Abstract
Cellular responses to many external stimuli are mediated by mitogen-activated protein kinases (MAPKs). We investigated whether dynamic intracellular movement contributes to the spatial and temporal characteristics of the responses elicited by a prototypic MAPK, Fus3, in the mating pheromone response pathway in budding yeast (Saccharomyces cerevisiae). Confining Fus3 in the nucleus, via fusion to a histone H2B, reduced MAPK activation and diminished all responses (pheromone-induced gene expression, cell cycle arrest, projection formation, and mating). Elimination of MAPK phosphatases restored more robust outputs for all responses, indicating that nuclear sequestration impedes full MAPK activation but does not abrogate its functional competence. Restricting Fus3 to the plasma membrane, via fusion to a lipid-modified CCaaX motif, led to MAPK hyperactivation yet severely impaired all response outputs. Fus3-CCaaX also caused aberrant cell morphology and a proliferation defect. Unlike similar phenotypes induced by pathway hyperactivation via upstream components, these deleterious effects were independent of the downstream transcription factor Ste12. Thus, appropriate cellular responses require free subcellular MAPK transit to disseminate MAPK activity optimally because preventing dynamic MAPK movement either markedly impaired signal-dependent activation and/or resulted in improper biological outputs.
Cellular adaptation to an extracellular stimulus often requires the action of mitogen-activated protein kinases (MAPKs) (58, 59). A MAPK serves as a nodal point in its signaling network, in the sense that it connects the relatively linear upstream pathway for its activation to a diverse set of downstream effectors. MAPK targets can include proteins that control quite different aspects of cell function, including cell morphology, cell cycle progression, and gene expression. MAPK-mediated phosphorylation of such substrates must occur in the correct spatial context and in the proper temporal order to elicit coherent and appropriately integrated responses by the cell. How a given MAPK-mediated signal is disseminated to its different cellular targets is a poorly understood aspect of intracellular signal transmission. In this study, we devised strategies to constrain a MAPK to a single subcellular compartment and examined the biochemical and phenotypic consequences of that restriction for the outputs normally elicited.
We used the MAPK of the mating pheromone response pathway of budding yeast (Saccharomyces cerevisiae), a well-established model system that has been helpful in elucidating many other basic principles of MAPK-dependent signal transduction (4, 17, 77). This pathway governs the part of the yeast life cycle in which a vegetatively growing haploid cell is induced to differentiate into a gamete-like cell. There are two haploid cell types, a and α, dictated by their genetic constitution at the mating-type locus (MATa and MATα, respectively). The α cells secrete a peptide (α-factor pheromone) that triggers the response pathway in a cells; a cells secrete a lipopeptide (a-factor pheromone) that evokes the response pathway in α cells. Exposure to pheromone induces new gene expression, arrests the cell cycle in the G1 phase, and promotes highly polarized growth to form a pronounced projection (shmoo formation). Once primed in this manner, an a and α cell mate at their projections, and after cell fusion, their nuclei congress, completing the formation of a MATa/MATα diploid cell.
A substantial amount is known about the molecular mechanisms by which a pheromone activates the MAPK Fus3. Each pheromone binds to a plasma membrane-localized G-protein-coupled receptor (GPCR); α-factor binds to GPCR Ste2 on a cells, and a-factor binds to GPCR Ste3 on α cells. Ligand-occupied Ste2 and Ste3 act as guanine nucleotide exchange factors (GEFs) to promote GTP-for-GDP exchange in the α subunit (Gpa1) of the heterotrimeric G protein that is coupled to these receptors. GTP binding to Gα causes conformational changes that expose a previously masked surface of the associated Gβγ (Ste4-Ste18) complex (40). Both Gpa1 and Gβγ remain anchored to the plasma membrane because each carries dual lipophilic substituents (47). The molecules of freed Gβγ now recruit via their previously buried face three crucial effectors: Far1 (12), Ste20 (43), and Ste5 (57, 81).
Far1 is a scaffold protein that carries the GEF (Cdc24) for the small (∼21-kDa) GTPase Cdc42, which is tethered to the plasma membrane via a C-terminal lipid modification, thereby stimulating formation of GTP-bound Cdc42 (52, 68, 82). One target activated by Cdc42-GTP is Ste20, a member of the p21-activated protein kinase (PAK) family (1, 42). Ste5 is another scaffold protein (19, 48, 56) that carries the components of the MAPK cascade of the mating pheromone response pathway—the MAPK kinase kinase (MAPKKK) Ste11, the MAPK kinase (MAPKK) Ste7, and the MAPK Fus3 (28, 86). One substrate of Ste20 is Ste11 (25, 60, 84). Thus, via mutual association with freed Gβγ complexes, Ste20 is placed in close proximity to both its upstream activator (Cdc42-GTP) and its downstream substrate (Ste11), triggering the cascade that generates active dually phosphorylated Fus3. This MAPK is primarily responsible for executing the array of cellular command decisions necessary for bringing about efficient mating. Pheromone response also stimulates MAPK Kss1, but it is activated only transiently (63) and seems to serve only an auxiliary function in pheromone response because efficacious mating occurs in a kss1Δ mutant (26, 29, 45).
In naïve cells, Fus3 is distributed throughout the cytoplasm and nucleus but is found predominantly in the latter compartment (74, 75). Upon pheromone stimulation, a fraction of the cellular content of Fus3 localizes to the tip of the mating projection (74, 75). Although the changes in subcellular distribution of Fus3 have been quantified carefully (7, 8, 46, 49, 69), it has never been established by direct experimental manipulation whether the dynamics of Fus3 localization contribute to any of its functions during pheromone response. This issue has become particularly pertinent, in light of the evidence that the yeast Hog1 MAPK (mammalian ortholog is p38/stress-activated protein kinase α [SAPKα]) is capable of supporting robust hyperosmotic stress resistance, even when excluded from the nucleus (50, 78), despite the fact that Hog1 normally translocates rapidly and nearly quantitatively into the nucleus upon hyperosmotic challenge (30, 79).
MATERIALS AND METHODS
Yeast strains and growth conditions.
S. cerevisiae strains used in this study are listed in Table 1 and were constructed using standard yeast genetic techniques (11). In the strains generated for this study, deletion alleles represent precise deletions of the corresponding open reading frames (ORFs), except for the ste12Δ mutation, which represents the same genomic deletion as that of the ste12Δ::LEU2 allele in JCY512 (6). The disruption markers KanMX4, CgHIS3, KlURA3, hphNT1, and natNT2 were amplified from pFA6a-KanMX4 (76), pCgH, pKlU, pRS306H, and pRS306N (71), respectively. In-frame fusions of the coding sequence for Aequoria victoria green fluorescent protein (GFP) to the 3′ end of the genomic FUS1 and PRM1 coding sequences were constructed using the method described by Longtine and colleagues (44). Strains carrying ura35-FOA were spontaneous 5-fluoroorotic acid-resistant Ura− derivatives of Ura+ parental strains. To make YJP73 and YJP131, YIplac128-based integration plasmids (35) were first constructed. These plasmids contain the promoters of FUS1 or STL1 (the 1-kb region 5′ of the open reading frame, cloned from YPH499) upstream of sequences coding for hemagglutinin-tagged enhanced GFP (HA-eGFP) or HA-td-Tomato (66), respectively. The plasmids were linearized within the FUS1 or STL1 promoter and integrated into YPH499 or YPH500, thus inserting the reporter construct immediately upstream of the normal FUS1 or STL1 locus, which remains intact. The LEU2 markers were subsequently replaced with ADE2, after which the reporters were combined into a single MATa strain by mating and sporulation. Unless otherwise indicated, strains were cultivated at 30°C in standard rich (YP) or defined (SC) medium (11) containing 2% glucose (dextrose [D] or Glc). For galactose induction experiments, cells were propagated in medium containing 2% raffinose and 0.2% sucrose (no glucose), and then galactose was added to a final concentration of 2%. When necessary, selection for plasmids was maintained by omission of appropriate nutrients.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype or description | Reference or sourcea |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 10 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | 10 |
| RCY9320 | BY4741 fus3Δ::KlURA3 kss1Δ::CgHIS3 | This study |
| RCY9335 | BY4741 fus3Δ::KanMX4 | Research Genetics, Inc. |
| RCY9336 | BY4741 ste7Δ::KanMX4 | Research Genetics, Inc. |
| RCY9337 | BY4741 ste5Δ::KanMX4 | Research Genetics, Inc. |
| RCY9338 | BY4741 rvs161Δ::KanMX4 | Research Genetics, Inc. |
| RCY9339 | BY4741 rvs167Δ::KanMX4 | Research Genetics, Inc. |
| RCY9340 | BY4741 ste2Δ::KanMX4 | Research Genetics, Inc. |
| RCY9341 | BY4741 far1Δ::KanMX4 | Research Genetics, Inc. |
| RCY9342 | BY4741 ste20Δ::KanMX4 | Research Genetics, Inc. |
| RCY9343 | BY4741 ste11Δ::KanMX4 | Research Genetics, Inc. |
| RCY9344 | BY4741 bni1Δ::KanMX4 | Research Genetics, Inc. |
| RCY9345 | BY4741 hog1Δ::KanMX4 | Research Genetics, Inc. |
| RCY9346 | BY4741 fus3Δ::KlURA3 kss1Δ::CgHIS3 ste5Δ::KanMX4 | This study |
| RCY9352 | BY4741 fus3Δ::Klura35-FOA kss1Δ::CgHIS3 | This study |
| RCY9354 | BY4741 fus3Δ::Klura35-FOA kss1Δ::CgHIS3 ste5Δ::KanMX4 | This study |
| RCY9358 | BY4741 fus3Δ::KlURA3 kss1Δ::CgHIS3 sst2Δ::KanMX4 | This study |
| RCY9366 | BY4741 ste12Δ::CgHIS3 | This study |
| RCY9367 | BY4741 ste12Δ::CgHIS3 | This study |
| RCY9368 | BY4741 ste4Δ::KanMX4 | Research Genetics, Inc. |
| RCY9380 | BY4741 FUS1-GFP::KanMX4 | This study |
| RCY9382 | BY4741 FUS1-GFP::KanMX4 fus3Δ::KlURA3 kss1Δ::CgHIS3 | This study |
| RCY9384 | BY4741 PRM1-GFP::KanMX4 | This study |
| RCY9386 | BY4741 fus3Δ::KlURA3 kss1Δ::CgHIS3 PRM1-GFP::KanMX4 | This study |
| RCY9395 | BY4741 fus3Δ::KlURA3 kss1Δ::CgHIS3 ste7Δ::KanMX4 | This study |
| RCY9401 | BY4741 fus3Δ::KlURA3 kss1Δ::CgHIS3 msg5Δ::natNT2 ptp2Δ::KanMX4 ptp3Δ::hphNT1 | This study |
| YJP73 | YPH499 PFUS1-HA-eGFP::leu2::ADE2 PSTL1-HA-tdtomato::leu2::ADE2 | This study |
| YJP131 | YPH499 PFUS1-HA-eGFP::leu2::ADE2 PSTL1-HA-tdtomato::leu2::ADE2 hog1Δ::hphNT1 | This study |
| YPH499 | MATaade2-101och his3-Δ200 leu2-Δ1 lys2-801a trp1-Δ63 ura3-52 | 67 |
| YPH500 | MATα ade2-101och his3-Δ200 leu2-Δ1 lys2-801a trp1-Δ63 ura3-52 | 67 |
Research Genetics, Inc., no longer exists. The deletion collections that this commercial enterprise sold are now marketed by Invitrogen Corp.
Plasmids and recombinant DNA methods.
Plasmids (Table 2) were constructed and propagated in Escherichia coli using standard recombinant DNA methods (64). The fidelity of all constructs generated in this study was verified by nucleotide sequence analysis. pRC192 was constructed by cloning the BY4741 FUS3 locus (the coding sequence and 500 bp of its promoter-containing 5′-flanking region) into pRS315 via SacI-ApaI (sites introduced into the primers); nucleotides encoding the sequence SGRIPSEPGLTDAS and including the restriction sites BamHI, XmaI, and MluI were included before the stop codon to facilitate subsequent introductions of C-terminal tags. pRC202 was constructed by introducing the GFP coding sequence into pRC192 as described by Westfall and coworkers (78). pRC205 and pRC206 were as described by Westfall et al. (78). pRC224 contains, as a SacI-ApaI insert in pRS315, the FUS3 coding sequence, including the native stop codon, preceded by the TPI1 promoter (the 500 nucleotides (nt) upstream of the BY4741 TPI1 coding sequence). The TPI1 promoter is followed by an XbaI site and an NdeI site that overlaps the start codon of the FUS3 coding sequence. pRC225, pRC226, and pRC227 are identical to pRC202, pRC205, and pRC206, respectively, except that the FUS3 promoter was replaced with the TPI1 promoter and junction sequence from pRC224. pRC249 and pRC250 are identical to pRC226 and pRC227, respectively, except that the TPI1 promoter was replaced with the GAL1 promoter amplified from pGS5-Cpr (57) as a SacI-NdeI fragment (sites introduced in the primers). pRC251 and pRC252 were constructed by cloning HTB2 from BY4741 into pRC202 and pRC225, respectively, via XmaI-ApaI (sites introduced in the primers); nucleotides encoding a (Gly)9 linker were included in the primers upstream of the HTB2 coding sequence. pRC254, pRC255, pRC259, pRC261, pRC284, and pRC288 contain a fus3(D137A) mutation but are otherwise identical to their FUS3 counterparts. pRC273 and pRC274 were constructed by subcloning the SacII-XmaI fragment of pRC192 into pRC226 and pRC227, respectively. pRC281 and pRC283 were constructed by subcloning the appropriate SacI-KpnI fragment (KpnI does not cut uniquely in these donor vectors) from pRC225 and pRC252, respectively, into pRS316. pRC285 and pRC286 are identical to pRC249 and pRC250, respectively, except that the FUS3 coding sequence was replaced by a sequence encoding Met-Lys-Leu-. pRC287 was constructed by subcloning the SacI-XmaI fragment of pRC274 into pRC252.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pRC192 | pRS315 PFUS3-FUS3 | This study |
| pRC202 | pRS315 PFUS3-FUS3-GFP | This study |
| pRC205 | pRS315 PFUS3-FUS3-GFP-CCaaX | This study |
| pRC206 | pRS315 PFUS3-FUS3-GFP-SSaaX | This study |
| pRC224 | pRS315 PTPI1-FUS3 | This study |
| pRC225 | pRS315 PTPI1-FUS3-GFP | This study |
| pRC226 | pRS315 PTPI1-FUS3-GFP-CCaaX | This study |
| pRC227 | pRS315 PTPI1-FUS3-GFP-SSaaX | This study |
| pRC249 | pRS315 PGAL1-FUS3-GFP-CCaaX | This study |
| pRC250 | pRS315 PGAL1-FUS3-GFP-SSaaX | This study |
| pRC251 | pRS315 PFUS3-FUS3-GFP-HTB2 | This study |
| pRC252 | pRS315 PTPI1-FUS3-GFP-HTB2 | This study |
| pRC254 | pRS315 PTPI1-fus3(D137A)-GFP-CCaaX | This study |
| pRC255 | pRS315 PFUS3-fus3(D137A)-GFP | This study |
| pRC259 | pRS315 PGAL1-fus3(D137A)-GFP-CCaaX | This study |
| pRC261 | pRS315 PGAL1-fus3(D137A)-GFP-SSaaX | This study |
| pRC273 | pRS315 PTPI1-FUS3-CCaaX | This study |
| pRC274 | pRS315 PTPI1-FUS3-SSaaX | This study |
| pRC281 | pRS316 PTPI1-FUS3-GFP | This study |
| pRC283 | pRS316 PTPI1-FUS3-GFP-HTB2 | This study |
| pRC284 | pRS315 PTPI1-fus3(D137A)-GFP-SSaaX | This study |
| pRC285 | pRS315 PGAL1-GFP-CCaaX | This study |
| pRC286 | pRS315 PGAL1-GFP-SSaaX | This study |
| pRC287 | pRS315 PTPI1-FUS3-HTB2 | This study |
| pRC288 | pRS315 PTPI1-fus3(D137A)-GFP | This study |
| pRS315 | CEN LEU2 | 67 |
| pRS316 | CEN URA3 | 67 |
| YCpDIG1-3HA | pRS316 PDIG1-DIG1-(HA)3 | Judith Zhu- Shimoni, Thorner laboratory |
Assays of mating and pheromone-induced growth arrest.
To quantitatively assess mating efficiency, cells from exponentially growing cultures of the MATa strain of interest and the tester strain BY4742 were combined at a 1:5 ratio in yeast extract-peptone-dextrose (YPD) medium. The resulting culture was shaken gently for 4 h at 30°C and plated on a medium selective for diploids; mating efficiency was calculated as the number of colonies arising divided by the number of input MATa cells. Relative mating efficiencies are normalizations, with respect to the appropriate wild-type (positive-control) strain assessed in parallel in each trial. Qualitative patch mating assays were performed as described previously (33). Assessment of pheromone-induced growth arrest and recovery by halo bioassay were performed as described previously (61).
Microscopy.
Cells were examined under the 100× or 60× lens objective of an epifluorescence microscope (model BH-2; Olympus America, Center Valley, PA) using a GFP band-pass filter (Chroma Technology Corp.), as appropriate. Images were collected with a charge-coupled-device camera (Olympus) and processed with Magnafire SP imaging software (Optronics) and Photoshop (Adobe).
Preparation of cell extracts and immunoblotting.
Preparation of yeast cell extracts by rapid alkaline lysis followed by trichloroacetic acid precipitation, SDS-PAGE, and immunoblotting were all performed as described previously (78). Primary antibodies used were anti-phospho-MAPK (Cell Signaling Technology; no. 9101), anti-GFP (Roche), anti-HA (Covance; HA.11, PRB-101P), anti-Cdc10 (Santa Cruz Biotechnology; sc-26296), and anti-Rvs167 (gift of Brenda Andrews, University of Toronto, Canada).
Quantification of pheromone response pathway reporter expression.
Yeast cultures were grown in YPD at 30°C overnight, diluted to an A600 of 0.025 and incubated at the indicated temperatures until an A600 of 0.5, diluted once again to an A600 of 0.025, and cultivated for an additional 5 h. Cultures were then supplemented with an equal volume of YPD containing α-factor, sorbitol, or neither and incubated for the indicated times. Cells were placed on an agarose pad and examined using the epifluorescence microscope (BH-2; Olympus) equipped with a 60× 1.40-numerical-aperture (NA) objective lens. For each field, GFP fluorescence was imaged using a 470-nm (40-nm-bandwidth) excitation filter and a 525-nm (50-nm-bandwidth) emission filter (Endow GFP 41007; Chroma), and autofluorescence was imaged using a 330- to 385-nm bandpass excitation filter (UG1; Olympus). Images were captured using the CCD camera (Olympus), and those acquired using the same filter set were captured with identical exposure times. Image analysis was performed using CellProfiler, version 1.0.5122 (13). Corrections for background and for variable illumination across the field were applied. Cells were identified using the autofluorescence images, and the average pixel intensity within each cell in the aligned GFP image was calculated. These values were averaged across cells to determine the extent of GFP expression in each condition.
RESULTS
Restricting subcellular localization of functional Fus3.
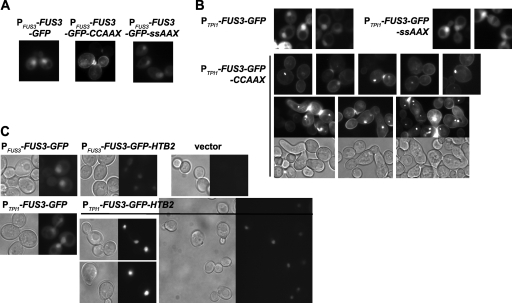
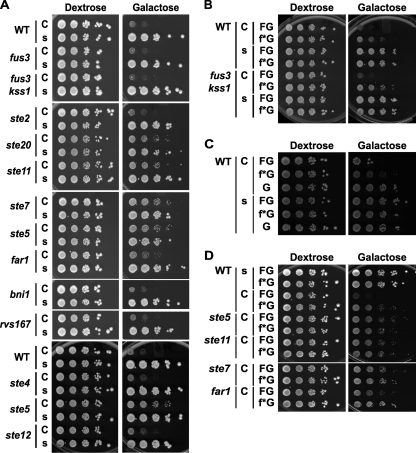
To examine whether Fus3 activation and its physiological consequences are dependent on dynamic changes in its subcellular localization, we constructed two Fus3 chimeras: one fused to Htb2, one of two S. cerevisiae histone H2B homologs; and another fused to the C-terminal CCaaX box of Ras2, one of two yeast RAS homologs. We anticipated that the former would be largely trapped on chromosomes in the nucleus and that the latter would be modified by S prenylation and S palmitoylation of the Cys residues in the CCaaX motif (32) and directed mainly to the plasma membrane. As a control for the latter, a derivative (SSaaX) was constructed that obviates lipid modification, preventing plasma membrane tethering. Each tag was attached in frame to the C terminus of a Fus3-GFP fusion to permit facile visualization, and these constructs were expressed in fus3Δ kss1Δ cells from a low-copy-number (CEN) vector at a near-endogenous level (from the FUS3 promoter), at a higher constitutive level (from the TPI1 promoter), or in certain cases, at a very high level (from the GAL1 promoter). Regardless of its expression level, each derivative exhibited the expected localization (Fig. 1).
FIG. 1.
Subcellular localization of plasma membrane- and nucleus-restricted alleles of Fus3. Exponentially growing cultures of strain RCY9320 (fus3 kss1) carrying the plasmids indicated below were examined by bright-field and/or fluorescence microscopy. (A) PFUS3-FUS3-GFP (pRC202) (left), PFUS3-FUS3-GFP-CCaaX (pRC205) (middle), or PFUS3-FUS3-GFP-SSaaX (pRC206) (right). (B) PTPI1-FUS3-GFP (pRC225) (top left), PTPI1-FUS3-GFP-CCaaX (pRC226) (bottom), or PTPI1-FUS3-GFP-SSaaX (pRC227) (top right). (C) PFUS3-FUS3-GFP (pRC202) (top left), PFUS3-FUS3-GFP-HTB2 (pRC251) (top middle), PTPI1-FUS3-GFP (pRC225) (bottom left), PTPI1-FUS3-GFP-HTB2 (pRC252) (bottom right), or empty vector (pRS315) (top right).
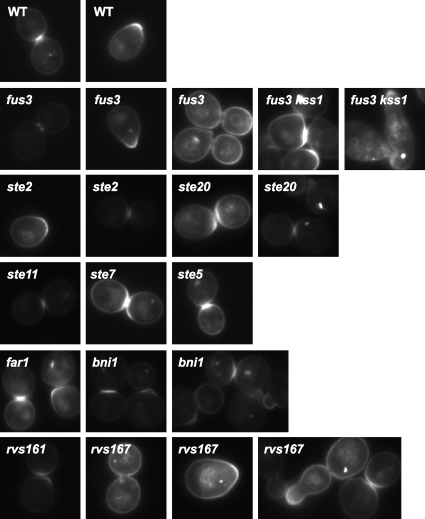
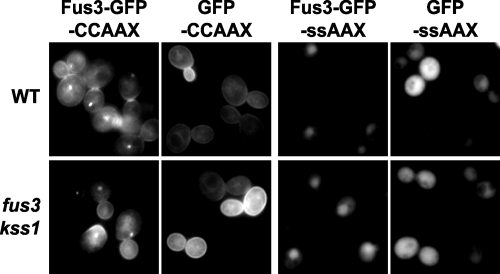
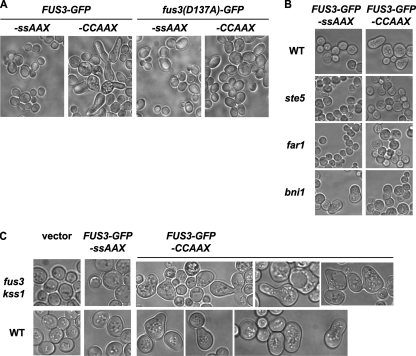
Although generally Fus3-GFP-CCaaX was observed along the entire plasma membrane, we noted that it was frequently enriched at sites of membrane growth or remodeling, including small buds, the bud neck, and polarized shmoo tips (Fig. 1 and 2). Additionally, it was often visible at intracellular membranes as well as a small number of unidentified intracellular foci, usually one per cell. The nature of the plasma membrane distribution of Fus3-GFP-CCaaX, including the bias for membrane growth sites, was unaltered even in cells lacking every other core component of the pheromone response pathway tested (Fig. 2) and was not observed for GFP-CCaaX (Fig. 3), indicating that this localization is a specific property conferred by the Fus3 polypeptide and is not a general feature of the trafficking of CCaaX-containing proteins.
FIG. 2.
Anisotropic membrane distribution of Fus3-GFP-CCaaX is independent of core pheromone response pathway components. Strains of the indicated genotypes (BY4741, RCY9320, RCY9335, RCY9336, RCY9337, RCY9338, RCY9339, RCY9340, RCY9341, RCY9342, RCY9343, and RCY9344) were transformed with a plasmid encoding Fus3-GFP-CCaaX (pRC226). Exponentially growing cultures were examined by fluorescence microscopy.
FIG. 3.
Membrane distribution of Fus3-GFP-CCaaX is conferred by the Fus3 sequence. Wild-type (WT) (BY4741) and fus3 kss1 (RCY9320) cells were transformed with galactose-inducible expression vectors encoding PGAL1-FUS3-GFP-CCaaX (pRC249), PGAL1-GFP-CCaaX (pRC285), PGAL1-FUS3-GFP-SSaaX (pRC250), or PGAL1-GFP-SSaaX (pRC286). Exponentially growing 26°C cultures were examined by fluorescence microscopy 2.5 h after addition of galactose.
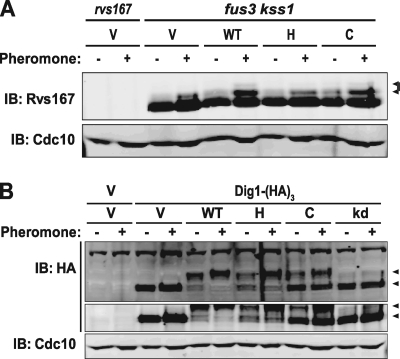
One plasma membrane-localized substrate of Fus3 is the amphiphysin ortholog Rvs167 (31), a portion of which becomes hyperphosphorylated (manifested as a band of lower mobility) in response to pheromone treatment. Pheromone-induced Rvs167 phosphorylation was observed but diminished in fus3Δ kss1Δ cells expressing Fus3-GFP-CCaaX, despite its tethering to the plasma membrane, compared to the same cells expressing wild-type Fus3-GFP (Fig. 4A ). Reassuringly, in cells expressing only Fus3-GFP-Htb2, the level of pheromone-induced Rvs167 phosphorylation was similar to that of a vector-only control. One nuclear substrate of Fus3 is the chromosome-localized Ste12-bound transcriptional repressor Dig1 (20, 72), and we previously observed (J. Zhu-Shimoni and J. Thorner, unpublished results) that Dig1 displays a dramatic and nearly quantitative electrophoretic mobility shift that requires Fus3 activity (Fig. 4B). This Dig1 phosphorylation was readily detectable but diminished in fus3Δ kss1Δ cells expressing Fus3-GFP-Htb2, despite its tethering to chromosomes, compared to the same cells expressing wild-type Fus3-GFP (Fig. 4B). Reassuringly, in cells expressing only Fus3-GFP-CCaaX, Dig1 phosphorylation was very inefficient, as judged by the substantial amount of unphosphorylated Dig1 present after pheromone treatment.
FIG. 4.
Restriction of Fus3 localization affects its phosphorylation of substrates. (A) rvs167 (RCY9339) and fus3 kss1 cells (RCY9320) were transformed with an empty vector (V; pRS315) or plasmids encoding Fus3-GFP (WT; pRC225), Fus3-GFP-Htb2 (H; pRC252), or Fus3-GFP-CCaaX (C; pRC226). Exponentially growing cultures were incubated in the absence (−) or presence (+) of 3 μM α-factor (pheromone) for 1.5 h, after which whole-cell extracts were prepared, resolved by SDS-PAGE, and analyzed by immunoblotting (IB) with anti-Rvs167 antibody and with anti-Cdc10 antibody to confirm equivalent sample loading. Arrowheads indicate phosphorylated forms of Rvs167. (B) fus3 kss1 cells (RCY9352) were transformed with plasmids encoding Dig1-(HA)3 (YCpDIG1-3HA), Fus3-GFP (WT; pRC225), Fus3-GFP-Htb2 (H; pRC252), Fus3-GFP-CCaaX (C; pRC226), Fus3(D137A)-GFP (kd; pRC288), and/or empty vectors (V; pRS315 or pRS316) as indicated. Exponentially growing cultures were incubated in the absence (−) or presence (+) of 3 μM α-factor (pheromone) for 1.5 h, after which whole-cell extracts were prepared, resolved by SDS-PAGE, and analyzed by immunoblotting with anti-HA antibody (two SDS-PAGE-immunoblots of the same samples are shown). Lower arrowhead, Dig1; upper arrowhead, phosphorylated Dig1. The anti-Cdc10 immunoblot serves as a loading control.
Plasma membrane localization is limiting for basal and pheromone-induced Fus3 activation.
As judged both by fluorescence intensity (Fig. 1) and by immunoblotting (Fig. 5), Fus3-GFP-Htb2 and Fus3-GFP-CCaaX both exhibited a lower steady-state level than either Fus3-GFP or Fus3-GFP-SSaaX. These differences were observed whether expression was driven by the native FUS3 promoter (PFUS3) or by the stronger TPI1 promoter (PTPI1), indicating that this effect was not due to loss of the positive feedback normally exerted on PFUS3 both basally and in response to pheromone (27, 62). These observations suggest that permanent residence in the nucleus and at the plasma membrane subjects Fus3 to degradative processes that this MAPK normally escapes. In any event, use of constructs driven by PFUS3 and by PTPI1 in all of our subsequent analyses allowed us to examine the effects of restricted localization at expression levels both lower and higher than that of endogenous FUS3 (or PFUS3-driven wild-type Fus3-GFP).
FIG. 5.
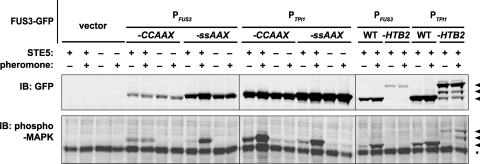
Basal and pheromone-induced Fus3 activation depend on its subcellular localization. Strains RCY9320 (fus3 kss1 STE5+) and RCY9346 (fus3 kss1 ste5) were transformed with an empty vector (pRS315) or plasmids encoding PFUS3-FUS3-GFP-CCaaX (pRC205), PFUS3-FUS3-GFP-SSaaX (pRC206), PTPI1-FUS3-GFP-CCaaX (pRC226), PTPI1-FUS3-GFP-SSaaX (pRC227), PFUS3-FUS3-GFP (pRC202), PFUS3-FUS3-GFP-HTB2 (pRC251), PTPI1-FUS3-GFP (pRC225), or PTPI1-FUS3-GFP-HTB2 (pRC252). Exponentially growing cultures were incubated in the absence (−) or presence (+) of 2 μM α-factor (pheromone) for 15 min, after which whole-cell extracts were prepared and resolved by SDS-PAGE. Total and activated levels of Fus3 were assessed by immunoblotting with anti-GFP and anti-phospho-MAPK antibodies, respectively. Arrowheads indicate corresponding bands in the anti-GFP and anti-phospho-MAPK immunoblots. The asterisk denotes a band present in all lanes, which likely corresponds to the phosphorylated form of the MAPK Slt2/Mpk1.
To assess the efficiency of initial activation, extracts of these cells were resolved by SDS-PAGE and analyzed by immunoblotting with an antibody that detects the activated (dually phosphorylated) form of this MAPK (2, 34) (Fig. 5). In comparison to wild-type Fus3-GFP (or Fus3-GFP-SSaaX), Fus3-GFP-Htb2 activation was barely detectable. As shown below, phosphatases that reside predominantly in the nucleus and that can remove phosphate from the phosphotyrosine (P-Tyr) in the activation loop of yeast MAPKs contribute to this pattern. In striking contrast, Fus3-GFP-CCaaX exhibited elevated basal activation (i.e., in the absence of pheromone stimulation) and further activation when exposed to pheromone, confirming our prior findings (78). Confining Fus3 to the plasma membrane may reduce its accessibility to deactivating phosphatases or increase its accessibility to the upstream activation machinery (or both). Normally, Fus3 encounters its immediate activators (MAPKK Ste7 and MAPKKK Ste11) in the context of the scaffold protein Ste5 (3, 19), and Ste5 is recruited to the plasma membrane in a manner that requires free Gβγ, as well as certain plasma membrane phospholipids (33, 83). To determine whether, due to its constitutive membrane tethering, Fus3-GFP-CCaaX is able to bypass the need for Ste5 in Fus3 activation, we compared STE5+ and ste5Δ cells. Absence of Ste5 almost completely abrogated both basal and pheromone-induced Fus3 activation (Fig. 5), in agreement with other cumulative evidence that Ste5 is not simply a passive platform to hold the MAPK components in close proximity but also exerts allosteric effects on these proteins that are necessary for signal propagation (36, 41, 65).
Restriction of Fus3 localization impairs mating.
Multiple, physiologically relevant phosphoacceptor substrates for Fus3 have been identified (18). Successful mating presumably requires the cumulative action of this MAPK on these targets. Hence, to determine whether nuclear or membrane restriction prevents Fus3 from performing its overall biological function, we assessed, first, the ability of Fus3-GFP-Htb2 and Fus3-GFP-CCaaX to support mating. Compared to the wild-type Fus3-GFP control, Fus3-GFP-Htb2 exhibited only about a 4- or 5-fold decrease in mating proficiency, whereas Fus3-GFP-CCaaX displayed more than a 10-fold decrease (Fig. 6A and B). Thus, the critical functions that Fus3 performs cannot be achieved by relying on the dynamic behavior of its targets (and/or regulators). Moreover, coexpression of Fus3-GFP-CCaaX and Fus3-GFP-Htb2 did not potentiate mating efficiency above the level afforded by either alone (Fig. 6B). Hence, effective Fus3 action in multiple compartments cannot be achieved by this MAPK simply being present in them. Therefore, optimal mating proficiency requires that Fus3 be free to actively shuttle between the nucleus, cytosol, and plasma membrane, unlike what has been observed for Hog1 MAPK and survival in response to hyperosmotic stress (78). Indeed, the fact that coexpression of Fus3-GFP-CCaaX with Fus3-GFP-Htb2 lowered mating efficiency to the level observed for Fus3-GFP-CCaaX alone (Fig. 6) indicates that prolonged residence of Fus3 at the plasma membrane is actually deleterious to the overall conjugation process. The dominance of Fus3-GFP-CCaaX over Fus3-GFP-Htb2 may be due to sequestration of activators (like Ste7 MAPKK) by the plasma membrane-tethered Fus3, thus reducing the already low frequency with which such activators can visit the nuclear compartment to activate the chromatin-tethered Fus3.
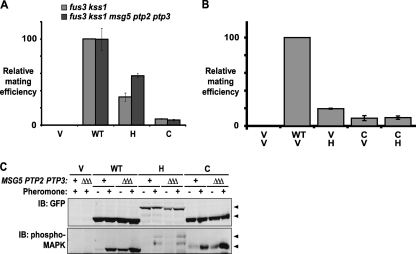
FIG. 6.
Restriction of Fus3 localization impairs mating. (A) fus3 kss1 (RCY9320) and fus3 kss1 msg5 ptp2 ptp3 (RCY9401) cells were transformed with an empty vector (V; pRS315) or plasmids encoding Fus3-GFP (WT; pRC225), Fus3-GFP-Htb2 (H; pRC252), or Fus3-GFP-CCaaX (C; pRC226). Mating proficiency was determined relative to fus3 kss1 cells carrying Fus3-GFP. Averages are shown; bars depict standard errors of the means (n = 2). (B) fus3 kss1 (RCY9352) cells were transformed with two plasmids as indicated: V, vector (pRS315 or pRS316); WT, Fus3-GFP (pRC225); H, Fus3-GFP-Htb2 (pRC283); and C, Fus3-GFP-CCaaX (pRC226). Mating proficiency was determined relative to cells carrying Fus3-GFP. Averages are shown; bars depict standard errors of the means (n ≥ 4). (C) fus3 kss1 (RCY9320) and fus3 kss1 msg5 ptp2 ptp3 (RCY9401) cells were transformed with an empty vector (V; pRS315) or plasmids encoding Fus3-GFP (WT; pRC225), Fus3-GFP-Htb2 (H; pRC252), or Fus3-GFP-CCaaX (C; pRC226). Exponentially growing cultures were incubated in the absence (−) or presence (+) of 2 μM α-factor (pheromone) for 15 min, after which whole-cell extracts were prepared and resolved by SDS-PAGE. Total and activated levels of Fus3 were assessed by immunoblotting with anti-GFP and anti-phospho-MAPK antibodies, respectively. Arrowheads indicate corresponding bands in the anti-GFP and anti-phospho-MAPK immunoblots.
MAPK dephosphorylation occurs primarily in the nucleus.
It was somewhat paradoxical that the apparent level of Fus3-GFP-Htb2 activation was much lower than that of Fus3-GFP-CCaaX (Fig. 5) yet was able to support significantly more efficacious mating (Fig. 6). These observations suggested that Fus3 activation can occur in the nucleus but is simply more transient, likely due to greater susceptibility to MAPK phosphatases known to be located in the nucleus. Consistent with this view, deletion of the genes encoding three MAPK-specific phosphatases, Msg5, Ptp2, and Ptp3 (23, 85), substantially improved mating by cells expressing Fus3-GFP-Htb2, whereas this tactic did not detectably ameliorate the mating defect of cells expressing Fus3-GFP-CCaaX (Fig. 6A), even though absence of these phosphatases increased the amount of activated Fus3-GFP-CCaaX (Fig. 6C).
Taken together, these results indicate, first, that the activation loop phosphorylation of this MAPK undergoes rapid turnover in the nuclear compartment, which largely explains the mating impairment displayed by Fus3-GFP-Htb2. Second, despite the observed translocation of Fus3 to the shmoo tip and the ensuing morphology and cell surface changes that haploids undergo in preparation for conjugation, our findings indicate that it is the actions executed by Fus3 in the nucleus that are especially important for successful mating. Third, the fact that the nuclear role of Fus3 MAPK is of primary importance for mating is in stark contrast to Hog1 MAPK, whose nuclear localization and subsequent effects on transcription are dispensable for its role in hyperosmotic stress response (50, 78).
Attempts to use a constitutively active allele of the MAPKKK STE11-4 (70) to stimulate even greater activation of Fus3-GFP-Htb2 and Fus3-GFP-CCaaX were problematic. In our hands, overexpression of the hyperactive MAPKKK allele STE11-4 from either the TPI1 promoter or the GAL1 promoter caused rather severe growth inhibition even in the control (fus3Δ kss1Δ) cells, so it was difficult to discern whether it had any salutary effect on the mating deficiency of cells expressing either Fus3-GFP-Htb2 or Fus3-GFP-CCaaX (R. E. Chen and J. Thorner, unpublished observations).
Subcellular location and the separation of MAPK functions.
The mating process requires coordination of MAPK-dependent changes in cell cycle progression, cell shape, and gene expression. Therefore, we examined which processes are able to occur and which are the most impaired when Fus3 is confined to the nucleus or to the plasma membrane.
Among the genes most highly induced in a Fus3-dependent manner (62) are FUS1 (73) and PRM1 (39). As expected, in cells expressing either wild-type Fus3-GFP or Fus3-GFP-SSaaX, the products of both of these genes (each marked with GFP) were induced upon pheromone treatment (Fig. 7A). Despite its presence on chromatin, but in keeping with its weaker apparent activation, Fus3-Htb2 supported induction of PRM1 expression at a reduced level and a barely detectable level of FUS1 expression. This observation suggests that different response genes require different levels of MAPK activity for their induction. Conversely, despite its high basal and pheromone-induced activation, but in keeping with its exclusion from the nucleus, Fus3-GFP-CCaaX was unable to support expression of either diagnostic pheromone-inducible gene (before or after pheromone treatment). The failure of Fus3-GFP-CCaaX to induce pheromone-regulated genes is likely a major contributing factor to its more severe mating deficiency. Moreover, the fact that Fus3-GFP-CCaaX is clearly active yet cannot support pheromone-induced gene expression rules out the possibility that Fus3 modifies transcriptional regulators outside the nucleus, which are then imported into the nucleus to influence the expression of pheromone-responsive genes.
FIG. 7.
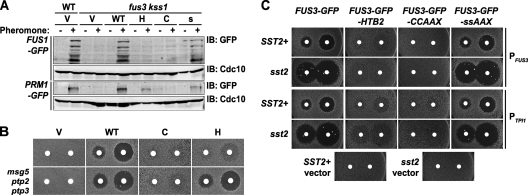
Pheromone-inducible gene expression and growth arrest depend on the localization of Fus3. (A) Strains RCY9380 (FUS3+ KSS1+ FUS1-GFP), RCY9382 (fus3 kss1 FUS1-GFP), RCY9384 (FUS1+ KSS1+ PRM1-GFP), and RCY9386 (fus3 kss1 PRM1-GFP) were transformed with plasmids encoding Fus3 (WT; pRC224), Fus3-Htb2 (H; pRC287), Fus3-CCaaX (C; pRC273), Fus3-SSaaX (s; pRC274), or an empty vector (V; pRS315). Exponentially growing cultures were incubated in the absence (−) or presence (+) of 2 μM α-factor (pheromone) for 1.5 h, after which whole-cell extracts were prepared and resolved by SDS-PAGE. Levels of Fus1-GFP, Prm1-GFP, and Cdc10 (loading control) were assessed by immunoblotting with anti-GFP and anti-Cdc10 antibodies. (B) The strains described above for panel A were assessed for pheromone-induced growth arrest by an agar diffusion (halo) assay. (C) fus3 kss1 (RCY9320) and fus3 kss1 sst2 (RCY9358) cells carrying plasmids encoding PFUS3-FUS3-GFP (pRC202), PFUS3-FUS3-GFP-HTB2 (pRC251), PFUS3-FUS3-GFP-CCaaX (pRC205), PFUS3-FUS3-GFP-SSaaX (pRC206), PTPI1-FUS3-GFP (pRC225), PTPI1-FUS3-GFP-HTB2 (pRC252), PTPI1-FUS3-GFP-CCaaX (pRC226), PTPI1-FUS3-GFP-SSaaX (pRC227), or an empty vector (pRS315) were assessed for pheromone-induced growth arrest by an agar diffusion (halo) assay.
Pheromone-imposed arrest of the cell division cycle in the G1 phase is a necessary prelude to mating (37) and can be conveniently assessed by a plate-based bioassay that measures the extent of growth inhibition (“halo”) in a lawn of MATa cells caused by α-factor diffusing from a filter disk (61). The fus3Δ kss1Δ cells carrying empty vector failed to respond to α-factor, whereas the same cells expressing Fus3-GFP or Fus3-GFP-SSaaX (Fig. 7B and C) displayed the prominent clear zone characteristic of pheromone-imposed growth arrest, as expected. By comparison, cells expressing Fus3-GFP-Htb2 exhibited a barely detectable retardation of growth, and cells expressing Fus3-GFP-CCaaX displayed an even weaker response. Despite their turbidity, the halos in the lawn of the cells expressing Fus3-GFP-Htb2 had the same diameter as those in the lawn of the cells expressing Fus3-GFP, suggesting either very inefficient imposition of G1 arrest or greatly accelerated recovery from it. To distinguish impaired initiation of cell cycle arrest from faster desensitization, we introduced an sst2Δ mutation into all of the strains (Fig. 7C). Sst2 is essential for resumption of cell cycle progression following pheromone-induced arrest, and therefore, its absence greatly increases pheromone sensitivity (and thus halo size) (14, 22). If the turbid halos displayed by Fus3-GFP-Htb2 cells are due to faster Sst2-promoted recovery, then loss of Sst2 should allow such cells to form clear halos. However, Fus3-GFP-Htb2 sst2Δ cells still formed turbid halos (Fig. 7C), suggesting that nucleus-restricted Fus3 is simply inefficient in imposing pheromone-induced cell cycle arrest. In further support of this conclusion, when Fus3-GFP-Htb2 was more robustly activated (in msg5Δ ptp2Δ ptp3Δ cells), its ability to impose G1 arrest was completely restored (Fig. 7C), indicating that nuclearly restricted Fus3 is fully competent to cause cell cycle arrest, provided that it is activated above a certain threshold. In marked contrast, neither removal of Sst2 nor elimination of the MAPK phosphatases rescued the G1 arrest defect of cells expressing Fus3-GFP-CCaaX (Fig. 7B and C), despite the fact that it is activated to a much greater extent than Fus3-GFP-Htb2 under all conditions examined.
The target of Fus3 necessary to impose pheromone-induced G1 arrest is Far1 (15), a protein with multiple functions. Far1 undergoes nucleocytoplasmic shuttling, thereby conveying its bound cargo (the Cdc24 GEF) from the nucleus to the cytoplasm (82). It acts as a scaffold to tether Cdc24 to the plasma membrane in a Gβγ-dependent manner (52), thereby locally producing Cdc42-GTP to activate Ste20 but also to stimulate polarized growth. Finally, the Fus3-phosphorylated form of Far1 is thought to act as a direct inhibitor of the G1 CDK (Cln-bound Cdc28) (55). Far1 contains a nuclear localization signal (NLS) and is located predominantly in the nucleus in the absence of pheromone, but its rate of export from the nucleus via the karyopherin Msn5 is stimulated by pheromone (9, 12, 53). Confining Far1 to the nucleus is necessary and sufficient for its role in pheromone-induced cell cycle arrest (9). Our observation that nuclearly restricted Fus3-GFP-Htb2 supports pheromone-induced G1 arrest confirms the conclusion that only the nuclear pool of Far1 can serve as a cell cycle inhibitor. Moreover, our results significantly extend our understanding by demonstrating that only the nuclear pool of Fus3 is capable of modifying Far1 to generate its CDK-inhibitory form. The fact that Fus3-GFP-CCaaX is clearly active yet cannot exert pheromone-induced G1 arrest rules out the possibility that Far1 performs its arrest function by being phosphorylated by Fus3 at the plasma membrane or near the cell cortex and then being imported back into the nucleus. Together, our results strongly suggest that Fus3-mediated phosphorylation of Far1 and its role as a CDK inhibitor occur exclusively and obligatorily in the nucleus.
Another function of Fus3 action is to promote the polarized growth required for projection extension. As expected, fus3Δ kss1Δ cells expressing either Fus3-GFP or Fus3-GFP-SSaaX efficiently supported shmoo formation (Fig. 8A ). Strains expressing Fus3-GFP-CCaaX exhibited a polarized cell shape even in the absence of pheromone (Fig. 1 and 9, discussed further below) and, compared to the control (Fus3-GFP and Fus3-GFP-SSaaX) cells, displayed grossly abnormal cell morphogenesis in the presence of pheromone (Fig. 8A). Although Fus3-GFP-Htb2 cells did not exhibit pheromone-induced cell polarization regardless of its level of expression (Fig. 8A), when its level of activation was enhanced (in msg5Δ ptp2Δ ptp3Δ cells), the ability to form pheromone-induced mating projections was restored (Fig. 8B). Thus, the only reason that Fus3-GFP-Htb2 is unable to stimulate polarized growth is because it is unable to sustain an adequate level of activation in the nuclear compartment. Indeed, we observed that, in the absence of the Msg5, Ptp2, and Ptp3 phosphatases, a significant fraction of the cells expressing wild-type Fus3-GFP (but not cells lacking Fus3) exhibited polarized morphology in the absence of pheromone, indicative of enhanced basal activation (Fig. 8B).
FIG. 8.
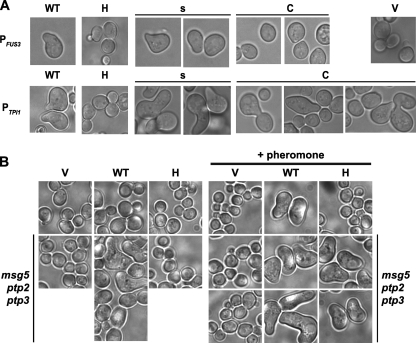
Pheromone-inducible polarized growth depends on the localization of Fus3. (A) Strain RCY9320 (fus3 kss1) was transformed with plasmids encoding Fus3-GFP (WT; pRC202, pRC225), Fus3-GFP-Htb2 (H; pRC251, pRC252), Fus3-GFP-SSaaX (s; pRC206, pRC227), Fus3-GFP-CCaaX (C; pRC205, pRC226), or empty vector (V; pRS315). Exponentially growing cultures were incubated with 2 μM α-factor for 1.5 to 2.5 h and examined by microscopy. (B) Strains RCY9320 (fus3 kss1) and RCY9401 (fus3 kss1 msg5 ptp2 ptp3) were transformed with plasmids encoding Fus3-GFP (WT; pRC225), Fus3-GFP-Htb2 (H; pRC252), or an empty vector (V; pRS315). Exponentially growing cultures were treated with or without 2 μM α-factor for 2 h and examined by microscopy.
FIG. 9.
Restriction of Fus3 to the plasma membrane causes aberrant cell morphology. (A) Exponentially growing cultures of fus3 kss1 (RCY9320) cells carrying plasmids encoding Fus3-GFP-SSaaX (pRC227), Fus3-GFP-CCaaX (pRC226), Fus3(D137A)-GFP-SSaaX (pRC284), or Fus3(D137A)-GFP-CCaaX (pRC254) were examined by microscopy. (B) Wild-type (BY4741) and isogenic ste5 (RCY9337), far1 (RCY9341), and bni1 (RCY9344) mutant cells were transformed with plasmids encoding PGAL1-FUS3-GFP-CCaaX (pRC249) or PGAL1-FUS3-GFP-SSaaX (pRC250), cultivated at 26°C in media containing 2% raffinose, 0.2% sucrose, and 2% galactose, and examined by microscopy. (C) Wild-type (BY4741) and isogenic fus3 kss1 (RCY9320) cells were transformed with plasmids encoding PGAL1-FUS3-GFP-CCaaX (pRC249), PGAL1-FUS3-GFP-SSaaX (pRC250), or an empty vector (pRS315), cultivated at 26°C in media containing 2% raffinose, 0.2% sucrose, and 2% galactose, and examined by microscopy.
Taken together, these observations indicate that gene induction and G1 arrest can be executed only by nuclear Fus3 and that membrane recruitment of Fus3 is not essential for stimulating polarized growth, provided that Fus3 activity is maintained above a certain threshold level. Thus, remarkably, achieving overall mating proficiency seems to rely more heavily on the nuclear functions of Fus3 than on the effects it has at the cell membrane. In fact, even though normally activated at the membrane in conjunction with the Ste5 scaffold and the rest of the upstream apparatus, the subsequent actions of Fus3 at the membrane must be only acute and rather transient because the chronic action of Fus3 at the membrane is quite deleterious, as described in the next two sections.
Plasma membrane-restricted Fus3 causes aberrant cell polarization.
In contrast to its lack of ability to support G1 arrest, gene expression, and overall mating proficiency, Fus3-GFP-CCaaX expressed in fus3Δ kssΔ1 cells had a unique property not displayed by any of the other constructs examined, namely, causing aberrant cell morphology in a readily detectable fraction of the population (∼2 to 10%), even in the absence of pheromone. This phenotype was more pronounced when Fus3-GFP-CCaaX was expressed from PTPI1 than from PFUS3 (Fig. 1) and greatly exacerbated when expression was driven by the galactose-inducible GAL1 promoter (Fig. 9). Introduction into Fus3-GFP-CCaaX of a point mutation (D137A) that renders Fus3 catalytically inactive and unable to support mating (data not shown) abolished the aberrant morphology (Fig. 9A). Induction of abnormal morphology by Fus3-GFP-CCaaX was also dependent on Ste4 (Gβ) and Ste5 (scaffold), but not Ste2 (pheromone receptor) (Fig. 9B and data not shown). The last result is probably explained by the fact that receptor-less cells display spontaneous stochastic dissociation of Gβγ (Ste4-Ste18) from Gpa1 (Gα) (38). Other pathway components located at the plasma membrane, including Far1 (scaffold) and Bni1 (formin), were also required for Fus3-GFP-CCaaX to affect morphology (Fig. 9B). All together, these results suggest that the aberrant polarized growth is due to the persistent and hyperactive effects of Fus3-GFP-CCaaX on the plasma membrane-located effectors and regulators normally involved in polarized morphogenesis. Indeed, this effect of Fus3-GFP-CCaaX behaved in a semirecessive manner because it was less pronounced in FUS3+ KSS1+ cells (Fig. 9C). Presumably, the presence of wild-type Fus3 and Kss1 coopts a portion of the upstream activation machinery and/or partially blocks access of the membrane-tethered Fus3 to the components required for polarized growth.
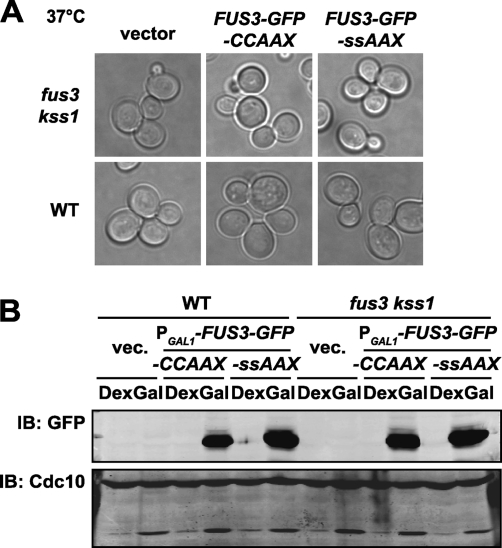
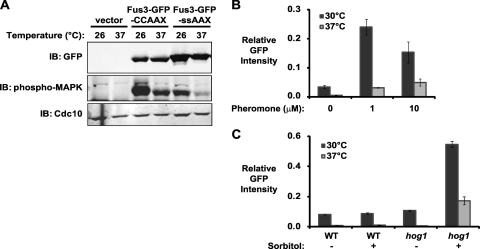
Interestingly, the aberrant morphology of FUS3-GFP-CCaaX cells was rescued at elevated temperature (Fig. 10A ), which was not due to any temperature-induced diminution in the level of Fus3-GFP-CCaaX expression (Fig. 10B) but might be explained, in part, by a reduction in the extent of activation at the higher temperature (Fig. 11A). Consistent with the notion that pheromone pathway activity is diminished at elevated temperature, we noted that at 37° (compared to 30°) there was reduced expression of a pathway reporter (PFUS1-GFP) during vegetative growth (basal expression) (Fig. 11B), following pheromone stimulation (Fig. 11B), and after sorbitol treatment of hog1Δ cells (due to “cross talk” arising from the fact that Ste11 activated by the osmosensors of the HOG pathway can escape in the absence of functional Hog1 and stimulate the pheromone response pathway [54]) (Fig. 11C).
FIG. 10.
The aberrant morphology caused by Fus3-GFP-CCaaX is rescued at elevated temperature. (A) An experiment otherwise identical to that described in the legend to Fig. 9C was performed using cultures cultivated in galactose-containing media at 37°C. (B) Wild-type (BY4741) and fus3 kss1 (RCY9320) cells were transformed with an empty vector (pRS315) or plasmids encoding PGAL1-FUS3-GFP-CCaaX (pRC249) or PGAL1-FUS3-GFP-SSaaX (pRC250) and cultivated at 37°C in media containing dextrose or galactose. Whole-cell extracts were prepared from exponentially growing cultures, and levels of Fus3 and Cdc10 (loading control) were assessed by immunoblotting with anti-GFP and anti-Cdc10 antibodies, respectively.
FIG. 11.
Pheromone pathway activation is reduced at elevated temperature. (A) Wild-type (BY4741) cells were transformed with plasmids encoding Fus3-GFP-CCaaX (pRC226), Fus3-GFP-SSaaX (pRC227), or an empty vector (pRS315) and cultivated at the indicated temperatures. Whole-cell extracts were prepared from exponentially growing cultures, and levels of total Fus3-GFP-CCaaX/-SSaaX, activated Fus3-GFP-CCaaX/-SSaaX, and Cdc10 (loading control) were assessed by immunoblotting with anti-GFP, anti-phospho-MAPK, and anti-Cdc10 antibodies, respectively. (B) PFUS1-HA-eGFP (YJP73) cells were grown at 30°C or 37°C and incubated with the indicated concentration of α-factor (pheromone) for 1 h. Cells were examined by microscopy, and GFP expression was quantified. The averages of three independent trials are shown; bars depict standard errors of the means across the three trials. In total, between 72 and 185 cells were examined for each condition. (C) PFUS1-HA-eGFP (YJP73) and hog1 PFUS1-HA-eGFP (YJP131) cells were grown at 30°C or 37°C and incubated in the absence (−) or presence (+) of 1 M sorbitol for 2 h. Cells were examined by microscopy, and GFP expression was quantified. The averages and standard errors of the means are shown (n = 28 to 82 cells).
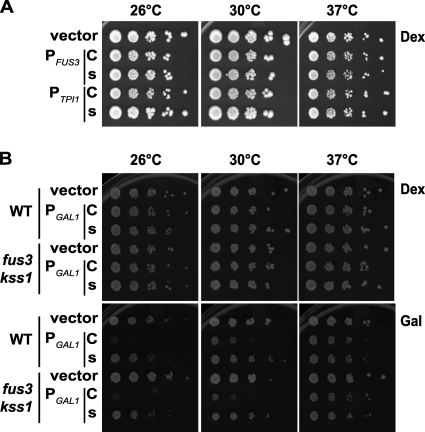
Plasma membrane-restricted Fus3 impairs cell proliferation.
When expressed from PTPI1, Fus3-GFP-CCaaX (but none of the other constructs examined) caused a detectable reduction in doubling time. This phenotype (Fig. 12A ) was not significant enough to adversely influence any of the phenotypic assays we conducted. However, expression of Fus3-GFP-CCaaX from the GAL1 promoter led to a pronounced growth defect (Fig. 13 and 12B). This growth inhibition was ameliorated at elevated temperature (Fig. 12B), as observed for the aberrant morphology caused by Fus3-GFP-CCaaX expression (Fig. 10). However, unlike the aberrant morphology, the growth defect caused by Fus3-GFP-CCaaX was not reduced by the presence of wild-type Fus3 (or of both Fus3 and Kss1) (Fig. 13A and B and 12B).
FIG. 12.
The cell proliferation defect caused by Fus3-GFP-CCaaX is dependent on expression strength and temperature. (A) fus3 kss1 (RCY9320) cells were transformed with an empty vector (pRS315) or plasmids encoding PFUS3-FUS3-GFP-CCaaX (C; pRC205), PFUS3-FUS3-GFP-SSaaX (s; pRC206), PTPI1-FUS3-GFP-CCaaX (C; pRC226), or PTPI1-FUS3-GFP-SSaaX (s; pRC227). Cultures were spotted in 10-fold dilution series on plates and assessed for growth at the indicated temperatures. (B) Wild-type (BY4741) and fus3 kss1 (RCY9320) cells were transformed with an empty vector (pRS315) or plasmids encoding PGAL1-FUS3-GFP-CCaaX (C; pRC249) or PGAL1-FUS3-GFP-SSaaX (s; pRC250). Cultures were spotted in 10-fold dilution series on plates containing dextrose (Dex) or galactose (Gal) as the carbon source and assessed for growth at the indicated temperatures.
FIG. 13.
Restriction of Fus3 to the plasma membrane impairs cell proliferation. (A) Strains BY4741, RCY9335, RCY9320, RCY9340, RCY9342, RCY9343, RCY9336, RCY9337, RCY9341, RCY9344, RCY9339, RCY9368, RCY9337, and RCY9366, with corresponding genotypes indicated, were transformed with plasmids encoding PGAL1-FUS3-GFP-CCaaX (C; pRC249) or PGAL1-FUS3-GFP-SSaaX (s; pRC250). Cultures were spotted in 10-fold dilution series on plates containing dextrose or galactose as the carbon source and assessed for growth at 26°C. (B to D) Strains BY4741, RCY9320, RCY9337, RCY9343, RCY9336, and RCY9341, with genotypes indicated, were transformed with plasmids encoding PGAL1-GFP-CCaaX (C, G; pRC285), PGAL1-FUS3-GFP-CCaaX (C, FG; pRC249), PGAL1-fus3(D137A)-GFP-CCaaX (C, f*G; pRC259), PGAL1-GFP-SSaaX (s, G; pRC286), PGAL1-FUS3-GFP-SSaaX (s, FG; pRC250), or PGAL1-fus3(D137A)-GFP-SSaaX (s, f*G; pRC261). Cultures were spotted in 10-fold dilution series on plates containing dextrose or galactose as the carbon source and assessed for growth at 26°C.
The ability of Fus3-GFP-CCaaX to impair growth required the PAK Ste20, the MAPKKK Ste11, the MAPKK Ste7, and the scaffolds Far1 and Ste5, but not Gβ (Ste4) or pheromone receptor (Ste2) (Fig. 13A). These results indicate that the Fus3-GFP-CCaaX-induced growth impairment depends on activation of this MAPK via the normal cascade and that, even in the absence of pheromone receptor-heterotrimeric G protein stimulation, the basal level of pathway activity at the membrane is high enough to support MAPK activation through the cascade once the concentration of Fus3 at the membrane is raised to a sufficiently high level via its tethering at this location.
The ability of Fus3-GFP-CCaaX to impair growth was not dependent on either of two known cytoplasmic substrates of this MAPK, Bni1 or Rvs167 (Fig. 13A). Nevertheless, the Fus3-GFP-CCaaX-dependent growth defect was largely eliminated when a catalytically inactive derivative (D137A) was used (Fig. 13B). However, growth of Fus3(D137A)-GFP-CCaaX cells was slightly less robust than that of Fus3-GFP-SSaaX or Fus3(D137A)-GFP-SSaaX cells. This residual effect could not be attributed to the nonspecific influence of its membrane-tethering per se because it was not caused by GFP-CCaaX (Fig. 13C). It also cannot be ascribed to the leaky activity of the Fus3(D137A) mutant, since rescue of the growth impairment afforded by Fus3(D137A)-GFP-CCaaX was not potentiated by deletions (ste5Δ, ste11Δ, and ste7Δ) that remove the upstream factors required for activation (Fig. 13D). This residual negative influence may be due to sequestration of (i.e., nondissociable interactions with) membrane proteins when catalytically inactive Fus3 is present at a high local concentration due to membrane anchoring. Nonetheless, the adverse effects caused by Fus3-GFP-CCaaX were largely dependent on the normal mechanisms of Fus3 activation and on the kinase activity of this MAPK.
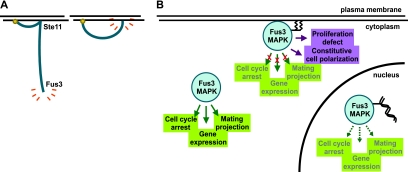
Others have documented that plasma membrane tethering of the MAPKKK Ste11 via a similar strategy (expression of Ste11-CCaaX) causes pheromone-independent growth arrest and that this arrest requires the downstream transcription factor Ste12 (83). In striking distinction, the growth defect caused by Fus3-GFP-CCaaX is not rescued by the absence of Ste12 (Fig. 13A). Thus, although membrane tethering of either the MAPKKK or the MAPK of this pathway impairs cell growth, different mechanisms are used. Restricting localization of an upstream pathway component, like Ste11, elevates pathway activation, but the resulting enhanced signal is then propagated via the normal downstream components, leaving Fus3 MAPK free to elicit its normal outputs, albeit more robustly (Fig. 14A ). In contrast, restricting localization of Fus3 MAPK also elevates its activation but prevents the enzyme from gaining access to some of its normal targets and apparently permits modification of inappropriate substrates, to the detriment of the cell (Fig. 14A).
FIG. 14.
MAPK dynamics and the induction of pathway responses. (A) The signal transduction cascade of the pheromone response pathway is depicted as a curve emanating from membrane-associated upstream components and terminating in the active MAPK. Restricting the localization of an intermediate pathway component (e.g., Ste11) does not prevent activation of normal downstream components or normal outputs (left), whereas restricting the localization of the terminal effector MAPK confines its activity to only some of its targets and biases its action toward inappropriate ones (right). (B) The activity of the upstream signal transduction pathway is directed to cellular processes according to the subcellular localization of Fus3. Only the free MAPK can access all of its relevant substrates efficiently.
DISCUSSION
Proper Fus3 localization is critical for switch-like pathway response.
Fus3 is inactive in the absence of pheromone and active in the presence of pheromone. We have shown that Fus3 trapped in the nucleus is relatively inactive under both conditions, whereas plasma membrane-tethered Fus3 has higher-than-normal basal activity yet can still respond further to pheromone stimulation. Thus, the ability of wild-type Fus3 to move between cellular compartments crucially affects the temporal and spatial characteristics, and the switch-like nature, of its activation and therefore the thresholds and kinetics with which the outputs of this signaling pathway are evoked upon exposure to pheromone.
Fus3 activation requires a cascade of sequential phosphorylations (Ste20 → Ste11 → Ste7 → Fus3). Ste20 associates via its N terminus with membrane-tethered Cdc42 and via its C terminus with membrane-anchored Gβγ. The other three kinases bind the Ste5 scaffold that is recruited to the membrane via interaction with Gβγ and with membrane phospholipids. Like artificial membrane tethering of Ste11, constitutive localization of Ste5 at the membrane also causes pheromone-independent pathway activation (57) because it places this MAPKKK in close proximity to its direct activator (Ste20). We showed here that membrane tethering of Fus3 also leads to its dual phosphorylation, even in the absence of pheromone, indicating that the propinquity of the upstream and downstream termini of the kinase cascade is sufficient to permit all of the interactions among them that are necessary for pathway activation, even in the absence of a pheromone. Thus, simply promoting congression of these elements is a major component of the mechanism by which the receptor-initiated stimulus triggers MAPK activation. The sensitivity of any given MAPK pathway to such effects is likely an emergent property dictated by its specific architecture. For example, in contrast to plasma membrane-restricted Fus3, membrane tethering of MAPK Hog1 (which is also activated via a Ste20-initiated kinase cascade) does not lead to significant basal Hog1 activation in the absence of the normal stimulus (hyperosmotic stress) (78).
Balanced activation of the pheromone response pathway avoids deleterious effects.
Pheromone-independent activation of the pheromone response pathway can be achieved in multiple ways, all of which lead to growth inhibition, such as deletion of the gene for the Gα subunit (Gpa1) that negatively regulates Gβγ (21, 51), overexpression of Gβγ (80), membrane tethering of either Ste5 (57) or Ste11 (83), use of the constitutively active alleles of Ste11 (70), and overexpression of Ste12 (24). In all cases, the observed growth retardation has been ascribed to the imposition of Fus3-initiated Ste12-dependent transcriptional responses and Far1-mediated cell cycle arrest.
As we have demonstrated here, membrane restriction of Fus3 abrogates Ste12-dependent transcription and prevents Far1-mediated cell cycle arrest (Fig. 7) yet still also impairs growth (Fig. 13). Consistent with the fact that Fus3-GFP-CCaaX is unable to access the nucleus, the growth defect that it induces is independent of Ste12 (Fig. 13). Thus, use of membrane-restricted Fus3 reveals that prolonged action of Fus3 at this location can cause a growth impairment that differs mechanistically from that thought to arise from hyperactivation via upstream components of the pathway (Fig. 14A). Thus, under the other situations previously studied, it is likely that the effects of chronic Fus3 activation that are exerted at the plasma membrane also contribute to impeding growth.
MAPK subcellular localization determines pathway behavior and signal responses.
Despite its hyperactivation, plasma membrane-bound Fus3 was largely defective for eliciting the normal panoply of pheromone-induced responses. In contrast, nuclearly restricted Fus3 was only poorly activated yet able to elicit, albeit weakly, pheromone-induced growth arrest and pheromone-induced gene expression. Furthermore, much of this apparent loss of function was restored (even the ability to form projections) by increasing its level of activation by eliminating counteracting phosphatases. Hence, and unlike Fus3 restricted to the plasma membrane, Fus3 restrained in the nucleus was much less compromised in evoking physiologically relevant responses. Thus, our analysis reveals how the function of Fus3 differs at distinct cellular locations. At the plasma membrane, Fus3 is competent to be activated and is spared from deactivation by phosphatases yet is unable to access some of its most critical effectors and therefore cannot generate the outputs most crucial for successful mating. If allowed to translocate into the nucleus, Fus3 encounters there its most critical effectors, but to achieve the necessary responses, it has to act in a certain window of opportunity before it becomes deactivated by the phosphatases resident in that compartment. Normally, in the presence of a continued stimulus and due to the positive feedback that arises from the pheromone-induced upregulation of both the FUS3 transcript (27) and its product (5), the effects of Fus3 in the nucleus, once initiated, are self-reinforcing. In the case of Fus3-GFP-Htb2, by contrast, its activation is weak and remains so because that process must rely on the nucleocytoplasmic shuttling of Ste5 and the amount of Ste7 MAPKK activated at the membrane that remains bound to the scaffold. Thus, restricted localization of Fus3 MAPK cannot be compensated for by the mobility of its upstream activators or downstream targets, further explaining why the ability of this MAPK to undergo dynamic changes in its localization is necessary for optimal pathway function. In other words, this MAPK must move to its targets, and not vice versa.
Our Fus3-GFP-CCaaX construct behaved like a neomorphic allele; although it was unable to elicit several pheromone responses, it acquired the ability to cause aberrant cell morphology and inhibit cell proliferation. Thus, the sustained presence of activated Fus3 at zones of growth is quite deleterious to cell physiology. Therefore, normally, the observed localization of Fus3 at the shmoo tip (49) must be transient, so that it modifies its substrates at this location in a kiss-and-run fashion. Consistent with this view, fluorescence recovery after photobleaching (FRAP) analysis suggests that Fus3-GFP at the shmoo tip does undergo rapid turnover (75). At the time, this behavior was interpreted as cycles of recruitment, activation, and release. However, our observations indicate that prolonged dwell time at the membrane is toxic. These considerations provide yet another rationale for why, even after its activation, MAPK mobility is crucial for successful biological outcomes in the mating process (Fig. 14B).
The fact that membrane tethering of Fus3 abrogates expression of pheromone-induced gene expression (Fig. 7A), whereas membrane tethering of Ste11 induces the expression of such genes even in the absence of pheromone (83), highlights two points. First, the untoward effects of Fus3-CCaaX are not merely the result of its hyperactive state because equivalent Fus3 hyperactivation is achieved by Ste11-CCaaX. Therefore, it matters what subcellular substrates activated Fus3 can access. A second and corollary point is that Fus3 hyperactivated by membrane-tethered MAPKKK is free to disseminate its activity to all of its normal targets (and is subject to deactivation by phosphatases in the nucleus); in contrast, although membrane tethering of Fus3 also leads to its hyperactivation, Fus3 in this case does not have access to any of its nuclearly localized substrates and is not subject to downregulation by nuclearly localized negative regulators (Fig. 14B).
Despite its much weaker activation (Fig. 5), Fus3-GFP-Htb2 supported mating 5- to 8-fold more efficiently than Fus3-GFP-CCaaX, suggesting either that the nuclear functions of this MAPK contribute more to mating proficiency than its cytoplasmic (and membrane) functions or that carrying out its nuclear functions requires significantly less active MAPK. The former seems more likely, given that enhancing the level of Fus3-GFP-Htb2 activity by eliminating its deactivating phosphatases promoted significantly more robust outputs and increased overall mating efficiency to 50 to 60% of that seen for cells expressing wild-type Fus3-GFP. Also consistent with the above conclusion, alleles of Fus3 that are catalytically inactive (but unrestricted in localization) are unable to support detectable mating, and Fus3-GFP-Htb2 promotes ≥106 times more efficient mating in STE7+ fus3 kss1 cells than in ste7 fus3 kss1 cells (16). The importance of the nuclear functions of Fus3 MAPK for mating stand in stark contrast with the dispensability of the nuclear functions of Hog1 MAPK for hyperosmotic stress resistance (78). In both cases, tethering of the cognate MAPK to the plasma membrane abolishes its capacity to induce gene expression, yet this restriction compromises mating but not growth and survival under hyperosmotic conditions. Thus, in the mating pheromone response pathway, encounter of the Fus3 MAPK with its activators, inactivators, and physiologically relevant substrates depends crucially on the dynamic intercompartmental movement of the MAPK itself.
Acknowledgments
This work was supported by a predoctoral fellowship from the UC Systemwide Cancer Research Coordinating Committee (to R.E.C.)., by NIH predoctoral traineeship GM07232 (to J.C.P.), and by NIH R01 research grant GM21841 (to J.T.).
We thank Michael A. McMurray for critical reading of the manuscript and other members of the Thorner laboratory for helpful discussions, especially Daniel R. Ballon and Lindsay S. Garrenton.
Footnotes
Published ahead of print on 28 June 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Ash, J., C. Wu, R. Larocque, M. Jamal, W. Stevens, M. Osborne, D. Y. Thomas, and M. Whiteway. 2003. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics 163:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballard, M. J., W. A. Tyndall, J. M. Shingle, D. J. Hall, and E. Winter. 1991. Tyrosine phosphorylation of a yeast 40 kDa protein occurs in response to mating pheromone. EMBO J. 10:3753-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, A. J., L. J. Flatauer, K. Matsukuma, J. Thorner, and L. Bardwell. 2001. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276:10374-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardwell, L. 2005. A walk-through of the yeast mating pheromone response pathway. Peptides 26:339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell, L., J. G. Cook, E. C. Chang, B. R. Cairns, and J. Thorner. 1996. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16:3637-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardwell, L., J. G. Cook, J. X. Zhu-Shimoni, D. Voora, and J. Thorner. 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. U. S. A. 95:15400-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackwell, E., I. M. Halatek, H. J. Kim, A. T. Ellicott, A. A. Obukhov, and D. E. Stone. 2003. Effect of the pheromone-responsive Gα and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase. Mol. Cell. Biol. 23:1135-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell, E., H. J. Kim, and D. E. Stone. 2007. The pheromone-induced nuclear accumulation of the Fus3 MAPK in yeast depends on its phosphorylation state and on Dig1 and Dig2. BMC Cell Biol. 8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blondel, M., P. M. Alepuz, L. S. Huang, S. Shaham, G. Ammerer, and M. Peter. 1999. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 13:2284-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 11.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter, A. E., T. R. Jones, M. R. Lamprecht, C. Clarke, I. H. Kang, O. Friman, D. A. Guertin, J. H. Chang, R. A. Lindquist, J. Moffat, P. Golland, and D. M. Sabatini. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan, R. K., and C. A. Otte. 1982. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol. Cell. Biol. 2:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, F., and I. Herskowitz. 1990. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell 63:999-1011. [DOI] [PubMed] [Google Scholar]
- 16.Chen, R. E. 2008. Function and regulation of mitogen-activated protein kinases in the yeast Saccharomyces cerevisiae. Ph.D. thesis. University of California, Berkeley, CA.
- 17.Chen, R. E., and J. Thorner. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773:1311-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherkasova, V. A. 2006. Measuring MAP kinase activity in immune complex assays. Methods 40:234-242. [DOI] [PubMed] [Google Scholar]
- 19.Choi, K. Y., B. Satterberg, D. M. Lyons, and E. A. Elion. 1994. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78:499-512. [DOI] [PubMed] [Google Scholar]
- 20.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831-2848. [DOI] [PubMed] [Google Scholar]
- 21.Dietzel, C., and J. Kurjan. 1987. The yeast SCG1 gene: a Gα-like protein implicated in the a- and α-factor response pathway. Cell 50:1001-1010. [DOI] [PubMed] [Google Scholar]
- 22.Dohlman, H. G., J. Song, D. Ma, W. E. Courchesne, and J. Thorner. 1996. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein α subunit). Mol. Cell. Biol. 16:5194-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doi, K., A. Gartner, G. Ammerer, B. Errede, H. Shinkawa, K. Sugimoto, and K. Matsumoto. 1994. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 13:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolan, J. W., and S. Fields. 1990. Overproduction of the yeast STE12 protein leads to constitutive transcriptional induction. Genes Dev. 4:492-502. [DOI] [PubMed] [Google Scholar]
- 25.Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 26.Elion, E. A., J. A. Brill, and G. R. Fink. 1991. Functional redundancy in the yeast cell cycle: FUS3 and KSS1 have both overlapping and unique functions. Cold Spring Harb. Symp. Quant. Biol. 56:41-49. [DOI] [PubMed] [Google Scholar]
- 27.Elion, E. A., P. L. Grisafi, and G. R. Fink. 1990. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell 60:649-664. [DOI] [PubMed] [Google Scholar]
- 28.Errede, B., A. Gartner, Z. Zhou, K. Nasmyth, and G. Ammerer. 1993. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature 362:261-264. [DOI] [PubMed] [Google Scholar]
- 29.Farley, F. W., B. Satterberg, E. J. Goldsmith, and E. A. Elion. 1999. Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics 151:1425-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrigno, P., F. Posas, D. Koepp, H. Saito, and P. A. Silver. 1998. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin-beta homologs NMD5 and XPO1. EMBO J. 17:5606-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friesen, H., K. Murphy, A. Breitkreutz, M. Tyers, and B. Andrews. 2003. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Mol. Biol. Cell 14:3027-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu, H. W., and P. J. Casey. 1999. Enzymology and biology of CaaX protein prenylation. Recent Prog. Horm. Res. 54:315-342. [PubMed] [Google Scholar]
- 33.Garrenton, L. S., S. L. Young, and J. Thorner. 2006. Function of the MAPK scaffold protein Ste5 requires a cryptic PH domain. Genes Dev. 20:1946-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gartner, A., K. Nasmyth, and G. Ammerer. 1992. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 6:1280-1292. [DOI] [PubMed] [Google Scholar]
- 35.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 36.Good, M., G. Tang, J. Singleton, A. Reményi, and W. A. Lim. 2009. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 136:1085-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartwell, L. H. 1980. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J. Cell Biol. 85:811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasson, M. S., D. Blinder, J. Thorner, and D. D. Jenness. 1994. Mutational activation of the STE5 gene product bypasses the requirement for G protein β and γ subunits in the yeast pheromone response pathway. Mol. Cell. Biol. 14:1054-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heiman, M. G., and P. Walter. 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151:719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein, S., H. Reuveni, and A. Levitzki. 2000. Signal transduction by a nondissociable heterotrimeric yeast G protein. Proc. Natl. Acad. Sci. U. S. A. 97:3219-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kusari, A. B., D. M. Molina, W. J. Sabbagh, C. S. Lau, and L. Bardwell. 2004. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Cell Biol. 164:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamson, R. E., M. J. Winters, and P. M. Pryciak. 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 22:2939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391:191-195. [DOI] [PubMed] [Google Scholar]
- 44.Longtine, M. S., A. R. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 45.Ma, D., J. G. Cook, and J. Thorner. 1995. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol. Biol. Cell 6:889-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maeder, C. I., M. A. Hink, A. Kinkhabwala, R. Mayr, P. I. Bastiaens, and M. Knop. 2007. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat. Cell Biol. 9:1319-1326. [DOI] [PubMed] [Google Scholar]
- 47.Manahan, C. L., M. Patnana, K. J. Blumer, and M. E. Linder. 2000. Dual lipid modification motifs in Gα and Gγ subunits are required for full activity of the pheromone response pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 11:957-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcus, A., A. Polverino, M. Barr, and M. Wigler. 1994. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc. Natl. Acad. Sci. U. S. A. 91:7762-7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metodiev, M. V., D. Matheos, M. D. Rose, and D. E. Stone. 2002. Regulation of MAPK function by direct interaction with the mating-specific Gα in yeast. Science 296:1483-1486. [DOI] [PubMed] [Google Scholar]
- 50.Mettetal, J. T., D. Muzzey, C. Gomez-Uribe, and A. van Oudenaarden. 2008. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319:482-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyajima, I., M. Nakafuku, N. Nakayama, C. Brenner, A. Miyajima, K. Kaibuchi, K. Arai, Y. Kaziro, and K. Matsumoto. 1987. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell 50:1011-1019. [DOI] [PubMed] [Google Scholar]
- 52.Nern, A., and R. A. Arkowitz. 1999. A Cdc24p-Far1p-Gβγ protein complex required for yeast orientation during mating. J. Cell Biol. 144:1187-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nern, A., and R. A. Arkowitz. 2000. Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J. Cell Biol. 148:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peter, M., and I. Herskowitz. 1994. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science 265:1228-1231. [DOI] [PubMed] [Google Scholar]
- 56.Printen, J. A., and G. F. Sprague, Jr. 1994. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138:609-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pryciak, P. M., and F. A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 12:2684-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qi, M., and E. A. Elion. 2005. MAP kinase pathways. J. Cell Sci. 118:3569-3572. [DOI] [PubMed] [Google Scholar]
- 59.Raman, M., W. Chen, and M. H. Cobb. 2007. Differential regulation and properties of MAPKs. Oncogene 26:3100-3112. [DOI] [PubMed] [Google Scholar]
- 60.Ramer, S. W., and R. W. Davis. 1993. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 90:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reneke, J. E., K. J. Blumer, W. E. Courchesne, and J. Thorner. 1988. The carboxy-terminal segment of the yeast α-factor receptor is a regulatory domain. Cell 55:221-234. [DOI] [PubMed] [Google Scholar]
- 62.Roberts, C., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 63.Sabbagh, W. J., L. J. Flatauer, A. J. Bardwell, and L. Bardwell. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1, 2, and 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 65.Sette, C., C. J. Inouye, S. L. Stroschein, P. J. Iaquinta, and J. Thorner. 2000. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell 11:4033-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheff, M. A., and K. S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21:661-670. [DOI] [PubMed] [Google Scholar]
- 67.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon, M. N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376:702-705. [DOI] [PubMed] [Google Scholar]
- 69.Slaughter, B. D., J. W. Schwartz, and R. Li. 2007. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc. Natl. Acad. Sci. U. S. A. 104:20320-20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevenson, B. J., N. Rhodes, B. Errede, and G. F. J. Sprague. 1992. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 6:1293-1304. [DOI] [PubMed] [Google Scholar]
- 71.Taxis, C., and M. Knop. 2006. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques 40:73-78. [DOI] [PubMed] [Google Scholar]
- 72.Tedford, K., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7:228-238. [DOI] [PubMed] [Google Scholar]
- 73.Trueheart, J., J. D. Boeke, and G. R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Drogen, F., and M. Peter. 2001. MAP kinase dynamics in yeast. Biol. Cell 93:63-70. [DOI] [PubMed] [Google Scholar]
- 75.van Drogen, F., V. M. Stucke, G. Jorritsma, and M. Peter. 2001. MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 3:1051-1059. [DOI] [PubMed] [Google Scholar]
- 76.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 77.Wang, Y., and H. G. Dohlman. 2004. Pheromone signaling mechanisms in yeast: a prototypical sex machine. Science 306:1508-1509. [DOI] [PubMed] [Google Scholar]
- 78.Westfall, P. J., J. C. Patterson, R. E. Chen, and J. Thorner. 2008. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc. Natl. Acad. Sci. U. S. A. 105:12212-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Westfall, P. J., and J. Thorner. 2006. Analysis of mitogen-activated protein kinase signaling specificity in response to hyperosmotic stress: use of an analog-sensitive HOG1 allele. Eukaryot. Cell 5:1215-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whiteway, M., L. Hougan, and D. Y. Thomas. 1990. Overexpression of the STE4 gene leads to mating response in haploid Saccharomyces cerevisiae. Mol. Cell. Biol. 10:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whiteway, M. S., C. Wu, T. Leeuw, K. Clark, A. Fourest-Lieuvin, D. Y. Thomas, and E. Leberer. 1995. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science 269:1572-1575. [DOI] [PubMed] [Google Scholar]
- 82.Wiget, P., Y. Shimada, A. C. Butty, E. Bi, and M. Peter. 2004. Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. EMBO J. 23:1063-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winters, M. J., R. E. Lamson, H. Nakanishi, A. M. Neiman, and P. M. Pryciak. 2005. A membrane binding domain in the Ste5 scaffold synergizes with Gβγ binding to control localization and signaling in pheromone response. Mol. Cell 20:21-32. [DOI] [PubMed] [Google Scholar]
- 84.Wu, C., M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270:15984-15992. [DOI] [PubMed] [Google Scholar]
- 85.Zhan, X. L., R. J. Deschenes, and K. L. Guan. 1997. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 11:1690-1702. [DOI] [PubMed] [Google Scholar]
- 86.Zhou, Z., A. Gartner, R. Cade, G. Ammerer, and B. Errede. 1993. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol. Cell. Biol. 13:2069-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]