Abstract
The major histocompatibility complex class II (MHC-II) locus includes a dense cluster of genes that function to initiate immune responses. Expression of insulator CCCTC binding factor (CTCF) was found to be required for expression of all MHC class II genes associated with antigen presentation. Ten CTCF sites that divide the MHC-II locus into apparent evolutionary domains were identified. To define the role of CTCF in mediating regulation of the MHC II genes, chromatin conformation capture assays, which provide an architectural assessment of a locus, were conducted across the MHC-II region. Depending on whether MHC-II genes and the class II transactivator (CIITA) were being expressed, two CTCF-dependent chromatin architectural states, each with multiple configurations and interactions, were observed. These states included the ability to express MHC-II gene promoter regions to interact with nearby CTCF sites and CTCF sites to interact with each other. Thus, CTCF organizes the MHC-II locus into a novel basal architecture of interacting foci and loop structures that rearranges in the presence of CIITA. Disruption of the rearranged states eradicated expression, suggesting that the formation of these structures is key to coregulation of MHC-II genes and the locus.
Spanning nearly 700 kb of the short arm of chromosome 6 at 6p21.31, the major histocompatibility complex class II (MHC-II) locus contains a dense cluster of highly polymorphic genes that encode the alpha and beta chains of the classical MHC-II heterodimeric molecules (reviewed in references 8, 21, and 59). Three MHC-II isotypes, HLA-DR, -DQ, and -DP, can be formed, and they present antigenic peptides to CD4 T lymphocytes in order to initiate, control, and/or maintain adaptive immune responses (20). This antigen presentation process is aided by two MHC-II region-encoded molecules, HLA-DM and HLA-DO, which are also alpha/beta heterodimers with sequence and structural homology with MHC-II proteins (55, 64). The locus includes several MHC-II homologous pseudogenes that represent defunct duplications of ancient MHC-II genes (29). Also within the locus are five genes that do not bear structural homology to MHC-II proteins, the TAP1 and TAP2 genes involved in peptide transport for MHC-I antigen presentation, the proteasome 20s core beta subunit genes PSMB8 and PSMB9, and the bromodomain and extra terminal domain transcriptional regulator gene BRD2 (25, 38, 67).
MHC-II genes are expressed in B lymphocytes, macrophages, and dendritic cells and certain cells within the thymus. Gamma interferon (IFN-γ) is a potent inducer of MHC-II gene expression in most cell types (12). The transcription factors RFX (regulatory factor X), CREB (cyclic AMP [cAMP] response element binding protein), and NF-Y (nuclear factor Y) are constitutively and ubiquitously expressed and bind to highly conserved MHC-II gene- and related gene promoter-proximal sequences termed the X1, X2, and Y boxes (9, 51, 59). CIITA (class II transactivator) interacts with the X-Y box-bound factors and mediates the interactions between the DNA-bound factors, the chromatin modification and remodeling factors, and the general transcription machinery (32, 51). CIITA expression is cell type specific and is induced upon exposure to IFN-γ (58). Each of these factors is required for MHC-II gene expression. While the genes involved in MHC-II antigen presentation are for the most part coregulated using the above-mentioned system, TAP1, TAP2, PSMB8, PSMB9, and BRD2 are not coexpressed or regulated with the classical MHC-II genes.
The mammalian protein CCCTC binding factor (CTCF) has been shown to demarcate and insulate regions of regulatory activity within the genome by functioning as an enhancer blocker (66) or by preventing the spread of heterochromatin into active genes (30). In vertebrates, all known insulator elements are associated with the function of CTCF (10, 18, 34). Using comparisons to model systems, CTCF has been proposed to function in part by forming the nexus of long-range chromatin loops, but the genes and regulatory-region targets of such loops and how the loops might coordinate gene expression are unknown. Recently, we identified a CTCF binding site (XL9) in the intergenic region between the HLA-DRB1 and HLA-DQA1 genes of the human MHC (35). Intriguingly, XL9 was found to serve as a focus for long-distance interactions with the promoters of the above-mentioned HLA genes (36). The interactions were dependent upon the presence of both CTCF and CIITA. Small interfering RNA (siRNA)-mediated CTCF knockdown resulted in a decrease in HLA-DRB1 and HLA-DQA1 expression, reduced histone acetylation at the promoters of the above-mentioned genes, and the loss of the long-distance interactions with the HLA-DRB1 and HLA-DQA1 promoter regions. CIITA and CTCF coimmunoprecipitated, suggesting that the two factors interacted directly or indirectly through a complex of factors. These observations demonstrated a role for CTCF in regulating HLA-DRB1 and HLA-DQB1 expression and suggested that CTCF may control gene expression by changing the spatial relationships and topological architecture of genes.
Here, CTCF activity was found to be required for expression of all MHC-II genes. This observation and our previous study (36) raised the question of whether XL9 regulated all MHC-II gene expression and whether it served as a hub for interactions with the promoter regions of the other MHC-II gene promoters or whether other CTCF elements were involved. Nine new CTCF sites were found in the MHC-II region. The sites separated the MHC-II subregions from each other in a manner that suggested that these regions might reflect borders of evolutionary duplications. Interactions between the CTCF sites and the MHC-II locus promoter regions were examined, and a complex network of transcription-associated and independent interactions was observed. Thus, these data describe novel CTCF interactions across a large and critical region of chromosome 6 and present two distinct modes of interaction, one that is associated with gene expression and another that is independent of expression. The former mode is required to express MHC-II genes and to provide a functional immune response.
MATERIALS AND METHODS
Cell culture.
The Burkitt's lymphoma B-cell line Raji (17) was used in this study as the wild-type MHC-II expressing cells and was obtained from the American Type Culture Collection (ATCC). The cell line RJ2.2.5 (provided by R. Accola, University of Insubria, Varese, Italy) is a derivative of the Raji line that does not express MHC-II genes due to mutations within the CIITA gene (1, 57). NCI-H929 (CRL-9068; ATCC), a plasmacytoma-derived cell line, does not express CIITA and MHC-II. These cells were grown in RPMI 1640 supplemented with 5% fetal bovine serum (HyClone, Inc., Logan, UT), 5% bovine calf serum (HyClone), 100 U/ml penicillin-streptomycin, 4.5 g/liter glucose, 1.0 mM sodium pyruvate, and 10 mM HEPES. A-431 cells, an epidermoid carcinoma cell line, obtained from the ATCC (CRL-1555), do not express MHC-II genes unless induced with IFN-γ (36). A-431 cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 100 U/ml penicillin-streptomycin. To induce MHC-II gene expression, cells were cultured with 500 U/ml of human IFN-γ (PeproTech Inc., NJ; catalog number 300-02) for the indicated time.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (6). Briefly, cells were cross-linked in 1% formaldehyde for 10 min, and a chromatin lysate was prepared and sonicated to generate fragments averaging 500 bp in length. For immunoprecipitations, 5 to 10 μg of anti-CTCF (Upstate; catalog number 06-917), cohesin (anti-Rad21; Upstate; catalog number 05-908), (anti-SMC3; Abcam; catalog number ab8263), anti-RFX5 (41), anti-CIITA (6), or anti-T-cell receptor (nonspecific control) antibody was used. Protein A beads (60 μl) were used to isolate the chromatin-antibody complexes. After extensive washing, the immunoprecipitated chromatin was eluted in 1% SDS and incubated overnight at 65°C to reverse the formaldehyde-induced cross-links. The DNA was purified and used as a template in real-time PCRs.
The immunoprecipitated DNA was quantitated by real-time PCR using a 5-point genomic DNA standard curve and an I-cycler (Bio-Rad Laboratories, Inc.). Quantitative-PCR mixtures contained 5% dimethyl sulfoxide (DMSO), 1× SYBR green (Bio Whittaker Molecular Applications), 0.04% gelatin, 0.3% Tween 20, 50 mM KCl, 20 mM Tris (pH 8.3), 3 mM MgCl2, 0.2 mM deoxynucleoside triphosphate(dNTP), and 100 nM each primer. Sequences for all primers used in the ChIP real-time PCR assays are available upon request. All ChIP experiments were performed at least three times from independent preparations of chromatin. The data were averaged and plotted with respect to the input chromatin.
EMSA.
Nuclear extracts were prepared from Raji cells as described previously (23, 53). The electrophoretic mobility shift assay (EMSA) probe was prepared by annealing complementary oligonucleotides of the chicken globin 5′ HS4 CTCF site (18, 34), end labeling them with [γ32P]ATP, and then purifying the probe by polyacrylamide gel electrophoresis. EMSA reaction mixtures (20 μl) contained 5% glycerol, 150 mM KCl, 20 mM HEPES, pH 7.9, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 0.5% Triton X-100, 400 ng poly(dG-dC), and 2 μg of nuclear extract. The reaction mixtures were incubated at room temperature for 10 min, and labeled probe and/or competitor DNA was added. The competitor DNAs corresponded to the core regions of the CTCF sites and were generated by PCR amplification of the primers (available upon request) used for ChIP assays.
Quantitative 3C assay.
A modified chromatin conformation capture (3C) assay protocol was performed in this experiment as described previously (36, 60). Cells (107) were washed in cold phosphate-buffered saline (PBS) buffer. Formaldehyde was added to the cells to a final concentration of 1%, and the cells were incubated for 10 min at room temperature. Glycine (final concentration, 125 mM) was added to stop the cross-linking reaction. Nuclei were collected from the cross-linked cells and digested overnight with either EcoRI, HindIII, or PstI, as indicated, at 37°C. The restriction enzymes were heat inactivated, diluted in ligation buffer at a ratio of ∼40:1, and then ligated overnight with T4 DNA ligase at 16°C. Proteinase K (final concentrations to 200 μg/ml) was added to the ligation reaction mixtures and incubated overnight at 65°C to reverse the cross-links and digest the proteins. The DNA was purified by phenol chloroform extraction and ethanol precipitated. Quantitation of the 3C products was performed by real-time PCR against a five-point standard curve as described previously (36). Standard-curve templates for the 3C products were generated in vitro by restriction enzyme cleaving and religating a bacterial artificial chromosome (BAC) containing the region being studied. The following BACs were used to cover the human MHC-II locus: RP11-974L24, RP11-257P24, RP11-544H13, RP11-10A19, RP11-1059J10, and RP11-699M5 (purchased from Children's Hospital Oakland Research Institute). All primer combinations were tested prior to use in the 3C assay to determine whether they could efficiently amplify a single product on the cleaved/religated BAC DNA. Primer sequences are available upon request. All primers had a greater-than-90% PCR efficiency and displayed only one product. Negative-control restriction fragments were chosen with the following bias. Control fragments had to be more than two restriction sites away from the test sequence and contained within a fragment that was <10 kb in length, as we have empirically found that very large fragments score poorly in these assays. Data are presented as cross-linked frequency and represent an average derived from three independent biological replicates. A standard error is provided for the data set. Student t tests were used to determine if observed differences were significant. Restriction enzyme site accessibility was tested in Raji, RJ2.2.5, A431, and H929 cells, as well as Raji cells depleted of CTCF. 3C chromatin was prepared and subjected to restriction digestion or left untreated according to the above-mentioned 3C protocol. After the cross-links were reversed and the DNA was purified, PCR amplification across the restriction site used was conducted, and the products were analyzed by agarose gel electrophoresis. Primer sequences for these reactions are available upon request.
RNAi and quantitative reverse transcription (RT)-PCR.
RNA interference (RNAi) using small interfering RNA (siRNA) was performed to knock down CTCF and HLA-DRA mRNA levels from Raji cells, using siRNA oligonucleotide SMART pools from Dharmacon Inc. (Lafayette, CO). These siRNAs were transfected into 4 × 106 Raji cells using a Nucleofection apparatus and transfecting reagents (Kit V) from Amaxa Biosystems. Following transfection, the cells were cultured for 72 h. Cells were then selected for the loss of HLA-DR surface expression using MACS colloidal superparamagnetic MicroBeads conjugated to anti-HLA-DR antibody (Miltenyi Biotech). HLA-DR surface expression is specifically lost following depletion of CTCF (36). Total RNA was isolated from either siRNA-treated or untreated cells using an RNeasy RNA isolation kit (Qiagen, Valencia, CA) according to company protocol. cDNA synthesis by Superscript II reverse transcriptase (Invitrogen, Inc., Carlsbad, CA) was performed using 2 μg of total RNA in a volume of 20 μl in PCR II buffer (Applied Biosystems, Foster City, CA) supplemented with MgCl2 to a final concentration of 5 mM. After reverse transcription, sample volumes were increased up to 200 μl with Tris-EDTA (TE) buffer. Three microliters of the cDNA produced was used for the subsequent quantitative-PCR assays. GAPDH primers were used in each of the experiments to normalize the data. The primers used in this experiment are available upon request. The data presented represent the average with standard deviation of three independent experiments. Statistical significance was calculated using a Student t test and specifically compared the values obtained using the HLA-DRA siRNAs to those using the CTCF siRNAs.
RESULTS
CTCF regulates MHC-II genes.
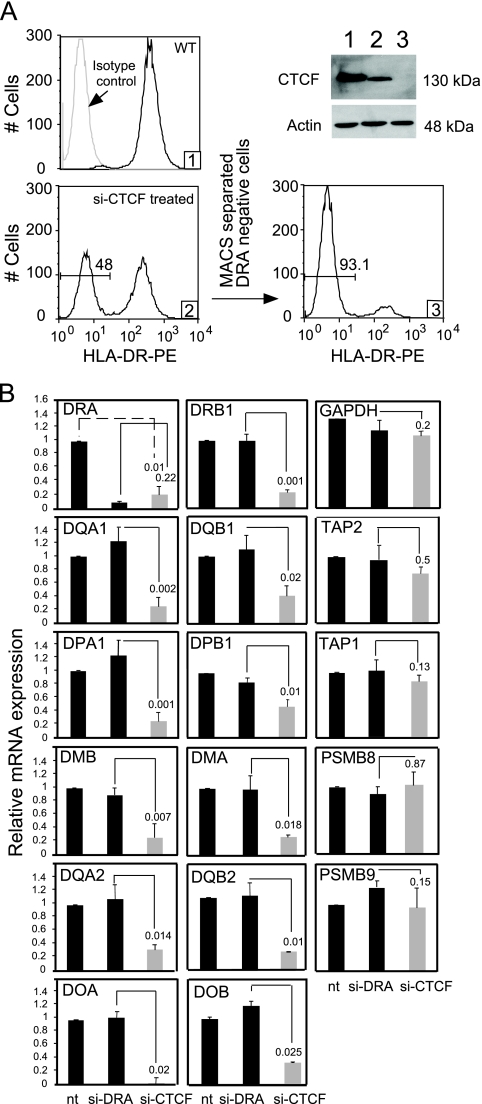
We showed recently that down modulation of CTCF through siRNA resulted in a reduction of the HLA-DRB1 and HLA-DQA1 mRNA transcripts and HLA-DR and -DQ surface expression (36). This result suggested that other MHC-II genes might also be regulated by CTCF. To determine if CTCF was required for the expression of all MHC-II genes, CTCF knockdown experiments in the MHC-II-expressing B-lymphoblastoid cell line Raji were conducted using CTCF-specific siRNA oligonucleotide SMART pools as previously described (36). Transient-transfection knockdown efficiency levels varied between 35 and 55% (Fig. 1A). As shown previously to be due to a loss of HLA-DRB1 mRNA, CTCF depletion results in a loss of HLA-DR surface expression, allowing the purification of CTCF-depleted cells by their failure to bind to HLA-DR antibody-coated paramagnetic MACS beads. This enrichment resulted in ∼90% of the cells being HLA-DR negative and depleted of CTCF (Fig. 1A). A similar population of HLA-DR-negative cells was generated using siRNAs to HLA-DRA transcripts.
FIG. 1.
Knockdown of CTCF specifically reduces the expression of human MHC-II genes. (A) Following siRNA knockdown in Raji cells using siRNAs specific to CTCF or HLA-DRA, the cells were purified by negative selection to HLA-DR antibody-coated MACS beads (36). Histograms 1, 2, and 3 display flow cytometry using phycoerythrin (PE)-conjugated antibodies for untreated, CTCF siRNA (si-CTCF)-treated, and negatively selected cells, respectively. The Western blot displays CTCF protein levels at each stage of purification. (B) MHC-II locus mRNA levels were determined by real-time RT-PCR using RNA prepared from untreated cells (nt) and cells transfected with si-CTCF or HLA-DRA siRNA (si-DRA), as purified in panel A. Specific reduc- tions in mRNA levels were observed for all genes involved in the MHC-II antigen presentation. No significant reduction was found with PSMB8, PSMB9, TAP1, TAP2, or GAPDH. These data were compiled from three biological replicates and are presented as relative mRNA expression with respect to the no-treatment (nt) lane. Standard errors are presented, along with Student t test comparisons between siRNAs to HLA-DRA and CTCF.
To determine the effects of CTCF knockdown on the expression of other genes within the MHC-II region, RNA isolated from untreated (not separated) and CTCF- and HLA-DRA siRNA-treated purified cells were analyzed by real-time RT-PCR (Fig. 1B). Whereas siRNA knockdown of HLA-DRA transcripts affected only the expression of the HLA-DRA gene, CTCF knockdown resulted in significant reductions in all the classical HLA class II genes. This reduction in gene expression extended to the MHC-II antigen-processing genes, HLA-DOA, -DOB, -DMA, and -DMB. The peptide transporter genes, TAP1 and TAP2, and the proteasome genes PSMB8 and PSMB9 (also included in the MHC-II locus) showed no reduction in expression, suggesting that their expression is not regulated by CTCF in these cells. GAPDH expression was also not affected. The finding that HLA-DRA mRNA knockdown did not affect HLA-DRB1 expression or expression of the other genes demonstrates specificity of the knockdown and that the loss of expression of one gene pair does not affect the expression of the other. We previously showed that similar siRNA depletion of CTCF had no effect on the expression of CIITA, RFX5, CARM1, Cox2, c-Myc, or PRMT1 mRNAs in these cells (36). Additionally, the in vivo binding of RFX5 or CIITA to the X-Y box of the HLA-DRB1 gene was unchanged following the reduction of CTCF in Raji cells (36). Together, these data indicate that CTCF is required for maximal expression of MHC-II and their related genes and that the effect is specific.
CTCF binds to 10 sites within the human MHC-II locus.
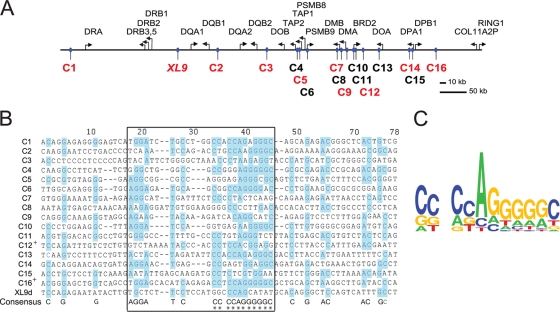
Using ChIP-chip and ChIP-seq approaches, two groups have mapped CTCF sites across the human genome (4, 27). More than 15,000 CTCF binding sites were identified in these studies, which involved human IMR90 fibroblasts and CD4+ T cells. Combining the data from these studies with a preliminary Chip-chip analysis of CTCF binding with Raji cells (data not shown) led to a compiled list with 16 additional CTCF sites (termed C1 to C16) across the human MHC-II locus (Fig. 2A). While it has been reported that CTCF recognizes long and diverse/different nucleotide sequences (19, 27, 28), DNA sequence alignment of the binding regions of these sites showed a low level of homology (Fig. 2B). A core consensus similar to that reported for CTCF (48) was deduced for these sequences, with several of the CTCF sites displaying a high degree of similarity within the core sequence (Fig. 2C).
FIG. 2.
Numerous putative CTCF sites sharing a consensus core sequence are located in the MHC-II locus. (A) Schematic diagram showing the relative positions of genes and CTCF binding sites (numbered C1 to C16) within the human MHC-II locus as identified by Barski et al. and Kim et al. (4, 27) and aligned using UCSC genome browser database tools (http://www.genome.ucsc.edu) (26). The red CTCF sites represent the confirmed sites as defined in the text. The black CTCF sites could not be confirmed. (B) The identified CTCF sites were aligned using vector NTI software. The region of greatest similarity is boxed. Blue highlights indicate conformation to a consensus sequence derived from the alignment, which is shown at the bottom of the alignment. The asterisks indicate similarities to published CTCF consensus sites (49). +, sequences in which the reverse complement was used in the alignment. (C) CTCF consensus sequence of core homology elements derived using a Web-based application (http://weblogo.berkeley.edu/ [13]).
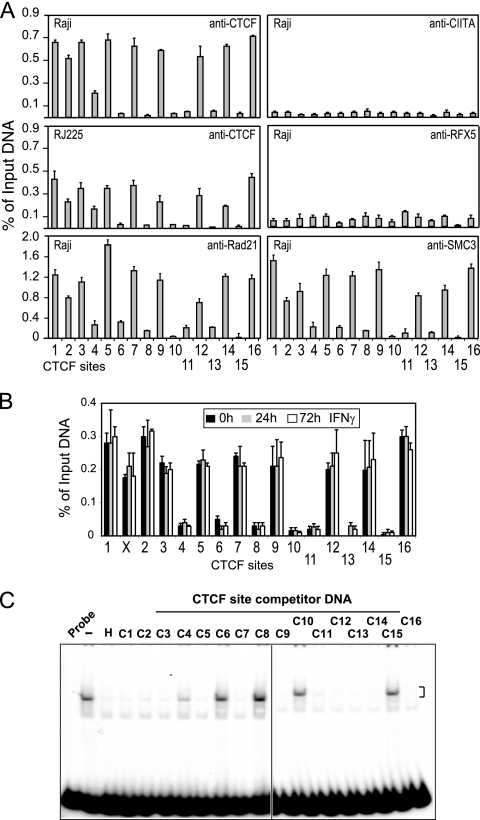
To determine whether any or all of these identified CTCF binding sites could bind CTCF in vivo, ChIP assays were employed on chromatin prepared from the MHC-II-expressing B-lymphoblastoid cell line Raji and the MHC-II-negative B-cell line RJ2.2.5 (Fig. 3A). Derived from Raji cells, RJ2.2.5 cells are mutated for CIITA and therefore MHC-II negative (1, 57). ChIP primers were designed to encompass the region of CTCF binding identified from whole-genome studies (4, 27). The results showed that 10 CTCF sites (C1, C2, C3, C4, C5, C7, C9, C12, C14, and C16) exhibited high levels of CTCF binding in both MHC-II-expressing and non-MHC-II-expressing cells (Raji and RJ2.2.5). While CTCF levels were consistently lower in RJ2.2.5 cells, the pattern of binding was similar. Low or near-background levels were found at sites C6, C8, C10, C11, C13, and C15. A similar pattern was also obtained for the plasma cell line H929, which is a CIITA- and MHC-II-negative plasmacytoma cell line (data not shown). Examination of the A-431 epidermoid carcinoma cell line showed similar binding of CTCF to the 10 sites (Fig. 3B), including the previously described XL9 site (36). Intriguingly, treatment of A-431 cells with IFN-γ showed no significant difference in the binding of CTCF to these sites (Fig. 3B), demonstrating that IFN-γ does not change the occupancy of CTCF at these sites, despite the fact that it induces MHC-II gene expression (36). CTCF antibodies did not immunoprecipitate HLA-DRA promoter region DNA, which does not contain a CTCF site (data not shown). ChIP experiments with anti-RFX5 or -CIITA antisera, which display robust binding at MHC-II promoter regions in Raji cells (data not shown and reference 22), showed no binding at any of the putative CTCF sites.
FIG. 3.
CTCF and cohesin bind strongly to nine new sites in the MHC-II locus. (A) Quantitative ChIP assays were conducted on Raji and RJ2.2.5 cells with the indicated antibodies to assess the in vivo occupancy of CTCF, CIITA, RFX5, and two cohesin components, Rad21 and SMC3, at the putative CTCF sites. Strong binding of CTCF was observed at 9 of the 16 sites, and intermediate binding was associated with 1 site (C4). Cohesin protein occupancy occurred only at the strong CTCF sites. No significant level of CIITA or RFX5 binding was detected at any of the CTCF sites. The error bars indicate standard errors. (B) Quantitative ChIP assays for CTCF binding to the 17 sites (including XL9) were conducted on A-431 cells, untreated or treated with IFN-γ for 24 and 72 h. All ChIP results were averaged from three independent chromatin preparations and are presented with respect to input chromatin. Standard errors are shown. (C) EMSA-based in vitro DNA competition using a 32P-labeled HS4 human β-globin insulator CTCF site probe was performed to assess the specificity of CTCF binding to the various CTCF sites. Competitor DNAs generated by PCR amplification of the CTCF sites are indicated. All strong CTCF binding sites competed efficiently for CTCF binding to the HS4 probe. Lanes: H, HS4 β-globin competitor DNA; −, no competitor DNA; C1 to C16, CTCF binding sites; and probe alone. The figure is representative of three independent experiments.
Competitor DNAs representing each of the CTCF sites were used to assess the ability of each putative site to compete for CTCF binding in an in vitro DNA binding EMSA. The CTCF site encoded in the chicken globin 5′HS4 insulator region was used as a probe (18), and Raji cell nuclear extracts were used as a source of CTCF. CTCF sites C1, C2, C3, C5, C7, C9, C11, C12, C13, C14, and C16 effectively competed for CTCF binding to the human 5′HS4 insulator CTCF site and demonstrated that these sites contain a CTCF binding site (Fig. 3C). Importantly C6, C8, C10, and C15 did not compete for CTCF binding, demonstrating that these regions do not encode CTCF sites. Unlike the ChIP results, which did not support CTCF binding at C11 and C13, the EMSA results suggest that these may be real sites that exhibit a cell type preference or bind CTCF weakly in vivo. C4 showed weak competitive activity, which corresponded with its weak binding activity as measured by ChIP in Raji and RJ2.2.5 cells, suggesting that C4 is at best a weak site in these cells. Background binding at C4 was observed in A-431 cells (Fig. 3B). Coupled with the previous results showing strong CTCF binding to XL9 (35), a total of 10 strong CTCF binding sites (C1, XL9, C2, C3, C5, C7, C9, C12, C14, and C16) are located within the MHC-II region (Fig. 2A, red letters).
MHC-II region CTCF sites bind cohesin.
Cohesin is expressed at high levels between the S and M phases of the cell cycle and is responsible for maintaining the pairing of sister chromatids during mitosis (37, 50, 63). Cohesin is composed of 4 subunits: Smc1, Smc3, Rad21/Scc1, and Scc3/SA1 (49). Recently, genome-wide ChIP-chip studies discovered that cohesin occupancy was enriched across the genome at CTCF sites (48, 56, 65). Additionally, transient knockdown of cohesin expression resulted in the loss of CTCF enhancer-blocking activity in a reporter assay, suggesting that cohesin was a contributor to insulator function (40). These results predict that strong CTCF site occupancy within the human MHC-II locus could be coincident with cohesin binding. To determine if this prediction is correct, ChIP was performed with antibodies to Smc3 and Rad21. The antibodies yielded similar results, which showed that the strong CTCF sites C1, C2, C3, C5, C7, C9, C12, C14, and C16 exhibited high levels of cohesin binding (Fig. 3A). XL9 also displayed cohesin binding that was comparable to that of the above-mentioned sites (data not shown). CTCF sites C4, C6, C11, and C13 exhibited 5- to 6-fold-lower levels than the average of the strong sites. Rad21 and Smc3 antibodies did not immunoprecipitate promoter region sequences of the HLA-DRA gene (data not shown). These data therefore indicate that cohesin is associated with the CTCF sites that reside within the MHC-II locus.
CTCF sites interact locally with the proximal promoter regions of MHC-II genes in a CIITA-dependent manner.
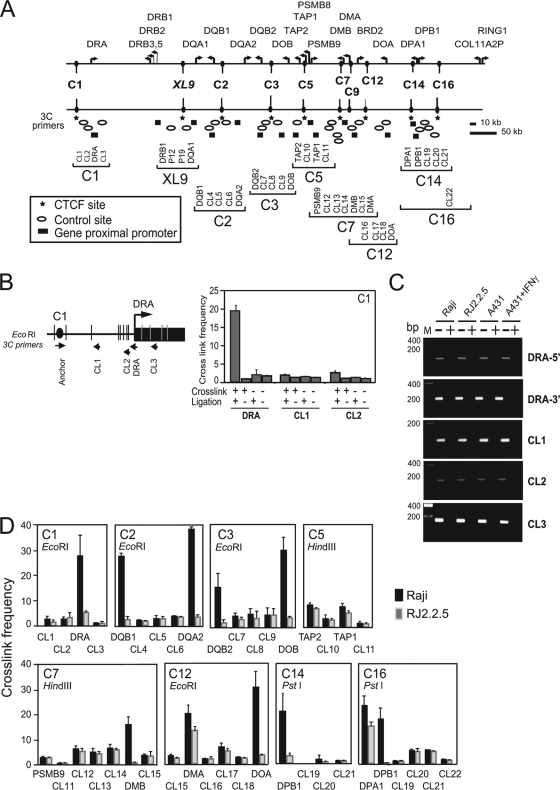
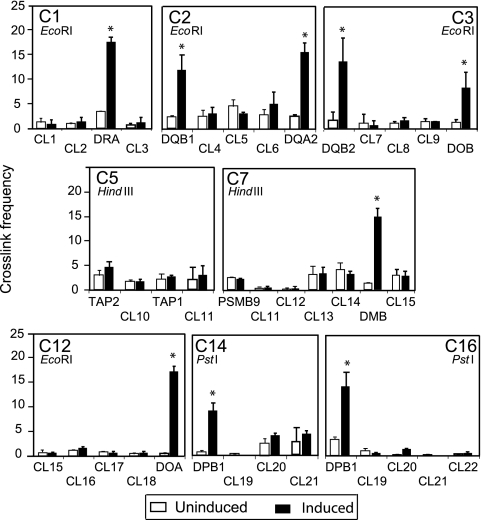
As described previously, XL9 interacted with the proximal promoter regions of the HLA-DRB1 and HLA-DQA1 genes (36), raising the question as to whether these CTCF sites also interact with MHC-II genes. To answer this question, quantitative 3C assays were performed (36). 3C assays allow one to capture and visualize interactions between cis-acting elements that are separated by long distances (14, 54). The assessment is carried out on formaldehyde-cross-linked chromatin that is subjected to complete restriction digestion, followed by dilution and subsequent ligation of the diluted DNA. Ligation conditions are such that only restriction fragments that remain in close proximity, due to the cross-linking, have a higher frequency of ligation than a random fragment. The novel sequence junction created by the ligation of the fragments is detected by quantitative PCR. BAC DNAs encoding the MHC-II locus were digested and ligated to provide a source DNA for the novel ligation junctions and served as a template for quantification. In this experimental series, interactions between each of the strong CTCF sites and the proximal promoter regions of the neighboring MHC-II genes, as well as a number of randomly chosen restriction fragments in the region, were assessed by 3C. Depending on the availability of restriction sites close to a cis element, assays were performed with EcoRI, HindIII, or PstI as the restriction enzyme. An overview schematic illustrating the locations of the primers and restriction sites used in these assays is presented in Fig. 4A. A detailed view of the C1 HLA-DRA region is shown in Fig. 4B. All primer pairs produced a single DNA band on the ligated BAC DNA but no bands on intact genomic DNA (data not shown).
FIG. 4.
CTCF sites interact with MHC-II gene promoter regions in a manner that is coincident with MHC-II gene activity. (A) Overview diagram of CTCF sites, MHC-II genes within the MHC-II locus, and the approximate positions of primer sets used to determine the relative frequencies of CTCF site interactions with the neighboring MHC-II gene promoters and control sites by 3C assay. The symbols represent the relative primer positions for CTCF sites, MHC-II gene promoter regions, and control restriction fragments, as indicated. (B) Detailed schematic of the HLA-DRA gene region displaying all EcoRI sites and the positions of the primers for CTCF site C1 and the HLA-DRA gene. In the histogram, control 3C reactions demonstrate that the formation of a 3C product between C1 and the restriction fragment containing the promoter region of HLA-DRA was dependent on cross-linking and DNA ligase. Interactions between C1 and the control restriction fragments CL1 and CL2 showed no 3C product. Relative cross-linked frequencies were determined by real-time PCR using 3C assay products. The error bars indicate standard errors. (C) Agarose gels of PCR assays across the restriction sites surrounding the HLA-DRA promoter, CL1, CL2, and CL3 sites are shown with (+) and without (−) restriction enzyme digestion of the formaldehyde-cross-linked 3C chromatin preparations. No differences were observed in the MHC-II-expressing and non-MHC-II-expressing cell lines shown for these sites or any other used in this study (data not shown). (D) A series of quantitative 3C assays determining the relative interaction frequencies between the indicated CTCF anchor sites and their neighboring MHC-II gene promoter region restriction fragments (as indicated) were conducted in Raji (MHC-II- and CIITA-positive) and RJ2.2.5 (MHC-II- and CIITA-negative) cell lines. Control regions are indicated as CL1 to CL22. The cross-linked frequency with standard error represents the relative average amount of 3C product (from three independent chromatin preparations) for each set of interactions divided by a nonspecific control fragment within each set of reactions.
To begin the analysis, interactions between CTCF site C1 and the HLA-DRA promoter restriction fragment were first assessed under conditions in which cross-linking and/or DNA ligase were not included (Fig. 4B). Interactions between C1 and the HLA-DRA promoter region restriction fragment were dependent on cross-linking and the inclusion of DNA ligase in the reaction. Importantly, interactions with nearby control region DNA sequences that reside between C1 and HLA-DRA showed no enhanced interactions. Thus, the experimental system showed specificity and an interaction between the C1 site and the HLA-DRA gene promoter region fragments. As assessed by PCR across the restriction site, no significant differences in restriction site accessibility to digestion for each site used in this study were observed among the different cell lines (Raji, RJ2.2.5, and A-431) or sites (Fig. 4C). Similar results were observed for all other restriction sites used in 3C in this study (data not shown).
Specific interactions between MHC-II gene promoters (HLA-DRA, DQB1, DQA2, DQB2, DOB, DMB, DOA, and DPB1) with the closest CTCF site analyzed were observed (Fig. 4D). For example, CTCF site C1 interacted at a high frequency with the HLA-DRA gene promoter region, C2 interacted with both HLA-DQB1 and HLA-DQA2 promoter regions, and C3 interacted with the promoter region fragments of HLA-DQB2 and HLA-DOB. With the exception of the C12-HLA-DMA and C16-HLA-DPA 3C assays, all interactions with MHC-II gene promoter regions occurred only in the CIITA-expressing cell line Raji and not in the CIITA mutant RJ2.2.5, demonstrating that CIITA was necessary for interactions with the CTCF sites. No interactions between the CTCF sites (C1, XL9, and C2) and HLA-DRA, -DRB1, or -DQA1 were observed in the plasma cell line H929, which lacks CIITA (data not shown). As specifically addressed below, the interactions observed between C12 and HLA-DMA, and C16 and HLA-DPA, suggested that another mechanism was at play for these experimental designs. Interactions between the CTCF sites and control restriction fragments were substantially lower for each region assayed, suggesting that there was no observable proximity bias in the assays. Importantly, the levels of the control interactions with the tested CTCF site were the same between Raji and RJ2.2.5 cells, suggesting that such signals represent background interactions that are commonly observed in 3C analyses (26, 43, 44, 61). Due to the absence of suitable restriction sites, specific roles for C9 could not be determined. TAP1, TAP2, and PSMB9 displayed low to near-background interaction levels with the CTCF sites tested (C5 and C7), and each displayed similar frequencies of interaction in Raji and RJ2.2.5 cells (Fig. 4D).
IFN-γ induces interactions between the CTCF sites and MHC-II promoters.
MHC-II genes and CIITA are not typically expressed in nonhematopoietic cells but can be induced upon exposure to IFN-γ (12, 16, 58). Exposure of cells to IFN-γ results in the induction of CIITA gene expression and assembly of CIITA at the MHC-II promoters (6, 57). To test whether the interactions between the CTCF sites and MHC-II promoter region restriction fragments were controlled by IFN-γ exposure and CIITA induction, 3C assays were performed on A-431 cells treated with IFN-γ (Fig. 5). In the absence of IFN-γ, 3C interactions were low or at background levels. This level of interaction likely represents a ground state for these genes in most cells that are CIITA and MHC-II negative. Following IFN-γ treatment, all MHC-II promoter regions assayed showed an increase in the frequency of 3C products associated with nearby CTCF sites. Interactions of HLA-DRB1 and HLA-DQA1 with XL9 were previously reported following IFN-γ induction (36). These interactions occurred after CIITA expression and were coincident with gene expression (36). HLA-DOB, which is specifically expressed in B lymphocytes and is not responsive to IFN-γ induction in A-431 cells (42), also showed a 3C interaction following IFN-γ treatment. Thus, while HLA-DOB expression is dependent on the presence of CIITA for its expression in B cells, an undefined mechanism controls its expression in non-B lymphocytes.
FIG. 5.
IFN-γ induces interactions between CTCF sites and MHC-II gene promoters. A-431 epithelial (MHC-II-negative) cells were treated with IFN-γ (500 U/ml) for 72 h to induce CIITA and MHC-II expression. 3C assays were carried out as described in the legend to Fig. 4 and represent the average of three independent experiments with standard errors. The asterisks indicate significant differences (P < 0.05) between control and IFN-γ-treated samples as determined by Student t tests. The anchor CTCF site is indicated at the top of each graph.
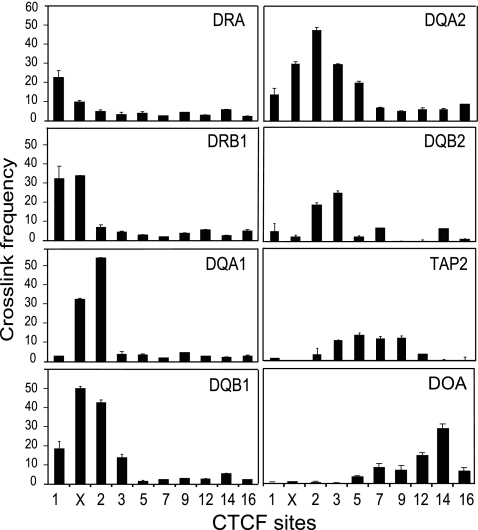
MHC-II gene promoter regions can interact with more than one CTCF site.
To determine if MHC-II gene promoter regions have a preference for one site over another or could interact with more than one site, 3C was performed across the locus using several of the MHC-II gene promoter regions as anchors (Fig. 6). HLA-DRA showed preferential interactions with C1 but could also interact to a lesser degree with XL9, which is ∼209 kb away. HLA-DRB1 interacted at the same frequency with C1 and XL9, the two CTCF sites that flank the gene. A similar observation was made for HLA-DQA1 and HLA-DQB1, which showed the highest-frequency interactions with XL9 and C2. HLA-DQB1 also interacted with moderate frequency with C1 and C3 but not with the CTCF sites located further down the locus. The pseudogenes HLA-DQA2 and HLA-DQB2, which are expressed (2, 3, 29, 45), interacted most strongly with C2 and C3, although HLA-DQA2 appears to form a broader spectrum of interactions across the locus. Because the same restriction enzyme (EcoRI) had to be used to assess interactions across the locus, some promoter interactions could not be assayed due to an inability to separate the promoter regions from CTCF sites by restriction digestion (e.g., HLA-DMA from C9, DPA/DPB from C16, and TAP1 from C5). As in the above-mentioned assays, TAP2 showed some interactions with neighboring CTCF sites. Overall, MHC-II promoter regions displayed a higher frequency for interaction with the closest CTCF sites but could also interact with multiple sites.
FIG. 6.
MHC-II promoter regions interact with multiple CTCF sites. 3C assays were performed using the indicated MHC-II locus gene promoters as anchors in Raji cells. To screen the greatest number of CTCF sites and MHC-II gene promoter regions, EcoRI was used as the restriction enzyme in this set of assays. 3C assays were carried out as described in the legend to Fig. 4 and represent the average of three independent experiments with standard errors. The MHC-II proximal promoter region used as the anchor for each series is indicated at the top of each graph.
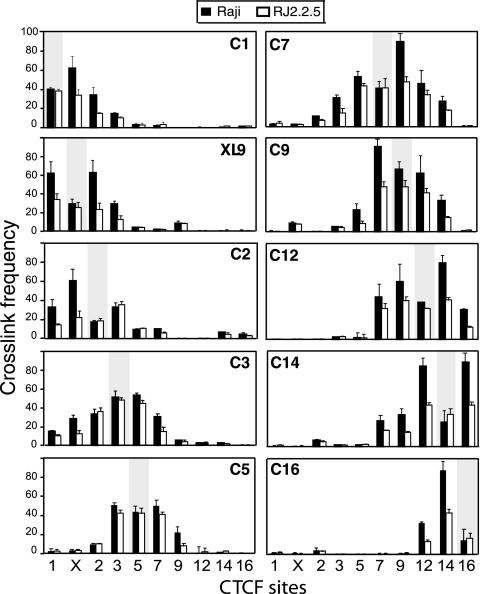
CTCF sites interact with each other, but such interactions are not associated with MHC-II gene expression.
Several cofactors have been identified as targeting CTCF (62, 68) or CTCF function (56, 65). Even without any cofactors, CTCF-bound DNA can self-interact and dimerize both in vivo and in vitro, thereby connecting two separate DNA fragments (5, 47). The large number of CTCF sites within the MHC-II region raises several questions. Do the CTCF sites interact with each other? If so, is there a preference for interactions, and do the interactions require a transcriptionally active MHC-II gene?
Using 3C assays, the relative frequencies of interactions between the strong CTCF sites (C1, XL9, C2, C3, C5, C7, C9, C12, C14, and C16) were determined in MHC-II-expressing Raji and non-MHC-II-expressing RJ2.2.5 cells. All of the strong CTCF sites interacted with multiple CTCF sites (Fig. 7). In general, the highest frequency of interaction was observed with the nearest upstream or downstream neighbor (Fig. 7). The frequencies decreased as the distances increased. Interactions were observed within approximately 250 kb of DNA across the region. In RJ2.2.5 cells, interactions between the CTCF sites were similar to those in Raji cells but were lower in frequency (Fig. 7), most likely reflecting the lower levels of CTCF present at these sites (Fig. 3A). Similar results for C1, XL9, and C2 were also observed in the H929 plasma cells (data not shown). Thus, CTCF-CTCF interactions occur across the locus irrespective of MHC-II expression, suggesting that these interactions form the foundation for this system's three-dimensional architecture. The self-ligated anchor fragments for each interaction set displayed equal ligation frequencies between Raji and RJ2.2.5 (Fig. 7), demonstrating that similar levels of chromatin were used in the assays and that the restriction sites surrounding each anchor primer were equally accessible between the cell lines.
FIG. 7.
Interactions between CTCF sites are not associated with MHC-II gene activity. 3C assays were conducted between CTCF sites to determine the frequencies of interactions between the sites. Raji (MHC-II-expressing) and RJ2.2.5 (MHC-II-negative) cells were compared. EcoRI was used as the enzyme that allowed the majority of sites to be assessed. 3C assays were carried out as described above. The data were derived from three independent preparations of cross-linked chromatin. The shaded areas represent self-ligating restriction fragments that contained the anchor primer set for each panel, as indicated. These self-ligating 3C fragments demonstrate that restriction site accessibilities for these fragments were similar in the two cell lines tested. The error bars indicate standard errors.
CTCF is required for all interactions with CTCF sites.
To determine a role for CTCF sites in the two sets of interactions described above, CTCF knockdown of Raji cells as performed in Fig. 1A was carried out to deplete CTCF from the system. ChIP for CTCF binding at the strong CTCF sites demonstrated that CTCF knockdown was effective in lowering the occupancy of CTCF at all CTCF binding sites within the MHC-II locus (Fig. 8A). Two sets of 3C assays were performed to examine the abilities of the CTCF sites to interact with the promoters of MHC-II genes and each other. In the absence of CTCF, interactions with MHC-II promoters were markedly reduced (Fig. 8B). Similarly, interactions between C1, XL9, and C2 and all other CTCF sites that could be assayed were also greatly reduced in the absence of CTCF (Fig. 8C). As determined by PCR across the restriction sites, CTCF depletion by RNAi did not affect the accessibility of the sites to restriction enzyme digestion during the 3C assay protocol (Fig. 8D and data not shown). Thus, CTCF is required for the formation of observed 3C interactions across the locus.
FIG. 8.
CTCF depletion results in loss of occupancy and 3C interactions across the locus. (A) ChIP assays for CTCF binding from control (siCONT) and CTCF si-RNA-treated HLA-DR-negative purified cells as described in the legend to Fig. 1 were carried out and analyzed for the indicated loci. The error bars indicate standard errors. (B) 3C was performed to assess interactions between the indicated CTCF sites and MHC-II promoter regions using chromatin preparations from control and si-CTCF-transfected cells, as in panel A. (C) 3C was performed to assess interactions between C1, XL9, and C2 and the other CTCF sites across the MHC-II region in the presence or absence of CTCF. The shaded bars indicate the self-ligating anchor 3C fragments, as in Fig. 7. The results of these assays represent an average derived from three independent siRNA replicates. (D) As in Fig. 4C, ethidium bromide-stained agarose gels of PCR assays were used to assess restriction enzyme accessibility in chromatin prepared from CTCF-depleted cells. CTCF depletion by RNAi did not prevent restriction enzyme access to the sites assayed by 3C region. All sites used were assessed and showed similar results (data not shown).
DISCUSSION
In this study, experiments were designed to identify the role and potential function of CTCF as it relates to MHC-II gene expression and chromatin organization. The results showed that CTCF is required for maximal expression of the classical MHC-II genes, as well as the MHC-II homologous accessory genes that are included within the locus. These data suggest that a common mechanism is used by CTCF to regulate the expression of this system and the immunological process of antigen presentation. While numerous CTCF sites were identified from ChIP-chip/seq studies (4, 24, 27), not all could be verified by either standard ChIP or EMSA in B lymphocytes. Such differences may reflect the cell types used (CD4 T cells and IMR90 fibroblasts versus B lymphocytes). There was also considerable sequence diversity among the sites, which may reflect the fact that CTCF contains 11 Zn fingers, only a few of which mediate DNA contact (19, 27). This would allow degenerate sites to evolve while still maintaining the function of the region. Nonetheless, a consensus sequence that matched the sequence derived from the genome studies was computationally derived from the MHC-II CTCF sites (4, 27).
The finding that the CTCF sites fell between subregions of the MHC-II regions, essentially separating/insulating MHC-II family genes from non-MHC-II family genes (Fig. 2A), was unexpected. For example, a pair of CTCF sites (C1 and XL9) surround the HLA-DR subregion, and a pair (XL9 and C2) surround the HLA-DQ region. C7 and C9 surround the HLA-DM genes, separating them from the proteosome and BRD2 genes. Intriguingly, this does not occur for HLA-DOB, which is within the same CTCF domain as TAP1, a gene that is ubiquitously expressed. This may contribute to or necessitate the unique regulation of HLA-DOB that dissociates it from the rest of the system (42). The occurrence of these domains, the fact that even the HLA-DQ pseudogenes are flanked by CTCF sites, and the reliance on CTCF for MHC-II expression suggests that such sites may mark the borders of ancient gene duplication events that occurred during the evolution of this locus. If cohesins and CTCF participate in events that mediate recombination, then it might be possible that misalignments at CTCF/cohesin sites within the MHC locus could facilitate duplication/deletion events.
3C assays record frequencies at which DNA fragments are in close spatial proximity to one another, and the formation of a 3C product between two DNA fragments is interpreted to demonstrate that those fragments bind factors that interact (15, 61, 68). While this assay provides a powerful approach to scoring interactions, there are some limitations (39). One limitation that was evident in this study was the paucity of convenient restriction sites that would allow separation of gene promoter regions from CTCF sites. Thus, some potential interactions could not be scored, despite the use of several enzymes. The assay also requires a significant number of controls (14). In addition to the biochemical controls provided here and previously (36), multiple control fragments were employed to demonstrate specificity of the interactions. Importantly, a genetic control could be employed in this analysis through the use of cells deficient for CIITA or cells depleted of CTCF. Additional specificity was provided by the observations that IFN-γ could induce CIITA and subsequent MHC-II promoter region-CTCF interactions. In a time course following IFN-γ induction in A-431 cells, CIITA is produced and detected within 4 to 6 h (36); however, MHC-II expression does not occur at that time point. At 12 h following IFN-γ treatment, both MHC-II expression and interactions between XL9 and HLA-DRB1 and HLA-DQA1 promoter regions could be detected (36). Such data support the notion that these interactions are critical to MHC-II gene expression.
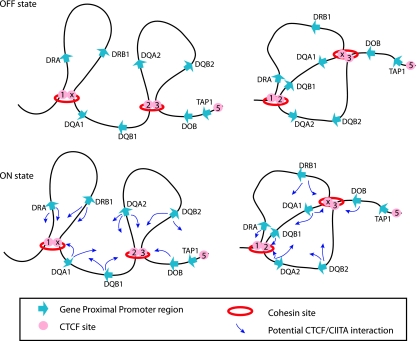
The results from this study define at least two organizational architectures that shape the three-dimensional structure of the human MHC-II locus (Fig. 9). In the transcriptionally “off” state for this system, where MHC-II genes are not expressed, the region is not a simple linear template, but rather a complex chromatin structure with inherent loops, with the foci of these loops occurring at interacting CTCF sites (Fig. 9, top). The 3C assay and the data do not allow determination of whether a single CTCF site interacts with only one other CTCF site at a time or whether it can interact with multiple sites at the same time. If CTCF site interactions occur through CTCF dimerization, as has been suggested (68), then such interactions may be limited to only a pair of sites. If this is the case, then multiple architectures can form across the locus, and the MHC-II architecture of one MHC chromosome will be different than that of its homolog. As such, the architectures within a population of B lymphocytes will be numerous. Two of the many potential architectures are depicted (Fig. 9, top) to emphasize the point that a CTCF site contained within one loop may also form interactions with other CTCF sites.
FIG. 9.
CTCF and MHC-II promoters form multiple architectures. Shown are schematic representations of CTCF-CTCF and CTCF-CIITA interactions for the HLA-DR and -DQ subregions. (Top) In the off state, CTCF sites within the MHC-II locus can interact with their neighboring sites in several different configurations, two of which are presented. These interactions are independent of MHC-II gene expression and represent a baseline architecture for the locus. (Bottom) In the transcriptionally on state, CIITA binds to the proximal promoter regions of MHC-II genes. CIITA-bound promoter regions can interact with one of several nearby CTCF sites, as indicated by the small arrows. The data suggest that many distinct configurations are possible, e.g., HLA-DRA can interact with either C1 or XL9 and DRB1 with C1 or XL9, etc.
The above model raises the question of how CTCF sites choose sites to interact with. One possibility could be that the sites choose to interact based on the relative occupancy of CTCF at a site. That is, sites with the highest CTCF occupancy frequency would interact with each other at the greatest frequency. While this may be true, the quantitative ChIP data presented showed a remarkably even pattern of CTCF binding across the locus (Fig. 3A). This suggests that distance/proximity and opportunity may be the determining factors. Thus, we propose that CTCF sites interact with the most proximal site at the time and do so in a stochastic manner. While the MHC-II region appears to display a distance-dependent effect on CTCF interactions, distance in the nucleus is complex, and our understanding of the three-dimensional architecture of the nucleus is in its infancy. In other gene systems, such as the Igf2-H19 and DM loci, CTCF sites were found to interact (52). In some cases, CTCF-CTCF site interactions were between chromosomes, such as in the Igf2-H19 locus in chromosome 7 and the Wsb1-Nf1 locus in chromosome 11 (33), arguing for spatial proximity or another mechanism as opposed to simple cis distance.
In B lymphocytes and IFN-γ-treated cells, CIITA is expressed, and by virtue of its ability to interact with CTCF either directly or in complexes (36), an additional set of interactions was identified. These interactions were between the CIITA-bound promoter fragments of the MHC-II genes and the CTCF-bound sites. Thus, in the transcriptionally “on,” or active, state, the CIITA-bound proximal promoter regions of MHC-II genes associate with the CTCF-bound foci of the loops (Fig. 9, bottom). The data suggest that MHC-II promoters do not have a single preferred CTCF site but can interact with multiple CTCF sites, and as above, we propose that spatial proximity/distance and opportunity are the determining features. It is not known how CTCF interacts with CIITA (if it does so directly), but because CTCF-CTCF interactions occur with similar frequencies irrespective of CIITA binding, it is likely that if CIITA binds directly to CTCF, it does so with a domain that is not required for CTCF-CTCF dimerization. Thus, it is not anticipated that a CTCF-CTCF interaction would prevent a CIITA-CTCF interaction. Previously, we observed an HLA-DRB1 and an HLA-DQA1 3C product. This suggests that both promoters can interact with a single CTCF focus: XL9 (36). In light of the current data, we suggest that because CTCF-CTCF site interactions orchestrate a background architecture, each member of the CTCF dimer could interact with a single MHC-II promoter. Thus, the data suggest that multiple configurations of the system are possible when the genes are expressed, with the principle that an expressed MHC-II gene does so by interacting with a CTCF site. In this model, it is likely that each MHC-II locus may adopt a distinct conformation within a single cell.
Genome-wide ChIP-chip studies recently found that cohesin binding sites and CTCF sites had a high degree of overlap, suggesting that the cohesin complex may play a role in the function activities associated with CTCF (48, 56, 65). Parelho et al. showed that CTCF binding is a prerequisite to target cohesin onto the DNA (48). Cohesin and CTCF can bind to the insulator of the chicken lysozyme locus and function as an upstream enhancer blocker (31). Here, it was observed that CTCF sites also bound cohesin subunits (Rad21 and Smc3). This may suggest that cohesin helps stabilize the foci of the loops and provides structural integrity to the architectures. Cohesin binding to the CTCF sites was independent of CIITA expression and binding to MHC-II promoters.
What is the role of CTCF in this system? Depletion of CTCF through siRNA demonstrated that CTCF was critical for MHC-II gene expression. Importantly, the TAP genes, which are expressed in these cells, were not affected by CTCF depletion, suggesting that the role of CTCF is specific to the MHC-II genes in this locus. Previous work showed that siRNA knockdown of CTCF resulted in a reduction of active histone modifications at the promoters of the HLA-DRB1 and HLA-DQA1 genes (36). Because of the possibility of forming a complex series of interactions between multiple CTCF sites and promoters, the architecture brings promoters into close proximity to each other and in a sense forms a domain of high transcriptional activity that is reminiscent of a transcription factory or chromatin hub (46). We suggest that the formation of the CTCF-CTCF-CIITA loops allows the coordinate expression of these antigen presentation and selection genes by bringing all of the regulatory components into the same three-dimensional space within the nucleus. In other systems, such as the human β-globin gene cluster, CTCF sites are within the locus control region flanking the entire locus (61). The CTCF sites also interact with one another, suggesting that this may be a common theme. Additional interactions occur with the β-globin locus control region (LCR) and the actively transcribed β-globin genes (11, 60). In the CFTR (cystic fibrosis transmembrane conductance regulator)-expressing primary epididymis, the CFTR promoter and 3′-end DNase hypersensitive region, which contains a CTCF binding site, interact with each other and form a chromatin loop, whereas no interaction was found in CFTR-negative skin fibroblasts (7). Each of these examples portrays a role for CTCF to organize a transcriptional domain. The MHC-II locus appears to have taken many of these features and used them to coordinate the expression of an entire locus by first establishing a baseline architecture that can reorganize itself into a transcriptional active domain when the signal (CIITA expression) is recruited into the locus.
Acknowledgments
We acknowledge R. Butler and J. Lee for excellent technical assistance and J. Lucchesi, P. Wade, and members of the laboratory for their critical comments on the manuscript. We also thank M. Krangel and E. Chan of Duke University for consultation on this work and G. Mooney for editorial assistance in the preparation of the manuscript.
This work was supported by Public Health Service grant GM47310 from the National Institute of General Medical Sciences.
We have no financial conflict of interest to report.
Footnotes
Published ahead of print on 28 June 2010.
REFERENCES
- 1.Accolla, R. S. 1983. Human B cell variants immunoselected against a single Ia antigen subset have lost expression of several Ia antigen subsets. J. Exp. Med. 157:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auffray, C., J. Kuo, R. DeMars, and J. L. Strominger. 1983. A minimum of four human class II alpha-chain genes are encoded in the HLA region of chromosome 6. Nature 304:174-177. [DOI] [PubMed] [Google Scholar]
- 3.Auffray, C., J. W. Lillie, A. J. Korman, J. M. Boss, N. Frechin, F. Guillemot, J. Cooper, R. C. Mulligan, and J. L. Strominger. 1987. Structure and expression of HLA-DQ alpha and -DX alpha genes: interallelic alternate splicing of the HLA-DQ alpha gene and functional splicing of the HLA-DQ alpha gene using a retroviral vector. Immunogenetics 26:63-73. [DOI] [PubMed] [Google Scholar]
- 4.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 5.Bartkuhn, M., and R. Renkawitz. 2008. Long range chromatin interactions involved in gene regulation. Biochim. Biophys. Acta 1783:2161-2166. [DOI] [PubMed] [Google Scholar]
- 6.Beresford, G. W., and J. M. Boss. 2001. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat. Immunol. 2:652-657. [DOI] [PubMed] [Google Scholar]
- 7.Blackledge, N. P., C. J. Ott, A. E. Gillen, and A. Harris. 2009. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 37:1086-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boss, J. M. 1997. Regulation of transcription of MHC class II genes. Curr. Opin. Immunol. 9:107-113. [DOI] [PubMed] [Google Scholar]
- 9.Boss, J. M., and P. E. Jensen. 2003. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr. Opin. Immunol. 15:105-111. [DOI] [PubMed] [Google Scholar]
- 10.Bushey, A. M., E. R. Dorman, and V. G. Corces. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell 32:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 12.Collins, T., A. J. Korman, C. T. Wake, J. M. Boss, D. J. Kappes, W. Fiers, K. A. Ault, M. A. Gimbrone, Jr., J. L. Strominger, and J. S. Pober. 1984. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 81:4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker, J. 2006. The three ′C's of chromosome conformation capture: controls, controls, controls. Nat. Methods 3:17-21. [DOI] [PubMed] [Google Scholar]
- 15.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 295:1306-1311. [DOI] [PubMed] [Google Scholar]
- 16.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein, M. A., B. G. Achong, Y. M. Barr, B. Zajac, G. Henle, and W. Henle. 1966. Morphological and virological investigations on cultured Burkitt tumor lymphoblasts (strain Raji). J. Natl. Cancer Inst. 37:547-559. [PubMed] [Google Scholar]
- 18.Farrell, C. M., A. G. West, and G. Felsenfeld. 2002. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol. 22:3820-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filippova, G. N., S. Fagerlie, E. M. Klenova, C. Myers, Y. Dehner, G. Goodwin, P. E. Neiman, S. J. Collins, and V. V. Lobanenkov. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germain, R. N., N. S. Braunstein, M. A. Brown, L. H. Glimcher, R. I. Lechler, J. McCluskey, D. H. Margulies, J. Miller, M. A. Norcross, W. E. Paul, et al. 1986. Structure and function of murine class II major histocompatibility complex genes. Mt. Sinai J. Med. 53:194-201. [PubMed] [Google Scholar]
- 21.Germain, R. N., and D. H. Margulies. 1993. The biochemistry and cell biology of antigen processing and presentation. Annu. Rev. Immunol. 11:403-450. [DOI] [PubMed] [Google Scholar]
- 22.Gomez, J. A., P. Majumder, U. M. Nagarajan, and J. M. Boss. 2005. X box-like sequences in the MHC class II region maintain regulatory function. J. Immunol. 175:1030-1040. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa, S. L., and J. M. Boss. 1991. Two B cell factors bind the HLA-DRA X box region and recognize different subsets of HLA class II promoters. Nucleic Acids Res. 19:6269-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jothi, R., S. Cuddapah, A. Barski, K. Cui, and K. Zhao. 2008. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 36:5221-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, A., S. H. Powis, L. A. Kerr, I. Mockridge, T. Elliott, J. Bastin, B. Uchanska-Ziegler, A. Ziegler, J. Trowsdale, and A. Townsend. 1992. Assembly and function of the two ABC transporter proteins encoded in the human major histocompatibility complex. Nature 355:641-644. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S. I., S. J. Bultman, C. M. Kiefer, A. Dean, and E. H. Bresnick. 2009. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl. Acad. Sci. U. S. A. 106:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, T. H., Z. K. Abdullaev, A. D. Smith, K. A. Ching, D. I. Loukinov, R. D. Green, M. Q. Zhang, V. V. Lobanenkov, and B. Ren. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klenova, E. M., R. H. Nicolas, H. F. Paterson, A. F. Carne, C. M. Heath, G. H. Goodwin, P. E. Neiman, and V. V. Lobanenkov. 1993. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 13:7612-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korman, A. J., J. M. Boss, T. Spies, R. Sorrentino, K. Okada, and J. L. Strominger. 1985. Genetic complexity and expression of human class II histocompatibility antigens. Immunol. Rev. 85:45-86. [DOI] [PubMed] [Google Scholar]
- 30.Labrador, M., and V. G. Corces. 2002. Setting the boundaries of chromatin domains and nuclear organization. Cell 111:151-154. [DOI] [PubMed] [Google Scholar]
- 31.Lefevre, P., J. Witham, C. E. Lacroix, P. N. Cockerill, and C. Bonifer. 2008. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol. Cell 32:129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeibundGut-Landmann, S., J. M. Waldburger, M. Krawczyk, L. A. Otten, T. Suter, A. Fontana, H. Acha-Orbea, and W. Reith. 2004. Mini-review: Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur. J. Immunol. 34:1513-1525. [DOI] [PubMed] [Google Scholar]
- 33.Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen, X. W. Qiu, A. M. Cherry, and A. R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312:269-272. [DOI] [PubMed] [Google Scholar]
- 34.Magdinier, F., T. M. Yusufzai, and G. Felsenfeld. 2004. Both CTCF-dependent and -independent insulators are found between the mouse T cell receptor alpha and Dad1 genes. J. Biol. Chem. 279:25381-25389. [DOI] [PubMed] [Google Scholar]
- 35.Majumder, P., J. A. Gomez, and J. M. Boss. 2006. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J. Biol. Chem. 281:18435-18443. [DOI] [PubMed] [Google Scholar]
- 36.Majumder, P., J. A. Gomez, B. P. Chadwick, and J. M. Boss. 2008. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 205:785-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marston, A. L., and A. Amon. 2004. Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 5:983-997. [DOI] [PubMed] [Google Scholar]
- 38.McCusker, D., T. Jones, D. Sheer, and J. Trowsdale. 1997. Genetic relationships of the genes encoding the human proteasome beta subunits and the proteasome PA28 complex. Genomics 45:362-367. [DOI] [PubMed] [Google Scholar]
- 39.Miele, A., and J. Dekker. 2008. Long-range chromosomal interactions and gene regulation. Mol. Biosyst. 4:1046-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishiro, T., K. Ishihara, S. Hino, S. Tsutsumi, H. Aburatani, K. Shirahige, Y. Kinoshita, and M. Nakao. 2009. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 28:1234-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno, C. S., E. M. Rogers, J. A. Brown, and J. M. Boss. 1997. Regulatory factor X, a bare lymphocyte syndrome transcription factor, is a multimeric phosphoprotein complex. J. Immunol. 158:5841-5848. [PubMed] [Google Scholar]
- 42.Nagarajan, U. M., J. Lochamy, X. Chen, G. W. Beresford, R. Nilsen, P. E. Jensen, and J. M. Boss. 2002. Class II transactivator is required for maximal expression of HLA-DOB in B cells. J. Immunol. 168:1780-1786. [DOI] [PubMed] [Google Scholar]
- 43.Ni, Z., M. Abou El Hassan, Z. Xu, T. Yu, and R. Bremner. 2008. The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat. Immunol. 9:785-793. [DOI] [PubMed] [Google Scholar]
- 44.Oestreich, K. J., R. M. Cobb, S. Pierce, J. Chen, P. Ferrier, and E. M. Oltz. 2006. Regulation of TCR beta gene assembly by a promoter/enhancer holocomplex. Immunity 24:381-391. [DOI] [PubMed] [Google Scholar]
- 45.Okada, K., H. L. Prentice, J. M. Boss, D. J. Levy, D. Kappes, T. Spies, R. Raghupathy, R. A. Mengler, C. Auffray, and J. L. Strominger. 1985. SB subregion of the human major histocompatibility complex: gene organization, allelic polymorphism and expression in transformed cells. EMBO J. 4:739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palstra, R. J., B. Tolhuis, E. Splinter, R. Nijmeijer, F. Grosveld, and W. de Laat. 2003. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35:190-194. [DOI] [PubMed] [Google Scholar]
- 47.Pant, V., S. Kurukuti, E. Pugacheva, S. Shamsuddin, P. Mariano, R. Renkawitz, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 24:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parelho, V., S. Hadjur, M. Spivakov, M. Leleu, S. Sauer, H. C. Gregson, A. Jarmuz, C. Canzonetta, Z. Webster, T. Nesterova, B. S. Cobb, K. Yokomori, N. Dillon, L. Aragon, A. G. Fisher, and M. Merkenschlager. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422-433. [DOI] [PubMed] [Google Scholar]
- 49.Peters, J. M., A. Tedeschi, and J. Schmitz. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev. 22:3089-3114. [DOI] [PubMed] [Google Scholar]
- 50.Petronczki, M., M. F. Siomos, and K. Nasmyth. 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423-440. [DOI] [PubMed] [Google Scholar]
- 51.Reith, W., and B. Mach. 2001. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 19:331-373. [DOI] [PubMed] [Google Scholar]
- 52.Rubio, E. D., D. J. Reiss, P. L. Welcsh, C. M. Disteche, G. N. Filippova, N. S. Baliga, R. Aebersold, J. A. Ranish, and A. Krumm. 2008. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. U. S. A. 105:8309-8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shapiro, D. J., P. A. Sharp, W. W. Wahli, and M. J. Keller. 1988. A high-efficiency HeLa cell nuclear transcription extract. DNA 7:47-55. [DOI] [PubMed] [Google Scholar]
- 54.Simonis, M., J. Kooren, and W. de Laat. 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods 4:895-901. [DOI] [PubMed] [Google Scholar]
- 55.Sloan, V. S., P. Cameron, G. Porter, M. Gammon, M. Amaya, E. Mellins, and D. M. Zaller. 1995. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature 375:802-806. [DOI] [PubMed] [Google Scholar]
- 56.Stedman, W., H. Kang, S. Lin, J. L. Kissil, M. S. Bartolomei, and P. M. Lieberman. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27:654-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steimle, V., L. A. Otten, M. Zufferey, and B. Mach. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 75:135-146. [PubMed] [Google Scholar]
- 58.Steimle, V., C. A. Siegrist, A. Mottet, B. Lisowska-Grospierre, and B. Mach. 1994. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science 265:106-109. [DOI] [PubMed] [Google Scholar]
- 59.Ting, J. P., and J. Trowsdale. 2002. Genetic control of MHC class II expression. Cell 109(Suppl.):S21-S33. [DOI] [PubMed] [Google Scholar]
- 60.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 61.Vakoc, C. R., D. L. Letting, N. Gheldof, T. Sawado, M. A. Bender, M. Groudine, M. J. Weiss, J. Dekker, and G. A. Blobel. 2005. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17:453-462. [DOI] [PubMed] [Google Scholar]
- 62.Wallace, J. A., and G. Felsenfeld. 2007. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17:400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watanabe, Y. 2004. Modifying sister chromatid cohesion for meiosis. J. Cell Sci. 117:4017-4023. [DOI] [PubMed] [Google Scholar]
- 64.Weber, D. A., B. D. Evavold, and P. E. Jensen. 1996. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science 274:618-620. [DOI] [PubMed] [Google Scholar]
- 65.Wendt, K. S., K. Yoshida, T. Itoh, M. Bando, B. Koch, E. Schirghuber, S. Tsutsumi, G. Nagae, K. Ishihara, T. Mishiro, K. Yahata, F. Imamoto, H. Aburatani, M. Nakao, N. Imamoto, K. Maeshima, K. Shirahige, and J. M. Peters. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796-801. [DOI] [PubMed] [Google Scholar]
- 66.West, A. G., S. Huang, M. Gaszner, M. D. Litt, and G. Felsenfeld. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453-463. [DOI] [PubMed] [Google Scholar]
- 67.York, I. A., and K. L. Rock. 1996. Antigen processing and presentation by the class I major histocompatibility complex. Annu. Rev. Immunol. 14:369-396. [DOI] [PubMed] [Google Scholar]
- 68.Yusufzai, T. M., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13:291-298. [DOI] [PubMed] [Google Scholar]