Abstract
Excessive and chronic alcohol intake leads to a lower hepatic vitamin A status by interfering with vitamin A metabolism. Dietary provitamin A carotenoids can be converted into vitamin A mainly by carotenoid 15,15’-monooxygenase 1 (CMO1) and, to a lesser degree, carotenoid 9′10’-monooxygenase 2 (CMO2). CMO1 has been shown to be regulated by several transcription factors, such as the PPAR, retinoid X receptor, and thyroid receptor (TR). The regulation of CMO2 has yet to be identified. The impact of chronic alcohol intake on hepatic expressions of CMO1 and CMO2 and their related transcription factors are unknown. In this study, Fischer 344 rats were pair-fed either a liquid ethanol Lieber-DeCarli diet (n = 10) or a control diet (n = 10) for 11 wk. Hepatic retinoid concentration and expressions of CMO1, CMO2, PPARγ, PPARα, and TRβ as well as plasma thyroid hormones levels were analyzed. We observed that administering alcohol decreased hepatic retinoid levels but increased mRNA concentrations of CMO1, CMO2, PPARγ, PPARα, and TRβ and upregulated protein levels of CMO2, PPARγ, and PPARα. There was a positive correlation of PPARγ with CMO1 (r = 0.89; P < 0.0001) and both PPARγ and PPARα with CMO2 (r = 0.72, P < 0.001 and r = 0.62, P < 0.01, respectively). Plasma thyroid hormone concentrations did not differ between the control rats and alcohol-fed rats. This study suggests that chronic alcohol intake significantly upregulates hepatic expression of CMO1 and, to a much lesser extent, CMO2. This process may be due to alcohol-induced PPARγ expression and lower vitamin A status in the liver.
Introduction
Excessive and chronic alcohol intake is known to interfere with endocrine system functions, creating a hormonal and metabolic imbalance (1–3). Low vitamin A nutritional status is one of the major alterations caused by chronic alcohol intake. Vitamin A is stored mainly in the liver in the form of retinyl esters, which can undergo hydrolysis to retinol, the circulating form in the body. Substantial work has been done investigating the mechanisms by which excessive alcohol intake interferes with retinoid metabolism and signaling (4). More specifically, alcohol acts as a competitive inhibitor of vitamin A oxidation to retinoic acid involving alcohol dehydrogenases and acetaldehyde dehydrogenases, induces cytochrome P450 enzymes [particularly cytochrome P450 2E1 (CYP2E1)5] that degrade retinol and retinoic acid, and alters retinoid homeostasis by increasing vitamin A mobilization from the liver to extrahepatic tissues. Nutritional interventions that restore normal vitamin A status may offer protection at the cellular level to modify alcohol-related illness in high-risk human populations.
Vitamin A can be consumed directly from the diet, usually in the form of retinol or retinyl esters from meat and dairy foods. It can be produced by enzymatic cleavage of provitamin A carotenoids (β-carotene, α-carotene, and β-cryptoxanthin), which can be absorbed through the intestines and accumulate in the liver and other tissues of the human body. Recently, 2 different carotenoid monooxygenases, carotenoid 15,15’-monooxygenase 1 (CMO1) and carotenoid 9′10’-monooxygenase 2 (CMO2), were molecularly identified (5, 6). Both belong to a family of structurally related nonheme iron oxygenases (7, 8). The most common carotenoid substrate for CMO1 is β-carotene, which is cleaved in the central carbon 15,15’-double bond to produce retinal (9). CMO2 catalyzes the excentric oxidative cleavage of β-carotene at the C9′, C10’ double bond (6, 10) to form β-apo–carotenals, which can be oxidized to β-apo–carotenoic acids and then further oxidized through a β-oxidation–like process to form retinoic acid (11). High expressions of CMO1 and CMO2 in the liver may be related to hepatic metabolism of both provitamin A carotenoids and nonprovitamin A carotenoids (5, 6, 10, 12). Although the significance of CMO2 in bioconversion of provitamin A carotenoids into vitamin A has not been well defined, a recent study demonstrates that hepatic vitamin A levels were significantly lower in cows with a CMO2 mutation. This indicates that CMO2 is a key regulator of β-carotene metabolism (13).
CMO1 has been identified as a PPARγ target gene in mice. This is determined by identifying a peroxisomal proliferator response element (PPRE) in the promoter region of the CMO1 gene, which is a recognition site for PPARγ (14). Some authors report that CMO1 is transcriptionally regulated by the action of PPAR and retinoid X receptors (RXR) in both mice and humans (14, 15). RXR plays a central role in diverse biologic pathways by serving as an obligate heterodimeric partner for multiple steroid hormone nuclear receptors. These include PPAR, thyroid hormone receptor (TR), vitamin D receptor, and others (16). In addition, Yamaguchi and Suruga (17) showed that CMO1 is upregulated by thyroid hormones in human small intestinal-like Caco-2 cells. This suggests that these hormones contribute to the absorption and metabolism of vitamin A. The regulation of the CMO2 gene has yet to be identified, but it is conceivable that nonprovitamin A carotenoids may play a critical role in the transcriptional regulation of CMO2 (10, 18).
Daily consumption of alcohol is related to reduced β-carotene levels (19, 20). Lower vitamin A levels in the liver during chronic alcohol consumption are also well documented (21). It is acknowledged that alcohol activates nuclear transcription factors involved in the regulation of hepatic gene expression (22); however, direct evidence regarding the effects of alcohol on carotenoid cleavage enzymes and ingested β-carotene to vitamin A is still lacking. Considering that CMO1 can be regulated by PPAR and TR and alcohol can modulate PPAR and TR, we investigated the impact of chronic alcohol intake on expressions of CMO1, CMO2, PPAR, and TRβ in the livers of rats.
Materials and Methods
Rats and experimental protocol.
Two-month-old male Fischer 344 rats (weighing ∼130–140 g) were obtained from Charles River Laboratories and were housed in individual cages in an American Association of Accreditation of Laboratory Animal Care-accredited animal facility at the Human Nutrition Research Center on Aging at Tufts University. Their room was under controlled temperature (20–22°C), humidity (45–55%), and lighting (12-h-light/-dark cycle). The rats were acclimatized over 1 wk to a nonpurified diet (23) and then randomly divided into 2 groups: control (n = 10) and alcohol (n = 10). The alcohol intake in rats (36% of total energy) was approximately equal to the consumption of 100 g/d of alcohol (7.1 kcal/g alcohol; 1 kcal = 4.184 kJ) in a 2000-kcal human diet. The rats were adapted to the nutritionally adequate Lieber-DeCarli liquid control and alcohol diets (Dyets) (24) over a 2-wk period. In the control diet, isocaloric maltodextrin was the substitute for ethanol. The Lieber-DeCarli liquid diet contains vitamin A (0.9 mg/1000 kcal), which is a sufficient amount for both the control and experimental groups (25). During the experimental period, the rats were group pair-fed to the alcohol group and were given ~70 mL/d of the diet. Body weight was recorded once per week. The diets were prepared twice per week and were stored at 4°C. After 11 wk of treatment, the rats were killed by terminal exsanguination under deep anesthesia. Blood and livers were collected and stored at −80°C until required for analysis. This experimental protocol was reviewed and approved by the Animal Care and Use Committee at the Human Nutrition Research Center on Aging at Tufts University.
HPLC analysis.
Liver sample extractions were done as described previously (26). The extracts were evaporated under N2 gas and resuspended in 50 μL ethanol for injection into the HPLC system, as described (27). Individual retinoids were identified by coelution with standards and absorption spectrum analysis and quantified by determining peak areas calibrated against known amounts of standards.
Total RNA isolation and RT.
Total RNA was extracted from liver tissue using the TriPure reagent (Roche Applied Science) following the manufacturer’s protocol. Then 400 ng of RNA was used for the synthesis of 20 μL of cDNA by Primer Random p(dN)6 and Moloney Murine Leukemia Virus RT (Invitrogen).
Real-time PCR.
CMO1, CMO2, PPARγ, PPARα, and TRβ mRNA levels were determined by real-time PCR. Primers were designed using the Primer Express version 2.0 (Applied Biosystems) software. The sequences for CMO1 (NCBI reference sequence: NM_053648.2) were: forward, 5′-GCCAACCTGAACAAGGACTTCG-3′ and reverse, 5′-AGCCCACTTCTGCATCCTTGTC-3′; for CMO2 (NCBI reference sequence: DQ083174.1): forward, 5′-CATGTCAAGGTTTGAGCCACCT-3′ and reverse, 5′-ACGAATTTGCTCCAGTCCACC-3′; for PPARγ (NCBI reference sequence: NM_013124.1): forward, 5′-GGAAAAAACCCTTGCATCCTTC-3′ and reverse, 5′-TTCAAACTCCCTCATGGCCA-3′; for PPARα (NCBI reference sequence: NM_013196.1): forward, 5′-ACTAGCAACAATCCGCCTTTTG-3′ and reverse, 5′-GGACCTCTGCCTCCTTGTTTTC-3′; and for TRβ (NCBI reference sequence: NM_012672.2): forward, 5′-CTACCTCTCTGCATTCGGTCTG-3′ and reverse, 5′-AGGTCTGTTGCCATGCCAA-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (NCBI reference sequence: NM_017008.2) was used as internal control and the sequences were: forward, 5′-AGTGCCAGCCTCGTCTCATAG-3′ and reverse, 5′-CCTTGACTGTGCCGTTGAACT-3′. Quantitative measurement was performed using the SYBR Green qPCR kit (Invitrogen) according to the manufacturer’s instructions on an Applied Biosystems 7000 sequence detection system. The real-time cycler conditions were as follows: enzyme activation at 50°C for 2 min and after denaturation at 95°C for 10 min, the cDNA products were amplified with 40 cycles, each cycle consisting of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. Product purity was confirmed by dissociation curve analysis. Gene expression was quantified relative to the values of the control group after adjusting for GAPDH by the 2−ΔΔCT method as described previously (28).
Western blotting.
Protein abundance of the CMO2, PPARγ, and PPARα were assessed by Western blotting. For CMO2, liver tissues were homogenized with ice-cold, whole-cell lysate buffer containing inhibitors. Nuclear extraction of liver tissue was used for PPARγ and PPARα. Protein concentration was quantified spectrophotometrically (Bio-Spec 1601) by using Coomassie Blue. Protein samples (50 μg/lane) were separated by PAGE using 10% SDS-polyacrylamide gels. Samples were transferred to nitrocellulose membranes (Immobilon-P transfer membrane, Millipore) and blocked with 5% milk. The membrane was incubated with a CMO2 primary antibody against ferret CMO2, which was developed in our laboratory and cross-reacts with human and rat CMO2 (10), for 1.5 h at room temperature followed by the secondary antibody (against rabbit) for 30 min at room temperature. PPARγ (1:250, Santa Cruz) and PPARα (1:250, Santa Cruz) membranes were incubated with a primary antibody overnight at 4°C, followed by secondary antibody for 1 h at room temperature. For Western-blot analysis, GAPDH (1:5000, Chemicon) was used as an internal loading control. Immunoreactive bands were visualized and quantified using an imaging densitometry (GS-710, Imaging Densitometer; Bio-Rad Laboratories).
Free triiodothyronine, free thyroxine, and thyroid stimulating hormone dosage.
Plasma concentrations of free triiodothyronine (fT3), free thyroxine (fT4), and thyroid stimulating hormone (TSH) were measured by ELISA kits (Bio-Quant) following the manufacturer’s protocol.
Statistical analysis.
Sigma Stat (version 3.5) software was used for statistical analysis. Student’s t test was used for data analysis. To evaluate the association among CMO1, CMO2, PPARγ, PPARα, and TRβ, Pearson correlation coefficient was determined. Differences between means were considered significant at P < 0.05. Results are expressed as means ± SEM.
Results
Body and liver weights.
After 11 wk of treatment, the alcohol-fed rats had lower body (372.8 ± 13.1 g) and liver weights (11.59 ± 0.14 g; P < 0.05) than the control groups (429.7 ± 5.5 g and 12.69 ± 0.35 g, respectively; P < 0.05), as previously documented (29). The ratio of liver weight:body weight did not differ between the groups (ranging from 0.029 to 0.031), indicating that the decreased liver weight associated with the decreased body weight in the alcohol-fed rats.
Hepatic concentrations of retinoids.
Compared with controls, the hepatic concentration of retinoids was significantly lower in the ethanol-fed rats (Table 1). Ethanol feeding of rats for 11 wk resulted in lower hepatic retinoic acid, retinol, and retinyl palmitate concentrations by ∼62, 33, and 42%, respectively, compared with rats not fed ethanol.
TABLE 1.
Hepatic concentrations of retinoic acid, retinol, and retinyl palmitate in rats fed control diet or 36% ethanol diet for 11 wk1
| Treatment group | Retinoic acid | Retinol | Retinyl palmitate |
| nmol/g | |||
| Control | 0.243 ± 0.074 | 34.1 ± 4.97 | 719 ± 87.7 |
| Ethanol | 0.092 ± 0.013* | 22.9 ± 3.27* | 423 ± 54.7* |
Values are means ± SD, = 10. *Different from control, P < 0.01.
Hepatic expressions of CMO1 and CMO2.
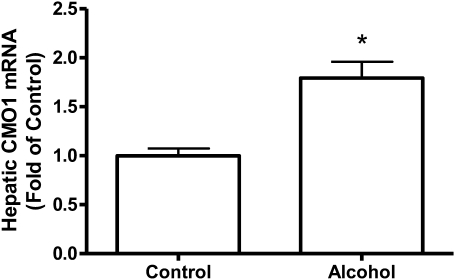
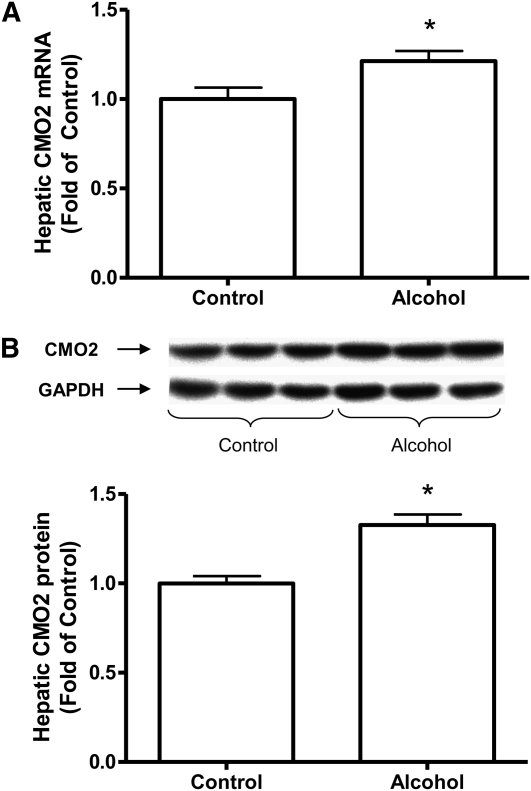
Chronic consumption of alcohol as 36% of total energy intake for 11 wk significantly increased the expression of CMO1 mRNA (P < 0.05) (Fig. 1). We were not able to measure the protein level of CMO1 due to lack of a specific antibody against CMO1. Ethanol also significantly increased the mRNA (Fig. 2A) and protein levels (Fig. 2B) for CMO2 (P < 0.05) but to a much lesser degree then CMO1. In the CMO2, the mRNA expression was 21% higher and the protein levels were 32% higher in the ethanol-fed rats compared with the nonalcohol-fed rats.
FIGURE 1.
Hepatic CMO1 mRNA expression in rats fed control diet or 36% ethanol diet for 11 wk. Values are means ± SEM, n = 10. *Different from control, P < 0.05.
FIGURE 2.
Hepatic CMO2 mRNA expression (A) and protein expression (B) in rats fed control diet or 36% ethanol diet for 11 wk. Values are means ± SEM, n = 10. *Different from control, P < 0.05.
Hepatic expressions of PPAR.
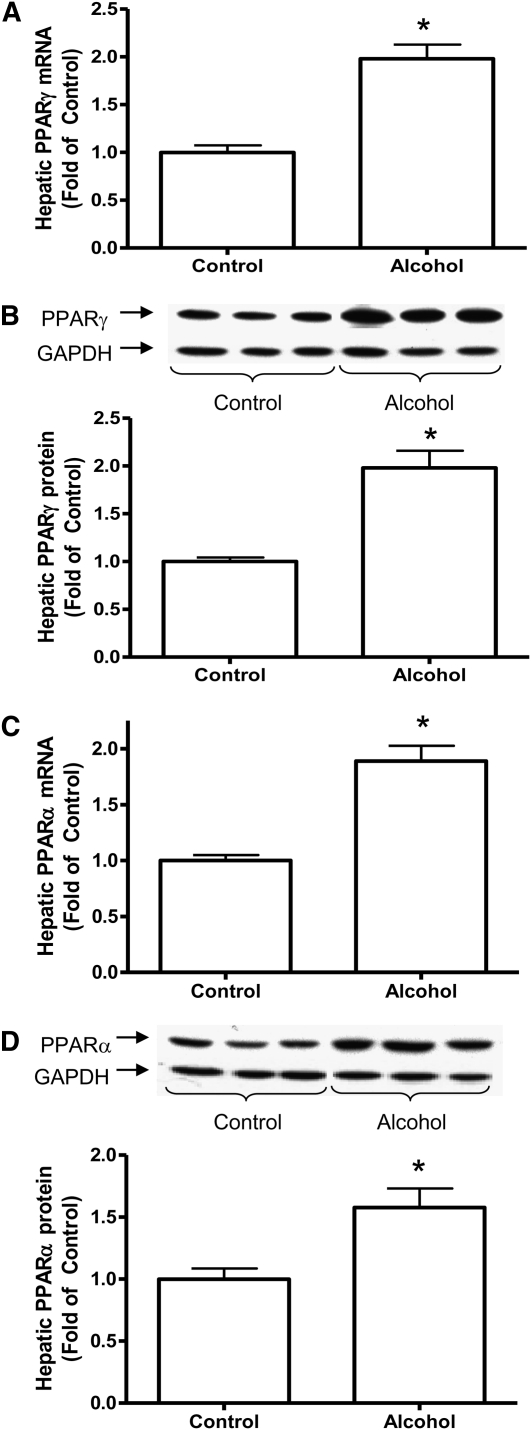
Given that CMO1 is regulated by PPAR, we then examined hepatic PPAR expressions in alcohol-fed rats. Alcohol intake significantly upregulated both mRNA (Fig. 3A) and protein expressions of PPARγ and PPARα (Fig. 3B) compared with the control group of rats.
FIGURE 3.
Hepatic PPARγ expressions [mRNA levels (A) and protein levels (B)] and PPARα expressions [mRNA levels (C) and protein levels (D)] in rats fed control diet or 36% ethanol diet for 11 wk. Inset: Representative Western blot for protein expression. Values are means ± SEM, n = 10. *Different from control, P < 0.05.
Plasma concentrations of fT3, fT4, and TSH and hepatic expression of TR#x03B2.
Because prolonged alcohol ingestion led to a significant reduction of plasma levels of fT3 and fT4 in rats compared with pair-fed controls (30, 31) and CMO1 was found to be upregulated by T3 (30), we examined plasma concentrations of fT3, fT4, and TSH as well as hepatic TRβ gene expression. Plasma concentrations of fT3 (2.87 ± 0.69 ng/L vs. 3.03 ± 0.57 ng/L), fT4 (0.303 ± 0.027 ng/L vs. 0.298 ± 0.025 ng/L), and TSH (1.25 ± 0.15 mIU/L vs. 1.44 ± 0.24 mIU/L; P = 0.06) did not differ significantly between the control and alcohol-fed groups. Alcohol modestly increased hepatic TRβ gene expression to 1.38-fold that of controls (P < 0.05).
Correlations.
We examined correlations between the studied genes using Pearson correlation coefficient analysis. We found a correlation between CMO1 and CMO2 (r = 0.65; P < 0.01) in the liver, suggesting that both genes were affected by alcohol consumption. We found a strong positive correlation for PPARγ compared with CMO1 (r = 0.89; P < 0.0001) but no correlation for PPARα compared with CMO1 (r = 0.36). Interestingly, we observed a correlation for both PPARγ compared with CMO2 (r = 0.72; P < 0.001) and PPARα compared with CMO2 (r = 0.62; P < 0.01) in the liver. In addition, hepatic TRβ expression correlated positively to CMO1 (r = 0.61; P < 0.05) but not to CMO2 (r = 0.27).
Discussion
Because the liver is the major organ that expresses high levels of CMO1 and accumulates both provitamin A carotenoids and vitamin A, we raised the question of whether the conversion of provitamin A carotenoids into vitamin A by CMO-1 is impaired by chronic alcohol intake. In this study, we show that chronic alcohol intake significantly upregulates hepatic expressions of CMO-1. Because PPRE has been identified in the promoter region of the CMO1 gene (14) and is transcriptionally regulated by the action of PPAR and RXR (14, 15), we evaluated the expressions of PPARα and PPARγ. Both share functions as well as have distinct activities in a variety of roles, including the regulation of lipid metabolism (32). Previous studies show that hepatic PPARγ expression remains unchanged in chronic alcohol-fed rats (33) and that PPAR expression is decreased by chronic alcohol feeding (34). However, there was no information regarding PPAR protein levels (33) and the specificity of PPAR subtype (34) in their report. Although our model was a little different from the previous one, e.g., we used Fisher rats rather than Sprague-Dawley rats, we found that both PPARα and PPARγ expressions at protein levels were upregulated in the livers of rats with alcohol feeding (Fig. 3). Interestingly, we observed that only PPARγ expression, not PPARα, was strongly correlated with CMO1 expression in alcohol-fed rats. This supports the previous observation that PPARγ is essential for CMO1 gene expression (14). In humans, PPARγ alone may not be sufficient to activate CMO1 expression. For example, the induction of CMO1 is dependent on the cooperation between PPARγ and myocyte enhancer factor 2 isoform (15).
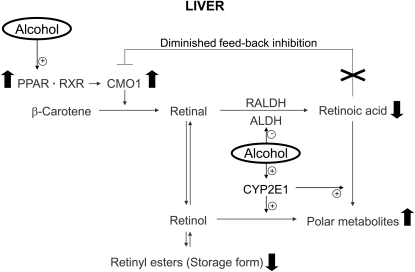
Our observation of the upregulation of CMO1 by alcohol intake is in agreement with previous observations that daily consumption of alcohol is related to reduced β-carotene levels (19, 20), but how can we explain lower vitamin A levels in the liver during chronic alcohol consumption? Although the Lieber-DeCarli liquid diet contains a sufficient amount of vitamin A for both the control and experimental groups, ethanol feeding of rats for 11 wk resulted in lower hepatic retinoic acid, retinol, and retinyl palmitate concentrations (by ∼62, 33, and 42%, respectively) compared with nonethanol-fed rats (Table 1). These data were in agreement with our previous studies showing that feeding rats with Lieber-DeCarli alcohol liquid diet for 1 mo significantly reduces hepatic vitamin A and retinoic acid levels in rats compared with rats pair-fed a diet matched for energy and vitamin A content (26, 35). It is shown that chronic alcohol intake increases catabolism of retinol and retinoic acid into more polar metabolites in the liver (36). Work from our laboratory showed that alcohol-reduced levels of retinol and retinoic acid were prevented by chlormethiazole, an inhibitor of cytochrome P4502E1, both in vitro and in vivo (27). This indicates that the alcohol-enhanced catabolism of retinoids in hepatic tissue after exposure to alcohol is a major mechanism for lowering vitamin A status (4). Previously, we conducted an in vitro incubation experiment with β-carotene using liver homogenates of alcohol-fed rats compared with controls (X.D. Wang, unpublished data). Although we did observe a significant loss of β-carotene after the incubation with the alcohol-fed liver homogenate, we could not detect either retinal or retinoic acid formation from β-carotene in the liver homogenates of the alcohol-fed rats compared with the control. We believe the strong induction of cytochrome P450 enzymes or certain nonspecific oxygenases by alcohol treatment degraded both β-carotene and newly formed retinol and retinoic acid. Therefore, even with CMO1 converting provitamin A carotenoids into vitamin A, the induction of hepatic CYP2E1 enzyme in chronic intermittent drinking continues to be a factor in destroying retinol and retinoic acid, even after the alcohol is cleared. In addition, the much lower level of retinoic acid, compared with retinol and retinyl palmitate (Table 1), could be due to a competitive inhibition of alcohol dehydrogenase and aldehyde dehydrogenase by alcohol, thereby inhibiting retinol oxidation into retinal and retinoic acid (37, 38). Takitani et al. (39) reported that hepatic CMO1 expression was suppressed by all-trans retinoic acid supplementation in the presence of vitamin A deficiency. It has also been shown that activity and expression of intestinal CMO1 were downregulated by retinoic acid treatment in chickens (40). It is an effective feedback regulatory mechanism of retinoic acid on CMO1 and has been suggested to be involved in vitamin A homeostasis (41). Therefore, the diminished feedback inhibition of CMO1 expression due to alcohol-reduced retinoic acid levels in the liver could contribute to the upregulation of CMO1 in the present study as well (Fig. 4). Interestingly, retinal, a metabolite of both retinol and β-carotene, suppresses PPARγ and inhibits adipogenesis (42); therefore, the alcohol-reduced retinoid levels in the liver could contribute to the upregulation of PPARγ-mediated CMO1 expression in the present study. In addition, fatty liver has been documented in this rat model using Lieber-Decarli alcoholic diet (29). The previous study in the CMO1 knockout mouse developed both vitamin A deficiency and a fatty liver (43). Therefore, in this study, the alcohol-reduced level of retinol and retinoic acid could be related to the upregulation of PPARγ and fatty liver as well. Future study examining CMO1 expression in alcohol-fed rats with vitamin A/retinoic acid supplementation will provide more evidence to support our conclusion.
FIGURE 4.
Proposed schematic representation of the effects of chronic alcohol intake (solid arrow) on CMO1 expression and in the hepatic metabolism of vitamin A. ALDH: aldehyde dehydrogenase; RALDH: retinal dehydrogenase.
Thyroid hormone can transcriptionally upregulate CMO1 (17). Because chronic alcohol intake is related to decreased thyroid axis (30, 31), we examined whether the upregulation of CMO1 by alcohol was due to its effects on circulating thyroid hormones, including fT3, fT4, and TSH, and hepatic TRβ expression. Although we did not detect any changes on the levels of fT3, fT4, and TSH, alcohol intake did increase hepatic TRβ expression. This correlated with the expression of CMO1. Because retinoic acid treatment decreases TR expression in a dose-dependent manner (41) and chronic alcohol intake results in lower levels of hepatic vitamin A and retinoic acid (Table 1), we proposed that alcohol treatment could indirectly increase hepatic TR expression by decreasing vitamin A levels in the livers. In addition, it has been reported that patients with hypothyroidism accumulated significant amounts of carotenoids in their body (41, 44) and that CMO1 was upregulated by T3 (17). Therefore, we speculate that the increase in TRβ could augment thyroid hormone’s effects and contribute to the upregulation of CMO1. Further studies are needed to evaluate this notion.
Previously, we showed that CMO2 catalyzes the excentric cleavage of all trans-β-carotene and cis-lycopene isomers at the 9′,10’ double bond (10). In the present study, alcohol intake caused only a slight increase in both mRNA expression and protein levels of CMO2 (Fig. 2), indicating that there was little effect of vitamin A status on CMO2 regulation in the liver. Although both PPARγ and PPARα were modestly correlated to CMO2 expression, the biochemical evaluation of the CMO2 promoter region has not been completed, and previous studies did not find PPRE in the CMO2 promoter region (45), we cannot affirm that PPARs are involved in CMO2 regulation. However, there is a difference between wild-type cows and those with a premature stop codon in CMO2, e.g., serum β-carotene concentrations were 48% higher and hepatic vitamin A levels were 33% lower in cows with premature stop codon in CMO (13). Therefore, the upregulation of CMO2 by alcohol may have certain biological significance in terms of potential interaction of alcohol with provitamin A carotenoid metabolism. However, some data show that β-carotene supplementation with concomitant alcohol consumption generates intrinsic hepatotoxicity (46). Recently, we demonstrated that lycopene supplementation at a higher dose significantly induces hepatic CYP2E1 protein and the incidence of inflammatory foci in the alcohol-fed rats but not in the control rats (29). The excessive formation of excentric cleavage products from carotenoids may produce detrimental effects in both smokers and alcohol drinkers (47). Clearly, determining whether CMO2 plays a role in those processes needs further investigation.
Taken together, these data indicate that chronic alcohol intake upregulates CMO1, PPARγ, and TRβ. This corroborates the idea that CMO1 is transcriptionally regulated by PPARγ and TRβ and that regulation of CMO1 acts by a negative feedback mechanism of vitamin A and retinoic acid. This involvement of multiple factors on the expression of carotenoid cleavage enzymes further indicates a complexity of the transcriptional regulatory mechanisms of carotenoid cleavage enzymes, and clearly more research is needed.
Acknowledgments
We thank Dr. Célia Regina Nogueira (Department of Clinical Medicine, Botucatu School of Medicine, UNESP, Brazil) for advice. We also thank Kang-Quan Hu for her technical assistance and Stephanie-Jo McGehee for her assistance in the preparation of this manuscript. X.D.W. designed research; R.A.M.L., A.F.N., S.V. and C.L. conducted research; A.F.N. analyzed data; R.A.M.L. and X.D.W. wrote the paper. All authors read and approved the final manuscript.
Footnotes
Supported by NIH grant R01CA104932, by USDA grant 1950-51000-064S, and by FAPESP (process no. 06/58177-0). Any opinions, findings, conclusion, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of NIH and the USDA.
Abbreviations used: CMO1, carotenoid 15,15’-monooxygenase 1; CMO2, carotenoid 9′10’-monooxygenase 2; CYP2E1, cytochrome P450 2E1; fT3, free triiodothyronine; fT4, free thyroxine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PPRE, peroxisomal proliferator response element; RXR, retinoid X receptor; TR, thyroid receptor; TSH, thyroid stimulating hormone.
Literature Cited
- 1.Richardson HN, Lee SY, O'Dell LE, Koob GF, Rivier CL. Alcohol self-administration acutely stimulates the hypothalamic-pituitary-adrenal axis, but alcohol dependence leads to a dampened neuroendocrine state. Eur J Neurosci. 2008;28:1641–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valeix P, Faure P, Bertrais S, Vergnaud AC, Dauchet L, Hercberg S. Effects of light to moderate alcohol consumption on thyroid volume and thyroid function. Clin Endocrinol (Oxf). 2008;68:988–95 [DOI] [PubMed] [Google Scholar]
- 3.Chaung WW, Jacob A, Ji Y, Wang P. Suppression of PGC-1alpha by ethanol: implications of its role in alcohol induced liver injury. Int J Clin Exp Med. 2008;1:161–70 [PMC free article] [PubMed] [Google Scholar]
- 4.Wang XD. Alcohol, vitamin A and cancer. Alcohol. 2005;35:251–8 [DOI] [PubMed] [Google Scholar]
- 5.von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem. 2000;275:11915–20 [DOI] [PubMed] [Google Scholar]
- 6.Kiefer C, Hessel S, Lampert JM, Vogt K, Lederer MO, Breithaupt DE, von Lintig J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J Biol Chem. 2001;276:14110–6 [DOI] [PubMed] [Google Scholar]
- 7.Moise AR, von Lintig J, Palczewski K. Related enzymes solve evolutionarily recurrent problems in the metabolism of carotenoids. Trends Plant Sci. 2005;10:178–86 [DOI] [PubMed] [Google Scholar]
- 8.Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308:267–9 [DOI] [PubMed] [Google Scholar]
- 9.Jirholt J, Lindqvist AK, Karlsson J, Andersson A, Holmdahl R. Identification of susceptibility genes for experimental autoimmune encephalomyelitis that overcome the effect of protective alleles at the EAE2 locus. Int Immunol. 2002;14:79–85 [DOI] [PubMed] [Google Scholar]
- 10.Hu KQ, Liu C, Ernst H, Krinsky NI, Russell RM, Wang XD. The biochemical characterization of ferret carotene-9′,10'-monooxygenase catalyzing cleavage of carotenoids in vitro and in vivo. J Biol Chem. 2006;281:19327–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebuterne X, Wang XD, Smith DE, Tang G, Russell RM. In vivo biosynthesis of retinoic acid from beta-carotene involves and excentric cleavage pathway in ferret intestine. J Lipid Res. 1996;37:482–92 [PubMed] [Google Scholar]
- 12.Redmond TM, Gentleman S, Duncan T, Yu S, Wiggert B, Gantt E, Cunningham FX., Jr Identification, expression, and substrate specificity of a mammalian beta-carotene 15,15'-dioxygenase. J Biol Chem. 2001;276:6560–5 [DOI] [PubMed] [Google Scholar]
- 13.Berry SD, Davis SR, Beattie EM, Thomas NL, Burrett AK, Ward HE, Stanfield AM, Biswas M, Ankersmit-Udy AE, et al. Mutation in bovine beta-carotene oxygenase 2 affects milk color. Genetics. 2009;182:923–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boulanger A, McLemore P, Copeland NG, Gilbert DJ, Jenkins NA, Yu SS, Gentleman S, Redmond TM. Identification of beta-carotene 15, 15'-monooxygenase as a peroxisome proliferator-activated receptor target gene. FASEB J. 2003;17:1304–6 [DOI] [PubMed] [Google Scholar]
- 15.Gong X, Tsai SW, Yan B, Rubin LP. Cooperation between MEF2 and PPARgamma in human intestinal beta,beta-carotene 15,15'-monooxygenase gene expression. BMC Mol Biol. 2006;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shulman AI, Mangelsdorf DJ. Retinoid X receptor heterodimers in the metabolic syndrome. N Engl J Med. 2005;353:604–15 [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi N, Suruga K. Triiodothyronine stimulates CMO1 gene expression in human intestinal Caco-2 BBe cells. Life Sci. 2008;82:789–96 [DOI] [PubMed] [Google Scholar]
- 18.Mein JR, Lian F, Wang XD. Biological activity of lycopene metabolites: implications for cancer prevention. Nutr Rev. 2008;66:667–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stryker WS, Kaplan LA, Stein EA, Stampfer MJ, Sober A, Willett WC. The relation of diet, cigarette smoking, and alcohol consumption to plasma beta-carotene and alpha-tocopherol levels. Am J Epidemiol. 1988;127:283–96 [DOI] [PubMed] [Google Scholar]
- 20.Fukao A, Tsubono Y, Kawamura M, Ido T, Akazawa N, Tsuji I, Komatsu S, Minami Y, Hisamichi S. The independent association of smoking and drinking with serum beta-carotene levels among males in Miyagi, Japan. Int J Epidemiol. 1996;25:300–6 [DOI] [PubMed] [Google Scholar]
- 21.Leo MA, Lieber CS. Alcohol, vitamin A, and beta-carotene: adverse interactions, including hepatotoxicity and carcinogenicity. Am J Clin Nutr. 1999;69:1071–85 [DOI] [PubMed] [Google Scholar]
- 22.Nagy LE. Molecular aspects of alcohol metabolism: transcription factors involved in early ethanol-induced liver injury. Annu Rev Nutr. 2004;24:55–78 [DOI] [PubMed] [Google Scholar]
- 23.Baez-Saldana A, Ortega E. Biotin deficiency blocks thymocyte maturation, accelerates thymus involution, and decreases nose-rump length in mice. J Nutr. 2004;134:1970–7 [DOI] [PubMed] [Google Scholar]
- 24.Kim CI, Leo MA, Lowe N, Lieber CS. Differential effects of retinoids and chronic ethanol consumption on membranes in rats. J Nutr. 1988;118:1097–103 [DOI] [PubMed] [Google Scholar]
- 25.Lieber CS, DeCarli LM. The feeding of alcohol in liquid diets: two decades of applications and 1982 update. Alcohol Clin Exp Res. 1982;6:523–31 [DOI] [PubMed] [Google Scholar]
- 26.Wang XD, Liu C, Chung J, Stickel F, Seitz HK, Russell RM. Chronic alcohol intake reduces retinoic acid concentration and enhances AP-1 (c-Jun and c-Fos) expression in rat liver. Hepatology. 1998;28:744–50 [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Chung J, Seitz HK, Russell RM, Wang XD. Chlormethiazole treatment prevents reduced hepatic vitamin A levels in ethanol-fed rats. Alcohol Clin Exp Res. 2002;26:1703–9 [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
- 29.Veeramachaneni S, Ausman LM, Choi SW, Russell RM, Wang XD. High dose lycopene supplementation increases hepatic cytochrome P4502E1 protein and inflammation in alcohol-fed rats. J Nutr. 2008;138:1329–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teschke R, Moreno F, Heinen E, Herrmann J, Kruskemper HL, Strohmeyer G. Hepatic thyroid hormone levels following chronic alcohol consumption: direct experimental evidence in rats against the existence of a hyperthyroid hepatic state. Hepatology. 1983;3:469–74 [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen DD, Sarkar DK, Roberts JL, Gore AC. Chronic daily ethanol and withdrawal. 4. Long-term changes in plasma testosterone regulation, but no effect on GNRH gene expression or plasma LH concentrations. Endocrine. 2003;22:143–50 [DOI] [PubMed] [Google Scholar]
- 32.Mello T, Polvani S, Galli A. Peroxisome proliferator-activated receptor and retinoic x receptor in alcoholic liver disease. PPAR Res. 2009;2009:748174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieber CS, Leo MA, Wang X, Decarli LM. Effect of chronic alcohol consumption on hepatic SIRT1 and PGC-1alpha in rats. Biochem Biophys Res Commun. 2008;370:44–8 [DOI] [PubMed] [Google Scholar]
- 34.Wan YJ, Morimoto M, Thurman RG, Bojes HK, French SW. Expression of the peroxisome proliferator-activated receptor gene is decreased in experimental alcoholic liver disease. Life Sci. 1995;56:307–17 [DOI] [PubMed] [Google Scholar]
- 35.Chung J, Veeramachaneni S, Liu C, Mernitz H, Russell RM, Wang XD. Vitamin E supplementation does not prevent ethanol-reduced hepatic retinoic acid levels in rats. Nutr Res. 2009;29:664–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Russell RM, Seitz HK, Wang XD. Ethanol enhances retinoic acid metabolism into polar metabolites in rat liver via induction of cytochrome P4502E1. Gastroenterology. 2001;120:179–89 [DOI] [PubMed] [Google Scholar]
- 37.Molotkov A, Duester G. Retinol/ethanol drug interaction during acute alcohol intoxication in mice involves inhibition of retinol metabolism to retinoic acid by alcohol dehydrogenase. J Biol Chem. 2002;277:22553–7 [DOI] [PubMed] [Google Scholar]
- 38.Khalighi M, Brzezinski MR, Chen H, Juchau MR. Inhibition of human prenatal biosynthesis of all-trans-retinoic acid by ethanol, ethanol metabolites, and products of lipid peroxidation reactions: a possible role for CYP2E1. Biochem Pharmacol. 1999;57:811–21 [DOI] [PubMed] [Google Scholar]
- 39.Takitani K, Zhu CL, Inoue A, Tamai H. Molecular cloning of the rat beta-carotene 15,15'-monooxygenase gene and its regulation by retinoic acid. Eur J Nutr. 2006;45:320–6 [DOI] [PubMed] [Google Scholar]
- 40.Bachmann H, Desbarats A, Pattison P, Sedgewick M, Riss G, Wyss A, Cardinault N, Duszka C, Goralczyk R, et al. Feedback regulation of beta,beta-carotene 15,15'-monooxygenase by retinoic acid in rats and chickens. J Nutr. 2002;132:3616–22 [DOI] [PubMed] [Google Scholar]
- 41.Perez P, Sanchez-Pacheco A, Pascual A, Aranda A. Retinoic acid decreases thyroid hormone receptor expression in pituitary GH1 cells. Biochem Biophys Res Commun. 1991;181:9–15 [DOI] [PubMed] [Google Scholar]
- 42.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, Tang G, Dolnikowski GG, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hessel S, Eichinger A, Isken A, Amengual J, Hunzelmann S, Hoeller U, Elste V, Hunziker W, Goralczyk R, et al. CMO1 deficiency abolishes vitamin A production from beta-carotene and alters lipid metabolism in mice. J Biol Chem. 2007;282:33553–61 [DOI] [PubMed] [Google Scholar]
- 44.Dotsch J, Zepf K, Schellmoser S, Rascher W, Dorr HG. Unmasking of childhood hypothyroidism by disseminated xanthomas. Pediatrics. 2001;108:E96. [DOI] [PubMed] [Google Scholar]
- 45.Zaripheh S, Nara TY, Nakamura MT, Erdman JW., Jr Dietary lycopene downregulates carotenoid 15,15'-monooxygenase and PPAR-gamma in selected rat tissues. J Nutr. 2006;136:932–8 [DOI] [PubMed] [Google Scholar]
- 46.Lieber CS. Alcohol: its metabolism and interaction with nutrients. Annu Rev Nutr. 2000;20:395–430 [DOI] [PubMed] [Google Scholar]
- 47.Mernitz H, Wang XD. Alcohol and tobacco smoke in retinoid metabolism and signaling: implications for carcinogenesis. : Cho S, Purohit V, Alcohol, tobacco and cancer. Basel: Karger AG; 2006. p. 140–59 [Google Scholar]