Abstract
Subjecting cardiomyocytes to mechanical stress or neurohumoral stimulation causes cardiac hypertrophy characterized in part by reactivation of the fetal cardiac gene program. Here we demonstrate a new common mechanism by which these stimuli are transduced to a signal activating the hypertrophic gene program. Mechanically stretching cardiomyocytes induced nuclear accumulation of myocardin-related transcription factor A (MRTF-A), a coactivator of serum response factor (SRF), in a Rho- and actin dynamics-dependent manner. Expression of brain natriuretic peptide (BNP) and other SRF-dependent fetal cardiac genes in response to acute mechanical stress was blunted in mice lacking MRTF-A. Hypertrophic responses to chronic pressure overload were also significantly attenuated in mice lacking MRTF-A. Mutation of a newly identified, conserved and functional SRF-binding site within the BNP promoter, or knockdown of MRTF-A, reduced the responsiveness of the BNP promoter to mechanical stretch. Nuclear translocation of MRTF-A was also involved in endothelin-1- and angiotensin-II-induced activation of the BNP promoter. Moreover, mice lacking MRTF-A showed significantly weaker hypertrophic responses to chronic angiotensin II infusion than wild-type mice. Collectively, these findings point to nuclear translocation of MRTF-A as a novel signaling mechanism mediating both mechanical stretch- and neurohumoral stimulation-induced BNP gene expression and hypertrophic responses in cardiac myocytes.
Hemodynamic overload, a combination of mechanical stress and neurohumoral stimulation, induces a hypertrophic response characterized in part by reactivation of the fetal gene program in cardiac myocytes (4, 15, 25, 45, 58). Though cardiac hypertrophy initially serves as an adaptive response to increased cardiac output, when sustained it leads to cardiac decompensation and heart failure, which is now a leading cause of morbidity and mortality around the world. Thus, elucidation of the molecular mechanisms underlying the development and progression of cardiac hypertrophy is an important issue when considering therapeutic intervention. To delineate the molecular pathways involved in the hypertrophic response to mechanical stress, in vitro stretching devices have been developed that enable stretch stress to be applied to cultured cardiac myocytes (51, 62). Using these devices, it was revealed that mechanical stress activates several signal transduction pathways involving mitogen-activated protein kinases (MAPKs), protein kinase C (PKC), Jak-STAT, and small G proteins (e.g., Rho, Rac, and Ras) in cultured cardiac myocytes (1, 16, 27, 47, 48, 51, 62). How these signaling molecules transduce mechanical stretch to a signal activating a set of transcription factors and ultimately the hypertrophic gene program, however, remains unclear.
In addition to mechanical stress, neurohumoral stimulation is also known to be a pivotal contributor to the chronic remodeling process in hearts (45). Angiotensin II (AngII), phenylephrine, and endothelin 1 (ET-1), which all act through G-protein-coupled receptors, have all been shown to induce cardiac hypertrophy. Clinical evidence showing the favorable effects of blocking AngII signaling on the course of heart failure and the ability of AngII blockade to repress cardiac hypertrophy supports the notion that neurohumoral factors play an important role in pathological cardiac remodeling (8). Among the variety of intracellular signaling molecules that have been shown to be activated following mechanical stretch or neurohumoral stimulation, Rho family small GTPases, especially Rho A and Rac1, have been highlighted as important regulators for cardiac hypertrophy (5, 24). The precise downstream mechanisms by which Rho GTPases activate the hypertrophic gene program remain obscure, however.
Serum response factor (SRF) is a MADS box transcription factor that regulates the expression of immediate-early genes and muscle-specific genes by binding to a conserved sequence [CC(A/T)6GG] known as the CArG box or serum response element. Moreover, several findings have confirmed the involvement of SRF in the induction of a subset of cardiac genes during adverse cardiac remodeling (23, 32, 34, 36, 57). Targeted deletion of SRF in the developing heart results in lethal cardiac defects, with reduced expression of many cardiac-specific genes (33, 49). In addition, overexpression of SRF in the postnatal heart leads to cardiomyopathy with increased fetal cardiac gene expression (63), while conditional deletion of SRF in isolated neonatal cardiac myocytes results in reduced expression of hypertrophic genes (43). Several fetal cardiac genes, including atrial natriuretic peptide (ANP), skeletal α-actin, smooth muscle α-actin, and smooth muscle 22α (SM22α), have been shown to contain a functionally important CArG box in their upstream transcription control region (53, 57). At least two signaling pathways are known to modulate SRF activity, one involving the phosphorylation of ternary complex factors in Ets domain family proteins and another controlled by Rho family small GTPases and actin dynamics (10, 12, 14, 56). It was recently shown in NIH 3T3 cells that stimulation of Rho- and actin dynamics-dependent signaling results in translocation of a novel SRF cofactor, myocardin-related transcription factor A (MRTF-A) (also called MAL or MKL1), from G-actin in the cytoplasm to the nucleus and in activation of SRF target genes (35).
In the present study, we investigated the role of MRTF-A in mediating prohypertrophic signaling evoked by mechanical stress and neurohumoral stimulation in cardiac myocytes. Our study defines Rho- and actin dynamics-dependent nuclear translocation of MRTF-A as a novel common mechanism transducing mechanical stretch and neurohumoral stimulation to activation of the hypertrophy gene program, including increased expression of the brain natriuretic peptide (BNP) gene, in cardiac myocytes.
MATERIALS AND METHODS
Plasmid construction.
A DNA fragment from the 5′ flanking region of the human BNP gene (bp −423 to +98 relative to the transcription start site) was isolated by PCR using human genomic DNA as a template with the primers 5-ACT CGG AAG ATC TGT CTT GGC CGG GGC TGT TTT CGC-3 (sense) and 5-GAT TCC CAA GCT TCA TGT CTC TGG AGG GAC TGC GG-3 (antisense), after which the fragment was inserted upstream of a luciferase gene in the pGL3 vector using the BglII/HindIII sites (−423hBNP-luc). In addition, the following primers were used to generate deletion mutants in which the BNP 5′-flanking region from bp −146 to +98 (−146hBNP-luc), −77 to +93 (−77hBNP-luc), or −25 to +93 (−25hBNP-luc) were deleted: −146hBNP, 5-GGC GGA AGA TCT CGG AGG GGC TCA TTC CCG −3; −77hBNP, 5-GGC GGA AGA TCT TGC ATG GCA GGG CAG GCC-3; and −25hBNP, 5-GGC GGA AGA TCT CCC GAG GAG CCA GGA GGA-3. −1823hBNP-luc, human ANP promoter-reporter gene (−452hANP-luc), SM22α-luc, and 4× CArG-luc were described previously (21, 44). To generate a mutation in the CArG-like sequences of the BNP 5′-flanking region, PCR-based mutagenesis was performed using the primers 5-GGC CCA TTT CTG TAC CCG GTC GGC TCT G-3 (mut CArG sense) and 5-CAG AGC CGA CCG GGT ACA GAA ATG GGC C-3 (mut CArG antisense).
Cell culture and transfection.
Primary neonatal rat ventricular myocytes were isolated and grown as described previously (37). Twenty-four hours after plating, the myocytes were transfected for 12 h with 200 ng of reporter plasmid and 200 ng of expression vector using the Gene Jammer reagent (Invitrogen) unless indicated otherwise. A Rous sarcoma virus (RSV)-driven lacZ expression vector was included in all transfections as an internal control. The transfectant cells were then incubated in serum-free medium for 6 h, after which ET-1 (100 nM), AngII (100 nM), or vehicle was added, and the cells were maintained for an additional 48 h.
Myocytes subjected to stretching were first transfected for 6 h with 300 ng of reporter plasmid using the Gene Jammer reagent (Invitrogen) unless indicated otherwise. The transfected cells were then incubated in serum-free medium for 12 h, after which they were subjected to 20% mechanical stretch for 4 h.
Recombinant adenovirus infection.
For adenovirus production, cDNAs encoding FLAG-tagged, full-length mouse MRTF-A were cloned into the pAC-CMV vector, and the resultant constructs were cotransfected into HEK 293 cells along with pJM17 using Fugene 6 (Roche Applied Science). Clonal populations of adenoviruses were amplified by reinfecting HEK 293 cells, after which titers of the viral preparations were determined using the agar overlay method.
Thirty-six hours after plating, cardiac myocytes grown on coverslips in 6-well dishes were infected for 6 h with recombinant adenovirus at a multiplicity of infection (MOI) of 5 and then maintained in serum-containing medium for 24 h. Thereafter, the growth medium was replaced with serum-free medium, and cells were incubated for an additional 12 h before being treated with 100 nM ET-1 or AngII for 1 h and fixed in 4% formaldehyde in phosphate-buffered saline (PBS).
Thirty-six hours after plating on silicone membranes, cardiac myocytes were infected for 6 h with recombinant adenovirus at a MOI of 5 and then maintained in serum-containing medium for 36 h. After changing to serum-free medium, the cells were incubated for an additional 12 h and then subjected to 20% mechanical stretch for 1 h. The cells were then fixed in 4% formaldehyde in PBS.
The subcellular distribution of MRTF-A was determined by immunostaining for the FLAG epitope (green). Cardiac myocytes were also positively stained with anti-ANP antibody to distinguish them from cardiac fibroblasts (red).
Luciferase assay.
Cells were harvested, and luciferase and control β-galactosidase activities were measured using a luciferase assay system (Promega) and a FluoReporter lacZ galactosidase quantitation kit (Invitrogen) according to the manufacturer's instructions. All assays were performed at least twice in triplicate.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed using double-stranded oligonucleotides corresponding to the SM22 CArG or BNP CArG-like sequence. The probe sequences were as follows: SM22 CArG, 5′-CCT GCC CAT AAA AGG TTT TTC CTG CCC ATA AAA GGT TTT T-3′; BNP CArG, 5′-CAT TTC TAT ACA AGG TCG GCC ATT TCT ATA CAA GGT CGG C-3′; and BNP mutCArG, 5′-CAT TTC TGT ACC CGG TCG GCC ATT TCT GTA CCC GGT CGG C-3′. For gel mobility shift assays utilizing SRF, 2 μl of a coupled in vitro translation reaction product (TNT kit; Promega) was incubated with the indicated 32P-labeled oligonucleotide probes in the presence of 1 μl of poly(dIdC) (1.5 μg/μl) for 20 min at room temperature, followed by nondenaturing electrophoresis. Unlabeled competitor oligonucleotides were added at a 10-, 100-, or 1,000-fold molar excess, and 2 μl of anti-SRF antibody (Santa-Cruz) was added for supershift experiments. The assay buffers and electrophoresis conditions were described previously (37).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were carried out according to the protocol supplied by the manufacturer (Upstate Biotechnology). Briefly, cultured ventricular myocytes fixed with 1% formaldehyde for 10 min at 37°C were collected, resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl [pH 8.1]) containing 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A, and sonicated seven times for 10 s each time. After clearing the lysate by centrifugation, one aliquot of the lysate (20 μl) was removed to serve as an input control. The remainder was incubated overnight at 4°C in ChIP dilution buffer (16.7 mM Tris [pH 8.1], 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, 0.01% SDS, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A) also containing 20 μl of anti-SRF antibody (Santa Cruz). Thereafter, 60 μl of salmon sperm DNA and a protein G-agarose bead suspension were added and incubated for an additional 1 h at 4°C. The beads were then sequentially washed for 5 min each in low-salt immune complex wash buffer (20 mM Tris-HCl [pH 8.1], 150 mM NACl, 2 mM EDTA, 0.1% SDS, and 1% Triton X-100), high-salt buffer (20 mM Tris-HCl [pH 8.1], 500 mM NACl, 2 mM EDTA, 0.1% SDS, and 1% Triton X-100), LiCl immune complex wash buffer (250 mM LiCl, 1% Igepal-CA630, 1% deoxycholic acid [sodium salt], 1 mM EDTA, 10 mM Tris [pH 8.1]), and TE buffer (10 mM Tris [pH 8.0] and 1 mM EDTA). The immune complexes were then eluted by incubating the beads with elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min at room temperature. After the addition of 5 M NaCl, the eluates were heated to 65°C for 12 h to reverse the protein-DNA cross-links. The DNA was then recovered through proteinase K treatment, phenol-chloroform extraction, and ethanol precipitation. The resultant pellets were resuspended in 20 μl of TE buffer. PCR was performed with 2-μl samples of DNA using the following primers: rBNP CArG FW, 5-CAA TAC GGG TGG GGC ACG GTA-3; rBNP CArG RV, 5-ACC CCT CTG TGC CTC TGT-3; rBNP distal FW, 5-TGG CAC CAA GCC ACA CTC TGA AGA-3; rBNP distal RV, 5-GAA TGC TGC ATT TAA CGG CCT GGA T-3; rSM22 CArG FW, 5-GGT CCT GCC CAT AAA AGG TTT-3; rSM22 CArGRV, 5-TGC CCA TGG AAG TCT GCT TGG-3; GAP FW, 5-GCT CTC TGC TCC TCC CTG TT-3; GAP RV, 5-CAT CCT CTG CAA TGC GGA GC-3.
RNA interference.
For analysis of MRTF-A using RNA interference (RNAi), a siGENOME Smart pool reagent against rat MRTF-A (M-081405-00-0010) with guaranteed minimum 75% mRNA knockdown was purchased from Dharmacon. A Block-It fluorescent oligomer (Invitrogen) was used as a nonspecific control. For luciferase assays, neonatal rat ventricular myocytes in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum were transfected with 100 pmol of small interfering RNA (siRNA) and 500 ng of luciferase reporter plasmid for 12 h using Fugene. An RSV-lacZ expression plasmid was included in all transfections as an internal control. The transfectant cells were then incubated in serum-free medium for 6 h, after which ET-1 (100 nM) or vehicle was added, and the cells were maintained for an additional 48 h.
For luciferase assays with myocytes subjected to mechanical stretch, neonatal rat ventricular myocytes in DMEM supplemented with 10% fetal bovine serum were transfected with 200 pmol of siRNA and 600 ng of luciferase reporter plasmid for 12 h using Fugene. The transfected cells were then incubated for 12 h in serum-free medium, after which they were subjected to mechanical stretch for 4 h.
To verify the efficiency of siRNA-mediated knockdown of MRTF-A expression, rat smooth muscle cells in 6-well dishes were transfected with 200 pmol of siRNA and 48 h later were harvested for real-time reverse transcriptase PCR (RT-PCR) analysis. In cells transfected with rat MRTF-A siRNA, we observed an 88% reduction in the endogenous expression of MRTF-A mRNA compared to results for cells transfected with control siRNA.
To assess the effect of knocking down MRTF-A expression on myocyte hypertrophy, cells were transfected for 24 h using Lipofectamine with On-Target plus siRNA reagent for rat MRTF-A (Dharmacon) or control scrambled siRNA 2000, as previously described (46).
Animal experiments.
MRTF-A−/− mice were kindly provided by E. N. Olson (University of Texas Southwestern Medical Center at Dallas) (26). The animal care and all experimental protocols were reviewed and approved by the Animal Research Committee at Kyoto University Graduate School of Medicine.
Thoracic aortic banding.
Six- to 8-week-old male mice either underwent a sham operation or were subjected to pressure overload induced by thoracic aortic banding (TAB), as described previously (13). A constriction created using a 27-gauge needle was placed in the transverse aorta between the innominate and left carotid arteries. We previously showed that constriction to a 27-gauge stenosis induces moderate hypertrophy without clinical signs of heart failure or malignant ventricular arrhythmia. The mice were sacrificed 1 h (for acute pressure overload) or 3 weeks (for chronic pressure overload) after TAB. At that time, we confirmed the integrity of the banding by inspecting the surgical constriction and noting the marked difference in the caliber of the right and left carotid arteries.
Chronic AngII administration.
AngII (dissolved in 10 mM acetic acid) was subcutaneously administered at the rate of 0.6 mg/kg of body weight/day for 2 weeks using an osmotic minipump (Alzet model 2002; Alza Corp.) implanted in each mouse at 10 weeks of age. After 1 week of AngII infusion, systolic blood pressure (SBP) was measured in conscious mice using a noninvasive computerized tail-cuff method (Muromachi).
Echocardiographic analysis.
Cardiac function was evaluated echocardiographically in conscious 10-week-old mice using a Hewlett Packard Sonos 5500 ultrasound system with a 12-MHz transducer, as described previously (9). Briefly, views were taken in planes that approximated the parasternal short-axis view (chordal level) and the apical long-axis view. Left ventricular internal diameters and wall thicknesses were measured (at least 3 cardiac cycles) at the end of systole and the end of diastole.
Real-time RT-PCR.
Total RNA was isolated from cultured neonatal ventricular myocytes or mouse hearts using Trizol and following the manufacturer's protocol. Real-time one-step RT-PCR was performed with 20 to 100 ng of total RNA using One-step RT-PCR master mix reagent (ABI). TaqMan primers and probes for mouse BNP, ANP, c-Fos, Egr-1, striated muscle activator of Rho signaling (STARS), skeletal α-actin, SM22α, and smooth muscle α-actin were purchased from ABI.
Statistical analysis.
Data are presented as means ± standard errors of the means (SEM). Unpaired t tests were used for comparison between two groups, and analysis of variance (ANOVA) with post hoc Fisher's test was used for comparison among groups. P values of <0.05 were considered significant.
RESULTS
Nuclear translocation of MRTF-A in response to mechanical stretch in cardiac myocytes.
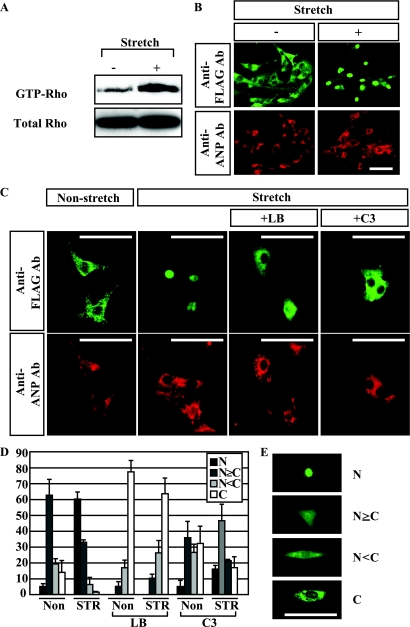
Using a fibroblast cell line, MRTF-A was previously shown to be translocated into the nucleus following sequential serum stimulation and Rho activation and to then activate SRF (35). In cardiac myocytes, SRF is known to be involved in hypertrophic gene reprogramming (23, 32, 57), while Rho is known to be activated by various hypertrophic stimuli, including mechanical stretch and neurohumoral stimulation (with ET-1 or AngII) (1, 2, 5, 20, 24, 48, 52, 59). We therefore hypothesized that nuclear translocation of MRTF-A mediates the hypertrophic signaling evoked by mechanical stretch, resulting in activation of the hypertrophic gene program. To test that idea, we first examined whether MRTF-A is translocated into the nucleus in response to mechanical stretch in cultured neonatal ventricular myocytes infected with an adenovirus encoding MRTF-A. We initially confirmed that stretching cardiac myocytes in our system rapidly leads to Rho activation, as previously reported by others (1) (Fig. 1A). When we stretched cardiac myocytes expressing FLAG-tagged MRTF-A, we observed accumulation of MRTF-A in the nucleus within 1 h after the initiation of stretch (Fig. 1B, D, and E). This translocation of MRTF-A was blocked in the presence of latrunculin B, an inhibitor of actin treadmilling, or C3 exoenzyme, an inhibitor of Rho, which suggests Rho-actin dynamics plays a critical role in the stretch-induced nuclear translocation of MRTF-A (Fig. 1C and D).
FIG. 1.
Mechanical stretch induces nuclear accumulation of MRTF-A in a Rho- and actin treadmilling-dependent manner. (A) Cardiomyocytes were subjected to 20% stretch for 5 min, after which GTP-RhoA was pulled down and visualized by Western blotting with anti-Rho antibody. (B) Cultured neonatal rat ventricular myocytes infected with adenovirus encoding FLAG-tagged MRTF-A (Ad-MRTF-A) were stretched by 20% for 1 h. The subcellular distribution of MRTF-A was determined by immunostaining for the FLAG epitope (green). Cardiac myocytes were positively stained with anti-ANP antibody (Ab) to distinguish them from cardiac fibroblasts (red). (C) Cultured neonatal rat ventricular myocytes infected with Ad-MRTF-A were stretched by 20% for 1 h in the presence or absence of an inhibitor of actin treadmilling (latrunculin B [LB]) or a Rho inhibitor (C3). The cells were then stained with anti-FLAG antibody (green) or anti-ANP antibody (red). (D) Graphs showing the percent MRTF-A localization in adenovirus-infected ventricular myocytes, with or without mechanical stretch, in the presence or absence of LB or C3. N, exclusive staining of MRTF-A in the nucleus; N≥C, nuclear staining of MRTF-A is greater than or equal to cytoplasmic staining; N<C, greater staining of MRTF-A in the cytoplasm than the nucleus; C, exclusive staining of MRTF-A in the cytoplasm. At least 100 infected cells were counted under each condition in three independent experiments. Values are means ± SEM. (E) Representative images of the four categories for MRTF-A localization are shown. In all images, bars represent 50 μm.
Expression of BNP and other SRF-dependent fetal genes in response to acute pressure overload is impaired in MRTF-A−/− mice.
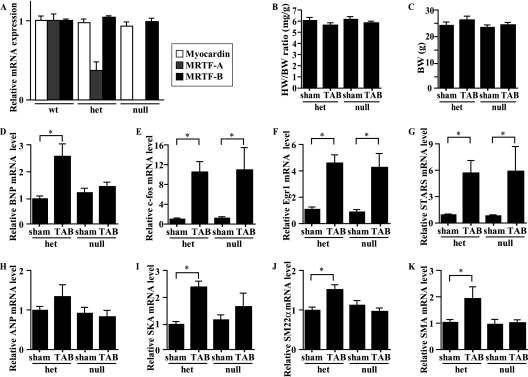
We next tested whether loss of MRTF-A diminishes activation of the hypertrophic gene program induced by mechanical load in vivo. To evaluate the contribution of MRTF-A to mechanical stress-induced genetic alterations separately from the effects of subsequent neurohumoral activation, we subjected MRTF-A−/− mice to acute pressure overload. Under basal conditions, male MRTF-A−/− mice have no obvious structural or physiological deficiencies (26). Consistent with that finding, echocardiographic analysis revealed parameters of cardiac function to be similar in MRTF-A+/+ (wild-type), MRTF-A+/−, and MRTF-A−/− mice (unpublished observation; the data for wild-type and MRTF-A−/− mice are shown in Table 1). We evaluated the expression of three myocardin family genes and confirmed that ablation of MRTF-A expression produces no significant change in myocardin or MRTF-B mRNA levels (Fig. 2A). When we compared the heart weight-to-body weight (HW/BW) ratios and BWs of control MRTF-A+/− and MRTF-A−/− mice subjected to a sham operation or acute mechanical overload caused by TAB for 1 h (13), we found no differences among the four groups of mice (Fig. 2B and C). We then measured the levels of brain natriuretic peptide (BNP) and c-fos mRNA as representative markers of fetal cardiac genes and early response genes, respectively, 1 h after TAB in MRTF-A−/− mice and MRTF-A+/− mice (11, 28, 31). The c-fos gene contains a CArG box and an Ets-binding site and is regulated by the formation of a phosphorylated Elk-1/SRF complex independently of myocardin family coactivators (54, 61). As shown in Fig. 2D, the increase in BNP mRNA expression following TAB was markedly smaller in MRTF-A−/− mice than in their MRTF-A+/− littermates, though there was no significant difference in the induction of c-fos mRNA expression, which is indicative of the similarity of the mechanical stresses applied (Fig. 2E). Consistent with that finding, Egr-1, another early response gene known to be regulated by SRF independently of MRTF-A, was also similarly induced in the two genotypes (Fig. 2F) (54). In addition, the gene encoding the cytoskeletal protein striated muscle activator of Rho signaling (STARS), a downstream target of MEF2 also rapidly induced by TAB, was similarly activated in both genotypes (Fig. 2G) (21).
TABLE 1.
Echocardiographic analysis of 11-week-old MRTF-A+/+ and MRTF-A−/− mice treated with AngII or left untreated
| Characteristica | Value for mouse groupb |
|||
|---|---|---|---|---|
| No treatment |
AngII treatment |
|||
| WT | MRTF-A−/− | WT | MRTF-A−/− | |
| HR (beats/min) | 687.0 ± 40.0 | 694.8 ± 20.1 | 687.0 ± 20.9 | 677.4 ± 18.4 |
| LVDd (mm) | 2.92 ± 0.14 | 2.82 ± 0.33 | 2.7 ± 0.22 | 2.94 ± 0.09 |
| LVDs (mm) | 1.12 ± 0.15 | 1.04 ± 0.19 | 1.23 ± 0.09* | 1.09 ± 0.12 |
| IVST (mm) | 0.95 ± 0.07 | 0.93 ± 0.01 | 1.03 ± 0.14 | 0.96 ± 0.06# |
| PWT (mm) | 0.90 ± 0.06 | 0.90 ± 0.04 | 1.30 ± 0.17* | 0.97 ± 0.05# |
| FS (%) | 62.4 ± 4.3 | 63.6 ± 3.7 | 63.1 ± 2.3 | 66.0 ± 2.3 |
| EF (%) | 93.6 ± 1.9 | 94.6 ± 1.6 | 94.8 ± 1.1 | 95.6 ± 0.8 |
| LVM (mg) | 74.7 ± 4.8 | 80.5 ± 10.3 | 122.1 ± 11.5* | 94.5 ± 4.7# |
HR, heart rate; LVDd, left ventricular end diastolic dimension; LVDs, left ventricular end systolic dimension; IVST, interventricular septal thickness; PWT, posterior wall thickness; FS, fractional shortening; EF, ejection fraction; LVM, left ventricular mass.
Values are means ± SEM. For the untreated groups, n = 5; for the AngII-treated groups, n = 8 (wild type [WT]) or 9 (MRTF-A−/−). *, P < 0.05 versus results for untreated (WT) mice. #, P < 0.05 versus results for WT mice treated with AngII.
FIG. 2.
The increased expression of BNP and SRF-dependent fetal cardiac genes induced by acute pressure overload is blunted in MRTF-A−/− mice. (A) Myocardin, MRTF-A, and MRTF-B mRNA levels evaluated by real-time RT-PCR in the ventricular myocardium of wild-type (wt), MRTF-A+/− (het), and MRTF-A−/− (null) mice. (B and C) HW/BW ratios (mg/g) (B) or BWs (g) (C) in 8- to 10-week-old MRTF-A+/− (het) and MRTF-A−/− (null) mice 1 h after TAB or sham operation (n = 8 each). (D to K) BNP (D), c-Fos (E), Egr-1 (F), STARS (G), ANP (H), skeletal α-actin (SKA) (I), SM22α (J), or smooth muscle α-actin (SMA) (K) gene expression was assessed by real-time RT-PCR using total RNA extracted from 8- to 10-week-old MRTF-A+/− (het) and MRTF-A−/− (null) mice 1 h after TAB or sham operation (n = 8 each). *, P < 0.05 versus results with sham. In all graphs, values are shown as means ± SEM.
We also examined the expression of other known SRF target genes potentially controlled by myocardin family proteins, including the genes for ANP, skeletal α-actin, SM22α, and smooth muscle α-actin (53, 57). Though the level of ANP mRNA expression was not significantly altered by 1 h of TAB, levels of skeletal α-actin, SM22α, and smooth muscle α-actin gene expression were significantly upregulated in wild-type mice (Fig. 2H to K), but those increases were significantly attenuated in MRTF-A−/− mice (Fig. 2I to K). Thus, MRTF-A is required to mediate the stretch-induced hypertrophic signaling that leads to the upregulation of several SRF-dependent fetal cardiac genes.
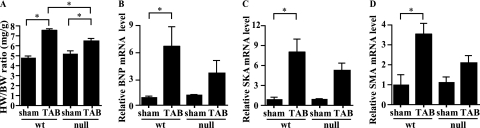
To evaluate the contribution of MRTF-A to mechanical stress-induced chronic hypertrophic responses, including genetic alterations, we next subjected wild-type and MRTF-A−/− mice to chronic pressure overload. When we compared the HW/BW ratios in wild-type and MRTF-A−/− mice subjected to a sham operation or TAB for 3 weeks (13), we found that the increase in HW/BW ratios in MRTF-A−/− mice subjected to TAB was significantly smaller than those seen in wild-type mice subjected to TAB (Fig. 3A). Consistent with that finding, TAB-induced increases in the expression of genes encoding BNP, skeletal α-actin, and smooth muscle α-actin were significantly smaller in MRTF-A−/− mice than in wild-type mice (Fig. 3B to D). These results further support the notion that MRTF-A is required to mediate the mechanical stress-induced hypertrophic signaling that leads to upregulation of several SRF-dependent fetal cardiac genes.
FIG. 3.
Hypertrophic response induced by chronic pressure overload is attenuated in MRTF-A−/− mice. (A) HW/BW ratios (mg/g) in wild type (wt) and MRTF-A−/− (null) mice 3 weeks after TAB or sham operation (n = 3 each). *, P < 0.05. (B to D) BNP (B), skeletal α-actin (SKA) (C), or smooth muscle α-actin (SMA) (D) gene expression was assessed with real-time RT-PCR using total RNA extracted from wild type and MRTF-A−/− mice 3 weeks after TAB or sham operation (n = 3 each).
BNP gene is a direct target of SRF.
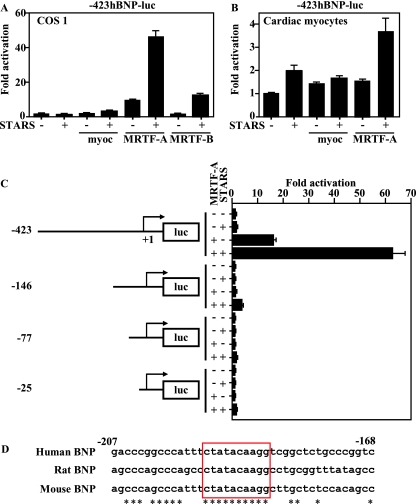
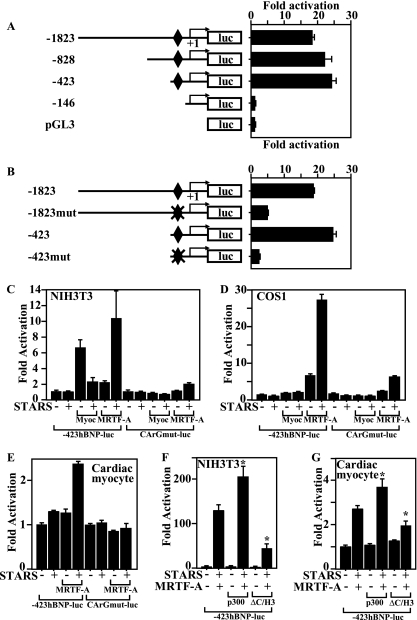
Although it has been suggested that BNP expression is under the control of SRF (43), a functional CArG box has yet to be identified in the BNP promoter, and it remains unclear whether BNP is a direct target of SRF. That said, the observed selective reduction of TAB-induced BNP expression in MRTF-A−/− mice suggests a direct involvement of SRF in BNP gene regulation. We previously showed that STARS induces nuclear translocation of MRTF-A and -B and activates SRF (19). To test whether BNP promoter activity is directly activated by SRF, we cotransfected COS1 cells with a BNP-luciferase gene and expression vectors encoding myocardin, MRTF-A, MRTF-B, or STARS. As shown in Fig. 4A, the BNP proximal promoter was activated by any of these SRF coactivators and was strongly activated by the combination of STARS and MRTF-A, clearly demonstrating that BNP is a MRTF-A-sensitive, direct downstream target of SRF. As in COS1 cells, the BNP promoter in cardiac myocytes was activated by myocardin, MRTF-A, and/or STARS and most strongly activated by the combination of STARS and MRTF-A (Fig. 4B).
FIG. 4.
Identification of the MRTF-A-responsive element in the BNP promoter. (A) MRTF-A and STARS activate −423BNP-luc in COS1 cells. COS1 cells were cotransfected with −423BNP-luc and expression vectors encoding STARS, myocardin (20 ng), MRTF-A (20 ng), or MRTF-B (20 ng). (B) Ventricular myocytes were cotransfected with −423BNP-luc and expression vectors encoding STARS, myocardin (20 ng), or MRTF-A (20 ng). (C) Ventricular myocytes were transfected with a luciferase gene driven by various DNA fragments from the BNP upstream region. Deletion from bp −423 to −146 abolished the response to MRTF-A and STARS in ventricular myocytes. A LacZ gene driven by the RSV promoter was also transfected as an internal control. (D) Sequences between bp −207 and −168 are shown. CArG box-like sequences completely conserved among humans, rats, and mice are shown in the red box.
We next used several BNP promoter deletion mutants to identify the response element for MRTF-A. We found that deletion from bp −423 to −146 significantly reduced the response of the BNP promoter to MRTF-A and STARS and that additional deletions did not reduce the activity further. This means the MRTF-A responsive element is located in a region between bp −423 and −146, within the BNP promoter (Fig. 4C). A search for a CArG box within this region revealed a sequence similar to a CArG box that extended from bp −184 to −193 (CTATACAAGG) and was completely conserved in humans, rats, and mice (Fig. 4D). We then performed electrophoretic mobility shift assays (EMSAs) using this sequence as a probe and found that SRF binds to the sequence but its affinity is weaker than that for the CArG box in the SM22α gene promoter (Fig. 5A). More importantly, we observed that endogenous SRF in cardiac myocytes binds to the sequence (Fig. 5A), and ChIP analysis confirmed the recruitment of SRF to this CArG-like sequence in the BNP gene in cardiac myocytes (Fig. 5B). Conversely, deletion or mutation of this element almost completely abolished activation of the BNP promoter by a SRF-VP16 fusion protein (Fig. 6A and B); moreover, mutations within this CArG element abolished the response of the BNP promoter to MRTF-A and STARS in both nonmuscle cells and cardiac myocytes (Fig. 6C to E), which makes this element the exclusive functional SRF binding site, at least within the 1,823-bp BNP promoter.
FIG. 5.
SRF binds to the CArG-like region in the BNP gene. (A) EMSAs were performed using in vitro-translated SRF (indicated as SRF) or nuclear extract from cultured cardiac ventricular myocytes (indicated as MC) and the BNP CArG-box-like region, with (mut) or without mutation, or the SM22 CArG-box (SM22) as a probe. The unlabeled BNP CArG-box-like region, mutated BNP CArG box-like region, or SM22 CArG box was used as a cold competitor (+, 10-fold; ++, 100-fold; and +++, 1,000-fold excess). Anti-SRF antibody was used for supershift assays. Arrows indicate bands representing a specific DNA-SRF complex. The lowest of the EMSA bands was markedly weakened when the wild-type and mutant BNP CArG probes were applied in the presence of the wild-type and mutant BNP CArG cold probes but not the cold SM22 CArG probe, indicating that these bands represent a complex between the BNP probe and a nonspecific protein binding to a sequence other than the core CArG-like sequence in the BNP probe. Note that this band is not affected by anti-SRF antibody. (B) ChIP assays were performed with ventricular myocytes using anti-SRF antibody. PCR was performed with chromatin from around the CArG box in the BNP promoter region, around the region 2 kbp distal to the CArG box in the BNP promoter, around the two CArG boxes in the SM22 promoter region, and around the first exon of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Aliquots of chromatin obtained before immunoprecipitation were also analyzed as input. Representative PCRs with identical results in three independent experiments are shown. Schematic representations of the location of the PCR primers in each gene are shown in the upper panel.
FIG. 6.
MRTF-A directly activates the BNP promoter via the SRF-binding site. (A) COS1 cells were cotransfected with a luciferase gene driven by various DNA fragments from the BNP upstream region and an expression vector encoding the SRF-VP16 fusion protein. The SRF-binding site is shown as a diamond. (B) COS1 cells were cotransfected with a luciferase gene driven by 1,823 bp or 423 bp of the BNP upstream region, with (mut) or without mutation of the identified SRF-binding site, and an expression vector encoding the SRF-VP16 fusion protein. The SRF-binding site is shown as a diamond. (C and D) NIH 3T3 (C) or COS1 (D) cells were cotransfected with a luciferase gene driven by 423 bp from the BNP upstream region, with (CArGmut-luc) or without (−423hBNP-luc) mutation of the SRF-binding site, and an expression vector encoding STARS, myocardin, or MRTF-A. (E) Cardiac myocytes were cotransfected with −423hBNP-luc or CArGmut-luc and an expression vector encoding STARS or MRTF-A. (F and G) p300 contributes to MRTF-A-mediated activation of SRF. NIH 3T3 cells (F) or cultured cardiac myocytes (G) were cotransfected with −423hBNP-luc and expression vectors encoding STARS, MRTF-A, and wild-type p300 or a dominant-negative p300 mutant (ΔC/H3). Values are shown as means ± SEM (n = 4 each). *, P < 0.05 versus results with −423hBNP-luc with STARS and MRTF-A in the absence of p300.
p300 is a transcriptional coactivator that possesses intrinsic histone acetyltransferase activity and reportedly participates in myocardin-mediated SRF activation (7). And because myocardin's transcription-activating domain is well conserved in MRTF-A, we tested whether p300 also participates in MRTF-A-induced activation of the BNP promoter. When we cotransfected cultured ventricular myocytes with a BNP-luciferase gene and expression vectors encoding MRTF-A, STARS, and/or wild type p300 or a dominant-negative p300 mutant (42), we found that wild-type p300 enhanced MRTF-A-mediated BNP promoter activation. In contrast, the dominant-negative p300 mutant significantly attenuated MRTF-A-induced activation of BNP gene transcription in both NIH 3T3 cells and cardiac myocytes (Fig. 6F and G). We also confirmed the physical interaction between MRTF-A and p300 (see Fig. S1A in the supplemental material). p300 thus appears to participate in MRTF-A-mediated activation of BNP gene transcription.
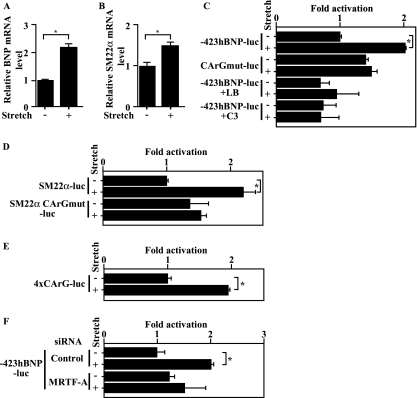
Mechanical stretch increases BNP promoter activity through SRF.
We hypothesized that the stretch-induced nuclear accumulation of MRTF-A we observed contributes to the stretch-induced increase in BNP gene transcription. To test that idea, we examined whether CArG in the BNP promoter is responsible for stretch-induced BNP gene expression. We initially confirmed that expression of endogenous BNP and SM22α mRNA is upregulated in an in vitro model of mechanical stretch, as reported previously (Fig. 7A and B) (18). Consistent with stretch-induced expression of the endogenous gene, the BNP promoter was activated by mechanical stretch in neonatal ventricular myocytes (Fig. 7C). Mechanical stretch also increased SM22α promoter activity in an SRF binding site-dependent manner (Fig. 7D) and activated a luciferase reporter gene controlled by tandem CArG boxes (4× CArG-luciferase), confirming that myocardial stretch activates SRF-dependent transcription (Fig. 7E). Mutation of CArG within the BNP promoter significantly reduced the response to mechanical stretch, suggesting the CArG element transduces the signal initiated by mechanical stretch (Fig. 7C). Stretch-induced BNP promoter activity was also diminished in the presence of latrunculin B or C3, which inhibit the nuclear translocation of MRTF-A (Fig. 7C and 1C), and in the presence of siRNA targeting MRTF-A, confirming that MRTF-A mediates stretch-induced BNP promoter activation (Fig. 7F).
FIG. 7.
Mechanical stretch activates the BNP promoter via the SRF-binding site. (A and B) Expression of BNP (A) or SM22α (B) mRNA was assessed using real-time RT-PCR in rat neonatal ventricular myocytes subjected to 20% mechanical stretch for 1 h. (C) Ventricular myocytes transfected with a luciferase gene driven by 423 bp from the BNP upstream region, with (CArGmut-luc) or without (−423hBNP-luc) mutation of the SRF-binding site, were subjected to mechanical stretch in the presence or absence of 0.5 μM latrunculin B (LB) or 2.5 μg/ml C3 exoenzyme (C3). (D) Ventricular myocytes transfected with a luciferase gene driven by the SM22 α upstream region, with (SM22α CArGmut) or without (SM22α-luc) mutation of SRF-binding sites, were subjected to mechanical stretch. (E) Ventricular myocytes transfected with 4× CArG-luc were subjected to mechanical stretch. (F) Ventricular myocytes transfected with −423hBNP-luc were subjected to mechanical stretch in the presence or absence of siRNA targeting MRTF-A. For all transfections, a LacZ gene driven by an RSV promoter was also transfected as an internal control. In all graphs, values are shown as means ± SEM; *, P < 0.05 versus results for the unstretched control.
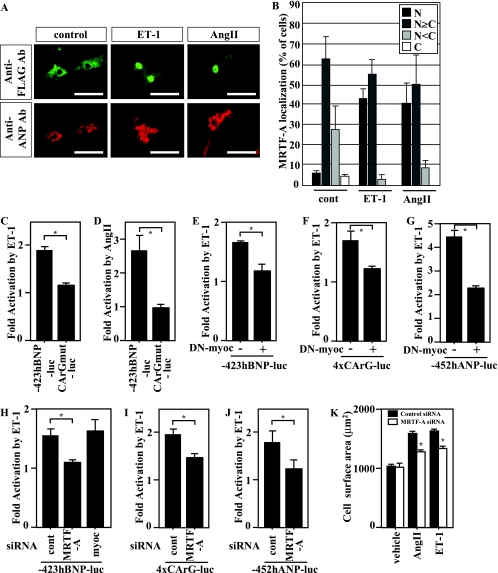
MRTF-A mediates ET-1- and AngII-induced activation of BNP gene transcription.
Since Rho activation is involved in cardiac hypertrophy induced by several humoral factors, including AngII, phenylephrine, and ET-1 (2, 20, 52), we also tested whether nuclear translocation of MRTF-A occurs following stimulation of cardiac myocytes with ET-1 or AngII. We found that both molecules enhanced nuclear accumulation of MRTF-A in ventricular myocytes (Fig. 8A and B) and that they activated the BNP promoter (Fig. 8C and D). Moreover, mutation of CArG significantly reduced the response, confirming that SRF activation plays an important role in ET-1- and AngII-induced activation of BNP gene transcription (Fig. 8C and D). ET-1-induced activation of the BNP promoter was blocked in the presence of a dominant-negative myocardin mutant that lacked a transcription-activating domain and inhibited the activities of all myocardin family members, including myocardin and MRTF-A and -B, which suggests their involvement in ET-1-induced, SRF-mediated activation of the BNP promoter (Fig. 8E). The dominant-negative myocardin mutant also inhibited ET-1-induced activation of a reporter gene controlled by tandem CArG boxes (4× CArG-luciferase) (Fig. 8F) and the ANP promoter (hANP-luciferase) (Fig. 8G), again suggesting the involvement of myocardin and MRTF family proteins. In addition, siRNA targeting MRTF-A but not myocardin inhibited ET-1-induced activation of the BNP promoter in ventricular myocytes, indicating that, as with mechanical stretch, nuclear translocation of MRTF-A mediates ET-1-induced hypertrophic signaling to activate BNP gene transcription (Fig. 8H). Finally, MRTF-A knockdown also inhibited ET-1-induced activation of 4× CArG-luciferase and hANP-luciferase (Fig. 8I and J), suggesting the general involvement of MRTF-A in G protein-coupled receptor-induced increases in SRF-mediated gene transcription. MRTF-A knockdown also significantly attenuated ET-1- and AngII-induced increases in the size of cardiac myocytes (Fig. 8K; see also Fig. S1B in the supplemental material).
FIG. 8.
ET-1 and AngII induce nuclear accumulation of MRTF-A. (A) Cultured neonatal rat ventricular myocytes infected with adenovirus encoding FLAG-tagged MRTF-A (Ad-MRTF-A) were treated with 100 nM ET-1 or 100 nM AngII for 1 h. The subcellular distribution of MRTF-A was determined by immunostaining of the FLAG epitope (green). Myocytes were positively stained with anti-ANP antibody (red). Bars represent 50 μm. (B) Graphs show the percent MRTF-A localization: N, exclusive staining of MRTF-A in the nucleus; N≥C, nuclear staining of MRTF-A was greater than or equal to the cytoplasmic staining; N<C, greater staining of MRTF-A in the cytoplasm than in the nucleus; C, exclusive staining of cytoplasmic MRTF-A (100 infected cells were counted). (C and D) Ventricular myocytes transfected with a luciferase gene driven by the human BNP upstream region, with (CArGmut) or without (−423hBNP-luc) mutation of SRF-binding sites, were treated with 100 nM ET-1 (C) or 100 nΜ AngII (D) for 48 h. (E to G) Ventricular myocytes cotransfected with a luciferase gene driven by the BNP upstream region (−423hBNP-luc) (E), by 4× CArG (4xCArG-luc) (F), or by the human ANP promoter (−452hANP-luc) (G), as well as an expression vector encoding a dominant-negative myocardin mutant (DN-myocardin), were treated with ET-1. (H) Ventricular myocytes cotransfected with −423hBNP-luc and siRNA targeting MRTF-A or myocardin were treated with ET-1. (I and J) Ventricular myocytes cotransfected with siRNA targeting MRTF-A and 4× CArG-luc (I) or −452hANP-luc (J) were treated with ET-1. In all luciferase experiments, a LacZ gene driven by the RSV promoter was also transfected as an internal control. The graphs show the fold activation by ET-1 in each group. In all graphs, values are shown as means ± SEM; *, P < 0.05. (K) Cultured ventricular myocytes in which MRTF-A expression was knocked down using siRNA were less susceptible to hypertrophic myocyte growth induced by ET-1 (100 nM) or AngII (100 nΜ) than control myocytes. Graphs show myocyte size (μm2) in each group. Values are shown as means ± SEM; *, P < 0.05 versus results for control siRNA in each group.
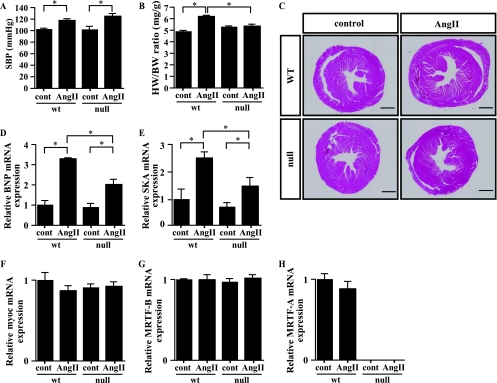
Reduced hypertrophic responses to chronic AngII treatment in MRTF-A−/− mice.
We next examined the role of MRTF-A in chronic cardiac remodeling, a process in which neurohumoral factors are known to play pivotal roles. When we subcutaneously administered AngII for 2 weeks, systolic blood pressure (SBP) was similarly increased in wild-type and MRTF-A−/− mice (Fig. 9A). Under these conditions, cardiac hypertrophy, indicated by significant increases in HW/BW ratios, was observed in wild-type mice but not in MRTF-A−/− mice (Fig. 9B and C). Moreover, the expression of hypertrophy-related genes, including BNP and skeletal α-actin, induced by chronic AngII treatment was significantly weaker in MRTF-A−/− mice than in wild-type mice (Fig. 9D and E). This supports the notion that MRTF-A is necessary for chronic AngII-induced cardiac hypertrophy. In addition, echocardiographic analysis showed that AngII-induced increases in the thickness of the interventricular septum and left ventricular posterior wall, as well as the calculated left ventricular mass, were all significantly attenuated in MRTF-A−/− mice compared to findings for wild-type mice (Table 1). Levels of myocardin and MRTF-B mRNA were not significantly altered in wild-type or MRTF-A−/− mice, with or without AngII treatment (Fig. 9F and G), nor were levels of MRTF-A mRNA in wild-type mice (Fig. 9H). Taken together, these results demonstrate that MRTF-A is a crucial participant in cardiac hypertrophy signaling during the cardiac remodeling induced by AngII.
FIG. 9.
MRTF-A participates in AngII-induced hypertrophic signaling in vivo. (A) SBP (mm Hg) in wild-type (wt) and MRTF-A−/− (null) mice, with or without AngII infusion for 2 weeks (n = 4 each for wild-type mice and 8 each for MRTF-A−/− mice). (B) HW/BW ratios (mg/g) in wt and MRTF-A−/− mice, with or without AngII infusion for 2 weeks (10 to 12 weeks of age; n = 4 each for wild-type mice and 8 each for MRTF-A−/− mice). (C) Hearts were sectioned coronally and stained with hematoxylin-eosin. Scale bar, 1 mm. (D to H) BNP (D), skeletal α-actin (SKA) (E), myocardin (myoc) (F), MRTF-B (G), or MRTF-A (H) gene expression was assessed by real-time RT-PCR using total RNA extracted from wt and MRTF-A−/− mice, with or without AngII for 2 weeks (n = 3 for wild-type mice without AngII, 4 for wild-type mice with AngII, 5 for MRTF-A−/− mice without AngII, and 6 for MRTF-A−/− mice with AngII). In all graphs, values are shown as means ± SEM; *, P < 0.05.
DISCUSSION
Mechanical stress is one of the earliest stimuli promoting the induction of cardiac hypertrophy, which is characterized in part by reactivation of the fetal cardiac gene program (e.g., ANP, BNP, skeletal α-actin, β-myosin heavy chain, SM22α, and smooth muscle α-actin) (4, 15, 25). Using an in vitro cardiac myocyte model, it has been shown that mechanical stretch activates a variety of intracellular signaling molecules, including PKC, MAPKs, p90 and p70 S6 kinases, Jak-STAT, and Rho family small G proteins (1, 16, 27, 47, 48, 51, 62). The precise molecular mechanism by which mechanical stretch is transduced to transcriptional activation remained unresolved, however. In the present study, we have shown that Rho- and actin treadmill-dependent nuclear accumulation of MRTF-A contributes to the transduction of mechanical stress to the transcriptional activation of SRF-dependent fetal cardiac genes in cardiac myocytes. In mice lacking MRTF-A, induction of BNP and other fetal cardiac genes in response to both acute and chronic pressure overload was significantly attenuated. We identified a functional SRF-responsive element in the 5′-flanking region of the BNP gene as a novel target of MRTF-A. In addition, we also showed the involvement of MRTF-A in chronic cardiac remodeling, a process in which neurohumoral factors play a pivotal role. Following stimulation with AngII or ET-1, MRTF-A was translocated into the nuclei of cardiac myocytes, where it activated SRF. Moreover, MRTF-A−/− mice showed significantly weaker hypertrophic responses than their wild-type littermates. Collectively, these findings indicate that MRTF-A is a common mediator of mechanical stress- and neurohumoral stimulation-induced prohypertrophic signaling.
There are two distinct pathways leading to SRF activation: one involves the phosphorylation of ternary complex factors in Ets domain family proteins, while the other is controlled by Rho family small GTPases and actin dynamics (10, 14, 40, 56). MRTF-A is involved in the latter (35, 50). In the regulation of some immediate-early genes, Ets domain family proteins, such as Elk-1, which is phosphorylated by extracellular signal-regulated kinase (ERK), associate with and activate SRF independently of MRTF-A (14, 22, 40, 54, 61). The fact that mechanical stretch activates ERKs therefore suggests that during mechanical stress, the ERKs-Elk1 pathway contributes to the increased expression of several immediate-early genes through activation of SRF. Thus, the genetic response to mechanical stretch involves both MRTF-A-dependent and -independent SRF activation. Moreover, SRF reportedly interacts with two other cardiac transcriptional factors, GATA and NKX2.5, and with transcriptional regulators, such as HOP, which do not bind to DNA. This suggests that full activation of the hypertrophic cardiac gene program may require SRF to also work with transcriptional factors situated downstream of signaling pathways other than Rho-actin dynamics-dependent and ERK-dependent pathways (3, 17, 55).
The expression of the BNP gene is rapidly and dramatically upregulated in both in vivo and in vitro models of cardiac hypertrophy in response to hypertrophic stimuli, including mechanical stress and neurohumoral stimulation (11, 18, 41). Indeed, plasma BNP levels are a clinical marker used to detect and manage cardiac hypertrophy and heart failure in humans (38, 39, 60). Although several signaling pathways and transcriptional factors are known to be involved in the stretch- and neurohumoral stimulation-induced activation of the BNP promoter (27, 30, 37, 44), the entire molecular process governing the transcriptional activation of BNP has not yet been characterized. In that regard, expression of BNP mRNA is reportedly altered in SRF−/− cardiac myocytes (43), though the functional SRF-binding site had not been identified in the BNP gene. In the present study, we identified a functional but atypical CArG element within the 1,823-bp BNP promoter region. Deletion or mutation of this CArG box almost completely abolished the increase in transcription induced by SRF-VP16, suggesting this region is functionally the most important SRF-binding site, at least within the 1,823-bp BNP promoter. Indeed, this CArG element is completely conserved among humans, rats, and mice. We further showed that mutation of CArG reduced the response of the BNP promoter to two hypertrophic stimuli, mechanical stretch and neurohumoral stimulation. That the ANP promoter also has two CArG boxes that have been implicated in hypertrophic induction of ANP gene transcription (57) suggests ANP and BNP may be coordinately regulated by SRF.
Among the variety of signaling molecules activated within cardiac myocytes following mechanical stretch or neurohumoral stimulation, Rho family small GTPases, especially Rho A and Rac1, are known to be important regulators of cardiac hypertrophy (5, 24). For instance, the inhibition of Rho or ROCK (Rho kinase), a downstream target of Rho, has been shown to ameliorate pathological cardiac hypertrophy (5, 20, 29). Our study defines MRTF-A as a critical downstream mediator of Rho- and actin dynamics-associated prohypertrophic signaling in cardiac myocytes, while others have shown that in epithelial cells MRTF-A is also activated downstream of Rac (6). Two important events that occur downstream of Rho and Rac activation are alteration of actin cytoskeletal organization and gene transcription. It appears that MRTF-A is a key mediator of the latter. Consequently, diminishing MRTF-A-mediated transcriptional activation by inhibiting its nuclear translocation and/or its coactivator function, which would selectively block transcriptional pathways activated downstream of Rho family small GTPases, could be a safer and more specific therapeutic approach to preventing pathological cardiac remodeling without the potential side effects caused by disruption of the physiological organization of the actin cytoskeleton.
Supplementary Material
Acknowledgments
We thank Yukari Kubo for her excellent secretarial work. We also thank E. N. Olson and R. Bassel-Duby for providing us with MRTF knockout mice and critically reading the manuscript.
This research was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to K. Kuwahara and K. Nakao), a grant from the Japanese Ministry of Health, Labor and Welfare (to K. Nakao), and grants from the Mitsubishi Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Uehara Memorial Foundation, the Japan Heart Foundation/Novartis Grant for Research Award on Molecular and Cellular Cardiology, the Japan Foundation for Applied Enzymology, the Mitsubishi Pharma Research Foundation, the Astellas Foundation for Research on Metabolic Disorders, the Kanae Foundation for the Promotion of Medical Science, the Ichiro Kanehara Foundation, the Suzuken Memorial Foundation, the Vehicle Racing Commemorative Foundation, the Japan Research Promotion Society for Cardiovascular Diseases, the Takeda Medical Research Foundation, and the Hoh-ansha Foundation (to K. Kuwahara).
We have no conflict of interest to report.
Footnotes
Published ahead of print on 6 July 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aikawa, R., I. Komuro, T. Yamazaki, Y. Zou, S. Kudoh, W. Zhu, T. Kadowaki, and Y. Yazaki. 1999. Rho family small G proteins play critical roles in mechanical stress-induced hypertrophic responses in cardiac myocytes. Circ. Res. 84:458-466. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, H., S. Izumo, and J. Sadoshima. 1998. Angiotensin II activates RhoA in cardiac myocytes: a critical role of RhoA in angiotensin II-induced premyofibril formation. Circ. Res. 82:666-676. [DOI] [PubMed] [Google Scholar]
- 3.Belaguli, N. S., J. L. Sepulveda, V. Nigam, F. Charron, M. Nemer, and R. J. Schwartz. 2000. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol. Cell. Biol. 20:7550-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, F. M., S. E. Packer, T. G. Parker, L. H. Michael, R. Roberts, R. J. Schwartz, and M. D. Schneider. 1991. The vascular smooth muscle alpha-actin gene is reactivated during cardiac hypertrophy provoked by load. J. Clin. Invest. 88:1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, J. H., D. P. Del Re, and M. A. Sussman. 2006. The Rac and Rho hall of fame: a decade of hypertrophic signaling hits. Circ. Res. 98:730-742. [DOI] [PubMed] [Google Scholar]
- 6.Busche, S., A. Descot, S. Julien, H. Genth, and G. Posern. 2008. Epithelial cell-cell contacts regulate SRF-mediated transcription via Rac-actin-MAL signalling. J. Cell Sci. 121:1025-1035. [DOI] [PubMed] [Google Scholar]
- 7.Cao, D., Z. Wang, C. L. Zhang, J. Oh, W. Xing, S. Li, J. A. Richardson, D. Z. Wang, and E. N. Olson. 2005. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol. Cell. Biol. 25:364-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn, J. N., R. Ferrari, and N. Sharpe. 2000. Cardiac remodeling—concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. J. Am. Coll. Cardiol. 35:569-582. [DOI] [PubMed] [Google Scholar]
- 9.Frey, N., T. Barrientos, J. M. Shelton, D. Frank, H. Rutten, D. Gehring, C. Kuhn, M. Lutz, B. Rothermel, R. Bassel-Duby, J. A. Richardson, H. A. Katus, J. A. Hill, and E. N. Olson. 2004. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 10:1336-1343. [DOI] [PubMed] [Google Scholar]
- 10.Gineitis, D., and R. Treisman. 2001. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 276:24531-24539. [DOI] [PubMed] [Google Scholar]
- 11.Harada, K., I. Komuro, Y. Zou, S. Kudoh, K. Kijima, H. Matsubara, T. Sugaya, K. Murakami, and Y. Yazaki. 1998. Acute pressure overload could induce hypertrophic responses in the heart of angiotensin II type 1a knockout mice. Circ. Res. 82:779-785. [DOI] [PubMed] [Google Scholar]
- 12.Hill, C. S., J. Wynne, and R. Treisman. 1995. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81:1159-1170. [DOI] [PubMed] [Google Scholar]
- 13.Hill, J. A., M. Karimi, W. Kutschke, R. L. Davisson, K. Zimmerman, Z. Wang, R. E. Kerber, and R. M. Weiss. 2000. Cardiac hypertrophy is not a required compensatory response to short-term pressure overload. Circulation 101:2863-2869. [DOI] [PubMed] [Google Scholar]
- 14.Janknecht, R., W. H. Ernst, V. Pingoud, and A. Nordheim. 1993. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 12:5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerkela, R., and T. Force. 2006. Recent insights into cardiac hypertrophy and left ventricular remodeling. Curr. Heart Fail. Rep. 3:14-18. [DOI] [PubMed] [Google Scholar]
- 16.Komuro, I. 2000. Molecular mechanism of mechanical stress-induced cardiac hypertrophy. Jpn. Heart J. 41:117-129. [DOI] [PubMed] [Google Scholar]
- 17.Kook, H., J. J. Lepore, A. D. Gitler, M. M. Lu, W. Wing-Man Yung, J. Mackay, R. Zhou, V. Ferrari, P. Gruber, and J. A. Epstein. 2003. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Invest. 112:863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudoh, S., H. Akazawa, H. Takano, Y. Zou, H. Toko, T. Nagai, and I. Komuro. 2003. Stretch-modulation of second messengers: effects on cardiomyocyte ion transport. Prog. Biophys. Mol. Biol. 82:57-66. [DOI] [PubMed] [Google Scholar]
- 19.Kuwahara, K., T. Barrientos, G. C. Pipes, S. Li, and E. N. Olson. 2005. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol. Cell. Biol. 25:3173-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuwahara, K., Y. Saito, O. Nakagawa, I. Kishimoto, M. Harada, E. Ogawa, Y. Miyamoto, I. Hamanaka, N. Kajiyama, N. Takahashi, T. Izumi, R. Kawakami, N. Tamura, Y. Ogawa, and K. Nakao. 1999. The effects of the selective ROCK inhibitor, Y27632, on ET-1-induced hypertrophic response in neonatal rat cardiac myocytes—possible involvement of Rho/ROCK pathway in cardiac muscle cell hypertrophy. FEBS Lett. 452:314-318. [DOI] [PubMed] [Google Scholar]
- 21.Kuwahara, K., G. C. Teg Pipes, J. McAnally, J. A. Richardson, J. A. Hill, R. Bassel-Duby, and E. N. Olson. 2007. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J. Clin. Invest. 117:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, S. M., M. Vasishtha, and R. Prywes. 12 May 2010. Activation and repression of cellular immediate early genes by SRF cofactors. J. Biol. Chem. [Epub ahead of print.] doi: 10.1074/jbc.M110.108878. [DOI] [PMC free article] [PubMed]
- 23.Lee, T. C., K. L. Chow, P. Fang, and R. J. Schwartz. 1991. Activation of skeletal alpha-actin gene transcription: the cooperative formation of serum response factor-binding complexes over positive cis-acting promoter serum response elements displaces a negative-acting nuclear factor enriched in replicating myoblasts and nonmyogenic cells. Mol. Cell. Biol. 11:5090-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lezoualc'h, F., M. Metrich, I. Hmitou, N. Duquesnes, and E. Morel. 2008. Small GTP-binding proteins and their regulators in cardiac hypertrophy. J. Mol. Cell Cardiol. 44:623-632. [DOI] [PubMed] [Google Scholar]
- 25.Li, L., J. M. Miano, P. Cserjesi, and E. N. Olson. 1996. SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ. Res. 78:188-195. [DOI] [PubMed] [Google Scholar]
- 26.Li, S., S. Chang, X. Qi, J. A. Richardson, and E. N. Olson. 2006. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol. Cell. Biol. 26:5797-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang, F., and D. G. Gardner. 1999. Mechanical strain activates BNP gene transcription through a p38/NF-kappaB-dependent mechanism. J. Clin. Invest. 104:1603-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang, F., J. Wu, M. Garami, and D. G. Gardner. 1997. Mechanical strain increases expression of the brain natriuretic peptide gene in rat cardiac myocytes. J. Biol. Chem. 272:28050-28056. [DOI] [PubMed] [Google Scholar]
- 29.Loirand, G., P. Guerin, and P. Pacaud. 2006. Rho kinases in cardiovascular physiology and pathophysiology. Circ. Res. 98:322-334. [DOI] [PubMed] [Google Scholar]
- 30.Ma, K. K., K. Banas, and A. J. de Bold. 2005. Determinants of inducible brain natriuretic peptide promoter activity. Regul. Pept. 128:169-176. [DOI] [PubMed] [Google Scholar]
- 31.Magga, J., O. Vuolteenaho, H. Tokola, M. Marttila, and H. Ruskoaho. 1997. Involvement of transcriptional and posttranscriptional mechanisms in cardiac overload-induced increase of B-type natriuretic peptide gene expression. Circ. Res. 81:694-702. [DOI] [PubMed] [Google Scholar]
- 32.Miano, J. M. 2003. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell Cardiol. 35:577-593. [DOI] [PubMed] [Google Scholar]
- 33.Miano, J. M., N. Ramanan, M. A. Georger, K. L. de Mesy Bentley, R. L. Emerson, R. O. Balza, Jr., Q. Xiao, H. Weiler, D. D. Ginty, and R. P. Misra. 2004. Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl. Acad. Sci. U. S. A. 101:17132-17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minty, A., and L. Kedes. 1986. Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells: presence of an evolutionarily conserved repeated motif. Mol. Cell. Biol. 6:2125-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miralles, F., G. Posern, A. I. Zaromytidou, and R. Treisman. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329-342. [DOI] [PubMed] [Google Scholar]
- 36.Molkentin, J. D., S. M. Jobe, and B. E. Markham. 1996. Alpha-myosin heavy chain gene regulation: delineation and characterization of the cardiac muscle-specific enhancer and muscle-specific promoter. J. Mol. Cell Cardiol. 28:1211-1225. [DOI] [PubMed] [Google Scholar]
- 37.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison, L. K., A. Harrison, P. Krishnaswamy, R. Kazanegra, P. Clopton, and A. Maisel. 2002. Utility of a rapid B-natriuretic peptide assay in differentiating congestive heart failure from lung disease in patients presenting with dyspnea. J. Am. Coll. Cardiol. 39:202-209. [DOI] [PubMed] [Google Scholar]
- 39.Mukoyama, M., K. Nakao, K. Hosoda, S. Suga, Y. Saito, Y. Ogawa, G. Shirakami, M. Jougasaki, K. Obata, H. Yasue, et al. 1991. Brain natriuretic peptide as a novel cardiac hormone in humans. Evidence for an exquisite dual natriuretic peptide system, atrial natriuretic peptide and brain natriuretic peptide. J. Clin. Invest. 87:1402-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murai, K., and R. Treisman. 2002. Interaction of serum response factor (SRF) with the Elk-1 B box inhibits RhoA-actin signaling to SRF and potentiates transcriptional activation by Elk-1. Mol. Cell. Biol. 22:7083-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa, O., Y. Ogawa, H. Itoh, S. Suga, Y. Komatsu, I. Kishimoto, K. Nishino, T. Yoshimasa, and K. Nakao. 1995. Rapid transcriptional activation and early mRNA turnover of brain natriuretic peptide in cardiocyte hypertrophy. Evidence for brain natriuretic peptide as an “emergency” cardiac hormone against ventricular overload. J. Clin. Invest. 96:1280-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa, Y., K. Kuwahara, G. Takemura, M. Akao, M. Kato, Y. Arai, M. Takano, M. Harada, M. Murakami, M. Nakanishi, S. Usami, S. Yasuno, H. Kinoshita, M. Fujiwara, K. Ueshima, and K. Nakao. 2009. p300 plays a critical role in maintaining cardiac mitochondrial function and cell survival in postnatal hearts. Circ. Res. 105:746-754. [DOI] [PubMed] [Google Scholar]
- 43.Nelson, T. J., R. Balza, Jr., Q. Xiao, and R. P. Misra. 2005. SRF-dependent gene expression in isolated cardiomyocytes: regulation of genes involved in cardiac hypertrophy. J. Mol. Cell Cardiol. 39:479-489. [DOI] [PubMed] [Google Scholar]
- 44.Ogawa, E., Y. Saito, K. Kuwahara, M. Harada, Y. Miyamoto, I. Hamanaka, N. Kajiyama, N. Takahashi, T. Izumi, R. Kawakami, I. Kishimoto, Y. Naruse, N. Mori, and K. Nakao. 2002. Fibronectin signaling stimulates BNP gene transcription by inhibiting neuron-restrictive silencer element-dependent repression. Cardiovasc. Res. 53:451-459. [DOI] [PubMed] [Google Scholar]
- 45.Olson, E. N. 2004. A decade of discoveries in cardiac biology. Nat. Med. 10:467-474. [DOI] [PubMed] [Google Scholar]
- 46.Onohara, N., M. Nishida, R. Inoue, H. Kobayashi, H. Sumimoto, Y. Sato, Y. Mori, T. Nagao, and H. Kurose. 2006. TRPC3 and TRPC6 are essential for angiotensin II-induced cardiac hypertrophy. EMBO J. 25:5305-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan, J., K. Fukuda, M. Saito, J. Matsuzaki, H. Kodama, M. Sano, T. Takahashi, T. Kato, and S. Ogawa. 1999. Mechanical stretch activates the JAK/STAT pathway in rat cardiomyocytes. Circ. Res. 84:1127-1136. [DOI] [PubMed] [Google Scholar]
- 48.Pan, J., U. S. Singh, T. Takahashi, Y. Oka, A. Palm-Leis, B. S. Herbelin, and K. M. Baker. 2005. PKC mediates cyclic stretch-induced cardiac hypertrophy through Rho family GTPases and mitogen-activated protein kinases in cardiomyocytes. J. Cell. Physiol. 202:536-553. [DOI] [PubMed] [Google Scholar]
- 49.Parlakian, A., D. Tuil, G. Hamard, G. Tavernier, D. Hentzen, J. P. Concordet, D. Paulin, Z. Li, and D. Daegelen. 2004. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol. Cell. Biol. 24:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parmacek, M. S. 2007. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circ. Res. 100:633-644. [DOI] [PubMed] [Google Scholar]
- 51.Sadoshima, J., and S. Izumo. 1997. The cellular and molecular response of cardiac myocytes to mechanical stress. Annu. Rev. Physiol. 59:551-571. [DOI] [PubMed] [Google Scholar]
- 52.Sah, V. P., M. Hoshijima, K. R. Chien, and J. H. Brown. 1996. Rho is required for Galphaq and alpha1-adrenergic receptor signaling in cardiomyocytes. Dissociation of Ras and Rho pathways. J. Biol. Chem. 271:31185-31190. [DOI] [PubMed] [Google Scholar]
- 53.Schneider, M. D., W. R. McLellan, F. M. Black, and T. G. Parker. 1992. Growth factors, growth factor response elements, and the cardiac phenotype. Basic Res. Cardiol. 87(Suppl. 2):33-48. [DOI] [PubMed] [Google Scholar]
- 54.Selvaraj, A., and R. Prywes. 2004. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol. Biol. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sepulveda, J. L., S. Vlahopoulos, D. Iyer, N. Belaguli, and R. J. Schwartz. 2002. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J. Biol. Chem. 277:25775-25782. [DOI] [PubMed] [Google Scholar]
- 56.Sotiropoulos, A., D. Gineitis, J. Copeland, and R. Treisman. 1999. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98:159-169. [DOI] [PubMed] [Google Scholar]
- 57.Sprenkle, A. B., S. F. Murray, and C. C. Glembotski. 1995. Involvement of multiple cis elements in basal- and alpha-adrenergic agonist-inducible atrial natriuretic factor transcription. Roles for serum response elements and an SP-1-like element. Circ. Res. 77:1060-1069. [DOI] [PubMed] [Google Scholar]
- 58.Tarone, G., and G. Lembo. 2003. Molecular interplay between mechanical and humoral signalling in cardiac hypertrophy. Trends Mol. Med. 9:376-382. [DOI] [PubMed] [Google Scholar]
- 59.Thorburn, J., S. Xu, and A. Thorburn. 1997. MAP kinase- and Rho-dependent signals interact to regulate gene expression but not actin morphology in cardiac muscle cells. EMBO J. 16:1888-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Troughton, R. W., C. M. Frampton, T. G. Yandle, E. A. Espiner, M. G. Nicholls, and A. M. Richards. 2000. Treatment of heart failure guided by plasma aminoterminal brain natriuretic peptide (N-BNP) concentrations. Lancet 355:1126-1130. [DOI] [PubMed] [Google Scholar]
- 61.Wang, Z., D. Z. Wang, D. Hockemeyer, J. McAnally, A. Nordheim, and E. N. Olson. 2004. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428:185-189. [DOI] [PubMed] [Google Scholar]
- 62.Yamazaki, T., I. Komuro, I. Shiojima, and Y. Yazaki. 1999. The molecular mechanism of cardiac hypertrophy and failure. Ann. N. Y. Acad. Sci. 874:38-48. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, X., G. Azhar, J. Chai, P. Sheridan, K. Nagano, T. Brown, J. Yang, K. Khrapko, A. M. Borras, J. Lawitts, R. P. Misra, and J. Y. Wei. 2001. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am. J. Physiol. Heart Circ. Physiol. 280:H1782-H1792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.