Abstract
Calcineurin is a widely expressed and highly conserved Ser/Thr phosphatase. Calcineurin is inhibited by the immunosuppressant drug cyclosporine A (CsA) or tacrolimus (FK506). The critical role of CsA/FK506 as an immunosuppressant following transplantation surgery provides a strong incentive to understand the phosphatase calcineurin. Here we uncover a novel regulatory pathway for cyclic AMP (cAMP) signaling by the phosphatase calcineurin which is also evolutionarily conserved in Caenorhabditis elegans. We found that calcineurin binds directly to and inhibits the proteosomal degradation of cAMP-hydrolyzing phosphodiesterase 4D (PDE4D). We show that ubiquitin conjugation and proteosomal degradation of PDE4D are controlled by a cullin 1-containing E3 ubiquitin ligase complex upon dual phosphorylation by casein kinase 1 (CK1) and glycogen synthase kinase 3β (GSK3β) in a phosphodegron motif. Our findings identify a novel signaling process governing G-protein-coupled cAMP signal transduction—opposing actions of the phosphatase calcineurin and the CK1/GSK3β protein kinases on the phosphodegron-dependent degradation of PDE4D. This novel signaling system also provides unique functional insights into the complications elicited by CsA in transplant patients.
Calcineurin (also known as protein phosphatase 2B) was first identified as a calcium/calmodulin-binding protein in brain extracts (40, 57). Further analysis demonstrated that calcineurin is a widely expressed and highly conserved phosphatase (5, 19, 21, 25, 53). Calcineurin is inhibited by the immunosuppressant drug cyclosporine A (CsA) or tacrolimus (FK506) (17, 24, 43). CsA (or FK506) forms complexes with cyclophilins (or FK506-binding proteins) and inhibits calcineurin. The critical role of CsA and FK506 as immunosuppressants following transplantation surgery (16, 45, 46, 48) provides a strong incentive to understand the phosphatase calcineurin.
At the molecular level, calcineurin contributes to immune regulation by inhibiting the transcription factor NFAT (nuclear factor of activated T cells). The transcription factor NFAT family was first identified as an important regulator of interleukin-2 (IL-2) gene expression (23). NFAT is located in the cytosol of resting cells (20, 30). Increased intracellular calcium activates the phosphatase calcineurin, which binds to a calcineurin docking motif (PXIXIT) (4, 6, 15) and dephosphorylates NFAT. Administration of CsA to transplant patients inhibits calcineurin and blocks the nuclear translocation of NFAT. Hence, NFAT remains in the cytosol and the expression of cytokine genes such as that for IL-2 is reduced. Since IL-2 is a critical mitogen of T cells, a reduced level of IL-2 abrogates T-cell differentiation and minimizes the incidence of host-versus-graft reactions in transplant patients.
Despite the tremendous advantages of CsA and FK506 inhibition of calcineurin after transplantation surgery, prolonged treatment with CsA or FK506 produces many side effects, including diabetes mellitus, hyperlipidemia, nephrotoxicity, neuronal damage, and cardiovascular disease (3, 26, 38, 48). The current model indicates that calcineurin-NFAT signaling regulates the growth and differentiation of beta cells in the pancreatic islets (27, 28). In the presence of CsA, changes in insulin signaling could account for the complications found in transplant patients.
Here we uncover a novel signaling system which centers on calcineurin controlling ubiquitin conjugation and proteosomal degradation of cyclic AMP (cAMP)-hydrolyzing phosphodiesterase 4D (PDE4D). Degradation of PDE4D is channeled through a cullin 1 (Cul1)-containing E3 ubiquitin ligase complex and is dependent on dual phosphorylation carried out by casein kinase 1 (CK1) and glycogen synthase kinase 3β (GSK3β). Integration of calcineurin, CK1, GSK3β, and Cul1 is also evolutionarily conserved in Caenorhabditis elegans. This new signaling system expands the repertoire of calcineurin substrates and indicates that calcineurin impinges on phosphodependent protein degradation. We propose that this novel calcineurin signaling system provides new insights into alternative therapeutic targets for a range of adverse pathological complications elicited by CsA in transplant patients.
MATERIALS AND METHODS
Mice.
Animal experiments were performed in accordance with the guidelines of the Albert Einstein College of Medicine Institute of Animal Studies. CnAβ−/− mice were generated as described previously, by the insertion of a neomycin resistance gene to disrupt exon 2 (12). CnAβ−/− mice were backcrossed with C57BL/6 mice at least 10 times before use. Backcrossed CnAβ−/− mice were also used for the isolation of mouse embryonic fibroblasts (MEFs).
Worm strains.
C. elegans strains were maintained under standard culture conditions on NGM agar plates with Escherichia coli OP50 as a food source (10). Wild-type animals were of Bristol strain N2 (a kind gift from Ji Ying Sze). Loss-of-function calcineurin tax-6(p675lf) (41) and PDE-4 pde-4(ok1290) mutants were a kind gift from Ji Ying Sze or obtained from the Caenorhabditis Genetics Center.
Reagents.
Mouse CnAβ was amplified by PCR from mouse mammary gland cDNA and hemagglutinin (HA) tagged using primers 5′-AGGAATTCGCCACCATGTACCCATACGATGTTCCAGATTA CGCTGCCGCCCCGGAGCCGGCC-3′ and 5′-CTCGAGTCACTGGGCACTATGGTT-3′. The PCR product was subcloned into the vector pCDNA3 using the EcoRI and XhoI sites. C. elegans PDE4 was amplified by PCR from plasmid KM#203 (a kind gift from Ken Miller) (13) and FLAG tagged using primers 5′-GATCGGATCCCGGCAGCCAACATGCCACGAAGACGCGGCTCGTCGTCGTCGTCGTCG-3′ and 5′-GATCGAATTCTCATTTGTCATCATCGTCCTTATAGTCTTTGTGCTCGTCATCTTCTGTTACAGT-3′. The PCR product was subcloned into the vector pCDNA3 using the BamHI and EcoRI sites. Expression vectors for VSV-G-tagged PDE4D3, VSV-G-tagged PDE4D5, FLAG-tagged GSK3β, HA-tagged CK1, FLAG-tagged dominant negative Cul1, HA-tagged ubiquitin, and C. elegans PDE4 (KM#203) have been described previously (13, 37, 44, 59, 60, 63). Point mutations were generated by the QuikChange protocol and sequenced. Recombinant PDE4D was expressed using the vector pGEX-5X1 by cloning into the BamHI and XhoI sites. Antibodies for PDE4D (sc25814 and sc25097), the VSV-G tag (sc66180), the HA tag (sc7392), the FLAG tag (F3185), calcineurin A (sc6124 and sc17808), and calcineurin B (sc6119) were obtained from Santa Cruz or Sigma. Polyclonal antibodies against phospho-Thr were obtained from Invitrogen (71-8200). Tubulin antibody (clone E7) was obtained from the monoclonal antibody facility at the University of Iowa. Polyclonal antibodies against PDE4D and C. elegans PDE4 were described previously (9, 13). Antibodies to phospho-Ser616 were generated by using the synthetic phosphopeptide Cys-Glu-Ser-Asp-Thr-Glu-Lys-Asp-Ser(P)-Gly-Ser-Gln-Val-Glu-Glu-Asp and conjugated to keyhole limpet hemocyanin. The phosphospecific antibody was affinity purified from rabbit serum as described previously (62). The PDE inhibitors IBMX and rolipram, the calcium ionophore ionomycin (Ion), the calcineurin inhibitor CsA, the GSK3β inhibitor lithium chloride (LiCl), the CK1 inhibitor D4476, the CK2 inhibitor 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT), and the proteosome inhibitor MG132 were obtained from Calbiochem, Sigma, and/or Fisher Thermo Scientific.
Cell culture.
MEFs were prepared from E13.5/14.5 embryos after trypsin digestion as described previously (61). MEFs and COS7 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). COS7 cells were transfected using Lipofectamine (Invitrogen) according to the manufacturer's protocol. Cells were harvested in Triton lysis buffer (20 mM Tris [pH 7.4], 134 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 2 mM NaPPi, 10% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 10 μg/ml leupeptin).
Semiquantitative RT-PCR.
Total RNA was isolated from MEFs using TRIzol reagent (Invitrogen). Five micrograms of RNA was reverse transcribed into cDNA using superscript II reverse transcriptase (Invitrogen). Semiquantitative reverse transcription (RT)-PCR was performed for each sample in triplicate with two different concentrations of cDNA to establish linear amplification. Twenty PCR cycles were amplified. The primers used for RT-PCR were as follows: for PDE4A, 5′-CCTGCCTGACAAGTTCCAAT-3′ and 5′-AGCAGAGATGACGGCAGAAT-3′; for PDE4B, 5′-ATCACCTTGCTGTGGGATTC-3′ and 5′-AACCAACCTGGGATTTTTCC-3′; for PDE4D, 5′-GTCCCATGTGTGACAAGCAC-3′ and 5′-CAAGTTTCAGGCTGGCTTTC-3′; for PDE7A, 5′-TGCTGCAGACGTTACTCAGG-3′ and 5′-AGCACATTTCAAGGCCATCT-3′.
Half-life determination.
The plasmid for VSV-G-tagged PDE4D3 expression was transfected into COS7 cells. Cells were treated with cycloheximide (CHX; 0.5 mM) in the presence or absence of CsA (5 μM) for 0, 1, 2, or 4 h before harvest. The effect of the transcriptional inhibitor actinomycin D (ActD; 0.5 μg/ml) was also examined.
Protein kinase assays.
Glutathione S-transferase (GST) recombinant protein (2 μg) encompassing either the NH2 or COOH terminus of the catalytic core of PDE4D was incubated with purified GSK3β or CK1 (5 U) in the presence of [γ-32P]ATP for 30 min. Phosphorylation of PDE4D was examined by sodium dodecyl sulfate-gel electrophoresis and autoradiography.
Binding assays.
The plasmid for VSV-G-tagged PDE4D3 expression was cotransfected with HA-tagged CnAβ into COS7 cells. Cell extracts prepared using Triton lysis buffer were incubated with protein G-agarose and HA antibody to immunoprecipitate CnAβ in coimmunoprecipitation assays. Binding assays were also performed by using GST or GST-PDE4D recombinant protein and purified calcineurin. After three washes with Triton lysis buffer, the bound proteins were detected by immunoblot analysis.
Statistical analysis.
The data shown are means ± standard errors of the means. The Student t test was performed, and P values of <0.05 were considered to be significant.
RESULTS
Posttranscriptional regulation of PDE4D expression by calcineurin.
We initially set out to determine whether calcineurin contributes to the regulation of cAMP-hydrolyzing PDE4D, given that targeted disruption of calcineurin led to metabolic complications with dysregulation in the β-adrenergic G-protein-coupled receptor (GPCR)/cAMP signaling pathway (data available on request). Among the 11 PDE families, PDE4 was chosen because of its widespread expression in different cells and tissues (18, 32). Indeed, PDE4 accounts for the bulk of the total cAMP-hydrolyzing PDE activity in primary MEFs (11). Furthermore, PDE4 can be regulated upon phosphorylation by protein kinase A (PKA) and/or ERK (29, 55), which are downstream effectors of the β-adrenergic receptor signaling pathway.
Unexpectedly, we found that the levels of PDE4D expression were potentiated upon coexpression with CnAβ (Fig. 1). Increased protein expression was found in at least two different members of the PDE4D family, including PDE4D3 and PDE4D5 (Fig. 1A). CnAβ, however, affected neither the level of expression of exogenous β-arrestin 1 (βArr-1) nor the levels of expression of endogenous tubulin and insulin growth factor receptor. These data indicate that CnAβ regulates PDE4D3 and PDE4D5 expression.
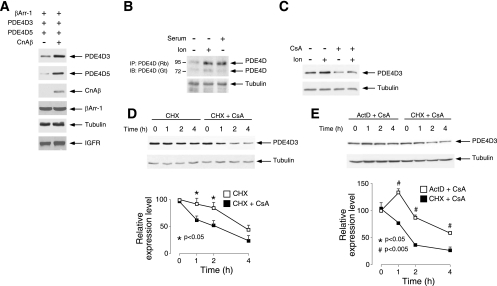
FIG. 1.
Posttranscriptional regulation of PDE4D expression by calcineurin. (A) Extracts prepared from COS7 cells transiently transfected with PDE4D3, PDE4D5, or βArr-1 were examined by immunoblot analysis. The effect of coexpression of CnAβ is also shown. Expression of endogenous IGF-1 receptor (IGF-1) and tubulin is shown as a control. (B) Expression of endogenous PDE4D in MEFs was detected by immunoprecipitation (IP) using rabbit (Rb) polyclonal PDE4D antibody (sc25814) and subsequent immunoblot analysis (IB) using goat (Gt) polyclonal PDE4D antibody (9). The effect of the calcium ionophore ionomycin and mitogenic stimuli (20% fetal calf serum) is also shown. (C) Extracts prepared from COS7 cells transiently transfected with PDE4D3 were treated with the calcium ionophore Ion (2 μM) and/or the calcineurin inhibitor CsA (5 μM). Expression of PDE4D3 was determined by immunoblot analysis. (D) COS7 cells were transiently transfected with PDE4D3. The half-life of PDE4D3 was determined in the presence CHX (0.5 mM). The effect of CsA (5 μM) is also shown. Expression of tubulin was used as a control. (E) COS7 cells were transiently transfected with PDE4D3. The half-life of PDE4D3 was determined in the presence of CsA (5 μM). The effect of the transcription inhibitor ActD (0.5 μg/ml) or the translational inhibitor CHX (0.5 mM) was examined. *, P < 0.05; #, P < 0.005.
We further ascertained calcineurin regulation of PDE4D expression by using the calcium ionophore ionomycin to activate calcineurin and the calcineurin inhibitor CsA (Fig. 1B and C). In the presence of ionomycin, the expression levels of endogenous PDE4D in MEFs were potentiated (Fig. 1B). The expression levels of endogenous PDE4D were also potentiated by serum stimulation, which induced a Ca2+ influx (Fig. 1B). Administration of CsA, however, reduced PDE4D3 expression (Fig. 1C). Notably, CsA abrogated the potentiation of PDE4D3 expression elicited by ionomycin (Fig. 1C). These data confirm that calcineurin regulates PDE4D expression.
To investigate the role of calcineurin in PDE4D protein expression, we determined the half-lives of PDE4D3 in the presence and absence of CsA (Fig. 1D). In the presence of the translational inhibitor CHX, we noted that the half-life of PDE4D3 was ∼4 h (Fig. 1D). Administration of CsA reduced the half-life of PDE4D3 to ∼1 h (Fig. 1D). These data indicate that calcineurin regulates the degradation of the PDE4D3 protein.
Given the prominent role of calcineurin in transcription regulation (20, 30), we asked whether the role of calcineurin in PDE4D3 potentiation was also transcription dependent (Fig. 1E). Unlike the effect of a translation inhibitor, administration of CsA in the presence of the transcription inhibitor ActD did not further reduce the half-life of PDE4D3 (Fig. 1E). These data indicate that calcineurin regulates the posttranscriptional degradation of the PDE4D3 protein.
Ubiquitin-mediated degradation of PDE4D via a phosphodegron motif.
Ubiquitin conjugation and subsequent proteosome-mediated degradation play a critical role in protein turnover (49, 50). We have identified a potential phosphodegron (DSGSQVEED623) located in the COOH-terminal conserved catalytic core of PDE4D (Fig. 2 A). In general, a phosphodegron (e.g., DpSGX2-4pS) promotes the polyubiquitin-mediated degradation of phosphoproteins via the Skp1-Cul-F box receptor (SCF) E3 ubiquitin ligase complex (34, 36). Phosphodependent degradation of PDE4D has also been implicated in a recent genome-wide identification of Cul1 substrates (65, 66). The location of the phosphodegron in the COOH-terminal conserved catalytic core further suggests that all PDE4D isoforms are regulated similarly.
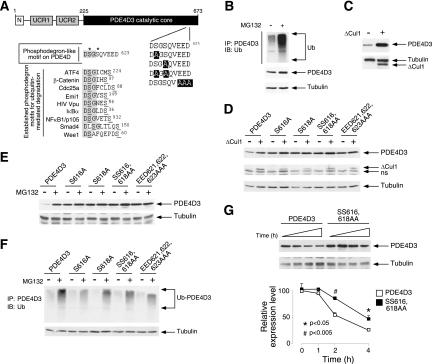
FIG. 2.
Ubiquitin-mediated degradation of PDE4D via a phosphodegron motif. (A) Schematic illustration of the location of the DSGSQVEED phosphodegron in PDE4D3. Sequences of known phosphodegrons are also shown. Conserved amino acid residues and phosphorylated Ser are highlighted and underlined, respectively. A phosphodegron-like motif (DSGSQVEED) and potential phosphorylation sites (Ser616 and Ser618) in PDE4D3 are also shown. (B) COS7 cells were transiently transfected with PDE4D3. Ubiquitin conjugation of PDE4D3 was determined by coimmunoprecipitation assays. The presence of ubiquitin in PDE4D3 immunoprecipitates (IP) was analyzed by immunoblot (IB) analysis. The effect of the proteosome inhibitor MG132 (10 μM) is also shown. (C) COS7 cells were transiently transfected with PDE4D3. The expression levels of PDE4D3 in the presence and absence of dominant negative Cul1 (ΔCul1) were determined by immunoblot analysis. (D) COS7 cells were transiently transfected with wild-type or phosphodegron-mutated (Ala616, Ala618, Ala616,618, or Ala621,622,623) PDE4D3. The expression levels of wild-type and phosphodegron-mutated PDE4D3 were determined in the presence and absence of ΔCul1. (E) COS7 cells were transiently transfected with wild-type or phosphodegron-mutated (Ala616, Ala618, Ala616,618, or Ala621,622,623) PDE4D3. The expression levels of wild-type and phosphodegron-mutated PDE4D3 were determined in the presence and absence of the proteosome inhibitor MG132 (10 μM). (F) COS7 cells were transiently transfected with wild-type or phosphodegron-mutated (Ala616, Ala618, Ala616,618, or Ala621,622,623) PDE4D3. Ubiquitin conjugation of wild-type and phosphorylation-defective PDE4D3 was determined by coimmunoprecipitation assays. The presence of ubiquitin in PDE4D3 immunoprecipitates was analyzed by immunoblot analysis. The effect of the proteosome inhibitor MG132 (10 μM) is also shown. (G) COS7 cells were transiently transfected with wild-type or phosphodegron-mutated (Ala616,618) PDE4D3. The half-lives of wild-type and phosphorylation-defective PDE4D3 were determined in the presence of CHX (0.5 mM) and CsA (5 μM). *, P < 0.05; #, P < 0.005.
Here we show that PDE4D degradation can be channeled through the polyubiquitin-dependent proteosome pathway (Fig. 2B). Indeed, in the presence of the proteosome inhibitor MG132, degradation was reduced while ubiquitin conjugation of PDE4D was increased (Fig. 2B). Notably, expression of dominant negative Cul1 (ΔCul1), which blocks recruitment and subsequent protein degradation mediated by the SCF E3 ubiquitin ligase complex, also reduced PDE4D degradation (Fig. 2C). These data indicate that the degradation of PDE4D is mediated by a Cul1-containing SCF E3 ubiquitin ligase complex.
Next, we determined the role of the phosphodegron in PDE4D degradation (Fig. 2D to F). Phosphodegron-defective PDE4D (Ala616, Ala618, Ala616,618, or Ala621-623) exhibited increased expression compared to the wild type (Fig. 2D to F). In the presence of CHX, the half-life of phosphodegron-defective PDE4D was potentiated (Fig. 2G). Expression of ΔCul1 reduced PDE4D degradation (Fig. 2D). The effect of ΔCul1 on PDE4D degradation, however, was abrogated in phosphodegron-defective PDE4D (Fig. 2D). These data indicate that degradation of PDE4D by the SCF E3 ubiquitin ligase complex is phosphodegron dependent.
Reduced degradation of phosphodegron-defective PDE4D in the presence of the proteosome inhibitor MG132 was also confirmed. In the presence of MG132, the expression and ubiquitin conjugation of PDE4D were potentiated (Fig. 2E and F). Phosphodegron-defective PDE4D, however, exhibited reduced ubiquitin conjugation despite potentiated expression (Fig. 2E and F). Together, these data indicate that the phosphodegron is required for ubiquitin-mediated degradation of PDE4D via a Cul1-containing SCF E3 ubiquitin ligase complex.
Calcineurin regulation of PDE4D degradation is phosphodegron dependent.
Given that calcineurin potentiates PDE4D expression (Fig. 1), we therefore asked whether calcineurin regulation is also phosphodegron dependent (Fig. 3). Expression of calcineurin reduced PDE4D degradation (Fig. 3). The effect of calcineurin on PDE4D degradation, however, was abrogated in phosphodegron-defective PDE4D (Fig. 3). Reduced degradation of phosphodegron-defective PDE4D in the presence of pharmacological agents that modulate endogenous calcineurin activity was also confirmed (data available on request). Administration of the calcium ionophore ionomycin reduced, while the calcineurin inhibitor CsA potentiated, PDE4D degradation (data available on request). The effect of ionomycin and CsA on PDE4D degradation, however, was abrogated in phosphodegron-defective PDE4D (data available on request). These data illuminate the role of calcineurin, via the phosphodegron, on PDE4D protein degradation.
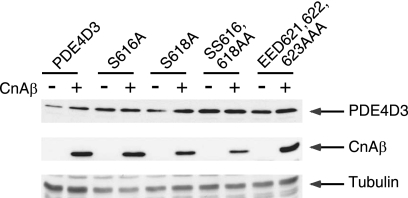
FIG. 3.
Calcineurin regulation of PDE4D degradation is phosphodegron dependent. Wild-type or phosphodegron-mutated (Ala616, Ala618, Ala616,618, or Ala621,622,623) PDE4D3 was coexpressed with CnAβ in COS7 cells. The expression levels of PDE4D3 and CnAβ were determined by immunoblot analysis. The expression level of tubulin was used as a control.
Mapping of functional PxIxIT-like calcineurin docking motifs in PDE4D.
Previous studies established that calcineurin interacts with its substrates via a PXIXIT docking motifs (4, 6, 15). For example, conserved PXIXIT docking motifs are found in all four calcineurin-regulated NFAT family members (Fig. 4 A), which play an important role in cytokine gene transcription and contribute significantly to the success of transplant surgery (30). Replacement of the PXIXIT motif with Ala attenuates the dephosphorylation of NFAT by calcineurin (4, 6, 15). In addition, the PXIXIT-containing peptide domain acts in a dominant negative fashion and interferes with calcineurin interaction and subsequent dephosphorylation (4, 6, 15), thereby inhibiting NFAT-mediated cytokine gene transcription. Similar PXIXIT motifs are found in calcineurin substrates in yeast (Fig. 4A) (21), indicating an evolutionarily conserved enzyme-substrate mechanism mediated by calcineurin docking via the PXIXIT motif.
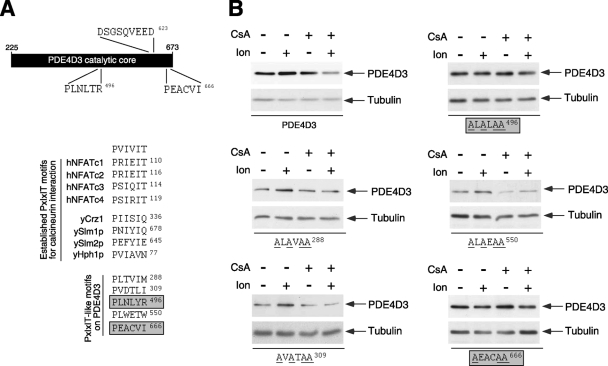
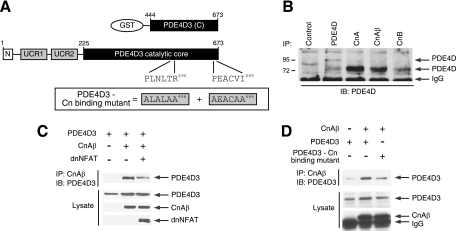
FIG. 4.
Mapping of functional PXIXIT-like calcineurin docking motifs in PDE4D. (A) Schematic illustration of the locations of the two functional PXIXIT-like motifs (PLNLYR496 and PEACVI666) in the catalytic core of PDE4D3. The location of the DSGSQVEED phosphodegron in PDE4D3 is also indicated. A sequence comparison of known calcineurin docking PXIXIT motifs found in human NFAT and yeast proteins is shown. Potential PXIXIT-like motifs in PDE4D3 and an optimized PXIXIT motif (PVIVIT) are also shown. Functional PXIXIT-like motifs in PDE4D3 are highlighted. (B) Wild-type and PXIXIT-mutated PDE4D3 were transiently transfected into COS7 cells. The expression levels of wild-type and PXIXIT-mutated PDE4D3 were determined in the presence and absence of Ion (2 μM) and/or CsA (5 μM). The expression of tubulin was used as a control. Replacements of critical amino acid residues with Ala in potential PXIXIT-like motifs of PDE4D3 are underlined. Functional PXIXIT-like motifs in PDE4D3 are highlighted.
We have identified several putative PXIXIT-like calcineurin docking motifs in the COOH-terminal conserved catalytic core of PDE4D (Fig. 4A). We surmised that some of these motifs are functional and are critical in mediating the effect of calcineurin on PDE4D degradation. Thus, we performed site-direct mutagenesis to replace consensus amino acids of the PXIXIT-like motif with Ala (AXAXAA) and tested whether the expression of the Ala-substituted PDE4Ds was regulated by ionomycin or CsA (Fig. 4B). Immunoblot analysis indicated that PLNLYR496 and PEACVI666 are functional PXIXIT-like calcineurin docking motifs in PDE4D (Fig. 4B). Replacement of PLNLYR496 or PEACVI666 with Ala abrogated the stabilization of PDE4D expression by ionomycin (Fig. 4B). Similarly, increased degradation of PDE4D in the presence of CsA was attenuated in Ala-substituted PLNLYR496 or PEACVI666 PDE4D (Fig. 4B). These data indicate that there are two functional PXIXIT-like calcineurin docking domains in PDE4D. The location of these functional calcineurin docking domains in the COOH-terminal conserved catalytic core of PDE4D implies that all PDE4D isoforms are subject to similar regulation.
Direct interaction between calcineurin and PDE4D.
Next, we examined whether calcineurin interacts with PDE4D and whether such interaction requires the two identified PXIXIT-like motifs (PLNLYR496 and PEACVI666) (Fig. 5 A). Coimmunoprecipitation assays showed that endogenous calcineurin interacts with endogenous PDE4D (Fig. 5B). Notably, the calcineurin-PDE4D interaction was attenuated by coexpression of a PXIXIT-containing calcineurin docking inhibitor, dnNFAT (6, 15) (Fig. 5C). Furthermore, the calcineurin-PDE4D interaction was impaired upon the replacement of critical residues in the PXIXIT-like motifs with Ala (ALALAA496 and AEACAA666) in PDE4D (Fig. 5D). Finally, binding assays using purified proteins supported the concept that calcineurin interacts directly with PDE4D (data available on request).
FIG. 5.
Direct interaction between calcineurin and PDE4D. (A) Schematic illustration of PDE4D3 and PDE4D3 GST fusion protein. Functional calcineurin docking motifs (PLNLTR496 and PEACVI666) are indicated. Calcineurin binding-defective PDE4D3 (ALALAA496 and AEACAA666) is also shown. (B) Binding of endogenous PDE4D and calcineurin was determined by coimmunoprecipitation assays. Endogenous PDE4D in COS7 cells was immunoprecipitated (IP) using goat polyclonal PDE4D antibody (sc25097), mouse monoclonal calcineurin A antibody (sc17808), goat polyclonal calcineurin Aβ antibody (sc6124), or goat polyclonal calcineurin B antibody (sc6119). The presence of endogenous PDE4D in calcineurin immunoprecipitates was analyzed by immunoblot (IB) analysis using rabbit polyclonal PDE4D antibody (sc25814). Mouse M2 monoclonal antibody was used as a control. (C) PDE4D3 was coexpressed with CnAβ in COS7 cells. Binding of PDE4D3 and CnAβ was determined by coimmunoprecipitation assays. The presence of PDE4D3 in CnAβ immunoprecipitates and in cell extracts was analyzed by immunoblot analysis. The effect of the PXIXIT-containing calcineurin docking inhibitor dnNFAT is also shown. (D) Wild-type and calcineurin binding-defective PDE4D3 were coexpressed in the presence and absence of CnAβ in COS7 cells. The presence of PDE4D3 in CnAβ immunoprecipitates and in cell extracts was analyzed by immunoblotting.
CK1 or GSK3β phosphorylates the PDE4D phosphodegron.
Interaction with and stabilization of PDE4D by calcineurin suggest that the phosphodegron motif (DSGSQVEED623) might be phosphorylated. We therefore sought protein kinases that might counterbalance the role of calcineurin in PDE4D stabilization (Fig. 6). Common Ser/Thr protein kinases that preferentially phosphorylate target sites encompassing acidic amino acid residues include CK1, CK2, and GSK3β. Phosphorylation mediated by GSK3β and/or CK1 can promote protein degradation (34, 60). Since integration of calcineurin, GSK3β, and CK1 regulates NFAT activation (7, 67), we asked whether GSK3β and/or CK1 phosphorylate PDE4D.
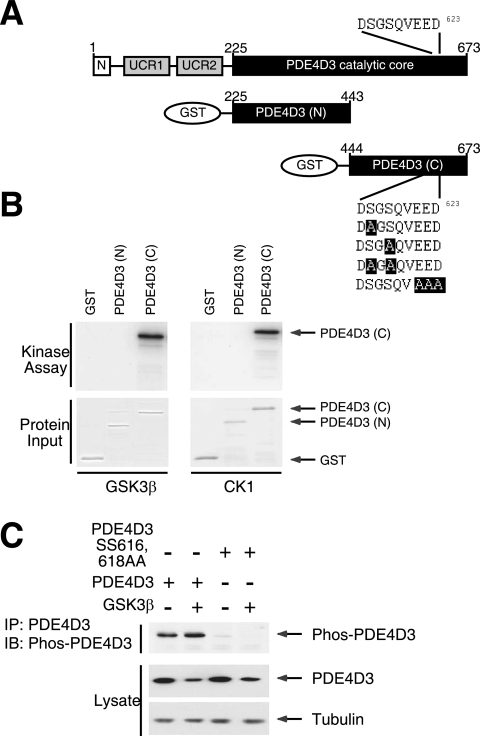
FIG. 6.
CK1 and GSK3β phosphorylate PDE4D phosphodegron. (A) Schematic illustration of PDE4D3 and PDE4D3 GST fusion protein. The location of the DSGSQVEED phosphodegron in PDE4D3 are also shown. Replacements of critical amino acid residues in the DSGSQVEED phosphodegron of PDE4D3 with Ala are highlighted. (B) GST recombinant proteins encompassing either the NH2-terminal or the COOH-terminal portion of the catalytic core of PDE4D3 were subjected to in vitro protein kinase assays in the presence of recombinant GSK3β or CK1 and [γ-32P]ATP. Autoradiography and Coomassie blue staining of recombinant PDE4D3 are shown. (C) COS7 cells transiently transfected with wild-type or Ala616,618 mutant PDE4D3. The effect of GSK3β was also examined. Phosphorylation of Ser616 of PDE4D3 was assessed by phospho-PDE4D antibody (Phos-PDE4D) in immunoblot analysis. Expression of PDE4D3 and tubulin is also shown.
In vitro protein kinase assays indicated that GSK3β and CK1 phosphorylated the COOH-terminal portion of the PDE4D catalytic core (Fig. 6A and B), which encompasses the DSGSQVEED phosphodegron. Replacement of Ser616 and/or Ser618 in the phosphodegron with Ala reduced PDE4D phosphorylation mediated by GSK3β or CK1 (data available on request). Replacement of Glu621, Glu622, and Asp623 with Ala also reduced GSK3β- or CK1-mediated PDE4D phosphorylation (data available on request), indicating that these acidic amino acid residues are critical for optimal phosphorylation. These data indicate that CK1 and GSK3β phosphorylate Ser616 and/or Ser618 at the phosphodegron in PDE4D.
To elucidate the phosphorylation of the phosphodegron of PDE4D in vivo, we developed phosphospecific antibodies to detect phospho-Ser616. Immunoblotting analyses demonstrated phosphorylation of PDE4D3, which increased in intensity upon the coexpression of GSK3β (Fig. 6C). Notably, GSK3β reduced the expression of PDE4D3, despite increased phosphorylation of PDE4D3. The replacement of Ser616,618 of PDE4D3 with Ala, however, abrogated the detection of PDE4D3 by the phosphospecific antibodies (Fig. 6C). In the presence of MG132 to inhibit proteosomal degradation, phosphorylated PDE4D3 accumulated (data available on request). Together, these data confirm phosphorylation at the phosphodegron of PDE4D3 in vivo. These data also support the concept that phosphorylation of PDE4D at the phosphodegron is a prerequisite for subsequent degradation via the proteosome.
CK1 or GSK3β promotes PDE4D degradation via the phosphodegron motif.
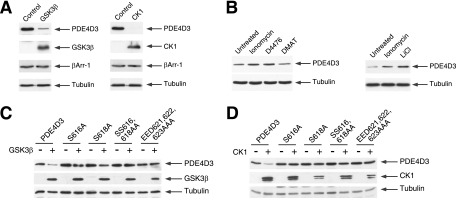
Given that calcineurin reduces PDE4D degradation, we asked whether GSK3β or CK1 mediates the opposing effect and potentiates PDE4D degradation (Fig. 7). Coexpression of either GSK3β or CK1 potentiated the degradation of PDE4D (Fig. 7A). Expression of exogenous βArr-1 and endogenous tubulin, however, was not affected by GSK3β or CK1 coexpression (Fig. 7A). We further ascertained that endogenous GSK3β or CK1 regulates PDE4D degradation using the protein kinase inhibitors LiCl and D4476, respectively (Fig. 7B). In the presence of LiCl or D4476, degradation of PDE4D was reduced (Fig. 7B). Inhibition of CK2 with DMAT did not affect the degradation of PDE4D (Fig. 7B). These data demonstrate that the endogenous protein kinase GSK3β or CK1 phosphorylates PDE4D and promotes its degradation.
FIG. 7.
CK1 and GSK3β promote PDE4D degradation. (A) PDE4D3 or βArr-1 was coexpressed with GSK3β or CK1 in COS7 cells. The expression levels of PDE4D3, βArr-1, GSK3β, and CK1 were determined by immunoblot analysis. The expression level of tubulin was used as a control. (B) PDE4D3 was transiently expressed in COS7 cells. The expression levels of PDE4D3 in the presence and absence of protein kinase inhibitors for 4 h were determined by immunoblot analysis. The effect of Ion (2 μM) is also shown. The protein kinase inhibitors administered included the GSK3 inhibitor LiCl (20 mM), the CK1 inhibitor D4476 (1 μM), and the CK2 inhibitor DMAT (400 nM). (C and D) Wild-type or phosphodegron-mutated (Ala616, Ala618, Ala616,618, or Ala621,622,623) PDE4D3 was coexpressed with GSK3β (C) or CK1 (D) in COS7 cells. The expression levels of PDE4D3, GSK3β, and CK1 were determined by immunoblot analysis. The expression level of tubulin was used as a control.
Next, we examined whether GSK3β or CK1 regulation of PDE4D is phosphodegron dependent (Fig. 7C and D). Expression of GSK3β or CK1 potentiated PDE4D degradation (Fig. 7C and D). The effect of GSK3β or CK1 on PDE4D degradation, however, was abrogated in phosphodegron-defective PDE4D (Fig. 7C and D). Conversely, administration of the inhibitor LiCl or D4476 to block endogenous protein kinase activity reduced PDE4D degradation (data available on request). The effect of LiCl or D4476 on PDE4D degradation, however, was abrogated in phosphodegron-defective PDE4D (data available on request). These data demonstrate that protein kinase GSK3β or CK1 phosphorylates PDE4D and promotes its degradation via the phosphodegron.
Evolutionarily conserved regulation of PDE4D degradation via calcineurin, CK1, and GSK3β.
Both PDE4 and calcineurin are highly conserved in evolution from C. elegans to mammals (18, 21, 41, 53) (Fig. 8 A). Indeed, we have identified similar phosphodegron and calcineurin docking motifs in the C. elegans PDE4 counterpart (cePDE4) (Fig. 8A). In addition, we have recently found that ablation of CnAβ led to sustained β-adrenergic receptor/PKA activation (data available on request). We found that sustained elevation of cAMP in the absence of CnAβ was due in part to impaired cAMP hydrolysis (data available on request).
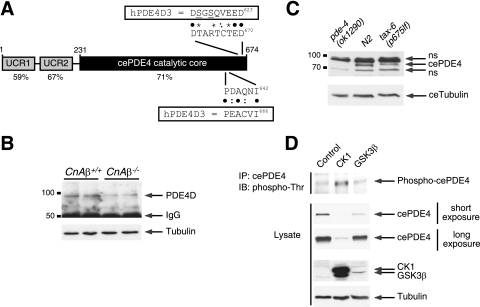
FIG. 8.
Evolutionarily conserved regulation of PDE4D degradation via calcineurin, CK1, and GSK3β. (A) Schematic illustration of C. elegans PDE4 (cePDE4). The locations of the potential phosphodegron and calcineurin docking motif are also shown. Sequence alignment and percent identity with human PDE4D (hPDE4D) are indicated. Bullets indicated conserved amino acid residues. Colons indicated similar amino acid residues. Asterisks indicated potential phosphorylation sites. (B) Extracts prepared from CnAβ−/− and CnAβ+/+ mouse skeletal muscle were subjected to immunoprecipitation and immunoblot analysis to determine the endogenous expression of PDE4D. The expression level of tubulin was used as a control. (C) Extracts prepared from N2 wild-type control C. elegans and loss-of-function calcineurin mutant worms [tax-6(p675lf)] were subjected to immunoblot analysis to determine the endogenous expression of cePDE4. Extracts prepared from PDE4 null worms [pde-4(ok1290)] were used to show the specificity of the cePDE4 antibody. The expression level of C. elegans tubulin was used as a control. (D) cePDE4 was transiently cotransfected with either CK1 or GSK3β into COS7 cells. Phosphorylation of cePDE4 was determined by immunoprecipitation (IP) using M2 monoclonal antibody against FLAG-tagged cePDE4 and subsequent immunoblotting (IB) analysis using phospho-Thr polyclonal antibodies. The expression levels of cePDE4, CK1, GSK3β, and tubulin are also shown.
Next, we asked whether, in analogy to calcineurin inhibition by CsA, the expression of PDE4D is altered upon ablation of CnAβ. Immunoblot analysis indicated reduced expression of PDE4D upon the ablation of CnAβ (Fig. 8B). Expression of PDE4D, as well as PDE4A and PDE4B, mRNA in CnAβ+/+ and CnAβ−/− cells, however, was indistinguishable (data available on request). These data indicate that, similar to calcineurin inhibition, ablation of CnAβ reduces PDE4D expression at the posttranscriptional level.
To further demonstrate the relevance of the calcineurin-PDE4D signaling cross talk, we used C. elegans as a model system. Immunoblot analysis demonstrated that the expression of endogenous cePDE4 was reduced in worms bearing a loss-of-function mutation in calcineurin [tax-6(p675lf)] (Fig. 8C) (41). Evolutionarily conserved regulation of PDE4 by calcineurin indicates the importance of this novel signaling cross talk.
At the molecular level, similar to that of mammalian PDE4D, degradation of cePDE4 was channeled through the Cul1-containing SCF E3 ubiquitin ligase complex (data available on request). Degradation of cePDE4 was reduced by either expression or activation of calcineurin (data available on request). Administration of CsA to inhibit calcineurin, however, potentiated cePDE4 protein degradation (data available on request). Expression of either CK1 or GSK3β protein kinase also potentiated cePDE4 degradation (Fig. 8D). Notably, expression of CK1 or GSK3β increased the phosphorylation of cePDE4, despite the reduced cePDE4 expression in the presence of these protein kinases (Fig. 8D). Together, these data demonstrate that regulation of PDE4 protein degradation by Cul, calcineurin, CK1, and GSK3β is evolutionarily conserved.
DISCUSSION
Calcineurin in cell signaling regulation.
In this report, we uncover a novel calcineurin signaling system (Fig. 9). We find that calcineurin regulates phosphodependent degradation of PDE4D, a widely expressed cAMP-degrading PDE that is well known to underpin key functions of cAMP signaling (18, 33). We demonstrate that calcineurin binds directly to PDE4D in order to oppose the novel action of the CK1 and GSK3β protein kinases in phosphorylating a phosphodegron motif in PDE4D. Such phosphorylation by CK1 and GSK3β allows the SCF E3 ubiquitin ligase to promote ubiquitin conjugation, which then targets PDE4D for proteosomal degradation. Intriguingly, our study of C. elegans indicates that calcineurin regulation of PDE4D degradation by a phosphodegron is evolutionarily conserved, supporting the importance of this novel calcineurin-PDE4D signaling system in GPCR/cAMP signal transduction. Thus, in addition to subcellular compartmentalization and phosphorylation-dependent activation (18, 33), controlling the stability of PDE4D by targeted ubiquitination involving CK1 and GSK3β phosphorylation and subsequent recruitment of the SCF E3 ubiquitin ligase complex provides a novel, evolutionarily conserved control point at which to regulate cAMP signaling in cells.
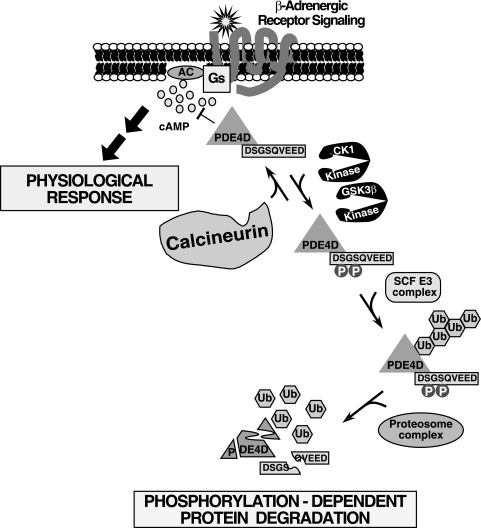
FIG. 9.
Model of the phosphatase calcineurin in the β-adrenergic signaling pathway via regulation of PDE4D degradation. PDE4D encompasses a DSGSQVEED phosphodegron. Phosphorylation at the phosphodegron by the protein kinases GSK3β and CK1 promotes PDE4D degradation, which is mediated by the SCF E3 ubiquitin (Ub) ligase complex. Ubiquitinated PDE4D was degraded by the proteosome complex. Conversely, dephosphorylation mediated by the phosphatase calcineurin opposes PDE4D degradation. Thus, phosphorylation-dependent degradation controls the expression level of PDE4D, which in turn regulates the intracellular concentration of the second messenger cAMP upon activation of the β-adrenergic receptor signaling pathway—a key component of the control of diverse physiological responses. The calcineurin/ubiquitin/PDE4D signaling axis is also evolutionarily conserved in C. elegans, indicating its important role in critical cellular processes to regulate cAMP duration. We propose that potentiation of the Gs-coupled signaling pathway due to sustained elevation of cAMP, which is a consequence of reduced PDE4D expression in the absence of calcineurin function, accounts for the metabolic complications (e.g., hyperlipidemia and hemodynamic dysregulations) found in transplant patients treated with CsA. AC, adenylyl cyclase.
Our studies also expand the repertoire of calcineurin function in regulating protein stability via phosphodegrons. Identification of substrates targeted for phosphodegron-mediated degradation has been proven difficult because of complex phosphorylation events for substrate recognition. These substrates are often phosphorylated at multiple sites and low in abundance, as they are rapidly degraded once phosphorylated. Therefore, identification of these substrates becomes a “catch-22” event, as phosphorylation induces binding to the SCF adaptors, which facilitate ubiquitin conjugation and promote degradation. Conversely, interaction with and subsequent dephosphorylation by calcineurin will increase the protein stability of some phosphodegron-mediated substrates. Future systematic investigations to identify such substrates will provide new insights for the pleiotropic effect of calcineurin.
The opposing actions of the phosphatase calcineurin and the CK1/GSK3β protein kinases on phosphodegron-dependent degradation of PDE4D are analogous to their roles in regulating the nucleocytoplasmic shuttling of NFAT transcription factors. Coordinated regulation by the phosphatase calcineurin (and CK1/GSK3β protein kinases) on NFAT-mediated transcription and on PDE4D protein stability would elicit intricate biological responses. For example, cAMP-dependent protein kinase (PKA) opposes calcineurin-mediated nuclear NFAT accumulation (7, 14). Stabilization of PDE4D, and hence dampening of PKA activation via increasing cAMP hydrolysis, would facilitate nuclear accumulation of NFAT upon activation of calcineurin. Induction of NFAT target genes (e.g., T-cell mitogens) and downregulation of cAMP concentration/localization in specific cellular compartments by calcineurin may also contribute to overall T-cell function and proliferation (1, 2, 56). A similar role for calcineurin in NFAT and PDE4D regulation may also be found in endocrine secreting cells, such as adipocytes and pancreatic β cells, where NFAT mediates the expression of secretory factors (e.g., adiponectin, resistin, and insulin) (28, 61), while PDE4D regulates exocytic release via compartmentalized cAMP signaling (58).
Calcineurin in posttransplantation complications.
Our findings on calcineurin regulation of the stability of PDE4D have clinical significance in providing a new perspective on the molecular pathology of posttransplantation complications elicited by the widely used calcineurin inhibitor and immunosuppressant CsA. Suppression of immune function by CsA during and after transplant surgery minimizes the incidence of host-versus-graft reactions in transplant patients and increases the success of organ transplantation. Many of these transplant patients, however, develop posttransplantation complications even without a history of such pathologies. Posttransplantation complications include a broad range of pathologies, including insulin resistance, hyperlipidemia, hypertension, and nephro- and neurotoxicity (3, 26, 38, 48). Their etiology, however, remains elusive.
Current models suggest that CsA disrupts the mitochondrial permeability transition pore function and causes cellular dysfunction in β cells (22). CsA also inhibits calcineurin (54), which dephosphorylates the transcription factor NFAT (17, 35). Calcineurin-NFAT signaling is critical for pancreatic β-cell growth and adipokine gene transcription (28, 61). Upon calcineurin inhibition by CsA, dysregulated pancreatic function and loss of insulin sensitivity can account for the metabolic complications found in transplant patients.
The effect of sustained elevation of cAMP upon the loss of calcineurin function in stabilizing PDE4D expression can also contribute to the adverse pathological complications found in CsA-treated transplant patients. The diverse functions of GPCR signal transduction and an array of effectors regulated by the second messenger cAMP, e.g., cAMP-dependent protein kinase (PKA) (39, 47), cAMP-gated ion channels (8), and cAMP-regulated guanine nucleotide exchange factors (Epac) (31), provide a wide base to account for the pathologies found in CsA-treated transplant patients. Notably, GPCR antagonists and calcium channel blockers, seemingly unrelated to calcineurin function, have shown certain success in attenuating pathological complications elicited by CsA (42, 51, 52, 64). We propose that metabolic complications found in transplant patients treated with CsA may be a consequence of not only pancreatic dysregulation via the insulin pathway but also of potentiation of the GPCR signaling pathway due to sustained elevation of cAMP.
Conclusions.
In conclusion, we have uncovered a novel, evolutionarily conserved calcineurin signaling system that regulates SCF-mediated ubiquitin conjugation and proteosomal degradation of PDE4D upon dual phosphorylation by CK1 and GSK3β. The calcineurin/ubiquitin/PDE4D signaling system impinges on diverse cellular processes by regulating cAMP responses. This new calcineurin signaling system provides new insights and points to novel therapeutic targets for a range of adverse pathological complications elicited by CsA in transplant patients.
Acknowledgments
H.Y.S. is a trainee sponsored by 5T32 GM07491. This research was supported, in part, by grants from the National Institutes of Health (C.-W.C., J.J., L.A.J., D.J.L., M.P.L., J.D.M., and P.E.S.), the American Diabetes Association (C.-W.C., D.J.L., and P.E.S.), and the Howard Hughes Medical Institute (J.D.M.).
Footnotes
Published ahead of print on 20 July 2010.
REFERENCES
- 1.Aandahl, E. M., W. J. Moretto, P. A. Haslett, T. Vang, T. Bryn, K. Tasken, and D. F. Nixon. 2002. Inhibition of antigen-specific T cell proliferation and cytokine production by protein kinase A type I. J. Immunol. 169:802-808. [DOI] [PubMed] [Google Scholar]
- 2.Abrahamsen, H., G. Baillie, J. Ngai, T. Vang, K. Nika, A. Ruppelt, T. Mustelin, M. Zaccolo, M. Houslay, and K. Tasken. 2004. TCR- and CD28-mediated recruitment of phosphodiesterase 4 to lipid rafts potentiates TCR signaling. J. Immunol. 173:4847-4858. [DOI] [PubMed] [Google Scholar]
- 3.Andoh, T. F., and W. M. Bennett. 1998. Chronic cyclosporine nephrotoxicity. Curr. Opin. Nephrol. Hypertens. 7:265-270. [DOI] [PubMed] [Google Scholar]
- 4.Aramburu, J., F. Garcia-Cozar, A. Raghavan, H. Okamura, A. Rao, and P. G. Hogan. 1998. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell 1:627-637. [DOI] [PubMed] [Google Scholar]
- 5.Aramburu, J., A. Rao, and C. B. Klee. 2000. Calcineurin: from structure to function. Curr. Top. Cell Regul. 36:237-295. [DOI] [PubMed] [Google Scholar]
- 6.Aramburu, J., M. B. Yaffe, C. Lopez-Rodriguez, L. C. Cantley, P. G. Hogan, and A. Rao. 1999. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285:2129-2133. [DOI] [PubMed] [Google Scholar]
- 7.Beals, C. R., C. M. Sheridan, C. W. Turck, P. Gardner, and G. R. Crabtree. 1997. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science 275:1930-1934. [DOI] [PubMed] [Google Scholar]
- 8.Biel, M., and S. Michalakis. 2009. Cyclic nucleotide-gated channels. Handb. Exp. Pharmacol. 191:111-136. [DOI] [PubMed] [Google Scholar]
- 9.Bolger, G. B., S. Erdogan, R. E. Jones, K. Loughney, G. Scotland, R. Hoffmann, I. Wilkinson, C. Farrell, and M. D. Houslay. 1997. Characterization of five different proteins produced by alternatively spliced mRNAs from the human cAMP-specific phosphodiesterase PDE4D gene. Biochem. J. 328(Pt. 2):539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruss, M. D., W. Richter, K. Horner, S.-L. C. Jin, and M. Conti. 2008. Critical role of PDE4D in {beta}2-adrenoceptor-dependent cAMP signaling in mouse embryonic fibroblasts. J. Biol. Chem. 283:22430-22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bueno, O. F., B. J. Wilkins, K. M. Tymitz, B. J. Glascock, T. F. Kimball, J. N. Lorenz, and J. D. Molkentin. 2002. Impaired cardiac hypertrophic response in calcineurin Abeta-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 99:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlie, N. K., A. M. Thomure, M. A. Schade, and K. G. Miller. 2006. The Dunce cAMP phosphodiesterase PDE-4 negatively regulates G{alpha}s-dependent and G{alpha}s-independent cAMP pools in the Caenorhabditis elegans synaptic signaling network. Genetics 173:111-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow, C. W., and R. J. Davis. 2000. Integration of calcium and cyclic AMP signaling pathways by 14-3-3. Mol. Cell. Biol. 20:702-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow, C. W., M. Rincon, and R. J. Davis. 1999. Requirement for transcription factor NFAT in interleukin-2 expression. Mol. Cell. Biol. 19:2300-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christians, U., and K. F. Sewing. 1993. Cyclosporin metabolism in transplant patients. Pharmacol. Ther. 57:291-345. [DOI] [PubMed] [Google Scholar]
- 17.Clipstone, N. A., and G. R. Crabtree. 1992. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 357:695-697. [DOI] [PubMed] [Google Scholar]
- 18.Conti, M., and J. Beavo. 2007. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 76:481-511. [DOI] [PubMed] [Google Scholar]
- 19.Crabtree, G. R. 2001. Calcium, calcineurin, and the control of transcription. J. Biol. Chem. 276:2313-2316. [DOI] [PubMed] [Google Scholar]
- 20.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.):S67-S79. [DOI] [PubMed] [Google Scholar]
- 21.Cyert, M. S. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143-1150. [DOI] [PubMed] [Google Scholar]
- 22.Düfer, M., P. Krippeit-Drews, N. Lembert, L. A. Idahl, and G. Drews. 2001. Diabetogenic effect of cyclosporin A is mediated by interference with mitochondrial function of pancreatic B-cells. Mol. Pharmacol. 60:873-879. [PubMed] [Google Scholar]
- 23.Durand, D. B., J. P. Shaw, M. R. Bush, R. E. Replogle, R. Belagaje, and G. R. Crabtree. 1988. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol. Cell. Biol. 8:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fruman, D. A., C. B. Klee, B. E. Bierer, and S. J. Burakoff. 1992. Calcineurin phosphatase activity in T lymphocytes is inhibited by FK 506 and cyclosporin A. Proc. Natl. Acad. Sci. U. S. A. 89:3686-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerini, D. 1997. Calcineurin: not just a simple protein phosphatase. Biochem. Biophys. Res. Commun. 235:271-275. [DOI] [PubMed] [Google Scholar]
- 26.Hauben, M. 1996. Cyclosporine neurotoxicity. Pharmacotherapy 16:576-583. [PubMed] [Google Scholar]
- 27.Heit, J. J. 2007. Calcineurin/NFAT signaling in the beta-cell: from diabetes to new therapeutics. Bioessays 29:1011-1021. [DOI] [PubMed] [Google Scholar]
- 28.Heit, J. J., A. A. Apelqvist, X. Gu, M. M. Winslow, J. R. Neilson, G. R. Crabtree, and S. K. Kim. 2006. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature 443:345-349. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann, R., G. S. Baillie, S. J. MacKenzie, S. J. Yarwood, and M. D. Houslay. 1999. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J. 18:893-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan, P. G., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205-2232. [DOI] [PubMed] [Google Scholar]
- 31.Holz, G. G., G. Kang, M. Harbeck, M. W. Roe, and O. G. Chepurny. 2006. Cell physiology of cAMP sensor Epac. J. Physiol. 577:5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houslay, M. D., and D. R. Adams. 2003. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 370:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houslay, M. D., G. S. Baillie, and D. H. Maurice. 2007. cAMP-specific phosphodiesterase-4 enzymes in the cardiovascular system: a molecular toolbox for generating compartmentalized cAMP signaling. Circ. Res. 100:950-966. [DOI] [PubMed] [Google Scholar]
- 34.Hunter, T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28:730-738. [DOI] [PubMed] [Google Scholar]
- 35.Jain, J., P. G. McCaffrey, Z. Miner, T. K. Kerppola, J. N. Lambert, G. L. Verdine, T. Curran, and A. Rao. 1993. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature 365:352-355. [DOI] [PubMed] [Google Scholar]
- 36.Jin, J., X. L. Ang, T. Shirogane, and J. Wade Harper. 2005. Identification of substrates for F-box proteins. Methods Enzymol. 399:287-309. [DOI] [PubMed] [Google Scholar]
- 37.Jin, J., T. Shirogane, L. Xu, G. Nalepa, J. Qin, S. J. Elledge, and J. W. Harper. 2003. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17:3062-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jindal, R. M., R. A. Sidner, and M. L. Milgrom. 1997. Post-transplant diabetes mellitus. The role of immunosuppression. Drug Saf. 16:242-257. [DOI] [PubMed] [Google Scholar]
- 39.Kim, C., D. Vigil, G. Anand, and S. S. Taylor. 2006. Structure and dynamics of PKA signaling proteins. Eur. J. Cell Biol. 85:651-654. [DOI] [PubMed] [Google Scholar]
- 40.Klee, C. B., T. H. Crouch, and M. H. Krinks. 1979. Calcineurin: a calcium- and calmodulin-binding protein of the nervous system. Proc. Natl. Acad. Sci. U. S. A. 76:6270-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhara, A., H. Inada, I. Katsura, and I. Mori. 2002. Negative regulation and gain control of sensory neurons by the C. elegans calcineurin TAX-6. Neuron 33:751-763. [DOI] [PubMed] [Google Scholar]
- 42.Lassila, M., P. Finckenberg, A.-K. Pere, L. Krogerus, J. Ahonen, H. Vapaatalo, and M.-L. Nurminen. 2000. Comparison of enalapril and valsartan in cyclosporine A-induced hypertension and nephrotoxicity in spontaneously hypertensive rats on high-sodium diet. Br. J. Pharmacol. 130:1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807-815. [DOI] [PubMed] [Google Scholar]
- 44.MacKenzie, S. J., G. S. Baillie, I. McPhee, G. B. Bolger, and M. D. Houslay. 2000. ERK2 mitogen-activated protein kinase binding, phosphorylation, and regulation of the PDE4D cAMP-specific phosphodiesterases. The involvement of COOH-terminal docking sites and NH2-terminal UCR regions. J. Biol. Chem. 275:16609-16617. [DOI] [PubMed] [Google Scholar]
- 45.Marchetti, P., and R. Navalesi. 2000. The metabolic effects of cyclosporin and tacrolimus. J. Endocrinol. Invest. 23:482-490. [DOI] [PubMed] [Google Scholar]
- 46.Matsuda, S., and S. Koyasu. 2000. Mechanisms of action of cyclosporine. Immunopharmacology 47:119-125. [DOI] [PubMed] [Google Scholar]
- 47.McConnachie, G., L. K. Langeberg, and J. D. Scott. 2006. AKAP signaling complexes: getting to the heart of the matter. Trends Mol. Med. 12:317-323. [DOI] [PubMed] [Google Scholar]
- 48.Mihatsch, M. J., M. Kyo, K. Morozumi, Y. Yamaguchi, V. Nickeleit, and B. Ryffel. 1998. The side-effects of cyclosporine-A and tacrolimus. Clin. Nephrol. 49:356-363. [PubMed] [Google Scholar]
- 49.Nalepa, G., M. Rolfe, and J. W. Harper. 2006. Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 5:596-613. [DOI] [PubMed] [Google Scholar]
- 50.O'Connell, B. C., and J. W. Harper. 2007. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr. Opin. Cell Biol. 19:206-214. [DOI] [PubMed] [Google Scholar]
- 51.Padi, S. S., and K. Chopra. 2002. Salvage of cyclosporine A-induced oxidative stress and renal dysfunction by carvedilol. Nephron 92:685-692. [DOI] [PubMed] [Google Scholar]
- 52.Padi, S. S., and K. Chopra. 2002. Selective angiotensin II type 1 receptor blockade ameliorates cyclosporine nephrotoxicity. Pharmacol. Res. 45:413-420. [DOI] [PubMed] [Google Scholar]
- 53.Rusnak, F., and P. Mertz. 2000. Calcineurin: form and function. Physiol. Rev. 80:1483-1521. [DOI] [PubMed] [Google Scholar]
- 54.Schreiber, S. L., and G. R. Crabtree. 1992. The mechanism of action of cyclosporin A and FK506. Immunol. Today 13:136-142. [DOI] [PubMed] [Google Scholar]
- 55.Sette, C., S. Iona, and M. Conti. 1994. The short-term activation of a rolipram-sensitive, cAMP-specific phosphodiesterase by thyroid-stimulating hormone in thyroid FRTL-5 cells is mediated by a cAMP-dependent phosphorylation. J. Biol. Chem. 269:9245-9252. [PubMed] [Google Scholar]
- 56.Skålhegg, B. S., B. F. Landmark, S. O. Doskeland, V. Hansson, T. Lea, and T. Jahnsen. 1992. Cyclic AMP-dependent protein kinase type I mediates the inhibitory effects of 3′,5′-cyclic adenosine monophosphate on cell replication in human T lymphocytes. J. Biol. Chem. 267:15707-15714. [PubMed] [Google Scholar]
- 57.Stewart, A. A., T. S. Ingebritsen, A. Manalan, C. B. Klee, and P. Cohen. 1982. Discovery of a Ca2+- and calmodulin-dependent protein phosphatase: probable identity with calcineurin (CaM-BP80). FEBS Lett. 137:80-84. [DOI] [PubMed] [Google Scholar]
- 58.Szaszák, M., F. Christian, W. Rosenthal, and E. Klussmann. 2008. Compartmentalized cAMP signalling in regulated exocytic processes in non-neuronal cells. Cell. Signal. 20:590-601. [DOI] [PubMed] [Google Scholar]
- 59.Thornton, T. M., G. Pedraza-Alva, B. Deng, C. D. Wood, A. Aronshtam, J. L. Clements, G. Sabio, R. J. Davis, D. E. Matthews, B. Doble, and M. Rincon. 2008. Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science 320:667-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei, W., J. Jin, S. Schlisio, J. W. Harper, and W. G. Kaelin, Jr. 2005. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8:25-33. [DOI] [PubMed] [Google Scholar]
- 61.Yang, T. T., H. Y. Suk, X. Yang, O. Olabisi, R. Y. Yu, J. Durand, L. A. Jelicks, J. Y. Kim, P. E. Scherer, Y. Wang, Y. Feng, L. Rossetti, I. A. Graef, G. R. Crabtree, and C. W. Chow. 2006. Role of transcription factor NFAT in glucose and insulin homeostasis. Mol. Cell. Biol. 26:7372-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, T. T., Q. Xiong, I. A. Graef, G. R. Crabtree, and C. W. Chow. 2005. Recruitment of the extracellular signal-regulated kinase/ribosomal S6 kinase signaling pathway to the NFATc4 transcription activation complex. Mol. Cell. Biol. 25:907-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yarwood, S. J., M. R. Steele, G. Scotland, M. D. Houslay, and G. B. Bolger. 1999. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J. Biol. Chem. 274:14909-14917. [DOI] [PubMed] [Google Scholar]
- 64.Yavuz, D. G., S. Tuglular, H. Koçak, A. Atakan, C. Ozener, E. Akoglu, and S. AkalIn. 2004. Angiotension converting enzyme inhibition and calcium channel blockage improves cyclosporine induced glucose intolerance in rats. Transplant. Proc. 36:171-174. [DOI] [PubMed] [Google Scholar]
- 65.Yen, H. C., and S. J. Elledge. 2008. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322:923-929. [DOI] [PubMed] [Google Scholar]
- 66.Yen, H. C., Q. Xu, D. M. Chou, Z. Zhao, and S. J. Elledge. 2008. Global protein stability profiling in mammalian cells. Science 322:918-923. [DOI] [PubMed] [Google Scholar]
- 67.Zhu, J., F. Shibasaki, R. Price, J. C. Guillemot, T. Yano, V. Dotsch, G. Wagner, P. Ferrara, and F. McKeon. 1998. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 93:851-861. [DOI] [PubMed] [Google Scholar]