Abstract
Both insulin and bone morphogenetic protein (BMP) signaling systems are important for adipocyte differentiation. Analysis of gene expression in BMP7-treated fibroblasts revealed a coordinated change in insulin signaling components by BMP7. To further investigate the cross talk between insulin and BMP signaling systems in brown adipogenesis, we examined the effect of BMP7 in insulin receptor substrate 1 (IRS-1)-deficient brown preadipocytes, which exhibit a severe defect in differentiation. Treatment of these cells with BMP7 for 3 days prior to adipogenic induction restored differentiation and expression of brown adipogenic markers. The high level of adipogenic inhibitor preadipocyte factor 1 (Pref-1) in IRS-1-null cells was markedly reduced by 3 days of BMP7 treatment, and analysis of the 1.3-kb pref-1 promoter revealed 9 putative Smad binding elements (SBEs), suggesting that BMP7 could directly suppress Pref-1 expression, thereby allowing the initiation of the adipogenic program. Using a series of sequential deletion mutants of the pref-1 promoter linked to the luciferase gene and chromatin immunoprecipitation, we demonstrate that the promoter-proximal SBE (−192/−184) was critical in mediating BMP7's suppressive effect on pref-1 transcription. Together, these data suggest cross talk between the insulin and BMP signaling systems by which BMP7 can rescue brown adipogenesis in cells with insulin resistance.
Obesity and insulin resistance are two hallmark features of a larger collection of abnormalities called the metabolic syndrome. It is estimated that over 40 million people in the United States have the metabolic syndrome, and it is this cluster of abnormalities that creates risk for many of our most common medical conditions, including glucose intolerance, dyslipidemias, nonalcoholic fatty liver, cardiovascular disease, renal failure, Alzheimer's disease, and even some cancers (7, 8, 21, 24). Adipose tissue plays an active role in systemic energy metabolism (14) and therefore contributes to the pathogenesis of the metabolic syndrome. There are two functionally different types of adipose tissue: white adipose tissue, which is specialized for the storage of excess energy in the form of triglycerides, and brown adipose tissue, which is completely dedicated to energy expenditure in response to cold or overfeeding. Given its tremendous capacity to dissipate chemical energy (4) and its recently demonstrated presence in adult humans (9, 53, 32, 40, 55, 62), promoting brown fat differentiation and function provides an attractive approach to the treatment of obesity and related metabolic complications.
Adipocyte differentiation is regulated by a network of hormones and growth factors (4, 14). Insulin signaling is known to play a pivotal role in both white and brown adipogenesis (38). Insulin receptor substrates (IRSs) function as the docking proteins coordinating hormone binding to the receptor and downstream signaling events (56). There are four members of the IRS protein family, and each has a unique and complementary function in different biological processes. We have previously demonstrated different roles for the different IRSs in brown adipocyte differentiation (48). While IRS-4-null cells show normal brown adipocyte differentiation, IRS-2 knockout (IRS-2KO) brown preadipocytes exhibit a mild defect in differentiation but have impaired insulin-stimulated glucose uptake (13). IRS-3-null cells also show a moderate defect in adipogenesis, and brown preadipocytes lacking IRS-1 display a severe defect in differentiation (11).
Emerging evidence suggests that the bone morphogenetic protein (BMP) family of developmental regulators plays an important role in the formation of different adipose depots (42). The effects of BMPs on adipogenesis appear to depend on the stage of cell development and the dosage of different BMP ligands. BMP2 can promote differentiation of 3T3-L1 white preadipocytes (37), and this process is enhanced by peroxisome proliferator-activated receptor γ (PPARγ) agonists (19, 46). Mice with genetic ablation of schnurri 2, a zinc finger-containing protein activated by BMP2, display reduced white, but not brown, fat mass, suggesting that BMP2 signaling is critical to white adipogenesis (22). BMP4 is required for the commitment step and subsequent differentiation of the multipotent C3H10T1/2 cells to the white adipocyte lineage (2, 3) through downregulating the expression of matrix metalloproteinase 3 (MMP-3) and MMP-13 (33). Recently, we showed that BMP7 specifically promotes brown adipogenesis in both committed brown preadipocytes and uncommitted multipotent mesenchymal precursors (49). BMP7 knockout embryos and newborn pups exhibit a marked paucity of interscapular brown fat, suggesting that BMP7 plays an indispensable role in the formation of brown adipose tissue during embryonic development (49). Whether BMP7 acts in concert with other hormonal or developmental signals to specify brown fat cell fate is currently unknown.
BMPs exert their biological activities through binding to a heterodimeric transmembrane complex composed of type I and II BMP receptors that possess serine/threonine kinase activity (20, 29). Binding of BMPs to this complex triggers phosphorylation of the type I BMP receptor (26, 27), which in turn phosphorylates downstream signaling mediator Smads (27). Phosphorylation of Smads decreases their affinity for cytoplasmic anchors and increases their affinity for nuclear factors, leading to their translocation into the nucleus, where they activate or repress target gene expression (44, 59). BMPs and transforming growth factor beta (TGF-β) signaling pathways are mediated by the transcriptional factors Smad1/5/8 and Smad2/3 (termed R-Smads, for receptor-specific Smads), respectively, which partner with the universal co-Smad, Smad4, to form heterodimers that target the Smad binding elements (SBEs) of gene promoters (28). In addition to the Smad pathway, BMPs also utilize the p38 mitogen-activated protein kinase (MAPK) pathway to regulate different biological functions (30). We previously showed that inhibition of the p38 MAPK pathway suppressed BMP7-induced UCP1 expression without affecting lipid accumulation, revealing an essential role of the p38MAPK pathway in the BMP7-induced thermogenic program, while this pathway appears to be dispensable for lipid accumulation (49).
Because both insulin and BMP signaling pathways are essential for brown adipogenesis, we hypothesized that cross talk between these two pathways is involved in driving the brown adipogenic program. In this study, we demonstrate that BMP7 treatment coordinately upregulates several components of insulin signaling with concomitant downregulation of the insulin signaling inhibitor suppressor of cytokine signaling 3 (SOCS3). Importantly, pretreatment of IRS-1KO cells with BMP7 prior to adipogenic induction rescued their differentiation defect, which was mediated, at least in part, by specifically inhibiting expression of preadipocyte factor 1 (Pref-1), a known suppressor of preadipocyte-adipocyte transition. We further show that downstream mediators of BMP signaling, Smad1/4, were able to bind to the pref-1 promoter to suppress its expression. These data reveal a heretofore unrecognized molecular mechanism utilized by BMP7 to regulate brown adipogenesis and highlight the fact that the cooperation of the BMP and insulin signaling systems plays a central role in the regulation of brown fat formation and function.
MATERIALS AND METHODS
Cell culture.
Establishment of wild-type (WT) and IRS-1KO brown preadipocytes isolated from newborn wild-type and IRS-1KO mice and their differentiation into brown adipocytes were described previously (11, 49-51), except that the cells were pretreated with 3.3 nM BMP7 or vehicle for 3 days before induction of differentiation, as indicated. Briefly, brown preadipocytes were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products). Adipocyte differentiation was induced by incubating 100% confluent cells in DMEM supplemented with 10% FBS, 0.5 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 20 nM insulin, 0.125 mM indomethacin, and 1 nM T3 for 2 days and then switching them to DMEM plus 10% FBS supplemented with 20 nM insulin and 1 nM triiodothyronine (T3), renewed every second day until the cells were harvested at the indicated time points.
Plasmids.
The mouse pref-1 promoter (−1271 to +110) and a series of deletion mutants were generated by PCR and cloned into pGLbasic4.1 vector to generate the pref-1 promoter-luciferase constructs [pGL(−1271), pGL(−809), pGL(−201), and pGL(−49)]. A point mutation in SBE9 [pGL(−201mut)] was generated by changing 2 nucleotides in the SBE9 sequence from 5′-GTCTAGCC-3′ to 5′-GACTACCC-3′ (underlining indicates SBE core sequences; italics indicate nucleotides substituted). pCS2-Flag-Smad1, pCS2-Flag-Smad4, and pCMV5-DPC4 (a dominant-negative form of Smad4) were kindly provided by Andrew Lassar, Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School.
Transfection and luciferase assay.
Transfections and luciferase assays were done using Lipofectamine 2000 (Invitrogen) and the Dual-Luciferase System (Promega), respectively, following the protocols recommended by the manufacturers. Briefly, WT and IRS-1KO cells were transfected with mouse pref-1 promoter constructs pGL(−201), pGL(−201mut), and pGL(−49). Six hours after transfection, the cells were treated with BMP7 for 24 h and then assayed for luciferase activity.
Microarray analysis.
C3H10T1/2 cells were cultured in DMEM supplemented with 10% FBS. At 60% confluence, the cells were treated with 3.3 nM BMP7 or vehicle for 3 days. RNA samples from four independent experiments were analyzed by microarray using the Affymetrix GeneChip mouse 430A. A total of 8 chips were used in this study. Intensity values were quantitated by MAS 5.0 software (Affymetrix). All chips were subjected to global scaling to a target intensity of 1,500 to take into account the inherent differences between the chips and their hybridization efficiencies. The significance of gene expression was determined by Student's t test using a P value of ≤0.05 as the threshold.
ChIP analysis.
WT and IRS-1KO cells were cotransfected with pGL(−201), pGL(−201mut), and pGL(−49) constructs, along with pCS2-Flag-Smad1/pCS2-Flag-Smad4. Eighteen hours after transfection, the cells were treated with vehicle or BMP7 for 1 h. Then, the cells were incubated in 1% formaldehyde at room temperature for 10 min for cross-linking, followed by addition of 1 M glycine to a final concentration of 0.125 M and incubation at room temperature for 10 min. The cells were washed twice with cold phosphate-buffered saline (PBS) and harvested in chromatin immunoprecipitation (ChIP) lysis buffer (0.1% SDS, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, and 20 mM Tris, pH 8). The samples were then subjected to sonication for 8 min to ensure that DNA was fragmented to the size range 100 bp to 1,000 bp. One hundred micrograms of sample, 20 μg protein G (Amersham), and 1 μg anti-Flag antibody (Sigma) or isogenic immunoglobulin (as a mock control) were used for each ChIP reaction to precipitate Smad1/4 protein. Coprecipitated DNA was subjected to a series of washes in buffers with different salt concentrations and then de-cross-linked in buffer (1% SDS, 0.1 M NaHCO3, 0.2 M NaCl) at 65°C for 4 h. After phenol chloroform extraction of the proteins, the DNA was precipitated and dissolved in nuclease-free water and then analyzed by quantitative RT-PCR (Q-RT-PCR) using primers specific for the transgene SBE9 that can distinguish the SBE9 sequence of the transfected pref-1 promoter construct from that of the endogenous pref-1 promoter. Ten percent of the ChIP input samples (without precipitation and washing) were also subjected to the same procedures and quantified by Q-RT-PCT as an internal control for normalization.
Western blotting and ELISA.
Cultured cells were harvested in protein sample buffer. Fifty micrograms of total proteins was loaded in each lane in SDS-PAGE for Western blotting. The antibodies used in Western blotting were UCP1 (Santa Cruz sc-6528), PPARγ (Santa Cruz sc-7273), CCAAT/enhancer binding protein α (C/EBPα) (Santa Cruz sc-61), Pref-1 (Santa Cruz sc-8625), and β-tubulin (Cell Signaling catalog no. 2146). Medium was harvested from cultures of WT and IRS-1KO cells overexpressing BMP7 and analyzed for the BMP7 expression level by enzyme-linked immunosorbent assay (ELISA) using a human BMP7 DuoSet ELISA Development kit from R&D Systems following the manufacturer-provided protocol.
Bioinformatics and statistical analyses.
pref-1 promoter sequences were analyzed by MatInspector (http://www.genomatix.de) for Smad binding elements. Student's t test was used in all Q-RT-PCR analyses.
Nucleotide sequence accession numbers.
The accession numbers of the pref-1 promoter sequences from different species are as follows: mouse, AC107681; rat, AB046763; human, AL132711; and chicken, GGU48890.
RESULTS
BMP7 induces expression of insulin signaling components in mouse embryonic fibroblasts.
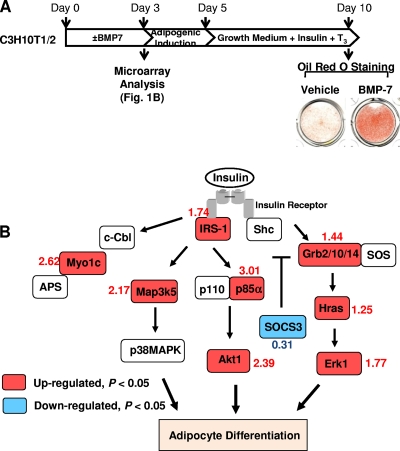
We previously reported that BMP7 triggers commitment of the multipotent mesenchymal C3H10T1/2 cells to a brown adipocyte lineage and that transplantation of BMP7-treated C3H10T1/2 cells into athymic nude mice results in formation of brown fat tissue in vivo (49). Treatment of these cells with BMP7 for 3 days, followed by standard adipogenic cocktail, led to a marked increase in lipid accumulation (Fig. 1 A) and expression of the brown fat-specific marker uncoupling protein 1 (UCP1) (49). To search for the potential molecular mechanisms mediating BMP7's brown adipogenic effect, we performed microarray analysis on C3H10T1/2 cells treated with BMP7 or vehicle for 3 days. Interestingly, microarray data revealed that BMP7 significantly regulated several key components of the insulin signaling pathway in these mesenchymal cells (Fig. 1B; also see Table S1 in the supplemental material). They include upregulation of IRS-1, the p85 subunit of phosphatidylinositol 3-kinase (PI3-kinase), thymoma viral proto-oncoprotein (Akt), growth factor receptor-bound protein 2 (Grb2), growth factor receptor-bound protein 10 (Grb10), and extracellular signal-regulated kinase 1 (Erk1) and downregulation of SOCS3, an inhibitor of insulin signaling. It has been shown that activation of the PI3K/Akt pathway is required for insulin-induced adipogenesis (1, 34, 58, 60), while adipocyte differentiation is associated with downregulation of SOCS3 expression (12). Thus, the coordinated regulation of insulin signaling components by BMP7 suggests that BMP7-induced adipogenesis is mediated at least partly by enhancing insulin signaling.
FIG. 1.
BMP7 alters the expression of insulin signaling components. C3H10T1/2 cells were cultured in DMEM supplemented with 10% FBS. At 60% confluence, the cells were treated with 3.3 nM BMP7 or vehicle for 3 days and then subjected to standard adipogenic differentiation. The cells were stained with Oil Red O on day 10 (A), and mRNAs were isolated on day 3 and subjected to microarray analysis (B). Panel B shows coordinated changes in several insulin signaling components after 3 days of BMP7 treatment analyzed by Genmapp (http://www.genmapp.org). The numbers beside the gene names indicate the fold change of a given gene in BMP7-treated cells relative to the control, which corresponds to the most significant P value among different probe sets.
BMP7 treatment rescues the differentiation defect of IRS-1KO brown preadipocytes.
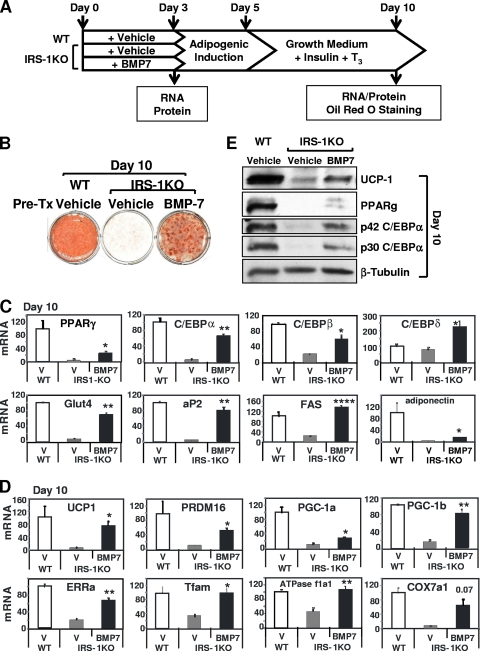
To further examine the effect of BMP7 on the insulin signaling pathway during brown adipogenesis, we treated IRS-1KO brown preadipocytes, which exhibited severe defects in differentiation, with BMP7 or vehicle for 3 days and subjected the cells to a standard protocol of brown adipocyte differentiation for an additional 7 days (Fig. 2 A). As a control, the WT brown preadipocytes were also treated with vehicle and then induced to undergo adipocyte differentiation (50). BMP7 treatment effectively restored the differentiation defect in IRS-1KO cells compared to that of wild-type cells achieved by a standard differentiation protocol and as judged by Oil Red O staining (Fig. 2B). Adipogenesis is regulated by a transcriptional network involving many transcriptional factors, with PPARγ and C/EBPs in the center and governing the whole terminal-differentiation process (10, 38). Q-RT-PCR analysis revealed that BMP7 treatment completely or partially restored the expression of the key adipogenic transcription factors PPARγ, C/EBPα, C/EBPβ, and C/EBPδ, as well as markers for mature adipocytes, including aP2, Glut4, FAS, and adiponectin, in IRS-1KO cells at day 10 of this program (Fig. 2C). Interestingly, expression of adiponectin, an adipokine that is highly associated with whole-body insulin sensitivity (6), was completely abolished in IRS-1KO cells, and BMP7 treatment could restore only 14% of the expression level in WT cells, suggesting that certain mechanisms for the regulation of adiponection gene expression were altered in IRS-1-deficient cells that cannot be rescued by BMP7 treatment.
FIG. 2.
BMP7 treatment rescued brown adipocyte differentiation of IRS-1KO cells. (A) Differentiation protocol. IRS-1KO cells were pretreated with vehicle or BMP7 for 3 days and then induced to differentiate, as described in Materials and Methods. As a control, WT cells were treated with vehicle only and then induced to differentiate. RNA and protein were isolated on day 3 and day 10 and analyzed by Q-RT-PCR and Western blotting, respectively. On day 10, the cells were fixed and stained with Oil Red O. (B) Oil Red O staining of cells at day 10. Pre-Tx, pretreatment. (C) Q-RT-PCR analysis of the expression of adipogenic markers at day 10. (D) Q-RT-PCR analysis of the expression of brown-fat-selective genes at day 10. V, vehicle. For Q-RT-PCR analysis, data are presented as the mean plus the standard error of the mean (SEM) from a representative of 4 independent experiments, with each performed in triplicate. The values for WT cells were set as 100 arbitrarily. Statistically significant differences between vehicle- and BMP7-treated IRS-1KO cells are indicated above the bars: *, P < 0.05; **, P < 0.01; ***, P < 0.0001. (E) Western blotting of general adipogenic and brown-fat-specific marker proteins. A representative of 4 independent experiments is shown.
Brown adipocytes are characterized by a high density of mitochondria and the unique expression of UCP1 (4). Importantly, BMP7 treatment in IRS-1KO cells and adipogenic induction led to nearly complete restoration of UCP1 expression at day 10 (Fig. 2D). This was accompanied by a significant increase in the expression of several key regulators of brown adipogenesis, including PR domain containing 16 (PRDM16) (43) and the nuclear coactivators peroxisome proliferative activated receptor gamma coactivator 1α (PGC-1α) (35, 57), PGC-1β (52), and the orphan nuclear receptor estrogen-related receptor α (ERRα) (41) (Fig. 2D). The transcriptional complex consisting of ERRα and PGC-1α has been shown to play an essential role in the regulation of mitochondrial gene expression and biogenesis (31, 36, 41). Thus, expression of genes involved in mitochondrial replication and oxidative phosphorylation was increased in cells treated with BMP7, including mitochondrial transcription factor A (Tfam), cytochrome c oxidase, subunit VIIa 1 (COX7a1), and ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1 (ATPase f1a1) (Fig. 2D). Western blotting further confirmed that BMP7 pretreatment restored protein expression of UCP1, PPARγ, and C/EBPα (Fig. 2E). These data suggest that BMP7 can reverse the differentiation defect in brown preadipocytes with impairment in insulin signaling.
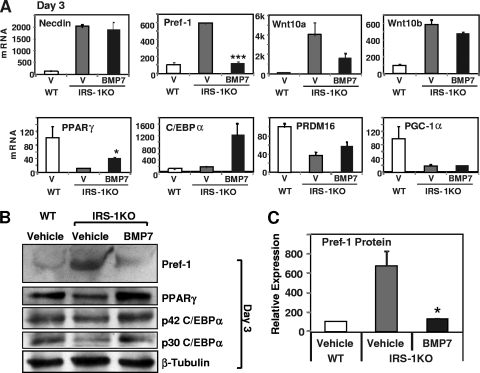
To further explore the molecular mechanisms underlying these processes, we examined mRNA and protein expression in IRS-1KO cells treated with BMP7 or vehicle for 3 days. It is known that prior to entering the adipogenic program, the preadipocytes need to be released from suppression controlled by a number of adipogenic inhibitors, such as necdin (17, 48), Wnt family proteins (39), and Pref-1 (47). Disruption of insulin signaling by knocking down IRS-1 led to increased expression of necdin, Wnt, and Pref-1 and resulted in blockage of the transcriptional cascade necessary for adipogenesis (48). Treatment of IRS-1KO cells with BMP7 for 3 days did not affect the expression of necdin and Wnt10b but tended to reduce the expression of Wnt10a, although it did not reach statistical significance. Importantly, BMP7 completely reversed the elevated expression of Pref-1 in IRS-1KO cells at both the mRNA and protein levels (Fig. 3 A, B, and C). Thus, the rescue of brown adipocyte differentiation by BMP7 in IRS-1KO cells is mediated, at least in part, by suppressing Pref-1 expression.
FIG. 3.
Regulation of early adipogenic markers and inhibitors by BMP7. WT and IRS-1KO cells were differentiated as described in the legend to Fig. 2. RNA and protein were isolated after 3 days of BMP7 pretreatment and analyzed for gene expression by Q-RT-PCR and Western blotting. (A) Q-RT-PCR analysis of adipogenic markers and inhibitors on day 3. V, vehicle. The data are presented as the mean plus SEM from a representative of 4 independent experiments, with each performed in triplicate. (B) Western blotting of early adipogenic inhibitor and marker proteins on day 3. A representative of 4 independent experiments is shown. (C) Quantification of the Pref-1 protein levels in Western blots by densitometry. The data represent the mean plus SEM from 4 independent experiments. The values for WT cells are set as 100 arbitrarily. The asterisks depict statistically significant differences between vehicle- and BMP7-treated IRS-1KO cells (*, P < 0.05; ***, P < 0.001).
In addition to removal of suppression, 3 days of BMP7 pretreatment also significantly induced mRNA and protein expression for the two key adipogenic transcription factors PPARγ and C/EBPα in IRS-1KO cells (Fig. 3A and B). Expression of the early brown adipogenic regulator PRDM16 was reduced by 50% in IRS-1KO cells, and this was partially rescued by BMP7 treatment (Fig. 3A). Interestingly, the reduced levels of PGC-1α in IRS-1KO cells were not altered at this early time point, but they were partially restored in BMP7-treated cells at the later time point (Fig. 2D).
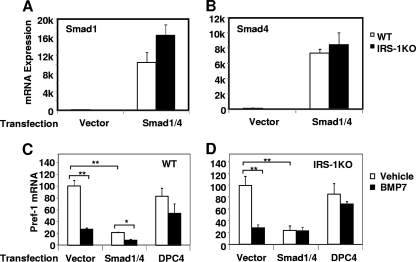
Overexpression of Smad1/4 suppresses Pref-1 expression.
Our observation that BMP pretreatment specifically downregulated Pref-1 expression in IRS-1KO cells prompted us to hypothesize that BMP signaling may directly target the pref-1 promoter to repress its expression at the early stage of differentiation. To determine whether inhibition of Pref-1 expression by BMP7 is mediated by the Smad signaling pathway, we transfected both WT and IRS-1KO cells with Smad1 and Smad4 expression vectors, a vector expressing a dominant-negative form of Smad4 (DPC4) (23), or the empty vector as a control. This resulted in a 70- to 160-fold increase in Smad1 and Smad4 expression over the basal levels (Fig. 4 A and B). After culturing the transfected cells in the presence or absence of BMP7 for 3 days, we isolated mRNA from these cells and analyzed Pref-1 gene expression. Consistent with the findings described above, BMP7 treatment resulted in a nearly 80% reduction in Pref-1 expression in both WT and IRS-1KO cells transfected with empty vector alone (Fig. 4C and D). Overexpression of Smad1/4 in these cells significantly suppressed Pref-1 expression in the basal state, and BMP7 treatment further decreased Pref-1 expression in WT cells, but not IRS-1KO cells. Importantly, when the dominant-negative form of Smad4, DPC4, was expressed in either WT or IRS-1KO cells, the inhibitory effect of BMP7 on Pref-1 expression was blunted, indicating that BMP7 worked through the Smad1/4 complex to inhibit Pref-1 expression.
FIG. 4.
Overexpression of Smad1/4 supresses Pref-1 expression. WT and IRS-1KO brown preadipocytes were transfected with plasmids expressing Smad1 and Smad4 or a dominant-negative form of Smad4, DPC4. Six hours after transfection, the cells were treated with vehicle or BMP7 for 72 h. mRNAs were isolated and analyzed for Smad1 (A), Smad4 (B), and Pref-1 expression in WT (C) and IRS-1KO (D) cells by Q-RT-PCR. The data represent means plus SEM from a representative of 4 independent experiments, with each performed in triplicate. The values for vector-transfected WT cells (A and B) or for vehicle-treated vector-transfected cells (C and D) are set as 100 arbitrarily. The asterisks indicate statistically significant differences (*, P < 0.05; **, P < 0.01).
Smad1/4 binds to the Smad binding element of the pref-1 promoter and inhibits its expression.
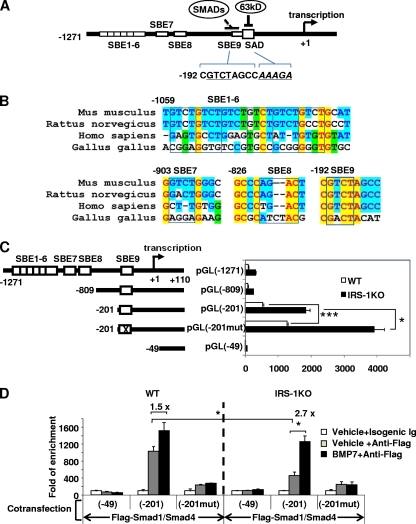
Based on the results described above, we reasoned that the Smad1/4 complex might directly suppress pref-1 promoter activity. Indeed, bioinformatics analysis revealed the presence of 9 putative SBEs, referred to here as SBE1 to SBE9, within the 1.3-kb pref-1 promoter region (from −1271 to + 110) (Fig. 5 A and B). The first 6 SBEs, consisting of the repeated core sequence 5′-GTCT-3′, overlapped each other, while SBE8 was in the opposite direction. Interestingly, the promoter-proximal SBE, SBE9, was located right next to the SAD (suppression in adipocyte differentiation) element, which was previously reported to bind a 63-kDa protein (p63) in response to dexamethasone treatment to repress pref-1 promoter activity (45) (Fig. 5A). Alignment of the putative SBE sequences in the pref-1 promoter from different species revealed that the SBE9 sequences shared 100% identity among mice, rats, and humans, while the other SBEs exhibited different degrees of variation (Fig. 5B). To determine whether these putative SBEs are required for regulation of Pref-1 expression, we generated luciferase reporter constructs harboring sequential deletions of the 1.3-kb pref-1 promoter and a point mutation in SBE9 (Fig. 5C) and transfected them into both WT and IRS-1KO cells. Given the inhibitory effect of BMP7/Smad on Pref-1 gene expression, we anticipated that deletion or mutation of the key Smad binding elements would lead to increased luciferase activity. Deletion of SBE1 to SBE8 slightly decreased reporter gene activity in both WT and IRS-1KO cells (Fig. 5C), suggesting that SBE1 to SBE8 do not play a role in suppression of Pref-1 expression by the Smad pathway. Further deletion of the sequence between −809 and −201 increased reporter gene activity in both WT and IRS-1KO cells (Fig. 5C), suggesting this fragment contains non-SBE elements that act to repress pref-1 promoter activity. Most importantly, introducing a mutation of 2 bases in SBE9 significantly increased the promoter activity by 2-fold in both WT and IRS-1KO cells (Fig. 5C), suggesting that SBE9 functions as an inhibitory element for Pref-1 expression. Further deletion of the promoter sequences from −201 to −49 completely abolished luciferase activity (Fig. 5C), indicating this region contains key regulatory elements essential for Pref-1 gene expression.
FIG. 5.
Smad1/4 interacts with SBE of the pref-1 promoter to repress Pref-1 expression. (A) Scheme of the ∼1,300-bp pref-1 promoter. The sequences of SBE9 and SAD are shown. Alignment was performed using the Align-X program in Vector NTI and then manually adjusted. Yellow highlighting indicates absolutely conserved nucleotides, cyan highlighting indicates conserved nucleotides, and green highlighting indicates similar nucleotides. (B) Alignment of the 9 SBE sequences of the pref-1 promoter from different species. The core sequences of SBEs are boxed. (C) Analysis of the pref-1 promoter. WT and IRS-1KO cells were transfected with the indicated mouse pref-1 promoter constructs and assayed for luciferase activity. The data represent the mean plus SEM from a representative of 3 independent experiments, with each performed in triplicate. The value for pGL(−1271) transfection of WT cells was set as 100 arbitrarily. (D) ChIP analysis of Smad1/4 binding to the pref-1 promoter. WT and IRS-1KO cells were cotransfected with the indicated plasmid constructs and analyzed by ChIP (see Materials and Methods). The pref-1 promoter SBE9 region enriched by ChIP was quantified by Q-RT-PCR using primers flanking SBE9. The values for the vehicle plus isogenic immunoglobulin were set as 1 arbitrarily. The data represent the mean plus SEM from a representative of 3 independent experiments, with each performed in triplicate. The asterisks depict statistically significant differences (*, P < 0.05; ***, P < 0.001).
Next, we sought to determine, using modified ChIP analysis, whether Smad1/4 can directly bind to SBE9. We cotransfected WT and IRS-1KO cells with Flag-tagged Smad1 and Smad4 expression vectors, along with the pref-1 promoter constructs pGL(−49), pGL(−201) (containing the wild-type SBE9), and pGL(−201mut) (containing the mutated SBE9). The cells were treated with BMP7 or vehicle and then subjected to ChIP analysis using the anti-Flag antibody to pull down protein-DNA complexes. As shown in Fig. 5D, the pref-1 promoter construct containing SBE9 appeared to bind to the Smad1/4 complex, as quantified by Q-RT-PCR using specific primers flanking SBE9 (Fig. 5D). Binding of the Smad1/4 complex to SBE9 [pGL(−201)] was increased by 1.5- and 2.7-fold by BMP7 treatment in WT and IRS-1KO cells, respectively. Importantly, this binding was almost completely abolished when SBE9 was mutated. Notably, in the basal state (vehicle treatment), the binding of Smad1/4 complex to SBE9 in IRS-1KO cells was about half of that in WT cells, indicating that Pref-1 expression is less suppressed in IRS-1KO cells than in WT cells. This may account for the high levels of Pref-1 in IRS-1KO cells, leading to the defect in differentiation (46). Together, these data suggest a critical role of the promoter-proximal SBE9 in direct binding of the Smad1/4 complex, which mediates BMP7's suppressive effect on Pref-1 gene expression.
Overexpression of BMP7 induces spontaneous differentiation in wild-type brown preadipocytes and rescues brown adipogenesis in IRS-1KO cells.
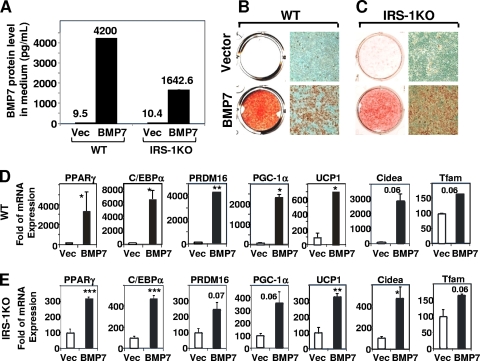
To further verify the findings that BMP7 rescued the differentiation defect of IRS-1KO cells, we stably expressed BMP7 in WT and IRS-1KO cells by transfecting these cells with a vector expressing human BMP7 (hBMP7) under the control of the adipocyte-specific aP2 promoter. ELISA analysis confirmed high levels of hBMP7 in the culture media collected on day 10 (Fig. 6 A). When subjected to continuous postconfluence culture, the WT cells overexpressing BMP7 underwent spontaneous brown adipocyte differentiation, as determined by lipid accumulation (Fig. 6B), and displayed increased expression of general and brown adipogenic marker genes (Fig. 6D). Overexpression of BMP7 in IRS-1KO cells did not result in spontaneous differentiation (data not shown). However, when put in the standard differentiation media, the BMP7-overexpessing IRS-1KO cells were able to differentiate efficiently into mature brown adipocytes (Fig. 6C) that expressed high levels of the key adipogenic transcription factors PPARγ and C/EBPα, as well as the brown-fat-selective markers PRDM16, PGC-1α, Cidea, and UCP1 (Fig. 6E). Given the fact that there is little to no aP2 expression in preadipocytes, it is conceivable that IRS-1KO cells transfected with the aP2 promoter-driven BMP7 vector still require induction cocktails to initiate differentiation but that once the cells enter the adipogenic program, they are able to produce BMP7 to further enhance differentiation.
FIG. 6.
Overexpression of BMP7 induced spontaneous differentiation of wild-type brown preadipocytes and rescued brown adipocyte differentiation of IRS-1KO preadipocytes. WT and IRS-1KO brown preadipocytes were stably transfected with an hBMP7 construct driven by the aP2 promoter or with empty vector (as a control). Pools of transfected clones were grown to confluence (defined as day 0) and proceeded to continuous culture in DMEM plus 10% FBS without any inducers (for WT cells) or with Dex-IBMX-insulin-indomethacinn-T3 (for IRS-1KO cells) (see Materials and Methods). On day 10, the analyses were performed. (A) hBMP7 levels in the medium supernatants from WT and IRS-1KO cells were determined by ELISA. (B and C) Oil Red O staining and microscopic views of WT cells (B) and IRS-1KO cells (C). (D and E) Expression of adipogenic markers and brown-fat-selective genes in WT cells (D) and IRS-1KO cells (E) was analyzed by Q-RT-PCR. The data are presented as the mean plus SEM from a representative of 3 independent experiments, with each performed in duplicate or triplicate. The values for vector are set as 100 arbitrarily. Statistically significant differences between vector- and hBMP7-transfected cells are indicated above the bars: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
It is well established that insulin signaling is required for adipogenesis. We previously showed that IRS-1KO brown preadipocytes had a severe defect in differentiation and expressed high levels of adipogenic inhibitors, including necdin, Wnts, and Pref-1. We also showed that knockdown of necdin restores brown adipogenesis with downregulation of Pref-1 and Wnt10a expression (48), suggesting that Pref-1 acts downstream of necdin to inhibit brown adipogenesis. In this study, we showed that when treated with BMP7 for 3 days, followed by a standard differentiation protocol, the defect of brown adipogenesis in IRS-1KO cells was nearly completely restored. This was mediated, at least in part, by suppressing Pref-1 expression. Using truncations of the pref-1 promoter and ChIP analysis, we demonstrated that the promoter-proximal Smad binding element directly binds Smad1/Smad4 to repress Pref-1 gene expression. These data describe a previously unrecognized mechanism for regulation of Pref-1 gene expression by the BMP signaling pathway and establish a new role for BMP7 in rescuing brown adipogenesis in cells with insulin resistance.
It has been well documented that the R-Smads/Smad4 heterodimer is able to both activate and inhibit gene expression by recruiting either transcriptional coactivators (e.g., p300, CBP, P/CAF, SMIF, and Swift) or transcriptional corepressors (e.g., ski, SnoN, p107, NCoR, mSin3, and histone deacetylases [HDACs]) to the promoters of target genes (28). Of the nine putative SBEs in the 1.3-kb pref-1 promoter, SBE9 exhibited 100% homology among different species, suggesting a functional conservation of SBE9 throughout evolution. Indeed, in this study, we demonstrated that SBE9 functions as an inhibitory element to repress Pref-1 expression by recruiting the Smad1/4 complex. The close proximity of SBE9 to the previously identified negative regulatory element SAD of the pref-1 promoter, which binds a 63-kDa suppressor (45), suggests that these factors may act synergistically to repress Pref-1 gene expression in response to BMP7 and other extracellular stimuli. While it is still unclear whether these transcription factors can directly interact with each other and recruit additional corepressors, such studies are warranted for future investigations.
Potential cross talk between insulin and BMP signaling systems have been described in different cell types, although the exact roles of these interactions in the regulation of different biological processes remain to be further defined. It has been reported that a combination of insulin and glucose induced BMP-9 expression and processing in rat nonparenchymal liver cells, while insulin resistance severely reduced this effect in vivo (5). In addition, insulin can potentiate BMP2-induced osteogenic differentiation in rat spinal ligament cells via activation of the PI3K/Akt pathway (25). Interestingly, BMPs have also been reported to modulate insulin gene expression, proinsulin processing, and insulin secretion in pancreatic beta-cells (18). It has also been reported that BMP2 induces osteogenesis by directly stimulating PI3K and Akt activation via BMP receptor threonine/serine kinase, which in turn modulates Smad5 nuclear translocation. This raises the possibility that BMPs could compensate for insulin in PI3K/Akt activation (15, 16). In this study, we demonstrated that in multipotent C3H10T1/2 cells, BMP7 coordinately regulates a panel of insulin signaling components, including upregulation of IRS-1, PI3K-p85, Akt, Gab2/10/14, Erk1, and Map3k5 and downregulation of SOCS3, an inhibitor of insulin signaling. More importantly, BMP7 is able to rescue brown adipogenesis in cells with defective insulin signaling, partly via directly targeting the pref-1 promoter to inhibit its expression, thereby permitting the brown preadipocytes to undergo terminal differentiation. Interestingly, one of the mechanisms utilized by Pref-1 to inhibit adipocyte differentiation is antagonizing insulin/IGF-1 signaling, leading to reduced activation of both the Erk1/2 and Akt pathways (54, 61). Thus, there exists a dynamic interaction among the BMP, insulin, and Pref-1 signaling systems in the regulation of brown adipocyte differentiation.
Obesity and insulin resistance are the two hallmark features of the metabolic syndrome, which has become an epidemic and has created huge medical and social burdens to society. Given the tremendous capacity of brown fat to burn stored energy, increasing brown fat mass and function provides an attractive approach to the treatment of obesity and the metabolic syndrome. Interestingly, we show in this study that BMP7 can induce the expression of several insulin signaling components and is able to restore brown adipogenesis in cells with impaired insulin signaling. These data suggest that, in addition to its previously described brown adipogenic effect (49), BMP7 may also have the potential to improve insulin resistance, providing a new avenue for the cure of the metabolic syndrome. Obviously, many studies need to be done before therapeutic interventions become possible, but these findings provide grounds for the use of BMP7 or its molecular mimetics in the treatment of the metabolic syndrome.
Supplementary Material
Acknowledgments
This work was supported in part by grants R01 DK077097, R21 DK070722, and UL1 RR 025758-01 from NIH (Harvard Catalyst/The Harvard Clinical and Translational Science Center) and research grants from the Eli Lilly Research Foundation and Harvard Stem Cell Institute (to Y.-H.T.), by the Joslin Diabetes and Endocrinology Research Center (P30DK036836 from the NIDDK), the Danish Natural Science Research Council, the Novo Nordisk Foundation, and the Carlsberg Foundation (to K.K.).
We acknowledge A. Lassar (Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School) for providing the Smad expression constructs and H. S. Sul for providing the pref-1 promoter vector. We thank C. R. Kahn for input on initiation of this project; S. Russell for helpful discussions on pref-1 promoter sequences; J. Schroeder, Y.-C. Lai, and J. Li for technical assistance; and K. Townsend and L. McDougall for critical reading of the manuscript.
We declare no competing financial interests.
Footnotes
Published ahead of print on 28 June 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baudry, A., Z. Z. Yang, and B. A. Hemmings. 2006. PKBalpha is required for adipose differentiation of mouse embryonic fibroblasts. J. Cell Sci. 119:889-897. [DOI] [PubMed] [Google Scholar]
- 2.Bowers, R. R., J. W. Kim, T. C. Otto, and M. D. Lane. 2006. Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proc. Natl. Acad. Sci. U. S. A. 103:13022-13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowers, R. R., and M. D. Lane. 2007. A role for bone morphogenetic protein-4 in adipocyte development. Cell Cycle 6:385-389. [DOI] [PubMed] [Google Scholar]
- 4.Cannon, B., and J. Nedergaard. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84:277-359. [DOI] [PubMed] [Google Scholar]
- 5.Caperuto, L. C., G. F. Anhe, T. D. Cambiaghi, E. H. Akamine, B. D. do Carmo, J. Cipolla-Neto, R. Curi, and S. Bordin. 2008. Modulation of bone morphogenetic protein-9 expression and processing by insulin, glucose, and glucocorticoids: possible candidate for hepatic insulin-sensitizing substance. Endocrinology 149:6326-6335. [DOI] [PubMed] [Google Scholar]
- 6.Cook, J. R., and R. K. Semple. 2010. Hypoadiponectinemia—cause or consequence of human “insulin resistance”? J. Clin. Endocrinol. Metab. 95:1544-1554. [DOI] [PubMed] [Google Scholar]
- 7.Cornier, M. A., D. Dabelea, T. L. Hernandez, R. C. Lindstrom, A. J. Steig, N. R. Stob, R. E. Van Pelt, H. Wang, and R. H. Eckel. 2008. The metabolic syndrome. Endocr. Rev. 29:777-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craft, S. 2007. Insulin resistance and Alzheimer's disease pathogenesis: potential mechanisms and implications for treatment. Curr. Alzheimer Res. 4:147-152. [DOI] [PubMed] [Google Scholar]
- 9.Cypess, A. M., S. Lehman, G. Williams, I. Tal, D. Rodman, A. B. Goldfine, F. C. Kuo, E. L. Palmer, Y. H. Tseng, A. Doria, G. M. Kolodny, and C. R. Kahn. 2009. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360:1509-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmer, S. R. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4:263-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fasshauer, M., J. Klein, K. M. Kriauciunas, K. Ueki, M. Benito, and C. R. Kahn. 2001. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol. Cell. Biol. 21:319-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fasshauer, M., J. Klein, U. Lossner, and R. Paschke. 2002. Isoproterenol is a positive regulator of the suppressor of cytokine signaling-3 gene expression in 3T3-L1 adipocytes. J. Endocrinol. 175:727-733. [DOI] [PubMed] [Google Scholar]
- 13.Fasshauer, M., J. Klein, K. Ueki, K. M. Kriauciunas, M. Benito, M. F. White, and C. R. Kahn. 2000. Essential role of insulin receptor substrate-2 in insulin stimulation of glut4 translocation and glucose uptake in brown adipocytes. J. Biol. Chem. 275:25494-25501. [DOI] [PubMed] [Google Scholar]
- 14.Gesta, S., Y. H. Tseng, and C. R. Kahn. 2007. Developmental origin of fat: tracking obesity to its source. Cell 131:242-256. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh-Choudhury, N., S. L. Abboud, B. Chandrasekar, and C. G. Ghosh. 2003. BMP-2 regulates cardiomyocyte contractility in a phosphatidylinositol 3 kinase-dependent manner. FEBS Lett. 544:181-184. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh-Choudhury, N., S. L. Abboud, R. Nishimura, A. Celeste, L. Mahimainathan, and G. G. Choudhury. 2002. Requirement of BMP-2-induced phosphatidylinositol 3-kinase and Akt serine/threonine kinase in osteoblast differentiation and Smad-dependent BMP-2 gene transcription. J. Biol. Chem. 277:33361-33368. [DOI] [PubMed] [Google Scholar]
- 17.Goldfine, A. B., S. Crunkhorn, M. Costello, H. Gami, E. J. Landaker, M. Niinobe, K. Yoshikawa, C. R. Kahn, D. Lo, A. Warren, J. Jimenez-Chillaron, and M. E. Patti. 2006. Necdin and E2F4 are modulated by rosiglitazone therapy in diabetic human adipose and muscle tissue. Diabetes 55:640-650. [DOI] [PubMed] [Google Scholar]
- 18.Goulley, J., U. Dahl, N. Baeza, Y. Mishina, and H. Edlund. 2007. BMP4-BMPR1A signaling in beta cells is required for and augments glucose-stimulated insulin secretion. Cell Metab. 5:207-219. [DOI] [PubMed] [Google Scholar]
- 19.Hata, K., R. Nishimura, F. Ikeda, K. Yamashita, T. Matsubara, T. Nokubi, and T. Yoneda. 2003. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor gamma during bone morphogenetic protein 2-induced adipogenesis. Mol. Biol. Cell 14:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogan, B. L. 1996. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10:1580-1594. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, I. R., S. P. Kim, M. Kabir, and R. N. Bergman. 2007. Metabolic syndrome, hyperinsulinemia, and cancer. Am. J. Clin. Nutr. 86:s867-s871. [DOI] [PubMed] [Google Scholar]
- 22.Jin, W., T. Takagi, S. N. Kanesashi, T. Kurahashi, T. Nomura, J. Harada, and S. Ishii. 2006. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev. Cell 10:461-471. [DOI] [PubMed] [Google Scholar]
- 23.Kim, D. W., and A. B. Lassar. 2003. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol. Cell. Biol. 23:8704-8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopelman, P. G. 2000. Obesity as a medical problem. Nature 404:635-643. [DOI] [PubMed] [Google Scholar]
- 25.Li, H., D. Liu, C. Q. Zhao, L. S. Jiang, and L. Y. Dai. 2008. Insulin potentiates the proliferation and bone morphogenetic protein-2-induced osteogenic differentiation of rat spinal ligament cells via extracellular signal-regulated kinase and phosphatidylinositol 3-kinase. Spine 33:2394-2402. [DOI] [PubMed] [Google Scholar]
- 26.Liu, F., F. Ventura, J. Doody, and J. Massague. 1995. Human type II receptor for bone morphogenic proteins (BMPs): extension of the two-kinase receptor model to the BMPs. Mol. Cell. Biol. 15:3479-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massagué, J., and Y. G. Chen. 2000. Controlling TGF-beta signaling. Genes Dev. 14:627-644. [PubMed] [Google Scholar]
- 28.Massague, J., J. Seoane, and D. Wotton. 2005. Smad transcription factors. Genes Dev. 19:2783-2810. [DOI] [PubMed] [Google Scholar]
- 29.Massague, J., and F. Weis-Garcia. 1996. Serine/threonine kinase receptors: mediators of transforming growth factor beta family signals. Cancer Surv. 27:41-64. [PubMed] [Google Scholar]
- 30.Miyazono, K., Y. Kamiya, and M. Morikawa. 2010. Bone morphogenetic protein receptors and signal transduction. J. Biochem. 147:35-51. [DOI] [PubMed] [Google Scholar]
- 31.Mootha, V. K., C. Handschin, D. Arlow, X. Xie, J. St Pierre, S. Sihag, W. Yang, D. Altshuler, P. Puigserver, N. Patterson, P. J. Willy, I. G. Schulman, R. A. Heyman, E. S. Lander, and B. M. Spiegelman. 2004. Err{alpha} and Gabpa/b specify PGC-1{alpha}-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U. S. A. 101:6570-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nedergaard, J., T. Bengtsson, and B. Cannon. 2007. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293:E444-E452. [DOI] [PubMed] [Google Scholar]
- 33.Otto, T. C., R. R. Bowers, and M. D. Lane. 2007. BMP-4 treatment of C3H10T1/2 stem cells blocks expression of MMP-3 and MMP-13. Biochem. Biophys. Res. Commun. 353:1097-1104. [DOI] [PubMed] [Google Scholar]
- 34.Peng, X.-D., P.-Z. Xu, M.-L. Chen, A. Hahn-Windgassen, J. Skeen, J. Jacobs, D. Sundararajan, W. S. Chen, S. E. Crawford, K. G. Coleman, and N. Hay. 2003. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 17:1352-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 36.Rangwala, S. M., X. Li, L. Lindsley, X. Wang, S. Shaughnessy, T. G. Daniels, J. Szustakowski, N. R. Nirmala, Z. Wu, and S. C. Stevenson. 2007. Estrogen-related receptor alpha is essential for the expression of antioxidant protection genes and mitochondrial function. Biochem. Biophys. Res. Commun. 357:231-236. [DOI] [PubMed] [Google Scholar]
- 37.Rebbapragada, A., H. Benchabane, J. L. Wrana, A. J. Celeste, and L. Attisano. 2003. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol. Cell. Biol. 23:7230-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosen, E. D., and O. A. MacDougald. 2006. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell. Biol. 7:885-896. [DOI] [PubMed] [Google Scholar]
- 39.Ross, S. E., N. Hemati, K. A. Longo, C. N. Bennett, P. C. Lucas, R. L. Erickson, and O. A. MacDougald. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289:950-953. [DOI] [PubMed] [Google Scholar]
- 40.Saito, M., Y. Okamatsu-Ogura, M. Matsushita, K. Watanabe, T. Yoneshiro, J. Nio-Kobayashi, T. Iwanaga, M. Miyagawa, T. Kameya, K. Nakada, Y. Kawai, and M. Tsujisaki. 2009. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreiber, S. N., R. Emter, M. B. Hock, D. Knutti, J. Cardenas, M. Podvinec, E. J. Oakeley, and A. Kralli. 2004. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. U. S. A. 101:6472-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz, T. J., and Y. H. Tseng. 2009. Emerging role of bone morphogenetic proteins in adipogenesis and energy metabolism. Cytokine Growth Factor Rev. 20:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seale, P., S. Kajimura, W. Yang, S. Chin, L. M. Rohas, M. Uldry, G. Tavernier, D. Langin, and B. M. Spiegelman. 2007. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 6:38-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi, Y., and J. Massague. 2003. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113:685-700. [DOI] [PubMed] [Google Scholar]
- 45.Smas, C. M., D. Kachinskas, C. M. Liu, X. Xie, L. K. Dircks, and H. S. Sul. 1998. Transcriptional control of the pref-1 gene in 3T3-L1 adipocyte differentiation. Sequence requirement for differentiation-dependent suppression. J. Biol. Chem. 273:31751-31758. [DOI] [PubMed] [Google Scholar]
- 46.Sottile, V., and K. Seuwen. 2000. Bone morphogenetic protein-2 stimulates adipogenic differentiation of mesenchymal precursor cells in synergy with BRL 49653 (rosiglitazone). FEBS Lett. 475:201-204. [DOI] [PubMed] [Google Scholar]
- 47.Sul, H. S. 2009. Minireview: Pref-1: role in adipogenesis and mesenchymal cell fate. Mol. Endocrinol. 23:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tseng, Y. H., A. J. Butte, E. Kokkotou, V. K. Yechoor, C. M. Taniguchi, K. M. Kriauciunas, A. M. Cypess, M. Niinobe, K. Yoshikawa, M. E. Patti, and C. R. Kahn. 2005. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat. Cell Biol. 7:601-611. [DOI] [PubMed] [Google Scholar]
- 49.Tseng, Y. H., E. Kokkotou, T. J. Schulz, T. L. Huang, J. N. Winnay, C. M. Taniguchi, T. T. Tran, R. Suzuki, D. O. Espinoza, Y. Yamamoto, M. J. Ahrens, A. T. Dudley, A. W. Norris, R. N. Kulkarni, and C. R. Kahn. 2008. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454:1000-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tseng, Y. H., K. M. Kriauciunas, E. Kokkotou, and C. R. Kahn. 2004. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol. Cell. Biol. 24:1918-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tseng, Y. H., K. Ueki, K. M. Kriauciunas, and C. R. Kahn. 2002. Differential roles of insulin receptor substrates in the anti-apoptotic function of insulin-like growth factor-1 and insulin. J. Biol. Chem. 277:31601-31611. [DOI] [PubMed] [Google Scholar]
- 52.Uldry, M., W. Yang, J. St Pierre, J. Lin, P. Seale, and B. M. Spiegelman. 2006. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3:333-341. [DOI] [PubMed] [Google Scholar]
- 53.van Marken Lichtenbelt, W. D., J. W. Vanhommerig, N. M. Smulders, J. M. Drossaerts, G. J. Kemerink, N. D. Bouvy, P. Schrauwen, and G. J. Teule. 2009. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360:1500-1508. [DOI] [PubMed] [Google Scholar]
- 54.Villena, J. A., C. S. Choi, Y. Wang, S. Kim, Y. J. Hwang, Y. B. Kim, G. Cline, G. I. Shulman, and H. S. Sul. 2008. Resistance to high-fat diet-induced obesity but exacerbated insulin resistance in mice overexpressing preadipocyte factor-1 (Pref-1): a new model of partial lipodystrophy. Diabetes 57:3258-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Virtanen, K. A., M. E. Lidell, J. Orava, M. Heglind, R. Westergren, T. Niemi, M. Taittonen, J. Laine, N. J. Savisto, S. Enerback, and P. Nuutila. 2009. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360:1518-1525. [DOI] [PubMed] [Google Scholar]
- 56.White, M. F. 1998. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell. Biochem. 182:3-11. [PubMed] [Google Scholar]
- 57.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115-124. [DOI] [PubMed] [Google Scholar]
- 58.Xu, J., and K. Liao. 2004. Protein kinase B/AKT 1 plays a pivotal role in insulin-like growth factor-1 receptor signaling induced 3T3-L1 adipocyte differentiation. J. Biol. Chem. 279:35914-35922. [DOI] [PubMed] [Google Scholar]
- 59.Xu, L., C. Alarcon, S. Col, and J. Massague. 2003. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J. Biol. Chem. 278:42569-42577. [DOI] [PubMed] [Google Scholar]
- 60.Yun, S. J., E. K. Kim, D. F. Tucker, C. D. Kim, M. J. Birnbaum, and S. S. Bae. 2008. Isoform-specific regulation of adipocyte differentiation by Akt/protein kinase Balpha. Biochem. Biophys. Res. Commun. 371:138-143. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, H., J. Noohr, C. H. Jensen, R. K. Petersen, E. Bachmann, B. Teisner, L. K. Larsen, S. Mandrup, and K. Kristiansen. 2003. Insulin-like growth factor-1/insulin bypasses Pref-1/FA1-mediated inhibition of adipocyte differentiation. J. Biol. Chem. 278:20906-20914. [DOI] [PubMed] [Google Scholar]
- 62.Zingaretti, M. C., F. Crosta, A. Vitali, M. Guerrieri, A. Frontini, B. Cannon, J. Nedergaard, and S. Cinti. 2009. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 23:3113-3120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.