Abstract
The small ubiquitin-related modifiers (SUMOs) are evolutionarily conserved polypeptides that are covalently conjugated to protein targets to modulate their subcellular localization, half-life, or activity. Steady-state SUMO conjugation levels increase in response to many different types of environmental stresses, but how the SUMO system is regulated in response to these insults is not well understood. Here, we characterize a novel mode of SUMO system control: in response to elevated alcohol levels, the Saccharomyces cerevisiae SUMO protease Ulp1 is disengaged from its usual location at the nuclear pore complex (NPC) and sequestered in the nucleolus. We further show that the Ulp1 region previously demonstrated to interact with the karyopherins Kap95 and Kap60 (amino acids 150 to 340) is necessary and sufficient for nucleolar targeting and that enforced sequestration of Ulp1 in the nucleolus significantly increases steady-state SUMO conjugate levels, even in the absence of alcohol. We have thus characterized a novel mechanism of SUMO system control in which the balance between SUMO-conjugating and -deconjugating activities at the NPC is altered in response to stress via relocalization of a SUMO-deconjugating enzyme.
The small ubiquitin-related modifiers (SUMOs) are a family of evolutionarily conserved polypeptides that are conjugated to protein targets via the concerted action of SUMO-specific E1 (activation), E2 (conjugation), and E3 (ligase) enzymes to effect changes in subcellular localization, half-life, or target activity. A family of SUMO-specific proteases act to remove the modifier from conjugates (8, 20). The SUMO system has been implicated in a variety of critical cellular functions, such as DNA repair and replication, RNA metabolism, and stress responses (8, 16, 20). Importantly, the SUMO system is highly dynamic and the SUMO pathway enzymes appear to work together to precisely control SUMO conjugate levels in the cell (8, 16, 20). However, how the SUMO system itself is regulated is poorly understood.
Localization of the SUMO pathway enzymes may play an important role in SUMO system function (21). For example, the budding yeast SUMO protease Ulp1 is tethered to the nuclear face of the nuclear pore complex (NPC) via an unconventional interaction with the karyopherin Kap121 and the heterodimeric Kap95/Kap60 complex (12, 13, 23). However, this SUMO protease is not maintained exclusively at the NPC but appears to be mobile, effecting desumoylation at diverse subcellular locations: e.g., during mitosis, Saccharomyces cerevisiae Ulp1 is recruited to the septin ring to desumoylate septins (15), Schizosaccharomyces pombe Ulp1 localization is regulated throughout the cell cycle (31), and a mammalian Ulp1 homolog, SENP2, is shuttled between the nucleus and the cytoplasm (7). Consistent with these observations, SUMO conjugate levels are significantly altered in yeast strains expressing mislocalized Ulp1 (13, 37).
Dramatic changes in SUMO conjugate populations have been noted in response to many different types of stresses in yeasts, mammals, and plants (9, 17, 27, 32, 38). For example, in S. cerevisiae, significantly increased steady-state SUMO conjugate levels are observed in response to elevated concentrations of ethanol (38). To better understand how the SUMO system is regulated in response to stress, we utilized alcohol as a model of a physiologically relevant stressor in yeast. Here, we demonstrate that alcohol stress results in a rapid, reversible nucleolar sequestration of Ulp1 and that enforced localization of Ulp1 in the nucleolus leads to a dramatic increase in steady-state SUMO conjugate levels. This is the first demonstration of regulated modulation of the intracellular localization of a SUMO enzyme in response to stress and thus represents a novel mechanism for SUMO system control.
MATERIALS AND METHODS
Yeast strains and microbiological techniques.
The S. cerevisiae strains used in this study were derivatives of BY4741 or DF5 haploid cells, unless otherwise specified, and are listed in Table 1. All yeast genetic manipulations were performed according to established procedures. Unless otherwise noted, yeast strains were grown at 30°C to mid-logarithmic phase in YPD or selective minimal (SM) medium supplemented with appropriate nutrients and 2% glucose. Temperature-sensitive mutants were grown at a permissive temperature of 24°C to mid-logarithmic phase in SM medium supplemented with appropriate nutrients and 2% glucose. Yeast cells were pelleted and transferred to prewarmed medium at 37°C for 3 h prior to treatment with alcohols. Transformations were performed as described previously (2).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Parent strain | Source or reference |
|---|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 3 | |
| Ulp1-TAP | MATaulp1::ULP1-TAP (His3MX6) | BY4741 | 15 |
| Aos1-TAP | MATaAos1-TAP (His3MX6) | BY4741 | 5 |
| Uba2-TAP | MATaUba2-TAP (His3MX6) | BY4741 | 5 |
| Ubc9-TAP | MATaUbc9-TAP (His3MX6) | BY4741 | 5 |
| Ulp1-HA | MATa pULP1-HA | BY4741 | This study |
| Ulp2-HA | MATa pULP2-HA | BY4741 | This study |
| Aos1-GFP | MATaAos1-GFP (His3MX6) | BY4741 | 6 |
| Uba2-GFP | MATaUba2-GFP (His3MX6) | BY4741 | 6 |
| Ubc9-GFP | MATaUbc9-GFP (His3MX6) | BY4741 | 6 |
| Ulp2-RFP | MATa pUlp2-RFP | BY4741 | This study |
| cNLS-RFP | MATa pcNLS-RFP | BY4741 | This study |
| Nup59-GFP | MATaNup59-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Nup100-GFP | MATaNup100-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Nop2-GFP | MATaNop2-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Noc3-GFP | MATaNoc3-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Nop56-GFP | MATaNop56-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Noc2-GFP | MATaNoc2-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Nop4-GFP | MATaNop4-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Nop6-GFP | MATaNop6-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Nop58-GFP | MATaNop58-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Nab2-GFP | MATaNab2-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Tom1-GFP | MATaTom1-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Hrb1-GFP | MATaHrb1-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Dbp5-GFP | MATaDbp5-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Mtr10-GFP | MATaMtr10-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Hrp1-GFP | MATaHrp1-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Gbp2-GFP | MATaGbp2-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Sub2-GFP | MATaSub2-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Yra1-GFP | MATaYra1-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Thp1-GFP | MATaThp1-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Sac3-GFP | MATaSac3-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Mex67-GFP | MATaMex67-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Mtr2-GFP | MATaMtr2-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Npl3-GFP | MATaNpl3-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Kap95-GFP | MATaKap95-GFP (His3MX6) pNop56-Dsred | BY47414 | This study |
| Kap60-GFP | MATaKap60-GFP (His3MX6) pNop56-Dsred | BY47414 | This study |
| Kap121-GFP | MATaKap121-GFP (His3MX6) pNop6-Dsred | BY47414 | This study |
| DF5 | MATaura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 | 4 | |
| Ulp1-GFP | MATaulp1Δ::KanMX4 CDC3-HA::HIS5 pULP1-GFP | DF5 | 15 |
| pULP1150-621-GFP | MATaulp1Δ::KanMX4 CDC3-HA::HIS5 pULP1150-621-GFP | DF5 | 15 |
| pULP1Δ150-340-GFP | MATaulp1Δ::KanMX4 CDC3-HA::HIS5 pULP1Δ150-340-GFP | DF5 | 15 |
| pULP11-150-GFP | MATa pULP11-150-GFP | BY4741 | This study |
| pULP1150-340-GFP | MATa pULP1150-340-GFP | BY4741 | This study |
| Nup60-GFP | MATaNup60-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Mlp1-GFP | MATaMlp1-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Mlp2-GFP | MATaMlp2-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Esc1-GFP | MATaEsc1-GFP (His3MX6) pULP1-RFP | BY47414 | This study |
| Nup2Δ pULP1-GFP | MATanup2Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Nup42Δ pULP1-GFP | MATanup42Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Nup53Δ pULP1-GFP | MATanup53Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Nup59Δ pULP1-GFP | MATanup59Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Nup60Δ pULP1-GFP | MATanup60Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Nup100Δ pULP1-GFP | MATanup100Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Nup133Δ pULP1-GFP | MATanup133Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Nup170Δ pULP1-GFP | MATanup170Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Nup188Δ pULP1-GFP | MATanup188Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Esc1Δ pULP1-GFP | MATaesc1Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Asr1Δ pULP1-GFP | MATaasr1Δ::KanMX4 pULP1-GFP | BY47416 | This study |
| Mlp1/2Δ pULP1-GFP | MATamlp1Δ::KanMX4 mlp2::natMX4 pULP1-GFP | BY47416 | This study |
| Nup59Δ pULP1150-621-GFP | MATanup59Δ::KanMX4 pULP1150-621-GFP | BY47416 | This study |
| Nup100Δ pULP1150-621-GFP | MATanup100Δ::KanMX4 pULP1150-621-GFP | BY47416 | This study |
| Nup2Δ pULP1150-621-GFP | MATanup2Δ::KanMX4 pULP1150-621-GFP | BY47416 | This study |
| Nup60Δ pULP1150-621-GFP | MATanup60Δ::KanMX4 pULP1150-621-GFP | BY47416 | This study |
| Esc1Δ pULP1150-621-GFP | MATaesc1Δ::KanMX4 pULP1150-621-GFP | BY47416 | This study |
| Mlp1/2Δ pULP1150-621-GFP | MATamlp1Δ::KanMX4 mlp2Δ::natMX4 pULP1150-621-GFP | BY47417 | This study |
| ulp2Δ | MATaulp2Δ::KanMX4 | BY47416 | 34 |
| ubc9-2 | MATα ubc9-2::NATR can1Δ::STE2pr-Sp_his5 lyp1Δ | TSQUERY370 | 16 |
| kap121-34 | MATakap121::ura3::HIS3 pkap121-34 | DF5 | 10 |
| kap95-14 | MATakap95::HIS5 pkap95-14 | DF5 | 1 |
| kap95-14 | MATα kap95::HIS5 pkap95-14 | DF5 | 1 |
| kap95-14 ULP1150-621-GFP | MATα kap95::HIS5 pkap95-14 ulp1Δ::KanMX4 CDC3-HA::HIS5 pULP1150-621-GFP | DF5 | This study |
| kap95-14 ULP1150-621-GFP | MATakap95::HIS5 pkap95-14 ulp1Δ::KanMX4 CDC3-HA::HIS5 pULP1150-621-GFP | DF5 | This study |
| kap95-14 cNLS-RFP | MATa pcNLS-RFP | DF510 | This study |
| kap121-34 pULP1150-340-GFP | MATa pULP1-GFP pNOP2-RFP | DF59 | This study |
| kap95-14 pULP1150-340-GFP | MATa pULP1-GFP pNOP2-RFP | DF510 | This study |
| ulp1-333SGG | MATα ulp1-333::NATR smt3ΔC::LEU2 can1Δ::STE2pr-Sp_his5 lyp1Δ | TSQUERY961 | 16 |
WCL preparation, affinity purification, SDS-PAGE, and Western blotting.
Whole-cell lysates (WCL) were prepared by alkaline lysis and trichloroacetic acid protein precipitation of cell pellets derived from 10-ml cultures. Protein pellets were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, sonicated for 10 s, and incubated at 90°C for 5 min. Affinity purification of tandem affinity purification (TAP)- or hemagglutinin (HA)-tagged proteins was performed essentially as described previously (26), except that cells were lysed by bead beating with 0.5-mm glass beads (BioSpec Products, Inc.) in 10% glycerol-50 mM HEPES (pH 8.0)-100 mM KCl-2 mM EDTA-0.1% Nonidet P-40. Lysates were cleared by centrifugation at 16,000 × g and incubated overnight with Dynabeads Pan Mouse IgG (Invitrogen). Beads were washed three times with phosphate-buffered saline. Proteins were eluted in Laemmli buffer and resolved on 10% or 4 to 12% SDS-polyacrylamide gels and transferred to 0.45-μm-pore-size nitrocellulose membrane (Pall Corp.) using standard methods. Specific rabbit polyclonal antibodies were used to detect yeast SUMO (Smt3p) and Swi6p (both from Rockland Immunochemicals, Inc.). Rabbit serum was used to detect the Staphylococcus aureus protein A moiety in the TAP and HA tags. A mouse monoclonal antibody was used for detection of beta-actin (EMD Chemicals, Inc.). Binding of primary antibodies was detected using horseradish peroxidase (HRP)-conjugated donkey anti-rabbit secondary antibodies (Bio-Rad Laboratories) or HRP-conjugated goat anti-mouse IgM (EMD Chemicals, Inc.) and visualized using ECL (Bio-Rad Laboratories) or ECL-plus (Millipore Corporation).
The alcohols used were methanol (99.8%; EMD Chemicals, Inc.), ethanol (95%; Commercial Alcohols, Brampton, Ontario, Canada), isopropanol (99.5%; BioShop Canada), butanol (99.8%; BioShop Canada), pentanol, and hexanol (98%; Fluka Chemie).
Live-cell confocal microscopy.
Mid-log-phase cells were collected from 1-ml cultures, washed briefly in H2O containing 2% glucose, and mounted on a glass slide. Cells were imaged with a 60×, 1.42 numerical aperture PlanApo objective on an Olympus IX80 inverted microscope (Olympus Canada) fitted with a Yokogawa CSU10 spinning-disk confocal scanner unit (Quorum Technologies Inc., Guelph, Ontario, Canada) and a 512-by-512-pixel electron multiplier charge-coupled device (CCD) camera (Hamamatsu, Japan). The system was controlled with Volocity 4.3.2 software (Improvision Ltd.). The CCD camera was operated at maximum resolution; the gain was set to 1, binning to ×1, and sensitivity to 100 (except for pUlp1-RFP, for which it was set to 130). Exposure times varied by strain, ranging from 0.2 to 10 s. Settings were maintained for all subsequent images of the same strain. Further processing of images was performed using Volocity and Adobe Photoshop CS2 (Adobe Systems Inc.).
RESULTS
Soluble Ulp1 levels are inversely correlated with steady-state sumoylation levels.
To better understand how alcohol stress modulates steady-state sumoylation levels, we characterized the effects of ethanol exposure on the proteins responsible for SUMO conjugation and deconjugation. Yeast strains expressing epitope-tagged SUMO pathway proteins were exposed to 10% ethanol for 60 min, and WCL (prepared by standard alkaline lysis and trichloroacetic acid protein precipitation) were analyzed via Western blotting. In WCL, no apparent changes in expression level or electrophoretic mobility were observed for any of the SUMO pathway proteins (Fig. 1 A). Interestingly, however, isolation of the same proteins from cell lysates via immunoprecipitation (affinity purification [AP] via IgG-Sepharose; see Materials and Methods) revealed a striking, rapid decrease in immunoprecipitable Ulp1 levels in response to alcohol stress (Fig. 1A). A smaller decrease was observed for Ulp2 (Fig. 1A). This effect did not appear to be related to a particular epitope tag, as the Ulp1-HA and Ulp1-TAP proteins behaved identically (compare Fig. 1A and B).
FIG. 1.
Ulp1 solubility is inversely correlated with steady-state SUMO conjugate levels. (A) Yeast strains expressing epitope-tagged versions of SUMO system proteins were grown in liquid culture and treated with 10% ethanol, and aliquots were removed at the indicated times. WCL were prepared, or AP was conducted for each protein, as described in Materials and Methods. Western blotting was conducted to monitor protein levels. Swi6 was used as a loading control. (B) Ulp1-TAP was affinity purified from cells treated with various concentrations of ethanol, as indicated. Purified material was subjected to SDS-PAGE and Western analysis. (C) The Ulp1-TAP strain was treated with 10% methanol, ethanol, isopropanol, or butanol, and apparent Ulp1 solubility was monitored by AP and Western analysis as described above. (D) A Ulp1-TAP culture was treated with 10% ethanol for 60 min, pelleted, washed, resuspended, and maintained in fresh culture medium for an additional 180 min. Aliquots were removed at the time points indicated. WCL and affinity-purified Ulp1-TAP were subjected to SDS-PAGE and Western analysis as indicated. Swi6 was used as a loading control. (E) A wild-type S. cerevisiae strain (BY4741) was exposed to 10% ethanol, aliquots were taken at the time points indicated, and WCL were subjected to SDS-PAGE and Western analysis. Steady-state SUMO conjugate levels (top, short exposure) and unconjugated (Free) SUMO protein levels (bottom, longer exposure) are shown. (F) A ulp2Δ mutant strain was subjected to 10% ethanol treatment as described above. Aliquots were removed at the time points indicated, and WCL were subjected to SDS-PAGE and Western analysis. WCL from an untreated wild-type (WT) strain is included for comparison.
The loss of Ulp1 from immunoprecipitates was ethanol dose dependent (Fig. 1B). While only a minor effect was noted within 60 min for 5% ethanol, 7.5% ethanol elicited Ulp1 disappearance within 60 min and higher concentrations led to a more rapid loss. To determine whether this effect was specific to ethanol, we tested a number of other simple alcohols. Exposure to methanol, isopropanol, butanol (Fig. 1C), pentanol, or hexanol (data not shown) also led to a loss of Ulp1 from immunoprecipitates. A loss of the apparent solubility of Ulp1 is therefore not limited to a specific alcohol but appears to be a general response to alcohol exposure.
Interestingly, steady-state SUMO conjugate levels were inversely correlated with soluble Ulp1 levels (Fig. 1D). The loss of Ulp1 from immunoprecipitates was also reversible: removal of alcohol from the culture medium was followed by a gradual increase in soluble Ulp1, accompanied by a concomitant decrease in SUMO conjugates (Fig. 1D, middle). As observed above, total Ulp1 levels measured in WCL were unchanged in response to alcohol exposure (Fig. 1D, bottom; Swi6 was used as a loading control). The increase in SUMO conjugates in alcohol-treated cells did not appear to be due to an effect on SUMO protein levels, as unconjugated (free) SUMO protein decreased in concert with increasing SUMO conjugates (Fig. 1E). Thus, while total Ulp1 protein levels are not affected by alcohol, the availability of Ulp1 for immunoprecipitation is dramatically decreased by alcohol stress and inversely correlated with steady-state SUMO conjugate levels.
Ulp2 is not required for the SUMO alcohol response.
While alcohol exposure had a rapid and dramatic effect on the apparent solubility of Ulp1, we also observed a smaller decrease in Ulp2 levels in immunoprecipitates in response to ethanol stress (Fig. 1A). However, ethanol-stimulated SUMO conjugate formation appeared to be unaffected in a ulp2Δ mutant strain (Fig. 1F). Similarly, cells overexpressing Ulp2 from a galactose-inducible promoter (data not shown) displayed no detectable change in ethanol-stimulated SUMO conjugate formation. Ulp2 thus does not appear to play a critical role in the alcohol-mediated increase in SUMO conjugate formation.
Ulp1 is recruited to the nucleolus in response to alcohol stress.
The fact that the apparent solubility of Ulp1 was affected by alcohol exposure (while total Ulp1 protein levels were not) suggested that it could be recruited to an intracellular compartment or structure that is insoluble under our standard lysis conditions. To explore this possibility, we characterized the localization of the SUMO conjugation and deconjugation enzymes in cells subjected to alcohol stress. No changes in intracellular localization were observed for Aos1-GFP, Uba2-GFP, Ubc9-GFP, or Ulp2-RFP in response to ethanol treatment (10% ethanol for 60 min; Fig. 2 A). However, Ulp1 localization was dramatically altered within minutes of alcohol exposure. As previously reported (13, 23), in untreated cells, Ulp1-GFP displays nuclear rim localization. However, following ethanol exposure, Ulp1 was also found in large nuclear foci (Fig. 2A). This effect was not restricted to ethanol; identical results were obtained with isopropanol, butanol, pentanol, and hexanol (data not shown).
FIG. 2.
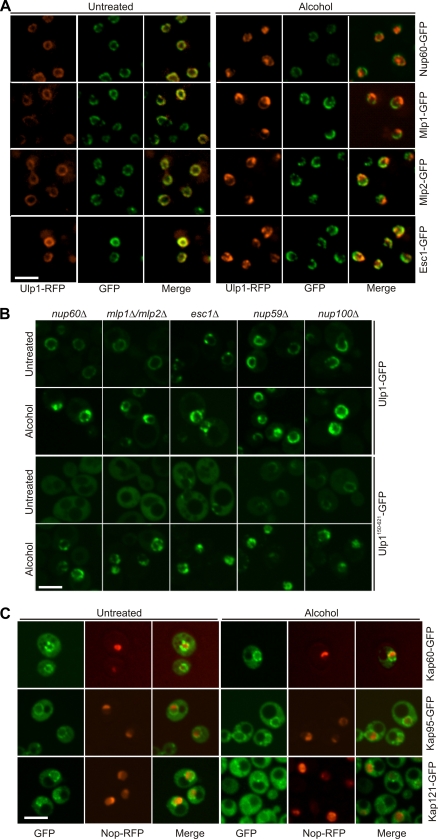
Ulp1 is recruited to the nucleolus in response to alcohol stress. (A) Confocal microscopy of untreated and ethanol-treated (10% for 60 min) yeast strains expressing GFP-tagged Aos1, Uba2, Ubc9, or Ulp1 or RFP-tagged Ulp2. (B) Cells coexpressing Ulp1-RFP and a GFP-tagged nucleoporin (Nup59-GFP or Nup100-GFP) or GFP-tagged nucleolar protein (Nop62-GFP or Noc3-GFP) were imaged before and after treatment with 10% ethanol (60 min). Scale bars, 5 μm.
To better define the location and composition of the Ulp1-containing foci, Ulp1-RFP was coexpressed with a series of green fluorescent protein (GFP)-tagged intracellular markers. As previously reported (13, 23), Ulp1-red fluorescent protein (RFP) colocalized with NPC components in untreated cells (e.g., Nup59-GFP and Nup100-GFP; Fig. 2B). Following alcohol treatment, Ulp1-RFP instead colocalized with Noc3, Nop62 (Fig. 2B), and several other nucleolar markers (data not shown). No colocalization of Ulp1 with these proteins was observed in untreated cells (Fig. 2B). Ulp1 is therefore recruited to the nucleolus in response to alcohol stress.
Many types of environmental stresses have been demonstrated to increase SUMO conjugate formation in yeasts, plants, and mammalian cells in culture (9, 17, 27, 32, 38). We therefore wished to determine if these other types of stress also effect Ulp1 nucleolar recruitment. Heat shock (42°C for 3 h), cold shock (16°C for 3 h), osmotic shock (1 M NaCl or 1 M sorbitol for 1 h), protein folding stress (5 μg/ml tunicamycin or 30 mM 2-mercaptoethanol for 1 h), and hydrogen peroxide (1, 10, or 100 mM for 1 h) did not affect Ulp1 localization (data not shown). Ulp1 nucleolar sequestration thus does not appear to be a general response to all stresses but may be a specific response to alcohol exposure.
A Ulp1 region required for nucleolar recruitment.
To better understand the structural requirements for Ulp1 nucleolar recruitment, we characterized the localization of a set of previously described Ulp1-GFP deletion mutant proteins (15) in the presence and absence of alcohol. Earlier studies demonstrated that Ulp1 is localized to the NPC via unconventional interactions with the karyopherin Kap121 and the heterodimer Kap95/Kap60 (12, 13, 23). The Kap121 binding region was localized to amino acids (aa) 1 to 150, and the Kap95/Kap60 binding site (along with a bipartite nuclear localization sequence) was demonstrated to reside within aa 150 to 340. Aa 340 to 403 comprise a coiled-coil motif of unknown function, and the catalytic region (aa 403 to 621) is located at the C terminus of the molecule (Fig. 3 A) (12, 13, 23).
FIG. 3.
The region encompassing Ulp1 aa 150 to 340 is necessary and sufficient for alcohol-mediated nucleolar localization. (A) Schematic representation of the Ulp1 deletion mutant proteins used in this study. Previously defined protein domains and motifs are indicated, along with the alcohol-induced nucleolar localization (this study) and NPC localization (previously demonstrated) properties of each deletion mutant (right). (B) Cells expressing GFP-tagged Ulp1 truncation mutants (as indicated) were imaged before and after treatment with 10% ethanol for 60 min.
Consistent with earlier reports (15), a Ulp11-150-GFP fusion protein displayed both cytoplasmic and nuclear localization in untreated cells (Fig. 3B). Ethanol treatment had no effect on its subcellular distribution (Fig. 3B). Also as previously observed (15), the N-terminal deletion mutant protein Ulp1150-621-GFP is partially mislocalized in untreated cells (presumably due to the absence of the Kap121 binding site), displaying both nuclear rim and cytoplasmic localization (Fig. 3B). However, like the full-length protein, this N-terminal deletion mutant was efficiently recruited to the nucleolus in response to ethanol treatment (Fig. 3B). A Ulp1150-340-GFP protein (lacking the C-terminal coiled-coil and catalytic regions) displayed the same properties (Fig. 3B), indicating that the coiled-coil and catalytic segments are not required for nucleolar localization. Finally, a Ulp1 mutant protein lacking only aa 150 to 340 (Ulp1Δ150-340-GFP) displayed both cytoplasmic and nuclear rim signals in untreated cells, and ethanol had no effect on its localization (Fig. 3B). The region encompassing aa 150 to 340 (containing a nuclear localization signal and the Kap95/Kap60 binding region) is therefore necessary and sufficient for Ulp1 nucleolar recruitment and sequestration in response to alcohol stress.
NPC-associated proteins do not appear to be involved in Ulp1 recruitment to the nucleolus.
Several NPC-associated proteins (Esc1, Mlp1, Mlp2, and Nup60) have been implicated in Ulp1 localization at the NPC (11, 22, 37). To determine whether these proteins also play a role in Ulp1 nucleolar recruitment and localization, we examined Esc1-GFP, Mlp1-GFP, Mlp2-GFP, and Nup60-GFP localization before and after ethanol treatment (Fig. 4 A). While Ulp1 was efficiently recruited to the nucleolus in all of these strains in response to ethanol treatment, no change in localization was observed for any of the NPC-associated proteins (Fig. 4A). We also examined Ulp1 nucleolar recruitment in strains with the esc1, mlp1 and mlp2, or nup60 gene disrupted. Ulp1-GFP and Ulp1150-621-GFP nucleolar targeting and sequestration were unaffected in these strains (Fig. 4B).
FIG. 4.
NPC-associated proteins are not required for Ulp1 nucleolar recruitment. (A) Proteins previously demonstrated to be required for proper Ulp1 localization at the NPC, Nup60-GFP, Mlp1-GFP, Mlp2-GFP, and Esc1-GFP, were coexpressed with Ulp1-RFP and imaged before and after treatment with 10% ethanol for 60 min. Scale bar, 5 μm. (B) Full-length Ulp1-GFP (top) or Ulp1150 −621-GFP (bottom) was expressed in cells with nup60, mlp1 and mlp2, esc1, nup59, or nup100 deleted and imaged before and after treatment with 10% ethanol for 60 min. Scale bar, 5 μm. (C) Kap60-GFP, Kap95-GFP, and Kap121-GFP were imaged before and after treatment with 10% ethanol for 60 min. Scale bar, 5 μm.
To examine the role of NPC structural proteins in Ulp1 nucleolar recruitment, Ulp1-GFP was expressed in strains lacking individual NPC components (nup59Δ, nup100Δ, nup42Δ, nup53Δ, nup170Δ, and nup188Δ). No defects in alcohol-mediated Ulp1 nucleolar targeting were observed in any of these strains (Fig. 4B and data not shown). The mechanism of recruitment of Ulp1 to the nucleolus in response to alcohol stress thus appears to differ dramatically from that of its targeting to the NPC.
Kap121 and the Kap95/Kap60 heterodimer are also critical for Ulp1 localization at the NPC (12, 13, 23). To determine whether these karyopherins play a direct role in Ulp1 nucleolar targeting, we examined the intracellular localization of Kap95-GFP, Kap60-GFP, and Kap121-GFP before and after ethanol treatment. No changes in the localization of the GFP-tagged protein populations were observed (Fig. 4C). Since these karyopherins do not appear to be specifically enriched in the nucleolus following alcohol exposure, the Kaps themselves are most likely not involved in targeting Ulp1 to the nucleolus.
Enforced Ulp1 nucleolar localization effects an alcohol-independent increase in steady-state SUMO conjugates.
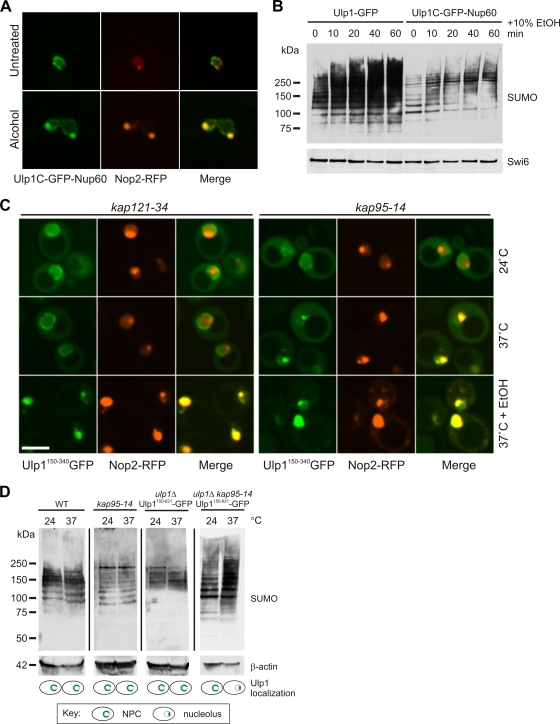
In an attempt to disrupt Ulp1 nucleolar recruitment, we utilized a yeast strain that expressed a previously described fusion protein, Ulp1C-GFP-Nup60, as its only source of Ulp1 (10). This protein is composed of the catalytic region of Ulp1 fused to Nup60, a nucleoporin localized to the nucleoplasmic face of the NPC. Surprisingly, we found that even this fusion protein was recruited to the nucleolus in response to ethanol treatment (Fig. 5 A). Consistent with this observation, steady-state SUMO conjugate formation, while somewhat blunted in this strain compared to that in the wild type, was increased after ethanol exposure (Fig. 5B).
FIG. 5.
Enforced Ulp1 nucleolar localization effects an increase in steady-state SUMO conjugates. (A) Ulp1C-GFP-Nup60 (in a ulp1Δ mutant strain) was coexpressed with the nucleolar marker Nop2-RFP, and cells were imaged before and after treatment with 10% ethanol for 60 min. (B) A ulp1Δ mutant strain expressing either Ulp1-GFP or Ulp1C-GFP-Nup60 (as the only source of Ulp1 in these strains) was exposed to 10% ethanol. Aliquots were taken at the time points indicated, and WCL were subjected to SDS-PAGE and Western analysis. Swi6 was used as a loading control. (C) Ulp1150-340-GFP and Nop2-RFP were coexpressed in kap121-34 and kap95-14 mutant strains. Cells were imaged at permissive (24°C) and restrictive (37°C for 3 h) temperatures, as well as at the restrictive temperature plus 10% ethanol (EtOH; 60 min). (D, top) WCL were prepared from the strains indicated at the permissive (24°C) and restrictive temperatures (37°C for 3 h) and subjected to SDS-PAGE and anti-SUMO Western analysis. Actin was used as a loading control. (D, bottom) Schematic representation of Ulp1 localization in each strain at the permissive and restrictive temperatures.
Finally, we characterized Ulp1 localization in kap95-14 and kap121-34 temperature-sensitive mutant yeast strains. Transport of cargos recognized specifically by these karyopherins is eliminated at the restrictive temperature in these strains (10, 18). As expected, inactivation of Kap121 function had no effect on Ulp1 nucleolar recruitment. The Ulp1150-340-GFP protein (which lacks the Kap121 binding region) displayed nuclear rim and cytoplasmic localization in the kap121-34 strain at both the permissive and restrictive temperatures, and ethanol treatment effected nucleolar sequestration at both temperatures (Fig. 5C, left). Ulp1150-340-GFP also displayed nuclear rim and cytoplasmic localization in the kap95-14 strain at the permissive temperature. However, shifting this strain to the restrictive temperature resulted in a constitutive, ethanol-independent localization of Ulp1150-340-GFP to the nucleolus (Fig. 5C, right). Treatment of these cells with ethanol caused no further change in Ulp1 localization (Fig. 5C, right). Identical results were obtained with the Ulp1150-621-GFP fusion protein (data not shown). Taken together, these data suggest that Ulp1 nucleolar localization is normally inhibited by a Kap95-dependent interaction at the NPC and that the region encompassing Ulp1 aa 150 to 340 contains a cryptic nucleolar targeting signal.
To establish a direct link between Ulp1 nucleolar sequestration and the increase in SUMO conjugate formation following alcohol exposure, Ulp1150-621-GFP was expressed (as the only source of Ulp1 protein) in a ulp1Δ mutant strain, as well as in a ulp1Δ kap95-14 double-mutant strain (14, 15). As expected, Ulp1150-621-GFP was localized to the nuclear rim and cytoplasm at the permissive temperature (24°C) in both strains. Shifting to the restrictive temperature (37°C for 3 h) effected a constitutive, ethanol-independent localization of Ulp1150-621-GFP to the nucleolus in the double-mutant strain (ulp1Δ kap95-14; as in Fig. 5C). Importantly, shifting to the restrictive temperature had no effect on SUMO conjugates in wild-type cells or either of the ulp1Δ or kap95-14 single-mutant strains (Fig. 5D). However, shifting the ulp1Δ kap95-14 double-mutant strain to the restrictive temperature to enforce nucleolar Ulp1 relocalization elicited a dramatic increase in SUMO conjugate formation (Fig. 5D).
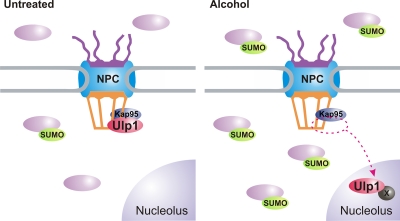
Taken together, our data strongly suggest that decreasing SUMO deconjugation at the NPC by sequestration of Ulp1 in the nucleolus alters the balance of SUMO conjugation and deconjugation in the cell and is responsible for the alcohol stress-mediated increase in steady-state SUMO conjugate levels (Fig. 6).
FIG. 6.
Model of the alcohol-mediated SUMO stress response. In untreated cells (left panel), Ulp1 is localized primarily to the nuclear face of the NPC in a Kap95-dependent manner. Steady-state SUMO conjugate levels are maintained at a relatively low level in this state. Following alcohol exposure (right panel), Ulp1 is sequestered in the nucleolus (presumably via a reversible interaction with an unknown nucleolar component [×]), resulting in an increase in steady-state SUMO conjugate levels.
DISCUSSION
Ulp1 is an essential SUMO protease localized primarily to the NPC via an unconventional interaction with the karyopherins Kap121 and Kap95/Kap60 (12, 13, 23). However, Ulp1 is not localized exclusively to this intracellular location. For example, during mitosis, Ulp1 is recruited to the septin ring to desumoylate the septin proteins (15). In S. pombe, Ulp1 localization is regulated throughout the cell cycle (31), and a mammalian Ulp1 homolog, SENP2, shuttles between the nucleus and cytoplasm (7). Here, we report that in S. cerevisiae, Ulp1 is sequestered in the nucleolus in response to alcohol stress. We further demonstrate that the previously defined Ulp1 Kap95/Kap60 binding region (aa 150 to 340) is necessary and sufficient for nucleolar localization and that enforced Ulp1 nucleolar localization (even in the absence of alcohol) results in a dramatic increase in steady-state SUMO conjugate levels.
The nucleolus and regulation of protein activity.
The nucleolus is a multifunctional subcompartment (25). In addition to its role in the assembly of ribosomal subunits, it plays important roles in a number of other critical cellular processes, in part via the sequestration of regulatory proteins. Compartmentalization provides a rapid and energy-efficient mechanism to modulate protein activity. For example, in mammalian cells, telomerase reverse transcriptase is sequestered in the nucleolus until telomere replication (35), the von Hippel-Lindau protein is sequestered in the nucleolus in response to hypoxia (19), and Mdm2, a ubiquitin E3 ligase involved in p53 degradation, is sequestered in the nucleolus in response to DNA damage (33). In S. cerevisiae, the cell cycle regulator Cdc14 is sequestered in the nucleolus until mitosis, when it is liberated to effect cell cycle progression (30). Our data suggest that the nucleolus can also play an important role in the regulation of the SUMO system.
A model of alcohol stress and SUMO system control.
We do not yet understand how Ulp1 is recruited to and maintained in the nucleolus. The process does appear to be ATP dependent, but it does not require any of the proteins previously demonstrated to be involved in Ulp1 localization at the NPC, i.e., Nup60, Mlp1, Mlp2, or Esc1 (6, 17, 31; Y. Sydorskyy et al., unpublished data). We also expressed Ulp1-GFP in strains lacking individual NPC components (nup42Δ, nup53Δ, nup59Δ, nup100Δ, nup170Δ, and nup188Δ mutants). No defects in alcohol-mediated nucleolar targeting were observed in any of these strains. Finally, we characterized Ulp1 nucleolar targeting in yeast strains lacking proteins previously reported to be recruited to the nucleus in response to alcohol treatment, i.e., Asr1 (2), Nmd5, and Ssa4 (24). Ulp1 was efficiently recruited to the nucleolus in an alcohol-dependent manner in all of these strains (Sydorskyy et al., unpublished). The mechanism of recruitment of Ulp1 to the nucleolus following alcohol stress thus appears to differ dramatically from that of its targeting to the NPC.
Our data suggest that, once released from the NPC in response to alcohol stress, Ulp1 can diffuse into the nucleus, and hence into the nucleolus, where it likely interacts with one or more nucleolar components to anchor it in this subnuclear space (Fig. 6). Removal of the SUMO-deconjugating enzyme from the NPC shifts the balance of SUMO conjugation/deconjugation in the cell, resulting in a buildup of SUMO conjugates. Note that no change in the activity of the SUMO-activating (E1) or -conjugating (E2) enzymes is required in such a model.
How might ethanol cause Ulp1 nucleolar targeting? Alcohols have been demonstrated to increase NPC permeability, most likely by interfering with hydrophobic interactions among the FG (phenylalanine and glycine-rich) nucleoporins that line the inner channel of the nuclear pore (28, 29). It is thus likely that weakening of Kap-Nup interactions at the NPC by alcohols leads to a release of Ulp1 from the nuclear pore. It is tempting to speculate that the Ulp1-Kap-Nup interactions at the NPC act as a sensor for environmental alcohol levels, such that above a certain alcohol threshold, a SUMO stress response is activated.
Importantly, however, NPC release alone cannot explain the behavior of Ulp1 in response to alcohol stress. A small but significant proportion of the Ulp1150-621-GFP and Ulp1150-340-GFP proteins is not localized to the NPC but is observed in the cytoplasm, yet these mutant proteins are only found in the nucleolus after alcohol exposure (Fig. 3B). Similarly, in yeast mutants lacking proteins previously demonstrated to be required for proper NPC localization (nup60Δ, mlp1Δ/mlp2Δ, and esc1Δ mutants), Ulp1150-621-GFP and Ulp1150-340-GFP are almost completely mislocalized to the cytoplasm yet are recruited to the nucleolus only after alcohol treatment (Fig. 4B). Further study is therefore required to identify the cellular factors required for Ulp1 nucleolar recruitment, to better characterize the effects of alcohol on the Ulp1-NPC interaction, and to identify the Ulp1 nucleolar anchor(s).
The mammalian SUMO system is more complex than that in S. cerevisiae, with six unique SUMO proteases (36). Two of these proteases, SENP3 and SENP5, are localized to the nucleolus (36). It would be interesting to examine whether other SENPs are recruited to the nucleolus, or perhaps whether these SUMO proteases are released from the nucleolus, under specific conditions. Additionally, while a steady-state increase in yeast SUMO conjugates is clearly observed in alcohol-treated cells, we cannot rule out a scenario in which Ulp1 also effects the desumoylation of specific nucleolar targets in response to alcohol stress.
Acknowledgments
We are grateful to C. Boone, B. Andrews, R. Wozniak, and M. Tyers for strains and plasmids.
Y.S. was funded by an Amgen/OCI postdoctoral fellowship. T.S. and S.M.J. were funded by Canadian Institutes of Health Research (CIHR) student fellowships, and S.W. was funded by an Ontario Graduate Student fellowship. We thank I. Jurisica for use of the confocal microscope, funded by the Canada Foundation for Innovation and Ontario Ministry of Innovation, grant 12301. A.C.G. holds the Canada Research Chair in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics and is supported by CIHR MOP-84314. B.R. holds the Canada Research Chair in Proteomics and Molecular Medicine and was supported by funding from the Canada Foundation for Innovation, the Ontario Ministry of Innovation, the Ontario Ministry of Health and Long Term Care, and CIHR grant MOP-81268.
Footnotes
Published ahead of print on 20 July 2010.
REFERENCES
- 1.Aitchison, J. D., G. Blobel, and M. P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624-627. [DOI] [PubMed] [Google Scholar]
- 2.Betz, C., G. Schlenstedt, and S. M. Bailer. 2004. Asr1p, a novel yeast ring/PHD finger protein, signals alcohol stress to the nucleus. J. Biol. Chem. 279:28174-28181. [DOI] [PubMed] [Google Scholar]
- 3.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 4.Finley, D., E. Ozkaynak, and A. Varshavsky. 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48:1035-1046. [DOI] [PubMed] [Google Scholar]
- 5.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 6.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 7.Itahana, Y., E. T. Yeh, and Y. Zhang. 2006. Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol. Cell. Biol. 26:4675-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 9.Kurepa, J., J. M. Walker, J. Smalle, M. M. Gosink, S. J. Davis, T. L. Durham, D. Y. Sung, and R. D. Vierstra. 2003. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278:6862-6872. [DOI] [PubMed] [Google Scholar]
- 10.Leslie, D. M., B. Grill, M. P. Rout, R. Wozniak, and J. D. Aitchison. 2002. Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol. Cell. Biol. 22:2544-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis, A., R. Felberbaum, and M. Hochstrasser. 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 178:813-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 13.Li, S. J., and M. Hochstrasser. 2003. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160:1069-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makhnevych, T., C. P. Lusk, A. M. Anderson, J. D. Aitchison, and R. W. Wozniak. 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell 115:813-823. [DOI] [PubMed] [Google Scholar]
- 15.Makhnevych, T., C. Ptak, C. P. Lusk, J. D. Aitchison, and R. W. Wozniak. 2007. The role of karyopherins in the regulated sumoylation of septins. J. Cell Biol. 177:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makhnevych, T., Y. Sydorskyy, X. Xin, T. Srikumar, F. J. Vizeacoumar, S. M. Jeram, Z. Li, S. Bahr, B. J. Andrews, C. Boone, and B. Raught. 2009. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell 33:124-135. [DOI] [PubMed] [Google Scholar]
- 17.Manza, L. L., S. G. Codreanu, S. L. Stamer, D. L. Smith, K. S. Wells, R. L. Roberts, and D. C. Liebler. 2004. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem. Res. Toxicol. 17:1706-1715. [DOI] [PubMed] [Google Scholar]
- 18.Marelli, M., D. J. Dilworth, R. Wozniak, and J. D. Aitchison. 2001. The dynamics of karyopherin-mediated nuclear transport. Biochem. Cell Biol. 79:603-612. [PubMed] [Google Scholar]
- 19.Mekhail, K., L. Gunaratnam, M. E. Bonicalzi, and S. Lee. 2004. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 6:642-647. [DOI] [PubMed] [Google Scholar]
- 20.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 21.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28:612-618. [DOI] [PubMed] [Google Scholar]
- 22.Palancade, B., X. Liu, M. Garcia-Rubio, A. Aguilera, X. Zhao, and V. Doye. 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell 18:2912-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panse, V. G., B. Kuster, T. Gerstberger, and E. Hurt. 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5:21-27. [DOI] [PubMed] [Google Scholar]
- 24.Quan, X., R. Rassadi, B. Rabie, N. Matusiewicz, and U. Stochaj. 2004. Regulated nuclear accumulation of the yeast hsp70 Ssa4p in ethanol-stressed cells is mediated by the N-terminal domain, requires the nuclear carrier Nmd5p and protein kinase C. FASEB J. 18:899-901. [DOI] [PubMed] [Google Scholar]
- 25.Raska, I., P. J. Shaw, and D. Cmarko. 2006. New insights into nucleolar architecture and activity. Int. Rev. Cytol. 255:177-235. [DOI] [PubMed] [Google Scholar]
- 26.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 27.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 28.Shulga, N., and D. S. Goldfarb. 2003. Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol. Cell. Biol. 23:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shulga, N., N. Mosammaparast, R. Wozniak, and D. S. Goldfarb. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149:1027-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stegmeier, F., and A. Amon. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203-232. [DOI] [PubMed] [Google Scholar]
- 31.Taylor, D. L., J. C. Ho, A. Oliver, and F. Z. Watts. 2002. Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease. J. Cell Sci. 115:1113-1122. [DOI] [PubMed] [Google Scholar]
- 32.Tempé, D., M. Piechaczyk, and G. Bossis. 2008. SUMO under stress. Biochem. Soc. Trans. 36:874-878. [DOI] [PubMed] [Google Scholar]
- 33.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 34.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 35.Wong, J. M., L. Kusdra, and K. Collins. 2002. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 4:731-736. [DOI] [PubMed] [Google Scholar]
- 36.Yeh, E. T. 2009. SUMOylation and de-SUMOylation: wrestling with life's processes. J. Biol. Chem. 284:8223-8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao, X., C. Y. Wu, and G. Blobel. 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167:605-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, W., J. J. Ryan, and H. Zhou. 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279:32262-32268. [DOI] [PMC free article] [PubMed] [Google Scholar]