Abstract
Biological methylation is a fundamental enzymatic reaction for a variety of substrates in multiple cellular processes. Mammalian N6amt1 was thought to be a homologue of bacterial N6-adenine DNA methyltransferases, but its substrate specificity and physiological importance remain elusive. Here, we demonstrate that N6amt1 functions as a protein methyltransferase for the translation termination factor eRF1 in mammalian cells both in vitro and in vivo. Mass spectrometry analysis indicated that about 70% of the endogenous eRF1 is methylated at the glutamine residue of the conserved GGQ motif. To address the physiological significance of eRF1 methylation, we disrupted the N6amt1 gene in the mouse. Loss of N6amt1 led to early embryonic lethality. The postimplantation development of mutant embryos was impaired, resulting in degeneration around embryonic day 6.5. This is in contrast to what occurs in Escherichia coli and Saccharomyces cerevisiae, which can survive without the N6amt1 homologues. Thus, N6amt1 is the first glutamine-specific protein methyltransferase characterized in vivo in mammals and methylation of eRF1 by N6amt1 might be essential for the viability of early embryos.
Nucleic acids, proteins, carbohydrates, and lipids, as well as a body of small molecules, are subject to methylation in a wide variety of biological contexts (3). The majority of methylation reactions are catalyzed by S-adenosylmethionine (AdoMet)-dependent methyltransferases (MTases). These enzymes ubiquitously exist in species from all three domains of life.
Methylation of DNA occurs on one of two bases: cytosine or adenine (19). In prokaryotes, adenine methylation is as widespread as cytosine methylation. In contrast, eukaryotic genomes are devoid of adenine methylation or this type of methylation is too rare to be detected (23, 26). Intriguingly, two putative N6-adenine DNA MTases, named N6amt1 and N6amt2, are encoded in the mouse and human genomes. In addition to the conserved AdoMet-binding signature motif GXGXG and other sequence elements, they possess the NPPY motif characteristic of the N6-adenine or N4-cytosine DNA MTases in bacteria (6, 14). N6amt1 was thus proposed as an AdoMet-dependent DNA MTase, although no evidence had been provided that N6amt1 could methylate DNA (23).
No functional clue for N6amt1 existed until two groups independently identified Escherichia coli HemK, distantly related to N6amt1, as a protein MTase for polypeptide release factors RF1 and RF2 (8, 17). The HemK gene was initially discovered in a genetic screen for heme biosynthesis mutants (18), although subsequent studies revealed no direct involvement in heme metabolism. The presence of an NPPY motif, thought to be restricted to members of the adenine and cytosine amino methyltransferases, led to the suggestion that HemK could be an AdoMet-dependent DNA MTase (2). However, a series of genetic and biochemical experiments finally revealed that HemK methylates the side-chain amide group of a glutamine residue in the universally conserved tripeptide motif GGQ of the two release factors in E. coli (8, 17). Methylation of the release factors ensures efficient translation termination and release of newly synthesized peptide from the ribosome (16). Similarly, the yeast HemK homologue, YDR140w (Mtq2p), was confirmed to methylate the eukaryotic release factor eRF1 on a corresponding glutamine residue (9, 22). More recently, the human homologue N6amt1 (HemK2) was reported to methylate release factor 1 (eRF1) in vitro (5).
We initially sought to characterize the function of N6amt1 as a potential DNA adenine MTase. Interestingly, the human N6amt1 gene is located on chromosome 21q21.3, a critical region for Down syndrome (1, 20). In this study, we report the identification of murine N6amt1 as a glutamine-specific MTase of eRF1 both in vitro and in vivo. Mammalian eRF1, the only mammalian release factor, is indeed methylated at the glutamine residue of the GGQ motif. Inactivation of the N6amt1 gene by targeted disruption led to embryonic lethality in the mouse. These data confirm that N6amt1 functions as a protein MTase in mammals and indicate that modulation of the eRF1 activity by N6amt1-mediated glutamine methylation might be essential for embryo viability.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain DH5α was used for plasmid extraction, and BL21 CodonPlus (DE3)-RP was used for protein expression. The mouse N6amt1 gene was amplified from the kidney cDNA pool (Clontech) using the sense primer 5′-ACCATGGCGGCGCCGAGTGTCCCCACGC-3′ (NcoI site underlined) and antisense primer 5′-AGCGGCCGCCTAGGACTTGCTGAACCTGAGGACT-3′ (NotI underlined), and the mouse Trm112 gene was amplified from embryonic stem (ES) cell cDNA using the sense primer 5′-GAAGATCTATGAAACTGCTCACCCAC-3′ (BglII underlined) and antisense primer 5′-TGTCGACGCCGTCTCAGTCTCCTCATC-3′ (SalI underlined). The N6amt1 and Trm112 open reading frames (ORFs) were inserted into the multiple cloning sites 1 (MCS1) and MCS2 of the coexpression vector pRSFDuet-1. pRSET-eRF1 and pGEX-2T-eRF3 plasmids were kindly provided by B. A. Hemmings and P. Cron. eRF1 mutant plasmids were constructed using QuikChange site-directed mutagenesis kit (Stratagene) according to the instruction manual and the following primers were used: Q185I sense, 5′-AAGAAACACGGTAGAGGAGGTATTTCAGCCTTGCGTTTTGCCC-3′, and antisense, 5′-GGGCAAAACGCAAGGCTGAAATACCTCCTCTACCGTGTTTCTT-3′; and Q185N sense, 5′-GAAACACGGTAGAGGAGGTAACTCAGCCTTGCGTTTTGC-3′, and antisense 5′-GCAAAACGCAAGGCTGAGTTACCTCCTCTACCGTGTTTC-3′. N6amt1, Trm112, eRF1, and eRF3 ORFs were also subcloned into pcDNA vectors for mammalian cell expression.
The N6AMT1 knockdown construct was prepared based on the drug-inducible system (25). Briefly, an N6AMT1 short hairpin RNA (shRNA) duplex containing the small interfering RNA (siRNA) sequence (5′-GTTGATCTTCTGGTGTTTAAT-3′) was inserted into pSUPER vector, and the confirmed H1-shN6AMT1 cassette was subcloned into pLVUT-tTRKRAB, resulting in pLVUT-shN6AMT1.
Protein purification.
N6amt1, Trm112, eRF1, and eRF3 (amino acids 136 to 633) were expressed from the E. coli BL21 CodonPlus(DE3)-RP strain. Ni-nitrilotriacetic acid (NTA) resin was used for copurification of 6×His-N6amt1 and FLAG-Trm112 and the purification of 6×His-eRF1. Glutathione S-transferase (GST)-eRF3 was purified on glutathione Sepharose 4B (Amersham Biosciences) according to the standard protocol.
Methylation assay.
Methylation assays were performed in buffer MET (10 mM Tris-HCl, pH 7.6, 50 mM KCl, 10 mM Mg acetate, 6 mM β-mercaptoethanol) and 7.7 μM [methyl-3H]AdoMet (0.80 Ci/mmol). GTP was added at a final concentration of 1 mM. Each reaction mixture contained 4 pmol of N6amt1, 4 pmol of Trm112, 10 pmol of eRF3, and 10 pmol of eRF1 or mutants. The reaction mixture was incubated at 37°C for 1 h before being terminated by adding an equal volume of 20% cold trichloroacetic acid (TCA) or 6× SDS loading buffer.
Scintillation counting.
After the incubation, the proteins were precipitated with cold TCA as described previously (17). Briefly, an equal volume of 20% cold TCA was added and the mixture was incubated on ice for 30 min, followed by centrifugation at 13,000 rpm for 15 min to pellet the proteins. The pellet was washed twice with 10% cold TCA and once with cold acetone. It was then dissolved in 50 μl of 100 mM Tris-HCl, pH 8.8, plus 100 mM NaCl. Three hundred microliters of the scintillation liquid was added to each tube, and the scintillation counts were read on a Beckman LS 500 scintillation counter.
Fluorography detection.
The methylation reaction terminated by the addition of 6× SDS loading buffer was separated by 15% SDS-PAGE. The proteins were visualized by Coomassie blue staining and then destained. The gel was agitated in En3Hance (NEN Life Science) and 10% cold polyethylene glycol (PEG) solution sequentially and then dried and exposed to Kodak Biomax MS film for 1 day at −80°C.
Generation of the N6AMT1 knockdown stable cell line in HEK-293T cells.
The N6AMT1 knockdown stable cell line was generated following the drug-inducible systems (25). Briefly, pLVUT-shN6 was cotransfected with VSVG and PAX2 vector into HEK-293T cells to produce the virus. HEK-293T cells were infected by the virus to generate the knockdown stable cell line. The N6AMT1 expression in the generated cell line N6-A9 and control cell line H1-B4 was verified by real-time PCR with the forward primer 5′-GGTCTGGTGTAGTATCTGC-3′ and reverse primer 5′-TTCGGTCAATCTTGGTAGC-3′. The RNA interference (RNAi) effect was examined by transfecting FLAG-N6AMT1 to the cells and testing N6AMT1 expression.
Immunoprecipitation.
Endogenous eRF1 was immunoprecipitated by anti-eRF1 polyclonal antibody (Abcam). Overexpressed eRF1-FLAG was immunoprecipitated by anti-FLAG M2 affinity gel (Sigma). All of the immunoprecipitation steps were performed on ice or at 4°C. A total of 2 × 107 HEK-293T cells were lysed with 1 ml of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NCl, 1 mM EDTA, 0.1% SDS, 0.5% deoxycholic acid, 1% Triton X-100) and then centrifuged at 12,000 rpm for 10 min. The supernatant was incubated with 5 μg of anti-eRF1 polyclonal antibody and 75 μl of protein A beads (GE Healthcare) for 4 h. The beads were washed in RIPA buffer twice, and the endogenous eRF1 was eluted with 2× SDS loading buffer without dithiothreitol (DTT) or β-mercaptoethanol. For immunoprecipitation of eRF1-FLAG, 2 × 107 transfected HEK-293T cells and 50 μl of anti-FLAG M2 affinity gel were used. The bound eRF1-FLAG was eluted with 75 μl of 10-mg/ml FLAG peptide. The eluted proteins were separated by 15% SDS-PAGE and visualized by Coomassie blue staining. The eRF1 band was excised from the gel for mass spectrometry (MS) analysis.
MALDI-TOF MS, LC-MS/MS, and data analysis.
All matrix-assisted laser desorption (MALDI) MS data were acquired in a positive-ion reflector mode on an ABI 4800 MALDI-tandem time of flight (TOF/TOF) mass spectrometry instrument. The matrix used for all mass spectra was a-cyano-4-hydroxycinnamic acid (CHCA), as a saturated solution in 1% trifluoroacetic acid (TFA) in acetonitrile-water (1:1 [vol/vol]). Samples for MS were prepared by using 1 μl of peptide solution mixed with 1 μl of the matrix solution. In the TOF/TOF experiments, a 2-kV MS/MS operating mode was used, collision-induced dissociation (CID) using air was turned off, and metastable suppression was enabled. External mass calibration was performed with a mixture of peptide standards provided by ABI.
The liquid chromatography (LC)-MS/MS was based on our previous work (13). Briefly, chromatography was performed using a Surveyor LC system (Thermo Finnigan) on an analytical C18 column (reverse phase [RP], 5 μm, 150 μm by 100 mm; Column Technology, Inc.). The pump flow rate was split to achieve a flow rate of 2 μl/min. The tryptic peptide mixtures were eluted using a gradient of 0 to 35% elution buffer (0.1% fluorescent antibody [FA]/acetonitrile [ACN]) over 120 min. The MS/MS was performed on a LTQ linear ion trap mass spectrometer (Thermo Finnigan). The mass spectrometer was set so that one full MS scan was followed by 10 MS/MS scans of the 10 most intense ions. Dynamic exclusion was set at a repeat count of 2, repeat duration of 30 s, and exclusion duration of 120 s. The acquired MS/MS spectra were automatically searched against the eRF1 protein sequence using the TurboSEQUEST program in the BioWorks 3.2 software package. Database searches were performed by applying the precursor ion m/z tolerances of ±3 Da and fragment ion m/z tolerances of ±1 Da. Cysteine residues were statically modified by 57 Da due to carboxyamidomethylation. Dynamic modifications were permitted to allow the detection of oxidized Met (+16) and methylated Gln (+14). The maximum number of internal cleavage sites was set to 1. Peptide identifications were filtered with the following criteria: Xcorr of ≥1.9 with charge state 1+, Xcorr of ≥2.2 with charge state 2+, and Xcorr of ≥3.75 with charge state 3+. In addition, ΔCn cutoff values were ≥0.1.
In-gel digestion.
Protein bands were excised from the one-dimensional (1D) gel and transferred to 1.5-ml siliconized centrifuge tubes. The excised gel pieces were destained in 30% acetonitrile (ACN)-100 mM NH4HCO3 (vol/vol). After being lyophilized, the gel pieces were reduced by dithiothreitol (DTT) and alkylated by iodoacetamide (IAA). The gel particles were washed with 100 mM NH4HCO3 and completely dried in a Speed Vac. A 12.5-ng/μl concentration of trypsin or chymotrypsin in 50 mM NH4HCO3 was added to cover the gel pieces. Digestion was performed overnight at 37°C. The peptides were extracted with 0.1% TFA-80% acetonitrile, dried by vacuum centrifugation, and stored at −80°C for further analysis by mass spectrometry.
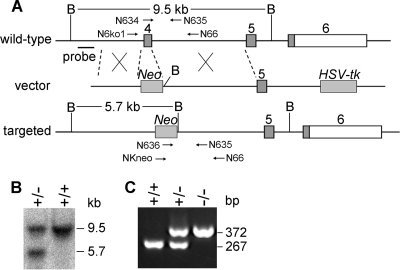
Targeting vector and generation of N6amt1 null mice.
To generate the N6amt1 targeting vector, DNA fragments for the 5′ and 3′ homology arms were amplified from mouse (129/SvEv) genomic DNA by PCR. The left arm consists of a 1.0-kb region immediately upstream of the fourth exon, and the right arm is a 3.7-kb fragment immediately downstream of the fourth exon. The two arms were cloned into the pPNT vector in NotI-XhoI and BamHI-KpnI sites, respectively, and confirmed by sequencing. The primers used for the amplification of the left and right arms are as follows: left arm, sense, 5′-ATAAGAATGCGGCCGCAAAGTTTGCAAGTCCAATGGGCCTC-3′ (NotI is underlined), and antisense 5′-CCGCTCGAGTGAGGGTAGCAATAATGCAGGTTAG-3′ (XhoI is underlined); right arm, sense 5′-CGGGATCCAGACTCACAACAGAACTCAGTCTTT-3′ (BamHI is underlined), and antisense, 5′-GGGGTACCTTTACGGTAACTAAGTAGAACAGCC-3′ (KpnI is underlined).
The targeting vector linearized with NotI was electroporated into CJ7 ES cells (129/S6/SVJ derived), which were subsequently cultured in the presence of G418 and ganciclovir on mitotically inactivated mouse embryonic fibroblast (MEF) cells. Resistant ES cell clones were picked 6 to 8 days after drug selection. Three targeted ES clones were identified by Southern blotting analysis using the 5′ external probe. The 5′ external probe was prepared by PCR (sense, 5′-GGTGGCCCTGAACTAAACTGATTTG-3′; antisense, 5′-ACACGGACTTTATTCCCACTGGGGC-3′). Positive ES clones identified by Southern blotting were microinjected into the C57BL/6J blastocysts and then transferred into pseudopregnant foster mothers. Highly chimeric male mice were bred with C57BL/6J females to generate heterozygous N6amt1+/− mice. Mouse genotypes were identified by Southern analysis of tail DNA using a 5′ external probe and by PCR using the following primers: N634, 5′-CTATGTAGTGACTCCGCCTGAAGAG-3′; N635, 5′-ATTTAGGGTAATGGACCCGTTTAAG-3′; and N636, 5′-GTACTGTGGTTTCCAAATGTGTCAG-3′. Nested PCR was used for the genotyping of blastocysts. Primers N6ko1, NKneo, and N66 were used for the 1st round of PCR, and primers N634, N635, and N636 were used for the 2nd round of PCR. The sequences of these primers are as follows: N6ko1, 5′-GATTATCTTTCGACCTCCATTTGTGTACAG-3′; NKneo, 5′-CAGGACATAGCGTTGGCTACCCGTG-3′; and N66, 5′-TTATTACTCTATGTCCCTTCACCTT-3′.
In vitro embryo culture.
Heterozygous N6amt1+/− mice were intercrossed, and females positive for the presence of the vaginal plug were sacrificed at 3.5 days postcoitus (dpc), and blastocysts were collected by uterine flush and grown on 0.1% gelatin-coated dishes in ES cell medium with antibiotics and leukemia inhibitory factor. Embryo morphology was observed by phase-contrast microscopy, and blastocyst outgrowths were photographed each day. After 9 days of culture in vitro, outgrowths were scraped off and lysed for genotyping as described above.
Generation of anti-methyl-eRF1 and anti-N6AMT1 antibodies.
Antibody against methyl-eRF1 was raised in rabbit with a biotinylated peptide, HGRGGQSALRC, in which the glutamine residue is monomethylated (Dragonfly Sciences, Inc). The antibody was affinity purified and tested for specificity by enzyme-linked immunosorbent assay (ELISA). Antibody against N6AMT1 was raised in rabbit with full-length human N6AMT1 protein.
Histological analysis.
Embryos from N6amt1+/− intercrossed mice were processed for histological analysis as described previously (12). Briefly, pregnant females of the indicated time were sacrificed. Whole deciduae were dissected, fixed in paraformaldehyde (PFA), dehydrated, oriented with respect to the antimesometrial-mesometrial axis, and embedded in paraffin. Sagittal sections were cut and stained with hematoxylin and eosin (H&E).
RESULTS
Identification of Gln185 methylation of eRF1 in HEK-293T cells.
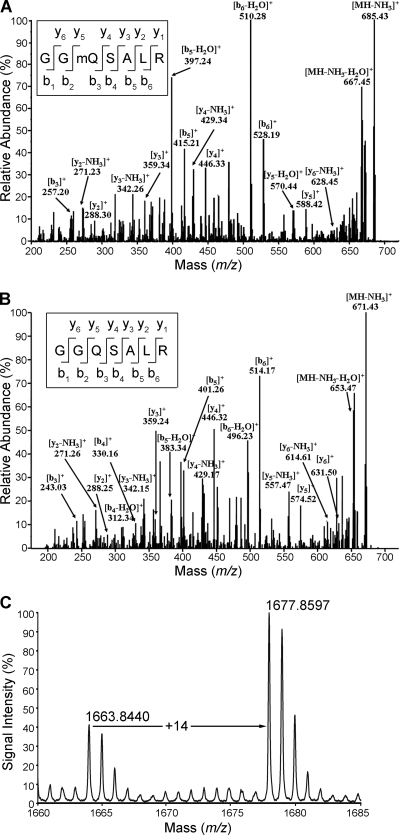
A recent report indicated that human recombinant N6amt1 can methylate release factor 1 (eRF1) in vitro (5). However, whether methylation is present on eRF1 in vivo in mammalian cells has not been demonstrated. Since in E. coli and Saccharomyces cerevisiae methylation of release factors occurs at the glutamine residue of the conserved GGQ motif (4, 9), we set out to examine the in vivo methylation status of Gln185 of the eRF1 GGQ motif in HEK-293T cells. To this end, endogenous eRF1 was isolated by immunoprecipitation using anti-eRF1 polyclonal antibody, separated by SDS-PAGE, excised from the gel, and cleaved with trypsin. A complete tryptic digest of eRF1 is predicted to produce a heptapeptide with the amino acid sequence GGQSALR (GGQ motif underlined) at m/z 702.34, if the Q is methylated. The predicted peak was indeed detected by the mass spectrometry identification. Fragmentation of the selected ions indicated that Gln185 was attached with a moiety of 14 Da, based on the mass difference of 142.09 m/z at Gln185 (Fig. 1 A) between the y4 and y5 ions with 14-Da shift from a normal Gln residue. To confirm that the 14-Da shift was caused by posttranslational methylation of the glutamine residue, recombinant eRF1 purified from E. coli was analyzed with electrospray ionization (ESI)-MS/MS as a control. The fragmentation ions of a heptapeptide at m/z 688.32 corresponding to the unmodified form of the heptapeptide were obtained (Fig. 1B). The difference (128.20 m/z) between the y4 and y5 ions was identical to the molecular weight of a glutamine residue expected of recombinant eRF1 lacking any conjugation. Further comparison of multiple b and y ions detected in the spectra of peptides from endogenous and recombinant sources substantiated the presence of monomethylation in endogenous eRF1 (Table 1). The m/z values of ions containing Gln185 (b3, b5, b6, y4, and y5) from the endogenous eRF1 have 14-m/z shift higher than that of their corresponding ions from the recombinant eRF1. To ascertain the assignment of glutamine methylation, we synthesized Gln185-methylated and Gln185-unmethylated heptapeptides. The MS/MS spectra of these two synthetic peptides were almost identical to those of the peptides derived from endogenous and recombinant eRF1, respectively (see Fig. S1 in the supplemental material). We therefore conclude that eRF1 is methylated at Gln185 in HEK-239T cells.
FIG. 1.
Identification of Gln185 methylation of eRF1 in vivo in mammalian cells. (A) ESI-MS/MS spectrum of methylated 702.34-Da fragments [heptapeptide GG(mQ)SALR, amino acids 183 to 189] from endogenous eRF1. eRF1 was purified by immunoprecipitation from the HEK-293T cell extract using anti-eRF1 polyclonal antibodies, separated by PAGE, and subjected to in-gel digestion with trypsin prior to ESI-MS/MS. The fragmentation pattern established the heptapeptide sequence indicated. (B) ESI-MS/MS spectrum of methylation-free 688.32-Da fragments from recombinant eRF1 purified from E. coli. (C) MALDI-TOF MS spectrum of endogenous eRF1 of HEK-293T cells. eRF1 was affinity purified with specific antibodies. The peak at m/z 1,663.84 is from the unmethylated peptide (sequence TVDLPKKHGRGGQSAL, amino acids 173 to 188) generated from chymotrypsin digestion. The peak at m/z 1,677.86 represents the methylated peptide.
TABLE 1.
Amino acid sequence of the GGQ-containing heptapeptide of eRF1
| Amino acid |
m/za |
|||
|---|---|---|---|---|
| Recombinant eRF1 |
Endogenous eRF1 |
|||
| b ion | y ion | b ion | y ion | |
| Gly | 58.03 | 58.03 | ||
| Gly | 115.05 | 631.50 | 115.05 | 645.37 |
| Glnb | 243.03 | 574.52 | 257.20 | 588.42 |
| Ser | 330.16 | 446.32 | 344.16 | 446.33 |
| Ala | 401.26 | 359.24 | 415.21 | 359.34 |
| Leu | 514.17 | 288.25 | 528.19 | 288.30 |
| Arg | 175.12 | 175.12 | ||
m/z values for the missing ions in the MS detection are in italics.
The residue is methylated.
To assess the methylation level of eRF1 in vivo, we performed matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) on immunoprecipitated eRF1 from HEK-293T cells. Since tryptic digestion produced another peptide (HNYVR) with the same m/z value (688.32) as the unmethylated heptapeptide GGQSALR, chymotrypsin digestion was performed to obtain Gln185-containing peptides with a unique mass peak. MALDI-TOF MS analysis revealed two peptide ions at m/z 1,663.84 and 1,677.86, expected to represent, respectively, the unmethylated and methylated forms with the TVDLPKKHGRGGQSAL sequence (Fig. 1C). The result of quantification by comparing the peak intensity indicated that the methylated form of eRF1 accounted for ∼70% in the total protein.
N6amt1 methylates Gln185 of eRF1 in vitro.
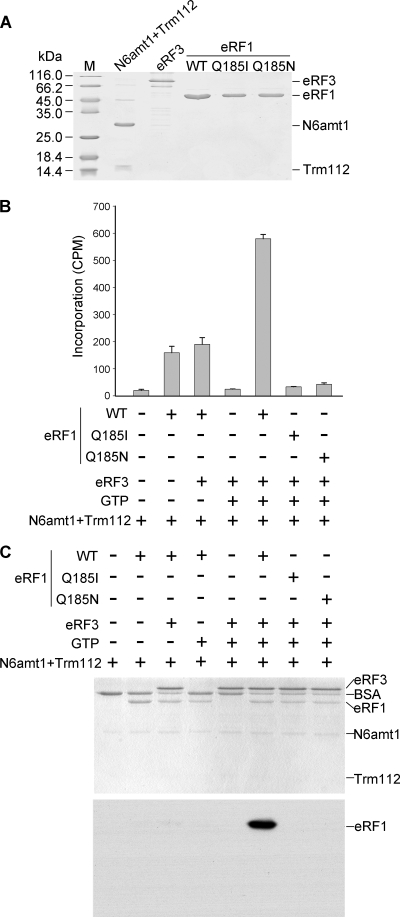
We then examined whether eRF1 can be methylated by murine N6amt1 in vitro. Purification of recombinant N6amt1 was achieved as a heterodimer with the interacting partner protein Trm112 coexpressed in E. coli (Fig. 2 A), a strategy used for the yeast homologous proteins (10). Two mutants (Q185I and Q185N) of eRF1 were prepared in addition to the wild type. Recombinant eRF3 was also prepared as it forms a complex with eRF1 (15) and is required for eRF1 methylation in yeast (9). An in vitro methylation assay using [methyl-3H]AdoMet as methyl donors indicated that the N6amt1-Trm112 complex displayed the maximal activity in the presence of both eRF3 and GTP (Fig. 2B). Leaving out either eRF3-GTP or GTP alone from the reaction led to a decrease of the methylation activity to about one-third of the maximal. This cofactor dependence was less strong than the yeast enzyme (9). Most importantly, no incorporation was detected on the two mutant eRF1 substrates with substitutions at glutamine 185, indicating that no other site could be methylated by N6amt1. To further confirm the methyl transfer reaction, we separated the methylation products on an SDS-PAGE gel and performed 3H fluorography. A predominant band with a mobility corresponding to eRF1 appeared when the wild-type eRF1 was incubated with N6amt1-Trm112 in the presence of all cofactors (Fig. 2C). These data together demonstrate that the murine N6amt1-Trm112 heterodimer functions as a highly specific protein MTase and its activity can be stimulated by eRF3 and GTP.
FIG. 2.
Characterization of the in vitro methylation activity of recombinant N6amt1. (A) Recombinant proteins used for the methylation assay. 6×His-tagged N6amt1 coexpressed FLAG-Trm112, GST-eRF3, and wild-type (WT) and mutant 6×His-eRF1 proteins were purified from E. coli and examined by Coomassie staining. (B) Methyltransferase activity detected by scintillation counting. eRF1 substrates were incubated with N6amt1 in the presence or absence of other factors using [methyl-3H]AdoMet as the methyl donor, and the proteins were then precipitated by trichloroacetic acid (TCA) for the measurement of methyl-3H incorporation. Averages from three independent experiments ± standard deviations are shown. (C) Methyl-transferase activity detected by fluorography. After the methylation reaction in the presence of radioactive [methyl-3H]AdoMet, the proteins were separated by SDS-PAGE and visualized by Coomassie staining (upper panel), and the same gel was subjected to fluorography (lower panel). Note that only the wild-type eRF1 was radioactively labeled.
N6amt1 methylates Gln185 of eRF1 in mammalian cells.
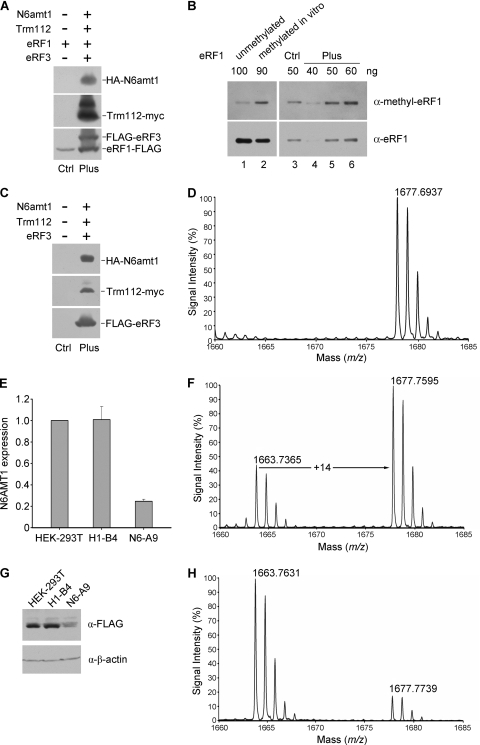
We then examined the function of N6amt1 in vivo in cultured mammalian cells. For the detection of methylated eRF1 by an approach independent of mass spectrometry, rabbit polyclonal antibody against a Gln185-methylated eRF1 peptide was generated and its specificity was validated by dot blotting and Western analysis (see Fig. S2 in the supplemental material). FLAG-tagged eRF1 was transfected into HEK-293T cells alone (the control sample) or with hemagglutinin (HA)-N6amt1, Trm112-myc, and FLAG-eRF3 (the “Plus” sample) (Fig. 3A). Western blot analysis with anti-methyl-eRF1 showed a stronger signal in the Plus sample than in the control in repeated experiments (Fig. 3B, compare lanes 5 and 3). To further verify the function of N6amt1 in vivo, we affinity purified the endogenous eRF1 from HEK-293T cells with and without the expression of transfected N6amt1, Trm112, and eRF3 (Fig. 3C) and digested it by chymotrypsin in gel prior to MALDI-TOF MS analysis. In the absence of ectopic expression of N6amt1, Trm112, and eRF3, both MS peaks with m/z values of 1,663.84 and 1,677.86 corresponding to unmethylated and methylated peptides appeared, similar to the result in Fig. 1C (see Fig. S3 in the supplemental material). However, the MS peak of unmethylated peptide (1,663.84) was missing upon the ectopic expression of N6amt1, Trm112, and eRF3 (Fig. 3D), suggesting that the endogenous eRF1 has become completely methylated. Therefore, N6amt1 is able to methylate Gln185 of the endogenous eRF1 in vivo in mammalian cells.
FIG. 3.
Characterization of the in vivo methylation activity of N6amt1 in transfected HEK-293T cells. (A) Detection of transfected proteins by Western analysis. HEK-293T cells were transiently transfected with the eRF1-FLAG expression construct alone (“Ctrl” sample) and with the constructs for HA-N6amt1, Trm112-myc, and FLAG-eRF3 (“Plus” sample). Epitope-specific antibodies were used in the detection. (B) Increased methylation of eRF1 when cotransfected with HA-N6amt1, Trm112-myc, and FLAG-eRF3. The ectopic eRF1 was immunoprecipitated from the cell extract using anti-FLAG M2 beads prior to Western analysis. Bacterial recombinant eRF1 protein samples, unmethylated and methylated in vitro, were loaded as a control to monitor preferential detection of the methylated form by the anti-methyl-eRF1 (α-methyl-eRF1) antibody (lanes 1 and 2). Different amounts (40, 50, and 60 ng) of the “Plus” eRF1 sample were loaded for quantification of the methylated eRF1. The antibodies used are indicated to the right. α-eRF1, anti-eRF1 antibody. (C) Western detection of N6amt1 and its cofactors transfected into HEK-293T cells for the methylation analysis on endogenous eRF1. Epitope-specific antibodies were used. (D) MALDI-TOF MS analysis for Gln185 methylation of endogenous eRF1 from HEK-293T cells transfected with N6amt1 and its cofactors (“Plus” sample). The endogenous eRF1 protein was immunopurified with anti-eRF1 polyclonal antibodies from transfected HEK-293T cells, separated, and in-gel digested with chymotrypsin prior to MALDI-TOF MS analysis. The peak at m/z 1,677.69 corresponds to methylated peptides containing Gln185 (Fig. 1C). The peak for unmethylated peptide was not detected. (E) N6AMT1 depletion in knockdown cell line N6-A9 verified by real-time PCR. siRNA expression was induced by addition of doxycycline. H1-B4 is a control cell line harboring an empty siRNA vector. (F) N6AMT1 depletion in knockdown cell line N6-A9 verified by Western blotting. (G) MALDI-TOF MS analysis for Gln185 methylation of FLAG-eRF1 in H1-B4 cells. α-FLAG, anti-FLAG antibody; α-β-actin, anti-β-actin antibody. (H) MALDI-TOF MS analysis for Gln185 methylation of FLAG-eRF1 in N6-A9 cells.
To further confirm the methyltransferase function of N6AMT1 in vivo, we generated an inducible knockdown stable cell line from HEK-293T cells. Reduced N6AMT1 expression in knockdown cells was verified by both real-time PCR and Western blotting (Fig. 3E and G). FLAG-tagged eRF1 was then transfected into the knockdown cells and purified for MALDI-TOF MS analysis. In the control cell line H1-B4, the eRF1 methylation level (∼70%) was similar to that of HEK-293T cells (compare Fig. 3F with 1C). In N6-A9 knockdown cells, the peak intensity of methylated peptide (1,677.77), however, dropped below 20% of that of unmethylated peptide (1,663.76) (Fig. 3H). This indicated that less than 17% of the eRF1 was methylated when N6AMT1 was depleted. These results strongly suggest that N6AMT1 methylates eRF1 in vivo.
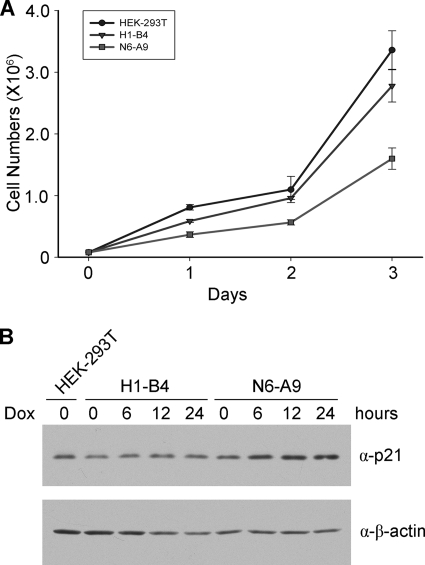
N6AMT1 depletion leads to reduced cell proliferation.
Since deletion of the N6amt1 homolog in E. coli and yeast result in growth defects, we examined whether depletion of N6AMT1 would affect mammalian cell proliferation in culture. Indeed, the knockdown N6-A9 cells had a lower growth rate (Fig. 4A). The growth rate of N6-A9 was about 60% of that of the control H1-B4 and 50% of that of the HEK-293T cells. Although there was no significant cell cycle difference between H1-B4 and N6-A9 (data not shown), expression of p21 was moderately elevated during the first 24 h upon doxycycline induction (Fig. 4B).
FIG. 4.
Reduced proliferation of N6AMT1 knockdown cells and modest upregulation of p21. (A) Growth curve of knockdown (N6-A9) and control (HEK-293T and H1-B4) cells in medium with doxycycline. (B) Expression of p21 upon doxycycline (Dox) induction in N6-A9 cells. Western blotting was carried out three times, and representative results are shown.
Disruption of the murine N6amt1 gene results in embryonic lethality.
To investigate the physiological significance of the N6amt1 methylation function, we set out to generate mutant mice by targeted gene disruption. The murine N6amt1 gene consists of 6 exons that form two transcript isoforms with the first 4 exons in common (23). Proteins encoded by both isoforms contain the conserved GXGXG motif essential for the binding of AdoMet and the NPPY motif essential for the binding of an amide side chain (27). To ensure functional loss of the N6amt1 gene, exon 4 encoding the NPPY motif was replaced by a Neo gene through homologous recombination in embryonic stem (ES) cells (Fig. 5A). Three independent heterozygous ES clones were identified by genomic PCR and Southern blot analysis (data not shown). Mouse chimeras were derived from the blastocysts injected with the N6amt1 heterozygous ES cells and were crossed with C57BL/6 mice. Heterozygote offspring were identified by Southern blot analysis (Fig. 5B) and PCR genotyping. They were normal in gross appearance and showed no discernible impairment of growth and fertility (data not shown). However, no homozygous littermates were obtained from intercrosses among heterozygous mice, while wild-type mice and heterozygotes were identified in a 1:2 ratio among the 95 born pups (Table 2). This clearly indicates that N6amt1 disruption led to embryonic lethality in the mouse.
FIG. 5.
Targeted disruption of the N6amt1 gene in mice. (A) Targeting strategy to generate the mutant N6amt1 allele. Numbered gray boxes represent coding exons, and open boxes represent the 3′ untranscribed region (UTR). B, BamHI restriction site; Neo, neomycin resistance gene; HSV-tk, herpes simplex virus thymidine kinase gene. The locations of the Southern probe and the expected BamHI fragments are indicated. PCR primers used for genotyping are indicated with arrows. (B) Genotyping of ES clones by Southern blot analysis. Genomic DNA was digested with BamHI. The 9.5-kb and 5.7-kb bands were derived from the wild-type and targeted alleles, respectively. (C) Genotyping of mouse embryos by PCR. Heterozygous mice were intercrossed, and E3.5 blastocysts were collected. The 267-bp and 372-bp PCR products were derived from the wild-type and targeted alleles, respectively.
TABLE 2.
Viability analysis of pups and embryos from N6amt1+/− intercrosses
| Embryonic stage | No. of pups/embryos with characteristic by: |
Total for genotype or phenotype | |||||
|---|---|---|---|---|---|---|---|
| Genotype |
Phenotype |
||||||
| +/+ | +/− | −/− | NDa | Normal | Abnormal | ||
| Newborn | 30 | 65 | 0 | 0 | 95 | 0 | 95 |
| E10.5 | 2 | 6 | 0 | 3 | 8 | 3 | 11 |
| E9.5 | 2 | 4 | 0 | 2 | 6 | 2 | 8 |
| E7.5 | 7 | 13 | 4 | 3 | 21 | 6 | 27 |
| E3.5 | 11 | 12 | 6 | 3 | 29 | 3 | 32 |
| Cultured blastocysts | 17 | 32 | 6 | 8 | 57 | 6 | 63 |
ND, genotype not determined due to failure of PCR.
N6amt1 is essential for early postimplantation development.
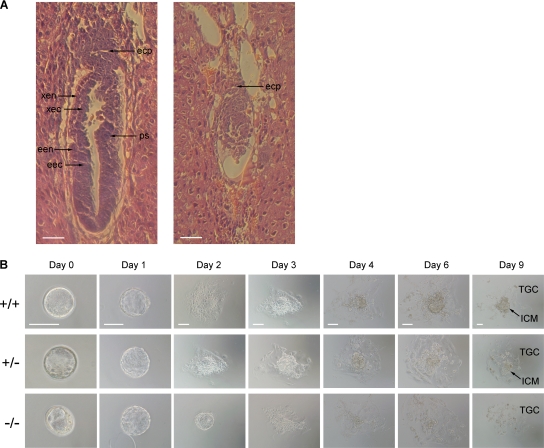
In order to identify the timing of developmental defects caused by N6amt1 deficiency, embryos arising from heterozygote intercrosses were collected at different stages. PCR genotyping of these embryos demonstrated that N6amt1 homozygotes could be recovered after implantation at embryonic day 7.5 (E7.5) but were not found afterwards (Fig. 5C and Table 2). Since the preimplantation development did not appear to be affected, we focused on the analysis of E6.5 and E7.5 embryos. Of 27 E7.5 embryos, 6 were much smaller in size, exhibiting a resorption phenotype, and 4 of them were successfully genotyped as homozygotes (Table 2). For further analysis of E6.5 embryos, sections of decidua embedded in paraffin were stained with hematoxylin and eosin. In the longitudinal section of normal embryos, a primitive streak appeared and endoderm and ectoderm could be readily distinguished for either the extraembryonic or embryonic region (Fig. 6A). In contrast, abnormal embryos were much smaller in overall size, and the ectoplacental cone was underdeveloped. These embryos had no typical embryonic structures. A round shape instead of an elongated egg cylinder was repeatedly observed.
FIG. 6.
Early embryonic lethality caused by deficiency in N6amt1. (A) H&E staining of longitudinal sections of E6.5 embryos from intercrosses of N6amt1+/− mice. Typical embryos with normal (left) and abnormal (right) morphologies are shown. The normal E6.5 embryo has initiated the formation of a primitive streak (ps) and developed an elongated egg cylinder. The abnormal embryo is much smaller in overall size. The ectoplacental cone (epc) is underdeveloped, and no primitive streak forms. Endoderm and ectoderm cannot be distinguished. xen, extraembryonic endoderm; xec, extraembryonic ectoderm; een, embryonic endoderm; eec, embryonic ectoderm. Scale bar, 50 μm. (B) Outgrowth of blastocysts from N6amt1+/− intercrosses in vitro. E3.5 blastocysts were collected and cultured in ES medium for 9 days. Genotypes were determined by genomic PCR. ICM and trophoblastic giant cells (TGC) in outgrowth at day 9 are pointed out. Scale bars, 100 μm.
For further determination of the consequence of N6amt1 loss, preimplatation E3.5 blastocysts were isolated. Six homozygotes were identified from 32 blastocysts. This number was slightly less than the expected 25% from the Mendelian principle, suggesting that N6amt1 inactivation was not sufficient to cause blastocyst lethality if there was any negative effect on preimplantation development. To further determine whether the blastocyst and later developmental stages could be affected, the isolated blastocysts were cultured in vitro for 9 days. For the first 3 days, no different phenotype was observed for the N6amt1−/− blastocysts (Fig. 6B). Outgrowths of the homozygous blastocysts showed normal inner cell mass (ICM) morphology above the trophectodermal cell layer. On days 4 and 6, however, the homozygous mutant exhibited a slower outgrowth, and on day 9, ICM cells were missing. Consistently, no ES cell lines could be established from mutant blastocysts homozygous for N6amt1. Taken together, the gene-targeting experiment indicates that the N6amt1 gene is essential for mouse early embryonic development and the survival of ES cells.
DISCUSSION
In this work, we demonstrate that a proposed mammalian DNA adenine methyltransferase, N6amt1, acts as a protein methyltransferase for the release factor eRF1 in vitro and in vivo. By gene targeting in mice, we establish the role of the N6amt1 gene for the viability of early embryos. Deficiency of N6amt1 led to early embryonic lethality at postimplantation stage. Mutant cells from inner cell mass (ICM) lacking N6amt1 failed to proliferate. The requirement of N6amt1 in mouse early development suggests the importance of modulation of the translation termination activity of eRF1 in protein synthesis. To date, N6amt1 is the only characterized glutamine methyltransferase in mammals as all other protein MTases are specific either for lysine or arginine residues.
The finding of N6amt1 as a bona fide glutamine-specific protein methyltransferase is not surprising considering that a similar mechanism can be utilized to methylate the exocyclic amide group of an adenine (N6) or cytosine (N4) base in DNA and the side-chain amide group of a glutamine residue in a polypeptide. The NPPY motif found in the active site of both glutamine- and adenine-specific methyltransferases makes major contacts with the target amide group in the substrate and the methyl donor AdoMet, in common to both types of the enzymes (27). This misidentification of N6amt1 resembles the case of Dnmt2, which was initially classified as a putative cytosine DNA methyltransferase based on its prominent sequence conservation but later turned out to be a robust tRNA cytosine methyltransferase (7). It is still under debate whether Dnmt2 can also methylate DNA in vivo despite its negligible activity.
One key question regarding the in vivo function of N6amt1 is how disruption of N6amt1 causes the mouse embryonic lethality. Although we cannot address the exact molecular mechanism in the early embryo, the knockdown experiment in cell culture suggests that deficiency in N6amt1 can lead to activation of the p53-p21 pathway, thus affecting cell proliferation (Fig. 4). Therefore, it is plausible that N6amt1-deficient embryos fail to develop due to cell cycle defects.
Although methylation at the glutamine residue in the active center of eRF1 enhances its function in promoting the release of newly synthesized polypeptides from the ribosome (4, 16), the biological significance of this modification has been poorly understood. Deletion of the HemK methyltransferase for the release factor in E. coli had no effect on cell growth in rich media but reduced growth considerably on poor carbon sources (16, 17); inactivation of the N6amt1 homologue YDR140w in S. cerevisiae was permissive to cell viability under standard conditions but resulted in decrease in growth rate and other phenotypes under a stress environment, including cold sensitivity and sensitivity to translation fidelity drugs paromomycin and Geneticin (22). The observed phenotypes in these two lower organisms are mild and can be only loosely related to inefficient termination of protein translation. In contrast, we found that mammalian eRF1 methyltransferase N6amt1 is indispensable for the embryo viability. Embryos deficient in the enzyme die soon after implantation, and embryonic cells from null blastocysts fail to proliferate. This severe consequence can be accounted for by higher sensitivity of mammalian cells to abnormal proteins arising from potentially more frequent read-through at stop codons. Conversely, dysfunction of unmethylated eRF1 in translation termination may result in ribosomal stress accompanying activation of the p53-p21 pathway (21, 24, 28). Nevertheless, an intriguing possibility is that the mammalian methyltransferase N6amt1 might have other unknown functions or substrate specificity in addition to glutamine-specific methylation of eRF1. In this connection, it is noteworthy that N6amt1 might bind DNA in a sequence-specific manner, according to a recent profiling of the human protein-DNA interactome (11). This finding warrants further investigation for potential functions of this protein in modification of DNA or RNA, in spite of the fact that adenine methylation was undetectable in total mouse genomic DNA even with a highly sensitive method (23).
In contrast to complete methylation of RF1 and RF2 in E. coli (4, 17), eRF1 in both yeast (9) and human cells (this work) is methylated to 60 to 75%. The fact of partial methylation raises interesting questions regarding the regulatory mechanism for the modification of eRF1 and the biological significance of eRF1 partition in two forms in eukaryotes. Since overexpression of N6amt1 in transfected cells led to complete methylation of eRF1, it would be of interest to examine if there is a change in the methylation level of eRF1 in Down syndrome patients with an extra copy of the N6AMT1 gene.
Supplementary Material
Acknowledgments
We thank B. A. Hemmings and P. Cron for pGEX-2T-eRF3 and pRSET-eRF1, C. Li and Y. Jin for the effort to derive the N6amt1−/− ES cell line, and G. F. Xu and J. Gao for technical assistance with embryonic culture.
This work was supported by the Ministry of Science and Technology of China (grants 2005CB522400 and 2007CB947503) and the National Science Foundation of China to G.L.X.
Footnotes
Published ahead of print on 6 July 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Antonarakis, S. E., R. Lyle, S. Deutsch, and A. Reymond. 2002. Chromosome 21: a small land of fascinating disorders with unknown pathophysiology. Int. J. Dev. Biol. 46:89-96. [PubMed] [Google Scholar]
- 2.Bujnicki, J. M., and M. Radlinska. 1999. Is the HemK family of putative S-adenosylmethionine-dependent methyltransferases a “missing” zeta subfamily of adenine methyltransferases? A hypothesis. IUBMB Life 48:247-249. [DOI] [PubMed] [Google Scholar]
- 3.Clarke, S. 1993. Protein methylation. Curr. Opin. Cell Biol. 5:977-983. [DOI] [PubMed] [Google Scholar]
- 4.Dincbas-Renqvist, V., A. Engstrom, L. Mora, V. Heurgue-Hamard, R. Buckingham, and M. Ehrenberg. 2000. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19:6900-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figaro, S., N. Scrima, R. H. Buckingham, and V. Heurgue-Hamard. 2008. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 582:2352-2356. [DOI] [PubMed] [Google Scholar]
- 6.Goedecke, K., M. Pignot, R. S. Goody, A. J. Scheidig, and E. Weinhold. 2001. Structure of the N6-adenine DNA methyltransferase M. TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 8:121-125. [DOI] [PubMed] [Google Scholar]
- 7.Goll, M. G., F. Kirpekar, K. A. Maggert, J. A. Yoder, C. L. Hsieh, X. Zhang, K. G. Golic, S. E. Jacobsen, and T. H. Bestor. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311:395-398. [DOI] [PubMed] [Google Scholar]
- 8.Heurgue-Hamard, V., S. Champ, A. Engstrom, M. Ehrenberg, and R. H. Buckingham. 2002. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heurgue-Hamard, V., S. Champ, L. Mora, T. Merkulova-Rainon, L. L. Kisselev, and R. H. Buckingham. 2005. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 280:2439-2445. [DOI] [PubMed] [Google Scholar]
- 10.Heurgue-Hamard, V., M. Graille, N. Scrima, N. Ulryck, S. Champ, H. van Tilbeurgh, and R. H. Buckingham. 2006. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 281:36140-36148. [DOI] [PubMed] [Google Scholar]
- 11.Hu, S., Z. Xie, A. Onishi, X. Yu, L. Jiang, J. Lin, H. S. Rho, C. Woodard, H. Wang, J. S. Jeong, S. Long, X. He, H. Wade, S. Blackshaw, J. Qian, and H. Zhu. 2009. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139:610-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufman, H. E., and E. C. Adams. 1954. Water-soluble chelates in histochemical staining. Science 120:723-724. [DOI] [PubMed] [Google Scholar]
- 13.Li, R. X., H. B. Chen, K. Tu, S. L. Zhao, H. Zhou, S. J. Li, J. Dai, Q. R. Li, S. Nie, Y. X. Li, W. P. Jia, R. Zeng, and J. R. Wu. 2008. Localized-statistical quantification of human serum proteome associated with type 2 diabetes. PLoS One 3:e3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 15.Merkulova, T. I., L. Y. Frolova, M. Lazar, J. Camonis, and L. L. Kisselev. 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443:41-47. [DOI] [PubMed] [Google Scholar]
- 16.Mora, L., V. Heurgue-Hamard, M. de Zamaroczy, S. Kervestin, and R. H. Buckingham. 2007. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 282:35638-35645. [DOI] [PubMed] [Google Scholar]
- 17.Nakahigashi, K., N. Kubo, S. Narita, T. Shimaoka, S. Goto, T. Oshima, H. Mori, M. Maeda, C. Wada, and H. Inokuchi. 2002. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl. Acad. Sci. U. S. A. 99:1473-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayashiki, T., K. Nishimura, and H. Inokuchi. 1995. Cloning and sequencing of a previously unidentified gene that is involved in the biosynthesis of heme in Escherichia coli. Gene 153:67-70. [DOI] [PubMed] [Google Scholar]
- 19.Noyer-Weidner, M., and T. A. Trautner. 1993. Methylation of DNA in prokaryotes, p. 39-108. In J. P. Jost and H. P. Saluz (ed.), DNA methylation: molecular biology and biological significance. Birkhauser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 20.Patterson, D. 2009. Molecular genetic analysis of Down syndrome. Hum. Genet. 126:195-214. [DOI] [PubMed] [Google Scholar]
- 21.Pestov, D. G., Z. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21:4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polevoda, B., L. Span, and F. Sherman. 2006. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 281:2562-2571. [DOI] [PubMed] [Google Scholar]
- 23.Ratel, D., J. L. Ravanat, M. P. Charles, N. Platet, L. Breuillaud, J. Lunardi, F. Berger, and D. Wion. 2006. Undetectable levels of N6-methyl adenine in mouse DNA: cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 580:3179-3184. [DOI] [PubMed] [Google Scholar]
- 24.Sulic, S., L. Panic, M. Barkic, M. Mercep, M. Uzelac, and S. Volarevic. 2005. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 19:3070-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szulc, J., M. Wiznerowicz, M. O. Sauvain, D. Trono, and P. Aebischer. 2006. A versatile tool for conditional gene expression and knockdown. Nat. Methods 3:109-116. [DOI] [PubMed] [Google Scholar]
- 26.Wion, D., and J. Casadesus. 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4:183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, Z., L. Shipman, M. Zhang, B. P. Anton, R. J. Roberts, and X. Cheng. 2004. Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase. J. Mol. Biol. 340:695-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, Y., and H. Lu. 2009. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16:369-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.