Abstract
The polypyrimidine tract near the 3′ splice site is important for pre-mRNA splicing. Using pseudouridine incorporation and in vivo RNA-guided RNA pseudouridylation, we have identified two important uridines in the polypyrimidine tract of adenovirus pre-mRNA. Conversion of either uridine into pseudouridine leads to a splicing defect in Xenopus oocytes. Using a variety of molecular biology methodologies, we show that the splicing defect is due to the failure of U2AF65 to recognize the pseudouridylated polypyrimidine tract. This negative impact on splicing is pseudouridine specific, as no effect is observed when the uridine is changed to other naturally occurring nucleotides. Given that pseudouridine favors a C-3′-endo structure, our results suggest that it is backbone flexibility that is key to U2AF binding. Indeed, locking the key uridine in the C-3′-endo configuration while maintaining its uridine identity blocks U2AF65 binding and splicing. This pseudouridine effect can also be applied to other pre-mRNA polypyrimidine tracts. Thus, our work demonstrates that in vivo binding of U2AF65 to a polypyrimidine tract requires a flexible RNA backbone.
In eukaryotic cells, the removal of intervening sequences (or introns) from mRNA precursors (pre-mRNA), a process known as splicing, is necessary for the production of mature mRNA (5, 13, 32, 40). Pre-mRNA possesses several key consensus sequence elements that are required for accurate splicing. Of these, the 5′ splice site, the 3′ splice site, and the branch site have been well documented (5). In higher eukaryotic pre-mRNA, there also exists a polypyrimidine tract (a sequence generally rich in uridines) adjacent to the 3′ splice site (1, 5, 12).
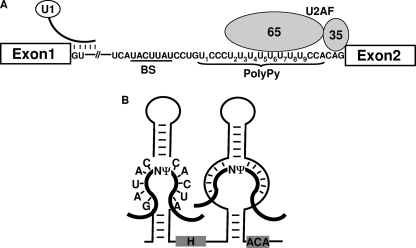
Pre-mRNA splicing occurs in the spliceosome, a multicomponent complex consisting of five small nuclear ribonucleoproteins (snRNPs)—U1, U2, U4, U5, and U6—and a large number of protein factors (5, 12, 13, 26, 32, 40). In test tubes, the spliceosome is assembled in a stepwise fashion. Early in spliceosome assembly, U1 recognizes the 5′ splice site, and U2AF (U2 auxiliary factor), a heterodimer with a 65-kDa subunit (U2AF65) and a 35-kDa subunit (U2AF35), recognizes the polypyrimidine tract/3′ splice site (Fig. 1A). U2AF65 also interacts with other splicing factors and helps recruit U2 to the branch site, thereby facilitating base-pairing interactions between U2 and the branch site. Finally, the U4/U5/U6 tri-snRNP joins the complex, converting it into a fully assembled spliceosome. Following a series of conformational changes, the spliceosome becomes activated, initiating splicing reactions.
FIG. 1.
Early stage of spliceosome assembly and box H/ACA RNA in directing RNA pseudouridylation. (A) At the early stage in spliceosome assembly, U1 recognizes the 5′ splice site of pre-mRNA, and U2AF65 recognizes the polypyrimidine tract. The branch site (BS) and polypyrimidine tract (PolyPy) of adenovirus pre-mRNA are shown. U1 and U2AF (both the U2AF65 and U2AF35 subunits) are also indicated. The uridines in the polypyrimidine tract are sequentially numbered. (B) The hairpin-hinge-hairpin-tail structure of a box H/ACA RNA is shown. The internal loops of the hairpins serve as guides (or pseudouridylation pockets) that base pair with their respective substrates. The thick lines represent substrate RNAs, and the thin lines stand for box H/ACA RNA. The target nucleotides to be pseudouridylated are indicated (Ψ). Box H and box ACA are also indicated. The guide sequence of the 5′ pseudouridylation pocket is taken from PugU2-34/44, a Xenopus box H/ACA guide RNA targeting U2 at positions 34 and 44.
The binding of U2AF65 to the polypyrimidine tract has been extensively studied (29-31, 36). Although polypyrimidine tracts vary widely in their nucleotide sequences, a pyrimidine (primarily uridine)-rich consensus sequence is sufficient for U2AF65 binding. Several years ago, the Kielkopf lab solved the crystal structure of the U2AF65 RNA binding domain bound to a polyuridine tract (29). The protein-RNA structure suggest that the side chains of U2AF65 are quite flexible in binding with RNA, thus offering a possible explanation as to how U2AF65 is able to bind to a variety of polypyrimidine tract sequences (29). However, relatively little is known about polypyrimidine tract requirements for binding.
Pseudouridine (Ψ) is found in stable eukaryotic RNAs, such as snRNAs and rRNAs (21, 25, 33). Ψ is derived from uridine via a posttranscriptional isomerization reaction known as pseudouridylation, in which the nitrogen-carbon bond linking the base to the sugar ring is broken and a new carbon-carbon bond forms, reestablishing the base-sugar linkage. The U-to-Ψ conversion frees the nitrogen on the base, thus creating an extra hydrogen bond donor in pseudouridine. In addition to a possible function in RNA-protein and RNA-RNA interactions, the extra hydrogen bond donor in Ψ can potentially add rigidity to the RNA backbone, locking the sugar pucker in a C-3′-endo conformation (2, 6, 15, 23).
Box H/ACA RNP-catalyzed (or RNA-guided) pseudouridylation is the primary mechanism responsible for most U-to-Ψ conversions in eukaryotes (25, 42). Box H/ACA RNP consists of one box H/ACA RNA and four core proteins, including Cbf5/Nap57/dyskerin (pseudouridylase), Nhp2, Gar1, and Nop10 (3, 7, 37, 42). The RNA component of the RNP folds into a unique structure known as the helix-hinge-helix-tail (Fig. 1B). The two internal loops each form a pseudouridylation pocket that base pairs with the RNA substrate, positioning the target uridine at the base of the upstem (Fig. 1B) (11, 24). Once the target uridine is identified and positioned, the pseudouridylase Cbf5/Nap57/dyskerin converts it into pseudouridine (10, 18). This modification scheme has been extensively studied and, consequently, verified in various organisms (3, 7, 11, 19, 24, 37, 38).
In the present work, we use U-to-Ψ conversion to identify uridines in pre-mRNA that are important for splicing in vivo. We show that conversion of uridine into pseudouridine (but not any other nucleotide) at either position 5 or position 7 in the polypyrimidine tract of adenovirus pre-mRNA (Fig. 1) blocks pre-mRNA splicing in Xenopus oocytes. Detailed analyses indicate that pseudouridylation at the critical uridine sites brings rigidity to the backbone of the polypyrimidine tract. Consequently, the pseudouridylated polypyrimidine tract loses its ability to bind to U2AF65. Our results thus demonstrate that in vivo binding of U2AF65 requires a flexible RNA backbone within the polypyrimidine tract.
MATERIALS AND METHODS
Construction of various adenovirus pre-mRNAs, U2 snRNA, and artificial box H/ACA guide RNAs.
T7 in vitro transcription was used to generate uniformly radiolabeled wild-type adenovirus pre-mRNA and mutant adenovirus pre-mRNA containing a U-to-G substitution at position 5 or 7 (U5 or U7) in the polypyrimidine tract, as previously described (39, 41). The full length of in vitro-transcribed adenovirus pre-mRNA is 415 nucleotides (nucleotides 1 to 66, exon 1; 67 to 309, intron; and 310 to 415, exon 2). T7 in vitro transcription was also used for generation of trace-radiolabeled U2 snRNA for splicing reconstitution experiments and trace-radiolabeled artificial box H/ACA guide RNAs for targeted pre-mRNA pseudouridylation. Based on PusU2-34/44, a naturally occurring Xenopus box H/ACA guide RNA (44), we constructed, via PCR, the T7-DNA templates for transcription of artificial box H/ACA guide RNA. The 5′ pseudouridylation pocket of PugU2-34/44 (Fig. 1B) was changed to fit the new target site, whereas the remaining nucleotide sequences remained unchanged.
Chimeric adenovirus pre-mRNAs, whose 5′ halves were derived from regularly transcribed pre-mRNA and whose 3′ halves were derived from pre-mRNA with pseudouridine substitution (or in reciprocal combination), were constructed according to a previously published protocol (Fig. 2B) (41). First, regular pre-mRNA and pre-mRNA with pseudouridine substitution were synthesized via T7 in vitro transcription. Specifically, while regular pre-mRNA was transcribed in the presence of regular nucleotides (nucleoside triphosphates [NTPs]), in vitro synthesis of pre-mRNA with pseudouridine substitution was carried out under similar conditions, except that UTP was replaced with ΨTP (Sierra Bioresearch, Tucson, AZ) in the transcription reaction. Second, regular pre-mRNA and pre-mRNA with pseudouridine substitution were subjected to site-specific RNase H cleavage directed by 2′-O-methyl RNA-DNA chimeras. Finally, by mixing and matching, the 5′ half of regular pre-mRNA and the 3′ half of pre-mRNA with pseudouridine substitution (or reciprocally, the 5′ half of pre-mRNA with pseudouridine substitution and the 3′ regular pre-mRNA) were aligned using an antisense bridging oligonucleotide and ligated by T4 DNA ligase (Fermentas). Three 2′-O-methyl RNA-DNA chimeras were used to direct the cleavage at three positions: the phosphodiester bond between nucleotides 2 and 3 with respect to the 5′ splice site (cleavage 1), the phosphodiester bond between nucleotides −19 and −20 with respect to the 3′ splice site (cleavage 2), and the phosphodiester bond between nucleotides −3 and −4 with respect to the 3′ splice site (cleavage 3). Upon ligation, three pairs of chimeric adenovirus pre-mRNAs were created.
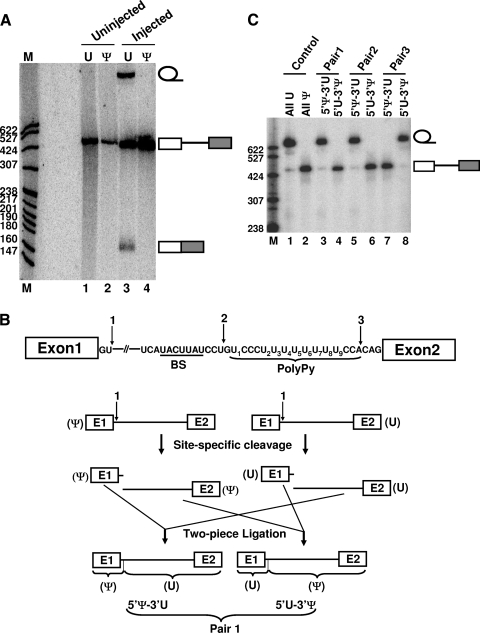
FIG. 2.
Mapping of the important uridines within adenovirus pre-mRNA. (A) pre-mRNA splicing in Xenopus oocytes. Lanes 1 and 2 are uninjected pre-mRNAs, regularly transcribed (U) and with pseudouridine substitution (Ψ), respectively. Lanes 3 and 4 are injected pre-mRNAs, regularly transcribed (U) and with pseudouridine substitution (Ψ), respectively. The RNA bands corresponding to the lariat intron, pre-mRNA, and mature mRNA are indicated. M, molecular size markers. (B) Construction of chimeric pre-mRNAs using RNase H site-specific cleavage directed by 2′-O-methyl RNA-DNA chimeras followed by mix-and-match two-piece ligation. The three cleavage sites are indicated, and the cleavage and ligation strategy, using cleavage site 1 as an example, is shown. Pre-mRNA with pseudouridine substitution is on the left, and the regularly transcribed (containing U) pre-mRNA is on the right. Upon cleavage at the same site (directed by the 2′-O-methyl RNA-DNA chimera), the 5′ and 3′ halves were mixed and matched. Two-piece ligation was then followed, generating a pair of chimeric pre-mRNAs. (C) Three pairs of chimeric pre-mRNAs (lanes 3 to 8) as well as regularly transcribed pre-mRNAs (lane 1; all U) and pre-mRNAs with pseudouridine substitution (lane 2; all Ψ) were assayed for splicing in Xenopus oocytes. 5′Ψ-3′U represents pre-mRNAs whose 5′ halves contained Ψ's and 3′ halves contained U's. 5′U-3′Ψ represents pre-mRNAs with reciprocal combinations.
To construct adenovirus pre-mRNA containing a 2′-F-uridine (IDT), 2′-H-uridine (IDT), or 2′-4′-locked uridine (C-3′-endo configuration) (14) at position 7 in the polypyrimidine tract, an in vitro-transcribed 5′ fragment (nucleotides 1 to 295), an in vitro-transcribed 3′ fragment (nucleotides 309 to 415), and a chemically synthesized middle-fragment oligoribonucleotide covering nucleotides 296 to 308 (5′-[32P]UUUUU[dU]UUCCACA-3′, where dU is 2′-deoxyuridine; 5′-[32P]UUUUU[FU]UUCCACA-3′, where FU is 2′-F-uridine; 5′-[32P]U[2′-4′U]UUUUUUCCACA-3′, where 2′-4′U is a uridine locked in the C-3′-endo configuration; or 5′-[32P]UUUUU[2′-4′U]UUCCACA-3′, where 2′-4′U is a uridine locked in the C-3′-endo configuration) were aligned using a bridging DNA oligonucleotide complementary to nucleotides 278 to 329 of adenovirus pre-mRNA (1:1:4:1 5′ fragment-3′ fragment-middle fragment-bridging oligonucleotide) and ligated together by T4 DNA ligase (Fermentas), essentially as described previously (39).
Adenovirus pre-mRNA containing a pseudouridine at position 7 (Ψ7) within the polypyrimidine tract was constructed by two-piece ligation. The 5′ piece (nucleotides 1 to 295) was in vitro transcribed, and the 3′ piece (nucleotides 296 to 319, where nucleotide 301 is equivalent to position 7 of the polypyrimidine tract and nucleotides 310 to 319 correspond to the first 10 nucleotides of exon 2) was chemically synthesized (Dharmacon) and 5′ phosphorylated with [γ-32P]ATP. Upon hybridization with the bridging oligonucleotide (complementary to nucleotides 278 to 329), the two pieces of adenovirus pre-mRNA were then ligated by T4 DNA ligase.
To produce a stable adenovirus polypyrimidine tract for the competition assay, a T7-polypyrimidine tract template (TAATACGACTCACTATAGGATGCAGGTAACTGCATGGAACCGCAGACCGAACGTCCCTTTTTTTTCACAGCACCATCTCGACGAGAGTCGAGAGAATT) was generated by overlapping PCR. T7 transcription with UTP or ΨTP generated an RNA containing, respectively, a regular polypyrimidine tract or polypyrimidine tract with pseudouridine substitution, flanked by a 5′ stem-loop structure and a 3′ stem-loop structure.
Xenopus oocyte microinjection and reconstitution of pre-mRNA splicing.
Microinjection and reconstitution were performed essentially as previously described (41). For checking the splicing activity of a pre-mRNA, a single injection was used. Specifically, 9 nl of 32P uniformly radiolabeled or 32P singly radiolabeled pre-mRNA was directly injected into the nuclei of Xenopus oocytes. After a 1-h incubation, the nuclei were isolated, RNA was recovered, and splicing was assayed on a 7% polyacrylamide-8 M urea gel (19:1 acrylamide-bisacrylamide; EM Science).
For the pseudouridylation-splicing reconstitution assay, multiple injections were used. Briefly, 46 nl of a mixture of 2 mg/ml antisense U2 DNA oligonucleotide (complementary to nucleotides 28 to 42) and 1 mg/ml in vitro-transcribed artificial box H/ACA guide RNA, targeting uridines of the adenovirus pre-mRNA polypyrimidine tract, were injected into the cytoplasm of Xenopus oocytes. After an overnight (or ∼4-h) incubation, 9 nl of radiolabeled adenovirus splicing substrate (500,000 cpm/μl) was injected into the nucleus of oocytes. Following another overnight (or ∼4-h) incubation, 40 nl of 50 ng/μl in vitro-transcribed U2 snRNA was injected into the cytoplasm of the oocytes to trigger splicing. After an overnight (or ∼1-h) reconstitution, nuclei were isolated. Upon proteinase K treatment, total nuclear RNA was extracted with perchloric acid (PCA) and then precipitated with ethanol. The recovered RNA was loaded on a 7% polyacrylamide-8 M urea gel (19:1 acrylamide-bisacrylamide; EM Science). Radiolabeled pre-mRNA and splicing products were visualized after autoradiography.
For the competition assay, the procedure was essentially the same, with one slight modification. Specifically, at the step of injecting radiolabeled adenovirus pre-mRNA, an excess amount (∼15- to 45-fold excess over labeled RNA) of trace-radiolabeled polypyrimidine tract (either regular or with pseudouridine substitution) was also injected.
Analysis of splicing complexes by native gel electrophoresis.
Spliceosome assembly was analyzed according to previous reports (16, 41). Briefly, 10 min after nuclear injection of 9 nl of radiolabeled adenovirus splicing substrate (500,000 cpm/μl), nuclei were isolated and broken by being pipetted up and down several times in 10 μl of loading dye containing 9 mM HEPES (pH 7.9), 22.5 mM KCl, 0.09 mM EDTA, 0.22 mM dithiothreitol (DTT), and 1 mg/ml heparin. The samples were then loaded onto a 4% polyacrylamide native gel (80:1 acrylamide-bisacrylamide). The splicing complexes were visualized by autoradiography.
Immunoprecipitation.
Anti-Sm (Y12) and anti-U2AF65 immunoprecipitations were carried out essentially as described previously (17, 41). Briefly, 9 nl of radiolabeled adenovirus splicing substrate (500,000 cpm/μl) was injected into the nuclei of Xenopus oocytes. After a 10 min-incubation, the oocyte nuclei were isolated and broken by pipetting up and down 20 times using P10 tips, followed by vigorous mixing in the Net-2 buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.05% NP-40). The sample was then clarified by centrifugation at 13,000 × g for 5 min, and the supernatant was mixed with anti-Sm antibodies (Y12) or anti-U2AF65 antibodies prebound to protein A-Sepharose. The mixture was then nutated for 2 h at 4°C. After a brief centrifugation, the beads were washed four times with Net-2 buffer and digested with proteinase K at 42°C in G50 buffer (20 mM Tris-HCl at pH 7.5, 300 mM sodium acetate, 2 mM EDTA, 0.3% SDS). Coprecipitated RNA was recovered by PCA extraction and ethanol precipitation and analyzed on a 7% denaturing gel.
U2AF65-polypyrimidine tract binding assay.
U2AF65 was expressed in Escherichia coli and purified by ion-exchange chromatography, as described previously (36, 43). The polypyrimidine tract of adenovirus pre-mRNA was synthesized in vitro (Dharmacon), and 5′ radiolabeled with [γ-32P]ATP and polynucleotide kinase (Promega). The binding assay was carried out essentially as described previously (22, 28). Briefly, a 10-μl reaction mixture containing 10 mM Tris-HCl, pH 7.5, 5% glycerol, 1 mM DTT, 50 mM KCl, 0.5 U/μl RNasin, 0.09 μg/μl bovine serum albumin (BSA), 0.15 μg/μl carrier tRNA, protein dilutions (0, 5, 10, or 23 ng/μl), and 1 fmol of 5′-radiolabeled RNA probe (polypyrimidine tract) was incubated at 30°C for 30 min. The reaction mixture was then immediately loaded onto a 5% native polyacrylamide gel (1:39 bisacrylamide-acrylamide). After electrophoresis, the polypyrimidine tract and polypyrimidine tract-UAF65 complex were visualized by autoradiography, and the dissociation constant (Kd) was calculated. The 5′-radiolabeled polypyrimidine tracts used in the binding assay were as follows: U7 probe, 5′-GUC CCU UUU UU7U UCC ACA GCT CGC GGT TG-3′; and Ψ7 probe, 5′-GUC CCU UUU UΨ7U UCC ACA GCT CGC GGT TG-3′.
Pseudouridylation assay.
To assess the efficiency of pseudouridylation, pre-mRNA containing a single 32P in the polypyrimidine tract (5′ of a test uridine), rather than a uniformly radiolabeled pre-mRNA, was injected into the U2-depleted Xenopus oocytes containing an artificial box H/ACA guide RNA. Following an overnight (or ∼4-h) incubation, nuclei were isolated and total nuclear RNA was recovered by PCA extraction and ethanol precipitation. The recovered RNA was then digested with nuclease P1 (final 200 μg/ml in 3 μl of sodium acetate at pH 5.2) for 1 h at 37°C. The digested sample was then dotted on thin-layer chromatography (TLC) PEI plates (EM Science) and chromatographed in HCl-H2O-isopropanol (15:15:70 [vol/vol/vol]) buffer for ∼6 to 7 h. 32PU and 32Pψ were separated and visualized by autoradiography. The ratio of 32PU to 32Pψ was determined using a PhosphorImager (Molecular Dynamics).
RESULTS
Replacement of uridines with pseudouridines in the polypyrimidine tract blocks pre-mRNA splicing.
To identify uridines within pre-mRNA that are required for splicing, we replaced uridines of the adenovirus pre-mRNA splicing substrate with pseudouridines via in vitro transcription (using ΨTP instead of UTP). The pre-mRNA was then injected into Xenopus oocytes, and in vivo splicing was assessed.
As shown in Fig. 2A, while regular pre-mRNA with uridines was efficiently spliced, pre-mRNA containing pseudouridines completely failed to splice. In this experiment, because all uridines were changed to pseudouridines in the pre-mRNA, the minimal U-to-Ψ changes needed to block splicing remained unclear.
To narrow down the list of possibly important uridine sites, we constructed three pairs of chimeric pre-mRNAs using a combination of site-specific RNase H cleavage directed by 2′-O-methyl RNA-DNA chimeras and two-piece ligation (Fig. 2B) (41). One RNA in each pair derived its 5′ sequence (exon 1 plus the first two nucleotides of the intron [arrow 1], exon 1 plus the 5′ intron sequence prior to the polypyrimidine tract [arrow 2], or exon 1 plus all but the last three nucleotides of the intron [arrow 3]) from pre-mRNA transcript with pseudouridine substitution, and its 3′ sequence (all but the first two nucleotides of the intron plus exon 2 [arrow 1], the intron sequence starting from the polypyrimidine tract plus exon 2 [arrow 2], or the last three nucleotides of the intron plus exon 2 [arrow 3]) from regular pre-mRNA transcript (containing uridines); conversely, in the other RNA, the 5′ sequence was derived from regular pre-mRNA transcript, and its 3′ sequence was from transcript with pseudouridine substitution. These chimeric pre-mRNAs were separately injected into the nuclei of Xenopus oocytes for splicing assays. As shown in Fig. 2C, pre-mRNAs containing pseudouridines in the 5′ portion of the molecule, including exon 1 and the first two nucleotides of intron (lane 3) or exon 1 plus the intron sequence until prior to the polypyrimidine tract (lane 5), spliced just as efficiently as did the control pre-mRNA containing no pseudouridines (lane 1). In contrast, the reciprocal pre-mRNAs containing pseudouridines in the 3′ portion of the molecule, starting with either the third nucleotide of the intron (lane 4) or the polypyrimidine tract (lane 6), completely failed to splice, just as the pre-mRNA with full seudouridine substitution did (lane 2). However, when the 5′ portion of the molecule was extended to include the polypyrimidine tract, the outcome became completely different. Specifically, pre-mRNA containing pseudouridines in the 5′ portion of the molecule completely failed to splice (lane 7); however, the reciprocal chimeric pre-mRNA containing pseudouridines in the 3′ portion of the molecule (lane 8) spliced as efficiently as did the control pre-mRNA (containing no pseudouridines) (lane 1). Together, these results indicated that uridine sites in the polypyrimidine tract (a total of 9) were sensitive to pseudouridine substitution.
Two of the nine uridines in the polypyrimidine tract are sensitive to U-to-Ψ change.
To further dissect the polypyrimidine tract and to pinpoint the important uridine(s) in this region, we used a new strategy, namely, RNA-guided RNA pseudouridylation, to examine each of the nine uridine sites. Using PugU2-34/44 (44), a known naturally-occurring Xenopus box H/ACA RNA, we constructed nine artificial box H/ACA guide RNAs, each targeting one of the nine uridines in the polypyrimidine tract. Specifically, for each artificial guide RNA, only the guide sequence in the 5′ pseudouridylation pocket of PugU2-34/44 was changed (to target the uridines in the polypyrimidine tract); all of the other PugU2-34/44 sequences remained unchanged (Fig. 1B).
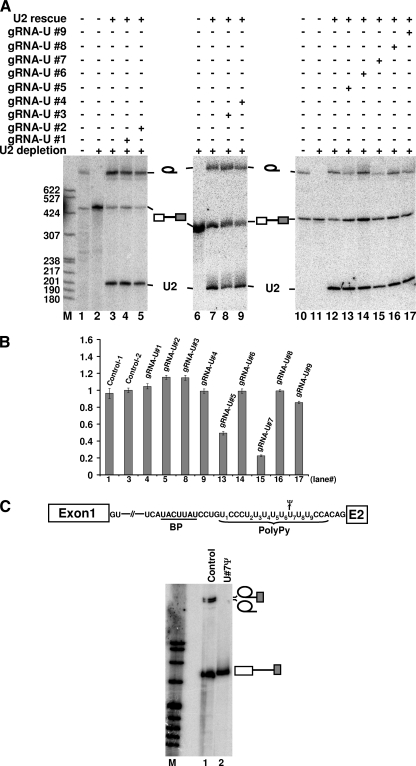
The artificial box H/ACA guide RNAs and pre-mRNA were sequentially injected into Xenopus oocytes depleted of U2. The purpose of using U2-depleted oocytes was to slow down (or completely pause) splicing, thus providing enough time for the guide RNA to direct pre-mRNA pseudouridylation in the polypyrimidine tract. After a 4-h incubation, U2 snRNA was then injected, thereby resuming splicing competence in the oocytes. As shown in Fig. 3A and B, while injection of seven of the nine guide RNAs had virtually no effect on pre-mRNA splicing, injection of guide RNA targeting U5 or U7 (lanes 13 and 15) resulted in greatly reduced splicing capability. To ensure that RNA-guided pseudouridylation occurred efficiently, we injected, under the same conditions, pre-mRNAs containing a single 32P at the target sites (rather than the uniformly labeled pre-mRNA). Upon a 4-h incubation, RNA was recovered, and pseudouridylation efficiency was assessed using RNase P1 digestion followed by TLC analysis. Our results indicated that RNA-guided pseudouridylation occurred almost equally efficiently (∼70 to 80%) at the target sites (data not shown). Thus, our results suggested that only two uridine sites, U5 and U7, were sensitive to U-to-Ψ substitution.
FIG. 3.
Identification of two important pseudouridines in the polypyrimidine tract by RNA-guided RNA pseudouridylation. (A) Nine artificial box H/ACA guide RNAs targeting the nine uridines in the polypyrimidine tract (gRNA-U#1 to gRNA-U#9) were separately injected into U2-depleted oocytes (lanes 4, 5, 8, 9, and 13 to 17). Labeled pre-mRNA was then injected. Following pre-mRNA pseudouridylation, U2 snRNA was injected to rescue splicing activity. Lanes 1 and 10 are controls in which radiolabeled pre-mRNA was directly injected into intact oocytes. Lanes 2, 6, and 11 are negative controls in which radiolabeled pre-mRNA was injected into U2-depleted oocytes and no rescuing U2 was injected later. Lanes 3, 7, and 12 are positive controls in which radiolabeled pre-mRNA was injected into U2-depleted oocytes and rescuing U2 was injected later. Bands corresponding to the lariat intron, pre-mRNA, and rescuing U2 are indicated. (B) Relative splicing efficiency (relative ratio of lariat intron to the sum of lariat intron and unspliced pre-mRNA) of each reaction was quantified based on three independent experiments, with the control reaction (U2 depletion and reconstitution) being set as 1 (control-2). The numbers on the x axis correspond to the lane numbers in panel A. (C) Pre-mRNA containing a wild-type U7 (lane 1) or a U7-to-Ψ7 change (lane 2) in the polypyrimidine tract was synthesized (also see the top panel), and splicing was examined in Xenopus oocytes. Bands corresponding to the 2/3 lariat intermediate, lariat intron product, and unspliced pre-mRNA are indicated. BP, branch point; M, molecular size markers.
To verify our observations, we constructed, using chemical synthesis and two-piece ligation, a pre-mRNA in which U7 was completely replaced with Ψ7 (see Materials and Methods). This pre-mRNA failed to splice, whereas the parallely synthesized control pre-mRNA (containing U7) underwent substantial splicing (Fig. 3C). Thus, our experiments demonstrated that U7 (and most likely U5 as well) was a key uridine that could not tolerate U-to-Ψ substitution.
The inhibitory effect on splicing is pseudouridine specific.
Although it is an isomerization reaction, strictly speaking, the U-to-Ψ conversion could be considered a mutation. To gain a better understanding of the effect of this U-to-Ψ conversion, we next carried out a comparison analysis in which U-to-Ψ conversion was compared with a uridine-to-other-nucleotide mutation.
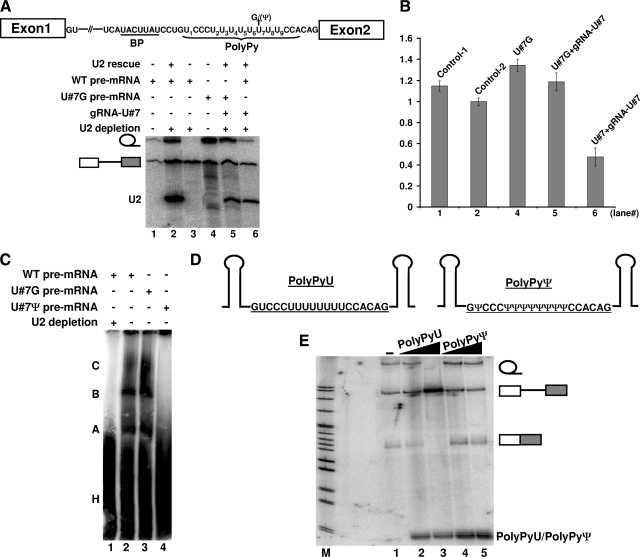
Specifically, we constructed pre-mRNAs containing either a U7-to-G7, U7-to-C7, or U7-to-A7 mutation in the polypyrimidine tract and tested their ability to splice in Xenopus oocytes. Interestingly, these mutant pre-mRNAs spliced nearly as well as the wild-type pre-mRNA did (Fig. 4A and B, lanes 4 and 5; data not shown), in either the presence or absence of the artificial guide RNA targeting U7. This result is in sharp contrast to the result shown in Fig. 3C, where pre-mRNA containing Ψ7 in the polypyrimidine tract completely failed to splice. Consistently, in the presence of the artificial guide RNA targeting U7, wild-type pre-mRNA splicing was greatly reduced (Fig. 4A and B, lane 6). Together, these results indicated that the inhibitory effect on pre-mRNA splicing was not merely due to a mutation. It appeared that the effect was pseudouridine specific.
FIG. 4.
Ψ-specific effect and its impact on the assembly of early splicing complexes. (A) Wild-type (WT) pre-mRNA (lanes 1, 2, 3, and 6) and a mutant pre-mRNA containing a U7-to-G7 change (lanes 4 and 5) were assayed for splicing in Xenopus oocytes. Lanes 1 and 4, splicing in intact oocytes; lane 3, splicing in U2-depleted oocytes; lane 2, splicing in oocytes in which U2 was depleted and later reconstituted; lanes 5 and 6, as in lane 2, with the exception that an artificial box H/ACA guide RNA, targeting position 7 of the polypyrimidine tract, was present. BP, branch point sequence. (B) Relative splicing efficiency (see legend to Fig. 3B). The numbers on the x axis correspond to the lane numbers in panel A. (C) Wild-type pre-mRNA (lanes 1 and 2) and mutant pre-mRNA containing a U7-to-G7 (lane 3) or a U7-to-Ψ7 (lane 4) change were assayed for spliceosome assembly in Xenopus oocytes. Lane 1 is a negative control in which spliceosome assembly was assayed in U2-depleted oocytes. Splicing complexes A, B, and C, as well as the heterogeneous complex H, are indicated. (D) Competitor adenovirus polypyrimidine tracts (with U [PolyPyU] on the left and with Ψ [PolyPyΨ] on the right) sandwiched by two stem structures are shown. (E) Wild-type pre-mRNA splicing was tested in the presence of competitor PolyPyU, with 15-fold (lane 2) and 45-fold (lane3) molar excess relative to pre-mRNA, or in the presence of competitor PolyPyΨ, with 15-fold (lane 4) and 45-fold (lane 5) molar excess. To monitor the stability of the competitors, a trace amount of radiolabeled PolyPyU or PolyPyΨ was mixed with the unlabeled competitor before injection. Lane 1, splicing without competitor. Bands corresponding to lariat intron, unspliced pre-mRNA, and spliced mRNA, as well as competitor PolyPyU or PolyPyΨ, are indicated.
The change of U to Ψ in the polypyrimidine tract affects binding of a splicing factor at an early stage in spliceosome assembly.
To identify the stage of spliceosome assembly or function affected by the U-to-Ψ change, we conducted native complex gel analysis (41). Wild-type pre-mRNAs containing U7 in the polypyrimidine tract, mutant pre-mRNAs containing G7 in the polypyrimidine tract, or altered pre-mRNAs containing Ψ7 in the polypyrimidine tract were injected into the oocyte nuclei under mineral oil. Ten minutes later, the nuclei were broken, mixed with native gel loading dye, and loaded on a native gel. As shown in Fig. 4C, both wild-type pre-mRNA and mutant pre-mRNA containing G7 in the polypyrimidine tract were able to assemble into splicing complexes A, B, and C (lanes 2 and 3). In contrast, pre-mRNA containing Ψ7 was unable to form any splicing-specific complexes (lane 4), suggesting that the effect manifested itself at an early stage during spliceosome assembly.
To gain some insights into the mechanisms by which the U-to-Ψ conversion in the polypyrimidine tract inhibited pre-mRNA splicing, we carried out a competition assay in Xenopus oocytes. Before injecting pre-mRNA, we injected a short RNA sequence corresponding to either the wild-type polypyrimidine tract or an altered polypyrimidine tract in which uridines were changed to pseudouridines (Fig. 4D). While a dose-dependent inhibitory effect was observed when the wild-type polypyrimidine tract was injected (Fig. 4E, compare lanes 2 and 3 with lane 1), no effect was observed when the polypyrimidine tract with pseudouridine substitution was injected (compare lanes 4 and 5 with lane 1). This result suggested that the polypyrimidine tract was competing in trans with pre-mRNA in binding with a splicing factor and that the replacement of uridines with pseudouridines abolished its ability to bind to this splicing factor.
U-to-Ψ change in the polypyrimidine tract affects U2AF binding.
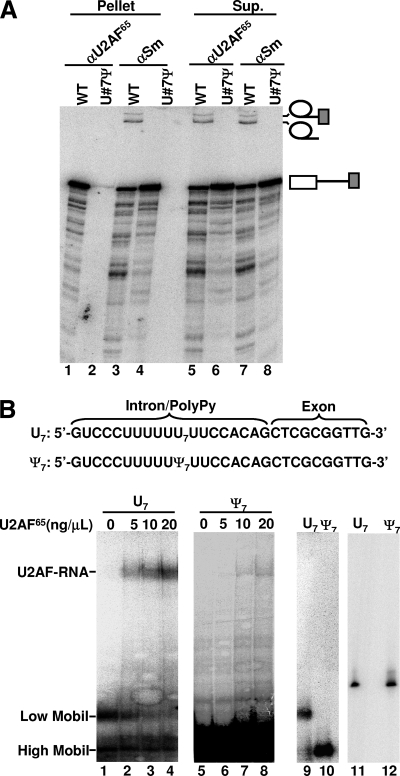
Given that the essential splicing factor U2AF65 is known to bind to the polypyrimidine tract at an early stage of splicing, we reasoned that the U-to-Ψ change in the polypyrimidine tract may give rise to a U2AF65-binding defect, thereby inhibiting splicing. To test this hypothesis, we carried out immunoprecipitation analyses. After injection of radiolabeled pre-mRNA, either the wild-type pre-mRNA or the pre-mRNA containing Ψ7 in the polypyrimidine tract, the oocyte nuclei were broken and immunoprecipitated with either anti-U2AF65 antibody or anti-Sm antibody (specific for snRNPs) as a control. The coprecipitated RNA was recovered and analyzed on a denaturing gel. As shown in Fig. 5A, while wild-type pre-mRNA was coprecipitated with U2AF65 (lane 1), virtually no altered pre-mRNA, containing Ψ7 in the polypyrimidine tract, was brought down by anti-U2AF65 antibody (lane 2). In contrast, both pre-mRNAs—unmodified wild-type pre-mRNA and pre-mRNA containing Ψ7 in the polypyrimidine tract—were precipitated by anti-Sm antibody (lanes 3 and 4). These results are consistent with the fact that both U1 snRNP and U2AF independently bind to pre-mRNA at early times during spliceosome assembly and indicate that U7-to-Ψ7 change blocked binding with U2AF but not with U1. Furthermore, in the case of the wild-type pre-mRNA, which was spliced, no lariat intermediate and product were coprecipitated with U2AF65 (lane 1); the lariat intermediate and product were only in the supernatant (lane 5). However, the lariat intermediate and product were precipitated by anti-Sm antibody. These results are in agreement with the fact that U2AF65 is released from pre-mRNA prior to functional spliceosome formation (4, 8, 34), whereas U2 and U5, both of which are targets of anti-Sm, remain bound to the lariat intermediate and product after the first and second steps of splicing (22, 28).
FIG. 5.
Effect of U7-to-Ψ7 change on U2AF65 binding. (A) Wild-type pre-mRNA (odd-numbered lanes) or pre-mRNA containing a Ψ7 in the polypyrimidine tract (even-numbered lanes) was injected into the nuclei of Xenopus oocytes. Nuclear extracts were subsequently prepared and immunoprecipitated with anti-U2AF65 (αU2AF65) (lanes 1, 2, 5, and 6), or anti-Sm (αSm) (lanes 3, 4, 7, and 8) antibodies. Lanes 1 to 4 are the precipitated fraction (pellet), and lanes 5 to 8 are the unbound fraction (supernatant [Sup.]). (B) In vitro U2AF65-polypyrimidine binding assay. The 5′-end radiolabeled U7-containing polypyrimidine tract (U7) (lanes 1 to 4) or Ψ7-containing polypyrimidine tract (Ψ7) (lanes 5 to 8; panel overexposed in order to visualize the shifted complex) was incubated with the indicated amount of pure U2AF65 (lanes 1 to 8). After the reaction, the RNA-protein complex (indicated as U2AF-RNA) and unbound RNA (indicated as low mobility [Low Mobil] and high mobility [High Mobil]) were resolved on the native gel. Lanes 9 and 10 are controls in which the U7 polypyrimidine tract and Ψ7 polypyrimidine tract alone (in the absence of U2AF65) were loaded, respectively, onto the native gel. Lanes 11 and 12 are controls in which the U7 polypyrimidine tract and Ψ7 polypyrimidine tract were analyzed on a denaturing gel, respectively. The sequences of U7 and Ψ7 are shown at the top.
To confirm our immunoprecipitation results, we carried out the in vitro binding assay (22, 28) using U2AF65 and the polypyrimidine tract of adenovirus pre-mRNA containing either U7 or Ψ7. As shown in Fig. 5B, while U2AF65 bound to the U7-containing polypyrimidine tract with high affinity (Kd = 1.8 × 10−7 M) (lanes 1 to 4), binding of U2AF65 to the Ψ7-containing polypyrimidine tract was greatly reduced (Kd = 8.1 × 10−6 M) (lanes 5 to 8). Interestingly, based on native gel analysis, the U7-containing polypyrimidine tract appeared to adopt two conformations, with the vast majority of the molecules in a low-mobility conformation and a small fraction in a high-mobility conformation (lanes 1 to 4 and 9). It appeared that only the low-mobility fraction shifted upon addition of U2AF65 (lanes 1 to 4), suggesting that this conformation was active in binding with U2AF65. In contrast, the Ψ7-containing polypyrimidine tract adopted only the high-mobility conformation (lanes 5 to 8 and 10), which was barely shifted by the addition of U2AF65 (lanes 5 to 8). Thus, these results have reinforced the notion that a single pseudouridine in the polypyrimidine tract can alter the folding (or conformation) of the polypyrimidine tract, impacting negatively on U2AF65 binding.
U2AF binding requires a flexible polypyrimidine tract backbone.
It is well established that through a bridging water molecule, pseudouridine can form a hydrogen bond with its own phosphate backbone, thus favoring the C-3′-endo sugar ring configuration and bringing rigidity to the backbone and the base (2, 6, 15, 23). Our observed pseudouridine-specific effect on U2AF binding to the polypyrimidine tract suggested that it was perhaps the inflexibility of the backbone that negatively impacted U2AF binding. To test this hypothesis, we synthesized a pre-mRNA in which the important uridine (U7) within the polypyrimidine tract was locked into the C-3′-endo sugar ring configuration (the nucleotide itself was kept as uridine) (14). This pre-mRNA was injected into Xenopus oocytes for the splicing assay.
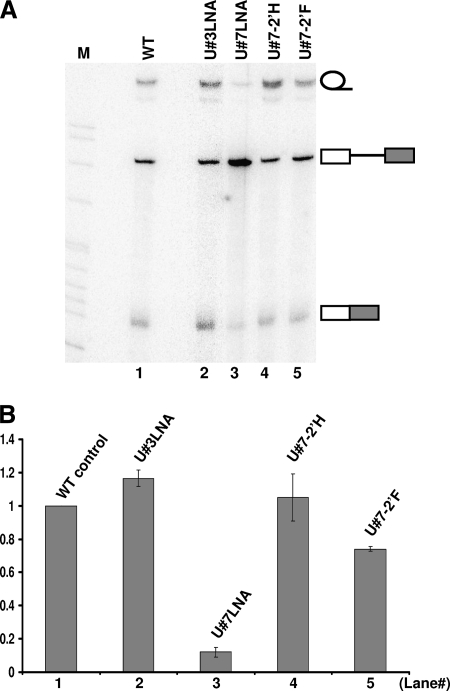
As shown in Fig. 6A and B, while the pre-mRNA containing a regular uridine (U7) was spliced efficiently (lane 1), splicing was almost completely blocked when U7 was locked up in the C3′-endo configuration (locked nucleic acid [LNA], in which its 2′-O was linked, through a carbon, to the 4′-C position) (lane 3). As a control, when an unimportant uridine (U3) was locked in the C-3′-endo configuration, splicing was not affected (lane 2).
FIG. 6.
U2AF65 binding requires RNA backbone flexibility within the polypyrimidine tract. (A) Splicing of pre-mRNA containing a modified nucleotide in the polypyrimidine tract. Wild-type pre-mRNA (lane 1) and pre-mRNA in which U3 (lane 2) or U7 (lane 3) in the polypyrimidine tract was locked in the C-3′-endo configuration (LNA), or the 2′-OH group of U7 was changed to 2′-H (lane 4) or 2′-F (lane 5) were assayed for splicing in Xenopus oocytes. (B) Relative splicing efficiency (relative ratio of the sum of lariat intron and spliced mRNA to the sum of lariat intron, spliced mRNA, and unspliced pre-mRNA) (see the legend to Fig. 3A). The numbers on the x axis correspond to the lane numbers in panel A.
To further confirm our observations and to rule out the possibility that the 2′-OH of U7 played a role in splicing (2′-OH was altered in LNA), we synthesized additional pre-mRNAs, in which the 2′-OH group of U7 was changed to 2′-F or 2′-H. It was reported that, in relative terms, 2′-F favors the C-3′-endo configuration, whereas 2′-H favors the C-2′-endo configuration (35). As shown in Fig. 6A and B, splicing occurred efficiently when pre-mRNA with a 2′-H at U7 was used (lane 4). In contrast, when pre-mRNA with a 2′-F at U7 was used, a reduction in splicing, albeit small, was evident (lane 5). These results, coupled with the results of conventional mutagenesis analyses (Fig. 4), suggest that it is the sugar backbone configuration (rather than the nucleotide identity or the 2′-OH moiety) at position 7 that is key to U2AF65 binding and splicing.
U-to-Ψ change also affects the splicing of pre-mRNA bearing a β-globin polypyrimidine tract.
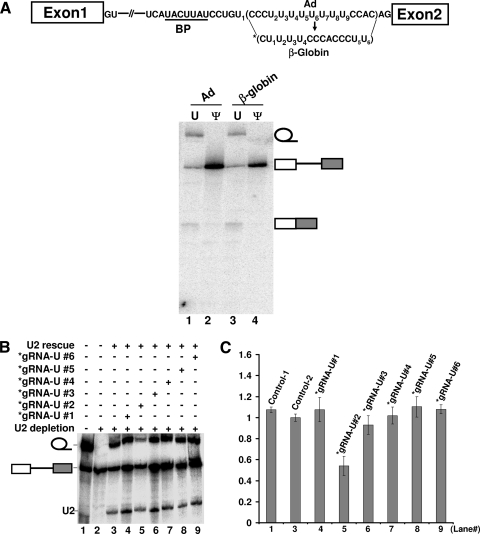
It is known that the polypyrimidine tract sequences in various mammalian pre-mRNAs, although pyrimidine rich, are quite different. To test whether the effect of U-to-Ψ change in the adenovirus polypyrimidine tract can be applied to other pre-mRNAs, we replaced the polypyrimidine tract of the adenovirus pre-mRNA (CCCUUUU5UU7UUCCAC) with that of β-globin pre-mRNA (CUU2UUCCCACCCUU). The new pre-mRNA was synthesized in vitro, injected into Xenopus oocytes, and assayed for splicing. As shown in Fig. 7A, while pre-mRNA containing uridines (lane 3) spliced as efficiently as its parental pre-mRNA (with an adenovirus polypyrimidine tract) did (lane 2), pre-mRNA containing pseudouridines completely failed to splice (lane 4), just as its parental pre-mRNA with pseudouridine substitution did (lane 2). Further analysis using targeted pseudouridylation indicated that the second-most 5′ uridine (U2) (out of six uridines) in the β-globin polypyrimidine tract could not tolerate a U-to-Ψ change (Fig. 7B and C, lane 5). These results suggested that the effects of the U-to-Ψ change could be applied to different polypyrimidine tracts, despite the fact that their sequences are quite different (see Discussion).
FIG. 7.
The Ψ effect can be applied to different polypyrimidine tracts. (A) Pre-mRNA containing an adenovirus (Ad) (lanes 1 and 2) or β-globin (lane 3 and 4) polypyrimidine tract was assayed for splicing in Xenopus oocytes. In lanes 1 and 3, uridines were incorporated into the polypyrimidine tracts, whereas in lanes 2 and 4, pseudouridines were incorporated into the polypyrimidine tracts. Bands corresponding to the lariat intron, unspliced pre-mRNA, and spliced mRNA are indicated. The diagram of pre-mRNA and the sequences of polypyrimidine tract (both adenovirus and β-globin), as well as the branch point (BP) sequence, are shown at the top of the figure. The asterisk represents the substituted β-globin polypyrimidine tract. (B) As in Fig. 3A, except that six (instead of nine) uridines (see panel A) were targeted for pseudouridylation and function. The asterisks represent guide RNAs that are specific for uridines in the substituted β-globin polypyrimidine tract (see panel A). (C) Relative splicing efficiency (relative ratio of lariat intron to the sum of the lariat intron and unspliced pre-mRNA) (as in Fig. 3A). The numbers on the x axis correspond to the lane numbers in panel B.
DISCUSSION
Using the Xenopus oocyte microinjection system, we have identified two uridines (U5 and U7) in the polypyrimidine tract region of adenovirus pre-mRNA that cannot tolerate U-to-Ψ change without compromising splicing. Indeed, U-to-Ψ conversion at each site halted the binding of U2AF65, an essential splicing factor in higher eukaryotes, thus suggesting that the two uridines are critical for U2AF65 binding. Further dissection indicated that a flexible backbone, which was rigidified upon U-to-Ψ conversion at either of the two sites (U5 and U7), is required for U2AF65 binding. Our current work has also raised some interesting questions with regard to the observed effects of the U-to-Ψ change.
U-to-Ψ conversion in the pre-mRNA polypyrimidine tract inhibits splicing: a Ψ-
specific backbone effect. According to our experimental data (Fig. 4), while a U-to-Ψ change at uridine position 7 of the polypyrimidine tract of adenovirus pre-mRNA resulted in a defect in U2AF65 binding and pre-mRNA splicing, U-to-other-nucleotide (G, C, or A) changes did not. These results indicate that the observed effect is due to U-to-Ψ isomerization, not to conventional mutations, and is therefore a Ψ-specific effect.
What is unique about Ψ? Isomerization of U to Ψ involves the breakage of a N-C bond that used to link the base with the sugar ring and the concurrent formation of a new C-C bond which reestablishes the linkage between the base and sugar. Consequently, this reaction frees a nitrogen (N-1), resulting in an extra hydrogen bond donor in the newly formed Ψ. It is possible that this hydrogen bond donor specifically forms a hydrogen bond with some other factor(s), thereby competing with U2AF65 for binding. Alternatively, the new hydrogen bond donor could form a hydrogen bond with a water molecule, which in turn would form another two hydrogen bonds with the phosphodiester backbone of the RNA (2, 6, 15, 23). Consequently, both the Ψ base and its sugar ring backbone would become rigid. Such a rigid configuration may disfavor binding with U2AF, giving rise to a splicing defect phenotype. Our experimental results strongly support the hypothesis that a flexible sugar backbone at position 7 (and most likely at position 5 as well) or a C-2′-endo sugar pucker (S form) (rather than the C-3′-endo [N form]) is required for U2AF65 binding in vivo. Specifically, we have shown that, when U7 is locked in the C-3′-endo configuration (LNA via 2′-O, 4′-C linkage), the polypyrimidine tract becomes inactive in binding with U2AF65 (Fig. 6). However, when the uracil base at position 7 is changed to other bases (Fig. 4) or the 2′-OH group at position 7 is changed to 2′-H (Fig. 6), splicing is not affected. Thus, our results indicate that the sugar ring backbone conformation, rather than the base identity or the 2′-OH moiety, is critical for U2AF65 binding. Along these lines, a recent report indicates that the rigid nature of the Ψ-containing Sm binding site leads to the defect in the binding of U7 snRNA with its Sm proteins (15).
With regard to the structural basis of polypyrimidine tract-U2AF65 binding, the crystal structure of a U2AF65 RNA binding domain bound to a seven-uridine tract (U1U2U3U4U5U6U7) has been solved (29). The structural data indicate that there is only one 2′-OH (the 2′-OH of U5) that interacts with U2AF65; there are no contacts between U2AF65 and the 2′-OH groups of the other six uridines. This structural information is consistent with our experimental results, which indicate that at least the 2′-OH group of U7 in the adenovirus polypyrimidine tract is not required for U2AF65 binding (Fig. 6). Although most interactions are through contacts between uridine bases and U2AF65 (29), the C-5 and C-6 atoms of the bases, which have changed in the pseudouridine context (N1 in pseudouridine is equivalent to C-5 in uridine), show no contact with U2AF65, suggesting that the N1 atom of the pseudouridine base does not contribute to the observed defect in U2AF65 binding (Fig. 5). Importantly, the crystal structure also indicates that the sugar puckers of the first three uridines adopt the C-2′-endo conformation (C. Kielkopf, personal communication), a structure often adopted by DNA rather than RNA. Notably, while binding with U2AF65, the seven-uridine tract forms a turn, with the third nucleotide being at the turn of the chain. Given the above structural information, it is tempting to align the seven-uridine tract with the adenovirus polypyrimidine tract starting with U5 (U5U6U7U8U9CC) (Fig. 1A). Such an alignment would suggest that the sugar puckers of U5, U6, and U7 (the first three uridines) of the adenovirus polypyrimidine tract adopt the C-2′-endo conformation when bound with U2AF65. This alignment would thus allow us to explain our experimental data, which show that locking U5 and U7 (the nucleotide at the turn) in the C-3′-endo conformation (by replacing them with either pseudouridines or LNA) leads to profound defects in U2AF65 binding and splicing (Fig. 5 and 6). However, our results still cannot explain U6, the second-most 5′ uridine of the seven-pyrimidine tract. Perhaps the C-2′-endo conformation of U6 is not as crucial in binding with U2AF65. More research is necessary.
Is the U-to-Ψ effect universal to all polypyrimidine tracts?
We have shown that U-to-Ψ change in the polypyrimidine tract of adenovirus pre-mRNA blocks U2AF binding and pre-mRNA splicing. It appears that this effect is not limited to the adenovirus polypyrimidine tract, as when the polypyrimidine tract of β-globin pre-mRNA was used, a similar effect was observed, suggesting that this effect is universally applicable.
Given the totally different polypyrimidine tract sequences that are known to exist in various pre-mRNAs, the commonality of determinants in different sequences is somewhat puzzling. Which uridine(s) (apparently with no commonality in sequence context) is critical: in other words, which uridine(s) cannot be changed to pseudouridine(s) without compromising U2AF binding and splicing? Taking the current experiments as an example, the uridines mapped in the adenovirus polypyrimidine tract are the fifth and seventh uridines (UCCCUUUU5UU7UUCC) or the first and third uridines (U5UU7UUCC) when aligned with the seven-uridine tract in the crystal, whereas the uridine mapped in the β-globin polypyrimidine tract (CUU2UUCCCACCCUU) is the second uridine (U2). However, if the pyrimidine tract begins with the 5′-most C, U2 of the β-globin pyrimidine tract (CUU2UUCC) becomes the third pyrimidine and would be equivalent to U3 of the seven-uridine tract, thus offering a possible explanation of why replacement of this uridine with a pseudouridine resulted in a splicing defect (Fig. 7B and C). Thus, we predict that the sugar puckers of the 5′-most C and the third pyrimidine (U2) prefer to take a C-2′-endo formation when complexed with U2AF65.
The polypyrimidine tract provides a possible target for gene silencing in vivo.
We have shown that a U-to-Ψ change at a specific site(s) in the polypyrimidine tract effectively blocks pre-mRNA splicing. Using artificial box H/ACA guide RNAs to target the critical uridines, we have also recapitulated this inhibitory effect in vivo. Thus, such an approach provides a way to regulate gene expression at the level of pre-mRNA splicing. However, it has long been noted that box H/ACA RNAs are localized to the nucleoli and Cajal bodies, where their natural target RNAs, rRNAs and snRNAs, are located. This raises an interesting question as to whether mRNA and pre-mRNA, which are known to be absent in the nucleoli and Cajal bodies, will be modified by the box H/ACA-guided mechanism.
Interestingly, recent work from several labs suggests that guide RNAs may be dispersed throughout the nucleoplasm. For instance, the Gall lab has demonstrated that Drosophila cells lacking Cajal bodies are capable of modifying snRNAs (9). Work from our lab and others suggests that some specific guide RNAs may indeed reside within the nucleoplasm (20, 44). More directly, it has been shown that an artificial guide RNA can direct nucleoplasmic pre-mRNA modification in yeast and mammalian cells (27, 45). Our current work also indicates that pseudouridylation can be introduced into pre-mRNA. All of these lines of evidence suggest that the approach may in principle be used for gene silencing and regulation. Given the well-known mechanisms of RNA-guided RNA modification and pre-mRNA splicing, it would be relatively simple and straightforward to design artificial guide RNAs to target important uridines in pre-mRNA, thereby regulating pre-mRNA splicing in vivo.
Acknowledgments
We thank Clara Kielkopf for kind gifts of anti-U2AF65 antibody and the U2AF65 plasmid. We also thank Clara Kielkopf for valuable discussions about the experimental results and information about the crystal structure of the U2AF65-uridine tract. We are grateful to Doug Turner for advice on LNA.
This work was supported by grant GM62937 from the National Institutes of Health.
Footnotes
Published ahead of print on 6 July 2010.
REFERENCES
- 1.Adams, M. D., D. Z. Rudner, and D. C. Rio. 1996. Biochemistry and regulation of pre-mRNA splicing. Curr. Opin. Cell Biol. 8:331-839. [DOI] [PubMed] [Google Scholar]
- 2.Arnez, J. G., and T. A. Steitz. 1994. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33:7560-7567. [DOI] [PubMed] [Google Scholar]
- 3.Baker, D. L., O. A. Youssef, M. I. Chastkofsky, D. A. Dy, R. M. Terns, and M. P. Terns. 2005. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 19:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, M., S. Michaud, J. Kingston, and R. Reed. 1992. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 6:1986-2000. [DOI] [PubMed] [Google Scholar]
- 5.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 6.Charette, M., and M. W. Gray. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49:341-351. [DOI] [PubMed] [Google Scholar]
- 7.Charpentier, B., S. Muller, and C. Branlant. 2005. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 33:3133-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiara, M. D., L. Palandjian, R. Feld Kramer, and R. Reed. 1997. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 16:4746-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deryusheva, S., and J. G. Gall. 2009. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol. Biol. Cell 20:5250-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan, J., L. Li, J. Lu, W. Wang, and K. Ye. 2009. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol. Cell 34:427-439. [DOI] [PubMed] [Google Scholar]
- 11.Ganot, P., M. L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89:799-809. [DOI] [PubMed] [Google Scholar]
- 12.Green, M. R. 1991. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu. Rev. Cell Biol. 7:559-599. [DOI] [PubMed] [Google Scholar]
- 13.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 14.Kierzek, E., A. Ciesielska, K. Pasternak, D. H. Mathews, D. H. Turner, and R. Kierzek. 2005. The influence of locked nucleic acid residues on the thermodynamic properties of 2′-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 33:5082-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolev, N. G., and J. A. Steitz. 2006. In vivo assembly of functional U7 snRNP requires RNA backbone flexibility within the Sm-binding site. Nat. Struct. Mol. Biol. 13:347-353. [DOI] [PubMed] [Google Scholar]
- 16.Konarska, M. M. 1989. Analysis of splicing complexes and small nuclear ribonucleoprotein particles by native gel electrophoresis. Methods Enzymol. 180:442-453. [DOI] [PubMed] [Google Scholar]
- 17.Lerner, E. A., M. R. Lerner, C. A. Janeway, Jr., and J. A. Steitz. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. U. S. A. 78:2737-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, B., J. Zhou, E. Kahen, R. M. Terns, M. P. Terns, and H. Li. 2009. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat. Struct. Mol. Biol. 16:740-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang, X. H., Q. Liu, and M. J. Fournier. 2007. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 28:965-977. [DOI] [PubMed] [Google Scholar]
- 20.Liang, X. H., Y. X. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 8:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massenet, S., A. Mougin, and C. Branlant. 1998. Posttranscriptional modifications in the U small nuclear RNAs, p. 201-228. In H. Grosjean (ed.), Modification and editing of RNA. ASM Press, Washington, DC.
- 22.Moore, M. J., C. C. Query, and P. A. Sharp. 1993. Splicing of precursors to mRNAs by the spliceosome, p. 303-357. In R. F. Gesteland and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Newby, M. I., and N. L. Greenbaum. 2002. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. U. S. A. 99:12697-12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565-573. [DOI] [PubMed] [Google Scholar]
- 25.Ofengand, J., and M. Fournier. 1998. The pseudouridine residues of rRNA: number, location, biosynthesis, and function., p. 229-253. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, DC.
- 26.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 27.Semenov, D. V., O. V. Vratskih, E. V. Kuligina, and V. A. Richter. 2008. Splicing by exon exclusion impaired by artificial box c/d RNA targeted to branch-point adenosine. Ann. N. Y. Acad. Sci. 1137:119-124. [DOI] [PubMed] [Google Scholar]
- 28.Sharp, P. A. 1994. Split genes and RNA splicing. Cell 77:805-815. [DOI] [PubMed] [Google Scholar]
- 29.Sickmier, E. A., K. E. Frato, H. Shen, S. R. Paranawithana, M. R. Green, and C. L. Kielkopf. 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell 23:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh, R., H. Banerjee, and M. R. Green. 2000. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA 6:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh, R., J. Valcarcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 32.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 33.Stephenson, D., J. Karijolich, and Y. T. Yu. 2008. Functional roles of spliceosomal snRNA modifications in pre-mRNA splicing, p. 175-189. In H. C. Smith (ed.), RNA and DNA editing. John Wiley & Sons, Hoboken, NJ.
- 34.Tisserant, A., and H. Konig. 2008. Signal-regulated pre-mRNA occupancy by the general splicing factor U2AF. PLoS One 3:e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uesugi, S., H. Miki, M. Ikehara, H. Iwahashi, and Y. Kyogoku. 1979. A linear relationship between electronegativity of 2′-substituents and conformation of adenine nucleosides. Tetrahedron Lett. 20:4073-4076. [Google Scholar]
- 36.Valcarcel, J., R. Singh, P. D. Zamore, and M. R. Green. 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362:171-175. [DOI] [PubMed] [Google Scholar]
- 37.Wang, C., and U. T. Meier. 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23:1857-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao, M., C. Yang, P. Schattner, and Y. T. Yu. 2009. Functionality and substrate specificity of human box H/ACA guide RNAs. RNA 15:176-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, Y. T. 2000. Site-specific 4-thiouridine incorporation into RNA molecules. Methods Enzymol. 318:71-88. [DOI] [PubMed] [Google Scholar]
- 40.Yu, Y. T., E. C. Scharl, C. M. Smith, and J. A. Steitz. 1999. The growing world of small nuclear ribonucleoproteins, p. 487-524. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Yu, Y. T., M. D. Shu, and J. A. Steitz. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17:5783-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, Y. T., R. M. Terns, and M. P. Terns. 2005. Mechanisms and functions of RNA-guided RNA modification. Top. Curr. Genet. 12:223-262. [Google Scholar]
- 43.Zamore, P. D., and M. R. Green. 1989. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl. Acad. Sci. U. S. A. 86:9243-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, X., Z. H. Li, R. M. Terns, M. P. Terns, and Y. T. Yu. 2002. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA 8:1515-1525. [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, X., and Y. T. Yu. 2008. Targeted pre-mRNA modification for gene silencing and regulation. Nat. Methods 5:95-100. [DOI] [PubMed] [Google Scholar]