Abstract
Limb-bud and heart (LBH) is a novel key transcriptional regulator of vertebrate development. However, the molecular mechanisms upstream of LBH and its role in adult development are unknown. Here we show that in epithelial development, LBH expression is tightly controlled by Wnt signaling. LBH is transcriptionally induced by the canonical Wnt pathway, as evident by the presence of conserved functional T-cell factor (TCF)/lymphoid enhancer-binding factor (LEF) binding sites in the LBH locus and rapid β-catenin-dependent upregulation of endogenous LBH by Wnt3a. In contrast, LBH induction by Wnt/β-catenin signaling is inhibited by Wnt7a, which in limb development signals through a noncanonical pathway involving Lmx1b. Furthermore, we show that LBH is aberrantly overexpressed in mammary tumors of mouse mammary tumor virus (MMTV)-Wnt1-transgenic mice and in aggressive basal subtype human breast cancers that display Wnt/β-catenin hyperactivation. Deregulation of LBH in human basal breast cancer appears to be Wnt/β-catenin dependent, as DKK1 and Wnt7a inhibit LBH expression in breast tumor cells. Overexpression studies indicate that LBH suppresses mammary epithelial cell differentiation, an effect that could contribute to Wnt-induced tumorigenesis. Taken together, our findings link LBH for the first time to the Wnt signaling pathway in both development and cancer and highlight LBH as a potential new marker for therapeutically challenging basal-like breast cancers.
Increasing evidence suggests that embryonic development and tumorigenesis share some of the same molecular mechanisms. In particular, aberrant reactivation of latent developmental signaling pathways and transcription factors in tumor cells has been associated with and shown to play causal roles in advanced-stage, invasive cancers (5, 6, 80).
Limb-bud and heart (LBH) is a highly conserved, novel tissue-specific transcription cofactor in vertebrates with important roles in embryonic development (7, 8, 19). We have previously identified Lbh as a novel mouse gene with unique spatiotemporal expression during early embryogenesis that reflects pattern formation in the developing limb buds and heart (8). Lbh encodes a small acidic protein (molecular mass, 12.3 kDa) that contains a conserved putative nuclear localization signal and a glutamate-rich putative transcriptional activation domain but lacks a DNA binding domain (DBD) (8). In mammalian reporter assays, LBH has both transcriptional activator and corepressor functions (7, 8). Recent biophysical analysis has revealed a high degree of structural disorder in LBH, suggesting that conformational plasticity may play a significant role in modulating LBH-dependent transcriptional processes (2).
Aberrant gain of LBH function is associated with partial trisomy 2p syndrome (7), a rare human autosomal disorder that is characterized by multiple congenital anomalies, including cardiovascular, skeletal, and postaxial limb defects (46). Partial trisomy 2p syndrome patients harbor a triplication of chromosomal region 2p23, where LBH maps, indicating that increased LBH gene dosage is pathological in humans (7). Transgenic misexpression of Lbh during embryonic heart development in mice phenocopies congenital heart disease observed in these patients (7) and indicates that Lbh functions to attenuate cardiac chamber differentiation through corepression of key cardiac transcription factors NKX2.5 and TBX5 (7). Interestingly, gain of function of LBH during mouse heart development also causes various growth defects, such as ventricular hyperplasia and increased cardiac valve formation, as well as the abnormally sustained self-renewal of cardiomyocytes after birth, suggesting that LBH has promitogenic activity (7). Consistent with these findings, retroviral overexpression of Lbh in chicken embryonic limbs leads to increased cell proliferation of immature chondrocytes and markedly delays bone differentiation (19). However, the regulatory pathways acting upstream of LBH and its role in adult development have remained obscure.
Wnt signaling plays a fundamental role in embryonic development by regulating pattern formation, cell proliferation, differentiation, and migration (44). Wnt ligands are secreted lipid-modified glycoproteins that act as morphogens and elicit different cell behaviors depending on whether receptor interaction activates a canonical β-catenin-dependent transduction pathway or other β-catenin-independent noncanonical pathways (39, 73). In adults, canonical Wnt signaling promotes the self-renewal and maintenance of stem cells required for normal tissue homeostasis (52), a function that becomes oncogenic when this pathway is deregulated (18). Activation of canonical Wnt signaling leads to the stabilization of cytoplasmic β-catenin and its subsequent translocation into the nucleus, where it forms a heteromeric complex with DNA-binding proteins of the T-cell factor (TCF)/lymphoid enhancer-binding factor (LEF) family to activate target gene transcription (18). In the absence of Wnt, TCF/LEF factors are bound to target gene promoters but act as transcriptional repressors by forming a complex with Groucho/Grg/TLE corepressors (18). The amplitude of canonical Wnt signaling is autoregulated via multiple positive and negative feedback mechanisms that include TCF/LEF factors themselves and the secreted antagonist Dickkopf 1 (DKK1), respectively (13, 18, 26, 51). In addition, Wnt ligands can activate multiple noncanonical signaling pathways, including the planar cell polarity (PCP) and Wnt/Ca2+ pathways and, in the case of Wnt7a in limb development, a pathway involving the homeodomain transcription factor Lmx1b (37, 62).
Genetic studies of mice first indicated that canonical Wnt signaling is oncogenic and implicated in breast cancer. Aberrant activation of Wnt1 through proviral integration of the mouse mammary tumor virus (MMTV) causes aggressive mammary tumors in mice (53, 71). Similarly, transgenic overexpression of stabilized β-catenin in mouse mammary glands results in formation of invasive mammary adenocarcinomas (33, 69). Abnormal activation of the canonical Wnt pathway is also associated with human breast cancer (41) and is most common in a highly aggressive subtype of breast cancers known as basal carcinomas (22, 28, 38, 64). This tumor subtype accounts for 15 to 20% of breast cancers and is characterized by early onset and a highly invasive, poorly differentiated (keratin 5/6-positive) tumor phenotype. Basal-like breast cancers have a poor clinical outcome and represent a challenge for therapeutic intervention due to their lack of expression of the therapeutic targets estrogen receptor (ER), progesterone receptor (PR), and the ERBB2 oncogene (57, 65, 66), thus emphasizing the need for identification of new tumor-specific markers.
The spatiotemporal expression pattern of Lbh during mouse embryogenesis (8) led us to hypothesize that Lbh may be controlled by morphogenic signaling pathways that orchestrate cell specification and pattern formation. Using a combination of molecular, mammalian tissue culture, mouse genetics, and in silico analyses, we set out to identify the molecular pathways operating upstream of LBH. In doing so, we discovered that LBH expression in epithelial development is tightly controlled by an antagonistic relationship between canonical Wnt/β-catenin and noncanonical Wnt7a signaling. Whereas LBH transcription is induced by Wnt/β-catenin signaling via four conserved TCF/LEF binding sites in the LBH gene locus, this induction is efficiently blocked by Wnt7a. Given the parallels between Wnt signaling in development and cancer, we hypothesized that LBH, as a canonical Wnt target gene, might be deregulated in breast cancer. Indeed, we found that LBH is aberrantly overexpressed in mammary tumors of MMTV-Wnt1 transgenic mice as well as in highly aggressive basal subtype human breast cancers. Overexpression of Lbh in HC11 mammary epithelial cells (MECs) further demonstrates that LBH suppresses terminal cell differentiation, an effect that could contribute to Wnt-induced tumorigenesis. Collectively, our data suggest that LBH is a direct Wnt target gene that is reactivated in a particularly lethal form of human breast cancer.
MATERIALS AND METHODS
Genomic DNA isolation and plasmids.
A Lambda gt11-129EV mouse genomic DNA library (Stratagene) was screened with Lbh-specific cDNA probes (8). Several overlapping genomic clones comprising approximately 30 kb of the murine Lbh gene locus were isolated and mapped by restriction analysis. A SexAI-NotI genomic fragment containing approximately 1.5 kb of Lbh promoter region and 283 bp downstream of the transcriptional start site, including exon 1 (−1469 to +283), was inserted into the XhoI-HindIII sites of a pGL3-luciferase vector (Pwt). Lbh enhancer regions 1 and 2 (−6365 to −6445 and +1240 to +2003, respectively) were PCR amplified and cloned individually into the KpnI site of the Pwt plasmid construct upstream of the Lbh promoter to generate constructs E1wt and E2wt. In vitro mutagenesis was performed to introduce mismatch mutations into Lbh-specific TCF/LEF binding elements (TBEs) T1 to T4 using the QuikChange II XL site-directed mutagenesis kit (Stratagene). The sequence of mutagenic primers is shown in Table S1 in the supplemental material. To generate a His-TCF4 expression vector, the TCF4 DNA binding domain (DBD; residues 265 to 496) was PCR amplified using pGST-TCF4 (51) (generously provided by Tetsu Akiyama, University of Tokyo, Japan) as a template and inserted into the BamHI-HindIII restriction sites of pET28A vector (Novagen). Recombinant His-TCF4 was expressed in Escherichia coli and purified with nickel beads (Novagen) according to the manufacturer's protocol.
Electrophoretic mobility shift assays (EMSAs).
Double-stranded DNA oligonucleotides (30-mers) containing the genomic TBE sites T1 to T4 with flanking sequences (see Table S1 in the supplemental material) were 5′ end labeled with 32P, and 5,000 cpm/μl of labeled probe was incubated with 1 μg of recombinant His-TCF4 protein in a total volume of 15 μl binding buffer (25). For competition and supershift experiments, His-TCF4 was preincubated with unlabeled DNA oligonucleotides at a 400-fold excess or with 1 to 5 μg of anti-6×His tag antibody (Abcam) for 10 min prior to addition of labeled probe. Samples were separated on 5% nondenaturing polyacrylamide gels for 1 h at 400 V. Protein-DNA complexes were detected by phosphorimaging on a Storm 840 scanner (Molecular Dynamics).
Luciferase reporter assays.
Luciferase reporter assays were performed as described in reference 7 with the following modifications: 1 day prior to transfection 2.0 × 105 cells were plated per well of a 12-well plate. Cells were cotransfected with 100 ng of different luciferase reporter plasmids (Pwt, E1wt, and E2wt or TOPFlash and FOPFlash) and 300 ng of pCDNA/β-cateninS37Y expression plasmid (kindly provided by Antonio Garcia de Herreros, Universitat Pompeu-Fabra, Spain) using Lipofectamine 2000 reagent (Invitrogen). The fold transactivation of each Lbh-luciferase construct represents the ratio between normalized luciferase values of β-cateninS37Y-cotransfected cells and of cells transfected with the respective Lbh-luciferase constructs alone. For TOPFlash reporter assays, fold activation represents the ratio between normalized TOPFlash and FOPFlash activities. All transfections were performed in duplicate, and results of at least three independent experiments were statistically analyzed using a paired Student t test.
Cell culture.
Human mammary epithelial cells (HMEC) were obtained from Clonetics; all other human breast epithelial tumor cell lines were from the American Type Culture Collection (ATCC) and grown per the recommendations of these distributors. 293T and L-Wnt3a cells (ATCC) were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and grown under standard conditions at 37°C in a 5% CO2 atmosphere. Wnt3a-conditioned medium was prepared according to the distributor's protocol. HC11 cells (kindly provided by Kermit Carraway) were grown in RPMI medium supplemented with 10% FBS, 10 ng/ml insulin (Sigma), and 5 μg/ml epidermal growth factor (EGF; Invitrogen). Stable polyclonal cell lines were established by Lipofectamine transfection of HC11 cells with linearized pCDNA3 empty vector or pCDNA3-NLbh plasmid (8) followed by selection in 200 μg/ml of G418 (Invitrogen).
Wnt induction and RNAi.
For time course experiments, 293T cells were cocultured with Wnt3a-conditioned medium for 0, 4, 8, 16, and 24 h. Inhibition experiments used 100 ng/ml of recombinant human DKK1, Wnt5a, or Wnt7a (R&D Systems), which was added either alone for the indicated time points or 8 h prior to an 8-h treatment of cells with Wnt3a-conditioned medium. For RNA interference (RNAi) studies, 4 × 105 293T cells were transfected with 100 nM synthetic small interfering RNA (siRNA) specific for CTNNB1/β-catenin or a scrambled control sequence using Dharmafect 1 reagent (Dharmacon). Approximately 65 h after siRNA transfection, 293T cells were trypsinized and transferred to a dish with twice the surface area to allow for growth. At 72 h posttransfection, Wnt3a-conditioned medium was added for an additional 16 h. After the cells were harvested, total RNA was isolated using TRIzol reagent (Invitrogen) and treated with Turbo DNase (Ambion).
qPCR.
cDNA was synthesized from 1 μg DNase-treated total RNA using the Transcriptor first-strand cDNA synthesis kit (Roche). Quantitative real-time PCRs (qPCRs) were carried out in 20 μl using SYBR green Master Mix (NEB) containing 10 nM 6-carboxyfluorescein (Sigma) as a reference dye, 50 to 100 ng of cDNA, and 2 μM primers. The reactions were performed in triplicate on a Bio-Rad iCycler and quantified using the iCycler iQ software. The relative quantities of LBH, DKK1, and β-catenin mRNA were determined for each sample based on the threshold cycle (CT) value normalized to the corresponding values for GAPDH. For sequences of the qPCR primers, see Table S1 in the supplemental material.
Mice, histology, in situ hybridization (ISH), and X-Gal staining.
MMTV-Wnt1 [B6SJL-Tg(Wnt1)1Hev/J] and TopGal [Tg(Fos-lacZ)34Efu/J] transgenic mice were purchased from Jackson Laboratories (Bar Harbor, ME). The Wnt7a−/− and Lmx1b−/− mice were generously provided by Andy McMahon, (Harvard University, Boston, MA) and Randy Johnson (Baylor University, Houston, TX). Mice were bred and maintained in accordance with the guidelines set forth by the National Institutes of Health Guide for the Care and Use of Laboratory Animals, published by the U.S. Public Health Service. All experimental protocols were approved by the Institutional Animal Care and Use Committee at the University of Miami. Whole-mount in situ RNA hybridization and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of embryos and sections were performed as previously described (8). Moreover, 14-μm cryosections of snap-frozen mouse mammary glands or 5-μm paraffin sections of MMTV-Wnt1 mammary tumors were hybridized with a mouse Lbh-specific antisense probe (8).
Western blot analysis.
Mammary epithelial cells (MECs) from mammary glands of wild-type mice were isolated via proteolytic digestion with 100 units/ml hyaluronidase (Sigma) and 2 mg/ml collagenase A (Roche) in 15 ml DMEM for 3 h at 37°C with gentle agitation followed by washing in DMEM plus 5% FBS. Tumors from MMTV-Wnt1 transgenic mice were snap-frozen in liquid nitrogen and mechanically pulverized. Isolated MECs and ground tumors were lysed in RIPA lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% SDS) containing protease inhibitors (Amresco). For Western blot analysis a total of 25 μg protein extract per sample was separated by SDS-PAGE, blotted on nitrocellulose membrane, and incubated with the following antibodies in TBST (20 mM Tris HCl, pH 7.5, 140 mM NaCl, 0.1% Tween 20) plus 5% nonfat dry milk: a rabbit polyclonal Lbh antibody raised against murine Lbh and purified by Melon Gel IgG purification (Pierce; 1:1,000), polyclonal keratin 5 (Covance; 1:10,000), polyclonal keratin 8/18 (Progen; 1:2,000), monoclonal β-actin (AC-15, Sigma A5441; 1:50,000), and anti-rabbit, anti-mouse, or anti-guinea pig horseradish peroxidase (HRP)-coupled secondary antibodies (Amersham, Sigma; 1:10,000).
Immunofluorescence.
Cells were grown overnight on BD Biosciences culture slides at a density of 2 × 105 cells per well and induced with Wnt3a-conditioned medium for 6 h. Cells were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature, followed by cell permeabilization in 0.3% Triton X-100 in PBS. Cells were blocked for 1.5 h in PBS plus 10% normal goat serum (NGS) and incubated with β-catenin antibody (BD Biosciences; 1:200) followed by subsequent incubation with anti-mouse Cy3 (Jackson ImmunoResearch; 1:400). Cells were mounted in Slowfade plus DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes) according to the manufacturer's protocol. Images were taken on a DMRI Leica inverted microscope.
Chromatin immunoprecipitation (ChIP).
HC11 cells were grown to 70% confluence prior to addition of Wnt3a-conditioned medium for 3 h. Cells were fixed in a final concentration of 1% formaldehyde for 10 min at room temperature followed by a quenching of fixative with 125 mM glycine. Cells were incubated for 10 min on ice in swelling buffer (5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 8.0, 85 mM KCl, 1% NP-40) at a concentration of 5 × 107 cells/ml followed by Dounce homogenization 15 times. Nuclei were pelleted at 2,500 rpm for 5 min and resuspended in sonication buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS) at a concentration of 1 × 108 cells/ml. Sonication of cells for 6 pulses of 15 s each on ice-water at 50% power on a Misonix sonicator resulted in chromatin fragments of an average length of 1 kb. Lysates were cleared for 10 min at top speed. For each IP reaction, 1 × 107 cell equivalents were diluted to a 1-ml total volume in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.1 mM EDTA, 16.7 mM Tris-HCl, pH 8, 167 mM NaCl) and precleared for 2 h with 40 μl protein A/G Sepharose beads (GE Healthcare). Cleared lysates were incubated overnight with 5 μg of normal rabbit IgG, anti-acetyl-histone 3, or anti-β-catenin antibodies (Upstate). Thereafter, precipitation of immunocomplexes was performed according to the Upstate EZ ChIP protocol. PCRs for 35 cycles were carried out using Phusion polymerase (Finnzymes).
Cell proliferation and differentiation assays.
For proliferation assays, 2,000 cells were seeded in triplicate on 96-well plates. Two hours postplating, 20 μl of CellTiter 96 AQueous One cell proliferation assay reagent (Promega) was added to wells containing cells and blank medium controls. Reagent was applied at the same time daily, and absorbance at 492 nm was measured 2 h later on a microplate reader for 7 days. Background was eliminated by subtracting values of medium controls. Differentiation of HC11 cells was carried out for 3 days according to the method in reference 3.
Gene expression analysis.
Affymetrix gene expression data representing a total of 1,107 primary breast tumors from six previously published microarray studies (16, 21, 34, 56, 68, 76) were integrated as described previously using a mean-batch centering method (5, 63). Centroid prediction (11) was used to assign the tumors from each data set to the five Norway/Stanford subtypes (Basal, Luminal A, Luminal B, ERBB2, and Normal-like [57, 65, 66]). Centered average linkage clustering of the integrated tumor data sets was performed using the Cluster (23) and TreeView programs as described previously (65).
RESULTS
Identification of functional Wnt-responsive elements in the Lbh gene locus.
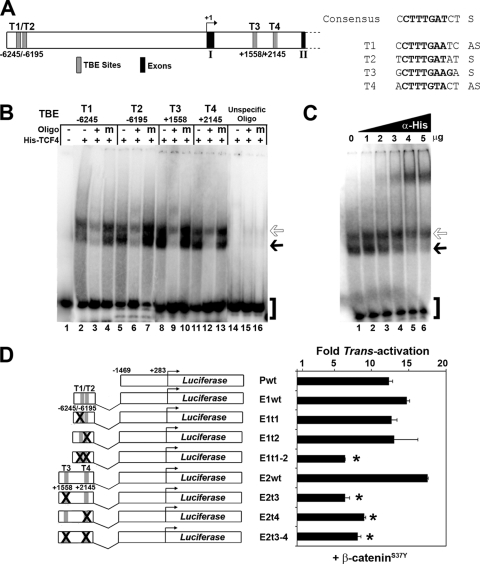
To elucidate the molecular pathways acting upstream of Lbh, we screened murine Lbh genomic sequences for potential transcription factor binding sites. This in silico search identified four conserved putative TCF/LEF binding elements (TBEs) in the Lbh gene locus. Two TBEs with the consensus motif 5′-CTTTG(A/T)(A/T)-3′ (75) were located within an enhancer region (E1) at bp −6245 (T1) and −6195 (T2) upstream of the Lbh transcriptional start site (Fig. 1 A). In addition, two consensus TBEs were found in an enhancer (E2) contained within the first intron of the Lbh gene at bp positions +1558 (T3) and +2145 (T4) (Fig. 1A). To directly assay for TCF binding to these sites, electrophoretic mobility shift assay (EMSA) was performed. Recombinant TCF4 protein bound with high affinity to all Lbh-specific TBEs (T1 to T4) but not to an unspecific oligonucleotide (Fig. 1B). TCF4 binding to these sites was efficiently competed by addition of a 400-fold excess of unlabeled wild-type (+) oligonucleotide but not by addition of a 400-fold excess of mutant (m) oligonucleotide (Fig. 1B). Furthermore, TCF4-specific protein-DNA complexes were supershifted with increasing amounts of an antibody against recombinant TCF4 (Fig. 1C).
FIG. 1.
Identification of functional TCF binding elements (TBEs) in the mouse Lbh gene locus. (A) Schematic of genomic Lbh promoter/enhancer sequences in the 5′ upstream region and the first intron between exons 1 and 2. The positions of four putative conserved TBE sites (T1 to T4) predicted by MatInspector (Genomatix) and/or rVista (http://rvista.dcode.org) computer software are shown in relationship to the transcriptional start site (+1). Sequences of T1 to T4 in comparison with the TBE consensus site (75) are indicated. S, sense strand; AS, antisense strand. (B) Electrophoretic mobility shift assay (EMSA) showing high-affinity binding of recombinant His-tagged TCF4 protein to T1 through T4. In oligonucleotide competition experiments, no competitor oligonucleotide (−), a 400-fold excess of unlabeled wild-type TBE oligonucleotide (+), or mutant TBE oligonucleotide (m) was added to gel shift reaction mixtures. Migration of free probe (brackets) and of TCF4 protein-DNA complexes (solid and open arrows) is indicated. (C) Supershift of His-TCF4 binding to the T3 (+1558) site with 1 to 5 μg of antibody to the his tag epitope (α-His). (D) Transient reporter assays in HC11 mouse mammary epithelial cells. Luciferase (Luc) reporter constructs containing different murine Lbh promoter/enhancer sequences (P, E1, and E2) with wild-type (wt) and mutant TBEs (t1 to t4) as shown schematically were cotransfected with a pCDNA construct expressing constitutively active β-catenin (β-cateninS37Y). Values represent relative fold increase of transcriptional activation for each construct (see Materials and Methods). *, P < 0.02.
Subsequently, cell-based reporter assays were performed to test whether the Lbh gene-specific TBE sites (T1 to T4) were functionally responsive to overexpression of β-catenin, which is the Wnt-inducible component of the TCF/β-catenin transcriptional complex. HC11 mouse mammary epithelial cells were used because this cell line abundantly expresses TCF4 but has low endogenous Wnt/β-catenin signaling activity (17). The Lbh enhancer regions E1 and E2 were cloned individually into a promoter-luciferase construct (Pwt) upstream of approximately 1.5 kb of murine Lbh gene promoter sequences that do not contain any apparent consensus binding sites for TCF/β-catenin. The three Lbh-luciferase reporter constructs (Pwt, E1wt, and E2wt) were cotransfected with a pCDNA plasmid vector expressing constitutively active β-catenin (β-cateninS37Y). As shown in Fig. 1D, Lbh-luciferase constructs containing wild-type TBE sites (E1wt and E2wt) were induced by β-cateninS37Y approximately 14- to 18-fold. The basal Lbh promoter-luciferase construct (Pwt) also showed transcriptional activation despite the lack of TBEs, indicating that β-catenin may also have indirect effects on Lbh promoter activity. Most importantly, however, mutations of both T1 and T2 together (E1t1-2) or of T3 and T4 either individually or in combination (E2t3, E2t4, and E2t3-4) significantly reduced (2.5- to 3-fold; P < 0.02) transcriptional activation of Lbh reporters by β-cateninS37Y (Fig. 1D). Mutation of either T1 or T2 alone had little effect, suggesting that binding of a TCF/β-catenin transcriptional complex to only one of these sites is sufficient for activity of this enhancer (E1). These data suggest that Lbh is activated by the canonical Wnt pathway at the transcriptional level via high-affinity TCF binding elements located within upstream and intronic enhancer regions of the Lbh gene.
Expression of endogenous LBH is upregulated by canonical Wnt signaling in 293T cells.
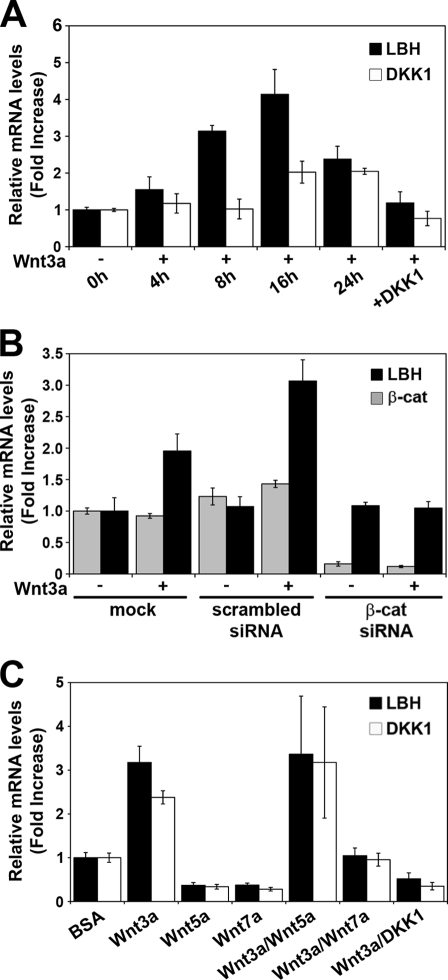
To further test whether LBH is a bona fide Wnt/β-catenin target gene, we examined whether endogenous LBH mRNA expression was responsive to Wnt. Human 293T embryonic kidney epithelial cells were cocultured with Wnt3a-conditioned medium (here referred to as Wnt3a), and mRNA levels of LBH, as well as of a known Wnt target gene, DKK1 (13, 26, 51), were assayed over a 24-h time course using quantitative real-time PCR (qPCR) analysis. Induction of LBH was detectable within 4 h of Wnt3a treatment and reached a maximum at 16 h (>4-fold increase) (Fig. 2 A). DKK1 was induced to a smaller degree, and its induction was delayed compared to that of LBH (Fig. 2A). Induction of both LBH and DKK1 mRNA expression by Wnt3a was efficiently blocked by recombinant DKK1 protein (Fig. 2A and C), a potent inhibitor of canonical Wnt/β-catenin signaling (50). Moreover, Wnt3a-mediated induction of LBH and DKK1 was abrogated by depletion of β-catenin expression using RNAi, while scrambled control siRNA had no effect (Fig. 2B). These results reinforce the notion that LBH is a direct transcriptional target of the canonical Wnt signaling pathway.
FIG. 2.
Regulation of endogenous LBH mRNA expression in human 293T cells by Wnt signaling. (A to C) qPCR analysis measuring relative mRNA levels of LBH, DKK1, and β-catenin normalized to GAPDH mRNA levels. All values represent means ± standard deviations (n = 3). (A) Time course analysis showing induction of LBH and DKK1 expression in response to Wnt3a. Cells were treated with Wnt3a-conditioned medium (Wnt3a) for the indicated time points. Wnt3a-mediated induction of both genes was inhibited by coadministration of recombinant Wnt inhibitor DKK1 (100 ng/ml), which was added 8 h prior to an 8-h treatment of cells with Wnt3a. (B) siRNA knockdown of β-catenin abolishes Wnt3a-induced upregulation of LBH, confirming activation of LBH expression by the canonical Wnt pathway. At 72 h posttransfection, 293T cells were treated with vehicle (−) or with Wnt3a-conditioned medium (+) for an additional 16 h. Note the reduction of β-catenin mRNA levels to less than 20% of endogenous expression levels in β-catenin (β-cat) siRNA-transfected cells. (C) Wnt7a, but not Wnt5a, inhibits Wnt3a-induced activation of LBH and DKK1. Cells were treated for 16 h with recombinant Wnt5a or Wnt7a proteins (100 ng/ml) individually or in conjunction with Wnt3a. As a control, cells were treated for 16 h with bovine serum albumin (BSA) vehicle.
To investigate whether Wnt ligands that signal through noncanonical pathways could also induce LBH gene expression, 293T cells were treated for 16 h with recombinant Wnt5a or Wnt7a (Fig. 2C). In contrast to Wnt3a, both Wnt5a and Wnt7a treatment alone did not induce LBH but modestly reduced baseline LBH and DKK1 expression (Fig. 2C). Since Wnt5a has previously been shown to inhibit Wnt3a-induced canonical Wnt signaling (48), we also examined LBH gene expression in cells treated with both Wnt3a and the individual noncanonical Wnt ligands. Surprisingly, Wnt7a strongly inhibited LBH and DKK1 induction by Wnt3a, whereas Wnt5a failed to block Wnt3a-mediated induction of these genes (Fig. 2C). Thus, LBH is specifically induced by canonical Wnt signaling, whereas noncanonical Wnt7a signaling has an antagonistic effect on LBH expression and its induction by Wnt3a.
Lbh is downstream of Wnt signaling in mouse embryonic limb development.
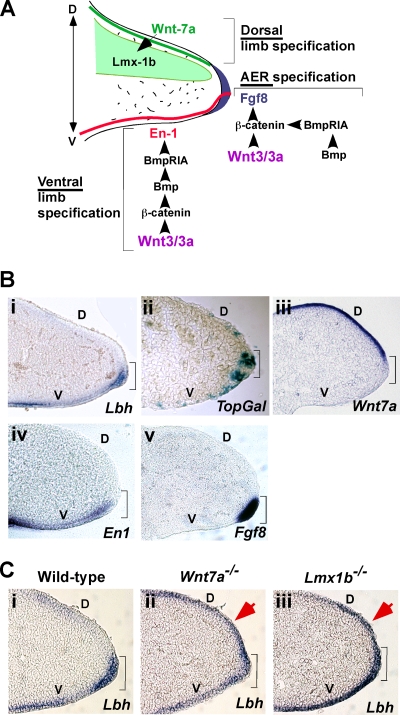
As Lbh was identified as a developmental regulatory gene (8), we next examined whether Wnt signaling might play a role in regulating Lbh expression during vertebrate embryonic development. We have previously shown that during early mouse limb development Lbh is expressed in the apical ectodermal ridge (AER) and in a ventral pattern in non-AER limb bud ectoderm (8). These ectodermal domains provide the cues for proximo-distal and ventral limb patterning, respectively (14, 70), and have recently been shown to be patterned by canonical Wnt/β-catenin signaling in concert with the Bmp pathway (Fig. 3 A) (4, 67). Conversely, Wnt7a, which is expressed and secreted from dorsal limb ectoderm, induces dorsal limb differentiation through a noncanonical pathway involving Lmx1b upregulation in the underlying dorsal limb mesenchyme (Fig. 3A and B, panel iii) (37, 55). Thus, we reexamined Lbh expression in relationship to the different Wnt signaling activities during crucial stages of mouse embryonic limb development.
FIG. 3.
Lbh expression during mouse embryonic limb bud formation is regulated by Wnt signaling. (A) Model for the genetic regulation of dorsoventral (D to V) limb patterning and apical ectodermal ridge (AER) formation. Wnt3 (Wnt3a in chicken embryo) and β-catenin act upstream of the Bmp pathway to induce En1 gene expression (red) in the ventral limb and AER ectoderm. En1 is required for ventral limb specification and restricts expression of Wnt7a (green) to the dorsal ectoderm. Wnt7a induces dorsal limb specification through induction of Lmx1b (light green) in the underlying dorsal mesoderm. Canonical Wnt/β-catenin signaling also directly induces expression of Fgf8 (blue) in the AER and is required for AER maintenance (4, 37, 45, 67). (B) Whole-mount RNA in situ hybridization analysis of E10.5 wild-type mouse embryonic limbs and transverse sections thereof showing that LBH expression in AER (brackets) and ventral limb ectoderm (i) overlaps with (ii) canonical Wnt activity, as detected by X-Gal staining of TopGal reporter mice at E10.5, as well as with expression of known Wnt/β-catenin-induced genes En1 (iv) and Fgf8 (v). In contrast, Wnt7a expression in dorsal limb ectoderm (iii) is mutually exclusive with expression of Lbh. (C) Lbh is ectopically expressed in the dorsal limb ectoderm of Wnt7a−/− (ii) and Lmx1b−/− (iii) mouse patterning mutants at E10.5 (red arrows), compared to wild-type limbs (i). D, dorsal; V, ventral.
As shown in Fig. 3B, panel i, Lbh expression was exclusively confined to the ventral limb and AER ectoderm of embryonic day 10.5 (E10.5) wild-type embryos as revealed by whole-mount in situ RNA hybridization analysis. Similarly, canonical Wnt activity was detected predominantly in the AER and ventral limb ectoderm, as evident by lacZ expression in TopGal embryonic limbs at the same stage, although some cells in the most distal dorsal ectoderm also expressed lacZ (Fig. 3B, panel ii). Moreover, genes regulated by canonical Wnt signaling, such as Fgf8 and En1 (37), were exclusively restricted to the AER and/or ventral limb ectoderm, respectively, and overlapped with Lbh expression in these ectodermal domains (Fig. 3B, panels i, iv, and v). The pattern of Lbh expression in embryonic limb ectodermal cells is also similar to that described for other Wnt/β-catenin target genes, including Dkk1 and axin2, which are expressed in the AER (1, 67). In contrast, expression of Lbh, as well as of En1 and Fgf8, was mutually exclusive with the Wnt7a expression domain in dorsal limb ectoderm (Fig. 3B, panels i and iii to v).
The complementary expression patterns of Wnt7a and Lbh (Fig. 3B, panels iii and i), as well as repression of LBH by Wnt7a in tissue culture cells (Fig. 2C), prompted us to test potential Wnt7a effects on Lbh expression in vivo. Lbh expression was examined in mouse mutants in which the noncanonical Wnt7a pathway was rendered inactive by gene targeting (15, 55). Remarkably, in Wnt7a−/− mutant mice and animals lacking the Wnt7a downstream transcriptional regulator Lmx1b (Lmx1b−/−), Lbh was ectopically expressed in the distal dorsal limb ectoderm (Fig. 3C, panels ii and iii). In contrast, Lbh exhibited a normal ventral expression pattern in wild-type littermates (Fig. 3C, panel i). Taken together, these developmental genetic studies provide the first functional evidence that Lbh expression in vivo coincides with canonical Wnt signaling activity and that it is repressed by noncanonical Wnt7a-Lmx1b signaling during embryonic limb development.
Lbh is expressed in postnatal mammary gland development, and levels are elevated in mammary tumors of MMTV-Wnt1 transgenic mice.
To test our hypothesis that Lbh might be implicated in Wnt-induced tumorigenesis, we examined Lbh expression in MMTV-Wnt1 transgenic mice, a mouse model for Wnt-induced breast cancer (Fig. 4) (71). Moreover, since the expression pattern of Lbh in normal adult breast tissue was not known, we analyzed Lbh expression during postnatal mouse mammary gland development using RNA in situ hybridization and Western blot analyses.
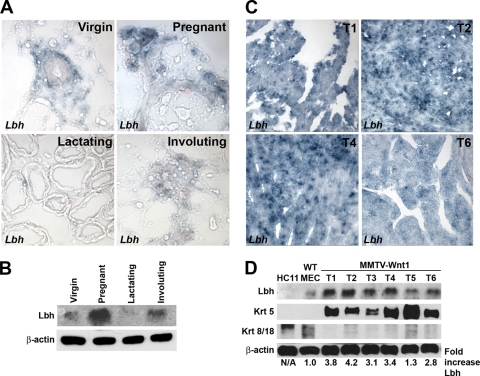
FIG. 4.
Lbh expression during normal mouse mammary gland development and overexpression in Wnt-induced mammary tumors. (A) RNA in situ hybridization analysis of sagittal cryosections of 7-week virgin, 13-day pregnant, 12-day lactating, and 4-day involuting normal mammary glands (original magnification, ×40). Lbh is expressed in basal-myoepithelial, terminal end bud, and stromal cells in virgin mammary glands, as well as in the lobulo-alveolar units during pregnancy and involution. Note that Lbh is not expressed in luminal epithelial cells or in lactating mammary glands. (B) Western blot analysis depicting Lbh protein levels during normal mammary gland development at the same stages as in panel A. (C and D) RNA in situ hybridization (C) and Western blot analysis (D) showing elevated Lbh expression levels in mammary tumors of MMTV-Wnt1 transgenic mice (T1 to T6) compared to HC11 and isolated wild-type (WT) mammary epithelial cells (MEC). Basal keratin 5 (Krt 5) and luminal keratin 8/18 (Krt 8/18) mammary epithelial markers, as well as a β-actin loading control, are shown. Quantification of Lbh protein levels by densitometry normalized to β-actin values is shown in the bottom panel. N/A, not applicable.
In postpubertal (7 weeks) virgin female mammary glands, expression of Lbh was restricted to stromal, basal-myoepithelial, and terminal end bud (TEB) mammary epithelial cells (Fig. 4A and data not shown). In contrast, Lbh was absent from ductal luminal mammary epithelial cells at all postnatal development stages analyzed (Fig. 4A). During pregnancy, Lbh levels drastically increased and Lbh transcripts were primarily detected in the proliferating lobulo-alveolar compartment, a pattern that was maintained during early involution (Fig. 4A and B). Notably, Lbh expression was virtually absent in lactating mammary glands, suggesting that Lbh is not expressed in terminally differentiated secretory mammary epithelial cells (Fig. 4A and B). Most remarkably, Lbh expression levels were significantly elevated (2.8- to 4.2-fold) in 9 out of 10 mammary tumors from different MMTV-Wnt1 transgenic mice compared to nonpregnant mammary glands, HC11 cells, and mammary epithelial cells isolated from equiparous wild-type littermates (Fig. 4A to D and data not shown). Moreover, in MMTV-Wnt1 tumors, which phenocopy human basal breast cancer (30), Lbh expression correlated with expression of the basal marker keratin 5, whereas it inversely correlated with expression of the luminal markers keratin 8 and 18 (Fig. 4C). Thus, Lbh is expressed at normal levels in basal and proliferative alveolar mammary epithelial cells during normal mammary gland development, whereas it is overexpressed in Wnt-induced basal breast epithelial tumors.
Lbh overexpression suppresses the differentiation of HC11 mammary epithelial cells.
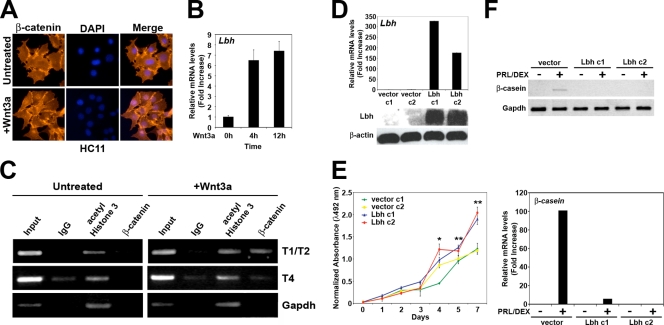
As we found Lbh expression specifically in cellular targets of canonical Wnt signaling during normal mammary gland tissue homeostasis (9, 69, 72) and Lbh over expression in Wnt-induced mammary tumors (Fig. 4), we further investigated the functional relationship between Wnt/β-catenin signaling and Lbh in a cell culture system for mammary epithelial development. HC11 was chosen because it is one of few existing nontransformed mammary epithelial cell lines that can be induced to differentiate in vitro with lactogenic hormones (3). Moreover, overexpression of different Wnt ligands has been shown to lead to cellular transformation of these cells (31, 32). To test whether Lbh could be downstream of canonical Wnt signaling in mammary epithelial cells, we treated HC11 cells, which do not express Lbh (Fig. 4), with Wnt3a. Wnt3a treatment resulted in nuclear localization of β-catenin as well as a rapid increase in Lbh mRNA levels (Fig. 5 A and B). In addition, ChIP analysis showed that the Lbh gene regulatory sequences T1 to T4 (Fig. 1A) were occupied by endogenous β-catenin in Wnt3a-treated cells but not in untreated control cells (Fig. 5C and data not shown).
FIG. 5.
Wnt/β-catenin-mediated induction and ectopic expression of Lbh in HC11 mammary epithelial cells. (A) Immunofluorescence analysis showing nuclear translocation of β-catenin in HC11 cells treated with Wnt3a-conditioned medium (+Wnt3a) for 6 h but not in untreated cells. (B) qPCR analysis measuring rapid induction of Lbh in cells treated with Wnt3a at the indicated time points. (C) ChIP analysis of β-catenin occupancy of endogenous Lbh gene regulatory sequences (T1/T2 and T4) in cells treated with Wnt3a for 3 h. DNA derived from sheared chromatin fragments from untreated and Wnt3a-treated cells immunoprecipitated with antibodies to β-catenin, acetyl-histone 3, and normal rabbit IgG was quantified by semiquantitative reverse transcription PCR (RT-PCR). As a control, <1% of input chromatin was used. (D) qPCR (top) and Western blot analyses (bottom) of two polyclonal cultures (c1 and c2) of HC11 cells stably expressing pCDNA3-Lbh or pCDNA3 vector alone, showing overexpression of Lbh in HC11-Lbh cultures. β-Actin was used as a loading control. (E) Ectopic expression of Lbh increases cell growth as assessed by CellTiter 96 AQueous One solution cell proliferation assay. Values represent the mean values; error bars represent the standard deviations (n = 3). Student's t test was used to evaluate significance: *, P < 0.01; **, P < 0.001. (F) Semiquantitative RT-PCR (top) and qPCR (bottom) analyses measuring induction of the terminal differentiation marker β-casein. Confluent cell cultures were treated for 3 days with normal growth medium or serum-free differentiation medium containing 5 μg/ml prolactin and 1 μM dexamethasone (PRL/DEX). qPCR values were normalized to Gapdh. All values represent means ± standard errors of the means (n = 3).
Having demonstrated that Lbh is a direct transcriptional target of Wnt/β-catenin in HC11 cells, we asked whether overexpression of Lbh elicits some of the same effects that have been reported for overexpression of Wnt ligands in this cell line (31, 32). Several polyclonal HC11 cell lines stably expressing Lbh (Lbh c1 and c2) were generated by transfection with a pCDNA3-Lbh plasmid, and Lbh overexpression was confirmed by qPCR and Western blot analyses (Fig. 5D). No Lbh expression was detectable in vector control-transfected cells or in the parental HC11 cells (Fig. 4D and 5D). Although ectopic Lbh expression did not result in cell transformation as determined by soft agar assays (data not shown), the growth rates of Lbh-expressing HC11 cells were significantly increased compared to those of vector control cells (Fig. 5E). Moreover, whereas differentiation induction with prolactin and dexamethasone increased mRNA expression of the milk protein β-casein in parental and vector control cells, induction of β-casein in response to these lactogenic hormones was lost in HC11-Lbh cells (Fig. 5F). Thus, overexpression of Lbh promotes cell proliferation and blocks terminal differentiation of HC11 mammary epithelial cells.
LBH is overexpressed in highly invasive ER-negative, basal subtype human breast cancers.
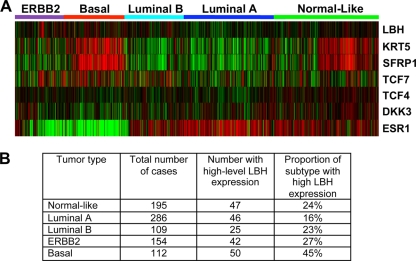
To further examine whether LBH might be deregulated in human breast cancer, meta-analysis of six Affymetrix gene expression data sets comprising 1,107 primary human breast cancers was performed as previously described (63). These data represent the five “intrinsic” breast tumor subtypes normal-like, luminal A, luminal B, ERBB2 positive, and basal-like (65), which can be distinguished by specific gene signatures and differences in clinical outcome, with basal-like breast cancers having the worst prognosis (Fig. 6) (57, 65, 66). Strikingly, LBH expression was significantly associated with aggressive, poorly differentiated basal type carcinomas. Almost half (45%) of the basal breast tumors had high LBH expression levels (Fig. 6B). In contrast, elevated LBH was observed in far smaller proportions of normal-like (24%), luminal A (16%), luminal B (23%), and ERBB2+ (27%) breast carcinomas (Fig. 6B). Moreover, a strong inverse correlation was observed between LBH and estrogen receptor alpha (ESR1) expression (Fig. 6A; see also Fig. S1 in the supplemental material) (R = −0.29, P < 0.0001), whereas no significant correlation existed with ERBB2 status (see Fig. S1) (R = −0.01). Most remarkably, however, LBH expression in breast tumors strongly correlated with the basal marker keratin 5 and canonical Wnt pathway genes, such as SFRP1, TCF4, TCF7, and DKK3 (Fig. 6A; see also Fig. S1) (P < 0.0001). These data highlight LBH as a novel molecular marker for difficult-to-treat ER-negative basal type breast cancer and suggest that LBH deregulation in breast cancer could be a consequence of oncogenic Wnt signaling.
FIG. 6.
LBH gene expression in human breast cancer correlates with ER-negative, basal-like tumor subtype. (A) Meta-analysis of 1,107 human primary breast carcinoma samples from six published Affymetrix data sets (63) showing a strong positive correlation of LBH with basal tumor type, as well as expression of keratin 5 (KRT5) and Wnt pathway genes SFRP1, TCF7, TCF4, and DKK3. In contrast, the LBH signature inversely correlates with estrogen receptor alpha (ESR1) expression. Clustering of tumor subtypes—basal (red), ERBB2 (purple), luminal A (dark blue), luminal B (light blue,) and normal-like (green)—was done according to reference 66. Red, high expression; green, low expression. (B) Table showing the proportion of breast cancer specimens with high levels (overall upper quartile) of LBH mRNA expression in individual tumor subtypes. P = 0.0000014 (chi-squared test).
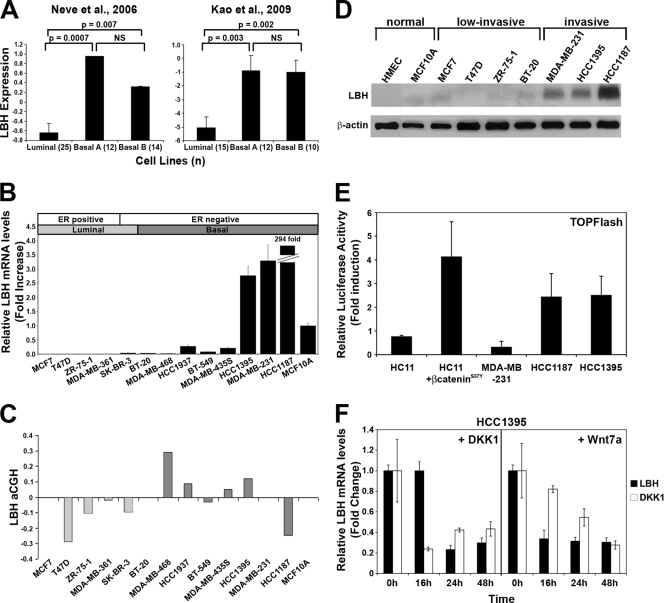
The lack of suitable antibodies currently precludes analysis of LBH protein expression in clinical specimens. Therefore, we analyzed human breast carcinoma cell lines to validate our findings. We first queried published Affymetrix gene expression data from 51 human breast cancer cell lines (49) to confirm the existence of a relationship between LBH expression and breast cancer subtype. Expression of LBH was significantly higher in both the basal A and basal B cell line subtypes than in those classified as luminal (P < 0.007) (Fig. 7 A). Specifically, 50% of the basal A (n = 12) and 29% of the basal B (n = 14) cell lines had high (upper quartile) expression of LBH, compared to only 12% of luminal (n = 25) cell lines (Fig. 7A). Similar results were observed in a more recent cDNA microarray study of breast cancer cell line gene expression (36) (Fig. 7A), which also demonstrated significantly higher expression of LBH in the basal A and basal B cell lines than in the luminal cell line (42%, 40%, and 12% of cells with high expression, respectively).
FIG. 7.
Validation of LBH expression and Wnt responsiveness in human breast tumor cell lines. (A) LBH mRNA expression is significantly higher in basal rather than in luminal breast carcinoma cell lines classified according to tumor subtype (36, 49). Values represent the means, and error bars represent the standard errors. (n), number of samples per tumor subtype; NS, not significant. (B) qPCR analysis of relative LBH mRNA expression in a panel of human breast tumor cell lines showing overexpression of LBH in HCC1395, MDA-MB-231, and HCC1187 tumor cells. Cell lines are arranged by tumor subtype (49). All measurements were performed in triplicate, and expression levels were normalized to mRNA levels of GAPDH. (C) Comparative genomic hybridization array (aCGH) analysis of the same breast tumor cell lines as those in panel B. (D) Western blot analysis detecting expression of LBH protein exclusively in invasive ER-negative basal type breast cancer lines but not in two nontransformed (normal) mammary epithelial cell lines or in low-invasive breast tumor cell lines. β-Actin was used as a loading control. (E) TOPFlash reporter assay detects Wnt signaling activity in LBH-expressing HCC1395 and HCC1187 cells but not in MDA-MB-231 cells. HC11 and HC11 transiently transfected with pCDNA3/β-cateninS37Y were used as negative and positive controls, respectively. Values represent the mean ratios of TOPFlash over FOPFlash activity ± standard deviations. (F) Administration of recombinant DKK1 and Wnt7a (100 μg/ml) for the indicated time points strongly inhibits LBH and DKK1 mRNA expression in HCC1395 cells as revealed by qPCR analysis. Values represent means ± standard errors of the means (n = 3).
We next examined LBH expression in a panel of 13 established human breast cancer cell lines by using qPCR and Western blot analysis. High levels of LBH expression were detected only in the ER-negative basal subtype breast tumor cell lines HCC1395, MDA-MB-231, and HCC1187 (Fig. 7B and D). In contrast, none of the ER-positive lines (MCF7, T47D, ZR-75-1, and MDA-MB-361) or ER-negative (SK-BR-3) luminal cell lines expressed LBH at detectable levels (Fig. 7B and D). Furthermore, LBH protein was not detected in normal human mammary epithelial cells (HMEC) or in nonmalignant MCF10A cells (Fig. 7D). Thus, consistent with our gene expression analysis in primary breast tumors, LBH expression in breast cancer-derived cell lines correlated with an invasive basal carcinoma phenotype and inversely correlated with expression of the good prognostic marker ER.
LBH deregulation in breast cancer may be due to aberrant Wnt/β-catenin pathway activation.
To begin to investigate the mechanisms underlying LBH deregulation in breast cancer, we queried comparative genomic hybridization array (aCGH) data that were available for these breast tumor cell lines. Only one of three LBH-overexpressing basal tumor cell lines (HCC1395) had a modest increase in LBH copy number (Fig. 7C). Moreover, aCGH analysis of primary breast tumor data sets did not show a significant correlation between increased LBH copy number and LBH overexpression in basal subtype tumors (data not shown), suggesting that changes in LBH gene dosage play a minor role in LBH dysregulation in basal breast carcinomas. To further test whether LBH overexpression may be a consequence of aberrant Wnt signaling, we measured endogenous Wnt signaling activity in LBH-positive breast tumor cell lines using TOPFlash reporter assays. Strikingly, 2 out of 3 of these cell lines (HCC1187 and HCC1395) displayed increased Wnt/β-catenin signaling activity, similar to HC11 cells transfected with pCDNA/β-cateninS37Y (Fig. 7E). Consistent with previous studies (74), no detectable Wnt activity was measured in MDA-MB-231 cells, nor in HC11 cells, which served as a negative control. Furthermore, treatment of HCC1395 cells with DKK1 inhibitor blocked LBH expression, indicating that expression of LBH in this breast tumor cell line is dependent on Wnt/β-catenin signaling (Fig. 7F). Finally, we explored whether Wnt7a could serve as a means to inhibit LBH expression in basal breast tumor cells. Remarkably, treatment of HCC1395 cells with Wnt7a efficiently suppressed mRNA expression of LBH as well as of DKK1 (Fig. 7F). Thus, aberrant canonical Wnt signaling, at least in part, is responsible for LBH overexpression in basal subtype breast carcinoma cells.
DISCUSSION
Previous overexpression studies with mice and chicks have clearly defined a pivotal role of the novel vertebrate transcriptional regulator LBH in embryonic development and congenital disease (7, 8, 19). However, the molecular mechanisms upstream of LBH and its role in adult development have remained obscure. The present study links LBH for the first time to the Wnt signaling pathway, an essential developmental and oncogenic signaling network, as well as implicating LBH overexpression in breast cancer.
We demonstrate through cell-based induction, RNAi knockdown of β-catenin, and reporter assays, as well as through molecular in vitro EMSA and in vivo ChIP analysis, that LBH is a direct transcriptional target of the canonical Wnt/β-catenin pathway. Specifically LBH is upregulated via binding of TCF/β-catenin transcriptional complexes to four functional conserved TCF/LEF binding sites (75) in two Lbh enhancer regions. A basal Lbh promoter-reporter construct (Pwt) containing no TBE sites also showed transcriptional activation by β-cateninS37Y, which could be due to association of β-catenin with p300 or CREB (29, 78), each of which has binding sites in the basal Lbh promoter (data not shown). Interestingly, LBH induction by Wnt3a in 293T cells consistently occurred to a greater extent (4-fold versus 2-fold induction) and with more rapid kinetics than for the known canonical Wnt target gene DKK1 (13, 26, 51). Given that Wnt ligands act as morphogens and that different concentrations of Wnts elicit distinct cellular responses (81), these results might indicate that differences in the numbers and affinities of TBE sites in LBH versus DKK1 may be a critical determinant of when and where these Wnt target genes are turned on. Most importantly, the placement of LBH downstream of Wnt appears to be “universal” in mammalian epithelial development, as it was observed in both embryonic (293T, limb ectoderm) and adult (postnatal mammary gland, HC11 cells) epithelial systems, as well as in epithelial neoplasia (MMTV-Wnt1 mammary tumors, human breast cancer cell lines). Such an intimate relationship between Wnt/β-catenin and other target embryonic transcription factors does not always exist. For instance, TWIST, a transcriptional regulator of mesenchymal cell fate and inducer of epithelial-mesenchymal transition (EMT) during breast metastasis, is upregulated in mouse Wnt-induced breast tumors (22, 31, 79) but unlike LBH is not expressed in normal epithelial development. Moreover, the Lbh expression pattern overlapped not only with Wnt/β-catenin signaling activity during embryonic limb ectoderm development but also with the expression patterns of several Wnt ligands (9, 10, 24) during postnatal mammary gland development. Wnt has been shown to stimulate the growth of TEB cells during mammary gland branching morphogenesis, the expansion of the lobulo-alveolar compartment during pregnancy, and the self-renewal of mammary epithelial stem cells, which are interspersed in the basal myoepithelial layer (9, 69, 72). Specific expression of Lbh in these cellular targets of canonical Wnt signaling during normal mammary gland development, as well as the promitogenic and differentiation-inhibiting effects of Lbh overexpression in HC11 cells, suggests a potential novel function of LBH in adult mammary gland tissue homeostasis downstream of Wnt. A positive relationship between canonical Wnt signaling and Lbh expression may also have relevance to embryonic heart development, where LBH plays a major role (7). This is suggested by a recent microarray study showing that Lbh induction during cardiac differentiation of mouse embryonic stem (ES) cells is abolished by inhibition of canonical Wnt signaling (43). Thus, LBH may act downstream of the canonical Wnt pathway in multiple aspects of embryonic and adult vertebrate development.
By studying the effects of different Wnt ligands on LBH expression in tissue culture, we discovered that Wnt7a efficiently blocked Wnt3a-mediated induction of LBH as well as of the known β-catenin target gene DKK1 in 293T cells. Moreover, Wnt7a, which is a tumor suppressor in lung cancer (77), strongly repressed LBH and DKK1 expression in human HCC1395 breast cancer cells. To our knowledge, this is the first evidence that Wnt7a can inhibit canonical Wnt signaling. One possible mechanism for the observed inhibitory effect could be that Wnt7a activates the noncanonical planar cell polarity (PCP) pathway in tissue culture cells (12, 40). However, given our developmental genetics studies and the fact that Wnt7a and Wnt5a, which can also activate the PCP pathway (58, 61), exerted different effects on Wnt3-induced gene expression, we propose that the inhibitory effect of Wnt7a on canonical Wnt3a signaling in tissue culture cells could be mediated by a poorly understood noncanonical pathway involving Lmx1b (37).
The antagonistic relationship between canonical Wnt/β-catenin and noncanonical Wnt7a signaling may have important implications for dorsoventral limb pattern formation during embryonic development. It has remained enigmatic how expression of canonical Wnt target genes gets restricted to ventral limb and AER ectoderm, especially since Wnt3 and its downstream signaling components are expressed throughout the limb ectoderm (4, 54, 67). Furthermore, canonical Wnt signaling activity is detected in some dorsal ectodermal cells as evident from our TopGal reporter assays. Thus, ectopic expression of Lbh in distal dorsal limb ectoderm of mouse mutants lacking Wnt7a pathway activity (Wnt7a−/− and Lmx1b−/−) together with the repressive effects of Wnt7a on canonical Wnt signaling in tissue culture cells suggests that Wnt7a-Lmx1b signaling may be an important repressive mechanism that blocks Wnt/β-catenin target gene expression, and consequently ventral differentiation, in dorsal limb ectoderm.
Most importantly, aberrant overexpression of LBH in MMTV-Wnt1 mammary tumors, as well as in human breast tumors and cell lines, provides the first indication that somatic gain of LBH function occurs in cancer. Notably, LBH is specifically deregulated in ER-negative breast tumors and correlates strongly with the most clinically aggressive basal-like tumor phenotype (66). Elevated expression of LBH mRNA is observed in approximately half of basal-like tumors but is present in only 16 to 23% of luminal breast tumors, which have a good prognosis (66). Since only a few distinct molecular markers have been identified to date that are uniquely associated with basal-like breast cancers (47, 59), LBH may prove to be a valuable diagnostic marker for this difficult-to-treat clinical subtype.
The strong correlation between expression of LBH and canonical Wnt pathway genes in basal breast tumors, as well as TOPFlash reporter and inhibition assays in breast tumor cell lines, furthermore suggests that dysregulation of LBH in breast cancer is due, at least in part, to aberrant Wnt/β-catenin signaling. We detected high Wnt signaling activity in all LBH-expressing basal breast carcinoma cell lines (HCC1187 and HCC1395) except for MDA-MB-231 with cell-based TOPFlash reporter assays. However, recent short hairpin RNA (shRNA) knockdown studies of the Wnt coreceptor LRP6 suggest that MDA-MB-231 cells do have some endogenous Wnt signaling activity (42), which may account for LBH expression in this cell line. Moreover, this result may suggest that other, as-yet-unidentified, signaling pathways may also contribute to LBH overexpression in breast cancer. In further support of a clinical association of LBH with Wnt/β-catenin signaling, additional meta-analysis showed that LBH overexpression also correlates with Wnt pathway gene expression in colon cancer (A. H. Sims, unpublished data), which is primarily driven by Wnt activating mutations (18). Although deregulation of LBH in congenital heart disease is associated with increased LBH gene dosage, aCGH analysis does not suggest that increased LBH gene copy number is the underlying cause for LBH deregulation in basal-like breast cancers. However, we noted that luminal tumor cell lines frequently displayed a decrease in LBH copy number, suggesting that a decrease in LBH gene dosage may play a role in inactivation of LBH in ER-positive luminal tumors. Genomic instability inherent to tumor cells may also lead to LBH overexpression in other human cancers, as karyotypic anomalies involving translocation or triplication of chromosomal region 2p23, where LBH maps, are frequently observed in hematopoietic malignancies, testicular cancers, and neuroblastomas (20, 27, 35, 60). Thus, deregulation of LBH may be a more general event in human cancer.
In summary, our findings raise the intriguing possibility that LBH may act as a downstream effector of canonical Wnt/β-catenin signaling in both normal and neoplastic epithelial development, which is under the tight control of antagonistic noncanonical Wnt7a signaling.
Supplementary Material
Acknowledgments
We thank Bert Vogelstein, Tetsu Akiyama, Alberto Muñoz, Antonio Garcia de Herreros, Kermit Carraway, Andy McMahon, and Randy Johnson for reagents and mice. We are grateful to Alexandra Joyner for initial support of these studies. We also thank Joyce Slingerland, David Robbins, Mike Xu, and Chaitanya Jain for suggestions on the manuscript, Feng Gong for equipment usage, Virneliz Fernandez-Vega for technical assistance, and the University of Miami Analytical Imaging Core Facility for imaging.
This work was supported by the James and Ester King Biomedical Program, Florida State Department of Health NIR05-01-5186, the Braman Family Breast Cancer Institute of the University of Miami Sylvester Cancer Center (K.J.B.), DOD predoctoral fellowship W81XWH-08-1-0253 (M.E.R.), and funding from Breakthrough Breast Cancer (A.H.S.).
Footnotes
Published ahead of print on 6 July 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adamska, M., B. T. MacDonald, Z. H. Sarmast, E. R. Oliver, and M. H. Meisler. 2004. En1 and Wnt7a interact with Dkk1 during limb development in the mouse. Dev. Biol. 272:134-144. [DOI] [PubMed] [Google Scholar]
- 2.Al-Ali, H., M. E. Rieger, K. L. Seldeen, T. K. Harris, A. Farooq, and K. J. Briegel. 2010. Biophysical characterization reveals structural disorder in the developmental transcriptional regulator LBH. Biochem. Biophys. Res. Commun. 391:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, R. K., R. R. Friis, C. A. Schoenenberger, W. Doppler, and B. Groner. 1988. Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 7:2089-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrow, J. R., K. R. Thomas, O. Boussadia-Zahui, R. Moore, R. Kemler, M. R. Capecchi, and A. P. McMahon. 2003. Ectodermal Wnt3/beta-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev. 17:394-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Porath, I., M. W. Thomson, V. J. Carey, R. Ge, G. W. Bell, A. Regev, and R. A. Weinberg. 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40:499-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briegel, K. J. 2006. Embryonic transcription factors in human breast cancer. IUBMB Life 58:123-132. [DOI] [PubMed] [Google Scholar]
- 7.Briegel, K. J., H. S. Baldwin, J. A. Epstein, and A. L. Joyner. 2005. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor Lbh. Development 132:3305-3316. [DOI] [PubMed] [Google Scholar]
- 8.Briegel, K. J., and A. L. Joyner. 2001. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev. Biol. 233:291-304. [DOI] [PubMed] [Google Scholar]
- 9.Brisken, C., A. Heineman, T. Chavarria, B. Elenbaas, J. Tan, S. K. Dey, J. A. McMahon, A. P. McMahon, and R. A. Weinberg. 2000. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 14:650-654. [PMC free article] [PubMed] [Google Scholar]
- 10.Buhler, T. A., T. C. Dale, C. Kieback, R. C. Humphreys, and J. M. Rosen. 1993. Localization and quantification of Wnt-2 gene expression in mouse mammary development. Dev. Biol. 155:87-96. [DOI] [PubMed] [Google Scholar]
- 11.Calza, S., P. Hall, G. Auer, J. Bjohle, S. Klaar, U. Kronenwett, E. T. Liu, L. Miller, A. Ploner, J. Smeds, J. Bergh, and Y. Pawitan. 2006. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 8:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carmon, K. S., and D. S. Loose. 2008. Secreted frizzled-related protein 4 regulates two Wnt7a signaling pathways and inhibits proliferation in endometrial cancer cells. Mol. Cancer Res. 6:1017-1028. [DOI] [PubMed] [Google Scholar]
- 13.Chamorro, M. N., D. R. Schwartz, A. Vonica, A. H. Brivanlou, K. R. Cho, and H. E. Varmus. 2005. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J. 24:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, H., and R. L. Johnson. 1999. Dorsoventral patterning of the vertebrate limb: a process governed by multiple events. Cell Tissue Res. 296:67-73. [DOI] [PubMed] [Google Scholar]
- 15.Chen, H., Y. Lun, D. Ovchinnikov, H. Kokubo, K. C. Oberg, C. V. Pepicelli, L. Gan, B. Lee, and R. L. Johnson. 1998. Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat. Genet. 19:51-55. [DOI] [PubMed] [Google Scholar]
- 16.Chin, K., S. DeVries, J. Fridlyand, P. T. Spellman, R. Roydasgupta, W. L. Kuo, A. Lapuk, R. M. Neve, Z. Qian, T. Ryder, F. Chen, H. Feiler, T. Tokuyasu, C. Kingsley, S. Dairkee, Z. Meng, K. Chew, D. Pinkel, A. Jain, B. M. Ljung, L. Esserman, D. G. Albertson, F. M. Waldman, and J. W. Gray. 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10:529-541. [DOI] [PubMed] [Google Scholar]
- 17.Civenni, G., T. Holbro, and N. E. Hynes. 2003. Wnt1 and Wnt5a induce cyclin D1 expression through ErbB1 transactivation in HC11 mammary epithelial cells. EMBO Rep. 4:166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clevers, H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127:469-480. [DOI] [PubMed] [Google Scholar]
- 19.Conen, K. L., S. Nishimori, S. Provot, and H. M. Kronenberg. 2009. The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development. Dev. Biol. 333:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crockford, G. P., R. Linger, S. Hockley, D. Dudakia, L. Johnson, R. Huddart, K. Tucker, M. Friedlander, K. A. Phillips, D. Hogg, M. A. Jewett, R. Lohynska, G. Daugaard, S. Richard, A. Chompret, C. Bonaiti-Pellie, A. Heidenreich, P. Albers, E. Olah, L. Geczi, I. Bodrogi, W. J. Ormiston, P. A. Daly, P. Guilford, S. D. Fossa, K. Heimdal, S. A. Tjulandin, L. Liubchenko, H. Stoll, W. Weber, D. Forman, T. Oliver, L. Einhorn, M. McMaster, J. Kramer, M. H. Greene, B. L. Weber, K. L. Nathanson, V. Cortessis, D. F. Easton, D. T. Bishop, M. R. Stratton, and E. A. Rapley. 2006. Genome-wide linkage screen for testicular germ cell tumour susceptibility loci. Hum. Mol. Genet. 15:443-451. [DOI] [PubMed] [Google Scholar]
- 21.Desmedt, C., F. Piette, S. Loi, Y. Wang, F. Lallemand, B. Haibe-Kains, G. Viale, M. Delorenzi, Y. Zhang, M. S. d'Assignies, J. Bergh, R. Lidereau, P. Ellis, A. L. Harris, J. G. Klijn, J. A. Foekens, F. Cardoso, M. J. Piccart, M. Buyse, and C. Sotiriou. 2007. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 13:3207-3214. [DOI] [PubMed] [Google Scholar]
- 22.DiMeo, T. A., K. Anderson, P. Phadke, C. Feng, C. M. Perou, S. Naber, and C. Kuperwasser. 2009. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 69:5364-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavin, B. J., and A. P. McMahon. 1992. Differential regulation of the Wnt gene family during pregnancy and lactation suggests a role in postnatal development of the mammary gland. Mol. Cell. Biol. 12:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giese, K., A. Amsterdam, and R. Grosschedl. 1991. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 5:2567-2578. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Sancho, J. M., O. Aguilera, J. M. Garcia, N. Pendas-Franco, C. Pena, S. Cal, A. Garcia de Herreros, F. Bonilla, and A. Munoz. 2005. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene 24:1098-1103. [DOI] [PubMed] [Google Scholar]
- 27.Griffin, C. A., A. L. Hawkins, C. Dvorak, C. Henkle, T. Ellingham, and E. J. Perlman. 1999. Recurrent involvement of 2p23 in inflammatory myofibroblastic tumors. Cancer Res. 59:2776-2780. [PubMed] [Google Scholar]
- 28.Hayes, M. J., D. Thomas, A. Emmons, T. J. Giordano, and C. G. Kleer. 2008. Genetic changes of Wnt pathway genes are common events in metaplastic carcinomas of the breast. Clin. Cancer Res. 14:4038-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herschkowitz, J. I., K. Simin, V. J. Weigman, I. Mikaelian, J. Usary, Z. Hu, K. E. Rasmussen, L. P. Jones, S. Assefnia, S. Chandrasekharan, M. G. Backlund, Y. Yin, A. I. Khramtsov, R. Bastein, J. Quackenbush, R. I. Glazer, P. H. Brown, J. E. Green, L. Kopelovich, P. A. Furth, J. P. Palazzo, O. I. Olopade, P. S. Bernard, G. A. Churchill, T. Van Dyke, and C. M. Perou. 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howe, L. R., O. Watanabe, J. Leonard, and A. M. Brown. 2003. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 63:1906-1913. [PubMed] [Google Scholar]
- 32.Humphreys, R. C., and J. M. Rosen. 1997. Stably transfected HC11 cells provide an in vitro and in vivo model system for studying Wnt gene function. Cell Growth Differ. 8:839-849. [PubMed] [Google Scholar]
- 33.Imbert, A., R. Eelkema, S. Jordan, H. Feiner, and P. Cowin. 2001. Delta N89 beta-catenin induces precocious development, differentiation, and neoplasia in mammary gland. J. Cell Biol. 153:555-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivshina, A. V., J. George, O. Senko, B. Mow, T. C. Putti, J. Smeds, T. Lindahl, Y. Pawitan, P. Hall, H. Nordgren, J. E. Wong, E. T. Liu, J. Bergh, V. A. Kuznetsov, and L. D. Miller. 2006. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 66:10292-10301. [DOI] [PubMed] [Google Scholar]
- 35.Kansal, R., S. N. Sait, A. W. Block, P. M. Ward, F. L. Kelly, R. T. Cheney, M. Czuczman, M. L. Brecher, and M. Barcos. 2005. Extra copies of chromosome 2 are a recurring aberration in ALK-negative lymphomas with anaplastic morphology. Mod. Pathol. 18:235-243. [DOI] [PubMed] [Google Scholar]
- 36.Kao, J., K. Salari, M. Bocanegra, Y. L. Choi, L. Girard, J. Gandhi, K. A. Kwei, T. Hernandez-Boussard, P. Wang, A. F. Gazdar, J. D. Minna, and J. R. Pollack. 2009. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One 4:e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kengaku, M., J. Capdevila, C. Rodriguez-Esteban, J. De La Pena, R. L. Johnson, J. C. I. Belmonte, and C. J. Tabin. 1998. Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science 280:1274-1277. [DOI] [PubMed] [Google Scholar]
- 38.Khramtsov, A. I., G. F. Khramtsova, M. Tretiakova, D. Huo, O. I. Olopade, and K. H. Goss. 2010. Wnt/{beta}-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am. J. Pathol. 176:2911-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komiya, Y., and R. Habas. 2008. Wnt signal transduction pathways. Organogenesis 4:68-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Grand, F., A. E. Jones, V. Seale, A. Scime, and M. A. Rudnicki. 2009. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell 4:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, S. Y., W. Xia, J. C. Wang, K. Y. Kwong, B. Spohn, Y. Wen, R. G. Pestell, and M. C. Hung. 2000. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. U. S. A. 97:4262-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, C. C., J. Prior, D. Piwnica-Worms, and G. Bu. 2010. LRP6 overexpression defines a class of breast cancer subtype and is a target for therapy. Proc. Natl. Acad. Sci. U. S. A. 107:5136-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, Y., M. Asakura, H. Inoue, T. Nakamura, M. Sano, Z. Niu, M. Chen, R. J. Schwartz, and M. D. Schneider. 2007. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 104:3859-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Logan, C. Y., and R. Nusse. 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20:781-810. [DOI] [PubMed] [Google Scholar]
- 45.Loomis, C. A., E. Harris, J. Michaud, W. Wurst, M. Hanks, and A. L. Joyner. 1996. The mouse Engrailed-1 gene and ventral limb patterning. Nature 382:360-363. [DOI] [PubMed] [Google Scholar]
- 46.Lurie, I. W., H. G. Ilyina, D. B. Gurevich, N. V. Rumyantseva, I. V. Naumchik, C. Castellan, A. Hoeller, and A. Schinzel. 1995. Trisomy 2p: analysis of unusual phenotypic findings. Am. J. Med. Genet. 55:229-236. [DOI] [PubMed] [Google Scholar]
- 47.Mani, S. A., J. Yang, M. Brooks, G. Schwaninger, A. Zhou, N. Miura, J. L. Kutok, K. Hartwell, A. L. Richardson, and R. A. Weinberg. 2007. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc. Natl. Acad. Sci. U. S. A. 104:10069-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mikels, A. J., and R. Nusse. 2006. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 4:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neve, R. M., K. Chin, J. Fridlyand, J. Yeh, F. L. Baehner, T. Fevr, L. Clark, N. Bayani, J. P. Coppe, F. Tong, T. Speed, P. T. Spellman, S. DeVries, A. Lapuk, N. J. Wang, W. L. Kuo, J. L. Stilwell, D. Pinkel, D. G. Albertson, F. M. Waldman, F. McCormick, R. B. Dickson, M. D. Johnson, M. Lippman, S. Ethier, A. Gazdar, and J. W. Gray. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niehrs, C. 2006. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 25:7469-7481. [DOI] [PubMed] [Google Scholar]
- 51.Niida, A., T. Hiroko, M. Kasai, Y. Furukawa, Y. Nakamura, Y. Suzuki, S. Sugano, and T. Akiyama. 2004. DKK1, a negative regulator of Wnt signaling, is target of the beta-catenin/TCF pathway. Oncogene 23:8520-8526. [DOI] [PubMed] [Google Scholar]
- 52.Nusse, R., C. Fuerer, W. Ching, K. Harnish, C. Logan, A. Zeng, D. Ten Berge, and Y. Kalani. 2008. Wnt signaling and stem cell control. Cold Spring Harbor Symp. Quant Biol. 73:59-66. [DOI] [PubMed] [Google Scholar]
- 53.Nusse, R., and H. E. Varmus. 1982. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31:99-109. [DOI] [PubMed] [Google Scholar]
- 54.Oosterwegel, M., M. van de Wetering, J. Timmerman, A. Kruisbeek, O. Destree, F. Meijlink, and H. Clevers. 1993. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development 118:439-448. [DOI] [PubMed] [Google Scholar]
- 55.Parr, B. A., and A. P. McMahon. 1995. Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature 374:350-353. [DOI] [PubMed] [Google Scholar]
- 56.Pawitan, Y., J. Bjohle, L. Amler, A. L. Borg, S. Egyhazi, P. Hall, X. Han, L. Holmberg, F. Huang, S. Klaar, E. T. Liu, L. Miller, H. Nordgren, A. Ploner, K. Sandelin, P. M. Shaw, J. Smeds, L. Skoog, S. Wedren, and J. Bergh. 2005. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 7:R953-R964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perou, C. M., T. Sorlie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, C. A. Rees, J. R. Pollack, D. T. Ross, H. Johnsen, L. A. Akslen, O. Fluge, A. Pergamenschikov, C. Williams, S. X. Zhu, P. E. Lonning, A. L. Borresen-Dale, P. O. Brown, and D. Botstein. 2000. Molecular portraits of human breast tumours. Nature 406:747-752. [DOI] [PubMed] [Google Scholar]
- 58.Qian, D., C. Jones, A. Rzadzinska, S. Mark, X. Zhang, K. P. Steel, X. Dai, and P. Chen. 2007. Wnt5a functions in planar cell polarity regulation in mice. Dev. Biol. 306:121-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodriguez-Pinilla, S. M., D. Sarrio, G. Moreno-Bueno, Y. Rodriguez-Gil, M. A. Martinez, L. Hernandez, D. Hardisson, J. S. Reis-Filho, and J. Palacios. 2007. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod. Pathol. 20:474-481. [DOI] [PubMed] [Google Scholar]
- 60.Satge, D., S. W. Moore, C. A. Stiller, F. K. Niggli, K. Pritchard-Jones, N. Bown, J. Benard, and D. Plantaz. 2003. Abnormal constitutional karyotypes in patients with neuroblastoma: a report of four new cases and review of 47 others in the literature. Cancer Genet. Cytogenet. 147:89-98. [DOI] [PubMed] [Google Scholar]
- 61.Seifert, J. R., and M. Mlodzik. 2007. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat. Rev. Genet. 8:126-138. [DOI] [PubMed] [Google Scholar]
- 62.Semenov, M. V., R. Habas, B. T. Macdonald, and X. He. 2007. SnapShot: noncanonical Wnt signaling pathways. Cell 131:1378. [DOI] [PubMed] [Google Scholar]
- 63.Sims, A. H., G. J. Smethurst, Y. Hey, M. J. Okoniewski, S. D. Pepper, A. Howell, C. J. Miller, and R. B. Clarke. 2008. The removal of multiplicative, systematic bias allows integration of breast cancer gene expression datasets—improving meta-analysis and prediction of prognosis. BMC Med. Genomics 1:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smid, M., Y. Wang, Y. Zhang, A. M. Sieuwerts, J. Yu, J. G. Klijn, J. A. Foekens, and J. W. Martens. 2008. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 68:3108-3114. [DOI] [PubMed] [Google Scholar]
- 65.Sorlie, T., C. M. Perou, R. Tibshirani, T. Aas, S. Geisler, H. Johnsen, T. Hastie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, T. Thorsen, H. Quist, J. C. Matese, P. O. Brown, D. Botstein, P. Eystein Lonning, and A. L. Borresen-Dale. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 98:10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sorlie, T., R. Tibshirani, J. Parker, T. Hastie, J. S. Marron, A. Nobel, S. Deng, H. Johnsen, R. Pesich, S. Geisler, J. Demeter, C. M. Perou, P. E. Lonning, P. O. Brown, A. L. Borresen-Dale, and D. Botstein. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U. S. A. 100:8418-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soshnikova, N., D. Zechner, J. Huelsken, Y. Mishina, R. R. Behringer, M. M. Taketo, E. B. Crenshaw III, and W. Birchmeier. 2003. Genetic interaction between Wnt/beta-catenin and BMP receptor signaling during formation of the AER and the dorsal-ventral axis in the limb. Genes Dev. 17:1963-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sotiriou, C., P. Wirapati, S. Loi, A. Harris, S. Fox, J. Smeds, H. Nordgren, P. Farmer, V. Praz, B. Haibe-Kains, C. Desmedt, D. Larsimont, F. Cardoso, H. Peterse, D. Nuyten, M. Buyse, M. J. Van de Vijver, J. Bergh, M. Piccart, and M. Delorenzi. 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98:262-272. [DOI] [PubMed] [Google Scholar]
- 69.Teuliere, J., M. M. Faraldo, M. A. Deugnier, M. Shtutman, A. Ben-Ze'ev, J. P. Thiery, and M. A. Glukhova. 2005. Targeted activation of beta-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development 132:267-277. [DOI] [PubMed] [Google Scholar]
- 70.Tickle, C. 1999. Morphogen gradients in vertebrate limb development. Semin. Cell Dev. Biol. 10:345-351. [DOI] [PubMed] [Google Scholar]
- 71.Tsukamoto, A. S., R. Grosschedl, R. C. Guzman, T. Parslow, and H. E. Varmus. 1988. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 55:619-625. [DOI] [PubMed] [Google Scholar]
- 72.Turashvili, G., J. Bouchal, G. Burkadze, and Z. Kolar. 2006. Wnt signaling pathway in mammary gland development and carcinogenesis. Pathobiology 73:213-223. [DOI] [PubMed] [Google Scholar]
- 73.van Amerongen, R., A. Mikels, and R. Nusse. 2008. Alternative wnt signaling is initiated by distinct receptors. Sci. Signal. 1:re9. [DOI] [PubMed] [Google Scholar]
- 74.van de Wetering, M., N. Barker, I. C. Harkes, M. van der Heyden, N. J. Dijk, A. Hollestelle, J. G. Klijn, H. Clevers, and M. Schutte. 2001. Mutant E-cadherin breast cancer cells do not display constitutive Wnt signaling. Cancer Res. 61:278-284. [PubMed] [Google Scholar]
- 75.van de Wetering, M., M. Oosterwegel, D. Dooijes, and H. Clevers. 1991. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 10:123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, Y., J. G. Klijn, Y. Zhang, A. M. Sieuwerts, M. P. Look, F. Yang, D. Talantov, M. Timmermans, M. E. Meijer-van Gelder, J. Yu, T. Jatkoe, E. M. Berns, D. Atkins, and J. A. Foekens. 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365:671-679. [DOI] [PubMed] [Google Scholar]
- 77.Winn, R. A., L. Marek, S. Y. Han, K. Rodriguez, N. Rodriguez, M. Hammond, M. Van Scoyk, H. Acosta, J. Mirus, N. Barry, Y. Bren-Mattison, T. J. Van Raay, R. A. Nemenoff, and L. E. Heasley. 2005. Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J. Biol. Chem. 280:19625-19634. [DOI] [PubMed] [Google Scholar]
- 78.Xu, L., R. B. Corcoran, J. W. Welsh, D. Pennica, and A. J. Levine. 2000. WISP-1 is a Wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 14:585-595. [PMC free article] [PubMed] [Google Scholar]
- 79.Yang, J., S. A. Mani, J. L. Donaher, S. Ramaswamy, R. A. Itzykson, C. Come, P. Savagner, I. Gitelman, A. Richardson, and R. A. Weinberg. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927-939. [DOI] [PubMed] [Google Scholar]
- 80.Yang, J., and R. A. Weinberg. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14:818-829. [DOI] [PubMed] [Google Scholar]
- 81.Zecca, M., K. Basler, and G. Struhl. 1996. Direct and long-range action of a wingless morphogen gradient. Cell 87:833-844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.