Abstract
RDS (retinal degeneration slow) is a photoreceptor-specific tetraspanin protein required for the biogenesis and maintenance of rod and cone outer segments. Mutations in the Rds gene are associated with multiple forms of rod- and cone-dominant retinal degeneration. To gain more insight into the mechanisms underlying the regulation of this gene the identification of regulatory sequences within the promoter of Rds was undertaken. A 3.5kb fragment of the 5′ flanking region of the mouse Rds gene was isolated and binding sites for Crx, Otx2, Nr2e3, RXR family members, Mef2C, Esrrb, NF1, AP1, and SP1 in addition to several E-boxes, GC-boxes and GAGA-boxes were identified. Crx binding sequences were conserved in all mammalian species examined. Truncation expression analysis of the Rds promoter region in Y-79 retinoblastoma cells showed maximal activity in the 350bp proximal promoter region. We also show that inclusion of more distal fragments reduced promoter activity to the basal level, and that the promoter activities are cell-type and direction specific. Co-transfection with Nrl increased promoter activity, suggesting that this gene positively regulates Rds expression. Based on these findings, a relatively small fragment of the Rds promoter may be useful in future gene transfer studies to drive gene expression in photoreceptors.
Keywords: Rds gene, promoter activity and specificity, cis-elements, in vitro transfection, gene therapy
Introduction
Rds is one of the most common ocular genes to carry pathogenic mutations. Over 80 different disease causing mutations in Rds have been identified and are responsible for a wide range of degenerative phenotypes including autosomal dominant retinitis pigmentosa (RP) and various forms of macular dystrophy (Berson, 1993, Keen and Inglehearn, 1996). We and others have successfully delivered wild type murine Rds to the retinas of mice with an Rds-associated haploinsufficiency RP phenotype (the rds+/-) and reported structural and functional rescue of the diseased retina (Ali, et al., 2000, Cai, et al., 2009b). For our studies we used both ubiquitous (chicken beta-actin, CBA) and photoreceptor-specific (human interphotoreceptor retinoid binding protein, IRBP) promoters and while they represented satisfactory preliminary choices, further knowledge of the native Rds promoter will be a great advantage for our future studies. However thus far, no reports have specifically addressed regulation of this gene. Characterization of the Rds promoter region will give a better understanding of the native regulation of the Rds gene and may enable us to enhance our gene therapy studies by incorporating critical regulatory elements in our vector design.
Gene therapy has been a popular and promising therapeutic approach for the treatment of inherited retinal degenerations in various animal models and patients [rodents (Ali, et al., 2000, Cai, et al., 2009b, Weber, et al., 2003), dogs (Acland, et al., 2005, Acland, et al., 2001, Le Meur, et al., 2006), primates (Jacobson, et al., 2006, Lotery, et al., 2003, Weber, et al., 2003) and humans (Bainbridge, et al., 2008, Cideciyan, et al., 2008, Hauswirth, et al., 2008)], and optimization of ocular gene therapy by expanding promoter choices is advantageous. Due to the prevalence of inherited retinal degenerations associated with photoreceptor and retinal pigment epithelial (RPE) defects, these cell types have often been targets of gene delivery studies. While tissue-specific promoters like vitelliform macular dystrophy 2 (VMD2) and rhodopsin (MOP) and ubiquitously expressed promoters like chicken beta-actin (CBA) have been successfully used to direct expression in the retina (Allocca, et al., 2007, Cai, et al., 2009a, Cai, et al., 2009b), strong promoters that can direct proper levels of gene expression in rods and cones have been lacking. The strongest currently used ocular promoter (the MOP promoter) is typically thought to be rod specific, or to drive very low levels of gene expression in cones (Glushakova, et al., 2006). For the treatment of rod-based diseases, this promoter is a good choice, however, many diseases target both cones and rods. The most commonly used promoters to target both rods and cones have been the promoter for the photoreceptor transcription factor Crx and the IRBP (interphotoreceptor retinoid binding protein) promoter (Nour, et al., 2004, Oh, et al., 2007) although other promoters such as the rhodopsin kinase promoter, have also been studied for this purpose (Khani, et al., 2007). To expand the available options for strong rod/cone promoters, we chose to characterize the promoter region for a gene that is expressed robustly in both photoreceptor types; Rds (retinal degeneration slow, also referred to as Peripherin/rds, P/rds, or Prph2). Our goals were first, to characterize a novel promoter that could be potentially used to direct high levels of expression of any gene (but particularly Rds) in rods and cones in gene therapy studies; and second, to study regulation of the Rds gene to better understand the expression and regulation of this key outer segment protein. We isolated a 3.5kb fragment of the 5′ flanking region of the mouse Rds gene from wild type C57BL/6 genomic DNA, identified regulatory factor binding sites in the promoter, and characterized the in vitro activity and cell-type specificity of various promoter fragments.
Materials and methods
Cloning of the 5′ flanking region and identification of regulatory sequences
3.5 kb of the 5′ flanking region of the murine Rds gene was isolated from C57BL/6 genomic DNA using the PromoterFinder™ DNA Walking kit (Clontech Laboratories, Inc., Palo Alto, CA, for details see Supplementary Methods and Supplementary Figure 1). Products were cloned into the pBluescriptKS+ vector and sequenced. Sequences were blasted against the ensembl database (www.ensembl.org). Analysis for the presence of known transcription factor binding sites (cis-elements) in the 3.5kb murine RDS 5′ flanking region was carried out by MatInspector version 7.0 using the Matrix Family library database version 7.0 (Genomatix, Munich, Germany). Similar assessment was carried out on 3.5kb of the flanking region of bovine, rat, Xenopus, and human RDS promoter (using the ensembl sequences). For comparison sake, the same 5′ flanking region was analyzed from the mouse HPRT housekeeping gene and the liver specific HSD17b gene. Identity calculations and Clustal alignment of sequences from multiple species was carried out using Vector NTI 11 (Invitrogen, Carlsbad, CA). Analysis of CpG islands was carried out according to the method by Gardiner-Garden and Frommer (Gardiner-Garden and Frommer, 1987). Briefly, CpG rich areas were defined as regions at least 200 bp in length (starting from the transcription initiation site-TSS) that had a GC content above 50% and an observed/expected CG ration of greater than 0.6. In all cases, the translation initiation site (ATG) was defined as +1.
Animal care and use
C57/Bl6 and Balb/C mice were maintained in the breeding colony under cyclic light (14L:10D) conditions; cage illumination was ∼7 foot-candles during the light cycle. All procedures were approved by the University of Oklahoma Health Science Center Institutional Animal Care and Use Committee (IACUC) and adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Visual Research (http://www.arvo.org/). Genomic DNA was harvested from tail cuts as described previously (Naash, et al., 1993).
Plasmid construction, cell transfection and luciferase assay
Rds promoter fragments were inserted upstream of the luciferase reporter gene in the multiple cloning site of the pGL2-Basic plasmid (Promega, Madison WI) using standard techniques. For cell transfections, plasmids were prepared using the Endofree Plasmid Maxi Kit (Qiagen, Chatsworth, CA). COS-1 cells, low passage (<40) human Y-79 retinoblastoma cells (Di Polo and Farber, 1995), and in some experiments NIH3T3, 661W, and MCF-7 cells were seeded in six well plates at 3.2×105 cells/well 24 hours prior to transfection. Y-79 cells were cultured/transfected in suspension, in RPMI-40 media containing 15% FBS (Invitrogen/Gibco, Carlsbad CA) while adherent 661W, COS, MCF-7 and NIH3T3 cells were cultured/transfected in DMEM containing 10% FBS. All media contained standard antibiotics (penicillin 100 U/ml and streptomycin 100 μg/ml-Invitrogen/Gibco). Adherent cells were ∼70% confluent at the time of transfection. 15 μg of each plasmid (pLUC468, pLUC468IV, pLUC1084, pLUC1439, pLUC2632, pLUC3304, pGL2-Basic, and pGL2-promoter) was used for standard CaPO4 transfection carried out according to the manufacturer procotocl using the CalMaximizer (Clontech) kit. As an internal control for transfection efficiency, 5 μg of pCH110 plasmid (Amersham GE, Piscataway NJ) was co-transfected. Cells were harvested 48 hours after transfection in 250 μl of 1× lysis buffer containing 5% (v/v) Triton X-100. Luciferase activity was assayed by mixing 20 μl of this lysate with 100 μl of luciferase assay reagent (Luciferase Assay System, Promega) and light emission was measured immediately in a spectrophotometer. As described previously (Rosenthal, 1987), β-gal activity was measured by ortho-Nitrophenyl-β-galactoside (ONPG) cleavage assay using the same cytosolic extract. The promoter activity of each construct in each cell line was defined as Relative Light Unit/β-gal activity. Each data point was the average of three readings and each experiment was repeated three times.
mRNA collection and real-time PCR
Murine eyes from P30 WT, CRX-NRL and nrl-/- animals were collected (5-6 eyes per group), and total RNA was isolated using Trizol (Invitrogen) as per the manufacturers instruction. DNase treatment and cDNA synthesis were performed as described previously (Cai, et al., 2009b, Farjo, et al., 2006). qRT-PCR for Rds was performed in triplicate for each sample using a Bio-rad iCycler single color system and the previously published primers (Farjo, et al., 2006). The HPRT housekeeping gene was used as an internal control as previously described (Farjo, et al., 2006).
Immunocytochemistry and western blotting
Cells were fixed with 4% paraformaldehyde at room temperature (RT) for 20 min. The monoclonal anti-RDS antibody 5H2 (1:2, a generous gift from Dr. Robert Molday, University of British Columbia) was applied overnight at room temperature. The cells were rinsed with PBS and covered with FITC-anti-rabbit IgG (1:100) for 30 min. After three washings in PBS, cells were mounted in Vectashield with DAPI (Vector Laboratory Inc, Burlingame CA) and viewed under an epifluorescent microscope (Zeiss USA, Thornwood NY). Western blotting was performed using the RDS-CT polyclonal antibody and HRP conjugated secondary (as previously described) (Farjo, et al., 2006) and actin-HRP (Sigma-Aldrich, St. Louis MO).
Statistical Analysis
One-way ANOVA with Bonferroni's post-hoc test was used to test the null hypothesis in cases where there were three or more different groups. Two-tailed Student's t-tests were used in cases where there were only two groups to compare (Fig. 4B and 5A).
Figure 4. Tissue and direction specificity of Rds promoter activity.
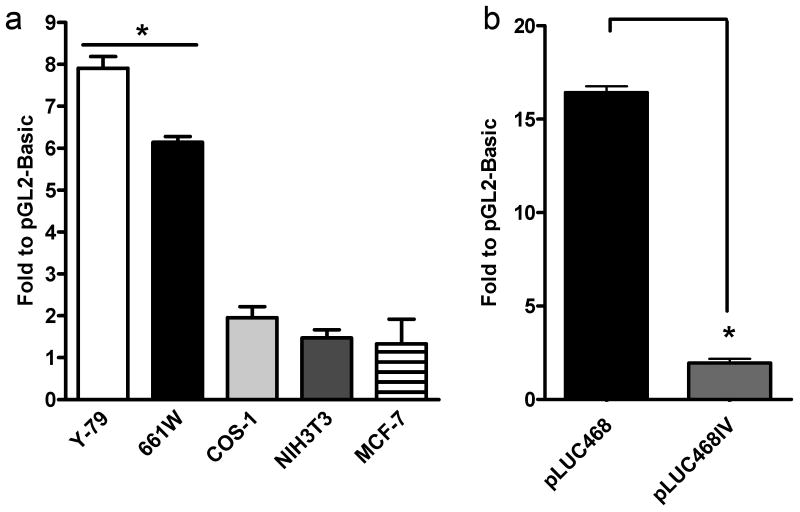
a. pLUC468 (with pCHIP110 as a control) was transfected into several different ocular and non-ocular cell lines. Significant expression (i.e. above levels detected in pGL2-Basic transfected cells) was only detected in Y-79 and 661W cells. b. pLUC468IV transfection did not direct significant promoter activity indicating that the Rds promoter is direction specific. The activity presented is an average of three independent experiments. * p<0.01
Figure 5. Nrl and Nr2e3 regulation of Rds expression.
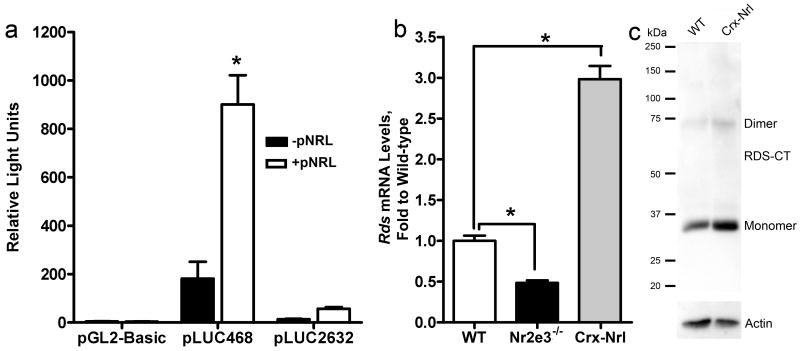
a. Y-79 cells were co-transfected with the luciferase construct (and pCHIP110) and, as indicated, pNrl. Expression of Nrl led to a significant increase in Rds gene expression. b. Rds mRNA levels (qRT-PCR) were significantly reduced in Nr2e3-/- mice and significantly increased in Crx-Nrl mice. * p<0.01. c. Western blots on 5 μg of total retinal extract from WT or Crx-Nrl mice were probed with polyclonal antibodies against the RDS C-terminal (RDS-CT) and with antibodies against actin. RDS protein levels (when normalized to actin) were increased by approximately 2-fold.
Results
Sequence analysis of the 5′ flanking region
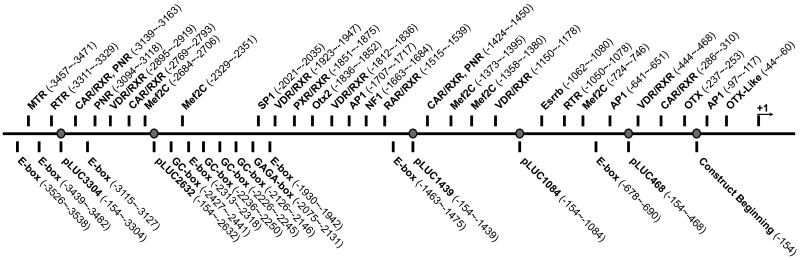
The 5′ flanking region of murine RDS was sequenced and analyzed as described in the methods (Supplementary Fig. 2). The GC content in the 400 bp immediately 5′ to the ATG initiation codon is 55%. The observed/expected CpG ratio was 0.49 indicating that the immediate upstream promoter region is not CpG rich. Sequence analysis of the flanking region showed the absence of typical TATA and CCAAT elements. Over 800 potential cis-regulatory elements were detected in the analyzed region. Those relevant to photoreceptor gene regulation are shown on the sequence in Supplementary Fig. 2, and schematically in Figure 1. At the -3 and +4 positions (see Supplementary Fig. 2), A and G residues were observed respectively, consistent with the proposed consensus sequence for initiation of translation in higher eukaryotes (Kozak, 1987). Transcription start sites are observed at -218 and -221. An OTX binding site was observed at position -237 to -253. AP1 elements were observed at -641 to -651 and -1707 to -1717. Several binding sites for RXR (Retinoid X receptor) family members including CAR, VDR, RAR, PXR and RTR were located throughout the promoter region as were putative Mef2C sites. Three Nr2e3 binding sites (PNR) were found, at -1424 to -1450, -3094 to -3118 and -3139 to -3163. Five E-boxes were located in the distal region of the fragment, with two additional E-boxes in the proximal region. Sites for Otx2 (-1836 to -1852), and NF1 (-1663 to -1684) were observed near the middle of the promoter region. Just upstream from these elements are one SP1 site (-2021 to -2035), one GAGA-box (-2075 to -2131) and four GC-boxes. One mammalian transcriptional repressor binding site was found located in the extreme distal part of the isolated flanking region (-3457 to -3471).
Figure 1. Schematic distribution of relevant regulatory sequences in the 5′ flanking region of Rds gene.
Relevant putative transcription factor binding sites or other regulatory regions are highlighted (as identified using the procedures found in the Materials and Methods section). The relative location of start/end sites for the in vitro expression constructs is also noted (grey circles).
On the base pair level, the 5′ flanking region is moderately conserved among the species we examined; 46% similarity between mouse and human, 46% between mouse and bovine, and 44% between mouse and Xenopus Tropicalis, and 39% between mouse and rat (Supplementary Fig. 3). Cis-element analysis of the 5′ regions of each of these species was then undertaken. The only regulatory element that appears in the same region of all the mammalian species examined is the CRX binding site (OTX) at position -237 suggesting a prime regulatory role for this transcription factor. As shown in Table 1, additional cis-elements were conserved across species within regions of the promoter (as opposed to in exactly the same place). Those conserved in the proximal (0 to -500), medial (-500 to -1500), and distal regions (-1500 to -3500) are shown in Table 1. Cis-element analysis of the HPRT (housekeeping) gene and the non-ocular HSD17B (hydroxysteroid 17β dehydrogenase) gene are included on table 1 as a reference for which cis-elements are found ubiquitously and which may be more retina-specific.
Table 1. Preservation of transcription factor binding sites across species.
Transcription factor binding sites in the 3.5kb promoter region of the Rds gene from multiple species (with a non-tissue specific [HPRT] and non-photoreceptor gene [HSD17b] used as controls) were identified as described in the methods. The analyzed regions were divided into proximal (within 500 base pairs of the translation start site), medial (from 500-1500 base pairs) and distal (1500-3500 bp). An X indicates that the listed binding site was found in that region.
| CREs (Trans factor) | Mouse Rds | Bovine Rds | Human Rds | Rat Rds | Xenopus Rds | Mouse HPRT | Mouse HSD17B | Role | References |
|---|---|---|---|---|---|---|---|---|---|
| Proximal | |||||||||
| AP1 | X | X | X | X | Not Nrl response element | (Rehemtulla, et al., 1996) | |||
| OTX (Crx) | X | X | X | X | Core activator of photoreceptor genes | (Chen, et al., 1997, Furukawa, et al., 1997, Kennedy, et al., 1998, Moritz, et al., 2002, Whitaker and Knox, 2004) | |||
| CAR/RXR (RXRγ) | X | X | X | X | Regulation of M- vs. S-cones | (Hennig, et al., 2008, Roberts, et al., 2006, Yoshida, et al., 2004) | |||
| Medial | |||||||||
| AP1 | X | X | X | X | As above | ||||
| Mef2c (Mef2c) | X | X | X | X | X | X | X | Rod enhanced | (Hsiau, et al., 2007) |
| Esrrb (Esrrb) | X | X | Rod-enhanced orphan nuclear receptor | (Hsiau, et al., 2007) | |||||
| VDR/RXR (RXRγ) | X | X | X | X | X | X | X | As above | |
| CAR/RXR (RXRγ) | X | X | X | X | As above | ||||
| PNR (Nr2E3) | X | X | X | X | X | Dual activity rod/cone transcription factor | (Cheng, et al., 2006, Cheng, et al., 2004, Peng, et al., 2005) | ||
| Distal | |||||||||
| OTX2 (OTX2) | X | X | X | Activator of Crx regulatory network | (Koike, et al., 2007, Rath, et al., 2006) | ||||
| PXR (RXRγ) | X | X | X | X | Regulation of M- vs. S-cones | (Hennig, et al., 2008, Roberts, et al., 2006, Yoshida, et al., 2004) | |||
| SP1 (SP1) | X | X | X | Ubiquitous transcription factor | (Hennig, et al., 2008) | ||||
| Mef2c (Mef2c) | X | X | X | X | X | X | X | As above | |
| CAR/RXR (RXRγ) | X | X | X | X | As above | ||||
| VDR/RXR (RXRγ) | X | X | X | X | X | X | X | As above | |
| PNR (Nr2e3) | X | X | X | X | X | X | As above | ||
| RTR (RXRγ) | X | X | X | X | As above | ||||
Promoter activity of the Rds gene in vitro
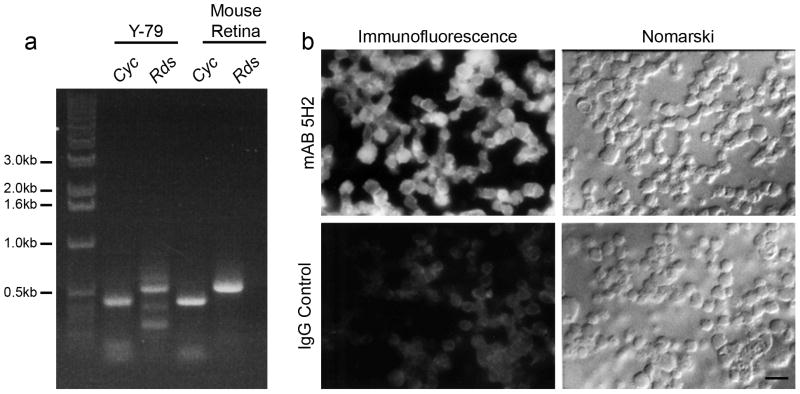
To test the promoter activity of the 5′ flanking region of the Rds gene, transient transfection experiments were carried out in vitro using human Y-79 retinoblastoma cells. Although these cells have been shown to express both rod and cone specific genes, (Di Polo and Farber, 1995) it was necessary to confirm the presence of all basal and tissue-specific factors required for Rds expression. The most straightforward way to confirm that the cells possess the ability to express Rds was to examine Rds expression. RT-PCR across intron 1 revealed that Y-79 cells express Rds transcripts: a PCR product of the expected size (387 bp) was detected in both Y-79 and the positive control (mouse retina) samples (Fig. 2a). Immunocytochemistry with mAB 5H2 (against RDS) was performed and substantial specific immunoreactivity was observed (Fig. 2b) confirming that RDS is expressed in these cells.
Figure 2. Expression of the mouse Rds gene in Y-79 cell lines.
a. Expression of the Rds gene detected by RT-PCR. Primers used for each PCR reaction are indicated on top of each lane (Rds, Cyc). The size of the products from Rds and cyclophilin gene (control) are also labeled. b. Expression of the Rds gene detected by immunocytochemistry using mAB 5H2 against RDS. Immunostaining was observed in the majority of the Y-79 cells (top left) while no staining was seen in IgG stained control cells. Scale bar 25 μm.
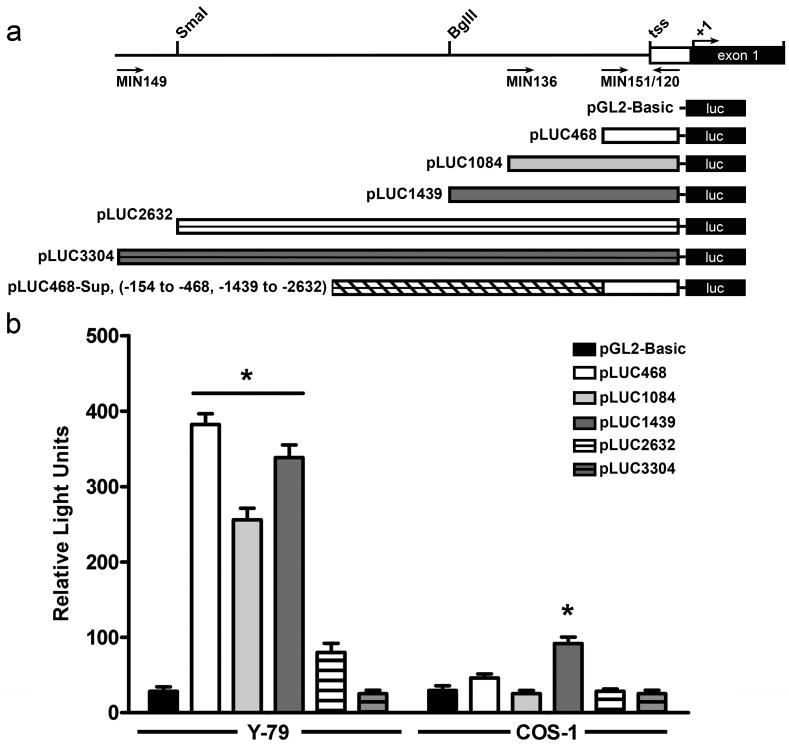
To evaluate the promoter activity and cell type specificity of the isolated flanking region, a series of deletion mutants (Fig. 3a) was generated by PCR or restriction digestion. Promoter fragments in the native orientation were inserted into the pGL2-Basic plasmid containing the luciferase reporter gene as follows: 1) pLUC468, [-154 to -468], 2) pLUC1084, [-154 to -1084], and 3) pLUC1439 [-154 to -1439], 4) pLUC2632 [-154 to -2632] and 5) pLUC3304 [-154 to -3304]. The tested fragments did not include the region downstream from positions -154 to +58. This region includes a modified start codon (ATG) and stop codon (TGA) and based on our initial tests it does not have any promoter activity (data not shown). The fragments were transfected into Y-79 retinoblastoma cells or COS-1 cells (as a non-ocular control) and luciferase activity was measured. Transfection with pGL2-Basic (no promoter) was used to determine the basal level of luciferase activity and transfection with pGL2-SV40 (pGL2-Basic with SV40 promoter) served as a positive control. As an internal control to evaluate the transfection efficiency, pCH110 containing the β-gal reporter gene driven by SV40 promoter was co-transfected with the luciferase vectors.
Figure 3. Promoter activity of the 5′ flanking region of the mouse Rds gene.
a. Schematic showing the construction of the luciferase reporter constructs. b-c. Luciferase activity (normalized to β-gal) after co-transfection of 15 μg of the indicated luciferase construct (containing different Rds promoter fragments) and 10 μg of pCH110 (used as an internal control) into Y-79 and COS-1 cells. b. Three of the constructs drove significant expression after transfection into human Y-79 retinoblastoma cells (left) while little expression was detected after transfection into COS-1 cells (right). The data shown is an average of three independent experiments, * p<0.01.
High levels of luciferase activity were observed when the pGL2-SV40 plasmid was delivered to Y-79 and COS-1 cells, indicating that these cells can be successfully transfected and the plasmid expressed (data not shown). In Y-79 cells, pLUC468, pLUC1084 and pLUC1439 drove significantly higher levels of luciferase expression than pGL2-Basic, although the level observed in pLUC1084 transfected cells was approximately 33% lower than that in pLUC468 (Fig 3b). The longer pLUC2632 construct promoted much lower level luciferase expression than other constructs; 79% reduction compared to the activity of pLUC468. This strongly suggests the presence of basic promoter activity in the 314 bp fragment (pLUC468), potential negative regulatory element(s) in the region from [-468 to -1084], and additional enhancer elements in the region between [-1084 and -1439]. pLUC2632 exhibited a modest but not statistically significant amount of activity, while pLUC3304 was virtually identical to non-expressing controls (pGL2-basic). The absence of detectable in vitro promoter activity in the longer pLUC3304 constructs suggests that additional elements in the distal regions of the 5′ flanking region may also negatively regulate transcription. In COS-1 cells, only pLUC1439 showed a low level of tissue non-specific promoter activity (Fig. 3b).
Cell type specificity of pLUC468 mediated gene expression
In Y-79 cells, pLUC468 showed the highest promoter activity of all the fragments (12 fold higher than the promoterless construct) but it directed no detectable expression in COS-1 cells. To further explore the cell-type specificity of this region, two additional non-photoreceptor cell lines, NIH3T3 (mouse embryonic fibroblast derived) and MCF-7 (human breast cancer derived) and one additional photoreceptor cell line, 661W (cone photoreceptor derived (Tan, et al., 2004)) were transfected with pLUC468. None of the non-photoreceptor cell lines exhibited any gene expression above baseline when transfected with pLUC468 (Fig. 4a), while transfection in 661W cells resulted in a significant level of luciferase expression.
In addition to the cell-type specificity of the promoter activity, we were also interested in its directional specificity. To that end, we tested the promoter activity of the 314 bp fragment in Y-79 cells when it was inverted (pLUC468IV). As can be seen in Figure 4b, inversion of the promoter fragment abolished its ability to drive luciferase gene expression confirming that the promoter activity is directional.
Effect of photoreceptor developmental transcription factors on Rds expression
Rds is expressed in both rod and cone photoreceptor cells. Sequence analysis of the 3.5kb 5′ flanking region of the mouse Rds gene revealed the presence of several cis-elements known to bind key transcription factors in the development of these two divergent cell types, specifically Crx and Nr2e3. Crx and Nr2e3 are known to regulate transcription with the aid of a third photoreceptor-specific factor, Nrl (Peng, et al., 2005) and some ubiquitous transcription factors such as the Sp zinc finger proteins (Hennig, et al., 2008). Crx and Nr2e3 are known to regulate Rds transcription. Chromatin immunoprecipitation assays demonstrate that Nr2e3 binds to the Rds promoter region and in a mouse model lacking Nr2e3 (rd7/rd7 mice), Rds transcript levels are several fold lower than in wild-type controls (Nystuen, et al., 2008). Similarly, Rds levels are decreased in mice lacking Crx (Crx-/-) (Furukawa, et al., 1997, Livesey, et al., 2000). In spite of the observation that these photoreceptor transcription factors usually work as a group, our cis element analysis did not identify any Nrl sites in the Rds promoter region. To determine whether Nrl might therefore have an indirect regulatory affect on Rds, we co-transfected Y-79 cells with pLUC2632 or pLUC468 and a plasmid containing the Nrl (pNrl) gene under the control of the adenovirus major late promoter (pMT3 vector, a generous gift from Dr. Anand Swaroop, NEI). Co-transfection of the pNrl and pLUC468 resulted in significantly enhanced (∼5 fold) promoter activity compared to single transfected controls (Fig. 5a). Co-transfection with pLUC2632 and pNrl also resulted in a ∼2-fold increase in promoter activity (Fig. 5a). To determine whether these transcription factors might affect Rds expression in vivo, we measured Rds message levels by qRT-PCR in mice that overexpress Nrl (Crx-Nrl transgenics (Oh, et al., 2007)). As can be seen in Figure 5, overexpression of Nrl leads to enhanced expression of Rds transcript (∼3 fold higher than in wild-type animals, Fig. 5b) and protein (higher in wild-type animals, actin shown to verify loading, Fig. 5c). Nr2e3 promotes rod development directly in concert with Nrl while simultaneously suppressing cone differentiation. In Figure 5b, we show that, consistent with previously published results (Nystuen, et al., 2008), the absence of Nr2e3 (Nr2e3-/- mice) yields significantly decreased steady-state Rds transcript levels (∼50% reduction compared to wild-type).
Discussion
In this study, we evaluated the 3.5kb flanking region of the Rds gene due to its structural importance in the photoreceptor outer segment, functional necessity for proper vision, and experimental value in the design of optimized gene therapy vectors. Unlike many other dual-photoreceptor specific genes, it lacks typical TATA and CAAT promoter elements but contains several other regulatory elements. This region is not CpG rich. Notably, several critical photoreceptor transcription factor binding sites were observed on the isolated 3.5kb fragment including OTX (Crx), PNR (Nr2e3), EsrrB, Mef2C and RXR family members. Additional, non-photoreceptor specific regulatory elements were detected including SP1 sites, several E-boxes, one GAGA-box, and several GC-boxes. In vitro analysis of the activity of various promoter fragments contained within the 3.5kb 5′ flanking region indicated that the region from [-154 to -468] possesses the basal, direction-specific promoter activity in retina-derived cell lines (Y-79/661W) but not in any other non-ocular cell lines tested. Although two of the other promoter fragments possessed some activity (pLUC1084, pLUC1439), expression of longer constructs was significantly suppressed in Y-79 cells.
Photoreceptor gene regulation is controlled by a highly conserved network of transcription factors centered on Crx (Hennig, et al., 2008). Crx regulates expression of the Nrl gene which, among other things, regulates expression of Nr2e3. Crx plays a critical role in the differentiation and maintenance of photoreceptor cells, (Chen, et al., 1997, Furukawa, et al., 1997), regulation of tissue specificity, and levels of rod and cone gene expression and photoreceptor development (Boatright, et al., 1997, Kennedy, et al., 1998, Kennedy, et al., 2001, Mani, et al., 2001, Martinez and Barnstable, 1998, Morabito, et al., 1991, Tao, et al., 1993, Whitaker and Knox, 2004, Yu, et al., 1996). Nrl and Nr2e3 work together to promote rod development although Nr2e3 is considered a dual action transcription factor. In the absence of Nr2e3, cone genes are de-repressed and rod genes are suppressed resulting in an enhanced S-cone phenotype (Haider, et al., 2006, Haider, et al., 2009). All three (Crx, Nr2e3, and Nrl) can act together along with ubiquitously expressed transcription factors such as the Sp zinc finger proteins to co-regulate photoreceptor gene expression (Cheng, et al., 2004, Hennig, et al., 2008).
Binding sites for Crx (OTX), Nr2e3 (PNR), and Sp1 were identified in the Rds promoter region however, the specifically modified AP1 sites (Rehemtulla, et al., 1996) to which Nrl binds were not found in the 5′ flanking region of the Rds gene. In spite of this, in vitro results shown here demonstrate that reporter gene expression is increased in cells co-transfected with our pLUC468 Rds construct and pNrl, supporting a role for Nrl in the regulation of Rds. Although Nrl could possibly exert regulatory effects by binding to the various non-modified AP1 sites identified in the 5′ flanking region, the absence of these sites in the region shown to be regulated by Nrl (pLUC468) suggests this is unlikely. It is possible that Nrl regulates Rds expression indirectly by enhancing expression of Nr2e3 or Crx, or by participating in the Crx regulatory complex. It is likewise possible that Nrl regulates Rds expression by binding to an as yet unidentified sequence. The lack of a predicted cognate binding site does not preclude transcription factor binding; indeed many photoreceptor gene regulatory elements have been identified after being missed by standard sequence comparisons (Hsiau, et al., 2007, Peng and Chen, 2005). Support for the idea that Nrl regulates Rds comes from our observation that in the Nrl over-expressing all-rod retina (Crx-Nrl) Rds message levels are significantly increased by approximately 3 fold. As the murine retina is only 3% cones, this dramatic increase in Rds message (and protein) cannot be accounted for by the small increase in Rds which would be expected to accompany the conversion of cones to rods.
Historically, Nr2e3 and Nrl were considered the only rod-specific transcription factors. However, recent evidence has suggested that two more photoreceptor transcription factors may be rod-enhanced: Mef2C and EsrrB (Hsiau, et al., 2007). Both of these transcription factors have binding sites in the 5′ flanking region of Rds and may play a role in Rds expression in rods. The transcription factor RxrG, which interacts with the RXR cis-element (found in the 5′ flanking region of Rds), is required for some cone gene transcription (Hennig, et al., 2008, Roberts, et al., 2006, Yoshida, et al., 2004), is upregulated in Nrl-/- mice (Hsiau, et al., 2007) and may positively regulate Rds expression in cones. However, RXR/RAR cis elements, and Mef2C binding sites were found in all promoter regions examined, so it is difficult to make conclusions on their role in the regulation of Rds specifically.
For most photoreceptor genes, the maximal promoter activity and cell type specificity resides within the first 500 bp of the 5′ flanking region (Fujimaki, et al., 2004, Mohamed, et al., 1998, Pittler, et al., 2004, Young, et al., 2003). This appears to be the case for the Rds promoter: we observed strongest activity with our pLUC468 construct. This region contains the only highly conserved cis-element among the species we examined, the OTX element (conserved in the X. laevis Rds promoter region (Moritz, et al., 2002)) which will likely be recognized by Crx thus transactivating the promoter activity. There appear to be suppressors in the regions between [-468 to -1084] and [-1439 to- 2632] and a positive regulator or enhancer between [-1084 to -1439]. The pLUC1084 construct (which showed 33% less activity than pLUC468) contains one AP1 site and one E-box. The increased promoter activity conferred by inclusion of the region between [-1084 and -1439] (i.e. pLUC1439 vs. pLUC1084) is likely due to positive regulation by Nr2e3 (PNR) and possibly Mef2C and CAR/RXR. However, additional PNR, RXR, and Mef2C sites are found more distally in regions of the 5′ flanking sequence which demonstrated suppressed transcription (e.g. within pLUC2632 and pLUC3304), so regulation is clearly not straightforward and additional interpretation will require further experiments. In some cases, gene expression is inversely correlated with plasmid size (Kreiss, et al., 1999, Yin, et al., 2005), however not always (Maucksch, et al., 2009). In this case, however, lack of expression with the larger constructs is not likely to be due primarily to increased plasmid size. In experiments designed specifically to test the effects of plasmid size on gene expression, gene expression decreased in a consistent, near-linear fashion over the range of plasmid sizes comparable to what we use here (Yin, et al., 2005). In contrast, expression from our constructs does not steadily decrease as the plasmid size increases. In fact, expression is higher for pLUC1439 than the smaller pLUC1084 and then drops precipitously for pLUC2632.
Gene therapy studies require a well-characterized promoter fragment to reliably drive high levels of tissue specific gene expression. Several photoreceptor specific promoters have been studied including those for rod opsin, cone opsin, Crx, and IRBP. However, the preponderance of inherited retinal diseases associated with mutations in Rds coupled with RDS' unique expression pattern (expressed at high levels in rods, and substantial but lesser levels in cones (Farjo, et al., 2006)) makes characterization of the endogenous Rds promoter a useful step for our progress in developing gene based therapeutics. Furthermore, our experiments demonstrating that co-delivery of constructs containing positive regulators of transcription (including Nrl) and therapeutic constructs containing the Rds promoter may help increase expression of the therapeutic gene. Since Nrl is thought to have a positive regulatory role on the expression of many rod genes, it is also possible that delivery of Nrl alone could improve the phenotype of rod-dominant eye diseases that are associated with haploinsufficiency in various genes. Development of a genetic therapy that is not specific to any particular gene has been an area of interest and would be a great advantage for the treatment of multiple retinal degenerative conditions.
Our results suggest that the 314 bp or 1,285 bp regions contained in pLUC468 and pLUC1439 drive the best expression in vitro and are likely the best choices for incorporation into photoreceptor-targeting vectors. Often promoter activity in vitro does not correlate well with activity in vivo, and future studies will involve the use of these constructs in vivo. Our data suggest that Rds is regulated by the same core Crx-based photoreceptor regulatory network as many other photoreceptor specific genes. The presence of binding sites for both cone and rod transcription factors may help explain the differential expression pattern of Rds in rods and cones.
Supplementary Material
Acknowledgments
This work was supported by the National Eye Institute (EY10609-MIN, EY018656-MIN, EY14052-MRA, EY018512-SMC), and the Foundation Fighting Blindness (MIN, MRA).
Abbreviations
- RDS
retinal degeneration slow
- RXR
retinoid X receptor
- RTR
Retinoid receptor-related testis-associated receptor
- Crx
cone-rod homeobox
- Nrl
neural retinal leucine zipper
- Nr2e3
orphan nuclear receptor subfamily 2, group E, member 3
- Otx2
orthodenticle homeobox 2
- RxrG
retinoid X receptor γ
- CAR
constitutive androstane receptor
- VDR
vitamin D receptor
- RAR
retinoic acid receptor
- PXR
pregnane X receptor
- PNR
photoreceptor cell-specific nuclear receptor
- OTX
orthodenticle homeobox
- Mef2c
myocyte enhancer factor 2C
- Esrrb
estrogen-related receptor β
- NF1
nuclear factor 1
- AP1
activator protein 1
- SP1
specificity protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acland GM, Aguirre GD, Bennett J, Aleman TS, Cideciyan AV, Bennicelli J, Dejneka NS, Pearce-Kelling SE, Maguire AM, Palczewski K, Hauswirth WW, Jacobson SG. Long-term restoration of rod and cone vision by single dose rAAV-mediated gene transfer to the retina in a canine model of childhood blindness. Mol Ther. 2005;12:1072–82. doi: 10.1016/j.ymthe.2005.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acland GM, Aguirre GD, Ray J, Zhang Q, Aleman TS, Cideciyan AV, Pearce-Kelling SE, Anand V, Zeng Y, Maguire AM, Jacobson SG, Hauswirth WW, Bennett J. Gene therapy restores vision in a canine model of childhood blindness. Nat Genet. 2001;28:92–5. doi: 10.1038/ng0501-92. [DOI] [PubMed] [Google Scholar]
- Ali RR, Sarra GM, Stephens C, Alwis MD, Bainbridge JW, Munro PM, Fauser S, Reichel MB, Kinnon C, Hunt DM, Bhattacharya SS, Thrasher AJ. Restoration of photoreceptor ultrastructure and function in retinal degeneration slow mice by gene therapy. Nat Genet. 2000;25:306–10. doi: 10.1038/77068. [DOI] [PubMed] [Google Scholar]
- Allocca M, Mussolino C, Garcia-Hoyos M, Sanges D, Iodice C, Petrillo M, Vandenberghe LH, Wilson JM, Marigo V, Surace EM, Auricchio A. Novel adeno-associated virus serotypes efficiently transduce murine photoreceptors. J Virol. 2007;81:11372–80. doi: 10.1128/JVI.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N, Petersen-Jones S, Bhattacharya SS, Thrasher AJ, Fitzke FW, Carter BJ, Rubin GS, Moore AT, Ali RR. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–9. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Berson EL. Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1993;34:1659–76. [PubMed] [Google Scholar]
- Boatright JH, Borst DE, Peoples JW, Bruno J, Edwards CL, Si JS, Nickerson JM. A major cis activator of the IRBP gene contains CRX-binding and Ret-1/PCE-I elements. Mol Vis. 1997;3:15. [PubMed] [Google Scholar]
- Cai X, Conley SM, Nash Z, Fliesler SJ, Cooper MJ, Naash MI. Gene delivery to mitotic and postmitotic photoreceptors via compacted DNA nanoparticles results in improved phenotype in a mouse model of retinitis pigmentosa. FASEB J. 2009a doi: 10.1096/fj.09-139147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Nash Z, Conley SM, Fliesler SJ, Cooper MJ, Naash MI. A partial structural and functional rescue of a retinitis pigmentosa model with compacted DNA nanoparticles. PLoS ONE. 2009b;4:e5290. doi: 10.1371/journal.pone.0005290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–30. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum Mol Genet. 2006;15:2588–602. doi: 10.1093/hmg/ddl185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–75. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Aleman TS, Boye SL, Schwartz SB, Kaushal S, Roman AJ, Pang JJ, Sumaroka A, Windsor EA, Wilson JM, Flotte TR, Fishman GA, Heon E, Stone EM, Byrne BJ, Jacobson SG, Hauswirth WW. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc Natl Acad Sci U S A. 2008;105:15112–7. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Polo A, Farber DB. Rod photoreceptor-specific gene expression in human retinoblastoma cells. Proc Natl Acad Sci U S A. 1995;92:4016–20. doi: 10.1073/pnas.92.9.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo R, Skaggs JS, Nagel BA, Quiambao AB, Nash ZA, Fliesler SJ, Naash MI. Retention of function without normal disc morphogenesis occurs in cone but not rod photoreceptors. J Cell Biol. 2006;173:59–68. doi: 10.1083/jcb.200509036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimaki T, Huang ZY, Kitagawa H, Sakuma H, Murakami A, Kanai A, McLaren MJ, Inana G. Truncation and mutagenesis analysis of the human X-arrestin gene promoter. Gene. 2004;339:139–47. doi: 10.1016/j.gene.2004.06.032. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–41. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Glushakova LG, Timmers AM, Issa TM, Cortez NG, Pang J, Teusner JT, Hauswirth WW. Does recombinant adeno-associated virus-vectored proximal region of mouse rhodopsin promoter support only rod-type specific expression in vivo? Mol Vis. 2006;12:298–309. [PubMed] [Google Scholar]
- Haider NB, Demarco P, Nystuen AM, Huang X, Smith RS, McCall MA, Naggert JK, Nishina PM. The transcription factor Nr2e3 functions in retinal progenitors to suppress cone cell generation. Vis Neurosci. 2006;23:917–29. doi: 10.1017/S095252380623027X. [DOI] [PubMed] [Google Scholar]
- Haider NB, Mollema N, Gaule M, Yuan Y, Sachs AJ, Nystuen AM, Naggert JK, Nishina PM. Nr2e3-directed transcriptional regulation of genes involved in photoreceptor development and cell-type specific phototransduction. Exp Eye Res. 2009 doi: 10.1016/j.exer.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L, Conlon TJ, Boye SL, Flotte TR, Byrne BJ, Jacobson SG. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19:979–90. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–33. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One. 2007;2:e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Boye SL, Aleman TS, Conlon TJ, Zeiss CJ, Roman AJ, Cideciyan AV, Schwartz SB, Komaromy AM, Doobrajh M, Cheung AY, Sumaroka A, Pearce-Kelling SE, Aguirre GD, Kaushal S, Maguire AM, Flotte TR, Hauswirth WW. Safety in nonhuman primates of ocular AAV2-RPE65, a candidate treatment for blindness in Leber congenital amaurosis. Hum Gene Ther. 2006;17:845–58. doi: 10.1089/hum.2006.17.845. [DOI] [PubMed] [Google Scholar]
- Keen TJ, Inglehearn CF. Mutations and polymorphisms in the human peripherin-RDS gene and their involvement in inherited retinal degeneration. Hum Mutat. 1996;8:297–303. doi: 10.1002/(SICI)1098-1004(1996)8:4<297::AID-HUMU1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kennedy BN, Goldflam S, Chang MA, Campochiaro P, Davis AA, Zack DJ, Crabb JW. Transcriptional regulation of cellular retinaldehyde-binding protein in the retinal pigment epithelium. A role for the photoreceptor consensus element. J Biol Chem. 1998;273:5591–8. doi: 10.1074/jbc.273.10.5591. [DOI] [PubMed] [Google Scholar]
- Kennedy BN, Vihtelic TS, Checkley L, Vaughan KT, Hyde DR. Isolation of a zebrafish rod opsin promoter to generate a transgenic zebrafish line expressing enhanced green fluorescent protein in rod photoreceptors. J Biol Chem. 2001;276:14037–43. doi: 10.1074/jbc.M010490200. [DOI] [PubMed] [Google Scholar]
- Khani SC, Pawlyk BS, Bulgakov OV, Kasperek E, Young JE, Adamian M, Sun X, Smith AJ, Ali RR, Li T. AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Invest Ophthalmol Vis Sci. 2007;48:3954–61. doi: 10.1167/iovs.07-0257. [DOI] [PubMed] [Google Scholar]
- Koike C, Nishida A, Ueno S, Saito H, Sanuki R, Sato S, Furukawa A, Aizawa S, Matsuo I, Suzuki N, Kondo M, Furukawa T. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007;27:8318–29. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–48. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiss P, Cameron B, Rangara R, Mailhe P, Aguerre-Charriol O, Airiau M, Scherman D, Crouzet J, Pitard B. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 1999;27:3792–8. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Meur G, Stieger K, Smith AJ, Weber M, Deschamps JY, Nivard D, Mendes-Madeira A, Provost N, Pereon Y, Cherel Y, Ali RR, Hamel C, Moullier P, Rolling F. Restoration of vision in RPE65-deficient Briard dogs using an AAV serotype 4 vector that specifically targets the retinal pigmented epithelium. Gene Ther. 2006 doi: 10.1038/sj.gt.3302861. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–10. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- Lotery AJ, Yang GS, Mullins RF, Russell SR, Schmidt M, Stone EM, Lindbloom JD, Chiorini JA, Kotin RM, Davidson BL. Adeno-associated virus type 5: transduction efficiency and cell-type specificity in the primate retina. Hum Gene Ther. 2003;14:1663–71. doi: 10.1089/104303403322542301. [DOI] [PubMed] [Google Scholar]
- Mani SS, Batni S, Whitaker L, Chen S, Engbretson G, Knox BE. Xenopus rhodopsin promoter. Identification of immediate upstream sequences necessary for high level, rod-specific transcription. J Biol Chem. 2001;276:36557–65. doi: 10.1074/jbc.M101685200. [DOI] [PubMed] [Google Scholar]
- Martinez JA, Barnstable CJ. Erx, a novel retina-specific homeodomain transcription factor, can interact with Ret 1/PCEI sites. Biochem Biophys Res Commun. 1998;250:175–80. doi: 10.1006/bbrc.1998.9261. [DOI] [PubMed] [Google Scholar]
- Maucksch C, Bohla A, Hoffmann F, Schleef M, Aneja MK, Elfinger M, Hartl D, Rudolph C. Transgene expression of transfected supercoiled plasmid DNA concatemers in mammalian cells. J Gene Med. 2009;11:444–53. doi: 10.1002/jgm.1310. [DOI] [PubMed] [Google Scholar]
- Mohamed MK, Taylor RE, Feinstein DS, Huang X, Pittler SJ. Structure and upstream region characterization of the human gene encoding rod photoreceptor cGMP phosphodiesterase alpha-subunit. J Mol Neurosci. 1998;10:235–50. doi: 10.1007/BF02761777. [DOI] [PubMed] [Google Scholar]
- Morabito MA, Yu X, Barnstable CJ. Characterization of developmentally regulated and retina-specific nuclear protein binding to a site in the upstream region of the rat opsin gene. J Biol Chem. 1991;266:9667–72. [PubMed] [Google Scholar]
- Moritz OL, Peck A, Tam BM. Xenopus laevis red cone opsin and Prph2 promoters allow transgene expression in amphibian cones, or both rods and cones. Gene. 2002;298:173–82. doi: 10.1016/s0378-1119(02)00923-x. [DOI] [PubMed] [Google Scholar]
- Naash MI, Hollyfield JG, al-Ubaidi MR, Baehr W. Simulation of human autosomal dominant retinitis pigmentosa in transgenic mice expressing a mutated murine opsin gene. Proc Natl Acad Sci U S A. 1993;90:5499–503. doi: 10.1073/pnas.90.12.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour M, Ding XQ, Stricker H, Fliesler SJ, Naash MI. Modulating expression of peripherin/rds in transgenic mice: critical levels and the effect of overexpression. Invest Ophthalmol Vis Sci. 2004;45:2514–21. doi: 10.1167/iovs.04-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystuen AM, Sachs AJ, Yuan Y, Heuermann L, Haider NB. A novel mutation in Prph2, a gene regulated by Nr2e3, causes retinal degeneration and outer-segment defects similar to Nr2e3 (rd7/rd7) retinas. Mamm Genome. 2008;19:623–33. doi: 10.1007/s00335-008-9138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh EC, Khan N, Novelli E, Khanna H, Strettoi E, Swaroop A. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci U S A. 2007;104:1679–84. doi: 10.1073/pnas.0605934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–64. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci. 2005;22:575–86. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- Pittler SJ, Zhang Y, Chen S, Mears AJ, Zack DJ, Ren Z, Swain PK, Yao S, Swaroop A, White JB. Functional analysis of the rod photoreceptor cGMP phosphodiesterase alpha-subunit gene promoter: Nrl and Crx are required for full transcriptional activity. J Biol Chem. 2004;279:19800–7. doi: 10.1074/jbc.M401864200. [DOI] [PubMed] [Google Scholar]
- Rath MF, Munoz E, Ganguly S, Morin F, Shi Q, Klein DC, Moller M. Expression of the Otx2 homeobox gene in the developing mammalian brain: embryonic and adult expression in the pineal gland. J Neurochem. 2006;97:556–66. doi: 10.1111/j.1471-4159.2006.03773.x. [DOI] [PubMed] [Google Scholar]
- Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci U S A. 1996;93:191–5. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA. Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci U S A. 2006;103:6218–23. doi: 10.1073/pnas.0509981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal N. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 1987;152:704–20. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- Tan E, Ding XQ, Saadi A, Agarwal N, Naash MI, Al-Ubaidi MR. Expression of cone-photoreceptor-specific antigens in a cell line derived from retinal tumors in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45:764–8. doi: 10.1167/iovs.03-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Pandey S, Simon MI, Fong HK. Structure of the bovine transducin gamma subunit gene and analysis of promoter function in transgenic mice. Exp Eye Res. 1993;56:497–507. doi: 10.1006/exer.1993.1063. [DOI] [PubMed] [Google Scholar]
- Weber M, Rabinowitz J, Provost N, Conrath H, Folliot S, Briot D, Cherel Y, Chenuaud P, Samulski J, Moullier P, Rolling F. Recombinant adeno-associated virus serotype 4 mediates unique and exclusive long-term transduction of retinal pigmented epithelium in rat, dog, and nonhuman primate after subretinal delivery. Mol Ther. 2003;7:774–81. doi: 10.1016/s1525-0016(03)00098-4. [DOI] [PubMed] [Google Scholar]
- Whitaker SL, Knox BE. Conserved transcriptional activators of the Xenopus rhodopsin gene. J Biol Chem. 2004;279:49010–8. doi: 10.1074/jbc.M406080200. [DOI] [PubMed] [Google Scholar]
- Yin W, Xiang P, Li Q. Investigations of the effect of DNA size in transient transfection assay using dual luciferase system. Anal Biochem. 2005;346:289–94. doi: 10.1016/j.ab.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, Hero AO, Swaroop A. Expression profiling of the developing and mature Nrl-/- mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet. 2004;13:1487–503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- Young JE, Vogt T, Gross KW, Khani SC. A short, highly active photoreceptor-specific enhancer/promoter region upstream of the human rhodopsin kinase gene. Invest Ophthalmol Vis Sci. 2003;44:4076–85. doi: 10.1167/iovs.03-0197. [DOI] [PubMed] [Google Scholar]
- Yu X, Leconte L, Martinez JA, Barnstable CJ. Ret 1, a cis-acting element of the rat opsin promoter, can direct gene expression in rod photoreceptors. J Neurochem. 1996;67:2494–504. doi: 10.1046/j.1471-4159.1996.67062494.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.