp1D333A strain compared to wild-type strain as determined using χ2 test. (B) Cell size at division of cells of the indicated strains grown in or on agar. Invasive growth: wild-type strain, n = 16; asp1H397A strain, n = 15. Surface growth: wild-type strain, n = 30; asp1H397A strain, n = 42; asp1D333A strain, n = 10. (C) Selection of growth site of invasively and surface-grown strains of the indicated strain or genotype. Top row of cells denotes “parental” cells, and the growth pattern of these cells up to cell division is indicated by arrows. Bottom row of cells depicts the progeny of the upper cells and their mode of growth following cytokinesis. Numbers given are cells in that particular category. (D) Following cytokinesis, the growth zones of the end-most cell and other cells in the pseudohyphal structure were determined.
Next, we analyzed growth zone selection of invasively growing cells. The majority of invasively growing cells defined the old end as the growth zone after cytokinesis (Fig. 5C, 70% of wild-type and 63% of Asp1H397A cells). Interestingly, all analyzed end-most cells of a pseudohyphal structure (1st cell, Fig. 5D) initiated growth at the old end. As this end is farthest away from the colony, it is possible that this growth zone is environmentally determined.
Microtubule and actin cytoskeleton in invasive growth.
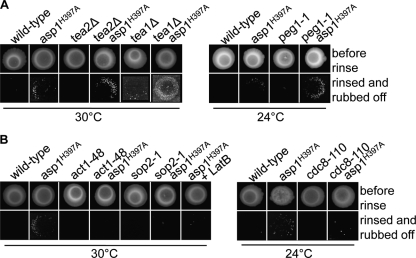
Proper polarized growth of surface- and/or liquid-grown S. pombe cells is dependent on functional actin and microtubule cytoskeletons (15, 44, 60). To determine the impact of these two cytoskeletal structures on invasive, pseudohyphal growth, we assayed the ability of a number of actin and microtubule cytoskeletal mutants to grow invasively. We tested invasive growth of mal3, tea1, and tea2 deletion strains (the mal3Δ, tea1Δ, tea2Δ strains) and the temperature-sensitive peg1-1 strain at the permissive temperature (6, 10, 12, 27). The Tea2 kinesin protein is required for the transport of the kelch repeat protein Tea1 to the cell ends, where morphogenesis is controlled via the Tea1-Tea4 complex (9, 45, 77). Mal3 and Peg1 are components of the microtubule +TIP complex that regulates microtubule dynamics (6, 12, 27). Mal3 is also required for Tea1 deposition at the cell end, while Peg1 is not (9, 12, 27).
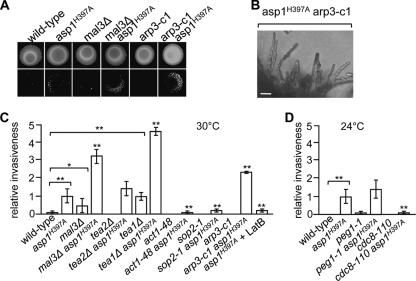
The mal3Δ and tea1Δ strains showed a higher number of agar-invading colonies than the wild-type strain, while the peg1-1 mutant strain had a frequency of agar invasion similar to that of the wild-type strain (Fig. 6 A and C and 7A). The tea2Δ strain did not show invasive growth after 20 days of incubation; however, a tea2Δ strain can grow invasively after prolonged incubation (Fig. 6C) (19). Double mutants carrying mal3Δ, tea1Δ, tea2Δ, peg1-1, or asp1H397A showed more invasively growing colonies than the single asp1H397A mutant strain (Fig. 6A and C). However, the colonies of the double mutant strains were, in general, smaller than those of the asp1H397A single mutant strain.
FIG. 6.
An altered microtubule or actin cytoskeleton increases the invasive growth of asp1H397A. (A) The wild-type strain and strains with the indicated genotypes were grown on YE5S for 20 days at 30°C (top), and the cells were washed/rubbed off (bottom). (B) Photomicrographs of invasively growing asp1H397A arp3-c1 cells. Bar, 10 μm. (C) The indicated strains were grown at 30°C or 24°C as described in the legend to panel A, and the number of invasive colonies was counted. Invasiveness of asp1H397A was set to 100%. At least 3 independent experiments were carried out. *, P < 0.05 for mal3Δ strain compared to wild-type strain as determined using Welch test. **, P < 0.01 for asp1H397A or tea1Δ strains compared to wild-type strain and for mal3Δ asp1H397A, tea1Δ asp1H397A, act1-48 asp1H397A, sop2-1 asp1H397A, arp3-c1 asp1H397A, asp1H397A + LatB, and cdc8-110 asp1H397A strains compared to asp1H397A strain as determined using Welch test.
FIG. 7.
Photomicrographs of invasive growth assays. (A and B) Wild-type strain and strains with the indicated genotypes were grown for 20 days at 30°C or 24°C on YE5S (top), and the cells were washed/rubbed off (bottom).
We then analyzed the impact of the actin cytoskeleton on invasive growth and found that cold-sensitive act1 (encodes actin), sop2 (encodes a subunit of the Arp2/3 complex), and arp3 (encodes the Arp3 protein) mutants and the temperature-sensitive cdc8 (encodes tropomyosin) mutant were unable to grow invasively under permissive growth conditions (Fig. 6A and C and 7B) (5, 16, 47, 48). The asp1H397A act1-48 double mutant strain showed no invasive growth (Fig. 6C). The asp1H397A sop2-1 strain was able to grow invasively, albeit at a reduced rate comparable to that of the asp1H397A strain grown in the presence of the actin polymerization inhibitor latrunculin B (Fig. 6C). Surprisingly, the asp1H397A arp3-c1 strain had an increased number of invasively growing cells compared to the asp1H397A single mutant strain (Fig. 6A and C). Microscopic analysis revealed that the asp1H397A arp3-c1 strain showed an increased cell separation defect, which results in very neat chains of invasively growing cells (Fig. 6B). This type of pseudohyphal growth might facilitate invasion into the agar.
Asp1-mediated invasive growth is dependent on the cyclic AMP (cAMP) protein kinase A (PKA) pathway.
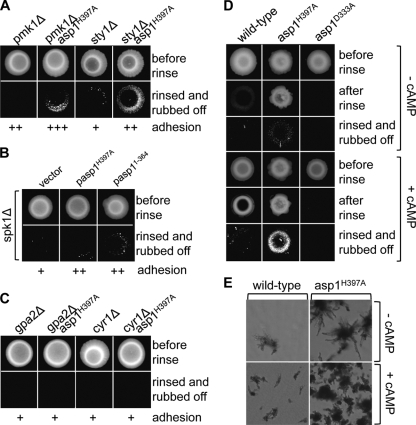
Environmental changes are detected and adapted to via a number of kinase signaling pathways (18). These include mitogen-activated protein (MAP) kinase pathways, such as the pheromone-responsive pathway, the Sty1/Spc1 stress response pathway, the Pmk1/Spm1 cell-integrity pathway, and the cAMP-dependent PKA pathway (26, 31, 78, 79, 84). To analyze which of these pathways were required for Asp1-mediated invasive growth, we made double mutants of asp1H397A and pmk1Δ (component of cell integrity pathway) or sty1Δ (component of stress response pathway). The double mutants could still grow invasively, thus indicating that these pathways are not required for Asp1-induced invasive growth (Fig. 8 A). Next, as the asp1H397A strain and a spk1 deletion strain (pheromone response pathway) could not mate, we transformed the spk1Δ strain with plasmids expressing Asp1H397A or Asp11−364. We found that production of these proteins could induce increased invasive growth (Fig. 8B). However, double mutants of asp1H397A and null alleles of genes encoding components of the protein kinase A pathway were unable to grow invasively, as shown for the asp1H397A gpa2Δ and asp1H397A cyr1Δ strains (Fig. 8C). gpa2+codes for part of the heterotrimeric G protein, while cyr1+ encodes the adenylate cyclase (31). These findings imply that the cAMP pathway is required for invasive growth. To determine if cAMP PKA acts upstream or downstream of Asp1 kinase function, we grew wild-type, asp1H397A, and asp1D333A strains on medium containing cAMP (Fig. 8D and E). We found that in the presence of cAMP, wild-type and asp1H397A strains showed increased invasive growth compared to growth of these strains on cAMP-less medium, but the asp1D333A strain was still unable to grow invasively. We conclude that the cAMP PKA pathway acts upstream of Asp1 kinase function (1, 31).
FIG. 8.
Asp1 function in invasive growth requires the cAMP PKA pathway. (A) Strains with the indicated genotypes were grown for 20 days at 30°C on YE5S (top), and the cells were washed/rubbed off (bottom). + to +++, ability, from weakest to strongest, respectively, to adhere to the agar. + represents wild-type adhesion (compare to Fig. 3A). (B) spk1Δ cells transformed with a vector control or plasmids expressing asp1H397A or asp11−364 were incubated on plasmid-selective agar for 20 days at 30°C and treated as described in the legend to panel A. (C) Strains with the indicated genotypes were grown and treated as described for panel A. (D) The indicated strains were grown for 13 days at 30°C on YE5S with or without 1 mM cAMP, and the cell spots were rinsed or were rinsed and rubbed off. (E) Photomicrographs of the invasively growing wild-type and asp1H397A colonies shown in panel D.
The Vip1 family also regulates invasive pseudohyphal growth in S. cerevisiae.
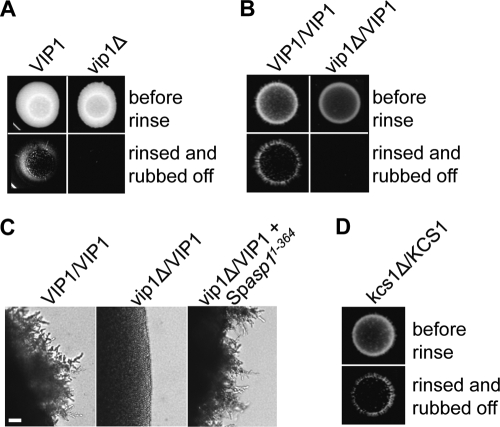
To determine if the Vip1 family function, with respect to invasive growth, was conserved, we generated S. cerevisiae haploid and diploid strains in which the VIP1 gene was deleted and tested these strains in invasive growth assays (25, 65). The haploid vip1Δ strain showed a moderately reduced ability to grow invasively compared to the isogenic wild-type strain, implying that Vip1 activity contributes to but is not essential for haploid invasive growth (Fig. 9 A). Next, we tested the requirement of Vip1 in diploid pseudohyphal growth. As a homozygous vip1Δ/vip1Δ strain showed growth defects on our test medium, we assayed the ability of the heterozygous vip1Δ/VIP1 strain to grow in an invasive fashion on the appropriate SLAHD medium (25). Loss of one VIP1 copy and thus a presumably 50% reduction in Vip1 protein levels severely reduced the ability of this strain to grow in a pseudohyphal manner (Fig. 9B and C). Expression of the S. pombe Asp11−364 variant in this strain restored the ability to grow invasively (Fig. 9C). Thus, the S. pombe Asp1 and S. cerevisiae Vip1 proteins are functional orthologs, and the role of the Vip1 family in dimorphic switching is conserved.
FIG. 9.
S. cerevisiae Vip1 is required for invasive growth. (A) A total of 105 haploid wild-type and vip1Δ cells were spotted on YPD plates and grown for 9 days at 30°C (top). Plates were rinsed, and the remaining cells rubbed off (bottom). (B) A total of 105 diploid wild-type and vip1Δ/VIP1 cells were incubated on SLAHD plates for 14 days at 30°C and treated as described in the legend to panel A. (C) Photomicrographs of diploid wild-type, vip1Δ/VIP1, and vip1Δ/VIP1 cells expressing S. pombe Asp11-364. Cells were grown on SLAHD agar for 5 days. Bar, 20 μm. (D) A diploid kcs1Δ/KCS1 strain was analyzed as described in the legend to panel B.
To determine if members of the IP6K family regulate dimorphic switching, we generated a homozygous kcs1Δ/ksc1Δ strain (70, 71). However, this strain could not be tested in our assay, due to massive growth defects (data not shown). We therefore assayed the ability of a heterozygous kcs1Δ/KCS1 strain to make pseudohyphae. Formation of pseudohyphae was comparable to that of the wild-type strain (Fig. 9D).
DISCUSSION
Vip1 kinases regulate fungal dimorphism.
We have used S. pombe to identify a key regulator of the switch from the single-cell yeast form to an invasive pseudohyphal growth form. Our analysis has shown that the kinase activity of the Vip1 member Asp1 is essential for the alternative growth form in S. pombe. Similarly, Vip1 plays a role in haploid invasive and diploid pseudohyphal growth of S. cerevisiae cells. As S. pombe cells and diploid and haploid S. cerevisiae cells change to an invasive filamentous type of growth in response to different environmental cues, our data suggest that Vip1-generated inositol pyrophosphates could per se play a role in fungal dimorphism (1, 24). Furthermore, as an increase in Asp1-generated inositol pyrophosphates increases the ability to switch to and grow in an invasive growth mode, we propose that these inositol molecules act as “second messengers” (Fig. 10).
FIG. 10.
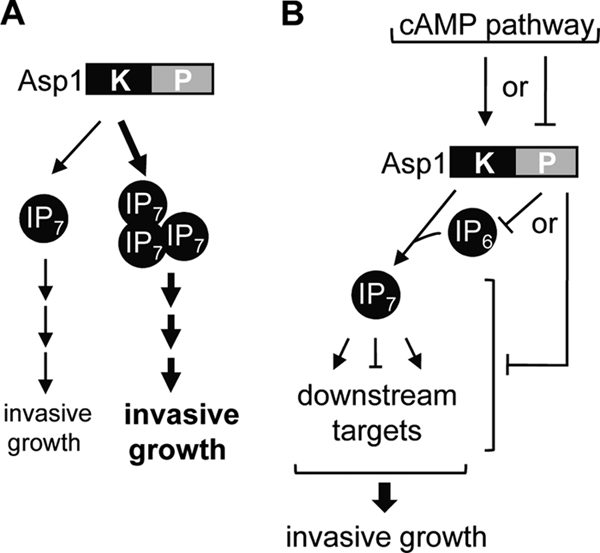
Model for the regulation of invasive growth by Asp1. (A) Increased Asp1 kinase activity leads to increased invasive growth phenotype. (B) Asp1 kinase activity is regulated negatively by the Asp1 phosphatase domain. The latter domain might have an enzymatic activity that uses the Asp1 kinase substrate. Alternatively, it might regulate downstream targets of Asp1 kinase. Active Asp1 kinase leads to the production of IP7 (or IP8), which regulates a number of pathways required for invasive growth.
Higher inositol phosphates have been discussed as sensors/transmitters for environmental conditions and have been shown to be upregulated in response to hyperosmotic stress and to mediate chemotaxis in Dictyostelium (43, 59). One of the very few studies that have described such a role specifically for members of the Vip1 family is the regulation of phosphate metabolism. In response to phosphate starvation, the S. cerevisiae Pho80-Pho85-Pho81 complex is inactivated via Vip1-generated IP7 molecules, leading to activation of the Pho4 transcription factor (39, 41). Here, we have identified a new function of inositol pyrophosphates generated by the Vip1 family as being part of the signaling network that leads to an altered growth mode in response to external cues.
Members of the Vip1 family generate a specific IP7 isomer, namely, 1/3 diphosphoinositol pentakisphosphate, and phosphorylate the IP7 isomer generated by members of the IP6K family to IP8 (17, 23, 42, 54, 57). We were unable to determine if Vip1-generated IP7 and/or Vip1- and IP6K-generated IP8 was required for pseudohyphal growth, as the S. pombe IP6K member SPCC970.08 is essential for growth (36) and the S. cerevisiae kcs1Δ/kcs1Δ diploid strain showed massive growth defects. However, in contrast to the diploid vip1Δ/VIP1 strain which has a haploinsufficient phenotype, the diploid S. cerevisiae strain heterozygous for KCS1 generated pseudohyphae similar to those of the wild-type strain. This implies that a presumably 50% reduction in Kcs1-generated IP7 does not affect the dimorphic transition and might suggest that the IP7 molecules generated by Vip1 kinases regulate the dimorphic switch. In this context, it has been shown recently that activation of the S. cerevisiae Pho4 transcription factor by Vip1-regulated inhibition of Pho85 kinase function results in the downregulation of cellular Kcs1 activity by the generation of antisense KCS1 RNAs (56). As the Kcs1 and Vip1 kinases both use IP6 as a substrate and Kcs1 is the major inositol polyphosphate kinase, such a regulation might enable Vip1 to utilize more of its substrate (54, 56, 83). Thus, reduction of Kcs1 protein levels in our diploid heterozygous strain could result in more IP6 substrate that can be utilized by Vip1.
Autoinhibition of Asp1 kinase activity?
The present analysis implies that certain environmental conditions induce an upregulation of Asp1 kinase activity via the PKA pathway. How this regulation is achieved at the molecular level is at present unclear. However, it has been proposed that the C-terminal part of Vip1 kinases, which includes a seemingly enzymatically inactive histidine acid phosphatase domain, might act as a regulatory domain (73). Our data strongly suggest that this is the case, as the presence of the wild-type Asp1 phosphatase domain has a negative impact on Asp1 function. In all our assays, we find that the phenotype of the wild-type strain is in between those of strains expressing Asp1 kinase-only variants (Asp1H397A or Asp11−364) and strains expressing the Asp1 kinase-dead variant (asp1D333A) or an asp1Δ strain. (i) The asp1H397A strain is more resistant to TBZ than the wild-type strain, which is more TBZ resistant than the asp1D333A strain (Fig. 1C). Similar data were obtained for the resistance to digestion by zymolyase. The asp1D333A and asp1Δ strains are more resistant to digestion by the enzyme than the wild-type strain, which is more resistant than the asp1H397A strain. (ii) The asp1H397A strain shows an increased ability to grow in an invasive, pseudohyphal manner compared to the wild-type strain, while the asp1D333A strain and the asp1Δ strain are unable to grow invasively. (iii) Overexpression of the Asp1 kinase-only variants (Asp11−364 and Asp1H397A) gives rise to massive flocculation of an asp1Δ strain, while overexpression of wild-type asp1+ does not.
These data imply that the C-terminal part of Asp1, which encompasses a putative histidine acid phosphatase domain, negatively regulates the Asp1 kinase function. How can this be brought about? Such a regulation could be mediated by influencing Asp1 kinase activity either directly or by modulating downstream effectors of the Asp1-mediated inositol pyrophosphate regulation. For example, the phosphatase domain could regulate the amount of Asp1-generated inositol pyrophosphates directly by competing with the Asp1 kinase for the substrate. However, it must be pointed out that no enzymatic activity has as yet been described for the Asp1 phosphatase domain (17, 23, 54). Alternatively, the C-terminal part of Asp1 could influence a pathway that is regulated in the opposite way by Asp1 kinase activity. As all scored phenotypes of the asp1Δ strain and the asp1D333A strain are very similar if not identical, we conclude that the putative phosphatase domain has a function that comes into play only in the presence of a wild-type kinase domain. Interestingly, in vitro analysis of human and yeast Vip1 kinase activities suggests that the kinase activity might be regulated directly by the phosphatase domain. Full-length human and yeast Vip1 proteins have significantly less specific activity than variants, which consist only of the kinase domain (23). These findings fit with our observation that plasmid-borne strong overexpression of asp1D333A or asp1365−920 (an Asp1 variant that consists only of the C-terminal phosphatase domain) massively reduces the ability of the asp1H397A strain to adhere to the agar and grow invasively. trans inhibition by massive overexpression of the inhibitory domain has been shown for a number of proteins regulated by autoinhibition (63).
Upstream regulators of Asp1 kinase function.
A number of MAP kinase pathways detect alterations in their surroundings, leading to cellular adaptation. By testing several kinase signaling pathways, we found that Asp1 kinase-mediated pseudohyphal invasive growth requires a functional cAMP PKA pathway. Mutations in this pathway abolish this type of growth, while exogenous cAMP enhances pseudohyphal growth in strains with a wild-type Asp1 kinase domain. Exogenous cAMP does not lead to invasive growth of the asp1D333A strain, indicating that cAMP regulates Asp1 kinase function and not vice versa. Both the S. pombe Asp1 and S. cerevisiae Vip1 proteins have multiple phosphorylation sites, and S. cerevisiae Vip1 is a target of Tpk1 and Tpk3, which represent two of the catalytic cAMP PKA catalytic subunits (62, 72, 82). Thus, we speculate that activation or enhancement of the kinase activity of Vip1 family members could be modulated directly by PKA.
Growth pattern of invasively growing S. pombe cells and the role of the cytoskeleton.
Filamentous invasive growth has been described for S. pombe (1, 19, 61), but we have analyzed for the first time the growth pattern of such cells. Invasive growth of wild-type and asp1H397A cells is a variation on the same theme that is a combination of different properties, such as cell-cell and cell-substrate adhesion, defects in cell separation, the ability to alter cell shape in response to environmental constraints, and an alteration of the normal growth pattern. A wild-type actin cytoskeleton is needed for this type of growth, as strains harboring mutations in genes coding for the actin cytoskeletal components Arp2/3, formin, tropomyosin, and actin are unable to grow invasively (19; our data).
The role of the interphase microtubule cytoskeleton and associated proteins in invasive growth is less clear-cut. We and others found that invasive pseudohyphal growth did not depend on the presence of the Tea1-Tea4 cell end marker complex or the microtubule regulators Mal3 and Peg1 (19; our data). Indeed in the absence of, for example, the Tea1 protein, the asp1H397A strain showed an increased number of invasively growing colonies. However, these colonies were, in general, smaller than those of the asp1H397A single mutant strain, possibly due to the aberrant microtubule cytoskeleton which led to an alteration of the cylindrical cell shape and bent or lemon-shaped cells (data not shown). A cylindrical cell form probably facilitates the formation of straight chains of cells required for efficient invasive pseudohyphal growth. Thus, a wild-type microtubule cytoskeleton and the Tea1-Tea4 complex are not essential for invasive pseudohyphal growth, similar to what has been described for mating S. pombe cells, in which growth is redirected by external pheromones (55). In both systems, extrinsic signals regulate/modify the normal internal polarization program that requires the microtubule-based delivery of the Tea1-Tea4 complex: mating S. pombe cells turn into the pheromone-induced shmooing growth mode (55), while invasively growing cells show an alteration of the normal growth pattern, as they do not initiate NETO and grow predominantly monopolarly. Interestingly, the absence of Tea1 led to a faster switch to pheromone-induced growth, indicating that factors that regulate the intrinsic growth mode need to be dismantled before extrinsically controlled growth can commence (55). We speculate that this might also be the case for the switch from the single-celled growth form to the pseudohyphal growth form, and thus, a defective microtubule cytoskeleton might aid in the switch. However, as the vast majority of wild-type and asp1H397A invasively growing cells were cylindrical, microtubules and associated proteins must play some role during invasive growth. Indeed, our preliminary analysis of Tea1-GFP localization in invasively growing cells indicates that Tea1 is present at one cell tip, with no preference for the growing or nongrowing tip.
Taken together, these data suggest that extrinsic signals modify the normal internal microtubule-dependent polarization program leading to the switch from bipolar single-celled growth to monopolar pseudohyphal growth. If Asp1 kinase is involved in growth zone definition in invasively growing cells is as yet unclear. However, Asp1 kinase function is needed for NETO in vegetatively growing cells.
Acknowledgments
We are very grateful to Shelley Sazer for the very careful reading of the manuscript. We thank Kathleen Gould, Iain Hagan, Johannes Hegemann, Jonathan Millar, Takashi Toda, Fulvia Verde, John York, and the Yeast Genetic Resource Center (Osaka, Japan) for strains and reagents.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 590).
Footnotes
Published ahead of print on 12 July 2010.
REFERENCES
- 1.Amoah-Buahin, E., N. Bone, and J. Armstrong. 2005. Hyphal growth in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 4:1287-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azevedo, C., A. Burton, E. Ruiz-Mateos, M. Marsh, and A. Saiardi. 2009. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. U. S. A. 106:21161-21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bähler, J. 2005. A transcriptional pathway for cell separation in fission yeast. Cell Cycle 4:39-41. [DOI] [PubMed] [Google Scholar]
- 4.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian, M. K., A. Feoktistova, D. McCollum, and K. L. Gould. 1996. Fission yeast Sop2p: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. EMBO J. 15:6426-6437. [PMC free article] [PubMed] [Google Scholar]
- 6.Beinhauer, J. D., I. M. Hagan, J. H. Hegemann, and U. Fleig. 1997. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 139:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman, J. 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 9:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhandari, R., A. Saiardi, Y. Ahmadibeni, A. M. Snowman, A. C. Resnick, T. Z. Kristiansen, H. Molina, A. Pandey, J. K. Werner, Jr., K. R. Juluri, Y. Xu, G. D. Prestwich, K. Parang, and S. H. Snyder. 2007. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. U. S. A. 104:15305-15310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browning, H., D. D. Hackney, and P. Nurse. 2003. Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5:812-818. [DOI] [PubMed] [Google Scholar]
- 10.Browning, H., J. Hayles, J. Mata, L. Aveline, P. Nurse, and J. R. McIntosh. 2000. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol. 151:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busch, K. E., and D. Brunner. 2004. The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14:548-559. [DOI] [PubMed] [Google Scholar]
- 12.Busch, K. E., J. Hayles, P. Nurse, and D. Brunner. 2004. Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev. Cell 6:831-843. [DOI] [PubMed] [Google Scholar]
- 13.Carthew, R. W. 2007. Pattern formation in the Drosophila eye. Curr. Opin. Genet. Dev. 17:309-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cartwright, J. L., and A. G. McLennan. 1999. The Saccharomyces cerevisiae YOR163w gene encodes a diadenosine 5′, 5′′′-P1,P6-hexaphosphate (Ap6A) hydrolase member of the MutT motif (Nudix hydrolase) family. J. Biol. Chem. 274:8604-8610. [DOI] [PubMed] [Google Scholar]
- 15.Chang, F. 2001. Establishment of a cellular axis in fission yeast. Trends Genet. 17:273-278. [DOI] [PubMed] [Google Scholar]
- 16.Chang, F., A. Woollard, and P. Nurse. 1996. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109:131-142. [DOI] [PubMed] [Google Scholar]
- 17.Choi, J. H., J. Williams, J. Cho, J. R. Falck, and S. B. Shears. 2007. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 282:30763-30775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowan, K. J., and K. B. Storey. 2003. Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. J. Exp. Biol. 206:1107-1115. [DOI] [PubMed] [Google Scholar]
- 19.Dodgson, J., H. Avula, K. L. Hoe, D. U. Kim, H. O. Park, J. Hayles, and J. Armstrong. 2009. Functional genomics of adhesion, invasion, and mycelial formation in Schizosaccharomyces pombe. Eukaryot. Cell 8:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Draskovic, P., A. Saiardi, R. Bhandari, A. Burton, G. Ilc, M. Kovacevic, S. H. Snyder, and M. Podobnik. 2008. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 15:274-286. [DOI] [PubMed] [Google Scholar]
- 21.Drummond, D. R., and R. A. Cross. 2000. Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr. Biol. 10:766-775. [DOI] [PubMed] [Google Scholar]
- 22.Feoktistova, A., D. McCollum, R. Ohi, and K. L. Gould. 1999. Identification and characterization of Schizosaccharomyces pombe asp1(+), a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics 152:895-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fridy, P. C., J. C. Otto, D. E. Dollins, and J. D. York. 2007. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 282:30754-30762. [DOI] [PubMed] [Google Scholar]
- 24.Gancedo, J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:107-123. [DOI] [PubMed] [Google Scholar]
- 25.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 26.Gotoh, Y., E. Nishida, M. Shimanuki, T. Toda, Y. Imai, and M. Yamamoto. 1993. Schizosaccharomyces pombe Spk1 is a tyrosine-phosphorylated protein functionally related to Xenopus mitogen-activated protein kinase. Mol. Cell. Biol. 13:6427-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grallert, A., C. Beuter, R. A. Craven, S. Bagley, D. Wilks, U. Fleig, and I. M. Hagan. 2006. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 20:2421-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grishin, N. V. 1999. Phosphatidylinositol phosphate kinase: a link between protein kinase and glutathione synthase folds. J. Mol. Biol. 291:239-247. [DOI] [PubMed] [Google Scholar]
- 29.Guldal, C. G., and J. Broach. 2006. Assay for adhesion and agar invasion in S. cerevisiae. J. Vis Exp. 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. U. S. A. 97:12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman, C. S. 2005. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 33:257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holbrook, E. D., and C. A. Rappleye. 2008. Histoplasma capsulatum pathogenesis: making a lifestyle switch. Curr. Opin. Microbiol. 11:318-324. [DOI] [PubMed] [Google Scholar]
- 33.Ingram, S. W., S. T. Safrany, and L. D. Barnes. 2003. Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene, and effects on growth rate, morphology and intracellular diadenosine 5′,5′′′-P1,P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem. J. 369:519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamiguchi, H. 2007. The role of cell adhesion molecules in axon growth and guidance. Adv. Exp. Med. Biol. 621:95-103. [DOI] [PubMed] [Google Scholar]
- 35.Kerres, A., C. Vietmeier-Decker, J. Ortiz, I. Karig, C. Beuter, J. Hegemann, J. Lechner, and U. Fleig. 2004. The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association. Mol. Biol. Cell 15:5255-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, D. U., J. Hayles, D. Kim, V. Wood, H. O. Park, M. Won, H. S. Yoo, T. Duhig, M. Nam, G. Palmer, S. Han, L. Jeffery, S. T. Baek, H. Lee, Y. S. Shim, M. Lee, L. Kim, K. S. Heo, E. J. Noh, A. R. Lee, Y. J. Jang, K. S. Chung, S. J. Choi, J. Y. Park, Y. Park, H. M. Kim, S. K. Park, H. J. Park, E. J. Kang, H. B. Kim, H. S. Kang, H. M. Park, K. Kim, K. Song, K. B. Song, P. Nurse, and K. L. Hoe. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knaus, M., P. Wiget, Y. Shimada, and M. Peter. 2005. Control of cell polarity in response to intra- and extracellular signals in budding yeast. Novartis Found. Symp. 269:47-58, 223-230. [PubMed] [Google Scholar]
- 38.La Carbona, S., C. Le Goff, and X. Le Goff. 2006. Fission yeast cytoskeletons and cell polarity factors: connecting at the cortex. Biol. Cell 98:619-631. [DOI] [PubMed] [Google Scholar]
- 39.Lee, Y. S., K. Huang, F. A. Quiocho, and E. K. O'Shea. 2008. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, Y. S., S. Mulugu, J. D. York, and E. K. O'Shea. 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316:109-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenburg, M. E., and E. K. O'Shea. 1996. Signaling phosphate starvation. Trends Biochem. Sci. 21:383-387. [PubMed] [Google Scholar]
- 42.Lin, H., P. C. Fridy, A. A. Ribeiro, J. H. Choi, D. K. Barma, G. Vogel, J. R. Falck, S. B. Shears, J. D. York, and G. W. Mayr. 2009. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 284:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo, H. R., Y. E. Huang, J. C. Chen, A. Saiardi, M. Iijima, K. Ye, Y. Huang, E. Nagata, P. Devreotes, and S. H. Snyder. 2003. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell 114:559-572. [DOI] [PubMed] [Google Scholar]
- 44.Martin, S. G. 2009. Microtubule-dependent cell morphogenesis in the fission yeast. Trends Cell Biol. 19:447-454. [DOI] [PubMed] [Google Scholar]
- 45.Martin, S. G., W. H. McDonald, J. R. Yates III, and F. Chang. 2005. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell 8:479-491. [DOI] [PubMed] [Google Scholar]
- 46.Martin, S. G., S. A. Rincon, R. Basu, P. Perez, and F. Chang. 2007. Regulation of the formin for3p by cdc42p and bud6p. Mol. Biol. Cell 18:4155-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCollum, D., M. Balasubramanian, and K. Gould. 1999. Identification of cold-sensitive mutations in the Schizosaccharomyces pombe actin locus. FEBS Lett. 451:321-326. [DOI] [PubMed] [Google Scholar]
- 48.McCollum, D., A. Feoktistova, M. Morphew, M. Balasubramanian, and K. L. Gould. 1996. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J. 15:6438-6446. [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchison, J. M., and P. Nurse. 1985. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75:357-376. [DOI] [PubMed] [Google Scholar]
- 50.Mitsuzawa, H. 2006. Ammonium transporter genes in the fission yeast Schizosaccharomyces pombe: role in ammonium uptake and a morphological transition. Genes Cells 11:1183-1195. [DOI] [PubMed] [Google Scholar]
- 51.Moreno, M. B., A. Duran, and J. C. Ribas. 2000. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast 16:861-872. [DOI] [PubMed] [Google Scholar]
- 52.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 53.Mullaney, E. J., and A. H. Ullah. 2003. The term phytase comprises several different classes of enzymes. Biochem. Biophys. Res. Commun. 312:179-184. [DOI] [PubMed] [Google Scholar]
- 54.Mulugu, S., W. Bai, P. C. Fridy, R. J. Bastidas, J. C. Otto, D. E. Dollins, T. A. Haystead, A. A. Ribeiro, and J. D. York. 2007. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316:106-109. [DOI] [PubMed] [Google Scholar]
- 55.Niccoli, T., and P. Nurse. 2002. Different mechanisms of cell polarisation in vegetative and shmooing growth in fission yeast. J. Cell Sci. 115:1651-1662. [DOI] [PubMed] [Google Scholar]
- 56.Nishizawa, M., T. Komai, Y. Katou, K. Shirahige, T. Ito, and E. A. Toh. 2008. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol. 6:2817-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Onnebo, S. M., and A. Saiardi. 2009. Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem. J. 423:109-118. [DOI] [PubMed] [Google Scholar]
- 58.Pérez, P., and J. C. Ribas. 2004. Cell wall analysis. Methods 33:245-251. [DOI] [PubMed] [Google Scholar]
- 59.Pesesse, X., K. Choi, T. Zhang, and S. B. Shears. 2004. Signaling by higher inositol polyphosphates. Synthesis of bisdiphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J. Biol. Chem. 279:43378-43381. [DOI] [PubMed] [Google Scholar]
- 60.Piel, M., and P. T. Tran. 2009. Cell shape and cell division in fission yeast. Curr. Biol. 19:R823-R827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prevorovský, M., J. Stanurova, F. Puta, and P. Folk. 2009. High environmental iron concentrations stimulate adhesion and invasive growth of Schizosaccharomyces pombe. FEMS Microbiol. Lett. 293:130-134. [DOI] [PubMed] [Google Scholar]
- 62.Ptacek, J., G. Devgan, G. Michaud, H. Zhu, X. Zhu, J. Fasolo, H. Guo, G. Jona, A. Breitkreutz, R. Sopko, R. R. McCartney, M. C. Schmidt, N. Rachidi, S. J. Lee, A. S. Mah, L. Meng, M. J. Stark, D. F. Stern, C. De Virgilio, M. Tyers, B. Andrews, M. Gerstein, B. Schweitzer, P. F. Predki, and M. Snyder. 2005. Global analysis of protein phosphorylation in yeast. Nature 438:679-684. [DOI] [PubMed] [Google Scholar]
- 63.Pufall, M. A., and B. J. Graves. 2002. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 18:421-462. [DOI] [PubMed] [Google Scholar]
- 64.Reynolds, T. B., and G. R. Fink. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878-881. [DOI] [PubMed] [Google Scholar]
- 65.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 66.Rohrschneider, M. R., and J. Nance. 2009. Polarity and cell fate specification in the control of Caenorhabditis elegans gastrulation. Dev. Dyn. 238:789-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Safrany, S. T., J. J. Caffrey, X. Yang, M. E. Bembenek, M. B. Moyer, W. A. Burkhart, and S. B. Shears. 1998. A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 17:6599-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Safrany, S. T., S. W. Ingram, J. L. Cartwright, J. R. Falck, A. G. McLennan, L. D. Barnes, and S. B. Shears. 1999. The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase. Overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 274:21735-21740. [DOI] [PubMed] [Google Scholar]
- 69.Saiardi, A., R. Bhandari, A. C. Resnick, A. M. Snowman, and S. H. Snyder. 2004. Phosphorylation of proteins by inositol pyrophosphates. Science 306:2101-2105. [DOI] [PubMed] [Google Scholar]
- 70.Saiardi, A., H. Erdjument-Bromage, A. M. Snowman, P. Tempst, and S. H. Snyder. 1999. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9:1323-1326. [DOI] [PubMed] [Google Scholar]
- 71.Saiardi, A., E. Nagata, H. R. Luo, A. M. Snowman, and S. H. Snyder. 2001. Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 276:39179-39185. [DOI] [PubMed] [Google Scholar]
- 72.Santangelo, G. M. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:253-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shears, S. B. 2009. Diphosphoinositol polyphosphates: metabolic messengers? Mol. Pharmacol. 76:236-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 75.Smukalla, S., M. Caldara, N. Pochet, A. Beauvais, S. Guadagnini, C. Yan, M. D. Vinces, A. Jansen, M. C. Prevost, J. P. Latge, G. R. Fink, K. R. Foster, and K. J. Verstrepen. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135:726-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tatebe, H., K. Nakano, R. Maximo, and K. Shiozaki. 2008. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 18:322-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tatebe, H., K. Shimada, S. Uzawa, S. Morigasaki, and K. Shiozaki. 2005. Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr. Biol. 15:1006-1015. [DOI] [PubMed] [Google Scholar]
- 78.Toda, T., S. Dhut, G. Superti-Furga, Y. Gotoh, E. Nishida, R. Sugiura, and T. Kuno. 1996. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 16:6752-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toone, W. M., and N. Jones. 1998. Stress-activated signalling pathways in yeast. Genes Cells 3:485-498. [DOI] [PubMed] [Google Scholar]
- 80.Verstrepen, K. J., and F. M. Klis. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60:5-15. [DOI] [PubMed] [Google Scholar]
- 81.Wang, Y. 2009. CDKs and the yeast-hyphal decision. Curr. Opin. Microbiol. 12:644-649. [DOI] [PubMed] [Google Scholar]
- 82.Wilson-Grady, J. T., J. Villen, and S. P. Gygi. 2008. Phosphoproteome analysis of fission yeast. J. Proteome Res. 7:1088-1097. [DOI] [PubMed] [Google Scholar]
- 83.York, S. J., B. N. Armbruster, P. Greenwell, T. D. Petes, and J. D. York. 2005. Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 280:4264-4269. [DOI] [PubMed] [Google Scholar]
- 84.Zaitsevskaya-Carter, T., and J. A. Cooper. 1997. Spm1, a stress-activated MAP kinase that regulates morphogenesis in S. pombe. EMBO J. 16:1318-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]